Abstract
PURPOSE
The response to cancer therapies is typically assessed with radiologic imaging 6-10 weeks after treatment initiation. Circulating tumor DNA (ctDNA), however, has a short half-life, and dynamic changes in ctDNA quantity may allow for earlier assessment of the therapeutic response.
METHODS
Patients with advanced solid tumors referred to the Department of Investigational Cancer Therapeutics at The University of Texas MD Anderson Cancer Center were invited to participate in a liquid biopsy protocol for which serial blood samples were collected before, during, and after systemic therapy. We isolated ctDNA from serially collected plasma samples at baseline, mid-treatment, and first restaging. Genomically informed droplet digital polymerase chain reaction (ddPCR) was performed, and ctDNA quantities were reported as aggregate variant allele frequencies for all detected molecular aberrations.
RESULTS
We included 204 patients receiving 260 systemic therapies. The ctDNA detection rate was higher in progressors (patients with progressive disease) compared with nonprogressors (patients with stable disease, partial responses, or complete responses) at all time points (P < .009). Moreover, ctDNA detection was associated with a shorter median time-to-treatment failure (P ≤ .001). Positive delta and slope values for changes in ctDNA quantity were more frequent in progressors (P ≤ .03 and P < .001, respectively) and were associated with a shorter median time-to-treatment failure (P ≤ .014 and P < .001, respectively). Increasing ctDNA quantity was predictive of clinical and/or radiologic progressive disease in 73% of patients (median lead time, 23 days).
CONCLUSION
Detection of ctDNA and early dynamic changes in its quantity can predict the clinical outcomes of systemic therapies in patients with advanced solid tumors.
INTRODUCTION
Molecular profiling of tumor tissue has led to substantial advances in personalized cancer therapy.1 However, tumor tissue is not always available, and repeated biopsies can increase treatment costs and the risk of complications.2 Therefore, interest in alternative sources of tumor DNA, such as cell-free DNA (cfDNA)—released into circulation from necrotic and apoptotic cells—has increased.3 The tumor fraction of cfDNA–circulating tumor DNA (ctDNA)–originates from primary and/or metastatic cancer lesions and can be used for molecular testing in lieu of cancer tissue. Different technologies, including polymerase chain reaction (PCR)–based and sequencing-based methods, can be used for molecular testing of ctDNA.4 PCR is sensitive and relatively straightforward, but it lacks the capacity for testing of multiple mutations. Sequencing can test for multiple alterations but at the price of increased cost and complexity.4 Advances in these technologies have considerably improved ctDNA detection rates and enabled the incorporation of ctDNA analysis workflows into different clinical settings, particularly those involving patients with advanced disease.5,6 For instance, US Food and Drug Administration approved the first PCR-based EGFR mutation ctDNA test for advanced/metastatic lung cancer in 2016 and targeted next-generation sequencing test for solid tumors in 2020.7-9
CONTEXT
Key Objective
Current clinical practice uses radiologic imaging to assess for tumor response to therapies 6-10 weeks after treatment initiation. We hypothesized that dynamic changes in the quantity of circulating tumor DNA (ctDNA), which has a short half-life, can be used as an early predictor of therapeutic response in patients with advanced solid tumors.
Knowledge Generated
We have shown that genomically informed longitudinal monitoring of ctDNA can be used to predict treatment outcomes as early as day 21 of first cycle. Moreover, we have demonstrated that progression in ctDNA quantity precedes or co-occurs with clinical progression in most patients. We propose ctDNA response criteria, which, if validated, can be used in lieu of or complement imaging-based assessment.
Relevance
Our study provides a proof on the feasibility and value of using ctDNA dynamics to monitor therapeutic response in patients with advanced cancers.
Unlike tumor biopsies, blood sample collections for ctDNA isolation are minimally invasive, can be repeated multiple times during therapy, and therefore can be used to study clonal evolution and adaptive resistance mechanisms.10 In addition, ctDNA has a relatively short half-life of 8-147 minutes.11 Consequently, dynamic changes in ctDNA quantity can be very early surrogate markers of therapeutic response.12-15 In clinical practice, therapy response is typically assessed with imaging performed at baseline and then every 6-10 weeks for the duration of therapy and with prespecified criteria (eg, RECIST version 1.1).16 Accordingly, many patients unlikely to benefit from cancer therapy are exposed to weeks of costly but futile and potentially toxic treatment.17 We hypothesized that, in patients with advanced solid tumors, dynamic changes in ctDNA quantity during the first few weeks of systemic cancer therapy can predict clinical outcomes.
METHODS
Patients
Patients with advanced solid tumors who were referred to the Department of Investigational Cancer Therapeutics at The University of Texas MD Anderson Cancer Center were invited to participate in an institutional review board–approved liquid biopsy protocol (LAB10-0334) allowing for serial blood sample collection before, during, and after therapy. Eligible patients had at least one known mutation detected in the tumor tissue with standard-of-care molecular testing. Informed consent was obtained from all participants before any study-related procedure.
All patients underwent imaging at baseline and at their first restaging, which was done between weeks 6 and 10 of therapy. Therapy response was assessed using RECIST 1.1.16 Patients were classified as responders (patients with a complete response [CR] or partial response [PR]) or nonresponders (patients with stable disease [SD] or progressive disease [PD]) and as progressors (patients with PD) or nonprogressors (patients with CR, PR, or SD).
ctDNA Assessment
Serial peripheral blood samples were collected into EDTA tubes at baseline (cycle 1, day −21 to day 8), mid-treatment (cycle 1, day 21 ± 2 weeks), first restaging (cycle 1, day of first restaging ± 2 weeks), and whenever possible throughout therapy. Plasma samples were obtained by two-step centrifugation, as previously described,18 and was stored at −80°C until processing. We extracted cfDNA from an average of 4 mL plasma using QIAamp Circulating Nucleic Acid Kit (QIAGEN, Germantown, MD) according to the manufacturer's protocol. Extracted cfDNA was stored at −20°C until further testing. DNA was quantified using a Quant-iT PicoGreen dsDNA Assay Kit (Thermo Fisher, Waltham, MA) on a SpectraMax M2 Microplate Reader (Molecular Devices, San Jose, CA). Serial dilutions were used to prepare reference standard curves for accurate cfDNA quantification (Data Supplement).
Using the known molecular profile of tumor tissue, we performed digital droplet PCR (ddPCR) ctDNA molecular testing with a Qx200 Droplet Digital PCR System (Bio-Rad, Hercules, CA) for plasma samples at all available time points, regardless of the baseline samples' ctDNA analysis results. Dual-labeled (HEX or FAM) fluorescent probes for wild-type loci and clinically relevant molecular alterations in PIK3CA, TP53, ESR1, NF1, KRAS, BRAF, KIT, NRAS, NOTCH1, AKT1, IDH1, and IDH2 were used. PCR mixtures were prepared using ddPCR Supermix for Probes (No dUTP; Bio-Rad) and added to 96-well plates according to the ddPCR protocol. Data were analyzed using QuantaSoft 1.7.4 (Bio-Rad), and ctDNA aggregate variant allele frequency (VAF) was quantified for all detected molecular alterations.
Because tumors can shed different amounts of ctDNA, we classified tumors as high shedders (cancer types with ≥ 75% of patients with detected ctDNA) or low shedders (cancer types with < 75% of patients with detected ctDNA) on the basis of their estimated ctDNA-shedding potential using a previously published stratification approach.19
Delta values (defined as the difference in VAF between two time points) and slope values (calculated using the formula , where x and y are the sample means) were calculated at mid-treatment and first restaging and tested for associations with clinical outcomes. Delta and slope value positivity was defined as any value above 0 and was used to determine the possibility of using longitudinal ctDNA monitoring to predict treatment outcomes.
We defined ctDNA progression as the first occurrence of a high ctDNA level after the ctDNA nadir. The date of ctDNA progression was recorded, and continuation of treatment beyond ctDNA progression was claimed when the difference between the timing of the ctDNA progression and treatment completion was more than 2 weeks. The time after progression was defined as the duration between the date of treatment completion and that of ctDNA progression.
Statistical Analysis
Statistical analysis was performed using IBM SPSS Statistics for Windows, version 26.0. In patients who received multiple lines of therapy, each therapy was analyzed separately. Descriptive data consisted of frequencies and percentages for categorical variables and means, medians, standard deviations, ranges, and interquartile ranges for continuous variables. Chi-square, Fisher exact, and Mann-Whitney tests were used to assess statistical inference, as appropriate. The Kaplan-Meier method was used to analyze the time to treatment failure (TTF), defined as the time from the date of treatment initiation to that of treatment completion. Log-rank test was used to assess differences between subgroups. Whenever applicable, all tests were two-sided. P values less than .05 were considered significant.
