Abstract
Stimuli‐responsive soft materials enable controlled release of loaded drug molecules and biomolecules. Controlled release of potent chemotherapeutic or immunotherapeutic agents is crucial to reduce unwanted side effects. In an effort to develop controlled release strategies that can be triggered by using Cerenkov luminescence, we have developed polymer hydrogels that can release bovine serum albumin and immunoglobulin G by using light (254 nm–375 nm) as a trigger. We describe the synthesis and photochemical characterization of two light sensitive phenacyl bis‐azide crosslinkers that are used to prepare transparent self‐supporting hydrogel patches. One crosslinker was designed to optimize the overlap with the Cerenkov luminescence emission window, bearing an π‐extended phenacyl core, resulting in a high quantum yield (14 %) of photocleavage when irradiated with 375 nm light. We used the extended phenacyl crosslinker for the preparation of protein‐loaded dextran hydrogel patches, which showed efficient and selective dosed release of bovine serum albumin or immunoglobulin G after irradiation with 375 nm light. Cerenkov‐triggered release is as yet inconclusive due to unexpected side‐reactivity. Based on the high quantum yield, efficient release and large overlap with the Cerenkov window, we envision application of these photosensitive soft materials in radiation targeted drug release.
Keywords: controlled release, dextran, phenacyl, photocages, polymers, hydrogels
Polymer phenacyl crosslinked hydrogels that can release doxorubicin, bovine serum albumin and immunoglobulin G by using light (254 nm–375 nm) as a trigger are reported. One crosslinker was designed to optimize the overlap with the Cerenkov luminescence emission window, bearing an π‐extended phenacyl core, resulting in a high quantum yield (QY, 14 %) of photocleavage when irradiated with 375 nm light.
Introduction
Stimuli responsive drug carrier systems have emerged as important devices in the field of biomedical applications and targeted drug delivery. General challenges for controlled release of drugs are timed or prolonged release, targeted or local release, and overcoming physicochemical barriers in the body. In order to release the cargo a phase transition or macroscopic change of the hydrogel network has to be triggered. Several triggers have been successfully applied, such as: pH, ionic strength, mechanical stress, redox potential and light. Light is of particular interest since it is spatio‐temporally controlled and can be harmless when clinically applied, depending on wavelength and intensity. [1] However, human tissue limits the use of light as the penetration depth is just a few centimeters depending on the applied wavelength, which inherently limits the use of external light sources for triggered release. [2] To overcome these limitations, we were excited to investigate the potential of in situ generated light, known as Cerenkov luminescence (CL), and employ it as a drug release trigger. CL is generated when a charged particle, generated by the decay of radionuclides or external radiation, travels faster than the speed of light in a dielectric medium such as water or tissue. CL has a broad emission spectrum with a maximum emission around 380 nm (UV‐A), and is usually generated with low intensity. [3] CL has been employed as a light source in anticancer therapies, where nanoparticles passively target solid tumors and generate reactive oxygen species (ROS). [4] Moreover, CL is found an effective trigger for decaging doxorubicin [5] and is employed to generate reactive singlet nitrene species, that subsequently react with extracellular proteins or lipids and ‘tag’ solid tumors with a drug or dye. [6] For future application with CL triggers, we aimed at developing hydrogels with light sensitive groups with an absorption maximum in the Cerenkov emission window and high quantum yield of photodegradation.
Light sensitive hydrogels have been employed in a broad variety of settings and rely on light sensitive functionalities incorporated into the hydrogel network. [7] Two well‐known light sensitive moieties are the ortho‐nitrobenzyl group (oNB) and its ortho‐nitroveratryl analog. [8] Although ortho‐nitrobenzyl and ortho‐nitroveratryl groups have been employed successfully many times in a variety of studies, they have low to very low quantum yields and during photocleavage toxic nitroso adducts are formed. [9] Therefore we are curious to employ phenacyl based photo cleavable hydrogel crosslinkers (Figure 1C and 1D), as these have a significantly higher quantum yield. [10] Photo responsive phenacyl moieties have been applied to photocage thiols, [11] control the degree of swelling in polymer coatings, [12] induced light triggered folding of polymers chains [13] and to photocage a drug such as chlorambucil. [14]
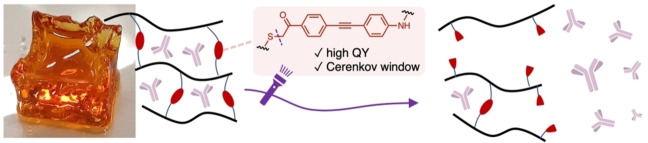
Figure 1.
