Abstract
GeneMatcher is a platform through which various stakeholders can connect with others interested in candidate gene findings. GeneDx, a diagnostic laboratory, has utilized GeneMatcher over the last seven years to successfully facilitate connections between clinicians and researchers, generating fruitful research collaborations. Our ultimate goal in reporting candidate gene findings is to amass sufficient evidence to establish novel disease–gene relationships (DGRs), thus providing diagnostic answers to families and clinicians. Our database of over 300,000 clinical exomes has been a major driver of DGR discovery. Our laboratory accounts for over 20% of total GeneMatcher submissions. Largely fueled by GeneMatcher matches, we have published over 200 articles involving new DGRs or expanded phenotypes for known disease‐causing genes in the past three years. These endeavors require commitments to sharing data and dedicating resources to investigate potential matches. Ultimately, GeneMatcher enables collaboration on a broad scale: we are grateful to the clinicians, researchers, patients, and caregivers who have partnered with us to accelerate the pace of DGR discovery. GeneMatcher opens the door to new partnerships, new discoveries, and families finding answers that otherwise may not have been possible.
Keywords: candidate gene, disease–gene relationships, exome sequencing, gene discovery, GeneDx, GeneMatcher, research
GeneDx GeneMatcher Connection Workflow
1. INTRODUCTION
GeneMatcher describes itself as “a freely accessible website designed to enable connections between clinicians and researchers from around the world who share an interest in the same gene or genes” (Sobriera et al., 2021b; https://genematcher.org). The use of GeneMatcher is ideal for candidate gene findings that, with further evidence, could lead to the identification of new Mendelian disease‐gene relationships (DGRs) (Sobreira et al., 2015). The presence of international contributors of different types (clinicians, researchers, clinical laboratories, and even families) increases the chances of matching what are often rare instances of individuals with variants in particular candidate genes.
As a clinical testing laboratory, GeneDx is strictly neither the “clinician” nor “researcher” described above but rather exists at a nexus for facilitating interactions between various stakeholders. These interactions have resulted in collaborations, ongoing research, publications in peer‐reviewed journals, and, most importantly, answers for families. While genetic testing laboratories are traditionally focused on test interpretation and reporting for known Mendelian conditions, GeneDx has focused on exome sequencing (ES). This inherently involves the identification of candidate gene findings, or variants in a gene without an established Mendelian disease relationship (Biesecker et al., 2021). These findings could also include novel mechanisms or modes of inheritance for established disease genes (e.g., a possible dominant gain of function mechanism with a distinct phenotype in a gene with only a previously established recessive loss of function disease). The ultimate goal of reporting candidate gene findings is to amass sufficient evidence to clearly establish novel DGRs and thus provide diagnostic answers to patients, caregivers, and ordering providers.
Through the course of diagnostic ES and, more recently, whole‐genome sequencing (WGS), GeneDx identifies candidate gene findings, such as de novo variants in constrained genes. While we routinely apply statistical meta‐analysis of the aggregated data across our cohort (Kaplanis et al., 2020), functional or animal model validation of candidate gene findings is beyond the scope of our work or capabilities as a diagnostic laboratory. The ability to identify cases beyond our referral cohort can also provide additional statistical and clinical evidence for novel DGRs. Thus, GeneDx has dedicated significant resources and effort, including three full‐time staff members, towards participation in GeneMatcher and support of the ensuing matches.
GeneDx has performed over 300,000 clinical exomes since 2012, consisting of 45% affected probands and the remainder sequenced for concurrent familial analysis. The resulting database has huge potential for gene discovery. Our participation in GeneMatcher has two closely linked purposes based on the aim of reclassifying candidate genes: to fine‐tune our ability to provide accurate results and to give answers to families even beyond those tested at GeneDx.
