Abstract
A granulovirus (GV) isolated from Epinotia aporema (Lepidoptera: Tortricidae)—a major soybean pest—was studied in terms of its main morphological, biochemical, and biological properties. The ovoidal occlusion bodies were 466 by 296 nm in size, and their most prominent protein had an apparent molecular mass of 29 kDa. Its amino-terminal sequence was remarkably homologous to that of the granulins of other GVs. The DNA genome size was estimated to be 120 kbp. The high specificity and pathogenicity of this newly described granulovirus (EpapGV) indicate that it is indeed a good candidate for the biological control of this pest.
The bean shoot borer, Epinotia aporema (Wals.), is a serious pest of soybean and other legume crops in South America. In Argentina, damages caused by this tortricid decrease soybean yields up to 40%, depending on population level and environmental conditions (18). Currently, broad-spectrum chemical insecticides are used to control this caterpillar, delaying or suppressing field colonization by natural enemies. This scenario compromises the implementation of a proper integrated pest management scheme for soybean crops, in areas where the incidence of E. aporema reaches high population levels (25).
Among pathogens, a granulovirus (GV) was detected as the main mortality factor (6, 26). In general, the potential of baculoviruses for pest control has been well documented and they have proven to be effective against many pests (9, 15, 24). In this regard, preliminary experiments showed that E. aporema GV (EpapGV) could be a safe alternative for inclusion in soybean pest management (25). However, former studies were directed towards evaluating its virulence, and none of the previously cited reports addressed the characterization of the virus. As an initial point in our research, we identified and characterized a GV isolate from E. aporema, indigenous to the main soybean area of Argentina.
Virus isolation and multiplication.
A colony of E. aporema caterpillars reared on artificial medium (12) was established in our laboratory. EpapGV was isolated from a single E. aporema larva collected in Oliveros (Santa Fe, Argentina). Viral amplification was carried out, allowing fourth-instar larvae to feed on formalin-free artificial diet, superficially contaminated with 4,000 granules per mm2. Granules were purified from homogenized larvae by two cycles of centrifugation on continuous 40 to 65% (wt/wt) sucrose gradients at 100,000 × g for 1 h. Virions were released by hydrolyzing granules in 0.1 M Na2CO3–0.17 M NaCl–0.01 M EDTA (pH 10.5) at 37°C for 30 min. Following the dissolution of the granules, the suspension was neutralized by addition of 1 M HCl, and undissolved material was removed by low-speed centrifugation.
Electron microscopy.
Tissues were dissected from infected and control larvae, fixed in 3% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.3)–0.25 M sucrose for 3 h, postfixed in 1% osmium tetroxide for 1 h, dehydrated through an ethanol-propylene oxide series, and embedded in Epon-Araldite resin. Ultrathin sections, stained in 2% uranyl acetate and lead citrate, were examined with a JEM-1200 EX II electron microscope.
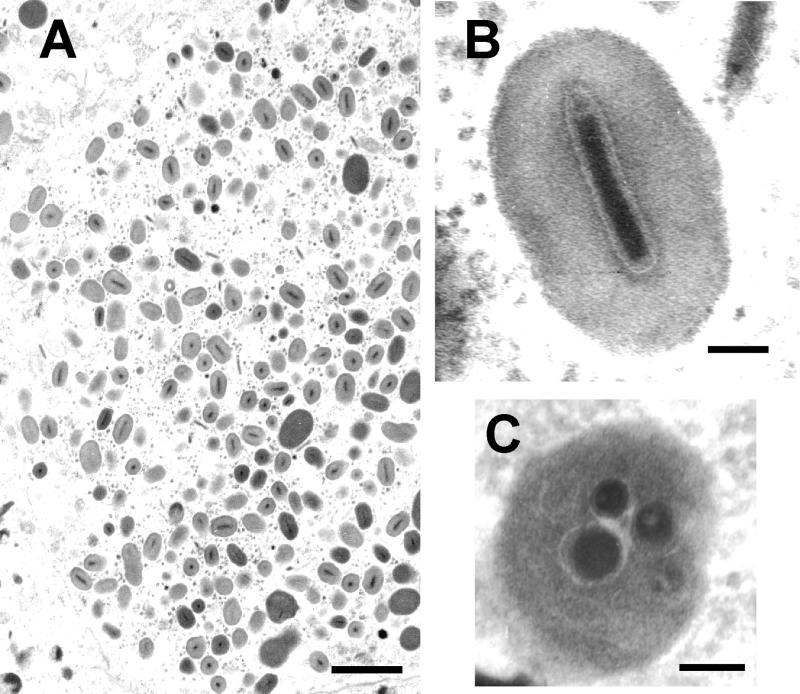
The occlusion bodies were ovoidal in shape, with a mean size of 466 (±8.9) by 296 (±6.0) nm. In general, each granule contained a single rod-shaped virion (290 [±10.9] × 61.2 [±1.8] nm) with one nucleocapsid (226 [±8.9] × 41.7 [±4.1] nm) (Fig. 1A and B). In rare cases, granules containing more than one virion were observed (Fig. 1C).
