Abstract
Freshwater habitats are under stress from agricultural land use, most notably the influx of neonicotinoid pesticides and increased nutrient pressure from fertilizer. Traditional studies investigating the effects of stressors on freshwater systems are often limited to a narrow range of taxa, depending heavily on morphological expertise. Additionally, disentanglement of multiple simultaneous stressors can be difficult in field studies, whereas controlled laboratory conditions do not accurately reflect natural conditions and food webs. To overcome these drawbacks, we investigated the impacts of two agricultural stressors (the neonicotinoid insecticide thiacloprid and fertilizer) in full‐factorial design in a semi‐natural research site, using environmental DNA sampling to study three different taxonomic groups representing three trophic levels: bacteria (decomposers), phytoplankton (primary producers), and chironomids (consumers). The results show considerable impact of both stressors across trophic levels, with an additive effect of fertilizer and thiacloprid on community composition at all levels. These findings suggest that agricultural stressors affect the entire food web, either directly or through cascade reactions. They are also consistent with morphological assessments that were performed in the same study site, even at a lower number of replicates. The study presented shows that the use of multimarker environmental DNA provides a more comprehensive assessment of stressor impacts across multiple trophic levels, at a higher taxonomic resolution than traditional surveys. Additionally, many putative novel bioindicators for both agricultural stressors were discovered. We encourage further investigations into stressors impacts at different trophic levels, which will lead to more effective monitoring and management of freshwater systems.
Keywords: ecotoxicology, environmental DNA, impact assessment, metabarcoding
1. INTRODUCTION
Freshwater ecosystems contain a rich diversity of both taxa and microhabitats, despite the fact that they cover <1% of the Earth’s surface. They are disproportionately affected by anthropogenic impacts, and seem to be under greater threat than terrestrial and marine systems (Dudgeon et al., 2006; WWF, 2014). Effective monitoring of biological quality of freshwater systems is essential for timely interventions, especially since freshwater is not only important for the management of aquatic flora and fauna, but also for the “ecosystem services” that are essential to people’s well‐being and health (Corvalan et al., 2005).
One of the most important stressors to freshwater systems is agricultural land use as many freshwater habitats are directly connected to agricultural areas. Next to the removal and fragmentation of habitat, pesticide and fertilizer use are the most prominent stressors here (Matson et al., 1997; Schreiner et al., 2016). While pesticides are used on agricultural land to prevent crop losses by pests, they may enter adjacent freshwater through spray drift, run‐off, and seepage. The widespread use of neonicotinoid insecticides in agriculture has been subject of debate as they are found to impact nontarget species, including many freshwater invertebrate species (Morrissey et al., 2015; Pisa et al., 2014; Raby et al., 2018), and have the potential to disrupt the entire food web (Yamamuro et al., 2019). Research has shown that neonicotinoid insecticides can negatively impact macroinvertebrate communities and have significant effects on food web structuring since invertebrates are critical in the transfer of nutrients from the primary producers to the consumers at the top of the food chain (Chagnon et al., 2015; Van Dijk et al., 2013; Schrama et al., 2017). The effects of neonicotinoids and the interaction with other common stressors such as increased influx of nutrients or fine sediments have been studied via morphological assessments in model systems (Barmentlo et al., 2019; Chará‐Serna et al., 2019), showing alternative impacts of neonicotinoids to macroinvertebrate communities in combination with other stressors.
Traditional morphological surveys, such as employed in the above‐mentioned studies, have several drawbacks which have implications on the quality and quantity of data that are collected. Morphological assessments of macroinvertebrate communities rely on skilled taxonomists, may be biased between assessors (Haase et al., 2010) and are labour‐intensive and therefore often expensive (Jones, 2008). The costs specifically affect decisions made on sampling frequency and intensity, and the time‐consuming nature can cause delays that prevent timely interventions into impacted systems (Keeley et al., 2018). Additionally, traditional morphological surveys are limited in accurately assessing many taxa that are likely to be affected by stressors, such as bacteria or planktonic organisms. Tools used to assess impact of pollutants on the aquatic ecosystem thus need to be refined (Schwarzenbach et al., 2006).
In the last decade, molecular tools, including environmental DNA (eDNA) metabarcoding, are being more routinely used for detecting and identifying taxa. Environmental DNA refers to any DNA collected from the environment without specifically collecting or isolating target specimens (Taberlet et al., 2012), and has become a popular tool in surveying freshwater metazoans (e.g., Hänfling et al., 2016; Shaw et al., 2016). The use of eDNA has also found its way into environmental impact studies, such as studies on the impact of aquaculture on benthic sediments (Pochon et al., 2015; Stoeck et al., 2018). eDNA enables the detection of other, potentially more informative, organism groups than those studied in traditional impact studies (Macher et al., 2018). The use of eDNA allows for the defining of new indicators to stressors (e.g., Chariton et al., 2014; Li et al., 2018), and metabarcoding techniques can lead to the creation of new molecular operational taxonomic unit (MOTU)‐based biotic indices (Apothéloz‐Perret‐Gentil et al., 2017). Despite their potential, most eDNA‐based impact assessments still focus on one or few taxonomic groups, and only recently have multimarker approaches been introduced to evaluate different taxonomic groups simultaneously (Andújar et al., 2018; Cordier et al., 2019; Keeley et al., 2018; Laroche et al., 2018; Li et al., 2018), and even some taxonomy‐free approaches (Apothéloz‐Perret‐Gentil et al., 2017; Armstrong & Verhoeven, 2020; Cordier et al., 2018).
Impact assessments are often performed directly in the field, where the myriad of simultaneous stressors make it difficult to identify the impact of individual stressors (Côté et al., 2016; Piggott et al., 2015). Multitrophic eDNA approaches have proven to provide stronger correlations with environmental variables than approaches that use a single guild (Keeley et al., 2018), but the possibility that different guilds respond differently to stressors make interpretation of novel multitrophic eDNA approaches in natural settings difficult. Due to a lack of multitrophic impact assessment studies where results gathered using eDNA and traditional approaches are combined, it remains unclear to what extent eDNA‐based assessments can accurately detect the impacts in such complex environments.
In this study, we assess the impact of two main agricultural stressors on multiple trophic levels in naturally colonized freshwater communities in outdoor experimental ditches. In a full factorial setup, we use eDNA to assess the single and combined impacts of fertilizer and pesticide (the neonicotinoid thiacloprid) application on the richness, taxonomic composition and community dissimilarity of three trophic levels: bacteria, representing decomposers; phytoplankton, representing primary producers; and chironomids, as representatives of the primary and secondary consumers, as well as a traditional indicator group for water quality. Using eDNA in this experimental impact assessment allows us to achieve the following aims: (1) to assess multitrophic impacts on taxon groups that may be sensitive to stressors, but are not traditionally used in freshwater impact assessments due to their difficulty in identification, using novel multimarker eDNA approaches; (2) to assess the impact of two agrochemicals on freshwater communities, while also being able to compare results with a concomitant traditional morphology‐based impact study (Barmentlo et al., 2019); and (3) to pinpoint potential new bioindicators for the health of freshwater ecosystems.
2. MATERIALS AND METHODS
2.1. Experimental setup
Environmental DNA sampling was performed in 20 experimental ditches located in the outdoor research facility the “Living Lab”. Ditches were 10 m long, with a width of 0.8 m and a depth of 0.3 m, and dug adjacent to an existing water level compensation reservoir in Oegstgeest (The Netherlands) that is connected to the Old‐Rhine watershed via canals (Figure S1). The experimental ditches were directly connected to the reservoir, but could be closed off with acrylic plates. The surrounding grassland has been uncultivated for the past 30 years, and there is no agricultural activity in close proximity (see Barmentlo et al., 2019) for a more detailed description of the site and treatments). Prior to the experiments, ditches were left connected to the adjacent reservoir for 6 months to allow for natural colonization of freshwater communities in the ditches. Before starting the experiment, ditches were hydrologically closed off using acrylic plates to isolate the ditches from the reservoir. Subsequently, the ditches were exposed to two different agrochemical stressors in a full factorial design (five ditches per treatment): (1) control, with no added substances; (2) addition of the insecticide thiacloprid (Sigma‐Aldrich) in two spikes (week 20 and 22) with a nominal time weighted average concentration for 1 month of 0.4 µg/L; (3) addition of nutrients in the form of three sachets with 75 g of slow‐releasing artificial fertilizer granulates (“Osmocote”; N:P:K = 15:9:11 combined with microelements) per ditch that were replaced every 6 weeks; and (4) a combination of thiacloprid and fertilizer in the same concentrations and application as described for the single‐treatment ditches. Thiacloprid is used as a model neonicotinoid insecticide, as it is known to be toxic to nontarget organisms, and abundantly present in surface waters in the Netherlands (Barmentlo et al., 2019; Vijver et al., 2008). Even though thiacloprid dissipates from surface waters quickly, it has been shown to be continually present in surface waters, most probably due to continued supply via runoff. To ensure thiacloprid was present in environmentally relevant concentrations, two biweekly spikes were applied. Before the application of the treatments, measurements found no thiacloprid in the water (see also Barmentlo et al., 2019).
