SUMMARY
Galactose toxicity (Gal‐Tox) is a widespread phenomenon ranging from Escherichia coli to mammals and plants. In plants, the predominant pathway for the conversion of galactose into UDP‐galactose (UDP‐Gal) and UDP‐glucose is catalyzed by the enzymes galactokinase, UDP‐sugar pyrophosphorylase (USP) and UDP‐galactose 4‐epimerase. Galactose is a major component of cell wall polymers, glycolipids and glycoproteins; therefore, it becomes surprising that exogenous addition of galactose leads to drastic root phenotypes including cessation of primary root growth and induction of lateral root formation. Currently, little is known about galactose‐mediated toxicity in plants. In this study, we investigated the role of galactose‐containing metabolites like galactose‐1‐phosphate (Gal‐1P) and UDP‐Gal in Gal‐Tox. Recently published data from mouse models suggest that a reduction of the Gal‐1P level via an mRNA‐based therapy helps to overcome Gal‐Tox. To test this hypothesis in plants, we created Arabidopsis thaliana lines overexpressing USP from Pisum sativum. USP enzyme assays confirmed a threefold higher enzyme activity in the overexpression lines leading to a significant reduction of the Gal‐1P level in roots. Interestingly, the overexpression lines are phenotypically more sensitive to the exogenous addition of galactose (0.5 mmol L−1 Gal). Nucleotide sugar analysis via high‐performance liquid chromatography‐mass spectrometry revealed highly elevated UDP‐Gal levels in roots of seedlings grown on 1.5 mmol L−1 galactose versus 1.5 mmol L−1 sucrose. Analysis of plant cell wall glycans by comprehensive microarray polymer profiling showed a high abundance of antibody binding recognizing arabinogalactanproteins and extensins under Gal‐feeding conditions, indicating that glycoproteins are a major target for elevated UDP‐Gal levels in plants.
Keywords: galactose toxicity, UDP‐sugar pyrophosphorylase, galactose‐1‐phosphate, nucleotide sugars, cell wall polymer profiling
Significance Statement
Studies on Gal‐Tox in different model organisms suggest that Gal‐1P plays a central role as toxic agent. Here we generated transgenic lines of Arabidopsis thaliana with significantly reduced Gal‐1P levels under Gal‐feeding conditions by pyrophosphorylase overexpression, which are surprisingly more sensitive towards galactose, indicating that other galactose‐containing metabolites like UDP‐Gal are much more crucial for the development of Gal‐Tox showing different mechanisms of Gal‐Tox in plants.
INTRODUCTION
Galactose is an abundant sugar in the pro‐ and eukaryotic kingdom representing a major component of various glycoproteins, glycolipids or polysaccharides. In mammals, fungi and some red algae, galactose is metabolized via the Leloir pathway into the products UDP‐galactose (UDP‐Gal) and glucose‐1‐phosphate (Glc‐1P), with contribution of the enzymes galactokinase (GALK), galactose‐1‐phosphate uridylyltransferase (GALT) and UDP‐galactose 4‐epimerase (UGE; Gross and Schnarrenberger, 1995; Paladini and Leloir, 1952; Siow et al., 2012). First, galactose is phosphorylated at the OH‐1 position to galactose‐1‐phosphate (Gal‐1P) by the action of GALK. After the transfer of an uridylyl‐residue from UDP‐glucose (UDP‐Glc) to Gal‐1P, the nucleotide sugar UDP‐Gal is formed via GALT and Glc‐1P as side‐product. The equilibrium between UDP‐Gal and UDP‐Glc can then be adjusted by the action of UGE. In humans, the deficiency of one of the Leloir enzymes results in different types of galactosemia representing a life‐threatening disease. Even though galactosemia has been known for over 100 years, the pathophysiology is not yet fully understood. Studies in animal and yeast models provide important insights into molecular mechanisms of galactose toxicity (Gal‐Tox); nevertheless, the only available treatment is the restriction of galactose in diet, which is insufficient to prevent chronic impairments (Delnoy et al., 2021). To advance the therapy approach, it is necessary to identify targets of Gal‐Tox.
Putative targets for Gal‐Tox
Patients suffering from severe galactosemia and GALT‐deficient mouse models exhibit highly elevated levels of Gal‐1P, leading to the hypothesis that Gal‐1P is a key agent in Gal‐Tox (Gitzelmann, 1995; Leslie et al., 1996). Innovative therapy approaches for classic galactosemia aim to alleviate Gal‐Tox by decreasing the Gal‐1P level with GALK inhibitors or mRNA‐based therapies; this suggests that there is a strong correlation between the concentration of Gal‐1P and the strength of the Gal‐Tox phenotype (Balakrishnan et al., 2020; Tang et al., 2012). Data from yeast suggest that Gal‐1P could act as an inhibitor of several enzymes like phosphoglucomutase or inositol‐monophosphatase (Bhat, 2003; de Jongh et al., 2008; Parthasarathy et al., 1997). Furthermore, it has been hypothesized that excess Gal‐1P competes with Glc‐1P for UDP‐hexose pyrophosphorylases resulting in a reduction of intracellular UDP‐hexose concentrations in human galactosemic cells (Lai et al., 2003).
Gal‐Tox in plants
The galactose metabolism in plants is similar to the Leloir pathway in animal and yeast models except that the conversion of Gal‐1P into UDP‐Gal is catalyzed by the enzyme UDP‐sugar pyrophosphorylase (USP) instead of the GALT enzyme. Arabidopsis thaliana contains a single copy gene encoding the USP enzyme (At5g52560) showing a broad substrate specificity towards various sugar‐1‐phosphates (Kotake et al., 2004; Litterer et al., 2006). The biological function of the USP enzyme has not been clearly identified, but a cross of heterozygous knockout usp‐mutants leads to collapsed non‐fertile pollen resulting in lethality (Schnurr et al., 2006). Silencing of the usp‐gene causes sterile seeds and dwarf plants, when the total activity of USP is below 10% of wild‐type (WT) level. Metabolite analysis of silenced USP‐plants mainly accumulates arabinose. The accumulation of arabinose is unlikely the cause of the dwarf phenotype, as knockouts in the gene of arabinokinase show no comparable phenotype (Behmuller et al., 2016). The acceptance of multiple sugar‐1‐phosphates by USP complicates the search for the relevant substrates of USP. Nevertheless, it confirms a role of USP in regular plant metabolism beyond the role in pollen development (Geserick and Tenhaken, 2013). The first description of Gal‐Tox in plants revealed drastic effects of galactose on the root system, including killing of primary root tips and a multi‐branched root system in Vicia villosa and Pisum sativum (Knudson, 1915). Interestingly, in plant models, the toxic effect of galactose can be observed in the WT, whereas the phenomenon of Gal‐Tox in animal and yeast models resulted from mutations in one of the enzymes of the Leloir pathway. There are many data available about the flux of galactose via different pathways into, for example, plant cell wall polymers (Sharples and Fry, 2007), but the targets of Gal‐Tox in plants remain elusive. We recently showed that Gal‐1P accumulates during Gal‐Tox, but the strong increase does not inhibit the enzyme phosphoglucomutase (Althammer et al., 2020) as observed in, for example, yeast.
The analysis of a T‐DNA insertion mutant in GALK showed highly resistant seedlings towards Gal up to 100 mmol L−1, indicating that phosphorylation of Gal is essential for the development of a Gal‐Tox phenotype (Egert et al., 2012). Here we test a strategy to reduce the concentration of Gal‐1P during Gal‐feeding by overexpression of USP. While higher USP activity indeed lowers the concentration of Gal‐1P, the phenotype of Gal‐Tox unexpectedly worsens.
RESULTS
Galactose toxicity in plants is characterized by a severe phenotype showing strong retardation in growth. The main root of Arabidopsis WT seedlings growing on MS plates supplemented with low Gal concentrations (1.5 mmol L−1 Gal) arrests growth. Instead, lateral roots take over the function of the main root. Knockout lines in galK are insensitive to the exogenous addition of Gal (Egert et al., 2012). Growth of WT plants and galK mutants on plates without sugar or on plates with sucrose reveals no visible phenotype; this changed strongly when both plants were grown on plates with Gal. The phosphorylation of Gal in WT plants is essential for the Gal‐Tox phenotype. Throughout our studies we often make use of the galK‐mutant as a control, because both lines grow on the same plates. Current studies in different model organisms suggest that Gal‐1P plays a key role as toxic agent in Gal‐Tox (Balakrishnan et al., 2020; Gibney et al., 2018). Gas chromatography‐mass spectrometry (GC‐MS) measurements revealed that feeding 5 mmol L−1 Gal leads to 20‐fold higher accumulation of Gal‐1P in roots of Arabidopsis WT seedlings (Althammer et al., 2020). To elucidate the role of Gal‐1P in Gal‐Tox in plants, we tested the hypothesis whether a reduction of the Gal‐1P level due to USP overexpression rescues roots from Gal‐Tox. The USP enzyme catalyzes the conversion of Gal‐1P with UTP as co‐substrate into UDP‐Gal and pyrophosphate (PPi) in higher plants, whereas the GALT enzyme uses Gal‐1P together with UDP‐Glc for the formation of UDP‐Gal and Glc‐1P in humans, yeast and red algae (Figure S1).
