Graphical abstract
Keywords: CRISPR, surfaceome, online tool, sgRNA
Abstract
CRISPR-based genome-editing tools have emerged as an efficient tool for functional genomics studies. Online tools and databases have been developed to facilitate the design and selection of CRISPR single guide RNA (sgRNA) for gene modifications. However, to the best of our knowledge, none of these tools or database are designated to cell surface proteins. In a previous study, we described the development and application of surfaceome CRISPR libraries targeting to cell surface proteins on human cells. Here, we present the design and construction of an online tool and database (https://crispr-surfaceome.siais.shanghaitech.edu.cn/home), named CRISPR-Surfaceome, for the design of highly efficient sgRNA targeting to the surface proteins on human cells. To show case and validate the efficiencies of sgRNAs designed by this online tool, we chose ICAM-1 gene for knockout studies and found that all the 10 designed ICAM-1 sgRNAs could efficiently generate knockout cells, with more than 80% gene disruption rates. These ICAM-1 knockout cells were found to be resistant to the infection of rhinovirus (RV), which utilizes ICAM-1 as the receptor. Therefore, CRISPR-Surfaceome can serve the research community for the functional genomics studies on cell surface proteins, such as identification of pathogen receptors and discovery of drug targets.
1. Introduction
The Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/CRISPR-associated endonuclease (Cas)9-mediated genome editing has revolutionized biomedical research [1]. In practical applications, chimeric single guide RNAs (sgRNAs) are designed to direct Cas9 nuclease to specific genomic locations, leading to double-stranded DNA breaks (DSBs). In eukaryotes, DSBs are repaired mainly by two competing pathways: error-prone nonhomologous end joining (NHEJ) and homologous recombination (HR). During CRISPR-mediated NHEJ, nucleotide insertions or deletions (indels) can be introduced into the DSBs that enabled the perturbation of the open reading frame of targeted genes. These features have made CRISPR-Cas9 one of the most widely used tools for loss of function analyses in functional genomics studies [2].
In addition to single sgRNA-mediated gene knockout, pooled sgRNA libraries can be constructed for performing large-scale, unbiased genetic screen [3]. This CRISPR-based genetic screen has become one of the most important technologies in functional genomics with a variety of applications, including discovery of cancer drug targets or therapies [4], [5], uncovering the mechanisms of neurological diseases [6], dissection of pathogen-host interactions [7] and others. Nevertheless, most of the conventional CRISPR screens rely on genome-wide sgRNA libraries [8], [9], [10], which may be prone to false positive signals [11], [12]. As an alternative approach, focused sgRNA libraries composing of certain subsets of genes have been constructed and employed, including those targeting to kinome [13], [14], epigenome [15], nuclear factors [16], and cancer-related genes [17], [18].
Among those focused subsets of genes, cell surface proteins represent particularly interesting targets. Surface proteins participate in the initial interactions between cells and external stimuli and constitute more than 60 % of the therapeutic targets [19]. Particularly, cell surface proteins are the most important targets for macromolecule drugs or cell-based therapeutics, which lack the access to intracellular components. We and others have constructed cell surface protein-focused CRISPR libraries and demonstrated the feasibility of targeting surface proteins for functional genomics studies [20], [21], [22], [23].
There are commercial or academic websites providing user-friendly tools to populate the application of CRISPR-based gene editing technologies [24], [25], [26]. However, these online tools typically provide sgRNA design methods at the genome-wide scale without the focus on subsets of genes such as cell surface proteins. In the present study, we describe the design, construction and validation of CRISPR-Surfaceome (https://crispr-surfaceome.siais.shanghaitech.edu.cn/home), an online tool and database for designing sgRNAs with a specific focus on cell surface proteins. CRISPR-Surfaceome can provide online queries for sgRNA design for a single gene and the genome-wide and surface protein-targeted sgRNA information as a downloadable file. We then validated the sgRNA design from this database using a rhinovirus (RV) infection model and sgRNAs targeting to RV receptor ICAM-1.
2. Materials and methods
2.1. Design of the website
Our website is constructed based on Dubbo, an open source remote procedure call (RPC) and microservice framework. The current architecture and the hardware system allow facile upgrade to support increased number of users and queries.
