Abstract
Dynamic contacts are formed between endoplasmic reticulum (ER) and mitochondria that enable the exchange of calcium and phospholipids. Disturbed contacts between ER and mitochondria impair mitochondrial dynamics and are a molecular hallmark of Parkinson’s disease, which is also characterized by impaired complex I activity and dopaminergic neuron degeneration. Here, we analyzed the role of cysteine-rich with EGF-like domain (Creld), a poorly characterized risk gene for Parkinson’s disease, in the regulation of mitochondrial dynamics and function. We found that loss of Creld leads to mitochondrial hyperfusion and reduced ROS signaling in Drosophila melanogaster, Xenopus tropicalis, and human cells. Creld fly mutants show differences in ER-mitochondria contacts and reduced respiratory complex I activity. The resulting low–hydrogen peroxide levels are linked to disturbed neuronal activity and lead to impaired locomotion, but not neurodegeneration, in Creld mutants. We conclude that Creld regulates ER-mitochondria communication and thereby hydrogen peroxide formation, which is required for normal neuron function.
Creld regulates reactive oxygen species formation required for locomotion and neuron activity.
INTRODUCTION
The role of cellular reactive oxygen species (ROS) is ambiguous: According to the free radical theory of aging, oxidative stress caused by excessive ROS seems to be the major driver of aging and occurs because of a decline in mitochondrial function. This theory was challenged by findings that long-lived species have normal or elevated ROS levels, and increasing ROS genetically does not shorten the life span (1). Moreover, ROS are now acknowledged as important signaling molecules; for example, hydrogen peroxide (H2O2) is required for the excitability of neurons (2), and certain types of ROS have been implicated in increasing longevity (3). Mitochondrial oxidative phosphorylation is the major cellular source of ROS. Superoxide anions (O2.−) are produced from small electron leaks mainly by the respiratory complexes I and III. They are cleared by superoxide dismutases, which convert them into H2O2. H2O2 can freely diffuse into the intermembrane space and into the cytosol but is mostly converted to H2O and oxygen by mitochondrial catalase. If the equilibrium between ROS production and scavenging is disturbed, then O2.− and H2O2 accumulate (4). Accumulation of ROS in the dopaminergic neurons of the substantia nigra as a result of dysfunctional mitochondria is considered as one of the main insults leading to degeneration of dopaminergic neurons in Parkinson’s disease (PD) (5, 6).
Mitochondria are highly dynamic and can form elongated tubes and networks or small, fragmented structures by undergoing cycles of fusion and fission. A key protein that coordinates mitochondrial quality control is phosphatase and tensin homolog–induced putative kinase 1 (PINK1), which is implicated in PD etiology (7). PINK1 directly interacts with proteins that determine mitochondrial dynamics (8, 9), among them the key player in mitochondrial fission, the large guanosine triphosphatase dynamin-related protein 1 (Drp1). PINK1 phosphorylates Drp1 at Ser616, which promotes its pro-fission activity. PINK1 further influences mitochondrial dynamics by inhibiting the pro-fusion factor mitofusin 2 (9).
Mitochondrial fragmentation takes place at endoplasmic reticulum (ER)–mitochondria contact sites. Drp1 binds at these contact sites, and the ER structurally contributes to mitochondrial fission by wrapping itself around the fission site (10). ER-mitochondria contacts are altered in PD patients and animal models, although reports on whether the contacts increase or decrease are sometimes contradictory (11–14). Besides a role in mitochondrial fission, ER-mitochondria contacts also influence mitochondrial activity by allowing the exchange of calcium and phospholipids between both compartments, which also plays a role in PD pathology (13).
Mitochondria produce energy by means of their electron transport chain: It contains four protein complexes that pump protons into the intermembrane space, thereby producing a proton gradient that drives an adenosine triphosphatase, which is sometimes referred to as complex V. The first and largest of the complexes is reduced form of nicotinamide adenine dinucleotide (NADH):ubiquinone oxidoreductase or complex I, which oxidizes NADH and reduces ubiquinone and transports protons from the inner mitochondrial membrane to the intermembrane space (15). Together with complex III, it is the main source of ROS production in eukaryotic cells. In neurons of patients with PD, complex I is functionally impaired and misassembled (7, 16), which is consistent with a role of ROS in PD pathology. Moreover, the complex I inhibitor rotenone, which has been used as a pesticide in agriculture, can induce PD in animal models, and PD is significantly more prevalent in people who were exposed to it (17). PINK1 is involved in the regulation of complex I: It mediates the phosphorylation of the complex I subunit NDUFA10 (NADH:ubiquinone oxidoreductase subunit A10) and thereby regulates its activity (18). The phospholipids cardiolipin, phosphatidylethanolamine (PE), and phosphatidylcholine (PC), which are exchanged at ER-mitochondria contacts, support complex I function (19); thus, ER-mitochondria communication might directly influence complex I activity. Notably, the functional relationship between ER-mitochondria contacts and complex I activity might be bidirectional: Inhibition of complex I with rotenone increases the expression of tether proteins in neuroblastoma cells (20).
Genetically, PD is heterogeneous: Risk genes for familial forms of PD comprise leucine rich repeat kinase 2 (LRRK), PINK1, and Parkin. PD is characterized by aggregates in neurons, the Lewy bodies, which contain α-synuclein folded into amyloid fibrils (21). A recent study shows that Lewy bodies mostly contain organelles and membrane fragments, highlighting the importance of organelle quality control and neuronal organelle trafficking in PD insurgence (22).
Proteins of the Creld family [cysteine-rich with epithelial growth factor (EGF)–like domains] are ER proteins that have been reported to regulate ER stress response and untranslated protein response (23, 24). CRELD1 is a risk gene for atrioventricular septal defects, and murine CRELD1 is required during heart development (25). A recent study links human CRELD1 to T cell survival (26). Mechanistic studies in Caenorhabditis elegans show that Creld1 is required for the maturation and assembly of acetylcholine receptors in the ER (27). CRELD1 is one of the 10 genes that was identified as differentially regulated on both protein and mRNA level in PD, in a combined proteomics and RNA sequencing approach in samples from the prefrontal cortex (28), suggesting a so far unknown connection to PD pathology.
RESULTS
Creld is conserved between fly, frog, and human
Creld proteins belong to a family that is evolutionary conserved across species. Mammalian Creld proteins contain a signaling peptide at the N terminus, a characteristic tryptophan and glutamic acid–rich (WE) domain, and two EGF-like and two calcium-binding domains (Fig. 1A) (29). Drosophila Creld shows high conservation to both the human CRELD1 and CRELD2: protein sequence identity is 44.1 and 43%, respectively, and 89 and 78% identity in the WE domain (fig. S1A). The Drosophila Creld protein contains a transmembrane domain like its human counterpart CRELD1, which, according to protein domain prediction, anchors it in the ER membrane, with the N terminus facing the lumen. The Drosophila genome contains only one gene for Creld, which results in two peptides, Creld-PA and Creld-PB, by alternative splicing. We raised an antibody against the conserved WE domain of Creld and found that it colocalized primarily with the ER marker KDEL (fig. S1B). We generated a Creld mutant (CreldΔ51) by homologous recombination that resulted in complete deletion of the gene locus (fig. S2A) and that it is a transcript null for both Creld transcripts (fig. S2B). In the following text, this strain is referred to as the Creld mutant, unless specifically stated otherwise. Creld mutants are homozygous viable but show slightly lower survival rates on a control diet, which is further reduced on a sugar-depleted diet (fig. S2C). The median life span of adult Creld mutants is shortened by ~30% (fig. S2D).
Fig. 1. Impaired locomotion, increased diastole, and mitochondrial accumulation in Creld mutants.
(A) Domain structure of human CRELD1 and CRELD2 and Drosophila Creld indicating protein identity of domains. (B) Single images from videos of w1118 and Creld mutants, showing their walking behavior. (C) Startle-induced negative geotaxis (SING) assay of 2-day-old w1118 and Creld mutants. (D) M-mode tracing of pixel movement of the adult heart. (E) Quantification of diastole, systole [not significant (ns), P = 0.43], and heart period. (F) Heart rate of w1118 and Creld mutant flies. (G) Transmission electron micrographs of abdominal tissue of w1118 and Creld mutant adults. Sections show myocardial cells. (H) Transmission electron micrographs of thorax tissue of w1118 and Creld mutant adults. Sections show indirect flight muscles (IFMs). Boxes represent the interquartile range and median; whiskers represent minimum and maximum. ***P < 0.001.
Loss of Creld leads to severe locomotion impairment
Adult Creld mutants show a strong locomotion deficit: They climb slowly and are almost unable to fly. Creld mutants show a distinct walking behavior: While wild-type flies show coordinated walking movements with an upright body position and a body axis of about 45° in relation to the surface, Creld mutants show uncoordinated walking movements and a horizontal body position, which causes them to fall on their heads frequently (Fig. 1B and movies S1 and S2). We used the startle-induced negative geotaxis (SING) assay to measure adult locomotion and found that Creld mutants are hardly able to climb (Fig. 1C). Ubiquitous expression of RNA interference (RNAi) directed against Creld also impairs the climbing performance (fig. S2E). We generated an independent allele by CRISPR-Cas9–mediated knockout (Creld12-2), which resulted in a point mutation after 90 bases and a frame shift after 30 amino acids (fig. S2F). We found that the Creld12-2 mutant and transheterozygous CreldΔ51/12-2 have reduced transcript levels of both transcripts and strongly reduced antibody signal (fig. S2, G to I). Climbing performance is equally reduced in both mutants and transheterozygotes (fig. S2J).
Prolonged heart resting phase shows energy deficit in Creld mutants
Mutations in human CRELD1 are associated with heart development defects (30), and mouse Creld1 mutants show defects in the proliferation of endocardial cells (31). The insect heart is morphologically different from the mammalian heart but contains valves and cardiac cells. Flies do not have a closed but an open circulatory system, and the heart pumps the hemolymph through the body to supply organs with nutrients and signaling molecules (32). We asked whether Creld mutants had heart defects and analyzed heart morphology and function. We found that heart development is normal, as is shown in third instar larvae, which show no morphological differences in the valve-like ostial cells (fig. S3A). Next, we analyzed heart function in adult flies. M-mode recordings from semi-intact Drosophila heart preparations and digital high-speed movie analysis show that the heart rate is regular but with a longer total heart period in Creld mutants compared to control flies (Fig. 1D). Upon quantification of the heart rate, we found that the total heart period is ~300 ms in wild-type flies and ~400 ms in Creld mutants and that this prolongation of the heart rate is due to a longer diastolic interval. The systole duration of the heart period is unaltered in Creld mutants, but the diastole duration increases from ~100 ms in wild types to ~250 ms in Creld mutants (Fig. 1E), and the heart rate is reduced from 3.5 to 2.5 Hz (Fig. 1F). We thus conclude that the contractility of the heart is not disturbed but that the resting phase between the contraction is delayed. Prolonged heart rates and an increased diastolic interval are associated with a deficiency in energy supply (33). We thus hypothesized that Creld mutants are not sufficiently supplied with adenosine triphosphate. Analysis of phosphorylated adenosine monophosphate–activated protein kinase (AMPK) confirmed that levels of phospho-AMPK are highly increased in protein extracts from adult Creld mutants compared to wild types (fig. S3, B and C). We conclude that heart development in Drosophila Creld mutants is normal, but that heart rate and diastolic interval are prolonged because of an energy deficit, which might also be involved in the locomotion defect displayed by Creld mutants.