About 6% of exome cases completed at GeneDx have a candidate gene variant as the sole finding, and 20% have a candidate gene finding in addition to another finding. Since candidate genes are of uncertain significance (sometimes called Genes of Uncertain Significance, or GUSs), variants in a candidate gene are classified as variants of uncertain significance (VUS) for clinical reporting. Communicating candidate gene findings can be challenging for clinicians, and patients who receive a VUS have been reported to experience: less reassurance from genetic counseling (Culver et al., 2013); sustained distress following result‐disclosure (O'Neill et al., 2009); behavioral and/or lifestyle intentions based on that VUS (Lawal et al., 2018); uncertainty stemming from the lack of information about the VUS, leading to dissatisfaction with the result (Lawal et al., 2018); and disappointment with the VUS result (Predham et al., 2016). Interestingly, patients who received a VUS result have also identified the potential future value of the result (Skinner et al., 2018), and some have expressed an expectation of reclassification over time (Solomon et al., 2017).
Candidate gene research has a positive impact on gene discovery and potentially contributes to a better clinical experience for the clinician and patient by reducing the number of candidate gene variants reported, thus making participation in GeneMatcher an obvious choice for GeneDx. It enables us to efficiently and consistently submit candidate genes from our database, as well as query for genes‐of‐interest.
1.1. Experience with GeneMatcher
In December 2014, GeneDx facilitated a mass submission of all previously reported candidate genes (1258 unique submissions and 812 unique genes) into GeneMatcher, effectively doubling the number of entries in the platform. Soon after, the number of matches also increased (Sobreira et al., 2015, fig. 2). Since then, GeneDx submits candidate gene entries immediately at the time of reporting. To protect patient privacy, only gene name and inheritance are submitted. As of 9/28/2021, GeneMatcher listed the following statistics (Sobriera et al., 2021a; https://genematcher.org/statistics/).
| Category | Counts |
|---|---|
| Submitters | 10,919 (94 Countries) |
| Submissions | 58,036 |
| Unique genes | 13,941 |
| Genes with matches | 8914 |
Of those, GeneDx has made 11,690 submissions (20.1% of total) in 3507 unique genes (25.2% of total). In the past year, GeneDx received an average of ten new GeneMatcher inquiries per day (i.e., direct email inquiries from GeneMatcher users who have an interest in a particular gene and see that they matched with a GeneDx entry), in addition to GeneMatcher‐generated match notifications and follow‐up inquiries and discussions regarding ongoing collaborations.
GeneDx submits all candidate genes included in its clinical diagnostic reports to GeneMatcher. A gene is considered a candidate gene if there is no established disease‐gene relationship (DGR) supported by publication(s) with functional studies and/or a cohort with multiple unrelated patients. A finding may be considered a candidate even if there is a known DGR; a different mechanism, inheritance, or substantially distinct phenotype would be considered a candidate finding for the purposes of reporting and submission to Genematcher.
GeneDx staffs a dedicated team of genetic counselors who field, investigate, and respond to GeneMatcher inquiries. Inquirers are generally clinicians, clinician/researchers, or researchers (human and otherwise). More rarely will a family/parent initiate contact via GeneMatcher. Most inquiries are for the purposes of discussion between clinicians about a case, cohort‐building, and/or functional work to supplement the clinical findings. GeneMatcher is also used by other clinical laboratories to aid in their variant classification. Upon receiving a match inquiry, the GeneDx team assesses our database to identify matches and provide responses to the inquiries, often soliciting additional search criteria from the inquirer to narrow our search (Figure 1). If the internal matches look promising given the nature of the inquiry (i.e., matching phenotype or variant type or inheritance), we offer to contact the ordering clinicians, which can involve multiple cases, some back‐and‐forth, and a significant amount of time. At this point, however, we do not share case details. Since the ordering clinician initiated testing of their patient and is an integral part of the clinical–laboratory–research loop, an introduction between the ordering clinician and inquirer is only made with the clinician's permission. Further discussion may occur regarding the purpose of the inquiry and the logistics of the research. Should the family not be interested in participating in a research cohort, the discussion ends. If the family is interested, the collaboration continues. Appropriate consent is obtained from participants. Some collaborations are focused on gathering clinical details for a cohort, while others do this in addition to functional studies. Often, GeneDx continues to identify new cases, both internally and via GeneMatcher, while the functional work and manuscript development is ongoing, thus facilitating a growing cohort in real‐time. In situations where publication is the goal, ordering clinicians, external researchers, and GeneDx work cohesively toward the final manuscript submission.
