Abstract
Campylobacter jejuni is a significant cause of human gastroenteritis worldwide and all strains express an N-glycan that is added to at least 80 different proteins. We characterized 98 C. jejuni isolates from infants from seven low- and middle-income countries and identified 4 isolates unreactive with our N-glycan-specific antiserum that was raised against the C. jejuni heptasaccharide composed of GalNAc-GalNAc-GalNAc(Glc)-GalNAc-GalNAc-diNAcBac. Mass spectrometric analyses indicated these isolates express a hexasaccharide lacking the glucose branch. Although all 4 strains encode the PglI glucosyltransferase (GTase), one aspartate in the DXDD motif was missing, an alteration also present in ~4% of all available PglI sequences. Deleting this residue from an active PglI resulted in a non-functional GTase when the protein glycosylation system was reconstituted in E. coli, while replacement with Glu/Ala was not deleterious. Molecular modelling proposed a mechanism for how the DXDD residues and the structure/length beyond the motif influences activity. Mouse vaccination with an E. coli strain expressing the full-length heptasaccharide produced N-glycan-specific antibodies and a corresponding reduction in Campylobacter colonization and weight loss following challenge. However, the antibodies did not recognize the hexasaccharide and were unable to opsonize C. jejuni isolates lacking glucose suggesting this should be considered when designing N-glycan based vaccines to prevent campylobacteriosis.
Keywords: Campylobacter, N-glycan, glycosyltransferase, PglI, immunogenicity, vaccine
Graphical Abstract
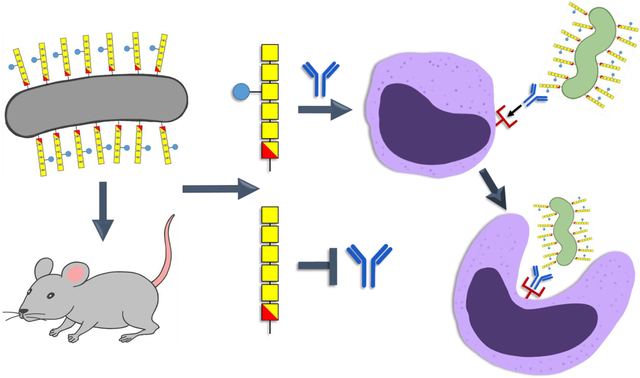
Immune responses to engineered Escherichia coli expressing the Campylobacter jejuni N-glycan heptasaccharide. The attenuated live E. coli vaccine (grey) was administered through intranasal delivery or intragastric gavage in mice. In both cases, IgG antibodies were generated that recognized the bacterial heptasaccharide (GalNAc-GalNAc-GalNAc(Glc)-GalNAc-GalNAc-diNAcBac), but did not react with the hexasaccharide lacking the glucose branch (blue circle). Mouse heptasaccharide antibodies were also active in C. jejuni (green) opsonophagocytosis assays (OPA). Schematic created by Bibi Zhou.
Introduction
Campylobacter jejuni is the major cause of diarrhea worldwide1 and among the most prevalent of enteropathogens in recent pediatric enteric disease investigations including the Global Enteric Multicenter (GEMS) and the Malnutrition and Enteric Diseases (MAL-ED) studies2–3. Infection with C. jejuni has also been associated with serious post-infectious sequelae such as Guillain-Barré syndrome, irritable bowel syndrome, reactive arthritis, and pediatric growth stunting in low-resource settings4–7. Although Campylobacter is generally susceptible to antibiotics, increasing resistance, particularly to fluoroquinolones, is a growing concern8. Multiple approaches have been used to target C. jejuni including phages9, probiotics10, general biosecurity measures11–12, and various vaccination strategies13–14. However, the development of an effective vaccine to prevent C. jejuni-associated disease is hindered by the variability of its surface-antigens including capsular polysaccharides, lipooligosaccharides (LOS) and O-linked flagellar glycans, that would otherwise be effective targets13. Despite this variability, all Campylobacter species produce a unique and conserved N-glycan structure, that in the case of C. jejuni is a heptasaccharide15–16 capable of inducing an immune response in humans, rabbits and mice17–18. In poultry, the major source attributed to foodborne Campylobacter infections, an N-glycan-based vaccine has been shown to induce an antigen-specific response and to significantly reduce the Campylobacter load19–20, while in human volunteers, antibodies formed against natural glycoproteins are not protective18.
The process of protein N-glycosylation in C. jejuni is well understood. An undecaprenyl-pyrophosphate-linked heptasaccharide is assembled on the cytosolic side of the inner membrane21–22 by the sequential action of five glycosyltransferases: the initiating GTase, PglC, adds UDP-diNAcBac (di-N-acetylbacillosamine or 2,4-diacetamido-2,4,6-trideoxyglucopyranose) to the lipid carrier undecaprenyl phosphate (Und-P) releasing UMP; PglA and PglJ transfer the second and the third N-acetylgalactosamine (GalNAc) residues; PglH adds another 3 GalNAcs; and PglI adds the final glucose (Glc) branch23–25. The heptasaccharide is then flipped to the periplasm by the ABC transporter PglK26 and transferred to the amino group of asparagine residues within the consensus sequence, [D/E-X1-N-X2-S/T], where X1 and X2 can be any amino acid except proline, by the oligosaccharyltransferase, PglB15, 27–28. At least 80 different proteins have been identified to be modified with the N-glycan29–30. The importance of this posttranslational modification is emphasized by the fact that complete loss of N-glycosylation (in a pglB mutant) and mutations in the biosynthetic pathway (e.g. pglH, pglJ, pglD, pglE or pglF mutants24, 31–34) result in similar phenotypes, despite the fact that in certain pathway mutants N-glycosylation still occurs (albeit with lower efficiency) and shorter N-glycans are added to proteins35–36. In addition, recent studies indicated that N-glycosylation of CmeA, of the CmeABC efflux pump, is not only required for bile salt and antibiotic resistance, but also for chicken colonization37–38, suggesting that a selective pressure exists to maintain this pathway. However, the relevance of the glucose on the N-glycan is intriguing. Loss of PglI in C. jejuni results in reduced chicken colonization24, 39, suggesting that the glucose branch is required, but not absolutely necessary. Moreover, in some instances, the glucose branch contributes to immunogenicity of the structure; it has been shown that rabbit antisera independently raised against the N-glycan either recognize (hR6) or do not recognize (R1) the N-glycan without the glucose branch16. In addition, other thermotolerant Campylobacter species including C. lari, lack pglI16, but can still reside in their avian hosts40–42. Therefore, the question of whether the Glc branch is required for survival and/or pathogenicity only in specific hosts remains to be answered.
Here we present an N-glycan survey of 98 C. jejuni and C. coli isolates and identify 4 strains lacking the Glc branch despite the presence of an intact pglI gene. Sequence comparisons showed that those strains are missing the terminal Asp in the 91DDDD94 sequence comprising the DXDD motif within the PglI GTase. Reconstitution of the pgl system in E. coli revealed that those PglIs indeed have no or very low GTase activity. Modelling and analyses of PglI point mutants showed that it is the length of the loop rather than the presence of Asp94 that is important for activity. Moreover, vaccination of mice with an engineered E. coli strain presenting the full-length N-glycan structure on its LOS significantly reduced the C. jejuni load, but the polyclonal N-glycan antibodies did not recognize the hexasaccharide, indicating that the Glc branch is part of the immunogenic epitope. These findings increase our understanding of this posttranslational modification and emphasize that variations in the N-glycan exist and should be taken into consideration when developing an effective vaccine against C. jejuni.
Results and Discussion
N-glycan survey of Campylobacter isolates.
All thermophilic Campylobacter species (including C. jejuni (Cj), C. coli, C. upsaliensis and C. lari) modify their glycoproteins with N-glycan structures. In Cj, C. coli and C. upsaliensis, the N-glycan consists of GalNAc-α1,4-GalNAc-α1,4-[Glcβ1,3-]GalNAc-α1,4-GalNAc-α1,4-GalNAcα1,3-diNAcBac-β1, where diNAcBac is di-N-acetylbacillosamine or 2,4-diacetamido-2,4,6-trideoxyglucopyranose15, 28, whereas C. lari and C. lari-related species synthesize a hexasaccharide lacking the Glc branch. In contrast, non-thermophilic Campylobacters (e. g. C. fetus and C. concisus) express a diverse range of N-glycan structures on their glycoproteins16, 43. To investigate the commonality of the Cj-N-glycan more broadly in settings where Campylobacter infections are most prevalent and have an impact on child development, we surveyed 98 Campylobacter strains that were isolated from infants <1 year of age in seven different low-to-middle-income countries (LMICs) from the GEMS study2 (Table S3).
We found that 94 isolates produce the C. jejuni N-glycan heptasaccharide on multiple proteins as evidenced by reactivity with our Cj-N-glycan specific (R1) antiserum (Figure 1A, Figure S1). Interestingly, 4 strains (1H2, 1J2, 1J5, and 1D7) did not react with R1 indicating that either no or structurally different N-glycans are attached to the proteins. Whole cell lysates of C. jejuni strain NCTC 11168 (WT), proven to N-glycosylate proteins with the heptasaccharide, and the isogenic pglB mutant, devoid of N-glycans, served as positive and negative controls, respectively. Coomassie-stained SDS-PAGE gels were run in parallel as loading controls.
Figure 1. The N-glycosylation status of various Campylobacter isolates.
