Abstract
We investigated antibiotic resistance of staphylococci isolated from 1128 samples of high-circulating RTE foods in Taiwan. A total of 111 Staphylococcus aureus and 709 coagulase-negative staphylococci (CoNS) comprising 23 species were isolated. The prevalence of S. aureus differed in various category of RTE foods, highest in fresh-cut fruits/vegetables (20.5%) and lowest in low-water activity (LWA) foods (0.7%). The overall staphylococcal contamination was highest in fresh-cut fruits/vegetables (62.2%), in which multiple isolates (up to 10) or species (up to 6) in single sample were frequently found. Distinct distribution of species contributed to unique feature in each category. Prevalence of antibiotic-resistant S. aureus was higher in fresh-cut fruits/vegetables samples (14.2% in 127) compared to other food categories (0–7.1%). A total of 4 MRSA carrying SCCmec type IV or VT were identified (3.6% in 111), in which 3 belonged to sequence type ST59 and one was ST5. Among CoNS, S. epidermidis and S. warneri exhibited higher non-intrinsic antibiotic resistance than other species. Of 41 methicillin-resistant CoNS (5.8% in 709) isolates, SCCmec type IV (n = 16) and type VT (n = 6) were most frequent. Isolates of S. saprophyticus, S. xylosus and S. sciuri displayed high rates of resistance to fusidic acid. Novel fusB-family determinants were identified in S. xylosus, S. sciuri and S. kloosii, which may contribute to their intrinsic resistance to fusidic acid. Compared to other food categories, fresh-cut fruits/vegetables were more contaminated by staphylococci carrying non-intrinsic resistance determinants including methicillin resistance. This nation-wide study demonstrated that some categories may have potential risk for transmitting antibiotic resistance, in which S. epidermidis and S. warneri should be gotten more attention.
Keywords: Antibiotic resistance determinants, Methicillin-resistant staphylococci, Ready-to-eat foods, Staphylococcal community
1. Introduction
Staphylococci are commonly distributed in the environment, animals and humans. Staphylococcus aureus is known as a food-borne pathogen producing heat-stable enterotoxins that cause food poisoning symptoms [1]. It displays high pathogenicity and often develops multiple drug resistance in clinical settings [2]. Coagulase-negative staphylococci (CoNS) are the major commensal microbes of human skin. Some of the CoNS species are recognized as opportunistic pathogens [3], and some are used as starters in meat fermentation or other food processing [4]. Rare cases of food poisoning have been caused by CoNS species, but CoNS may serve as reservoirs of antibiotic resistance genes for S. aureus [5]. Until now, many measures have been applied to avoid transmission of S. aureus in clinical settings, food processing and the livestock industry. Several official programs of long-term systemic surveillance have been implemented to monitor the prevalence of antibiotic resistance from farm to table [6,7], and most of them focus on food-borne pathogens such as S. aureus. The risks of CoNS in food safety have not been taken as seriously.
It was found that food-isolated S. aureus and CoNS carrying mobile elements such as the staphylococcal cassette chromosome mec (SCCmec) conferring methicillin resistance and reduced susceptibility to multiple antimicrobial agents [8], suggesting that the food production chain could provide a substantial transmission route for resistance genes [9]. Apart from the studies focused on livestock and animal-based products that reflect the need to improve policies regarding animal drug usage, an increasing number of reports on ready-to-eat (RTE) foods directly demonstrate the risks of food contamination [10]. Previous studies about staphylococci in RTE foods mainly focused on animal-based foods such as products of meat and dairy [10,11]. Other RTE categories were also investigated in some studies [12,13], most of which concentrated on characteristics of S. aureus.
Discussion of the role of staphylococci in food safety is ongoing. This study was aimed not only animal-based products but also other RTE food categories with high consumption in Taiwan, including fresh-cut fruits/vegetables, ice desserts, rice products and low-water activity (LWA) foods, and each RTE food category had over 100 samples for analysis. By the full view of staphylococcal species found in highly consumed RTE foods in Taiwan, we tried to discover the unique characteristics of staphylococcal species with antibiotic resistance genes in various RTE food categories to reveal the potential risks and to further explore the feasible strategies to enhance food safety.
2. Methods
2.1. Sample collection, isolation and identification of staphylococci
In consideration of the consumption habits and wide acceptance of RTE food products in Taiwan, only the most popular, high-circulating RTE food categories were collected. A total of 1128 samples were collected from convenience stores, supermarkets, hypermarkets, traditional markets, delis, restaurants and other food-selling sites in 16 densely populated cities and counties in Taiwan from February to November in 2014, including animal-based products (n = 492), fresh-cut fruits/vegetables (without dressing) (n = 127), ice desserts (n = 187), rice products (n = 183) and LWA foods (n = 139) (Supplementary Table 1). Food sampling, preparation of sample homogenate and detection of staphylococci were followed by the U.S. Food and Drug Administration’s Bacteriological Analytical Manual (BAM-FDA) [14]. Species identification was conducted by Matrix-Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry (MALDI-TOF MS) (Biotyper, Bruker Daltonics, Bremen, Germany) and/or dnaJ genes PCR-RFLP or 16S rRNA gene sequencing [15].
2.2. Antimicrobial susceptibility testing
Antimicrobial susceptibility testing (AST) was initially performed by VITEK 2 Compact (Biomérieux, Marcy l’Etoile, France) using AST card P592 and double-checked by the disk diffusion method. The minimal inhibitory concentrations (MICs) for antimicrobial drugs were determined by agar dilution methods described by the Clinical and Laboratory Standards Institute (CLSI) [16]. The breakpoints used to indicate antibiotic resistance were according to the guidelines of the CLSI except fusidic acid. Isolates displaying MIC >1 μg/mL for fusidic acid were considered resistant as recommended by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) [17]. S. aureus ATCC 29213 was used as the reference strain.