Figure 1.
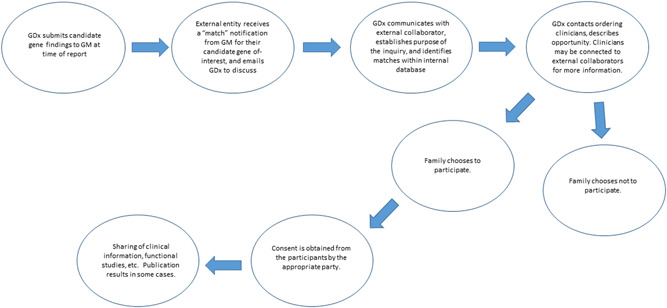
GeneDx GeneMatcher connection workflow
Involving our ordering clinicians in the GeneMatcher process is vital to the success of the individual collaborations. They are the key to robust clinical information and often the source of additional insights into the patient. Given this, many clinicians are co‐authors of the resulting manuscripts. In the first seven months of 2021 alone, 94 institutions and 254 clinician authorship spots (218 unique clinician co‐authors) were associated with published GeneDx collaborations. The laboratory scientists who were involved in the analysis and reporting of GeneDx cases or have expertise in a particular gene family or condition also often participate in the manuscript review process and are named co‐authors.
Ultimately, the work of facilitating GeneMatcher inquiries, collaborating, cohort‐building, and publishing better informs our analysis of candidate gene findings. When a GeneDx‐affiliated paper is published, a separate team within GeneDx promptly reviews the gene to determine if it can be reclassified from candidate gene to disease‐associated gene, based on the criteria described earlier. For cases already in our database, each variant is re‐classified and updated reports are issued as appropriate, providing answers for families previously faced with a VUS. Reclassification of the candidate genes also affects future cases, avoiding VUSs in favor of diagnostic results. As a result, 908 out of 3974 unique genes (22.8%) with diagnostic findings reported by our laboratory from January 1, 2021 to September 28, 2021 were originally identified as candidate genes and submitted to GeneMatcher before sufficient evidence existed to categorize them as definitive DGRs. This rate has increased by a mean of 18.7% annually since we began submitting to GeneMatcher. Of the 908 genes that shifted from candidate to validated DGR status during this time, at least 375 were pursued as active collaborations by our laboratory with GeneMatcher as the primary facilitator of the initial connections.
1.2. Outcomes of GeneMatcher participation
In general, our participation in GeneMatcher has been a very positive experience. Patients tested at GeneDx who receive a candidate gene variant, as well as their ordering clinicians, benefit from GeneMatcher both individually and collectively. Having access to the GeneMatcher research hub exposes us to a diversity of collaborators that we would otherwise not be able to access: clinicians writing up case series, laboratory researchers, other major commercial laboratories, and international collaborators.
In the last 3 years, at least 200 publications on which GeneDx has collaborated have involved either new DGRs (~50%) or expanded phenotypes for known disease‐causing genes (~26%). Expanded phenotype publications involve genes known to cause human disease, but a new cohort produces further information about additional features or a new mechanism of disease (e.g., a disease gene with established autosomal recessive inheritance is newly determined to have an autosomal dominant mechanism). To different degrees, these publications were facilitated by GeneMatcher connections, either new connections or ongoing partnerships with external collaborators originally connected to us via GeneMatcher. In PubMed, of the 142 articles published in the year September 2020 to September 2021 that cited GeneMatcher, 41/142 (28.9%) have GeneDx co‐authors. In the first seven months of 2021, GeneDx co‐authored publications on 26 novel DGRs, 24 of which had sufficient evidence to be upgraded from candidate to validated DGR status internally. During that same time, we also co‐authored publications on 18 additional genes with expanded phenotypes when compared to established DGRs.