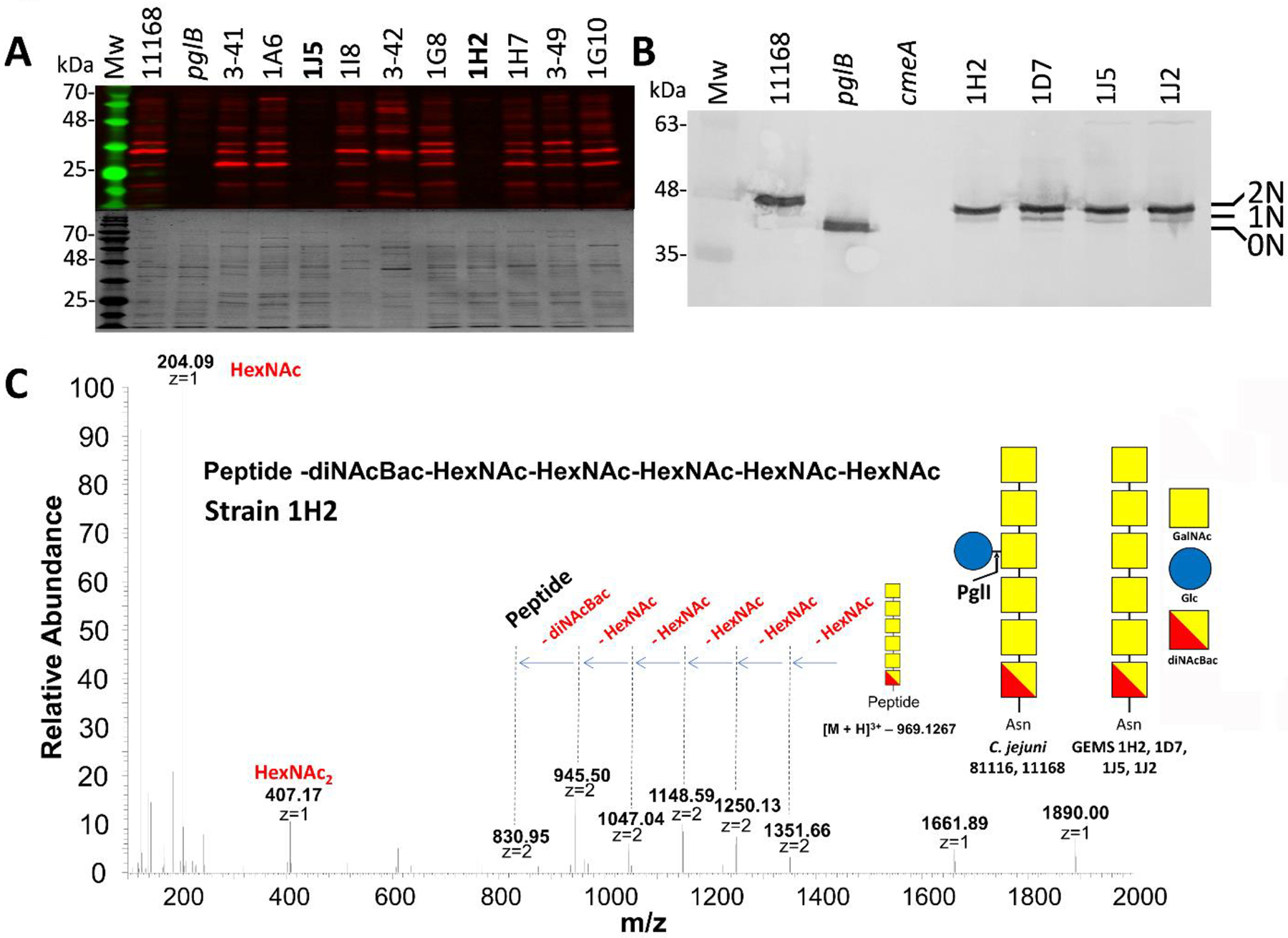
(A) Western blot (upper panel) with Cj-N-glycan specific R1 antiserum of equal amounts (3 μg) of whole cell lysates of C. jejuni WT (NCTC 11168), the isogenic pglB mutant (pglB) and other C. jejuni and C. coli isolates. Coomassie blue-stained gels were run in parallel as loading controls (lower panel). Western blots and Coomassie blue-stained gels for all the other GEMS isolates are provided in Figure S1. The full list of GEMS strains is provided in Table S3. (B) Western blot with Cj-CmeA-specific antiserum of C. jejuni WT (NCTC 11168), the isogenic pglB mutant (pglB), the Cj-cmeA mutant (cmeA, background control) and the GEMS strains (as indicated) that did not react with R1 serum. Non-, mono- and di-glycosylated CmeA proteins are indicated as 0N, 1N and 2N, respectively. Note that mono-glycosylated CmeA cannot be detected in the Cj-WT since it produces only di-glycosylated (2N) CmeA while the N-glycan is absent in the pglB mutant (0N). Molecular weight markers (Mw in kDa) are indicated on the left. A full top-to-bottom scan is provided in Figure S2. (C) Liquid chromatography-mass spectrometry (LC-MS)-based analyses of CmeA glycopeptides. HCD MS2 spectrum showing the fragmentation pattern of the carbohydrate portion on the CmeA peptide resulted in the identification of a HexNAc5-diNAcBac N-glycan attached to CmeA of C. jejuni 1H2. Spectra for N-glycopeptides from strains 1J2, 1J5 and 1D7 are provided in Figure S3. Since the genome and/or proteome of the GEMS strains was not available for databank searches, only the attached glycan structure was sequenced. The symbolic representation of the glucose-containing N-glycan from C. jejuni strain NCTC11168 is depicted for comparison; analysis of the full-length C. jejuni heptasaccharide N-glycan (HexNAc-HexNAc-HexNAc(Hex)-HexNAc-HexNAc-diNAcBac) attached to CmeA of 11168 WT is in Figure S3.
GEMS strain CmeA proteins unreactive with N-glycan sera are still glycosylated.
To investigate the R1-negative phenotype in the 4 GEMS strains in more detail, the glycosylation status of the CmeA protein, known be N-glycosylated at two sites in C. jejuni strain NCTC 11168 was analyzed by western blotting with CmeA-specific antiserum (Figure 1B, Figure S2). The signal corresponding to CmeA in the 4 R1-negative GEMS strains migrated at a higher molecular weight when compared to the signal of the CmeA from Cj-pglB mutant (non-glycosylated, negative control), but at a slightly lower molecular weight when compared to the Cj-WT. This indicated that CmeA in the GEMS strains might still be N-glycosylated, but with a shorter N-glycan which is not recognized by the heptasaccharide specific antibody. In addition, the presence of a signal potentially corresponding to mono-glycosylated CmeA in the GEMS strains (that is not visible in the Cj-WT) could potentially result from a reduced transfer activity for the GEMS N-glycan by the oligosaccharyltransferase PglB or just be a natural feature in these strains. No signal was present in whole cell lysates of the Cj-cmeA mutant that was included as a negative control.
Further investigation by liquid chromatography-mass spectrometry (LC-MS) of CmeA-derived glycopeptides from all 4 GEMS strains revealed that their CmeA was modified exclusively with a hexasaccharide (Figure 1C, Figure S3) comprised of HexNAc5-diNAcBac. This strongly indicated that those 4 GEMS strains glycosylate CmeA with an N-glycan similar to the C. jejuni heptasaccharide, but lack the glucose branch, the product of the C. jejuni PglI GTase.
PglI proteins from GEMS strains unreactive with the N-glycan-specific antiserum are lacking a terminal aspartate (Asp94) in the DXDD motif.
Loss of reactivity with the N-glycan-specific antiserum and the presence of only a hexasaccharide lacking Glc suggested that these specific C. jejuni isolates are either lacking the pglI gene or do not produce a functional PglI GTase.
We confirmed the presence of the pglI gene in all 4 R1-negative strains by PCR with pglI specific primers followed by sequencing and aligning the deduced amino acid (aa) sequences with PglIs from two reference strains C. jejuni 11168 and 81116 (Figure 2). Sequence comparisons revealed 99.03% (for GEMS strains 1D7 and 1H2) and 98.06% (for GEMS strains 1J2 and 1J5) similarity to PglI of Cj-81116; however, all 4 GEMS PglIs are one amino acid shorter, lacking D94 within the sequence 88TFLDDDDE95. Cj-PglI belongs to the GT-A-fold, GTase2 (GT-2) family (http://www.cazy.org/GT2.html) transferring Glc from the UDP-α-Glc donor to the 3rd GalNAc of the N-glycan acceptor, creating the β-linked product and completing the C. jejuni heptasaccharide24, 44. Despite their low sequence similarity, members of the GT2 family are structurally similar45 and the DXD or the extended DXDD motif that is predominantly found in these 3-glycosyl-and galactosyltransferases (reviewed by46) is required for coordinating a divalent cation necessary for function and interaction with the nucleotide phosphodiester of the donor45, 47–48.
Figure 2. Multiple sequence alignment of Campylobacter GEMS PglI proteins.
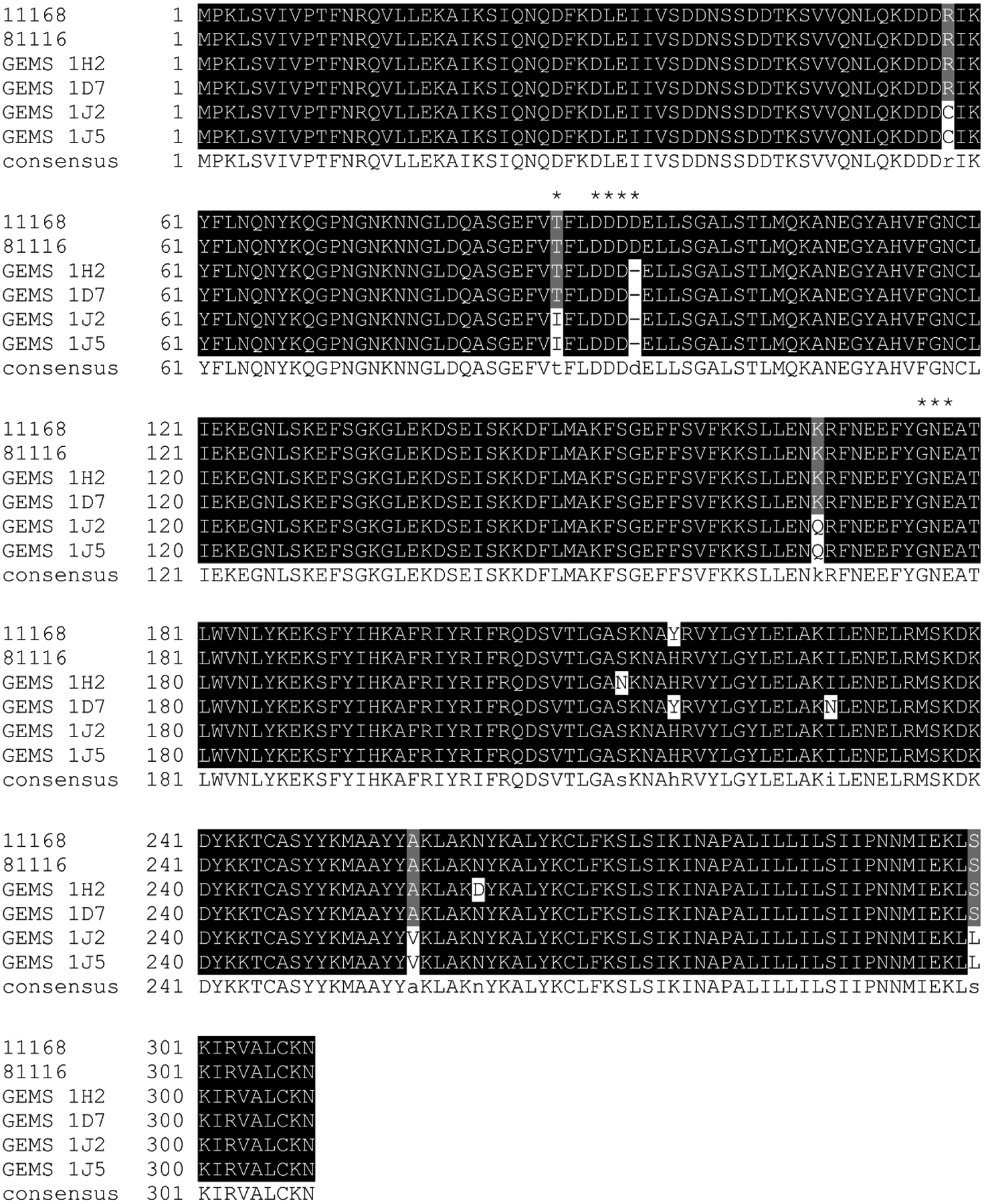
The DXDD motif as found in the reference strains Cj-11168 and Cj-81116 (where glycosylation of the N-glycan takes place to form the full length heptasaccharide) as well as the 177Gly-Asn-Glu179 motif that corresponds to the x-Glu-Asp motif containing the catalytic base in other related GTases are highlighted by asterisks. Highlighted in black are amino acids that are invariable, gray highlighting indicates amino acid conservation in at least 50% of the sequences, white indicates a non-conserved amino acid, and (−) marks a deletion at this position.
