Highlights
-
•
Scutellaria baicalensis has reversal effect on radio-resistance of colorectal cancer (CRC).
-
•
Wogonin is the main active component of Scutellaria baicalensis.
-
•
SULT2B1 is highly expressed in CRC and associated with poor prognosis.
-
•
Wogonin could increase the radio-resistance sensitivity of CRC.
-
•
Wogonin reverses radio-resistance in CRC by targeting SULT2B1.
Keywords: Colorectal cancer, Radio-resistance, Scutellaria baicalensis, SULT2B1, Wogonin
Abstract
Scutellaria baicalensis (SB) has been shown to improve the therapeutic effects of colorectal cancer (CRC) and perform well for reversing radio-resistance in different cancers. However, its potential function and mechanism related to radio-resistance in CRC has not been explored. A radio-resistant human CRC cell line (HCT116R) was applied. A network pharmacological analysis was performed to reveal the potential mechanism of SB for reversing radio-resistance in CRC, and computational pathological analysis was applied to indicate the clinicopathological significance of the key targets. Then, our hypothesis was further verified by molecular docking. The network pharmacology analysis showed that wogonin is the key compound of SB for reversing the radio-resistance of CRC. A Kyoto Encyclopedia of Genes and Genomes analysis showed that the genes for SB that reverse radio-resistance in CRC are mainly involved in steroid hormone biosynthesis. An enrichment analysis pointed out that Sulfotransferase family 2B member 1 (SULT2B1) is a potentially vital gene. SULT2B1 was demonstrated as being highly expressed in CRC and upregulated in radio-resistant rectal tissues or cell lines. A CCK-8 and clone formation test showed that the viability and clone formation ability of HCT116R were significantly decreased by wogonin combined with radiotherapy, compared to radiotherapy alone. By contrast, flow cytometry revealed that the apoptosis of HCT116R was significantly increased when wogonin treatment combined with radiotherapy, compared with radiotherapy alone. Molecular docking verification indicated that SULT2B1 and wogonin have a good binding ability. Taken together, SULT2B1 may be the potential drug target in treating radio-resistant CRC. Wogonin may be the core compound of SB for reversing radio-resistance in CRC by targeting SULT2B1.
Introduction
Colorectal cancer (CRC) is one of the most common cancers around the world, and the expected numbers of new cases, and the probability of cancer, are increasing every year [1]. According to the Global Cancer Statistics, CRC ranks third in terms of incidence, but second in terms of mortality [2]. The vast majority of patients with CRC at the time of diagnosis are in the middle or late stage, making simple surgery difficult to bring about complete resection, and the metastasis and recurrence rate is extremely high, leading to poor prognoses. Radiotherapy is one method used for the treatment of locally advanced cancers. It mainly relies on radiation energy to cause structural cellular changes and produce oxygen free radicals. As one of the most important therapeutic strategies for cancer treatment, radiotherapy plays an important role in the treatment of CRC [3,4]. However, due to side effects such as the low sensitivity of radiotherapy and damage to normal tissues, the problem of radio-resistance has become a key factor limiting the effectiveness of CRC treatments [5]. With this in mind, finding ways to reverse radiotherapy resistance and improve radiotherapy sensitivity is of great clinical value in the treatment of CRC.
Traditional Chinese Medicine (TCM) has been demonstrated to have a good synergistic and detoxifying effect in the adjuvant treatment of tumors [6,7]. Scutellaria baicalensis (SB, or huangqin in Chinese) is a major TCM herb. SB is often used for clearing away internal heat and keeping a proper temperature, stopping bleeding, and the prevention of miscarriage in clinical settings. Studies have shown that a good number of active components in SB have antiviral [8], antibacterial [9], anti-injury [10], anti-inflammatory [11], anti-tumor [12], [13], [14], and other pharmacological effects. In particular, a number of SB's active components have been shown to have good antagonistic effects in CRC [15], [16], [17]. Recently, studies have reported that baicalin, the active ingredient in SB, has a good effect on reversing the radio-resistance of nasopharyngeal carcinoma [18], and that the baicalein in SB can increase the radiotherapy sensitivity of breast cancers [19]. SB seems to have a certain reversal effect on the radio-resistance of various tumors; however, its underlying mechanism and promising use as a drug for targeting CRC radio-resistance have not been confirmed. Therefore, it is of great value and clinical significance to investigate potential core targets and mechanisms of SB related to radio-resistance in CRC.
In this study, we aimed to explore the potential molecular mechanism of SB and to excavate a promising drug target for antagonizing radio-resistance in CRC via network pharmacology and molecular docking, combined with computational pathology analysis. We then verified our hypotheses by a series of experiments. It is hoped that this research will provide theoretical support for the application of TCM to radiotherapy sensitization in CRC, further the exploration of treatment strategies for tumor radio-resistance, and promote the clinical application of TCM. A flowchart of our study is shown in Fig. 1.
Fig. 1.
The flow chart of SB for treatment radio-resistance in CRC.
Materials and methods
Analysis of network pharmacology
Screening of ingredients and potential targets
The potential targets of SB were searched in the Traditional Chinese Medicine Systems Pharmacy database (TCMSP, https://tcmspw.com/tcmsp.php) and the Encyclopedia of Traditional Chinese Medicine database (ETCM, http://www.tcmip.cn/ETCM/index.php/Home/Index/), which can complement the TCMSP in finding potential targets. The screening criteria for active ingredients in the TCMSP were the compounds having good oral bioavailability (OB≥30%) and drug-like properties (DL≥0.18); the potential targets of each compound were investigated. The screening criterion of targets for SB in the ETCM was Tanimoto>0.8. Studies have also shown that baicalin, wogonin, and baicalein [18], [19], [20], which are the main active ingredients of SB, have a good reversal effect on radio-resistance in cancers. Considering the high reliability of the compounds verified in the literature, the TCMSP, Traditional Chinese Medicine Integrated Database (TCMID, http://www.megabionet.org/tcmid/), HERB (http://herb.ac.cn/), and ETCM databases were used to conduct a wide, comprehensive search for the targets of the three compounds, and to supplement the targets of SB. Finally, all potential targets of SB were sorted, the duplicates were deleted, the union was obtained, and the potential targets of SB were identified. All active ingredients of SB related to the intersection genes were searched using the above-mentioned ETCM and TCMSP databases.
Acquisition of radio-resistance gene profiles in CRC
To obtain the genes related to CRC radio-resistance, cell lines with acquired radio-resistance characteristic were established, and in-house RNA-seq of CRC cells was employed. In brief, human CRC cell lines CX-1 and HCT-116 were routinely cultured and exposed to X-ray radiation intermittently, as previously reported [21]. The radio-resistant cell lines, namely CX-1-R and HCT-116-R, together with their parental cells (CX-1 and HCT-116) were subjected to RNA sequencing at Lc-Bio Technologies (Hangzhou, China) according to the instructions of the manufacturer, and the differentially expressed genes were computed.