2.2. Cell culture
H1-Hela and HEK293T cells were obtained from the American Type Culture Collection (ATCC) and Cell Bank of Shanghai Institutes for Biological Science (SIBS) respectively. Authentication of cell lines were performed by VivaCell Biosciences (Shanghai, China) using the short tandem repeats (STR) method. H1-Hela and HEK293T were grown in Dulbecco’s modified Eagle’s medium (DMEM, Thermo Fisher Scientific, Waltham, MA, USA) containing 10 % fetal bovine serum (FBS, Thermo) and 1 % penicillin–streptomycin (Thermo) at 37 °C in a fully humidified incubator containing 5 % CO2. H1-Hela cells and HEK293T cells were passaged at a subcultivation ratio of 1:3 and passaged every 2–3 days. The H1-Hela and HEK293T cells in this study were used under 10 passages. All cells were confirmed by PCR to be free of mycoplasma contamination.
2.3. Construction of lentiviral plasmids for CRISPR/sgRNA
To clone sgRNAs into the lentiviral vector pLeniCRISPR-v2, the vector was first treated with Esp3I (Thermo) to generate a sticky end. Forward and reverse oligonucleotides (Table S1) encoding the 20 bp sgRNA target sequence were annealed to form double-stranded DNA with overhangs that matched the sticky ends in the vector. Annealed sgRNA products were ligated into vector and transformed into DH5α E. coli (Tsingke, Beijing, China).
2.4. Lentivirus (LV) production
For LV production, HEK293T cells were seeded on to 6-well plates with a cell density of 2 × 106 cells per well. At 24 h after seeding, HEK293T cells at a confluence of approximately 70 %were transfected with 1,100 ng LV packaging plasmid pMD2.G, 1670 ng envelope plasmid psPAX and 2,200 ng transfer plasmid pLentiCRISPR-v2-sgRNA (1:1.5:2) that carries ICAM-1 sgRNA using Lipofectamine 3000 (Thermo). At 6 h after transfection, the medium was removed and fresh DMEM medium was supplemented. At 48 h post transfection, LVs were collected from the medium supernatant by centrifugation at 2000 rpm for 10 min, purified by filtration through a 0.45-μm filter (Merck, Darmstadt, Germany) and then stored at −80 °C for subsequent studies.
2.5. Generation of CRISPR-Cas9 knockout cells
H1-Hela cells were transduced with CRISPR/sgRNA LVs using spinfection. Briefly, 1 × 106 H1-Hela cells were incubated with 500 μL LVs in 500 μL DMEM supplemented with 10 % FBS in the presence of 10 μg/mL polybrene (Merck) under centrifugation at 2,000 rpm for 2 h. Upon completion of spinfection, the medium containing LVs was removed and fresh DMEM medium containing 10 % FBS and 2 μg/mL puromycin (Thermo) was supplemented. After 2 to 3 days’ selection with puromycin, survived cells were collected.
The genomic DNA of edited cells were extracted using Quick Extraction kit (Lucigen, Middleton, WI, USA). The target sites carrying edited sequences were PCR amplified using specific primers (Table S2) and then Sanger sequenced. The knockout efficiency was determined by TIDE (https://tide.nki.nl/) [27].
2.6. RV infection
RVs were produced as previously described [23]. For RV infection, H1-Hela cells were seeded on to 96- or 12-well plates with a density of 2.0 × 104 cells per well. At 24 h after seeding, cells were infected with RVs at an MOI of 2. After RV infection for 3 h, the medium containing RVs was removed and the cells washed with PBS for three times. After washing, fresh DMEM (Thermo) containing 10 % fetal bovine serum (FBS, Thermo) and 1 % penicillin–streptomycin (Thermo) was supplemented and cells were maintained for another 24 h before further analysis.
2.7. Cell viability assay
H1-Hela cells were infected with RV as described above and the cell images of bright field were acquired by Evos (Thermo). Cell viability was determined using Cell Counting Kit-8 (CCK-8, Dojindo, Kumamoto, Japan) following manufacturer’s instructions. The absorbance at 450 nm was measured using Enspire multimode plate reader (PerkinElmer, Waltham, MA, USA).