In addition, strains 1J5 and 1J2 (that are 100% identical at the aa level) contained a T88I mutation, and 4 additional aa changes were found at positions 58, 168, 257 and 300 relative to the WT sequence. In contrast, we found (in addition to the Asp deletion) aa 217 and 229 altered in strain 1D7 and aa 213 and 262 altered in strain 1H2 (Figure 2). To shed more light on the distribution of the active site Asp stretch and the underlying coding sequence, a survey of >700 publicly available pglI nucleotide sequences from C. jejuni and C. coli showed that in C. coli the predominant sequence is GAT-(GAT/GAC)-GAT-GAT-GAG (DDDDE); the sequence encoding DDDDE in C. jejuni is slightly different (GAT/GAC)-GAT-(GAT/GAC)-GAT-(GAG/GAA) (Figure S4). Moreover, 14 out of 291 C. jejuni strains (4.8%) and 2/109 C. coli strains (1.8%) were missing the same aspartate residue that is missing in the GEMS strains, and two strains have a 5 Asp stretch (Figure S5). Interestingly, the alleles encoding 3 Asp all have the sequence (GAT-GAT-GAT), followed by GAG (for Glu) in C. coli and by either GAG or GAA (for Glu) in C. jejuni; the 2 strains that have an additional Asp (5Ds) have an additional GAT (Figure S4). The fact that this variation is located around a repeat (GAT stretch) suggests that this tract could be hypervariable like the homo-polymeric G or C tracts conferring phase variation of surface antigens by slipped-strand mispairing in Campylobacters49–51. Such a mechanism would require that some cells within a population have +/− 1 GAT and regulate PglI functionality dependent on environmental pressure; however, sequencing of the GEMS pglI PCR products did not reveal such a variation. Moreover, the fact that truncated N-glycans (without the Glc branch) have not been observed during proteomics studies16, 29–30, 52 and that the 4 GEMS strains exclusively produce the hexasaccharide (at least on CmeA), suggests that this variation most likely reflects a “standard” non-hypervariable mutation centered at a repeat motif that is, once manifested in this location, a stable variant.
Overall, we found that the sequence variation observed within the GEMS strains (lack of one Asp) is more widespread among previously-sequenced isolates with the potential that in all those strains the functionality of the PglI GTase could be affected. In addition, PSI BLAST, including 1000 GTase-A/GT2 proteins, revealed conservation of the DXD(D) motif at this position of the PglI protein; however, other sequence motifs identified and described for GTase-A/GT2 family members, including the 179x-Glu-Asp180 motif containing the catalytic base53 display less conservation (xED) corresponds to 177Gly-Asn-Glu179 in PglI) and/or were absent in Cj-PglI (Figure 2, Figure S6).
Lack of Asp94 in the GEMS PglI impacts GTase activity.
Functional analysis of the PglI proteins was carried out by reconstituting the C. jejuni N-glycosylation system in the E. coli 3-plasmid system by complementing the pglI gene mutation on plasmid pACYC184pgl/ΔpglI (as described54). Plasmid pACYC184pgl (expressing all components of the N-glycosylation pathway for biosynthesis of the full-length Cj-N-glycan/heptasaccharide) and pACYC184pgl/ΔpglI complemented with pglI from Cj-81116 were used as positive controls. Similar to the native CmeA protein in whole cell lysates of C. jejuni, glycosylation is observed by an increase in mass of CmeA-His6 that was used as the N-glycan acceptor. 0N, 1N and 2N correspond to non-, mono- and double-glycosylated CmeA-His6, respectively (Figure 3, Figure S7). Although the lack of pglI upon co-expression of pACYC184pgl/ΔpglI and CmeA-His6 also resulted in 3 bands with anti-His serum, only low/background reactivity with R1 was observed since CmeA-His6 is glycosylated with only the hexasaccharide and R1 does not react when the N-glycan lacks glucose. As expected, expression of the GEMS PglIs in the pACYC184pgl/ΔpglI, CmeA-His6 background resulted in no (GEMS strains 1J1 and 1J5) or very low (above background) reactivity (GEMS strains 1D7 and 1H2) with R1, the latter case indicating the presence of minor amounts of the full length Cj-N-glycan. However, in the GEMS strains no heptasaccharide was detected in 1D7 and 1H2 and the effect could either be a result of overexpressing the PglI proteins in the E. coli system and/or due to additional variations in the amino acid sequence beyond the variation in the DxDD motif which could also affect activity.
Figure 3. Functional analysis of PglI GTases in the heterologous E. coli glycosylation system.
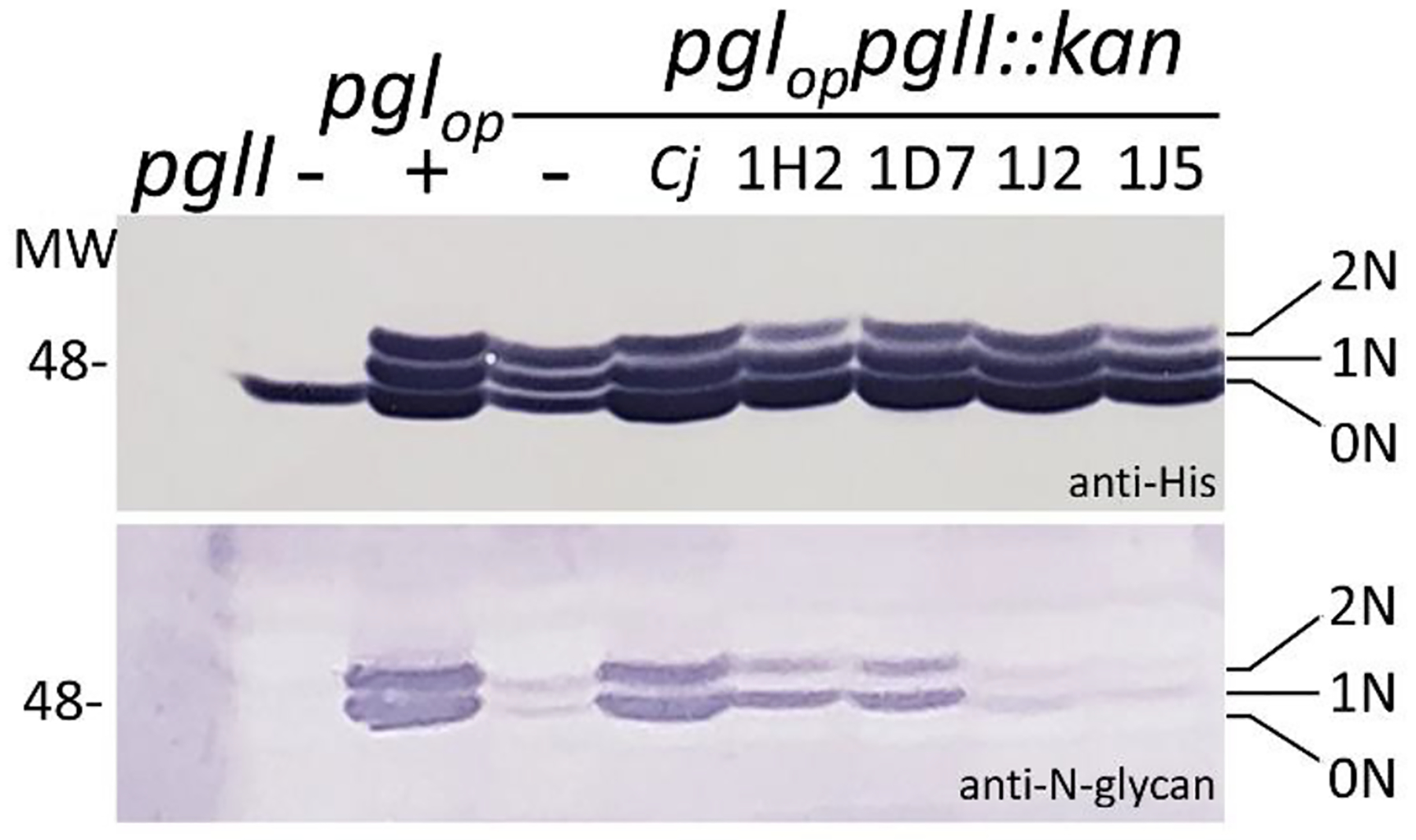
GTase-activities of GEMS PglIs in the E. coli 3-plasmid complementation system were analysed in whole cell lysates of E. coli CLM24 by western blotting; 5 μg of total protein were loaded in each lane. His-tag specific antibodies (upper panel) detect the glycosylation status of the glycan acceptor protein CmeA-His6; N-glycan-specific antibodies (R1, lower panel) detect the full-length Cj-N-glycan on CmeA-His6 that is only formed upon expression of an active PglI protein. Gene/plasmid combinations present in the underlying strains are indicated on top of each lane. Non, -mono, -and di-glycosylated CmeA-His6 proteins are indicated by 0N, 1N, and 2N, respectively. Molecular weight markers (Mw) in kDa are indicated on the left. Top-to-bottom scans are provided in the Figure S7.
Asp94 is required but not sufficient for GEMS strain PglI activity.
Since all 4 GEMS strains showed an amino acid deletion at exactly the same position (Asp94), we investigated whether the loss of Asp94 within in the aa sequence 88TFLDDDDE95 contributes to the loss and/or reduction of PglI activity. Mutants were created in Cj-81116 PglI and in the 1D7 and 1J2 PglIs as representatives, since 1J5 and 1J2 are 100% identical and 1D7 and 1H2 share the same low-activity phenotype when expressed in E. coli. PglI mediated Glc transfer (i.e. the formation of the full-length Cj-N-glycan) was analyzed with R1 antiserum after western blotting of whole cell lysates of the respective E. coli strains as shown in Figure 4A (and Figure S8A). Indeed, deletion of Asp94 in Cj-81116 (ΔD PglI) resulted in an inactive PglI protein since the produced N-glycan showed no reactivity with R1). Interestingly, previous studies investigating the DXDD motif in other GT-2 proteins, showed that amino acid changes (D94E or D94A) had no obvious impact on PglI activity indicating that PglI Asp94 does not have a catalytic, but rather a structural function55–59. However, to the best of our knowledge, none of those functional analyses describe a deletion in one of the active site Asp residues. The T88I mutation (the other residue near the active site that varied in two of the GEMS strains) did not have an effect on PglI activity and surprisingly neither did T88I in combination with ΔAsp94 which resembles the inactive 1J2 phenotype, thus indicating that other variation in the GEMS PglIs most likely contribute to their loss/reduction in activity. As expected, inserting Asp94 in 1D7 resulted in a functional PglI (Figure 4B), whereas the same insertion in 1J2 did not, nor did the I88T exchange alone; however, I88T in combination with the Asp94 insertion restored the activity of the 1J2 PglI. In all cases, reactivity of the R1 serum was indicative of PglI activity and formation of the full-length N-glycan with the Glc branch (Figure 4A/S8A, 4B/S8B, and Figure 4C). Glycosylation of CmeA-His6, independent of the presence of the glucose, was confirmed in parallel using anti-His antiserum (Figures 4A and 4B, Figure S8A and S8B).
