Abstract
Background
Genome-wide association studies have identified six genetic variants associated with severe COVID-19, yet the mechanisms through which they may affect disease remains unclear. We investigated proteomic signatures related to COVID-19 risk variants rs657152 (ABO), rs10735079 (OAS1/OAS2/OAS3), rs2109069 (DPP9), rs74956615 (TYK2), rs2236757 (IFNAR2) and rs11385942 (SLC6A20/LZTFL1/CCR9/FYCO1/CXCR6/XCR1) as well as their corresponding downstream pathways that may promote severe COVID-19 in risk allele carriers and their potential relevancies to other infection outcomes.
Methods
A DNA aptamer-based array measured 4870 plasma proteins among 11 471 participants. Linear regression estimated associations between the COVID-19 risk variants and proteins with correction for multiple comparisons, and canonical pathway analysis was conducted. Cox regression assessed associations between proteins identified in the main analysis and risk of incident hospitalized respiratory infections (2570 events) over a 20.7-year follow-up.
Results
The ABO variant rs657152 was associated with 84 proteins in 7241 white participants with 24 replicated in 1671 Black participants. The TYK2 variant rs74956615 was associated with ICAM-1 and -5 in white participants with ICAM-5 replicated in Black participants. Of the 84 proteins identified in the main analysis, seven were significantly associated with incident hospitalized respiratory infections including Ephrin type-A receptor 4 (hazard ratio (HR): 0.87; P = 2.3 × 10−11) and von Willebrand factor type A (HR: 1.17; P = 1.6x10−13).
Conclusions
Novel proteomics signatures and pathways for COVID-19-related risk variants TYK2 and ABO were identified. A subset of these proteins predicted greater risk of incident hospitalized pneumonia and respiratory infections. Further studies to examine these proteins in COVID-19 patients are warranted.
Introduction
Coronavirus disease 19 (COVID-19) is characterized by asymptomatic to moderate presentations in most individuals but crippling illness and death in others. Patients who are male, elderly, or have comorbidities including obesity, diabetes and underlying respiratory or cardiovascular disease have been shown to have greater risks for more severe outcomes (1,2). And yet, critical illnesses are observed in young or otherwise healthy individuals resulting in hospitalization, cytokine storm, microemboli formation, respiratory failure or death (3–5). It is therefore likely that other factors are involved in COVID-19 pathogenicity and severity—among these, genetic liability.
To date, two genome-wide association studies (GWASs) have identified six independent, candidate gene variants associated with greater risk of severe COVID-19. An initial GWAS conducted in a European sample population reported two gene variants independently associated with greater likelihoods of developing COVID-19 respiratory failure: rs657152 on chromosome 9 (proximal to the ABO gene) and rs11385942 on chromosome 3 (proximal to gene cluster SLC6A20, LZTFL1, CCR9, FYCO1, CXCR6 and XCR1) (6). A subsequent GWAS further identified four novel and independent variants proximal to genes TYK2 (rs74956615), IFNAR2 (rs2236757), DPP9 (rs2109069) and gene cluster OAS1, OAS2, OAS3 (rs10735079) (7). Collectively, these six loci represent a critical step in our understanding of COVID-19 liability, and examination of their associated plasma proteomic signatures and pathway analysis may elucidate the mechanisms through which they influence COVID-19 severity.
The present analysis aimed to determine the proteomic signatures of these six GWAS-identified variants using a multiplexed aptamer-based platform comprised of over 4600 unique protein measurements in a sample of 8914 Atherosclerosis Risk in Communities Study (ARIC) participants. Pathway analysis was performed to examine whether variant–protein association patterns suggest suppression or activation of known canonical pathways. An additional prospective analysis was conducted to test whether proteins identified in the main analysis may be associated with the risks of incident hospitalized respiratory infections and pneumonia. Using this approach, it was hypothesized that identified proteins and pathways may have implications for development of severe COVID-19 and other respiratory infections.
Results
Information on the previously identified top COVID-19 variants, including their chromosome positions, proximal genes, risk allele frequencies and imputation accuracy indices are shown in Table 1. All variants are intronic with the exception of the rs74956615 single nucleotide polymorphism (SNP), which is located in the three prime untranslated region of the TYK2 gene.
Table 1.
Chromosome positions, nearest genes, allele frequencies and imputation accuracies of COVID-19 variants related to risk of respiratory failure among Black and white ARIC participants
| rsID | Chr:position* | Proximal gene(s) | Non-coded | Risk | Black freq | Black R2 | White freq | White R2 | OR** |
|---|---|---|---|---|---|---|---|---|---|
| rs11385942 | 3:45834967 | SLC6A20, LZTFL1, CCR9, FYCO1, CXCR6, XCR1 | G | GA; A | 0.052 | 0.99 | 0.081 | 1.00 | 1.77 |
| rs657152 | 9:133263862 | ABO | C | A | 0.431 | 0.98 | 0.378 | 0.96 | 1.48 |
| rs10735079 | 12:112942203 | OAS1, OAS2, OAS3 | A | G | 0.226 | N/A | 0.350 | N/A | 1.3 |
| rs2109069 | 19:4719431 | DPP9 | G | A | 0.213 | 0.90 | 0.304 | 0.80 | 1.4 |
| rs74956615 | 19:10317045 | TYK2 | A | T | 0.009 | 0.88 | 0.056 | 0.88 | 1.6 |
| rs2236757 | 21:33252612 | IFNAR2 | G | A | 0.215 | 0.98 | 0.299 | 0.98 | 1.3 |
Associations between variants and plasma protein levels are shown in Table 2. Following Bonferroni correction, 84 significant associations between ABO variant rs657152 and plasma proteins were observed in white participants (top 20 shown in Table 2; remaining associations shown in Supplementary Material, Table S3). Strong associations in terms of significances and magnitudes of association were detected, and ABO and DC-SIGN proteins were among the top associations. Twenty-four of these associations were replicated in Black participants and, of the 84 total associations, seventy were directionally consistent but generally showed weaker magnitudes of association than in white participants.
Table 2.
