Abstract
Most circulating tumor DNA (ctDNA) assays are designed to detect recurrent mutations. Pediatric sarcomas share few recurrent mutations but rather are characterized by translocations and copy-number changes. We applied Cancer Personalized Profiling by deep Sequencing (CAPP-Seq) for detection of translocations found in the most common pediatric sarcomas. We also applied ichorCNA to the combined off-target reads from our hybrid capture to simultaneously detect copy-number alterations (CNA). We analyzed 64 prospectively collected plasma samples from 17 patients with pediatric sarcoma. Translocations were detected in the pretreatment plasma of 13 patients and were confirmed by tumor sequencing in 12 patients. Two of these patients had evidence of complex chromosomal rearrangements in their ctDNA. We also detected copy-number changes in the pretreatment plasma of 7 patients. We found that ctDNA levels correlated with metastatic status and clinical response. Furthermore, we detected rising ctDNA levels before relapse was clinically apparent, demonstrating the high sensitivity of our assay. This assay can be utilized for simultaneous detection of translocations and CNAs in the plasma of patients with pediatric sarcoma. While we describe our experience in pediatric sarcomas, this approach can be applied to other tumors that are driven by structural variants.
Introduction
Pediatric sarcomas consist of diverse subtypes that are rare and often difficult to diagnose. Diagnosis is currently performed using a combination of radiologic features, histologic analysis, and identification of molecular markers, including in some subtypes disease-defining translocations (1). In routine clinical practice, methods for translocation detection rely primarily on FISH of the tumor specimen. Most often, these are “break-apart” probes that identify an alteration in one translocation partner, but do not definitively identify the second partner, a practice that can sometimes lead to an erroneous diagnosis.
While some translocations are relatively common, many rare variants exist, often making diagnosis difficult using standard approaches. For example, in Ewing sarcoma, EWSR1-FLI1 is the most common translocation and occurs in 85% of tumors, while EWSR1-ERG occurs in 10% of tumors (2, 3). However, a small subset of Ewing sarcoma tumors do not have a translocation involving EWSR1. FISH break apart probes, which target EWSR1, fail to identify these non-EWSR1 rearranged tumors. Many other tumor subtypes are also characterized by EWSR1 translocations (4) and have distinct clinical courses and treatment regimens, necessitating accurate diagnosis. Although FISH may aid in the diagnosis of EWSR1-translocated tumors in some cases, the failure to identify the translocation partner can lead to inappropriate management.
Similarly, the diagnosis and management of rhabdomyosarcoma requires assessment for the presence or absence of a PAX3- or PAX7-FOXO1 translocation. Molecular analysis has shown that a subset of rhabdomyosarcoma tumors are characterized by fusions involving PAX3 or PAX7 and FOXO1 and the presence of these fusions portends a worse prognosis (5). Proper identification of fusion status and specific fusion partners is therefore paramount, as it can have therapeutic and prognostic implications (6–9).
Even for patients in whom the diagnosis can be accurately made using current approaches, there is a lack of sensitive and specific methods for monitoring disease burden during and after treatment. Imaging modalities can be misleading because tumor size does not always accurately reflect the presence of viable tumor, as some sarcomas can demonstrate a persistent mass despite adequate tumor necrosis (10–13). Furthermore, imaging approaches are expensive, may require sedation in young children, often lack the sensitivity to detect early relapse, and may contribute to the risk of future malignancies due to exposure to ionizing radiation (14).
One approach for noninvasive, sensitive, and specific monitoring of cancer is to evaluate circulating tumor DNA (ctDNA) (15–17). However, ctDNA assays have primarily been optimized to detect single-nucleotide variants. While detection of pediatric sarcoma translocations in ctDNA has been described, this has primarily relied on PCR-based amplification (18–20). Such methods require prior sequencing analysis of a tumor specimen for identification of the patient-specific translocation breakpoint followed by design of a customized assay for that patient. Thus, their clinical utility is limited, particularly for cases where the tumor is not available for analysis to define the translocation breakpoint. Additionally, PCR-based assays cannot simultaneously detect copy-number alterations (CNA), which are also highly prevalent in many pediatric sarcomas. While copy-number changes are not typically used for diagnostic purposes, they have increasingly significant prognostic and therapeutic implications (21–23). Recognizing these limitations, some groups have utilized next-generation sequencing (NGS) approaches for identification of translocations or CNAs in pediatric sarcomas (24, 25), however, a single assay that simultaneously detects these alterations may be advantageous. To develop a sarcoma-specific ctDNA assay capable of concurrently detecting translocations and copy-number changes, we used the Cancer Personalized Profiling by deep-Sequencing (CAPP-Seq; refs. 26, 27) approach to design a “pan-sarcoma” ctDNA assay that could be used “off-the-shelf”, without requiring a primary tumor specimen for analysis. Here we describe the use of this assay to diagnose and characterize pediatric sarcomas noninvasively and to monitor disease during the course of therapy.
Materials and Methods
Patient selection
Written informed consent for participation in this study was obtained from newly diagnosed or newly relapsed patients treated at Lucile Packard Children's Hospital Stanford or UCSF Benioff Children's Hospital San Francisco. The study was approved by the institutional review board (IRB) in accordance with established ethical guidelines (Belmont Report) Patients with a pathology-confirmed diagnosis of Ewing sarcoma, osteosarcoma, rhabdomyosarcoma, or synovial sarcoma were included if a plasma sample was available for collection prior to initiation of any curative intervention, including excisional biopsy. Clinical data, including histologic diagnosis, metastatic status at diagnosis, FISH results, clinical gene-panel testing results through Foundation Medicine (Cambridge, MA) or UCSF500 (San Francisco, CA), treatments received, and response to therapy was retrospectively collected (Supplementary Table S1). All radiographic examinations and laboratory analyses, including FISH, RT-PCR for translocations, and clinical gene-panel testing, were obtained as part of routine clinical care and at the discretion of the treating physician. Standardized tumor volumes from imaging studies were calculated using TeraRecon Aquarius reconstruction using manual circumscription, free region of interest demarcation, and volumetric exclusion (28).
Sample collection and processing
Pretreatment peripheral blood samples were prospectively collected from all patients prior to initiation of treatment for patients with newly diagnosed or newly relapsed pediatric sarcoma. When feasible, on-therapy samples of peripheral blood were also collected over the course of disease treatment. These time points generally coincided with surveillance scans and before/after surgical resection or radiation therapy courses. For a subset of patients, serial samples were also collected after completion of therapy, during surveillance visits. Plasma samples were also collected if there was concern for relapse. Peripheral blood (5–15 mL) was collected in K2EDTA tubes and processed within 2 hours of collection. Plasma was isolated by centrifugation at 1800 × g for 10 minutes and aliquoted into 2 mL cryovial tubes. The remaining plasma-depleted whole blood was mixed and aliquoted into 2 mL cryovial tubes. Plasma and whole-blood samples were stored at −80°C. Cell-free DNA (cfDNA) was extracted from plasma using the QIAamp Circulating Nucleic Acid Kit (Qiagen, catalog no. 55114) according to the manufacturer's instructions. Germline DNA was extracted from peripheral blood mononuclear cells using the DNeasy Blood & Tissue Kit (Qiagen, catalog no. 69504) according to the manufacturer's instructions.