To verify the radio-resistant genes from in-house RNA-seq, the radio-treated RNA-arrays and RNA sequence data of CRC were browsed and enrolled in the Gene Express Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/), Sequence Read Archive (SRA, https://www.ncbi.nlm.nih.gov/sra), and ArrayExpress (https://www.ebi.ac.uk/arrayexpress/) databases. To gather as much data as possible, the retrieval strategies were: ((colon) OR (rectal) OR (colorectal)) AND ((carcinoma) OR (cancer) OR (malignant) OR (tumor) OR (adenoma)) AND ((radio) OR (radiotherapy) OR (radiation)).
To ensure the availability of the included data, the inclusion criteria were: (1) human CRC tissue or cell lines and (2) the sample size of experiment groups and control groups were greater than three. The exclusion criteria were: (1) non-homo sapiens, (2) a sample size less than three, and (3) no control groups. The samples of each dataset were divided into radio-sensitive and radio-resistant groups, according to tumor regression grade (TRG) [22]; a score between 0 and 1 was regarded as radio-sensitive, while a score between 2 and 4 was regarded as radio-resistant. The limma and limma voom algorithms were used to calculate the differently expressed genes in radio-treated RNA-array and RNA-seq data. The genes highly expressed in the radio-resistant group were considered “radio-resistant genes.” A total of eight datasets from four detection platforms were obtained: GPL570(GSE 35452, GSE119409); GPL6244(GSE43206, GSE46862); GPL13497(GSE97543, GSE150082); GSE20298; and GSE60331.
Acquisition of intersection genes and network construction
Venny 2.1.1 (https://bioinfogp.cnb.csic.es/tools/venny/) was used to obtain the intersection genes of SB and radio-resistance in CRC. These genes were considered potential targets of SB against radio-resistance in CRC and named the “disease-related targets.” Then, we used Cytoscape 3.7.2 software (http://cytoscape.org/)to establish the drug ingredient disease-related-targets network for SB to treat CRC radio-resistance. In the network diagram, nodes represent compounds, target proteins, or drugs, while the edges represent the interactions among node molecules. The importance of each node in the network is evaluated by degree. The degree of a node represents the number of edges connected to it. The larger the degree, the more biological functions it participates in, indicating that the node is more important in the network.
Biological function and pathway enrichment analysis
To indicate the main pharmacological molecular mechanism of SB against radio-resistance in CRC, a biological function and enrichment analysis, including GO and KEGG, of the intersecting genes (11) of SB with radio-resistance in CRC was carried out. The RStudio software (https://www.rstudio.com/) package Cluster Profiler (4.0.2) was used for GO and KEGG function enrichment analysis and graphing. The Cluster Profiler can perform statistical and visual analyses of functional clusterings in gene clusters or collections. We set the threshold as adjusted P < 0.05 and statistically screened out the biological processes and pathways with significant statistical differences before drawing a bubble chart.
Pathological significance analysis of SULT2B1
Data mining of SULT2B1 expression using public databases
We made full use of public data resources. SULT2B1 expression in CRC data were acquired from TCGA (https://portal.gdc.cancer.gov) through August 31, 2020. Gene microarray data related to SULT2B1 in CRC were searched from the GEO, SRA, and ArrayExpress databases through August 31, 2020. Our search strategy was as follows: ((colon) OR (rectal) OR (colorectal)) AND ((carcinoma) OR (cancer) OR (malignant) OR (tumor)). The filtering conditions were 1) Series [entry type] and 2) Homo sapiens [organism]. A total of 24 datasets from 17 detection platforms relevant to SULT2B1 were obtained: GPL96 (GSE24514, GSE49355, GSE68468, GSE77953, GSE110223); GPL6480 (GSE21815, GSE35279); GPL10558 (GSE54986, GSE75548, GSE106582); GPL15207 (GSE81558, GSE113513); GSE15781; GSE20842; GSE28000; GSE156355; GSE141174; GSE113513; GSE103512; GSE87211; GSE47063; GSE44076; GSE41011; and GSE25071 [23], [24], [25], [26], [27], [28], [29], [30], [31]. SULT2B1 protein variations in colorectal tissues were explored by immunohistochemistry in the Human Protein Atlas (THPA) (https://www.proteinatlas.org/), an open-source database that provides data on the expression of proteins in a variety of human tissues.
Differential expression analysis of SULT2B1
The data of SULT2B1 expression obtained from different databases were log2 normalized, and the criteria for SULT2B1 to be considered statistically significant was P < 0.05. The differential expression levels of SULT2B1 in CRC tissues versus normal colon tissue samples and radio-resistant CRC samples versus radio-sensitive CRC samples were visualized using box plots and Receiver operating characteristic curve (ROC) made using the RStudio software package ggplot2. Considering that some datasets were actually from the same detection platforms, to avoid batch effects, the data from the same platform were merged and subjected to the subsequent analyses. The integrated computation of SMD was conducted to evaluate SULT2B1 expression from a holistic perspective. The χ2 and I2 tests were computed to assess heterogeneity within the meta‑analysis. A random-effects model was applied if I2>50%; otherwise, the fixed-effects model was adopted. The heterogeneity source was detected using a sensitivity analysis, as described previously [32]. Subsequently, to determine whether SULT2B1 was a suitable gene that discriminates radio-resistant from radio-sensitive samples, a diagnostic meta-analysis was conducted. The above comprehensive analysis was conducted using Stata 12.0 (Stata Corp LP, College Station, TX, USA). To evaluate the prognostic value of SULT2B1, Gene Expression Profiling Interactive Analysis (GEPIA2), a web server for gene expression analysis based on data from the TCGA database, was employed following the database's instructions [33]. The impact of SULT2B1 expression on CRC outcomes was estimated in terms of disease-free survival (DFS) and overall survival (OS), and the corresponding Kaplan-Meier (KM) survival curves were generated.
Experimental verification
Reagents and cell culture
Wogonin (purity>95%) was purchased from Chengdu Herbpurify Co., Ltd. and dissolved using dimethyl sulfoxide, and 5-fluorouracil (5-Fu, purity>95%, a positive control drug) was purchased from Shanghai Macklin Biochemical Technology Co., Ltd. (“Macklin Macklin”). HCT116R, a radio-resistant human colorectal cancer cell line, was established and cultured in the Experimental Center of the School of Pharmacy at Guangxi Medical University via long-term intermittent cumulative induction (cell irradiation condition was via X-ray beam (Varian Clinac iX, Varian Medical Systems Inc, Palo Alto, CA) at a dose rate of 522cGy/min, the accumulated total irradiation dose up to about 80Gy). Cell culture conditions were maintained in McCoy's 5A medium, supplemented with 10% fetal bovine serum.
Irradiation conditions
Cell line irradiation was performed using an X-ray beam (Varian Clinac iX, Varian Medical Systems Inc, Palo Alto, CA, USA) at a single dose rate of 265cGy/min(2Gy) or 522cGy/min(4Gy) for the experiments reported below.
Cell proliferation assays
Cell proliferation was measured using CCK8 assay. The HCT116 cells in the logarithmic phase were seeded at a density of 5 × 103cells/well into a 96-well plate. Then, the cells were cultured for 48 h in an incubator, after which the cells adhered to the wall and were pretreated with various concentrations (0, 2, 4, 8, 16, 32, 64 u mol/L) of wogonin. Then, CCK8 was added to the plates. The absorbances at 450 nm after 1 h were recorded under a microplate reader. After exploring the IC50 of wogonin on HCT116R cells, 10 μmol/L was selected as the administration concentration for subsequent experiments. Three replicate wells were evaluated in each group, and three independent experiments were performed. Cell survival was calculated using the following formula: Cell viability rate (%) = (OD of experimental group - OD of the blank control group) / (OD of control group - OD of the blank control group).
