Abstract
Collective migration is important to embryonic development and cancer metastasis, but migratory and nonmigratory cell fate discrimination by differential activity of signal pathways remains elusive. In Drosophila oogenesis, Jak/Stat signaling patterns the epithelial cell fates in early egg chambers but later renders motility to clustered border cells. How Jak/Stat signal spatiotemporally switches static epithelia to motile cells is largely unknown. We report that a nuclear protein, Dysfusion, resides on the inner nuclear membrane and interacts with importin α/β and Nup153 to modulate Jak/Stat signal by attenuating Stat nuclear import. Dysfusion is ubiquitously expressed in oogenesis but specifically down-regulated in border cells when migrating. Increase of nuclear Stat by Dysfusion down-regulation triggers invasive cell behavior and maintains persistent motility. Mammalian homolog of Dysfusion (NPAS4) also negatively regulates the nuclear accumulation of STAT3 and cancer cell migration. Thus, our finding demonstrates that Dysfusion-dependent gating mechanism is conserved and may serve as a therapeutic target for Stat-mediated cancer metastasis.
Drosophila Npas4 regulates Jak/Stat in collective invasion.
INTRODUCTION
Tissue morphogenesis during animal development and homeostatic wound healing relies on collective cell migration. Impaired cell migration causes devastating pathologies such as chronic wounds, birth defects, and immune deficiencies (1). Notably, cancer metastasis, whereby tumor cells invade neighboring tissues and migrate to distal organs, is a leading cause of patient death (2). However, it remains unclear how the population size of migratory cells is restricted by signaling pathways and how extracellular signals are ranked and integrated at the nuclear structure by gating the nuclear import of transcriptional factors, which is crucial to not only morphogenesis in animal development but also collective cancer metastasis. An increasing number of studies indicate that nucleocytoplasmic components or nuclear pore complexes (NPCs) are not just structural proteins gating macromolecules transport by size; some NPC subunits or karyopherin family proteins have different stoichiometry at the pore or differential expression during development (3, 4), which acts as a scaffold to recruit transcription factors for neuroprogenitor differentiation or for muscle development (5). Defects in nuclear transport not only impairs development, gametogenesis, or immune deficit due to insufficient nuclear accumulation of Smad, Ci, β-catenin, Sox9, NF-κB (nuclear factor κB), etc., but also affects the function and survival of motor neurons because of pathological aggregate of the nuclear RNA binding protein, TDP-43, in the cytoplasm (4, 6, 7). Those lines of evidence suggest that multilevel signaling may be controlled by timely delivery of nuclear protein into the nucleus, yet the molecular mechanism remains elusive.
The Janus kinase (Jak)/signal transducer and activator of transcription (Stat) pathway is a key signal perpetuated via the level of nuclear accumulation of phosphorylated Stat (p-Stat) (8–10). Upon ligand binding, the Jak/Stat receptor induces Jaks to auto-phosphorylate and trans-phosphorylate at select tyrosine residues of the receptor, which creates Stat docking sites. The Stats can then be phosphorylated by Jak, enabling dimerization with the importin α/β complex for nuclear entry (8, 11). During passage through the NPC, Stat is unloaded from the importin α/β complex upon binding to RanGTP, a small guanosine triphosphatase (GTPase) that regulates nuclear transport via guanosine triphosphate (GTP)/guanosine diphosphate (GDP) cycling (11, 12). Given the diverse functions of Jak/Stat signaling, insufficient or excessive activity can induce severe defects in animal development or tumorigenesis (10, 13–15). How different steps of Jak/Stat signaling are regulated has been investigated previously (16). PIAS (protein inhibitor of activated Stat) and Socs (suppressor of cytokine signaling) are conserved from Drosophila to mammals and negatively regulate Stat and Jak to maintain balanced signaling activity (17–23). Apart from phosphorylation of Stat resulting in its nuclear transport, the role of the NPC in regulating differential nuclear accumulation of Stat to level Jak/Stat activity remains elusive. It is unlikely that Stat nuclear entry is controlled by the importin α/β complex alone for differential activation of downstream genes, which is an essential process confining migratory and nonmigratory cells in epithelial, stem, and differentiated daughter cells (10, 16, 24–26). Furthermore, although tyrosine phosphorylation is a conserved mechanism for nuclear accumulation of Stats among vertebrates, nucleocytoplasmic shuttling varies among Stat family members (11). Moreover, although Jak/Stat-dependent cancer progression has been reported for ~70% of hematological and solid tumors (27), attempts to develop drugs that target Stat without severe side effects have been unsuccessful. Thus, uncovering the molecular mechanism by which nucleocytoplasmic Stat shuttling is regulated may be crucial to designing intervention strategies for Jak/Stat-mediated cancer metastasis.
In this study, we used border cells derived from Drosophila follicular epithelia of egg chambers as a model to investigate how Stat shuttling is regulated at the nuclear membrane, thereby controlling the size of migratory cell clusters. Border cells migrate as a group, with two polar cells being hauled by six to eight motile cells. Cell number in the cluster is determined by the Jak/Stat activity triggered by Upd (Unpaired), which is the Drosophila homolog of interleukin-6 (IL-6) and is secreted from the polar cells (24). Once Upd binds to its receptor, Domeless, graded Jak/Stat activity patterns the anterior-posterior axis of egg chamber development and instructs follicular epithelia cell fates (28–30). At stage 9 of oogenesis, Jak/Stat activity renders only six to eight follicular cells motile, forming a border cell cluster. Defects in Jak/Stat components hamper recruitment of migratory cohorts, whereas Jak/Stat hyperactivity leads to addition of extra invasive border cells (24). Apart from governing migratory cell fate, Jak/Sat activity is also required for maintaining persistent migration, because temporary blockage of the signaling impedes border cell movement by shifting statts, a temperature-sensitive allele of stat, to nonpermissive temperature for 30 min (25). Global analyses for border cell migration further reveal that slow border (slbo), the downstream target of Jak/Stat, spatially increases mRNA levels of several cytoskeleton-associated proteins by 2- to 18-fold in border cells (31, 32). Collectively, acquisition of motility for border cells demands a higher level of Jak/Stat signaling. Because the induction level of Jak/Stat signaling is the pivotal step in inducing border cell motility, spatiotemporal control of Jak/Stat to dampen Stat activity in nonmigratory follicle cells becomes critical (33, 34). In that regard, most previous studies have focused on Jak/Stat phosphorylation, transcriptional control of Stat, or Stat RNA/protein stability (10, 33, 34). However, how nuclear transport of Stat is modulated has not been explored in detail, although it could be vital to how Jak/Stat signaling is balanced during tissue morphogenesis and in cancer cell migration (16).
Here, we demonstrate that a basic helix-loop-helix (bHLH) nuclear protein [Dysfusion (Dysf)] acts as a negative regulator of Stat nuclear import in border cell migration. Overexpression of Dysf impairs border cell recruitment and persistent migration. Consistently, dysf loss-of-function mutation ectopically activated Jak/Stat signaling, leading to extra migrating border cells. Dysf is ubiquitously expressed on the nuclear membrane of Drosophila follicle cells, but its expression progressively declines in border cells and becomes undetectable as they reach the oocyte. In a biochemical screen, we identified Drosophila importin α2, Pendulin (Pen), and the importin β family member karyopherin β3 (Karyβ3) in the pull-down complex. Mechanically, Dysf binds to the importin β binding (IBB) domain of Pen, which blocks its interaction with Karyβ3 and impairs Stat nuclear import. A forward genetic screen further identified Nup153 genetically interacting with Dysf to regulate border cell migration. Evolutionarily, we show that the mammalian homolog of Dysf, neuronal PAS domain protein 4 (Npas4) (35), plays a similar role in Jak/Stat-mediated cancer cell migration. Our work provides insights into how Dysf spatiotemporally regulates Jak/Stat signaling by acting as a gatekeeper at the inner nuclear membrane to modulate nuclear translocation of Stat, explaining the long-standing mystery of why Jak/Stat signaling displays surge-like behavior during border cell migration. Moreover, our discovery of a novel role for Npas4 in suppressing Jak/Stat signaling–mediated cancer cell migration may represent a new treatment strategy for targeting Stat-mediated metastases.
RESULTS
Identification of Dysf as a suppressor of Jak/Stat signaling
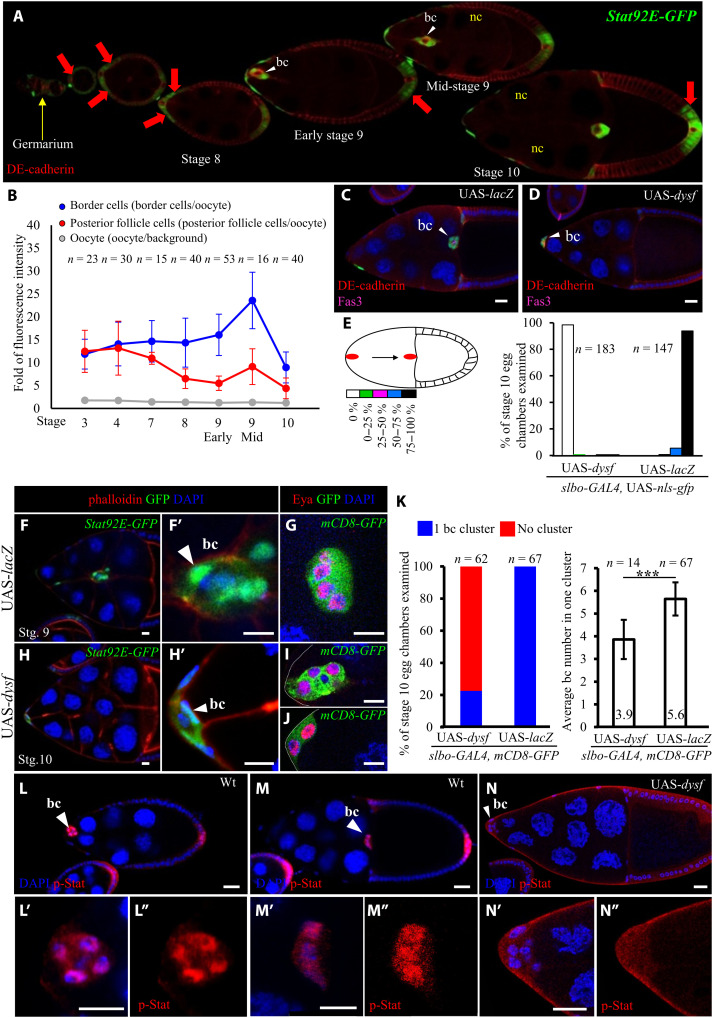
Jak/Stat signaling in early oogenesis is required for stalk cell formation. Germ line–derived Delta signaling induces polar cell formation, and the Jak/Stat ligand Upd is secreted from polar cells to promote stalk cell fate (fig. S1A) (29, 30). However, border cells are not recruited until stage 9 of oogenesis, although Jak overexpression in early oogenesis cannot induce precocious cluster formation. The only way to induce precocious migration of border cells is coactivating Jak/Stat and ecdysone signaling, the latter of which determines the timing of border cell detachment (36). Thus, we hypothesized that there might be two levels of Jak/Stat signaling, by stage 9, with a lower level harnessing follicle cell differentiation and a higher one recruiting border cells to initiate migration upon ecdysone signaling activation. To test that hypothesis, we assayed fluorescence signal of Stat92E–green fluorescent protein (GFP) to represent Jak/Stat activity during Drosophila oogenesis (37). The GFP reporter was expressed in the follicular precursors of germaria, with expression increasing gradually as the egg chambers developed, before ultimately being restricted to the anterior/posterior follicles and border cells (Fig. 1A), which is consistent with previous studies (28, 30, 38). Quantitative analysis revealed a significant increase in Stat92E-GFP expression after stage 8, with maximal levels in the migrating border cells. A similar pattern was observed during stages 9 to 10 in posterior follicle cells, but the expression levels were lower than that of border cells (Fig. 1B). Consequently, we hypothesize a temporal switch requiring a suppressor or activator to regulate Jak/Stat signaling activity in border cells. In a forward genetic screen, Dysf overexpression via the GAL4/UAS system (39, 40) impeded border cell movement, with 99% of border cells in 183 stage 10 egg chambers not migrating (Fig. 1, D and E). Notably, nearly 98% of those cells did not even detach from the anterior epithelium (Fig. 1D), unlike for control egg chambers (Fig. 1, C and E). Dysf overexpression also greatly reduced the number of cells expressing Stat92E-GFP in the anterior terminals of egg chambers (Fig. 1, F, F′, H, and H′). After carefully scrutinizing border cells marked with slbo-GAL4–driven UAS-mCD8-gfp and Eyes absent (Eya) staining (41), we found that only 22.58% of egg chambers contained a shrunk cluster with a decrease in border cell number from 5.6 to 3.9, and the rest (77.42%) failed to form a cluster (Fig. 1, G to K). To explore how Dysf affects Jak/Stat signaling, we randomly induced UAS-dysf by actin-GAL4 in individual border cells by means of a Flip-Out technique (42) to analyze its effect on Stat92E-GFP signal. Overexpressing Dysf in single border cells resulted in a 30% reduction in Stat92E-GFP signal (fig. S1, B to H) and delayed group cell migration (fig. S1I). We further assessed the effect of dysf on individual cell motility by quantifying clone position in the migrating cluster. Similar to the previous report, the control clones expressing UAS-lacZ randomly rotated within the cluster during migration (fig. S1J) (43). However, 88.2% of dysf-expressing cells were found to stay in the trailing part (fig. S1, B to D and J), and some of them even lagged behind the main cluster (fig. S1, E to G). These results indicate that up-regulation of dysf suppresses Jak/Stat-dependent motility and specification in border cells.