Mitochondria accumulate and elongate in Creld mutants
Mitochondria are the major source of energy in eukaryotic cells. To investigate the energy crisis that provokes impaired locomotion and heart rate prolongation in Creld mutants, we analyzed mitochondrial morphology by transmission electron microscopy (TEM) for ultrastructural analysis. Since the energy deficit seemed to affect heart function and locomotion, we analyzed both cardiac cells and musculature, which are rich in mitochondria. We found that in myocardial cells, mitochondria are more abundant and elongated in Creld mutants compared to wild type (Fig. 1G and fig. S3D). We also used TEM for ultrastructural analysis of mitochondria in indirect flight muscles (IFMs) and found that cristae structure is unchanged in mitochondria of Creld mutants but that mitochondria are more abundant and enlarged (Fig. 1H). Both transversal and longitudinal sections of IFM show that in Creld mutants, mitochondria occupy all the space between the muscle fibers, whereas single mitochondria are well discernible in the wild type. Our results indicate that in Creld mutants, there is a defect in mitochondrial network regulation and an imbalance toward mitochondrial fusion.
Mitochondrial hyperfusion upon Creld dysfunction is conserved
Members of the Creld protein family have not been associated with mitochondrial dynamics before. To assess whether mitochondrial fusion under Creld1 dysfunction can also be observed in vertebrates, we used the claw frog Xenopus tropicalis as a model. We injected X. tropicalis embryos with control or Creld1 morpholinos and a mito-eGFP (enhanced green fluorescent protein) construct in the two-cell stage and analyzed the mitochondrial morphology in bodywall muscles at stage 40. We found that muscles of animals injected with the control morpholino showed a balance between fused and fragmented mitochondria, while in animals injected with the Creld1 morpholino, mitochondria are mostly present as elongated and fused [Fig. 2A; morpholino-mediated knockdown efficiency is shown in fig. S4 (A and B)]. We calculated the mitochondrial fragmentation index and found that it is significantly reduced in muscle cells of animals injected with the Creld1 morpholino (Fig. 2B). To test whether a similar phenotype can be observed in mammalian cells, we used a double-stranded RNA (dsRNA) construct against CRELD1 in HeLa cells. We stained control and dsRNA-treated cells with an antibody against TOMM20 (translocase of outer mitochondrial membrane 20), a receptor in the outer mitochondrial membrane (34), and found that, also in HeLa cells, mitochondria are mostly fragmented in controls (35) but are fused, elongated, and network-like in cells that knock down CRELD1 (Fig. 2, C and D, and fig. S4C). Our results indicate that loss of Creld leads to defects in mitochondrial network regulation and an imbalance toward mitochondrial fusion in various tissues and species.
Fig. 2. Creld interaction with Pink1 and Parkin.
(A) X. tropicalis stage 40 bodywall muscle cells labeled with mito-eGFP and close-ups. (B) Quantification of the mitochondrial fragmentation index as the number of mitochondria divided by the total mitochondrial area of cells from animals injected with control and Creld1 morpholino (MO). (C) Untreated (control) or transfected with dsRNA against CRELD1 HeLa cells and close-ups. Mitochondria are stained with anti-TOMM20. (D) Quantification of the mitochondrial fragmentation index based on TOMM20 (C). (E) SING assay of w1118 and Creld mutants untreated or fed with 20 μM CsA in 10% sucrose solution. Analysis of variance (ANOVA) with F = 34.662 and 77 degrees of freedom. (F) SING assay of flies expressing a constitutively active form of Pink1. Genotypes are Creld−/−; tubulin Gal4/+, Creld/+ or −/−; and tubulin Gal4/UAS Pink1.C. ANOVA with F = 8.023 and 59 degrees of freedom. (G) SING assay of Pink1 mutants expressing UAS-Creld-HA. Genotypes are as indicated. ANOVA with F = 39.562 and 122 degrees of freedom. ns, P = 0.86. (H) SING assay of Creld-Parkin double mutants. Genotypes are as indicated. ANOVA with F = 56.584 and 204 degrees of freedom. ns, P = 0.29. Boxes represent the interquartile range and median; whiskers represent minimum and maximum. *P < 0.05, **P < 0.01, and *** P < 0.001.
Reduced association of the pro-fission factor Drp1 with mitochondria in Creld mutants
Mitochondrial fission is regulated by Drp1, which itself is regulated by several kinases and phosphatases. Drp1 localizes to mitochondria when activated to induce the fission process. To assess whether Drp1 locates to mitochondria in Creld mutants, we expressed a Drp1-hemagglutinin (HA) fusion protein in Creld mutants and compared the presence of Drp1-HA in the cytoplasmic and the mitochondrial fraction. After normalization to the protein content of the fractions, we found that 80% of Drp1-HA is found in the mitochondrial fraction in heterozygous controls. In Creld mutants, the mitochondrial portion of Drp1 is reduced, and only ~60% associate with mitochondria, while ~40% is found in the cytoplasmic fraction (fig. S4, D to G). This shows that, as a result of missing Creld function, Drp1 locates less to mitochondria, consistent with decreased fission activity.
Creld is required for the activation of the Drp1-activating phosphatase calcineurin in mammals (31, 36). To elucidate the Creld-calcineurin interaction in flies, we fed adult control flies and Creld mutants with the calcineurin inhibitor cyclosporin (CsA). Treatment with CsA strongly reduces the climbing ability of wild types and thus mimics the phenotype of Creld mutants (Fig. 2E). However, CsA treatment further reduces the climbing performance of Creld mutants, suggesting that an additional function of Creld, apart from activating calcineurin, is responsible for the locomotion phenotype. Drp1 can also be activated by phosphorylation by PINK1 (8). We overexpressed a constitutively active form of Pink1 (Pink1.C) under the control of the ubiquitous tubulin-Gal4 driver and found that it rescues the climbing performance of homozygous Creld mutants (Fig. 2F). This Pink1-mediated rescue was maintained after treatment with CsA (Fig. 2F). This suggests that Creld interacts genetically with Pink1, in addition to calcineurin, to regulate mitochondrial dynamics, and that both Pink1 and calcineurin act downstream and/or in parallel with Creld in the activation of Drp1 and mitochondrial fission.
Creld rescues the locomotion deficit of PINK1 mutants
Human CRELD1 was identified as a PD pathology gene (28), and the severe locomotion deficit that we observe is reminiscent of the PD phenotype in fly models. Moreover, expression of constitutively active PINK1 rescues the climbing performance of Creld mutants. We asked whether overexpression of Creld would rescue the locomotion deficit of PINK1 mutants. We generated a genetic rescue construct that allows for the expression of the full-length protein of Creld under the control of the binary Gal4/UAS transcription activator/enhancer system. We expressed the rescue construct under the control of the ubiquitous actin-Gal4 driver in Pink1 mutants and found that it significantly increases the climbing ability of Pink1 mutants (Fig. 2G). Pink1 recruits Parkin to mitochondria in the regulation of mitochondrial quality control. Heterozygous Creld and Parkin mutants show similar climbing performance. Heterozygous double mutants for Creld and Parkin show heterogeneous, but not overall reduced, climbing ability. Double mutants heterozygous for Creld and homozygous for Parkin show no further reduction of the climbing ability compared to homozygous Parkin mutants. By contrast, double mutants homozygous for Creld and heterozygous for Parkin show significantly worsened climbing behavior compared to Creld mutants (Fig. 2H). We did not find flies homozygous for both mutations and assume that the combination is lethal. Our results suggest that Creld modulates the Pink1-Parkin pathway of mitochondrial quality control.
Creld is required in neurons for normal locomotion
To explore the connection between the defects in mitochondrial fission, locomotion, and heart, we reintroduced Creld function genetically in specific tissues. We used the Creld rescue construct under the control of various tissue-specific Gal4 drivers. Homozygous Creld mutants that only carry the rescue construct, but no Gal4 driver, show impaired climbing similar to homozygous Creld mutants. Heterozygous Creld mutants show wild-type climbing performance. We found that expression in the heart with the handC-Gal4 driver (37) fails to rescue the climbing ability of Creld mutants. Expression in muscles with the Mef2-Gal4 driver partially rescues Creld mutants climbing. Similarly, ubiquitous expression of the genetic rescue construct with tubulin-Gal4 slightly but significantly rescues the climbing performance of homozygous Creld mutants (Fig. 3A). Expression of the genetic rescue construct in the nervous system by using the postmitotic neuron driver elav-Gal4 results in a strong rescue and markedly improves the climbing ability of homozygous Creld mutants (Fig. 3A). To further dissect the rescue in neurons, we used drivers specific for distinct neuron types. Expression of the rescue construct restricted to the insulin-producing cells (dilp2,3-Gal4), a group of median neurosecretory cells organized in two clusters in the pars intercerebralis of the adult brain that secrete the insulin-like peptides 2, 3, and 5 (38), fails to rescue the climbing deficit of Creld mutants (Fig. 3B). By contrast, use of the D42-Gal4 driver, which directs the expression of the rescue constructs to motor neurons (39), resulted in a complete rescue of the climbing deficit. Dopaminergic neurons are expressed in distinct clusters in the larval and adult brain (40) and integrate motor programs (41). In humans, dopaminergic neurons modulate locomotion by innervating the brain stem, and locomotion defects in PD are caused by degeneration of these neurons (42). This mechanism is strongly conserved, and in addition, flies present impaired locomotion and other PD symptoms upon loss of dopaminergic neurons (43). We used tyrosine hydroxylase (TH)–Gal4 to direct the expression of the rescue construct to dopaminergic neurons and found that it rescues the locomotion impairment of homozygous Creld mutants (Fig. 3B). Our findings indicate that loss of Creld affects locomotion because its function is required in motor neurons and dopaminergic neurons, not the heart, and only to a minor degree in muscles.
Fig. 3. Creld is required in neurons for normal locomotion.