Clone formation experiments
The HCT116R cells in the logarithmic growth phase were digested and blow as a single-cell suspension to perform the colony formation assay. The cells were seeded in 6-well plates (400 cells/well). After the cells adhered to the wall and were treated with wogonin (10 umol/L), the corresponding radiation was given. After 48h, the drug-free medium was replaced, and the culture continued to grow for 14 days. Then, the colonies were stained with 0.1% crystal violet solution and the number of colonies with more than 50 cells was counted. The rate of colony formation was calculated as follows: Colony formation rate (%) = Number of colonies/Number of cell inoculation. This experiment was independently repeated three times.
Apoptosis assay
Apoptosis assay was used to detect the effects of wogonin on the apoptosis of HCT116R cells. HCT116R cells were seeded in 6-well plates (1 × 105cells/well). After the cells adhered to the growth, they were immediately given wogonin (10 umol/L) and a corresponding radiation treatment (2 or 4Gy). The cells were incubated for 48 h and then harvested and stained using the annexin V-FITC/PI apoptosis detection kit (MultiSciences Lianke Biotech Co., Ltd.), according to the manufacturer's instructions. The resulting fluorescence was detected using flow cytometry. This experiment was independently repeated three times.
Statistical analysis
SPSS 22.0 statistical software (https://www.ibm.com/products/spss-statistics) was used to analyze the experiment data. Measurement data are presented as mean ± standard deviation (SD). The one-way ANOVA method was used for comparisons among groups. A P-value < 0.05 was considered statistically significant. Histograms of the results were made by using GraphPad Prism 6.0 software. Flowjo 10.0 software (https://www.flowjo.com/solutions/flowjo) was used to analyze data from the flow cytometric apoptosis assays.
Molecular docking verification
Molecular docking verification was made to explore the interaction between the key target gene and its related compounds of SB. We downloaded the 2D structure of small molecule compounds (SDF format) from the PubChem database(https://pubchem.ncbi.nlm.nih.gov/), and then OpenBabel software (http://openbabel.org/wiki/Main_Pageb) was used to convert and save the SDF format of small molecule compounds into mol2 format. We also downloaded the 3D structure (PDB format) of the protein receptor (key target genes) form the Research Collaboratory for Structural Bioinformatics’ Protein Database (PDB, http://www1.rcsb.org/) and ran the Surflex-Dock program in Sybyl-X2.2.1 software (https://sybyl.com/) to analyze the protein, including removing water and ligand molecules, hydrogenation, and the addition of polar and nonpolar hydrogen. Then, the key target genes and their related compounds were docked. The evaluation of molecular docking results was based on the similarity and affinity between the active pocket of protein and the structure of the small molecule compound. Generally speaking, the total score was greater than or equal to 5 [34], and the affinity between small molecules and receptors was considered reliable.
Results
Common targets of SB with radio-resistance in CRC
After removing duplicates, a total of 586 potential targets of SB were collected and screened. Using in-house high-throughput sequencing data and chips (data from GEO, SRA, and ArrayExpress) to construct the radio-resistant gene profiles of CRC, 155 radio-resistant genes were obtained. Finally, by taking an intersection of the SB target genes and disease-related genes, 11 common target genes were obtained after the intersection (Fig. 2A). These genes may be potential targets of SB against radio-resistance in CRC.
Fig. 2.
Network pharmacology analysis. (A) Common part of SB targets and radio-resistance genes of CRC (B). Ingredient-disease related target network (red nodes represent target genes of SB for treating radio-resistance in CRC, and blue nodes represent ingredients of SB). (C) GO enrichment of 11 insertion genes. (D) KEGG enrichment of 11 insertion genes.
Ingredient-disease-related targets network for SB in the treatment of radio-resistant CRC
We searched for active ingredients related to intersection genes (11 genes) from the above ETCM and TCMSP databases, as shown in Table 1. Then, the ingredient-disease-related targets network was constructed, which consisted of 24 nodes (13 compound nodes and 11 target nodes) and included 23 connections (Fig. 2B). Wogonin was at the core position in the network, which may be the key component of SB against radio resistance in CRC.
Table 1.
Active compounds of SB corresponding to intersecting genes.
| Genes | Compounds |
|---|---|
| SULT2B1 | wogonin,Sitosterol,Stigmasterol,Campesterol |
| NR1H4 | Palmitic Acid |
| AKR1C1 | Campesterol,Stigmasterol,Sitosterol,Norwogonin,Isoscutellarein,5,7,4′-Trihydroxy-6-Methoxyflavanone,5,7,2′,6′-Tetrahydroxyflavone,Eriodictyol,Dihydrooroxylin A,Baicalein,Chrysin |
| DPEP1 | wogonin |
| TMPRSS3 | wogonin |
| PRSS22 | wogonin |
| KLK6 | wogonin |
| KLK11 | wogonin |
| TMPRSS13 | wogonin |
| SH2D4A | wogonin |
| RAB25 | Wogonin |
GO and KEGG enrichment analyses
To further understand the molecular mechanism of SB against radio resistance in CRC, GO and KEGG enrichment analyses were performed on 11 potential common targets. The results of the GO analyses showed that these targets of molecular functions mainly involved serine type endopeptidase activity (GO: 0004252), serine type peptidase activity (GO: 0008236), serine hydrolase activity (GO: 0017171), endopeptidase activity (GO: 0004175), and so on (Fig. 2C). This indicated that these biological processes are mainly related to metabolic enzymes.
KEGG analyses showed that these potential genes were mainly enriched in steroid hormone biosynthesis (hsa00140), Bill secretion (hsa04976), and the metabolism of xenobiology by cytochrome P450 (hsa00980, Fig. 2D). Among these pathways, the most significant one is the steroid hormone biosynthesis pathway (hsa00140), in which SULT2B1 and AKR1C1 genes are enriched (Table 2). Interestingly, a recent study confirmed that overexpression of SULT2B1 has been shown to be an independent prognostic indicator for cell growth and invasion [35], and another study has revealed that steroid hormone biosynthesis plays an important role in the susceptibility and survival rate of colorectal cancer [36]. and another study has revealed that steroid hormone biosynthesis plays an important role in the susceptibility and survival rate of colorectal cancer.
Table 2.