Figure 4. Functional analysis of PglI variants and active site point mutants in the heterologous E. coli glycosylation system.
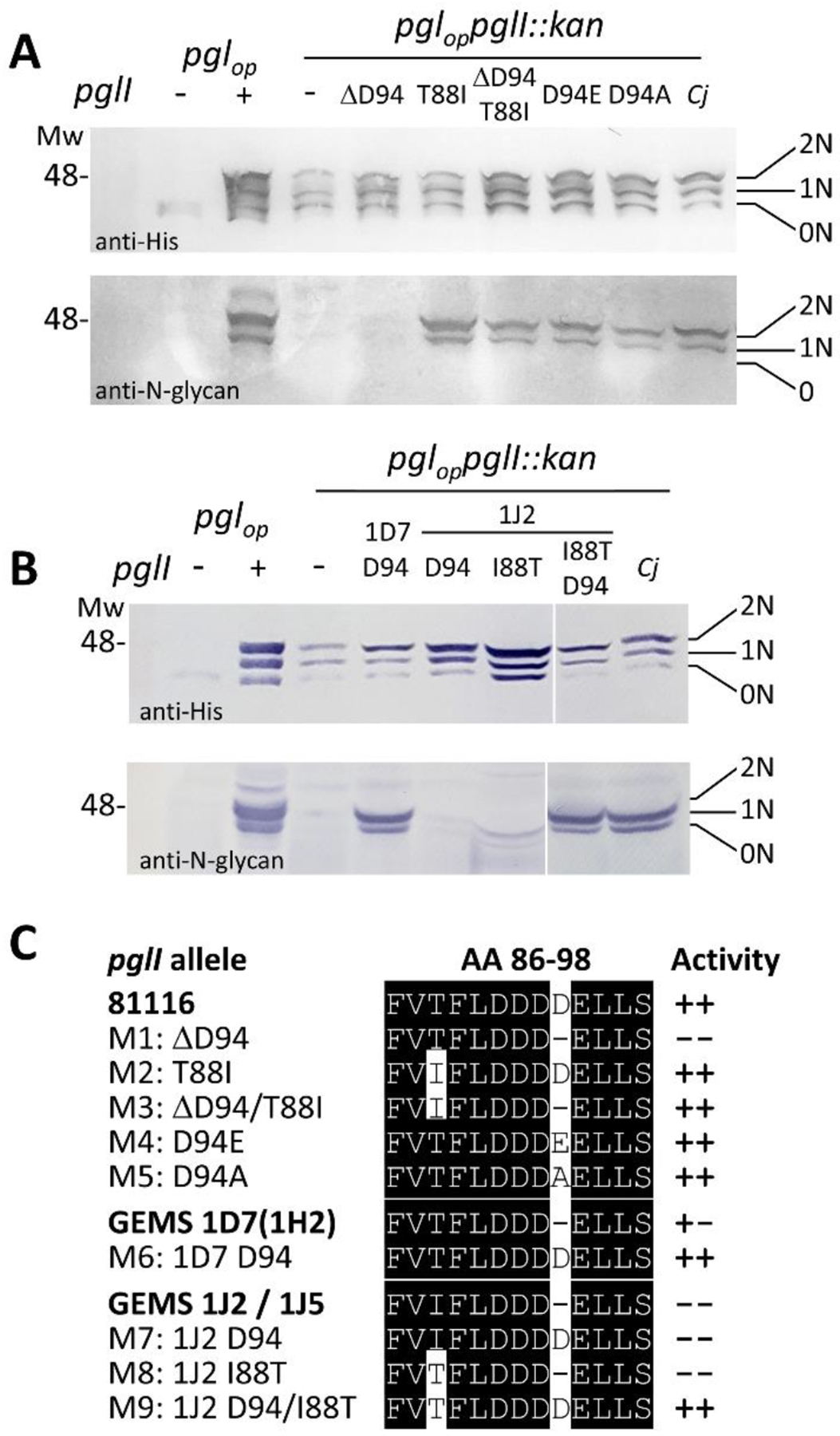
(A) The GTase-activities of active site point mutants in the PglI of 81116 and (B) in select GEMS PglIs were analyzed by western blotting. The glycan acceptor, CmeA-His6, was detected with His6-tag antibodies, the formation of the full length Cj-N-glycan was followed with R1 serum. Whole cell extracts of E. coli CLM24 (5 μg) containing and expressing the indicated gene/plasmid combinations are indicated on top of each lane. Non, -mono, -and di-glycosylated CmeA-His6 proteins are labelled with 0N, 1N, and 2N, respectively. Molecular weight markers (Mw) in kDa are indicated on the left. Top-to-bottom scans are provided in Figure S8. (C) Sequence alignment of the active site DXDD motif of Cj-81116, GEMS PglIs and point mutants. Activities as determined by western blotting in the 3-plasmid E. coli system are indicated by (−−) no activity (−+) low activity, and (++) activity comparable to Cj-81116 PglI.
PglI structure modelling.
To characterize the DXDD-containing structure in Cj-PglI, homology modelling and sequence alignments revealed that the structure of the N-terminal ~200 residues of PglI (up to approximately the middle of the long central loop shown in yellow, Figure 5) shows ~20–25% sequence similarity with several members of the GT-2 family and is predicted to share the canonical GT-A fold by sequence alignment and threading. The structure of the C-terminal ~100 residues of PglI following the central loop is predicted to be mostly helical and may share some similarities in tertiary structure with other bacterial glycosyltransferases like TarS (PDB ID 5TZE)56 and TarP (PDB ID 6H1J)60 from Staphylococcus aureus. The highly conserved positions of the DXDD motif and preceding β-strand in the GT-A fold further allowed for the prediction of the position of the catalytically important divalent metal cation and UDP-Glc donor53. In addition, the model strongly suggested that Asp91 and Asp93 occupy the two most functionally important residues that directly coordinate to the divalent metal ion in the DXDD motif of all members of the GT-A family. The deletion of Asp94 is thus expected to shorten the subsequent “90’s loop” (residues 95–99) and may affect the positioning of the helix at residues 100–110. Although the 90’s loop and the following helix are not directly involved with substrate binding or catalysis, they both contain residues that interact with two surface loops that are likely involved with acceptor binding interactions and possibly catalysis. Similarly, although the location of residue 88 is predicted to be somewhat distant from parts of the structure that are directly involved with substrate-binding and catalysis, the side chain of residue 88 is expected to directly interact with residues in the 90’s loop.
Figure 5. Structural model of PglI.
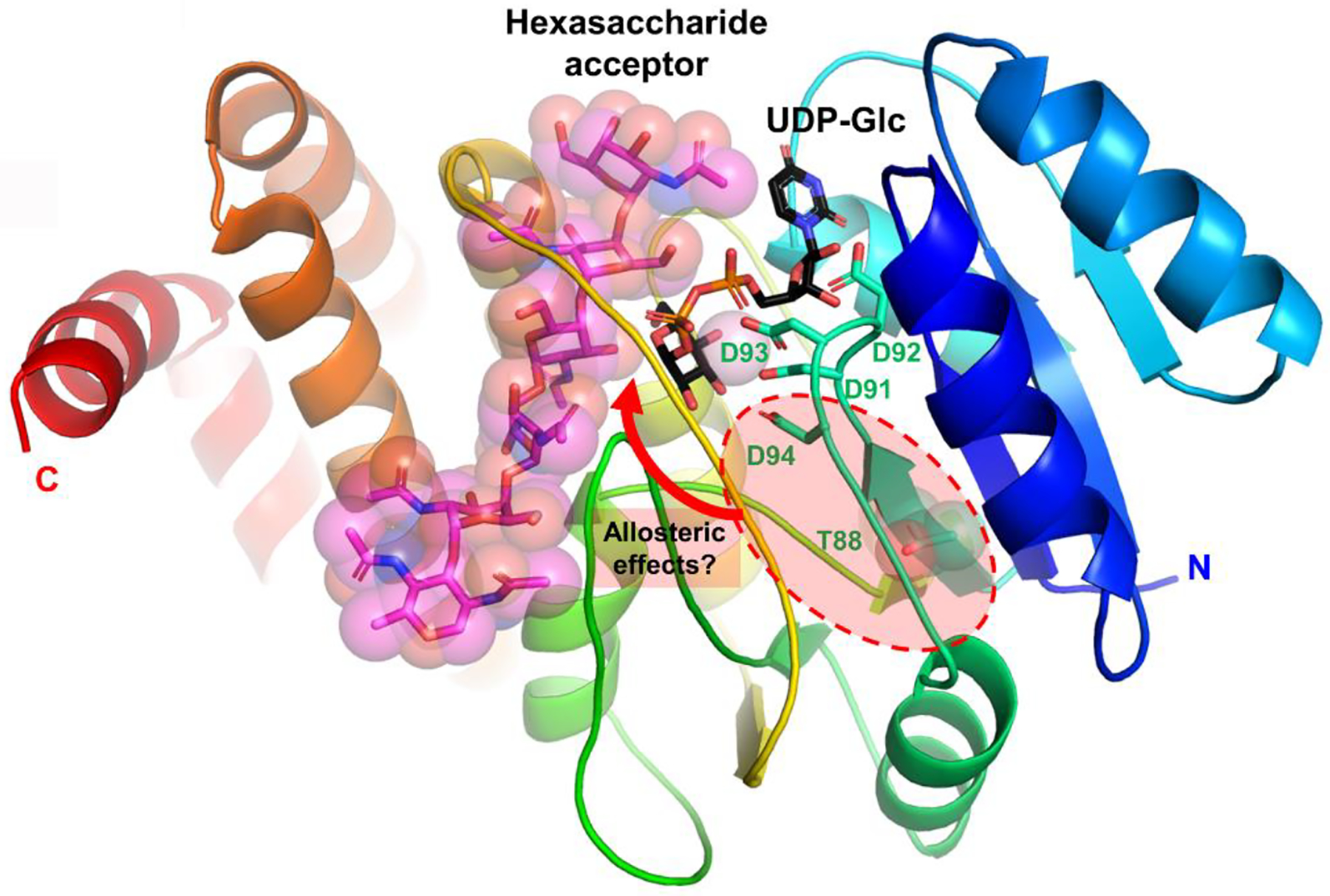
Cartoon representation of secondary structure elements with rainbow coloring starting with blue at the N-terminus and red at the C-terminus. UDP-Glc donor (black), hexasaccharide acceptor (magenta), the Asp91-X-Asp93 motif, Asp94 and Thr88 are all drawn in stick representation.