When fresh, unfixed tumor samples were available for analysis, samples were snap frozen, embedded in Optimal Cutting Temperature (OCT) compound, and sectioned. Cryosections were cut with a section depth of 5 μm on a cryostat and stained with hematoxylin and eosin (H&E). Sections were evaluated by a pediatric pathologist (F.K. Hazard or S.J. Cho) and samples with more than 70% tumor content were macro-dissected from the OCT block to a depth of up to 5 mm. Using a mortar and pestle under liquid nitrogen, fresh frozen tumor samples were disrupted and homogenized with a QIAshredder (QIAgen, catalog no. 79654), and DNA was extracted using the AllPrep kit (QIAgen, catalog no. 80204). For formalin-fixed, paraffin-embedded (FFPE) tumor tissue, 5-μm slides were obtained and a representative slide was stained with H&E and evaluated. Areas high in tumor content were scraped off adjacent slides for extraction. DNA from FFPE samples was extracted using the AllPrep DNA/RNA FFPE kit (Qiagen, catalog no. 80234).
cfDNA, tumor, and germline DNA were quantified using the Qubit High-Sensitivity double-stranded DNA (dsDNA) assay (Thermo Fisher). Tumor and germline DNA were also quantified using the Nanodrop 2000c (Thermo Fisher). Tumor and germline nucleic acid quality was determined using the Genomic DNA Analysis ScreenTape (Agilent, catalog no. 5067-5365) on the TapeStation 4200 (Agilent). Tumor-genomic DNA used for CAPP-Seq library preparation was sonicated to approximately 170 bp fragments using the S2 ultrasonicator (Covaris). Sheared DNA and cfDNA fragment size was determined using the 2100 Bioanalyzer (Agilent) or the Fragment Analyzer (AATI).
CAPP-Seq selector design
The CAPP-Seq targeted gene panel was designed by reviewing the Catalog of Somatic Mutations in Cancer (COSMIC; ref. 29) and the literature (30–37) to identify intronic regions involved in translocation breakpoints. For some translocations, both gene partners were not included in the selector design because the intronic regions were too large and would saturate the selector size. For example, the intronic regions of FOXO1 involved in PAX-FOXO1 translocations span approximately 130 kb. The final selector covers 150 kb and includes 25 intronic regions from 11 genes (Table 1). These intronic regions were used as input for the NimbleDesign software (Roche) to design a custom SeqCap EZ Choice capture panel (Supplementary Table S2).
Table 1.
Intronic regions included in the CAPP-seq selector.
| Gene | Introns | Size (bp) | Disease |
|---|---|---|---|
| EWSR1 | 7–10 | 5,100 | EWS |
| FLI1 | 4–8 | 37,229 | EWS |
| FUS | 5–8 | 4,422 | EWS |
| PAX3 | 6–7 | 19,562 | RMS |
| PAX7 | 7 | 32,334 | RMS |
| TP53 | 1, 9 | 13,574 | OS |
| ATRX | 12, 15 | 18,413 | OS |
| SS18 | 10 | 4,700 | SS |
| SSX1 | 4 | 3,134 | SS |
| SSX2 | 4 | 1,865 | SS |
| SSX4 | 1, 4 | 2,333 | SS |
| Total | 150,602 |
Abbreviations: EWS, Ewing sarcoma; RMS, rhabdomyosarcoma; OS, osteosarcoma; SS, synovial sarcoma.
CAPP-Seq library preparation, hybrid capture, and sequencing
A minimum of 32 ng of DNA was used as standard input for cfDNA library preparation, with up to 60 ng used if significant genomic DNA contamination was present. Genomic contamination was considered significant if more than 50% of DNA fragments were derived from high-molecular weight DNA as determined by Bioanalyzer or Fragment Analyzer. If less than 32 ng of cfDNA was obtained, the maximum amount available was used for library preparation (Supplementary Table S3). Plasma DNA was not amplified prior to library generation. A maximum of 100 ng of sheared genomic DNA was used as input for CAPP-Seq tumor-DNA library preparation. Libraries were prepared using the KAPA Hyper Prep Kit (Kapa Biosystems, catalog no. KK8502) and custom unique molecular identifiers, as described previously (26, 27). Pooled sequencing libraries subsequently underwent hybridization with the CAPP-Seq selector. Posthybridization pooled libraries were sequenced on an Illumina HiSeq4000 instrument using 2 × 150 paired-end reads with an 8-base indexing read. Median sequencing depth for cfDNA and tumor were 1,744X and 2,729X, respectively.
CAPP-Seq data analysis
Sequencing data was processed using a previously described custom bioinformatics pipeline (26, 27). Briefly, samples were demultiplexed and paired-end reads were mapped to the hg19 reference genome using Burrows–Wheeler Aligner (BWA) 0.6.2 with default parameters (38). Samples were sorted and indexed using SAMtools (39).
Structural-variant detection was performed using FACTERA (40), a highly sensitive and specific method for structural-variant detection in cfDNA. FACTERA takes as input a .bam alignment file of paired-end reads produced by BWA (38), exon coordinates in .bed format (hg19 RefSeq coordinates utilized), and a .2bit reference genome to enable fast sequence retrieval (hg19). In addition, the analysis was restricted to reads that overlap genomic regions in the CAPP-Seq selector used in this work. FACTERA processes the input in 3 sequential phases: identification of discordant reads, detection of breakpoints at base pair-resolution, and in silico validation of candidate fusions. For structural variants identified in tumor or in pretreatment cfDNA, monitoring in subsequent plasma samples was performed using the sequence across the breakpoint and submitting BLAST queries (41) for all reads (properly paired, improperly paired, and not paired) mapping to the genes involved in the fusion region. If even 1 read detected a structural variant, this was used to make a call, as translocations, unlike mutations, are unlikely to be attributed to sequencing errors. Allelic frequencies (AF) for structural variants not formally called by FACTERA were computed by dividing the average depth at the positions on each end of the structural variant (SV) by the number of reads containing the templated fusion sequence.
Off-target reads were combined to obtain low-depth coverage across the genome (median depth of 6.4X). Copy-number detection was performed using ichorCNA (17) and manual inspection.
