Abstract
The human choroid is a heterogeneous, highly vascular connective tissue that dysfunctions in age-related macular degeneration (AMD). In this study, we performed single-cell RNA sequencing on 21 human choroids, 11 of which were derived from donors with early atrophic or neovascular AMD. Using this large donor cohort, we identified new gene expression signatures and immunohistochemically characterized discrete populations of resident macrophages, monocytes/inflammatory macrophages and dendritic cells. These three immune populations demonstrated unique expression patterns for AMD genetic risk factors, with dendritic cells possessing the highest expression of the neovascular AMD-associated MMP9 gene. Additionally, we performed trajectory analysis to model transcriptomic changes across the choroidal vasculature, and we identified expression signatures for endothelial cells from choroidal arterioles and venules. Finally, we performed differential expression analysis between control, early atrophic AMD, and neovascular AMD samples, and we observed that early atrophic AMD samples had high expression of SPARCL1, a gene that has been shown to increase in response to endothelial damage. Likewise, neovascular endothelial cells harbored gene expression changes consistent with endothelial cell damage and demonstrated increased expression of the sialomucins CD34 and ENCM, which were also observed at the protein level within neovascular membranes. Overall, this study characterizes the molecular features of new populations of choroidal endothelial cells and mononuclear phagocytes in a large cohort of AMD and control human donors.
Graphical Abstract
Choroidal endothelial and immune cells are implicated in the pathogenesis of age-related macular degeneration (AMD). In a cohort of 11 AMD and 10 control human donors, Voigt et al. used single-cell RNA sequencing to identify gene expression changes across the choroidal endothelial cell and immune subpopulations. The background depicts a whole mount of human choroidal endothelial cells (red) and IBA1-expressing immune cells (green).
Introduction
Age-related macular degeneration (AMD) is a common cause of irreversible blindness in which photoreceptors degenerate within the central retina (termed the macula), resulting in a loss of central, detailed vision. Although the pathogenesis of AMD is multifactorial, choroidal endothelial cell degeneration is strongly linked with disease development. The choroid is a heterogeneous, highly vascular connective tissue that supports outer retinal physiology. Residing beneath the neural retina and retinal pigment epithelium (RPE), the choroid supplies oxygen, glucose and retinoids to its overlying tissues and clears metabolic waste through its numerous fenestrae and caveolae (1,2). This choroidal vasculature, which is part of the peripheral circulatory system, is arranged in three layers. Most superficially, the choriocapillaris is arranged into a dense network of large diameter capillaries organized into lobules (3). Beneath the choriocapillaris, arteries and veins are organized in layers of intermediate (Sattler’s layer) and large (Haller’s layer) caliber vessels. In addition to this rich vascular network, the choroid houses many other cell types found in connective tissues, including smooth muscle cells and pericytes, multiple classes of leukocytes, Schwann cells, melanocytes, and fibroblasts.
A central component of AMD pathophysiology is vascular degeneration, and choroidal endothelial cells degenerate very early in the disease course (4). Both histological (5,6) and optical coherence tomography imaging studies (7–11) have demonstrated that choriocapillaris endothelial cells are the first cells lost in AMD, and this degeneration often occurs under a confluent layer of RPE cells and is likely a consequence of complement injury (6). In addition, chronic inflammation has been described in association with AMD. For example, AMD patients have elevated circulating activated monocytes compared to control patients (12). Likewise, histological experiments have shown that AMD donor choroids have more white blood cells compared to control donors (5,13–16), and macrophages from AMD donors are smaller and have a more spherical morphology, suggestive of inflammatory activation (17).
Considering the multiple interwoven pathogenic mechanisms of AMD, it can be helpful to study individual populations of choroidal cells independently. We and others have used single-cell RNA sequencing (scRNA-seq) to study choroidal physiology in human donors (18–20). These studies have characterized the major choroidal cell populations and identified distinct clusters of choroidal arteries, veins and choriocapillaris endothelial cells. However, a limitation of these prior studies is that they have collectively profiled only two human donors with AMD (one atrophic, one neovascular). As such, the degree of gene expression differences between control and AMD choroids has been largely unquantified. Likewise, subsets of microglia (the resident macrophages of the retina) have been widely investigated with scRNA-seq (21–23), yet only one study has characterized transcriptomes of choroidal macrophage subpopulations (24). To further elucidate AMD pathogenesis mechanisms of these important cells (14), continued investigation of choroidal macrophage subpopulations in large AMD and control cohorts is warranted.
In this study, we perform scRNA-seq on 21 adult human donor choroids, 11 of which had clinical and/or histological evidence of atrophic (n = 9) or neovascular (n = 2) macular degeneration. We identified new gene expression signatures for several populations of immune cells within the choroid, including distinct clusters of resident macrophages, monocytes/inflammatory macrophages, and dendritic cells, which we validated with immunohistochemistry. In addition, we identified gene expression changes as choroidal endothelial cells transition from large caliber arteries to narrow capillaries and back to large caliber veins. Finally, we compared gene expression between early atrophic AMD, neovascular AMD, and control donors. We identified that the damage-associated gene SPARCL1 was enriched in choriocapillaris endothelial cells in early atrophic AMD donors and validated its protein-level expression in the choriocapillaris. Likewise, CD34 and ENCM were enriched in the neovascular AMD donors, and we immunohistochemically observed these gene products within choroidal neovascular membranes. Collectively, this study characterizes gene expression of several newly described choroidal cell populations in a large cohort of both AMD and control donors.
Results
scRNA-seq experiment design
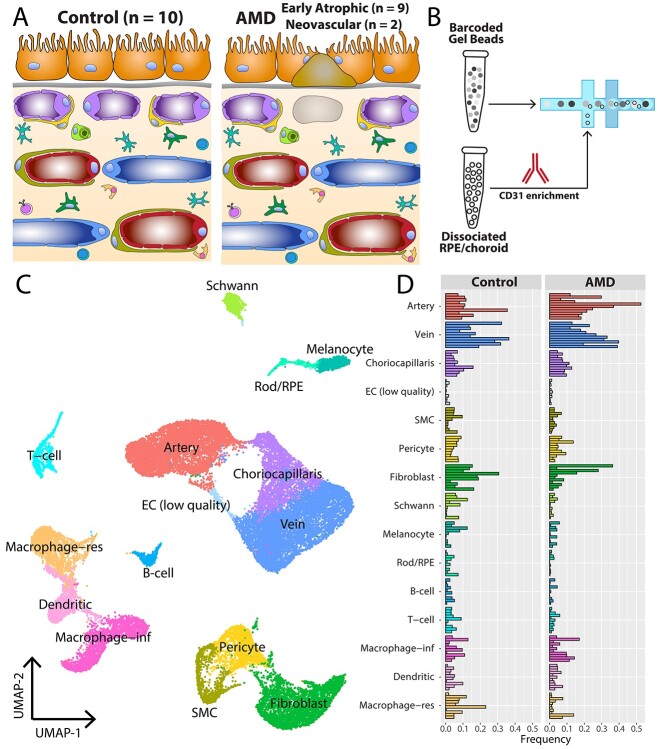
Gene expression changes in AMD choroidal cell populations have been largely unquantified. To better understand the AMD transcriptome, we collected 21 human macular RPE/choroid punch biopsies for scRNA-seq. Nine of these donors had histological evidence of early atrophic (non-neovascular or ‘dry’) AMD, demonstrating numerous soft drusen and/or basal laminar deposits (Fig. 1). In addition, two donors (donor 11, donor 18) had a clinical diagnosis of neovascular AMD (Table 1). In contrast, the remaining 10 donors had no histological nor clinical evidence of AMD and comprised a control cohort (Fig. 1).
Figure 1.
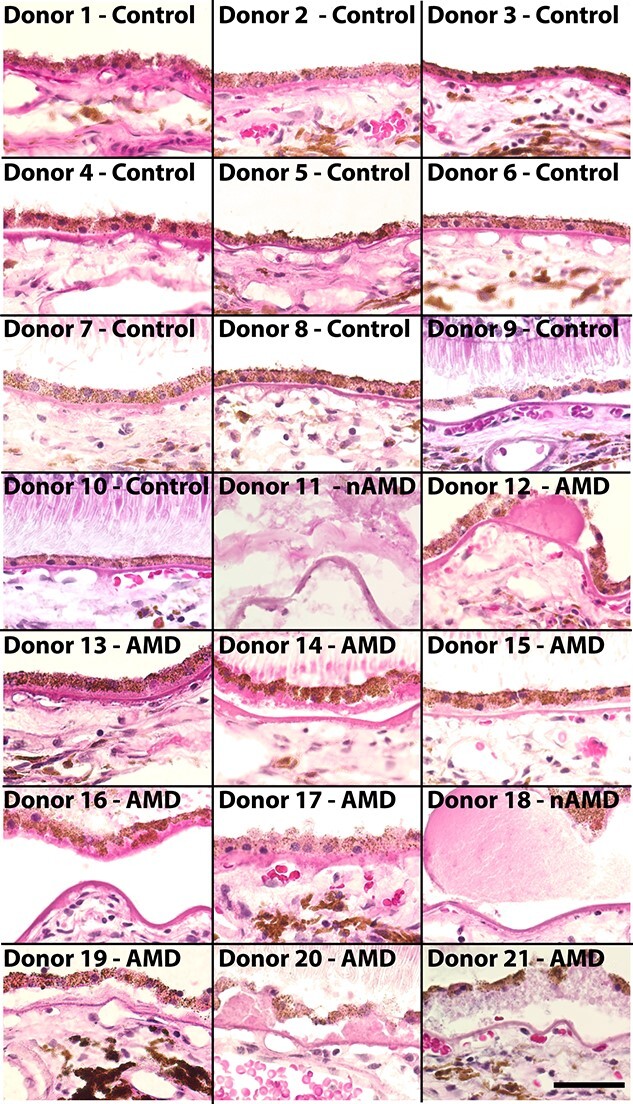
Hematoxylin and eosin staining of human donor eyes. Images from the submacular choroid of control donors (donors 1–10) versus AMD donors (donors 11–21). Donor 11 was diagnosed with neovascular AMD OU, and a neovascular membrane is shown. Donor 18 was diagnosed with neovascular AMD in OS (the eye used for scRNA-seq) while the histological image (large confluent drusen) was acquired from OD. All other AMD donors had histological evidence of early atrophic AMD. Scalebar (donor 21) = 50 μm. OS = oculus sinister (left eye); OD = oculus dexter (right eye); OU = oculus uterque (both eyes).
