Abstract
Our recent data demonstrates a critical role of the RIG-I-like receptor (RLR) family in regulating antifungal immunity against Aspergillus fumigatus in a murine model. However, the importance of this pathway in humans and the cell type(s) which utilize this innate immune receptor to detect A. fumigatus remains unresolved. Here using patients who underwent hematopoietic stem cell transplantation (HSCT), we demonstrate that a polymorphism in human MAVS present in the donor genome was associated with the incidence of invasive pulmonary aspergillosis (IPA). Moreover, in a separate cohort of confirmed IPA patients, polymorphisms in the IFIH1 gene alter the inflammatory response, including interferon-responsive chemokines. Returning to our murine model, we now demonstrate that CD11c+ SiglecF+ alveolar macrophages require Mavs expression to maintain host resistance against A. fumigatus. Our data support the role of MAVS signaling in mediating antifungal immunity in both mice and human at least in part through the role of MAVS-dependent signaling in alveolar macrophages.
INTRODUCTION
Aspergillus fumigatus is a ubiquitous environmental mold that humans inhale on a daily basis. Individuals with normal immune systems readily clear A. fumigatus conidia from their airways without complications. In contrast, immunocompromised individuals are at significantly greater risk of developing invasive pulmonary aspergillosis (IPA), including those receiving chemotherapy treatments for cancer and patients receiving immunosuppressive regimens to prevent GVHD following hematopoietic stem cell or solid organ transplantation (1–5). However, only a small proportion of immunosuppressed patients develop invasive fungal infections indicating that additional risk factors must exist. Numerous human studies have identified genetic polymorphisms in key antifungal pattern recognition receptors and inflammatory cytokines associated with fungal infections (reviewed in (6–8)). Thus, it is important to understand how these responses are coordinated in response to fungal infection of the lungs.
Recently, both type I and type III interferons have been shown to be essential for host resistance against pulmonary A. fumigatus challenge (9). Induction of the type I and type III interferon response following A. fumigatus challenge in the mouse model is dependent on both Dectin 1 (Clec7a) and MDA5 (Ifih1) (10, 11). Engagement of Dectin 1 leads to Syk activation which drive IRF5 activation for the production of IFNβ following Candida albicans challenge (12). MDA5 engagement by dsRNA leads to its interaction with MAVS resulting in the recruitment of IKKε and TBK1 that activate NFκB and IRF3 and IRF7, respectively, for the production of early cytokines and type I interferons (13). Genetic polymorphisms within CLEC7A have been associated with increased risk of developing IPA (14–16), while the role for genetic polymorphisms within IFIH1 or MAVS regarding susceptibility to IPA has not been explored to date.
Herein, our data demonstrates that genetic polymorphisms within IFIH1 and MAVS alter the production of interferon-dependent chemokines and risk for HSCT patients in developing IPA, respectively. Interestingly, increased risk for developing IPA was associated with genetic variation within MAVS only in the donor/hematopoietic compartment. Using our murine model, we identify alveolar macrophages as a key hematopoietic cell in the induction of the MDA5/MAVS-dependent interferon response which is necessary for host resistance against A. fumigatus. Overall, our study reveals a critical role for MDA5/MAVS in host anti-fungal immunity in both mice and humans.
MATERIALS & METHODS
Mice and Aspergillus fumigatus challenge model.
Ifih1(−/−) (Jackson Laboratory, Stock #015812), Mavs(fl/fl) (17), and Mavs(fl/fl) x Itgax-Cre mice (17) were bred in-house at Geisel School of Medicine at Dartmouth. C57BL/6J mice were purchased from Jackson Laboratory. All mice were 8–16 weeks of age at the time of challenge. Both female and male mice were used in these studies with no biological difference between the sexes. Animals were not co-housed to equilibrate their microbiota, which is a limitation of this study.
Preparation of Aspergillus fumigatus conidia and murine challenge model.
A. fumigatus CEA10 strains were used for this study. A. fumigatus was grown on glucose minimal media (GMM) agar plates for 3 days at 37°C. Conidia were harvested by adding 0.01% Tween 80 to plates and gently scraping conidia from the plates using a cell scraper. Conidia were then filtered through sterile Miracloth, were washed, and resuspended in phosphate buffered saline (PBS), and counted on a hemocytometer.
Mice were challenged with A. fumigatus conidia by the intratracheal (i.t.) route. Mice were anesthetized by inhalation of isoflurane; subsequently, mice were challenged i.t. with ~4 × 107 A. fumigatus conidia in a volume of 100 μl PBS. At the indicated time after A. fumigatus challenge, mice were euthanized using carbon dioxide. Bronchoalveolar lavage fluid (BALF) was collected by washing the lungs with 2 ml of PBS containing 0.05M EDTA. BALF was clarified by centrifugation and stored at −20°C until analysis. After centrifugation, the cellular component of the BALF was resuspended in 200 μl of PBS and total BAL cells were determined by hemocytometer count. BALF cells were subsequently spun onto glass slides using a Cytospin4 cytocentrifuge (Thermo Scientific) and stained with the Hema 3™ Stat Pack (Fisher Scientific) stain set for differential counting. For histological analysis lungs were filled with and stored in 10% buffered formalin phosphate for at least 24 hours. Lungs were then embedded in paraffin and sectioned into 5-micron sections. Sections were stained with Grocott-Gomori methenamine silver (GMS) using standard histological techniques to assess lung inflammatory infiltrates and fungal germination, respectively. Representative pictures of lung sections were taken using an Olympus BX50WI microscope with a QImaging Retiga 2000R camera.
Alveolar macrophage isolation and adoptive transfer.
Lungs from naïve Ifih1(−/−), Mavs(fl/fl) and Mavs(fl/fl) x Itgax-Cre were perfused with 20–30 ml of PBS. Lungs were then removed, injected with 700 μl of collagenase buffer [10ml RPMI, 1ml FBS, 25 μl 0.5M CaCl2, 25 μl 0.5M MgCl2, 50 μl HEPES, 100 μl L-glutamine, 100 μl Penn-Strep, 2 μl gentamycin, 25 μl DNase (VWR, Catalog # 77001–900), and 2 ml Liberase (Sigma-Aldrich)], and finally placed in 15 ml conical tube containing 2 ml of collagenase buffer. Lungs were digested or 30min at 37°C while shaking at 160–180 rpm. After which, 5 ml of ice cold stop buffer [25 ml RPMI, 1.25 ml FBS, 50 μl 0.5M EDTA] was added to each tube. Lungs were pushed through a 70 μm filter to generate a single cell suspension. Single cell lung suspensions were labeled with anti-Siglec-F MicroBeads (Miltenyi Biotec, Cat. No. 130-118-513) and selected for using LS Columns (Miltenyi Biotec, Cat. No. 130-042-401). Siglec F+ were eluted from the column, spun down, and resuspended in DMEM at 5×106 cells per ml. Subsequently 5×105 Siglec F+ alveolar macrophages were transferred i.t. to Mavs(fl/fl) x Itgax-Cre. Twenty-four hours later mice were challenge with A. fumigatus as described above.
Human HSCT cohort for SNP analysis.
A total of 460 hematologic patients undergoing allogeneic hematopoietic stem cell transplantation (HSCT) at the Hospital of Santa Maria, Lisbon and Instituto Português de Oncologia (IPO), Porto, between 2009 and 2015 were included in the study. Cases of IPA were identified and classified as ‘probable’ or ‘proven’ according to the 2008 criteria from the European Organization for Research and Treatment of Cancer/Mycology Study Group (EORTC/MSG) (18). Exclusion criteria included diagnosis of ‘possible’ IPA, infection with invasive molds other than Aspergillus spp. or history of pre-transplant mold infection. In accordance with institutional policies, all patients and donors (or their legal representative) provided informed written consent for data collection, DNA and cell storage, and their use for diagnostic and/or research purposes at the time of referral to transplantation. Study approval was obtained from the institutional review boards (SECVS-125/2014, HSM-632/14 and CES.26/015) and from the National Data Protection Commission (CNPD, 1950/2015) and was in compliance with all local relevant ethical regulations.