Significant associations of rs11385942, rs74956615 and rs657152 with plasma proteins among 1671 Black and 7241 white ARIC participants (top 20 for white participants shown for rs657152)
| White participants | Protein | Black participants | Protein | |||||
|---|---|---|---|---|---|---|---|---|
| Variant | Protein | Estimate* (95% CI) | P-value | SD | Estimate* (95% CI) | P-value | SD | |
| rs657152 | ABO system transferase | 2.85 (2.79, 2.92) | <1.0E-300 | 2.71 | 2.13 (1.96, 2.30) | 2.2E-109 | 2.89 | |
| DC-SIGN | 0.33 (0.32, 0.35) | <1.0E-300 | 0.50 | 0.24 (0.21, 0.28) | 2.2E-36 | 0.55 | ||
| Sulfhydryl oxidase 2 | 0.23 (0.22, 0.24) | 1.2E-299 | 0.38 | 0.10 (0.07, 0.12) | 5.0E-15 | 0.36 | ||
| Protein FAM3D | 0.46 (0.44, 0.49) | 9.9E-251 | 0.82 | 0.16 (0.11, 0.21) | 1.4E-10 | 0.74 | ||
| E-selectin | −0.28 (−0.29, −0.26) | 1.9E-223 | 0.52 | −0.14 (−0.18, −0.11) | 9.4E-18 | 0.48 | ||
| D-glucuronyl C5-epimerase | 0.14 (0.13, 0.15) | 2.6E-175 | 0.32 | 0.05 (0.03, 0.07) | 6.6E-06 | 0.32 | ||
| Golgi membrane protein 1 | 0.25 (0.23, 0.27) | 8.1E-144 | 0.45 | 0.09 (0.05, 0.12) | 4.9E-07 | 0.52 | ||
| Golgi membrane protein 1 | 0.19 (0.18, 0.20) | 2.4E-134 | 0.62 | 0.10 (0.06, 0.14) | 1.8E-06 | 0.66 | ||
| Platelet glycoprotein 4 | 0.19 (0.18, 0.21) | 2.4E-109 | 0.53 | 0.17 (0.12, 0.22) | 2.8E-10 | 0.95 | ||
| Protein CASC4 | 0.09 (0.08, 0.10) | 1.1E-86 | 0.26 | 0.04 (0.02, 0.05) | 6.4E-06 | 0.29 | ||
| sTie-1 | 0.09 (0.08, 0.10) | 2.9E-85 | 0.27 | 0.13 (0.11, 0.16) | 3.9E-30 | 0.33 | ||
| Coagulation factor VIII | 0.18 (0.16, 0.19) | 3.8E-81 | 0.54 | 0.16 (0.12, 0.20) | 1.3E-16 | 0.55 | ||
| C1GALT1-specific chaperone 1 | 0.06 (0.06, 0.07) | 3.2E-79 | 0.24 | 0.02 (0.01, 0.03) | 2.4E-03 | 0.20 | ||
| * * Plexin-D1 | −0.09 (−0.10, −0.08) | 3.1E-76 | 0.43 | −0.001 (−0.03, 0.02) | 9.7E-01 | 0.43 | ||
| THSD1 | −0.08 (−0.09, −0.07) | 2.6E-65 | 0.29 | −0.02 (−0.04, −0.001) | 3.5E-02 | 0.27 | ||
| Insulin receptor | −0.11 (−0.12, −0.10) | 2.7E-65 | 0.37 | −0.03 (−0.06, −0.003) | 2.9E-02 | 0.39 | ||
| * * IL-3 receptor subunit alpha | −0.13 (−0.14, −0.11) | 4.4E-63 | 0.47 | 0.02 (−0.008, 0.05) | 1.5E-01 | 0.50 | ||
| P-selectin | −0.10 (−0.11, −0.09) | 5.4E-59 | 0.37 | −0.05 (−0.08, −0.03) | 6.9E-05 | 0.37 | ||
| VEGF receptor 2 | −0.07 (−0.07, −0.06) | 1.7E-56 | 0.24 | −0.02 (−0.04, −0.003) | 1.9E-02 | 0.25 | ||
| LRC-32 | 0.07 (0.06, 0.08) | 2.2E-56 | 0.29 | 0.06 (0.04, 0.08) | 3.9E-09 | 0.32 | ||
| rs74956615 | ICAM-5 | −0.30 (−0.33, −0.27) | 1.4E-77 | 0.42 | −0.31 (−0.49, −0.14) | 5.2E-04 | 0.45 | |
| ICAM-5 | −0.30 (−0.32, −0.26) | 2.2E-70 | 0.43 | −0.30 (−0.48, 0.12) | 8.6E-04 | 0.48 | ||
| ICAM-1 | 0.25 (0.21, 0.29) | 7.3E-34 | 0.53 | 0.18 (0.01, 0.36) | 4.1E-02 | 0.45 | ||
| rs11385942 | * * TDGF-1 | 0.26 (0.19, 0.34) | 6.1E-13 | 1.21 | −0.02 (−0.15, 0.10) | 7.3E-1 | 0.84 | |
| High TDGF-1 | 0.04 (0.00, 0.08) | 2.6E-02 | 0.02 (−0.17, 0.20) | 8.7E-1 | ||||
| Low TDGF-1 | 0.01 (−0.01, 0.03) | 3.3E-01 | 0.01 (−0.04, 0.05) | 8.0E-1 | ||||
Model: General linear regression model adjusted for age, sex, field center, 10 principal components of ancestry and eGFR. Bonferroni correction for multiple testing in white participants, significant associations at P < 1.71E-6. Replication in Black participants for the 84 proteins identified in white participants stipulated a multiple testing corrected threshold of P < 5.95E-4.
*Change in log2 protein associated with one copy increment of at-risk allele for severe COVID-19, e.g. each copy of the rs657152 risk allele was associated with a 2.85 log2 greater ABO system transferase.
* * Variant protein associations that were not replicated in Black participant sample.
ICAM-5 levels were determined by two different DNA aptamers (Supplementary Material, Table S1).
ICAM = intercellular adhesion molecule; DC-SIGN=Dendritic Cell-Specific Intercellular adhesion molecule-3-Grabbing Non-integrin; sTie-1 = Soluble tyrosine-protein kinase receptor Tie-1, THSD1 = Thrombospondin type-1 domain-containing protein; IL = Interleukin; VEGF=Vascular endothelial growth factor; LRC-32 = Leucine-rich repeat-containing protein 32
Upon further investigation, it was found that the ABO protein has a distinct bimodal distribution. Since the linear regression model assumes a normal distribution—particularly for the dependent variable—the association between rs657152 and ABO protein levels became dubious. To better assess whether rs657152 is associated with the bimodally-distributed ABO protein, we first dichotomized protein levels into high and low values (using the 11.5 as the cutpoint); next, we used linear regression to examine the relationship between rs657152 and ABO levels in Black and white participants with adjustments for age, sex, field center, ten principal components of ancestry, and estimated glomerular filtration rate. Significant associations were observed between rs657152 and plasma levels of the ABO protein, but only for those in the `high' category in Black (β=0.323; p=3.0E-17) and white participants (β=0.484; p=5.0E-172).
The chromosome 19 variant rs74956615 was significantly associated with ICAM-1 (P = 7.3 × 10−34) and two independent aptamer measures of ICAM-5 in white participants (P = 1.4 × 10−77; P = 2.2 × 10−70). The former association with ICAM-1 was suggestive in Black participants but did not reach statistical significance following correction for multiple comparisons (P = 0.04), whereas the associations with ICAM-5 were replicated (P = 5.2 × 10−4; P = 8.6 × 10−4). Directionalities of associations for ICAM-1 and ICAM-5 were consistent between Black and white participants.