Table 1.
Demographic information of control and AMD donors
| Donor | Disease | Age | Sex | PMI | Eye | Cause of Death | Ophthalmologic History | MMP9 (rs4810482) |
|---|---|---|---|---|---|---|---|---|
| 1 | Control | 75 | F | 6:44 | OS | COPD | Glaucoma suspect OU, history of BRVO OU with 20/20 vision | C/C |
| 2 | Control | 77 | M | 4:48 | OD | Lung cancer with metastasis | Records negative for AMD, histologically normal Bruch’s in OS | C/T |
| 3 | Control | 61 | M | 3:21 | OS | Tumor lysis syndrome | No records received, histologically normal Bruch’s in OD | C/T |
| 4 | Control | 77 | F | 6:04 | OD | Cardiac arrest | Records negative for AMD, histologically normal Bruch’s in OS | T/T |
| 5 | Control | 63 | M | 4:44 | OD | Respiratory failure | Myopia OU, nuclear sclerosis OU, pigmentary dispersion syndrome, and glaucoma suspect OU | T/T |
| 6 | Control | 79 | F | 7:21 | OS | Respiratory failure secondary to COPD | No records received, histologically normal Bruch’s in OD | T/T |
| 7 | Control | 64 | M | 6:54 | OS | Cardiac shock | Type 2 diabetes without retinopathy | C/C |
| 8 | Control | 76 | F | 7:41 | OS | Cardiac failure | Records negative for AMD, histologically normal Bruch’s in OD | T/T |
| 9 | Control | 81 | M | 5:10 | OS | Septic shock | Records negative for AMD, histologically normal Bruch’s in OD | C/C |
| 10 | Control | 83 | M | 5:18 | OD | Cardiopulmonary arrest | Glaucoma/OHT OU | T/T |
| 11 | AMD (neovascular) | 92 | F | 5:04 | OD | Respiratory distress | Neovascular AMD OU | C/T |
| 12 | AMD | 80 | M | 5:50 | OS | Cardiac arrest | Early dry non-exudative AMD OU | T/T |
| 13 | AMD | 90 | M | 5:17 | OS | Aspiration pneumonia | Dry AMD with macular drusen/mottling | T/T |
| 14 | AMD | 90 | F | 5:33 | OS | Ovarian cancer with metastasis | Early dry AMD OU | T/T |
| 15 | AMD | 90 | M | 5:21 | OS | Septic shock | No records received, histologically observed basal laminar deposits in OD | T/T |
| 16 | AMD | 84 | M | 5:22 | OD | Stage 4 prostate cancer | Intermediate stage dry AMD OU, type 2 diabetes without retinopathy OU | C/T |
| 17 | AMD | 77 | F | 6:52 | OS | Sepsis | Mild non-exudative AMD OU | C/C |
| 18 | AMD (neovascular) | 78 | M | 6:42 | OS | Chronic respiratory failure | Exudative age-related macular degeneration with active choroidal neovascularization OS | T/T |
| 19 | AMD | 86 | F | 6:10 | OD | Lung cancer with metastasis | Retinoschisis OD, type 2 diabetes with retinopathy OU | C/C |
| 20 | AMD | 91 | F | 4:59 | OS | Urosepsis | No records received, histologically observed numerous soft drusen in OD | C/C |
| 21 | AMD | 92 | F | 6:37 | OS | Subfalcine herniation secondary to left subdural hematoma | No records received, histologically observed numerous soft drusen in OD | C/C |
OD, oculus dexter (right eye); OS, oculus sinister (left eye). PMI = post mortem interval; COPD = chronic obstructive pulmonary disease; BRVO = branched retinal vein occlusion.
Both histologic and optical coherence tomography angiography imaging data suggest that degeneration of choroidal endothelial cells, particularly choriocapillaris loss, is central to AMD pathogenesis. However, since endothelial cells represent a minority of unselected choroidal cells (19), we set out to recover large populations of endothelial cells from each donor by performing a CD31 enrichment of dissociated cell samples prior to scRNA-seq (Fig. 2A and B). After barcoding, library preparation, mapping and filtering low-quality cells, we recovered a total of 30 416 cells (17 097 from AMD donors, 13 319 from control donors), which were sequenced to a mean depth of 184 359 reads/cell. After normalization and graph-based clustering, we classified each cluster of cells into choroidal cell types based on previously published expression profiles (18–20,25) (Fig. 2C, Supplementary Material, Fig. S1).
Figure 2.
scRNA-seq experiment design. (A) Cartoon of the submacular RPE/choroid in health (left) and AMD (right). (B) After dissociation and cryopreservation, cell suspensions from the 21 donors were thawed before anti-CD31 (PECAM1) magnetic enrichment. CD31-enriched fractions from each donor underwent scRNA-seq. (C) Uniform manifold approximation and projection (UMAP) dimensionality reduction of the 30 416 recovered cells. Cell clusters were classified into cell types based on previously published expression profiles (Supplementary Material, Fig. S1). (D) Comparison of recovered cells from control and AMD samples. Each bar represents the proportion of recovered cells for one donor. SMC = smooth muscle cell. EC = endothelial cell. RPE = retinal pigment epithelium. Macrophage-res = resident macrophage. Macrophage-inf = inflammatory macrophage/monocyte.
Comparing cellularity of the choroid between AMD and control donors
After classifying all clusters into cell types, we compared cell type proportions between the AMD and control donors (Fig. 2D). By performing the CD31 enrichment, we expected to recover many choroidal endothelial cells which abundantly express this gene product. In addition, several immune populations also express CD31, including strong expression in monocytes/macrophages (26) and moderate expression in lymphoid cells (27,28). Unsurprisingly, choroidal veins and arteries were the most abundantly recovered cell types in this experiment, followed by fibroblasts, inflammatory and resident macrophages (see below), and choriocapillaris endothelial cells. In the AMD donors, we recovered a slightly higher proportion of large caliber veins (26% of AMD cells vs. 21% of control cells) and arteries (25% of AMD cells vs. 13% of control cells). Conversely, control donors had slightly more resident macrophages (3% of AMD cells vs. 9% of control cells) and fibroblasts (9% of AMD cells versus 15% of control cells). All other cell populations were recovered at a similar proportion (±3%) between AMD and control donors.
Identification of resident macrophages, inflammatory monocytes/macrophages and dendritic cells in the choroid
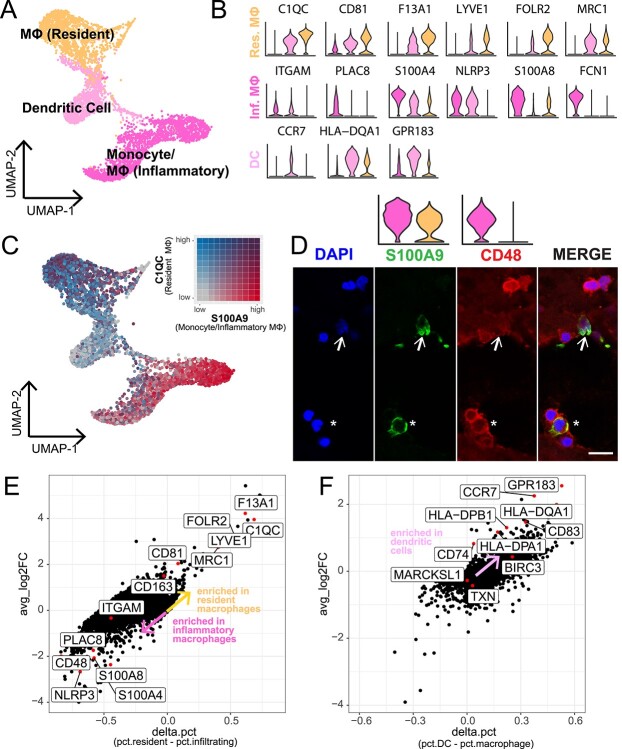
Interestingly, we identified three distinct clusters of mononuclear phagocytes with previously uncharacterized transcriptomes in the choroid (Fig. 3A). To classify potential macrophage and dendritic subtypes, we performed differential expression to identify genes unique to each of these clusters (Supplementary Material, Table S3). One cluster demonstrated high expression of common macrophage genes such as CD14 and AIF1 (which encodes the calcium-binding adaptor molecule IBA1). In addition, this cluster showed high expression of genes found in ‘resident’ tissue macrophages, including C1QC, CD81 (29), F13A1, LYVEI, FOLR2 and MRC1 (30). A second related cluster also expressed common macrophage genes, yet this cluster demonstrated specific expression of classic monocyte genes such as S100A8 (29) as well as ‘infiltrating’ and ‘inflammatory’ monocyte/macrophage genes, including ITGAM, PLAC8 (29), NLRP3 (30), S100A4, S100A8 and FCN1 (31) (Fig. 3B). Thus, we classified this second cluster as ‘monocyte/inflammatory macrophages.’ These two monocyte/macrophage clusters are well separated (Fig. 3C) and differential expression analysis recapitulates the enrichment of previously reported ‘resident’ versus ‘inflammatory’ macrophage genes (Fig. 3E).
Figure 3.