Information of the KEGG enrichment analysis of 11 intersection genes.
| Gene ID | Description | P Value | P Adjust | gene |
|---|---|---|---|---|
| hsa00140 | Steroid hormone biosynthesis | 0.00016709 | 0.000167095 | SULT2B1/ AKR1C1 |
| has00980 | Metabolism of xenobiotics by cytochrome P450 | 0.02866065 | 0.0330207710 | NR1H4 |
| hsa04976 | Bile secretion | 0.0330207 | 0.033020771 | AKR1C1 |
Differential expression and prognostic evaluation of SULT2B1
Upregulated SULT2B1 expression and its diagnostic value in radio-resistant CRC
SULT2B1 is not only the target of SB, it is also among the radio-resistant genes. We determined the expression of SULT2B1 in rectal tissues or cell lines with differential radiosensitivity. SULT2B1 was upregulated in radio-resistant samples, in comparison with those sensitive to radiotherapy (Fig. 3A–E). An integrated computation of all five datasets was conducted, to get a more precise assessment of SULT2B1 expression in a differential radiosensitivity sample of CRC. As shown in Fig. S1-A, and the heterogeneity test was I-Squard=2.1%, the integrated computation of SMD showed statistically significant differences, P = 0.394 > 0.05, which revealed there was no significant heterogeneity, so a fixed effects model was used. The SMD value of the pooled analysis of each study was 0.31>0, which indicated that SULT2B1 is highly expressed in radio-resistant cell lines. Tests for publication bias were performed (Begg's test was 0.221, Egger's test was 0.063), and the results showed no publication bias.
Fig. 3.
The box plots of SULT2B1 mRNA expression in rectal tissues or cells with differential radio-sensitivity.
ROC analysis was also used to evaluate the ability of SULT2B1 to distinguish radio-resistant samples from radio-sensitive samples. The corresponding ROC curves are shown in Fig. 4. As we can see, the area under curve (AUCs) were not satisfactorily high. The AUC of the sROC was 0.66 (95%CI: 0.61–0.70), and the combined sensitivity and specificity were 0.53 (95%CI: 0.41–0.65) and 0.77 (95%CI: 0.61–0.70; Fig. S1-I), respectively, indicating that SULT2B1 is not competent in discriminating radio-resistant samples from radio-sensitive samples.
Fig. 4.
The corresponding ROC curves for SULT2B1 expression in rectal tissues or cells with differential radio-sensitivity. (A) GPL570, (B) GPL6244, (C) GPL13497, (D) GSE20298, (E) GSE60331.
Overexpression of SULT2B1 and its diagnostic value in CRC
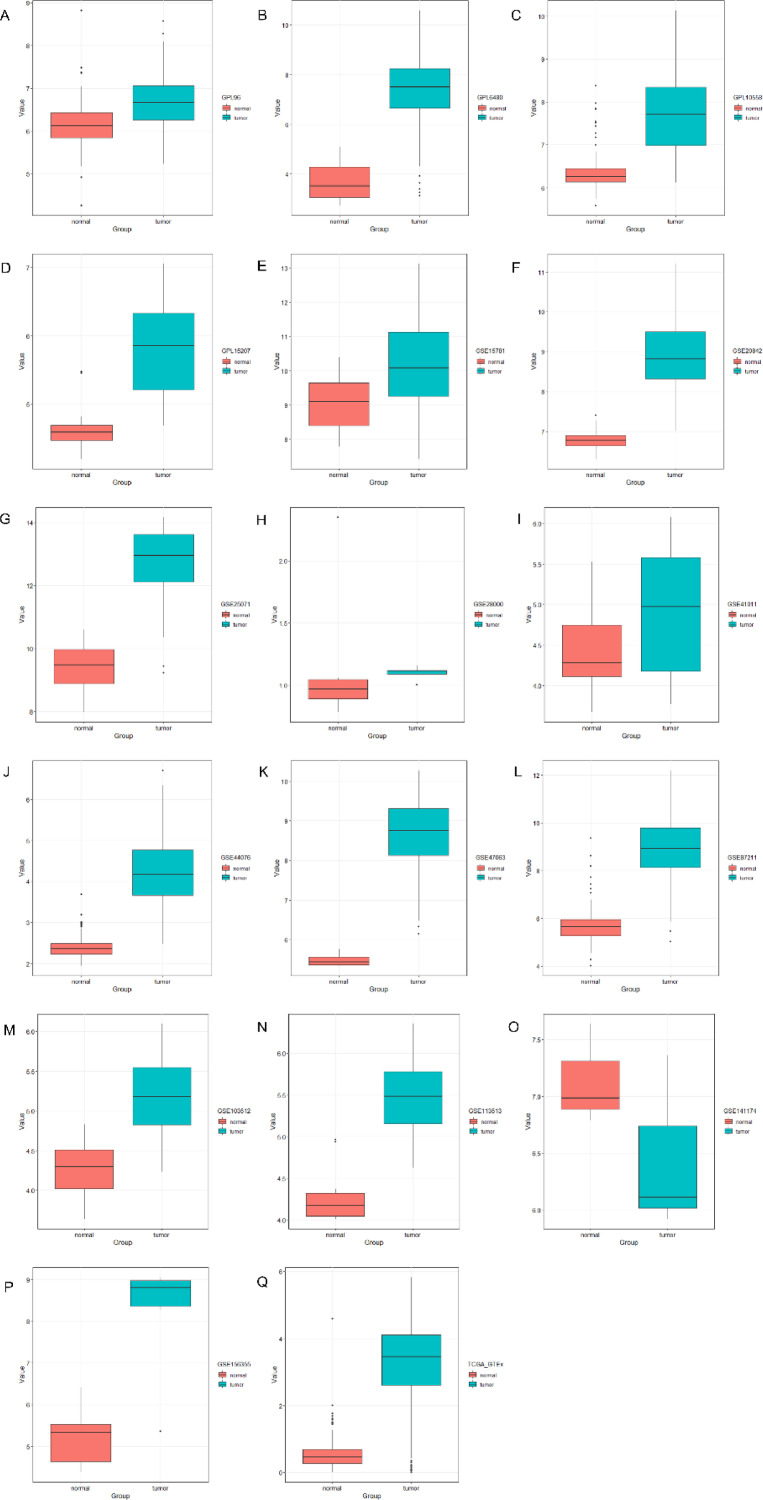
SULT2B1 is not only upregulated in radio-resistant CRC tissues, it is also highly expressed in CRC tissues. We mined data from three sources (the TCGA, GEO, and ArrayExpress databases, including 16 datasets and TCGA_GTEx). The expression value of SULT2B1 in CRC was calculated. The results of the expression analysis of these datasets revealed that SULT2B1 expression levels in CRC tissues were greater, compared with normal colon tissues (P < 0.05, Fig. 5 A–N, P,Q), although one of these datasets had P-values >0.05 (Fig. 5O, GSE141174). The overall pooled SMD of SULT2B1 was 2.00 (P ≤ 0.01; 95%CI: 1.5–2.51; Fig. S2-A), which indicated that SULT2B1 is highly expressed in CRC, and the test for heterogeneity was P < 0.05 (I2=94.4%). Random effect models were used to reduce the impact of heterogeneity. Furthermore, the publication bias was evaluated by a funnel chart: Begg's test was 0.174, and Egger's test was 0.471, which revealed that there was no conspicuous publication bias among these datasets. In general, we verified the overexpression of SULT2B1 mRNA in CRC tissue.
Fig. 5.
The expression of SULT2B1 mRNA in CRC tissues and normal colon tissues.
An ROC curve analysis of SULT2B1 in CRC was also performed, and the results are shown in Fig. 6A–Q. The AUC of the sROC was 0.96 (95%CI: 0.94–0.98), and the combined sensitivity and specificity were 0.89 (95%CI: 0.83–0.93) and 0.93 (95%CI: 0.87–0.96, Fig. S2-I), respectively. These results indicated that SULT2B1 may be a good biomarker for discriminating CRC tissues from normal colon tissues. The integrated computation also provided more specific knowledge of SULT2B1 expression and its diagnostic value.