2.3. Detection of antimicrobial resistance genes by PCR
Presence of resistance genes in staphylococci was detected by PCR. Bacterial DNA was extracted using QIAGEN DNeasy Blood and Tissue Kits (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions. The resistance determinants included those to penicillin (blaZ) [18], tetracycline (tetK, tetL, tetM, tetO, tetS, tetW) [19], oxacillin (mecA) [18], erythromycin (ermA, ermB, ermC, msrA/B, ermT) [20], aminoglycoside (aacA-aphD) [21], fusidic acid (fusB, fusC, fusD, fusF) [22] and trimethoprim (dfrA, dfrD, dfrG, dfrK) [18]. Staphylococcus strains with known antimicrobial resistance genes previously isolated in our laboratory were used as positive controls.
2.4. Molecular typing of S. aureus
The spa repeat region was amplified and sequenced using Sa0122-F and Sa0122-R [23]. Multilocus sequence typing (MLST) was conducted to determine the sequence types (STs) of S. aureus isolates. STs were assigned using the S. aureus MLST database (www.mlst.net) [24].
2.5. Identification of SCCmec in methicillin-resistant staphylococcal isolates
Multiplex PCR of SCCmec was applied for methicillin-resistant S. aureus [25]. SCCmec types in methicillin-resistant CoNS were determined by mec and ccr gene complexes described in the previous study [26].
3. Results
3.1. Staphylococcal contamination in each RTE food category
From 1128 RTE food samples, a total of 111 S. aureus (9.8%) and 709 CoNS comprising 23 species were isolated (Supplementary Table 2). The prevalence of S. aureus was highest in fresh-cut fruits/vegetables (20.5%, 26/127) followed by rice products (11.5%, 21/183), animal-based products (10.0%, 49/492), ice desserts (7.5%, 14/187) and LWA foods (0.7%, 1/139). The overall staphylococcal contamination was highest in fresh-cut fruits/vegetables (62.2% in 127), relatively low (10.8% in 139) in LWA foods and 38.0–45.9% in others (Table 1). Two or more (up to 10) isolates may be identified in each sample, which was relatively prominent in fresh-cut fruits/vegetables samples (31.5%) compared to other categories (2.9–21.3%). Contamination of multiple (3 or more, up to 6) species in single sample was obviously higher in fresh-cut fruits/vegetables (15.7% in 127) compared to others (0.7–8.7%). These data presented characteristics of staphylococcal contamination in each food category. Polymicrobial or multispecies contamination implied high degree of complexity in composition of staphylococcal community in RTE foods.
Table 1.
Characteristics of staphylococcal contamination on each RTE food category.
| Food categories | No. of samples | Staphylococcal contamination rate % (no. of samples) | Rate of samples containing 2 or more isolates % (no. of samples) | Percentage of samples containing 3 or more species % (no. of samples) | Top 3 CoNS species (prevalence) |
|---|---|---|---|---|---|
| Animal-based products | 492 | 42.5 (209) | 16.1 (79) | 3.7 (18) |
S. saprophyticus (25.4%) S. sciuri (9.6%) S. xylosus (7.9%) |
| Fresh-cut fruits/vegetables | 127 | 62.2 (79) | 31.5 (40) | 15.7 (20) |
S. warneri (29.9%) S. epidermidis (21.3%) S. saprophyticus (15.0%) |
| Ice desserts | 187 | 38.0 (71) | 12.3 (23) | 3.2 (6) |
S. warneri (21.4%) S. epidermidus (9.1%) S. saprophyticus (8.6%) |
| Rice products | 183 | 45.9 (84) | 21.3 (39) | 8.7 (16) |
S. saprophyticus (20.2%) S. warneri (12.0%) S. epidermidis (11.5%) |
| Low water activity foods | 139 | 10.8 (15) | 2.9 (4) | 0.7 (1) |
S. epidermidis (4.3%) S. saprophyticus (3.6%) S. captis (2.2%) |
| Total | 1128 | 40.6 (458) | 16.4 (185) | 5.4 (61) |
S. saprophyticus (17.9%) S. warneri (10.0%) S. epidermidis (8.1%) |
The prevalence of staphylococcal species was distinct in RTE foods (Table 1) (Supplementary Table 2). Except LWA foods, more than 10 species were identified in each RTE foods, in which S. saprophyticus, S. warneri, S. epidermidis, S. xylosus and S. carnosus appeared in all categories. The overall top 5 CoNS species were S. saprophyticus (17.9%), followed by S. warneri (10.0%), S. epidermidis (8.1%), S. sciuri (7.4%) and S. xylosus (7.4%) (Supplementary Table 2). S. saprophyticus was dominant in animal-based products (25.4%) and rice products (20.2%) but less frequent in ice products (8.6%). Conversely, S. epidermidis contamination was significant in fresh-cut fruits/vegetables (21.3%) but uncommon in animal-based products (4.1%). S. warneri was prominent in fresh-cut fruits/vegetables (29.9%) and ice desserts (21.4%) but rare in animal-based products (2.4%).