2.8. Real-time quantitative PCR (RT-qPCR)
To determine viral loads, H1-Hela cells were infected with RVs at an MOI of 1 unless noted otherwise. The medium supernatant or cell lysate containing RVs were harvested at 24 h after infection. The viral RNA (vRNA) in the supernatant and cell lysate was extracted using Trizol (Thermo), followed by purification using chloroform (Titan, Shanghai, China) extraction and isopropanol precipitation. Purified vRNA was reverse transcribed into cDNA using PrimeScript RT reagent Kit with gDNA Eraser (Takara Bio Inc., Shiga, Japan). Viral loads in medium supernatant was determined using RT-qPCR with Taqman probes and primers (Table S3) on an Applied Biosystems Q6 Real-Time PCR cycler (Thermo). The viral copy numbers were calculated using a standard curve of RV genome with known TCID50 and the R square of curve-fitting was guaranteed to be more than 0.99. VRNA in cell lysate was determined using RT-qPCR with SYBR green dye (Thermo) and specific primers (Table S3) on an Applied Biosystems Q6 Real-Time PCR cycler. The quality of SYBR Green primers was verified with the dissociation curves. The expression of vRNA in cell lysate was normalized to ribosomal gene RPLP0 (36b4).
2.9. Statistical analyses
Three biological replicates were performed for each experiment and the results are shown as mean ± SD. Statistical analyses and graphing were performed with GraphPad Prism8.0. Significant difference was determined using two-tailed, unpaired Student’s t-test.
3. Results and discussion
3.1. Design of surfaceome sgRNAs
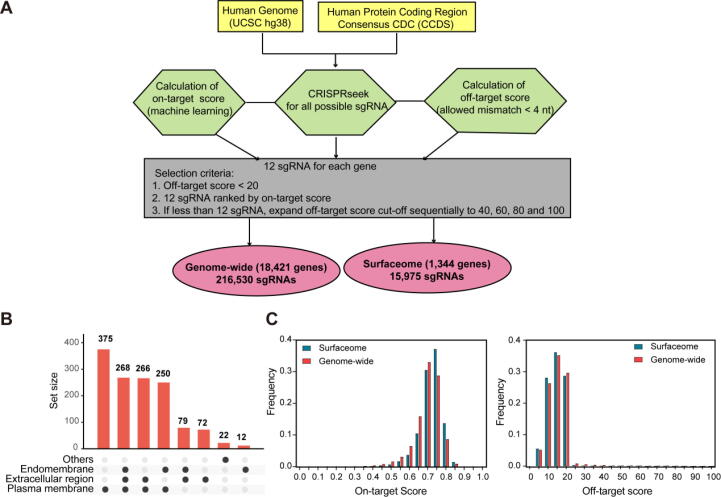
SgRNAs were designed to target to protein-coding regions (NCBI CCDS data) [28]. The on– and off-target scores of sgRNAs were calculated using Rule Set 2 [29] and Feng Zhang Lab’s algorithm [30] respectively. The sgRNAs were first ranked according to on-target scores and the top 12 sgRNAs with off-target scores of<20 were selected to disply on the webiste. If<12 sgRNAs met the above criteria, the cutoff of off-target scores were gradually increased to 40, 60, 80 and 100 until 12 sgRNAs were obtained (Fig. 1A).
Fig. 1.
Design of sgRNA in genome-wide and surfaceome-wide libraries. A. Schematic illustration of the design of sgRNA in genome-wide and surfaceome-wide libraries. B. Bar plot showing the composition of selected genes in surfaceome library. C. The distribution of sgRNA on-target and off-target scores in genome-wide and surfaceome libraries.
With the above criteria, the CRISPR-Surfaceome website contains 216,530 sgRNAs spanning the 18,421 genes in the genome with an average of 12 sgRNAs per gene. The 1,344 surface proteins are defined using the information from a mass spectrometry database [31] as previously described [23]. Gene ontology analysis of the cellular components of the surface proteins shows that the majority is located on plasma membrane and/or extracellular matrix, rather than intracellular membranes (Fig. 1B). The sgRNAs targeting to surface proteins are extracted from the genome-wide sgRNA pool (Fig. 1A). The on-target scores of most surface protein-targeted sgRNAs are around 0.75 in comparision with 0.70 in the genome-wide sgRNA pool. Importantly, the off-target scores of most sgRNAs in CRISPR-Surfaceome server are<20 (Fig. 1C).