Fig. 6.
The ROC curves of SULT2B1 expression in CRC tissues and normal colon tissues.
We also made use of the THPA database, to explore the expression of SULT2B1 at the protein level. As shown in Fig. 7, in accordance with the expression of SULT2B1 at the mRNA level data, SULT2B1 protein expression was higher in CRC tissue than in normal tissue.
Fig. 7.
The SULT2B1 expression in normal tissues and colorectal cancer tissues from the human Protein Atlas database [antibody HPA041724]. (A) Male, age 66. Patient id 4202. Small intestine(T-65000) normal tissues, NOS(M-00100), moderate expression for SULT2B1. The relative results of glandular cell are as follows: staining: medium; intensity: moderate; quantity: 75%–25%; location: cytoplasmic/membranous. (B) Male, age 65. Patient id 4910. Colon(T-67000) normal tissues, NOS(M-0010), deficient expression for SULT2B1. The relative results of glandular cell are as follows: staining: not detected; intensity: negative; quantity: none; location: none. (C) Female, age 68. Patient id 4207. Rectum(T-68000) normal tissues, NOS(M-00100), deficient expression for SULT2B1. The relative results of glandular cell are as follows: staining: not detected; intensity: negative; quantity: none; location: none. (D) Male, age 84. Patient id 291. Rectum(T-68000) adenocarcinoma, NOS(M-81403), high expression for SULT2B1. The relative results of tumor cell are as follows: staining: high; intensity: strong; quantity: 75%–25%; location: cytoplasmic/membranous.
Prognostic evaluation of SULT2B1 in CRC
The survival analysis results revealed that the DFS of patients with higher levels of SULT2B1 may survived shorter than those with lower expression, although the difference did not reach statistical significance (log rank P > 0.05; Fig. 8A). The OS of patients with high levels of SULT2B1 was a shorter time. The HR of overexpression SULT2B1 in DFS and OS were 1.3 (P > 0.05) and 1.6 (P > 0.05), but these differences did not reach statistical significance, despite indicating that SULT2B1 may be associated in some way with a poor prognosis.
Fig. 8.
Survival analysis in patients with CRC in terms of DFS and OS.
Effect of wogonin in the proliferation of the HCT116R cell line
The SB network pharmacology analysis showed that wogonin may be the core component of SB for reversing the radiotherapy resistance of CRC. To verify our assumption, we conducted several cell experiments. The cytotoxicity study of wogonin on the HCT116R cell line showed that wogonin inhibited the proliferation of HCT116R in a dose-dependent manner within a certain range, and the IC50 values of wogonin on the proliferation of HCT116R cell lines for 48h was 22.71 µmol/L (Fig. 9A). The effects of wogonin combined with radiotherapy on the cell viability of CRC radio-resistant cells (HCT116R) was then investigated. As shown in Fig. 9B, compared with the control group, the cell viability of each experimental group decreased (P < 0.05). Compared with the radiotherapy group (2 or 4Gy), the cell viability of radiotherapy combined with wogonin or 5-Fu decreased, and the difference was statistically significant (P < 0.05). The cell viability in the positive control (5-Fu) combined with radiotherapy group was lower than that in the wogonin combined with radiotherapy group (P < 0.01). The cell viability of the control, wogonin, 2Gy, 4Gy, 2Gy+wogonin, 4Gy+wogonin, 2Gy+5-Fu, and 4Gy+5-Fu groups were as follows: 100 ± 0.00%, 69.7 ± 1.91%, 74.4 ± 3.31%, 73.8 ± 0.95%, 61.23 ± 2.19%, 54.77 ± 0.15%, 38.6 ± 0.85%, and 38.6 ± 0.17%. In addition, the results of the clone formation experiment showed that, compared with the radiotherapy group, the cell clone formation rate in the wogonin combined with radiotherapy group was significantly decreased (P < 0.05, Fig. 9D). The above results reveal, to a certain extent, that wogonin can increase the radiosensitivity of radiation-resistant HCT116R cells.
Fig. 9.
Effect of wogonin in the proliferation of the HCT116R cell line. (A) The IC50 values of wogonin on the proliferation of HCT116R cell lines after 48h after drug treatment. (B) Effects of wogonin combined with radiotherapy on the cell viability of CRC radio-resistant cells (HCT116R). (C) Comparison of clone formation ability between no treatment with wogonin and treatment with wogonin. (D) Effects of wogonin combined with radiotherapy on the cloning efficiency of CRC radio-resistant cells (HCT116R). *P < 0.05, **P < 0.01 and ***P < 0.001 were contrasted with the control group; aP < 0.05 and aaP < 0.01 were contrasted with the 2Gy group; ##P < 0.01 was contrasted with the 4Gy group.
Effect of wogonin in apoptosis of the HCT116R cell line
Apoptosis was analyzed by flow cytometry to further investigate the apoptotic potential of wogonin (Fig. 10). Histogram data were presented as the mean ± standard deviation (Fig. 10A). Compared with the control group, the apoptosis rate of each experimental group increased to varying degrees (P < 0.05). Compared with the radiotherapy group, the apoptosis rate of the wogonin combined with radiotherapy group increased, and the difference was statistically significant (P < 0.05). There was no significant difference in the apoptosis rate between the wogonin combined with radiotherapy group and the 5-Fu combined with radiotherapy group. The apoptosis rates of the control, wogonin, 2Gy, 4Gy, 2Gy+wogonin, 4Gy+wogonin, 2Gy+5-Fu, and 4Gy+5-Fu groups were as follows: 8.56 ± 1.03%, 13.689 ± 0.76%, 15.19 ± 2.26%, 22.83 ± 1.59%, 19.96 ± 1.59%, 31.50 ± 3.58%,22.92 ± 3.00%, and 28.81 ± 3.52%.
Fig. 10.
Effect of wogonin in apoptosis of HCT116R cell line. (A) Effects of wogonin combined with radiotherapy on the apoptosis rate of CRC radio-resistant cells (HCT116R). (B) apoptosis of cells in each group after administration of wogonin. *P < 0.05, **P < 0.01 and ***P < 0.001 were contrasted with the control group; aP < 0.05 and aaP < 0.01 were contrasted with the 2Gy group; ##P < 0.01 and ###P < 0.001 were contrasted with the 4Gy group.
Molecular docking verification of wogonin and SULT2B1 protein
The above analysis showed that SULT2B1 may be a key gene in radio-resistant CRC, and a potential target of SB for radio-resistant CRC. Wogonin can effectively increase the radiosensitivity of CRC-resistant cells (HCT116R) by inhibiting their proliferation and increasing apoptosis. To verify whether wogonin can reverse the radio-resistance of CRC by targeting SULT2B1 and further understand their binding model, molecular docking using the unique structure of SULT2B1 (PDB, code: 1q22, ligands ID:AND) was made. As shown in Fig. 11 and Table 3, wogonin could easily enter and bind the active pocket of the SULT2B1, and it can form stable hydrogen bonds with THR73, LYS70, PHE272, LEU273, and ARG274 of the SULT2B1 protein. Generally speaking, the total score was greater than or equal to 5, and the affinity between small molecules and receptors could be considered reliable. Our results showed that SULT2B1 has a good binding force with wogonin (the total score for wogonin was 5.7439).