3.2. Antibiotic resistance in S. aureus
Among 111 S. aureus isolates, 65 (58.6%) were resistant to at least one antibiotic, of which 15 (13.5%) were multidrug resistant (resistant to 3 or more antibiotic class) (Table 2) (Supplementary Table 3). Penicillin resistance was common in S. aureus (55.0%), followed by tetracycline (31.5%). Resistance to erythromycin (10.8%), gentamicin (9.0%), oxacillin (3.6%), fusidic acid (1.8%) and trimethoprim/sulfamethoxazole (0.9%) was relatively low. MRSA isolates were identified from animal-based products (n = 2) and fresh-cut fruits/vegetables (n = 2) (Table 3). No vancomycin resistance was found.
Table 2.
Antibiotic resistance and major determinants in staphylococci from RTE foods.
| Resistance rates in Staphylococcus speciesb (Major determinant)c | |||||||
|---|---|---|---|---|---|---|---|
|
|
|||||||
| S. aureus (n = 111) | S. saprophyticus (n = 202) | S. warneri (n = 113) | S. epidermidis (n = 91) | S. sciuri (n = 84) | S. xylosus (n = 83) | S. pasteuri (n = 41) | |
| Antibiotics (%) | |||||||
| Oxacillin | 3.6 (mecA) | 5.0 (mecA) | 3.5 (mecA) | 23.1 (mecA) | 2.4 (mecA) | 0 | 2.4 (mecA) |
| Penicillin | 55.0 (blaZ) | 5.9 (blaZ) | 67.3 (blaZ) | 78.0 (blaZ) | 2.4 (N.D.)d | 3.6 (blaZ) | 61.0 (blaZ) |
| Tetracycline | 31.5 (tetK) | 22.8 (tetK) | 18.6 (tetK) | 24.2 (tetK) | 15.5 (tetK) | 27.7 (tetK) | 17.1 (tetK) |
| Erythromycin | 10.8 (msrA/B) | 6.9 (msrA/B) | 26.5 (msrA/B) | 31.9 (msrA/B) | 1.2 (ermA) | 4.8 (msrA/B) | 17.1 (msrA/B) |
| Gentamicin | 9.0 (aacA-aphD) | 0 | 20.4 (aacA-aphD) | 20.9 (aacA-aphD) | 0 | 1.2 (aacA-aphD) | 7.3 (aacA-aphD) |
| Fusidic acid | 1.8 (fusC) | 93.6 (fusD) | 0.9 (fusB) | 14.3 (fusB) | 98.8 (novel) | 78.3 (novel) | 0 |
| Trimethoprim/sulfamethoxazole | 0.9 (dfrG) | 3.5 (dfrG) | 0 | 12.1 (dfrA) | 1.2 (dfrG) | 4.8 (dfrG) | 0 |
| Rate of resistant to at least one antibiotic (%) | 58.6 | 96.5 | 87.6 | 84.6 | 100.0 | 84.3 | 61.0 |
| Multidrug resistant ratea (%) | 13.5 | 5.4 | 9.7 | 26.4 | 2.4 | 6.0 | 4.9 |
Isolates resistant to 3 or more antibiotic class.
Only CoNS species with > 10% of prevalence in any of RTE food categories were listed.
The details of antibiotic resistance determinants were listed in Supplementary Table 4.
Two isolates displayed resistant to both penicillin and oxacillin, but only mecA was found.
Table 3.
Characteristics of MRSA isolates.
| Isolate | Source | MICs of oxacillin (μg/mL) | Molecular typing | Antibiotic resistancea | |||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| STb | spa | SCCmec type | Profiles | Genes | |||
| M-01 | Roasted pork | 128 | ST59 | t437 | IV | OX-P | mecA, blaZ |
| #163 | Braised dishes | 256 | ST5 | t002 | IV | OX-P-TE-E-SXT | mecA, blaZ, tetM, ermC, dfrG |
| S-125 | Vegetable salad | 32 | ST59 | t437 | VT | OX-P-TE-E | mecA, blaZ, tetK, ermB |
| #181 | Vegetable salad | 128 | ST59 | t437 | IV | OX-P-E-CN | mecA, blaZ, ermB, aacA-aphD |
OX, oxacillin; P, penicillin; TE, tetracycline; E, erythromycin; CN, gentamicin; SXT, trimethoprim/sulfamethoxazole.
Sequence type.
Fresh-cut fruits/vegetables samples had slightly higher contamination rate of antibiotic-resistant S. aureus (14.2%, 18/127) followed by rice products (7.1%, 13/183), ice desserts (6.4%, 12/187) and animal-based products (4.5%, 22/492). The rate of multidrug-resistant S. aureus was also higher in fresh-cut fruits/vegetables (3.9%, 5/127) compared to rice products (2.2%, 4/183), animal-based products (1.0%, 5/492) and ice desserts (0.5%, 1/187). According to these data, we suggested greater risks of fresh-cut fruits/vegetables in transmitting antibiotic-resistant S. aureus compared to other food categories.