Whole-genome sequencing library preparation, sequencing, and data analysis
Whole-genome sequencing (WGS) libraries were made using the TruSeq Nano kit (Illumina) with a 350-bp insert as per the manufacturer's instructions. Tumor libraries were made using 400 ng of input DNA and sequenced to a depth of 60X. Germline libraries were made using 200 ng of input DNA and sequenced to a depth of 30X. Pooled libraries were sequenced on an Illumina HiSeq system with paired-end 2 × 150 bp reads. For 2 patients (042 and 005), germline and tumor were sequenced to a depth of 50X on an Illumina HiSeq system with paired-end 2 × 100 bp reads by Illumina, Inc. Raw DNA FASTQ data were preprocessed with NGSUtils (42) and aligned to the sex-specific GRCh38 reference genomes using BWA. Structural variants and CNAs were determined as previously described (23).
Statistical analysis
To test the hypothesis that patients that were metastatic had higher pretreatment ctDNA levels than patients that were nonmetastatic, we utilized the two-tailed Mann–Whitney U test. Kaplan–Meier estimates of progression-free survival and overall survival stratified by median pretreatment ctDNA level were performed using GraphPad Prism, version 8.2.1 for MacOS (GraphPad Software). HR and P value were calculated by the log–rank test.
Results
Patients and specimens
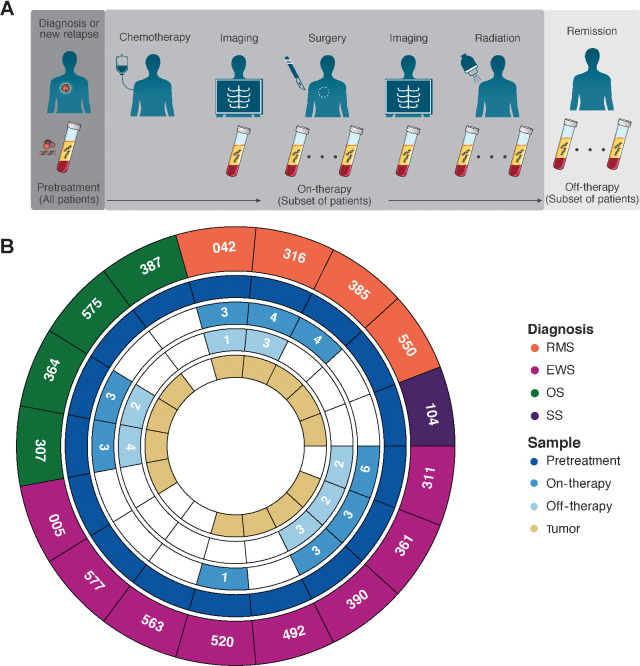
Samples were prospectively collected at the time points indicated in Fig. 1A. Samples available are summarized in Fig. 1B. The CAPP-Seq selector was applied to pretreatment plasma samples from 17 newly diagnosed or newly relapsed patients. Serial samples over the course of therapy were available from 9 patients. Of these, 7 patients also had plasma samples collected after completion of therapy. Matched tumor samples were available for genomic analysis by CAPP-Seq and/or research-grade WGS for 13 of 17 patients (Supplementary Table S4). Sequencing reports from clinical gene-panel testing were available for comparison for 4 patients (Supplementary Table S4).
Figure 1.
Sample collection schema and samples analyzed. A, All patients had plasma samples collected prior to initiation of treatment. When feasible, additional samples were obtained over the course of a patient's therapy and during off-therapy surveillance. B, Summary of plasma samples analyzed. Number of samples indicated if more than 1 sample was available for analysis. A subset of patients also had tumor specimens available for analysis using WGS and/or CAPP-Seq.
Recurrent sarcoma translocations detected in pretreatment plasma
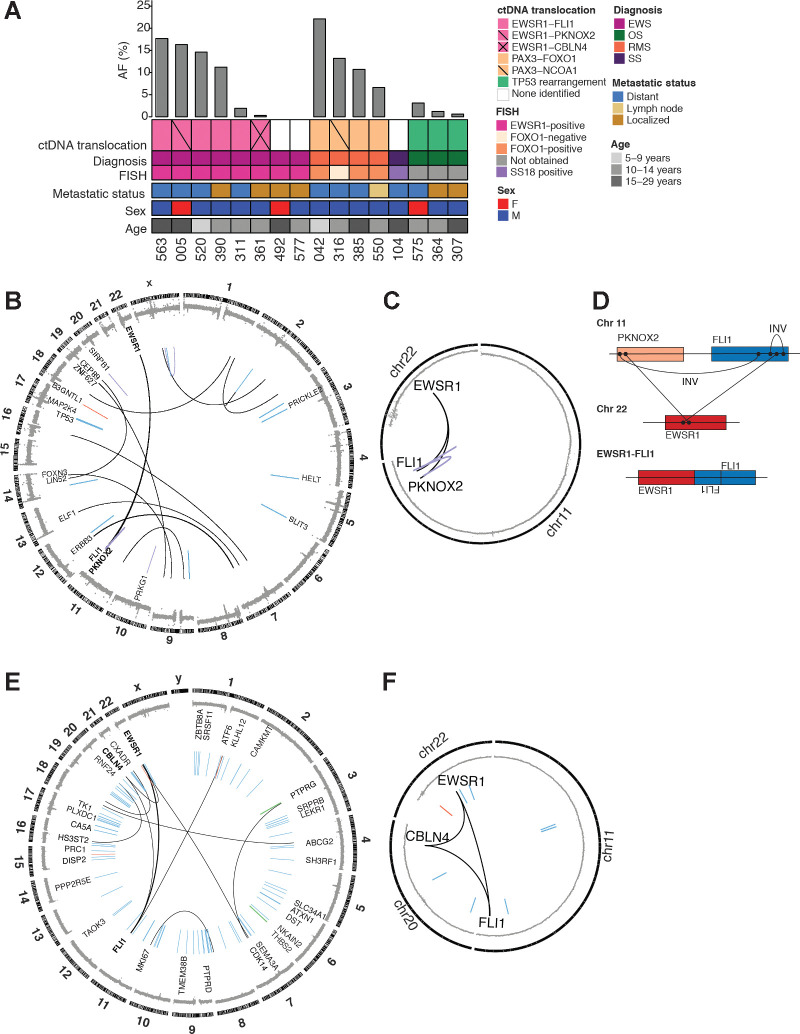
We first analyzed the ability of CAPP-Seq to detect translocations in pretreatment plasma samples. Sixteen patients had tumors with translocations identified by FISH or tumor sequencing. Translocations were identified in ctDNA with 81.3% sensitivity (13/16 patients) at AFs ranging from 0.31 to 22.1% (median 10.7%; Fig. 2A). The same translocations were also identified by sequencing of the tumor for the 12 patients that had tumor available for analysis (Supplementary Table S4). EWSR1-FLI1 translocations were detected in 4 patients with Ewing sarcoma, with a median AF of 12.9%. We did not identify the canonical EWSR1 rearrangements in the pretreatment plasma of 2 patients with Ewing sarcoma. Patient 492 had tumor WGS that demonstrated an EWSR1-FLI1 translocation that should have been detectable with our selector. Patient 577 had EWSR1 rearranged by break-apart probe analysis, but did not have any tumor available for sequencing, thus we were unable to confirm if this EWSR1 rearrangement should have been detected using our assay.