2.2. Sampling and DNA extraction
Environmental DNA sampling was performed in five replicate ditches for each treatment (20 in total) at four time points: 2 weeks prior to the start of the treatment (1 May 2017; week 18), and 2 weeks (31 May 31, 2017; week 22), 4 weeks (13 June 13, 2017; week 24) and 7 weeks (6 July 2017; week 27) after the start of the treatments. Surface water samples were collected in the morning from the centre of each ditch using sterilized bottles and filtered within 2 h in the laboratory. Filtration was performed using 0.2 μm polyethersulphone (PES) filter membranes (Sartorius) placed in sterilized Nalgene filter units (Thermo Fisher) attached to a vacuum pump. Up to 300 ml of water (until membrane clogged) was filtered for each of the 20 ditches. After filtration, membranes were stored in 700 µl CTAB at –20°C until extraction. A modified CTAB extraction protocol adapted from Turner et al. (2014) was used for DNA extraction (Beentjes, Speksnijder, Schilthuizen, Hoogeveen, & van der Hoorn, 2019). DNA precipitation was performed overnight at –20°C on 500 µl aqueous phase, with 1000 µl 96% ethanol, 360 µl 3 M sodium acetate and 3.0 µl glycogen.
2.3. DNA amplification and MiSeq sequencing
Three different markers for three different taxa groups were analysed separately: a ±400 bp fragment of 18S rRNA V4 subregion for diatoms and other phytoplankton (Zimmermann et al., 2011), a 273 bp fragment of the 16S rRNA for bacteria (Klindworth et al., 2013) and a 235 bp fragment of COI for chironomids (Bista et al., 2017) (for primers, see Table S1). The primers for the 18S rRNA V4 region were originally designed for diatom studies, but (unpublished) comparisons of various phytoplankton primers showed these primers had a broad coverage of multiple phytoplankton groups, and the subregion has relatively good coverage in NCBI Genbank. For each of the PCRs, all of the 80 reactions for each marker (20 replicate ditches, four time points) were performed in duplicate. The chironomid PCR contained two samples of DNA extracted from two chironomid specimens unlikely to occur in the setup (Einfeldia pagana and Tanytarsus brundini), which were used to estimate cross‐contamination between samples during the amplification.
Dual‐indexed Illumina amplicon libraries were prepared using a two‐step PCR protocol, in which the first PCR used primers with 5′ Illumina tails. Initial PCRs were performed in 25 µl reactions containing 1x Phire Green Reaction Buffer, 0.5 mM dNTPs, 0.5 µl Phire Hot Start II DNA Polymerase (Thermo Fisher), 0.5 µM of each primer and 2.0 µl of template DNA. Initial denaturation was performed at 98°C for 30 s, followed by 35 cycles at 98°C for 5 s, 50°C for 5 s and 72°C for 15 s, followed by final elongation at 72°C for 5 min. PCR products were checked on E‐Gel 96 pre‐cast agarose gel (Thermo Fisher) and cleaned with a one‐sided size selection using NucleoMag NGS‐Beads (Macherey‐Nagel), in a 1:0.9 ratio. Dual‐index PCRs were performed using 2.0 μl of PCR product from the first round in a 20 μl reaction containing 1x TaqMan Environmental Master Mix 2.0 (Thermo Fisher) and 1.0 μM of each primer. Initial denaturation was performed at 95°C for 10 min, followed by 10 cycles at 95°C for 30 s, 55°C for 60 s and 72°C for 30 s, followed by final elongation at 72°C for 7 min. These PCR products were quantified on the QIAxcel (Qiagen) and each replicate of each marker was pooled equimolarly separately. Pools were cleaned with a one‐sided size selection using NucleoMag NGS‐Beads, ratio 1:0.9, then quantified on the Bioanalyzer 2100 (Agilent Technologies) with the DNA High Sensitivity Kit. The pools for the bacteria and chironomids were combined equimolarly and sequenced on one run of Illumina MiSeq (v3 Kit, 2 × 300 paired‐end), the pools for the phytoplankton were combined equimolarly and sequenced on a separate run (v3 Kit, 2 × 300 paired‐end), both at BaseClear BV.
2.4. Bioinformatics
Quality filtering and clustering of all data was performed in a custom pipeline on the OpenStack environment of Naturalis Biodiversity Centre through a Galaxy instance (Afgan et al., 2018). Raw data was filtered with Sickle (quality threshold 20, minimum length 100 bp) (Joshi & Fass, 2011) and merged with flash v1.2.11 (minimum overlap 10, mismatch ratio 0.25) (Magoč & Salzberg, 2011), nonmerged reads were discarded. Primers were trimmed from both ends using cutadapt v1.16 (minimum match 5 bp, maximum error rate 0.2) (Martin, 2011) and any read without both primers present and anchored was discarded. prinseq v0.20.4 (Schmieder & Edwards, 2011) was used to filter reads on length, based on sequence length histogram data (390–420 bp for phytoplankton, 248–254 bp for bacteria, 230–250 bp for chironomids). Sequences were dereplicated and clustered into MOTUs using vsearch v2.10.3 (Rognes et al., 2016) with a cluster identity of 98% and a minimal accepted abundance of 2. MOTU tables were corrected using the occurrence of control chironomids in field samples with a tool based on the unspread.py script by Larsson et al. (2018) (https://github.com/sandberg‐lab/Spreading‐Correction) (rate of spread 0.003, cutoff value 5 reads). This tool calculates and corrects for a rate of spread, based on the abundance of MOTUs and the position of a sample in the PCR plate (using the Illumina index labels). Rate of spread was assumed to be identical for all markers. PCR replicates were combined, including all MOTUs that were present in at least one replicate.
Molecular operational taxonomic unit sequences were compared to custom reference databases using blast+ (Camacho et al., 2009). Phytoplankton MOTUs were compared to a data set that included all 18S rRNA sequences from GenBank (Release 227, Benson et al., 2005) (sequences downloaded 21 August 2018), bacteria were compared to Silva SSUParc (Release 132, Quast et al., 2013), chironomids were compared to a custom reference database (Beentjes, Speksnijder, Schilthuizen, Hoogeveen, Pastoor, et al., 2019) based on specimens collected in the Netherlands as part of a national DNA barcoding campaign (Beentjes et al., 2015), supplemented with sequences obtained from BOLD (Ratnasingham & Hebert, 2007).
2.5. Taxonomic assignment and diversity analysis
A 98% cutoff was used for species‐level identification, and a custom lowest common ancestor (LCA) script was used to identify MOTUs in those cases where no direct hits above 98% with the reference database were found (Beentjes, Speksnijder, Schilthuizen, Hoogeveen, Pastoor, et al., 2019). The same settings were used for all three data sets to keep the analyses consistent. LCA was performed on the top 10% hits with bitscore >170, a minimum identity of 85% and a minimum coverage of 90% (90% identity and 100% coverage for the bacteria). The LCA was set to identify MOTUs no further than genus level. Chironomid identifications above species level were cross‐checked against a blast against NCBI Genbank to rule out potential false positives due to the limited reference (only Chironomidae sequences) that could bias the LCA. Nontarget MOTUs were removed prior to subsequent analyses. For bacteria these were nonbacterial MOTUs (such as chloroplast and mitochondria), for phytoplankton these were any MOTUs that were not assigned to a phytoplankton group on at least phylum level, and for chironomids any MOTU’s not assigned to at least genus level and nonchironomids). Read data was normalized per sample to account for differences in read depth between samples. Differences in relative abundances in time, relative to the control samples, are assumed to be caused by the treatments (Barmentlo et al., 2019; Beermann et al., 2018).
2.6. Statistical analyses
Potential effects of the agrochemicals and time were assessed on the three different communities (bacteria, phytoplankton and chironomids). The effects of both fertilizer, thiacloprid, time, and all possible interactions were investigated on the normalized MOTU abundances using permutational analysis of variance (PERMANOVA, function adonis, R package vegan (Oksanen et al., 2019)). Bray‐Curtis was used as measure for dissimilarity, with 999 permutations. We accounted for the repeated measure design by including ditch number as a random variable. Differences in richness were analysed with ANOVA (R package stats). PCoA plots were created using function pcoa (R package ape (Paradis & Schliep, 2018)). Distances between centroids were calculated using centroid coordinates obtained via the betadisper function (R package vegan). Potential effects on beta dispersion were investigated by using distance‐based dispersion tests (function betadisper, R package vegan). Correlation between the distance matrices for the three communities analyzed in this study and the morphological assessment was investigated using a Mantel test (function mantel.rtest, R package ade4 (Dray & Dufour, 2007), 999 permutations). Indicative MOTUs for each of the treatments independently were identified using the multipatt function (R package indicspecies (De Cáceres & Legendre, 2009), 999 permutations), which uses the specificity (the probability that a ditch belongs to the treatment group given that a MOTU has been observed) and fidelity (the probability of finding a MOTU in ditches belonging to the treatment) to calculate in the indicator value (IndVal) of each MOTU (De Cáceres et al., 2010). All MOTUs with either specificity or fidelity scores below 0.5 were omitted, even if the indicator value was significant.
Morphological assessment in the original study by Barmentlo et al. (2019) was performed at three moments: before treatment, 1 month after treatment (June) and 4 months after treatment (September). The assessment in June was performed at the same time (directly after eDNA sampling) as the “week 24” measurement presented in this study. Data from the morphological assessment in June was compared directly to eDNA results from the same week.