3.2. Online querying and downloadable files
The current web server can support up to 100,000 users and 5,000 independent queries per day. The main functions of CRISPR-Surfaceome web server include querying, downloading and data uploading. Users can also choose species and library versions for sgRNA origins.
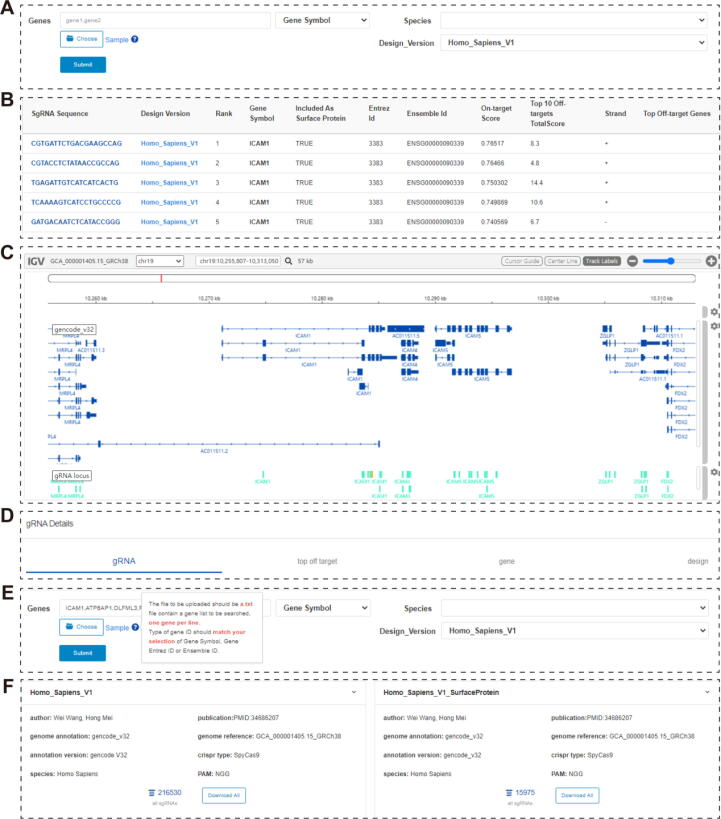
For querying function, CRISPR-Surfaceome provides two modes: single and batch queries. The single queries for sgRNAs of interest can be achieved using the searching keys “Gene symbol” (by inputing gene name), “Ensemble ID” (inputing Ensemble ID number) or “Entrez ID” (by inputing Entrez ID number) (Fig. 2A). The searching results will be displayed in a table, containing the information about sgRNA sequence, rank, on-target scores, off-target scores and targeting strand. Most importantly, the display page will indicate whether the related genes are defined as cell surface proteins in our surfaceome library V1 (Fig. 2B). Clicking on the sequence of the sgRNAs will direct the users to Intergrative Genomics Viewer (IGV) [32] and the selected sgRNAs will be labeled orange and placed under corresponding genomic loci (Fig. 2C). Under IGV presentation, details about sgRNAs, gense, off-target sites and sgRNA design are exhibited (Fig. 2D).
Fig. 2.
Online querying on CRISPR-Surfaceome. A. “Search” page. B. “Display” page. C. IGV page. D. Displayed details of sgRNAs in IGV page. E. Batch mode of gene querying. F. “Document” page for downloading the editable file for genome-wide and surfaceome libraries.
The batch queries are achieved by uploading gene list files of the.txt format or inputing the list of genes that are seperated by comma (Fig. 2E). The searching results will be downloadable.txt files containing the sgRNAs targeting the designated genes. This batch mode will be particularly useful for users who wish to design customized sgRNA libraries. In the “Document” page, the full list of the sgRNAs for each libraries can be downloaded (Fig. 2F). The downloading function is granted to registered users only to prevent improper use of the function that may disable the server.