This suggests that there is a requirement for a certain length and/or rigidity/flexibility of the catalytic site loop most likely required for proper substrate binding or release as it has been described for other members of this protein family61 (and reviewed by46). Moreover, the indirect connection linking the sequence differences at residues 88 and 94 to the elements of the structure that are directly involved with substrate binding and catalysis suggests that the loss of Glc transferase activity associated with the deletion of Asp94 and the suppression of the effects of the deletion by the T88I mutation in PglI of 81116 are most likely mediated by indirect allosteric effects on either substrate-binding or catalysis. In addition to indirect long-range structural effects within a single subunit of PglI, another explanation is that mutations distant from the active site of one subunit may affect either more directly or indirectly residues in the active site or substrate-binding site of an adjacent subunit in the mature quaternary structure formed by PglI. Unfortunately, the quaternary structure of PglI is not well-defined at present. Additional structural studies of WT and mutant PglI in complexes with donor and acceptor substrates, including measurements of substrate binding affinity and more detailed kinetics measurements, are expected to shed additional light on the structure-function relationships related to these sequence differences.
Vaccination with a C. jejuni N-glycan-presenting E. coli strain results in reduced loss of weight gain and significantly reduces Campylobacter shedding in mice.
Despite the fact that the 4 GEMS strains identified in this study originated from asymptomatic infants (Table S3), those isolates are still capable of colonizing their host and could lead to post-infectious sequelae warranting their consideration in vaccination efforts to eliminate C. jejuni. In particular, 3 of the 4 isolates express GM1-mimicking LOS (Patry, R. unpublished observation), a structure associated with the induction of Guillain-Barré syndrome. However, what are the implications of our findings for use of the N-glycan as a vaccine antigen? To investigate the antigenic properties of the N-glycan in a mouse model, we first engineered an E. coli strain for use in mammals that presents the full-length Cj-N-glycan on its surface (as a lipid A-core fusion). To do so, we integrated the Cj-pgl locus into the chromosome of the ACE vaccine subcomponent strain E.coli ACAM2025 and confirmed that the N-glycan structure is expressed on the LOS (Figure S9) similar to that expressed by our E. coli K12-based poultry vaccine strain19. Attaching the N-glycan to lipid A-core results in a compound that resembles a non-repeat lipopolysaccharide (LPS) structure, and which has been successfully employed to generate a whole cell vaccine against Shigella62. In general, attenuated whole cell vaccines e.g. against Vibrio cholerae rely on the production of anti-LPS antibodies to exert a protective effect against the pathogen63. Therefore, the presentation of the Cj-N-glycan on the cell surface could be an inexpensive and effective method to generate an anti-Campylobacter/E. coli-based whole cell vaccine for mammals.
Next, we employed a mouse model designed to mimic human disease to evaluate the potential of the vaccine strain as a means to reduce C. jejuni infection (Figure S10). Since the model was designed to induce Campylobacter-associated growth-stunting, we also followed the weight of each animal post-challenge. We found that non-vaccinated, but Cj-infected mice, displayed a significant reduction in body weight when compared to the uninfected controls. In the vaccinated groups (either intragastric (I.G.) or intranasal (I.N.) administration), we observed increased body weights when compared to the unvaccinated controls (Figure 6A). In particular, statistics indicate that the mouse group given the vaccine orally I.G. showed weights statistically different from the unvaccinated Cj control. In addition, we observed a significant reduction (1.5–2 log) in C. jejuni shedding in both I.N. and I.G. groups 5 days post-challenge (Figure 6B). After 9 days post challenge, the I.N. group still showed a significant reduction (p=0.051) of C. jejuni when compared to the non-vaccinated but challenged Cj group, whereas the reduction in the I.G. group did not reach statistical significance. These results indicate that vaccination with the E. coli live vaccine is partially protective when given I.G or I.N. in C57BL/6 mice. Protection after vaccination with a live E. coli vaccine strain has only been observed previously in our chicken model although in the latter model, birds were either fully protected (responders) or showed increases in Campylobacter levels that eventually were comparable to the non-vaccinated Cj challenge control (non –responders, Nothaft et al., submitted and19–20).
Figure 6. Mouse vaccination and antibody characterization.
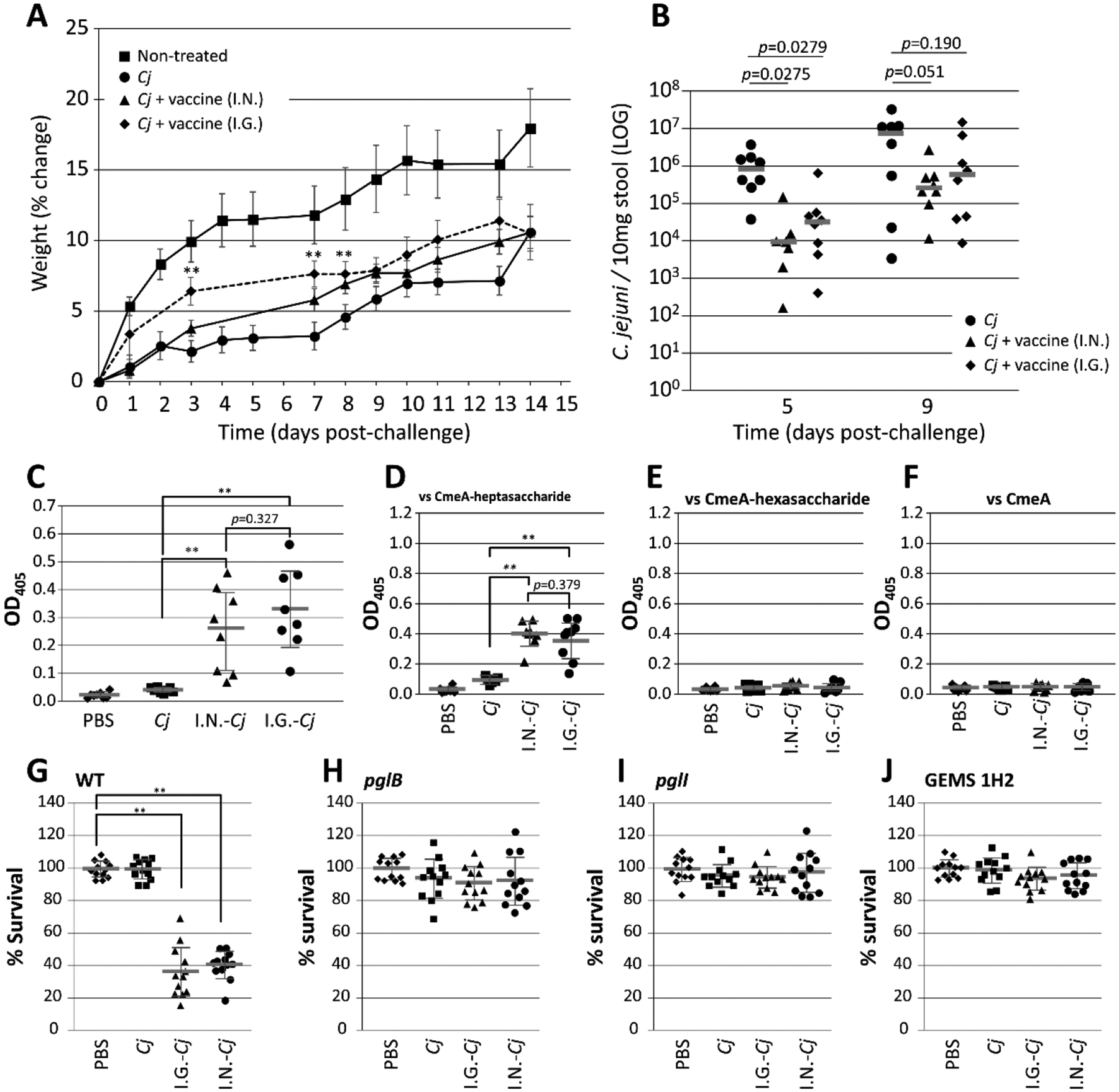
(A) Weight gain (in %) per day post-challenge was determined for each individual animal and plotted as the average for each group (as indicated). Error bars represent the standard error of the mean. (B) Campylobacter shedding on days 5 and 9 post-challenge presented as C. jejuni CFU per 10 mg stool sample. Each data point represents one animal from each group (as indicated). Bars represent averages for each group. (C) ELISAs were performed to confirm the presence and reactivity of N-glycan-specific antibodies for each mouse from the indicated groups using BSA conjugated to the Cj-heptasaccharide as the capture antigen. Individual sera were used at a 1:100 dilution. The specificity of the antibodies raised against variations in the Cj-N-glycan structure was determined in ELISAs using 1:10 dilutions of individually cross-adsorbed mouse sera (as indicated). Each point represents the average reactivity obtained from three biological replicates (assayed as triplicates) against: (D) the full length Cj-N-glycan; (E) the N-glycan, lacking the glucose branch; and (F) non-glycosylated CmeA-His6 (background control). Opsonophagocytosis assay: the percent survival of (G) the C. jejuni WT strain; (H) the pglB mutant (control); (I) the pglI mutant; and (J) one select GEMS strain (1H2) in the presence of pooled and Cj-pglB cross-adsorbed sera from each mouse group is shown. Each data point represents the average from three biological replicates (assayed as quadruplicates). Lines represent the median for each group, standard deviations are indicated by error bars. P-values (t-test) are given for each comparison or indicated by ** for p >0.005. I.N.; intranasal; I.G. intragastric; naïve mice (non-infected, PBS); Cj-infected, challenge-only mice (Cj).
Vaccination induces an N-glycan-specific immune-response.
Next, we tested if N-glycan-specific IgG antibodies are generated in mice after vaccination with the E. coli vaccine strain. ELISA results showed a statistically significant increase in N-glycan-specific antibody titers in both vaccinated groups when compared to the control groups (Figure 6C). No significant difference in antibody levels was observed between I.N.- and I.G-vaccinated mice while the slight increase in reactivity in the challenge only group (Cj) can be attributed to low levels of Cj-N-glycan-specific antibodies produced after challenge with C. jejuni.
The hexasaccharide is not recognized by mouse sera raised against the full-length N-glycan.