FIG. 1.
Electron micrographs illustrating the major morphological properties of EpapGV. (A) Section through an infected epidermal cell showing virions and granules (bar, 1 μm). (B) Longitudinal section through an occlusion body with a single embedded virion. Note the “nipple” and the “claw” ends of the nucleocapsid (bar, 100 nm). (C) Granule with multiple embedded virions (bar, 100 nm).
Major occlusion body protein.
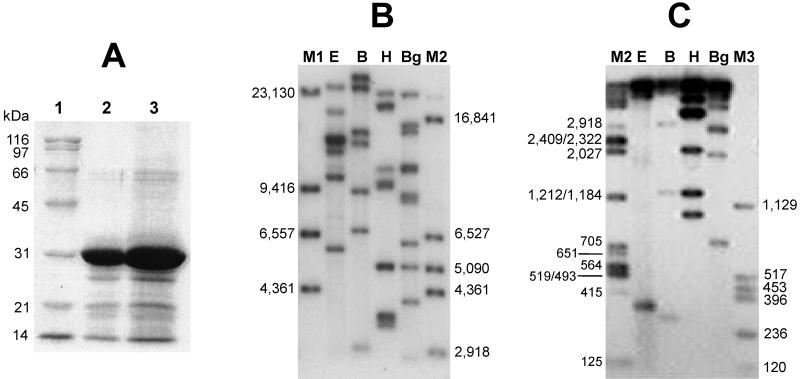
The major protein components of the occlusion bodies were analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (21). Proteins were visualized after staining with Coomassie brilliant blue, and sizes were estimated using molecular weight markers (Sigma, St. Louis, Mo.). The estimated molecular mass of the EpapGV major protein band (28.5 ± 0.5 kDa) (Fig. 2A) falls within the 27- to 31-kDa range, characteristic for granulins (27, 36).
FIG. 2.
(A) SDS-polyacrylamide gel electrophoresis of EpapGV proteins. Granules were purified from infected larvae as described in the text and run on a 12% polyacrylamide gel. Lane 1, molecular mass markers; lane 2, granules from EpapGV; lane 3, granules from CpGV. (B) Autoradiography of EpapGV DNA restriction fragments. Viral DNA was digested with EcoRI (E), BamHI (B), HindIII (H), and BglII (Bg); labeled with [α-32P]dATP; and resolved by electrophoresis on an 0.4% agarose gel. Lambda DNA was digested with HindIII (M1). (C) The same samples as in panel B separated on a 1.5% agarose gel. Lambda DNA digested with BamHI, HindIII, and BglII (M2) and pcDNA II DNA (Invitrogen) digested with HindIII and HinfI (M3) were used as molecular size markers (indicated in base pairs).
For amino acid sequencing, the proteins were resolved in a Tricine-polyacrylamide gel system (30) and transferred to a polyvinylidene difluoride membrane for 2 h at 0.25 A, and the granulin was visualized by Ponceau red staining (1% in water for 15 min and destaining with water). The section of membrane containing granulin (approximately 20 μg) was cut out and processed for protein sequencing by a modified Edman degradation method (23) at the LANAIS-PRO laboratory (Buenos Aires, Argentina).