(A) SING assay of animals expressing UAS-Creld-HA in specific tissues. Climbed distance in millimeters was measured after 6 s. Genotypes are (from left to right) Creld−/−; UAS-Creld-HA/+, Creld/+, or −/− as indicated; tubulin-Gal4/UAS Creld-HA, Creld+/−, or −/−; mef-Gal4; UAS-Creld-HA, Creld/+, or −/−; handC-Gal4; UAS-Creld-HA, Creld/+, or −/−; elav-Gal4; and UAS-Creld-HA. ANOVA with F = 46.1 and 195 degrees of freedom. (B) SING assay of animals expressing UAS-Creld-HA in specific neurons. Climbed distance in millimeters was measured after 6 s. Genotypes are Creld−/−; UAS-Creld-HA/+, Creld/+, or −/− as indicated; elav-Gal4/UAS Creld-HA, Creld/+, or −/−; dilp2,3-Gal4; UAS-Creld-HA, Creld/+, or −/−; D42-Gal4; UAS-Creld-HA, Creld/+, or −/−; TH-Gal4; and UAS-Creld-HA. ANOVA with F = 38.167 and 168 degrees of freedom. (C) Airyscan confocal imaging of ER in PPL1 cluster of dopaminergic neurons labeled by GFP KDEL (left) and mitochondria labeled by mCherry-mitoOMM (right) under the control of TH-Gal4. Genotypes are Creld/+; TH Gal4/UAS GFP KDEL or mCherry-mitoOMM and Creld−/−; and TH Gal4/UAS GFP KDEL or mCherry-mitoOMM. mCherry was detected with a DsRed antibody. (D) Mitochondrial fragmentation index in somata of dopaminergic neurons. (E and F) Calcium imaging in PPL2 clusters of dopaminergic neurons with UAS CaMPARI2. Images were taken after 30-s ultraviolet light induction and 4-min photoconversion. Genotypes are Creld/+ or −/− and TH Gal4/UAS CaMPARI2. Boxes represent the interquartile range and median; whiskers represent minimum and maximum. *P < 0.05, **P < 0.01, and ***P < 0.001.
Dopaminergic neurons are inactive in Creld mutants
Locomotion deficits in PD patients and PD fly models are elicited by degeneration of dopaminergic neurons. However, we found no loss of dopaminergic neurons in Creld mutants (fig. S5, A to C). We did not detect any enrichment of Creld protein in specific brain regions or neurons (fig. S5D). Histological sections did not show any sign of neurodegeneration (fig. S5E). The number of apoptotic cells in the brain is similar in wild-type flies and Creld mutants (fig. S5F). We first evaluated ER morphology in dopaminergic neurons by expressing GFP with the ER retention signal KDEL under the control of the TH-Gal4 driver. We analyzed dopaminergic neurons in control and Creld mutants with superresolution microscopy using Airyscan confocal imaging. We did not observe any differences in ER morphology (Fig. 3C). To evaluate mitochondrial morphology in dopaminergic neurons, we expressed a construct that carries the transmembrane domain of the mitochondrial protein Miro tagged with mCherry and thus labels the outer mitochondrial membrane (mCherry-mitoOMM), under the control of TH-Gal4. We found that mitochondria are round and fragmented in heterozygous Creld mutants but that the mCherry signal accumulates and shows mitochondria fused and elongated in homozygous Creld mutants [Airyscan confocal images in Fig. 3 (C and D); confocal images in fig. S6A]. Next, we asked whether dopaminergic neurons are less active in Creld mutants. We used the calcium-modulated photoactivatable ratiometric integrator CaMPARI2 (44) for calcium imaging of active dopaminergic neurons. We found that upon stimulation with ultraviolet light for 30 s and 4 min after stimulation, dopaminergic neuron somata show conversion of green to red in wild-type controls but less conversion in Creld mutants (Fig. 3, E and F). This indicates that dopaminergic neurons are less active, rather than degenerated, in Creld mutants.
ROS homeostasis is impaired in Creld mutants
Dopaminergic neuron loss in PD fly models is induced by oxidative stress, elicited by old and dysfunctional mitochondria that accumulate as a result of defective mitochondrial quality control. However, it is important to note that ROS are also important signaling molecules and indispensable for cell differentiation and neuronal activity, including motor neurons (45). With these considerations in mind, we asked whether ROS are altered in Creld mutants. We tested whether the observed mitochondrial phenotype led to reduced ROS levels. We used the superoxide indicator dihydroethidium (DHE) and the mitochondria-specific superoxide dye MitoSOX to assess superoxide levels in Creld mutants. We found that overall superoxide was reduced in both adult gut and brain (Fig. 4, A and B, and fig. S6B). By contrast, mitochondrial superoxide levels as stained by MitoSOX were increased (Fig. 4A). Superoxide anions are relatively stable and remain in the mitochondrial matrix and intermembrane space, while hydrogen peroxide can diffuse freely into the cytoplasm (4). Thus, we next measured hydrogen peroxide, a product of superoxide dismutation by Sod 1 and 2, with Amplex Red, and found that hydrogen peroxide is markedly reduced in Creld mutants (Fig. 4C and fig. S6C). We concluded that in mitochondria of Creld mutants, superoxide accumulates but is not degraded to hydrogen peroxide. Hydrogen peroxide is also reduced in extracts of X. tropicalis embryos that knock down Creld1 (fig. S6D). We measured the transcript levels of ROS-scavenging enzymes in flies and found that catalase and nitric oxide synthase (NOS) are expressed at wild-type levels, while superoxide dismutase 1 and 2 are significantly down-regulated (Fig. 4D), consistent with decreased hydrogen peroxide levels and, therefore, retention of ROS in mitochondria. To induce hydrogen peroxide formation in Creld mutants, we expressed superoxide dismutase 1 (Sod1) under the control of the ubiquitous arm-Gal4 driver and found that it rescues the climbing performance of Creld mutants (Fig. 4E), and ubiquitous expression of Pink1 and Sod1 and down-regulation of the ROS scavenger catalase increase hydrogen peroxide to control levels in Creld mutants. By contrast, down-regulation of Duox, which produces ROS by oxidation of cytoplasmic reduced form of nicotinamide adenine dinucleotide phosphate, does not restore hydrogen peroxide levels in Creld mutants (Fig. 4F). We tested whether modulation of hydrogen peroxide in dopaminergic neurons was sufficient to restore the climbing of Creld mutants. Upon induction of hydrogen peroxide formation by expression Pink1, Sod1, or down-regulation of catalase, the locomotion deficit was abolished. When hydrogen peroxide formation was repressed by down-regulation of Duox, climbing was not restored (Fig. 4G). Our results show that aberrant ROS signaling underlies the locomotion deficit in Creld mutants and suggest that reduced hydrogen peroxide exported from mitochondria leads to inactivity of dopaminergic neurons.
Fig. 4. Low H2O2 impairs dopaminergic neuron function.
(A and B) Adult gut tissue was stained with DHE to label superoxide anions. Adult gut was stained with MitoSOX to label mitochondrial superoxide anions. a.u., arbitrary units. (C) Hydrogen peroxide was measured in extracts of whole adults with Amplex Red (n = 8). (D) Transcript levels of ROS-scavenging enzymes in Creld mutants. Fold change was normalized to w1118 (n = 8). (E) SING assay of Creld mutants ubiquitously expressing UAS Sod1. Genotypes are Creld−/−; armadillo Gal4/+, Creld/+, or −/−; and armadillo Gal4/UAS Sod1. (F) Hydrogen peroxide was measured in extracts of whole adults with Amplex Red. Genotypes are Creld−/−; armadillo Gal4/+, Creld/+ or −/−; armadillo Gal4/UAS Pink1.C, Creld/+ or −/−; armadillo Gal4/UAS Sod1, Creld/+ or −/−; armadillo Gal4/UAS catalase-RNAi, Creld/+ or −/−; and armadillo Gal4/UAS Duox RNAi. (G) SING assay of Creld mutants expressing PINK1, Sod1, catalase-RNAi, or Duox-RNAi in dopaminergic neurons. Genotypes are Creld−/−; TH-Gal4;+; Creld/+ or −/−; TH-Gal4/UAS Pink1.C, Creld/+, or −/−; TH Gal4/UAS Sod1, Creld/+, or −/−; TH-Gal4/UAS catalase-RNAi, Creld/+, or −/−; and TH-Gal4/UAS Duox-RNAi. ANOVA with F = 33.627 and 145 degrees of freedom. (H) Oxygen consumption of larval bodywall preparations of w1118 and Creld mutants treated with pyruvate and malate and rotenone. (I) SING assay of Creld mutants expressing ND-39 in dopaminergic neurons. Genotypes are Creld−/−, TH-Gal4;+, Creld/+ or −/−, and TH-Gal4/UAS ND-39. Boxes represent the interquartile range and median; whiskers represent minimum and maximum. ***P < 0.001.
Mitochondrial activity is impaired at the level of respiratory complex I
A major source of superoxide and hydrogen peroxide in mitochondria is the respiratory complex I, which is regulated by Pink1 via phosphorylation of subunit ND-42 (46). We analyzed mitochondrial respiration in larval bodywall muscles using a Seahorse analyzer (Agilent). We found that both wild-type and Creld mutant tissue react to injection of pyruvate and malate as substrates for complex I by increasing mitochondrial respiration but that only oxygen consumption in wild-type tissue can be blocked with rotenone, a complex I inhibitor. Oxygen consumption in Creld mutant tissue remains at high levels upon rotenone treatment, which indicates that complex I is already inactive (Fig. 4H) and bypassed by other oxygen-consuming processes. Feeding wild-type adults with sublethal levels of rotenone (100 μM) decreases their climbing performance but not to the extent of Creld loss of function (fig. S6E). By contrast, the poor climbing performance of Creld mutants is not further affected by rotenone and even ameliorates slightly. Rotenone treatment lowers the levels of hydrogen peroxide in wild-type flies (although its mechanism of action leads to increased electron leakage, indicating a complex effect on ROS metabolism and transport) but not in Creld mutants, which already have low levels of hydrogen peroxide (fig. S6F). This shows that the climbing deficit of Creld mutants can be attributed to low complex I activity and concomitant changes in ROS metabolism and transport. Knockdown of complex I subunit ND-39 has been shown to prevent ROS formation by decreasing complex I levels (3). Expression of the complex I subunit ND-39 in dopaminergic neurons rescues the Creld mutant climbing deficit (Fig. 4I), and ubiquitous expression increases hydrogen peroxide levels (fig. S6G). This suggests that complex I activity and its impact on ROS production and signaling is indispensable for dopaminergic neuron function and locomotion, specifically.
Creld expression is up-regulated upon complex I inhibition
Complex 1 activity is altered in Creld mutants, and complex I defects are observed in PD. Creld was differentially expressed in PD prefrontal cortex samples (28). We therefore asked whether Creld reacts to conditions of low complex I activity and measured the transcript levels of Creld under different conditions in adult wild-type flies. We found that while normal under reduction of dietary sugar or upon starvation, Creld transcript levels increase upon feeding with rotenone (fig. S7A). We expressed an UAS-Creld construct tagged with HA in dopaminergic neurons and costained with an antibody against endogenous Creld to observe Creld expression on the protein level. We found that upon rotenone feeding, the signal intensity of both the ectopically expressed Creld and endogenous Creld markedly increases (fig. S7B). As this also affects the ectopically expressed UAS-Creld-HA, Creld accumulation upon complex I inhibition has to involve posttranscriptional mechanisms and transcriptional regulation. Our results show that Creld expression reacts to conditions of low complex I activity by up-regulation.