CRISPR-Surfaceome has a straightforward and relatively simple approach for administers to feed new data. The information that need to be provided by administers include name of libraries, species, genome annotation (.gtf files to be provided to server if not already in the file directory), genome information (genome fasta files to be uploaded to server if not already in the file directory) and type of CRISPR technologies. The information for sgRNAs such as nucleotide sequence, on-target score, off-targets, genomic location and others can be uploaded as a single.csv file. Once the feeding process is completed, the querying and downloading functions will be immediately activated without further validation process.
3.3. Validation of the top 10 sgRNAs of ICAM-1 from CRISPR-Surfaceome
To validate the efficiency of sgRNA design and the applicability of CRISPR-Surfaceome in loss-of-function functional genomics studies, we searched for the sgRNAs targeting to rhinovirus (RV) receptor [33] intercellular adhesion molecule-1 (ICAM-1) and constructed ICAM-1 knockout H1-Hela cells for investigation of the protection from RV infection [23]. The top 10 sgRNAs of ICAM-1 from CRISPR-Surfaceome database were selected for further analysis.
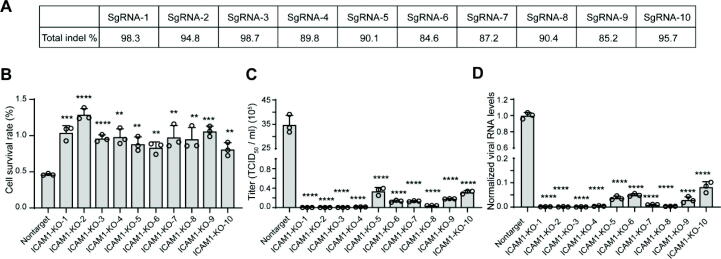
The sgRNA sequences were cloned to lentiviral plasmids and high-titer LVs were prepared. H1-Hela cells were then transduced with ICAM-1-targeting LVs. Genomic DNA was extracted from transduced cells and sgRNA-targeted sites were amplified for analysis of gene disruption efficiencies. It was found that all the 10 sgRNAs could knock out ICAM-1 with indel frequencies of more than 80 % (Fig. 3A and Figure S1), suggesting that CRISPR-Surfaceome is capable of generating highly efficient sgRNAs.
Fig. 3.
Validation of the top 10 sgRNAs of ICAM-1. A. Indel frequencies of the top 10 ICAM-1 sgRNAs, determined byTIDE analysis. B. Cell viability of ICAM-1 knockout cells upon RV challenge, in comparison with non-targeting sgRNA-treated cells. C-D. Viral loads in medium supernatant (C) or cell lysates (D) of non-targeting sgRNA-treated and ICAM-1 knockout cells after RV challenge. Viral RNA in cell lysates is normalized to RPLP0 expression. The significance of difference between ICAM-1 knockout and non-targeting sgRNA-treated cells are determined using two-tailed, unpaired Student’s t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ***, P < 0.0001.
To investigate if these ICAM-1 knockout could protect cells from RV infection, we adopted a previously reported live-and-dead model of RV infection [23]. It was found that all the 10 ICAM-1 knockout cells exhibited significantly increased cell viability upon RV-B14 challenge in comparison with non-targeting sgRNA-treated cells (Fig. 3B and Figure S2). In addition, these knockout cells showed markedly reduced viral loads in medium supernatant (Fig. 3C) and cell lysates (Fig. 3D). These results suggested that CRISPR-Surfaceome database is capable of generating sgRNAs for loss-of-function functional genomics studies.
3.4. Future update of CRISPR-Surfaceome database
It has been reported that up to 50 % of proteome has mixed localization and that surfaceome diversity depends on cell type and cell states [34]. The current database is generated using the experimental results of 41 human cells from a public resource Cell Surface Protein Atlas, or abbreviated as CSPA [31]. CSPA covers a limited number of cell types and cell states without the consideration of cellular perturbation, and is thus unlikely to encompass all possible human surface proteins. A recent study developed a machine-learning algorithm SURFY and predicted a human surfaceome of 2886 proteins [19]. Comparison of CSPA with SURFY revealed a low degree of overlap (Figure S3). In addition, a recent spatiotemporal proteomic profiling study has uncovered a series of proteins with altered subcellular localization under cellular perturbation [35]. Our analysis showed that CSPA only contained a small fraction of the proteins with dynamic cellular localization (Figure S3). These results suggest that it is possible to update the collection of cell surface proteins in CRISPR-Surfaceome once further experimental results are available. Therefore, our web server is designed to contain flexible updating and tracking ports for updating the protein database in the future.