Genomic DNA was isolated from whole blood of patients using the QIAcube automated system (Qiagen). SNPs were selected based on their previous association with infection or with putative functional consequences to the gene (19–21). Genotyping was performed using KASPar assays (LGC Genomics) in an Applied Biosystems 7500 Fast Real-Time PCR system (Thermo Fisher Scientific), according to the manufacturer’s instructions.
Cytokine analysis of BALF from human IPA patients.
BALF and blood samples were collected from hospitalized adult patients (≥18 years of age) at the Leuven University Hospitals, Leuven, Belgium, as previously described (22). All patients (or their legal representative) provided informed written consent for data collection, DNA and cell storage, and their use for diagnostic and/or research purposes at hospital admission. This study was approved and carried out in accordance with recommendations of the Ethics Subcommittee for Life and Health Sciences of the University of Minho, Portugal (SECVS-125/2014), and the Ethics Committee of the University Hospitals of Leuven, Belgium. Written informed consent was obtained from all subjects in accordance with the Declaration of Helsinki.
For this cytokine analysis, twenty-three cases of “probable” or “proven” IPA were identified according to the standard criteria from the European Organization for Research and Treatment of Cancer/Mycology Study Group (EORTC/MSG) (18) and included for cytokine analysis. Genomic DNA was isolated from EDTA venous blood and genotyped for the IFIH1 rs1990760 polymorphism. Cytokine levels were determined using the Cytokine & Chemokine 34-Plex Human ProcartaPlex and stratified based on their IFIH1 rs1990760 genotype.
Statistical analysis.
Statistical significance for in vitro and ex vivo data was determined by a Mann-Whitney U test, one-way ANOVA using a Bonferroni post-test, or Kruskal-Wallis one-way ANOVA with Dunn’s post-test through the GraphPad Prism 7 software as outlined in the figure legends. Mouse survival data were analyzed with the Mantel-Cox log rank test using GraphPad Prism. For the human HSCT cohort, the probability of IPA resulting from IFIH1 and MAVS SNPs was analyzed using the cumulative incidence method and compared using Gray’s test (23). The Gray’s test was selected for the analysis because, in contrast with the commonly used Kaplan-Meier and Cox models, it allows accounting for multiple competing risks and provides a better estimation for the risk of the main outcome of interest, in this case the development of IPA (24). Cumulative incidences were computed with the cmprsk package for R version 2.10.1 (25), with censoring of data at the date of last follow-up visit and defining relapse and death as competing events. A period of 24 months after transplant was chosen to include all cases of IPA.
Study approvals.
All animal experiments were approved by the Dartmouth College Institutional Animal Care and Use Committee under protocol number 00002168. For our human studies approval was obtained from the institutional review boards (SECVS-125/2014, HSM-632/14 and CES.26/015) and from the National Data Protection Commission (CNPD, 1950/2015) and was in compliance with all local relevant ethical regulations.
RESULTS
Single Nucleotide Polymorphisms (SNPs) in IFIH1 and MAVS influence the risk for developing invasive pulmonary aspergillosis.
Our recent mouse data suggests that MDA5/MAVS signaling is critical in maintaining host resistance against pulmonary challenge with A. fumigatus (11). Therefore, we wanted to explore the role of these molecules in a human cohort comprised of 460 HSCT patients (Supplemental Table 1). Missense SNPs within the coding region of IFIH1 (rs1990760 and rs3747517) are associated with autoimmune conditions, particularly interferonopathies (19). Moreover, a missense SNP in MAVS (rs17857295) is associated with altered type I interferon regulation (21). While an individual genotype for each SNP was not associated with risk of developing IPA (Supplemental Figure 1), we further examined these SNPs using a recessive genetic model because the autoimmune risk allele in IFIH1 rs1990760 has been shown to work in a dominant fashion (19). Notably, we found a significant association between the CC genotype at rs1990760 in IFIH1 and the risk of developing IPA in HSCT recipients (Figure 1A). This increased risk occurred when the variant was carried by the recipient in a recessive genetic model, but not when it was carried by the donor. In contrast, no association between the IFIH1 rs3747517 SNP and the risk of developing IPA was found (Figure 1B). Additionally, we also found a significant association between the MAVS rs17857295 SNP and the risk of developing IPA in HSCT recipients (Figure 1C). This increased risk occurred when the variant was carried by the donor in a recessive genetic model, but not when it was carried by the recipient. Moreover, the risk from the donor MAVS rs17857295 SNP held up to multivariate analysis accounting for relevant clinical risk factors (Table 1).
Figure 1. IFIH1 and MAVS polymorphisms are associated with invasive pulmonary aspergillosis in human transplant patients using a recessive allele model.
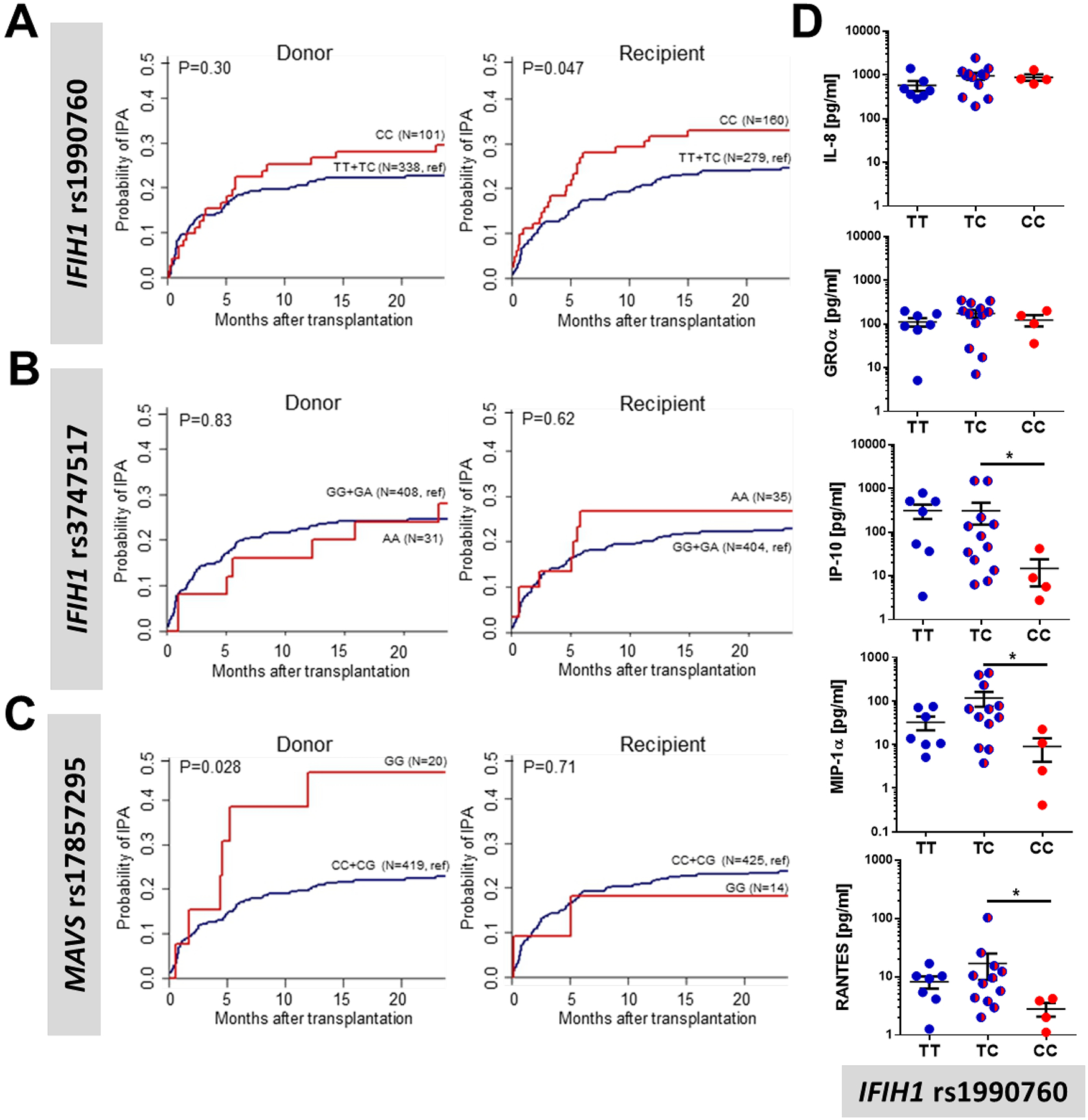
Cumulative incidence analysis of invasive aspergillosis after transplantation according to donor or recipient genotypes at rs1990760 in IFIH1 (A), rs3747517 in IFIH1 (B), or rs17857295 in MAVS (C) over 24 months after HSCT. Data were analyzed by two-sided Gray’s test. (D) Inflammatory cytokine levels in the bronchoalveolar lavage fluid from 23 patients with invasive pulmonary aspergillosis were measured using a 32-plex ProCarta Luminex assay and plotted based on their IFIH1 rs1990760 genotype. Data were analyzed using a Mann-Whitney U-test (* p < 0.05).