3. RESULTS
3.1. Sequence run statistics and taxonomic assignment
The two chironomid species used as cross‐contamination control contributed to a maximum of 0.25% of read data, although this was inflated in a few cases due to low number of total reads in a sample. After applying the filter to correct for cross‐contamination, none of the samples contained any reads for the control species. Similarly, all MOTUs present in the negative controls (PCR blanks) were filtered out by the correction (before correction, negative controls had an average of 153.8 reads vs. 14,608.4 on average in the samples). After merging and filtering, 6,751,664 reads were retained for bacteria, 4,553,770 for phytoplankton and 2,396,387 reads for chironomids (File S1). The chironomid data set also had a large proportion of reads (2,781,001) that did not conform to the expected length, this “by‐catch” was mostly identified as oomycetes and discarded from the data set prior to analysis. After clustering, and with the correction for cross‐contamination applied, the replicates combined and nontarget MOTUs omitted, there was a total of 5383 MOTUs for bacteria, 2819 for phytoplankton and 692 for chironomids. The bacteria data contained 4011 MOTUs (74.5% of total MOTUs, representing 61.0% of reads) that could be identified at least at phylum level, with the largest groups being Gammaproteobacteria (30.1%) and Bacteroidetes (20.9%). In the phytoplankton data, 1773 MOTUs (62.9% of total MOTUs, representing 54.2% of reads) could be identified to at least phylum level of relevant taxa, mostly Chlorophyta (45.4%) and Stramenopiles (34.5%). For the chironomid data set, 368 MOTUs (53.2% of total MOTUs, representing 44.8% of reads) could be identified as Chironomidae at genus or species level, representing 64 species from 35 genera; 207 MOTUs were only identified to genus level. One sample (a sample from a ditch with a fertilizer treatment from week 18) did not contain any chironomid reads. The morphological study by Barmentlo et al. (2019) confirmed the presence of chironomids in this ditch, which meant the non‐detection could not be translated in to an absence of chironomids; the sample was therefore omitted from the analyses presented here. Both MOTU richness and richness based on aggregated taxon data show no relevant significant differences between treatments (Figure S2), neither was there any correlation between the filtered volume and the MOTU and taxon richness (data not shown).
3.2. Effects on community dissimilarity and composition
Before the application of any agrochemical, there were no statistically significant differences between community species compositions of the prospective treatments. After application of the agrochemicals, fertilizer and thiacloprid addition showed a significant interaction, irrespective of time, leading to dissimilar communities relative to the control for all three communities (p = .001 for all comparisons) (Table 1). The interaction between thiacloprid and fertilizer was most pronounced in the weeks directly after application of thiacloprid (Figure 1). Two weeks after the start of the treatments, the impact of thiacloprid addition was more pronounced than the addition of fertilizer, with the former having a significant impact on the dissimilarity in all groups (p = .001, Table 2), while the impact of fertilizer was only significant for bacteria and phytoplankton (p = .021 and .001, respectively). Thiacloprid centroids were more distant from the control than the fertilizer centroids (visual inspection) for all three groups in week 22 and 24 (2 and 4 weeks after treatment), indicating that thiacloprid had a greater effect on the community composition in the short‐term than fertilizer. This reversed after the addition of fresh fertilizer pellets in week 25 as the effects of fertilizer became more pronounced compared to those of the thiacloprid addition (Figure 1). There was one sample in the control ditches prior to application of treatments, where we found only a single MOTU that was identified as a chironomid. This formed an outlier in the analysis of the chironomid data, and caused the centroid of the control samples in this measurement (week 18) to shift relative to the centroids of the other sets of ditches, explaining why the distances between centroids in week 18 were already elevated prior to start of treatment (Figure 1c).
TABLE 1.
PERMANOVA results (F‐statistic, R 2 and p‐values) for the different treatments and the combined effects, including the three‐way interaction with time, for data from all measurements combined. Significant p‐values are shown in bold
| Bacteria | Phytoplankton | Chironomidae | |||||||
|---|---|---|---|---|---|---|---|---|---|
| F | R 2 | p‐Value | F | R 2 | p‐Value | F | R 2 | P‐Value | |
| Thiacloprid | 6.823 | .034 | .001 | 2.605 | .017 | .001 | 4.068 | .038 | .001 |
| Fertilizer | 12.329 | .061 | .001 | 6.751 | .044 | .001 | 1.192 | .011 | .001 |
| Time | 29.331 | .436 | .001 | 19.850 | .387 | .001 | 8.950 | .254 | .001 |
| Fertilizer:Thiacloprid | 2.170 | .011 | .001 | 1.893 | .012 | .001 | 1.017 | .010 | .001 |
| Thiacloprid:Time | 3.100 | .046 | .002 | 2.115 | .041 | .016 | 1.531 | .043 | .041 |
| Fertilizer:Time | 4.785 | .071 | .001 | 2.911 | .057 | .002 | 0.872 | .025 | .695 |
| Fertilizer:Thiacloprid:Time | 1.543 | .023 | .195 | 1.287 | .025 | .269 | 0.846 | .024 | .755 |
FIGURE 1.
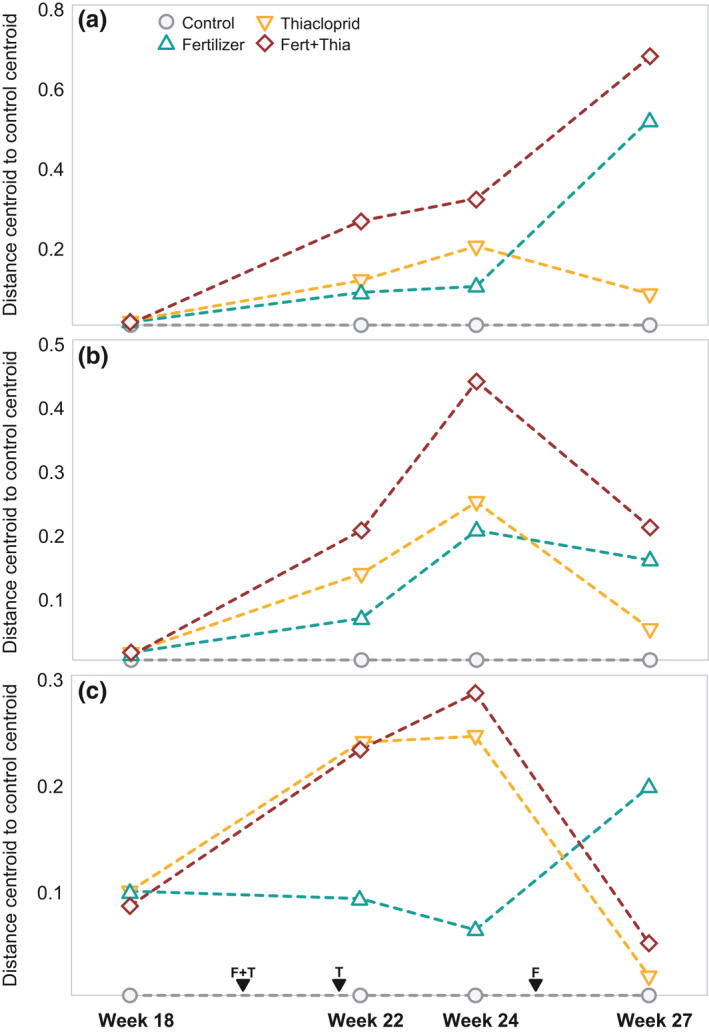
Average distance from centroid to the control centroid, for each of the taxonomic groups. (a) bacteria, (b) phytoplankton, and (c) chironomids. Moments of treatment application for thiacloprid (T) and fertilizer (F) are provided on the x‐axis of panel (c)
TABLE 2.
PERMANOVA results (F, R 2 and p‐values) for the different treatments for each of the time points evaluated separately
| Bacteria | Phytoplankton | Chironomidae | |||||||
|---|---|---|---|---|---|---|---|---|---|
| F | R 2 | p‐Value | F | R 2 | p‐Value | F | R 2 | p‐Value | |
| Week 18 | |||||||||
| Thiacloprid | 0.675 | .036 | .809 | 0.502 | .028 | .711 | 1.032 | .060 | .352 |
| Fertilizer | 1.329 | .071 | .201 | 1.190 | .066 | .287 | 0.316 | .018 | .990 |
| Fertilizer:Thiacloprid | 0.728 | .039 | .757 | 0.392 | .022 | .803 | 0.964 | .056 | .392 |
| Week 22 | |||||||||
| Thiacloprid | 6.428 | .234 | .001 | 3.520 | .144 | .001 | 4.087 | .181 | .001 |
| Fertilizer | 3.044 | .111 | .021 | 3.553 | .145 | .001 | 1.460 | .064 | .152 |
| Fertilizer:Thiacloprid | 2.005 | .073 | .069 | 1.348 | .055 | .179 | 1.093 | .048 | .335 |
| Week 24 | |||||||||
| Thiacloprid | 4.318 | .167 | .001 | 3.007 | .117 | .004 | 3.006 | .145 | .001 |
| Fertilizer | 3.806 | .147 | .003 | 4.210 | .164 | .001 | 0.857 | .041 | .662 |
| Fertilizer:Thiacloprid | 1.740 | .067 | .114 | 2.514 | .098 | .010 | 0.860 | .042 | .636 |
| Week 27 | |||||||||
| Thiacloprid | 2.027 | .061 | .070 | 0.892 | .041 | .550 | 1.269 | .066 | .173 |
| Fertilizer | 13.598 | .410 | .001 | 4.119 | .189 | .001 | 1.121 | .059 | .311 |
| Fertilizer:Thiacloprid | 1.514 | .046 | .149 | 0.750 | .034 | .764 | 0.728 | .038 | .782 |
Significant values are shown in bold
The effect of time on dissimilarity was prominent, being larger than most effects of the agrochemicals, indicating that continued species turnover occurred. Two‐way interactions of time with both fertilizer and thiacloprid were significant for all three communities studied (Table 1). There were no significant three‐way interactions for any of the three groups, indicating that the interaction between the effects of fertilizer and thiacloprid occurred irrespective of the time point sampled. Studying the individual weeks separately, there was a significant effect of thiacloprid addition on community dissimilarity compared to control ditches in all three groups in weeks 22 and 24. Fertilizer had a significant effect on the composition of phytoplankton and bacteria in all 3 weeks after the start of the treatments (Table 2).