Complex I inhibition enhances ER-mitochondria contacts
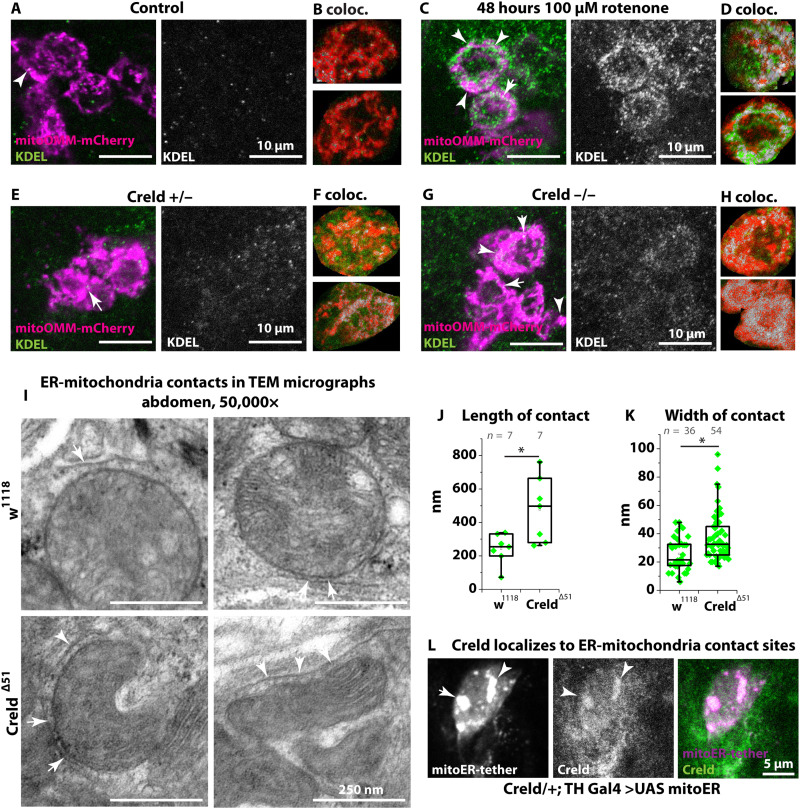
ER-mitochondria contacts regulate mitochondrial dynamics and are sites of material exchange (47). Drp1 binds at ER-mitochondria contacts to induce fission, and Pink1 and Parkin accumulate at contact sites in the regulation of mitophagy. Transfer of phospholipids and calcium ions (Ca2+) between ER and mitochondria takes place at their contact sites and is required for mitochondrial function: For example, phospholipids support the function of respiratory complex I (19). Human CRELD1 locates primarily in the ER (26), and Drosophila Creld also localizes mostly to the ER (fig. S1B). We thus asked how Creld affects the function of respiratory complex I from its ER position by assessing ER-mitochondria contacts in dependence on complex I activity. We expressed the mCherry-mitoOMM construct in dopaminergic neurons, costained with an antibody against the ER marker KDEL, and performed colocalization analysis on single confocal sections of single soma of dopaminergic neurons. In wild types, KDEL signal intensity is low and colocalization of ER and mitochondria markers is rare (Fig. 5, A and B). To inhibit complex I activity, we treated the flies for 48 hours with sublethal concentrations of rotenone. Upon rotenone treatment, KDEL signal intensity increases in the somata of dopaminergic neurons, resulting in enhanced colocalization of ER and mitochondria markers (Fig. 5, C and D, and fig. S7D). Similarly, the ER marker GFP-KDEL enhances in signal upon rotenone treatment (fig. S7C). We therefore suggest that complex I activity modulates ER-mitochondria contacts.
Fig. 5. Creld and complex I activity modulate ER-mitochondria contact.
(A) PPL1 cluster of dopaminergic neurons expressing mCherry-mitoOMM, stained with anti-DsRed and anti-KDEL in +/+; TH-Gal4/UAS mitoOMM. (B, D, F, and H) Regions of interest of single soma of dopaminergic neurons after colocalization analysis with ImageJ colocalization threshold tool. Colocalization is shown in gray. (C) PPL1 cluster of dopaminergic neurons expressing mCherry-mitoOMM, stained with anti-DsRed and anti-KDEL in +/+; TH-Gal4/UAS mitoOMM after treatment with rotenone. (E and G) PPL1 cluster of dopaminergic neurons expressing mCherry-mitoOMM, stained with anti-DsRed and anti-KDEL in Creld/+; TH-Gal4/UAS mitoOMM (A) and Creld−/−; TH-Gal4/UAS mitoOMM. Arrowheads show colocalization of ER and mitochondria. (I) TEM micrographs of adult abdomen showing ER-mitochondria contact sites. Scale bars are as indicated. (J) Quantification of ER-mitochondria contact length. (K) Quantification of ER-mitochondria contact width. (L) PPL1 cluster of dopaminergic neurons expressing an artificial tether construct (mitoER-tether-RFP) stained with α-Creld. Boxes represent the interquartile range and median; whiskers represent minimum and maximum. *P < 0.05 and ***P < 0.001.
Creld localizes to ER-mitochondria contacts
Complex I and mitochondrial dynamics are inhibited in Creld mutants, and complex I inhibition enhances ER-mitochondria contacts and induces Creld expression. To investigate ER-mitochondria contacts in Creld mutants, we used again the mCherry-mitoOMM construct in dopaminergic neurons and costained with an antibody against the ER marker KDEL. We found that in heterozygous controls, ER and mitochondrial markers show some few points of colocalization (Fig. 5, E and F, and fig. S7E). In homozygous Creld mutants, the KDEL signal is more intense in the somata of the dopaminergic neurons and colocalization between ER and mitochondrial markers is increased, similar to the situation in rotenone-treated wild types (Fig. 5, G and H). Since ER-mitochondria contact sites are beyond the resolution of confocal microscopy, we analyzed them in TEM micrographs. We confirmed that ER-mitochondria contacts stretch over longer parts of mitochondria in Creld mutants but that the gap between ER and mitochondrion is wider (Fig. 5, I to K), which poses the question of their functionality in terms of material exchange. To address whether endogenous Creld is located at ER-mitochondria contact sites, we expressed a synthetic mito-ER tether (11) in dopaminergic neurons. Tethering sites are marked with red fluorescent protein (RFP) in this line, and we found large spots of the RFP signal in the somata of dopaminergic neurons. The endogenous Creld signal colocalizes with these spots (Fig. 5L). Our results show that contacts between ER and mitochondria increase upon loss of Creld and upon complex I inhibition and that Creld localizes to ER-mitochondria contact sites that are induced by a synthetic mito-ER tether. Taking these observations together, it appears likely that Creld also localizes to endogenous ER-mitochondria contact sites in wild types.
Phospholipids accumulate at mitochondria-associated membranes (MAMs) in Creld mutants.
ER-mitochondria contacts have been identified as so-called MAMs: Crude mitochondrial fractions contain fragments of ER membranes (48). These sites function as important signaling hubs in a number of physiological and pathological processes (49). Phospholipids are transferred at ER-mitochondria contacts (47), and the phospholipids, PE, and PC are required for the function and stability of the respiratory complex I (19). We thus asked whether phospholipids can be transferred to mitochondria in Creld mutants. We isolated the cytoplasmic fraction, containing also plasma membrane, ER, and lysosomes; the crude mitochondrial fraction comprising mitochondria and MAMs; and the MAM fraction by ultracentrifugation and subjected the fractions to lipidomics analysis. We measured a technical sample containing only the buffers for each fraction and normalized the obtained data to the corresponding technical sample. We found that the total lipid content is by trend, albeit not quite significantly, in the cytoplasmic fraction in Creld mutants, while phospholipids are not significantly altered. Total lipids increase in the mitochondrial fraction, suggesting lipid allocation to mitochondria, but the phospholipid PE is reduced in Creld mutants. Total phospholipids accumulate in the MAM fraction, and when we looked at individual phospholipid species, we observed that PE and PC increase in the MAM fraction of Creld mutants (Fig. 6, A to C). Together, the data from our lipidomics analysis show that phospholipid distribution between ER and mitochondrial compartments is disturbed. While we do not have direct evidence for altered phospholipid transport in Creld mutants, it appears likely that transfer of phospholipids from the ER to mitochondria is impaired, as this is a known function of MAMs affecting complex I function. PE and PC, which are required for complex I activity, seem to get stuck at ER-mitochondria contacts. This hypothesis is consistent with the observation of wider gaps at MAMS in electromicrographs of Creld mutant tissues, as distance is one important factor in the exchange of lipids at MAMs (47). ER-mitochondria contacts in Creld mutants could be increased as a compensatory measure (Fig. 6, D to F), as this is also the case upon rotenone treatment and resulting complex I deficiency. We therefore propose that Creld normally functions under conditions of low complex I activity to facilitate phospholipid flux at ER-mitochondria contacts.
Fig. 6. Phospholipid transfer at ER-mitochondria contacts is impaired in Creld mutants.
(A) Total lipid, phospholipid, PE, and PC content of fractions containing the cytoplasm, ER, plasma membrane, and lysosomes. Fold change of extracts of Creld mutant flies normalized to w111. (B) Lipids as indicated in fractions containing mitochondria and MAM (crude mitochondria). (C) Lipids as indicated in fractions containing MAM. (D to F) Model of Creld action under (D) wild-type, (E) complex I–inhibited, and (F) Creld loss-of-function conditions. Bar charts show means and SD, and dots represent single data points. *P < 0.05. ns, not significant or P as indicated.
DISCUSSION
PD is characterized by motor function defects as a result of dopaminergic neuron loss. It is believed that one of the main pathology-driving factors is oxidative stress induced by failure of mitochondrial quality control, leading to neuron loss. In accordance with this theory, PINK1, one of the commonest risk genes for PD, not only induces clearance of old mitochondria but also directly regulates the machinery of mitochondrial fusion and fission. However, more recent data implicates PINK1 also in a role as a direct regulator of mitochondrial function via phosphorylation of the respiratory complex I subunit NDUFA10 (18), although independent of the Pink1 mutant locomotion deficit (46). Moreover, complex I dysfunction has been observed in tissue of patients with PD (16). Here, we present data on an unexplored PD risk gene, Creld, and show that Creld mutants exhibit a strong, PD-like locomotion deficit and mitochondrial hyperfusion. We found that Creld interacts genetically with Pink1. In contrast to canonical PD models, dopaminergic neurons do not degenerate in Creld mutants, and oxidative stress in the form of hydrogen peroxide is absent. Consistently, mitochondrial ultrastructure appeared undamaged, while mitochondrial abundance and fusion increase. Notably, increasing hydrogen peroxide production in dopaminergic neurons rescues the Creld locomotion defect, implicating disturbed ROS signaling rather than oxidative stress as driving the locomotion defect of Creld mutants. Further studies are required to reveal Creld-interacting proteins or Creld-dependent signaling that regulates mitophagy and mitochondrial biogenesis.