The N-terminal amino acid sequence of the protein (GYNKSLRYSRHEGTT) was compared with that of the homologous proteins from other GVs and in most cases showed no more difference than 1 or 2 out of 15 residues. The N-terminal sequences of polyhedrins are less conserved and clearly distinct from the granulin consensus sequence (Table 1). The lack of a methionine residue at the amino terminus probably reflects posttranslational processing. This is consistent with another granulin sequence (Agrotis segetum GV) that was also obtained by protein sequencing instead of deduction of the data from the DNA sequence (20).
TABLE 1.
Amino-terminal sequences of granulins
| Baculovirus or sequence | Insect host
|
Amino acid sequencea | Experimental determination | GenBank accession no. | |
|---|---|---|---|---|---|
| Species | Family | ||||
| AgseGV | Agrotis segetum | Noctuidae | GYNKSLRYSRHAGTS | Protein | P31035 |
| CpGV | Cydia pomonella | Tortricidae | MGYNKSLRYSRHDGTS | DNA | Y09478 |
| CrleGV | Cryptophlebia leucotreta | Tortricidae | MGYNKSLRYSRHDGTT | DNA | X79569 |
| ChfuGV | Choristoneura fumiferana | Tortricidae | MGYNKALRYSRHDGTS | DNA | U87621 |
| HbGV | Harrisinia brillians | Zygaenidae | MGYNKSLRYSRHEGTT | DNA | AF142425 |
| PbGV | Pieris brassicae | Pieridae | MGYNRALRYSKHEGTT | DNA | P06502 |
| PxGV | Plutella xylostella | Plutellidae | MGYNKSLRYSRHDGST | DNA | AB030174 |
| TnGV | Trichoplusia ni | Noctuidae | MGYNKSLRYSRHNGTT | DNA | P06503 |
| XcGV | Xestia c-nigrum | Noctuidae | MGYNKSLRYSRHNGTT | DNA | U70069 |
| EpapGV | Epinotia aporema | Tortricidae | GYNKSLRYSRHEGTT | Protein | |
| Consensus for granulin | MGYNKSLRYSRH.GTT | DNA | |||
| Consensus for polyhedrin | M...YSY.P..GRTYV | DNA | |||
The differences from the consensus sequence are indicated in boldface. The consensus resulted from applying the majority rule, i.e., the most abundant residue in each position is recorded as the consensus amino acid.
Genome.
Virion suspensions were adjusted to a final concentration of 0.5% SDS and incubated with proteinase K (0.25 mg/ml at 37°C for 3 h), and the DNA was isolated by phenol extraction and ethanol precipitation (29). The DNA fragments obtained after digestion with BamHI, BglII, EcoRI, and HindIII endonucleases were separated by electrophoresis on 0.4 and 1.5% (wt/vol) agarose gels, stained with ethidium bromide, and photographed under UV light. Alternatively, DNA digests were labeled with [α-32P]dATP, separated by electrophoresis, and visualized by autoradiography (29). The restriction patterns of EpapGV DNA were distinct from those published for other GVs (3, 4, 5, 7, 11, 14, 31, 33, 37, 39) (Fig. 2B and C). The existence of multiple fragments of similar mobilities was assessed by densitometry of the autoradiographs. To minimize the inaccuracy associated with the determination of the sizes of very large restriction fragments, double digests using combinations of restriction endonucleases were also analyzed (data not shown). A size of 120 kbp for the EpapGV genome was estimated from the sum of the fragments (Fig. 2B and C). This result is consistent with the known genomes of different GVs, which range from 89 kbp for Adoxophyes orana GV (22) to 179 kbp for Xestia c-nigrum GV (11). Detailed genome analysis is under way to complete the physical restriction map and sequencing of EpapGV, which would confirm the EpapGV species taxonomic status.
Since no submolar fragments were apparent in the restriction endonuclease analyses, the EpapGV isolate studied in our laboratory seems to be homogeneous. However, further studies on the viral genome and the comparison of DNA digests from other geographical and temporal isolates of EpapGV will allow us to determine if there are variants of this virus, as previously described for other GV isolates (3, 5, 32, 37).
Histopathology.