Beta‐dispersion was significantly higher in treatments containing fertilizer for both bacteria and phytoplankton (p < .001 and p = .005, respectively), meaning that communities diverged when fertilizer was added to the system. Thiacloprid addition had a significant effect on chironomids, leading to convergence of the communities across the replicate ditches (p = .002) (Figure S3). Combined with the patterns observed in the PCoA, this indicates that both community composition and dispersion are responsible for the significance in the PERMANOVA. There were moderate, but significant correlations between all three Bray Curtis distance matrices of the three taxon groups. The correlation between bacteria and phytoplankton was stronger (Pearson r = .820, p = .001) than correlations of bacteria and phytoplankton with chironomid data (r = .447 and r = .465, respectively, p = .001). This indicates that community dissimilarities caused comparable patterns for both bacteria and phytoplankton (Figure S4).
While the treatments had no apparent effect on the observed richness compared to control ditches (Figure S1), there were considerable shifts in the relative abundance of different taxa for all three communities analysed in this study (Figure 2). For bacteria, most notable were the proportional decline of Actinobacteria in ditches treated with fertilizer, to the point where they almost disappeared in the combined treatment in week 27 (representing 0.8% of the total reads in combined agrochemical ditches compared to 32.2% in the control), and the higher abundance of Bacteriodetes in ditches with added thiacloprid (representing 48.5% of the total reads vs. 30.3% in the control ditches).
FIGURE 2.
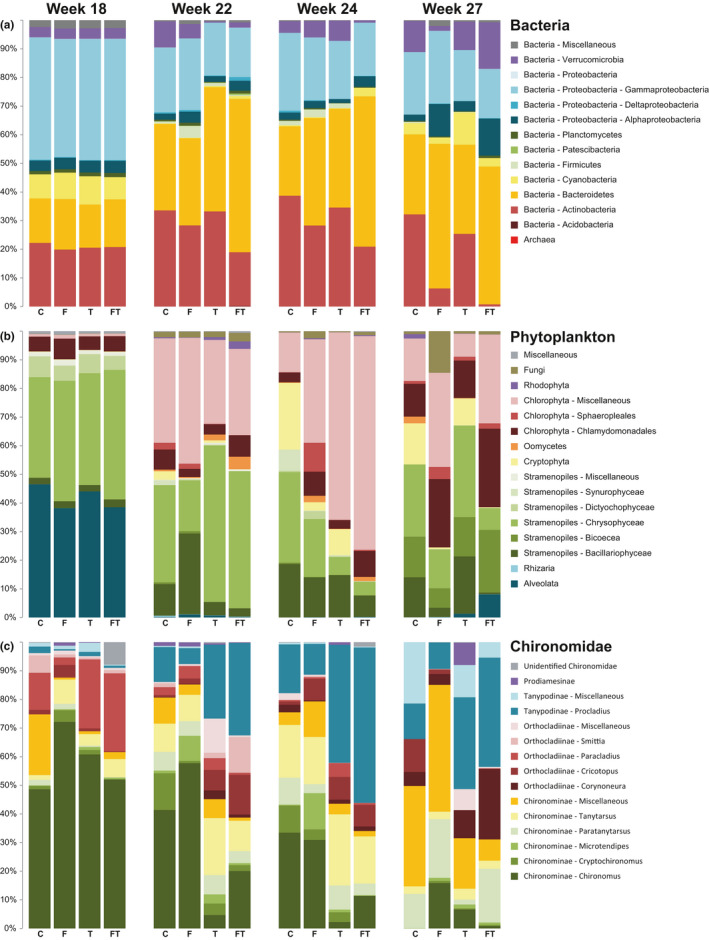
Read distributions observed for each of the different treatments and control both prior to (week 18) and after application of treatments (weeks 22–27) for each of the taxonomic groups. (a) Bacteria, (b) phytoplankton and (c) chironomids, in control situation (C), and with added fertilizer (F), thiacloprid (T) and combined treatments (FT)
The phytoplankton community compositions changed considerably as well, and were significantly affected by both fertilizer and thiacloprid (Table 2). The read distribution (Figure 2b) reflected these changes, with the thiacloprid treatment initially showing higher proportions of chrysophyte reads (average of 51.3% vs. 25.8% in control ditches), and later showing larger relative abundances of chlorophytes (76.3% vs. 36.0% in control ditches). After the addition of fresh fertilizer pellets in week 25, treated ditches were dominated by various groups of chlorophytes (60.6% vs. 24.9% in control ditches), whereas control and thiacloprid‐only ditches were dominated by the various stramenopile groups (60.0% vs. 27.0% in ditches with fertilzer). Cryptophytes represented 11.9% of the reads in treatments without fertilizer, but went nearly undetected in the treatments with fertilizer (0.4% of the reads). At this point in time, thiacloprid no longer showed a significant effect on the community composition (Table 2).
For the chironomids, the most notable differences were observed between ditches with and without added thiacloprid (Figure 2c). With thiacloprid addition, the genus Chironomus was no longer the most abundant and declined strongly in read abundance (12.4% in ditches with thiacloprid vs. 50.0% in control ditches), also compared to week 18 (before the start of treatments), where on average this genus represented 57.7% of the reads. Thiacloprid shifted the community composition towards genera outside of the subfamily Chironominae, such as Procladius (subfamily Tanypodinae) (29.0% vs. 8.9% in control ditches) and Cricotopus (subfamily Orthocladiinae) (10.5% vs. 1.3%). This shift continued in week 24, where thiacloprid ditches became dominated by Procladius (47.7% vs. 13.9%), at the expense of Chironomus (6.8% vs. 32.2%). In week 27, Procladius remained more abundant in the thiacloprid ditches, although not as pronounced as in week 24.
3.3. Indicator taxa
Indicator analysis on the three assessments after start of treatments separately identified 637 bacterial MOTUs, 472 phytoplankton MOTUs and 46 chironomid MOTUs that were indicative for either absence or presence of either of the two added agrochemicals in one or more of the three post‐treatment measurements (File S2, summarized in Table 3). With the observations of the three assessments combined (week 22, 24, 27), the indicator analysis identified 157 bacterial MOTUs, 28 of which acted as indicators for both agrochemicals. The majority were indicators for the absence of both fertilizer and thiacloprid (26), one MOTU (identified as a Cylindrospermum) was indicative for the presence of both agrochemicals. For phytoplankton, we found 115 indicators for the three sampling moments combined, in which nine acted as indicators for both agrochemicals, either for the absence (2) or presence (7) of both treatments. In the combined chironomid data there were 12 indicative MOTUs, with no MOTUs that acted as indicator for both agrochemicals. We did observe MOTUs with low fidelity values for the combined measurements, due to the fact that indicative MOTUs for all three groups were not observed in the ditches in each of the three assessments after the introduction of agrochemicals, but these were omitted from the data set.
TABLE 3.
Summarized indicator species analysis results, with the number of indicative MOTUs found for each of the three taxonomic groups: indicators for absence (F−) and presence (F+) of fertilizer, and absence (T−) and presence (T+) of thiacloprid. Analysis was performed on data from each post‐treatment measurement (weeks 22, 24 and 27), and combined data of the three measurements. An overview of all indicator MOTUs is provided in File S2
| Bacteria | Phytoplankton | Chironomidae | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F− | F+ | T− | T+ | F− | F+ | T− | T+ | F− | F+ | T− | T+ | |
| Week 22 | 52 | 54 | 128 | 50 | 60 | 63 | 39 | 56 | 1 | 8 | 13 | 10 |
| Week 24 | 43 | 46 | 110 | 39 | 12 | 127 | 40 | 48 | 3 | 4 | 5 | 11 |
| Week 27 | 194 | 109 | 25 | 4 | 90 | 91 | 9 | 2 | 3 | 0 | 0 | 3 |
| Weeks 22–27 | 79 | 25 | 73 | 8 | 42 | 53 | 15 | 14 | 3 | 0 | 1 | 8 |
3.4. Comparison to morphological assessment
Patterns observed in stressor responses as measured by distances between centroids in week 24 were similar for all three taxonomic groups assessed in this study, as well as the morphological assessment of macroinvertebrates assessed by Barmentlo et al. (2019) at the same sampling timepoint. The thiacloprid treatment showed more distance relative to the control than the fertilizer treatment, whereas the combined treatment showing the largest deviation for all four assessments (Figure 3), although the morphological assessment made use of nine replicate ditches instead of the five replicates that were used for eDNA evaluation. The distances between control centroids varied when using fewer replicates, but in all three eDNA assessments the pattern described above was observed with as little as three replicate ditches (out of five) (Figure S5B–D). For morphological data, at least seven (out of nine) replicate ditches were needed to reveal this pattern (Figure S5A). There were, however, no significant correlations with any of the eDNA‐based distance matrices and the distance matrix of the morphological assessment. The morphological assessments also showed no significant treatment effect on richness nor abundance of macroinvertebrates (Barmentlo et al., 2019).