ROS, especially mitochondrial hydrogen peroxide, have been recognized as important signaling molecules in recent years: Hydrogen peroxide is produced as a consequence of dopaminergic neuron activity but is also required for their activity We argue that mitochondrial ROS signaling, especially hydrogen peroxide resulting from complex I activity, is required in dopaminergic neurons to support their function. This notion is supported by a study showing that ROS resulting from reverse electron transport (RET) at complex I is not detrimental but instead delays aging (3), highlighting that ROS (as an umbrella term) can act differentially, depending on molecule, context, and location in the cell. In the same line of evidence, the PD-causing pesticide rotenone is a specific and well-studied complex I inhibitor that specifically blocks RET at complex I and reduces the formation of hydrogen peroxide (3), linking PD not only to complex I function in general but also specifically to complex I RET-dependent ROS production. Notably, we found that rotenone treatment provokes increased expression of Creld. It appears therefore likely that cells require Creld under conditions of low complex I activity and/or low levels of hydrogen peroxide signaling from mitochondria.
Creld is an ER protein that, in C. elegans, has been shown to act as a chaperone for the assembly of acetylcholine receptors (27). Misassembly of acetylcholine receptors might account for a muscle-induced climbing deficit also in fly Creld mutants but does not sufficiently explain mitochondrial hyperfusion and low complex I activity or the marked improvement of locomotion behavior upon expression of Sod1 or Creld in dopaminergic neurons. Instead, while the exact molecular mechanism of Creld in facilitating mitochondrial/complex I function is still elusive, our data implicate ER-mitochondria contacts as the site of Creld action. We could show that Creld accumulates at ER-mitochondria contacts that are artificially induced by expression of a synthetic tether. Both complex I inhibition by rotenone and Creld loss of function lead to increased regions of contact between ER and mitochondria. TEM imaging of Creld mutant tissue reveals broader gaps between the two compartments at the contact sites than in wild type, suggesting that they are functionally impaired as a result of Creld loss of function and implicating Creld in their function, maintenance, or regulation. Furthermore, the lipid composition of MAMs, ER, and mitochondria are altered, arguing for disturbed phospholipid exchange between the compartments in Creld mutants. This is an intriguing observation since MAMs are known to play a role in phospholipid transport between ER and mitochondria (47) and phospholipids regulate complex I activity (19). Our findings suggest that complex I dysfunction in Creld mutants could therefore be a direct consequence of impaired lipid exchange at ER-mitochondria contact sites and, in turn, affect neuron function due to impaired ROS signaling from complex I. However, the resulting locomotion defect is not caused by dopaminergic neuron loss, nor do we observe other signs of neurodegeneration. Nevertheless, we cannot rule out that dopaminergic neurons might degenerate eventually as a result of their inactivity in old Creld mutants. Determination of direct interacting proteins of Creld might reveal how Creld participates in the regulation of ER-mitochondria contacts.
To date, it is not entirely clear whether loose or tight contacts between ER and mitochondria better support the function of the cell: The formation of the contacts is important for mitochondrial dynamics quality control, but increased contacts have been associated with depletion of phospholipids from the ER (13). Furthermore, reduced contacts in flies as a result of knockdown of the tether protein Pdzd8 promote mitochondrial turnover in neurons and decelerate age-related locomotion deficits (50). On the basis of our data, we hypothesize that Creld is required for the formation of functional contacts between ER and mitochondria to provide phospholipids for the support of complex I function and thus allowing the formation of hydrogen peroxide as a signaling molecule for the activity of dopaminergic neurons. Complex I turns to its inactive state when oxygen is limiting (15), but phases of low complex I activity also occur during development and in dependence of nutrition (51). We suggest that Creld is required for dynamic ER-mitochondria contacts that enable the transfer of phospholipids to support or regulate complex I function and thus allows the formation of hydrogen peroxide as a signaling molecule for the activity of dopaminergic neurons. While direct interaction partners of Creld in the modulation of ER-mitochondrial contact dynamics are not yet known, it is conceivable that Creld might be required for the assembly of subunits of the respiratory complex I, a large protein complex comprising several subunits, or proteins that form the tether between ER and mitochondria. Further studies will reveal the molecular role of Creld and interaction in modulation of Pink1/Parkin pathway, ER-mitochondrial contact dynamics, and complex I activity.
MATERIALS AND METHODS
Fly husbandry
Flies were reared on standard cornmeal food (130 g of yarn agar, 248 g of Baker’s yeast, 1223 g of cornmeal, and 1.5 liters of sugar beet syrup in 20 liters of distilled water) and kept in a 25°C incubator with light-dark cycle. Fly stocks were obtained from the Bloomington Drosophila Stock Center (BDSC) and the Vienna Drosophila Resource Center. For stainings and SING assays, 2- to 5-day-old female flies were kept on control diet or on filter paper soaked with 10% sucrose solution and 50 or 100 μM rotenone or 20 μM CsA. We obtained the UAS-CaMPARI2 and the TH-Gal4 line from A. Schoofs and the UAS mito-ER tether lines from E. Ziviani. Other fly lines used in this study were w1118 (BDSC, no. 6326), elav-Gal4 (BDSC, no. 8760), tubulin-Gal4 (BDSC, no. 5138), mef-Gal4 (BDSC, no. 27390), handC-Gal4 (37), actin-Gal4 (BDSC, no. 4414), armadillo-Gal4 (BDSC, no. 1561), UAS-Creld-HA (this study), UAS-mCherry-mitoOMM (BDSC, no. 66533), Drp1-HA (BDSC, no. 42208), UAS Pink1.C (BDSC, no. 51648), UAS catalase-RNAi (BDSC, no. 34020), UAS Duox-RNAi (BDSC, no. 33975), UAS-Sod1 (BDSC, no. 24750), UAS ND-39 (BDSC, no. 15821), UAS GFP-KDEL (BDSC, no. 9899), Pink15 (BDSC, no. 51649), and park25 (52).
Creld mutant generation
The CreldΔ51 mutant was generated by homologous recombination. 5′ and 3′ homology arms were cloned into the pGX-attP vector (53) and injected into w1118 embryos. Primers were 5′ homology arm forward, GCGGCCGCCTTATTCGTATAGAACTTTGCCCG; 5′ homology arm reverse, GCGGCCGCTTTAACCGGCAAACTATCTGAG (both including Not I restriction sites); 3′ homology arm forward, TTGGCGCGCCAAATGATACGGCAACTCTCC; and 3′ homology arm reverse, TTGGCGCGCCAGTCCGCAGTGATTCAAACTC (both including Asc I restriction sites). Creld was deleted by homologous recombination and replaced by the mini-white open reading frame (ORF) and an attP site. The white gene was removed by Cre recombinase. The resulting mutants are transcript null. Expression of the neighboring genes zir and nhe1 was tested and found to be at wild-type levels. We generated a CRISPR-Cas9–mediated mutant allele (Creld12-2) by nonhomologous end joining. Primers were Creld_fwd, CTTCGCCTGTTCCGCCGCCCTGCA and Creld_rev, AAACTGCAGGGCGGCGGAACAGGC. Annealed oligos were cloned into the pU6-BbsI vector and sent to BestGene Inc. (USA) for injection into vas-Cas9 (BDSC, no. 55821). Mutants were identified by high-resolution melting analysis and sequencing. For the rescue construct, we designed a gblock containing a UAS coding sequence, the ORF for the Creld gene, and an HA tag (Integrated DNA Technologies). Cloning of the gblock construction was done by using Eco RI and Not I, followed by ligation into the pGE-attBGMR vector (53). Clones were sequenced to confirm successful integration, and the plasmid was sent to BestGene Inc. (USA) for injection.
Determination of life span
For determination of the life span of w− and Creld mutants, isogenized [backcrossed for six generations (54)] were separated by sex and transferred to longevity food [37.5 g of Baker’s yeast, 10 g of Agar Kobe I, 300 ml of dH2O, 3ml of 10 % Nipagin (in 70% ethanol) and 37.5 g of glucose were added after autoclavation] with 20 flies per tube. The number of viable and dead flies was controlled every day, and longevity food was changed every 2 days.
Semi-intact Drosophila heart preparation and digital high-speed movie analysis
All dissection steps were done in artificial hemolymph [108 mM NaCl2, 5 mM KCL2, 2 mM CaCl2 2xH2O, 8 mM MgCl2 6xH2O, 1 mM NaH2PO4, 4 mM NaHCO3, 15 mM Hepes, 1 mM sucrose, and 0.5 mM trehalose (pH 7.1)]. The flies were anesthetized with fly-nap, followed by transfer to a petri dish coated with Vaseline for dissection. After dissection, the submerged hearts were kept in oxygenated hemolymph for 15 min at room temperature for equilibration. The heart movements were recorded with a digital high-speed camera (connected to a Leica DMLB microscope with a 10× water immersion lens) and Fire capture Red 1.2 software. Movie analysis of the heart activity is then carried using the MATLAB R2010a software. Statistical analysis was done using Microsoft Excel. Images of morphology of semidissected fly hearts were taken by screenshots of the videos described above.
SING assay
For the analysis of the adult climbing performance, we used the SING assay. Flies show an innate escape response after being tapped to the bottom of a cylinder (negative geotaxis) and by connecting several tubes that allow simultaneous recording of several genotypes or conditions, the climbing performance can be monitored reproducibly (48). Five to 10 female flies were collected and allowed to recover from CO2 anesthesia for 24 hours. On the day of the experiment, flies were transferred into glass tubes for climbing. After 5 min of conditioning, flies were tapped to the bottom of the tubes and their climbing was recorded. The climbing apparatus was millimetrically marked to measure the climbing of the flies. Mp4 files were converted to JPEG using VideoProc video converter or VirtualDub. The height climbed by each individual fly was measured after 3, 6, and 10 s with ImageJ.
Transmission electron microscopy
Adult female flies were aged to 10 days on standard food. Heart tubes were dissected with the semidissection method described in the “Semi-intact Drosophila heart preparation and digital high-speed movie analysis” section, above. For the imaging of IFM, the thorax of the flies was cut open. Dissection was carried on in artificial hemolymph on ice. The tissue was fixed in TEM fixation buffer [2.0% paraformaldehyde (PFA) and 2.5% glutaraldehyde (both electron microscopy grade) in 0.1 M Na-cacodylate buffer (pH 7.4)] for 4 hours. Fixed specimens were further processed as previously described (55, 56). Briefly, samples were postfixed in 1% osmium tetroxide in phosphate-buffered saline (PBS) for 1 hour, dehydrated in a graded ethanol series up to 100% ethanol, and transferred into mixture of 100% ethanol and propylene oxide (1:1) for 10 min. Afterward, samples were rinsed in mixture of propylene oxide and Epon 812 and lastly embedded in Epon 812. Polymerization was carried out at 60°C for 72 hours. For TEM, ultrathin sections of 70 nm were prepared with a diamond knife on a Leica UC6 microtome. Mounted sections were contrasted with 2% uranyl acetate and lead citrate and lastly investigated with a Zeiss 902 TEM. ER-mitochondria contact sites were analyzed as previously described (57): A blinded investigator measured the length and width of the entire contact region in high-magnification images (50,000×).