Fig. 1. Dysf represses Jak/Stat signaling during Drosophila oogenesis.
Confocal micrographs of Drosophila egg chambers showing border cells (bc; arrowheads) migrating through nurse cells (nc). Anti–DE-cadherin staining [red in (A), (C), and (D)] labels cell margins, and DAPI (4′,6-diamidino-2-phenylindole; blue) marks nuclei. All UAS transgenes were driven by slbo-GAL4. (A) The Stat92E-GFP reporter (green) reveals Stat activity during Drosophila oogenesis from the germarium (yellow arrow) to egg chambers (red arrow). (B) Fluorescence intensity of Stat92E-GFP in Drosophila oogenesis. (C and D) Dysf overexpression impairs border cell migration. Fasciclin 3 (Fas3; magenta) staining labels polar cells. (E) Quantification of migration defect caused by dysf overexpression. The migration path is divided into five sections to quantify the extent of border cell migration. (F and H) Dysf overexpression reduced border cell number in the cluster. Stat92E-GFP signal marks border cells (white arrowheads), and phalloidin staining (red) labels cell margins. (G, I, and J) slbo-GAL4>UAS-mCD8gfp marks the cluster morphology, and anti-Eya labels anterior follicle cells. (K) Quantification of Dysf effect on border cells. (L and M) Anti–p-Stat staining (red) displays nuclear accumulation in border cells. Wt, wild type. (N) Dysf overexpression reduced p-Stat accumulation in border cell nuclei. n represents the number of egg chambers examined; ***P < 0.001, two-tailed t test. Error bars indicate SD. Scale bars, 20 μm.
To trigger signal activation, Stat proteins must first be phosphorylated by Jak to undergo dimerization and then translocate into the nucleus to bind the consensus DNA binding site (44, 45). Therefore, we examined the phosphorylation status of Stat in border cells by means of anti–p-Stat staining (Fig. 1, L to N). Consistent with the expression pattern of Stat92E-GFP, p-Stat staining is increased in border cell nuclei during migration (Fig. 1, L and M, white arrowheads, and fig. S2, A to C). The fluorescence intensity of p-Stat staining remains stable until stage 7 and then gradually increases and reaches the peak at early stage 9 (from 2.6- to 39-fold; fig. S2E, red plot). The nucleus/cytoplasm (N/C) ratio of the p-Stat signal starts to rise at stage 5 and keeps at a similar level (1.65-fold) from stages 6B to 10 (fig. S2E, blue plot). However, overexpressing Dysf in border cells reduced the nuclear staining of p-Stat (Fig. 1N and fig. S2D) with a decline in the fluorescence intensity (fig. S2F) and the N/C ratio (fig. S2G). To gain insights into the link between nuclear accumulation of p-Stat and cell motility, we analyzed the N/C ratio of p-Stat in Jak/Stat hyperactivation that can induce extra border cells (24). We ectopically expressed hopscotch (hop; the Drosophila homolog of Jak) and found that the N/C ratio of p-Stat was 1.44 in extra border cells, 1.56 in normal border cells, the front cluster that can reach oocyte, and 1.49 in wild-type border cells (fig. S2H, I and L). This result demonstrates the small fluctuation of p-Stat N/C ratio in Jak/Stat signaling–dependent migration.
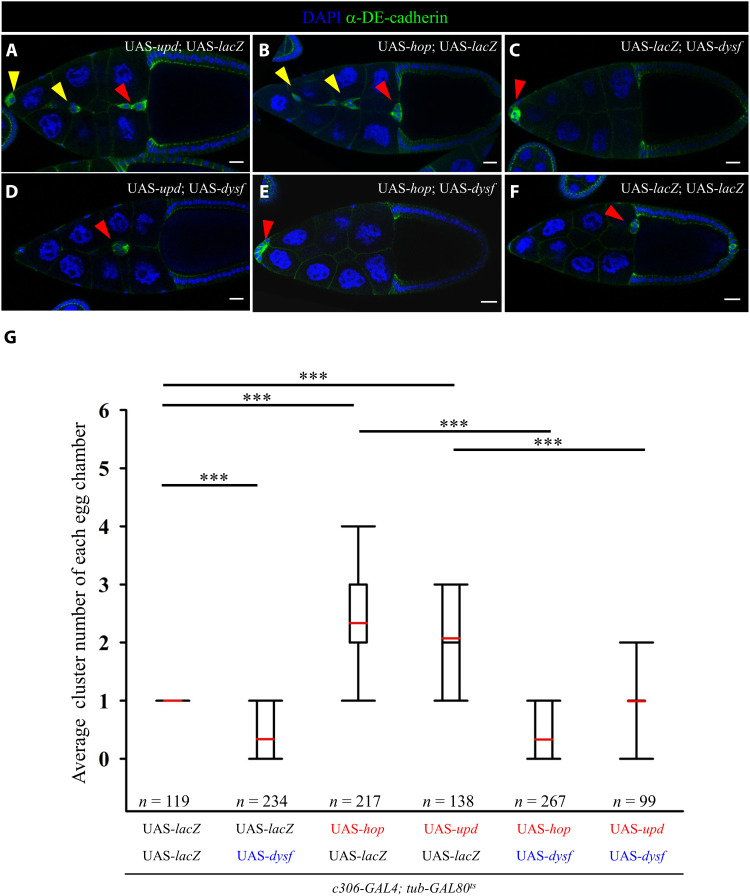
Dysf is a transcription factor that dimerizes with Tango (Tgo; a bHLH nuclear protein) to control gene expression for tracheal fusion in Drosophila embryogenesis, and it acts downstream of Notch signaling to regulate leg development (46–48). Accordingly, we wanted to ascertain whether Dysf functions as a repressor of Jak/Stat signaling by suppressing stat transcription. We determined expression of the enhancer trap line stat06346 (stat-lacZ; fig. S3, A to D), acting as a transcriptional reporter of a subset of stat expression during oogenesis and eye development (24, 49). There was no significant difference in stat-lacZ signal between wild-type and Dysf-overexpressing border cells (fig. S3E), indicating that Dysf does not transcriptionally control stat expression. To further probe epistasis between Dysf and Stat, we assessed whether Dysf overexpression suppresses the multi-cluster phenotype caused by Jak/Stat hyperactivity. As documented previously (24), up-regulation of upd or hop resulted in two to four border cell clusters within egg chambers, counted with the morphology of migratory cells stained with DAPI (4′,6-diamidino-2-phenylindole) and anti–DE-cadherin (Fig. 2, A, B, F, and G). Coexpression of upd or hop with dysf restored the cluster number to one and impeded border cell movement (Fig. 2, C to E and G). Notably, the impact of dysf almost outweighed the function of hop in terms of cluster formation and cell migration, suggesting that Dysf works downstream or in parallel with Jak. Thus, we conclude that Dysf suppresses extra border cell specification before high STAT signaling.
Fig. 2. Dysf suppresses the hyperactive Jak/Stat phenotype in border cells.
(A to F) Confocal micrographs showing border cell clusters (red arrowheads) in stage 10 egg chambers of the indicated genotypes. Anti–DE-cadherin staining (green) and DAPI (blue) mark cell margins and nuclei, respectively. Overexpression of upd or hop induces extra border cell clusters (A and B, yellow arrowheads). Egg chambers coexpressing dysf with upd or hop have one border cell cluster displaying migration delay (D and E), similar to the phenotype arising from dysf expression (C). Double UAS-lacZ serves as the control (F). (G) Quantitative assessment of the number of migrating clusters with indicated genotypes. n represents the total number of egg chambers examined; ***P < 0.001, two-tailed t test. The box plot shows the medians (black lines), means (red lines), the 25th and 75th range (boxes), and the 5th and 95th percentiles (whiskers). Scale bars, 20 μm.
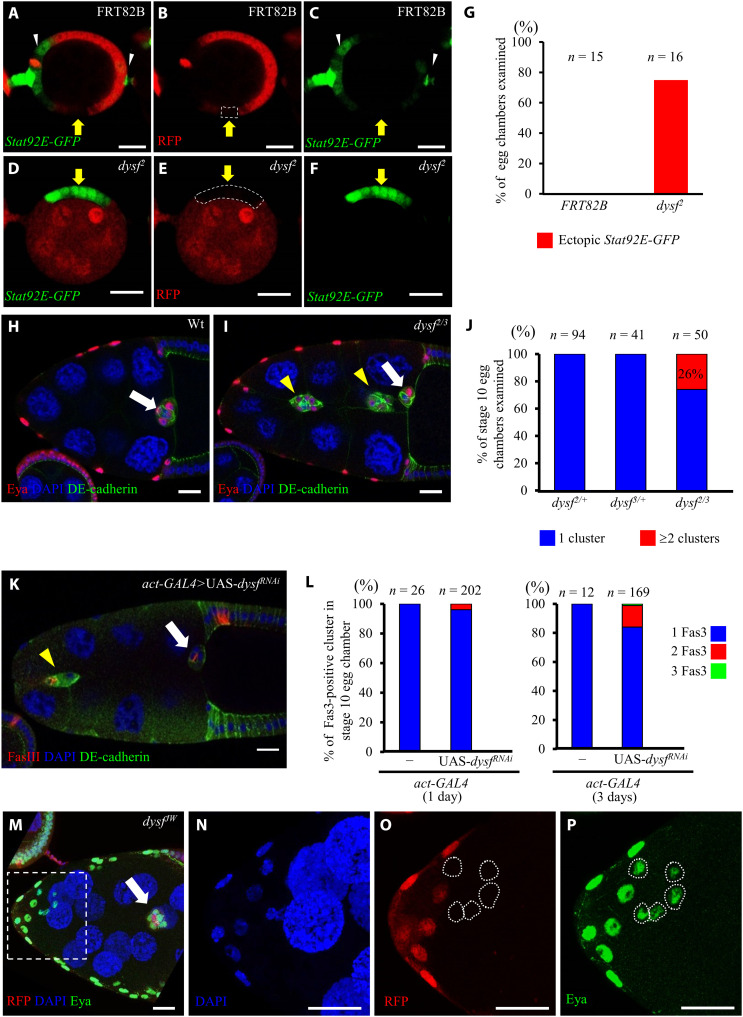
To further elucidate the role of Dysf in restricting Jak/Stat signaling, we tested whether dysf loss-of-function mutation resulted in ectopic Stat92E-GFP expression or additional border cells. Because of lethality, the regular treatment for clone induction led to no dysf2-homozygous mutant cells reaching the stage 9 of oogenesis to form border cells. We found that 75% of early-stage egg chambers carrying dysf2 mutant clones displayed ectopic Stat92E-GFP expression (Fig. 3, D to F, yellow arrows, and fig. S4A), which was not observed in the control FRT82B line (Fig. 3, A to C, yellow arrows, and G). However, this phenotype showed incomplete penetrance, meaning that not all dysf2 mutant clones were able to express ectopic Stat92E-GFP (Fig. 3G and fig. S4B, white arrowheads). In addition, some dysf2 clones showed follicle epithelial defects in earlier stages, including condensed DAPI staining (fig. S4C) and gaps between twin spots (fig. S4D), but the germline mutation did not delay border cell movement or cause any apparent defect (fig. S4E). Homozygous cells of dysfJW, a null allele generated in-house (fig. S5A), also encountered lethality issue. Therefore, we analyzed egg chambers dissected from dysf2/3 trans-heterozygotes and found that 26% of them had additional migrating clusters (Fig. 3, I and J), which was not the case for wild-type, dysf2/+, or dysf3/+ lines (Fig. 3, H and J) and resembled the phenotypes resulting from Jak/Stat hyperactivation. Similarly, RNA interference (RNAi)–mediated knockdown of dysf also phenocopied the excess in border cell clusters (Fig. 3K), with penetrance increasing from 4 to 16% in response to a longer expression time of dysfRNAi (Fig. 3L). The N/C ratio of p-Stat staining of ectopic border cells induced by dysf knockdown (1.48) was close to that in the wild-type control (1.51; fig. S2, J, K, and M). To distinguish whether the extra border cells were autonomously induced by dysf down-regulation or recruited by excessive polar cells, 1- to 2-day clones were generated to avoid lethality and ectopic polar cell formation. Under this condition, we retrieved very few egg chambers containing multiple dysfJW clones that invaded and migrated through the nurse cell cluster (Fig. 3, M to P). Together, we conclude that Dysf is a negative regulator constraining Jak/Stat signaling to control border cell recruitment, with loss of Dysf elevating Jak/Stat activity and inducing extra migrating cells.
Fig. 3. Down-regulation of dysf results in recruitment of extra border cells.