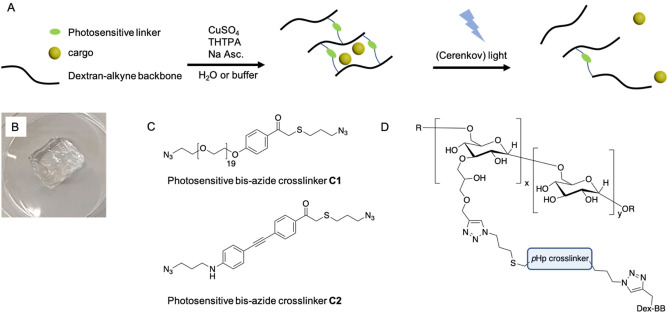
A) Schematic representation of dextran based photosensitive hydrogels. Alkyne modified dextran is crosslinked with a bis‐azide crosslinker using standard Cu‐click conditions (CuSO4, sodium ascorbate and activating ligand: Tris(3‐hydroxypropyltriazolylmethyl)amine (THTPA)). UV‐A light (including Cerenkov luminescence) triggers the photocleavage of the crosslinker resulting in free diffusion of the loaded cargo. B) Photograph of centimeter sized transparent self‐supporting dextran hydrogel. C) Bis‐azide crosslinkers C1 and C2, synthesized and investigated in this research. D) bis‐azide phenacyl crosslinked dextran structure.
Chemotherapy is one of the most important strategies to treat cancer. However, the administered dose of any particular drug is often low due to its toxicity and accompanying side effects. To mitigate these limiting factors and improve the efficacy of chemotherapeutic drugs, smart drug delivery systems are employed. Responsive hydrogel networks can be used for controlled delivery. Hydrogels are able to covalently or non‐covalently store a variety of anticancer drugs and show triggered release in the human body. [15] Alternatively, monoclonal antibodies or antibody‐drug conjugates (ADC) are employed to selectively target malignant tumor cells and induce cell death. [16] Controlled antibody release can improve therapy as the total administered dose is low, on target drug concentrations are high and systemic drug exposure is low which helps to reduce side effects. [17] Release mechanisms rely on a variety of internal and external triggers, including hydrolysis, [18] temperature, [19] ultrasound, [20] (UV) light [21] or enzymatic activity. [22] Also, passive release mechanisms are employed to deliver antibodies from hydrogels, were the degree of hydrogel branching and crosslink density determine the release profile of the loaded proteins. [23] Here, we present light cleavable dextran based hydrogels loaded with either doxorubicin, bovine serum albumin (BSA) or human Immunoglobulin G (IgG), which release their cargo when triggered by (UV‐A/C) light. Photosensitivity in the Cerenkov irradiation window is introduced by crosslinking alkyne modified dextran with bis‐azide phenacyl C1 or C2 using standard Cu‐click conditions (Figure 1).
Results and Discussion
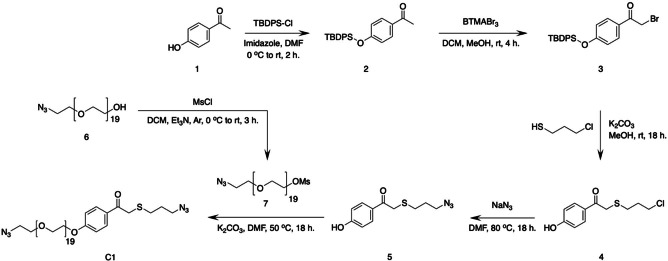
We synthesized bis‐azide crosslinker C1 starting with the protection of commercially available para‐hydroxyphenacyl 1 (pHp) using tert‐butyldiphenyl(chloro)silane (TBDPS−Cl), affording silyl ether 2 (Scheme 1). Next, bromination of compound 2, using benzyltrimethylammonium tribromide (BTMABr3), afforded compound 3. In reaction step 3, using alkaline conditions, 3‐chloro propane thiol was coupled to bromine 3. The combination of prolonged reaction time and alkaline conditions also resulted in the deprotection of the slightly acid hydroxyl in bromine 3, affording deprotected pHp 4. [24] Treating pHp 4 with NaN3 afforded azide 5, and in the final reaction step azide 5 was coupled to mesylated polyethylene