Table 1.
Multivariate analysis of the association of IFIH1 and MAVS SNPs with the risk of invasive pulmonary aspergillosis among transplant recipients.
| Genetic/clinical variables | Adjusted HR† (95% CI) | P value |
|---|---|---|
| Donor rs17857295 in MAVS | 2.39 (1.07 – 5.32) | 0.033 |
| Recipient rs1990760 in IFIH1 | 1.46 (0.89 – 2.40) | 0.121 |
| Acute GVHD grades III-IV | 1.73 (0.94 – 3.19) | 0.047 |
HR, hazard ratio; CI, confidence interval. Multivariate analyses were based on the subdistribution regression model of Fine and Gray.
Hazard ratios were adjusted for patient age and gender, and clinical variables with a P<0.15 in the univariate analyses. Only the clinical variables remaining significant after adjustment are shown.
To demonstrate a functional effect of the rs1990760 SNP in IFIH1 and the associated amino acid substitution on the function of MDA5, we analyzed the inflammatory response in the bronchoalveolar lavage fluid (BALF) of 23 patients with IPA after stratification by genotypes at rs1990760 in IFIH1 (Supplemental Table 2). We found that individuals with the CC genotype, which is associated with increased risk of developing IPA (Figure 1A), had a significant reduction in IP-10 (CXCL10), RANTES (CCL5), and MIP-1α (CCL3), but not IL-8 and GROα (CXCL1) (Figure 1D). IP-10 (CXCL10), RANTES (CCL5), and MIP-1α (CCL3) are known to be interferon-responsive genes, while IL-8 and GROα (CXCL1) are not (26). In the BALF samples from this patient cohort, the levels of IFN-α were below the detection limit (data not shown). Given the relatively rare frequency of the rs17857295 genotype in MAVS, it was not possible to analyze cytokine production according to MAVS genotype in the BAL samples (data not shown). Overall, these data support an important role for MDA5/MAVS signaling in regulating the inflammatory response and host resistance against Aspergillus spp. in humans.
Deletion of Mavs in CD11c+ cells result in increased susceptibility to Aspergillus fumigatus in mice.
Given the observation that patients in the HSCT cohort with the MAVS rs17857295 polymorphism within the donor population had altered risk for developing IPA, we next wanted to assess which hematopoietic cell population(s) in our invasive aspergillosis murine model relies on Mavs to maintain host resistance against A. fumigatus. To directly address which cells must express Mavs we utilized the recently developed Mavs(fl/fl) x Itgax-Cre conditional knock-out model, hereafter referred to as Mavs(Cd11c/Cd11c), that will delete Mavs in all CD11c-expressing cells (17), including both dendritic cells and alveolar macrophages. To test of role of Mavs expression in CD11c-expressing cells for the maintenance of host resistance against A. fumigatus, we challenged C57BL/6J, Mavs(fl/fl), and Mavs(Cd11c/Cd11c) mice with 4×107 conidia of the CEA10 strain and monitored survival over nine days. Mavs(Cd11c/Cd11c) mice were more susceptible to pulmonary challenge with A. fumigatus than either C57BL/6J or Mavs(fl/fl) mice (Figure 2A; Mantel-Cox log rank test, p < 0.0001). Mice completely lacking either Ifih1 or Mavs have decreased neutrophil recruitment, which was associated with increased fungal germination after pulmonary challenge with A. fumigatus (11). Thus, we assessed fungal germination in the lung by histological analysis at 48 h after conidial instillation. Strikingly, GMS staining of lung tissue from Mavs(Cd11c/Cd11c) mice revealed high levels of A. fumigatus germination at this time compared with control Mavs(fl/fl) mice (Figure 2B). When the percentage of germinated A. fumigatus was quantified, Mavs(fl/fl) mice displayed low levels of fungal germination (23.5% ± 9.1) compared with Mavs(Cd11c/Cd11c) mice (60.1% ± 8.2). Finally, we assessed inflammatory cell accumulation in the airways. The increased susceptibility of Mavs(Cd11c/Cd11c) mice to A. fumigatus challenge was associated with decreased accumulation of neutrophils in the airways compared with Mavs(fl/fl) mice (Figure 2C). These data match our prior observations with mice completely lacking either Ifih1 or Mavs having increased IPA susceptibility (11), suggesting that Mavs expression within CD11c-expressing cells is critical for host resistance against A. fumigatus challenge.
Figure 2. Mavs-dependent responses are essential in CD11c+ cells for host resistance against Aspergillus fumigatus.
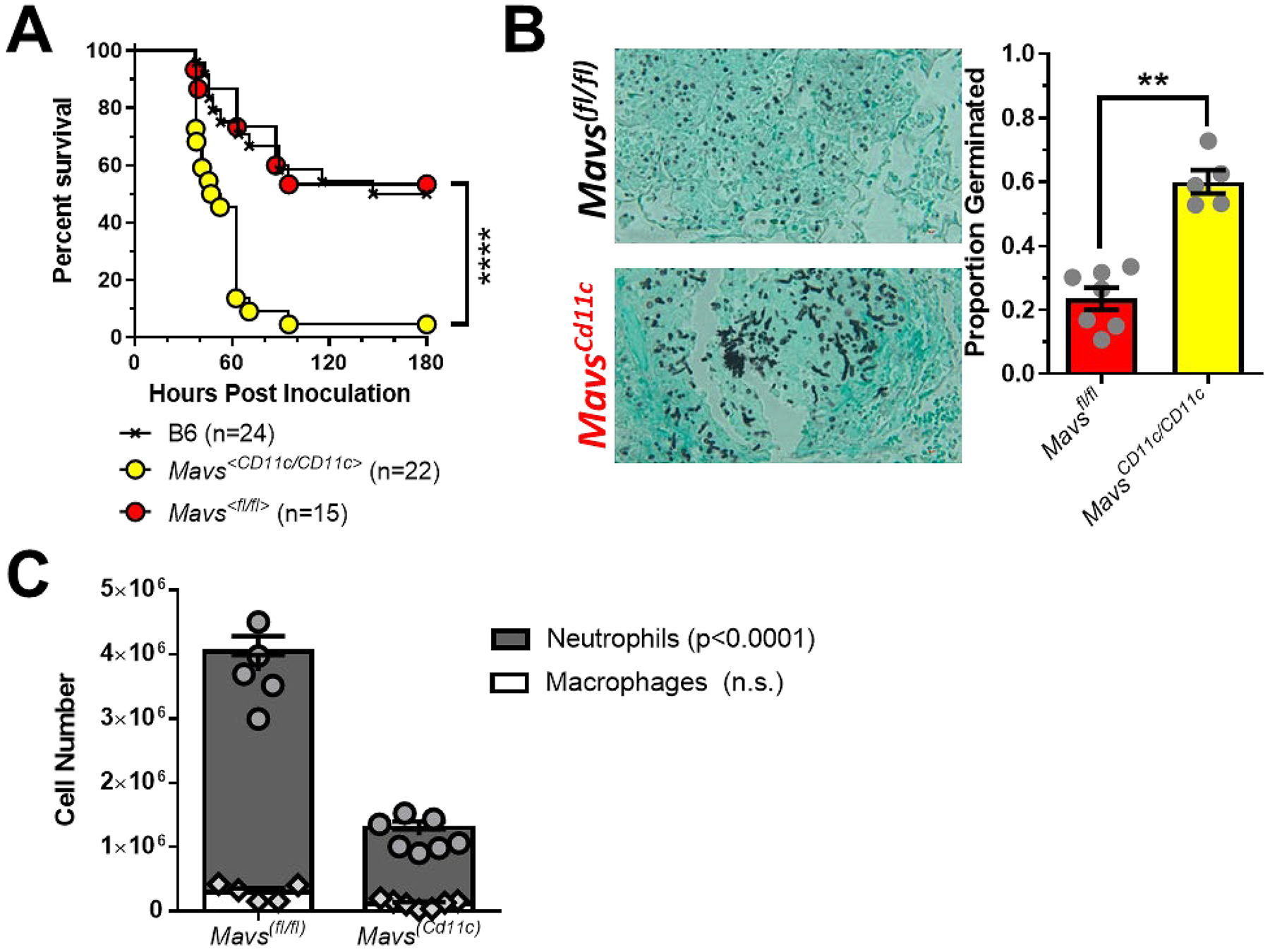
Mavs<Cd11c/Cd11c>, Mavs<fl/fl> and C57BL/6J mice were challenged i.t. with 4×107 resting conidia of the CEA10 isolate of Aspergillus fumigatus. (A) Survival analysis in immune-competent wild-type and knock-out mice were tracked over the first 9 days. ****, P < 0.0001 by Mantel-Cox log rank test. (B) Forty hours after A. fumigatus challenge mice were euthanized and fungal germination was assessed in the lungs by GMS staining. **, P < 0.01 by Mann-Whitney U-test. (C) At the same time point, cell differentials in the lung airways was determined by differential staining of BALF cytospins. Statistical significance was determined using a two-way ANOVA with a Sidak’s post-test.