The fact that a subset of Cj-strains does not decorate their proteins with the full-length glycan, but rather with an N-glycan lacking the glucose, led us to further characterize the N-glycan-specific IgG antibodies. First, we compared the serum reactivities against the full-length heptasaccharide versus the hexasaccharide on the Cj-CmeA glycoprotein produced and purified from E. coli. A statistically significant increase in reactivity was observed only when N-glycan antibody containing sera were probed against CmeA glycosylated with the full length Cj-heptasaccharide (Figure 6D). No reactivity was observed when testing CmeA glycosylated with the hexasaccharide (Figure 6E) or non-glycosylated CmeA as the capture antigen (Figure 6F), or when the naïve sera obtained from the PBS control group was applied. The slight increase in the reactivity (although not statistically significant compared to naïve sera) with sera obtained from non-vaccinated, but challenged mice can be attributed to very low levels of N-glycan specific antibodies produced after C. jejuni challenge. Based on these observations, we conclude that the Cj-N-glycan presented on the surface of E. coli ACAM2025 induces an N-glycan specific immune-response that likely contributes to reduced Campylobacter colonization levels after challenge and reduced weight loss compared to non-vaccinated, but challenged animals.
Next, we determined the opsonizing activity of the N-glycan-specific antibodies. Sera from each group were pooled and cross-absorbed against the C. jejuni pglB oligosaccharyltransferase mutant unable to N-glycosylate its proteins. We observed statistically significant N-glycan mediated bacterial killing of the Cj-WT for both I.G. and I.N. vaccinated groups (Figure 6G) that was abolished in the pglB mutant (Figure 6H). N-glycan mediated killing was not observed for the isogenic glucose mutant (pglI, (Figure 6I)) or the representative GEMS 1H2 strain naturally lacking glucose with the pooled I.G. serum (Figure 6J).
Therefore, vaccinated mice produce polyclonal antibodies recognizing N-glycan epitopes containing the Glc branch, similar to what we observed with our immune sera used for screening that was generated in rabbits16. This suggests that Cj-strains expressing N-glycans without the Glc could potentially evade immune-recognition; The importance of the Glc branch for immunodetection is consistent with other studies showing that branch points/epitopes in an oligosaccharide are predominantly recognized by the immune system64–65 resulting in altered antibody reactivities, structural changes in the LPS66–68, or major impacts on pathogenicity69.
Due to the potential importance of the branch, any mechanism that would allow C. jejuni to turn off PglI might provide a competitive advantage. A mechanism that “actively” eliminates one Asp from the active site DXDD would be supported by the underlying allele pattern (as described above); however, without knowledge of the original sequence, it is difficult to determine the exact mechanism resulting in loss of the Asp. Another argument against “active” immune evasion would be that loss of the glucose branch does not occur in chickens vaccinated with the N-glycan heptasaccharide expressed on our live E. coli vaccine strain19 and would most likely result (as previously observed24) in lower colonization levels. Nevertheless, the existence of C. jejuni strains that are missing the glucose due to the described variation in PglI should be taken into consideration when designing a broad range vaccine against all C. jejuni isolates, especially for human use in low resource settings.
Experimental Methods
Construction and verification of PglI expression plasmids.
The pglI expression plasmid from Cj strain 81116 has been previously54. GEMS strain pglIs were PCR amplified (in triplicate) from chromosomal DNA (cDNA) from strains 1J2, 1J5, 1H2 and 1D7 with oligonucleotides (pglI-81116-BamHI-F) and (81116-pglI-XhoI-R_short). Amplicons products were digested with BamHI and XhoI and inserted into plasmid pCE111–28 cut with the same enzymes. Candidate colonies obtained after ligation and transformation of E. coli DH5α were confirmed by restriction digest analyses, and the pglI sequence for each strain was determined for one candidate from each PCR product ligation. Point mutations in PglI were introduced using a 3-fragment ligation strategy as described in Supporting Information.
The E. coli 3-plasmid system to screen and analyze PglI activities.
The previously described54 E. coli 3-plasmid system was employed to follow PglI activity. Here, the N-glycan acceptor protein CmeA-His6 is constitutively expressed from plasmid pHI18b Cj-pgl genes are constitutively expressed from either plasmid pACYC184pgl (positive control)28 or plasmid pACYC184pgl/pglI::kan54. The individual pglI alleles are constitutively expressed from pCE111–28 derivatives. The glycosylation status of the CmeA-His6 protein in whole cell lysates of the respective strains was determined by western blotting using His6-tag (Rockland) and rabbit-derived N-glycan specific (R1)16 antisera.
Mouse vaccination and challenge.
To evaluate the immunogenicity of the N-glycan antigen when expressed on the surface of an E. coli strain specifically designed for the use in mammals (described in Supporting Information) and the impact of the vaccine on Cj-colonization a mouse model mimicking human disease was employed as described70 (Figure S10). The study consisted of four groups and each group consisted of eight animals. Mice in the control groups remained either untreated (non-vaccinated, non-challenged) or only received the Cj-challenge (non-vaccinated but challenged, Cj-group). Mice in the vaccine groups were given 100 μl containing 1×109 CFU (prepared as described20) live E. coli ACAM2025 expressing the C. jejuni N-glycan on its surface in combination with 2.5 μg/dose dmLT71 via intranasal administration (I.N.) or by oral gavage (I.G.) once a week for three consecutive weeks (days 7, 14 and 21). The oral C. jejuni challenge consisted of 100 μl PBS containing 1×106 CFU (1×107 CFU/ml) of C. jejuni 81–176 prepared as described20. Body weights were recorded after challenge until day 14; Campylobacter shedding in stool samples collected on day 5 and day 9 post-challenge was determined by qPCR based on the cadF probe of colony-counted controls as described70. Mice were euthanized at day 15 post-infection and blood samples were collected. All procedures were performed in accordance to the protocol approved by the Committee on the Ethics of Animal Experiments of the University of Virginia (Protocol Number: 3315).
Enzyme-linked immunosorbent assay (ELISA).
ELISA using fOS-BSA as the capture antigen were carried out as described19 with individual sera from all four groups of mice (N=8). Alkaline phosphatase-labelled anti-mouse serum (Santa Cruz, 1: 2500) served as the secondary antibody. To discriminate between serum reactivity against C. jejuni WT (full length heptasaccharide) and the pglI mutant hexasaccharide, purified CmeA-His6 proteins (100 μl, 50 ng/μl) glycosylated with either the full-length Cj-N-glycan (expressed and purified from E. coli CLM24 [pACYC184pgl, pIH18b]) or the Cj-N-glycan lacking the glucose (expressed and purified from E. coli CLM24 [pACYC184pgl/pglI::kan, pIH18b]) were used as capture antigens. Non-glycosylated CmeA purified from E. coli CLM24 pACYC184pglmut served as a control. CmeA-His6-derivatives were overexpressed and purified as described72. Prior to application CmeA-specific background reactivity (induced by the challenge strain) was eliminated by cross-adsorbing the individual mouse sera against non-glycosylated CmeA-His6 as follows: 10 μg of purified non-glycosylated CmeA-His6 protein (500 ng/μl in PBS) was mixed with 200 μl (slurry) of Ni-NTA agarose (Qiagen) and washed twice with 1 ml of PBS prior to loading. The mixture was transferred into individual PCR purification columns (Thermo Fisher) and washed twice with 500 μl PBS by centrifugation for 2 min at 2,000 rpm. 200 μl of each serum (diluted 1:10 in PBS) was loaded onto the columns and incubated with the Ni-NTA-CmeA6 mixture for 5 min at RT to allow CmeA-specific antibodies to bind. After centrifugation for 2 min at 2,000 rpm, the flow-through was collected and transferred into a fresh tube. The obtained cross-adsorbed sera were directly used for ELISA experiments or stored at 4 °C until further use.
Opsonophagocytosis Assay (OPA).
OPA were carried out as described73. Briefly, C. jejuni strain were grown on Mueller-Hinton (MH) agar for 16 h, harvested with 4 ml ice cold PBS, centrifuged for 1 min, (20,000 × g, 4 °C) and adjusted to an OD600 of 1.0 corresponding to 3×109 CFU/ml. Cells were diluted 1:10,000 in assay buffer and 10 μl containing 3×104 CFU were used to analyze serum antibodies for their ability to induce bacterial killing. Heparinized mouse blood (Cedarlane) served as a source for leukocytes, and pooled mouse sera (equal volumes of serum from mice within the respective group) were the source for the N-glycan specific antibodies. Pooled sera from naïve animals and non-vaccinated animal served as controls. To eliminate non-N-glycan-specific background activity, pooled sera from all groups were cross-adsorbed against whole cells of the Cj-pglB mutant. Cells of the Cj-pglB mutant were grown on MH, harvested with 4 ml of ice-cold PBS and set to an of OD600 of 1.0. One ml of these cells was pelleted for 1 min (20,000 × g, 4 °C), resuspended in 200 μl of pooled serum from each group and incubated on ice for 15 min. Cells were spun down and the supernatant was used for three additional rounds of cross-adsorption using fresh pglB mutant cells. Absorbed sera were stored at 4 °C until further use.
Supplementary Material
Acknowledgements
We would like to thank S. Tennant (University of Maryland School of Medicine) for the GEMS isolates, N. Kegley for help with western blotting, B. Zhou for providing the illustration for the graphical abstract and Figure S10, and PATH for the E. coli strain and invaluable input for the generation of the vaccine strain described in this study. PATH also provided dmLT with support from an award from the Bill and Melinda Gates Foundation (OPP1118786). We would also like to thank K. Moremen, the 2020 recipient of the Society for Glycobiology Karl Meyer award, for his helpful discussions on this study and his endless enthusiasm for glycosyltransferases. Work performed by PA and AS was supported by NIH R24GM137782.
Footnotes
Supporting Information
The following Supporting Information is available free of charge at the ACS website: Experimental Methods include: details on plasmid and strain construction, methods for protein and carbohydrate (N-glycan) analyses, and for PglI sequence analyses and homology modelling; Tables (S1–S3) summarizing oligonucleotides, plasmids and strain used in this study; Figures S1–S10, include full scans of all western blots, PglI sequence analyses/alignments, mass spectrometry and additional N-glycan western blots of GEMS isolates and an overview schematic for the mouse vaccination study.
Data availability
All raw MS data has been deposited into the Glycopost repository and is available at the following link: https://glycopost.glycosmos.org/preview/178377258560ccd67e60fe5; PIN CODE: 8293. All remaining data are presented in the manuscript or in the supporting information.