We tested the hypothesis whether reduction of Gal‐1P due to USP overexpression rescues roots from Gal‐Tox by transforming Arabidopsis with an Ubq10::PsUSP overexpressing construct. Initially, a larger number of transgenic lines were pre‐tested by reverse transcriptase‐polymerase chain reaction (RT‐PCR) for USP expression levels, and the three best candidate lines were selected for further experiments. The analysis of USP transcript levels by RT‐PCR revealed a 10‐fold higher level of transgenic PsUSP mRNA in overexpression line 4 and line 20 compared with the endogenous Arabidopsis USP levels (Figure 1a). The expression level of transgenic PsUSP in the weak overexpression line 23 showed similar levels like endogenous AtUSP level. In general, the expression level of Arabidopsis USP did not alter between WT and the three PsUSP overexpression lines.
Figure 1.
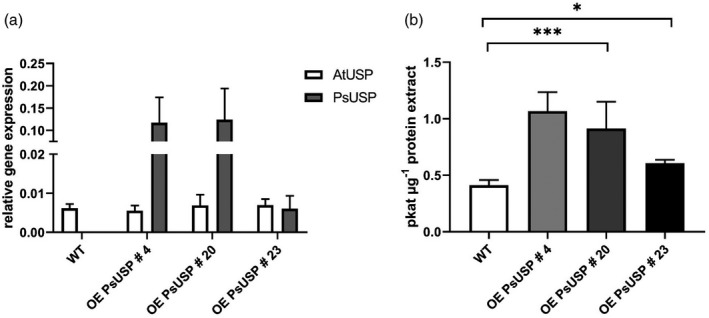
Characterization of UDP‐sugar pyrophosphorylase (USP) overexpression lines.
(a) Transcript expression level in leaves of five individual plants were analyzed by reverse transcriptase‐polymerase chain reaction (RT‐PCR).
(b) USP enzyme activity was measured in leaf extracts of six individual plants by adding galactose‐1‐phosphate (Gal‐1P) as a substrate. UDP‐galactose (UDP‐Gal) formation was determined by high‐performance liquid chromatography (HPLC). Mean values are depicted (±SD). Data analysis was performed using one‐way anova and Tukey’s multiple comparisons test. ns, not significant; ***P < 0.001.
The three selected overexpression lines were then investigated for increased USP enzyme activity. Therefore, crude leaf extracts from six individual plants were used for USP enzyme assay measuring the activity in the forward reaction by adding Gal‐1P as a substrate. Product formation was analyzed by high‐performance liquid chromatography (HPLC). Quantification of product formation showed elevated USP activities of 1.068 pkat µg−1 protein for overexpression line 4, 0.914 pkat µg−1 protein for overexpression line 20, and 0.607 pkat µg−1 protein for overexpression line 23 compared with the WT (0.412 pkat µg−1 protein), i.e. the overexpression lines 4 and 20 exhibited a two–threefold higher USP activity (Figure 1b), whereas the overexpression line 23 exhibited a 1.5‐fold higher USP activity.
To investigate the effect of USP overexpression on the Gal‐1P level in vivo, seedlings of transgenic lines and WT were grown on MS plates supplemented with 1.5 mmol L−1 Gal for 3 weeks, the roots were harvested, and metabolite extracts were analyzed via GC‐MS. The concentration of Gal‐1P in roots of the overexpression lines 4 and 20 was significantly reduced [22.1 nmol g−1 fresh weight (FW) or 19.7 nmol g−1 FW] when compared with the WT (33.0 nmol g−1 FW). The Gal‐1P level in roots of overexpression line 23 did not differ from WT level (30.5 nmol g−1 FW). The Gal‐1P concentrations we found in this study were much lower than the recently published data for Gal‐1P (Althammer et al., 2020), because of downscaling of Gal‐feeding from 5 mmol L−1 Gal to 1.5 mmol L−1 Gal resulting in a fivefold reduction of the Gal‐1P level in roots. Nevertheless, the characteristic phenotype of Gal‐Tox including growth cessation of the primary root can also be observed on 1.5 mmol L−1 Gal. Overexpression of USP leads to an additional significant reduction of Gal‐1P but, surprisingly, we did not find a rescue of the Gal‐Tox in transgenic lines, though the concentration of Gal‐1P is nearly down to 50% of WT levels (Figure 2).
Figure 2.
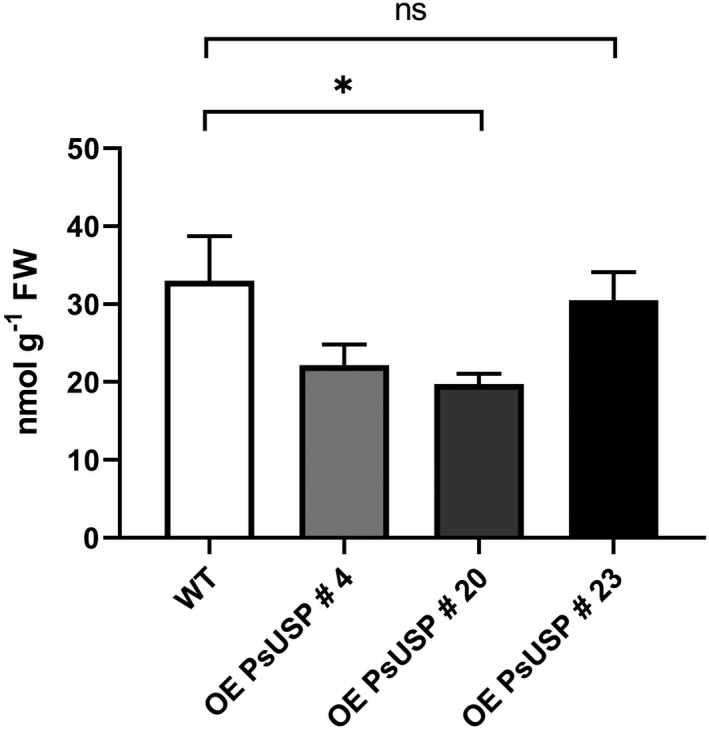
Galactose‐1‐phosphate (Gal‐1P) level in roots under Gal‐feeding conditions.
Gas chromatography‐mass spectrometry (GC‐MS) measurements of Gal‐1P level in roots of seedlings growing on 0.5 × MS plates supplemented with 1.5 mmol L−1 Gal. Root samples of the respective genotype growing on one plate were pooled representing one biological replicate. Mean values of three biological replicates with three technical replicates each are shown here (±SD). Data analysis was performed using one‐way anova and Dunnett’s multiple comparisons test; *P < 0.1.
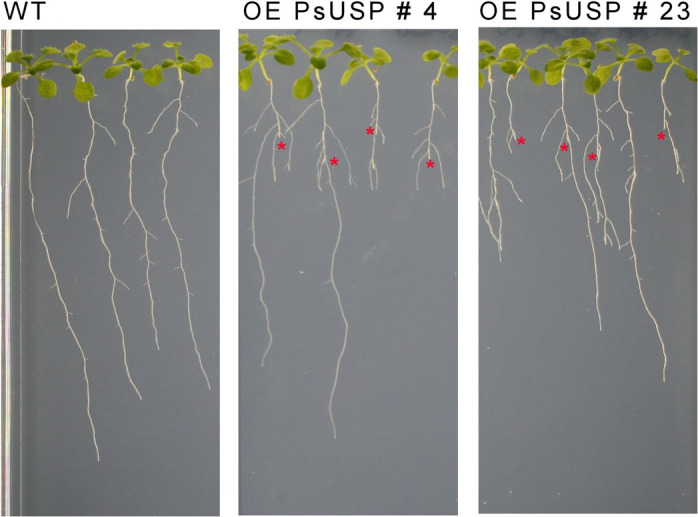
To investigate consequences of Gal‐1P reduction for the Gal‐Tox phenotype, seedlings of transgenic lines and WT were grown on MS plates supplemented with 0.5 mmol L−1 Gal. After 2 weeks of growth, most of the seedlings of the USP overexpression lines displayed the characteristic Gal‐Tox phenotype including a cessation of primary root growth and an overgrowth of lateral roots, whereas only one seedling of the WT line developed the Gal‐Tox phenotype (Figure 3a).
Figure 3.