Characterization of multiple mononuclear phagocyte cell populations in the human choroid. (A) Three clusters of mononuclear phagocytes were identified in the scRNA-seq study. (B) Violin plots show the expression of genes previously reported to localize to resident macrophages (orange), monocytes/inflammatory macrophages (dark pink) and dendritic cells (light pink). (C) Expression of C1QC (a marker for tissue-resident macrophages, colored grey-to-blue) and S100A8 (a marker for monocytes/inflammatory macrophages, colored grey-to-red) are colored on a bivariate color scale. The two populations are largely distinct. (D) Immunohistochemical labeling of CD48 (a gene expressed by monocytes/inflammatory macrophages) and S100A8/9 (expressed by both resident macrophages and monocytes/inflammatory macrophages) in the human choroidal stroma. Asterisks indicate cells that co-express CD48 and S100A9 and represent monocytes/inflammatory macrophages. Arrows indicate cells that express S100A9 but do not express CD48, representing resident macrophages. Overlying violin plots highlight gene expression of S100A9 and CD48 across the two macrophage populations. Scalebar = 10 μm. (E) Differential expression was completed between the resident macrophage versus the monocyte/inflammatory macrophage clusters. Each dot represents the differential expression results for one gene. Labeled genes have been previously reported to be enriched in resident or inflammatory macrophages. (F) Like (E), differential expression was performed between the dendritic cell cluster versus both macrophage clusters. Labeled genes have been previously reported to be enriched in dendritic cells from other tissues.
The third mononuclear phagocyte cluster demonstrated minimal expression of monocyte- and macrophage-specific genes and instead exhibited a dendritic cell expression signature. This cluster of cells demonstrated high expression of the dendritic cell-enriched genes CCR7, HLA-DQA1 and GPR183 (Fig. 3B) (32). In addition, we performed differential expression between the proposed dendritic cells versus the resident/inflammatory macrophage clusters, and the majority of previously reported dendritic cell genes (32) were enriched in this dendritic cell cluster (Fig. 3F).
Immunohistochemical labeling showed that CD48, which was enriched in the monocyte/inflammatory macrophage cluster, labeled several round cells in the choroidal stroma (Fig. 3D, red). In contrast, S100A8 was highly expressed by both the resident macrophage and monocyte/inflammatory macrophage cluster (Fig. 3D, green). We identified mononuclear cells that only expressed S100A8/9, characteristic of resident macrophages (Fig. 3D, arrow). Other S100A8/9-positive cells co-expressed CD48, matching the gene expression of monocytes/inflammatory macrophages (Fig. 3D, asterisk). We observed other round CD48-positive cells that did not co-label with S100A8/9. These CD48-expressing cells could represent either S100A8/9-negative macrophages or T cells (which express some CD48 mRNA but do not express S100A8/9).
Characterization of the continuous choroidal vascular tree
We identified distinct gene expression patterns for the major endothelial cell populations (arteries, capillaries, and veins) (Fig. 4A). While grouping cells into these discrete populations is helpful for studying endothelial biology, the mature choroidal vascular tree is continuous. Large caliber (and deep) arteries bifurcate into smaller caliber arterioles and even narrower (superficial) capillaries. These capillaries then drain blood into small venules, which eventually merge into large caliber (and deeper) veins.
Figure 4.
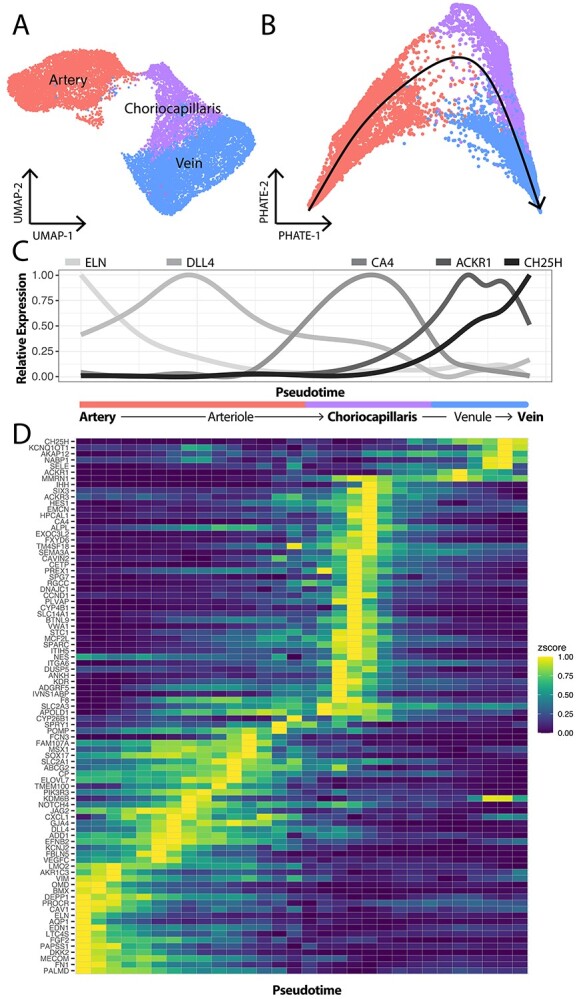
Trajectory analysis recapitulates the anatomic structure of the choroidal vascular tree. (A) Three distinct endothelial clusters (arteries, choriocapillaris and veins) were identified by scRNA-seq. (B) The PHATE dimensionality reduction method was applied to the endothelial expression data to better visualize progression from one cell state to another. Trajectory analysis was performed using slingshot (black line) to model expression changes from arteries to capillaries to veins. (C) Expression changes of five genes were visualized across this trajectory. The pseudotime trajectory is visualized along the x-axis; values on the left correspond to cells mapped to the arterial end of the trajectory while values on the right correspond to cells from the venous end. Previously reported arteriole (DLL4)- and venule (ACKR1)-specific genes demonstrated maximal expression at the artery–choriocapillaris and choriocapillaris–vein interface. (D) Differential expression was performed to identify genes enriched in arteries, arterioles, choriocapillaris, venules and veins. The expression of each gene was scaled from zero (dark blue) to one (yellow) and expression was visualized across pseudotime (x-axis) in a heatmap.
We investigated if these transitionary endothelial states were reflected in the gene expression data. First, we applied the dimensionality reduction method PHATE, which better preserves continuous progressions and branches in scRNA-seq data, to the choroidal endothelial cell clusters (33) (Fig. 4B). Next, we performed trajectory inference using slingshot to model gene expression changes from the arteries to the veins through the capillaries (Fig. 4B, black line). We visualized the expression of previously published markers of large caliber arteries (ELN), arterioles (DLL4), capillaries (CA4), venules (ACKR1), and large caliber veins (CH25H) across this trajectory (34) (Fig. 4C). In support of the trajectory analysis, arteriole gene expression peaked at the transition between arteries and capillaries, and venule gene expression peaked at the transition between capillaries and veins.
Based on pseudotime values, we manually classified cells into populations of arteries, arterioles, capillaries, venules and veins, and performed differential expression analysis to identify what genes are unique to each endothelial subpopulation (Fig. 4C, Supplementary Material, Table S4). This analysis suggested that AQP1 and PALMD are large artery-specific genes, FCN3 and GJA4 are arteriole genes, PLVAP and RGCC are choriocapillaris-specific genes, PKHD1L1 is a venule-specific gene, and TGFBR3 is a vein-specific gene (Supplementary Material, Fig. S2). In addition, some genes such as MGP and VCAM1 were restricted to large caliber choroidal arteries and veins (Supplementary Material, Fig. S2). In summary, this trajectory analysis suggests that endothelial cell transcriptomes can be ordered to recapitulate the anatomy of the choroidal vasculature.
Gene expression changes in early atrophic AMD
After characterizing novel choroidal cell mononuclear phagocyte and endothelial cell populations, we performed differential expression between early atrophic AMD (n = 9) and control (n = 10) donors across each cell population (Supplementary Material, Table S5). Conventionally, most scRNA-seq analysis packages treat all cells as independent observations in differential expression analyses. However, in a typical scRNA-seq experiment the number of recovered cells is orders of magnitude higher than the number of biological replicates. Hence, when comparing expression between early atrophic AMD versus control donors, a ‘pseudobulk’ differential expression analysis is most appropriate (35). In this strategy, the raw counts are summed for each donor across each independent cell type before differential expression. This more accurately captures the number of biological replicates in a study and avoids artificially significant P-values.
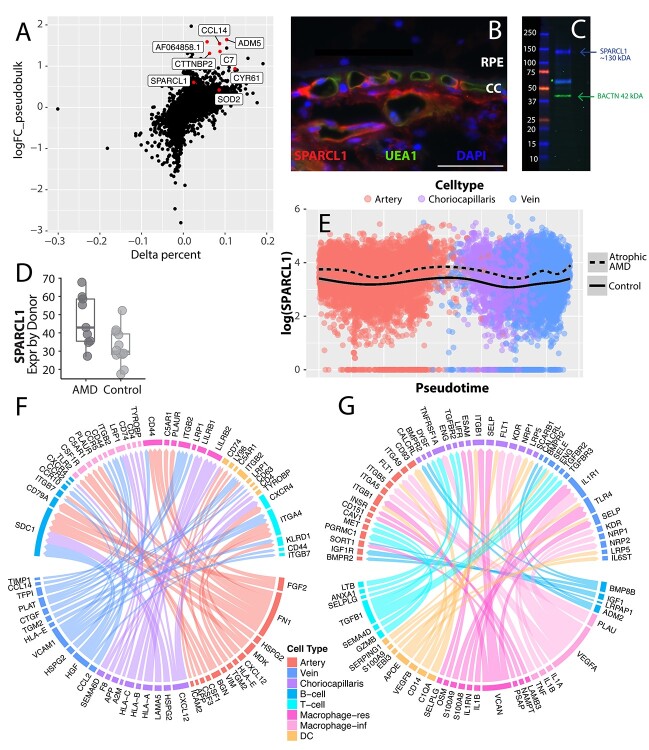
In the choriocapillaris, 98 genes demonstrated absolute log-fold changes greater than 0.5, although no genes yielded a false discovery rate (FDR) of less than 0.05 after correction for multiple hypotheses. While this lack of statistical significance is perhaps unsurprising (see Discussion), we performed a literature review of genes with the highest AMD enrichment (assessed by log-fold change) for potential hypothesis-generating insight into endothelial cell dysfunction in AMD (Fig. 5A). Genes enriched in early atrophic AMD samples included GALNT15, which catalyzes the synthesis of O-linked oligosaccharides (36), CYR61, which promotes endothelial migration and angiogenesis (37), and superoxide dismutase 2 (SOD2), which reduces the accumulation of reactive oxidative species. In addition, SPARCL1 is a secreted glycoprotein that is expressed in response to cell injury (38), and this gene was enriched in early atrophic AMD samples (Fig. 5D). Expression of SPARCL1 has been shown to decrease endothelial cell adhesion to the basement membrane and increase the spreading of endothelial cells in vitro (39). In sections of the human choroid, SPARCL1 localizes to choroidal endothelial cells (including the choriocapillaris) (Fig. 5B) and SPARCL1 protein was detected by western blot in human choroidal donor tissue (Fig. 5C). We examined SPARCL1 expression across pseudotime in early atrophic AMD and control samples (Fig. 5E), and SPARCL1 expression is higher in AMD cells across the entire vascular tree.