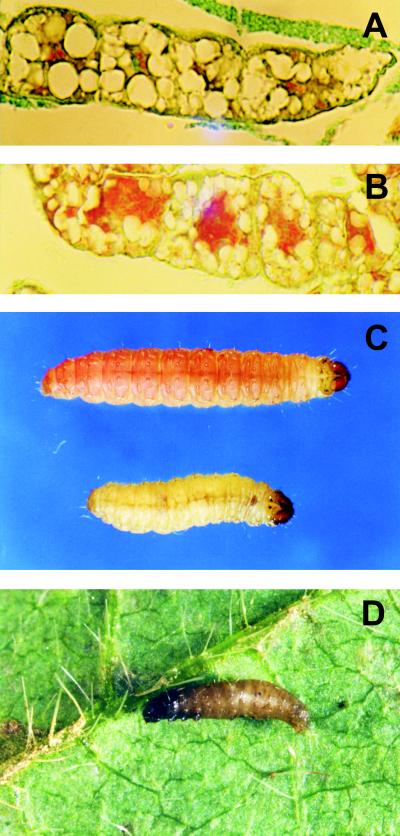
For light microscopy, larvae were fixed in aqueous Bouin's solution for 48 h, dehydrated in a tertiary-butyl alcohol series, and embedded in Paraplast, and 5-μm-thick sections were cut and stained using the modified Azan technique (13). EpapGV infection was polyorganotropic; the infected tissues included fatty tissue, tracheal matrix, and epidermis. Microscope observations of fatty tissue sections showed various stages of nuclear and cell disintegration (Fig. 3A and B).
FIG. 3.
Healthy (A) and infected (B) fatty tissue sections; the infected tissue shows an intense red color throughout the cells due to the presence of granules and a high degree of disintegration. By 3 days postinfection, the symptoms of the disease are evident; the last-instar control and infected larvae show differences in growth, development, and color (C). The infected larvae turn yellowish and swollen, and their development is interrupted, while the control larvae exhibit the characteristic pink color of the prepupal stage. Shortly after death, the larvae become brownish, and their tegument can be easily ruptured (D).
Biological activity.
The larvae infected by EpapGV showed the typical symptoms of a GV infection at a late stage of the disease, i.e., loss of appetite, decrease in mobility, and change of color due to the accumulation of occlusion bodies in the infected tissues (Fig. 3C). Shortly after death, the larvae became swollen and turned brownish, and the tegument was easily disrupted (Fig. 3D).
In order to estimate the virulence of the isolate, bioassays were conducted using the droplet feeding technique (17). The granules were suspended in a solution of 1% sucrose–0.1% Coomassie brilliant blue (1). The mean ingested volume per larva was estimated for neonates (0.016 ± 0.001 μl) and fourth-instar larvae (2.4 ± 0.2 μl). On the basis of these measurements, dilutions were adjusted to obtain 5 doses of between 2 and 32 granules per larva for neonates and between 2 × 102 and 1.6 × 104 granules for fourth-instar larvae in the 50% lethal dose (LD50; dose that produces a lethal infection in 50% of the larvae in 7 days) assays. Three replicate assays of 32 larvae each were carried out for each dose. An additional 32 larvae fed on the colored sucrose solution were used as controls.
Those larvae which had ingested the virus suspension were transferred individually to multiwell dishes containing formalin-free diet and held at 25 ± 1°C and 50% rH and in a 14-h-light/10-h-dark cycle. Mortality was recorded every 6 h (neonates) or daily (fourth-instar larvae). The time-mortality response was determined using the data obtained with the lowest dose that yielded 100% mortality. Dose and time-mortality response data were analyzed using ViStat software (Cornell University, Ithaca, N.Y.).
The bioassays carried out with neonates and fourth-instar larvae yielded LD50 estimates of 5.6 (range, 3.64 to 7.34) and 4,000 (range, 3,880 to 5,200) granules per larva, respectively. The median survival time using the lowest dose that yields 100% mortality was 90 (±4.4) h for neonates and 105 (±16) h for fourth-instar larvae. As expected, the time-mortality response was dose dependent (data not shown).
Host specificity was studied using the percentage of mortality and development of the disease as the criteria for susceptibility. The assays were performed on the major regional soybean pests in addition to E. aporema, i.e., Anticarsia gemmatalis, Rachiplusia nu, and Spodoptera frugiperda (Lepidoptera: Noctuidae). Two other species, one distantly and one closely related, Diatraea saccharalis (Lepidoptera: Pyralidae) and Cydia pomonella (Lepidoptera: Tortricidae), respectively, were also included in the assays.