FIGURE 3.
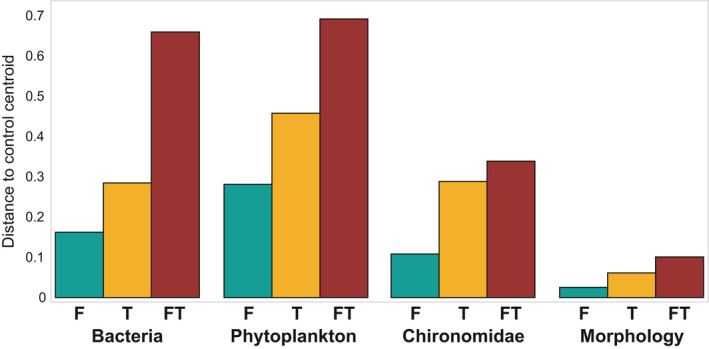
Centroid distance to the control centroid in week 24 (1 month after application of the agrochemicals), for the bacteria, phytoplankton, and chironomids assessed with environmental DNA, as well as the macroinvertebrates assessed with morphological methods (see Barmentlo et al., 2019), exposed to fertilizer (F), thiacloprid (T), and combined agrochemical addition (FT)
4. DISCUSSION
We aimed to investigate the applicability of eDNA for multitrophic impact assessments across multiple taxonomic groups for common agricultural stressors, compare findings to a concomitant morphological assessment, and uncover potential novel bioindicators. Clear impacts of both agricultural stressors were observed for all three taxonomic groups. Additionally, the simultaneous introduction of both agrochemicals in the ditches had strong additive effects on the three trophic levels analysed. We show that eDNA‐based impact assessments can provide useful insights into stressor responses in taxa that are usually not included in traditional assessments, with noninvasive sampling methods. Moreover, the responses measured are comparable and much more pronounced to the morphological assessments of macroinvertebrates conducted simultaneously during the same experiment (Barmentlo et al., 2019) and with the previously reported effects of neonicotinoids on macroinvertebrates and zooplankton (e.g., Yamamuro et al., 2019). Additionally, the three groups evaluated in this study have been observed to contain numerous putative indicative taxa that have potential as novel bioindicators for agricultural stressors.
Fertilizer addition caused significant changes in community composition of both bacteria and phytoplankton, with replicate ditches showing higher dissimilarity (divergence) compared to control ditches (Figure S3). Community dissimilarities showed comparable patterns (Figure S4) and strong correlations for bacteria and phytoplankton. This was expected as eutrophication has long been known to be associated with increased growth in phytoplankton (Heisler et al., 2008), and interactions such as nutrient cycling between phytoplankton and bacteria at the base of the food web (Seymour et al., 2017) render bacterial communities sensitive to changes in phytoplankton communities (and vice versa). Chironomids were also sensitive to the addition of fertilizer, although these fertilized communities were generally more similar to the control than to the thiacloprid treatment (Figure S4). Nutrient pressure has been shown to have effects on freshwater macroinvertebrates in previous research (e.g., Donohue et al., 2009), since eutrophication can lead to oxygen depletion and changes in food availability, which could explain the current findings.
The thiacloprid concentration used in this study (a nominal time weighted average of 0.4 µg/L) is considered a realistic concentration, based on commonly observed surface water concentrations in the Netherlands (Vijver et al., 2008). Previous research has already shown that freshwater macroinvertebrates are affected by neonicotinoids at concentrations observed in surface water (e.g., Morrissey et al., 2015; Sánchez‐Bayo et al., 2016). Indeed, thiacloprid addition had a much larger impact on the chironomid community composition than fertilizer addition and resulted in a significant convergence (Figure S3). Even after thiacloprid had dissipated from the water column after only a few weeks due to its rapid adsorption to the sediment (DT90 = 11.1 days; Barmentlo et al., 2019), the legacy effect of thiacloprid was still larger than the effect of the fertilizer (Table 2). That thiacloprid has such a pronounced effect on the macrofaunal community is not surprising, given that it is an insecticide, but our data suggest a single spike can have a lasting impact. There was an additive effect of both agrochemicals, as the impact of a combined treatment effect of fertilizer and thiacloprid was greater than that of each treatment separately, and communities under a combined treatment were more dissimilar relative to the control than communities exposed to a single agrochemical (Figure 1, Tables 1 and 2). Most two‐way interactions between fertilizer and thiacloprid were not significant, however, suggesting the effect was additive, rather than synergistic (Table 2).
Addition of agrochemicals strongly affected the community compositions. Changes in composition where most notable for chironomids, for which most MOTUs could be identified at species or genus level. Subfamilies Tanypodinae and Orthocladiinae seemed less susceptible to the presence of thiacloprid, consistent with findings from previous studies that showed significant effects on Chironominae in response to neonicotinoid insecticides (Langer‐Jaesrich et al., 2010; Williams & Sweetman, 2019). This pronounced effect is despite the chironomid data set being the one with the most discarded nontarget MOTUs. It is also not unexpected that the chironomid data set seems more patchy than other data sets, since this data is derived from actual eDNA, contrary to the phytoplankton and bacteria most probably caught as whole organisms, leading to a more heterogenous distribution in the water. The fact that the effects of the treatments are so pronounced in the chironomid data shows they are an excellent indicator. Optimization of sampling methods specific for arthropod eDNA or of the primers designed by Bista et al. (2017) may improve the use of chironomids in eDNA studies such as the one presented in this manuscript. Whilst the direct effects of fertilizer on bacterial and phytoplankton communities have been studied before (Carvalho et al., 2013), there is little research on the effects of neonicotinoid insecticides on those communities. One study suggests that algal blooms appear to increase in size under stress from the neonicotinoid imidacloprid (Sumon et al., 2018). The neonicotinoid insecticide thiacloprid, meant to target pest insects, also seemed to affect bacterial and algal community composition in the present study. Our data suggests that thiacloprid has an important impact on the structuring of the communities (Tables 1 and 2, Figure 2). It is likely that some of these effects on phytoplankton and bacteria communities have been caused by food web cascades. Indeed, previous research showed that even under stress from pesticide mixtures, biotic interactions played a major role in the structuring of plankton communities (Pereira et al., 2018). Similarly, responses to nutrient pressure by fertilizer in macroinvertebrates may also partly be caused by cascade reactions, such as the aforementioned changes in food availability. Processes such as eutrophication can have a significant impact on total community composition and food web structure via trophic cascades (Davis et al., 2010; Suikkanen et al., 2013), and a recent study evaluating anthropogenic stressors on freshwater food webs showed that macroinvertebrates had different reactions to fertilizer, herbicide and insecticide, depending on their food source (Schrama et al., 2017). The authors also noted, however, that cascading effects in the food web were hard to explain, and found some suggestions of shifts in diet induced by stressors.
Results from the morphological assessment matched the observed patterns presented in this study regarding dissimilarity relative to the control; there was an increase in effect size from fertilizer to thiacloprid to the mixture treatment for all communities investigated, although no significant effects were detected on the beta dispersion of the community in the morphological assessment (Barmentlo et al., 2019). In this study, however, we observed these stressor impact patterns at a lower number of replicates compared to the traditional assessment (Figure S3), and with a higher resolution (83 morphological taxa vs. 4011, 1773 and 368 MOTUs for bacteria, phytoplankton and chironomids, respectively). The morphological assessment of the macroinvertebrates revealed patterns similar to the eDNA assessment, with the thiacloprid treatment showing more distance relative to the control than the fertilizer treatment and the combined treatment showing the largest deviation (Figure 3), irrespective of the biota that were sampled. This indicates how strongly interconnected the different trophic levels are and that potential cascading food web responses to stressors can occur even in non‐target biota, and also shows the great potential of using eDNA in impact assessments.
Our analyses revealed a large number of indicative MOTUs for all three trophic levels assessed (Table 3), suggesting that many potential new bioindicators are hidden in taxon groups that are either difficult to identify (e.g., chironomids) or are underutilized in traditional bioassessments of water impacted by anthropogenic stressors due to a lack in legislative frameworks (e.g., bacteria and phytoplankton). Previous research has shown that MOTU‐based approaches can provide better resolution in impact assessments, such as with undescribed cryptic diversity demonstrating contrasting responses to stressors (Macher et al., 2016), or reference databases being unable to identify all the encountered molecular variation (Beermann et al., 2018). Several studies have shown that MOTU‐based assessment methods can accurately predict stressor impact on water systems (e.g., Andújar et al., 2018; Li et al., 2018). However, the inability to identify all MOTUs to species or even genus level complicates the ecological interpretations of shifts in communities caused by external stressors. Taxonomic hiatuses in the reference database are large, especially for microorganisms such as the freshwater bacteria and phytoplankton studied in this study. Accumulating MOTUs based on the higher‐level taxonomic assignments could be possible, in order to assign some ecological value to such indicators. The MOTUs, however, may represent a wide variety of ecological groups, and accumulating them into a single entity would decrease the sensitivity of any such bioindicators (Jones, 2008). While it may be difficult to link ecological information to unidentified MOTUs, they can still be of use in comparative studies, such as impact assessments (Li et al., 2018).