RESULTS
Patient Characteristics
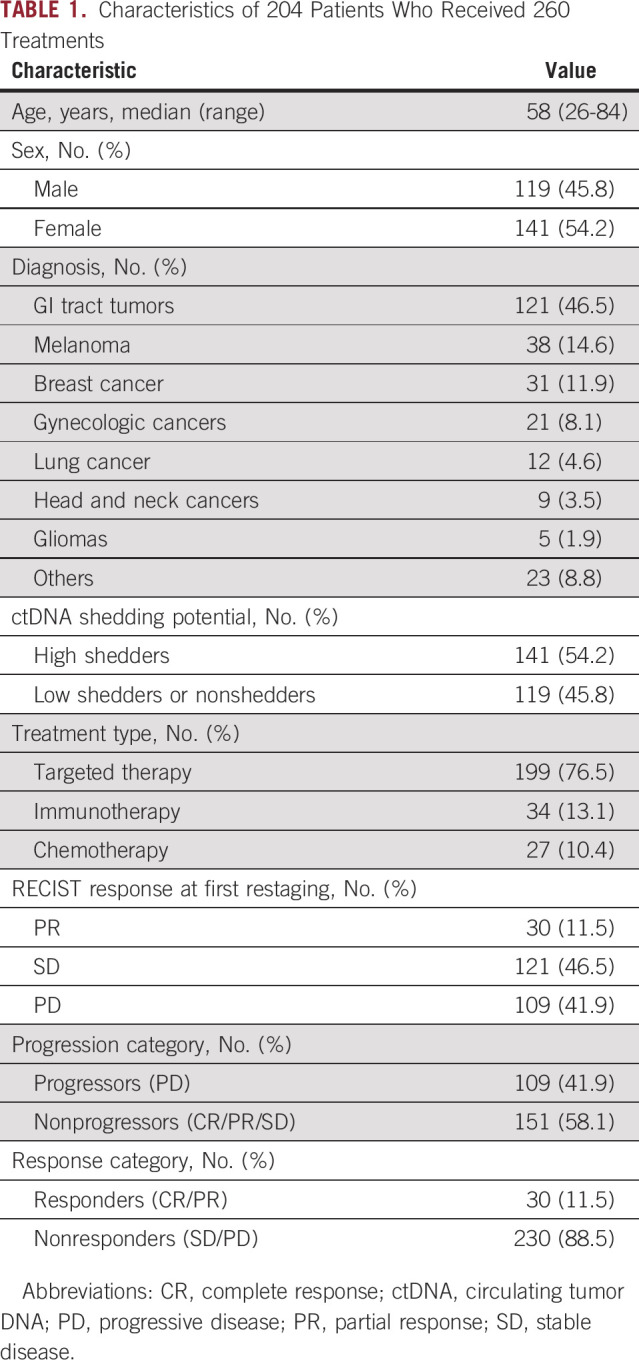
This study included 260 treatment courses administered to 204 patients between January 2012 and December 2019. Patients' median age at treatment initiation was 58 years. Detailed patient characteristics are provided in Table 1.
TABLE 1.
Characteristics of 204 Patients Who Received 260 Treatments
ctDNA Detection and Response to Therapy
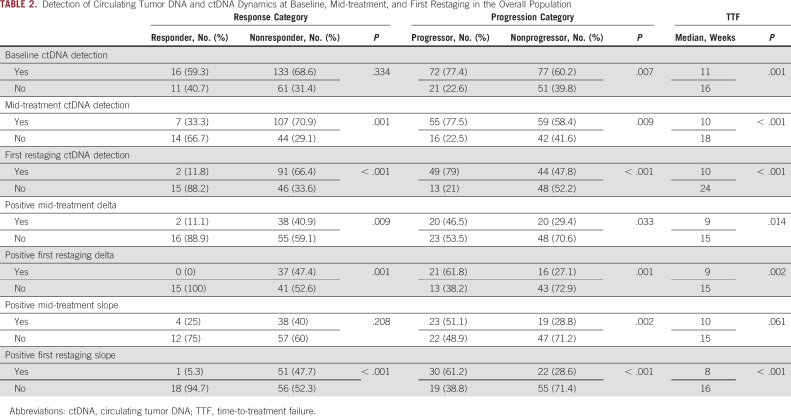
We detected ctDNA in 149 patients (67.4%) at baseline (approximately day 1 of cycle 1), in 114 (66.3%) at mid-treatment (approximately day 21 of cycle 1), and in 93 (60.4%) at first restaging (after approximately 6-9 weeks of therapy). Progressors (patients with PD at first restaging) had a higher ctDNA detection rate than nonprogressors (patients with SD, PR, or CR at first restaging) at baseline (72/93 [77.4%] v 77/128 [60.2%]; P = .007), mid-treatment (55/71 [77.5%] v 59/101 [58.4%]; P = .009), and first restaging (49/62 [79%] v 44/92 [47.8%]; P < .001). Furthermore, nonresponders (patients with PD or SD at first restaging) had a higher ctDNA detection rate than responders (patients with PR or CR at first restaging) at mid-treatment (107/148 [70.9%] v 7/21 [33.3%]; P = .001) and first restaging (91/137 [66.4%] v 2/17 [11.8%]; P < .001) but not baseline (133/194 [68.6%] v. 16/27 [59.3%]; P = .334; Table 2). Quantitatively, the median aggregate VAF, which represented the sum of the VAFs for all detected somatic alterations, was higher in progressors than in nonprogressors at baseline (2.1% v 0.3%; P = .001), mid-treatment (3.9% v 3.5%; P < .001), and first restaging (5.8% v 0%; P < .001). Moreover, the median aggregate VAF was significantly higher in nonresponders than in responders at mid-treatment (1.2% v 0%; P < .001) and first restaging (1.8% v 0%; P < .001) but not baseline (0.9% v 0.2%; P = .105; Data Supplement).
TABLE 2.
Detection of Circulating Tumor DNA and ctDNA Dynamics at Baseline, Mid-treatment, and First Restaging in the Overall Population
Subgroup analyses showed no significant differences in the response outcomes in patients with tumors of high versus low ctDNA shedding potential or among different treatment types. Details are shown in the Data Supplement.
ctDNA Detection and TTF
The presence of detectable ctDNA was associated with a shorter median TTF than the absence of detectable ctDNA at baseline (11 weeks; 95% CI, 9.1 to 12.9 v 16 weeks; 95% CI, 12.3 to 19.7; P = .001), mid-treatment (10 weeks; 95% CI, 7.9 to 12.1 v 18 weeks; 95% CI, 10.5 to 25.5; P < .001), and first restaging (10 weeks; 95% CI, 8.5 to 11.5 v 24 weeks; 95% CI, 17.3 to 30.7; P < .001; Table 2, Fig 1).
FIG 1.
