Abstract
Background
Lens development is orchestrated by transcription factors. Disease-causing variants in transcription factors and their developmental target genes are associated with congenital cataracts and other eye anomalies.
Methods
Using whole exome sequencing, we identified disease-causing variants in two large British families and one isolated case with autosomal dominant congenital cataract. Bioinformatics analysis confirmed these disease-causing mutations as rare or novel variants, with a moderate to damaging pathogenicity score, with testing for segregation within the families using direct Sanger sequencing.
Results
Family A had a missense variant (c.184 G>A; p.V62M) in PAX6 and affected individuals presented with nuclear cataract. Family B had a frameshift variant (c.470–477dup; p.A160R*) in PITX3 that was also associated with nuclear cataract. A recurrent missense variant in HSF4 (c.341 T>C; p.L114P) was associated with congenital cataract in a single isolated case.
Conclusions
We have therefore identified novel variants in PAX6 and PITX3 that cause autosomal dominant congenital cataract.
Subject terms: Genomics, Genetics
Introduction
Cataract the opacification of the eye lens is the most common, but treatable cause of blindness in the world (https://www.who.int/publications-detail/world-report-on-vision). Congenital cataracts are detected at birth or during the first decade of life. Hereditary cataract can be isolated or be a part of other ocular defects like anterior segment mesenchymal dysgenesis, glaucoma, microcornea, or aniridia; and systemic disorders such as heart disease, diabetes, deafness and Wolframin disease [1, 2]. Congenital cataract is usually autosomal dominant, followed by autosomal recessive and X linked inheritances. Congenital cataract are clinically and genetically heterogeneous, displaying various phenotypes [3].
So far nearly 50 genes have been found causing mostly isolated congenital cataracts broadly including genes encoding lens soluble proteins –crystallins; membrane proteins- gap junctions, aquaporins, receptor tyrosine kinase gene EPH receptor A2, an endoplasmic reticulum membrane-embedded protein, Wolframin, chromatin modifying protein-4B, lens integral membrane protein; AQP0, Connexin 50 and LIM2 cytoskeletal proteins- filensin, phakinin, vimentin and genes encoding transcription or developmental factors EYA1, MAF, FOXE3, VSX2, PAX6, PITX3 and HSF4 (https://cat-map.wustl.edu/) [4–6].
Transcription factors and developmental genes play a key role spatio-temporally in the embryonic development of ocular and other embryonic tissues [7, 8]. Disease-causing variants in these genes can be devastating for the developing eye, even causing anophthalmia [9, 10]. They display a spectrum of eye anomalies in the anterior segment of eye, but nuclear cataract phenotypes are consistent with early developmental effects as would be anticipated for PAX6 and PITX3 transcription factors. Recently, we have found two novel mutations in the transcription factors PAX6, PITX3 and one recurrent variant in HSF4.
PAX6, a paired-box and homeobox domain gene is one of the principal regulators in eye development, first described as a candidate for human aniridia [11]. PAX6 plays an important role in the early development of the lens including the interaction between the embryonic surface ectoderm and the budding optic vesicle. This interaction is critical for normal lens induction [12–14]. PAX6 is also expressed in the central nervous system, olfactory system and pancreas [15–17]. PAX6, a transcriptional regulator gene on chromosome 11p13, consists of 14 exons spanning 22 kb genomic region, encodes 422 amino acid residues. PAX6 consists of two highly conserved DNA-binding domains: at N-terminus a paired domain (PD) with N-terminal (NTS) and C-terminal (CTS) subdomains and the middle homeodomain (HD) connected by 79-amino-acid linker region. The C-terminal end of protein is a transregulatory region enriched in proline, serine and threonine called the PST domain [18, 19]. The human PAX6 gene produces two alternative splice isoforms due to the insertion of 14 additional amino acids encoded by exon 5a into the NTS subdomain of PD, which abolishes the DNA-binding activity of the NTS and unmasks the DNA-binding ability of the CTS subdomain [20]. Pathogenic variants in PAX6 cause severe lenticular and non-lenticular defects.