Fig. 11.
Molecular docking between the wogonin and protein 1q22 (encoded by SULT2B1).
Table 3.
Results of the molecular docking calculations.
| Target | PDB ID | Compound | Total Score | Crash | Polar |
|---|---|---|---|---|---|
| SULT2B1 | 1q22 | wogonin | 5.7439 | -2.3906 | 1.2041 |
Discussion
Radiotherapy plays a pivotal role in the treatment of a variety of cancers; however, the radio-resistance of tumors has become a key factor restricting the treatment effects and prognoses of cancer patients [37,38]. At present, TCM plays an important role in the adjuvant treatment of tumor radiotherapy and chemotherapy, and the synergistic and detoxification effects of TCM have been acknowledgd by a growing number of people. SB is a common TCM and is widely used in the clinical setting. In the treatment of CRC, it is mainly combined with other traditional Chinese medicines, such as Huangqin decoction [39], Gegenqinlian decoction [40,41], peony soup [42], and so on. Many studies have shown that these SB-related Chinese medicine compounds also have a good effect in the clinical application of CRC [43], [44], [45]. However, the effect of SB on reversing CRC radio-resistance and its possible mechanisms have not been confirmed. In the present study, we collected and screened SB targets (586) through multiple Chinese medicine databases. At the same time, based on evidence-based concepts, we collected and screened CRC radiotherapy chip data from the SRA, GEO, and ArrayExpress databases. Combined with in-house high-throughput sequencing data, we obtained 155 specific genes of radio-resistance in CRC through pathological integration calculations and then took the common drug disease genes (11) to establish the SB ingredient disease-related-targets network. The network revealed that wogonin may be the core component of SB for exerting anti-radiotherapy resistance in CRC. Then, we performed GO and KEGG enrichment analyses on these 11 intersection genes to reveal how SB antagonizes the potential mechanism of radio-resistance in CRC. Based on the enrichment analysis results, we can suggest that SULT2B1 may be a key gene in the potential pathway, and a series of analyses on differential expressions verification proved that SULT2B1 is highly expressed in CRC tissues and upregulated in radio-resistant CRC tissues or cell lines. The prognostic correlation analysis demonstrated that SULT2B1 may be associated with a poor prognosis, as all of our analyses proved the key role of SULT2B1 in the radio-resistance of CRC. Furthermore, through experiments. we confirmed the sensitizing effect of wogonin on CRC radio-resistance. Finally, we indicated that SULT2B1 may be a potential target for wogonin by reversing the radio-resistance of CRC via a molecular docking method.
SULT2B1, a member of sulfotransferase family, is mainly enriched in the steroid hormone biosynthesis pathway and plays a key role in the biosynthesis of steroids. At present, the abnormal expression of SULT2B1 has been reported in a variety of malignant tumors, and it is mostly related to tumor proliferation, invasion, migration, and poor prognoses; for example, Yang et al. [46] indicated that the expression of SULT2B1 in human liver cancer tumor tissues is relatively higher than that in its neighboring tissues, and it promotes the proliferation of hepatocellular carcinoma cells, both in vivo and in vitro. Hong et al. [47] showed that SULT2B1 affects gastric epithelial function and carcinogenesis induced by a carcinogenic agent. Hu et al. [48] discovered that SULT2B1 is associated with the metastatic invasion and poor prognosis of non-small cell lung cancer. In CRC, reports have also shown that SULT2B1 is related to the migration and invasion of CRC, and SULT2B1 may be an independent prognostic biomarker and potential therapeutic target [35]. Research has proven that single-nucleotide polymorphisms in estrogen metabolism pathway genes affect the susceptibility and survival rate of CRC, and SULT2B1, one of the genes in this pathway, has important significance for the prevalence and prognosis of CRC [36]. Our results further showed that SULT2B1 is not only related to CRC migration, invasion, and poor prognosis, but as a specific and vital gene in radio-resistant CRC, it plays a critical role in the formation of radio-resistance and may be a potential therapeutic target.
The results of our experimental verification and the molecular docking demonstrated that wogonin could increase the radiosensitivity of CRC-resistant HCT116R cells by inhibiting proliferation and promoting apoptosis, and it has a good affinity with SULT2B1. Studies have also shown that wogonin can reduce cholesterol levels in different ways. For instance, Chen et al. [49] investigated the effect of wogonin on the formation of macrophage foam cells from mouse J774A1 macrophages and found that wogonin attenuated oxidized low-density lipoprotein (oxLDL)-induced cholesterol accumulation in macrophages while increasing cholesterol efflux by increasing protein phosphatase 2B-dependent dephosphorylation at ATP-binding cassette transporter-A1 in macrophages. Bak et al. [50] found that the cholesterol levels in experimental group mice were significantly lower than those of control group mice after the oral administration of wogonin. While a large number of studies have found that cholesterol may be positively associated with the risk of CRC [51], [52], [53], interestingly, SULT2B1, as an important gene in the pathway of steroid hormone biosynthesis, is responsible for the sulfation of cholesterol and catalyzes sulfation of the 3-beta-hydroxyl groups of steroids, and it plays a role in epidermal cholesterol metabolism. Therefore, it can be seen that wogonin may inhibit the metabolism of cholesterol in the steroid hormone biosynthesis pathway by targeting SULT2B1 to antagonize the radio-resistance in CRC. However, these conjectures need to be verified by further experiments.
Despite our findings, some limitations of this study must be acknowledgd. We recognize that our paper is a mainly network pharmacology and computer pathology analysis, and our hypotheses from the present analysis have not been fully confirmed. First, to further verify the binding of wogonin and SULT2B1, related experiments should be conducted. Second, to further understand the role of the enzyme SULT2B1 in the radiotolerance of CRC, there may be some ways to assess the activity of SULT2B1 in radioresistant and radiosensitive tissue.
Conclusion
In summary, we revealed the potential mechanism of SB for antagonizing radio-resistance in CRC using network pharmacology and molecular docking. Combined with the analysis of computational pathology and cell experiments, we determined that SULT2B1 may be a key target for reversing radio-resistance and increasing the radiosensitivity of CRC, and SULT2B1 may be a potential drug target for treating radio-resistant CRC. Wogonin of SB may be the core compound of SB for reversing radio-resistance in CRC.
Data availability
All the data during the current study are included in the article or uploaded as supplementary information.
Funding
This research was supported by Guangxi Key Laboratory for Bioactive Molecules Research Evaluation and Wuhan Health Research Fund Number WZ21Q23.
CRediT authorship contribution statement
Jinmei Huang: Writing – original draft, Writing – review & editing, Methodology. Ming Zhou: Writing – original draft, Writing – review & editing, Methodology, Funding acquisition. Huan Zhang: Writing – original draft, Writing – review & editing. Yeying Fang: Data curation, Visualization. Gang Chen: Conceptualization, Validation, Supervision. Jiaying Wen: Visualization, Data curation. LiMin Liu: Conceptualization, Funding acquisition, Validation, Supervision.