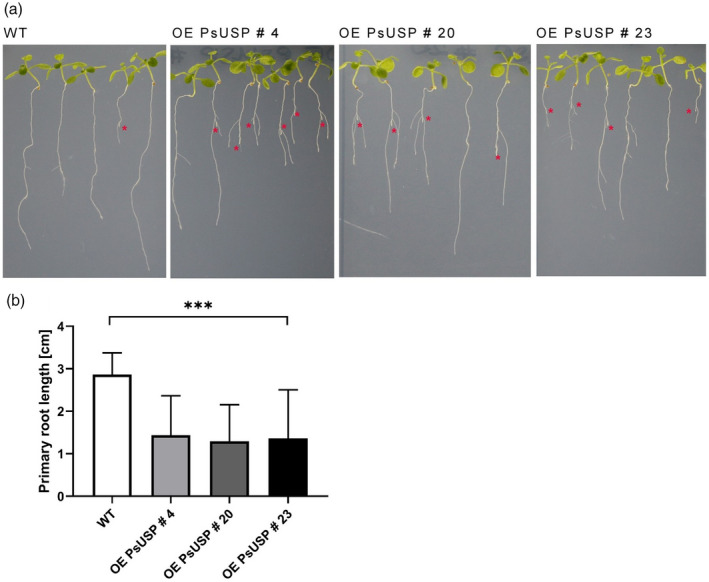
Phenotype of UDP‐sugar pyrophosphorylase (USP) overexpression lines.
Two‐week‐old seedlings of wild‐type (WT) and the two USP overexpression lines growing on 0.5 × MS plates with 0.5 mmol L−1 galactose (Gal) are shown (a). The tips of the primary roots, which were overgrown by lateral roots indicating the formation of the galactose toxicity (Gal‐Tox) phenotype, are marked by red asterisks.
(b) The reduction of primary root length in response to USP overexpression under Gal‐feeding conditions is shown. Each column represents mean values (±SD) of the measurement of primary root lengths after 2 weeks growth on 0.5 × MS plates supplemented with 0.5 mmol L−1 Gal using the ImageJ Software (n = 12 seedlings). Data analysis was performed using one‐way anova and Dunnett’s multiple comparisons test; ***P < 0.001.
The USP overexpression lines grown on MS plates with 0.5 mmol L−1 Gal showed a significant reduction of primary root lengths when compared with the WT, indicating an exacerbation of the Gal‐Tox phenotype (Figure 3b).
Therefore, we conclude that USP overexpression makes plants more susceptible to the exogenous addition of Gal, indicating that USP overexpression leads to higher Gal sensitivity.
Next, we examined how Gal‐feeding affects the pool of nucleotide sugars. For this experiment, we used WT seedlings and seedlings of transgenic line 4 and transgenic line 23 grown on 1.5 mmol L−1 Suc versus 1.5 mmol L−1 Gal to investigate levels of UDP‐Gal and UDP‐Glc. Extraction of metabolites was performed as described above followed by analysis by HPLC‐MS. We found highly elevated levels of UDP‐Gal in WT (93.02 pmol mg−1 FW) and USP overexpression line 4 (171.56 pmol mg−1 FW) and overexpression line 23 (180.52 pmol mg−1 FW) under Gal‐feeding conditions (Figure 4a,c) compared with Suc‐feeding conditions (WT: 17.75 pmol mg−1 FW; OE PsUSP 4: 28.56 pmol mg−1 FW; and OE PsUSP 23: 27.32 pmol mg−1 FW). The level of UDP‐Glc decreased significantly under Gal‐feeding from 93.78 pmol mg−1 FW to 74.10 pmol g−1 FW for the WT, from 142.97 pmol mg−1 FW to 106.03 pmol mg−1 FW in the overexpression line 4, and from 136.73 pmol mg−1 FW to 121.18 pmol mg−1 FW in the overexpression line 23 (Figure 4a,c). In general, overexpression of USP leads to higher levels of nucleotide sugars as suggested by the significant increase of UDP‐Gal under Gal‐feeding conditions (Figure 4c) or the significant increase of UDP‐Glc under Suc‐feeding conditions (Figure 4a). Data for nucleotide sugar levels in plants show similar values for UDP‐Glc and UDP‐Gal content in leaves of Arabidopsis WT (Dormann and Benning, 1998). Furthermore, the ratio of UDP‐Gal to UDP‐Glc shifted dramatically from 0.19 to 1.25 in WT, from 0.20 to 1.62 in the USP overexpression line 4, and from 0.20 to 1.45 in the USP overexpression line 23 under Gal‐feeding conditions due to five–sixfold accumulation of UDP‐Gal. Interestingly, the addition of 0.5 mmol L−1 Suc to 1.5 mmol L−1 Gal‐containing plates leads to significant reduction of the UDP‐Gal level from 93.02 pmol mg−1 FW to 52.07 pmol mg−1 FW in the WT (Figure 4b). Overexpression of USP resulted in highly elevated UDP‐Gal levels of 139.18 pmol mg−1 FW in line 4 and 130.74 pmol mg−1 FW in line 23 even under supplementary feeding conditions with the addition of 0.5 mmol L−1 Suc to 1.5 mmol L−1 Gal (Figure 4b).
Figure 4.
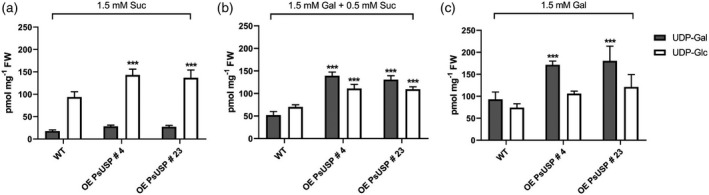
Nucleotide sugar analysis.
High‐performance liquid chromatography‐mass spectrometry (HPLC‐MS) measurements of UDP‐galactose (UDP‐Gal) and UDP‐glucose (UDP‐Glc) in roots of seedlings growing on 1.5 mmol L−1 Suc (a), 1.5 mmol L−1 Gal + 0.5 mmol L−1 Suc (b) and 1.5 mmol L−1 Gal (c) are shown. Values are averages of three biological and three technical replicates each (±SD). Data analysis was performed using one‐way anova and Dunnett’s multiple comparisons test; ***P < 0.001 comparing wild‐type (WT) with the two UDP‐sugar pyrophosphorylase (USP) overexpression lines.
The effect of Suc addition on the growth and development of the root system on Gal‐containing plates can be observed in Figure 5, indicating that the reduction of the UDP‐Gal level in WT helped to overcome Gal toxicity, whereas overexpression of USP prevented the removal of the Gal‐Tox phenotype due to significantly increased UDP‐Gal levels.
Figure 5.
Supplementary feeding of Suc to Gal‐containing plates.
Seedlings of wild‐type (WT) and UDP‐sugar pyrophosphorylase (USP) overexpression lines 4 and 23 grown on 0.5 × MS plates with 1.5 mmol L−1 Gal + 0.5 mmol L−1 Suc are depicted (2 weeks old). Defects in primary root growth and development are marked by red asterisk.
Nucleotide sugars are donor substrates for the biosynthesis of cell wall polymers by the action of glycosyltransferases (Gibeaut, 2000; Mohnen, 2008). To investigate the role of elevated UDP‐Gal levels on cell wall biosynthesis, we compared the cell wall composition of WT and galK seedlings growing on 1.5 mmol L−1 Gal versus 1.5 mmol L−1 Suc. Therefore, the roots were harvested after 4 weeks of growth, cell walls were extracted, hydrolyzed in trifluoroacetic acid (TFA), and monosaccharides determined by HPLC.
The results in Figure S2 confirm a significant higher Gal content in root cell walls of WT seedlings under Gal feeding conditions compared with the root cell walls of galK grown under the same conditions. In general, the galactose content of root cell walls did not alter between WT seedlings grown on Suc‐containing plates and galK seedlings grown on Gal‐containing plates, indicating that galK serves as a good control for cell wall analysis.
To get a deeper insight into alterations of cell wall polymers, the binding of monoclonal antibodies to more than 40 cell wall epitopes was analyzed by carbohydrate microarray profiling (Figure S3).