Alveolar macrophage transfer to Mavs(Cd11c/Cd11c) conditional knock-out mice reduces their susceptibility to Aspergillus fumigatus in mice.
Multiple cells in the lungs can express Igtax (CD11c) including alveolar macrophages, CD103+ dendritic cells, CD11b+ dendritic cells, monocyte-derived dendritic cells, and plasmacytoid dendritic cells, all of which have be implicated in the antifungal immune response against A. fumigatus (27–33). Utilizing the publicly available data assembled by the ImmGen Consortium (34), in the lungs we see that Mavs is highly expressed in Siglec F+ alveolar macrophages, but also moderately expressed in both monocytes and dendritic cells (data not shown). Interestingly, following RSV infection alveolar macrophages have been shown to be the key cell type for initiating the type I interferon through a MAVS-dependent mechanism (35, 36). While alveolar macrophages largely populate the lungs during embryogenesis and are maintained by local proliferation during homeostasis (37, 38) following conditioning for HSC transplantation alveolar macrophage can be repopulated from the donor HSC compartment (39–42). Therefore, to specifically address the role of Mavs in alveolar macrophages, we purified Siglec F+ cells from the lungs of naïve Mavs(fl/fl) or Mavs(Cd11c/Cd11c) mice and transferred 5×105 Siglec F+ cells intratracheally into Mavs(Cd11c/Cd11c) mice one day prior to challenging with 4×107 conidia of the CEA10 strain (Figure 3A). Forty-eight hours after challenge, we examined the lungs by GMS staining of lung tissue from Mavs(Cd11c/Cd11c) mice receiving the Mavs(Cd11c/Cd11c) Siglec F+ cells and revealed the presence of high levels of germinated A. fumigatus (68.1% ± 12.6), while that was not observed to the same extent in Mavs(Cd11c/Cd11c) mice receiving the Mavs(fl/fl) Siglec F+ cells (31.4% ± 6.7) (Figure 3B). This is in line with control Mavs(Cd11c/Cd11c) mice or Mavs(fl/fl) mice receiving DMEM (vehicle) administered intratracheally. When we examined the accumulation of leukocytes in the airways by differential cytospin analysis we found that Mavs(Cd11c/Cd11c) mice receiving Mavs(Cd11c/Cd11c) Siglec F+ cells had reduced numbers of neutrophils when compared with Mavs(Cd11c/Cd11c) mice receiving Mavs(fl/fl) Siglec F+ cells (Figure 3C). Strikingly, when we examined macrophage numbers in the airways, we found Mavs(Cd11c/Cd11c) mice receiving Mavs(fl/fl) Siglec F+ cells had many macrophages present, but Mavs(Cd11c/Cd11c) mice receiving Mavs(Cd11c/Cd11c) Siglec F+ cells had few macrophages in their airways at 48 hours post-inoculation with A. fumigatus (Figure 3D). Importantly, the decrease in macrophages in the airways is not observed in naïve Mavs(Cd11c/Cd11c) mice (Supplemental Figure 2A). Additionally, Mavs(Cd11c/Cd11c) mice receiving an i.t. transfer of Mavs(Cd11c/Cd11c) Siglec F+ cells 24h prior only had a moderate decrease in alveolar macrophage numbers (Supplemental Figure 2B).
Figure 3. Alveolar macrophages require a Mavs- and Ifih1-dependent response to maintain host resistance against Aspergillus fumigatus.
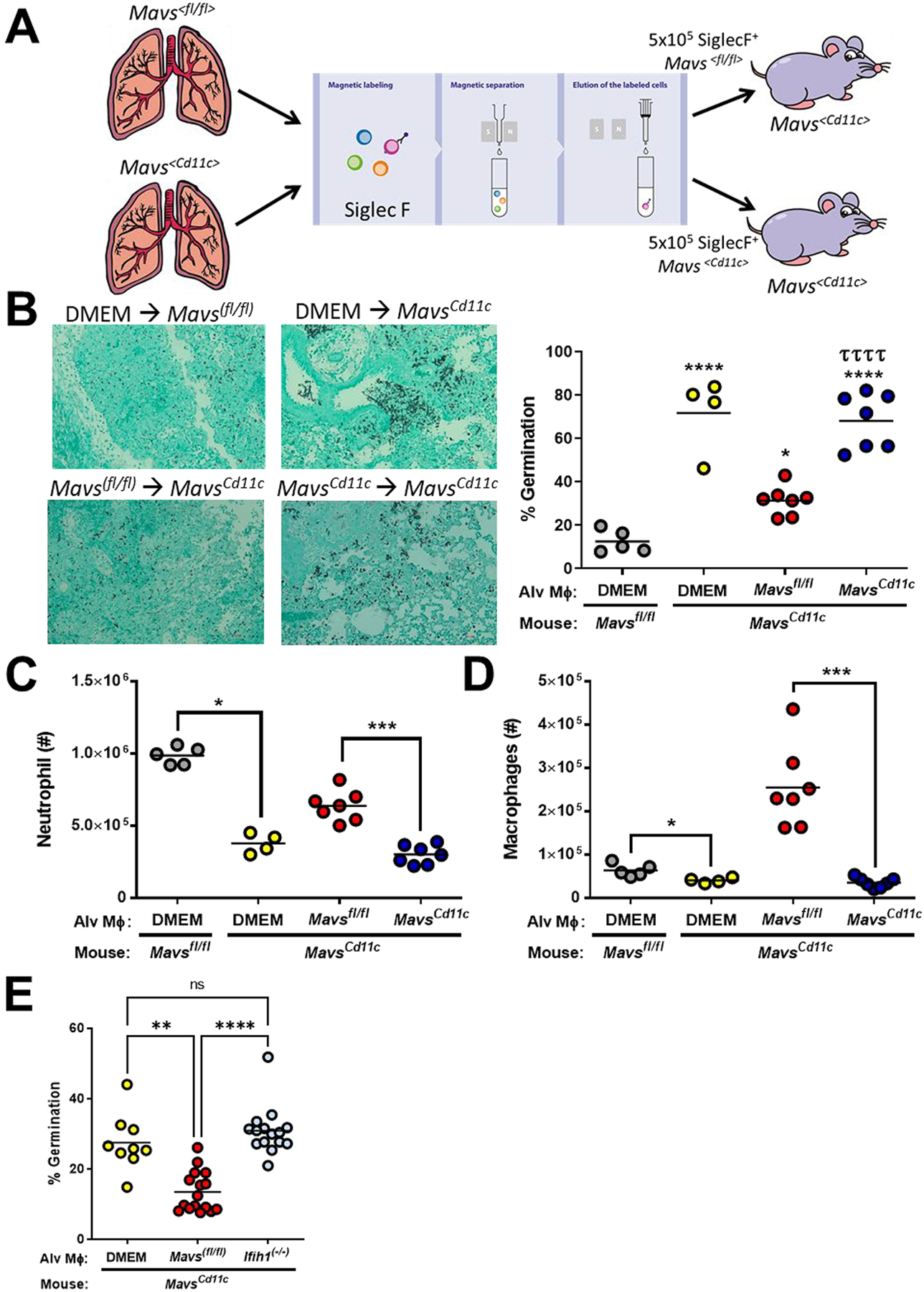
(A) Mavs<Cd11c/Cd11c> mice were complemented with 5×105 Mavs<fl/fl> or Mavs<Cd11c/Cd11c> alveolar macrophages (Siglec F+) given intranasally. One day later mice were challenged i.t. with 4×107 resting conidia of the CEA10 isolate of Aspergillus fumigatus. Forty hours after A. fumigatus challenge mice were euthanized. (B Fungal germination was assessed in the lungs by GMS staining. Representative 20x GMS images are shown. Statistically significant different were determined using a one-way ANOVA with a Tukey’s post-test: *p<0.05 (vs. DMEM → Mavs(fl//fl)), ****p<0.0001 (vs. DMEM → Mavs(fl//fl)), and ττττ p<0.0005 (vs. Mavs(fl//fl) → Mavs(Cd11c)). Neutrophils (C) and macrophages (D) in the lung airways was determined by differential staining of BALF cytospins. (E) Mavs<Cd11c/Cd11c> mice were complemented with 5×105 Mavs<fl/fl> or Ifih1(−/−) alveolar macrophages (Siglec F+) given intranasally. One day later mice were challenged i.t. with 4×107 resting conidia of the CEA10 isolate of A. fumigatus. Forty hours after A. fumigatus challenge mice were euthanized. Fungal germination was assessed in the lungs by GMS staining. Data are pooled from 2 independent experiments. Each dot represents an individual mouse. Statistically significant different were determined using a one-way ANOVA with a Kruskal-Wallis post-test: ****p<0.0001.