3.3. Antibiotic resistance in CoNS
Except for S. condimenti, S. vitulinus and S. succinus, 599 isolates of 20 species were found resistant to at least one antibiotic (84.5%, 599/709), in which 65 isolates (9.2%, 65/709) of 11 CoNS species displaying multidrug resistance. Resistance to fusidic acid was the most common (51.5%), followed by penicillin (31.3%), tetracycline (20.3%) and erythromycin (14.7%). Resistance to gentamicin (7.2%), oxacillin (5.8%) and trimethoprim/sulfamethoxazole (3.2%) was relatively low. MR-S. epidermidis was prominent (n = 21) followed by S. saprophyticus (n = 10), S. warneri (n = 4), S. sciuri (n = 2), S. cohnii (n = 2), S. haemolyticus (n = 1) and S. pasteuri (n = 1) (Table 4). MR-S. epidermidis had a broad range of susceptibility (2–128 μg/mL), but other species displayed moderate or high resistance to oxacillin (64–>256 μg/mL). No vancomycin-resistant CoNS were found. Among species contributing to the major staphylococcal community (over 10% of prevalence in any of RTE food categories), all S. sciuri, over 90% of S. saprophyticus and over half portion of S. warneri, S. epidermidis, S. xylosus and S. pasteuri isolates displayed antibiotic resistant, in which S. epidermidis (26.4%) had significantly high prevalence of multidrug resistance (Table 2). High rates of resistance to fusidic acid were found in S. sciuri (98.8%), S. saprophyticus (93.6%) and S. xylosus (78.3%). Penicillin resistance was common in S. epidermidis (78.0%), S. warneri (67.3%), and S. pasteuri (61.0%). Tetracycline resistance was frequent in S. xylosus (27.7%), S. epidermidis (24.2%) and S. saprophyticus (22.8%). Erythromycin and gentamicin were frequently found in S. epidermidis (31.9%, 20.9%) and S. warneri (26.5%, 20.4%) compared to other species. Few isolates displayed trimethoprim/sulfamethoxazole (0–12.1%), which were highest in S. epidermidis. Moreover, MR-CoNS appeared in various RTE foods, whereas the prevalence was higher in fresh-cut fruits/vegetables (11.8%, 15/127) compared to rice products (6.6%, 12/183), ice desserts (4.8%, 9/187) and animal-based products (1.0%, 5/492) (Table 4). Our data demonstrated that each species had unique patterns of antibiotic resistance. Of them, S. epidermidis and S. warneri exhibited higher resistance rates of various antibiotic resistance.
Table 4.
Profile of MR-CoNS isolates.
| Species (no. of isolates) | MIC (μg/mL) | mec Complex class | ccr Gene | SCCmec type | No. of isolates | Food source (No. of isolate) |
|---|---|---|---|---|---|---|
| S. epidermidis (21) | 2–128 | A | ccrA4B4 | VIII | 1 | Ice desserts (1) |
| B | ccrA2B2 | IV | 13 | Animal-based products (2) | ||
| Fresh-cut fruits/vegetables (5) | ||||||
| Ice desserts (3) | ||||||
| Rice products (3) | ||||||
| C2 | ccrC1 | VT | 4 | Fresh-cut fruits/vegetables (1) | ||
| Ice desserts (1) | ||||||
| Rice products (2) | ||||||
| C2 | ccrC1, ccrA2B2 | VT | 1 | Fresh-cut fruits/vegetables (1) | ||
| A | NTa | NT | 2 | Fresh-cut fruits/vegetables (1) | ||
| Ice desserts (1) | ||||||
| S. saprophyticus (10) | 64–>256 | A | ccrA3B3 | III | 3 | Animal-based products (1) |
| Fresh-cut fruits/vegetables (1) | ||||||
| Rice products (1) | ||||||
| A | ccrA1B1 | NT | 1 | Animal-based products (1) | ||
| NT | ccrC1 | NT | 2 | Fresh-cut fruits/vegetables (1) | ||
| Rice products (1) | ||||||
| A | NT | NT | 4 | Fresh-cut fruits/vegetables (2) | ||
| Rice products (2) | ||||||
| S. warneri (4) | 64–256 | B | ccrA2B2 | IV | 2 | Ice desserts (1) |
| Rice products (1) | ||||||
| C2 | ccrA1B1 | IX | 2 | Fresh-cut fruits/vegetables (1) | ||
| Rice products (1) | ||||||
| S. sciuri (2) | >256 | A | NT | NT | 2 | Animal-based products (1) |
| Fresh-cut fruits/vegetables (1) | ||||||
| S. cohnii (2) | 128 | A | NT | NT | 2 | Ice desserts (1) |
| Rice products (1) | ||||||
| S. haemolyticus (1) | 128 | C2 | ccrC1 | VT | 1 | Fresh-cut fruits/vegetables (1) |
| S. pasteuri (1) | 64 | B | ccrA2B2 | IV | 1 | Ice desserts (1) |
| Total (41) | 2–>256 | – | – | III | 3 | Animal-based products (5) |
| IV | 16 | Fresh-cut fruits/vegetables (15) | ||||
| VT | 6 | Ice products (9) | ||||
| VIII | 1 | Rice products (12) | ||||
| IX | 2 | |||||
| NT | 13 |
NT, untypable.
3.4. Antibiotic resistance determinants
The blaZ was detected in most penicillin-resistant staphylococcal isolates (272/283) (Supplementary Table 4). All 45 oxacillin-resistant isolates (4 MRSA and 41 MR-CoNS) carried mecA. Isolates resistant to gentamicin (n = 61) carried aacA-aphD. The genes tetK, tetL and tetM were identified in tetracycline resistant isolates, with tetK (157/179) the most frequent. The msrA/B was found in large portion of erythromycin-resistant isolates (92/116), and the ermA, ermB, ermC and ermT were also found in few isolates. The dfrG (18/24) and dfrA (11/24) were identified in isolates resistant to trimethoprim/sulfamethoxazole.
However, distribution of resistance determinants was diverse among staphylococcal species (Supplementary Table 4). Compared to other species, S. epidermidis more frequently carried msrA/B, mecA, aacA-aphD, fusB and dfrA. S. warneri and S. aureus frequently harbored msrA/B and aacA-aphD as well.