Therefore, seedlings of the WT and galk mutant were grown on 0.5 × MS plates supplemented with 1.5 mmol L−1 Gal for 4 weeks. The alcohol‐insoluble residue (AIR) was prepared from roots, sequentially extracted with diamino‐cyclo‐hexane‐tetra‐acetic acid (CDTA) and NaOH, and then probed. The galK mutant allowed us to include seedlings that are insensitive to the exogenous galactose up to 100 mmol L−1 due to the inability of phosphorylation of galactose (Egert et al., 2012). In the CDTA fraction, LM5 and LM6, which recognize galactose‐ and arabinose oligomers in, for example, pectic RGI, produced more signal in the WT than in galK mutant (Figure 6). Binding of INRA‐RU1 and INRA‐RU2 (RGI backbone) was higher in the NaOH fraction of galK; this alteration in extractible polymers suggests stronger physical interactions of RGI with hemicellulosic cell wall polymers, which predominated in the alkali fraction. This might be due to a reduced RGI decoration with galactose and/or arabinose (lower LM5 and LM6 signal), allowing for more interactions of RGI with hemicellulose backbones. The NaOH fraction revealed strong binding of BS‐400‐2 (callose) in the WT. Most interestingly, in both the NaOH and CDTA fractions, binding of probes recognizing glycoproteins was significantly higher in the WT than in the galK mutant. For example, the CDTA fraction yielded abundant binding of LM3, JIM11, JIM12 and JIM20, detecting extensin epitopes (Figure 6). The NaOH fraction yielded high binding signals for antibodies against arabinogalactan proteins (AGPs; JIM13, LM2, LM14 and MAC207).
Figure 6.
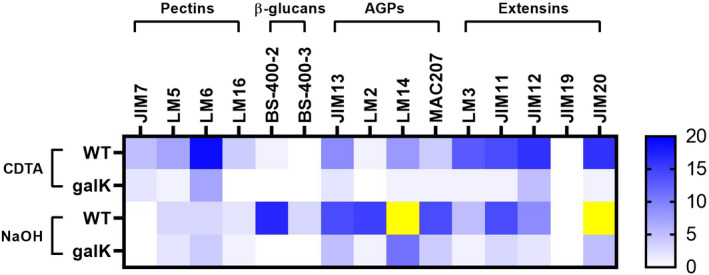
Comprehensive microarray polymer profiling (CoMPP).
Analysis of cell wall composition by carbohydrate profiling is shown. Alcohol‐insoluble residue (AIR) samples were isolated from roots of wild‐type (WT) and galK mutant after 1 month growth on 0.5 × MS plates supplemented with 1.5 mmol L−1 Gal. Cell wall components were extracted from AIR samples of WT and galK mutant with diamino‐cyclo‐hexane‐tetra‐acetic acid (CDTA) and NaOH. Blue colors indicate increased intensity of respective antibody binding. Yellow reveals an intensity higher than 20.
Hemicellulose, which predominated in the NaOH fraction, showed high signals for mannans (LM21), xyloglucans (LM15, LM25, CCRC‐M58) and xylans (LM28, CCRC‐M39), but there was no difference between the WT and galK mutant, with the exception of LM21 showing a higher signal in the galK mutant. There was no signal difference between WT and galK mutant for cellulose probes (CBM3a and CBM2a).
DISCUSSION
Reduction of Gal‐1P does not solve the problem of Gal‐Tox in plants
The phenomenon of Gal‐Tox has been known for over 100 years, but the mechanisms of Gal‐Tox are still under debate. Galactose‐containing metabolites like Gal‐1P or UDP‐Gal are often hypothesized to function as toxic agents. Currently published data from the mouse model show that reduction of Gal‐1P level in red blood cells and liver by mRNA‐based therapy helps to overcome galactose sensitivity (Balakrishnan et al., 2020). This prompted us to examine the role of Gal‐1P in the mechanisms of Gal‐Tox in plants. Studies in plant models reveal that phosphorylation of Gal into Gal‐1P is essential for development of the Gal‐Tox phenotype (Egert et al., 2012). Accumulation of Gal‐1P under Gal‐feeding conditions was also proven by GC‐MS measurements in roots of Arabidopsis, but it was shown that Gal‐1P does not act as an inhibitor of the enzyme phosphoglucomutase (Althammer et al., 2020) as, for example, suggested for yeast. To investigate the role of Gal‐1P in plants, we tried to lower the Gal‐1P level by overexpression of the enzyme USP in Arabidopsis. Here, we used the USP gene from P. sativum (Kotake et al., 2004) sharing 80% identity in nucleotide sequence to the open reading frame of At5g52560 from A. thaliana to avoid effects of silencing in transgenic lines. USP activity assays confirmed a two–threefold higher enzyme activity of USP in the two selected transgenic lines. GC‐MS measurements revealed a significant reduction of Gal‐1P in the roots of seedlings under Gal‐feeding due to USP overexpression. Contrary to mouse models, the reduction of Gal‐1P does not help to overcome Gal‐Tox in plants. We cannot detect any alleviation of the Gal‐Tox phenotype due to Gal‐1P reduction. Measurements of primary root lengths of USP overexpression lines under Gal‐feeding conditions rather indicate a higher sensitivity towards Gal. Therefore, we conclude that Gal‐1P is negatively correlated with Gal‐Tox phenotype in contrast to animals and humans, where a positive correlation is typically observed. Overexpression of USP cause no visible phenotype in leaves or roots. We start to see a higher sensitivity in USP‐overexpressor lines only in the presence of Gal.
The level of UDP‐Gal and the balance of nucleotide sugars represent a key checkpoint in Gal‐Tox
Nucleotide sugars like UDP‐Gal or UDP‐Glc are essential precursors for the biosynthesis of cell wall polymers. It was shown that synthesis of UDP‐Gal by UDP‐glucose epimerase 4 (UGE4) is required for organization and function of endomembrane systems, especially the trans‐Golgi network and early endosomes (Wang et al., 2015). Studies in yeast models showed that deficiency in UGE caused an accumulation of UDP‐Gal levels, and dramatically aggravated the sensitivity of yeast strains towards galactose when compared with GALT‐deficient strains (Mumma et al., 2008; Ross et al., 2004). Data from studies in Drosophila melanogaster also revealed that imbalance in nucleotide sugars due to UGE deficiency rather than Gal‐1P resulted in galactose‐sensitivity, showing the importance of nucleotide sugar homeostasis (Daenzer et al., 2012). To sum up, the results of these studies all clearly showed an exacerbation of Gal‐Tox due to UGE deficiency, probably by an imbalance in nucleotide sugars. In plants, the Gal‐Tox phenotype can be observed without any deficiency in UGE or any other enzyme of the Leloir pathway. Arabidopsis thaliana has five isoforms of UGE (UGE1–UGE5) displaying enzyme activities with K m values of 0.1–0.3 mmol L−1 and a turnover rate of 23–128 sec−1 (k cat) for UDP‐Gal (Barber et al., 2006). In comparison, characterization of UGE from Escherichia coli showed a k cat value of 500 sec−1, indicating a four to 20 times higher activity of E. coli UGE with UDP‐Gal as substrate (Chen et al., 1999; Wilson and Hogness, 1964). In this study, we determined a fivefold accumulation of UDP‐Gal and a slight decrease in UDP‐Glc under Gal‐feeding conditions in roots of Arabidopsis WT seedlings via HPLC‐MS measurements (Figure 4). The equilibrium between UDP‐Gal and UDP‐Glc with a usual ratio of approximately 0.33 cannot be achieved due to extremely accumulated levels of UDP‐Gal, which might result from Gal‐feeding together with a low turnover rate of Arabidopsis UGEs. Studies in Arabidopsis suggest that overexpression of UGE isoforms suppresses toxic effects of galactose, indicating the importance of UGE activity for Gal‐Tox in plants (Barber et al., 2006; Dormann and Benning, 1998). In this study, we showed an aggravation of the Gal‐Tox phenotype in USP overexpression lines by measurements of primary root lengths after 2 weeks of growth on 0.5 mmol L−1 Gal (Figure 3b). This observation can be explained by a 1.8‐fold higher accumulation of UDP‐Gal under Gal‐feeding conditions compared with WT due to USP overexpression (Figure 4c). Supplementary feeding of Suc to Gal‐containing plates leads to significant reduction of the UDP‐Gal level in WT seedlings resulting in a complete loss of the Gal‐Tox phenotype (Figure 5). The increase in root length uses more UDP‐Gal for polymer synthesis than UDP‐Glc, and thus lowers the amount of UDP‐Gal as well as the ratio of UDP‐Gal to UDP‐Glc as observed in Figure 4. Both changes occur at the same time. In contrast, USP overexpression produces highly elevated UDP‐Gal levels even under supplementary feeding conditions leading to growth defects with cessation of primary root growth (Figure 5). Therefore, we conclude that mainly the UDP‐Gal level is critical for normal root development especially under Gal‐feeding conditions.
UDP‐Gal is also a precursor for galactolipids in chloroplasts. The elevated levels observed in USP‐overexpressor lines could thus modify the amount of galactolipids in plants. However, studies from cold acclimated rye (Uemura and Steponkus, 1997) or phosphate‐starved Arabidopsis cells (Jouhet et al., 2004) indicate variability in the galactolipid levels as a response of changing the environment. This makes it rather unlikely that the increase in UDP‐Gal causes changes in galactolipids, leading to the root phenotypes that we observed.