REFERENCES
- 1.Havelaar AH; Kirk MD; Torgerson PR; Gibb HJ; Hald T; Lake RJ; Praet N; Bellinger DC; de Silva NR; Gargouri N; Speybroeck N; Cawthorne A; Mathers C; Stein C; Angulo FJ; Devleesschauwer B; World Health Organization Foodborne Disease Burden Epidemiology Reference, G., World Health Organization Global Estimates and Regional Comparisons of the Burden of Foodborne Disease in 2010. PLoS Med 2015, 12 (12), e1001923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kotloff KL; Nataro JP; Blackwelder WC; Nasrin D; Farag TH; Panchalingam S; Wu Y; Sow SO; Sur D; Breiman RF; Faruque AS; Zaidi AK; Saha D; Alonso PL; Tamboura B; Sanogo D; Onwuchekwa U; Manna B; Ramamurthy T; Kanungo S; Ochieng JB; Omore R; Oundo JO; Hossain A; Das SK; Ahmed S; Qureshi S; Quadri F; Adegbola RA; Antonio M; Hossain MJ; Akinsola A; Mandomando I; Nhampossa T; Acacio S; Biswas K; O’Reilly CE; Mintz ED; Berkeley LY; Muhsen K; Sommerfelt H; Robins-Browne RM; Levine MM, Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 2013, 382 (9888), 209–22. [DOI] [PubMed] [Google Scholar]
- 3.Platts-Mills JA; Babji S; Bodhidatta L; Gratz J; Haque R; Havt A; McCormick BJ; McGrath M; Olortegui MP; Samie A; Shakoor S; Mondal D; Lima IF; Hariraju D; Rayamajhi BB; Qureshi S; Kabir F; Yori PP; Mufamadi B; Amour C; Carreon JD; Richard SA; Lang D; Bessong P; Mduma E; Ahmed T; Lima AA; Mason CJ; Zaidi AK; Bhutta ZA; Kosek M; Guerrant RL; Gottlieb M; Miller M; Kang G; Houpt ER; Investigators M-EN, Pathogen-specific burdens of community diarrhoea in developing countries: a multisite birth cohort study (MAL-ED). Lancet Glob Health 2015, 3 (9), e564–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kirkpatrick BD; Tribble DR, Update on human Campylobacter jejuni infections. Curr Opin Gastroenterol 2011, 27 (1), 1–7. [DOI] [PubMed] [Google Scholar]
- 5.Lee G; Pan W; Penataro Yori P; Paredes Olortegui M; Tilley D; Gregory M; Oberhelman R; Burga R; Chavez CB; Kosek M, Symptomatic and asymptomatic Campylobacter infections associated with reduced growth in Peruvian children. PLoS neglected tropical diseases 2013, 7 (1), e2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dinh DM; Ramadass B; Kattula D; Sarkar R; Braunstein P; Tai A; Wanke CA; Hassoun S; Kane AV; Naumova EN; Kang G; Ward HD, Longitudinal Analysis of the Intestinal Microbiota in Persistently Stunted Young Children in South India. PloS one 2016, 11 (5), e0155405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amour C; Gratz J; Mduma E; Svensen E; Rogawski ET; McGrath M; Seidman JC; McCormick BJ; Shrestha S; Samie A; Mahfuz M; Qureshi S; Hotwani A; Babji S; Trigoso DR; Lima AA; Bodhidatta L; Bessong P; Ahmed T; Shakoor S; Kang G; Kosek M; Guerrant RL; Lang D; Gottlieb M; Houpt ER; Platts-Mills JA; Etiology RF; Interactions of Enteric, I.; Malnutrition; the Consequences for Child, H.; Development Project Network, I., Epidemiology and Impact of Campylobacter Infection in Children in 8 Low-Resource Settings: Results From the MAL-ED Study. Clin Infect Dis 2016, 63 (9), 1171–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.World Health Organization. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics; https://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf (accessed 2017).
- 9.Kittler S; Steffan S; Peh E; Plotz M, Phage Biocontrol of Campylobacter: A One Health Approach. Curr Top Microbiol Immunol 2021, 431, 127–168. [DOI] [PubMed] [Google Scholar]
- 10.Deng W; Dittoe DK; Pavilidis HO; Chaney WE; Yang Y; Ricke SC, Current Perspectives and Potential of Probiotics to Limit Foodborne Campylobacter in Poultry. Frontiers in microbiology 2020, 11, 583429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abd El-Hack ME; El-Saadony MT; Shehata AM; Arif M; Paswan VK; Batiha GE; Khafaga AF; Elbestawy AR, Approaches to prevent and control Campylobacter spp. colonization in broiler chickens: a review. Environ Sci Pollut Res Int 2021, 28 (5), 4989–5004. [DOI] [PubMed] [Google Scholar]
- 12.Meunier M; Guyard-Nicodeme M; Dory D; Chemaly M, Control strategies against Campylobacter at the poultry production level: biosecurity measures, feed additives and vaccination. Journal of applied microbiology 2016, 120 (5), 1139–73. [DOI] [PubMed] [Google Scholar]
- 13.Riddle MS; Guerry P, Status of vaccine research and development for Campylobacter jejuni. Vaccine 2016, 34 (26), 2903–2906. [DOI] [PubMed] [Google Scholar]
- 14.Pumtang-On P; Mahony TJ; Hill RA; Vanniasinkam T, A Systematic Review of Campylobacter jejuni Vaccine Candidates for Chickens. Microorganisms 2021, 9 (2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Young NM; Brisson JR; Kelly J; Watson DC; Tessier L; Lanthier PH; Jarrell HC; Cadotte N; St Michael F; Aberg E; Szymanski CM, Structure of the N-linked glycan present on multiple glycoproteins in the Gram-negative bacterium, Campylobacter jejuni. The Journal of biological chemistry 2002, 277 (45), 42530–9. [DOI] [PubMed] [Google Scholar]
- 16.Nothaft H; Scott NE; Vinogradov E; Liu X; Hu R; Beadle B; Fodor C; Miller WG; Li J; Cordwell SJ; Szymanski CM, Diversity in the protein N-glycosylation pathways within the Campylobacter genus. Mol Cell Proteomics 2012, 11 (11), 1203–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nothaft H; Szymanski CM, Bacterial protein N-glycosylation: new perspectives and applications. The Journal of biological chemistry 2013, 288 (10), 6912–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szymanski CM; Goon S; Allan B; Guerry P, Protein glycosylation in Campylobacter. In Campylobacter: Molecular and Cellular Biology, Ketley JM; Konkel ME, Eds. Horizon Bioscience: Norwich, United Kingdom, 2005; pp 259–273. [Google Scholar]
- 19.Nothaft H; Davis B; Lock YY; Perez-Munoz ME; Vinogradov E; Walter J; Coros C; Szymanski CM, Engineering the Campylobacter jejuni N-glycan to create an effective chicken vaccine. Scientific reports 2016, 6, 26511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nothaft H; Perez-Munoz ME; Gouveia GJ; Duar RM; Wanford JJ; Lango-Scholey L; Panagos CG; Srithayakumar V; Plastow GS; Coros C; Bayliss CD; Edison AS; Walter J; Szymanski CM, Coadministration of the Campylobacter jejuni N-Glycan-Based Vaccine with Probiotics Improves Vaccine Performance in Broiler Chickens. Applied and environmental microbiology 2017, 83 (23), 1523–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szymanski CM; Logan SM; Linton D; Wren BW, Campylobacter - a tale of two protein glycosylation systems. Trends in microbiology 2003, 11 (5), 233–8. [DOI] [PubMed] [Google Scholar]
- 22.Szymanski CM; Michael FS; Jarrell HC; Li J; Gilbert M; Larocque S; Vinogradov E; Brisson JR, Detection of conserved N-linked glycans and phase-variable lipooligosaccharides and capsules from campylobacter cells by mass spectrometry and high resolution magic angle spinning NMR spectroscopy. The Journal of biological chemistry 2003, 278 (27), 24509–20. [DOI] [PubMed] [Google Scholar]
- 23.Larkin A; Imperiali B, The expanding horizons of asparagine-linked glycosylation. Biochemistry 2011, 50 (21), 4411–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelly J; Jarrell H; Millar L; Tessier L; Fiori LM; Lau PC; Allan B; Szymanski CM, Biosynthesis of the N-linked glycan in Campylobacter jejuni and addition onto protein through block transfer. Journal of bacteriology 2006, 188 (7), 2427–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nothaft H; Szymanski CM, Protein glycosylation in bacteria: sweeter than ever. Nature reviews 2010, 8 (11), 765–78. [DOI] [PubMed] [Google Scholar]
- 26.Alaimo C; Catrein I; Morf L; Marolda CL; Callewaert N; Valvano MA; Feldman MF; Aebi M, Two distinct but interchangeable mechanisms for flipping of lipid-linked oligosaccharides. EMBO J 2006, 25 (5), 967–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kowarik M; Young NM; Numao S; Schulz BL; Hug I; Callewaert N; Mills DC; Watson DC; Hernandez M; Kelly JF; Wacker M; Aebi M, Definition of the bacterial N-glycosylation site consensus sequence. EMBO J 2006, 25 (9), 1957–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wacker M; Linton D; Hitchen PG; Nita-Lazar M; Haslam SM; North SJ; Panico M; Morris HR; Dell A; Wren BW; Aebi M, N-linked glycosylation in Campylobacter jejuni and its functional transfer into E. coli. Science (New York, N.Y 2002, 298 (5599), 1790–3. [DOI] [PubMed] [Google Scholar]
- 29.Cain JA; Dale AL; Niewold P; Klare WP; Man L; White MY; Scott NE; Cordwell SJ, Proteomics Reveals Multiple Phenotypes Associated with N-linked Glycosylation in Campylobacter jejuni. Mol Cell Proteomics 2019, 18 (4), 715–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scott NE; Parker BL; Connolly AM; Paulech J; Edwards AV; Crossett B; Falconer L; Kolarich D; Djordjevic SP; Hojrup P; Packer NH; Larsen MR; Cordwell SJ, Simultaneous glycan-peptide characterization using hydrophilic interaction chromatography and parallel fragmentation by CID, higher energy collisional dissociation, and electron transfer dissociation MS applied to the N-linked glycoproteome of Campylobacter jejuni. Mol Cell Proteomics 2011, 10 (2), M000031–MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hendrixson DR; DiRita VJ, Identification of Campylobacter jejuni genes involved in commensal colonization of the chick gastrointestinal tract. Molecular microbiology 2004, 52 (2), 471–84. [DOI] [PubMed] [Google Scholar]
- 32.Jones MA; Marston KL; Woodall CA; Maskell DJ; Linton D; Karlyshev AV; Dorrell N; Wren BW; Barrow PA, Adaptation of Campylobacter jejuni NCTC11168 to high-level colonization of the avian gastrointestinal tract. Infection and immunity 2004, 72 (7), 3769–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karlyshev AV; Everest P; Linton D; Cawthraw S; Newell DG; Wren BW, The Campylobacter jejuni general glycosylation system is important for attachment to human epithelial cells and in the colonization of chicks. Microbiology (Reading) 2004, 150 (Pt 6), 1957–1964. [DOI] [PubMed] [Google Scholar]
- 34.Vijayakumar S; Merkx-Jacques A; Ratnayake DB; Gryski I; Obhi RK; Houle S; Dozois CM; Creuzenet C, Cj1121c, a novel UDP-4-keto-6-deoxy-GlcNAc C-4 aminotransferase essential for protein glycosylation and virulence in Campylobacter jejuni. The Journal of biological chemistry 2006, 281 (38), 27733–43. [DOI] [PubMed] [Google Scholar]