Figure 2.
Translocations detected in pretreatment ctDNA. A, Oncoprint of pretreatment ctDNA translocations. Each column represents a specific patient. A bar plot at the top details the AF of a given translocation. Key clinical information is depicted below. B, Circos plot for patient 005. Copy-number changes are depicted in the outermost circle. Structural alterations are shown in the inner portion of the plot (blue, deletion; red, duplication; black, translocation; purple, inversion) and the weight of the line corresponds to the number of supporting reads. C, Circos plot focusing on rearrangements that form a loop for same sample as in (B). D, Schematic showing EWSR1-FLI1 fusion generated from complex loop rearrangement. E, Circos plot for patient 361. F, Circos plot focusing on looped rearrangements for same sample as in (E). INV, Inversion.
Three patients had detectable PAX3-FOXO1 translocations, with a median AF of 10.7%. We did not identify any patients with PAX7-FOXO1 translocations in their plasma or tumor. Patient 316 had been clinically diagnosed with fusion-negative alveolar rhabdomyosarcoma, based on FISH testing of the tumor for the FOXO1 gene rearrangement. However, analysis of the plasma by CAPP-Seq revealed a PAX3-NCOA1 translocation, a rare translocation that has been described in a subset of patients with rhabdomyosarcoma (30). This fusion results in a chimeric transcription factor activator that has potent PAX3 transactivation activity, resulting in a gene-expression profile similar to that of the canonical fusion (22, 43). Identification of the PAX3-NCOA1 fusion is of clinical value, as these tumors are associated with a worse prognosis (5).
In contrast to Ewing sarcoma and rhabdomyosarcoma, osteosarcoma is not associated with a canonical gene fusion event. However, approximately 50% of these tumors have structural variants in TP53, most commonly in intron 1 (34). In addition, structural variants in introns 12 and 15 of ATRX are seen in 14% of tumors (34). We detected TP53 translocations at a median AF of 1.2% in 3 of 3 patients with osteosarcoma with TP53 rearrangements identified from sequencing of their primary tumor. ATRX translocations were not identified in the tumor or plasma of any patients in this cohort.
Our initial cohort included only 1 patient with synovial sarcoma. We were unable to detect the canonical SSX1-SS18 translocation in the pretreatment plasma of this patient. The patient had a solitary lung nodule measuring 0.2 cm3 and we were able to detect the SSX1-SS18 translocation in this nodule using WGS and CAPP-Seq. We hypothesize that the small tumor size may have led to very low levels of ctDNA. Despite the 3 negative cases, our results support the utility of the CAPP-Seq selector to identify fusions in the majority of pediatric sarcomas.
Complex chromosomal rearrangements detected in ctDNA
Patient 005 was diagnosed with Ewing sarcoma after biopsy revealed a small round blue-cell tumor with an EWSR1 gene rearrangement by FISH. We detected an EWSR1-PKNOX2 translocation in the patient's plasma at the time of relapse, with breakpoints at chr11:125215841 and chr22:29684555 (hg19), at an AF of 16.3%. To our knowledge, this translocation has not previously been described. WGS of the matched primary tumor revealed multiple structural variants (Fig. 2B), including the same EWSR1-PKNOX2 translocation with translocation breakpoints identical to those identified in the ctDNA. Additionally, an inversion of PKNOX2 and FLI1, an intronic inversion of FLI1, and an EWSR1-FLI1 translocation, were detected by WGS (Fig. 2B and C). Recent findings show that 40% of EWS-ETS (E26 transformation-specific) fusions are generated via chromoplexy rearrangements (44). These complex “loop” rearrangements are caused by chain-like events that start at EWSR1 and end with the ETS partner but involve several intervening genes. The EWSR1-PKNOX2 translocation identified in this case likely represents an example of such a complex, loop-like rearrangement. Because the breakpoint for the EWSR1-FLI1 rearrangement occurs within the inverted FLI1 intron, we may have been unable to detect this EWSR1-FLI1 translocation in the cfDNA (Fig. 2D). Tumors harboring chromoplexy are associated with more aggressive disease and may have contributed to this patient's relapse. Similarly, we detected an EWSR1-CBLN4 translocation at an AF of 0.31% in the ctDNA of patient 361. Tumor WGS suggests that this patient may also have a complex loop rearrangement involving EWSR1, CBLN4, and FLI1 (Fig. 2E and F). To our knowledge, these complex chromoplectic events have not been previously identified in plasma and represent a potentially clinically useful aspect of the assay described here.
ctDNA levels correlate with clinical features
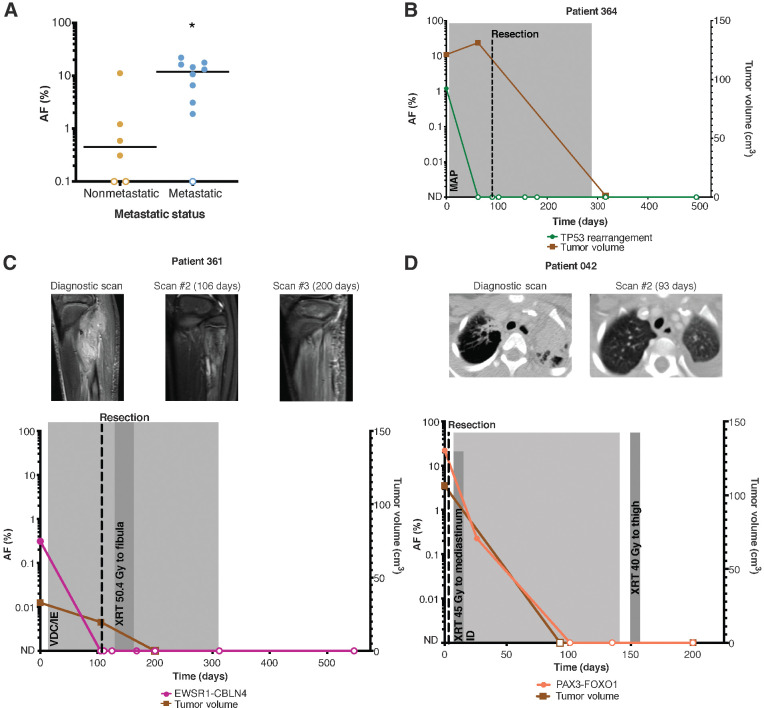
We sought to determine whether pretreatment ctDNA levels were correlated with clinical features. Patients with lymph node involvement or distant metastatic disease had higher absolute pretreatment ctDNA levels (mean AF = 10.6%, median AF = 11.95%) compared with patients with localized disease (mean AF = 2.2%, median AF = 0.45%; Fig. 3A). We also evaluated pretreatment ctDNA levels to determine their potential prognostic value. We defined a “ctDNA-low” and “ctDNA-high” group, with the cutoff determined by the median pretreatment ctDNA level of 4.85% across our cohort of patients. Pretreatment ctDNA levels did not correlate with risk of relapse (Supplementary Fig. S1A). However, patients in the “ctDNA-high” group had a significantly increased risk of disease-related death (HR 7.2, P = 0.0245; Supplementary Fig. S1B), though this may be attributable to metastatic status, as all patients in the “ctDNA-high” group had metastatic disease.