Glycoproteins are the target of elevated UDP‐Gal levels
In this study, we showed that Gal‐feeding leads to a fivefold accumulation of UDP‐Gal in the roots of Arabidopsis WT seedlings. The cell wall is a major Gal sink in plants; therefore, we investigated whether UDP‐Gal levels affect the cell wall composition via comprehensive microarray polymer profiling (CoMPP), allowing to test for the relative abundance of more than 40 cell wall epitopes. CoMPP revealed (Figure 6) a much higher signal for glycoproteins (AGPs, extensins) in the WT when compared with the galK mutant under Gal‐feeding conditions. This indicates higher glycosylation of AGPs and extensins in response to elevated UDP‐Gal levels. AGPs have various functions in plant growth and development, and strongly affect the cell wall properties and architecture (Hromadova et al., 2021; Silva et al., 2020). Disruption of the AGP function via complexation with the Yariv reagent causes reduced root elongation, swelling of epidermal cells and altered the organization of cytosolic microtubules (Nguema‐Ona et al., 2007; Willats and Knox, 1996). GH43 loss‐of‐function mutants lacking β‐1,3‐galactosidase activity revealed altered glycosylation of AGPs with increased AGP cell wall association; authors suggested that this impaired cell wall stiffening during cell expansion led to root swelling and stunted root growth (Nibbering et al., 2020). Therefore, we hypothesize that Gal‐feeding led to higher glycosylation of AGPs, which may have increased their physical interactions with other cell wall components, altering the cell wall’s architecture and extensibility. Moreover, when growing roots on 1.5 mmol L−1 Gal, we found the majority of AGP epitopes in the NaOH fraction, underpinning their strong cell wall association. In contrast, investigating normally grown Arabidopsis seedlings, Moller et al. (2007) and Nibbering et al. (2020) found a majority of AGP epitopes (e.g. LM2, JIM13) to be readily extractable by CaCl2 and/or CDTA. It is possible that an altered glycosylation of AGPs increases their chance to become covalently linked to pectin and/or arabinoxylan (Tan et al., 2013). Correspondingly, we found relatively high amounts of homogalacturonan epitopes (LM19) in the NaOH fraction of the Gal‐fed WT. Our hypothesis that an altered AGP structure is a major contributor to the Gal‐Tox phenotype (i.e. reduced root elongation and swelling of epidermal root cells) is further supported by the observation that treatment with the Yariv reagent or mutants with altered glycosylation of AGPs produce similar phenotypes (Nibbering et al., 2020; Willats and Knox, 1996). Besides AGPs, we found significantly more extensin epitopes in the Gal‐grown WT than in galK plants. Extensins can self‐assemble to highly regular networks (Lamport et al., 2011), and might have a role in mediating the assembly of pectin‐rich structures such as the cell plate (Hall and Cannon, 2002). Consequently, alteration of their glycosylation pattern as recognized by antibodies has strong consequences for the plant cell wall structure (Velasquez et al., 2011), and might help explaining the WT Gal‐Tox phenotype on 1.5 mmol L−1 Gal. Furthermore, the absence of normally glycosylated extensins in WT (very low CoMPP signals; Figure 6) in combination with a strong Gal‐Tox phenotype in WT roots supports the idea that expansin glycosylation strongly influences root growth, expansion and morphology.
In conclusion, overexpression of USP in A. thaliana resulted in significantly decreased Gal‐1P levels under Gal‐feeding conditions, with a concomitant higher sensitivity towards galactose. Analysis of cell wall polymers indicated alteration of glycosylation pattern of glycoproteins like AGPs and extensins probably due to elevated UDP‐Gal levels. However, further studies are required to investigate structure and localization of glycoproteins to understand how AGPs and extensins contribute to the Gal‐Tox phenotype.
EXPERIMENTAL PROCEDURES
Chemicals
Deionized water was produced in‐house with a MilliQ® Integral 3 instrument (Millipore, Billerica, MA, USA). Acetonitrile (ACN; ≥99.9%) and methanol (MeOH; ≥99.9%) were purchased from VWR International (Vienna, Austria). n‐Hexane (≥99.0%), trimethylamine (≥99.0%), acetic acid (glacial; ≥99.0%), N‐methyl‐N‐trimethylsilyltrifluoroacetamide (MSTFA; ≥98.5%), NaOH (≥97.0%) and CDTA (≥97.0%) were obtained from Merck (Darmstadt, Germany); helium (≥99.9999) came from SIAD (St Pantaleon, Austria).
Plasmid construction
The USP gene from P. sativum was amplified by PCR from plasmid pET32‐Pisum‐USP (Kotake et al., 2004), (fwd GAGTTTTTCTGATTAACCATGGCTTCCTCCCTCGG) (rev TTTGCGGACTCTAGAGGAGTCAAGCCTTGAAGTCAAATTTGC). The PCR product was inserted into a pMAA‐Ubq10 vector (Ali et al., 2012), cut with NcoI and BamHI, by hot fusion technology (Fu et al., 2014). The sequence was verified by sequencing the construct.
Plant transformation and growth conditions
The pMAA‐Ubq10‐PsUSP construct was introduced into competent Agrobacterium tumefaciens GV3101 cells, and A. thaliana (ecotype Columbia; NASC N60000) transformation was performed by the floral dipping method (Davis et al., 2009). Transformed seeds were selected based on their expression of the dsRed reporter in the seed coat and confirmed by PCR. Plants were grown in standard soil in a growth chamber under short (long) day conditions at 23°C with 10 h (–16 h) light (approximately 150 μE m−2 sec−1) and 18°C in the dark. Successful Agrobacterium‐transformed seeds were selected by checking the fluorescence under a Leica Stereo‐Fluorescence System (LEICA MZFLIIITM; Filter G: excitation filter 546/10 nm and barrier filter 590 nm) and sown out. The offspring was tested for PsUSP overexpression via RT‐PCR. After selection and expression analysis, we obtained a total of four independent PsUSP‐overexpressing lines, using the three best candidate lines for further experiments. The selected USP overexpression lines were heterozygous and still segregating. Therefore, transgenic seeds of the respective USP overexpression line were selected by their fluorescence prior to every experiment. For metabolite analysis, seeds were surface sterilized with ethanol and incubated on 0.5 × MS plates (Basal Salt Mixture, Duchefa #M0245), pH 5.7 (KOH) containing 0.80% plant agar (Duchefa) and the respective sugar. The roots of the seedlings were harvested after 2–4 weeks of growth on 0.5 x MS plates with respective sugar, frozen in liquid nitrogen and stored at −80°C.
RT‐PCR
Total RNA was extracted from A. thaliana leaves by TriReagent buffer combined with silica spin purification. cDNA was synthesized from 2.0 µg of total RNA following the protocol of RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). To investigate the transcript expression level, RT‐PCR was performed on an Agilent Aria MX1.2 real‐time cycler using SYBR Green as dye. The primers 5ʹ‐GATACCCTGATGGTGATGTC‐3ʹ and 5ʹ‐GCATCATACATTCAAGTCGAG‐3ʹ were used for quantification of transgenic PsUSP level, and primers 5ʹ‐CGTGCTAACAGCCTCATTCTT‐3ʹ and 5ʹ‐GGAACCGATGTGTCCTTGTAA‐3ʹ were used for analysis of endogenous AtUSP expression level. The EF1α gene was used as internal housekeeping gene control (5ʹ‐GACCAACTCTTCTTGAGGCTCTTGAC‐3ʹ and 5ʹ‐GGCACCGTTCCAATACCACCAATC‐3ʹ). The PCR reaction mixture consisted of 2× SYBR‐PCR‐Mix [40 mmol L−1 Tris‐Cl pH 8.4, 100 mmol L−1 KCl, 6 mmol L−1 MgCl2, 8% glycerol, 1% bovine serum albumin solution, 10 mg ml−1, 320 μmol L−1 dNTP, Sybr‐Green 1:25.000‐dilution of a commercial 10.000 ×‐SYBR‐Green‐stock (Roche)], 200 nmol L−1 primers each, 1 U Taq polymerase and 1.5 µl cDNA in a total volume of 20 µl. The PCR program cycles through 1 × 3 min at 94°C, and for 40 × 5 sec at 95°C, 10 sec at the specific primer pair annealing temperature, and 10 sec at 72°C, and in the end additional 30 sec at 95°C, 72°C and 95°C once, followed by a conclusive melting curve indicating identity and homogeneity of the product.