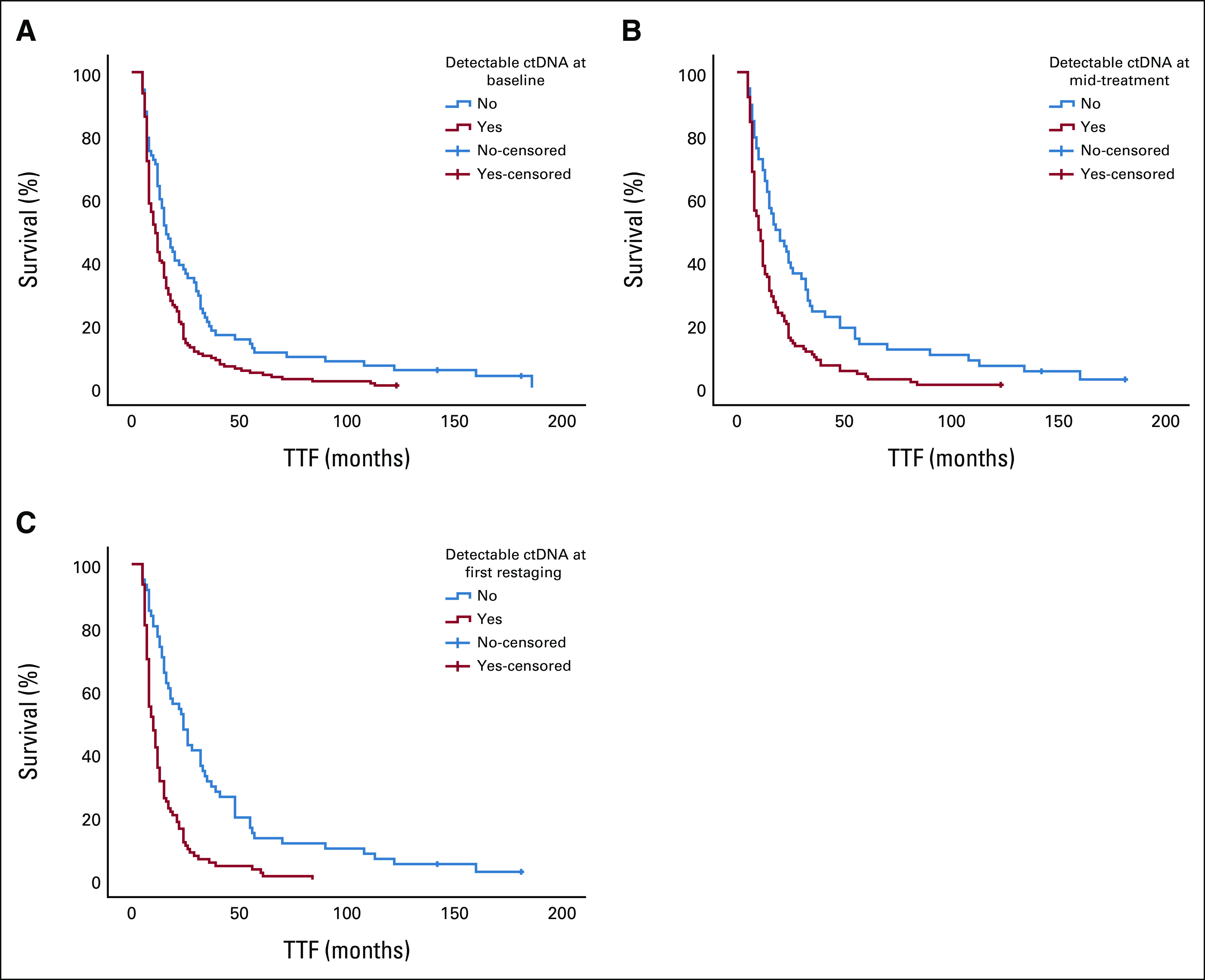
Kaplan-Meier curves for the differences in median TTF between patients with detectable and undetectable ctDNA at (A) baseline, (B) mid-treatment, and (C) first restaging. Detectable ctDNA was associated with a shorter median TTF than undetectable ctDNA at baseline (11 weeks; 95% CI, 9.1 to 12.9 v 16 weeks; 95% CI, 12.3 to 19.7, respectively; P = .001), mid-treatment (10 weeks; 95% CI, 7.9 to 12.1 v 18 weeks; 95% CI, 10.5 to 25.5, respectively; P < .001), and first restaging (10 weeks; 95% CI, 8.5 to 11.5 v 24 weeks; 95% CI, 17.3 to 30.7, respectively; P < .001). ctDNA, circulating tumor DNA; TTF, time-to-treatment failure.
Next, we tested whether the ctDNA quantity was associated with TTF by using different cutoffs to separate two groups (ie, median, 5% trimmed mean), which eliminated extreme high and low values. These analyses showed that patients with ctDNA quantities above the median or 5% trimmed mean had shorter TTF. Details are presented in the Data Supplement.
Dynamic Changes in ctDNA and Clinical Outcomes
Positive delta values for changes in ctDNA quantity from baseline to mid-treatment and first restaging (Fig 2) were more frequently observed in progressors (29/55, 52.7% v 32/92, 34.8%; P = .033) than in nonprogressors (30/50, 60% v 23/79, 29.1%; P = .001) and in nonresponders (17/92, 18.5% v 2/55, 3.6%; P = .009) than in responders (16/79, 20.3% v 0/50, 0%; P = .001; Table 2). Moreover, patients with positive delta values at mid-treatment and first restaging had a shorter median TTF than patients with nonpositive delta values (9 weeks, 95% CI, 7.4 to 10.6 v 15 weeks, 95% CI, 12.4 to 17.6; P = .014; Fig 3A; and 9 weeks, 95% CI, 7.1 to 10.9 v 15 weeks, 95% CI, 12.4 to 17.6, P = .002; Fig 3B). Subgroup analyses according to high versus low shedding potential demonstrated similar trends in the overall population. Details are presented in the Data Supplement.
FIG 2.
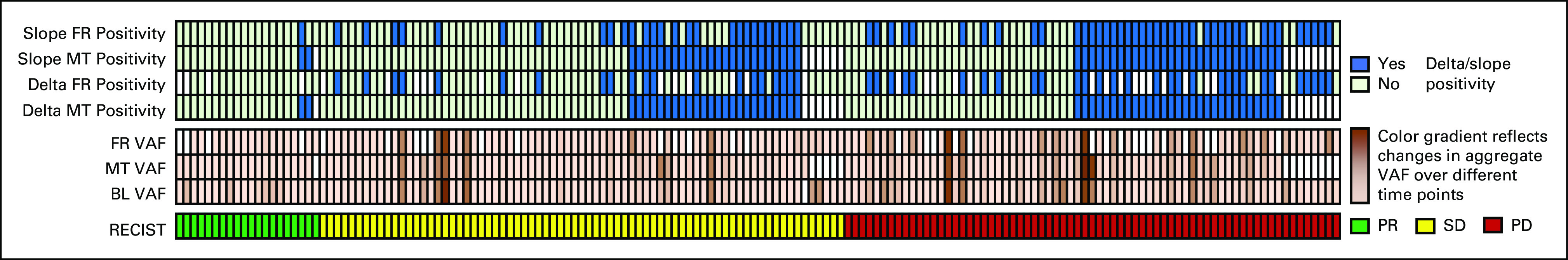
Studied variables and treatment response as assessed with the RECIST, version 1.1. Positive delta and slope values are highlighted in blue. Color gradients in the middle of the figure show the VAF changes at different time points. BL, baseline; FR, first restaging; MT, mid-treatment; PD, progressive disease; PR, partial response; SD, stable disease; VAF, variant allele frequency.
FIG 3.
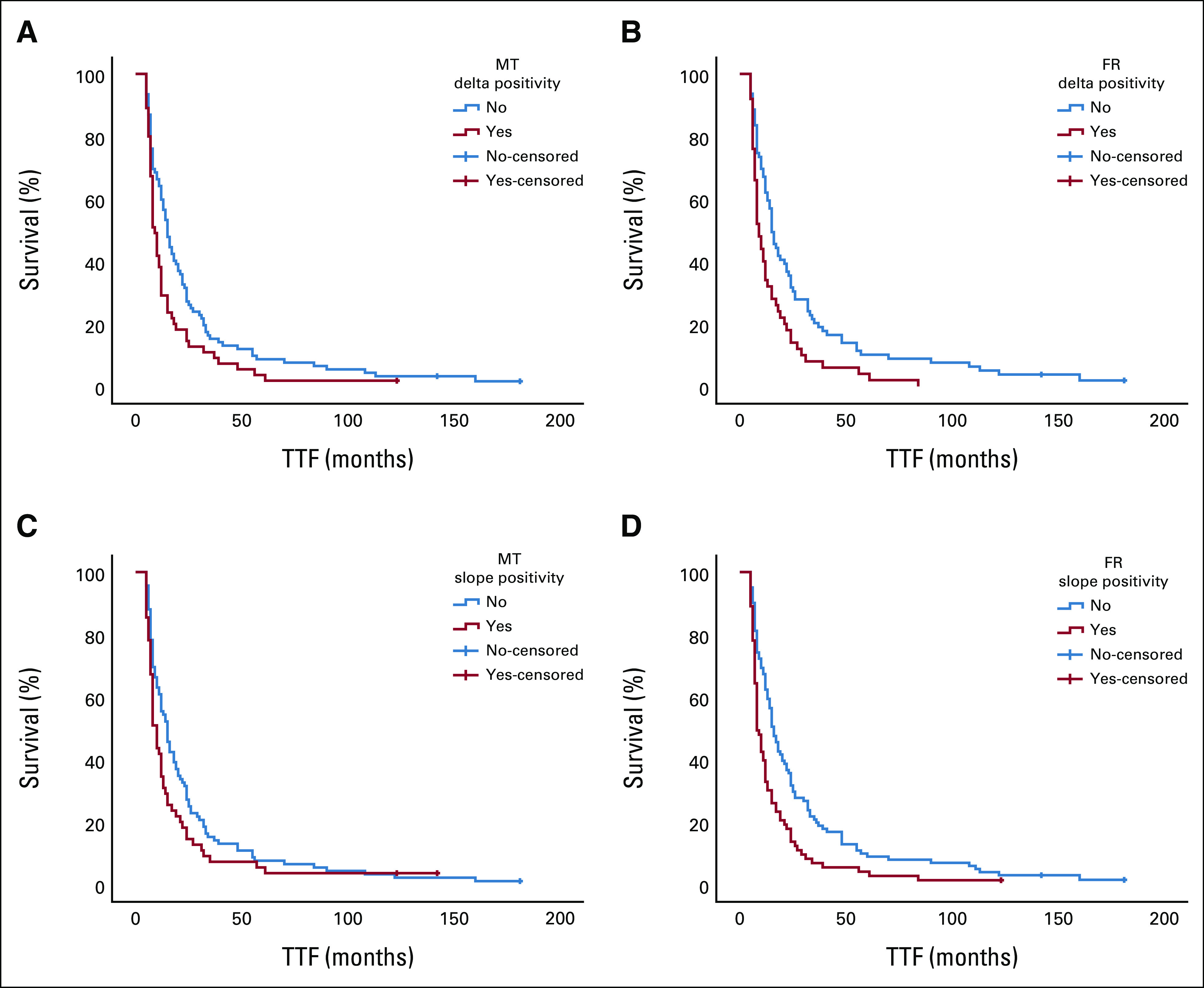
Kaplan-Meier curves demonstrate the differences in TTF between patients with positive and nonpositive delta values at (A) MT and (B) FR and the differences in TTF between patients with positive and nonpositive slope values at (C) MT and (D) FR. FR, first restaging; MT, mid-treatment; TTF, time-to-treatment failure.