glycol (PEG) 7 affording bis‐azide crosslinker C1 (Figure S1). C1 has an absorption maximum at 283 nm, which is not ideal for CL triggering. To gain insight in the photodegradation process of crosslinker C1, we performed an irradiation experiment in which one sample was irradiated with UV‐C light (254 nm, 1.02 mW/cm2) and one sample was kept in the dark as a control. We found that the absorption (283 nm) of the light irradiated sample decreases and shifts with increasing irradiation time, indicating photocleavage of C1 (Figures 2A and B (green data)), while the non‐irradiated sample remained stable over time (black data, Figure 2B). Using standard Cu‐click conditions (CuSO4 (0.47 mM), sodium ascorbate (3.0 mM), activating ligand: Tris(3‐hydroxypropyltriazolylmethyl)amine (THTPA) (0.24 mM)), crosslinker C1 and alkyne modified dextran (500 kDa, DS=36 %, Figure S3) were reacted to prepare transparent self‐supporting dextran hydrogels (Figure 2C), having a storage modulus (G’) of 4.3 ⋅ 102 Pa and tan δ (G’’/G’) of 6.7 ⋅ 10−3 (Figure S11). These hydrogels were loaded with doxorubicin (Figure 2D) and the washed hydrogels were then placed in fresh H2O (25 mL) and irradiated with UV‐C light (254 nm, 1.02 mW/cm2) (Figure 2F). Aliquots were collected at selected timepoints over a period of 24 h (Figure S8B) and UV/Vis analysis showed a linear increase of absorbance at 480 nm, which is doxorubicin's absorption maximum. Unfortunately, the observed increase in absorbance was very small, which is likely caused by the simultaneous photo degradation of doxorubicin. [25] In the same experiment, a 5‐fold increase in absorbance at 283 nm was observed which is contributed to degradation products of doxorubicin (Figure S8A). Degradation products of doxorubicin were not identified or quantified as degradation products of C1 also absorb in the 283 nm region. Although the photodegradation of doxorubicin is undesirable, the experiment does demonstrate that C1 phenacyl works as a photolabile crosslinker as the hydrogels completely disintegrated upon UV‐C irradiation (Figure 2G), in contrast with the control hydrogels that remained intact and retained the loaded doxorubicin within the hydrogel matrix (Figure 2H).
Scheme 1.
Synthetic pathway for the 6‐step synthesis of bis‐azide crosslinker C1, starting from parahydroxy phenacyl (pHp) 1.
Figure 2.
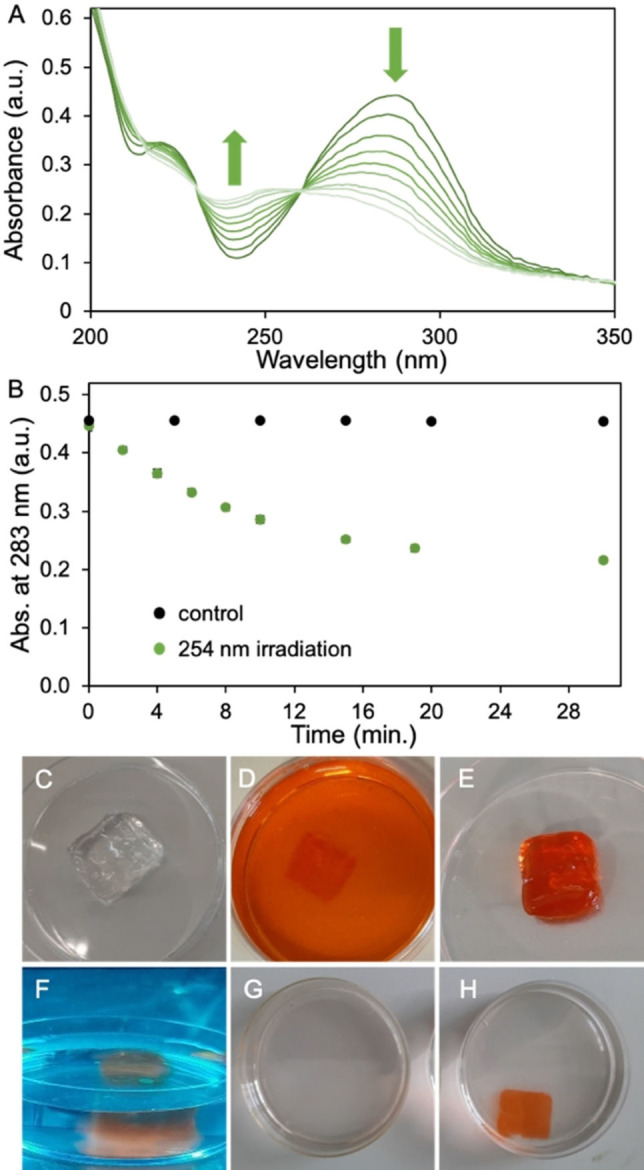
A) UV/Vis data recorded during 254 nm light (1.02 mW/cm2) irradiation experiment of crosslinker C1 (40 μM in H2O). B) Absorbance at 283 nm over time. Black data is a non‐irradiated control sample, green data is irradiated sample. C)–H) series of photographs taken during doxorubicin load and release process. C) H2O washed and swollen gel. D) Hydrogel submerged in doxorubicin loading solution (0.18 mM in H2O after 148 h incubation. E) Doxorubicin loaded hydrogel taken out from the doxorubicin loading solution. F) Doxorubicin loaded gel during 254 nm light irradiation. G) After irradiation, the hydrogel object has fully disintegrated. H) Non‐irradiated control hydrogel after 24 h in H2O.