Figure 5.
Genes enriched in early atrophic AMD endothelial cells. (A) Differential expression between early atrophic AMD (n = 9) and control (n = 10) choriocapillaris endothelial cells. The y-axis depicts a (pseudobulk) log-fold change, with positive values reflecting increased expression in early atrophic AMD donors (see Materials and Methods). The x-axis depicts a variable termed delta percentage, which is calculated by: (percentage of AMD cells that express the gene) − (percentage of control cells that express the gene). Genes with a positive delta percentage are expressed by a higher proportion of AMD cells. (B) SPARCL1 (red) labels the choroidal endothelial cells, including the choriocapillaris, of a human donor choroid. The endothelial-specific lectin UEA-1 (green) co-labels choroidal endothelial cells. (C) A western blot of protein isolated from the human choroid. A SPARCL1 band is visible at the molecular weight of 130 kDa (40) in addition to a smaller, possibly proteolytic fragment ~60 kDa (41). (D) Comparison of SPARCL1 choriocapillaris expression between early atrophic AMD and control donors. The average SPARCL1 expression is depicted on a donor-by-donor basis (each dot represents the average SPARCL1 expression of one donor). (E) SPARCL1 expression is compared across pseudotime (see Fig. 4) stratified by AMD status. There is an increase in SPARCL1 expression across the choroidal vascular tree in early atrophic AMD. (F, G) Ligand–receptor interactions are visualized between endothelial and immune subpopulations. Ligand–receptor pairs are only visualized if at least one member of the pair is enriched in early atrophic AMD cells above a (pseudobulk) log-fold change of 0.5. In (F), endothelial-expressed ligands are visualized with their immune cell targets. In (G), immune-expressed ligands are visualized with their endothelial targets.
In addition, we performed a connectome analysis to visualize signaling network interactions between choroidal endothelial cells and immune populations (Fig. 5F and G). We restricted our analysis to genes with an AMD enrichment (pseudobulk logFC > 0.5, see Materials and Methods) in at least one member of the ligand–receptor pair. We visualized endothelial-expressed ligands (Fig. 5F) as well as ligands expressed by immune cells (Fig. 5G) along with their respective receptor targets. Several of the mononuclear phagocyte populations demonstrated AMD-enriched receptor targets that promote inflammation. For example, the monocyte/inflammatory macrophage population expressed the adhesion molecules ITGB2 (which encodes CD18), which along with CD11b can bind to the complement component C3 and ICAM-1 (42). In addition, the monocyte/inflammatory macrophages demonstrated expression of C5AR1, which serves as a receptor for the complement component C5a that, when activated, drives inflammation (42–44). Finally, both dendritic cells and resident macrophages demonstrated S100A8/A9 enrichment in AMD. S100A8/A9 are calcium-binding proteins that promote leukocyte recruitment and have been implicated as drivers of inflammatory diseases (45).
Expression of genes associated with AMD risk
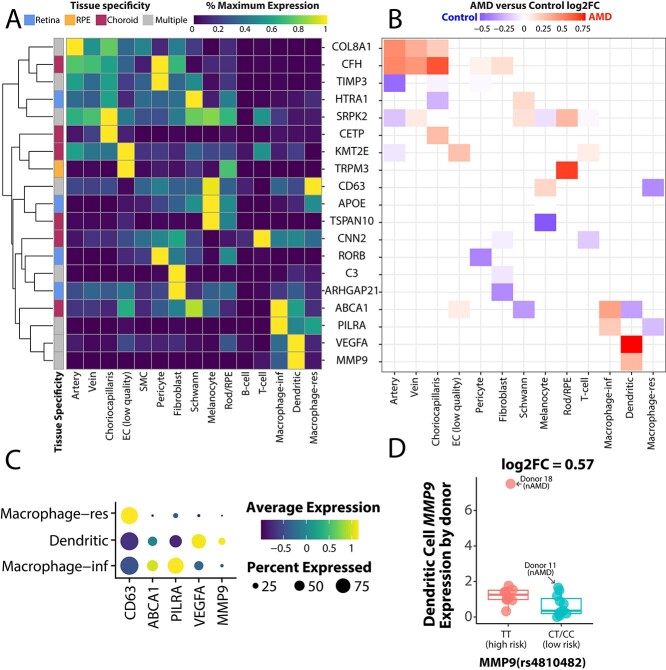
Next, we visualized the cell-specific expression patterns of AMD-associated genes identified by genome wide association studies (46). First, we used scRNA-seq data to classify if each AMD-associated gene was expressed more in the retina, RPE, or choroid. Of the 46 AMD-associated GWAS genes, only 24 had an expression level greater than 1.0 transcripts per 10 000 in the highest retinal, RPE, or choroidal cell population. Seven genes demonstrated maximal expression in the retina, six genes had maximal expression in the choroid, and three genes had maximal expression in the RPE. The remaining eight genes were expressed at comparable levels between two or more tissues.
We visualized the expression patterns of these AMD-associated genes across all choroidal cell types (Fig. 6A). Of note, three retinal-specific genes (VTN, TMEM97, COL4A3) and two RPE-specific genes (SLC16A8, RDH5) had very low (<1.0 transcript per 10 000) expression in this dataset and were thus excluded from this visualization. A group of genes (COL8A1, CFH, TIMP3, CETP and SPRK2) were most highly expressed by choroidal endothelial cells or their associating pericytes. A second set of genes (including CD63, ABCA1, PILRA, VEGFA and MMP9) were most highly expressed by one of the choroidal mononuclear phagocyte populations.
Figure 6.
Expression of AMD risk genes in the choroid. (A) A heatmap depicts cell-specific expression patterns of genes with AMD-associated genetic variants (46). Each gene is classified as expressed mostly in the retina, RPE, choroid or in multiple tissues (see Materials and Methods). For each gene, we scale expression according to this value from 0% to 100% of the maximally expressing cell type. (B) For all genes in (A), we compare expression between AMD and control cells. (C) A dot plot depicts the relative expression level (blue-to-yellow gradient) and percentage of expressing cells (circle size) of five genes maximally expressed by mononuclear phagocytes. (D) MMP9 expression was compared between donors genotyped at the rs4810482 SNP, which is associated with neovascular AMD. Each dot represents the average MMP9 expression of one donor.
We also compared the expression of these AMD-associated genes between our early atrophic AMD and control cohort (Fig. 6B). However, when two cell populations both have low expression of a gene, differential expression results (and log-fold changes) can be artificially inflated. Therefore, differential expression results are only visualized for cell types that have at least 50% of the maximal expression for a given gene. Genes that showed modest enrichment in AMD donors included COL8A1 and CFH in choroidal endothelial cells as well as VEGFA and MMP9 in choroidal dendritic cells (Fig. 6B).
The mononuclear phagocytes demonstrated interesting cell-specific expression patterns of AMD-associated transcripts (Fig. 6A). Thus, we investigated expression characteristics of the mononuclear phagocyte localized genes (CD63, ABCA1, PILRA, VEGFA and MMP9) in more detail (Fig. 6C). The resident macrophage cluster demonstrated high expression of CD63 while the monocyte/inflammatory macrophage cluster specifically expressed ABCA1 and PILRA. In contrast, choroidal dendritic cells showed most of the VEGFA and MMP9 expression. Recently, our group validated a genetic association between three single-nucleotide polymorphisms (SNPs) in the MMP9 locus and neovascular AMD (46,47). Thus, we genotyped each of the 21 donors at the MMP9 SNP rs4810482. Donors with the high-risk (TT) genotype at this locus demonstrated slightly higher MMP9 expression (log2FC = 0.57) (Fig. 6D).