There have also been unexpected results that have grown from GeneMatcher collaborations. Some families who have participated in research cohorts expressed their desire to be connected with other families and formed informal and formal family support groups. For families who perhaps thought they were the only one in the world with a particular candidate gene finding, these connections have reportedly been valuable. The development of collaborative relationships with colleagues around the world has also been beneficial. Through the successful GeneMatcher collaborations, GeneDx has identified many external clinicians and scientists with a mutual interest in candidate gene validation.
One example of a successful collaboration aided by GeneMatcher involves FAR1 and a newly described autosomal dominant condition. In 2014, autosomal recessive FAR1 deficiency was reported in a few cases. Over the next few years, GeneDx analysts noted this as an internal gene‐of‐interest based on several heterozygous cases with de novo variants at the p.Arg480 residue and overlapping clinical features. Data‐mining (reviewing our data set with a specific gene/variant in mind) identified eight cases from a cohort of 42,983 exome trios and 9205 exome‐based “Xpanded” panels including FAR1 analysis. GeneDx scientists presented a poster with our internal data at the American Society of Human Genetics meeting in 2017 and, in the meantime, GeneMatcher inquiries/matches led to the addition of several more patients to our cohort, at least one of which was submitted by the Undiagnosed Diseases Network (https://undiagnosed.hms.harvard.edu/). As our cohort grew and we collected clinical information, another GeneMatcher match led to a partnership with lipid synthesis researchers at the University of Amsterdam who were doing functional work regarding FAR1. Patients consented to the appropriate research protocols, GeneDx led the gathering of clinical information from our ordering clinicians and external clinical collaborators, and the functional work was completed and that data added to the manuscript. Our collaborative paper was published in Genetics in Medicine in April 2021 (PMID 33239752; Ferdinandusse et al., 2021). In all, we gathered twelve individuals (eight tested at GeneDx) with heterozygous de novo missense variants at the same residue. The individuals exhibited a common phenotype distinct from the clinical phenotype, and opposite from the biochemical phenotype, associated with autosomal recessive FAR1 deficiency. During the consenting process, parents were amazed to learn that there were other families with the same FAR1 variants (“We thought we were the only ones!”). Several parents expressed interest in connecting with other parents and, with permission of the families, GeneDx facilitated these connections; the parents subsequently formed a Facebook support group that remains active.
1.2.1. Challenges
GeneMatcher is a useful platform. The challenges associated with its use largely stem from a single issue: resources. Internal resources for identifying and developing collaboration opportunities are finite, particularly in the setting of a commercial laboratory. This issue became particularly apparent during the COVID‐19 pandemic. As did everyone, GeneDx shifted, reassessed, and developed workarounds as needed to adapt to unprecedented circumstances. Perhaps not surprisingly, we noticed an increase in GeneMatcher activity. Internal tracking showed that in 2019 we received approximately 8.5 new inquiries per day from GeneMatcher matches (2078 for the year). In 2020, new inquiries from GeneMatcher matches jumped to approximately 10 daily, or 2322 for the year. With stay‐at‐home orders, perhaps many clinicians and researchers suddenly had time they had lacked previously and decided to work on research projects. This increase in volume may also be driven by the snowball effect of accumulating GeneMatcher entries. As a result, our response time also increased, and we are no longer able to respond to GeneMatcher inquiries in real‐time. We have also noticed a number of commercial laboratories using GeneMatcher to classify the variants identified in their cases. During the first two weeks of September 2021, 15% of the GeneMatcher inquiries we received were from other diagnostic laboratories seeking information for variant classification purposes. This further impacts our turnaround times for GeneMatcher responses, as resources are used to review all inquiries.