Next, to examine the role of Ifih1 in alveolar macrophages, we purified Siglec F+ cells from the lungs of naïve Mavs(fl/fl) or Ifih1(−/−) mice and transferred 5×105 Siglec F+ cells intratracheally into Mavs(Cd11c/Cd11c) mice one day prior to challenging with 4×107 conidia of the CEA10 strain (Figure 3E). Forty-eight hours after challenge, we examined the lungs by GMS staining of lung tissue from Mavs(Cd11c/Cd11c) mice receiving either DMEM or Ifih1(−/−) Siglec F+ cells, revealing the presence of high levels of germinated A. fumigatus, which was not observed to the same extent in Mavs(Cd11c/Cd11c) mice receiving the Mavs(fl/fl) Siglec F+ cells (Figure 3E). Taken all together, these data demonstrate that Ifih1 and Mavs expression within Siglec F+ alveolar macrophages is critical for host resistance against A. fumigatus.
DISCUSSION
Humans inhale Aspergillus fumigatus spores on a daily basis, but individuals with healthy immune systems readily clear A. fumigatus conidia from their airways without problems. One lung sentinel cell that can be critical in the clearance of A. fumigatus is the alveolar macrophage. Alveolar macrophages are known to phagocytose and destroy A. fumigatus (31), but their role in host resistance has remained controversial (33, 43). Patients undergoing HSCT are at increased risk of developing IPA (1–5). In early studies from patients undergoing HSCT, alveolar macrophages are dysfunctional (41) and found at decreased in numbers (40–42) for at least 50 days post-transplantation. However, we currently do not understand the effects of modern HSCT conditioning regiments on alveolar macrophages function and numbers, which is a limitation of our study. Either way it is important to understand the role of alveolar macrophages in the initial inflammatory response induced by A. fumigatus. These lung sentinel alveolar macrophages are also known to participate in the inflammatory response induced by A. fumigatus (44), but their role in driving the interferon response following A. fumigatus challenge was not addressed. During respiratory syncytial virus infection alveolar macrophages have been demonstrated to be key drivers of the type I interferon response through a MAVS-dependent mechanism (35, 36). We recently describe a critical role for MDA5/MAVS-dependent induction of interferons following A. fumigatus challenge (11), but the cellular localization of MAVS-dependent signaling was not elucidated. Here we demonstrated that Mavs expression in alveolar macrophages was at least partially necessary for host resistance against A. fumigatus, through the regulation of neutrophil accumulation in the lung for the prevention of fungal growth. While alveolar macrophage transfer to Mavs(Cd11c/Cd11c) mice largely restores host resistance, it was not absolute suggesting additional CD11c+ cells might require Mavs expression for resistance. One potential candidate would be plasmacytoid dendritic cells that are known to produce type I interferons after fungal challenge (27) and are essential for host resistance A. fumigatus (45), but the importance of MAVS signaling in plasmacytoid dendritic cells in driving the interferon response remains controversial. Additionally, in a dendritic cell vaccination model, A. fumigatus conidial RNA was shown to be sufficient for the activation of dendritic cells to preferentially prime a Th1 immune response (46), which was at least partially dependent on TLR3 (47). The role of MAVS signaling in other CD11c+ cell populations will be explored in future studies. Alveolar macrophages also participate in the inflammatory response to other fungal pathogens including Cryptococcus neoformans (48), Pneumocystis spp. (49), and Rhizopus spp. (50). Interestingly, heterogeneity within the alveolar macrophage population has been demonstrated particularly in regard to CXCL2 expression (48), which could be important in our findings showing that the mice lacking Ifih1 or Mavs have decreased neutrophil accumulation following A. fumigatus challenge (11). Additionally, Jakubzick and colleagues have identified significant heterogeneity within human alveolar macrophages using single cell RNA-Seq, which include the identification of novel IFN responsive alveolar macrophage populations and a number of chemokine producing populations (51). Understanding the specific role of these alveolar macrophage populations in host resistance is the goal of future studies.
In addition to the role of alveolar macrophages in the interferon-dependent antifungal immune response that we describe herein, it has previously been shown that in vitro human bronchial epithelial cells can secrete IFNβ and CXCL10 in response to A. fumigatus spore challenge (52). Following respiratory virus infections, the lung epithelium can be a critical source of antiviral interferons, which can be driven by both TLRs and RLRs (53, 54). In the human bronchial epithelial cell model, Bals and colleagues demonstrated that fungal dsRNA which was transfected into the human bronchial epithelial cells was sufficient for inducing IFNβ and CXCL10 secretion (52), which is similar to our previous findings using in murine fibroblast (11). However, while Bals and colleagues demonstrated that A. fumigatus dsRNA was sufficient for the activation of TLR3-TRIF signaling, they did not examine the necessity of this pathway in driving IFNβ and CXCL10 secretion by the human bronchial epithelial cells (52). Interestingly, in our preliminary studies, cytosolic delivery of A. fumigatus dsRNA drives both MDA5/MAVS-dependent and TRIF-dependent IFNα and CXCL10 responses in murine fibroblasts (data not shown). Moreover, Romani and colleagues have found both Tlr3- and Ticam1 (TRIF)-deficient mice are highly susceptible to pulmonary A. fumigatus challenge (47, 55), and this was due entirely to TRIF-dependent response in epithelial cells (55). Interestingly, in our HSCT patients, the IFIH1 SNP association was found in the host genotype. Taken together, these data suggest the lung epithelial cells could be another potential source of IFNs after A. fumigatus challenge which needs to be explored in future studies.