Declaration of Competing Interest
The authors declare that they have no conflicts of interest.
Acknowledgment
Not applicable.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2022.101488.
Contributor Information
Jinmei Huang, Email: 2813440309@qq.com.
Ming Zhou, Email: 42974497@qq.com.
Huan Zhang, Email: zhanghuan0226@126.com.
Yeying Fang, Email: fangyeying2010@163.com.
Gang Chen, Email: chengang@gxmu.edu.cn.
Jiaying Wen, Email: wiskvot@163.com.
LiMin Liu, Email: liulimin126@163.com.
Appendix. Supplementary materials
References
- 1.Siegel R.L., Miller K.D., Fuchs H.E., et al. Cancer Statistics, 2021. CA Cancer J. Clin. 2021;71(1):7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 2.Bray F., Ferlay J., Soerjomataram I., et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Kim B.M., Hong Y., Lee S., et al. Therapeutic implications for overcoming radiation resistance in cancer therapy. Int. J. Mol. Sci. 2015;16(11):26880–26913. doi: 10.3390/ijms161125991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tam S.Y., Wu V.W.C. A review on the special radiotherapy techniques of colorectal cancer. Front. Oncol. 2019;9:208. doi: 10.3389/fonc.2019.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee Y.J., Cho J.M., Sai S., et al. 5-Fluorouracil as a tumor-treating field-sensitizer in colon cancer therapy. Cancers. 2019;11(12) doi: 10.3390/cancers11121999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liao Y.H., Li C.I., Lin C.C., et al. Traditional Chinese medicine as adjunctive therapy improves the long-term survival of lung cancer patients. J. Cancer Res. Clin. Oncol. 2017;143(12):2425–2435. doi: 10.1007/s00432-017-2491-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiang Y., Guo Z., Zhu P., et al. Traditional Chinese medicine as a cancer treatment: modern perspectives of ancient but advanced science. Cancer Med. 2019;8(5):1958–1975. doi: 10.1002/cam4.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pang P., Zheng K., Wu S., et al. Baicalin downregulates RLRs Signaling pathway to control influenza a virus infection and improve the prognosis. Evid. Based Complement. Alternat. Med. 2018;2018 doi: 10.1155/2018/4923062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qiu J., Niu X., Dong J., et al. Baicalin protects mice from Staphylococcus aureus pneumonia via inhibition of the cytolytic activity of α-hemolysin. J. Infect. Dis. 2012;206(2):292–301. doi: 10.1093/infdis/jis336. [DOI] [PubMed] [Google Scholar]
- 10.Shi L., Hao Z., Zhang S., et al. Baicalein and baicalin alleviate acetaminophen-induced liver injury by activating Nrf2 antioxidative pathway: the involvement of ERK1/2 and PKC. Biochem. Pharmacol. 2018;150:9–23. doi: 10.1016/j.bcp.2018.01.026. [DOI] [PubMed] [Google Scholar]
- 11.Wu Z., Fan Q., Miao Y., et al. Baicalin inhibits inflammation caused by coinfection of Mycoplasma gallisepticum and Escherichia coli involving IL-17 signaling pathway. Poult. Sci. 2020;99(11):5472–5480. doi: 10.1016/j.bcp.2018.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh S., Meena A., Luqman S. Baicalin mediated regulation of key signaling pathways in cancer. Pharmacol. Res. 2020 doi: 10.1016/j.phrs.2020.105387. [DOI] [PubMed] [Google Scholar]
- 13.Duan X., Guo G., Pei X., et al. Baicalin inhibits cell viability, migration and invasion in breast cancer by regulating miR-338-3p and MORC4. OncoTargets Ther. 2019;12:11183–11193. doi: 10.2147/ott.s217101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu Y., Fang J., Wang H., et al. Baicalin suppresses proliferation, migration, and invasion in human glioblastoma cells via Ca2+-dependent pathway. Drug Des. Dev. Ther. 2018;12:3247–3261. doi: 10.2147/dddt.s176403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tao Y., Zhan S., Wang Y., et al. Baicalin, the major component of traditional Chinese medicine Scutellaria baicalensis induces colon cancer cell apoptosis through inhibition of oncomiRNAs. Sci. Rep. 2018;8(1):14477. doi: 10.1038/s41598-018-32734-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang B., Bai H., Sa Y., et al. Inhibiting EMT, stemness and cell cycle involved in baicalin-induced growth inhibition and apoptosis in colorectal cancer cells. J. Cancer. 2020;11(8):2303–2317. doi: 10.7150/jca.37242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng Q., Wang H., Pang J., et al. Prevention of wogonin on colorectal cancer tumorigenesis by regulating p53 nuclear translocation. Front. Pharmacol. 2018;9:1356. doi: 10.3389/fphar.2018.01356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang C., Yang Y., Sun L., et al. Baicalin reverses radioresistance in nasopharyngeal carcinoma by downregulating autophagy. Cancer Cell Int. 2020;20:35. doi: 10.1186/s12935-020-1107-4axccs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yae K.S., Yong M.J., Tatsuya U., et al. Baicalein Suppresses stem cell-like characteristics in radio- and chemoresistant MDA-MB-231 human breast cancer cells through up-regulation of IFIT2. Nutrients. 2019;11(3) doi: 10.3390/nu11030624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng C.S., Chen J., Tan H.Y., et al. Scutellaria baicalensis and cancer treatment: recent progress and perspectives in biomedical and clinical studies. Am. J. Chin. Med. 2018;46(1):25–54. doi: 10.1142/s0192415x18500027. [DOI] [PubMed] [Google Scholar]
- 21.Park S.Y., Lee S.J., Cho H.J., et al. Epsilon-globin hbe1 enhances radiotherapy resistance by down-regulating BCL11A in colorectal cancer cells. Cancers. 2019;11(4) doi: 10.3390/cancers11040498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benson A.B., Venook A.P., Al-Hawary M.M., et al. Rectal cancer, version 2.2018, NCCN clinical practice guidelines in oncology. J. Natl. Compr. Cancer Netw. 2018;16(7):874–901. doi: 10.6004/jnccn.2018.0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uribe-Lewis S., Stark R., Carroll T., et al. 5-hydroxymethylcytosine marks promoters in colon that resist DNA hypermethylation in cancer. Genome Biol. 2015;16:69. doi: 10.1186/s13059-015-0605-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Snipstad K., Fenton C.G., Kjaeve J., et al. New specific molecular targets for radio-chemotherapy of rectal cancer. Mol. Oncol. 2010;4(1):52–64. doi: 10.1016/j.molonc.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olsson L., Hammarstrom M.L., Israelsson A., et al. Allocating colorectal cancer patients to different risk categories by using a five-biomarker mRNA combination in lymph node analysis. PLoS One. 2020;15(2) doi: 10.1371/journal.pone.0229007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moreno V., Alonso M.H., Closa A., et al. Colon-specific eQTL analysis to inform on functional SNPs. Br. J. Cancer. 