Gene expression changes in neovascular AMD
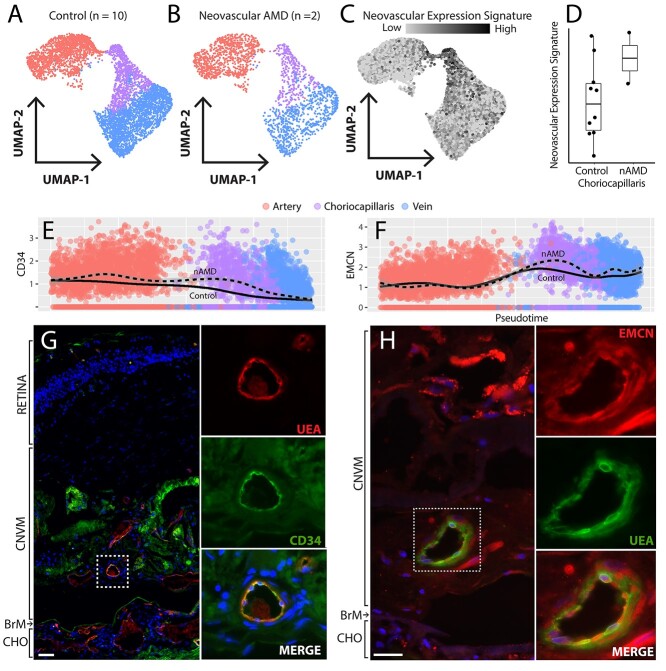
Recently, scRNA-seq has compared gene expression between healthy and laser-injured choroidal endothelial cells in a mouse model of choroidal neovascularization (CNV) (18). As two donors in our cohort were diagnosed with nAMD (Fig. 7A and B, Table 1), we investigated if the laser-injured mice and the nAMD donors shared gene expression changes. Laser injury to endothelial cells results in very acute damage opposed to the longstanding, chronic choroidal insults observed in human AMD. However, identifying shared expression perturbations between the laser injury model and human AMD may highlight biological changes specific to neovascular endothelial cells. Thus, we created a CNV gene expression signature from the top 50 genes enriched in laser-injured murine endothelial cells (excluding ribosomal genes). This gene expression signature predominately localized to the choriocapillaris in our human cohort (Fig. 7C). Although the small sample size of our neovascular cohort precludes statistical analysis, one nAMD donor (donor 11) had the highest CNV expression score while the other (donor 18) had the fifth-highest CNV expression score (Fig. 7D).
Figure 7.
Genes enriched in neovascular AMD endothelial cells. Choroidal endothelial cells (arteries = red, choriocapillaris = purple, veins = blue) from the 10 control donors (A) and the two neovascular AMD donors (B). (C) A neovascular expression signature was created by analyzing the top 50 upregulated genes in a laser injury model of choroidal endothelial cells (18). This expression signature was applied to endothelial cells in the current study, and choriocapillaris endothelial cells most highly expressed these signature genes. (D) The average neovascular expression score was computed between control and nAMD donors. (E, F) CD34 (E) and EMCN (F) expression was higher across the choroidal vascular tree in nAMD donors (dashed line). (G, H) CD34 (G) and EMCN (F) are expressed at the protein level in neovascular membranes in an independent CNV donor cohort. Scale bars = 50 μm.
Next, we performed differential expression between control (n = 10) and neovascular AMD (n = 2) donors across all cell populations (Supplementary Material, Table S6). Two neovascular-enriched genes of interest were CD34 and EMCN, two sialomucins that demonstrated high expression in the choriocapillaris of the nAMD donors (Fig. 7E and F). Both gene products were observed at the protein level in neovascular membranes in an independent CNV cohort (Fig. 7G and H).
Discussion
scRNA-seq has been used to study molecular features of the mouse (18,25) and human (18–20) choroid. However, previous scRNA-seq investigations have only included a limited number of donor choroids with documented AMD. In our group’s previous investigations of the human choroidal transcriptome at the single-cell level, we sequenced a total of two donors with phenotyped AMD (including one neovascular AMD donor, one atrophic AMD donor) (19,20). Rohlenova and colleagues have also performed scRNA-seq on one healthy human choroid, but AMD donors were not included in this study (18). This lack of biological replicates makes it difficult to quantify the degree of expression differences between AMD and control choroids. In this study, we set out to address this limitation by performing scRNA-seq on 21 human donor choroids, 11 of which had clinical and/or histological evidence of atrophic or neovascular AMD (nAMD).
Choriocapillaris cells degenerate early in AMD and are central to disease pathogenesis (6). Several genes demonstrated moderate log-fold change enrichments in early atrophic AMD that provide hypothesis-generating insight into this disease process. One such gene with an early atrophic AMD enrichment was SPARCL1 (pseudobulk log2FC = 0.61, absolute fold change = 1.53) (Fig. 5A–E). SPARCL1 encodes a matricellular protein that promotes cellular quiescence (48) and stimulates the detachment of cells from basal lamina through focal adhesion disassembly (49,50). SPARCL1 is additionally cleaved by matrix metalloproteinases and ADAMTS4 (51), generating a shorter C-terminal fragment that is associated with neovascularization in a murine glioma model (52). Likewise, the related protein SPARC is uniquely upregulated in the setting of cell injury (38). We verified that SPARCL1 was expressed at the protein level in a perivascular pattern around choroidal endothelial cells, including the choriocapillaris (Fig. 5B). Although we did not detect differences in SPARCL1 protein expression in an independent AMD and control cohort, the high choriocapillaris expression of this gene in several AMD donors is nonetheless interesting considering the pattern of endothelial loss observed in early AMD.
By including 21 human donor choroids in this study and sequencing cells very deeply (to an average depth of ~ 184 000 reads/cell), we were able to identify several human choroidal cell populations with previously uncharacterized transcriptomes. First, we were able to model gene expression as a function of pseudotime along the continuous choroidal vascular tree and identify transitionary populations of arterioles and venules (Fig. 4). We verified that previously reported arteriole- and venule-specific genes were maximally expressed at the pre- and post-capillary interface, validating this trajectory in the choroid (34). The expression signatures of post-capillary venules are of particular interest, as this is the location of maximal leukocyte adhesion and extravasation through the endothelium (53). Using the pseudotime models, we verified that endothelial adhesion molecules ICAM1, VCAM1, SELE and SELP reach peak RNA expression in the choroidal post-capillary venules (Supplementary Material, Fig. S3). An expression signature for all choroidal endothelial cell populations is reported in Supplementary Material, Table S4.
Likewise, we identified gene expression signatures for distinct populations of macrophages and dendritic cells (Fig. 3). While different subsets of microglia and monocyte-derived macrophages have been described in the retina (21,22), specialized subsets of choroidal macrophages have been largely uncharacterized. Conventionally, macrophages have been characterized as having proinflammatory (M1) or anti-inflammatory (M2) phenotypes, but this binary classification scheme does not accurately capture the degree of macrophage heterogeneity (29,54). Instead, we have identified populations of resident macrophages, which locally proliferate within the tissue, and recruited monocyte/inflammatory macrophages. Resident macrophages contribute to tissue homeostasis by phagocytizing cellular debris and helping resolve inflammation (55). In contrast, bone marrow-derived monocytes recruited to the choroid (which are referred to as macrophages once they extravasate out of the blood vessel) can drive inflammation by producing reactive oxygen species and cytokines (56). Dendritic cells are also phagocytic, but mainly serve to stimulate T cells (which are also found within the choroid) (15). Our previous immunohistochemical experiments suggest that approximately 75% of CD45-expressing immune cells reside within the choroidal stroma, while the remaining 25% were found within vascular lumens. Thus, it is possible that some of the sequenced immune cells had not migrated into the choroidal stroma and instead were recovered from the choroidal circulation.
Interestingly, several AMD-associated genes specifically localize to one of these three populations of mononuclear phagocytes. For example, the classical complement components C1QA/B/C are all highly expressed by tissue-resident macrophages while the complement receptors ITGB2, ITGAX, C3AR1 are specifically expressed by monocytes/proinflammatory macrophages (Supplementary Material, Fig. S4). Lastly, dendritic cells specifically express the neovascular AMD-associated gene MMP9 (Fig. 3). Characterizing gene expression in these subtypes of mononuclear phagocytes can help us better understand pathogenesis mechanisms for these important AMD-associated genes.
There are several limitations to this study. First, as discussed above, only a small handful of genes reached statistically significant enrichment in AMD or control donors after correction for multiple hypotheses. Properly accounting for biological variability in differential expression analysis is important to minimize false discoveries (57). In a recent experiment, scRNA-seq was used to compare gene expression between severe spinal cord contusion (n = 3) and control (n = 3) mice from the same genetic background (57). Endothelial cells demonstrated the greatest expression changes in the context of this spinal cord injury model, yet only 16 genes were observed to be differentially expressed after completion of a pseudobulk analysis. Considering these results, it is perhaps unsurprising that in our study, most cell populations do not have any AMD-enriched genes with statistically significant FDR-corrected P-values. Compared to mouse models, the human donor choroids in this study are from individuals with divergent ages, genetic backgrounds, environmental risk factors. Likewise, while all donors in the control cohort did not have clinical or histological signs on AMD, some control donors had other eye or systemic diseases that may influence gene expression. While our cohort of 21 donors is considered large for a scRNA-seq study, many more donors would be required to achieve genome-wide statistical confidence and correct for the intrinsic biological variability of human donors. Despite this lack of statistical significance, we observe many genes with moderate enrichment in early atrophic or neovascular AMD. We propose that the enclosed differential expression results are extremely valuable for hypothesis generation, especially considering the rarity of phenotyped human donor eyes with AMD. Second, while our group is most interested in studying choroidal endothelial cells (as these cells degenerate very early in AMD), RPE cells are also very important in retinal physiology and undergo dysfunction in AMD. The CD31 selection method prior to scRNA-seq, while necessary to analyze sufficient numbers of endothelial cells, precluded recovery of most RPE cells, and as such we were unable to quantify expression changes in AMD for this important cell population. Third, AMD is associated with the loss of some choroidal cell types (5,11). While we compared the proportion of each cell population in AMD versus control donors (Fig. 2D), the CD31 enrichment prevents an accurate quantification of the choroidal cellularity in AMD versus control donors.
This study provides new insight into the choroidal transcriptome in a large cohort of 10 healthy and 11 AMD human donors. With this dataset, we characterized the gene expression of multiple populations of choroidal macrophages/dendritic cells and we identified gene expression changes along the choroidal vascular tree. All raw data generated in this study have been deposited in the Gene Expression Omnibus database (GSE183320). In addition, processed data have been made fully available for interactive exploration with Spectacle (this dataset is labeled ‘choroid_amd_control’), which is freely accessible at https://singlecell-eye.org (58).