While our previous studies have shown the importance of MDA5/MAVS signaling (11) and type I/III interferons (9) in host resistance against A. fumigatus in the murine model, the importance of these pathways in humans has not been described. Herein, we have found that antifungal role of MDA5/MAVS signaling is also critical in humans. Our data from a cohort of HSCT patients demonstrate that SNPs in IFIH (rs1990760) and MAVS (rs17857295) are associated with an altered risk of developing IPA. Interestingly, SNPs within IFIH1 have been associated with numerous autoimmune diseases, particularly those that are highly dependent on type I interferons for their progression (19, 56). Recent evidence suggests that the autoimmune risk genotype of the rs1990760 SNP in IFIH1 (TT) has functional consequences in human PBMCs, specifically it is associated with elevated basal levels of IFNB1 (19). Knock-in of the IFIH1 rs1990760 autoimmune risk genotype (TT) into mice not only recapitulates the susceptibility to autoimmune diseases, but also results in increased host resistance against encephalomyocarditis virus (ECMV), a picornavirus specifically recognized by MDA5 (57), and elevated Ifnb1 expression (19). Moreover, in human pancreatic islets it has been shown that the IFIH1 rs1990760 TC genotype also drove enhanced interferon signaling after Coxsackievirus infection (20). This enhanced interferon response during Coxsackievirus infection was associated with greater signaling from the peroxisomes by the IFIH1 rs1990760 TC genotype (20), which has previously been shown to preferentially induce a type III interferon response (58, 59). Interestingly, our SNP analysis of a human HSCT cohort found that the T allele of IFIH1 rs1990760, which as just mentioned is associated with an increased interferon response (19, 20), was associated with a reduced risk for the development of IPA. In addition to the IFIH1 rs1990760 SNP, we also found a clinical association with the rs17857295 SNP in MAVS with the risk of developing IPA after HSCT. Much less is known about the functional outcome of the rs17857295 SNP in MAVS, but one report suggests overexpression of the GG genotype in 293T cells stably knocked-down for the endogenous MAVS allele resulted in an inability to induce Ifnb1 expression following PolyI:C stimulation (21). Overall, our analysis suggests that HSCT patients harboring these SNPs display a reduced ability to trigger an interferon response and are at greater risk of developing IPA.
In contrast to our clinical data from HSCT patients who develop IPA, the T allele at rs1990760 in IFIH1 is associated with an increased risk for the development of chronic mucocutaneous candidiasis (60). Again, the T allele at rs1990760 is associated with an increased interferon response (19, 20), but the lack of IFNAR-dependent signaling leads to increased susceptibility to invasive candidiasis in mice (12). In their study Netea and colleagues did not observe altered Ifnb expression in the absence of MDA5 after stimulation with Candida albicans hyphae (60), but a role for MDA5 in regulating other interferons was not explored. Our data with A. fumigatus suggests that MDA5/MAVS-signaling are more important in the regulation of type III interferons (IFNλ/IL-28), rather than type I interferons (IFNα/β). Excessive inflammation and tissue pathology can be critical in mediating disease during invasive candidiasis (61). Rivera and colleagues demonstrated that type III interferon (IFNλ/IL-28) signaling enhances the production of reactive oxygen species by neutrophils following A. fumigatus (9), which could enhance disease during invasive candidiasis. Thus, there appears to be an interesting dichotomy in MDA5 signaling that is crucial for tuning host resistance against different fungal pathogens and warrants further research.
Supplementary Material
Key Points/Findings:
Mavs expression in alveolar macrophages maintains resistance against A. fumigatus
A SNP in MAVS is associated with a risk of developing invasive aspergillosis
ACKNOWLEDGEMENTS
Thank you to Drs. Robert Cramer, Claudia Jakubzick, and David Leib (Geisel School of Medicine at Dartmouth) for helpful discussion on this project.
Footnotes
Research in this study was supported in part by institutional startup funds to JJO in part through the Dartmouth Lung Biology Center for Molecular, Cellular, and Translational Research grant P30 GM106394 (PI: Bruce A. Stanton) and Center for Molecular, Cellular and Translational Immunological Research grant P30 GM103415 (PI: William R. Green). JJO was partially supported by a Munck-Pfefferkorn Award from Dartmouth College and NIH R01 AI139133 grant. SMB is supported by the Division of Intramural Research, National Institutes of Health, National Institute of Allergy and Infectious Diseases. AC and CC were supported by the Fundação para a Ciência e a Tecnologia (FCT) (PTDC/SAU-SER/29635/2017, PTDC/MED-GEN/28778/2017, UIDB/50026/2020, UIDP/50026/2020, and CEECIND/04058/2018), the Northern Portugal Regional Operational Programme (NORTE 2020), under the Portugal 2020 Partnership Agreement, through the European Regional Development Fund (ERDF) (NORTE-01–0145-FEDER-000039), the European Union’s Horizon 2020 research and innovation programme under grant agreement no. 847507, and the “la Caixa” Foundation (ID 100010434) and FCT under the agreement LCF/PR/HR17/52190003. The funders had no role in the preparation or publication of the manuscript.
Abbrevations: IPA = invasive pulmonary aspergillosis; HCST = hematopoietic stem cell transplantation; GMM = glucose minimal media; PBS = phosphate buffered saline; i.t. = intratracheal; BALF = bronchoalveolar lavage fluid; GMS = Grocott-Gomori methenamine silver; SNP = single nucleotide polymorphism
Conflict of Interests: The authors have declared that no conflict of interest exists.
REFERENCES
- 1.Baddley JW, Andes DR, Marr KA, Kontoyiannis DP, Alexander BD, Kauffman CA, Oster RA, Anaissie EJ, Walsh TJ, Schuster MG, Wingard JR, Patterson TF, Ito JI, Williams OD, Chiller T, and Pappas PG. 2010. Factors associated with mortality in transplant patients with invasive aspergillosis. Clin Infect Dis 50: 1559–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steinbach WJ, Marr KA, Anaissie EJ, Azie N, Quan SP, Meier-Kriesche HU, Apewokin S, and Horn DL. 2012. Clinical epidemiology of 960 patients with invasive aspergillosis from the PATH Alliance registry. J Infect 65: 453–464. [DOI] [PubMed] [Google Scholar]
- 3.Garcia-Vidal C, Upton A, Kirby KA, and Marr KA. 2008. Epidemiology of invasive mold infections in allogeneic stem cell transplant recipients: biological risk factors for infection according to time after transplantation. Clin Infect Dis 47: 1041–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Upton A, Kirby KA, Carpenter P, Boeckh M, and Marr KA. 2007. Invasive aspergillosis following hematopoietic cell transplantation: outcomes and prognostic factors associated with mortality. Clin Infect Dis 44: 531–540. [DOI] [PubMed] [Google Scholar]
- 5.Thompson GR 3rd and Patterson TF. 2008. Pulmonary aspergillosis. Semin Respir Crit Care Med 29: 103–110. [DOI] [PubMed] [Google Scholar]
- 6.Romani L 2011. Immunity to fungal infections. Nat Rev Immunol 11: 275–288. [DOI] [PubMed] [Google Scholar]
- 7.Cunha C, Aversa F, Romani L, and Carvalho A. 2013. Human genetic susceptibility to invasive aspergillosis. PLoS Pathog 9: e1003434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campos CF, van de Veerdonk FL, Goncalves SM, Cunha C, Netea MG, and Carvalho A. 2019. Host Genetic Signatures of Susceptibility to Fungal Disease. Curr Top Microbiol Immunol 422: 237–263. [DOI] [PubMed] [Google Scholar]