4. Conclusion
In this study, we present CRISPR-Surfaceome as an online tool and database for designing highly efficient sgRNAs to generate surface protein knockout cells. The current surfaceome library consists of 1,344 surface proteins on human cells and is expandable in the future to a larger set of genes or to other organisms. CRISPR-Surfaceome can expedite loss-of-function analysis for functional genomics studies and may serve as an important tool for understanding cell–cell or cell-pathogen interactions and discovery of novel targets for macromolecule drugs and cell-based therapeutics.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
We thank Ms. Zhuojia (Julie) Liu at ShanghaiTech University for the graphic design of the web server and Discovery Technology Platform for the support of RV infection and cell viability assay.
Funding
This work is supported by Shanghai Frontiers Science Center for Biomacromolecules and Precision Medicine at ShanghaiTech University, the Joint Fund of Ninth People's Hospital Affiliated to Shanghai Jiao Tong University School of Medicine with ShanghaiTech University (JYJC202126 to J.L. and L.J.), ShanghaiTech University Startup Fund (2019F0301-000-01 to J.L.) and China Postdoctoral Science Foundation (2017M621551 to H.M).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2022.07.026.
Contributor Information
Lichun Jiang, Email: jianglch@shanghaitech.edu.cn.
Jia Liu, Email: liujia@shanghaitech.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Zhang H., et al. Application of the CRISPR/Cas9-based gene editing technique in basic research, diagnosis, and therapy of cancer. Mol Cancer. 2021;20(1):126. doi: 10.1186/s12943-021-01431-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang S.W., et al. Current applications and future perspective of CRISPR/Cas9 gene editing in cancer. Mol Cancer. 2022;21(1):57. doi: 10.1186/s12943-022-01518-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Przybyla L., Gilbert L.A. A new era in functional genomics screens. Nat Rev Genet. 2022;23(2):89–103. doi: 10.1038/s41576-021-00409-w. [DOI] [PubMed] [Google Scholar]
- 4.Haley B., Roudnicky F. Functional genomics for cancer drug target discovery. Cancer Cell. 2020;38(1):31–43. doi: 10.1016/j.ccell.2020.04.006. [DOI] [PubMed] [Google Scholar]
- 5.Buquicchio F.A., Satpathy A.T. Interrogating immune cells and cancer with CRISPR-Cas9. Trends Immunol. 2021;42(5):432–446. doi: 10.1016/j.it.2021.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kampmann M. CRISPR-based functional genomics for neurological disease. Nat Rev Neurol. 2020;16(9):465–480. doi: 10.1038/s41582-020-0373-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Puschnik A.S., et al. A CRISPR toolbox to study virus-host interactions. Nat Rev Microbiol. 2017;15(6):351–364. doi: 10.1038/nrmicro.2017.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han K., et al. CRISPR screens in cancer spheroids identify 3D growth-specific vulnerabilities. Nature. 2020;580(7801):136–141. doi: 10.1038/s41586-020-2099-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang W., et al. A genome-wide CRISPR-based screen identifies KAT7 as a driver of cellular senescence. Sci Transl Med. 2021;13(575) doi: 10.1126/scitranslmed.abd2655. [DOI] [PubMed] [Google Scholar]
- 10.Wei J., et al. Genome-wide CRISPR Screens Reveal Host Factors Critical for SARS-CoV-2 Infection. Cell. 2021;184(1):76–91.e13. doi: 10.1016/j.cell.2020.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haapaniemi E., et al. CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat Med. 2018;24(7):927–930. doi: 10.1038/s41591-018-0049-z. [DOI] [PubMed] [Google Scholar]
- 12.Ihry R.J., et al. p53 inhibits CRISPR-Cas9 engineering in human pluripotent stem cells. Nat Med. 2018;24(7):939–946. doi: 10.1038/s41591-018-0050-6. [DOI] [PubMed] [Google Scholar]
- 13.Tian R., et al. CRISPR interference-based platform for multimodal genetic screens in human iPSC-derived neurons. Neuron. 2019;104(2):239–255.e12. doi: 10.1016/j.neuron.2019.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang C., et al. CDK12 inhibition mediates DNA damage and is synergistic with sorafenib treatment in hepatocellular carcinoma. Gut. 2020;69(4):727–736. doi: 10.1136/gutjnl-2019-318506. [DOI] [PubMed] [Google Scholar]