The rs11385942 variant was associated with a single protein, teratocarcinoma-derived growth factor (TDGF-1; P = 6.1 × 10−13) but was non-significant in Black participants (P = 0.73). Upon inspection of TDGF-1, it was found that the protein has a distinct bimodal distribution in white participants (Supplementary Material, Fig. S1) and to a lesser degree in Black participants (Supplementary Material, Fig. S2). Upon dichotomizing the protein into ‘high’ and ‘low’ subgroups (cutoff threshold = 9.75 fluorescence units), no significant associations with rs11385942 were observed in either TDGF-1 subgroup (Table 2). Sex, race or age did not account for the bimodal distribution of TDGF-1 based on an examination of residuals of regression analyses with these covariates. The remaining three variants related to severe COVID-19 were not significantly associated with any plasma proteins.
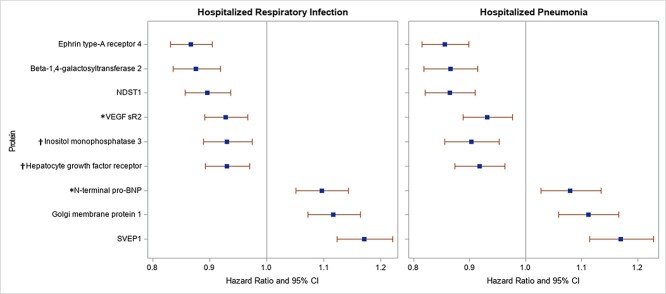
We next evaluated the relation of each of the 84 identified proteins and incident hospitalized respiratory infection and pneumonia. In total, we observed 2570 incident hospitalized respiratory infection events and 2087 incident hospitalized pneumonia events among the 10 775 participants. The median follow-up times were 20.7 years for incident respiratory infection events and 21.5 years for incident pneumonia. Ephrin type-A receptor 4 (P = 2.3 × 10−11), beta-1,4-galactosyltransferase 2 (P = 4.5 × 10−8), bifunctional heparan sulfate N-deacetylase/N-sulfotransferase 1 (NDST1) (P = 1.2 × 10−6) and vascular endothelial growth factor receptor 2 (P = 3.1 × 10−4) were significantly and inversely related to the risk of incident respiratory infection, whereas Sushi, von Willebrand factor type A (SVEP1) (P = 1.6 × 10−13), Golgi membrane protein 1 (P = 1.6 × 10−7) and N-terminal pro-BNP (P = 1.7 × 10−5) showed significant positive associations (Fig. 1, left panel). Results were similar for incident hospitalized pneumonia (Fig. 1, right panel); five significant protein observations were shared with respiratory infections and the associations were directionally consistent: ephrin type-A receptor 4 (P = 3.1 × 10−10), beta-1,4-galactosyltransferase 2 (P = 3.3 × 10−7), NDST1 (P = 3.2 × 10−8), Golgi membrane protein 1 (P = 1.9 × 10−5) and SVEP1 (P = 3.2 × 10−10). Additional significant associations were observed for pneumonia with inositol monophosphatase 3 (P = 1.9 × 10−4) and hepatocyte growth factor receptor (P = 5.3 × 10−4); these two proteins were directionally consistent with hospitalized respiratory infections. All of the above nine proteins identified for either infection outcomes were related to the ABO variant. TYK2-related proteins ICAM-1 and ICAM-5 were not significantly associated with either infection outcome after Bonferroni correction; however, associations for ICAM-5 were suggestive for risks of incident respiratory infection (P = 2.2 × 10−3) and pneumonia (P = 7.4 × 10−3).
Figure 1.
Cox regression analysis of significant proteins and risks of incident hospitalized respiratory infection (n for events = 2570) and hospitalized pneumonia (n for events = 2087) among ARIC participants over median 20.7- and 21.5-year follow-ups, respectively. Model: the Cox proportional hazards regression model assessed associations of proteins (per SD) with risks of incident infection outcomes with adjustments for age, gender, race, field center, eGFR, cigarette smoking status, BMI, prevalent diabetes, alcohol drinking status, estimated ethanol intake (grams/week), total cholesterol, prevalent cardiovascular disease, and prevalent stroke. Protein values outside of 6 SDs from the mean were excluded for each protein, and Bonferroni correction stipulated a significance level of P ≤ 5.9 × 10−4.
*VEGF sR2 and N-terminal BNP were significantly associated with incident respiratory infection but did not reach the Bonferroni corrected significance threshold for the pneumonia outcome. Inositol monophosphatase 3 and hepatocyte growth factor receptor were significantly associated with incident pneumonia but did not reach the Bonferroni-corrected significance threshold for the respiratory infection outcome. VEGF sR2 = vascular endothelial growth factor receptor 2.
Sensitivity analyses were conducted for both infection outcomes. Excluding participants with an incident hospitalized respiratory infection (n = 167 events) or incident pneumonia (n = 133 events) within 3 years of the Visit 3 baseline did not materially change most of these results; however, associations with vascular endothelial growth factor receptor 2 (P = 1.9 × 10−3) and hepatocyte growth factor receptor (P = 1.9 × 10−3) were rendered non-significant.
Ingenuity Pathway Analysis (IPA) was next conducted. Selected pathways and their corresponding proteins showing enrichment among carriers of the ABO risk variant are presented in Table 3 for white participants, and results were similar among Black participants (data not shown). Suppressions of IL-15 production (P = 2.5 × 10−10), signal transducer and activator of transcription 3 (STAT3) signaling (P = 6.1 × 10−6) and NF-κB signaling (P = 3.0 × 10−3) were identified. These pathways included proteins that were prospectively associated with incident hospitalized respiratory infections or pneumonia: hepatocyte growth factor receptor (IL-15 suppression), ephrin type-A receptor 4 (IL-15 suppression) and vascular endothelial growth factor receptor 2 (IL-15 suppression, STAT3 signaling and NF-κB signaling).
Table 3.
Enrichment of select canonical pathways and corresponding protein components associated with the ABO rs657152 variant
| Pathway | Pathway components | Symbol | Expression Log ratio | Expression P-value |
|---|---|---|---|---|
| IL-15 suppression | ||||
| Macrophage colony-stimulating factor 1 receptor | CSF1R | 0.033 | 2.2E-07 | |
| Insulin-like growth factor 1 receptor | IGF1R | -0.03 | 6.9E-11 | |
| Ephrin type-B receptor 4 | EPHB4 | -0.034 | 2.0E-11 | |
| Vascular endothelial growth factor receptor 3 | FLT4 | -0.074 | 1.2E-15 | |
| * Hepatocyte growth factor receptor | MET | -0.056 | 1.2E-35 | |
| * Ephrin type-A receptor 4 | EPHA4 | -0.058 | 1.0E-52 | |
| Vascular endothelial growth factor receptor 2 | KDR | -0.065 | 1.7E-56 | |
| Insulin receptor | INSR | -0.108 | 2.7E-65 | |
| Tyrosine-protein kinase receptor Tie-1 | TIE1 | 0.09 | 2.9E-85 | |
| STAT3 signaling | ||||
| Interleukin-6 receptor subunit beta | IL6ST | -0.019 | 2.9E-09 | |
| Insulin-like growth factor 1 receptor | IGF1R | -0.03 | 6.9E-11 | |
| Vascular endothelial growth factor receptor 3 | FLT4 | -0.074 | 1.2E-15 | |
| Vascular endothelial growth factor receptor 2 | KDR | -0.065 | 1.7E-56 | |
| Interleukin-3 receptor subunit alpha | IL3RA | -0.127 | 4.4E-63 | |
| Insulin receptor | INSR | -0.108 | 2.7E-65 | |
| NF-κB signaling | ||||
| Insulin-like growth factor 1 receptor | IGF1R | -0.03 | 6.9E-11 | |
| Vascular endothelial growth factor receptor 3 | FLT4 | -0.074 | 1.2E-15 | |
| Vascular endothelial growth factor receptor 2 | KDR | -0.065 | 1.7E-56 | |
| Insulin receptor | INSR | -0.108 | 2.7E-65 |
Discussion
Of the six GWAS-identified variants that have so far been related to severe COVID-19, two were significantly associated with 84 plasma proteins, many with immune-related functions. The TYK2 rs74956615 variant was strongly related to endothelial activation markers ICAM-5 and ICAM-1, whereas the ABO rs657152 variant was associated with dozens of protein targets. Prospective examination of these proteins revealed that nine showed consistent associations with incidence of hospitalized respiratory infections and pneumonia during over 20 years of follow-up, with five being statistically significant for both outcomes. Finally, pathway analysis showed that carriers of the ABO risk allele exhibit plasma proteomics patterns that are consistent with lower antiviral IL-15 activity as well as suppressed signaling of NF-κB and STAT3.