- 9.Espinosa V, Dutta O, McElrath C, Du P, Chang YJ, Cicciarelli B, Pitler A, Whitehead I, Obar JJ, Durbin JE, Kotenko SV, and Rivera A. 2017. Type III interferon is a critical regulator of innate antifungal immunity Sci Immunol 2: eaan5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dutta O, Espinosa V, Wang K, Avina S, and Rivera A. 2020. Dectin-1 Promotes Type I and III Interferon Expression to Support Optimal Antifungal Immunity in the Lung. Front Cell Infect Microbiol 10: 321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, Caffrey-Carr AK, Liu KW, Espinosa V, Croteau W, Dhingra S, Rivera A, Cramer RA, and Obar JJ. 2020. MDA5 Is an Essential Sensor of a Pathogen-Associated Molecular Pattern Associated with Vitality That Is Necessary for Host Resistance against Aspergillus fumigatus. J Immunol 205: 3058–3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.del Fresno C, Soulat D, Roth S, Blazek K, Udalova I, Sancho D, Ruland J, and Ardavin C. 2013. Interferon-beta production via Dectin-1-Syk-IRF5 signaling in dendritic cells is crucial for immunity to C. albicans. Immunity 38: 1176–1186. [DOI] [PubMed] [Google Scholar]
- 13.Loo YM and Gale M Jr. 2011. Immune signaling by RIG-I-like receptors. Immunity 34: 680–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cunha C, Di Ianni M, Bozza S, Giovannini G, Zagarella S, Zelante T, D’Angelo C, Pierini A, Pitzurra L, Falzetti F, Carotti A, Perruccio K, Latge JP, Rodrigues F, Velardi A, Aversa F, Romani L, and Carvalho A. 2010. Dectin-1 Y238X polymorphism associates with susceptibility to invasive aspergillosis in hematopoietic transplantation through impairment of both recipient- and donor-dependent mechanisms of antifungal immunity. Blood 116: 5394–5402. [DOI] [PubMed] [Google Scholar]
- 15.Sainz J, Lupianez CB, Segura-Catena J, Vazquez L, Rios R, Oyonarte S, Hemminki K, Forsti A, and Jurado M. 2012. Dectin-1 and DC-SIGN polymorphisms associated with invasive pulmonary Aspergillosis infection. PLoS One 7: e32273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fisher CE, Hohl TM, Fan W, Storer BE, Levine DM, Zhao LP, Martin PJ, Warren EH, Boeckh M, and Hansen JA. 2017. Validation of single nucleotide polymorphisms in invasive aspergillosis following hematopoietic cell transplantation. Blood 129: 2693–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dutta M, Robertson SJ, Okumura A, Scott DP, Chang J, Weiss JM, Sturdevant GL, Feldmann F, Haddock E, Chiramel AI, Ponia SS, Dougherty JD, Katze MG, Rasmussen AL, and Best SM. 2017. A Systems Approach Reveals MAVS Signaling in Myeloid Cells as Critical for Resistance to Ebola Virus in Murine Models of Infection. Cell Rep 18: 816–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Pauw B, Walsh TJ, Donnelly JP, Stevens DA, Edwards JE, Calandra T, Pappas PG, Maertens J, Lortholary O, Kauffman CA, Denning DW, Patterson TF, Maschmeyer G, Bille J, Dismukes WE, Herbrecht R, Hope WW, Kibbler CC, Kullberg BJ, Marr KA, Munoz P, Odds FC, Perfect JR, Restrepo A, Ruhnke M, Segal BH, Sobel JD, Sorrell TC, Viscoli C, Wingard JR, Zaoutis T, Bennett JE, R. European Organization for, G. Treatment of Cancer/Invasive Fungal Infections Cooperative, A. National Institute of, and G. Infectious Diseases Mycoses Study Group Consensus. 2008. Revised definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group. Clin Infect Dis 46: 1813–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gorman JA, Hundhausen C, Errett JS, Stone AE, Allenspach EJ, Ge Y, Arkatkar T, Clough C, Dai X, Khim S, Pestal K, Liggitt D, Cerosaletti K, Stetson DB, James RG, Oukka M, Concannon P, Gale M Jr., Buckner JH, and Rawlings DJ. 2017. The A946T variant of the RNA sensor IFIH1 mediates an interferon program that limits viral infection but increases the risk for autoimmunity. Nat Immunol 18: 744–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Domsgen E, Lind K, Kong L, Huhn MH, Rasool O, van Kuppeveld F, Korsgren O, Lahesmaa R, and Flodstrom-Tullberg M. 2016. An IFIH1 gene polymorphism associated with risk for autoimmunity regulates canonical antiviral defence pathways in Coxsackievirus infected human pancreatic islets. Sci Rep 6: 39378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xing F, Matsumiya T, Hayakari R, Yoshida H, Kawaguchi S, Takahashi I, Nakaji S, and Imaizumi T. 2016. Alteration of Antiviral Signalling by Single Nucleotide Polymorphisms (SNPs) of Mitochondrial Antiviral Signalling Protein (MAVS). PLoS One 11: e0151173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goncalves SM, Lagrou K, Rodrigues CS, Campos CF, Bernal-Martinez L, Rodrigues F, Silvestre R, Alcazar-Fuoli L, Maertens JA, Cunha C, and Carvalho A. 2017. Evaluation of Bronchoalveolar Lavage Fluid Cytokines as Biomarkers for Invasive Pulmonary Aspergillosis in At-Risk Patients. Front Microbiol 8: 2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gray RJ 1988. A Class of K-Sample Tests for Comparing the Cumulative Incidence of a Competing Risk. The Annals of Statistics 16: 1141–1154. [Google Scholar]
- 24.Dignam JJ and Kocherginsky MN. 2008. Choice and interpretation of statistical tests used when competing risks are present. J Clin Oncol 26: 4027–4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scrucca L, Santucci A, and Aversa F. 2007. Competing risk analysis using R: an easy guide for clinicians. Bone Marrow Transplant 40: 381–387. [DOI] [PubMed] [Google Scholar]
- 26.Bauer JW, Baechler EC, Petri M, Batliwalla FM, Crawford D, Ortmann WA, Espe KJ, Li W, Patel DD, Gregersen PK, and Behrens TW. 2006. Elevated serum levels of interferon-regulated chemokines are biomarkers for active human systemic lupus erythematosus. PLoS Med 3: e491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramirez-Ortiz ZG, Lee CK, Wang JP, Boon L, Specht CA, and Levitz SM. 2011. A nonredundant role for plasmacytoid dendritic cells in host defense against the human fungal pathogen Aspergillus fumigatus. Cell Host Microbe 9: 415–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Espinosa V, Jhingran A, Dutta O, Kasahara S, Donnelly R, Du P, Rosenfeld J, Leiner I, Chen CC, Ron Y, Hohl TM, and Rivera A. 2014. Inflammatory monocytes orchestrate innate antifungal immunity in the lung. PLoS Pathog 10: e1003940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hohl TM, Rivera A, Lipuma L, Gallegos A, Shi C, Mack M, and Pamer EG. 2009. Inflammatory monocytes facilitate adaptive CD4 T cell responses during respiratory fungal infection. Cell Host Microbe 6: 470–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zelante T, Wong AY, Ping TJ, Chen J, Sumatoh HR, Vigano E, Hong Bing Y, Lee B, Zolezzi F, Fric J, Newell EW, Mortellaro A, Poidinger M, Puccetti P, and Ricciardi-Castagnoli P. 2015. CD103(+) Dendritic Cells Control Th17 Cell Function in the Lung. Cell Rep 12: 1789–1801. [DOI] [PubMed] [Google Scholar]
- 31.Ibrahim-Granet O, Philippe B, Boleti H, Boisvieux-Ulrich E, Grenet D, Stern M, and Latge JP. 2003. Phagocytosis and intracellular fate of Aspergillus fumigatus conidia in alveolar macrophages. Infect Immun 71: 891–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grimm MJ, Vethanayagam RR, Almyroudis NG, Dennis CG, Khan AN, D’Auria AC, Singel KL, Davidson BA, Knight PR, Blackwell TS, Hohl TM, Mansour MK, Vyas JM, Rohm M, Urban CF, Kelkka T, Holmdahl R, and Segal BH. 2013. Monocyte- and macrophage-targeted NADPH oxidase mediates antifungal host defense and regulation of acute inflammation in mice. J Immunol 190: 4175–4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhatia S, Fei M, Yarlagadda M, Qi Z, Akira S, Saijo S, Iwakura Y, van Rooijen N, Gibson GA, St Croix CM, Ray A, and Ray P. 2011. Rapid host defense against Aspergillus fumigatus involves alveolar macrophages with a predominance of alternatively activated phenotype. PLoS One 6: e15943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heng TS, Painter MW, and C. Immunological Genome Project. 2008. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol 9: 1091–1094. [DOI] [PubMed] [Google Scholar]