One key limitation for assigning indicator taxa for freshwater communities is the large fluctuations in community composition over time. The large community turnover caused low fidelity scores for many indicator MOTUs observed in the indicator analysis on the combined data for the three post‐treatment measurements, due to the fact that many MOTUs do not occur in all time points (Supporting Information S2). Moreover, indicator MOTUs might not only be specific to a certain time frame, but can also be spatially limited, as it was previously observed that indicator taxa for the impact of offshore oil and gas drilling (Laroche et al., 2018) or nutrient loading (Clark et al., 2020) were highly site specific. Impact assessments based on novel indicators, or even based on MOTUs, should preferably be time‐ and location‐independent, to make their application on a broader scale feasible. This could prove challenging, especially when looking at microorganisms such as bacteria or phytoplankton taxa observed in the current study, as these groups tend to have a large turnover in their community composition on a relatively small time scale (Beentjes, Speksnijder, Schilthuizen, Hoogeveen, & van der Hoorn, 2019). However, the huge potential for these novel bio‐indicators in large‐scale impact assessments would make any efforts into a better understanding of their occurrence and behavior worthwhile.
5. CONCLUSIONS
We have shown that eDNA metabarcoding at multiple trophic levels provides insights into changes in freshwater communities under pressure of agricultural stressors. The full‐factorial design of the mostly natural study site allowed us to observe the impact of single stressors. We found an additive (but not synergistic) effect of artificial fertilizer and the insecticide thiacloprid on community composition at the level of decomposers (bacteria), primary producers (phytoplankton), and consumers (chironomids). This effect of multiple stressors was consistent and more pronounced with observations reported in traditional morphological assessments of the same experimental setup. These effects were even detected with a lower number of treatment replicates than the traditional morphological study, indicating the robustness of using eDNA metabarcoding in impact assessments. While both agrochemicals directly influenced different taxa at different trophic levels, the neonicotinoid insecticide thiacloprid, meant to target pest insects, also affected bacterial and algal community composition, be it directly or through cascade reaction through the food web. We encourage the use of multimarker eDNA for impact assessment across trophic levels in freshwater ecosystems, as it (1) provides a more comprehensive assessment of impacts on the entire food web, (2) provides more information at a higher taxonomic resolution compared to traditional morphological surveys, even if MOTUs are not all assigned to species level, and (3) allows for discovery of novel indicator taxa. The incorporation of eDNA methodology contributes to ecosystem understanding and would allow for more effective monitoring and management of freshwater systems, and help safeguard the ecosystem services they contribute to humanity.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
All authors contributed ideas and to the writing of the manuscript.
Supporting information
Supplementary Material
Supplementary Material
Supplementary Material
ACKNOWLEDGEMENTS
Experiments were performed in the outdoor experimental laboratory “Levend Lab” established by crowdfunding by Maarten Schrama and Martina Vijver at Leiden University. We thank Sam Boerlijst for his assistance with DNA extractions, and André van Nieuwenhuijzen for his critical look at the identifications of chironomids obtained from the eDNA metabarcoding. We thank Jelle Dercksen for supplying a drone photo of the field setup. This study was part of the DNA Waterscan project, funded by the Gieskes‐Strijbis Fonds.
Beentjes, K. K. , Barmentlo, S. H. , Cieraad, E. , Schilthuizen, M. , van der Hoorn, B. B. , Speksnijder, A. G. C. L. , & Trimbos, K. B. (2022). Environmental DNA metabarcoding reveals comparable responses to agricultural stressors on different trophic levels of a freshwater community. Molecular Ecology, 31, 1430–1443. 10.1111/mec.16326
DATA AVAILABILITY STATEMENT
Raw sequence data has been made available from the NCBI Sequence Read Archive (Bioproject accession PRJNA780792).
REFERENCES
- Afgan, E. , Baker, D. , Batut, B. , Van Den Beek, M. , Bouvier, D. , Ech, M. , Chilton, J. , Clements, D. , Coraor, N. , Grüning, B. A. , Guerler, A. , Hillman‐Jackson, J. , Hiltemann, S. , Jalili, V. , Rasche, H. , Soranzo, N. , Goecks, J. , Taylor, J. , Nekrutenko, A. , & Blankenberg, D. (2018). The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Research, 46, W537–W544. 10.1093/nar/gky379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andújar, C. , Arribas, P. , Gray, C. , Bruce, C. , Woodward, G. , Yu, D. W. , & Vogler, A. P. (2018). Metabarcoding of freshwater invertebrates to detect the effects of a pesticide spill. Molecular Ecology, 27, 146–166. 10.1111/mec.14410 [DOI] [PubMed] [Google Scholar]
- Apothéloz‐Perret‐Gentil, L. , Cordonier, A. , Straub, F. , Iseli, J. , Esling, P. , & Pawlowski, J. (2017). Taxonomy‐free molecular diatom index for high‐throughput eDNA biomonitoring. Molecular Ecology Resources, 17, 1231–1242. 10.1111/1755-0998.12668 [DOI] [PubMed] [Google Scholar]
- Armstrong, E. , & Verhoeven, J. P. (2020). Machine learning analyses of bacterial oligonucleotide frequencies to assess the benthic impact of aquaculture. Aquaculture Environment Interactions, 12, 131–137. 10.3354/aei00353 [DOI] [Google Scholar]
- Barmentlo, S. H. , Schrama, M. , van Bodegom, P. M. , de Snoo, G. R. , Musters, C. J. M. , & Vijver, M. G. (2019). Neonicotinoids and fertilizers jointly structure naturally assembled freshwater macroinvertebrate communities. Science of the Total Environment, 691, 36–44. 10.1016/j.scitotenv.2019.07.110 [DOI] [PubMed] [Google Scholar]
- Beentjes, K. K. , Speksnijder, A. G. C. L. , Schilthuizen, M. , Hoogeveen, M. , Pastoor, R. , & van der Hoorn, B. B. (2019). Increased performance of DNA metabarcoding of macroinvertebrates by taxonomic sorting. PLoS One, 14, e0226527. 10.1371/journal.pone.0226527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beentjes, K. K. , Speksnijder, A. G. C. L. , Schilthuizen, M. , Hoogeveen, M. , & van der Hoorn, B. B. (2019). The effects of spatial and temporal replicate sampling on eDNA metabarcoding. PeerJ, 7, e7335. 10.7717/peerj.7335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beentjes, K. K. , Speksnijder, A. G. C. L. , van der Hoorn, B. B. , & van Tol, J. (2015). DNA barcoding program at Naturalis Biodiversity Center, the Netherlands. Genome, 58, 193. 10.1139/gen-2015-0087 [DOI] [Google Scholar]
- Beermann, A. J. , Zizka, V. M. A. , Elbrecht, V. , Baranov, V. , & Leese, F. (2018). DNA metabarcoding reveals the complex and hidden responses of chironomids to multiple stressors. Environmental Sciences Europe, 30, 26. 10.1186/s12302-018-0157-x [DOI] [Google Scholar]
- Benson, D. A. , Karsch‐Mizrachi, I. , Lipman, D. J. , Ostell, J. , & Wheeler, D. L. (2005). GenBank. Nucleic Acids Research, 33, D34–D38. 10.1093/nar/gki063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bista, I. , Carvalho, G. R. , Walsh, K. , Seymour, M. , Hajibabaei, M. , Lallias, D. , Christmas, M. , & Creer, S. (2017). Annual time‐series analysis of aqueous eDNA reveals ecologically relevant dynamics of lake ecosystem biodiversity. Nature Communications, 8, 14087. 10.1038/ncomms14087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho, C. , Coulouris, G. , Avagyan, V. , Ma, N. , Papadopoulos, J. , Bealer, K. , & Madden, T. L. (2009). BLAST+: Architecture and applications. BMC Bioinformatics, 10, 421. 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho, L. , Poikane, S. , Lyche Solheim, A. , Phillips, G. , Borics, G. , Catalan, J. , De Hoyos, C. , Drakare, S. , Dudley, B. J. , Järvinen, M. , Laplace‐Treyture, C. , Maileht, K. , McDonald, C. , Mischke, U. , Moe, J. , Morabito, G. , Nõges, P. , Nõges, T. , Ott, I. , … Thackeray, S. J. (2013). Strength and uncertainty of phytoplankton metrics for assessing eutrophication impacts in lakes. Hydrobiologia, 704, 127–140. 10.1007/s10750-012-1344-1 [DOI] [Google Scholar]
- Chagnon, M. , Kreutzweiser, D. , Mitchell, E. A. D. , Morrissey, C. A. , Noome, D. A. , & Van Der Sluijs, J. P. (2015). Risks of large‐scale use of systemic insecticides to ecosystem functioning and services. Environmental Science and Pollution Research, 22, 119–134. 10.1007/s11356-014-3277-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chará‐Serna, A. M. , Epele, L. B. , Morrissey, C. A. , & Richardson, J. S. (2019). Nutrients and sediment modify the impacts of a neonicotinoid insecticide on freshwater community structure and ecosystem functioning. Science of the Total Environment, 692, 1291–1303. 10.1016/j.scitotenv.2019.06.301 [DOI] [PubMed] [Google Scholar]