A total of 367 isolates were resistant to fusidic acid. But only S. aureus, S. epidermidis, S. lugdunensis and S. warneri carried acquired (non-intrinsic) resistant gene fusB or fusC. S. saprophyticus and S. cohnii were known as intrinsically resistant to fusidic acid due to the presence of fusD and fusF, respectively. Most S. sciuri (83/84) and S. xylosus (65/83) and all S. kloosii (n = 5) isolates displayed low-level resistance but lacked known fusB family genes (fusB, fusC, fusD or fusF), which was also seen in previous studies [11,27,28]. For this, we conducted PCR amplification using a pair of degenerate primers of fusB family conserved region designed by our laboratory [22] and obtained PCR products from S. xylosus and S. kloosii isolates. Sequence of the above products matched (92–97% identity) to the gene encoding fibronectin-binding protein in S. xylosus (Sx-FBP, GenBank: CP007208.1) and the gene encoding elongation factor G-binding protein in S. kloosii (Sk-EFGBP, GenBank: CP027846.1), respectively. The genes show 75% and 67% amino acid similarity to FusD (GenBank: AP008934.1) (Supplementary Table 5). From the published genome sequence of S. sciuri (GenBank: CP022046.2), we found the gene encoding elongation factor G-binding protein S. sciuri (Sc-EFGBP) displaying 46% amino acid similarity to FusC (GenBank: NC_002953.3). Conserved domains such as the cysteines residues were shared among Sx-FBP, Sk-EFGBP, Sc-EFGBP and known FusB family protein [29] (Supplementary Fig. 1). From the above data, we suggested that these novel determinants from S. xylosus, S. kloosii, and S. sciuri could confer intrinsic resistance of fusidic acid, but may not contribute to the risks of transferring resistance.
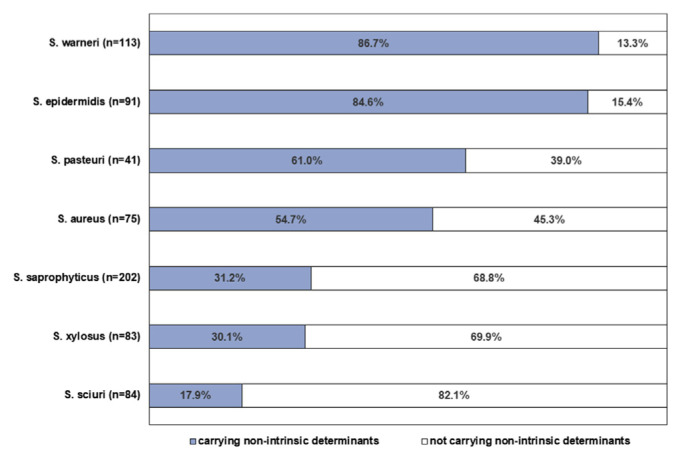
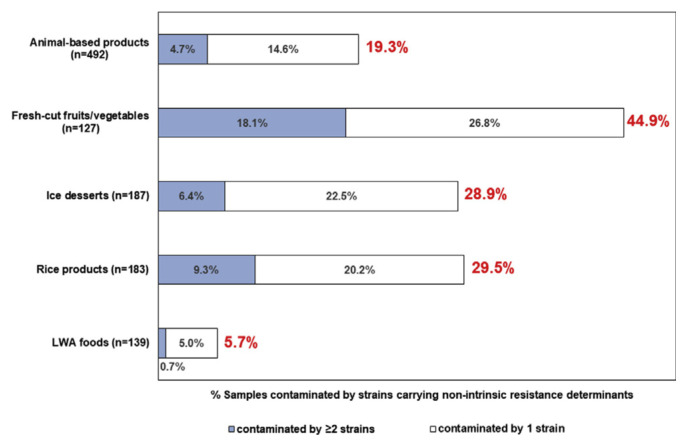
Ratio of non-intrinsic determinants in each species was listed in Fig. 1. The non-intrinsic determinants were carried highest in S. warneri (86.7%), followed by S. epidermidis (84.6%), S. pasteuri (61.0%) and S. aureus (54.7%). Owing to unique features of staphylococcal community in each food category, we suggested that the food category with high prevalence of S. warneri, S. epidermidis, S. pasteuri and S. aureus could be riskier in transmitting antibiotic resistance. Therefore, we analyzed prevalence of non-intrinsic determinants by food category. Non-intrinsic determinants were found higher in fresh-cut fruits/vegetables (44.9%), rice products (29.5%) and ice desserts (28.9%) than other categories (Fig. 2). In addition, the ratio of samples contaminated by 2 or more staphylococci carrying non-intrinsic determinants was significantly higher in fresh-cut fruits/vegetables (18.1%).
Fig. 1.
Percentage of isolates carrying (dark bar) or not carrying (light bar) non-intrinsic resistance determinants in the species contributing the majority of staphylococcal community in RTE foods (species had > 10% of prevalence in any of RTE food categories).
Fig. 2.
Percentage of samples contaminated staphylococci carrying non-intrinsic resistance determinants in each food category, including samples contaminated by 2 or more stains (dark bar) or by only 1 strain (light bar) carrying non-intrinsic resistance determinants.
3.5. Molecular profiles of methicillin-resistant staphylococci
The molecular profiles of MRSA were listed in Table 3. Three MRSA carried SCCmec type IV and displayed high MIC to oxacillin (MIC: 128–256 μg/mL), belonging to ST59/t437 (n = 2) or ST5/t002 (n = 1). One carried SCCmec type VT (MIC: 32 μg/mL) and belonged to ST59/t437.