External resources can also be a challenge, particularly limited research laboratory resources. In our anecdotal experience, the number of clinicians seeking answers for their patients exceeds the number of researchers with the ability to engage in functional studies. Therefore, some collaborations are limited to phenotypic characterizations of a small number of patients, which can be difficult to publish. It can be challenging to find a research laboratory with the ability to perform functional work. We choose to frame that challenge as an opportunity: Genematcher generates a high volume of matches, allowing us to identify possible research cohorts, which leads us to search for external groups who can perform functional studies and benefit from partnership. In fact, GeneMatcher has played a role in many of our ongoing partnerships with research laboratories, enabling us to have a “go‐to” list of researchers who are potentially interested in future collaborations.
Overall, given the large number of GeneMatcher submissions made by GeneDx, it is not surprising that we consistently receive a large number of inquiries. Triaging these inquiries as to purpose and urgency, then searching our database for matches and facilitating connections takes a considerable amount of time and effort. Ultimately, that effort does result in improved gene classifications and updated results for families, which makes it meaningful. However, it is not unusual for collaborations to span many months or years, and the impact may not be felt in a timely manner. As researchers know, many factors influence the success of a cohort project, not least of which include clinicians and or families moving, changing care or employment, or otherwise not engaging. GeneMatcher does not create these challenges, but our many simultaneous collaborations make these challenges more visible.
2. CONCLUSION
Genomic matchmaking has become common practice for many over the last five years, since the 2015 publication by Sobreira et al. Indeed, a number of those initial collaborations have proven themselves fruitful beyond the original match – the relationships and partnerships facilitated by GeneMatcher have continued to result in successful cohort‐building, case‐comparison, publication, and candidate gene reclassification. As a clinical testing laboratory, GeneDx is fortunate to view the evolution of GeneMatcher from a unique vantage point, one from which we see the interest in hundreds of genes and the work of multiple stakeholders. This is reflected in a collaborative publication record of which we are very proud: hundreds of publications in peer‐reviewed journals, a large proportion of which identify new DGRs.
We also embrace the benefits of GeneMatcher for our ordering clinicians, both for those seeking answers for their patients and for those interested in academic development. Participation in GeneMatcher requires a relatively small investment of time on the clinician's part but potentially enables them to join a broader study, resulting in relationship‐building within the scientific community, involvement in publications, and answers for current and future patients.
Our successes with GeneMatcher are built upon a commitment to data sharing, embracing the critical role of the ordering clinician in any collaboration and, ultimately, our commitment to helping patients and their families. As a diagnostic laboratory, we are grateful to the clinicians, researchers, patients, and caregivers who have participated with us in using GeneMatcher to accelerate the pace of DGR discovery, thus providing more definitive diagnoses to our referring physicians and patients. GeneMatcher is a platform that opens the door to new partnerships, new discoveries, and families finding answers that otherwise may not have been possible.
CONFLICT OF INTERESTS
The authors are employees of GeneDx, Inc.
ACKNOWLEDGMENTS
We acknowledge the valuable contributions of our ordering clinicians, external research collaborators, and especially the families who rely on GeneDx to continue to search for answers.
McWalter, K. , Torti, E. , Morrow, M. , Juusola, J. , & Retterer, K. (2022). Discovery of over 200 new and expanded genetic conditions using GeneMatcher. Human Mutation, 43, 760–764. 10.1002/humu.24351
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no data sets were generated or analyzed during the current study.