Previous studies
So far, one previous proteomics study by Katz et al. (8) has examined two variants associated with severe COVID-19 using a similar aptamer-based platform among Jackson Heart, Framingham Heart and Malmo Diet cohorts. Ten associations were reported between the rs657152 ABO variant and circulating proteins among 1813 Black participants of the Jackson Heart Study. Of these, we replicated eight associations in Black participants. Among the 3046 white participants, 21 protein associations were observed. Of these, we replicated 19 in white participants and detected an additional 66 associations—likely due our larger sample size and larger proteomics array (1305 versus 4870 protein analytes). A prominent finding by Katz et al., confirmed here, was the strong association of the rs657152 ABO variant with DC-SIGN, aka CD209, and these findings are consistent with a recent Mendelian randomization (MR) study showing that genetically higher levels of ABO (associated with the rs8176719 variant) are related to greater DC-SIGN levels (9). Functionally, DC-SIGN is a pathogen-recognition receptor but has been shown to facilitate viral cell entry for SAR-CoV-1 and HIV (10,11). DC-SIGN has similarly been shown to serve as a receptor for SARS-CoV-2 (12), and it may be speculated that genetically higher DC-SIGN levels result in greater DC-SIGN-mediated cell entry, resulting in more severe COVID-19 outcomes.
In contrast to the putative liability of DC-SIGN, a Neanderthal isoform of 2′–5′ oligoadenylate synthetase-1 (OAS1) may offer protection against COVID-19 based on a recent MR study (13). Higher circulating OAS1 protein levels were reported to reduce COVID-19 susceptibility, hospitalization and ventilation outcomes as determined through the instrument variable, rs4767027. This variant is in linkage disequilibrium with the variant tested in our study (rs10735079 proximal to OAS1/OAS2/OAS3, r2 = 0.89), and we found that the risk allele of rs10735079 is associated with lower OAS1 levels in white ARIC participants (beta: −0.014; P = 0.019)—supporting an inverse association between OAS1 protein and severe COVID outcomes. We were unable to show a prospective association between circulating OAS1 levels and respiratory infection [HR: 0.98; 95% confidence interval (CI): 0.92, 1.03] or pneumonia hospitalizations (HR: 1.02; 95% CI: 0.95, 1.08); however, this was not unexpected since the protective function of OAS1 may be limited to RNA viruses. Additional studies are needed to examine whether OAS1 reduces risk of infection and or virulence of SARS-Cov2 and other RNA viruses.
Pathway analysis was used to characterize ABO variant–protein associations more thoroughly. We observed enrichment of signaling pathways related to immune functions including suppressions of IL-15 production and both STAT3 and NF-κB signaling. Critically, these findings are supported by previous studies and align with the antiviral role of IL-15 and its proposed efficacy as a COVID-19 immunotherapy (14). Physiologically, IL-15 activates JAK/STAT and NF-κB signaling cascades, which induce NK cell and CD8+ cell activation for a sustained cytotoxic immune response (15) and clearance of viral-infected cells (16). Indeed, a clinical trial of an IL-15 superagonist is currently underway to determine its efficacy in treating COVID-19 patients (17). Taken together, proteomics coupled with pathway analyses suggest a genetically lower baseline immune status of IL-15-mediated pathways in carriers of the ABO risk variant.
Prospective analysis showed that, of the 84 proteins associated with the severe COVID-19 risk variants, nine significantly predicted the incidence of hospitalized respiratory infections or pneumonia, with five protein exposures being shared between these two outcomes. While most of the nine proteins have varied functions, immune-modulatory roles have been identified among them including mediation of cell adhesion, inflammation, cytotoxicity and host immune response to viral infection (Supplementary Material, Table S4). These observations provide some confirmatory evidence that a subset of identified proteins are involved in infection incidence and support a biological link between the ABO risk variant, identified proteins and risk of disease development.
The TYK2 rs74956615 risk variant was strongly associated with lower ICAM-5 and higher ICAM-1 levels in white participants. ICAM-5 is a neuron- and somadendritic-specific membrane glycoprotein, and its association with the TYK2 variant here remains unclear. By contrast, ICAM-1 is an endothelial activation marker expressed in the vasculature and alveolar epithelium, and elevated levels have been reported in plasma (18,19) and postmortem lung tissues of COVID-19 patients (20). Functionally, ICAM-1 is involved in endothelial activation, acute and chronic inflammation and hemostasis (21–24). With severe COVID-19, these processes become dysregulated and may induce the life-threatening features of disease including, but not limited to, cytokine storm, pulmonary vascular leakage, disseminated microemboli and acute respiratory distress syndrome (3–5). And while we cannot conclude that the lower plasma ICAM-5 and higher ICAM-1 levels in TYK2 variant carriers account for these phenomena and the genetic susceptibility of severe outcomes, ICAM-1 is a strong and biologically plausible candidate that is localized to tissues affected by COVID-19, is proximal to TYK2, and should be further investigated.