Among 41 MR-CoNS, SCCmec types were successfully identified in 28 isolates, in which SCCmec type IV was most prominent (n = 16) followed by type VT (n = 6), III (n = 3), IX (n = 2) and VIII (1). S. epidermidis harboring SCCmec type IV was significantly dominant (13/41). SCCmec types were not identified in most MR-S. saprophyticus (7/10) and all of MR-S. sciuri (n = 2) and MR-S. cohnii (n = 2) isolates (Table 4).
4. Discussion
In the present study, we conducted nation-wide and systemic surveillance of RTE foods in Taiwan, focusing on the incidence of staphylococcal isolates carrying non-intrinsic resistance determinants. Compared to previous studies in Taiwan [26,30], sample size (1128 samples, over 100 samples for each food category) and sample volume (50 g for each food sample) in the present study were greater and food items were more diverse. Our data successfully demonstrated that composition of different staphylococcal species in various RTE food categories contributed to diverse risks of potentially transmitting antibiotic resistance. Considering transmission of antibiotic resistance is getting more attentions from the public, we believe our findings would be helpful to food safety assessment.
Critical points of RTE food safety are mainly determined by sanitary quality of ingredients, the way of food processing and hygiene during transport and marketing. Factors such as water activity largely determine microbial survival, explaining lower rates of microbial contamination found in LWA foods compared to other categories. Unlike other countries [10,11], popular animal-based RTE products in Taiwan are usually well-cooked or sold at high temperatures, which could confine the composition of staphylococcal community. On the contrary, fresh-cut fruits/vegetables are composed of multiple, uncooked ingredients from various sources and processed by frequent hand contact, indicating high risk of being contaminated by microbes from human and environmental sources. Some reports indicated that S. aureus was frequently isolated from salad vegetables, fruits and sprouts [31,32]. We have tested several cleaning methods usually utilized by retailers and housewives. For leafy vegetables and sprouts, only a limited bacterial volume of S. aureus was removed (data not shown). In the present study, we found that fresh-cut fruits/vegetables were significant in staphylococcal contamination by S. aureus, multispecies, methicillin-resistant or other kinds of antibiotic-resistant staphylococci. The species frequently carrying various non-intrinsic antibiotic resistance such as S. warneri and S. epidermidis composed the majority of staphylococcal community in fresh-cut fruits/vegetables. These data indicated fresh-cut vegetables/fruits had higher risk in transmitting antibiotic resistance. To improve sanitation of fresh-cut fruits/vegetables, it is need to call for better control in the selection of raw materials, storage and processing steps. Furthermore, we recommend conducting routine monitoring of RTE foods to track the level of staphylococcal species with high potential for antibiotic resistance to enhance food safety.
Compared to S. aureus, the main hazards from food-related CoNS were considered limited to the presence of antibiotic resistance owing to their significantly lower toxigenic potentials [3]. Previous studies showed that the incidence and the kinds of antibiotic resistance were usually species-specific but not source-dependent [11]. Among CoNS, S. epidermidis is known as frequently isolated in infection cases and has ability to transmit resistance determinants to S. aureus [33]. In agreement with previous studies, we found S. epidermidis from RTE foods significantly displayed resistance to various antibiotics and carrying corresponding determinants. Furthermore, S. warneri was commonly isolated from various RTE foods, and its rate of non-intrinsic antibiotic resistance was as high as that of S. epidermidis. S. warneri is phenotypically similar to S. pasteuri [34] belonging to S. epidermidis-like group based on phylogenetic studies [3]. It frequently appeared in foods and human skin, animals, environments [35,36]. The ability of S. warneri to produce biofilm and enterotoxin has been reported [37]. Multidrug resistance has been reported in S. warneri isolated from many sources, but little was known about its genomic structure conferring resistance [38]. More studies are needed to explore the characteristics of S. warneri concerning antibiotic resistance and its correlation with food safety.
Molecular typing of four MRSA in the present study revealed three ST59 and one ST5. ST59/SCCmec type IV or VT are the major community-acquired MRSA in Taiwan, whereas ST5 is frequently found in hospital-acquired MRSA in many Asia countries [39]. MRSA is the major concern of public health. The MRSA contamination rate in RTE foods in the present study was 3.6% (4/111), similar to the data from China [13], Korea [32], Malaysia [40] and Japan [12]. CoNS is recognized as the origin and reservoirs of SCCmec. Our data showed that SCCmec types IV and VT were also identified in MR-CoNS. Therefore, we believe that continuous surveillance of both MRSA and MR-CoNS from RTE foods is important.
5. Conclusion
This study provides the systemic profile of prevalence and antibiotic resistance of staphylococci in various RTE foods popular in Taiwan. Composition of the staphylococci community in RTE foods was category-specific. Among them, fresh-cut fruits/vegetable were more contaminated by staphylococci carrying non-intrinsic resistance determinants including methicillin resistance. Therefore, it was recommended to routinely trace resistance characteristics of Staphylococcus species in RTE foods to make a complete assessment of the impacts of staphylococci on RTE food safety to review hazards and critical control points during food processing. Another new finding is that high rates of resistance to fusidic acid in S. xylosus and S. sciuri were due to carriage of novel intrinsic determinants, thus possibly will not contribute to the risk of transferring resistance.
Acknowledgments
This work was supported by the Ministry of Science and Technology of Taiwan [grant no. MOST 106-2320-B-002-039-MY3].
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jfda.2019.05.003.
Funding Statement
This work was supported by the Ministry of Science and Technology of Taiwan [grant no. MOST 106-2320-B-002-039-MY3].
Footnotes
Conflicts of interest
The authors declare no conflict of interest.