To eliminate the impact of fluctuating ctDNA levels and address the overall trend of ctDNA dynamic changes, we calculated slope values and reanalyzed the data using slope positivity/negativity as a reflection of trends (Fig 2). Similar to the results of the delta value analysis, the results of this analysis showed that patients with positive slope values were more frequently progressors than nonprogressors (43/73, 58.9% v 30/101, 29.7%; P < .001) and more frequently nonresponders than responders (19/101, 18.8% v 1/73, 1.4%; P < .001). A positive slope value was also associated with a shorter median TTF than was a nonpositive slope value (8 weeks, 95% CI, 6.4 to 9.6 v 16 weeks, 95% CI, 13.2 to 18.8; P < .001; Table 2).
Next, we analyzed the relationship between the timing of ctDNA progression and clinical or radiologic progression. Of the 91 patients (35% of the overall population) in whom ctDNA levels were monitored throughout the course of treatment until the end of therapy (± 2 weeks), ctDNA progression preceded or occurred with PD in 66 (72.5%) with a median lead time of 23 days (range, 0-406 days; Data Supplement). Patients who continued treatment beyond ctDNA progression had clinical or radiologic progression within a median time of 5 weeks.
ctDNA Response Criteria
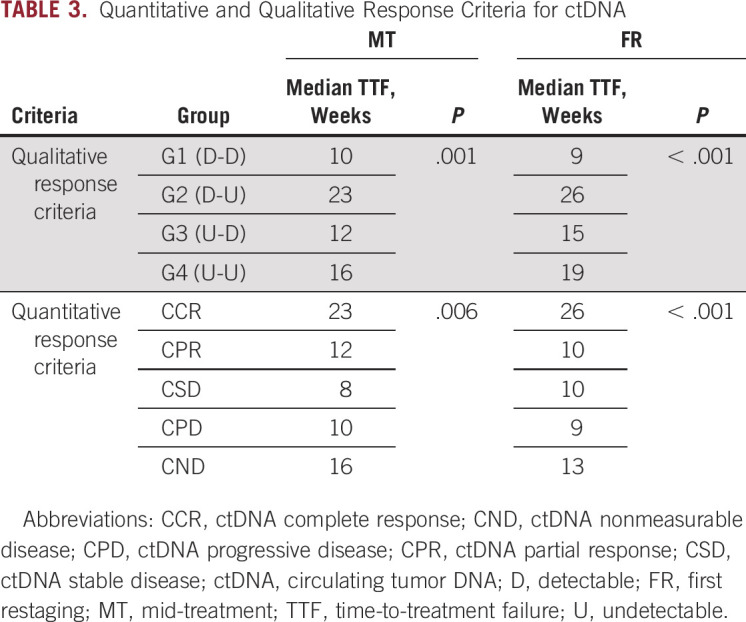
We propose ctDNA response criteria potentially applicable to clinical practice by separating patients into distinctive qualitative and quantitative groups that differed by their median TTFs. To evaluate the qualitative response criteria, patients were divided into four groups: G1 (patients with detectable ctDNA remaining detectable during therapy), G2 (those with detectable ctDNA becoming undetectable during therapy), G3 (those with undetectable ctDNA becoming detectable on therapy), and G4 (those with undetectable ctDNA remaining undetectable on therapy). The quantitative response criteria depended on the percent change of aggregate VAF and included five patient groups: ctDNA CR (ctDNA clearance after initial detectability), ctDNA PR (decrease of more than 10% in VAF), ctDNA PD (increase of more than 10% in VAF or de novo ctDNA detection), ctDNA SD (no increase or < 10% increase or decrease in VAF), and ctDNA nonmeasurable disease (undetectable ctDNA before and after treatment). Using the qualitative response criteria, we found that the patients in the G2 group had the longest TTF among all of the four qualitative groups we identified. Those who had detectable baseline ctDNA that remained detectable at mid-treatment and first restaging had the worst outcomes. Using the quantitative response criteria, we determined that patients with ctDNA CRs at mid-treatment or first restaging had longer TTFs (P = .006 and < .001, respectively) than patients with ctDNA PRs or, progressive, stable, and nonmeasurable disease (Table 3).
TABLE 3.
Quantitative and Qualitative Response Criteria for ctDNA
DISCUSSION
In our study, we systematically investigated ctDNA detection and dynamic changes in ctDNA quantity as potential surrogate markers for clinical outcomes in 204 patients with advanced cancers who were treated with 260 systemic therapies. The detection and high quantity of ctDNA at any prespecified time point (baseline, mid-treatment, and first restaging) were associated with a lack of both treatment response and disease control.
Multiple studies have reported that ctDNA detection in patients with advanced cancers may be associated with shorter progression-free survival (PFS) and overall survival (OS) durations.20-24 However, there is much less evidence supporting an association between ctDNA detection and response to therapy. A study involving the detection of circulating exosomal nucleic acids and ctDNA before therapy demonstrated an association between increased quantities of mutant exosomal content and lack of response or durable disease control per RECIST.18 Another study that used targeted methylation sequencing to detect ctDNA demonstrated a correlation between the magnitude of treatment response per RECIST and baseline ctDNA quantity as measured by methylation score (r = 0.32, P = .03).25 Our study also demonstrated that ctDNA quantity during treatment provided valuable insight into the treatment response at first restaging.
We also determined that patients with presence and/or high quantity of ctDNA collected at prespecified time points (baseline, mid-treatment, and first restaging) demonstrated shorter median TTFs than patients without or with a low quantity of ctDNA. For samples collected at baseline, this was not unexpected. For instance, Santiago-Walker et al,22 in a study of patients with BRAF V600–positive melanoma treated with dabrafenib or trametinib, found that baseline detection of BRAF mutations in ctDNA was an independent prognostic factor for PFS and that patients with BRAF-negative ctDNA had longer PFS and OS. Another study exploring the potential of testing baseline ctDNA for BRAF mutations to predict treatment outcomes in patients with advanced cancers suggested that higher BRAF V600E VAFs were associated with shorter OS.21 In patients with BRAF V600 mutations in tissue, the presence of BRAF V600 mutant ctDNA was also associated with a shorter median TTF.21 Another study by Zhang et al26 investigated the prognostic and predictive ability of ctDNA in advanced cancers treated with immune checkpoint inhibitors and found that a higher pretreatment VAF was associated with poorer OS. Whereas all these studies explored the potential to predict outcomes using ctDNA collected before therapy, our study demonstrated that detectable ctDNA at mid-treatment and first restaging was also associated with worse clinical outcomes. These data suggest that ctDNA detection can be used to supplement conventional imaging as a means of assessing treatment responses, especially if imaging response assessments are inconclusive.
Because ctDNA has a short half-life,11 dynamic changes in ctDNA quantity can be used as an early indicator of clinical outcomes. We demonstrated that dynamic changes measured by delta or slope values were associated with treatment responses and TTF. For instance, compared with nonpositive delta and slope values, positive delta and slope values were associated with a higher frequency of disease progression (defined by the number of progressors) and a shorter TTF. Therefore, our findings suggest that ctDNA changes can indicate treatment efficacy even at the mid-treatment time point. Furthermore, we noticed that ctDNA could be used to detect disease progression before or at the time of imaging in 72.5% of patients (median lead time, 23 days). It is not immediately clear whether this lead time was related to the underlying biology of ctDNA or whether it was affected by uncertainties in the protocols and the timing of sample collection. Our results, however, are consistent with data available from prior studies that suggested that ctDNA is a promising predictive biomarker in patients with advanced solid tumors.14,15 For example, Zhang and colleagues26 suggested that ctDNA dynamics during treatment are predictive of immunotherapy outcomes. Specifically, early ctDNA reduction correlated with improved PFS, OS, and responses to therapy. In alignment with our subgroup analysis of immunotherapy-treated cohort, other studies have also shown ctDNA's potential in discriminating between true and pseudoprogression. In addition, ctDNA clearance has been proposed as a potential tool for selecting patients for elective immunotherapy discontinuation.27-29 Studies in patients treated with other modalities also have been consistent regarding the value of detecting ctDNA changes. For example, two studies suggested that early ctDNA changes can be used as surrogates for clinical outcomes in patients with metastatic colorectal cancer and in patients with advanced breast cancer.12,30
We were encouraged to find that ctDNA dynamic changes at mid-treatment and at first restaging were associated with qualitative and quantitative clinical outcomes, which suggests that ctDNA analysis can be used in lieu of radiologic imaging to evaluate treatment outcomes. Similar to our results, a study by Thompson et al31 showed that molecular responders (> 50% decrease in VAF) had significantly longer PFS and OS compared with molecular nonresponders. Moreover, Zhang et al26 proposed that the ctDNA molecular response can predict long-term survival. Therefore, we proposed quantitative and qualitative ctDNA response criteria that can be used to separate patients into distinctive groups that differ by a median TTF. These criteria, if validated, may be used as an alternative to imaging and would be of substantial clinical usefulness, especially in scenarios in which disease is hard to measure or evaluate.