(A to C) Immunofluorescence micrographs showing Stat92E-GFP (green) expression patterns in egg chambers. White arrowheads indicate egg chamber terminals. (D to F) The dysf2 mutant clone [red fluorescence protein (RFP)–negative; dotted circle] presents ectopic Stat92E-GFP signal (yellow arrows) relative to control clones [yellow arrows in (A) to (C)]. (G) Quantification of ectopic Stat92E-GFP in indicated genotypes. (H and I) dysf2/3 displays multiple border cell clusters (yellow arrowheads). (J) Quantification of extra border cells in dysf2/3. (K) Knocking down dysf induced extra border cell clusters (yellow arrowheads). Anti-FasIII labels polar cells. (L) Quantification of ectopic cluster formation by dysf RNAi knockdown. Colors indicate the border cell cluster number. (M to P) Three-dimensional projection exhibits freely migrating border cells (white arrow) under dysfJW mutation (RFP-negative; white outline). All white arrows indicate border cells reaching the oocyte border. Enlargements of the boxed region are shown in (N) to (P). Anti-Eya stains follicle cells and border cells [red in (H) and (I); green in (M) and (P)]. DE-cadherin staining (green) labels cell margins (H, I, and K). DAPI (blue) marks all nuclei. Scale bars, 20 μm.
Dysf is located on the inner nuclear membrane where it gates nuclear entry of Stat
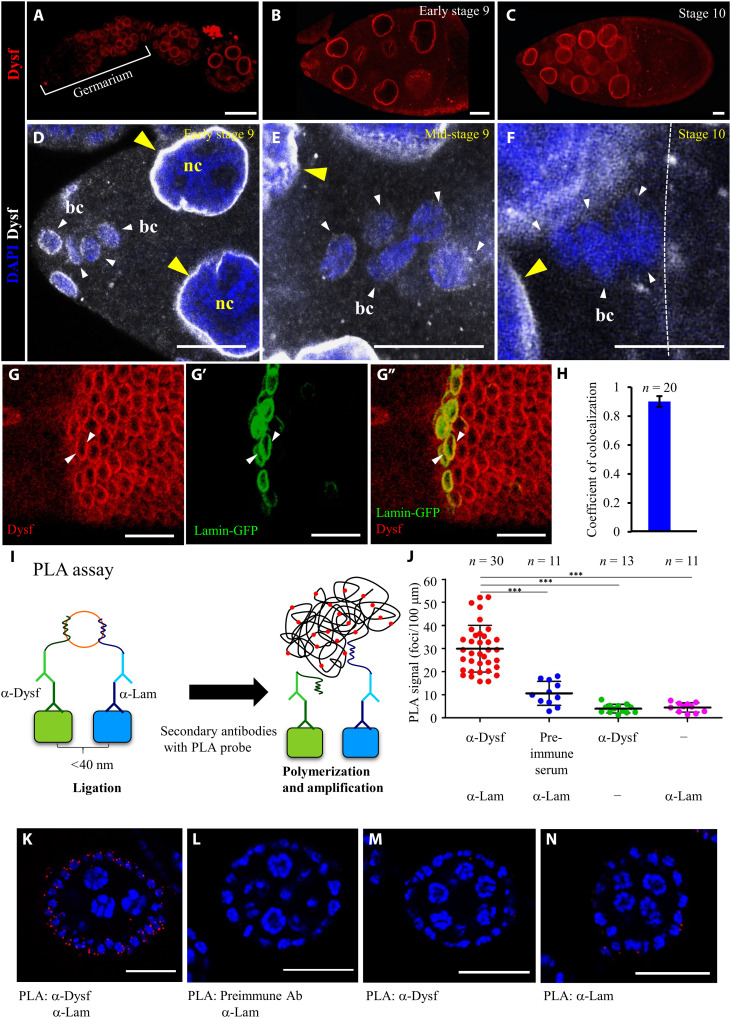
Dysf has been identified as a bHLH nuclear protein that is expressed in fusion cells to regulate Drosophila trachea development (50). Unexpectedly, rather than the nucleus-wide staining pattern reported previously for Dysf (50), we observed that Dysf expression was restricted to the nuclear membrane throughout oogenesis, including for germline and follicle cells (Fig. 4, A to F). Specificity of the anti-Dysf staining was confirmed with mutant clone analysis (fig. S5, B to D). As border cells began to migrate, staining signal of Dysf at the nuclear membrane gradually diminished (Fig. 4, D and E), becoming undetectable at stage 10 when the cluster came into contact with the oocyte (Fig. 4F). To depict the spatial relationship between Dysf and the nuclear membrane, we compared patterns of anti-Dysf staining with expression signal of GFP-tagged Lamin, a fibrous protein that lines the inner nuclear membrane (Fig. 4, G to G″). Confocal microscopy and Zen software analyses revealed a high colocalization coefficient (0.901) between these two proteins (Fig. 4H). Unlike border cells, the nuclear membrane staining of Dysf remains clear in the anterior follicle cells nearby border cells and the posterior follicles (fig. S6, A to C). We quantified the fold change in fluorescence intensity and found that anti-Dysf staining in border cells was significantly reduced from 2.58 to 1.19 during stages 8 to 10 (fig. S6D). A gradual decrease from 2.68 to 1.82 in Dysf staining was also observed in posterior follicle cells (fig. S6D). To further analyze the anti-Dysf distribution, we measured and plotted the fluorescence intensity across the nucleus of border cells and posterior follicles (fig. S6E). In both cell types, the fluorescence intensity profile of Dysf staining displays twin peaks at stage 8, indicating enriched signals on the nuclear membrane (arrows, fig. S6, F and G); however, in later stages, border cell profile turns wavy, similar to that of DAPI without specific accumulation (fig. S6F). In contrast, the posterior follicle cells kept the two-peak pattern all the way to stage 10 (arrows, fig. S6G). This observation concludes a spatiotemporal down-regulation of Dysf on the nuclear membrane of border cells when border cells start to move at stage 9. To further clarify whether Dysf resides on the inner nuclear membrane or in the perinuclear space, we conducted a Duolink proximity ligation assay (PLA), enabling us to examine intramolecular distances to within 40 nm (Fig. 4I) (51, 52). Confocal fluorescence microscopy revealed accumulated PLA foci upon coincubation of anti-Dysf and anti-Lamin antibody (Fig. 4, J and K) but not in negative controls containing preimmune serum or single primary antibody alone (Fig. 4, J and L to N). Moreover, anti-Dysf antibodies paired with antibodies against major components of the NPC, such as mAb414 that detects FG repeat–containing nucleoporins and Nup107, interacted in PLA (fig. S7, A to C). In contrast, no proximity was detected by PLA for Dysf and outer nuclear membrane proteins, including MSP-300 and Klarsicht (fig. S7, A, D and E) (53, 54). Quantitative analyses also demonstrate significant interactions of Dysf with Nup107 and central FG repeat–containing nucleoporins in the follicular epithelium (fig. S7, F to H). These PLA results demonstrate that Dysf is located on the inner nuclear membrane and, more precisely, near the nuclear lamina and NPC.
Fig. 4. The nuclear membrane of border cells displays a gradual decline in Dysf expression.
(A to G) Immunofluorescence micrographs showing anti-Dysf staining during Drosophila oogenesis [red in (A) to (C), (G), and (G″); white in (D) to (F)]. Arrowheads indicate nuclear membrane staining of Dysf in border cells (white) and nurse cells (yellow) in early stage 9 (D), mid-stage 9 (E), and stage 10 (F) egg chambers. The dotted white line denotes the oocyte border. (G to G″) Colocalization of anti-Dysf staining (red) and Lamin-GFP (green) signals on nuclear membrane in main-body follicles (arrowheads). (H) Colocalization coefficient for Dysf and Lamin-GFP. n indicates the number of nuclei examined. (I) Schematic illustrating detection of the Dysf-Lamin interaction by proximity ligation assay (PLA). (J) Quantifications of PLA signals using the indicated antibodies. The line boundaries show the SD of signal distributions, with the midline marking the average. (K to N) Confocal micrographs of egg chambers showing PLA reactions (red) with indicated antibodies. DAPI (blue) labels nuclei. n represents total sample size; ***P < 0.001, two-tailed t test. Error bars indicate SD. Scale bars, 25 μm (A to F) and 20 μm (G and K to N).
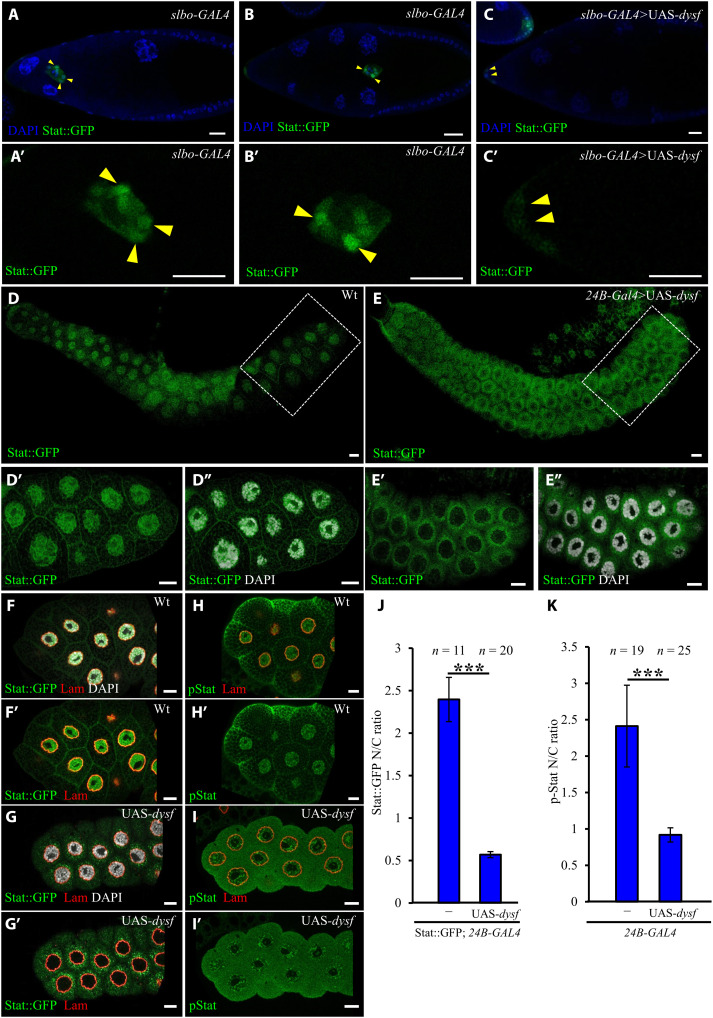
Given its subcellular localization, we wondered whether Dysf localizes close to the nuclear lamina and NPC to regulate nucleocytoplasmic transport of Stat. To address that possibility, we examined the expression pattern of stat92e-stat::gfp that encodes a Stat protein fused with GFP under control of its endogenous promoter (55). Dissimilar to anti–p-Stat staining, visualization of Stat::GFP distribution does not require a complicated fixation process or phosphatase inhibitor treatment. In addition, previous investigation revealed p-Stat staining as a useful tool for high-threshold Jak/Stat activity in stages 9 to 10 of oogenesis, but for signaling activity below the threshold, p-Stat antibody is not sensitive enough to detect the signal fluctuation (56). Thus, we applied stat92e-stat::gfp to detect the nuclear translocation of Stat. In the early stages of oogenesis, the intensity of Stat::GFP was relatively low and gradually increased from 6.7- to 14.3-fold during stage 8 to mid-stage 9 (fig. S8, A to E). Higher Stat::GFP signal was observed in border cell nuclei during migration (Fig. 5, A and B, yellow arrowheads), and the N/C ratio of fluorescence signal increased from 1.9 to 2.2, while border cells start to move at stage 9 (fig. S8F, red plot). By contrast, the N/C ratio of nonmigratory follicle cells remained lower than the anterior ones or border cells, staying at 1.53 to 1.61 during stage 6b to mid-stage 9 but rising to 1.77 at stage 10 (fig. S8F, blue plot). Overexpression of Dysf affected the cytoplasmic and nuclear distribution of Stat::GFP (Fig. 5, A to C), with the N/C ratio dropping to 1.19 (fig. S9E). To further examine the effect of Dysf on the cellular distribution of Stat::GFP and to test its conservation across cell types, we scrutinized Stat::GFP in the salivary glands of Drosophila larvae because of the larger cell size that can improve nucleocytoplasmic visualization (Fig. 5, D to G). In wild-type salivary glands, Stat::GFP was found predominantly in nuclei (Fig. 5, D to D″). However, under the condition of Dysf overexpression, Stat::GFP also accumulated at the periphery of the nuclear membrane (Fig. 5, E to E″) and outside the lamin meshwork (Fig. 5, G and G′). To quantify this effect, we measured the N/C ratio of Stat::GFP signal in the salivary gland cells and observed a reduction from 2.40 to 0.57 upon Dysf overexpression (Fig. 5J). Similarly, the ratio of p-Stat staining also decreased from 2.41 to 0.92 by Dysf up-regulation (Fig. 5, H, I, and K). We next examined the Stat::GFP signal in extra recruited border cells induced by overexpression of hop or dysf RNAi, and found that N/C ratios of Stat::GFP in these migrating cells were between 1.95 and 1.76, respectively, close to 1.83 in wild-type stage 10 border cells (fig. S9, A to E), but higher than 1.6 and 1.54 in nonmigratory cells at stage 9 to mid-stage 9 (fig. S8, A″ to D″ and F, blue plot). Observations here demonstrate that up-regulation of Dysf reduced the amount of nuclear Stat, but conversely, down-regulation of it led to an increase in Stat N/C and induced extra border cells, suggesting a role for Dysf in modulating Stat nuclear import to control border cell migration.