- Chariton, A. A. , Ho, K. T. , Proestou, D. , Bik, H. , Simpson, S. L. , Portis, L. M. , Cantwell, M. G. , Baguley, J. G. , Burgess, R. M. , Pelletier, M. M. , Perron, M. , Gunsch, C. , & Matthews, R. A. (2014). A molecular‐based approach for examining responses of eukaryotes in microcosms to contaminant‐spiked estuarine sediments. Environmental Toxicology and Chemistry, 33, 359–369. 10.1002/etc.2450 [DOI] [PubMed] [Google Scholar]
- Clark, D. E. , Pilditch, C. A. , Pearman, J. K. , Ellis, J. I. , & Zaiko, A. (2020). Environmental DNA metabarcoding reveals estuarine benthic community response to nutrient enrichment – Evidence from an in‐situ experiment. Environmental Pollution, 267, 115472. 10.1016/j.envpol.2020.115472 [DOI] [PubMed] [Google Scholar]
- Cordier, T. , Forster, D. , Dufresne, Y. , Martins, C. I. , Stoeck, T. , & Pawlowski, J. (2018). Supervised machine learning outperforms taxonomy‐based environmental DNA metabarcoding applied to biomonitoring. Molecular Ecology Resources, 18(6), 1381–1391. 10.1111/1755-0998.12926 [DOI] [PubMed] [Google Scholar]
- Cordier, T. , Frontalini, F. , Cermakova, K. , Apothéloz‐Perret‐Gentil, L. , Treglia, M. , Scantamburlo, E. , Bonamin, V. , & Pawlowski, J. (2019). Multi‐marker eDNA metabarcoding survey to assess the environmental impact of three offshore gas platforms in the North Adriatic Sea (Italy). Marine Environmental Research, 146, 24–34. 10.1016/j.marenvres.2018.12.009 [DOI] [PubMed] [Google Scholar]
- Corvalan, C. , Hales, S. , & McMichael, A. (2005). Ecosystems and human well‐being: Health synthesis. Island Press. [Google Scholar]
- Côté, I. M. , Darling, E. S. , & Brown, C. J. (2016). Interactions among ecosystem stressors and their importance in conservation. Proceedings of the Royal Society B: Biological Sciences, 283, 1–9. 10.1098/rspb.2015.2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, J. M. , Rosemond, A. D. , Eggert, S. L. , Cross, W. F. , & Wallace, J. B. (2010). Long‐term nutrient enrichment decouples predator and prey production. Proceedings of the National Academy of Sciences of the United States of America, 107(1), 121–126. 10.1073/pnas.0908497107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cáceres, M. , & Legendre, P. (2009). Associations between species and groups of sites: Indices and statistical inference. Ecology, 90, 3566–3574. 10.1890/08-1823.1 [DOI] [PubMed] [Google Scholar]
- De Cáceres, M. , Legendre, P. , & Moretti, M. (2010). Improving indicator species analysis by combining groups of sites. Oikos, 119, 1674–1684. 10.1111/j.1600-0706.2010.18334.x [DOI] [Google Scholar]
- Donohue, I. , Donohue, L. A. , Ainín, B. N. , & Irvine, K. (2009). Assessment of eutrophication pressure on lakes using littoral invertebrates. Hydrobiologia, 633, 105–122. 10.1007/s10750-009-9868-8 [DOI] [Google Scholar]
- Dray, S. , & Dufour, A. (2007). The ade4 package: Implementing the duality diagram for ecologists. Journal of Statistical Software, 22, 1–20. 10.18637/jss.v022.i04 [DOI] [Google Scholar]
- Dudgeon, D. , Arthington, A. H. , Gessner, M. O. , Kawabata, Z.‐I. , Knowler, D. J. , Lévêque, C. , Naiman, R. J. , Prieur‐Richard, A.‐H. , Soto, D. , Stiassny, M. L. J. , & Sullivan, C. A. (2006). Freshwater biodiversity: Importance, threats, status and conservation challenges. Biological Reviews of the Cambridge Philosophical Society, 81, 163–182. 10.1017/S1464793105006950 [DOI] [PubMed] [Google Scholar]
- Haase, P. , Pauls, S. U. , Schindehütte, K. , & Sundermann, A. (2010). First audit of macroinvertebrate samples from an EU Water Framework Directive monitoring program: Human error greatly lowers precision of assessment results. Journal of the North American Benthological Society, 29(4), 1279–1291. 10.1899/09-183.1 [DOI] [Google Scholar]
- Hänfling, B. , Handley, L. L. , Read, D. S. , Hahn, C. , Li, J. , Nichols, P. , Blackman, R. C. , Oliver, A. , & Winfield, I. J. (2016). Environmental DNA metabarcoding of lake fish communities reflects long‐term data from established survey methods. Molecular Ecology, 25, 3101–3119. 10.1111/mec.13660 [DOI] [PubMed] [Google Scholar]
- Heisler, J. , Glibert, P. M. , Burkholder, J. M. , Anderson, D. M. , Cochlan, W. , Dennison, W. C. , Dortch, Q. , Gobler, C. J. , Heil, C. A. , Humphries, E. , Lewitus, A. , Magnien, R. , Marshall, H. G. , Sellner, K. , Stockwell, D. A. , Stoecker, D. K. , & Suddleson, M. (2008). Eutrophication and harmful algal blooms: A scientific consensus. Harmful Algae, 8, 3–13. 10.1016/j.hal.2008.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, F. C. (2008). Taxonomic sufficiency: The influence of taxonomic resolution on freshwater bioassessments using benthic macroinvertebrates. Environmental Reviews, 16, 45–69. 10.1139/A07-010 [DOI] [Google Scholar]
- Joshi, N. A. , & Fass, J. N. (2011) Sickle: A sliding‐window, adaptive, quality‐based trimming tool for FastQ files (Version 1.33). https://github.com/najoshi/sickle
- Keeley, N. , Wood, S. A. , & Pochon, X. (2018). Development and preliminary validation of a multi‐trophic metabarcoding biotic index for monitoring benthic organic enrichment. Ecological Indicators, 85, 1044–1057. 10.1016/j.ecolind.2017.11.014 [DOI] [Google Scholar]
- Klindworth, A. , Pruesse, E. , Schweer, T. , Peplies, J. , Quast, C. , Horn, M. , & Glöckner, F. O. (2013). Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next‐generation sequencing‐based diversity studies. Nucleic Acids Research, 41, e1. 10.1093/nar/gks808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer‐Jaesrich, M. , Köhler, H. R. , & Gerhardt, A. (2010). Assessing toxicity of the insecticide thiacloprid on chironomus riparius (Insecta: Diptera) using multiple end points. Archives of Environmental Contamination and Toxicology, 58, 963–972. 10.1007/s00244-009-9420-x [DOI] [PubMed] [Google Scholar]
- Laroche, O. , Wood, S. A. , Tremblay, L. A. , Ellis, J. I. , Lear, G. , & Pochon, X. (2018). A cross‐taxa study using environmental DNA/RNA metabarcoding to measure biological impacts of offshore oil and gas drilling and production operations. Marine Pollution Bulletin, 127, 97–107. 10.1016/j.marpolbul.2017.11.042 [DOI] [PubMed] [Google Scholar]
- Larsson, A. J. M. , Stanley, G. , Sinha, R. , Weissman, I. L. , & Sandberg, R. (2018). Computational correction of index switching in multiplexed sequencing libraries. Nature Methods, 15, 305–307. 10.1038/nmeth.4666 [DOI] [PubMed] [Google Scholar]
- Li, F. , Peng, Y. , Fang, W. , Altermatt, F. , Xie, Y. , Yang, J. , & Zhang, X. (2018). Application of environmental DNA metabarcoding for predicting anthropogenic pollution in rivers. Environmental Science and Technology, 52, 11708–11719. 10.1021/acs.est.8b03869 [DOI] [PubMed] [Google Scholar]
- Macher, J. N. , Salis, R. K. , Blakemore, K. S. , Tollrian, R. , Matthaei, C. D. , & Leese, F. (2016). Multiple‐stressor effects on stream invertebrates: DNA barcoding reveals contrasting responses of cryptic mayfly species. Ecological Indicators, 61, 159–169. 10.1016/j.ecolind.2015.08.024 [DOI] [Google Scholar]
- Macher, J. N. , Vivancos, A. , Piggott, J. J. , Centeno, F. C. , Matthaei, C. D. , & Leese, F. (2018). Comparison of environmental DNA and bulk‐sample metabarcoding using highly degenerate cytochrome c oxidase I primers. Molecular Ecology Resources, 18, 1456–1468. 10.1111/1755-0998.12940 [DOI] [PubMed] [Google Scholar]
- Magoč, T. , & Salzberg, S. L. (2011). FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics, 27, 2957–2963. 10.1093/bioinformatics/btr507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, M. (2011). Cutadapt removes adapter sequences from high‐throughput sequencing reads. EMBnet.journal, 17, 10. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Matson, P. A. , Parton, W. J. , Power, A. G. , & Swift, M. J. (1997). Agricultural intensification and ecosystem properties. Science, 277, 504–509. 10.1126/science.277.5325.504 [DOI] [PubMed] [Google Scholar]
- Morrissey, C. A. , Mineau, P. , Devries, J. H. , Sanchez‐Bayo, F. , Liess, M. , Cavallaro, M. C. , & Liber, K. (2015). Neonicotinoid contamination of global surface waters and associated risk to aquatic invertebrates: A review. Environment International, 74, 291–303. 10.1016/j.envint.2014.10.024 [DOI] [PubMed] [Google Scholar]
- Oksanen, J. , Blanchet, F. G. , Friendly, M. , Kindt, R. , Legendre, P. , McGlinn, D. , Minchin, P. R. , O’Hara, R. B. , Simpson, G. L. , Solymos, P. , Stevens, M. H. H. , Szoecs, E. , & Wagner, H. (2019) vegan: Community Ecology Package. R package version 2.5‐6.