Figure 3.
ctDNA levels correlate with clinical features. A, Metastatic patients (n = 10) had higher levels of pretreatment ctDNA compared with nonmetastatic patients (n = 6). P value = 0.0224 (calculated by the Mann–Whitney U test). Serial ctDNA levels detected by CAPP-seq reflect response to therapy in osteosarcoma (B), Ewing sarcoma (C), and alveolar rhabdomyosarcoma (D). ND, not detected; M, methotrexate; A, adriamycin; P, cisplatinum; V, vincristine; D, doxorubicin; C, cyclophosphamide; I, ifosfamide; E, etoposide; XRT, radiotherapy.
Utility of ctDNA for longitudinal analysis in sarcomas
For the subset of patients with longitudinal samples, ctDNA levels were analyzed and compared with clinical status and imaging studies. Although pretreatment ctDNA levels did not correlate with primary tumor volume at presentation (Supplementary Fig. S1C), we observed a decline in ctDNA levels after initiation of therapy in all patients, in concordance with their clinical response to therapy (Supplementary Table S5). One patient (Fig. 3D) presented with relapsed alveolar rhabdomyosarcoma and multiple distant metastases. We detected a PAX3-FOXO1 translocation at an AF of 22%. After surgical excision of a lower extremity tumor, systemic chemotherapy, and radiation therapy to a metastatic site, ctDNA levels fell and were no longer detectable within 4 months, in concordance with radiologic findings. ctDNA remained undetectable and the patient remains in remission at last follow-up. Similar sustained declines in ctDNA levels after initiation of treatment were seen in osteosarcoma and Ewing sarcoma (Fig. 3B and C; Supplementary Table S5).
ctDNA can be detected by CAPP-Seq prior to clinically evident relapse
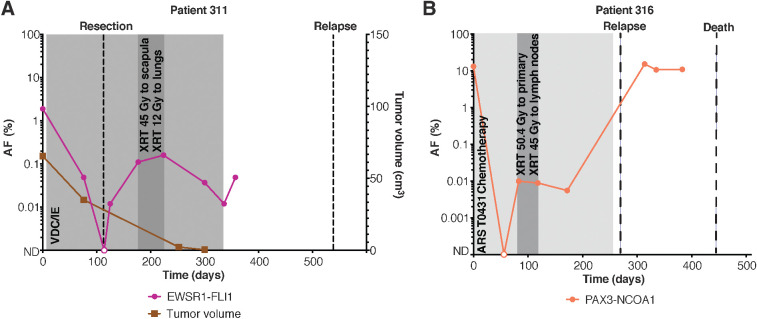
ctDNA was measurable at AFs as low as 0.003%, demonstrating the ultra-sensitive detection capabilities of CAPP-Seq. Patient 311 had an EWSR1-FLI1 translocation detected in the pretreatment plasma at an AF of 1.9%. The patient underwent surgical resection with positive margins and had detectable ctDNA postoperatively. He received radiotherapy at his positive margins and was considered to be in remission, however, his ctDNA levels remained elevated throughout his treatment. Plasma samples were obtained during off-therapy surveillance visits and ctDNA levels persisted (Fig. 4A). The patient ultimately presented with pulmonary relapse 6 months after completing therapy. The patient with a PAX3-NCOA1 translocation had ctDNA detected at a plasma AF of 13.2% at diagnosis. ctDNA levels initially declined to undetectable with chemotherapy, but rose to 0.008% 118 days after the pretreatment blood draw (Fig. 4B). The patient experienced clinical relapse 176 days later and ultimately died of disease.
Figure 4.
CAPP-seq can detect ctDNA before relapse is clinically evident in patients with pediatric sarcoma. A, Example of a patient with detectable pretreatment ctDNA that remained elevated after initiation of therapy. The patient ultimately relapsed 6 months after completion of therapy. B, Example of a patient with detectable pretreatment ctDNA who experienced an initial decline after starting chemotherapy. ctDNA was detected again 1 month later and remained elevated. The patient ultimately relapsed and died of disease.
CNAs detected using CAPP-Seq
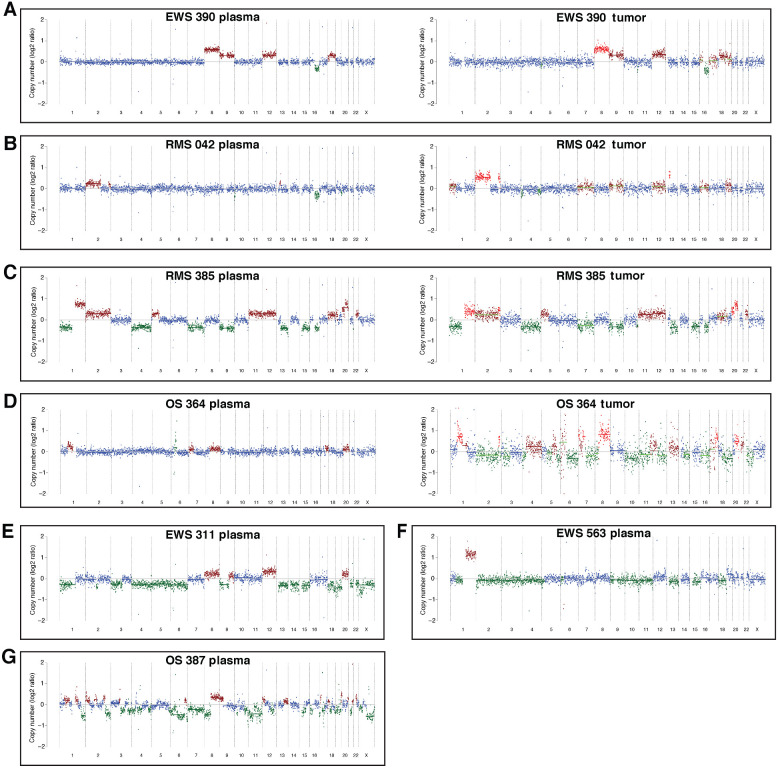
Seven patients had copy-number changes that were detectable in the plasma using off-target reads from CAPP-Seq (Fig. 5A–G). Four of these patients had tumor specimen available for copy-number analysis with CAPP-Seq (Fig. 5A–D) and 3 of the 4 patients also had tumor available for copy-number analysis by WGS (Supplementary Fig. S2). Copy-number analysis of the tumor using off-target reads was concordant with WGS and copy-number changes detected in the plasma were concordant with those found in the tumor (Supplementary Fig. S4). Two patients with Ewing sarcoma had gains in chromosomes 8 and 12 identified in their plasma. These CNAs have been previously described in Ewing sarcoma, with chromosome-8 gains being the most common (45, 46). These copy-number changes have also been shown to be coassociated, but do not necessarily bear a prognostic significance (46). Gain of 1q, which has been associated with poor survival (45, 46), was identified in a patient with relapsed Ewing sarcoma. CNAs were also identified in the plasma of 2 patients with rhabdomyosarcoma. Widespread copy-number changes were identified in 2 patients with osteosarcoma, including a gain in chromosome 8, which harbors MYC. While 6 of the patients with detectable copy-number changes also had translocations identified in their plasma, 1 of the osteosarcoma patients (387) did not have a TP53 rearrangement detected by sequencing of the tumor or plasma.