Fig. 5. Suppression of Stat nuclear import upon Dysf overexpression.
(A to C) Confocal images of GFP-tagged Stat distribution (green) in stage 9 or 10 egg chambers stained with DAPI (blue) to mark nuclei. Border cells indicated by arrowheads are shown below at higher magnification (A′ to C′). Stat::GFP expression (green) (D to G) and anti–p-Stat staining (H and I) in the salivary glands of the indicated genotypes, as revealed by immunofluorescence staining, and boxed regions are enlarged and displayed below. DAPI labels nuclei (white), and anti-Lamin (red) staining defines the boundary between the nucleus and cytoplasm. Quantitative assessment of the impact of Dysf on Stat nuclear localization by examining the nucleus:cytoplasm ratio of Stat::GFP (J) or anti–p-Stat (K) fluorescence signal. n indicates the number of salivary glands examined; ***P < 0.001, two-tailed t test. Error bars indicate SD. Scale bars, 20 μm.
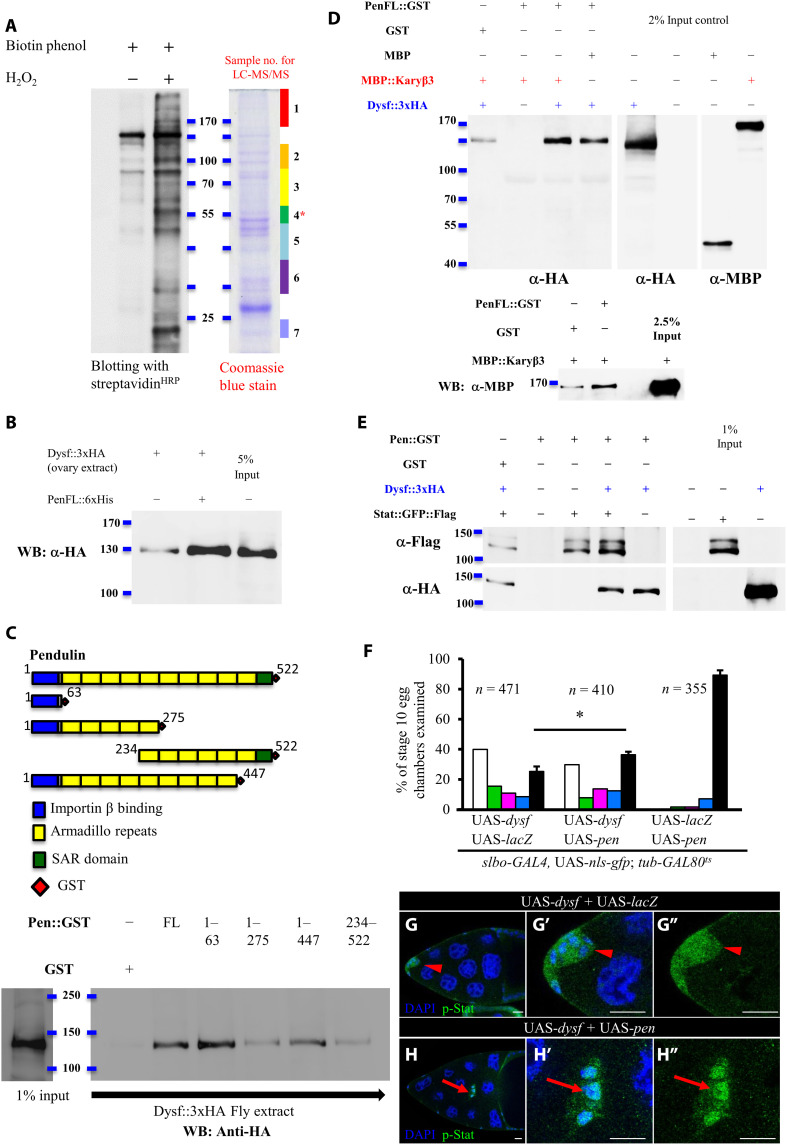
To uncover the molecular mechanism by which Dysf regulates Stat activity in situ and in vivo, it is critical to purify Dysf-associated proteins under native conditions. Thus, we generated the N-APEX2-dysf knock-in fly line that harbors an engineered ascorbate peroxidase (APEX2) inserted into the dysf locus to form a fused gene under endogenous promoter control (fig. S5A). APEX2 is one of the most efficient enzymes for proximity labeling, with proteins of interest fused with APEX2 tagging nearby proteins with biotin, enabling subcellular protein compartments to be mapped or the interacting partners of target proteins to be purified by means of streptavidin beads (52, 57, 58). We compared protein expression in fly ovaries treated with or without biotin labeling by blotting with streptavidinHRP. Seven slices were excised from the protein gel in alignment with the blot and then analyzed by liquid chromatography with tandem mass spectrometry (LC-MS/MS; Fig. 6A). By doing so, we identified 34 candidate genes, including histone binding proteins, nuclear transporters, a GTPase, ribosomal proteins, and RNA binding proteins, among others. Notable among them were Drosophila importin α2, Pen, and Karyβ3 (of the importin β family), indicating that they might participate in Dysf-mediated nucleocytoplasmic Stat translocation. We prioritized analysis of the interaction between Pen and Dysf. Given that APEX2-mediated biotin labeling requires proximity between two proteins, we used a pull-down assay to determine whether Pen and Dysf interact. Consistent with our blotting result (Fig. 6A), we found that hemagglutinin-tagged Dysf (Dysf::HA) bound to bacterially expressed Pen fused with a 6xHis::MBP tag (His, histidine; MBP, maltose binding protein) (Fig. 6B). Stat binds the armadillo repeats of importin α and forms a complex with importin β for nuclear transport (11). Given that Dysf also binds Pen and negatively regulates the nuclear localization of Stat, we postulated that the association between Dysf and Pen might interfere with the formation of the trimeric complex comprising importin α/β and Stat. Accordingly, we constructed various truncation mutants of Pen protein to determine which domain of Pen is responsible for interacting with Dysf. Glutathione S-transferase (GST) pull-down assays revealed that the IBB domain (covering amino acids 1 to 63) of Pen showed the strongest binding affinity for Dysf (Fig. 6C), relative to its armadillo repeats and the SAR domain (Fig. 6C). Our domain analysis indicates that rather than disrupting the Stat-Pen association, Dysf may modulate Stat translocation across the nuclear membrane by binding to Pen to interfere with Pen/Karyβ3 complex formation. To test this hypothesis, we carried out a competition assay to appraise the binding affinity of Pen for Dysf and Karyβ3. A pull-down assay showed that Dysf-Pen binding was not affected in the presence of abundant MBP::Karyβ3 recombinant protein (Fig. 6D, top panel). Moreover, we found that MBP::Karyβ3 interacts with Pen::GST (Fig. 6D, bottom panel). Next, we tested whether the Dysf-Pen interaction leads to dissociation of the Stat-Pen complex. We observed that Stat bound to Pen and formed a stable complex with Dysf, demonstrating that Dysf does not hamper the Stat-Pen association to attenuate Stat nuclear entry (Fig. 6E). These results suggest that the higher affinity of Dysf for the IBB domain of Pen may influence importin β–dependent nucleocytoplasmic transport. In a prevailing model without cargoes, importin α undergoes internal binding between its IBB domain and armadillo repeats (59). However, upon importin α binding to cargo, its IBB domain is exposed to importin β, which carries the transport complex through the NPC by interacting with the FG domain of Nups (60–63). Combining this conventional model with our findings, Dysf may occupy the IBB domain of Pen to restrict interaction between Karyβ3 and the Pen-Stat complex, thereby impairing delivery of Stat into the nucleus. If our model is correct, elevating Pen expression should counteract the effect of dysf up-regulation to increase the flow of Stat into the nucleus. In comparison to the control of UAS-dysf with 25.3% of border cells reaching the oocyte border, 36.3% of clusters that coexpressed Pen and Dysf completed the migration path, indicating a partial rescue of Dysf-induced migration defect by UAS-pen (Fig. 6F). Moreover, Pen overexpression also suppressed the phenotype of nuclear down-regulation of p-Stat caused by Dysf (Fig. 6, G and H, and fig. S2N). To reveal how Nup proteins participate in Dysf-mediated Stat transport, we performed a modified screening for any NPC component that could be linked to the Dysf-induced migration defect when overexpressed or down-regulated. After systematically examining 28 Nup genes, we noted that down-regulation of nup153 by RNAi knockdown or incorporation of a transposon-inserted mutant allele not only significantly suppressed the migration delay caused by Dysf (fig. S10, A and B) but also restored nuclear accumulation of p-Stat (fig. S10, C and D). Together, based on our findings, we hypothesize that Dysf resides on the nuclear lamina to attenuate passage of Stat through the NPC by binding to the IBB domain of Pen, thereby promoting dissociation of Karyβ3 from the Pen-Stat complex, which results in cargo being retained at the inner nuclear membrane (Fig. 7Q). The spatial decline in Dysf protein within stage 9 border cells increases Stat inflow into the nuclei, escalating Jak/Stat signaling to induce formation of a cell cluster and prompting persistent cell migration.
Fig. 6. Dysf-Pen interaction regulates Stat nuclear import.
(A) Biotinylated proteins were pulled down by streptavidin beads and detected by streptavidinHRP. SDS–polyacrylamide gel electrophoresis (PAGE) gels subjected to LC-MS/MS analysis were stained with Coomassie blue. (B) Pull-down assay validated the interaction between Dysf and Pen. (C) Schematic showing full-length and truncated Pen proteins with known domains. GST pull-down assay (bottom) demonstrated interaction of Dysf with Pen variant proteins. (D and E) GST pull-down assay revealed the impacts of Karyβ3 (D) or Stat (E) on Dysf-Pen association. (F) Quantification of border cell migration defects in indicated genotypes. Coexpression of pen partially rescues dysf-induced migration defect. (G and H) Fluorescence micrographs of stage 10 egg chambers stained with anti–p-Stat (green) and DAPI (blue). Arrowheads indicate an even distribution of p-Stat under the condition of Dysf overexpression (G to G″). Arrows highlight nuclear accumulation of p-Stat in cells coexpressing dysf and pen (H to H″). n is the total number of egg chambers examined; *P < 0.05, two-tailed t test. Error bars indicate SD. Scale bars, 20 μm.
Fig. 7. The human homolog of Dysf, Npas4, impairs Stat3 nuclear localization and cancer cell migration.
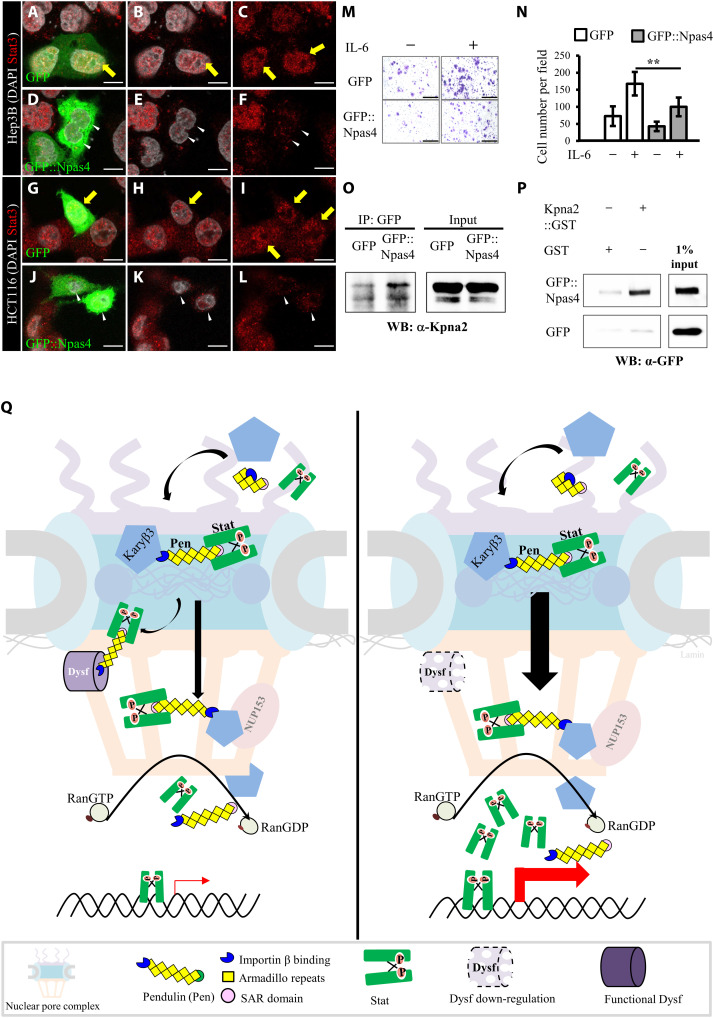
Confocal images of cancer cells stained with anti-Stat3 antibodies (red) and DAPI (white). After IL-6 stimulation, control cells transfected with GFP (green) revealed an accumulation of Stat3 in nuclei [yellow arrows in (A) to (C) and (G) to (I)]. Transfection of GFP::Npas4 (green) led to a lack of Stat3 signal in nuclei [white arrowheads in (D) to (F) and (J) to (L)]. (M) Crystal violet–stained (purple) Hep3B cells that crossed the transwell chamber membrane. (N) Quantification of our transwell migration assay (**P < 0.01, two-tailed t test; error bars indicate SEM). (O) Coimmunoprecipitation analysis demonstrated Npas4-Kpna2 interaction. (P) GST pull-down assay confirmed Npas4-Kpna2 interaction. (Q) Model of how Dysf gates nuclear import of Stat. Dysf protein resides on the inner nuclear membrane and binds to Pen via its IBB domain. Left: By stage 9, Dysf-Pen interaction reduces nuclear translocation of Stat. Right: In border cells, the gradual decrease of Dysf releases the constraint on Stat nuclear influx, inducing higher signaling activity and leading to cluster formation that maintains persistent migration. Nup153 serves as an additional gatekeeper modulating nuclear shuttling of Stat in border cells. Scale bars, 10 μm (A to L) and 500 μm (M).