USP enzyme assay
To investigate USP enzyme activity in transgenic lines, leaf extracts from 4‐week‐old plants were prepared in the cold with extraction buffer consisting of 30 mmol L−1 Tris/Cl pH 8, 2 mmol L−1 MgCl2, 10% glycerol and 0.5 mmol L−1 phenylmethylsulfonyl fluoride (PMSF). After centrifugation at 13 000 rpm for 3 min, 500 µl of the supernatant was desalted in the same buffer (but without PMSF) using a NAP5 size‐exclusion column (GE Healthcare, http://www.gehealthcare.com). USP enzyme activity was measured in the forward reaction using Gal‐1P as a substrate. The enzyme assay consisted of 50 µl desalted protein extract, 0.8 mmol L−1 UTP, 1 mmol L−1 Gal‐1P and 1 µl PPi (1:15 dilution in assay buffer from enzyme stock; Roche #14156423) with a total volume of 100 µl. After incubation at 30°C for 20 min, the reaction was stopped by heat (95°C for 5 min). The product formation of UDP‐Gal was measured by HPLC using a Partisil 10 SAX HPLC column (3.0 × 150 mm) with 100 mmol L−1 Na‐phosphate pH 2.8 as solvent A and 750 mmol L−1 Na‐phosphate pH 3.7 as solvent B. Separations of the metabolites were performed by the following gradient program: 0 min 2% B, 12 min 10% B, 19 min 10% B, 25 min 36% B, 35 min 60% B.
Metabolite analysis and derivatization
Sugar metabolites were extracted from roots by using the methanol/chloroform method described in Lunn et al. (2006) and Arrivault et al. (2009), with the addition of 400 µl instead of 200 µl water. To investigate the Gal‐1P level in roots, liquid samples were transferred into glass inlets. After adding 13C‐D‐Glc as an internal standard, the samples were evaporated to dryness using a centrifugal vacuum dryer. For derivatization, 25 µl pyridine and 25 µl MSTFA were added to the dried samples and incubated at 90°C for 30 min. After 1:2 dilution in n‐hexane, the samples were analyzed by GC‐MS. To investigate nucleotide sugars, the aqueous phases were evaporated to dryness and re‐dissolved in 1 ml water. In order to reduce the sample complexity, solid‐phase extraction was performed with Supelclean ENVI‐Carb SPE Tubes (1.0 ml, 0.1 g, particle size: 120–400 mesh) consisting of graphitized non‐porous carbon. The solid‐phase extraction was performed by the method described in Behmuller et al. (2014). Briefly, the SPE tubes were equilibrated 3× with 1 ml 60% ACN in water containing 0.3% formic acid adjusted to pH 9.0 with ammonia and 1× with 1 ml water. Afterwards, the re‐dissolved sample was added to an SPE column. Washing steps were performed with 1 ml water and 500 µl 60% ACN in water. Sample elution was done with 2.0 ml 60% ACN in water containing 0.3% formic acid adjusted to pH 9.0 with ammonia. After vaporization, the samples were re‐dissolved in 100 µl water and analyzed by HPLC‐MS.
HPLC
Chromatographic separation of 2.5 µl injected sample was carried out according to Rautengarten et al. (2011) on a Thermo Scientific™ UltiMate™ 3000 Rapid Separation system (Thermo Fisher Scientific, Germering, Germany) at a flow rate of 200 μl min−1 employing a Phenomenex® Synergi™ Hydro‐RP column (150 × 2.0 mm i.d., 4 µm particle size, 80 Å pore size; Phenomenex, Aschaffenburg, Germany) with a security guard cartridge C18 (4.0 × 3.0 mm i.d.; Phenomenex, Aschaffenburg, Germany) operated at a temperature of 28°C. The mobile phase A was composed of 20 mmol L−1 buffered trimethylamine/acetic acid pH 6.0 (TEAA) in water, mobile phase B of ACN. The separation started isocratic at 100.0% A for 15 min followed by a linear gradient of 0−5.0% B in 5 min, a second isocratic phase at 90.0% B for 3 min, and re‐equilibration of the column at 100% A for 7 min. The total runtime for one sample was 30 min. UV‐detection was carried out at 262 nm.
Mass spectrometry
Mass spectrometry was conducted by parallel reaction monitoring on a Thermo Scientific™ Q Exactive™ Hybrid Quadrupole‐Orbitrap™ mass spectrometer equipped with a Thermo Scientific™ Ion Max™ ion source with a heated electrospray ionization (HESI) probe, both from Thermo Fisher Scientific (Bremen, Germany). The source heater temperature was set to 250°C, spray voltage to −4.0 kV, sheath gas flow to 35 arbitrary units, auxiliary gas flow of 5 arbitrary units, capillary temperature to 350°C, and S‐lens RF level to 60.0. The m/z values set on the inclusion list were 535.060 (UDP‐xylose and UDP‐arabinose), 565.050 (UDP‐Glc and UDP‐Gal), 579.050 (UDP‐glucuronate and UDP‐galacturonate) and 482.980 (UTP) with an isolation window of 1.0 m/z. The resolution was set to 17 500 at m/z 200, the automatic gain control target was set to 1e6 charges with a maximum injection time of 150 msec, higher‐energy collisional dissociation was done with a collision energy of 23 eV.
GC‐MS
Gas chromatography‐mass spectrometry was carried out according to Althammer et al. (2020) on a Focus™ GC instrument equipped with an AI3000 autosampler and a DSQ™ II quadrupole mass spectrometer (all from Thermo Fisher Scientific, Dreieich, Germany). The separation was performed employing a TRACE™ TR‐5 column (5%‐phenyl‐methylpolysiloxane, 15 m × 0.25 mm i.d., 0.25 µm film thickness) using a constant carrier gas flow rate of 1.5 ml min−1 helium. A 1‐µl sample was injected in splitless injection mode at an inlet temperature of 250°C. First, the analytes were focused at the head of the column with an isothermal phase at 120°C for 1 min, followed by a gradient of 1.5°C min−1 from 120°C to 195°C, a second gradient of 20°C min−1 from 195°C to 300°C, and a final isothermal phase at 300°C for 2 min. The MS transfer line was set to 200°C, ionization was carried out in positive mode applying electron ionization at 70 eV at a temperature of 200°C. The data were acquired in selected ion monitoring mode for the m/z values 204.02, 205.99, 257.00, 298.93, 314.97, 387.04 and 428.95 with an isolation width of m/z ± 0.5 and a dwell time of 10 msec.
Data evaluation
Data acquisition and evaluation were conducted using Thermo Scientific™ Chromeleon™ 7.2 CDS (Thermo Fisher Scientific, Germering, Germany). The GC‐MS system was operated with the Xcalibur™ 3.0.63 software (Thermo Fisher Scientific, Waltham, MA, USA).
Cell wall extraction and glycan microarray analysis
The composition of root cell walls from seedlings grown on MS‐plates with 3.0 mmol L−1 Gal or 3.0 mmol L−1 Suc were homogenized in a Retsch ball mill. The samples were extracted twice with 75% ethanol followed by an acetone extraction. The resulting cell wall material was hydrolysed in 2 mol L−1 TFA at 121°C. After hydrolysis, samples were dried in a vacuum centrifuge. The dry pellet was redissolved in H2O and analyzed on a Thermo Dionex ICS 3000 system using a Carbopac PA20 (150 × 3.0 mm; flow rate 0.45 ml min−1) column. Neutral sugars were separated with 15 mmol L−1 NaOH, and uronic acids eluted with 30 mmol L−1 NaOH and 200 mmol L−1 Na‐acetate. The sugars were quantified by pulsed amperometric detection using authentic standards.
Roots of Arabidopsis seedlings (WT Col‐O or galK mutants) grown for 4 weeks on 0.5 × MS plates containing 1.5 mmol L−1 Gal were collected, homogenized frozen and their cell walls (AIR), were extracted in 75% ethanol until the supernatant was transparent. Cell wall polysaccharides and glycoproteins were sequentially extracted from AIR in 50 mmol L−1 CDTA (solubilizing pectins, glycoproteins) and 4 mol L−1 NaOH (solubilizing hemicelluloses, glycoproteins). The following glycan microarray analysis (Moller et al., 2007) was performed as described in detail before (Kracun et al., 2017). Arrays were printed as distinct dots onto nitrocellulose membranes with an ArrayJet Sprint microarray printer (ArrayJet, Roslin, UK) and probed with ~40 cell wall probes, including antibodies and carbohydrate binding modules (Table S1). For controls, primary antibodies were heat‐inactivated prior to use. Seedling growth, cell wall extraction and probing were performed in triplicates, whereas the WT (Col‐O) root FW was ~30–45 mg (giving ~2–4 mg AIR), and the galK root FW was ~35–45 mg (~8 mg AIR).