Figure 5.
A–G, CNAs can be detected utilizing off-target CAPP-seq reads in the plasma of Ewing sarcoma, rhabdomyosarcoma, and osteosarcoma. These findings are confirmed by CAPP-seq of the tumor in a subset of patients (right side of A–D).
Discussion
A major challenge in the clinical care of patients with pediatric sarcoma is the lack of sensitive, noninvasive methods to accurately diagnose and monitor disease. Currently, biopsy serves as the gold standard for diagnosis, but tumor purity, tumor heterogeneity, and inadequate sampling can limit detection of somatic variants. Tumor specimens often require sophisticated molecular assays for gene-rearrangement detection in order to make a definitive diagnosis. However, there is variability in the sensitivity and specificity of these molecular assays. Identification of these translocations is important for diagnosis and can also be relevant for prognosis. An even larger challenge is finding highly sensitive and specific biomarkers to monitor disease response.
To address these issues, we designed and tested a NGS assay for detection of recurrent translocations and CNAs in the plasma of patients with pediatric sarcoma. A key goal was to develop an “off-the-shelf” assay that could be rapidly applied to all patients without requiring primary tumor to be available for analysis. The CAPP-Seq selector tiles across the intronic regions most commonly involved in translocation events. While initially designed to detect a subset of key translocations found in pediatric sarcomas, this assay could easily be expanded to include other less common translocations relevant to the diagnosis and clinical care of pediatric solid tumors. With the current assay, we detected recurrent translocations in the cfDNA of patients with Ewing sarcoma, rhabdomyosarcoma, and osteosarcoma, with an overall sensitivity of 81% for translocation detection. We were able to detect a prognostically important PAX3-NCOA1 translocation that was not detected by standard FISH testing in a patient with rhabdomyosarcoma who was presumed to have fusion-negative disease. Furthermore, in 2 patients with Ewing sarcoma, we detected EWSR1 structural variants that may represent components of complex chromosomal loop rearrangements, which are associated with more aggressive disease. Additionally, by leveraging off-target reads from the hybrid capture, we were able to obtain low-depth coverage across the genome and detect copy-number changes in the cfDNA of a subset of patients with Ewing sarcoma, rhabdomyosarcoma, and osteosarcoma. One of these patients did not have a detectable translocation in the tumor, demonstrating that the added ability to detect copy-number changes can increase our overall sensitivity in detecting ctDNA. Although these copy-number changes are not diagnostic, they are of clinical significance, as they can have prognostic and therapeutic relevance. Utilizing a single assay that detects translocations and copy-number changes may be beneficial, especially when implementing such assays in the clinical setting, where the costs of specimen preparation, sequencing, and data analysis, as well as turn-around time, are paramount considerations.
The CAPP-Seq selector used here did not detect expected translocations in the cfDNA of 3 evaluated patients. For 1 patient, the SSX1-SS18 translocation was detected by CAPP-Seq in the primary tumor, demonstrating that the translocation-breakpoint region is detectable with our assay. The inability to detect this translocation in plasma could be due to low tumor burden. We were unable to detect the pretreatment EWSR1 rearrangements in 2 patients with Ewing sarcoma for unclear reasons. Additional studies will be needed to define the sensitivity of our assay in a larger patient population.
Our results suggest that pretreatment ctDNA levels detected by CAPP-Seq could be used to determine the likelihood of lung metastases in cases deemed indeterminate by imaging alone. ctDNA levels also correlated with clinical response to therapy, demonstrating its utility as a biomarker of disease. Importantly, our assay detected ctDNA several months before relapse was clinically detectable by imaging in Ewing sarcoma and alveolar rhabdomyosarcoma.
In summary, we have developed an ultrasensitive, “off-the-shelf” ctDNA assay that can simultaneously detect a wide variety of pediatric sarcoma translocations and CNAs. This can aid in diagnosis, monitoring, and early detection of relapse. This assay will need to be evaluated in a larger cohort of patients with pediatric cancer and, in particular, those with multiple plasma samples available across their disease course.
Authors' Disclosures
D.M. Kurtz reports personal fees from Roche Molecular Diagnostics, Genentech; and other support from Foresight Diagnostics outside the submitted work; in addition, D.M. Kurtz has a patent for Blood-based early detection of cancer pending and a patent for Ultrasensitive Cancer Monitoring by Phased variant Enrichment Sequencing pending. F.K. Hazard reports grants from United Therapeutics Corporation outside the submitted work. S.L. Spunt reports grants from Bayer AG, LOXO Oncology; and grants from Bayer Healthcare Pharmaceuticals outside the submitted work. M. Diehn reports personal fees from Roche Sequencing Solutions, Genentech, Novartis, Gritstone Oncology, BioNTech; grants and personal fees from AstraZeneca; personal fees and nonfinancial support from Illumina; other support from Cibermed, Foresight Diagnostics; and grants from Varian outside the submitted work; in addition, M. Diehn has a patent for ctDNA detection issued, licensed, and with royalties paid from Roche and a patent for ctDNA detection pending, licensed, and with royalties paid from Foresight Diagnostics. A.A. Alizadeh reports grants from BMS, personal fees and other support from Gliead, other support from Roche, other support from Genentech, personal fees and other support from Chugai, grants and other support from Celgene, other support from FortySeven, Foresight Dx, and other support from CiberMed outside the submitted work; in addition, A.A. Alizadeh has a patent for ctDNA diagnostics issued and licensed to Roche. No disclosures were reported by the other authors.