X. tropicalis husbandry, morpholino-mediated gene silencing, and mitochondria labeling
Two-cell stage embryos of X. tropicalis were obtained by natural mating using wild-type mature male and female frogs. Embryos were raised in 1/9× Modified Frog Ringer’s (1/9× MR) at 25°C. The stage of the embryos was defined according to Nieuwkoop and Faber (NF) (58). For knockdown of Creld1, a morpholino complementary to blocking the translation start of Creld1 in X. tropicalis was designed (5′-3′ sequence: TGACATACCCATAGCTGAGTGACTC) and tagged with Lissamine by Gene Tools. A morpholino standard control (5′-3′ sequence: CCTCTTACCTCAGTTACAATTTATA) tagged with Lissamine was used as control. Five nanograms of morpholino control or morpholino-Creld1 was injected in one- or two-cell stage embryos of X. tropicalis for hydrogen peroxide and mitochondria fragmentation analysis. For Western blot analysis of knockdown efficiency, injection of morpholinos took place at the one-cell stage, and protein extracts were taken from NF stage 9 embryos that are characterized by a homeostatic peak of Creld1 expression according to Xenbase (www.xenbase.org). To label mitochondria in the somatic muscle of X. tropicalis, two-cell stage embryos (NF stage 2) were injected with 4 nl of mito-eGFP reporter construct (100 ng/μl) and were injected into one blastomere. The embryo was raised in 1/9× MR buffer and kept at 25°C. Screening of eGFP+ injected embryos was performed under an epifluorescence microscope 24 hours after the injection. Half-body fluorescent animals were selected and kept in propylthiouracil buffer [phenylthiourea (0.03 g/liter) in 1/9× MR buffer] at 25°C. Larvae at NF stage 40 were anesthetized and fixed in 4% PFA overnight at 4°C. After PBS washes, the nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI; 1:5000). Mitochondria fragmentation index (MFI) was calculated blind using ImageJ software. Briefly, images were first thresholded and converted to binary images; eGFP+ muscle cells were delimitated, and then, the particle number was quantified. The MFI was calculated by dividing the number of particles by the total area of mitochondria.
Cell culture
HeLa cells were grown in T75cm2 flasks using Dulbecco’s modified Eagle’s medium (Thermo Fisher Scientific) and 1× penicillin-streptomycin (Thermo Fisher Scientific), supplemented with 10% fetal bovine serum, in an atmosphere of 37°C and 5% CO2. HeLa cells were subcultured three times a week at a ratio of 1:3 and 1:5 depending on the confluence. Cell density was determined using a hemocytometer. Cells were detached using 2.5 ml of trypsin-EDTA solution (Thermo Fisher Scientific). Experiments were conducted between passages 5 and 20. Dicer-substrate small interfering RNAs (DsiRNA) were resuspended as described by the manufacturer. On the day of the experiment, 10 nM DsiRNA (combination of 1-hs-CRELD1 and 2-hs-CRELD1) was reverse-transfected into 10,000 HeLa cells per well of an eight-well tissue culture chamber (SARSTEDT) using Lipofectamine RNAiMAX as transfection reagent. Cells used for transfection were grown in media without antibiotics and imaged after 48 hours. Validated Dicer substrate for hypoxanthine phosphoribosyltransferase 1 (HPRT-S1) DsiRNA was used as positive control, and a nontargeting DsiRNA was used as negative control and further used as imaging control. DsiRNA sequences are as follows: 1-hs-CRELD1, 5′-AAAAGGCAUCAGUCUUACACAGGUAGUAAGACUGAU-3′, and 2-hs-CRELD1, 1 5′-GCUGACCAAUUCUGCGUGCAGUGUUCACGCAGAAUU-3′.
Oxygen consumption assay
We used an Agilent Seahorse XFe96 to measure the oxygen consumption rate of larval bodywall tissue. Cartridges were watered and equilibrated according to the manufacturer’s instruction. Plates were coated with poly-l-lysine. Bodywall muscles of third instar larvae were dissected in HL3A buffer (115 mM sucrose, 70 mM NaCl, 20 mM MgCl2, 10 mM NaHCO3, 5 mM KCl, 5 mM Hepes, and 5 mM trehalose) by inside-out preparation and attached to the bottom of the well (n = 6). Tissue was covered with 180 μl of HL3A buffer. Assay was performed at 25°C. For the assay, three measurements without injection (baseline), three with pyruvate and malate (end concentration, 10 and 1 mM, respectively), and three with rotenone (end concentration, 5 μM) were taken.
Quantitative real-time PCR
Whole RNA of five individuals or ~300,000 HeLa cells were isolated using TRIzol reagent (Invitrogen, catalog no. 15596026). Tissue was homogenized using a Precellys 24 homogenizer (PEQLAB). Transcription to cDNA was performed using the Luna Universal One-Step RT-qPCR Kit (New England Biolabs, catalog no. E3005). Quantitative polymerase chain reaction (PCR) was performed with a CFX Connect cycler (Bio-Rad). A minus reverse transcriptase was analyzed in a PCR for each cDNA. Quantitative PCR was performed with a CFX Connect cycler (Bio-Rad) using Luna Universal qPCR Master Mix (New England Biolabs, catalog no. E3003). Values were normalized against two housekeeping genes (actin5c and rp49) and against control (w1118, e.g., female brain to female body, ΔΔCq). Each experiment was repeated at least four times. Primers were actin-5C, 5′-GGCCATCTCCTGCTCAAAGTC-3′ and 5′-GATCTGGCTGGTCGCGATT-3′; rp49, 5′-TCCTACCAGCTTCAAGATGAC-3′ and 5′-CACGTTGTGCACCAGGAACT-3′; Creld, 5′-GGCAGGACTCGAAAGGACAA-3′ and 5′-CAGGTTGTGGCAGTGATCCT-3′; Creld-RA, 5′-GCCACCTGTGTCATCTTCCA-3′ and 5′-TCTATTTCGGGTTGGTGGCC-3′; Creld-RB, 5′-AGTTGCCTGGAGTGTGATCG-3′ and 5′-TCTTCGTGCGTGGACTCTTC-3′; Sod1, 5′-GGTCAACATCACCGACTCCA-3′ and 5′-CGCTTAGACCTTGGCAATGC-3′; Sod2, 5′-ATCACCAGAAGCACCACCAG-3′ and 5′-CTTGTTGGGCGAGAGGTTCT-3′; NOS, 5′-TCGGAGTCGTCCCATTATTC-3′ and 5′-CAGCCAAAGAAGAGCCACA-3′; and catalase, 5′-GATGCGGCTTCCAATCAGTTG-3′ and 5′-GCAGCAGGATAGGTCCTCG-3′. Relative expression of human CRELD1 knockdown was analyzed as described above and normalized to hs–β-actin using nontargeting negative control as baseline. Primers were as follows: hs–β-actin, 5′-GCATCCACGAAACTACCTTCAA-3′ and 5′-TCAGGAGGAGCAATGATCTTGAT-3′; HPRT, 5′-CATTATGCTGAGGATTTGGAAAGG-3′ and 5′-CTTGAGCACACAGAGGGCTACA-3′; and CRELD1, 5′-CTATGAGTGCCGAGACTGTG-3′ and 5′-ATCCACATCGAGACACTTGG-3′.
Western blot
For the detection of p-AMPK, 15 μl of 1× PBS was added and animals were homogenized. Laemmli buffer (1×) was added, and the lysate was boiled at 99°C for 3 min to destroy all proteases. For the detection of Drp1-HA, 1× Laemmli buffer was added to the cytoplasmic and mitochondrial fraction, and the fractions were boiled at 99°C for 3 min. Electrophoresis was performed at 150 V in 1× SDS running buffer. Semidry blotting was performed to transfer the proteins onto a polyvinylidene difluoride membrane. Blotting was done at 100 V for 1.5 hours on a prior in methanol-activated membrane. The membrane was blocked in 5% milk in tris-buffered saline with Tween 20 (TBS-T) buffer (1 M) for 20 min followed by incubation with anti–p-AMPK (1:1000; Cell Signaling Technology), anti-HA (1:400; Roche), or anti-Creld (1:100; Davids Biotechnologie) antibody in 5% milk/TBS-T buffer at 4°C overnight. For immunodetection of the antibody, membrane was incubated with horseradish peroxidase (HRP)–coupled secondary antibody (1:7500; Santa Cruz Biotechnology) in 5% milk/TBS-T buffer in the dark at room temperature for 2 hours. Detection of antibodies was done with SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific) incubated for 1 min and exposed for 30 s. Signal acquisition was performed with Curix 60 (AGFA).
Histology
For semithin sections, heads of Creld and w− adult Drosophila melanogaster were fixed in 4% formaldehyde in 1× PBS overnight at 4°C. Following dehydration of tissue in a graded ethanol series (70, 80, 90, 96, and 100%) for half an hour per step, the tissue was embedded in JB-4 solution (Polysciences Inc.). Embedded tissue was cut into 5-μm sections with an ultramicrotome by a glass knife and transferred to a coated microscope slide (Superfrost Plus, Thermo Fisher Scientific). Furthermore, cut tissue was stained with hematoxylin for 3 min, then washed in ddH2O for 3 min, and stained with eosin for 3 min, followed by washing in ddH2O for 30 s. Sections were mounted in Entellan New (Merck).
Hydrogen peroxide detection
H2O2 was measured in extracts of three adult female flies. Three flies were homogenized in ice-cold PBS and centrifuged for 15 min at 4°C at 12,000g. One milliliter of working reagent contained 970 μl of PBS, 20 μl of HRP, and 10 μl of Amplex Red. Fifty microliters of supernatant of each sample was measured in triplicates. Fluorescence was measured at 544/590 nm excitation/emission using a TECAN plate reader. A 3 μM dilution of H2O2 was used for the standard curve.