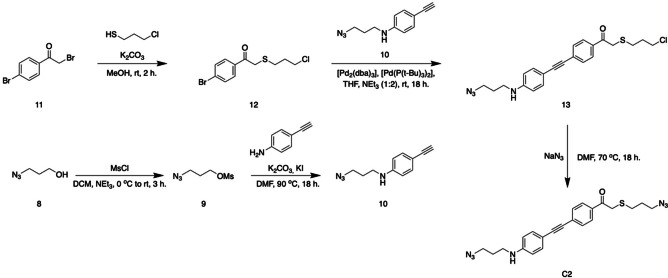
In this stage of the research we opted to develop a π‐extended analogue of crosslinker C1, which would absorb light in the UV‐A region to trigger its photocleavage. This would enable the release of proteins using our hydrogels. Since many proteins often do not absorb light above 300 nm, we expected no interference during light irradiation experiments. Also, the emission maximum of CL is centered around 380 nm light, and the increased overlap of the CL window with the absorbance spectrum of the new crosslinker would increase the efficiency of our system. [3] We synthesized crosslinker C2 which has an extended conjugated π‐system compared to crosslinker C1 and has a maximum absorbance at 383 nm, instead of 283 nm. The synthesis of bis‐azide crosslinker C2 was started with the mesylation of 3‐azido‐propanol (Scheme 2, compound 8) and the subsequent coupling with 4‐ethynyl‐aniline afforded alkyne 10 (Scheme 2). In parallel, reacting 3‐chloro‐propane thiol with bromide 11 yielded bromide 12. The Sonogashira coupling of alkyne 10 and bromine 12, catalyzed by [Pd2(dba)3] and [Pd(P(t‐Bu)3)2], afforded extended pHp 13, which in the final reaction step was treated with NaN3 to yield bis‐azide crosslinker C2 as a yellow powder.
Scheme 2.
Synthetic pathway for the 5‐step synthesis of bis‐azide crosslinker C2.
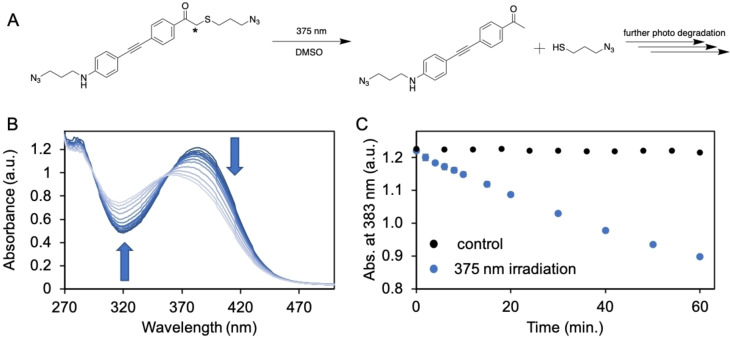
To examine the photochemical properties of C2 a solution was prepared and irradiated with 375 nm light (1240 mW, incident intensity=2.93 ± 0.08 ⋅ 1015 determined by ferrioxalate actinometry [26] (details in Supporting Information)). During a period of 60 minutes, aliquots were collected and analyzed using UV/Vis spectroscopy (Figure 3B). We observed a linear decrease of absorbance (383 nm), indicating photodegradation of C2 (Figure 3C, blue data), while the absorbance data collected for the non‐irradiated control sample remained stable over time (Figure 3C, black data). Photodegradation was confirmed by ESI‐MS analysis were we found m/z=319.12 ([M+H+]) (Figure S4), corresponding with the mass of the ketone photoproduct generated in the photodegradation process of C2 (Figure 3A). [27] The photodegradation rate of C2 was calculated (C2 extinction coefficient=ϵ383 27210 M−1 cm−1 (Figure S6)) according the collected UV/Vis (Figures 3B and C) data and equals 6.8 ⋅ 10−10 mol s−1. Alternatively, the photodegradation process of C2 was monitored by 1H NMR analyses. Here we observed a linear decrease of the resonance at 4.03 ppm (in DMSO‐d6) belonging to the α‐carbon in C2 (indicated with asterisk, Figures 3A and S5), indicating the photocleavage of the thioether. Additionally, the photodegradation rate was determined according the collected 1H NMR data (Figure S5) and equals 6.5 ⋅ 10−10 mol s−1, which is in line with the degradation rate determined according the UV/Vis data. The quantum yield (ϕ) of photodegradation for crosslinker C2 equals ϕ=0.14 calculated using UV/Vis data or ϕ=0.13 calculated using 1H NMR data, which is in line with reported values of comparable thioether phenacyl structures,[ 27b , 28 ] and is higher than typical o‐nitrobenzyl or o‐nitroveratryl cages. [10] Chromophore extension brings the absorption into the visible range, similar to other core‐extended photocages such as nitrobenzyls, acridines and some coumarins. [29]
Figure 3.