Materials and Methods
Human donor eyes
Enucleated human donor eyes were acquired from the Iowa Lions Eye Bank with full consent from the donor’s next of kin in accordance with the Declaration of Helsinki. A total of 21 human RPE/choroid samples were included in this study (Table 1). Six of these donors (control donors 1–3, 5; AMD donor 11–12) have been previously sequenced (19,20) and were integrated with newly acquired data from 15 donors. Supplementary Material, Table S1 contains GSE accession information for these six previously sequenced donors.
AMD classification
Samples were assigned to early atrophic AMD/drusen, neovascular AMD, or control groups based on chart review when medical records were available. Donors with advanced dry AMD/geographic atrophy were excluded from this study. In all cases, the contralateral macula was sectioned and evaluated histologically for confluent basal laminar deposits, pigment changes and/or large drusen. In cases without medical records, the histological appearance of the sections was used to populate the study. Samples that were ambiguous or unable to be read were not included in scRNA-seq experiments.
Sample preparation for scRNA-seq
Macular RPE/choroidal tissue was isolated from donors with 8- or 12-mm trephine punch biopsies centered on the fovea. Overlying retinal tissue was removed before the RPE/choroid was minced into approximately 1 mm × 1 mm squares with a razor blade. Cells were next placed in 2 mg/ml of collagenase II (Gibco, Grand Island, NY) resuspended in Hanks’ Balanced Salt Solution containing calcium chloride and magnesium chloride (Life Technologies, Grand Island, NY). Each sample was incubated at 37°C on a shaker for 1 h. The dissociated cells were gently centrifuged at 400 g before resuspension in DMSO-based Recovery Cell Culture Freezing Media (Life Technologies). Samples were subsequently frozen at −80°C in a CoolCell LX container (Corning, Corning, NY) to cool at 1°C/min for 3–12 h, after which samples were transferred to liquid nitrogen for long term storage.
Cryopreserved samples were rapidly thawed in parallel at 37°C and passed through a 70 μm filter to remove cell aggregates. Next, dissociated cells were incubated with human anti-CD31 microbeads (Miltenyi Biotec, Bergisch Gladbach, Germany) for 15 min according to the manufacturer’s instructions. Each cell suspension underwent an automated enrichment of the CD31-positive fraction with the autoMACS magnetic separator (Miltenyi Biotec) using the positive-selection (possel) program. The CD31-enriched fraction was immediately counted and microfluidically barcoded with the Chromium system v3.1 chemistry kit (10X Genomics, Pleasanton, CA). The resulting libraries were pooled and sequenced on the NovaSeq6000 platform (Illumina, San Diego, CA), generating 100-bp paired-end reads.
Genotyping
SNPs in the MMP9 locus have been associated with increased risk for neovascular AMD (46,47). All samples included in this study were genotyped at the MMP9 AMD risk locus rs4810482. From each of the 21 donors, genomic DNA was purified from flash-frozen peripheral retina trephine punch biopsies (DNeasy Blood & Tissue Kit, Qiagen). Samples were genotyped using the rhAMP SNP Genotyping system (Integrated DNA Technologies) following the manufacturer’s instructions. Briefly, 5 μl reactions were run on a 384-well plate with each reaction containing 3–10 ng genomic DNA. Predesigned assays from IDT were used (MMP9 rs4810482—Hs.GT.rs4810482.C.1). Assays were run in duplicate on the QuantStudio 6 Flex instrument (Applied Biosystems) on the Genotyping setting and genotype calls (Table 1) were generated automatically.
Immunohistochemistry
Immunohistochemistry experiments were performed on flash-frozen, paraformaldehyde-fixed human tissues sections as previously described (20). Each section was blocked with 1 mg/ml of bovine serum albumin for 15 min before incubation with primary antibody. Before blocking, sections for SPARCL1 analysis were treated with TrueBlack autofluorescence quencher (Biotinium, Hayward, CA) for 60 s followed by three 5-min washes in phosphate buffered saline (PBS) to quench autofluorescence in RPE cells. Sections were subsequently washed and incubated with secondary antibody resuspended in PBS supplemented with diamidino-2-phenylindole (DAPI) (Sigma, St. Louis, MO) for 30 min. Sections were next washed three times in PBS before being coverslipped. All sections were imaged alongside negative controls without primary antibodies. Cases and controls were treated identically (e.g. same exposure times). Primary and secondary antibody information is provided in Supplementary Material, Table S2.
Computational analysis of scRNA-seq data
The bcl2fastq software (Illumina) was used to generate FASTQ files from base calls. FASTQ files were mapped to the human GRCh38 reference with CellRanger (v6.0.0). Cells were removed from downstream analysis based on the following filtering criteria: <500 unique genes per cell (likely low quality), >7000 unique genes per cell (likely doublets), >15% of reads mapping to mitochondrial genes or >25% of reads mapping to ribosomal genes (to eliminate partially lysed cells from the dataset (59)). The filtered libraries were normalized and integrated with canonical correlation analysis using the Seurat package (v3.2.3) (60). A total of 2000 variable features were identified with the variance stabilizing transformation (VST) selection method.
Differential expression analysis was performed to identify genes enriched in early atrophic AMD (n = 9) versus control (n = 10) samples or neovascular AMD (n = 2) versus control (n = 10) samples. Differential expression analysis can treat every individual cell as an independent observation (as is default in Seurat), however, this can artificially inflate P-values when comparing gene expression between biological variables (such as AMD vs. control samples). Therefore, we additionally computed a ‘pseudobulk’ differential expression by summing the counts for all reads in each independent library (e.g. biological replicate) before differential expression (35). This pseudobulk comparison more accurately reflects the number of biological replicates between these disease states and genotypes (61). Lowly expressed genes (expressed by fewer than 10% of cells in a given population or with normalized expression values less than 1.0) were excluded from pseudobulk differential expression analysis. All raw and processed data are deposited in the Gene Expression Omnibus under the accession number GSE183320. In addition, fully processed scRNA-seq data are hosted online at https://singlecell-eye.org under the dataset ‘choroid_amd_vs_control’ for interactive visualization (58).
Characterization of gene expression along the choroidal vascular tree
To better visualize the transitions between endothelial cell types, the PHATE dimensionality reduction method (33) was applied to the integrated, scaled gene expression data from arteries, veins and choriocapillaris endothelial cells with the phateR package (v1.0.7). For each cell, a total of 10 nearest neighbors were used to build the kernel with a decay factor of 75. Next, a slingshot (62) trajectory was fit to the PHATE embedding with the slingshot R package (v2.0.0), with the trajectory starting at the arteries. Cells were then manually classified as large arteries, arterioles, choriocapillaris, venules or large veins based on pseudotime values.
Interactome
We visualized ligand–receptor interactions with the Connectome R package (63). Briefly, genes with an AMD enrichment (pseudobulk logFC of greater than 0.5) were identified for endothelial (artery, choriocapillaris or vein) or immune (B-cell, T-cell, dendritic cell, resident macrophage or monocyte/inflammatory macrophage) clusters. Next, this AMD-enriched gene list was intersected with the NicheNet database (64) which contains known ligand–receptor interactions. Only ligand–receptor pairs with at least one AMD-enriched member were included in the downstream analysis. Connectome visualizations were generated with both endothelial cells and immune cells as ‘senders’ of ligands.
Classification of retinal, RPE or choroid specificity
For each AMD-associated gene (Fig. 6) (46), we calculated the expression level in each choroidal cell population (using data from the current study), each retinal cell population (using data from Voigt et al. (65)), and in the RPE (using data from Voigt et al. (19)). Next, we compared the expression value in the highest individual cell population across these three tissues. Genes were categorized as ‘mostly choroid’ if they were expressed at least twice the level in the choroid compared to the retina or RPE. Genes were categorized as ‘mostly retina’ if they were expressed at least twice the level in the retina compared to the choroid or RPE. Genes were categorized as ‘mostly RPE’ if they were expressed at least twice the level in the RPE compared to the retina or choroid. All other genes were deemed to be expressed at comparable levels by multiple tissues.
Supplementary Material
Acknowledgements
The authors gratefully acknowledge the eye donors and their families who make so many advances in biomedical research possible.
Conflict of Interest statement. The authors declare that they have no conflict of interest.
Contributor Information
Andrew P Voigt, Department of Ophthalmology and Visual Sciences, The University of Iowa Carver College of Medicine, Iowa City, IA 52242, USA; Institute for Vision Research, The University of Iowa, Iowa City, IA 52242, USA.
Nathaniel K Mullin, Department of Ophthalmology and Visual Sciences, The University of Iowa Carver College of Medicine, Iowa City, IA 52242, USA; Institute for Vision Research, The University of Iowa, Iowa City, IA 52242, USA.
Kelly Mulfaul, Department of Ophthalmology and Visual Sciences, The University of Iowa Carver College of Medicine, Iowa City, IA 52242, USA; Institute for Vision Research, The University of Iowa, Iowa City, IA 52242, USA.
Lola P Lozano, Department of Ophthalmology and Visual Sciences, The University of Iowa Carver College of Medicine, Iowa City, IA 52242, USA; Institute for Vision Research, The University of Iowa, Iowa City, IA 52242, USA.
Luke A Wiley, Department of Ophthalmology and Visual Sciences, The University of Iowa Carver College of Medicine, Iowa City, IA 52242, USA; Institute for Vision Research, The University of Iowa, Iowa City, IA 52242, USA.
Miles J Flamme-Wiese, Department of Ophthalmology and Visual Sciences, The University of Iowa Carver College of Medicine, Iowa City, IA 52242, USA; Institute for Vision Research, The University of Iowa, Iowa City, IA 52242, USA.
Erin A Boese, Department of Ophthalmology and Visual Sciences, The University of Iowa Carver College of Medicine, Iowa City, IA 52242, USA; Institute for Vision Research, The University of Iowa, Iowa City, IA 52242, USA.