Our previous study demonstrates that ecdysone serves as a temporal signal to conduct border cell detachment and coactivation of ecdysone and Jak/Stat signaling induces border cell migration 12 hours in advance (36). Therefore, we wanted to test whether this precocious movement can be achieved by ecdysone hyperactivation with dysf down-regulation instead. In many cases, coexpression of UAS-dysfRNAi and UAS-tai(ΔB), the constitutive-active form of the ecdysone receptor coactivator taiman, led to egg chamber degeneration, but we still observed five early-detached border cell clusters in total 48 stage 8 egg chambers examined (fig. S11, A, B, E, and F). However, this early migration phenotype was not found in overexpression of UAS-tai(ΔB) or UAS-dysfRNAi alone (fig. S11, C to F). These data showed that dysf hypoactivity can replace Jak/Stat hyperactivity to trigger precocious border cell movement under excessive ecdysone activity, namely, dysf functions as a temporal regulator that controls Jak/Stat-dependent migration in border cells.
Suppression of Stat3 nuclear import and cancer cell migration by mammalian Npas4
Dysregulation of Stat proteins has been reported previously for cancer cell lines and patient-derived tumor tissues, with most cancer treatment strategies targeting Jak/Stat signaling designed to inhibit phosphorylation of Jak or Stat (64–66). Inhibitors to suppress general nuclear translocation are undergoing clinical trials for the treatment of hematological or solid tumors, but to date, they have displayed significant toxicity in patients (67). Therefore, selective approaches that block Stat nuclear transport have been proposed to reduce side effects (11). However, an interaction molecule that can prevent Stat nuclear localization in cancer cells has yet to be developed. Moreover, several genes homologous to those involved in Drosophila border cell migration have been implicated in regulating cancer progression (1, 16, 68, 69). In search of the correlation between Stat and the human homolog of Dysf (35), Npas4, in cancer development, The Cancer Genome Atlas (TCGA) data mining was preformed and revealed an inverse gene profile of these two genes (70, 71). Npas4 belongs to the bHLH-Per-Arnt-Sim (PAS) family and participates in memory formation (72). The Pas1 domain of Dysf shares 54% identity with the Pas domain of Npas4, and overall sequence identity in amino acid is 25%. In Stat-hyperactivated cancer cells, Npas4 expression is very low or barely detected (70). Accordingly, we extended our study to explore whether Npas4 operates via a similar mechanism to suppress Jak/Stat signaling. To investigate the effect of Npas4 on nuclear Stat3 translocation and invasive behavior, we transfected it into the cancer cell lines Hep3B and HCT116, both of which show high level of Stat3 expression. Following IL stimulation, we observed that anti-Stat3 signal was enriched in the nuclei of both sets of control cancer cell lines (Fig. 7, A to C and G to I), but Stat3 signals were reduced in GFP::Npas4-transfected cancer cell lines (Fig. 7, D to F and J to L). The nuclear staining of p-Stat3 was also reduced by Npas4 overexpression (fig. S12). Npas4 significantly impaired IL-6/Stat3–mediated migration in a transwell assay (Fig. 7, M and N). Moreover, GFP::Npas4 can be coimmunoprecipitated with the human homolog of Pen, karyopherin α 2 (Kpna2) (Fig. 7O), and the direct interaction was further confirmed by GST pull-down assay (Fig. 7P). These results provide functional evidence supporting the idea that the human homolog of Dysf, Npas4, can also antagonize Jak/Stat signaling in cancer metastases.
DISCUSSION
In response to a variety of signals such as morphogens, cytokines, hormones, or stress signals derived from pathogens, tissues must integrate all these inputs and react differentially by modulating expression of various genes. To do so, an array of positive and negative regulators must be orchestrated to define expression boundaries, which is the key to segregating stem cells and differentiated daughter cells in the niche, tissue patterning during embryogenesis, as well as the static and migratory cells of tissue morphogenesis and cancer metastases. Jak/Stat signaling is an example of precise control of gene activation, although its ligand emanates from a very localized source without significant differences in concentration. The Jak/Stat ligand Upd is secreted from anterior polar cells to frame the anterior epithelial pattern of egg chambers during Drosophila oogenesis, but only six to eight cells adopt a mesenchymal cell fate because of higher Jak/Stat signaling (24). Thus, suppression of excessive Stat activity in nonmigratory cells is a critical mechanism by which additional recruitment of extra follicle cells is prevented, as also observed for tissue morphogenesis and cancer cell invasion (73). A previous study showed that apontic (apt), a downstream gene of Stat, suppresses Jak/Stat signaling via auto-feedback to limit border cell number (33). apt down-regulates stat via its downstream target, miR-279, which targets the 3′ untranslated region (3′UTR) of stat to suppress gene expression (34). The negative feedback regulation established by apt/miR-279 accounts for how Stat activity is attenuated in a broad domain of anterior follicle cells to constrain the transmitted effect of Upd secreted from polar cells. However, in the initial stage, anti-Apt and anti-Stat staining signal overlaps in border cell precursors (33), indicating that the feedback loop cannot suppress Stat expression or fully account for border cell fate determination. Our study unveils an additional mechanism by which Stat activity can be differentially controlled by the degree of Stat nuclear import to determine cell fate, motile border cells, or static follicles (Fig. 7Q). Our APEX2-based screen uncovered a physical association between Dysf and Pen, and biochemical and genetic analyses show that Dysf gates nuclear import of Stat. Upon interacting with Dysf, the IBB domain of Pen dissociates from importin β, thereby impairing Stat transport across the nuclear membrane. In the NPC, the FG-rich repeat Nup proteins form a hydrogel-like molecular sieve that prevents diffusion of nonspecific cargoes (74). During nuclear import, the transport complexes carry cargoes across the nuclear membrane by interacting with Nup proteins via importin β, rendering importin α/β part of the molecular sieve modulating nucleocytoplasmic transport (75). In our screen, we also identified the NPC component, Nup153, as part of the regulatory system. Suppression of nuclear Stat translocation by Dysf is dosage dependent because cell motility and nuclear Stat import are both rescued in a genetic background of pen overexpression or nup153 down-regulation. This scenario is consistent with a previous observation that Ran-GTP, importin β, and Nup153 form a higher-order complex to modulate permeability in the nuclear basket (76). Moreover, when Nup153 dosage is reduced, more cargoes such as Stat proteins tend to be translocated into the nucleus. Before stage 9 of oogenesis, endogenous Dysf and Nup153 partially retain the importin α/Stat complex at the nuclear face of Nup to prevent Stat from binding its downstream genes (Fig. 7Q). We found that 26% of egg chambers carrying a dysf mutation had extra migratory clusters, suggesting that Dysf fine-tunes the threshold of Stat activity. Persistent expression of Dysf in nonmigratory follicle cells ensures cell fate stability by preventing perturbations of Jak/Stat activity surpassing the threshold to ectopically induce cell motility. However, once Jak/Stat suppression is lost, extra border cells are induced. Progressive degradation of Dysf at the nuclear membrane of border cells unleashes importin α/Stat complexes associated with Dysf proteins up to stage 9. This temporal regulation, the decline of Dysf at stage 9, is a critical step for border cell fate specification. In combination with ecdysone hyperactivation, precocious down-regulation of dysf resulted in early detachment and formation of migrating clusters. This finding demonstrates that Dysf takes part in the spatiotemporal circuit integrated with Jak/Stat and ecdysone signaling to schedule border cell migration. In addition, a previous study has shown that the IBB domain of importin α displays auto-inhibitory activity by binding to its own NLS (Nuclear Localization Signals)–cargo binding pocket, resulting in cargo dissociation in the nucleus (77). Thus, newly unleashed Pen/Stat complexes may disassemble via this auto-inhibitory mechanism. Alternatively, the released Pen/Stat complexes may rebind to importin β, follow the conventional pathway to transit the NPC, and then disassemble by interacting with Ran-GTP (61).
Furthermore, quantitative analyses reveal that slight alternation of the N/C ratio of p-Stat staining or Stat::GFP can significantly affect border cell migration in hop hyperactivation or dysf dysregulation by RNAi knockdown or overexpression. This result is also consistent with that of the previous investigation, in which a 30-min shift to nonpermissive temperature hindered border cell motility under statts background (25). Despite a 114% rise in Stat::GFP expression from stage 8 to mid-stage 9 (fig. S8E), the gradual decline of Dysf from stage 8 to early stage 9 followed by sequential increase of Stat::GFP nuclear import, a 16% increase in the N/C ratio (fig. S8F), boosts Jak/Stat signaling activity to the highest level at mid-stage 9 of oogenesis (Fig. 1B). However, we also observed a significant down-regulation of Stat92E-GFP in stage 10 border cells when the N/C ratio of Stat::GFP declined and the Dysf staining was almost undetectable. This observation implies an additional mechanism beyond Dysf to attenuate Stat activity, by which Stat may undergo nuclear export and degradation after mid-stage 9. Identification of the novel attenuation mechanism would further fully elucidate the differential control of Stat. In addition, GFP::Npas4 transfection reduced anti–p-Stat or anti-Stat staining not only in the nucleus but also in the cytoplasm in cancer cells. It is likely that the cytoplasmic p-Stat proteins may be dephosphorylated by phosphatases or subjected to proteasome degradation as reported in previous studies (9, 78). However, we cannot exclude the possibility that Npas4 may suppress Stat expression apart from its function on nucleocytoplasmic transport. All of these questions left unaddressed are of great interest to investigate in the future.
Jak/Stat signaling is a key process in several cellular functions, including cell proliferation, angiogenesis, differentiation, survival, and apoptosis, with Jak/Stat dysregulation contributing to diverse pathological defects such as solid tumors and cancer metastases (27). Aberrant activation of Stat3 is estimated to occur in more than 70% of cancers (73, 79). Our findings demonstrate that the human homolog of Dysf, Npas4, plays a similar role in negatively regulating Stat-dependent cancer cell migration. Inhibitors or monoclonal antibodies targeting the IL-6/Jak/Stat3 axis are currently under development and hold promise as antitumor proliferation therapeutics (80). The major limitation of these therapies is the immunosuppression resulting from general inhibition of Jak/Stat-dependent immune responses, which can offset the antitumor effects of these treatments (81). Thus, tissue-specific biomarkers beyond the Jak/Stat core components are needed for more targeted treatments, with Npas4 revealed by our work potentially representing a novel target for therapeutic development.
MATERIALS AND METHODS
Drosophila strains and fly genetics
The following fly strains were used in this study [obtained from the Bloomington Drosophila Stock Center (BDSC)]: w1118 or UAS-lacZ served as wild-type controls, dysf2 (BDSC 9590), dysf3 (BDSC 9591), UAS-dysf (BDSC 9592), UAS-lam::gfp (BDSC, 7378), UASp-dsRNA-dysf (BDSC 35010), stat92e-stat92e::gfp/Cyo (BDSC 38670), gfp-nup107 (BDSC 35514), msp300-gfp (BDSC 59757), stat-lacZ (BDSC 11681), and UAS-tai(ΔB) (BDSC 28273). UAS-nup153RNAi (VDRC 47155) and nup153NP2104 (Kyoto 104091) were obtained from Vienna Drosophila Resource Center and Kyoto Stock Center (DGRC), respectively. UAS-dysf-3xHA was obtained from FlyORF stock (F001839). The Jak/Stat signaling reporter Stat92E-GFP was obtained from E. Bach (37). Transgene expression in follicle cells was induced by slbo-GAL4 (40), c306-GAL4 (82), or actin-GAL4/Cyo (BDSC 4414) at 29°C for 12 to 14 hours. UAS-upd (83) and UAS-hop (84) were obtained from Yu-Chen Tsai. UAS-pen (UASp-impα2-3xMyc) was a gift from S. Kobayashi (85). hsFLP; FRT82B, ubi-rfp was a gift from Chi Kuang Yao. UAS-dysf expression in salivary glands was induced by 24B-GAL4 obtained from Tzu-Yang Lin. To prevent lethality caused by transgene expression during development, we combined tub-GAL80ts (BDSC 7019) with a GAL4 system to suppress its effect in embryogenesis. Therefore, flies were raised below or at 25°C until nonpermissive temperature treatment at 31°C for 12 hours to 3 days (86). In the precocious migration experiment, transgenes were induced by act-GAL4, tub-GAL80ts and incubated flies at 31°C for 2 days.