AUTHOR CONTRIBUTIONS
The study was designed by all authors. CR, CB and CGH developed the analytical GC‐MS and HPLC‐MS procedures. KH was responsible for the analysis of cell wall polymers by CoMPP. MA and EV performed the enzyme assays. Data analysis was performed by MA, KH, CR, RT. The manuscript was mainly written by MA and RT with contributions of all authors.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
Supporting information
Figure S1. Metabolism of galactose in higher plants compared with humans, yeast and red algae. Alternative routes for the conversion of Gal‐1P by the USP enzyme in higher plants and by the GALT enzyme in humans, yeast and red algae, where the USP enzyme is lacking. Gal, galactose; Gal‐1P, galactose‐1‐phosphate; Glc‐1P, glucose‐1‐phosphate; UDP, uridine diphosphate; PPi, pyrophosphate; UTP, uridine triphosphate; GALK, galactokinase; GALT, galactose‐1‐phosphate uridylyltransferase; USP, UDP‐sugar pyrophosphorylase.
Figure S2. Cell wall sugar composition. Cell walls were extracted from roots of 4‐week‐old WT and galK seedlings growing under Suc‐feeding (1.5 mmol L−1 Suc) versus Gal‐feeding (1.5 mmol L−1 Gal) conditions. After hydrolysis in TFA, monosaccharides were separated on HPLC and quantified by pulsed amperometric detection. Values are averages of four biological replicates each (±SD).
Figure S3. Cell wall polymer profiling (CoMPP analysis). Detection of 42 cell wall epitopes. The ratio of WT to galK mutant for both the CDTA and NaOH fraction is shown. Blue colors indicate increasing abundance of antibody binding in the WT.
Table S1. Cell wall probes used in the present study.
ACKNOWLEDGEMENTS
The authors would like to thank Doris Wittmann for her excellent technical help. K.H. was supported by an NNF Emerging Investigator grant (no. NNF21OC0067180).
DATA AVAILABILITY STATEMENT
All relevant data can be found within the manuscript and its supporting materials.
REFERENCES
- Ali, M.A. , Shah, K.H. & Bohlmann, H. (2012) pMAA‐Red: a new pPZP‐derived vector for fast visual screening of transgenic Arabidopsis plants at the seed stage. BMC Biotechnology, 12, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Althammer, M. , Blochl, C. , Reischl, R. , Huber, C.G. & Tenhaken, R. (2020) Phosphoglucomutase is not the target for galactose toxicity in plants. Frontiers in Plant Science, 11, 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrivault, S. , Guenther, M. , Ivakov, A. , Feil, R. , Vosloh, D. , van Dongen, J.T. et al. (2009) Use of reverse‐phase liquid chromatography, linked to tandem mass spectrometry, to profile the Calvin cycle and other metabolic intermediates in Arabidopsis rosettes at different carbon dioxide concentrations. The Plant Journal, 59, 826–839. [DOI] [PubMed] [Google Scholar]
- Balakrishnan, B. , An, D. , Nguyen, V. , DeAntonis, C. , Martini, P.G.V. & Lai, K. (2020) Novel mRNA‐based therapy reduces toxic galactose metabolites and overcomes galactose sensitivity in a mouse model of classic galactosemia. Molecular Therapy, 28, 304–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber, C. , Rosti, J. , Rawat, A. , Findlay, K. , Roberts, K. & Seifert, G.J. (2006) Distinct properties of the five UDP‐D‐glucose/UDP‐D‐galactose 4‐epimerase isoforms of Arabidopsis thaliana. Journal of Biological Chemistry, 281, 17276–17285. [DOI] [PubMed] [Google Scholar]
- Behmuller, R. , Forstenlehner, I.C. , Tenhaken, R. & Huber, C.G. (2014) Quantitative HPLC‐MS analysis of nucleotide sugars in plant cells following off‐line SPE sample preparation. Analytical and Bioanalytical Chemistry, 406, 3229–3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behmuller, R. , Kavkova, E. , Duh, S. , Huber, C.G. & Tenhaken, R. (2016) The role of arabinokinase in arabinose toxicity in plants. Plant Journal, 87, 376–390. [DOI] [PubMed] [Google Scholar]
- Bhat, P.J. (2003) Galactose‐1‐phosphate is a regulator of inositol monophosphatase: a fact or a fiction? Medical Hypotheses, 60, 123–128. [DOI] [PubMed] [Google Scholar]
- Chen, X. , Kowal, P. , Hamad, S. , Fan, H.N. & Wang, P.G. (1999) Cloning, expression and characterization of a UDP‐galactose 4‐epimerase from Escherichia coli. Biotechnology Letters, 21, 1131–1135. [Google Scholar]
- Daenzer, J.M. , Sanders, R.D. , Hang, D. & Fridovich‐Keil, J.L. (2012) UDP‐galactose 4'‐epimerase activities toward UDP‐Gal and UDP‐GalNAc play different roles in the development of Drosophila melanogaster. PLoS Genetics, 8, e1002721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, A.M. , Hall, A. , Millar, A.J. , Darrah, C. & Davis, S.J. (2009) Protocol: streamlined sub‐protocols for floral‐dip transformation and selection of transformants in Arabidopsis thaliana. Plant Methods, 5, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jongh, W.A. , Bro, C. , Ostergaard, S. , Regenberg, B. , Olsson, L. & Nielsen, J. (2008) The roles of galactitol, galactose‐1‐phosphate, and phosphoglucomutase in galactose‐induced toxicity in Saccharomyces cerevisiae. Biotechnology and Bioengineering, 101, 317–326. [DOI] [PubMed] [Google Scholar]
- Delnoy, B. , Coelho, A.I. & Rubio‐Gozalbo, M.E. (2021) Current and future treatments for classic galactosemia. Journal of Personalized Medicine, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormann, P. & Benning, C. (1998) The role of UDP‐glucose epimerase in carbohydrate metabolism of Arabidopsis. The Plant Journal, 13, 641–652. [DOI] [PubMed] [Google Scholar]
- Egert, A. , Peters, S. , Guyot, C. , Stieger, B. & Keller, F. (2012) An Arabidopsis T‐DNA insertion mutant for galactokinase (AtGALK, At3g06580) hyperaccumulates free galactose and is insensitive to exogenous galactose. Plant and Cell Physiology, 53, 921–929. [DOI] [PubMed] [Google Scholar]
- Fu, C. , Donovan, W.P. , Shikapwashya‐Hasser, O. , Ye, X. & Cole, R.H. (2014) Hot Fusion: an efficient method to clone multiple DNA fragments as well as inverted repeats without ligase. PLoS One, 9, e115318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geserick, C. & Tenhaken, R. (2013) UDP‐sugar pyrophosphorylase is essential for arabinose and xylose recycling, and is required during vegetative and reproductive growth in Arabidopsis. The Plant Journal, 74, 239–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibeaut, D.M. (2000) Nucleotide sugars and glycosyltransferases for synthesis of cell wall matrix polysaccharides. Plant Physiology and Biochemistry, 38, 69–80. [Google Scholar]
- Gibney, P.A. , Schieler, A. , Chen, J.C. , Bacha‐Hummel, J.M. , Botstein, M. , Volpe, M. et al. (2018) Common and divergent features of galactose‐1‐phosphate and fructose‐1‐phosphate toxicity in yeast. Molecular Biology of the Cell, 29, 897–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitzelmann, R. (1995) Galactose‐1‐phosphate in the pathophysiology of galactosemia. European Journal of Pediatrics, 154, S45–S49. [DOI] [PubMed] [Google Scholar]
- Gross, W. & Schnarrenberger, C. (1995) Purification and characterization of a galactose‐1‐phosphate: UDP‐glucose uridyltransferase from the red alga Galdieria sulphuraria. European Journal of Biochemistry, 234, 258–263. [DOI] [PubMed] [Google Scholar]
- Hall, Q. & Cannon, M.C. (2002) The cell wall hydroxyproline‐rich glycoprotein RSH is essential for normal embryo development in Arabidopsis. The Plant Cell, 14, 1161–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hromadova, D. , Soukup, A. & Tylova, E. (2021) Arabinogalactan proteins in plant roots—an update on possible functions. Frontiers in Plant Science, 12, 674010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouhet, J. , Marechal, E. , Baldan, B. , Bligny, R. , Joyard, J. & Block, M.A. (2004) Phosphate deprivation induces transfer of DGDG galactolipid from chloroplast to mitochondria. Journal of Cell Biology, 167, 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudson, L. (1915) Toxicity of galactose for certain of the higher plants. Annals of the Missouri Botanical Garden, 2, 659–666. [Google Scholar]