REFERENCES
- 1. Pinchuk IV, Beswick EJ, Reyes VE. Staphylococcal enterotoxins. Toxins (Basel) 2010;2:2177–97. doi: 10.3390/toxins2082177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. DeLeo FR, Chambers HF. Reemergence of antibiotic-resistant Staphylococcus aureus in the genomics era. J Clin Investig. 2009;119:2464–74. doi: 10.1172/JCI38226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Becker K, Heilmann C, Peters G. Coagulase-negative staphylococci. Clin Microbiol Rev. 2014;27:870–926. doi: 10.1128/CMR.00109-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Resch M, Nagel V, Hertel C. Antibiotic resistance of coagulase-negative staphylococci associated with food and used in starter cultures. Int J Food Microbiol. 2008;127:99–104. doi: 10.1016/j.ijfoodmicro.2008.06.013. [DOI] [PubMed] [Google Scholar]
- 5. Otto M. Coagulase-negative staphylococci as reservoirs of genes facilitating MRSA infection: staphylococcal commensal species such as Staphylococcus epidermidis are being recognized as important sources of genes promoting MRSA colonization and virulence. Bioessays. 2013;35:4–11. doi: 10.1002/bies.201200112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Aidara-Kane A, Andremont A, Collignon P. Antimicrobial resistance in the food chain and the AGISAR initiative. J Infect Public Health. 2013;6:162–5. doi: 10.1016/j.jiph.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 7. Bager F. DANMAP: monitoring antimicrobial resistance in Denmark. Int J Antimicrob Agents. 2000;14:271–4. doi: 10.1016/s0924-8579(00)00135-7. [DOI] [PubMed] [Google Scholar]
- 8. Bhargava K, Zhang Y. Characterization of methicillin-resistant coagulase-negative staphylococci (MRCoNS) in retail meat. Food Microbiol. 2014;42:56–60. doi: 10.1016/j.fm.2014.02.019. [DOI] [PubMed] [Google Scholar]
- 9. Founou LL, Founou RC, Essack SY. Antibiotic resistance in the food chain: a developing country-perspective. Front Microbiol. 2016;7:1881. doi: 10.3389/fmicb.2016.01881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chajecka-Wierzchowska W, Zadernowska A, Nalepa B, Sierpinska M, Laniewska-Trokenheim L. Coagulase-negative staphylococci (CoNS) isolated from ready-to-eat food of animal origin - phenotypic and genotypic antibiotic resistance. Food Microbiol. 2015;46:222–6. doi: 10.1016/j.fm.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 11. Fijalkowski K, Peitler D, Karakulska J. Staphylococci isolated from ready-to-eat meat - identification, antibiotic resistance and toxin gene profile. Int J Food Microbiol. 2016;238:113–20. doi: 10.1016/j.ijfoodmicro.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 12. Hammad AM, Watanabe W, Fujii T, Shimamoto T. Occurrence and characteristics of methicillin-resistant and -susceptible Staphylococcus aureus and methicillin-resistant coagulase-negative staphylococci from Japanese retail ready-to-eat raw fish. Int J Food Microbiol. 2012;156:286–9. doi: 10.1016/j.ijfoodmicro.2012.03.022. [DOI] [PubMed] [Google Scholar]
- 13. Yang X, Zhang J, Yu S, Wu Q, Guo W, Huang J, et al. Prevalence of Staphylococcus aureus and methicillin-resistant Staphylococcus aureus in retail ready-to-eat foods in China. Front Microbiol. 2016;7:816. doi: 10.3389/fmicb.2016.00816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.US Food and Drug Administration. Bacteriological Analytical Manual (BAM) Chapter 12: Staphylococcus aureus. Silver Spring MD; U.S. FDA: 2016. [accessed 10, 03, 18]. Available at: https://www.fda.gov/food/laboratory-methods-food/bamstaphylococcus-aureus. [Google Scholar]
- 15. Shah MM, Iihara H, Noda M, Song SX, Nhung PH, Ohkusu K, et al. dnaJ gene sequence-based assay for species identification and phylogenetic grouping in the genus Staphylococcus. Int J Syst Evol Microbiol. 2007;57:25–30. doi: 10.1099/ijs.0.64205-0. [DOI] [PubMed] [Google Scholar]
- 16.Clinical and Laboratory Standards Institute. M100-S27: performance standards for antimicrobial susceptibility testing; 27th informational supplement. Wayne, PA: CLSI; 2017. [Google Scholar]
- 17.European Committee on Antimicrobial Susceptibility Testing (EUCAST) 2018. [accessed 21.12.18]. Available at: http://www.eucast.org.