The dysf2 (BDSC 9590) or dysfJW alleles were recombined with FRT82B (BDSC 2035) to establish the dysf2, FRT82B and dysfJW, FRT82B lines, which were then crossed to hsFLP; FRT82B, ubi-gfp/TM3, Sb1 or hsFLP; FRT82B, ubi-rfpTM3, Sb1 to generate mutant clones. The female flies were heat-shocked at 37°C for 1 hour twice per day for one or three consecutive days and then raised at 25°C for 2 or 5 days on dry yeast and standard fly food before dissection. For the dysf knockdown experiment, female flies carrying actin-GAL4 and UASp-dsRNA-dysf were raised at 29°C for 1 to 3 days before dissection. For clonal induction of UAS-dysf by Flip-Out technique (87), hsFLP/UAS-dysf; AyGAL4, UAS-lacZ/Stat92E-GFP flies were heat-shocked twice at 37°C for 1 hour and then fed with dry yeast and maintained at 29°C for 1 day before dissection.
Generation of an APEX2 knock-in tag at the dysf locus
The dysfJW allele was generated by CRISPR-Cas9–mediated genome editing, which was performed by WellGenetics (Taiwan). In brief, the guide RNA (ACGTGGCCTAGACAAGGTGACGG) designed to create a double-strand break at the 5′UTR of the dysf locus was cloned into the CRISPR-Cas9 vector pDCC6. APEX2 (a gift from T.-Y. Lin) (88), homology arms, a Piggy Bac terminal repeat, the 3xP3-hsp70 promoter, and DsRed2 were cloned into pUC57-Kan donor plasmid. By microinjection, the transgene, APEX2-PBacDsRed, was inserted into the 5′ end of dysf coding region by homology-dependent repair. To kick out the Ds-Red marker and generate N-APEX2-dysf, dysfJW (fig. S5A) was crossed with P{Tub-PBac\T2/wgSp-1 (BL8285) to excise the PBacDsRed fragment (fig. S5A).
Production of anti–phospho-Drosophila Stat92E (Tyr704) antibody
The phospho-peptide around residue Tyr704 of Stat92E was synthesized and conjugated with a KLH peptide, resulting in H-Cys-Glu-Pro-Leu-Val-Leu-Asp-Pro-Val-Thr-Gly-{pTyr}-Val-Lys-Ser {KLH-peptide via Cys822828822 82-OH (89). A rabbit was then immunized with the peptide to generate antibodies.
Immunohistochemistry
Before dissection, flies or larvae were fattened on wet or dry yeast for 14 to 16 hours and maintained at 29°C. Egg chambers or salivary glands were dissected and fixed in buffer [4% paraformaldehyde (Electron Microscopy Sciences), 1× phosphate-buffered saline (PBS; Sigma-Aldrich), and 1 mM calcium chloride (Sigma-Aldrich)] on ice for 30 min. For anti-Dysf staining, the fixation time was reduced to 20 min. Samples were then washed three times with 1× PBS buffer containing 0.3% Triton X-100. Washing buffer with 0.5% Triton X-100 was applied for anti-Dysf staining. The following primary antibodies used for immunostaining were obtained from the Developmental Studies Hybridoma Bank (DSHB): mouse anti-FasIII (7G10; 1:10), mouse anti-Eya (eya10H6; 1:10), mouse anti–β-galactosidase (40-1a; 1:10), mouse anti-Lamin (ADL67.10; 1:10), and rat anti–DE-cadherin (DCAD2; 1:10). Rabbit anti-Dysf (1:500) was a gift from L. Jiang (Oakland University, USA). Mouse anti-Stat3 (124H6; 1:500) and rabbit anti–p-Stat3 (D3A7; 1:20) were purchased from Cell Signaling Technology. Mouse anti–Lamin A/C (636; 1:100) was ordered from Santa Cruz Biotechnology. Phalloidin (P5282; 1:5000) was purchased from Sigma-Aldrich. For anti–p-Stat staining, flies were dissected in S2 medium containing 10% fetal bovine serum (FBS) and 1× phosphatase inhibitors (PhosSTOP, Sigma-Aldrich) and followed by fixation buffer (4% paraformaldehyde, 1× PBS, and 1× phosphatase inhibitors) at 4°C for 2 hours. Samples were then washed three times with 0.5% PBT (PBS containing 0.5% Triton X-100). The anti–p-Stat antibodies (1:400) were diluted with 0.3% PBT (PBS containing 0.3% Triton X-100) and incubated with samples overnight at 4°C. The following secondary antibodies conjugated with fluorescent dye were used at 1:400 dilutions: Alexa Fluor 488, Alexa Fluor 568, or Alexa Fluor 647 (Invitrogen, Molecular Probes). Nuclei shown in all confocal micrographs were stained with DAPI (Invitrogen; 1:10,000). Fluorescence images were obtained by Zeiss LSM-780 confocal microscopy (Instrument Development Center, NCKU, Taiwan) and processed by ZEN software.
Quantitative assessment of Stat92E-GFP expression and anti–p-Stat staining during oogenesis
Micrographs of Stat92E-GFP reporter, Stat::GFP signals, and anti–p-Stat staining were obtained by Zeiss LSM 780 confocal microscopy. To ensure that egg chambers at the same stage were used for analysis, we measured their axes along the anterior to posterior direction (90, 91). Stat92E-GFP, Stat::GFP expression/anti–p-Stat staining was defined quantitatively by the ratio of fluorescence intensity in anterior follicle cells over that of oocytes at different stages, except for stages 9 to 10 when measurements were taken in border cells. We targeted the nuclear edges of border cells or follicle cells to measure fluorescence intensity with ZEN software. The fluorescence intensity of oocytes served as an internal control to normalize values of Stat92E-GFP, Stat::GFP, and anti–p-Stat.
APEX2 proximity labeling to map protein interactions
The APEX2 labeling protocol was a modification from a previous publication (58). We dissected 120 pairs of fly ovaries in Drosophila S2 cell medium and then treated them with 0.3% PBT for 15 min to enhance tissue penetrance, before applying 500 μM Biotin-Phenol (Iris Biotech) for 15 min. After this pretreatment, 1 mM H2O2 was added and incubated for 1 min to induce APEX2 to label nearby proteins. The reaction was stopped by means of quencher buffer [5 mM Trolox (Sigma-Aldrich), 10 mM sodium azide (Sigma-Aldrich), and 10 mM sodium ascorbate (Sigma-Aldrich) in PBS]. After biotin labeling, samples were lysed with lysis buffer (50 mM tris-Cl, 150 mM NaCl, 0.1% SDS, 1% Triton X-100, and 1× protease inhibitor cocktail), before being passed through streptavidin magnetic beads (Thermo Fisher Scientific) to enrich for biotinylated proteins. After purification, samples were subjected to SDS–polyacrylamide gel electrophoresis (SDS-PAGE) and blotted with anti-streptavidin–horseradish peroxidase (HRP; 1:5000; Invitrogen). Fourteen slices of protein interest in gel that include control and experiments were extracted from the PAGE and identified by LC-MS/MS.
Plasmid DNA construction
DNA fragments encoding GST were cloned into the Hind III/Xho I sites of the pET24a vector to generate pET24a-GST, which served as vector for Pen-truncated constructs. Full-length pen was amplified by polymerase chain reaction (PCR) using forward (5′-GAATTCATGAGTAAGGCGGATTCTAA-3′) and reverse (5′-AAGCTTGAACGTGTAGCCACCTTC-3′) primers from an expressed sequence tag (EST) clone (LD24935) and then subcloned into the Eco RI/Hind III sites of pET24a-GST. Other subdomain deletion constructs were amplified by (i) forward primer (5′-GAATTCATGAGTAAGGCGGATTCTAA-3′) and reverse primer (5′-AAGCTTGCCATTGAGCTCTTTGAGC-3′) for pET24a-Pen (1–63)-GST, (ii) forward primer (5′-GAATTCATGAGTAAGGCGGATTCTAA-3′) and reverse primer (5′-AAGCTTGATCTTGGTGTTATCGTCG-3′) for pET24a-Pen (1–275)-GST, (iii) forward primer (5′-GAATTCATGAGTAAGGCGGATTCTAA-3′) and reverse primer (5′-AAGCTTGGTGCCACCAAGTTTCTC-3′) for pET24a-Pen (1–447)-GST, and (iv) forward primer (5′-GAATTCATGCCATTCGATCAGGTGAAG-3′) and reverse primer (5′-AAGCTTGAACGTGTAGCCACCTTC-3′) for pET24a-Pen (234–552)-GST. Full-length karyopherin β3 was amplified from EST clone FI07923 by PCR with forward primer (5′-GGTACCGAGCTCATGGGTAAGCCTATCCCTAACCCTCTCCTCGGTCTCGATTCTACGATGGCAGCGGATCAGGCC-3′) and reverse primer (5′-GCGGCCGCGGCGGGAGCCACGTTTGC-3′), before being subcloned into the Sac I/Not I site of pDB-6xHis-MBP. Human Npas4 was amplified by PCR with forward primer (5′-AGATCTATGTACCGCTCCACCAAG-3′) and reverse primer (5′-CTGCAGAAACGTTGGTTCCCCTCC-3′) from cDNA of a HeLa cell line and subcloned into the Bgl II/Pst I sites of pEGFP-C1.
Human Kpna2 was amplified by PCR with forward primer (5′-AATGGGTCGCGGATCCATGGATTACAAGGACGACGATGACAAGATGTCCACCAACGAGAATGC-3′) and reverse primer (5′-TAGGGGACATAAGCTTAAAGTTAAAGGTCCCAGGAGCCC-3′) from cDNA of a Hep3B cell line and subcloned into the Eco RI/Hind III sites of pET24a-GST.
Recombinant protein expression
The plasmids encoding GST, Pen (FL)-GST, Pen (1-63)-GST, Pen (1-275)-GST, Pen (1-447)-GST, Pen (234-552)-GST, human Kpna2-GST, MBP, and MBP-Karyβ3 were transformed into BL21(DE3)-competent cells (Invitrogen) for protein expression. Fusion protein expression was induced by treatment with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 24 hours at 18°C. After induction, Escherichia coli were resuspended in ice-cold lysis buffer [1× PBS (pH 7.4), 0.3% Triton, 150 mM NaCl, 10% glycerol, 10 mg/ml lysozyme, and 5 mM EDTA] and then sonicated using a ChromTech UP-500 system (10 cycles at 30% amplitude for 5 s on/5 s off). After sonication, lysates were centrifuged at 12,000g at 4°C for 20 min. Supernatants were subjected to affinity purification using Ni2+ resin (Invitrogen) or Glutathione Sepharose 4B (GE Healthcare) to collect recombinant proteins. Before pull-down assays, beads binding to recombinant proteins were stored in binding buffer (50 mM tris-Cl, 150 mM NaCl, 0.05% SDS, 1% Triton X-100, and 1× Roche EDTA-free protease inhibitor).
Tissue extract preparation
Dysf-3xHA (UAS-dysf-3xHA, tubGAL80ts) was overexpressed by slbo-GAL4 in follicle cells. Female flies were kept at 25°C for 4 days and then shifted to 31°C for 1 day before dissection. We dissected 400 pairs of ovaries from female flies overexpressing UAS-dysf-3xHA in Drosophila S2 cell medium (Thermo Fisher Scientific) and then lysed them in 200 μl of radioimmunoprecipitation assay buffer (50 mM tris-Cl, 150 mM NaCl, 0.1% SDS, 1% Triton X-100, and 1× EDTA-free protease inhibitor). The ovary lysate was kept on ice and sonicated (ChromTech UP-500) for 10 cycles at 30% amplitude for 5 s on/10 s off. After sonication, lysates were centrifuged at 15,000g at 4°C for 20 min. The supernatant was applied for pull-down assay. Tissue was harvested from 24B-GAL4>UAS-dysf-3xHA, tubGAL80ts larvae maintained at 25°C for 4 days and then shifted to 31°C for 1 day before dissection and extraction of Dysf-3xHA. Tissue was harvested from stat92e-stat92e::gfp larvae kept at 25°C for 5 days before dissection and extraction of Stat::Flag. Salivary glands and imaginal discs from 200 larvae were dissected in S2 cell medium and lysed as described above for ovary extract preparation. All lysates were incubated with deoxyribonuclease I (6 U/ml; NEB) at 4°C for 30 min before undergoing pull-down assay.