Our study had several limitations. First, it was retrospective, and the time points for blood sample collection varied. Additionally, we enrolled patients with diverse cancer types treated with various therapies, which added to the study heterogeneity. Shedding of ctDNA can differ among different cancer types, which could have affected our results although we performed separate subanalyses for tumors with high and low ctDNA shedding potential and obtained consistent results. Finally, we used unamplified ddPCR and the tumor-informed approach for ctDNA detection. More comprehensive approaches, such as targeted next-generation sequencing, which can test dozens or hundreds of genes, can better reflect clonal evolution and result in different reported quantities of ctDNA.5,32 Nevertheless, our results offer a proof of concept for the development of ctDNA-based approaches for predicting cancer therapy outcomes. Our results should be validated in future prospective studies.
ACKNOWLEDGMENT
The authors would like to thank Laura L Russel from The Research Medical Library at The University of Texas MD Anderson Cancer Center for manuscript editing.
Sarina A. Piha-Paul
Consulting or Advisory Role: CRC Oncology
Research Funding: AbbVie (Inst), Aminex (Inst), Biomarin (Inst), Boehringer Ingelheim (Inst), Bristol Myers Squib (Inst), Cerulean Pharma (Inst), Chugai Pharma (Inst), Curis (Inst), Five Prime Therapeutics (Inst), Genmab (Inst), GlaxoSmithKline (Inst), Helix BioPharma (Inst), Incyte (Inst), Jacobio (Inst), MedImmune (Inst), Medivation (Inst), Merck Sharp and Dohme Corp (Inst), Novartis (Inst), Pieris Pharmaceuticals (Inst), Pfizer (Inst), Principa Biopharma (Inst), Puma Biotechnology (Inst), RAPT Therapeutics (Inst), Seattle Genetics (Inst), Taiho Oncology (Inst), Tesaro (Inst), TransThera Biosciences (Inst), Amphivena Therapetics, Inc (Inst), Alkermes (Inst), Daichi Sanko (Inst), Lilly (Inst), ABM (Inst), Acepodia (Inst), ENB Therapeutics (Inst), Gene Quantum (Inst), Silverback Therapeutics (Inst), NIH/NCI (Inst), Cyclacel (Inst), F-Star Beta Limited (Inst), F-Star Therapeutics, Ltd (Inst), HiberCell (Inst), Immunomedics (Inst), Lytix Biopharma (Inst), Synologic Therapeutics (Inst), Gilead Sciences (Inst), Phanes Therapeutics (Inst), Purinomia Biotech, Inc (Inst), ZielBio, Inc (Inst)
S. Greg Call
Employment: Tempus, AstraZeneca
Daniel D. Karp
Research Funding: Phosplatin Therapeutics (Inst)
Travel, Accommodations, Expenses: Phosplatin Therapeutics
Siqing Fu
Research Funding: Novartis (Inst), NeuPharma, Inc (Inst), BeiGene (Inst), MacroGenics (Inst), BioAtla (Inst), Parexel International, LLC (Inst), Boehringer Ingelheim (Inst), Abbisko (Inst), Lilly (Inst), Hookipa Pharma (Inst), IMV (Inst), Innovent Biologics (Inst), Lyvgen Biopharma (Inst), Millennium (Inst), Nerviano Medical Sciences (Inst), NIH/NCI (Inst), Sellas Life Sciences (Inst), Soricimed (Inst), NovoCure (Inst), Turnstone Bio (Inst), Taiho Oncology (Inst), NCCN (Inst), Exelis (Inst), K-Group Beta (Inst), NextCure (Inst), Ningbo NewBay Medical Technology (Inst), PureTech (Inst), SQZ Biotech (Inst), Sumitomo Dainippon Pharma Oncology (Inst), Treadwell Therapeutics (Inst), Tyligand Bioscience (Inst), Vaccibody (Inst), Greenfire Bio (Inst)
Aung Naing
Consulting or Advisory Role: Novartis, CytomX Therapeutics, OncoSec, STCube Pharmaceuticals Inc, Kymab, Takeda (I), CSL Behring (I), Horizon Pharma (I), Genome & Company, Deka Biosciences, Pharming NV (I)
Research Funding: NCI, EMD Serono, MedImmune, Atterocor, Amplimmune, ARMO BioSciences, Karyopharm Therapeutics, Incyte, Novartis, Regeneron, Merck, Bristol Myers Squibb, Pfizer, CytomX Therapeutics, Neon Therapeutics, Calithera Biosciences, TopAlliance BioSciences Inc, Healios, Lilly, Kymab, PsiOxus Therapeutics, Immune Deficiency Foundation (I), Arcus Biosciences, NeoImmuneTech, ImmuneOncia, Surface Oncology, Baxalta (I), Jeffrey Modell Foundation (I), Chao Physician-Scientist Awards (I), Monopteros Therapeutics, BioNTech SE, Seven and Eight Biopharmaceuticals, Sotio
Travel, Accommodations, Expenses: ARMO BioSciences
Vivek Subbiah
Consulting or Advisory Role: MedImmune, Helsinn Therapeutics, Loxo, R-Pharm, QED Therapeutics
Research Funding: Novartis (Inst), GlaxoSmithKline (Inst), NanoCarrier (Inst), Northwest Biotherapeutics (Inst), Genentech/Roche (Inst), Berg Pharma (Inst), Bayer (Inst), Incyte (Inst), Fujifilm (Inst), PharmaMar (Inst), D3 Oncology Solutions (Inst), Pfizer (Inst), AbbVie (Inst), Multivir (Inst), Blueprint Medicines (Inst), LOXO (Inst), Vegenics (Inst), Takeda (Inst), Alfasigma (Inst), Agensys (Inst), Idera (Inst), Boston Biomedical (Inst), Inhibrx (Inst), Exelixis (Inst), Amgen (Inst), Turning Point Therapeutics (Inst)
Travel, Accommodations, Expenses: PharmaMar, Bayer, Novartis, Helsinn Therapeutics
Other Relationship: Medscape
Shubham Pant
Honoraria: 4D Pharma
Consulting or Advisory Role: Xencor, Zymeworks, Ipsen, 4D Pharma, Novartis, Janssen
Research Funding: Mirati Therapeutics (Inst), Lilly (Inst), RedHill Biopharma (Inst), Xencor (Inst), Five Prime Therapeutics (Inst), Novartis (Inst), Rgenix (Inst), Sanofi/Aventis (Inst), ArQule (Inst), Bristol Myers Squibb (Inst), Onco Response (Inst), GlaxoSmithKline (Inst), Ipsen (Inst), Astellas Pharma (Inst), Purple Biotech (Inst), 4D Pharma (Inst), Boehringer Ingelheim (Inst), NGM Biopharmaceuticals (Inst), Janssen (Inst), Arcus Biosciences (Inst), Elicio Therapeutics (Inst)
Apostolia M. Tsimberidou