REFERENCES
- Biesecker, L. G. , Adam, M. P. , Alkuraya, F. S. , Amemiya, A. R. , Bamshad, M. J. , Beck, A. E. , Bennett, J. T. , Bird, L. M. , Carey, J. C. , Chung, B. , Clark, R. D. , Cox, T. C. , Curry, C. , Palko Dinulos, M. B. , Dobyns, W. B. , Giampietro, P. F. , Girisha, K. M. , Glass, I. A. , Graham Jr., J. M. , … Zarate, Y. A. (2021). A dyadic approach to the delineation of diagnostic entities in clinical genomics. American Journal of Human Genetics, 108(1), 8–15. 10.1016/j.ajhg.2020.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culver, J. O. , Brinkerhoff, C. D. , Clague, J. , Yang, K. , Singh, K. E. , Sand, S. R. , & Weitzelm, J. N. (2013). Variants of uncertain significance in BRCA testing: Evaluation of surgical decisions, risk perception, and cancer distress. Clinical Genetics, 84(5), 464–472. 10.1111/cge.12097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferdinandusse, S. , McWalter, K. , Te Brinke, H. , IJlst, L. , Mooijer, P. M. , Ruiter, J. P. N. , van Lint, A. E. M. , Pras‐Raves, M. , Wever, E. , Millan, F. , Guillen Sacoto, M. J. , Begtrup, A. , Tarnopolsky, M. , Brady, L. , Ladda, R. L. , Sell, S. L. , Nowak, C. B. , Douglas, J. , Tian, C. , … Vaz, F. M. (2021). An autosomal dominant neurological disorder caused by de novo variants in FAR1 resulting in uncontrolled synthesis of ether lipids. Genetics in Medicine, 23(4), 740–750. 10.1038/s41436-020-01027-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplanis, J. , Samocha, K. E. , Wiel, L. , Zhang, K. , Arvai, K. J. , Eberhardt, R. Y. , Gallone, G. , Lelieveld, S. H. , Martin, H. C. , McRae, J. F. , Short, P. J. , Torene, R. I. , de Boer, E. , Danecek, P. , Gardner, E. J. , Huang, N. , Lord, J. , Martincorena, I. , Pfundt, R. , … Retterer, K. (2020). Evidence for 28 genetic disorders discovered by combining healthcare and research data. Nature, 586(7831), 757–762. 10.1038/s41586-020-2832-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawal, T. A. , Lewis, K. L. , Johnston, J. J. , Heidlebaugh, A. R. , Ng, D. , Gaston‐Johansson, F. G. , Klein, W. M. P. , Biesecker, B. B. , & Biesecker, L. G. (2018). Disclosure of cardiac variants of uncertain significance results in an exome cohort. Clinical Genetics, 93(5), 1022–1029. 10.1111/cge.13220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill, S. C. , Rini, C. , Goldsmith, R. E. , Valdimarsdottir, H. , Cohen, L. H. , & Schwartz, M. D. (2009). Distress among women receiving uninformative BRCA1/2 results: 12‐month outcomes. Psycho‐Oncology, 18(10), 1088–1096. 10.1002/pon.1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Predham, S. , Hathaway, J. , Hulait, G. , Arbour, L. , & Lehman, A. (2016). Patient recall, interpretation, and perspective of an inconclusive long QT syndrome genetic test result. Journal of Genetic Counseling, 26, 150–158. 10.1007/s10897-016-9991-4 [DOI] [PubMed] [Google Scholar]
- Skinner, D. , Roche, M. I. , Weck, K. E. , Raspberry, K. A. , Foreman, A. K. M. , Strande, N. T. , Berg, J. S. , Evans, J. P. , & Henderson, G. E. (2018). “Possibly positive or certainly uncertain?”: Participants’ responses to uncertain diagnostic results from exome sequencing. Genetics in Medicine, 20(3), 313–319. 10.1038/gim.2017.135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobreira, N. , Schiettecatte, F. , Valle, D. , & Hamosh, A. (2015). GeneMatcher: A matching tool for connecting investigators with an interest in the same gene. Human Mutation, 36(No. 10), 928–930. 10.1002/humu.22844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobreira, N. , Schiettecatte, F. , Valle, D. , & Hamosh, A. (2021a). Statistics September 28, 2021. https://genematcher.org/statistics/
- Sobreira, N. , Schiettecatte, F. , Valle, D. , & Hamosh, A. (2021b). Retrieved from https://genematcher.org
- Solomon, I. , Harrington, E. , Hooker, G. , Erby, L. , Axilbund, J. , Hampel, H. , Semotiuk, K. , Blanco, A. , Klein, W. M. P. , Giardiello, F. , & Leonard, L. (2017). Lynch syndrome limbo: Patient understanding of variants of uncertain significance. Journal of Genetic Counseling, 26(4), 866–877. 10.1007/s10897-017-0066-y [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no data sets were generated or analyzed during the current study.