A) Reaction scheme showing the 375 nm light triggered photocleavage of crosslinker C2, generating the ketone photoproduct of C2 and 3‐azido‐propanethiol. B) UV/Vis data recorded during the 375 nm light (photon flux=2.93±0.08 ⋅ 1015 ⋅ s−1) irradiation experiment of crosslinker C2 (40 μM in DMSO). C) Absorbance at 383 nm over time. Black data is non‐irradiated control sample, blue data is irradiated sample.
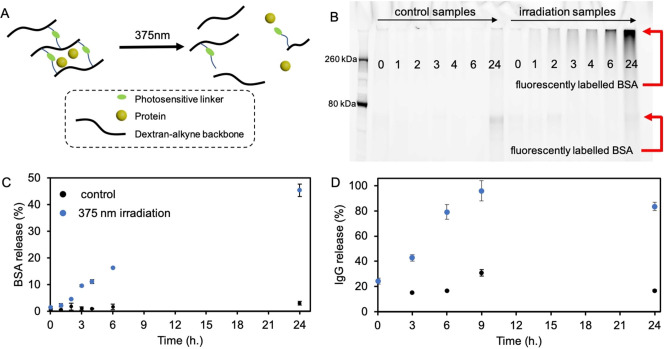
Next, we prepared hydrogel patches (4×4 cm, thickness ∼ 2 mm) loaded with BSA as a model protein (Figure 4A). First, a BSA stock solution (60 μl, 0.63 mg/mL, phosphate buffer (PB) (100 mM, pH 7.4)) was prepared and mixed with a dextran‐alkyne solution (10 wt% in DMSO), which subsequently was mixed with a crosslinker C2 solution (400 μl, 4.61 μM, DMSO) containing Cu‐click reactants (CuI 3.2 μM, DIPEA 8.6 μM, DMSO) and transferred to a quartz cuvette (4×4×4 cm) to gel overnight. After washing the hydrogel patches (details in Supporting Information), one hydrogel was irradiated with 375 nm light in order to degrade the hydrogel matrix and release the loaded BSA (66.5 kDa). Another hydrogel was kept in the dark as a control. During irradiation, aliquots were collected and subsequently analyzed for their protein content by SDS‐page gel analyses (sample mixture fluorescently labelled with NHS−Cy5, details in Supporting Information) (Figure 4B, right lanes). Here, we observed an increasing trend in fluorescence intensity as irradiation time increases, indicating the release of BSA protein. In contrast, barely any fluorescent signal is observed in the non‐irradiated control hydrogel after 24 h (Figure 4B, left lanes), indicating that the mesh size of the hydrogel matrix is small enough to efficiently trap the BSA protein. The fluorescent protein band is located high (approximately at 500 kDa range) in the SDS‐page gel, which is likely the result of resolubilized dextran polymer (500 kDa) during light irradiation, resulting in clogging of the SDS‐page gel which prevents BSA to migrate freely through the SDS‐page gel material. [30] Since NHS‐fluorescent labelling is specific towards protein amines, we are confident that we are observing the light triggered release of BSA protein from the dextran hydrogel matrix. This is supported by accompanying UV/Vis (NanoDrop) data we collected, which shows a linear increase of protein release reaching a 45 % release efficiency after 24 h of 375 nm light irradiation (Figure 4C). Encouraged by these results we decided to load human IgG in our hydrogel patches. Human IgG is an interesting model protein as antibodies and antibody drug conjugates (ADC) are potent anticancer agents. [31] The molecular weight of human IgG is approximately 150 kDa which is 2.3 times heavier than BSA (66.5 kDa). Larger molecules typically diffuse slower from a hydrogel matrix. [23b] This allowed us to reduce the crosslinker C2 loading (25 % lower initial C2 concentration) in our hydrogels while still hampering protein diffusion from the hydrogel matrix. To test antibody release, we prepared hydrogel patches as described before, and loaded human IgG (0.5 mg/mL). After gelation was complete, 375 nm light irradiation was started and the IgG release was quantified according a Bradford assay. We found a linear increase in IgG concentration with increasing irradiation time, reaching a release efficiency of 96 % after 9 h of 375 nm light irradiation (Figure 4D).