Consulting or Advisory Role: Vincerx Pharma, Diaccurate
Research Funding: IMMATICS (Inst), Karus Therapeutics (Inst), OBI Pharma (Inst), Tempus (Inst), Parker Institute for Cancer Immunotherapy (Inst), Agenus (Inst), Novocure (Inst), Tvardi Therapeutics (Inst)
David S. Hong
Stock and Other Ownership Interests: OncoResponse, Telperian, MolecularMatch
Consulting or Advisory Role: Bayer, Guidepoint Global, Gerson Lehrman Group, Alphasights, Axiom Biotechnologies, Medscape, Numab, Pfizer, Takeda, Trieza Therapeutics, WebMD, Infinity Pharmaceuticals, Amgen, Adaptimmune, Boxer Capital, EcoR1 Capital, Tavistock Life Sciences, Baxter, COG, Genentech, GroupH, Janssen, Acuta, HCW Precision, Prime Oncology, ST Cube, Alkermes, AUM Biosciences, Bridgebio, Cor2Ed, Gilead Sciences, Immunogen, Liberum, Oncologia Brasil, Pharma Intelligence, Precision Oncology Experimental Therapeutics, Turning Point Therapeutics, ZIOPHARM Oncology, Cowen, Gennao Bio, MedaCorp, YingLing Pharma, RAIN
Research Funding: Genentech (Inst), Amgen (Inst), Daiichi Sankyo (Inst), Adaptimmune (Inst), AbbVie (Inst), Bayer (Inst), Infinity Pharmaceuticals (Inst), Kite, a Gilead company (Inst), MedImmune (Inst), National Cancer Institute (Inst), Fate Therapeutics (Inst), Pfizer (Inst), Novartis (Inst), Numab (Inst), Turning Point Therapeutics (Inst), Kyowa (Inst), Loxo (Inst), Merck (Inst), Eisai (Inst), Genmab (Inst), Mirati Therapeutics (Inst), Mologen (Inst), Takeda (Inst), AstraZeneca (Inst), Navire (Inst), VM Pharma (Inst), Erasca, Inc (Inst), Bristol Myers Squibb (Inst), Adlai Nortye (Inst), Seattle Genetics (Inst), Deciphera (Inst), Pyramid Biosciences (Inst), Lilly (Inst), Endeavor BioMedicines (Inst), F. Hoffmann LaRoche (Inst), Ignyta (Inst), Teckro (Inst), TCR2 Therapeutics (Inst)
Travel, Accommodations, Expenses: Genmab, Society for Immunotherapy of Cancer, Bayer Schering Pharma, ASCO, AACR, Telperian
Jordi Rodon
Consulting or Advisory Role: Peptomyc, Kelun Pharmaceuticals/Klus Pharma, Ellipses Pharma, Molecular Partners, iOnctura
Research Funding: Blueprint Medicines (Inst), Black Diamond Therapeutics (Inst), Merck Sharp & Dohme (Inst), Hummingbird (Inst), Yingli Pharma (Inst), Vall d'Hebron Institute of Oncology/Cancer Core Europe (Inst), Novartis (Inst), Spectrum Pharmaceuticals (Inst), Symphogen (Inst), BioAtla (Inst), Pfizer (Inst), Genmab (Inst), CytomX Therapeutics (Inst), Kelun (Inst), Takeda/Millennium (Inst), GlaxoSmithKline (Inst), Taiho Pharmaceutical (Inst), Roche (Inst), Bicycle Therapeutics (Inst), Merus (Inst), Curis (Inst), Bayer (Inst), AADi (Inst), Nuvation Bio (Inst), Fore Biotherapeutics (Inst), BioMed Valley Discoveries (Inst), Loxo (Inst), Hutchison MediPharma (Inst), Cellestia Biotech (Inst), Deciphera (Inst), IDEAYA Biosciences (Inst), Amgen (Inst), Tango Therapeutics (Inst), Mirati Therapeutics (Inst), Linnaeus Therapeutics (Inst)
Travel, Accommodations, Expenses: ESMO
Other Relationship: Vall d'Hebron Institute of Oncology/Ministerio De Empleo Y Seguridad Social, Chinese University of Hong Kong, Boxer Capital, Tang Advisors
Funda Meric-Bernstam
Employment: MD Anderson Cancer Center
Consulting or Advisory Role: Xencor, Debiopharm Group, Roche, PACT Pharma, eFFECTOR Therapeutics, Kolon Life Sciences, Tyra Biosciences, Zymeworks, Zentalis, Infinity Pharmaceuticals, AbbVie, Black Diamond Therapeutics, Eisai, OnCusp Therapeutics, Lengo Therapeutics, Tallac Therapeutics, Karyopharm Therapeutics, Biovica, AstraZeneca, Seattle Genetics, Loxo, Silverback Therapeutics
Research Funding: Novartis (Inst), AstraZeneca (Inst), Taiho Pharmaceutical (Inst), Genentech (Inst), Calithera Biosciences (Inst), Debiopharm Group (Inst), Bayer (Inst), Aileron Therapeutics (Inst), PUMA Biotechnology (Inst), CytomX Therapeutics (Inst), Jounce Therapeutics (Inst), Zymeworks (Inst), Curis (Inst), Pfizer (Inst), eFFECTOR Therapeutics (Inst), AbbVie (Inst), Boehringer Ingelheim (Inst), Guardant Health (Inst), Daiichi Sankyo (Inst), GlaxoSmithKline (Inst), Seattle Genetics (Inst), Klus Pharma (Inst), Takeda (Inst)
Filip Janku
Employment: Monte Rosa Therapeutics
Leadership: Monte Rosa Therapeutics
Stock and Other Ownership Interests: Cardiff Oncology, Monte Rosa Therapeutics
Consulting or Advisory Role: Deciphera, Novartis, Sequenom, Foundation Medicine, Guardant Health, Synlogic, Valeant/Dendreon, IFM Therapeutics, Sotio, PureTech, Jazz Pharmaceuticals, Immunomet, IDEAYA Biosciences, Cardiff Oncology, Fore Biotherapeutics
Research Funding: Novartis (Inst), BioMed Valley Discoveries (Inst), Roche (Inst), Agios (Inst), Astellas Pharma (Inst), Deciphera (Inst), Plexxikon (Inst), Piqur (Inst), Fujifilm (Inst), Symphogen (Inst), Bristol Myers Squibb (Inst), Asana Biosciences (Inst), Astex Pharmaceuticals (Inst), Genentech (Inst), Proximagen (Inst)
Other Relationship: Bio-Rad
No other potential conflicts of interest were reported.
PRIOR PRESENTATION
Presented at the ASCO Virtual Scientific Program, Virtual, May 29-31, 2020; and the 32nd EORTC-NCI-AACR Symposium on Molecular Targets and Cancer Therapeutics, virtual, October 24-25, 2020.
SUPPORT
Supported by the National Center for Advancing Translational Sciences (UL1 TR000371); the National Institutes of Health through MD Anderson Cancer Center’s Support Grant (P30CA016672); the National Cancer Institute (CA235620-01A1); the Rising Tide Foundation (CR-18-600); and the Andrew Sabin Family Foundation (F.J.).
DATA SHARING STATEMENT
Data will be available upon reasonable request from authors.