2018;119(8):971–977. doi: 10.1038/s41416-018-0018-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jovov B., Araujo-Perez F., Sigel C.S., et al. Differential gene expression between African American and European American colorectal cancer patients. PLoS One. 2012;7(1):e30168. doi: 10.1371/journal.pone.0030168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu Y., Gaedcke J., Emons G., et al. Colorectal cancer susceptibility loci as predictive markers of rectal cancer prognosis after surgery. Genes Chromosom. Cancer. 2018;57(3):140–149. doi: 10.1002/gcc.22512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gaedcke J., Grade M., Jung K., et al. Mutated KRAS results in overexpression of DUSP4, a MAP-kinase phosphatase, and SMYD3, a histone methyltransferase, in rectal carcinomas. Genes Chromosom. Cancer. 2010;49(11):1024–1034. doi: 10.1002/gcc.20811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Danielsen S.A., Cekaite L., Agesen T.H., et al. Phospholipase C isozymes are deregulated in colorectal cancer–insights gained from gene set enrichment analysis of the transcriptome. PLoS One. 2011;6(9):e24419. doi: 10.1371/journal.pone.0024419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brouwer-Visser J., Cheng W.Y., Bauer-Mehren A., et al. Regulatory T-cell genes drive altered immune microenvironment in adult solid cancers and allow for immune contextual patient subtyping. Cancer Epidemiol. Biomark. Prev. 2018;27(1):103–112. doi: 10.1158/1055-9965.EPI-17-0461. [DOI] [PubMed] [Google Scholar]
- 32.Higgins J.P., Thompson S.G., Deeks J.J., et al. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang Z., Kang B., Li C., et al. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucl. Acids Res. 2019;47(W1):W556–W560. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feng S., Wang T., Fan L., et al. Exploring the potential therapeutic effect of Eucommia ulmoides-Dipsaci Radix herbal pair on osteoporosis based on network pharmacology and molecular docking technology. RSC Adv. 2022;12(4):2181–2195. doi: 10.1039/d1ra05799e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu L., Yang G.Z., Zhang Y., et al. Overexpression of SULT2B1b is an independent prognostic indicator and promotes cell growth and invasion in colorectal carcinoma. Lab. Invest. 2015;95(9):1005–1018. doi: 10.1038/labinvest.2015.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li S., Xie L., Du M., et al. Association study of genetic variants in estrogen metabolic pathway genes and colorectal cancer risk and survival. Arch. Toxicol. 2018;92(6):1991–1999. doi: 10.1007/s00204-018-2195-y. [DOI] [PubMed] [Google Scholar]
- 37.Kim H.S., Kim N.K. Challenges and shifting treatment strategies in the surgical treatment of locally advanced rectal cancer. Ann. Gastroenterol. Surg. 2020;4(4):379–385. doi: 10.1002/ags3.12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park I.J., Yu C.S. Current issues in locally advanced colorectal cancer treated by preoperative chemoradiotherapy. World J. Gastroenterol. 2014;20(8):2023–2029. doi: 10.3748/wjg.v20.i8.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song Z.M., Song H.Q., Song P.P. Clinical study on huangqin decoction combined with chemotherapy for colon cancer. J. Tradit. Chin. Med. 2020;52(17):25–27. [Google Scholar]
- 40.Zhong F.Q., Bi L., Yan H.J., et al. Clinical observation of modified gegen qinlian decoction in treating damp-heat diarrhea after surgery of colorectal cancer. Guid. China Med. 2020;18(02):166–167. [Google Scholar]
- 41.Sharifi-Rad J., Herrera-Bravo J., Salazar L.A., et al. The therapeutic potential of wogonin observed in preclinical studies. Evid. Based Complement. Alternat. Med. 2021;2021 doi: 10.1155/2021/9935451. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Song X.P. Clinical observation of modified shaoyao decoction combined with conventional chemotherapy in the treatment of advanced colorectal cancer. China's Naturop. 2020;28(08):74–76. [Google Scholar]
- 43.Lv J., Jia Y., Li J., et al. Gegen Qinlian decoction enhances the effect of PD-1 blockade in colorectal cancer with microsatellite stability by remodelling the gut microbiota and the tumour microenvironment. Cell Death Dis. 2019;10(6):415. doi: 10.1038/s41419-019-1638-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li M.Y., Li M.X., Xu N., et al. Effects of Huangqin Decoction on ulcerative colitis by targeting estrogen receptor alpha and ameliorating endothelial dysfunction based on system pharmacology. J. Ethnopharmacol. 2021;271 doi: 10.1016/j.jep.2021.113886. [DOI] [PubMed] [Google Scholar]
- 45.Wang X., Saud S.M., Zhang X., et al. Protective effect of Shaoyao Decoction against colorectal cancer via the Keap1-Nrf2-ARE signaling pathway. J. Ethnopharmacol. 2019;241 doi: 10.1016/j.jep.2019.111981. [DOI] [PubMed] [Google Scholar]
- 46.Yang X., Xu Y., Guo F., et al. Hydroxysteroid sulfotransferase SULT2B1b promotes hepatocellular carcinoma cells proliferation in vitro and in vivo. PLoS One. 2013;8(4):e60853. doi: 10.1371/journal.pone.0060853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hong W., Guo F., Yang M., et al. Hydroxysteroid sulfotransferase 2B1 affects gastric epithelial function and carcinogenesis induced by a carcinogenic agent. Lipids Health Dis. 2019;18(1):203. doi: 10.1186/s12944-019-1149-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu R., Huffman K.E., Chu M., et al. Quantitative secretomic analysis identifies extracellular protein factors that modulate the metastatic phenotype of non-small cell lung cancer. J. Proteome Res. 2016;15(2):477–486. doi: 10.1021/acs.jproteome.5b00819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen C.Y., Shyue S.K., Ching L.C., et al. Wogonin promotes cholesterol efflux by increasing protein phosphatase 2B-dependent dephosphorylation at ATP-binding cassette transporter-A1 in macrophages. J. Nutr. Biochem. 2011;22(11):1015–1021. doi: 10.1016/j.jnutbio.2010.08.014. [DOI] [PubMed] [Google Scholar]
- 50.Bak E.J., Kim J., Choi Y.H., et al. Wogonin ameliorates hyperglycemia and dyslipidemia via PPARα activation in db/db mice. Clin. Nutr. 2014;33(1):156–163. doi: 10.1016/j.clnu.2013.03.013. [DOI] [PubMed] [Google Scholar]
- 51.Azeem S., Gillani S.W., Siddiqui A., et al. Diet and Colorectal cancer risk in Asia-a systematic review. Asian Pac. J. Cancer Prev. 2015;16(13):5389–5396. doi: 10.7314/apjcp.2015.16.13.5389. [DOI] [PubMed] [Google Scholar]
- 52.van Duijnhoven F.J., Bueno-De-Mesquita H.B., Calligaro M., et al. Blood lipid and lipoprotein concentrations and colorectal cancer risk in the European prospective investigation into cancer and nutrition. Gut. 2011;60(8):1094–1102. doi: 10.7314/apjcp.2015.16.13.5389. [DOI] [PubMed] [Google Scholar]
- 53.Jacobs R.J., Voorneveld P.W., Kodach L.L., et al. Cholesterol metabolism and colorectal cancers. Curr. Opin. Pharmacol. 2012;12(6):690–695. doi: 10.1016/j.coph.2012.07.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data during the current study are included in the article or uploaded as supplementary information.