- Paradis, E. , & Schliep, K. (2018). ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics, 35, 526–528. 10.1093/bioinformatics/bty633 [DOI] [PubMed] [Google Scholar]
- Pereira, A. S. , Dâmaso‐Rodrigues, M. L. , Amorim, A. , Daam, M. A. , & Cerejeira, M. J. (2018). Aquatic community structure in Mediterranean edge‐of‐field waterbodies as explained by environmental factors and the presence of pesticide mixtures. Ecotoxicology, 27, 661–674. 10.1007/s10646-018-1944-2 [DOI] [PubMed] [Google Scholar]
- Piggott, J. J. , Townsend, C. R. , & Matthaei, C. D. (2015). Reconceptualizing synergism and antagonism among multiple stressors. Ecology and Evolution, 5, 1538–1547. 10.1002/ece3.1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisa, L. W. , Amaral‐Rogers, V. , Belzunces, L. P. , Bonmatin, J. M. , Downs, C. A. , Goulson, D. , Kreutzweiser, D. P. , Krupke, C. , Liess, M. , Mcfield, M. , Morrissey, C. A. , Noome, D. A. , Settele, J. , Simon‐Delso, N. , Stark, J. D. , Van Der Sluijs, J. P. , Van Dyck, H. , & Wiemers, M. (2014). Effects of neonicotinoids and fipronil on non‐target invertebrates. Environmental Science and Pollution Research, 22, 68–102. 10.1007/s11356-014-3471-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pochon, X. , Wood, S. A. , Keeley, N. B. , Lejzerowicz, F. , Esling, P. , Drew, J. , & Pawlowski, J. (2015). Accurate assessment of the impact of salmon farming on benthic sediment enrichment using foraminiferal metabarcoding. Marine Pollution Bulletin, 100, 370–382. 10.1016/j.marpolbul.2015.08.022 [DOI] [PubMed] [Google Scholar]
- Quast, C. , Pruesse, E. , Yilmaz, P. , Gerken, J. , Schweer, T. , Yarza, P. , Peplies, J. , & Glöckner, F. O. (2013). The SILVA ribosomal RNA gene database project: Improved data processing and web‐based tools. Nucleic Acids Research, 41, D590–D596. 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raby, M. , Nowierski, M. , Perlov, D. , Zhao, X. , Hao, C. , Poirier, D. G. , & Sibley, P. K. (2018). Acute toxicity of 6 neonicotinoid insecticides to freshwater invertebrates. Environmental Toxicology and Chemistry, 37, 1430–1445. 10.1002/etc.4088 [DOI] [PubMed] [Google Scholar]
- Ratnasingham, S. , & Hebert, P. D. N. (2007). BOLD: The Barcode of Life Data System (www.barcodinglife.org). Molecular Ecology Notes, 7, 355–364. 10.1111/j.1471-8286.2006.01678.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rognes, T. , Flouri, T. , Nichols, B. , Quince, C. , & Mahé, F. (2016). VSEARCH: A versatile open source tool for metagenomics. PeerJ, 4, e2584. 10.7717/peerj.2584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez‐Bayo, F. , Goka, K. , & Hayasaka, D. (2016). Contamination of the aquatic environment with neonicotinoids and its implication for ecosystems. Frontiers in Environmental Science, 4, 71. 10.3389/fenvs.2016.00071 [DOI] [Google Scholar]
- Schmieder, R. , & Edwards, R. (2011). Quality control and preprocessing of metagenomic datasets. Bioinformatics, 27, 863–864. 10.1093/bioinformatics/btr026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrama, M. , Barmentlo, S. H. , Hunting, E. R. , van Logtestijn, R. S. P. , Vijver, M. G. , & van Bodegom, P. M. (2017). Pressure‐induced shifts in trophic linkages in a simplified aquatic food web. Frontiers in Environmental Science, 5, 75. 10.3389/fenvs.2017.00075 [DOI] [Google Scholar]
- Schreiner, V. C. , Szöcs, E. , Bhowmik, A. K. , Vijver, M. G. , & Schäfer, R. B. (2016). Pesticide mixtures in streams of several European countries and the USA. Science of the Total Environment, 573, 680–689. 10.1016/j.scitotenv.2016.08.163 [DOI] [PubMed] [Google Scholar]
- Schwarzenbach, R. P. , Escher, B. I. , Fenner, K. , Hofstetter, T. B. , Johnson, C. A. , Von Gunten, U. , & Wehrli, B. (2006). The challenge of micropollutants in aquatic systems. Science, 313, 1072–1077. 10.1126/science.1127291 [DOI] [PubMed] [Google Scholar]
- Seymour, J. R. , Amin, S. A. , Raina, J. B. , & Stocker, R. (2017). Zooming in on the phycosphere: The ecological interface for phytoplankton‐bacteria relationships. Nature Microbiology, 2, 17065. 10.1038/nmicrobiol.2017.65 [DOI] [PubMed] [Google Scholar]
- Shaw, J. L. A. , Clarke, L. J. , Wedderburn, S. D. , Barnes, T. C. , Weyrich, L. S. , & Cooper, A. (2016). Comparison of environmental DNA metabarcoding and conventional fish survey methods in a river system. Biological Conservation, 197, 131–138. 10.1016/j.biocon.2016.03.010 [DOI] [Google Scholar]
- Stoeck, T. , Frühe, L. , Forster, D. , Cordier, T. , Martins, C. I. M. , & Pawlowski, J. (2018). Environmental DNA metabarcoding of benthic bacterial communities indicates the benthic footprint of salmon aquaculture. Marine Pollution Bulletin, 127, 139–149. 10.1016/j.marpolbul.2017.11.065 [DOI] [PubMed] [Google Scholar]
- Suikkanen, S. , Pulina, S. , Engström‐Öst, J. , Lehtiniemi, M. , Lehtinen, S. , & Brutemark, A. (2013). Climate change and eutrophication induced shifts in northern summer plankton communities. PLoS One, 8, e66475. 10.1371/journal.pone.0066475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumon, K. A. , Ritika, A. K. , Peeters, E. T. H. M. , Rashid, H. , Bosma, R. H. , Rahman, M. S. , Fatema, M. K. , & Van den Brink, P. J. (2018). Effects of imidacloprid on the ecology of sub‐tropical freshwater microcosms. Environmental Pollution, 236, 432–441. 10.1016/j.envpol.2018.01.102 [DOI] [PubMed] [Google Scholar]
- Taberlet, P. , Coissac, E. , Hajibabaei, M. , & Rieseberg, L. H. (2012). Environmental DNA. Molecular Ecology, 21, 1789–1793. 10.1111/j.1365-294X.2012.05542.x [DOI] [PubMed] [Google Scholar]
- Turner, C. R. , Miller, D. J. , Coyne, K. J. , & Corush, J. (2014) Improved methods for capture, extraction, and quantitative assay of environmental DNA from Asian bigheaded carp (Hypophthalmichthys spp.). PLoS One, 9, e114329. 10.1371/journal.pone.0114329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dijk, T. C. , Van Staalduinen, M. A. , & Van der Sluijs, J. P. (2013). Macro‐invertebrate decline in surface water polluted with imidacloprid. PLoS One, 8, e62374. 10.1371/journal.pone.0062374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijver, M. G. , Van ‘T Zelfde, M. , Tamis, W. L. M. , Musters, K. J. M. , & De Snoo, G. R. (2008). Spatial and temporal analysis of pesticides concentrations in surface water: Pesticides atlas. Journal of Environmental Science and Health ‐ Part B Pesticides, Food Contaminants, and Agricultural Wastes, 43, 665–674. 10.1080/03601230802388728 [DOI] [PubMed] [Google Scholar]
- Williams, N. , & Sweetman, J. (2019). Effects of neonicotinoids on the emergence and composition of chironomids in the Prairie Pothole Region. Environmental Science and Pollution Research, 26, 3862–3868. 10.1007/s11356-018-3683-6 [DOI] [PubMed] [Google Scholar]
- WWF (2014). The Living Planet Report 2014. 10.1016/j.edurev.2013.01.001 [DOI]
- Yamamuro, M. , Komuro, T. , Kamiya, H. , Kato, T. , Hasegawa, H. , & Kameda, Y. (2019). Neonicotinoids disrupt aquatic food webs and decrease fishery yields. Science, 366, 620–623. 10.1126/science.aax3442 [DOI] [PubMed] [Google Scholar]
- Zimmermann, J. , Jahn, R. , & Gemeinholzer, B. (2011). Barcoding diatoms: Evaluation of the V4 subregion on the 18S rRNA gene, including new primers and protocols. Organisms Diversity and Evolution, 11, 173–192. 10.1007/s13127-011-0050-6 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Supplementary Material
Supplementary Material
Data Availability Statement
Raw sequence data has been made available from the NCBI Sequence Read Archive (Bioproject accession PRJNA780792).