Figure 4.
A) Schematic representation of a dextran‐based protein loaded hydrogel, which shows light triggered protein release. B) Fluorescent SDS‐page gel analysis of aliquots collected during 375 nm light irradiation experiments of BSA loaded C2 crosslinked hydrogel patches. The collected aliquots where incubated with NHS−Cy5 fluorophore prior to SDS‐page gel analysis. Fluorescent intensity of the entire lane was analyzed. Control samples show no fluorescent signal except the 24‐hour timepoint which shows low intensity fluorescence. An increase in fluorescence is observed over a time span of 24 h. C) 375 nm light triggered release of BSA from C2 crosslinked hydrogel patches. BSA concentration was determined by UV/Vis analysis (NanoDrop). Initial BSA hydrogel loading=1.8 μg/mL, maximal detected BSA concentration after 24 h of light irradiation=0.8 μg/mL (45 % release efficiency). SD determined by n=2 experiments. D) 375 nm light triggered release of IgG from C2 crosslinked hydrogel patches. IgG concentration was determined by a Bradford assay. Initial IgG hydrogel loading=0.5 mg/mL, maximal detected IgG concentration after 9 h of light irradiation=0.48 mg/mL (96 % release efficiency). SD determined by n=2 experiments.
Encouraged by our obtained results on light triggered protein release, we were curious if we could photodegrade our hydrogel using real CL, which would result in protein release. To test this, we prepared protein containing hydrogel patches of which one was placed in a 60Co‐source and one was kept aside as a control. The γ‐radiation emitted by a 60Co source (0.6 kGy/h) generates low intensity light due to the Cerenkov effect, which could potentially trigger the photocleavage of crosslinker C2, resulting in the triggered release of the loaded proteins. However, we did not detect a significant amount of protein in the hydrogel's supernatant aqueous solution compared to the control hydrogel, indicating that the triggered release had failed. When visually inspecting the γ‐irradiated hydrogels we observed a remarkable contraction effect where the hydrogels consistently showed a macroscopic contraction of the hydrogel volume. Inspired by these results we started to investigate this contraction effect and found that the residual, unreacted alkyne moieties after initial Cu‐click mediated crosslinking, form new alkyne‐alkyne crosslinks. These secondary γ‐irradiation induced crosslinks result in an increased hydrogel stiffness and a significantly reduced hydrogel volume. [32] Moreover, the diffusivity of the hydrogel loaded proteins is thus further restrained due to the increased crosslink density, which explains why no protein release was observed. All together, we were not able to perform proper CL triggered release experiments, due to the inference caused by the residual alkyne groups. For future hydrogel design we will have to carefully design the polymer backbone structure and avoid residual reactive groups that potentially could interfere with γ‐radiation. At present, CL‐triggered release using phenacyl crosslinkers remains inconclusive.
Conclusion
In conclusion, we have synthesized two phenacyl bis‐azide crosslinkers, which have been incorporated in dextran‐based hydrogels to enable photosensitivity in the Cerenkov emission window. Crosslinker C1 photodegrades upon 254 nm light irradiation resulting in the complete dissolution of C1 crosslinked dextran hydrogels releasing loaded doxorubicin. However, the absorption spectrum of C1 has a too small overlap with the CL spectrum, and UV‐A irradiation leads to drug disintegration. A redesign led to extended crosslinker C2. C2 crosslinked dextran hydrogels were loaded with BSA protein or human IgG antibody which was released upon 375 nm light irradiation. The release efficiency for BSA was 45 % after 24 h irradiation, and 96 % for the IgG antibody after 9 h of 375 nm light irradiation. With a quantum yield of photodegradation of 0.14 and an absorption maximum at 380 nm, crosslinker C2 is more suited for application with CL triggers. Although we show promising photosensitivity of our protein loaded hydrogels we could not yet demonstrate protein release triggered by Cerenkov light. Development of CL triggered drug release materials is currently being pursued in our lab.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Acknowledgements
This work was supported by the European Research Council (ERC Consolidator Grant 726381).
T. G. Brevé, M. Filius, S. Weerdenburg, S. J. van der Griend, T. P. Groeneveld, A. G. Denkova, R. Eelkema, Chem. Eur. J. 2022, 28, e202103523.
A previous version of this manuscript has been deposited on a preprint server (https://chemrxiv.org/engage/chemrxiv/article‐details/60d5ec01c6229572e61b08e6).