ABO and COVID-19
ABO blood group phenotypes and ABO variants have previously been associated with other viral, bacterial and parasitic infections including cholera, malaria, noroviruses, SARS-Cov-1, and Helicobacter pylori (25–29), but there remains an ongoing controversy as to whether they may be related to COVID-19. In one of the first GWASs in COVID-19 patients, the rs657152 variant at the ABO locus was shown to be associated with the risk of severe COVID-19 among 1610 European patients and 2205 controls [odds ratio (OR) for the A allele: 1.32; 95% CI: 1.20–1.47; P = 4.95 × 10−8] (6). In a subsequent GWAS, the ABO signal did not reach genome-wide significance in discovery phase but was close to the significance threshold in their combined meta-analysis (7). Significant associations with COVID-19 infection and hospitalization were reported in a subsequent meta-analysis of three GWASs (30). Phenotype studies of blood type and COVID-19 infection or severe outcomes (e.g. hospitalization or death) have reported lower risks in those with blood type O and a corresponding greater risk for those with blood type A (31–34) as well as null findings (35–38). Finally, Hernandez et al. (39) showed that ABO gene expression co-localized with susceptibility and severity of COVID-19 in a multi-omics Bayesian co-localization and MR study. The investigators further showed evidence for a causal relationship between plasma ABO protein levels and COVID-19 severity using MR, which coincides with our finding of a strong association between the rs657152 risk variant and ABO protein levels (Table 2). While the above evidence is not definitive, the mixed findings coupled with the MR results suggest a modest effect of the ABO variant on COVID-19 outcomes, which may be mediated by pleiotropic effects of the ABO gene/protein.
Strengths and limitations
This study was composed of a large cohort, and protein levels were obtained using an aptamer-based platform that has undergone extensive quality control and reproducibility testing. To limit the probability of type I errors, Bonferroni corrections for multiple comparisons were applied in statistical analyses. For proteins identified in the main analysis, secondary analyses with incident hospitalized respiratory infections and pneumonia suggested that these protein targets may have clinically meaningful effects in addition to their biologically plausible roles in immune function.
In terms of limitations, we were unable to replicate all associations in Black participants, possibly because of the smaller sample size; however, previous GWASs were mostly conducted in European populations, and thus, the generalizability of our findings to other ancestry groups may be a limitation as well. Genetic risk variants were selected from previous GWASs but may not represent definitive risk factors for severe COVID-19. It must also be acknowledged that no proteins were associated with four of the six COVID-related variants, and it is possible that our proteomics array may not have had the coverage to assess all relevant proteins. Additionally, our approach is based on carriers of these genetic risk variants, absent of infection, to show proteomics signatures associated with genetic susceptibility of more severe COVID-19 outcomes. We cannot evaluate the possibility that some variant-related differences in protein levels are only detectable following development of COVID-19. And yet, numerous potential mediators consistent with worse COVID-19 outcomes were observed and coincide with disease pathophysiology. Finally, an MR analysis can be useful to evaluate whether identified proteins may be causally related to hospitalized infection outcomes. Since there are limited GWAS reports of the identified proteins in the literature, we conducted a GWAS for these proteins in ARIC European Americans (n = 9345) and attempted to replicate significant independent SNPs in the INTERVAL cohort (40). While we identified and replicated a few significant SNPs for each protein, these SNPs were found to explain a small % of protein variance (<5% for most proteins). Coupled with the relatively modest magnitudes of association of the proteins with the infection outcomes of interest, it was deemed unlikely that we would have sufficient statistical power to detect evidence of a causal relationship between the identified proteins and risk of either infection outcome using this approach.
Conclusions
Taken together, we used a large-scale proteomics approach to provide novel evidence of the genetic susceptibility to severe COVID-19 in carriers of the TYK2 and ABO risk variants. We showed that a subset of identified proteins are related to risk of other hospitalized respiratory infections events over an extended > 20-year follow-up, which provides supporting evidence and biological plausibility that they may be involved in SARS-CoV-2 infection or COVID-19 severity. Targeted research of proteomic profiles and pathway characteristics in COVID-19 patient carriers and non-carriers of these variants may confirm these findings and provide other evidence of their downstream mechanisms in severe COVID-19 respiratory syndrome.
Materials and Methods
Study population
The ARIC prospective cohort study was designed to identify cardiovascular disease and atherosclerosis risk factors (41). From 1987 to 1989, male and female participants (N = 15 792) between ages 45 and 64 were recruited among four US communities (Washington County, Maryland; the northwest suburbs of Minneapolis, Minnesota; Jackson, Mississippi and Forsyth County, North Carolina). The Jackson center only enrolled Black participants. Participants received annual follow-up calls (semi-annual after 2012) to report cardiovascular events and hospitalizations. Risk factor information was collected at baseline and across several follow-up examinations. The present study used proteomic biomarker data measured in specimens collected at Visit 3 (1993–95).
Genotyping
Information on genotyping, quality control and imputation procedures has been detailed previously (42). Whole blood genomic DNA was genotyped using the Affymetrix Genome-Wide Human SNP array 6.0 (Affymetrix, Santa Clara, CA). To expand the number of genetic markers, race-specific imputation of variant dosages to the TopMed reference panel were conducted (43). Race-specific principal components of genetic ancestry or population substructure based on the GWAS array data were generated by EIGENSTRAT 7 (44) in ARIC participants.
Assessment of kidney function
Estimated glomerular filtration rate (eGFR; ml/min/ 1.73 m2) at Visit 3 was calculated using the Chronic Kidney Disease Epidemiology Collaboration combined creatinine-cystatin C equation (45–47).
Proteomics measurement and quality control
EDTA-plasma collected at Visit 3 was analyzed using a modified aptamer-based capture array (SOMAscan version 4.0, Somalogic, Inc., Boulder, CO) as described previously (48–51). Plasma samples were transferred to the SomaLogic laboratory and incubated with proprietary reagents. Protein levels were measured using single-stranded DNA-based modified aptamers that bind specific protein epitopes and reported as relative fluorescence units.
Proof of principle and assay validations of specificity and intra- and inter-assay variability have been published (49–51). Briefly, protein measurements were standardized and normalized (51). Hybridization control normalization was applied to each sample based on a set of hybridization control sequences to correct for systematic biases during hybridization. Metrics of assay reproducibility have been previously reported (51) with a median coefficient of variance (quartile 1, quartile 3) of 5.0 (4.1, 6.9) and a median intraclass correlation (quartile 1, quartile 3) of 0.96 (0.92, 0.98). Each assay plate contained calibrators for each aptamer reagent to correct for plate-to-plate variation based on global reference materials.
Log base 2 transformation to all protein measures were applied to correct for skewness. A total of 422 blind duplicate plasma aliquots were included, and the median inter-assay Bland–Altman coefficient of variation was 6.3%. The median split sample reliability coefficient was 0.85 after excluding the following quality control outliers: of the 5284 available aptamer measurements, 94 were excluded because of a Bland–Altman coefficient of variation > 50% or a variance of < 0.01 on the log scale; an additional 313 measurements were excluded because of non-specific binding to non-proteins. After all quality control measures were completed, 4870 aptamer measurements were included that corresponded to 4697 unique human proteins or protein complexes. Proteins identified in the present analysis and their corresponding aptamer sequence identification numbers are shown in Supplementary Material, Table S1.
Ascertainment of respiratory infection and pneumonia hospitalizations
Hospitalizations and deaths in the ARIC cohort were identified through annual telephone interviews with participants, surveillance of local hospital discharge lists and linkage to the National Death Index. Medical records and death certificates were obtained, and International Classification of Diseases, Ninth Revision (ICD-9) and Tenth Revision (ICD-10) codes recorded. Respiratory infections and pneumonia were defined based on ICD-9 and ICD-10 codes (Supplementary Material, Table S2). In an effort to exclude infections that were not an important cause for hospitalization, events were restricted to those in positions 1 through 5 in medical records.