AUTHOR CONTRIBUTIONS
Conception and design: Mohamed A. Gouda, Helen J. Huang, S. Greg Call, Aung Naing, David S. Hong, Filip Janku
Financial support: David S. Hong
Administrative support: Vivek Subbiah, David S. Hong
Provision of study materials or patients: Sarina A. Piha-Paul, Daniel D. Karp, Siqing Fu, Aung Naing, Vivek Subbiah, Apostolia M. Tsimberidou, David S. Hong, Jordi Rodon, Funda Meric-Bernstam, Filip Janku
Collection and assembly of data: Mohamed A. Gouda, Helen J. Huang, Daniel D. Karp, Siqing Fu, Aung Naing, Vivek Subbiah, Apostolia M. Tsimberidou, David S. Hong, Jordi Rodon, Funda Meric-Bernstam, Filip Janku
Data analysis and interpretation: Mohamed A. Gouda, Helen J. Huang, Sarina A. Piha-Paul, Siqing Fu, Aung Naing, Vivek Subbiah, Shubham Pant, Derek J. Dustin, David S. Hong, Funda Meric-Bernstam, Filip Janku
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Sarina A. Piha-Paul
Consulting or Advisory Role: CRC Oncology
Research Funding: AbbVie (Inst), Aminex (Inst), Biomarin (Inst), Boehringer Ingelheim (Inst), Bristol Myers Squib (Inst), Cerulean Pharma (Inst), Chugai Pharma (Inst), Curis (Inst), Five Prime Therapeutics (Inst), Genmab (Inst), GlaxoSmithKline (Inst), Helix BioPharma (Inst), Incyte (Inst), Jacobio (Inst), MedImmune (Inst), Medivation (Inst), Merck Sharp and Dohme Corp (Inst), Novartis (Inst), Pieris Pharmaceuticals (Inst), Pfizer (Inst), Principa Biopharma (Inst), Puma Biotechnology (Inst), RAPT Therapeutics (Inst), Seattle Genetics (Inst), Taiho Oncology (Inst), Tesaro (Inst), TransThera Biosciences (Inst), Amphivena Therapetics, Inc (Inst), Alkermes (Inst), Daichi Sanko (Inst), Lilly (Inst), ABM (Inst), Acepodia (Inst), ENB Therapeutics (Inst), Gene Quantum (Inst), Silverback Therapeutics (Inst), NIH/NCI (Inst), Cyclacel (Inst), F-Star Beta Limited (Inst), F-Star Therapeutics, Ltd (Inst), HiberCell (Inst), Immunomedics (Inst), Lytix Biopharma (Inst), Synologic Therapeutics (Inst), Gilead Sciences (Inst), Phanes Therapeutics (Inst), Purinomia Biotech, Inc (Inst), ZielBio, Inc (Inst)
S. Greg Call
Employment: Tempus, AstraZeneca
Daniel D. Karp
Research Funding: Phosplatin Therapeutics (Inst)
Travel, Accommodations, Expenses: Phosplatin Therapeutics
Siqing Fu
Research Funding: Novartis (Inst), NeuPharma, Inc (Inst), BeiGene (Inst), MacroGenics (Inst), BioAtla (Inst), Parexel International, LLC (Inst), Boehringer Ingelheim (Inst), Abbisko (Inst), Lilly (Inst), Hookipa Pharma (Inst), IMV (Inst), Innovent Biologics (Inst), Lyvgen Biopharma (Inst), Millennium (Inst), Nerviano Medical Sciences (Inst), NIH/NCI (Inst), Sellas Life Sciences (Inst), Soricimed (Inst), NovoCure (Inst), Turnstone Bio (Inst), Taiho Oncology (Inst), NCCN (Inst), Exelis (Inst), K-Group Beta (Inst), NextCure (Inst), Ningbo NewBay Medical Technology (Inst), PureTech (Inst), SQZ Biotech (Inst), Sumitomo Dainippon Pharma Oncology (Inst), Treadwell Therapeutics (Inst), Tyligand Bioscience (Inst), Vaccibody (Inst), Greenfire Bio (Inst)
Aung Naing
Consulting or Advisory Role: Novartis, CytomX Therapeutics, OncoSec, STCube Pharmaceuticals Inc, Kymab, Takeda (I), CSL Behring (I), Horizon Pharma (I), Genome & Company, Deka Biosciences, Pharming NV (I)
Research Funding: NCI, EMD Serono, MedImmune, Atterocor, Amplimmune, ARMO BioSciences, Karyopharm Therapeutics, Incyte, Novartis, Regeneron, Merck, Bristol Myers Squibb, Pfizer, CytomX Therapeutics, Neon Therapeutics, Calithera Biosciences, TopAlliance BioSciences Inc, Healios, Lilly, Kymab, PsiOxus Therapeutics, Immune Deficiency Foundation (I), Arcus Biosciences, NeoImmuneTech, ImmuneOncia, Surface Oncology, Baxalta (I), Jeffrey Modell Foundation (I), Chao Physician-Scientist Awards (I), Monopteros Therapeutics, BioNTech SE, Seven and Eight Biopharmaceuticals, Sotio
Travel, Accommodations, Expenses: ARMO BioSciences
Vivek Subbiah
Consulting or Advisory Role: MedImmune, Helsinn Therapeutics, Loxo, R-Pharm, QED Therapeutics
Research Funding: Novartis (Inst), GlaxoSmithKline (Inst), NanoCarrier (Inst), Northwest Biotherapeutics (Inst), Genentech/Roche (Inst), Berg Pharma (Inst), Bayer (Inst), Incyte (Inst), Fujifilm (Inst), PharmaMar (Inst), D3 Oncology Solutions (Inst), Pfizer (Inst), AbbVie (Inst), Multivir (Inst), Blueprint Medicines (Inst), LOXO (Inst), Vegenics (Inst), Takeda (Inst), Alfasigma (Inst), Agensys (Inst), Idera (Inst), Boston Biomedical (Inst), Inhibrx (Inst), Exelixis (Inst), Amgen (Inst), Turning Point Therapeutics (Inst)
Travel, Accommodations, Expenses: PharmaMar, Bayer, Novartis, Helsinn Therapeutics
Other Relationship: Medscape
Shubham Pant
Honoraria: 4D Pharma
Consulting or Advisory Role: Xencor, Zymeworks, Ipsen, 4D Pharma, Novartis, Janssen
Research Funding: Mirati Therapeutics (Inst), Lilly (Inst), RedHill Biopharma (Inst), Xencor (Inst), Five Prime Therapeutics (Inst), Novartis (Inst), Rgenix (Inst), Sanofi/Aventis (Inst), ArQule (Inst), Bristol Myers Squibb (Inst), Onco Response (Inst), GlaxoSmithKline (Inst), Ipsen (Inst), Astellas Pharma (Inst), Purple Biotech (Inst), 4D Pharma (Inst), Boehringer Ingelheim (Inst), NGM Biopharmaceuticals (Inst), Janssen (Inst), Arcus Biosciences (Inst), Elicio Therapeutics (Inst)
Apostolia M. Tsimberidou
Consulting or Advisory Role: Vincerx Pharma, Diaccurate
Research Funding: IMMATICS (Inst), Karus Therapeutics (Inst), OBI Pharma (Inst), Tempus (Inst), Parker Institute for Cancer Immunotherapy (Inst), Agenus (Inst), Novocure (Inst), Tvardi Therapeutics (Inst)
David S. Hong
Stock and Other Ownership Interests: OncoResponse, Telperian, MolecularMatch
Consulting or Advisory Role: Bayer, Guidepoint Global, Gerson Lehrman Group, Alphasights, Axiom Biotechnologies, Medscape, Numab, Pfizer, Takeda, Trieza Therapeutics, WebMD, Infinity Pharmaceuticals, Amgen, Adaptimmune, Boxer Capital, EcoR1 Capital, Tavistock Life Sciences, Baxter, COG, Genentech, GroupH, Janssen, Acuta, HCW Precision, Prime Oncology, ST Cube, Alkermes, AUM Biosciences, Bridgebio, Cor2Ed, Gilead Sciences, Immunogen, Liberum, Oncologia Brasil, Pharma Intelligence, Precision Oncology Experimental Therapeutics, Turning Point Therapeutics, ZIOPHARM Oncology, Cowen, Gennao Bio, MedaCorp, YingLing Pharma, RAIN
Research Funding: Genentech (Inst), Amgen (Inst), Daiichi Sankyo (Inst), Adaptimmune (Inst), AbbVie (Inst), Bayer (Inst), Infinity Pharmaceuticals (Inst), Kite, a Gilead company (Inst), MedImmune (Inst), National Cancer Institute (Inst), Fate Therapeutics (Inst), Pfizer (Inst), Novartis (Inst), Numab (Inst), Turning Point Therapeutics (Inst), Kyowa (Inst), Loxo (Inst), Merck (Inst), Eisai (Inst), Genmab (Inst), Mirati Therapeutics (Inst), Mologen (Inst), Takeda (Inst), AstraZeneca (Inst), Navire (Inst), VM Pharma (Inst), Erasca, Inc (Inst), Bristol Myers Squibb (Inst), Adlai Nortye (Inst), Seattle Genetics (Inst), Deciphera (Inst), Pyramid Biosciences (Inst), Lilly (Inst), Endeavor BioMedicines (Inst), F. Hoffmann LaRoche (Inst), Ignyta (Inst), Teckro (Inst), TCR2 Therapeutics (Inst)
Travel, Accommodations, Expenses: Genmab, Society for Immunotherapy of Cancer, Bayer Schering Pharma, ASCO, AACR, Telperian
Jordi Rodon
Consulting or Advisory Role: Peptomyc, Kelun Pharmaceuticals/Klus Pharma, Ellipses Pharma, Molecular Partners, iOnctura
Research Funding: Blueprint Medicines (Inst), Black Diamond Therapeutics (Inst), Merck Sharp & Dohme (Inst), Hummingbird (Inst), Yingli Pharma (Inst), Vall d'Hebron Institute of Oncology/Cancer Core Europe (Inst), Novartis (Inst), Spectrum Pharmaceuticals (Inst), Symphogen (Inst), BioAtla (Inst), Pfizer (Inst), Genmab (Inst), CytomX Therapeutics (Inst), Kelun (Inst), Takeda/Millennium (Inst), GlaxoSmithKline (Inst), Taiho Pharmaceutical (Inst), Roche (Inst), Bicycle Therapeutics (Inst), Merus (Inst), Curis (Inst), Bayer (Inst), AADi (Inst), Nuvation Bio (Inst), Fore Biotherapeutics (Inst), BioMed Valley Discoveries (Inst), Loxo (Inst), Hutchison MediPharma (Inst), Cellestia Biotech (Inst), Deciphera (Inst), IDEAYA Biosciences (Inst), Amgen (Inst), Tango Therapeutics (Inst), Mirati Therapeutics (Inst), Linnaeus Therapeutics (Inst)
Travel, Accommodations, Expenses: ESMO
Other Relationship: Vall d'Hebron Institute of Oncology/Ministerio De Empleo Y Seguridad Social, Chinese University of Hong Kong, Boxer Capital, Tang Advisors
Funda Meric-Bernstam
Employment: MD Anderson Cancer Center
Consulting or Advisory Role: Xencor, Debiopharm Group, Roche, PACT Pharma, eFFECTOR Therapeutics, Kolon Life Sciences, Tyra Biosciences, Zymeworks, Zentalis, Infinity Pharmaceuticals, AbbVie, Black Diamond Therapeutics, Eisai, OnCusp Therapeutics, Lengo Therapeutics, Tallac Therapeutics, Karyopharm Therapeutics, Biovica, AstraZeneca, Seattle Genetics, Loxo, Silverback Therapeutics
Research Funding: Novartis (Inst), AstraZeneca (Inst), Taiho Pharmaceutical (Inst), Genentech (Inst), Calithera Biosciences (Inst), Debiopharm Group (Inst), Bayer (Inst), Aileron Therapeutics (Inst), PUMA Biotechnology (Inst), CytomX Therapeutics (Inst), Jounce Therapeutics (Inst), Zymeworks (Inst), Curis (Inst), Pfizer (Inst), eFFECTOR Therapeutics (Inst), AbbVie (Inst), Boehringer Ingelheim (Inst), Guardant Health (Inst), Daiichi Sankyo (Inst), GlaxoSmithKline (Inst), Seattle Genetics (Inst), Klus Pharma (Inst), Takeda (Inst)
Filip Janku
Employment: Monte Rosa Therapeutics
Leadership: Monte Rosa Therapeutics
Stock and Other Ownership Interests: Cardiff Oncology, Monte Rosa Therapeutics
Consulting or Advisory Role: Deciphera, Novartis, Sequenom, Foundation Medicine, Guardant Health, Synlogic, Valeant/Dendreon, IFM Therapeutics, Sotio, PureTech, Jazz Pharmaceuticals, Immunomet, IDEAYA Biosciences, Cardiff Oncology, Fore Biotherapeutics
Research Funding: Novartis (Inst), BioMed Valley Discoveries (Inst), Roche (Inst), Agios (Inst), Astellas Pharma (Inst), Deciphera (Inst), Plexxikon (Inst), Piqur (Inst), Fujifilm (Inst), Symphogen (Inst), Bristol Myers Squibb (Inst), Asana Biosciences (Inst), Astex Pharmaceuticals (Inst), Genentech (Inst), Proximagen (Inst)
Other Relationship: Bio-Rad
No other potential conflicts of interest were reported.