References
- 1. Fomina N., Sankaranarayanan J., Almutairi A., Adv. Drug Delivery Rev. 2012, 64, 1005–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ash C., Dubec M., Donne K., Bashford T., Lasers Med. Sci. 2017, 32, 1909–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Thorek D. L., Robertson R., Bacchus W. A., Hahn J., Rothberg J., Beattie B. J., Grimm J., Am. J. Nucl. Med. Mol. Imaging 2012, 2, 163–173. [PMC free article] [PubMed] [Google Scholar]
- 4.
- 4a. Duan D., Liu H., Xu Y., Han Y., Xu M., Zhang Z., Liu Z., ACS Appl. Mater. Interfaces 2018, 10, 5278–5286; [DOI] [PubMed] [Google Scholar]
- 4b. Kotagiri N., Cooper M. L., Rettig M., Egbulefu C., Prior J., Cui G., Karmakar P., Zhou M., Yang X., Sudlow G., Marsala L., Chanswangphuwana C., Lu L., Habimana-Griffin L., Shokeen M., Xu X., Weilbaecher K., Tomasson M., Lanza G., DiPersio J. F., Achilefu S., Nat. Commun. 2018, 9, 275; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4c. Yu B., Ni D., Rosenkrans Z. T., Barnhart T. E., Wei H., Ferreira C. A., Lan X., Engle J. W., He Q., Yu F., Cai W., Adv. Mater. 2019, 31, 1904894; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4d. Kamkaew A., Cheng L., Goel S., Valdovinos H. F., Barnhart T. E., Liu Z., Cai W., ACS Appl. Mater. Interfaces 2016, 8, 26630–26637; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4e. Ni D., Ferreira C. A., Barnhart T. E., Quach V., Yu B., Jiang D., Wei W., Liu H., Engle J. W., Hu P., Cai W., J. Am. Chem. Soc. 2018, 140, 14971–14979; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4f. Denkova A. G., Liu H., Men Y., Eelkema R., Adv. Ther. 2020, 3, 1900177; [Google Scholar]
- 4g. Liu H., Laan A. C., Plomp J., Parnell S. R., Men Y., Dalgliesh R. M., Eelkema R., Denkova A. G., ACS Appl. Polym. Mater. 2021, 3, 968–975. [Google Scholar]
- 5. Yao C., Li J., Cao X., Gunn J. R., Wu M., Jiang S., Pogue B. W., ACS Appl. Mater. Interfaces 2020, 12, 44383–44392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Das S., Haedicke K., Grimm J., J. Nucl. Med. 2018, 59, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li L., Scheiger J. M., Levkin P. A., Adv. Mater. 2019, 31, 1807333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.
- 8a. Zhao H., Sterner E. S., Coughlin E. B., Theato P., Macromolecules 2012, 45, 1723–1736; [Google Scholar]
- 8b. Men Y., Breve T., Liu H., Denkova A. G., Eelkema R., Polym. Chem. 2021, 12, 3612–3618; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8c. LeValley P. J., Neelarapu R., Sutherland B. P., Dasgupta S., Kloxin C. J., Kloxin A. M., J. Am. Chem. Soc. 2020, 142, 4671–4679; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8d. LeValley P. J., Sutherland B. P., Jaje J., Gibbs S., Jones R. M., Gala R. P., Kloxin C. J., Kiick K. L., Kloxin A. M., ACS Biomater. Sci. Eng. 2020, 3, 6944–6958; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8e. Kloxin A. M., Kasko A. M., Salinas C. N., Anseth K. S., Science 2009, 324, 59–63; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8f. Azagarsamy M. A., Alge D. L., Radhakrishnan S. J., Tibbitt M. W., Anseth K. S., Biomacromolecules 2012, 13, 2219–2224; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8g. Ruskowitz E. R., DeForest C. A., Nat. Rev. Mater. 2018, 3, 17087. [Google Scholar]
- 9.
- 9a. Givens R. S., Rubina M., Wirz J., Photochem. Photobiol. 2012, 11, 472–488; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9b. Du X., Frei H., Kim S. H., Biopolymers 2001, 62, 147–149. [DOI] [PubMed] [Google Scholar]
- 10. Klán P., Šolomek T., Bochet C. G., Blanc A., Givens R., Rubina M., Popik V., Kostikov A., Wirz J., Chem. Rev. 2013, 113, 119–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.