Cell extract preparation
The Hep3B cells were incubated in coimmunoprecipitation (co-IP) buffer [50 mM tris-HCl, 120 mM NaCl (pH 8.0), 0.05 mM EDTA, 0.5% Trixon-100, and 1× protease inhibitor] and rocked at 200 rpm for 15 min on ice. Then, cells were transferred to Eppendorf tubes and centrifuged at 12,000 rpm for 5 min at 4°C. The supernatant was collected before pull-down assay and co-IP.
Pull-down assay
Beads bound to recombinant proteins were incubated with tissue lysates at 4°C for 3 hours to overnight and then washed three times with wash buffer I (50 mM tris-Cl, 150 mM NaCl, 1% Triton X-100, and 1× Roche EDTA-free protease inhibitor). Last, high-salt wash buffer II (50 mM tris-Cl, 500 mM NaCl, 1% Triton X-100, and 1× EDTA-free protease inhibitor) was used to wash beads three times to avoid nonspecific binding, before being subjected to SDS-PAGE and Western blot detection. Anti-HA (1:3000; Ab9110, Abcam), anti-Flag (1:1000; M2, Sigma-Aldrich), and anti-MBP (1:1000; 2A1, DSHB) were used to detect transgenic proteins fused with indicated tags.
Co-IP assay
The GFP primary antibody was added into the cell extract and incubated with rotation at 4°C for 2 to 3 hours. About 35 to 40 μl of protein A/G beads that were prewashed with co-IP buffer were then added and incubated with rotation at 4°C for 1 hour. Captured immunoprecipitates were washed three times with lysis washing buffer and two times with 1× PBS. Immunoprecipitated proteins were denatured and subjected to immunoblot analysis using the following antibodies: anti-GFP (1:5000; JL-8, Clontech) and anti-Kpna2 (1:10,000; ab70160, Abcam).
Analysis of Dysf subcellular localization by PLAs
Our PLA protocol was modified from the Duolink In Situ Red Starter Kit Mouse/Rabbit (Sigma-Aldrich) protocol. The following primary antibodies were used: rabbit anti-Dysf (1:1000; in-house), mouse anti-Lamin (1:100; ADL67.10, DSHB), mouse anti-GFP (1:1000; 6AT316, Abcam), mouse anti-mab414 (1:1000; ab24609, Abcam), and KLAR-C (1:10; 9C10, DSHB).
Ovarioles were dissected in S2 cell medium after fattening for 2 days on dry yeast. Tissues were then fixed on ice with 4% paraformaldehyde in PBS buffer for 30 min. After fixation, the egg chambers were rinsed with 0.5% PBT for 10 min before being transferred to a 4°C refrigerator with gentle shaking overnight. The samples were then incubated overnight with primary antibody in 0.5% PBT at 4°C. The egg chambers were washed three times for 40 min with 0.5% PBT before being incubated in PLA probe solution (plus and minus probe diluted 1:5 in 0.5% PBT) at room temperature. After a 1-hour reaction in PLA solution, the samples were washed twice with wash buffer A from kit for 5 min, followed by 30-min ligation at 37°C. Samples were further washed twice with buffer A for 5 min and then incubated in amplification solution for 100 min at 37°C. After sequential rinsing with buffer B and then 0.01× buffer B from kit for 5 min each, samples were transferred to mounting medium containing DAPI and stored at 4°C. Stage 2 to 3 egg chambers were imaged by means of Zeiss LSM 780 confocal microscopy, and PLA signals were assessed quantitatively using Fiji ImageJ software. Pixel values less than or greater than that of the negative control (lacking anti-Dysf) were deemed as displaying no interaction or positive interaction, respectively.
Cell culture, transfection, and IL-6 treatment
Hep3B and HCT116 cells were purchased from BCRC (Food Industry Research and Development Institute, Taiwan). Hep3B cells were maintained in Dulbecco’s modified Eagle’s medium (high glucose, 1954626, Gibco). HCT116 cells were maintained in RPMI 1640 medium (31800022, Gibco). GFP and GFP::Npas4 were transfected into Hep3B and HCT116 by PolyJet and Lipofectamine 2000, respectively, according to the manufacturers’ instructions. For IL-6 treatment, cells were starved for 18 hours by maintaining them on serum-free medium before being subjected to IL-6 (25 ng/ml; GF338, Millipore) treatment at the indicated time points.
Migration assay
We seeded 8 × 104 cells suspended in FBS-free medium in the upper chamber of a 24-well transwell system (pore size 8 μm, BD Biosciences). Culture medium containing 10% FBS or conditioned medium was added to the lower chamber. After 48-hour incubation, migrated cells were fixed with methanol for 20 min and then stained with 0.1% crystal violet. Numbers of migrated cells were counted under optical microscopy in five random fields, and an average number is presented as mean ± SEM.
Acknowledgments
We thank Y. Henry Sun, Lan Jiang, T.-Y. Lin, C.-K. Yao, Min-Lang Huang, S. Kobayashi, and the Taiwan Fly Core for providing fly stocks. We thank the Bloomington Drosophila Stock Center, FlyORF, Vienna Drosophila Resource Center, and Kyoto Stock Center. We also thank Tzong-Yueh Chen and the technical support team of the Instrument Development Center of NCKU for providing critical assistance with Carl Zeiss LSM 780 laser-scanning micrograph imaging.
Funding: This research was supported by MOST (Ministry of Science and Technology, R.O.C.) grants to A.C.-C.J., MOST 110-2311-B-006-005 and MOST 107-2311-B-006-002-MY3, and to Y.-C.C., MOST 110-2311-B-110-004-MY3.
Author contributions: J.-W.W. conducted most experiments. C.-W.W. performed PLA and fluorescence intensity analyses. R.-Y.C. and L.-Y.H. contributed to Fig. 7. Y.-C.T. generated p-Stat antibody. Y.-T.C. carried out the modifier screen. A.C.-C.J. and Y.-C.C. designed and performed experiments, interpreted the data, and wrote the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S12
Table S1
REFERENCES AND NOTES
- 1.Stuelten C. H., Parent C. A., Montell D. J., Cell motility in cancer invasion and metastasis: Insights from simple model organisms. Nat. Rev. Cancer 18, 296–312 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Welch D. R., Hurst D. R., Defining the hallmarks of metastasis. Cancer Res. 79, 3011–3027 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guglielmi V., Sakuma S., D’Angelo M. A., Nuclear pore complexes in development and tissue homeostasis. Development 147, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hogarth C., Itman C., Jans D. A., Loveland K. L., Regulated nucleocytoplasmic transport in spermatogenesis: A driver of cellular differentiation? Bioessays 27, 1011–1025 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Raices M., Bukata L., Sakuma S., Borlido J., Hernandez L. S., Hart D. O., D’Angelo M. A., Nuclear pores regulate muscle development and maintenance by assembling a localized Mef2C complex. Dev. Cell 41, 540–554.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fallini C., Khalil B., Smith C. L., Rossoll W., Traffic jam at the nuclear pore: All roads lead to nucleocytoplasmic transport defects in ALS/FTD. Neurobiol. Dis. 140, 104835 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Satomura A., Brickner J. H., Nuclear pore complexes: A scaffold regulating developmental transcription? Trends Cell Biol. 27, 621–622 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levy D. E., Darnell J. E. Jr., Stats: Transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol. 3, 651–662 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Hu X., Li J., Fu M., Zhao X., Wang W., The JAK/STAT signaling pathway: From bench to clinic. Signal Transduct. Target. Ther. 6, 402 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herrera S. C., Bach E. A., JAK/STAT signaling in stem cells and regeneration: From Drosophila to vertebrates. Development 146, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ernst S., Muller-Newen G., Nucleocytoplasmic shuttling of STATs. A target for intervention? Cancers (Basel) 11, 1815 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ushijima R., Sakaguchi N., Kano A., Maruyama A., Miyamoto Y., Sekimoto T., Yoneda Y., Ogino K., Tachibana T., Extracellular signal-dependent nuclear import of STAT3 is mediated by various importin alphas. Biochem. Biophys. Res. Commun. 330, 880–886 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Owen K. L., Brockwell N. K., Parker B. S., JAK-STAT signaling: A double-edged sword of immune regulation and cancer progression. Cancers 11, 2002 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeidler M. P., Bausek N., The Drosophila JAK-STAT pathway. JAKSTAT 2, e25353 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu H., Pardoll D., Jove R., STATs in cancer inflammation and immunity: A leading role for STAT3. Nat. Rev. Cancer 9, 798–809 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trivedi S., Starz-Gaiano M., Drosophila Jak/STAT signaling: Regulation and relevance in human cancer and metastasis. Int. J. Mol. Sci. 19, 4056 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Betz A., Lampen N., Martinek S., Young M. W., Darnell J. E. Jr., A Drosophila PIAS homologue negatively regulates stat92E. Proc. Natl. Acad. Sci. U.S.A. 98, 9563–9568 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Starr R., Hilton D. J., Negative regulation of the JAK/STAT pathway. Bioessays 21, 47–52 (1999). [DOI] [PubMed] [Google Scholar]
- 19.Alexander W. S., Hilton D. J., The role of suppressors of cytokine signaling (SOCS) proteins in regulation of the immune response. Annu. Rev. Immunol. 22, 503–529 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Rawlings J. S., Rennebeck G., Harrison S. M., Xi R., Harrison D. A., Two Drosophila suppressors of cytokine signaling (SOCS) differentially regulate JAK and EGFR pathway activities. BMC Cell Biol. 5, 38 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karsten P., Hader S., Zeidler M. P., Cloning and expression of Drosophila SOCS36E and its potential regulation by the JAK/STAT pathway. Mech. Dev. 117, 343–346 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Callus B. A., Mathey-Prevot B., SOCS36E, a novel Drosophila SOCS protein, suppresses JAK/STAT and EGF-R signalling in the imaginal wing disc. Oncogene 21, 4812–4821 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Monahan A. J., Starz-Gaiano M., Socs36E attenuates STAT signaling to optimize motile cell specification in the Drosophila ovary. Dev. Biol. 379, 152–166 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silver D. L., Montell D. J., Paracrine signaling through the JAK/STAT pathway activates invasive behavior of ovarian epithelial cells in Drosophila. Cell 107, 831–841 (2001). [DOI] [PubMed] [Google Scholar]
- 25.Silver D. L., Geisbrecht E. R., Montell D. J., Requirement for JAK/STAT signaling throughout border cell migration in Drosophila. Development 132, 3483–3492 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Jiang H., Patel P. H., Kohlmaier A., Grenley M. O., McEwen D. G., Edgar B. A., Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell 137, 1343–1355 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verhoeven Y., Tilborghs S., Jacobs J., de Waele J., Quatannens D., Deben C., Prenen H., Pauwels P., Trinh X. B., Wouters A., Smits E. L. J., Lardon F., van Dam P. A., The potential and controversy of targeting STAT family members in cancer. Semin. Cancer Biol. 60, 41–56 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Xi R., McGregor J. R., Harrison D. A., A gradient of JAK pathway activity patterns the anterior-posterior axis of the follicular epithelium. Dev. Cell 4, 167–177 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Torres I. L., Lopez-Schier H., Johnston D. S., A Notch/Delta-dependent relay mechanism establishes anterior-posterior polarity in Drosophila. Dev. Cell 5, 547–558 (2003). [DOI] [PubMed] [Google Scholar]
- 30.Assa-Kunik E., Torres I. L., Schejter E. D., Johnston D. S., Shilo B. Z., Drosophila follicle cells are patterned by multiple levels of Notch signaling and antagonism between the Notch and JAK/STAT pathways. Development 134, 1161–1169 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Wang X., Bo J., Bridges T., Dugan K. D., Pan T. C., Chodosh L. A., Montell D. J., Analysis of cell migration using whole-genome expression profiling of migratory cells in the Drosophila ovary. Dev. Cell 10, 483–495 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Borghese L., Fletcher G., Mathieu J., Atzberger A., Eades W. C., Cagan R. L., Rørth P., Systematic analysis of the transcriptional switch inducing migration of border cells. Dev. Cell 10, 497–508 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Starz-Gaiano M., Melani M., Wang X., Meinhardt H., Montell D. J., Feedback inhibition of Jak/STAT signaling by apontic is required to limit an invasive cell population. Dev. Cell 14, 726–738 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Yoon W. H., Meinhardt H., Montell D. J., miRNA-mediated feedback inhibition of JAK/STAT morphogen signalling establishes a cell fate threshold. Nat. Cell Biol. 13, 1062–1069 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu Y., Flockhart I., Vinayagam A., Bergwitz C., Berger B., Perrimon N., Mohr S. E., An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinformatics 12, 357 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jang A. C., Chang Y. C., Bai J., Montell D., Border-cell migration requires integration of spatial and temporal signals by the BTB protein Abrupt. Nat. Cell Biol. 11, 569–579 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bach E. A., Ekas L. A., Ayala-Camargo A., Flaherty M. S., Lee H., Perrimon N., Baeg G. H., GFP reporters detect the activation of the Drosophila JAK/STAT pathway in vivo. Gene Expr. Patterns 7, 323–331 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Denef N., Schupbach T., Patterning: JAK-STAT signalling in the Drosophila follicular epithelium. Curr. Biol. 13, R388–R390 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Brand A. H., Perrimon N., Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415 (1993). [DOI] [PubMed] [Google Scholar]