Statistical analysis
Analyses included participants who attended Visit 3 when plasma samples for protein measurements were collected and had data on SNPs and covariates. Those missing genotyping or imputation data (n = 2737), protein measures (n = 1041) or other covariate data (n = 197) were excluded, resulting in a sample of 1671 Black and 7241 white participants. Race-specific multiple linear regression evaluated associations between variant dosages and log base 2 transformed proteins, adjusting for age, sex, eGFR, field center and 10 principal components of ancestry. Bonferroni correction adjusted for multiple testing, stipulating a significance threshold of P ≤ 1.71 × 10−6 in white participants (6 variants*4870 aptamer measurements). Significant associations were then tested for replication in Black participants, with Bonferroni corrections for the number of significant proteins tested for each variant. Distributions of protein values were inspected for those demonstrating significant associations. For TDGF-1, a bimodal distribution was observed. To determine whether this may be because of demographic factors of sex, age or race, protein levels were regressed against these variables, and residual plots were then inspected to determine whether any patterns could be observed.
Proteins identified in the above analysis were tested for associations with risks of incident hospitalized respiratory infections and pneumonia events. We defined incident hospitalized respiratory infection and pneumonia as those that occurred for the first time following the Visit 3 baseline in participants who did not have any such events up to three years before Visit 3. Accordingly, we excluded individuals with a hospitalized respiratory infection event up to 3 years prior to Visit 3 (N = 178). We used the relatively wide window to exclude prevalent events because a recent respiratory infection may also influence the plasma proteome. Cox proportional hazards regression analysis was performed among 10 775 participants with Visit 3 plasma proteins and covariates. Participants were followed until incident hospitalization with the outcome of interest, loss-to-follow-up, death or 31 December 2018. Proteins were analyzed as continuous variables [per standard deviation (SD)], and protein values outside of 6 SDs from the mean were excluded. We adjusted for age, gender, race, field center, eGFR, cigarette smoking status, body mass index (BMI), prevalent diabetes, alcohol drinking status, estimated ethanol intake (grams/week), total cholesterol, prevalent cardiovascular disease and prevalent stroke in Cox regression. We used Bonferroni correction to adjust for multiple testing, corresponding to a significance threshold of P ≤ 5.9 × 10−4 for testing of 85 aptamer measures corresponding to 84 proteins. Lastly, a sensitivity analysis was conducted in which participants who had an infection event within 3 years following the Visit 3 baseline were excluded. The purpose of this was to exclude the possibility of a mild infection that occurred around the Visit 3 baseline (affecting the plasma proteome), which then developed in to a more severe hospitalized infection in the subsequent years. Using this approach resulted in the exclusion of 167 incident respiratory infection and 133 incident pneumonia events from their respective analyses.
Pathway analysis
To identify signaling mechanisms and canonical pathway enrichment through which variant-associated proteins may promote susceptibility COVID-19, network pathway analysis was conducted using the IPA (QIAGEN Inc.) (52). Protein identifiers and estimates from SNP–protein analysis were uploaded, and analyses were restricted by applying a Benjamini–Hochberg false discovery rate adjustment for multiple comparisons whereby q-value ≤ 0.05 was deemed significant. Core pathway analyses were conducted using the uploaded data as the reference set and direct and indirect experimentally confirmed relationships across species were examined.
Supplementary Material
Acknowledgements
The authors thank the staff and participants of the ARIC study for their important contributions.
Conflict of Interest statement. Authors have no conflicts.
Contributor Information
Brian T Steffen, Division of Epidemiology and Community Health, University of Minnesota, Minneapolis, MN 55455, USA.
James S Pankow, Division of Epidemiology and Community Health, University of Minnesota, Minneapolis, MN 55455, USA.
Pamela L Lutsey, Division of Epidemiology and Community Health, University of Minnesota, Minneapolis, MN 55455, USA.
Ryan T Demmer, Division of Epidemiology and Community Health, University of Minnesota, Minneapolis, MN 55455, USA; Department of Epidemiology, Mailman School of Public Health, Columbia University, New York, NY 10032, USA.
Jeffrey R Misialek, Division of Epidemiology and Community Health, University of Minnesota, Minneapolis, MN 55455, USA.
Weihua Guan, Division of Biostatistics, University of Minnesota School of Public Health, Minneapolis, MN 55455, USA.
Logan T Cowan, Department of Biostatistics, Epidemiology, and Environmental Health Sciences, Jiann Ping-Hsu College of Public Health, Statesboro, GA 30458, USA.
Josef Coresh, Department of Epidemiology, Johns Hopkins Bloomberg School of Public Health, Welch Center for Prevention, Epidemiology and Clinical Research, Johns Hopkins Medical Institutions, Baltimore, MD 21218, USA.
Faye L Norby, Division of Epidemiology and Community Health, University of Minnesota, Minneapolis, MN 55455, USA; Department of Cardiology, Smidt Heart Institute, Cedars-Sinai Health System, Los Angeles 90048, CA.
Weihong Tang, Division of Epidemiology and Community Health, University of Minnesota, Minneapolis, MN 55455, USA.
Funding
This work was supported by the National Heart, Lung, and Blood Institute [grant number 2T32HL007779-26] and by an award from the Hawley Foundation to [BTS]. The Atherosclerosis Risk in Communities study has been funded in whole or in part with Federal funds from the National Heart, Lung, and Blood Institute, National Institutes of Health, Department of Health and Human Services [grant numbers HHSN268201700001I, HHSN268201700002I, HHSN268201700003I, HHSN268201700004I and HSN268201700005I, R01HL087641, R01HL059367 and R01HL086694]. Funding was also supported by R01HL087641, R01HL059367 and R01HL086694; National Human Genome Research Institute contract U01HG004402; and National Institutes of Health contract HHSN268200625226C. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research. SomaLogic Inc. conducted the SomaScan assays in exchange for use of ARIC data. This work was supported in part by the National Heart, Lung, and Blood Institute [grant R01 HL134320].