- 11a. Glassner M., Oehlenschlaeger K. K., Welle A., Bruns M., Barner-Kowollik C., Chem. Commun. 2013, 49, 633–635; [DOI] [PubMed] [Google Scholar]
- 11b. Tischer T., Claus T. K., Oehlenschlaeger K. K., Trouillet V., Bruns M., Welle A., Linkert K., Goldmann A. S., Börner H. G., Barner-Kowollik C., Macromol. Rapid Commun. 2014, 35, 1121–1127. [DOI] [PubMed] [Google Scholar]
- 12. Carias V., Wang J., Toomey R., Langmuir 2014, 30, 4105–4110. [DOI] [PubMed] [Google Scholar]
- 13. Claus Tanja K., Zhang J., Martin L., Hartlieb M., Mutlu H., Perrier S., Delaittre G., Barner-Kowollik C., Macromol. Rapid Commun. 2017, 38, 1700264. [DOI] [PubMed] [Google Scholar]
- 14. Gangopadhyay M., Jana A., Rajesh Y., Bera M., Biswas S., Chowdhury N., Zhao Y., Mandal M., Singh N. D. P., ChemistrySelect 2016, 1, 6523–6531. [Google Scholar]
- 15. Fu X., Hosta-Rigau L., Chandrawati R., Cui J., Chem 2018, 4, 2084–2107. [Google Scholar]
- 16. Diamantis N., Banerji U., Br. J. Cancer 2016, 114, 362–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huynh V., Jesmer A. H., Shoaib M. M., D′Angelo A. D., Rullo A. F., Wylie R. G., ChemBioChem 2019, 20, 747–753. [DOI] [PubMed] [Google Scholar]
- 18. Ashley G. W., Henise J., Reid R., Santi D. V., Proc. Natl. Acad. Sci. USA 2013, 110, 2318–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.
- 19a. Gregoritza M., Messmann V., Abstiens K., Brandl F. P., Goepferich A. M., Biomacromolecules 2017, 18, 2410–2418; [DOI] [PubMed] [Google Scholar]
- 19b. Chung C. K., Fransen M. F., van der Maaden K., Campos Y., García-Couce J., Kralisch D., Chan A., Ossendorp F., Cruz L. J., J. Controlled Release 2020, 323, 1–11. [DOI] [PubMed] [Google Scholar]
- 20. Huebsch N., Kearney C. J., Zhao X., Kim J., Cezar C. A., Suo Z., Mooney D. J., Proc. Natl. Acad. Sci. USA 2014, 111, 9762–9767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Azagarsamy M. A., Anseth K. S., Angew. Chem. Int. Ed. 2013, 52, 13803–13807; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2013, 125, 14048–14052. [Google Scholar]
- 22. Franssen O., Vos O. P., Hennink W. E., J. Controlled Release 1997, 44, 237–245. [Google Scholar]
- 23.
- 23a. Kim J., Francis D. M., Thomas S. N., Nanomaterials 2021, 11, 471;33673289 [Google Scholar]
- 23b. Hennink W. E., Talsma H., Borchert J. C. H., De Smedt S. C., Demeester J., J. Controlled Release 1996, 39, 47–55; [Google Scholar]
- 23c. Sun G., Chu C.-C., Carbohydr. Polym. 2006, 65, 273–287. [Google Scholar]
- 24.
- 24a. Wilson N. S., Keay B. A., Tetrahedron Lett. 1997, 38, 187–190; [Google Scholar]
- 24b. Prakash C., Saleh S., Blair I. A., Tetrahedron Lett. 1994, 35, 7565–7568. [Google Scholar]
- 25. Nawara K., Krysinski P., Blanchard G. J., J. Phys. Chem. A 2012, 116, 4330–4337. [DOI] [PubMed] [Google Scholar]
- 26.
- 26a. Hatchard C. G., Parker C. A., Bowen E. J., Proc. Roy. Soc. A 1956, 235, 518–536; [Google Scholar]
- 26b. Lehóczki T., Józsa É., Ősz K., J. Photochem. Photobiol. A 2013, 251, 63–68. [Google Scholar]
- 27.
- 27a. Sheehan J. C., Umezawa K., J. Org. Chem. 1973, 38, 3771–3774; [Google Scholar]
- 27b. Specht A., Loudwig S., Peng L., Goeldner M., Tetrahedron Lett. 2002, 43, 8947–8950. [Google Scholar]
- 28. Parthiban C., Pavithra M., Reddy L. V. K., Sen D., Samuel M., Singh N. D. P., Org. Biomol. Chem. 2018, 16, 7903–7909. [DOI] [PubMed] [Google Scholar]
- 29. Weinstain R., Slanina T., Kand D., Klán P., Chem. Rev. 2020, 120, 13135–13272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shaw M. M., Riederer B. M., Proteomics 2003, 3, 1408–1417. [DOI] [PubMed] [Google Scholar]
- 31. Hamilton G. S., Biologicals 2015, 43, 318–332. [DOI] [PubMed] [Google Scholar]
- 32. Breve T. G., Liu H., Denkova A. G., Eelkema R., Macromol. Mater. Eng. 2021, 10.1002/mame.202100623. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information