Using the above-described method, doses from 4 to 256 granules per larva were fed to neonates of each of these species. Doses of up to 40 LD50 for E. aporema neonates did not affect development of the heterologous species. In all cases, with the exception of C. pomonella, larvae fed on EpapGV-contaminated diet pupated simultaneously with the controls. At the highest dose tested, mortality produced by a GV was observed only for C. pomonella. Subsequent restriction enzyme analysis of the viral DNA, isolated from the granules found in dead larvae, indicated that the infection was actually produced by a C. pomonella GV (CpGV) (data not shown) and was possibly due to the activation of a latent virus present in the C. pomonella colony. This observation was previously reported for other baculoviral infections (16).
EpapGV is a potential control agent for E. aporema.
EpapGV has been considered as a tentative species in the genus Granulovirus (Baculoviridae) based on its pathology, although direct morphological and biochemical characterization was lacking (38). Here, we present electron microscopy evidence and biochemical data that confirm its generic identity. The biological properties and DNA restriction patterns are consistent with a new GV that seems to be different from others previously described in the literature.
According to the work of Federici (10), three different types of GVs can be distinguished by their unique pathobiological pattern. In this regard, our results showed that EpapGV exhibits the typical broad tissue tropism of the type 2 GVs. This type includes CpGV, which is the GV most successfully used for pest control (9, 15). On the basis of the estimates for LD50 and median survival time using the lowest dose that yields 100% mortality obtained for neonate larvae, it can be concluded that EpapGV is a highly virulent virus and, indeed, a good candidate for the microbial control of E. aporema larvae. Its potential is not limited to soybean—a crop for which it could be a complementary tool to be used in combination with other biotechnological or biological strategies–but applies to the pest management of other important regional crops, such as beans, peanuts, and alfalfa. However, in order to develop a viral insecticide for this pest, further studies on its efficacy under field conditions are necessary.
A significant decrease of the susceptibility with larval age was apparent, a phenomenon that was observed elsewhere for several other baculoviruses (2, 8, 19, 28, 34, 35). In this context, it is also important to establish complementary methods to monitor the population dynamics of E. aporema, in order to determine the proper timing of application that would ensure control during the early instars of the larvae, when they are more exposed and susceptible to the virus.
In summary, the results presented here provide the preliminary information required both for further detailed characterization of EpapGV and for the development of a new alternative for the biocontrol of E. aporema. In addition, it is interesting that, while environmentally sound pest management strategies are available for other insects that affect soybean, E. aporema is currently controlled only with broad-spectrum chemical insecticides.
Acknowledgments
This research was supported by BID-SECyT 802 OC/AR PID 410 (V.R. and A.S.-C.), UNLP X120 and X198 (V.R. and P.D.G.), and CIC BA (V.R.). V.R. holds a research career award from CONICET (Consejo Nacional de Investigaciones Científicas y Técnicas, Argentina).
We thank S. Lorenzatti de Diez and J. C. Gamundi (E.E.A. Oliveros INTA, Argentina) for supplying the material from which the virus was isolated and M. Rivas, D. Moreyra, and J. Cap for their collaboration in insect rearing and bioassays.