- 15.Li F., et al. In vivo epigenetic CRISPR screen identifies Asf1a as an immunotherapeutic target in kras-mutant lung adenocarcinoma. Cancer Discov. 2020;10(2):270–287. doi: 10.1158/2159-8290.CD-19-0780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cortez J.T., et al. CRISPR screen in regulatory T cells reveals modulators of Foxp3. Nature. 2020;582(7812):416–420. doi: 10.1038/s41586-020-2246-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inoue D., et al. Spliceosomal disruption of the non-canonical BAF complex in cancer. Nature. 2019;574(7778):432–436. doi: 10.1038/s41586-019-1646-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bersuker K., et al. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature. 2019;575(7784):688–692. doi: 10.1038/s41586-019-1705-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bausch-Fluck D., et al. The in silico human surfaceome. Proc Natl Acad Sci U S A. 2018;115(46):E10988–E10997. doi: 10.1073/pnas.1808790115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chong Z.S., et al. Pooled extracellular receptor-ligand interaction screening using CRISPR activation. Genome Biol. 2018;19(1):205. doi: 10.1186/s13059-018-1581-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ye L., et al. In vivo CRISPR screening in CD8 T cells with AAV-Sleeping Beauty hybrid vectors identifies membrane targets for improving immunotherapy for glioblastoma. Nat Biotechnol. 2019;37(11):1302–1313. doi: 10.1038/s41587-019-0246-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao X., et al. Human neonatal Fc receptor is the cellular uncoating receptor for enterovirus B. Cell. 2019;177(6):1553–1565.e16. doi: 10.1016/j.cell.2019.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mei H., et al. Surfaceome CRISPR screen identifies OLFML3 as a rhinovirus-inducible IFN antagonist. Genome Biol. 2021;22(1):297. doi: 10.1186/s13059-021-02513-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Concordet J.P., Haeussler M. CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 2018;46(W1):W242–W245. doi: 10.1093/nar/gky354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lei Y., et al. CRISPR-P: a web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol Plant. 2014;7(9):1494–1496. doi: 10.1093/mp/ssu044. [DOI] [PubMed] [Google Scholar]
- 26.Haeussler M., et al. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 2016;17(1):148. doi: 10.1186/s13059-016-1012-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brinkman E.K., et al. Easy quantification of template-directed CRISPR/Cas9 editing. Nucleic Acids Res. 2018;46(10):e58. doi: 10.1093/nar/gky164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pruitt K.D., et al. The consensus coding sequence (CCDS) project: Identifying a common protein-coding gene set for the human and mouse genomes. Genome Res. 2009;19(7):1316–1323. doi: 10.1101/gr.080531.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hsu P.D., et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31(9):827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doench J.G., et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34(2):184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bausch-Fluck D., et al. A mass spectrometric-derived cell surface protein atlas. PLoS ONE. 2015;10(3):e0121314. doi: 10.1371/journal.pone.0121314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robinson J.T., et al. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Greve J.M., et al. The major human rhinovirus receptor is ICAM-1. Cell. 1989;56(5):839–847. doi: 10.1016/0092-8674(89)90688-0. [DOI] [PubMed] [Google Scholar]
- 34.Min K.W., Lee S.H., Baek S.J. Moonlighting proteins in cancer. Cancer Lett. 2016;370(1):108–116. doi: 10.1016/j.canlet.2015.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mulvey C.M., et al. Spatiotemporal proteomic profiling of the pro-inflammatory response to lipopolysaccharide in the THP-1 human leukaemia cell line. Nat Commun. 2021;12(1):5773. doi: 10.1038/s41467-021-26000-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.