Imaging
Antibodies used in this study were α-GFP, α-KDEL (both Santa Cruz Biotechnology), α-DsRed (Takara), α-HA (Invitrogen), and α-TOMM20 (Sigma-Aldrich). α-Creld was generated by immunization with a peptide targeting the conserved WE domain (TAWEEEKLRSYKNSE, Davids Biotechnologie). For immunohistochemistry, brains from adult female flies were dissected in PBS and fixed for 1 hour in 0.5% PBS–Tween 20 and 4% formaldehyde. Tissue was washed with 0.5% PBS–Tween 20 and blocked with donkey serum before incubation with the primary antibody (overnight at 4°C). Tissue was washed in 0.1% PBS–Tween 20 before incubation with the secondary antibody at room temperature in the dark for 1 hour. Secondary antibodies coupled to Alexa or cyanine dyes were from Molecular Probes. Tissue was washed and incubated for 5 min with DAPI. For colocalization analysis of KDEL and mitoOMM-mCherry, adult brains were stained with α-KDEL–mouse and α-DsRed–rabbit. Secondary antibodies were donkey-α-mouse-Alexa488 and donkey-α-rabbit-Cy3. Single confocal sections of dopaminergic neuron PPL1(protocerebral posterior lateral 1), PPL2, and PAL (protocerebral anterior lateral) clusters were analyzed with the ImageJ colocalization threshold tool. For live staining with MitoSOX (Thermo Fisher Scientific), adult gut tissue was used because its large cells are suitable for live imaging of subcellular structures. Guts were dissected in cold PBS and incubated with 5 μM MitoSOX for 10 min. Tissue was washed in PBS, mounted in Fluoromount (Thermo Fisher Scientific), and analyzed immediately. For staining of superoxide with DHE (Sigma-Aldrich), tissue was dissected as described above and incubated for 15 min in 30 μM DHE and fixed for 15 min in 4% formaldehyde. Tissue was washed and incubated with DAPI, mounted in Fluoromount, and analyzed immediately. For imaging, we used a Zeiss LSM 710 with a 25× water lens (Plan-Neofluar, Zeiss), 40× water lens (C-Apochromat, Zeiss), and 63× water lens (Plan-Apochromat, Zeiss) and a Zeiss LSM 880 with Airyscan detector.
Subcellular fractionation
For the analysis of cytoplasmic and mitochondrial protein, the mitochondrial fraction was enriched from extracts of 10 female flies by homogenization in 200 μl of mitochondria isolation buffer [250 mM sucrose, 10 mM tris (pH 7.4), and 0.15 mM MgCl2]. Homogenates were filtered with cotton by centrifugation at 4000g for 15 min, and the cleared homogenate was transferred to a fresh cup. Mitochondria and cytoplasm were fractionated by centrifugation at 16,000g for 30 min at 4°C. The cytoplasmic fraction in the supernatant was transferred to fresh cups, and the mitochondrial pellet was dissolved in 200 μl of mitochondria isolation buffer. For isolation of MAMs, we followed the protocol by Wieckowski et al. (59). Briefly, 200 flies (100 male and 100 female) were homogenized in isolation buffer 1, 10 μl per fly, using a Precellys (PEQLAB) homogenizer. The homogenate was centrifuged twice at 1000g to remove chitin and other bulk material. The cleared homogenate was centrifuged at 7000g for 10 min and contained cytosol, ER, plasma membrane, and lysosomes in the supernatant and crude mitochondria in the pellet. The pellet was resuspended in 200 μl of isolation buffer 2 and pelleted again at 10,000g. The pellet was resuspended in 2 ml of mitochondria resuspension buffer and ultracentrifuged in a Beckman Optima L-60 Ultracentrifuge on Percoll medium to isolate mitochondria from MAMs. Lipidomics analysis of cytoplasmic fraction, crude mitochondria, and MAM was performed. For each isolation step, we included a technical sample containing only buffer. A minimum of five replicates were measured for each fraction. Lipidomics analysis was performed in a Thermo Scientific Q Exactive Plus Hybrid Quadrupole-Orbitrap mass spectrometer as described previously (60). Reads were normalized to protein content of the fraction, and reads from the technical samples were subtracted.
Statistical analysis
Bar charts represent mean and SD. Boxes in box plots represent the interquartile range and median, and whiskers represent minimum and maximum. Green squares in box plots represent single data points. We used Microsoft Excel for bar charts and Origin Pro 8G for box plots. We used the software GraphPad InStat for our statistical analyses. Two-sided Student’s t test was applied for normally distributed data in single comparisons, assuming heteroscedasticity. One-way analysis of variance (ANOVA) with Tukey-Kramer posttest was used for multiple comparisons. The Kolmogorov-Smirnov test was applied to test normality, and Bartlett’s method was used to test for equal SDs within groups. *P < 0.05, **P < 0.01, ***P < 0.001. A minimum of three biological replicates was used for each analysis.
Acknowledgments
We thank A. Schoofs for sharing reagents and for his support with the CaMPARI experiments and X. P. Garmendia for help with the subcellular fractionation. We thank A. Teleman and M. Pankratz for valuable comments on the manuscript. We thank D. Schmucker for supporting the project and all Schmucker Lab members for helpful discussions.
Funding: This work was supported by German Research Foundation BU 3731/1-1 (to M.H.B.); New Frontiers in Research Fund NFRFE-2019-00007 (to M.H.B.); Fonds Wetenschappelijk Onderzoek junior postdoctoral fellow grant 1214420N (to G.E.F.); and German Research Foundation PA517/13-1, PA517/15-1, PA517/16-1, SFB 944-TP7, and SFB 944 Z-Project (to A.P.).
Author contributions: Conceptualization: J.S. and M.H.B. Methodology: M.H.B., J.S., M.P., N.K., C.T., and A.P. Investigation: M.H.B., M.P., N.K., G.E.F., S.J.M.P., B.S., C.M., J.K., and I.J. Writing (original draft): M.H.B. Resources: M.H.B. and R.B. Writing (review and editing): M.H.B. and J.S. Visualization: M.H.B., S.J.M.P., and J.S. Funding acquisition: M.H.B. Supervision: M.H.B., J.S., A.P., C.T., and R.B.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Raw data are deposited on Mendeley data under the title “Creld.” https://data.mendeley.com/datasets/vm4hxk8v6r/1.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
Other Supplementary Material for this manuscript includes the following:
Movies S1 and S2
REFERENCES AND NOTES
- 1.Lapointe J., Hekimi S., When a theory of aging ages badly. Cell. Mol. Life Sci. 67, 1–8 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ohashi M., Hirano T., Watanabe K., Shoji H., Ohashi N., Baba H., Endo N., Kohno T., Hydrogen peroxide modulates neuronal excitability and membrane properties in ventral horn neurons of the rat spinal cord. Neuroscience 331, 206–220 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Scialò F., Sriram A., Fernández-Ayala D., Gubina N., Lõhmus M., Nelson G., Logan A., Cooper H. M., Navas P., Enríquez J. A., Murphy M. P., Sanz A., Mitochondrial ROS produced via reverse electron transport extend animal lifespan. Cell Metab. 23, 725–734 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morán M., Moreno-Lastres D., Marín-Buera L., Arenas J., Martín M. A., Ugalde C., Mitochondrial respiratory chain dysfunction: Implications in neurodegeneration. Free Radic. Biol. Med. 53, 595–609 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Trist B. G., Hare D. J., Double K. L., Oxidative stress in the aging substantia nigra and the etiology of Parkinson’s disease. Aging Cell 18, e13031 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wei Z., Li X., Li X., Liu Q., Cheng Y., Oxidative stress in Parkinson’s Disease: A systematic review and meta-analysis. Front. Mol. Neurosci. 11, 236 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haelterman N. A., Yoon W. H., Sandoval H., Jaiswal M., Shulman J. M., Bellen H. J., A mitocentric view of Parkinson’s disease. Annu. Rev. Neurosci. 37, 137–159 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han H., Tan J., Wang R., Wan H., He Y., Yan X., Guo J., Gao Q., Li J., Shang S., Chen F., Tian R., Liu W., Liao L., Tang B., Zhang Z., PINK1 phosphorylates Drp1 S616 to regulate mitophagy-independent mitochondrial dynamics. EMBO Rep. 21, e48686 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Y., Ouyang Y., Yang L., Beal M. F., McQuibban A., Vogel H., Lu B., Pink1 regulates mitochondrial dynamics through interaction with the fission/fusion machinery. Proc. Natl. Acad. Sci. U.S.A. 105, 7070–7075 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kraus F., Roy K., Pucadyil T. J., Ryan M. T., Function and regulation of the divisome for mitochondrial fission. Nature 590, 57–66 (2021). [DOI] [PubMed] [Google Scholar]
- 11.Basso V., Marchesan E., Peggion C., Chakraborty J., von Stockum S., Giacomello M., Ottolini D., Debattisti V., Caicci F., Tasca E., Pegoraro V., Angelini C., Antonini A., Bertoli A., Brini M., Ziviani E., Regulation of ER-mitochondria contacts by Parkin via Mfn2. Pharmacol. Res. 138, 43–56 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Gómez-Suaga P., Bravo-San Pedro J. M., González-Polo R. A., Fuentes J. M., Niso-Santano M., ER–mitochondria signaling in Parkinson’s disease. Cell Death Dis. 9, 337 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valadas J. S., Esposito G., Vandekerkhove D., Miskiewicz K., Deaulmerie L., Raitano S., Seibler P., Klein C., Verstreken P., ER lipid defects in neuropeptidergic neurons impair sleep patterns in Parkinson’s disease. Neuron 98, 1155–1169.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Wilson E. L., Metzakopian E., ER-mitochondria contact sites in neurodegeneration: Genetic screening approaches to investigate novel disease mechanisms. Cell Death Differ. 28, 1804–1821 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Babot M., Birch A., Labarbuta P., Galkin A., Characterisation of the active/de-active transition of mitochondrial complex I. Biochim. Biophys. Acta 1837, 1083–1092 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keeney P. M., Xie J., Capaldi R. A., Bennett J. P. Jr., Parkinson’s disease brain mitochondrial complex I has oxidatively damaged subunits and is functionally impaired and misassembled. J. Neurosci. 26, 5256–5264 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pouchieu C., Piel C., Carles C., Gruber A., Helmer C., Tual S., Marcotullio E., Lebailly P., Baldi I., Pesticide use in agriculture and Parkinson’s disease in the AGRICAN cohort study. Int. J. Epidemiol. 47, 299–310 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Morais V. A., Haddad D., Craessaerts K., De Bock P.-J., Swerts J., Vilain S., Aerts L., Overbergh L., Grünewald A., Seibler P., Klein C., Gevaert K., Verstreken P., De Strooper B., PINK1 loss-of-function mutations affect mitochondrial complex I activity via NdufA10 ubiquinone uncoupling. Science 344, 203–207 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Sharpley M. S., Shannon R. J., Draghi F., Hirst J., Interactions between phospholipids and NADH: Ubiquinone oxidoreductase (Complex I) from bovine mitochondria. Biochemistry 45, 241–248 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Ramalingam M., Huh Y. J., Lee Y. I., The impairments of α-synuclein and mechanistic target of rapamycin in rotenone-induced SH-SY5Y cells and mice model of Parkinson’s disease. Front. Neurosci. 13, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahul-Mellier A.-L., Burtscher J., Maharjan N., Weerens L., Croisier M., Kuttler F., Leleu M., Knott G. W., Lashuel H. A., The process of Lewy body formation, rather than simply α-synuclein fibrillization, is one of the major drivers of neurodegeneration. Proc. Natl. Acad. Sci. U.S.A. 117, 4971–4982 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shahmoradian S. H., Lewis A. J., Genoud C., Hench J., Moors T. E., Navarro P. P., Castaño-Díez D., Schweighauser G., Graff-Meyer A., Goldie K. N., Sütterlin R., Huisman E., Ingrassia A., de Gier Y., Rozemuller A. J. M., Wang J., De Paepe A., Erny J., Staempfli A., Hoernschemeyer J., Großerüschkamp F., Niedieker D., El-Mashtoly S. F., Quadri M., Van IJcken W. F. J., Bonifati V., Gerwert K., Bohrmann B., Frank S., Britschgi M., Stahlberg H., Van de Berg W. D. J., Lauer M. E., Lewy pathology in Parkinson’s disease consists of crowded organelles and lipid membranes. Nat. Neurosci. 22, 1099–1109 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Oh-hashi K., Koga H., Ikeda S., Shimada K., Hirata Y., Kiuchi K., CRELD2 is a novel endoplasmic reticulum stress-inducible gene. Biochem. Biophys. Res. Commun. 387, 504–510 (2009). [DOI] [PubMed] [Google Scholar]