REFERENCES
- 1.Begon M, Haji Daud K B, Young P, Howells R E. Invasion and replication of a granulosis virus in the Indian moth, Plodia interpunctella: an electron microscope study. J Invertebr Pathol. 1993;61:281–295. [Google Scholar]
- 2.Bourcias D G, Johnson D W, Allen G E. Effects of host age, virus dosage, and temperature on the infectivity of a nuclear polyhedrosis virus against velvetbean caterpillar, Anticarsia gemmatalis. Environ Entomol. 1980;9:59–61. [Google Scholar]
- 3.Crook N E. Restriction enzymes analysis of granulosis virus isolated from Artogeia rapae and Pieris brassicae. J Gen Virol. 1986;67:781–787. [Google Scholar]
- 4.Crook N E, James J D, Smith I R L, Winstanley D. Comprehensive physical map of the Cydia pomonella granulovirus genome and sequence analysis of the granulin gene region. J Gen Virol. 1997;78:965–974. doi: 10.1099/0022-1317-78-4-965. [DOI] [PubMed] [Google Scholar]
- 5.Crook N E, Spencer R A, Payne C C, Leisy D J. Variation in Cydia pomonella granulosis virus isolates and physical map of the DNA from three variants. J Gen Virol. 1985;66:2433–2440. [Google Scholar]
- 6.Diaz B, Diez S. IV Conferencia Mundial de Investigación en soja, Buenos Aires, Argentina, vol. III. 1989. Presencia de un virus de la granulosis en larvas de Epinotia aporema (Wals.), en cultivos de soja; pp. 1588–1592. [Google Scholar]
- 7.Dwyer K G, Granados R R. A physical map of the Pieris rapae granulosis virus genome. J Gen Virol. 1987;68:1471–1476. [Google Scholar]
- 8.Engelhard E K, Volkman L E. Developmental resistance in fourth instar Trichoplusia ni orally inoculated with Autographa californica M nuclear polyhedrosis virus. Virology. 1995;209:384–389. doi: 10.1006/viro.1995.1270. [DOI] [PubMed] [Google Scholar]
- 9.Entwistle P F. A world survey of virus control of insect pests. In: Hunter-Fujita F R, Entwistle P F, Evans H F, Crook N E, editors. Insect viruses and pest management. Chichester, England: John Wiley & Sons Ltd.; 1998. pp. 189–200. [Google Scholar]
- 10.Federici B A. Viral pathobiology in relation to insect control. In: Beckage N, Federici B A, Thompson N, editors. Parasites and pathogens of insects. Vol. 2. San Diego, Calif: Academic Press; 1993. pp. 81–101. [Google Scholar]
- 11.Goto C, Minobe Y, Iizuka T. Restriction endonuclease analysis and mapping of the genomes of granulosis viruses isolated from Xestia c-nigrum and five other noctuid species. J Gen Virol. 1992;73:1491–1497. doi: 10.1099/0022-1317-73-6-1491. [DOI] [PubMed] [Google Scholar]
- 12.Greene G L, Leppla N C, Dickerson W A. Velvetbean caterpillar: a rearing procedure and artificial medium. J Econ Entomol. 1976;69:487–488. [Google Scholar]
- 13.Hamm J. A modified Azan technique for inclusion body viruses. J Invertebr Pathol. 1966;8:125–126. doi: 10.1016/0022-2011(66)90113-3. [DOI] [PubMed] [Google Scholar]
- 14.Hashimoto Y, Hayashi K, Okuno Y, Hayakawa T, Saimoto A, Granados R R, Matsumoto T. Physical mapping and identification of interspersed homologous sequences in the Trichoplusia ni granulosis virus genome. J Gen Virol. 1996;77:555–563. doi: 10.1099/0022-1317-77-3-555. [DOI] [PubMed] [Google Scholar]
- 15.Huber J. Use of baculoviruses in pest management programs. In: Granados R R, Federici B A, editors. The biology of baculoviruses. I. Boca Raton, Fla: CRC Press, Inc.; 1986. pp. 181–202. [Google Scholar]
- 16.Hughes D, Possee R, King L A. Activation and detection of a latent baculovirus resembling Mamestra brassicae nuclear polyhedrovirus in Mamestra brassicae insects. Virology. 1993;194:608–618. doi: 10.1006/viro.1993.1300. [DOI] [PubMed] [Google Scholar]
- 17.Hughes P, Wood H A. A synchronous technique for the bioassay of insect viruses. J Invertebr Pathol. 1981;37:154–159. [Google Scholar]
- 18.Ianonne N, Parisi R, Dagoberto E. Incidencia del “barrenador de los brotes” Epinotia aporema (Wals.), en soja. EEA-INTA Pergamino Informe Técnico. 1987;209:1–24. [Google Scholar]