Ian C Han, Department of Ophthalmology and Visual Sciences, The University of Iowa Carver College of Medicine, Iowa City, IA 52242, USA; Institute for Vision Research, The University of Iowa, Iowa City, IA 52242, USA.
Todd E Scheetz, Department of Ophthalmology and Visual Sciences, The University of Iowa Carver College of Medicine, Iowa City, IA 52242, USA; Institute for Vision Research, The University of Iowa, Iowa City, IA 52242, USA.
Edwin M Stone, Department of Ophthalmology and Visual Sciences, The University of Iowa Carver College of Medicine, Iowa City, IA 52242, USA; Institute for Vision Research, The University of Iowa, Iowa City, IA 52242, USA.
Budd A Tucker, Department of Ophthalmology and Visual Sciences, The University of Iowa Carver College of Medicine, Iowa City, IA 52242, USA; Institute for Vision Research, The University of Iowa, Iowa City, IA 52242, USA.
Robert F Mullins, Department of Ophthalmology and Visual Sciences, The University of Iowa Carver College of Medicine, Iowa City, IA 52242, USA; Institute for Vision Research, The University of Iowa, Iowa City, IA 52242, USA.
Funding
National Institute of Health (grant nos EY031923, EY024605, EY025580, EY033308 and T32GM008629); the Elmer and Sylvia Sramek Charitable Trust; Research to Prevent Blindness and the Edward N. & Della L. Thome Memorial Foundation.
References
- 1. Nickla, D.L. and Wallman, J. (2010) The multifunctional choroid. Prog. Retin. Eye Res., 29, 144–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nakanishi, M., Grebe, R., Bhutto, I.A., Edwards, M., McLeod, D.S. and Lutty, G.A. (2016) Albumen transport to Bruch's membrane and RPE by choriocapillaris caveolae. Invest. Ophthalmol. Vis. Sci., 57, 2213–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hayreh, S.S. (1974) Vascular pattern of the choriocapillaris. Exp. Eye Res., 19, 101–104. [DOI] [PubMed] [Google Scholar]
- 4. Biesemeier, A., Taubitz, T., Julien, S., Yoeruek, E. and Schraermeyer, U. (2014) Choriocapillaris breakdown precedes retinal degeneration in age-related macular degeneration. Neurobiol. Aging, 35, 2562–2573. [DOI] [PubMed] [Google Scholar]
- 5. Mullins, R.F., Johnson, M.N., Faidley, E.A., Skeie, J.M. and Huang, J. (2011) Choriocapillaris vascular dropout related to density of drusen in human eyes with early age-related macular degeneration. Invest. Ophthalmol. Vis. Sci., 52, 1606–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Whitmore, S.S., Sohn, E.H., Chirco, K.R., Drack, A.V., Stone, E.M., Tucker, B.A. and Mullins, R.F. (2015) Complement activation and choriocapillaris loss in early AMD: implications for pathophysiology and therapy. Prog. Retin. Eye Res., 45, 1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kim, D.Y., Fingler, J., Zawadzki, R.J., Park, S.S., Morse, L.S., Schwartz, D.M., Fraser, S.E. and Werner, J.S. (2013) Optical imaging of the chorioretinal vasculature in the living human eye. Proc. Natl. Acad. Sci. U. S. A., 110, 14354–14359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Waheed, N.K., Moult, E.M., Fujimoto, J.G. and Rosenfeld, P.J. (2016) Optical coherence tomography angiography of dry age-related macular degeneration. Dev. Ophthalmol., 56, 91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Choi, W., Moult, E.M., Waheed, N.K., Adhi, M., Lee, B., Lu, C.D., deCarlo, T.E., Jayaraman, V., Rosenfeld, P.J., Duker, J.S.et al. (2015) Ultrahigh-speed, swept-source optical coherence tomography angiography in nonexudative age-related macular degeneration with geographic atrophy. Ophthalmology, 122, 2532–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Spaide, R.F. (2016) Choriocapillaris flow features follow a power law distribution: implications for characterization and mechanisms of disease progression. Am J. Ophthalmol., 170, 58–67. [DOI] [PubMed] [Google Scholar]
- 11. Lutty, G.A., McLeod, D.S., Bhutto, I.A., Edwards, M.M. and Seddon, J.M. (2020) Choriocapillaris dropout in early age-related macular degeneration. Exp. Eye Res., 192, 107939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cousins, S.W., Espinosa-Heidmann, D.G. and Csaky, K.G. (2004) Monocyte activation in patients with age-related macular degeneration: a biomarker of risk for choroidal neovascularization. Arch. Ophthalmol. (Chicago, Ill.: 1960), 122, 1013–1018. [DOI] [PubMed] [Google Scholar]
- 13. Penfold, P.L., Killingsworth, M.C. and Sarks, S.H. (1985) Senile macular degeneration: the involvement of immunocompetent cells. Graefes Arch. Clin. Exp. Ophthalmol., 223, 69–76. [DOI] [PubMed] [Google Scholar]
- 14. Skeie, J.M. and Mullins, R.F. (2009) Macrophages in neovascular age-related macular degeneration: friends or foes? Eye (London, England), 23, 747–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ezzat, M.K., Hann, C.R., Vuk-Pavlovic, S. and Pulido, J.S. (2008) Immune cells in the human choroid. Br. J. Ophthalmol., 92, 976–980. [DOI] [PubMed] [Google Scholar]
- 16. Cherepanoff, S., McMenamin, P., Gillies, M.C., Kettle, E. and Sarks, S.H. (2010) Bruch's membrane and choroidal macrophages in early and advanced age-related macular degeneration. Br. J. Ophthalmol., 94, 918–925. [DOI] [PubMed] [Google Scholar]
- 17. McLeod, D.S., Bhutto, I., Edwards, M.M., Silver, R.E., Seddon, J.M. and Lutty, G.A. (2016) Distribution and quantification of choroidal macrophages in human eyes with age-related macular degeneration. Invest. Ophthalmol. Vis. Sci., 57, 5843–5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rohlenova, K., Goveia, J., García-Caballero, M., Subramanian, A., Kalucka, J., Treps, L., Falkenberg, K.D., deRooij, L., Zheng, Y., Lin, L.et al. (2020) Single-cell RNA sequencing maps endothelial metabolic plasticity in pathological angiogenesis. Cell Metab., 31, 862–877.e814. [DOI] [PubMed] [Google Scholar]
- 19. Voigt, A.P., Mulfaul, K., Mullin, N.K., Flamme-Wiese, M.J., Giacalone, J.C., Stone, E.M., Tucker, B.A., Scheetz, T.E. and Mullins, R.F. (2019) Single-cell transcriptomics of the human retinal pigment epithelium and choroid in health and macular degeneration. Proc. Natl. Acad. Sci. U. S. A., 116, 24100–24107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Voigt, A.P., Whitmore, S.S., Mulfaul, K., Chirco, K.R., Giacalone, J.C., Flamme-Wiese, M.J., Stockman, A., Stone, E.M., Tucker, B.A., Scheetz, T.E.et al. (2020) Bulk and single-cell gene expression analyses reveal aging human choriocapillaris has pro-inflammatory phenotype. Microvasc Res., 131, 104031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ronning, K.E., Karlen, S.J., Miller, E.B. and Burns, M.E. (2019) Molecular profiling of resident and infiltrating mononuclear phagocytes during rapid adult retinal degeneration using single-cell RNA sequencing. Sci. Rep., 9, 4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. O'Koren, E.G., Yu, C., Klingeborn, M., Wong, A.Y.W., Prigge, C.L., Mathew, R., Kalnitsky, J., Msallam, R.A., Silvin, A., Kay, J.N.et al. (2019) Microglial function is distinct in different anatomical locations during retinal homeostasis and degeneration. Immunity, 50, 723–737.e727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Heng, J.S., Hackett, S.F., Stein-O'Brien, G.L., Winer, B.L., Williams, J., Goff, L.A. and Nathans, J. (2019) Comprehensive analysis of a mouse model of spontaneous uveoretinitis using single-cell RNA sequencing. Proc. Natl. Acad. Sci. U. S. A., 116, 26734–26744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Droho, S., Thomson, B.R., Makinde, H.M., Cuda, C.M., Perlman, H. and Lavine, J.A. (2020) Ocular macrophage origin and heterogeneity during steady state and experimental choroidal neovascularization. J. Neuroinflammation, 17, 341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lehmann, G.L., Hanke-Gogokhia, C., Hu, Y., Bareja, R., Salfati, Z., Ginsberg, M., Nolan, D.J., Mendez-Huergo, S.P., Dalotto-Moreno, T., Wojcinski, A.et al. (2020) Single-cell profiling reveals an endothelium-mediated immunomodulatory pathway in the eye choroid. J. Exp. Med., 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu, L. and Shi, G.P. (2012) CD31: beyond a marker for endothelial cells. Cardiovasc. Res., 94, 3–5. [DOI] [PubMed] [Google Scholar]
- 27. Stacchini, A., Chiarle, R., Antinoro, V., Demurtas, A., Novero, D. and Palestro, G. (2003) Expression of the CD31 antigen in normal B-cells and non Hodgkin's lymphomas. J. Biol. Regul. Homeost. Agents, 17, 308–315. [PubMed] [Google Scholar]
- 28. Tanaka, Y., Albelda, S.M., Horgan, K.J., vanSeventer, G.A., Shimizu, Y., Newman, W., Hallam, J., Newman, P.J., Buck, C.A. and Shaw, S. (1992) CD31 expressed on distinctive T cell subsets is a preferential amplifier of beta 1 integrin-mediated adhesion. J. Exp. Med., 176, 245–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mulder, K., Patel, A.A., Kong, W.T., Piot, C., Halitzki, E., Dunsmore, G., Khalilnezhad, S., Irac, S.E., Dubuisson, A., Chevrier, M.et al. (2021) Cross-tissue single-cell landscape of human monocytes and macrophages in health and disease. Immunity, 54, 1883–1900.e1885. [DOI] [PubMed] [Google Scholar]