- 24.Kern P., Balzer N. R., Blank N., Cygon C., Wunderling K., Bender F., Frolov A., Sowa J. P., Bonaguro L., Ulas T., Homrich M., Kiermaier E., Thiele C., Schultze J. L., Canbay A., Bauer R., Mass E., Creld2 function during unfolded protein response is essential for liver metabolism homeostasis. FASEB J. 35, e21939 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beckert V., Rassmann S., Kayvanjoo A. H., Klausen C., Bonaguro L., Botermann D. S., Krause M., Moreth K., Spielmann N., da Silva-Buttkus P., Fuchs H., Gailus-Durner V., de Angelis M. H., Händler K., Ulas T., Aschenbrenner A. C., Mass E., Wachten D., Creld1 regulates myocardial development and function. J. Mol. Cell. Cardiol. 156, 45–56 (2021). [DOI] [PubMed] [Google Scholar]
- 26.Bonaguro L., Köhne M., Schmidleithner L., Schulte-Schrepping J., Warnat-Herresthal S., Horne A., Kern P., Günther P., ter Horst R., Jaeger M., Rahmouni S., Georges M., Falk C. S., Li Y., Mass E., Beyer M., Joosten L. A. B., Netea M. G., Ulas T., Schultze J. L., Aschenbrenner A. C., CRELD1 modulates homeostasis of the immune system in mice and humans. Nat. Immunol. 21, 1517–1527 (2020). [DOI] [PubMed] [Google Scholar]
- 27.D’Alessandro M., Richard M., Stigloher C., Gache V., Boulin T., Richmond J. E., Bessereau J.-L., CRELD1 is an evolutionarily-conserved maturational enhancer of ionotropic acetylcholine receptors. eLife 7, e39649 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dumitriu A., Golji J., Labadorf A. T., Gao B., Beach T. G., Myers R. H., Longo K. A., Latourelle J. C., Integrative analyses of proteomics and RNA transcriptomics implicate mitochondrial processes, protein folding pathways and GWAS loci in Parkinson disease. BMC Med. Genomics 9, 5 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rupp P. A., Fouad G. T., Egelston C. A., Reifsteck C. A., Olson S. B., Knosp W. M., Glanville R. W., Thornburg K. L., Robinson S. W., Maslen C. L., Identification, genomic organization and mRNA expression of CRELD1, the founding member of a unique family of matricellular proteins. Gene 293, 47–57 (2002). [DOI] [PubMed] [Google Scholar]
- 30.Asim A., Agarwal S., Panigrahi I., Sarangi A. N., Muthuswamy S., Kapoor A., CRELD1 gene variants and atrioventricular septal defects in Down syndrome. Gene 641, 180–185 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Mass E., Wachten D., Aschenbrenner A. C., Voelzmann A., Hoch M., Murine creld1 controls cardiac development through activation of calcineurin/NFATc1 signaling. Dev. Cell 28, 711–726 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Rotstein B., Paululat A., On the morphology of the drosophila heart. J. Cardiovasc. Dev. Dis. 3, 15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kass D. A., Bronzwaer J. G. F., Paulus W. J., What mechanisms underlie diastolic dysfunction in heart failure? Circ. Res. 94, 1533–1542 (2004). [DOI] [PubMed] [Google Scholar]
- 34.De Miranda B. R., Rocha E. M., Castro S. L., Greenamyre J. T., Protection from α-Synuclein induced dopaminergic neurodegeneration by overexpression of the mitochondrial import receptor TOM20. npj Parkinsons Dis. 6, 38 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Collins T. J., Berridge M. J., Lipp P., Bootman M. D., Mitochondria are morphologically and functionally heterogeneous within cells. EMBO J. 21, 1616–1627 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu R., Liu T., Ning C., Tan F., Jin S.-B., Lendahl U., Zhao J., Nistér M., The phosphorylation status of Ser-637 in dynamin-related protein 1 (Drp1) does not determine Drp1 recruitment to mitochondria. J. Biol. Chem. 294, 17262–17277 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sellin J., Albrecht S., Kölsch V., Paululat A., Dynamics of heart differentiation, visualized utilizing heart enhancer elements of the Drosophila melanogaster bHLH transcription factor Hand. Gene Expr. Patterns 6, 360–375 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Luo J., Becnel J., Nichols C. D., Nässel D. R., Insulin-producing cells in the brain of adult Drosophila are regulated by the serotonin 5-HT1A receptor. Cell. Mol. Life Sci. 69, 471–484 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanyal S., Genomic mapping and expression patterns of C380, OK6 and D42 enhancer trap lines in the larval nervous system of Drosophila. Gene Expr. Patterns 9, 371–380 (2009). [DOI] [PubMed] [Google Scholar]
- 40.Niens J., Reh F., Çoban B., Cichewicz K., Eckardt J., Liu Y.-T., Hirsh J., Riemensperger T. D., Dopamine modulates serotonin innervation in the drosophila brain. Front. Syst. Neurosci. 11, 76 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fuenzalida-Uribe N., Campusano J. M., Unveiling the dual role of the dopaminergic system on locomotion and the innate value for an aversive olfactory stimulus in drosophila. Neuroscience 371, 433–444 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Ryczko D., Dubuc R., Dopamine and the brainstem locomotor networks: From lamprey to human. Front. Neurosci. 11, 295 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun J., Xu A. Q., Giraud J., Poppinga H., Riemensperger T., Fiala A., Birman S., Neural control of startle-induced locomotion by the mushroom bodies and associated neurons in drosophila. Front. Syst. Neurosci. 12, 6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moeyaert B., Holt G., Madangopal R., Perez-Alvarez A., Fearey B. C., Trojanowski N. F., Ledderose J., Zolnik T. A., Das A., Patel D., Brown T. A., Sachdev R. N. S., Eickholt B. J., Larkum M. E., Turrigiano G. G., Dana H., Gee C. E., Oertner T. G., Hope B. T., Schreiter E. R., Improved methods for marking active neuron populations. Nat. Commun. 9, 4440 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oswald M. C., Brooks P. S., Zwart M. F., Mukherjee A., West R. J. H., Giachello C. N. G., Morarach K., Baines R. A., Sweeney S. T., Landgraf M., Reactive oxygen species regulate activity-dependent neuronal plasticity in Drosophila. eLife 7, e39393 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pogson J. H., Ivatt R. M., Sanchez-Martinez A., Tufi R., Wilson E., Mortiboys H., Whitworth A. J., The complex I subunit NDUFA10 selectively rescues Drosophila pink1 mutants through a mechanism independent of mitophagy. PLOS Genet. 10, e1004815 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Flis V. V., Daum G., Lipid transport between the endoplasmic reticulum and mitochondria. Cold Spring Harb. Perspect. Biol. 5, a013235 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gargano J. W., Martin I., Bhandari P., Grotewiel M. S., Rapid iterative negative geotaxis (RING): A new method for assessing age-related locomotor decline in Drosophila. Exp. Gerontol. 40, 386–395 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Rodríguez-Arribas M., Yakhine-Diop S. M. S., Pedro J. M. B. S., Gómez-Suaga P., Gómez-Sánchez R., Martínez-Chacón G., Fuentes J. M., González-Polo R. A., Niso-Santano M., Mitochondria-associated membranes (MAMs): Overview and its role in Parkinson’s disease. Mol. Neurobiol. 54, 6287–6303 (2017). [DOI] [PubMed] [Google Scholar]
- 50.Hewitt V. L., Miller-Fleming L., Andreazza S., Mattedi F., Prudent J., Polleux F., Vagnoni A., Whitworth A. J., Decreasing pdzd8-mediated mitochondrial-ER contacts in neurons improves fitness by increasing mitophagy. bioRxiv 2020.11.14.382861 (2020). [Google Scholar]
- 51.Perez-Gomez R., Magnin V., Mihajlovic Z., Slaninova V., Krejci A., Downregulation of respiratory complex I mediates major signalling changes triggered by TOR activation. Sci. Rep. 10, 4401 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Greene J. C., Whitworth A. J., Kuo I., Andrews L. A., Feany M. B., Pallanck L. J., Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc. Natl. Acad. Sci. U.S.A. 100, 4078–4083 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang J., Zhou W., Dong W., Watson A. M., Hong Y., Directed, efficient, and versatile modifications of the Drosophila genome by genomic engineering. Proc. Natl. Acad. Sci. U.S.A. 106, 8284–8289 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Piper M. D. W., Partridge L., Protocols to study aging in drosophila. Methods Mol. Biol. 291–302 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dehnen L., Janz M., Verma J. K., Psathaki O. E., Langemeyer L., Fröhlich F., Heinisch J. J., Meyer H., Ungermann C., Paululat A., A trimeric metazoan Rab7 GEF complex is crucial for endocytosis and scavenger function. J. Cell Sci. 133, jcs247080 (2020). [DOI] [PubMed] [Google Scholar]
- 56.Lehmacher C., Abeln B., Paululat A., The ultrastructure of Drosophila heart cells. Arthropod Struct. Dev. 41, 459–474 (2012). [DOI] [PubMed] [Google Scholar]
- 57.Dietrich M. O., Liu Z.-W., Horvath T. L., Mitochondrial dynamics controlled by mitofusins regulate Agrp neuronal activity and diet-induced obesity. Cell 155, 188–199 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.J. Gerhart, M. Kirschner, Normal Table of Xenopus Laevis (Daudin), P. D. Nieuwkoop, J. Faber, Eds. (Garland Science, 2020). [Google Scholar]
- 59.Wieckowski M. R., Giorgi C., Lebiedzinska M., Duszynski J., Pinton P., Isolation of mitochondria-associated membranes and mitochondria from animal tissues and cells. Nat. Protoc. 4, 1582–1590 (2009). [DOI] [PubMed] [Google Scholar]
- 60.Thiele C., Wunderling K., Leyendecker P., Multiplexed and single cell tracing of lipid metabolism. Nat. Methods 16, 1123–1130 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Movies S1 and S2