References
- 1. Yang, J., Zheng, Y., Gou, X., Pu, K., Chen, Z., Guo, Q., Ji, R., Wang, H., Wang, Y. and Zhou, Y. (2020) Prevalence of comorbidities and its effects in patients infected with SARS-CoV-2: a systematic review and meta-analysis. Int. J. Infect. Dis., 94, 91–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. LoPresti, M., Beck, D.B., Duggal, P., Cummings, D.A.T. and Solomon, B.D. (2020) The role of host genetic factors in coronavirus susceptibility: review of animal and systematic review of human literature. Am. J. Hum. Genet., 107, 381–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ye, Q., Wang, B. and Mao, J. (2020) The pathogenesis and treatment of the `Cytokine Storm' in COVID-19. J. Inf. Secur., 80, 607–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Roberts, K.A., Colley, L., Agbaedeng, T.A., Ellison-Hughes, G.M. and Ross, M.D. (2020) Vascular manifestations of COVID-19 - thromboembolism and microvascular dysfunction. Front Cardiovasc Med, 7, 598400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang, H. and Ma, S. (2008) The cytokine storm and factors determining the sequence and severity of organ dysfunction in multiple organ dysfunction syndrome. Am. J. Emerg. Med., 26, 711–715. [DOI] [PubMed] [Google Scholar]
- 6. Ellinghaus, D., Degenhardt, F., Bujanda, L., Buti, M., Albillos, A., Invernizzi, P., Fernández, J., Prati, D., Baselli, G., Asselta, R. et al. (2020) Genomewide association study of severe Covid-19 with respiratory failure. N. Engl. J. Med., 383, 1522–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pairo-Castineira, E., Clohisey, S., Klaric, L., Bretherick, A.D., Rawlik, K., Pasko, D., Walker, S., Parkinson, N., Fourman, M.H., Russell, C.D. et al. (2021) Genetic mechanisms of critical illness in COVID-19. Nature, 591, 92–98. [DOI] [PubMed] [Google Scholar]
- 8. Katz, D.H., Tahir, U.A., Ngo, D., Benson, M.D., Bick, A.G., Pampana, A., Gao, Y., Keyes, M.J., Correa, A., Sinha, S. et al. (2020) Proteomic profiling in biracial cohorts implicates DC-SIGN as a mediator of genetic risk in COVID-19. medRxiv, in press. [Google Scholar]
- 9. Anisul, M., Shilts, J., Schwartzentruber, J., Hayhurst, J., Buniello, A., Mohammed, S.E., E., Zheng, J., Holmes, M., Ochoa, D., Carmona, M. et al. (2021) A proteome-wide genetic investigation identifies several SARS-CoV-2-exploited host targets of clinical relevance. elife, 10, e69719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jeffers, S.A., Tusell, S.M., Gillim-Ross, L., Hemmila, E.M., Achenbach, J.E., Babcock, G.J., Thomas, W.D., Jr., Thackray, L.B., Young, M.D., Mason, R.J. et al. (2004) CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. U. S. A., 101, 15748–15753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pustylnikov, S., Dave, R.S., Khan, Z.K., Porkolab, V., Rashad, A.A., Hutchinson, M., Fieschi, F., Chaiken, I. and Jain, P. (2016) Short communication: inhibition of DC-SIGN-mediated HIV-1 infection by complementary actions of dendritic cell receptor antagonists and Env-targeting virus inactivators. AIDS Res. Hum. Retrovir., 32, 93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Amraei, R., Yin, W., Napoleon, M.A., Suder, E.L., Berrigan, J., Zhao, Q., Olejnik, J., Chandler, K.B., Xia, C., Feldman, J. et al. (2021) CD209L/L-SIGN and CD209/DC-SIGN act as receptors for SARS-CoV-2. bioRxiv. in press. [DOI] [PMC free article] [PubMed]
- 13. Zhou, S., Butler-Laporte, G., Nakanishi, T., Morrison, D.R., Afilalo, J., Afilalo, M., Laurent, L., Pietzner, M., Kerrison, N., Zhao, K. et al. (2021) A Neanderthal OAS1 isoform protects individuals of European ancestry against COVID-19 susceptibility and severity. Nat. Med., 27, 659–667. [DOI] [PubMed] [Google Scholar]
- 14. Kandikattu, H.K., Venkateshaiah, S.U., Kumar, S. and Mishra, A. (2020) IL-15 immunotherapy is a viable strategy for COVID-19. Cytokine Growth Factor Rev., 54, 24–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Verbist, K.C. and Klonowski, K.D. (2012) Functions of IL-15 in anti-viral immunity: multiplicity and variety. Cytokine, 59, 467–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Adib-Conquy, M., Scott-Algara, D., Cavaillon, J.M. and Souza-Fonseca-Guimaraes, F. (2014) TLR-mediated activation of NK cells and their role in bacterial/viral immune responses in mammals. Immunol. Cell Biol., 92, 256–262. [DOI] [PubMed] [Google Scholar]
- 17. National Library of Medicine (U.S.) (2020) A Phase I/II Study of Universal Off-the-shelf NKG2D-ACE2 CAR-NK Cells for Therapy of COVID-19. Identifier NCT04324996. https://clinicaltrials.gov/ct2/show/NCT04324996.
- 18. Tong, M., Jiang, Y., Xia, D., Xiong, Y., Zheng, Q., Chen, F., Zou, L., Xiao, W. and Zhu, Y. (2020) Elevated expression of serum endothelial cell adhesion molecules in COVID-19 patients. J. Infect. Dis., 222, 894–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Escher, R., Breakey, N. and Lämmle, B. (2020) Severe COVID-19 infection associated with endothelial activation. Thromb. Res., 190, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nagashima, S., Mendes, M.C., Camargo Martins, A.P., Borges, N.H., Godoy, T.M., Miggiolaro, A., da Silva Dezidério, F., Machado-Souza, C. and de Noronha, L. (2020) Endothelial dysfunction and thrombosis in patients with COVID-19-brief report. Arterioscler. Thromb. Vasc. Biol., 40, 2404–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. van Hinsbergh, V.W. (2012) Endothelium--role in regulation of coagulation and inflammation. Semin. Immunopathol., 34, 93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhu, G.D., Arendsen, D.L., Gunawardana, I.W., Boyd, S.A., Stewart, A.O., Fry, D.G., Cool, B.L., Kifle, L., Schaefer, V., Meuth, J. et al. (2001) Selective inhibition of ICAM-1 and E-selectin expression in human endothelial cells. 2. Aryl modifications of 4-(aryloxy)thieno[2,3-c]pyridines with fine-tuning at C-2 carbamides. J. Med. Chem., 44, 3469–3487. [DOI] [PubMed] [Google Scholar]
- 23. Perico, L., Benigni, A., Casiraghi, F., Ng, L.F.P., Renia, L. and Remuzzi, G. (2021) Immunity, endothelial injury and complement-induced coagulopathy in COVID-19. Nat Rev Nephrol, 17, 46–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Greineder, C.F., Johnston, I.H., Villa, C.H., Gollomp, K., Esmon, C.T., Cines, D.B., Poncz, M. and Muzykantov, V.R. (2017) ICAM-1-targeted thrombomodulin mitigates tissue factor-driven inflammatory thrombosis in a human endothelialized microfluidic model. Blood Adv, 1, 1452–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cheng, Y., Cheng, G., Chui, C.H., Lau, F.Y., Chan, P.K., Ng, M.H., Sung, J.J. and Wong, R.S. (2005) ABO blood group and susceptibility to severe acute respiratory syndrome. JAMA, 293, 1450–1451. [DOI] [PubMed] [Google Scholar]
- 26. Lindesmith, L., Moe, C., Marionneau, S., Ruvoen, N., Jiang, X., Lindblad, L., Stewart, P., LePendu, J. and Baric, R. (2003) Human susceptibility and resistance to Norwalk virus infection. Nat. Med., 9, 548–553. [DOI] [PubMed] [Google Scholar]