- 19.Kirkpatrick B A, Washburn J O, Volkman L E. AcMNPV pathogenesis and developmental resistance in fifth instar Heliothis virescens. J Invertebr Pathol. 1998;72:63–72. doi: 10.1006/jipa.1997.4752. [DOI] [PubMed] [Google Scholar]
- 20.Kozlov E A, Rodnin N V, Levitina T L, Gusak M M, Radomskij N F, Palchikovskaya L J. The amino acid sequence determination of a granulin and polyhedrin from two baculoviruses infecting Agrotis segetum. Virology. 1992;189:320–323. doi: 10.1016/0042-6822(92)90708-w. [DOI] [PubMed] [Google Scholar]
- 21.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 22.Li X, Benz G. Restriction endonuclease analysis of the granulosis virus of Adoxophyes orana F.V.R. (Lep., Tortricidae) Bull OILB/SROP. 1994;17:244–247. [Google Scholar]
- 23.Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987;262:10035–10038. [PubMed] [Google Scholar]
- 24.Moscardi F. Assessment of the application of baculoviruses for control of Lepidoptera. Annu Rev Entomol. 1999;44:257–489. doi: 10.1146/annurev.ento.44.1.257. [DOI] [PubMed] [Google Scholar]
- 25.Moscardi F, Sosa Gomez D. Use of viruses against soybean caterpillars in Brazil. In: Copping L G, Green M B, Rees R T, editors. Pest management in soybean. London, United Kingdom: Elsevier Applied Science; 1992. pp. 98–109. [Google Scholar]
- 26.Ripa R. Presencia de dos nuevos patógenos en Epinotia aporema Wals. Agric Tec (Chile) 1982;41:55–56. [Google Scholar]
- 27.Rohrmann G R. Baculovirus structural proteins. J Gen Virol. 1992;73:749–761. doi: 10.1099/0022-1317-73-4-749. [DOI] [PubMed] [Google Scholar]
- 28.Sait S M, Begon M, Thompson D J. The influence of larval age on the response of Plodia interpunctella to a granulosis virus. J Invertebr Pathol. 1994;63:107–110. [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Schägger H, Von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 31.Singaravelu B, Ramakrishnan N. Characterization of a granulosis virus from the castor semilooper, Archaea janata L. J Invertebr Pathol. 1998;71:227–235. doi: 10.1006/jipa.1997.4735. [DOI] [PubMed] [Google Scholar]
- 32.Smith I R L, Crook N E. Physical maps of the genomes for variants of Artogeia rapae granulosis virus. J Gen Virol. 1988;69:1741–1747. [Google Scholar]
- 33.Smith I R L, Crook N E. Characterization of new baculovirus genotypes arising from inoculation of Pieris brassicae with a granulosis virus. J Gen Virol. 1993;74:415–424. doi: 10.1099/0022-1317-74-3-415. [DOI] [PubMed] [Google Scholar]
- 34.Smith P H, Vlak J M. Biological activity of Spodoptera exigua nuclear polyhedrosis virus against S. exigua larvae. J Invertebr Pathol. 1988;51:107–114. [Google Scholar]
- 35.Teackle R E, Jensen J M, Giles J B. Age-related susceptibility of Heliothis punctiger to a commercial formulation of nuclear polyhedrosis virus. J Invertebr Pathol. 1986;47:82–92. [Google Scholar]
- 36.Tweeten K A, Bulla L A, Consigli R A. Applied and molecular aspects of insect granulosis viruses. Microbiol Rev. 1981;45:379–408. doi: 10.1128/mr.45.3.379-408.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vickers J M, Cory J S, Entwistle P F. DNA characterization of eight geographic isolates of granulosis virus from the potato tuber moth (Phtorimaea operculella) (Lepidoptera, Gelechiidae) J Invertebr Pathol. 1991;57:334–342. [Google Scholar]
- 38.Volkman L E, Blissard G W, Friesen P, Keddie B A, Possee R, Theilmann D A. Baculoviridae. In: Murphy F A, Fauquet C M, Bishop D H L, Ghabrial S A, Jarvis A W, Martelli G P, Mayo M A, Summers M D, editors. Virus taxonomy. Sixth report of the International Committee on Taxonomy of Viruses. New York, N.Y: Springer-Verlag; 1995. pp. 104–111. [Google Scholar]
- 39.Zeddam J L, Pollet A, Mangoendiharjo S, Ramadhan T H, López Ferber M. Occurrence and virulence of a granulosis virus in Phtorimaea operculella (Lep., Gelechiidae) populations in Indonesia. J Invertebr Pathol. 1999;74:48–54. doi: 10.1006/jipa.1998.4840. [DOI] [PubMed] [Google Scholar]