REFERENCES
- 1. Gambardella V, Tarazona N, Cejalvo JM, et al. Personalized medicine: Recent progress in cancer therapy. Cancers (Basel) 2020;12:1009. doi: 10.3390/cancers12041009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Overman MJ, Modak J, Kopetz S, et al. Use of research biopsies in clinical trials: Are risks and benefits adequately discussed? J Clin Oncol 3117–222013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Polivka J, Jr, Pesta M, Janku F.Testing for oncogenic molecular aberrations in cell-free DNA-based liquid biopsies in the clinic: Are we there yet? Expert Rev Mol Diagn 151631–16442015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lim JSJ, Janku F, Yap TA.Circulating tumor DNA-From bench to bedside Curr Probl Cancer 41212–2212017 [DOI] [PubMed] [Google Scholar]
- 5.Rolfo C, Mack P, Scagliotti GV, et al. Liquid biopsy for advanced NSCLC: A consensus statement from the International Association for the Study of Lung Cancer Res Social Adm Pharm 161647–16622021 [DOI] [PubMed] [Google Scholar]
- 6.Dasari A, Morris V, Allegra CJ, et al. ctDNA applications and integration in colorectal cancer: an NCI Colon and Rectal-Anal Task Forces whitepaper Nat Rev Clin Oncol 17757–7702020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.FDA Approval: cobas EGFR Mutation Test v2. 2016. [Google Scholar]
- 8.FDA Approval: FoundationOne Liquid CDx. 2020. [Google Scholar]
- 9.FDA Approval: Guardant360 CDx. 2020. [Google Scholar]
- 10.Crowley E, Di Nicolantonio F, Loupakis F, et al. Liquid biopsy: Monitoring cancer-genetics in the blood Nat Rev Clin Oncol 10472–4842013 [DOI] [PubMed] [Google Scholar]
- 11.Chen K, Zhao H, Shi Y, et al. Perioperative dynamic changes in circulating tumor DNA in patients with lung cancer (DYNAMIC) Clin Cancer Res 257058–70672019 [DOI] [PubMed] [Google Scholar]
- 12.Tie J, Kinde I, Wang Y, et al. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer Ann Oncol 261715–17222015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Husain H, Melnikova VO, Kosco K, et al. Monitoring daily dynamics of early tumor response to targeted therapy by detecting circulating tumor DNA in urine Clin Cancer Res 234716–47232017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dumbrava EE, Call SG, Huang HJ, et al. PIK3CA mutations in plasma circulating tumor DNA predict survival and treatment outcomes in patients with advanced cancers. ESMO Open. 2021;6:100230. doi: 10.1016/j.esmoop.2021.100230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lapin M, Huang HJ, Chagani S, et al. Monitoring of dynamic changes and clonal evolution in circulating tumor DNA from patients with IDH-mutated cholangiocarcinoma treated with isocitrate dehydrogenase inhibitors. JCO Precis Oncol. 2022;6:e2100197. doi: 10.1200/PO.21.00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1) Eur J Cancer 45228–2472009 [DOI] [PubMed] [Google Scholar]
- 17.Grothey A, Van Cutsem E, Sobrero A, et al. Regorafenib monotherapy for previously treated metastatic colorectal cancer (CORRECT): An international, multicentre, randomised, placebo-controlled, phase 3 trial Lancet 381303–3122013 [DOI] [PubMed] [Google Scholar]
- 18.Mohrmann L, Huang HJ, Hong DS, et al. Liquid biopsies using plasma exosomal nucleic acids and plasma cell-free DNA compared with clinical outcomes of patients with advanced cancers Clin Cancer Res 24181–1882018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janku F, Angenendt P, Tsimberidou AM, et al. Actionable mutations in plasma cell-free DNA in patients with advanced cancers referred for experimental targeted therapies Oncotarget 612809–128212015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janku F, Huang HJ, Claes B, et al. BRAF mutation testing in cell-free DNA from the plasma of patients with advanced cancers using a rapid, automated molecular diagnostics System Mol Cancer Ther 151397–14042016 [DOI] [PubMed] [Google Scholar]
- 22.Santiago-Walker A, Gagnon R, Mazumdar J, et al. Correlation of BRAF mutation status in circulating-free DNA and tumor and association with clinical outcome across four BRAFi and MEKi clinical trials Clin Cancer Res 22567–5742016 [DOI] [PubMed] [Google Scholar]
- 23.Schwaederle MC, Patel SP, Husain H, et al. Utility of genomic assessment of blood-derived circulating tumor DNA (ctDNA) in patients with advanced lung adenocarcinoma Clin Cancer Res 235101–51112017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pairawan S, Hess KR, Janku F, et al. Cell-free circulating tumor DNA variant allele frequency associates with survival in metastatic cancer Clin Cancer Res 261924–19312020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu L, Toung JM, Jassowicz AF, et al. Targeted methylation sequencing of plasma cell-free DNA for cancer detection and classification Ann Oncol 291445–14532018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Q, Luo J, Wu S, et al. Prognostic and predictive impact of circulating tumor DNA in patients with advanced cancers treated with immune checkpoint blockade Cancer Discov 101842–18532020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anagnostou V, Forde PM, White JR, et al. Dynamics of tumor and immune responses during immune checkpoint blockade in non-small cell lung cancer Cancer Res 791214–12252019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hellmann MD, Nabet BY, Rizvi H, et al. Circulating tumor DNA analysis to assess risk of progression after long-term response to PD-(L)1 blockade in NSCLC Clin Cancer Res 262849–28582020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goldberg SB, Narayan A, Kole AJ, et al. Early assessment of lung cancer immunotherapy response via circulating tumor DNA Clin Cancer Res 241872–18802018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hrebien S, Citi V, Garcia-Murillas I, et al. Early ctDNA dynamics as a surrogate for progression-free survival in advanced breast cancer in the BEECH trial Ann Oncol 30945–9522019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson JC, Carpenter EL, Silva BA, et al. Serial monitoring of circulating tumor DNA by next-generation gene sequencing as a biomarker of response and survival in patients with advanced NSCLC receiving pembrolizumab-based therapy JCO Precis Oncol 5510–5242021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mezquita L, Swalduz A, Jovelet C, et al. Clinical relevance of an amplicon-based liquid biopsy for detecting ALK and ROS1 fusion and resistance mutations in patients with non-small-cell lung cancer JCO Precis Oncol 4272–2822020 [DOI] [PMC free article] [PubMed] [Google Scholar]