- 35.Goritzka M, Makris S, Kausar F, Durant LR, Pereira C, Kumagai Y, Culley FJ, Mack M, Akira S, and Johansson C. 2015. Alveolar macrophage-derived type I interferons orchestrate innate immunity to RSV through recruitment of antiviral monocytes. J Exp Med 212: 699–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kirsebom FCM, Kausar F, Nuriev R, Makris S, and Johansson C. 2019. Neutrophil recruitment and activation are differentially dependent on MyD88/TRIF and MAVS signaling during RSV infection. Mucosal Immunol 12: 1244–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guilliams M, De Kleer I, Henri S, Post S, Vanhoutte L, De Prijck S, Deswarte K, Malissen B, Hammad H, and Lambrecht BN. 2013. Alveolar macrophages develop from fetal monocytes that differentiate into long-lived cells in the first week of life via GM-CSF. J Exp Med 210: 1977–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Svedberg FR, Brown SL, Krauss MZ, Campbell L, Sharpe C, Clausen M, Howell GJ, Clark H, Madsen J, Evans CM, Sutherland TE, Ivens AC, Thornton DJ, Grencis RK, Hussell T, Cunoosamy DM, Cook PC, and MacDonald AS. 2019. The lung environment controls alveolar macrophage metabolism and responsiveness in type 2 inflammation. Nat Immunol 20: 571–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zinter MS and Hume JR. 2021. Effects of Hematopoietic Cell Transplantation on the Pulmonary Immune Response to Infection. Front Pediatr 9: 634566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thomas ED, Ramberg RE, Sale GE, Sparkes RS, and Golde DW. 1976. Direct evidence for a bone marrow origin of the alveolar macrophage in man. Science 192: 1016–1018. [DOI] [PubMed] [Google Scholar]
- 41.Winston DJ, Territo MC, Ho WG, Miller MJ, Gale RP, and Golde DW. 1982. Alveolar macrophage dysfunction in human bone marrow transplant recipients. Am J Med 73: 859–866. [DOI] [PubMed] [Google Scholar]
- 42.Nakata K, Gotoh H, Watanabe J, Uetake T, Komuro I, Yuasa K, Watanabe S, Ieki R, Sakamaki H, Akiyama H, Kudoh S, Naitoh M, Satoh H, and Shimada K. 1999. Augmented proliferation of human alveolar macrophages after allogeneic bone marrow transplantation. Blood 93: 667–673. [PubMed] [Google Scholar]
- 43.Mircescu MM, Lipuma L, van Rooijen N, Pamer EG, and Hohl TM. 2009. Essential role for neutrophils but not alveolar macrophages at early time points following Aspergillus fumigatus infection. J Infect Dis 200: 647–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dubourdeau M, Athman R, Balloy V, Philippe B, Sengmanivong L, Chignard M, Philpott DJ, Latge JP, and Ibrahim-Granet O. 2006. Interaction of Aspergillus fumigatus with the alveolar macrophage. Med Mycol 44: S213–S217. [DOI] [PubMed] [Google Scholar]
- 45.Guo Y, Kasahara S, Jhingran A, Tosini NL, Zhai B, Aufiero MA, Mills KAM, Gjonbalaj M, Espinosa V, Rivera A, Luster AD, and Hohl TM. 2020. During Aspergillus Infection, Monocyte-Derived DCs, Neutrophils, and Plasmacytoid DCs Enhance Innate Immune Defense through CXCR3-Dependent Crosstalk. Cell Host Microbe doi: 10.1016/j.chom.2020.1005.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bozza S, Perruccio K, Montagnoli C, Gaziano R, Bellocchio S, Burchielli E, Nkwanyuo G, Pitzurra L, Velardi A, and Romani L. 2003. A dendritic cell vaccine against invasive aspergillosis in allogeneic hematopoietic transplantation. Blood 102: 3807–3814. [DOI] [PubMed] [Google Scholar]
- 47.Carvalho A, De Luca A, Bozza S, Cunha C, D’Angelo C, Moretti S, Perruccio K, Iannitti RG, Fallarino F, Pierini A, Latge JP, Velardi A, Aversa F, and Romani L. 2012. TLR3 essentially promotes protective class I-restricted memory CD8(+) T-cell responses to Aspergillus fumigatus in hematopoietic transplanted patients. Blood 119: 967–977. [DOI] [PubMed] [Google Scholar]
- 48.Xu-Vanpala S, Deerhake ME, Wheaton JD, Parker ME, Juvvadi PR, MacIver N, Ciofani M, and Shinohara ML. 2020. Functional heterogeneity of alveolar macrophage population based on expression of CXCL2. Sci Immunol 5: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bhagwat SP, Gigliotti F, Wang J, Wang Z, Notter RH, Murphy PS, Rivera-Escalera F, Malone J, Jordan MB, Elliott MR, and Wright TW. 2018. Intrinsic Programming of Alveolar Macrophages for Protective Antifungal Innate Immunity Against Pneumocystis Infection. Front Immunol 9: 2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Andrianaki AM, Kyrmizi I, Thanopoulou K, Baldin C, Drakos E, Soliman SSM, Shetty AC, McCracken C, Akoumianaki T, Stylianou K, Ioannou P, Pontikoglou C, Papadaki HA, Tzardi M, Belle V, Etienne E, Beauvais A, Samonis G, Kontoyiannis DP, Andreakos E, Bruno VM, Ibrahim AS, and Chamilos G. 2018. Iron restriction inside macrophages regulates pulmonary host defense against Rhizopus species. Nat Commun 9: 3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li X, Kolling FW, Aridgides D, Mellinger D, Ashare A, and Jakubzick CV. 2022. ScRna-seq expression of IFI27and APOC2 identifies four alveolar macrophage superclusters in healthy BALF. bioRxiv [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beisswenger C, Hess C, and Bals R. 2012. Aspergillus fumigatus conidia induce interferon-beta signalling in respiratory epithelial cells. Eur Respir J 39: 411–418. [DOI] [PubMed] [Google Scholar]
- 53.Ioannidis I, Ye F, McNally B, Willette M, and Flano E. 2013. Toll-like receptor expression and induction of type I and type III interferons in primary airway epithelial cells. J Virol 87: 3261–3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Crotta S, Davidson S, Mahlakoiv T, Desmet CJ, Buckwalter MR, Albert ML, Staeheli P, and Wack A. 2013. Type I and type III interferons drive redundant amplification loops to induce a transcriptional signature in influenza-infected airway epithelia. PLoS Pathog 9: e1003773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de Luca A, Bozza S, Zelante T, Zagarella S, D’Angelo C, Perruccio K, Vacca C, Carvalho A, Cunha C, Aversa F, and Romani L. 2010. Non-hematopoietic cells contribute to protective tolerance to Aspergillus fumigatus via a TRIF pathway converging on IDO. Cell Mol Immunol 7: 459–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cen H, Wang W, Leng RX, Wang TY, Pan HF, Fan YG, Wang B, and Ye DQ. 2013. Association of IFIH1 rs1990760 polymorphism with susceptibility to autoimmune diseases: a meta-analysis. Autoimmunity 46: 455–462. [DOI] [PubMed] [Google Scholar]
- 57.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, Yamaguchi O, Otsu K, Tsujimura T, Koh CS, Reis e Sousa C, Matsuura Y, Fujita T, and Akira S. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441: 101–105. [DOI] [PubMed] [Google Scholar]
- 58.Odendall C, Dixit E, Stavru F, Bierne H, Franz KM, Durbin AF, Boulant S, Gehrke L, Cossart P, and Kagan JC. 2014. Diverse intracellular pathogens activate type III interferon expression from peroxisomes. Nat Immunol 15: 717–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dixit E, Boulant S, Zhang Y, Lee AS, Odendall C, Shum B, Hacohen N, Chen ZJ, Whelan SP, Fransen M, Nibert ML, Superti-Furga G, and Kagan JC. 2010. Peroxisomes are signaling platforms for antiviral innate immunity. Cell 141: 668–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jaeger M, van der Lee R, Cheng SC, Johnson MD, Kumar V, Ng A, Plantinga TS, Smeekens SP, Oosting M, Wang X, Barchet W, Fitzgerald K, Joosten LA, Perfect JR, Wijmenga C, van de Veerdonk FL, Huynen MA, Xavier RJ, Kullberg BJ, and Netea MG. 2015. The RIG-I-like helicase receptor MDA5 (IFIH1) is involved in the host defense against Candida infections. Eur J Clin Microbiol Infect Dis 34: 963–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pappas PG, Lionakis MS, Arendrup MC, Ostrosky-Zeichner L, and Kullberg BJ. 2018. Invasive candidiasis. Nat Rev Dis Primers 4: 18026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