PITX3 a paired-like homeodomain transcription factor gene is a member of the REIG/PITX family of homeobox transcription factors, including PITX1 and PITX2 [21]. PITX2 and PITX3 participate in eye development and express in most developing ocular tissues, including the retina, lens and cornea [22]. Mutations in PITX2 have been linked to Rieger syndrome causing glaucoma and mild craniofacial dysmorphism in humans [23]. In the aphakia mouse mutant, two deletions in the promoter of the homeobox transcription factor Pitx3 lead to loss of its function and to arrest of eye development at the lens stalk stage [24].
HSF4 belongs to the heat-shock transcription factors family that regulate the expression of heat-shock proteins in response to various stresses, such as high temperature, infection, and free radicals, and also in development. In the lens HSF4 is expressed in both cell types (epithelial cells and fibre cells) at an early stage of development and is required for normal cell growth and differentiation of these two cell types [25] and for transcription regulation of αB-crystallin in the lens [26]. Disease causing variants in HSF4 cause both dominant and recessive cataracts.
Herein we report two novel and one known variant(s) in PAX6, PITX3 and HSF4 genes.
Methods
Phenotyping
Patients were identified via the proband attending the Genetic Service at Moorfields Eye Hospital, London, UK. The study protocol adhered to the Tenets of the Declaration of Helsinki and was approved by UCL research ethics committee, (project ID − 4817/001). All the family members participating in this study gave written informed consent and underwent full ophthalmic examination, including slit lamp examination. All affected individuals from two families and one isolated case were diagnosed as having an isolated congenital cataract as described below.
Whole exome sequencing (WES) and bioinformatics analysis
Genomic DNA was extracted from EDTA treated blood samples using the Nucleon II DNA Extraction Kit (Scotlab Bioscience, Strathclyde, Scotland, UK). The DNA samples were sequenced at Macrogen Europe. Exon capture and target enrichment was performed using the SureSelectXT Human All Exon V6 post, (Agilent, Santa Rosa, CA, USA). Paired-end sequencing was performed on an Illumina Hiseq 2500 high-throughput sequencer, generating mean exome coverage of 50x. Raw data in fastq format was aligned to the UCSC Genome Browser GRCh37/hg19 human reference sequence and analysed using the Phenopolis bioinformatics platform as before [27, 28]. We used tiered approach to prioritised rare coding variants using Kaviar (http://db.systemsbiology.net/kaviar/) [29] and Genome Aggregation Database (GnomAD http://gnomad.broadinstitute.org/) or rare variants (GnomAD allele frequency <0.0001) in all the known cataract genes (https://cat-map.wustl.edu/). The variant were then filtered using CADD score, predicted to be moderately or highly damaging (CADD>15) with the highest at the top for both known and unknown genes for cataracts. Further bioinformatic validations were done on the varsome platform (varsome.com).
Sanger sequencing
Sanger sequencing was performed to validate the variant identified by whole exome sequencing. Genomic DNA was amplified by PCR using GoTaq 2X master mix (AB gene; Thermo Scientific, Epsom, UK) and PAX6, PITX3 and HSF4 -specific primers designed with http://bioinfo.ut.ee/primer3-0.4.0/ PCR conditions were as follows: 94 °C for 5 min of initial denaturation followed by 30 cycles of amplification of 30 s at 94 °C denaturing, 30 s at 60 °C annealing, and 45 s at 72 °C for extending. After cleaning, the PCR products were reacted with BigDye Terminator v3.1, they were run on ABI 3730 Genetic Analyzer (both from Applied Biosystems, Foster City, CA, USA) and analysed using SeqMan Pro (version 8.0.2 from DNASTAR) sequence analysis. After validating the variant, segregation was performed in all the available family members.