- 30. Cochain, C., Vafadarnejad, E., Arampatzi, P., Pelisek, J., Winkels, H., Ley, K., Wolf, D., Saliba, A.E. and Zernecke, A. (2018) Single-cell RNA-seq reveals the transcriptional landscape and heterogeneity of aortic macrophages in murine atherosclerosis. Circ. Res., 122, 1661–1674. [DOI] [PubMed] [Google Scholar]
- 31. Liao, M., Liu, Y., Yuan, J., Wen, Y., Xu, G., Zhao, J., Cheng, L., Li, J., Wang, X., Wang, F.et al. (2020) Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med., 26, 842–844. [DOI] [PubMed] [Google Scholar]
- 32. Kang, H.M., Subramaniam, M., Targ, S., Nguyen, M., Maliskova, L., McCarthy, E., Wan, E., Wong, S., Byrnes, L., Lanata, C.M.et al. (2018) Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. Nat. Biotechnol., 36, 89–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Moon, K.R., vanDijk, D., Wang, Z., Gigante, S., Burkhardt, D.B., Chen, W.S., Yim, K., Elzen, A.V.D., Hirn, M.J., Coifman, R.R.et al. (2019) Visualizing structure and transitions in high-dimensional biological data. Nat. Biotechnol., 37, 1482–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kalucka, J., deRooij, L., Goveia, J., Rohlenova, K., Dumas, S.J., Meta, E., Conchinha, N.V., Taverna, F., Teuwen, L.A., Veys, K.et al. (2020) Single-cell transcriptome atlas of murine endothelial cells. Cell, 180, 764, e720–779. [DOI] [PubMed] [Google Scholar]
- 35. Lun, A.T.L. and Marioni, J.C. (2017) Overcoming confounding plate effects in differential expression analyses of single-cell RNA-seq data. Biostatistics, 18, 451–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bennett, E.P., Mandel, U., Clausen, H., Gerken, T.A., Fritz, T.A. and Tabak, L.A. (2012) Control of mucin-type O-glycosylation: a classification of the polypeptide GalNAc-transferase gene family. Glycobiology, 22, 736–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kifuji, K. (1989) Study of the mechanisms of histamine release from human leukocytes by challenge of mycoplasma. Arerugi, 38, 1226–1235. [PubMed] [Google Scholar]
- 38. Bradshaw, A.D. and Sage, E.H. (2001) SPARC, a matricellular protein that functions in cellular differentiation and tissue response to injury. J. Clin. Invest., 107, 1049–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Girard, J.P. and Springer, T.A. (1996) Modulation of endothelial cell adhesion by hevin, an acidic protein associated with high endothelial venules. J. Biol. Chem., 271, 4511–4517. [DOI] [PubMed] [Google Scholar]
- 40. Brekken, R.A., Sullivan, M.M., Workman, G., Bradshaw, A.D., Carbon, J., Siadak, A., Murri, C., Framson, P.E. and Sage, E.H. (2004) Expression and characterization of murine hevin (SC1), a member of the SPARC family of matricellular proteins. J. Histochem. Cytochem., 52, 735–748. [DOI] [PubMed] [Google Scholar]
- 41. Girard, J.P. and Springer, T.A. (1995) Cloning from purified high endothelial venule cells of hevin, a close relative of the antiadhesive extracellular matrix protein SPARC. Immunity, 2, 113–123. [DOI] [PubMed] [Google Scholar]
- 42. Sándor, N., Lukácsi, S., Ungai-Salánki, R., Orgován, N., Szabó, B., Horváth, R., Erdei, A. and Bajtay, Z. (2016) CD11c/CD18 dominates adhesion of human monocytes, macrophages and dendritic cells over CD11b/CD18. PLoS One, 11, e0163120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nozaki, M., Raisler, B.J., Sakurai, E., Sarma, J.V., Barnum, S.R., Lambris, J.D., Chen, Y., Zhang, K., Ambati, B.K., Baffi, J.Z.et al. (2006) Drusen complement components C3a and C5a promote choroidal neovascularization. Proc. Natl. Acad. Sci. U. S. A., 103, 2328–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Skeie, J.M., Fingert, J.H., Russell, S.R., Stone, E.M. and Mullins, R.F. (2010) Complement component C5a activates ICAM-1 expression on human choroidal endothelial cells. Invest. Ophthalmol. Vis. Sci., 51, 5336–5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang, S., Song, R., Wang, Z., Jing, Z., Wang, S. and Ma, J. (2018) S100A8/A9 in inflammation. Front. Immunol., 9, 1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fritsche, L.G., Igl, W., Bailey, J.N., Grassmann, F., Sengupta, S., Bragg-Gresham, J.L., Burdon, K.P., Hebbring, S.J., Wen, C., Gorski, M.et al. (2016) A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat. Genet., 48, 134–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sohn, E.H., Han, I.C., Roos, B.R., Faga, B., Luse, M.A., Binkley, E.M., Boldt, H.C., Folk, J.C., Russell, S.R., Mullins, R.F.et al. (2021) Genetic association between MMP9 and choroidal neovascularization in age-related macular degeneration. Ophthalmology Science, 1, 100002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Naschberger, E., Liebl, A., Schellerer, V.S., Schütz, M., Britzen-Laurent, N., Kölbel, P., Schaal, U., Haep, L., Regensburger, D., Wittmann, T.et al. (2016) Matricellular protein SPARCL1 regulates tumor microenvironment-dependent endothelial cell heterogeneity in colorectal carcinoma. J. Clin. Invest., 126, 4187–4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Murphy-Ullrich, J.E., Lane, T.F., Pallero, M.A. and Sage, E.H. (1995) SPARC mediates focal adhesion disassembly in endothelial cells through a follistatin-like region and the Ca2+-binding EF-hand. J. Cell. Biochem., 57, 341–350. [DOI] [PubMed] [Google Scholar]
- 50. Wang, Y., Liu, S., Yan, Y., Li, S. and Tong, H. (2019) SPARCL1 promotes C2C12 cell differentiation via BMP7-mediated BMP/TGF-β cell signaling pathway. Cell Death Dis., 10, 852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Weaver, M.S., Workman, G., Cardo-Vila, M., Arap, W., Pasqualini, R. and Sage, E.H. (2010) Processing of the matricellular protein hevin in mouse brain is dependent on ADAMTS4. J. Biol. Chem., 285, 5868–5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Weaver, M., Workman, G., Schultz, C.R., Lemke, N., Rempel, S.A. and Sage, E.H. (2011) Proteolysis of the matricellular protein hevin by matrix metalloproteinase-3 produces a SPARC-like fragment (SLF) associated with neovasculature in a murine glioma model. J. Cell. Biochem., 112, 3093–3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. House, S.D. and Lipowsky, H.H. (1987) Leukocyte-endothelium adhesion: microhemodynamics in mesentery of the cat. Microvasc. Res., 34, 363–379. [DOI] [PubMed] [Google Scholar]
- 54. Mould, K.J., Jackson, N.D., Henson, P.M., Seibold, M. and Janssen, W.J. (2019) Single cell RNA sequencing identifies unique inflammatory airspace macrophage subsets. JCI Insight, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Honold, L. and Nahrendorf, M. (2018) Resident and monocyte-derived macrophages in cardiovascular disease. Circ. Res., 122, 113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Italiani, P. and Boraschi, D. (2014) From monocytes to M1/M2 macrophages: phenotypical vs. functional differentiation. Front. Immunol., 5, 514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Squair, J.W., Gautier, M., Kathe, C., Anderson, M.A., James, N.D., Hutson, T.H., Hudelle, R., Qaiser, T., Matson, K.J.E., Barraud, Q.et al. (2021) Confronting false discoveries in single-cell differential expression. Nat. Commun., 12, 5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Voigt, A.P., Whitmore, S.S., Lessing, N.D., DeLuca, A.P., Tucker, B.A., Stone, E.M., Mullins, R.F. and Scheetz, T.E. (2020) Spectacle: an interactive resource for ocular single-cell RNA sequencing data analysis. Exp. Eye Res., 200, 108204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Luecken, M.D. and Theis, F.J. (2019) Current best practices in single-cell RNA-seq analysis: a tutorial. Mol. Syst. Biol., 15, e8746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Butler, A., Hoffman, P., Smibert, P., Papalexi, E. and Satija, R. (2018) Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol., 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Thurman, A.L., Ratcliff, J.A., Chimenti, M.S. and Pezzulo, A.A. (2021) Differential gene expression analysis for multi-subject single cell RNA sequencing studies with aggregateBioVar. Bioinformatics, 37, 3243–3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Street, K., Risso, D., Fletcher, R.B., Das, D., Ngai, J., Yosef, N., Purdom, E. and Dudoit, S. (2018) Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics, 19, 477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Raredon, M.S.B., Yang, J., Garritano, J., Wang, M., Kushnir, D., Schupp, J.C., Adams, T.S., Greaney, A.M., Leiby, K.L., Kaminski, N.et al. (2021) Connectome: computation and visualization of cell-cell signaling topologies in single-cell systems data. bioRxiv, in press., 2021.2001.2021.427529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Browaeys, R., Saelens, W. and Saeys, Y. (2020) NicheNet: modeling intercellular communication by linking ligands to target genes. Nat. Methods, 17, 159–162. [DOI] [PubMed] [Google Scholar]
- 65. Voigt, A.P., Mullin, N.K., Whitmore, S.S., DeLuca, A.P., Burnight, E.R., Liu, X., Tucker, B.A., Scheetz, T.E., Stone, E.M. and Mullins, R.F. (2021) Human photoreceptor cells from different macular subregions have distinct transcriptional profiles. Hum. Mol. Genet., 30, 1543–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.