Acknowledgments
A.T. Shah was a Debra and Andrew Rachleff endowed fellow and was also funded by a St. Baldrick's Research Fellowship Grant, the Alex's Lemonade Stand Foundation, and the Frank A. Campini Foundation. T.D. Azad was funded by a Doris Duke Clinical Research Mentorship Award. H.E. Daldrup-Link was funded by a grant from the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD, R01 HD081123A). E.A. Sweet-Cordero was funded by a grant from the Alex's Lemonade Stand Foundation, the Child Health Research Institute (CHRI, Stanford), and the Children's Oncology Group Solid Malignancy-Integrated Translational Science Center. Additional funding was provided by the Lucile Packard Children's Hospital (Stanford University) and the Helen Diller Family Comprehensive Cancer Center (University of California San Francisco). This work used resources provided by the Stanford Genetics Bioinformatics Service Center and the Stanford Functional Genomics Facility, including those funded with support from an NIH S10 Shared Instrumentation Grant (S10OD018220). This work also used resources provided by the UCSF Center for Advanced Technology. The artwork for Fig. 1A was designed by Sarah Pyle. We thank the Stanford Tissue Bank and UCSF Tissue Bank for help collecting patient samples. We also thank all members of the Sweet-Cordero lab for their input and thoughtful discussion. We are especially grateful to all the patients and their families, who generously agreed to share blood and tumor samples to make this work possible.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Therapeutics Online (http://mct.aacrjournals.org/).
Authors' Contributions
A.T. Shah: Conceptualization, data curation, formal analysis, investigation, visualization, methodology, writing–original draft. T.D. Azad: Formal analysis, methodology. M.R. Breese: Data curation, formal analysis, visualization, methodology. J.J. Chabon: Data curation, formal analysis. E.G. Hamilton: Investigation. K. Straessler: Data curation, investigation. D.M. Kurtz: Investigation. S.G. Leung: Data curation. A. Spillinger: Data curation. H.-Y. Liu: Data curation. I.H. Behroozfard: Data curation. F.M. Wittber: Methodology. F.K. Hazard: Validation. S.-J. Cho: Validation. H.E. Daldrup-Link: Resources. K.T. Vo: Resources. A. Rangaswami: Resources. A. Pribnow: Resources, data curation. S.L. Spunt: Resources, data curation. N.J. Lacayo: Resources, data curation. M. Diehn: Conceptualization, writing–original draft, project administration, writing–review and editing. A.A. Alizadeh: Conceptualization, methodology, writing–original draft. E.A. Sweet-Cordero: Conceptualization, resources, formal analysis, funding acquisition, investigation, writing–original draft, project administration, writing–review and editing.
References
- 1. Lazar A, Abruzzo LV, Pollock RE, Lee S, Czerniak B. Molecular diagnosis of sarcomas: chromosomal translocations in sarcomas. Arch Pathol Lab Med 2006;130:1199–207. [DOI] [PubMed] [Google Scholar]
- 2. Fisher C. The diversity of soft tissue tumours with EWSR1 gene rearrangements: a review. Histopathology 2014;64:134–50. [DOI] [PubMed] [Google Scholar]
- 3. Chen S, Deniz K, Sung Y-S, Zhang L, Dry S, Antonescu CR. Ewing sarcoma with ERG gene rearrangements: A molecular study focusing on the prevalence of FUS-ERG and common pitfalls in detecting EWSR1-ERG fusions by FISH. Genes Chromosomes Cancer 2016;55:340–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cantile M, Marra L, Franco R, Ascierto P, Liguori G, De Chiara A, et al. Molecular detection and targeting of EWSR1 fusion transcripts in soft tissue tumors. Med Oncol 2013;30:412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Williamson D, Missiaglia E, de Reyniès A, Pierron G, Thuille B, Palenzuela G, et al. Fusion gene-negative alveolar rhabdomyosarcoma is clinically and molecularly indistinguishable from embryonal rhabdomyosarcoma. J Clin Oncol 2010;28:2151–8. [DOI] [PubMed] [Google Scholar]
- 6. Skapek SX, Anderson J, Barr FG, Bridge JA, Gastier-Foster JM, Parham DM, et al. PAX-FOXO1 fusion status drives unfavorable outcome for children with rhabdomyosarcoma: a children's oncology group report. Pediatr Blood Cancer 2013;60:1411–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Arnold MA, Anderson JR, Gastier-Foster JM, Barr FG, Skapek SX, Hawkins DS, et al. Histology, Fusion Status, and Outcome in Alveolar Rhabdomyosarcoma With Low-Risk Clinical Features: A Report From the Children's Oncology Group. Pediatr Blood Cancer 2016;63:634–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rudzinski ER, Anderson JR, Chi Y-Y, Gastier-Foster JM, Astbury C, Barr FG, et al. Histology, fusion status, and outcome in metastatic rhabdomyosarcoma: a report from the Children's Oncology Group. Pediatr Blood Cancer 2017;64:10.1002/pbc.26645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sorensen PHB, Lynch JC, Qualman SJ, Tirabosco R, Lim JF, Maurer HM, et al. PAX3-FKHR and PAX7-FKHR gene fusions are prognostic indicators in alveolar rhabdomyosarcoma: a report from the children's oncology group. J Clin Oncol 2002;20:2672–9. [DOI] [PubMed] [Google Scholar]
- 10. Erlemann R, Sciuk J, Bosse A, Ritter J, Kusnierz-Glaz CR, Peters PE, et al. Response of osteosarcoma and Ewing sarcoma to preoperative chemotherapy: assessment with dynamic and static MR imaging and skeletal scintigraphy. Radiology 1990;175:791–6. [DOI] [PubMed] [Google Scholar]
- 11. Hawkins DS, Rajendran JG, Conrad EU, Bruckner JD, Eary JF. Evaluation of chemotherapy response in pediatric bone sarcomas by [F-18]-fluorodeoxy-D-glucose positron emission tomography. Cancer 2002;94:3277–84. [DOI] [PubMed] [Google Scholar]
- 12. Rosenberg AR, Anderson JR, Lyden E, Rodeberg DA, Wolden SL, Kao SC, et al. Early response as assessed by anatomic imaging does not predict failure-free survival among patients with Group III rhabdomyosarcoma: a report from the Children's Oncology Group. Eur J Cancer 2014;50:816–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rodeberg DA, Stoner JA, Hayes-Jordan A, Kao SC, Wolden SL, Qualman SJ, et al. Prognostic significance of tumor response at the end of therapy in group III rhabdomyosarcoma: a report from the Children's Oncology Group. J Clin Oncol 2009;27:3705–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Inskip PD, Curtis RE. New malignancies following childhood cancer in the United States, 1973–2002. Int J cancer 2007;121:2233–40. [DOI] [PubMed] [Google Scholar]
- 15. Dawson S-J, Tsui DWY, Murtaza M, Biggs H, Rueda OM, Chin S-F, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med 2013;368:1199–209. [DOI] [PubMed] [Google Scholar]