- 27. Rowe, J.A., Handel, I.G., Thera, M.A., Deans, A.M., Lyke, K.E., Koné, A., Diallo, D.A., Raza, A., Kai, O., Marsh, K. et al. (2007) Blood group O protects against severe Plasmodium falciparum malaria through the mechanism of reduced rosetting. Proc. Natl. Acad. Sci. U. S. A., 104, 17471–17476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Barua, D. and Paguio, A.S. (1977) ABO blood groups and cholera. Ann. Hum. Biol., 4, 489–492. [DOI] [PubMed] [Google Scholar]
- 29. Borén, T., Falk, P., Roth, K.A., Larson, G. and Normark, S. (1993) Attachment of Helicobacter pylori to human gastric epithelium mediated by blood group antigens. Science, 262, 1892–1895. [DOI] [PubMed] [Google Scholar]
- 30. COVID-19 Host Genetics Initiative . (2021) Mapping the human genetic architecture of COVID-19. Nature, 600, 472–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Barnkob, M.B., Pottegård, A., Støvring, H., Haunstrup, T.M., Homburg, K., Larsen, R., Hansen, M.B., Titlestad, K., Aagaard, B., Møller, B.K. et al. (2020) Reduced prevalence of SARS-CoV-2 infection in ABO blood group O. Blood Adv, 4, 4990–4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Göker, H., Aladağ Karakulak, E., Demiroğlu, H., Ayaz Ceylan, Ç.M., Büyükaşik, Y., Inkaya, A., Aksu, S., Sayinalp, N., Haznedaroğlu, I.C., Uzun, Ö. et al. (2020) The effects of blood group types on the risk of COVID-19 infection and its clinical outcome. Turk J Med Sci, 50, 679–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kibler, M., Dietrich, L., Kanso, M., Carmona, A., Marchandot, B., Matsushita, K., Trimaille, A., How-Choong, C., Odier, A., Gennesseaux, G. et al. (2020) Risk and severity of COVID-19 and ABO blood group in transcatheter aortic valve patients. J. Clin. Med., 9, 3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wu, Y., Feng, Z., Li, P. and Yu, Q. (2020) Relationship between ABO blood group distribution and clinical characteristics in patients with COVID-19. Clin. Chim. Acta, 509, 220–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Latz, C.A., DeCarlo, C., Boitano, L., Png, C.Y.M., Patell, R., Conrad, M.F., Eagleton, M. and Dua, A. (2020) Blood type and outcomes in patients with COVID-19. Ann. Hematol., 99, 2113–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lehrer, S. and Rheinstein, P.H. (2021) ABO blood groups, COVID-19 infection and mortality. Blood Cells Mol. Dis., 89, 102571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Boudin, L., Janvier, F., Bylicki, O. and Dutasta, F. (2020) ABO blood groups are not associated with risk of acquiring the SARS-CoV-2 infection in young adults. Haematologica, 105, 2841–2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Anderson, J.L., May, H.T., Knight, S., Bair, T.L., Muhlestein, J.B., Knowlton, K.U. and Horne, B.D. (2021) Association of sociodemographic factors and blood group type with risk of COVID-19 in a US population. JAMA Netw. Open, 4, e217429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hernández Cordero, A.I., Li, X., Milne, S., Yang, C.X., Bossé, Y., Joubert, P., Timens, W., van den Berge, M., Nickle, D., Hao, K. et al. (2021) Multi-omics highlights ABO plasma protein as a causal risk factor for COVID-19. Hum. Genet., 140, 969–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sun, B.B., Maranville, J.C., Peters, J.E., Stacey, D., Staley, J.R., Blackshaw, J., Burgess, S., Jiang, T., Paige, E., Surendran, P. et al. (2018) Genomic atlas of the human plasma proteome. Nature, 558, 73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. The ARIC investigators (1989) The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. Am. J. Epidemiol., 129, 687–702. [PubMed] [Google Scholar]
- 42. Pankow, J.S., Tang, W., Pankratz, N., Guan, W., Weng, L.C., Cushman, M., Boerwinkle, E. and Folsom, A.R. (2017) Identification of genetic variants linking protein C and lipoprotein metabolism: the ARIC study (Atherosclerosis Risk in Communities). Arterioscler. Thromb. Vasc. Biol., 37, 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Taliun, D., Harris, D.N., Kessler, M.D., Carlson, J., Szpiech, Z.A., Torres, R., Taliun, S.A.G., Corvelo, A., Gogarten, S.M., Kang, H.M. et al. (2021) Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature, 590, 290–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Byun, J., Han, Y., Gorlov, I.P., Busam, J.A., Seldin, M.F. and Amos, C.I. (2017) Ancestry inference using principal component analysis and spatial analysis: a distance-based analysis to account for population substructure. BMC Genomics, 18, 789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Inker, L.A., Schmid, C.H., Tighiouart, H., Eckfeldt, J.H., Feldman, H.I., Greene, T., Kusek, J.W., Manzi, J., Van Lente, F., Zhang, Y.L. et al. (2012) Estimating glomerular filtration rate from serum creatinine and cystatin C. N. Engl. J. Med., 367, 20–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ballew, S.H., Chen, Y., Daya, N.R., Godino, J.G., Windham, B.G., McAdams-DeMarco, M., Coresh, J., Selvin, E. and Grams, M.E. (2017) Frailty, kidney function, and polypharmacy: the Atherosclerosis Risk in Communities (ARIC) study. Am. J. Kidney Dis., 69, 228–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Grubb, A., Blirup-Jensen, S., Lindström, V., Schmidt, C., Althaus, H. and Zegers, I. (2010) First certified reference material for cystatin C in human serum ERM-DA471/IFCC. Clin. Chem. Lab. Med., 48, 1619–1621. [DOI] [PubMed] [Google Scholar]
- 48. Gold, L., Walker, J.J., Wilcox, S.K. and Williams, S. (2012) Advances in human proteomics at high scale with the SOMAscan proteomics platform. New Biotechnol., 29, 543–549. [DOI] [PubMed] [Google Scholar]
- 49. Han, Z., Xiao, Z., Kalantar-Zadeh, K., Moradi, H., Shafi, T., Waikar, S.S., Quarles, L.D., Yu, Z., Tin, A., Coresh, J. et al. (2018) Validation of a novel modified aptamer-based array proteomic platform in patients with end-stage renal disease. Diagnostics (Basel), 8, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kim, C.H., Tworoger, S.S., Stampfer, M.J., Dillon, S.T., Gu, X., Sawyer, S.J., Chan, A.T., Libermann, T.A. and Eliassen, A.H. (2018) Stability and reproducibility of proteomic profiles measured with an aptamer-based platform. Sci. Rep., 8, 8382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tin, A., Yu, B., Ma, J., Masushita, K., Daya, N., Hoogeveen, R.C., Ballantyne, C.M., Couper, D., Rebholz, C.M., Grams, M.E. et al. (2019) Reproducibility and variability of protein analytes measured using a multiplexed modified aptamer assay. J Appl Lab Med, 4, 30–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Krämer, A., Green, J., Pollard, J., Jr. and Tugendreich, S. (2014) Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics, 30, 523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.