- 40.Rorth P., Szabo K., Bailey A., Laverty T., Rehm J., Rubin G. M., Weigmann K., Milan M., Benes V., Ansorge W., Cohen S. M., Systematic gain-of-function genetics in Drosophila. Development 125, 1049–1057 (1998). [DOI] [PubMed] [Google Scholar]
- 41.Bai J., Montell D., Eyes absent, a key repressor of polar cell fate during Drosophila oogenesis. Development 129, 5377–5388 (2002). [DOI] [PubMed] [Google Scholar]
- 42.Ito K., Sass H., Urban J., Hofbauer A., Schneuwly S., GAL4-responsive UAS-tau as a tool for studying the anatomy and development of the Drosophila central nervous system. Cell Tissue Res. 290, 1–10 (1997). [DOI] [PubMed] [Google Scholar]
- 43.Chang Y. C., Wu J. W., Hsieh Y. C., Huang T. H., Liao Z. M., Huang Y. S., Mondo J. A., Montell D., Jang A. C. C., Rap1 negatively regulates the hippo pathway to polarize directional protrusions in collective cell migration. Cell Rep. 22, 2160–2175 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yan R., Small S., Desplan C., Dearolf C. R., Darnell J. E. Jr., Identification of a Stat gene that functions in Drosophila development. Cell 84, 421–430 (1996). [DOI] [PubMed] [Google Scholar]
- 45.Schindler C., Darnell J. E. Jr., Transcriptional responses to polypeptide ligands: The JAK-STAT pathway. Annu. Rev. Biochem. 64, 621–652 (1995). [DOI] [PubMed] [Google Scholar]
- 46.Jiang L., Crews S. T., Dysfusion transcriptional control of Drosophila tracheal migration, adhesion, and fusion. Mol. Cell. Biol. 26, 6547–6556 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang L., Crews S. T., Transcriptional specificity of Drosophila dysfusion and the control of tracheal fusion cell gene expression. J. Biol. Chem. 282, 28659–28668 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cordoba S., Estella C., The bHLH-PAS transcription factor dysfusion regulates tarsal joint formation in response to Notch activity during drosophila leg development. PLOS Genet. 10, e1004621 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zeidler M. P., Perrimon N., Strutt D. I., Polarity determination in the Drosophila eye: A novel role for unpaired and JAK/STAT signaling. Genes Dev. 13, 1342–1353 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jiang L., Crews S. T., The Drosophila dysfusion basic helix-loop-helix (bHLH)-PAS gene controls tracheal fusion and levels of the trachealess bHLH-PAS protein. Mol. Cell. Biol. 23, 5625–5637 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Söderberg O., Gullberg M., Jarvius M., Ridderstråle K., Leuchowius K.-J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L.-G., Landegren U., Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods 3, 995–1000 (2006). [DOI] [PubMed] [Google Scholar]
- 52.Mannix K. M., Starble R. M., Kaufman R. S., Cooley L., Proximity labeling reveals novel interactomes in live Drosophila tissue. Development 146, dev176644 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Becker R., Leone M., Engel F. B., Microtubule organization in striated muscle cells. Cell 9, 1395 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Palka M., Tomczak A., Grabowska K., Machowska M., Piekarowicz K., Rzepecka D., Rzepecki R., Laminopathies: What can humans learn from fruit flies. Cell. Mol. Biol. Lett. 23, 32 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sotillos S., Krahn M., Espinosa-Vazquez J. M., Hombría J. C., Src kinases mediate the interaction of the apical determinant Bazooka/PAR3 with STAT92E and increase signalling efficiency in Drosophila ectodermal cells. Development 140, 1507–1516 (2013). [DOI] [PubMed] [Google Scholar]
- 56.Hayashi Y., Sexton T. R., Dejima K., Perry D. W., Takemura M., Kobayashi S., Nakato H., Harrison D. A., Glypicans regulate JAK/STAT signaling and distribution of the Unpaired morphogen. Development 139, 4162–4171 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen C. L., Hu Y., Udeshi N. D., Lau T. Y., Wirtz-Peitz F., He L., Ting A. Y., Carr S. A., Perrimon N., Proteomic mapping in live Drosophila tissues using an engineered ascorbate peroxidase. Proc. Natl. Acad. Sci. U.S.A. 112, 12093–12098 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hung V., Udeshi N. D., Lam S. S., Loh K. H., Cox K. J., Pedram K., Carr S. A., Ting A. Y., Spatially resolved proteomic mapping in living cells with the engineered peroxidase APEX2. Nat. Protoc. 11, 456–475 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kobe B., Autoinhibition by an internal nuclear localization signal revealed by the crystal structure of mammalian importin α. Nat. Struct. Biol. 6, 388–397 (1999). [DOI] [PubMed] [Google Scholar]
- 60.Gorlich D., Henklein P., Laskey R. A., Hartmann E., A 41 amino acid motif in importin-alpha confers binding to importin-beta and hence transit into the nucleus. EMBO J. 15, 1810–1817 (1996). [PMC free article] [PubMed] [Google Scholar]
- 61.Lee S. J., Matsuura Y., Liu S. M., Stewart M., Structural basis for nuclear import complex dissociation by RanGTP. Nature 435, 693–696 (2005). [DOI] [PubMed] [Google Scholar]
- 62.Dickmanns A., Kehlenbach R. H., Fahrenkrog B., Nuclear pore complexes and nucleocytoplasmic transport: From structure to function to disease. Int. Rev. Cell Mol. Biol. 320, 171–233 (2015). [DOI] [PubMed] [Google Scholar]
- 63.Davis L. K., Saric A., Hoogenboom B. W., Zilman A., Physical modeling of multivalent interactions in the nuclear pore complex. Biophys. J. 120, 1565–1577 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu H., Lee H., Herrmann A., Buettner R., Jove R., Revisiting STAT3 signalling in cancer: New and unexpected biological functions. Nat. Rev. Cancer 14, 736–746 (2014). [DOI] [PubMed] [Google Scholar]
- 65.Bromberg J. F., Wrzeszczynska M. H., Devgan G., Zhao Y., Pestell R. G., Albanese C., Darnell J. E. Jr., Stat3 as an oncogene. Cell 98, 295–303 (1999). [DOI] [PubMed] [Google Scholar]
- 66.Pilati C., Zucman-Rossi J., Mutations leading to constitutive active gp130/JAK1/STAT3 pathway. Cytokine Growth Factor Rev. 26, 499–506 (2015). [DOI] [PubMed] [Google Scholar]
- 67.Turner J. G., Dawson J., Cubitt C. L., Baz R., Sullivan D. M., Inhibition of CRM1-dependent nuclear export sensitizes malignant cells to cytotoxic and targeted agents. Semin. Cancer Biol. 27, 62–73 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chang Y.-C., Wu J.-W., Wang C.-W., Jang A. C.-C., Hippo Signaling-mediated Mechanotransduction in cell movement and cancer metastasis. Front. Mol. Biosci. 6, 157 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jang A. C., Starz-Gaiano M., Montell D. J., Modeling migration and metastasis in Drosophila. J. Mammary Gland Biol. Neoplasia 12, 103–114 (2007). [DOI] [PubMed] [Google Scholar]
- 70.Ghandi M., Huang F. W., Jané-Valbuena J., Kryukov G. V., Lo C. C., McDonald E. R. III, Barretina J., Gelfand E. T., Bielski C. M., Li H., Hu K., Andreev-Drakhlin A. Y., Kim J., Hess J. M., Haas B. J., Aguet F., Weir B. A., Rothberg M. V., Paolella B. R., Lawrence M. S., Akbani R., Lu Y., Tiv H. L., Gokhale P. C., de Weck A., Mansour A. A., Oh C., Shih J., Hadi K., Rosen Y., Bistline J., Venkatesan K., Reddy A., Sonkin D., Liu M., Lehar J., Korn J. M., Porter D. A., Jones M. D., Golji J., Caponigro G., Taylor J. E., Dunning C. M., Creech A. L., Warren A. C., McFarland J. M., Zamanighomi M., Kauffmann A., Stransky N., Imielinski M., Maruvka Y. E., Cherniack A. D., Tsherniak A., Vazquez F., Jaffe J. D., Lane A. A., Weinstock D. M., Johannessen C. M., Morrissey M. P., Stegmeier F., Schlegel R., Hahn W. C., Getz G., Mills G. B., Boehm J. S., Golub T. R., Garraway L. A., Sellers W. R., Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 569, 503–508 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nusinow D. P., Szpyt J., Ghandi M., Rose C. M., McDonald E. R. III, Kalocsay M., Jané-Valbuena J., Gelfand E., Schweppe D. K., Jedrychowski M., Golji J., Porter D. A., Rejtar T., Wang Y. K., Kryukov G. V., Stegmeier F., Erickson B. K., Garraway L. A., Sellers W. R., Gygi S. P., Quantitative proteomics of the cancer cell line encyclopedia. Cell 180, 387–402.e16 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sun X., Lin Y., Npas4: Linking neuronal activity to memory. Trends Neurosci. 39, 264–275 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Teng Y., Ross J. L., Cowell J. K., The involvement of JAK-STAT3 in cell motility, invasion, and metastasis. JAKSTAT 3, e28086 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Frey S., Richter R. P., Gorlich D., FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science 314, 815–817 (2006). [DOI] [PubMed] [Google Scholar]
- 75.Kapinos L. E., Huang B., Rencurel C., Lim R. Y. H., Karyopherins regulate nuclear pore complex barrier and transport function. J. Cell Biol. 216, 3609–3624 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lowe A. R., Tang J. H., Yassif J., Graf M., Huang W. Y. C., Groves J. T., Weis K., Liphardt J. T., Importin-β modulates the permeability of the nuclear pore complex in a Ran-dependent manner. eLife 4, e04052 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Harreman M. T., Hodel M. R., Fanara P., Hodel A. E., Corbett A. H., The auto-inhibitory function of importin alpha is essential in vivo. J. Biol. Chem. 278, 5854–5863 (2003). [DOI] [PubMed] [Google Scholar]
- 78.Butturini E., Carcereri de Prati A., Mariotto S., Redox regulation of STAT1 and STAT3 signaling. Int. J. Mol. Sci. 21, 7034 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Frank D. A., STAT3 as a central mediator of neoplastic cellular transformation. Cancer Lett. 251, 199–210 (2007). [DOI] [PubMed] [Google Scholar]
- 80.Johnson D. E., O’Keefe R. A., Grandis J. R., Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat. Rev. Clin. Oncol. 15, 234–248 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Qureshy Z., Johnson D. E., Grandis J. R., Targeting the JAK/STAT pathway in solid tumors. J. Cancer Metastasis Treat. 6, 27 (2020). [PMC free article] [PubMed] [Google Scholar]
- 82.Manseau L., Baradaran A., Brower D., Budhu A., Elefant F., Phan H., Philp A. V., Yang M., Glover D., Kaiser K., Palter K., Selleck S., GAL4 enhancer traps expressed in the embryo, larval brain, imaginal discs, and ovary of Drosophila. Dev. Dyn. 209, 310–322 (1997). [DOI] [PubMed] [Google Scholar]
- 83.Tsai Y. C., Sun Y. H., Long-range effect of upd, a ligand for Jak/STAT pathway, on cell cycle in Drosophila eye development. Genesis 39, 141–153 (2004). [DOI] [PubMed] [Google Scholar]
- 84.Harrison D. A., Binari R., Nahreini T. S., Gilman M., Perrimon N., Activation of a Drosophila Janus kinase (JAK) causes hematopoietic neoplasia and developmental defects. EMBO J. 14, 2857–2865 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Asaoka M., Hanyu-Nakamura K., Nakamura A., Kobayashi S., Maternal Nanos inhibits Importin-α2/Pendulin-dependent nuclear import to prevent somatic gene expression in the Drosophila germline. PLOS Genet. 15, e1008090 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McGuire S. E., Mao Z., Davis R. L., Spatiotemporal gene expression targeting with the TARGET and gene-switch systems in Drosophila. Sci. STKE 2004, pl6 (2004). [DOI] [PubMed] [Google Scholar]
- 87.Ito K., Awano W., Suzuki K., Hiromi Y., Yamamoto D., The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development 124, 761–771 (1997). [DOI] [PubMed] [Google Scholar]
- 88.Lam S. S., Martell J. D., Kamer K. J., Deerinck T. J., Ellisman M. H., Mootha V. K., Ting A. Y., Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat. Methods 12, 51–54 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li J., Li W., Calhoun H. C., Xia F., Gao F. B., Li W. X., Patterns and functions of STAT activation during Drosophila embryogenesis. Mech. Dev. 120, 1455–1468 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.A. C. Spradling, Developmental Genetics of Oogenesis, in The Development of Drosophila melanogaster (CSH Laboratory Press, 1993), pp. 1–70. [Google Scholar]
- 91.Chen D. Y., Crest J., Streichan S. J., Bilder D., Extracellular matrix stiffness cues junctional remodeling for 3D tissue elongation. Nat. Commun. 10, 3339 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S12
Table S1