- 18.Loncaric I, Tichy A, Handler S, Szostak MP, Tickert M, Diab-Elschahawi M, et al. Antibiotics (Basel) 2019. Prevalence of methicillin-resistant Staphylococcus sp (MRS) in different companion animals and determination of risk factors for colonization with MRS; p. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Belen Florez A, Alegria A, Rossi F, Delgado S, Felis GE, Torriani S, et al. Molecular identification and quantification of tetracycline and erythromycin resistance genes in Spanish and Italian retail cheeses. Biomed Res Int. 2014;2014:746859. doi: 10.1155/2014/746859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wan TW, Hung WC, Tsai JC, Lin YT, Lee H, Hsueh PR, et al. Novel structure of Enterococcus faecium-originated ermB-positive Tn1546-like element in Staphylococcus aureus. Antimicrob Agents Chemother. 2016;60:6108–14. doi: 10.1128/AAC.01096-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Strommenger B, Kettlitz C, Werner G, Witte W. Multiplex PCR assay for simultaneous detection of nine clinically relevant antibiotic resistance genes in Staphylococcus aureus. J Clin Microbiol. 2003;41:4089–94. doi: 10.1128/JCM.41.9.4089-4094.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen HJ, Hung WC, Lin YT, Tsai JC, Chiu HC, Hsueh PR, et al. A novel fusidic acid resistance determinant, fusF, in Staphylococcus cohnii. J Antimicrob Chemother. 2015;70:416–9. doi: 10.1093/jac/dku408. [DOI] [PubMed] [Google Scholar]
- 23. Shopsin B, Gomez M, Montgomery SO, Smith DH, Waddington M, Dodge DE, et al. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J Clin Microbiol. 1999;37:3556–63. doi: 10.1128/jcm.37.11.3556-3563.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38:1008–15. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen HJ, Lin YT, Hung WC, Tsai JC, Hsueh PR, Teng LJ. Distribution of staphylococcal cassette chromosome (SCC) mec element types in fusidic acid-resistant Staphylococcus epidermidis and identification of a novel SCC7684 element. Antimicrob Agents Chemother. 2016;60:5006–9. doi: 10.1128/AAC.00231-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yang TY, Hung WW, Lin L, Hung WC, Tseng SP. mecA-related structure in methicillin-resistant coagulase-negative staphylococci from street food in Taiwan. Sci Rep. 2017;7:42205. doi: 10.1038/srep42205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Castanheira M, Watters AA, Bell JM, Turnidge JD, Jones RN. Fusidic acid resistance rates and prevalence of resistance mechanisms among Staphylococcus spp. isolated in North America and Australia, 2007–2008. Antimicrob Agents Chemother. 2010;54:3614–7. doi: 10.1128/AAC.01390-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cirkovic I, Trajkovic J, Hauschild T, Andersen PS, Shittu A, Larsen AR. Nasal and pharyngeal carriage of methicillin-resistant Staphylococcus sciuri among hospitalised patients and healthcare workers in a Serbian university hospital. PLoS One. 2017;12:e0185181. doi: 10.1371/journal.pone.0185181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Guo X, Peisker K, Backbro K, Chen Y, Koripella RK, Mandava CS, et al. Structure and function of FusB: an elongation factor G-binding fusidic acid resistance protein active in ribosomal translocation and recycling. Open Biol. 2012;2:120016. doi: 10.1098/rsob.120016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fang TJ, Wei QK, Liao CW, Hung MJ, Wang TH. Microbiological quality of 18 degrees C ready-to-eat food products sold in Taiwan. Int J Food Microbiol. 2003;80:241–50. doi: 10.1016/s0168-1605(02)00172-1. [DOI] [PubMed] [Google Scholar]
- 31. Viswanathan P, Kaur R. Prevalence and growth of pathogens on salad vegetables, fruits and sprouts. Int J Hyg Environ Health. 2001;203:205–13. doi: 10.1078/S1438-4639(04)70030-9. [DOI] [PubMed] [Google Scholar]
- 32. Seo Y-H, Jang J-H, Moon K-D. Occurrence and characterization of enterotoxigenic Staphylococcus aureus isolated from minimally processed vegetables and sprouts in Korea. Food Sci Biotechnol. 2010;19:313–9. [Google Scholar]
- 33. Bloemendaal AL, Brouwer EC, Fluit AC. Methicillin resistance transfer from Staphylocccus epidermidis to methicillin-susceptible Staphylococcus aureus in a patient during antibiotic therapy. PLoS One. 2010;5:e11841. doi: 10.1371/journal.pone.0011841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vandenesch F, Perrier-Gros-Claude JD, Bes M, Fuhrmann C, Delorme V, Mouren C, et al. Staphylococcus pasteuri-specific oligonucleotide probes derived from a random amplified DNA fragment. FEMS Microbiol Lett. 1995;132:147–52. doi: 10.1111/j.1574-6968.1995.tb07824.x. [DOI] [PubMed] [Google Scholar]
- 35. Irlinger F. Safety assessment of dairy microorganisms: coagulase-negative staphylococci. Int J Food Microbiol. 2008;126:302–10. doi: 10.1016/j.ijfoodmicro.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 36. Phukon M, Sahu P, Srinath R, Nithya A, Babu S. Unusual occurrence of Staphylococcus warneri as endophyte in fresh fruits along with usual Bacillus spp. J Food Saf. 2013;33:102–6. [Google Scholar]
- 37. Szczuka E, Krzyminska S, Kaznowski A. Clonality, virulence and the occurrence of genes encoding antibiotic resistance among Staphylococcus warneri isolates from bloodstream infections. J Med Microbiol. 2016;65:828–36. doi: 10.1099/jmm.0.000287. [DOI] [PubMed] [Google Scholar]
- 38. Hung WC, Chen HJ, Lin YT, Tsai JC, Chen CW, Lu HH, et al. Skin commensal staphylococci may act as reservoir for fusidic acid resistance genes. PLoS One. 2015;10:e0143106. doi: 10.1371/journal.pone.0143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chen CJ, Huang YC. New epidemiology of Staphylococcus aureus infection in Asia. Clin Microbiol Infect. 2014;20:605–23. doi: 10.1111/1469-0691.12705. [DOI] [PubMed] [Google Scholar]
- 40. Puah SM, Chua KH, Tan JA. Virulence factors and antibiotic susceptibility of Staphylococcus aureus isolates in ready-to-eat foods: detection of S. aureus contamination and a high prevalence of virulence genes. Int J Environ Res Public Health. 2016;13:199. doi: 10.3390/ijerph13020199. [DOI] [PMC free article] [PubMed] [Google Scholar]