- Kotake, T. , Yamaguchi, D. , Ohzono, H. , Hojo, S. , Kaneko, S. , Ishida, H.K. et al. (2004) UDP‐sugar pyrophosphorylase with broad substrate specificity toward various monosaccharide 1‐phosphates from pea sprouts. Journal of Biological Chemistry, 279, 45728–45736. [DOI] [PubMed] [Google Scholar]
- Kracun, S.K. , Fangel, J.U. , Rydahl, M.G. , Pedersen, H.L. , Vidal‐Melgosa, S. & Willats, W.G.T. (2017) Carbohydrate microarray technology applied to high‐throughput mapping of plant cell wall glycans using comprehensive microarray polymer profiling (CoMPP). Methods in Molecular Biology, 1503, 147–165. [DOI] [PubMed] [Google Scholar]
- Lai, K. , Langley, S.D. , Khwaja, F.W. , Schmitt, E.W. & Elsas, L.J. (2003) GALT deficiency causes UDP‐hexose deficit in human galactosemic cells. Glycobiology, 13, 285–294. [DOI] [PubMed] [Google Scholar]
- Lamport, D.T. , Kieliszewski, M.J. , Chen, Y. & Cannon, M.C. (2011) Role of the extensin superfamily in primary cell wall architecture. Plant Physiology, 156, 11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie, N.D. , Yager, K.L. , McNamara, P.D. & Segal, S. (1996) A mouse model of galactose‐1‐phosphate uridyl transferase deficiency. Biochemical and Molecular Medicine, 59, 7–12. [DOI] [PubMed] [Google Scholar]
- Litterer, L.A. , Schnurr, J.A. , Plaisance, K.L. , Storey, K.K. , Gronwald, J.W. & Somers, D.A. (2006) Characterization and expression of Arabidopsis UDP‐sugar pyrophosphorylase. Plant Physiology and Biochemistry, 44, 171–180. [DOI] [PubMed] [Google Scholar]
- Lunn, J.E. , Feil, R. , Hendriks, J.H. , Gibon, Y. , Morcuende, R. , Osuna, D. et al. (2006) Sugar‐induced increases in trehalose 6‐phosphate are correlated with redox activation of ADPglucose pyrophosphorylase and higher rates of starch synthesis in Arabidopsis thaliana. The Biochemical Journal, 397, 139–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohnen, D. (2008) Pectin structure and biosynthesis. Current Opinion in Plant Biology, 11, 266–277. [DOI] [PubMed] [Google Scholar]
- Moller, I. , Sorensen, I. , Bernal, A.J. , Blaukopf, C. , Lee, K. , Obro, J. et al. (2007) High‐throughput mapping of cell‐wall polymers within and between plants using novel microarrays. The Plant Journal, 50, 1118–1128. [DOI] [PubMed] [Google Scholar]
- Mumma, J.O. , Chhay, J.S. , Ross, K.L. , Eaton, J.S. , Newell‐Litwa, K.A. & Fridovich‐Keil, J.L. (2008) Distinct roles of galactose‐1P in galactose‐mediated growth arrest of yeast deficient in galactose‐1P uridylyltransferase (GALT) and UDP‐galactose 4'‐epimerase (GALE). Molecular Genetics and Metabolism, 93, 160–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguema‐Ona, E. , Bannigan, A. , Chevalier, L. , Baskin, T.I. & Driouich, A. (2007) Disruption of arabinogalactan proteins disorganizes cortical microtubules in the root of Arabidopsis thaliana. The Plant Journal, 52, 240–251. [DOI] [PubMed] [Google Scholar]
- Nibbering, P. , Petersen, B.L. , Motawia, M.S. , Jorgensen, B. , Ulvskov, P. & Niittyla, T. (2020) Golgi‐localized exo‐beta1,3‐galactosidases involved in cell expansion and root growth in Arabidopsis. Journal of Biological Chemistry, 295, 10581–10592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paladini, A.C. & Leloir, L.F. (1952) Studies on uridine‐diphosphate‐glucose. The Biochemical Journal, 51, 426–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parthasarathy, R. , Parthasarathy, L. & Vadnal, R. (1997) Brain inositol monophosphatase identified as a galactose 1‐phosphatase. Brain Research, 778, 99–106. [DOI] [PubMed] [Google Scholar]
- Rautengarten, C. , Ebert, B. , Herter, T. , Petzold, C.J. , Ishii, T. , Mukhopadhyay, A. et al. (2011) The interconversion of UDP‐arabinopyranose and UDP‐arabinofuranose is indispensable for plant development in Arabidopsis. The Plant Cell, 23, 1373–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, K.L. , Davis, C.N. & Fridovich‐Keil, J.L. (2004) Differential roles of the Leloir pathway enzymes and metabolites in defining galactose sensitivity in yeast. Molecular Genetics and Metabolism, 83, 103–116. [DOI] [PubMed] [Google Scholar]
- Schnurr, J.A. , Storey, K.K. , Jung, H.J. , Somers, D.A. & Gronwald, J.W. (2006) UDP‐sugar pyrophosphorylase is essential for pollen development in Arabidopsis. Planta, 224, 520–532. [DOI] [PubMed] [Google Scholar]
- Sharples, S.C. & Fry, S.C. (2007) Radioisotope ratios discriminate between competing pathways of cell wall polysaccharide and RNA biosynthesis in living plant cells. Plant Journal, 52, 252–262. [DOI] [PubMed] [Google Scholar]
- Silva, J. , Ferraz, R. , Dupree, P. , Showalter, A.M. & Coimbra, S. (2020) Three decades of advances in arabinogalactan‐protein biosynthesis. Frontiers in Plant Science, 11, 610377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siow, R.S. , Teo, S.S. , Ho, W.Y. , Shukor, M.Y. , Phang, S.M. & Ho, C.L. (2012) Molecular cloning and biochemical characterization of galactose‐1‐phosphate Uridylyltransferase from Gracilaria Changii (Rhodophyta)(1). Journal of Phycology, 48, 155–162. [DOI] [PubMed] [Google Scholar]
- Tan, L. , Eberhard, S. , Pattathil, S. , Warder, C. , Glushka, J. , Yuan, C. et al. (2013) An Arabidopsis cell wall proteoglycan consists of pectin and arabinoxylan covalently linked to an arabinogalactan protein. The Plant Cell, 25, 270–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, M. , Odejinmi, S.I. , Vankayalapati, H. , Wierenga, K.J. & Lai, K. (2012) Innovative therapy for classic galactosemia—tale of two HTS. Molecular Genetics and Metabolism, 105, 44–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura, M. & Steponkus, P.L. (1997) Effect of cold acclimation on the lipid composition of the inner and outer membrane of the chloroplast envelope isolated from rye leaves. Plant Physiology, 114, 1493–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasquez, S.M. , Ricardi, M.M. , Dorosz, J.G. , Fernandez, P.V. , Nadra, A.D. , Pol‐Fachin, L. et al. (2011) O‐Glycosylated cell wall proteins are essential in root hair growth. Science, 332, 1401–1403. [DOI] [PubMed] [Google Scholar]
- Wang, S. , Ito, T. , Uehara, M. , Naito, S. & Takano, J. (2015) UDP‐D‐galactose synthesis by UDP‐glucose 4‐epimerase 4 is required for organization of the trans‐Golgi network/early endosome in Arabidopsis thaliana root epidermal cells. Journal of Plant Research, 128, 863–873. [DOI] [PubMed] [Google Scholar]
- Willats, W.G. & Knox, J.P. (1996) A role for arabinogalactan‐proteins in plant cell expansion: evidence from studies on the interaction of beta‐glucosyl Yariv reagent with seedlings of Arabidopsis thaliana. The Plant Journal, 9, 919–925. [DOI] [PubMed] [Google Scholar]
- Wilson, D.B. & Hogness, D.S. (1964) The enzymes of the galactose operon in Escherichia Coli. I. Purification and characterization of uridine diphosphogalactose 4‐epimerase. Journal of Biological Chemistry, 239, 2469–2481. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Metabolism of galactose in higher plants compared with humans, yeast and red algae. Alternative routes for the conversion of Gal‐1P by the USP enzyme in higher plants and by the GALT enzyme in humans, yeast and red algae, where the USP enzyme is lacking. Gal, galactose; Gal‐1P, galactose‐1‐phosphate; Glc‐1P, glucose‐1‐phosphate; UDP, uridine diphosphate; PPi, pyrophosphate; UTP, uridine triphosphate; GALK, galactokinase; GALT, galactose‐1‐phosphate uridylyltransferase; USP, UDP‐sugar pyrophosphorylase.
Figure S2. Cell wall sugar composition. Cell walls were extracted from roots of 4‐week‐old WT and galK seedlings growing under Suc‐feeding (1.5 mmol L−1 Suc) versus Gal‐feeding (1.5 mmol L−1 Gal) conditions. After hydrolysis in TFA, monosaccharides were separated on HPLC and quantified by pulsed amperometric detection. Values are averages of four biological replicates each (±SD).
Figure S3. Cell wall polymer profiling (CoMPP analysis). Detection of 42 cell wall epitopes. The ratio of WT to galK mutant for both the CDTA and NaOH fraction is shown. Blue colors indicate increasing abundance of antibody binding in the WT.
Table S1. Cell wall probes used in the present study.
Data Availability Statement
All relevant data can be found within the manuscript and its supporting materials.