- 16. Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014;6:224ra24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Adalsteinsson VA, Ha G, Freeman SS, Choudhury AD, Stover DG, Parsons HA, et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat Commun 2017;8:1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hayashi M, Chu D, Meyer CF, Llosa NJ, McCarty G, Morris CD, et al. Highly personalized detection of minimal Ewing sarcoma disease burden from plasma tumor DNA. Cancer 2016;122:3015–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Krumbholz M, Hellberg J, Steif B, Bäuerle T, Gillmann C, Fritscher T, et al. Genomic EWSR1 fusion sequence as highly sensitive and dynamic plasma tumor marker in Ewing Sarcoma. Clin Cancer Res 2016;22:4356–65. [DOI] [PubMed] [Google Scholar]
- 20. Shukla NN, Patel JA, Magnan H, Zehir A, You D, Tang J, et al. Plasma DNA-based molecular diagnosis, prognostication, and monitoring of patients with EWSR1 fusion-positive Sarcomas. JCO Precis Oncol 2017;2017:PO.16.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brohl AS, Solomon DA, Chang W, Wang J, Song Y, Sindiri S, et al. The genomic landscape of the Ewing Sarcoma family of tumors reveals recurrent STAG2 mutation. PLoS Genet 2014;10:e1004475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shern JF, Chen L, Chmielecki J, Wei JS, Patidar R, Rosenberg M, et al. Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion-positive and fusion-negative tumors. Cancer Discov 2014;4:216–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sayles LC, Breese MR, Koehne AL, Leung SG, Lee AG, Liu H-Y, et al. Genome-informed targeted therapy for Osteosarcoma. Cancer Discov 2019;9:46–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Klega K, Imamovic-Tuco A, Ha G, Clapp AN, Meyer S, Ward A, et al. Detection of somatic structural variants enables quantification and characterization of circulating tumor DNA in children with solid tumors. JCO Precis Oncol 2018;2018:PO.17.00285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Barris DM, Weiner SB, Dubin RA, Fremed M, Zhang X, Piperdi S, et al. Detection of circulating tumor DNA in patients with osteosarcoma. Oncotarget 2018;9:12695–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Newman AM, Bratman SV, To J, Wynne JF, Eclov NCW, Modlin LA, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014;20:548–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Newman AM, Lovejoy AF, Klass DM, Kurtz DM, Chabon JJ, Scherer F, et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol 2016;34:547–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rkein AM, Harrigal C, Friedman AC, Persky D, Krupinski E. Comparison of the accuracy of CT volume calculated by circumscription to prolate ellipsoid volume (bidimensional measurement multiplied by coronal long axis). Acad Radiol 2009;16:181–6. [DOI] [PubMed] [Google Scholar]
- 29. Forbes SA, Tang G, Bindal N, Bamford S, Dawson E, Cole C, et al. COSMIC (the Catalogue of Somatic Mutations in Cancer): a resource to investigate acquired mutations in human cancer. Nucleic Acids Res 2010;38:D652–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sumegi J, Streblow R, Frayer RW, Dal Cin P, Rosenberg A, Meloni-Ehrig A, et al. Recurrent t(2;2) and t(2;8) translocations in rhabdomyosarcoma without the canonical PAX-FOXO1 fuse PAX3 to members of the nuclear receptor transcriptional coactivator family. Genes Chromosomes Cancer 2010;49:224–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Barr FG. Gene fusions involving PAX and FOX family members in alveolar rhabdomyosarcoma. Oncogene 2001;20:5736–46. [DOI] [PubMed] [Google Scholar]
- 32. Berger M, Dirksen U, Braeuninger A, Koehler G, Juergens H, Krumbholz M, et al. Genomic EWS-FLI1 fusion sequences in Ewing sarcoma resemble breakpoint characteristics of immature lymphoid malignancies. PLoS One 2013;8:e56408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sankar S, Lessnick SL. Promiscuous partnerships in Ewing's sarcoma. Cancer Genet 2011;204:351–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen X, Bahrami A, Pappo A, Easton J, Dalton J, Hedlund E, et al. Recurrent somatic structural variations contribute to tumorigenesis in pediatric osteosarcoma. Cell Rep 2014;7:104–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Agus V, Tamborini E, Mezzelani A, Pierotti MA, Pilotti S. Re: A novel fusion gene, SYT-SSX4, in synovial sarcoma. J Natl Cancer Inst 2001;93:1347–9. [DOI] [PubMed] [Google Scholar]
- 36. Ladanyi M. Fusions of the SYT and SSX genes in synovial sarcoma. Oncogene 2001;20:5755–62. [DOI] [PubMed] [Google Scholar]
- 37. Wei Y, Sun M, Nilsson G, Dwight T, Xie Y, Wang J, et al. Characteristic sequence motifs located at the genomic breakpoints of the translocation t(X;18) in synovial sarcomas. Oncogene 2003;22:2215–22. [DOI] [PubMed] [Google Scholar]
- 38. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence Alignment/Map format and SAMtools. Bioinformatics 2009;25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Newman AM, Bratman SV, Stehr H, Lee LJ, Liu CL, Diehn M, et al. FACTERA: a practical method for the discovery of genomic rearrangements at breakpoint resolution. Bioinformatics 2014;30:3390–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol 1990;215:403–10. [DOI] [PubMed] [Google Scholar]
- 42. Breese MR, Liu Y. NGSUtils: a software suite for analyzing and manipulating next-generation sequencing datasets. Bioinformatics 2013;29:494–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wachtel M, Dettling M, Koscielniak E, Stegmaier S, Treuner J, Simon-Klingenstein K, et al. Gene expression signatures identify rhabdomyosarcoma subtypes and detect a novel t(2;2)(q35;p23) translocation fusing PAX3 to NCOA1. Cancer Res 2004;64:5539–45. [DOI] [PubMed] [Google Scholar]
- 44. Anderson ND, de Borja R, Young MD, Fuligni F, Rosic A, Roberts ND, et al. Rearrangement bursts generate canonical gene fusions in bone and soft tissue tumors. Science 2018;361:eaam8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Armengol G, Tarkkanen M, Virolainen M, Forus A, Valle J, Böhling T, et al. Recurrent gains of 1q, 8 and 12 in the Ewing family of tumours by comparative genomic hybridization. Br J Cancer 1997;75:1403–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tirode F, Surdez D, Ma X, Parker M, Le Deley MC, Bahrami A, et al. Genomic landscape of Ewing sarcoma defines an aggressive subtype with co-association of STAG2 and TP53 mutations. Cancer Discov 2014;4:1342–53. [DOI] [PMC free article] [PubMed] [Google Scholar]