- 35.Nothaft H; Liu X; McNally DJ; Li J; Szymanski CM, Study of free oligosaccharides derived from the bacterial N-glycosylation pathway. Proc Natl Acad Sci U S A 2009, 106 (35), 15019–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nothaft H; Liu X; Li J; Szymanski CM, Campylobacter jejuni free oligosaccharides: function and fate. Virulence 2010, 1 (6), 546–50. [DOI] [PubMed] [Google Scholar]
- 37.Dubb RK; Nothaft H; Beadle B; Richards MR; Szymanski CM, N-glycosylation of the CmeABC multidrug efflux pump is needed for optimal function in Campylobacter jejuni. Glycobiology 2020, 30 (2), 105–119. [DOI] [PubMed] [Google Scholar]
- 38.Abouelhadid S; North SJ; Hitchen P; Vohra P; Chintoan-Uta C; Stevens M; Dell A; Cuccui J; Wren BW, Quantitative Analyses Reveal Novel Roles for N-Glycosylation in a Major Enteric Bacterial Pathogen. mBio 2019, 10 (2), e00297–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Vries SP; Gupta S; Baig A; Wright E; Wedley A; Jensen AN; Lora LL; Humphrey S; Skovgard H; Macleod K; Pont E; Wolanska DP; L’Heureux J; Mobegi FM; Smith DGE; Everest P; Zomer A; Williams N; Wigley P; Humphrey T; Maskell DJ; Grant AJ, Genome-wide fitness analyses of the foodborne pathogen Campylobacter jejuni in in vitro and in vivo models. Scientific reports 2017, 7 (1), 1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller WG; Yee E; Chapman MH; Smith TP; Bono JL; Huynh S; Parker CT; Vandamme P; Luong K; Korlach J, Comparative genomics of the Campylobacter lari group. Genome Biol Evol 2014, 6 (12), 3252–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Skirrow MB; Benjamin J, ‘1001’ Campylobacters: cultural characteristics of intestinal campylobacters from man and animals. The Journal of hygiene 1980, 85 (3), 427–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benjamin J; Leaper S; Owen RJ; Skirrow MB, Description of Campylobacter-Laridis, a New Species Comprising the Nalidixic-Acid Resistant Thermophilic Campylobacter (NARTC) Group. Current microbiology 1983, 8 (4), 231–238. [Google Scholar]
- 43.Jervis AJ; Butler JA; Lawson AJ; Langdon R; Wren BW; Linton D, Characterization of the structurally diverse N-linked glycans of Campylobacter species. Journal of bacteriology 2012, 194 (9), 2355–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Glover KJ; Weerapana E; Imperiali B, In vitro assembly of the undecaprenylpyrophosphate-linked heptasaccharide for prokaryotic N-linked glycosylation. Proc Natl Acad Sci U S A 2005, 102 (40), 14255–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Breton C; Snajdrova L; Jeanneau C; Koca J; Imberty A, Structures and mechanisms of glycosyltransferases. Glycobiology 2006, 16 (2), 29R–37R. [DOI] [PubMed] [Google Scholar]
- 46.Brockhausen I, Crossroads between Bacterial and Mammalian Glycosyltransferases. Front Immunol 2014, 5, 492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Breton C; Fournel-Gigleux S; Palcic MM, Recent structures, evolution and mechanisms of glycosyltransferases. Current opinion in structural biology 2012, 22 (5), 540–9. [DOI] [PubMed] [Google Scholar]
- 48.Unligil UM; Rini JM, Glycosyltransferase structure and mechanism. Current opinion in structural biology 2000, 10 (5), 510–7. [DOI] [PubMed] [Google Scholar]
- 49.Bacon DJ; Szymanski CM; Burr DH; Silver RP; Alm RA; Guerry P, A phase-variable capsule is involved in virulence of Campylobacter jejuni 81–176. Molecular microbiology 2001, 40 (3), 769–77. [DOI] [PubMed] [Google Scholar]
- 50.Gilbert M; Karwaski MF; Bernatchez S; Young NM; Taboada E; Michniewicz J; Cunningham AM; Wakarchuk WW, The genetic bases for the variation in the lipo-oligosaccharide of the mucosal pathogen, Campylobacter jejuni. Biosynthesis of sialylated ganglioside mimics in the core oligosaccharide. The Journal of biological chemistry 2002, 277 (1), 327–37. [DOI] [PubMed] [Google Scholar]
- 51.Guerry P; Szymanski CM; Prendergast MM; Hickey TE; Ewing CP; Pattarini DL; Moran AP, Phase variation of Campylobacter jejuni 81–176 lipooligosaccharide affects ganglioside mimicry and invasiveness in vitro. Infection and immunity 2002, 70 (2), 787–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scott NE; Marzook NB; Cain JA; Solis N; Thaysen-Andersen M; Djordjevic SP; Packer NH; Larsen MR; Cordwell SJ, Comparative proteomics and glycoproteomics reveal increased N-linked glycosylation and relaxed sequon specificity in Campylobacter jejuni NCTC11168 O. Journal of proteome research 2014, 13 (11), 5136–50. [DOI] [PubMed] [Google Scholar]
- 53.Taujale R; Venkat A; Huang LC; Zhou Z; Yeung W; Rasheed KM; Li S; Edison AS; Moremen KW; Kannan N, Deep evolutionary analysis reveals the design principles of fold A glycosyltransferases. Elife 2020, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Duma J; Nothaft H; Weaver D; Fodor C; Beadle B; Linton D; Benoit SL; Scott NE; Maier RJ; Szymanski CM, Influence of Protein Glycosylation on Campylobacter fetus Physiology. Frontiers in microbiology 2020, 11, 1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang S; Czuchry D; Liu B; Vinnikova AN; Gao Y; Vlahakis JZ; Szarek WA; Wang L; Feng L; Brockhausen I, Characterization of two UDP-Gal:GalNAc-diphosphate-lipid beta1,3-galactosyltransferases WbwC from Escherichia coli serotypes O104 and O5. Journal of bacteriology 2014, 196 (17), 3122–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sobhanifar S; Worrall LJ; King DT; Wasney GA; Baumann L; Gale RT; Nosella M; Brown ED; Withers SG; Strynadka NC, Structure and Mechanism of Staphylococcus aureus TarS, the Wall Teichoic Acid beta-glycosyltransferase Involved in Methicillin Resistance. PLoS pathogens 2016, 12 (12), e1006067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Keenleyside WJ; Clarke AJ; Whitfield C, Identification of residues involved in catalytic activity of the inverting glycosyl transferase WbbE from Salmonella enterica serovar borreze. Journal of bacteriology 2001, 183 (1), 77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jank T; Belyi Y; Aktories K, Bacterial glycosyltransferase toxins. Cellular microbiology 2015, 17 (12), 1752–65. [DOI] [PubMed] [Google Scholar]
- 59.Jank T; Bogdanovic X; Wirth C; Haaf E; Spoerner M; Bohmer KE; Steinemann M; Orth JH; Kalbitzer HR; Warscheid B; Hunte C; Aktories K, A bacterial toxin catalyzing tyrosine glycosylation of Rho and deamidation of Gq and Gi proteins. Nat Struct Mol Biol 2013, 20 (11), 1273–80. [DOI] [PubMed] [Google Scholar]
- 60.Gerlach D; Guo Y; De Castro C; Kim SH; Schlatterer K; Xu FF; Pereira C; Seeberger PH; Ali S; Codee J; Sirisarn W; Schulte B; Wolz C; Larsen J; Molinaro A; Lee BL; Xia G; Stehle T; Peschel A, Methicillin-resistant Staphylococcus aureus alters cell wall glycosylation to evade immunity. Nature 2018, 563 (7733), 705–709. [DOI] [PubMed] [Google Scholar]
- 61.Qasba PK; Ramakrishnan B; Boeggeman E, Structure and function of beta −1,4-galactosyltransferase. Curr Drug Targets 2008, 9 (4), 292–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim MJ; Moon YH; Kim H; Rho S; Shin YK; Song M; Walker R; Czerkinsky C; Kim DW; Kim JO, Cross-Protective Shigella Whole-Cell Vaccine With a Truncated O-Polysaccharide Chain. Frontiers in microbiology 2018, 9, 2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Holmgren J, An Update on Cholera Immunity and Current and Future. Trop Med Infect Dis 2021, 6 (2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nnalue NA, All accessible epitopes in the Salmonella lipopolysaccharide core are associated with branch residues. Infection and immunity 1999, 67 (2), 998–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kong Q; Yang J; Liu Q; Alamuri P; Roland KL; Curtiss R 3rd, Effect of deletion of genes involved in lipopolysaccharide core and O-antigen synthesis on virulence and immunogenicity of Salmonella enterica serovar typhimurium. Infection and immunity 2011, 79 (10), 4227–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wright A, Mechanism of conversion of the Salmonella O-antigen by bacteriophage epsilon 34. Journal of bacteriology 1971, 105 (3), 927–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huan PT; Whittle BL; Bastin DA; Lindberg AA; Verma NK, Shigella flexneri type-specific antigen V: cloning, sequencing and characterization of the glucosyl transferase gene of temperate bacteriophage SfV. Gene 1997, 195 (2), 207–16. [DOI] [PubMed] [Google Scholar]
- 68.Mann E; Ovchinnikova OG; King JD; Whitfield C, Bacteriophage-mediated Glucosylation Can Modify Lipopolysaccharide O-Antigens Synthesized by an ATP-binding Cassette (ABC) Transporter-dependent Assembly Mechanism. The Journal of biological chemistry 2015, 290 (42), 25561–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Talyansky Y; Nielsen TB; Yan J; Carlino-Macdonald U; Di Venanzio G; Chakravorty S; Ulhaq A; Feldman MF; Russo TA; Vinogradov E; Luna B; Wright MS; Adams MD; Spellberg B, Capsule carbohydrate structure determines virulence in Acinetobacter baumannii. PLoS pathogens 2021, 17 (2), e1009291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Giallourou N; Medlock GL; Bolick DT; Medeiros PH; Ledwaba SE; Kolling GL; Tung K; Guerry P; Swann JR; Guerrant RL, A novel mouse model of Campylobacter jejuni enteropathy and diarrhea. PLoS pathogens 2018, 14 (3), e1007083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Norton EB; Lawson LB; Freytag LC; Clements JD, Characterization of a mutant Escherichia coli heat-labile toxin, LT(R192G/L211A), as a safe and effective oral adjuvant. Clin Vaccine Immunol 2011, 18 (4), 546–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Barre Y; Nothaft H; Thomas C; Liu X; Li J; Ng KKS; Szymanski CM, A conserved DGGK motif is essential for the function of the PglB oligosaccharyltransferase from Campylobacter jejuni. Glycobiology 2017, 27 (10), 978–989. [DOI] [PubMed] [Google Scholar]
- 73.Harding CM; Nasr MA; Scott NE; Goyette-Desjardins G; Nothaft H; Mayer AE; Chavez SM; Huynh JP; Kinsella RL; Szymanski CM; Stallings CL; Segura M; Feldman MF, A platform for glycoengineering a polyvalent pneumococcal bioconjugate vaccine using E. coli as a host. Nat Commun 2019, 10 (1), 891. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw MS data has been deposited into the Glycopost repository and is available at the following link: https://glycopost.glycosmos.org/preview/178377258560ccd67e60fe5; PIN CODE: 8293. All remaining data are presented in the manuscript or in the supporting information.