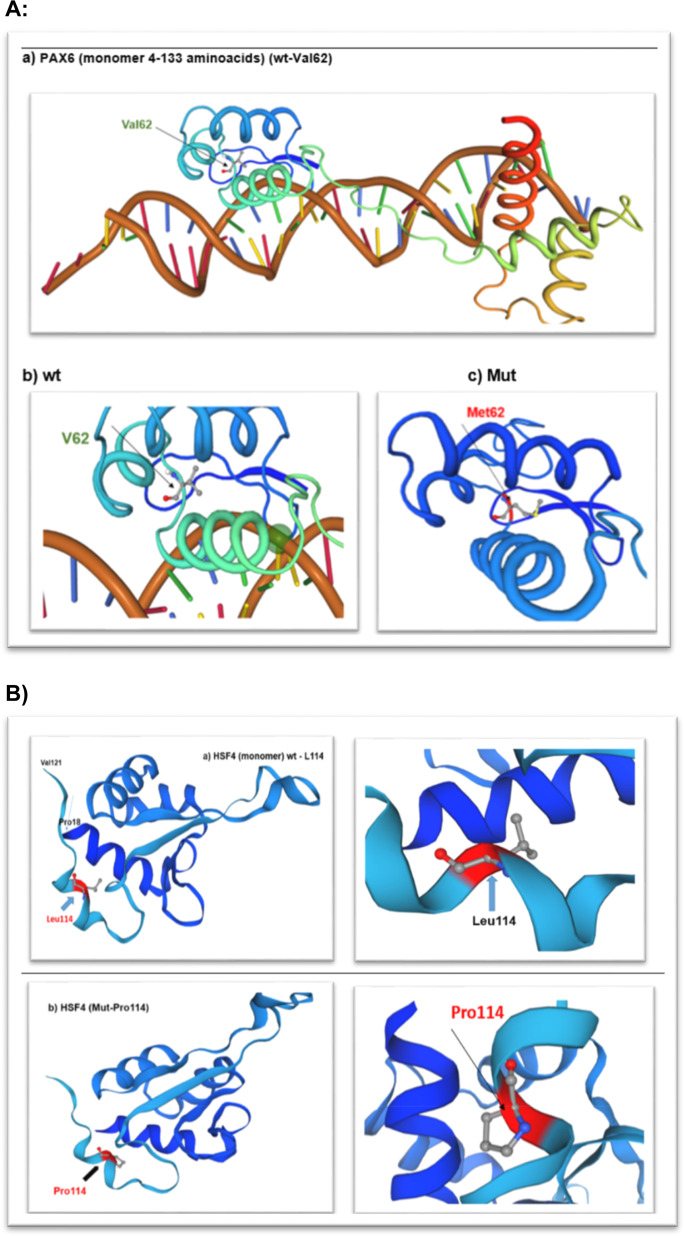
The protein structure of PAX6 and HSF4 were analysed using SWISSMODEL (Fig. 2: A, B).
Fig. 2. Structural view of PAX6 and HSF4 proteins.
A Crystal structure of the human PAX-6 paired domain-dna complex reveals a general model for PAX protein-dna interactions- X-ray diffraction, 2.50 A monomer (4-133 aminoacids). Wt -V62 PAX6: https://swissmodel.expasy.org/repository/uniprot/P26367 Mut - M62-PAX6: https://swissmodel.expasy.org/interactive/1hpd5p/models/. B Structure of the DNA-binding domain of HSF2 with sequence homology to HSF4 at aminoacids position (pro18-Val121). Wt: L114 https://swissmodel.expasy.org/interactive/0QmfH7/models/ Heat shock factor protein 2-Human HSF2 DNA Binding Domain in complex with 3-site HSE DNA at 2.1 Angstroms Resolution; Mut: P114 https://swissmodel.expasy.org/interactive/Fx4QGZ/models/.
Wt-PAX6 (https://swissmodel.expasy.org/repository/uniprot/P26367)
Mut-PAX6 (https://swissmodel.expasy.org/interactive/1hpd5p/models/)
Wt-HSF4 (https://swissmodel.expasy.org/interactive/0QmfH7/models/)
Mut-HSF4 (https://swissmodel.expasy.org/interactive/Fx4QGZ/models/)
Results
Cataract families
In this study, we have investigated two families A and B and an isolated individual C with autosomal dominant congenital cataract.
Family A- novel missense variant (c.184 G>A; p.V62M) in PAX6
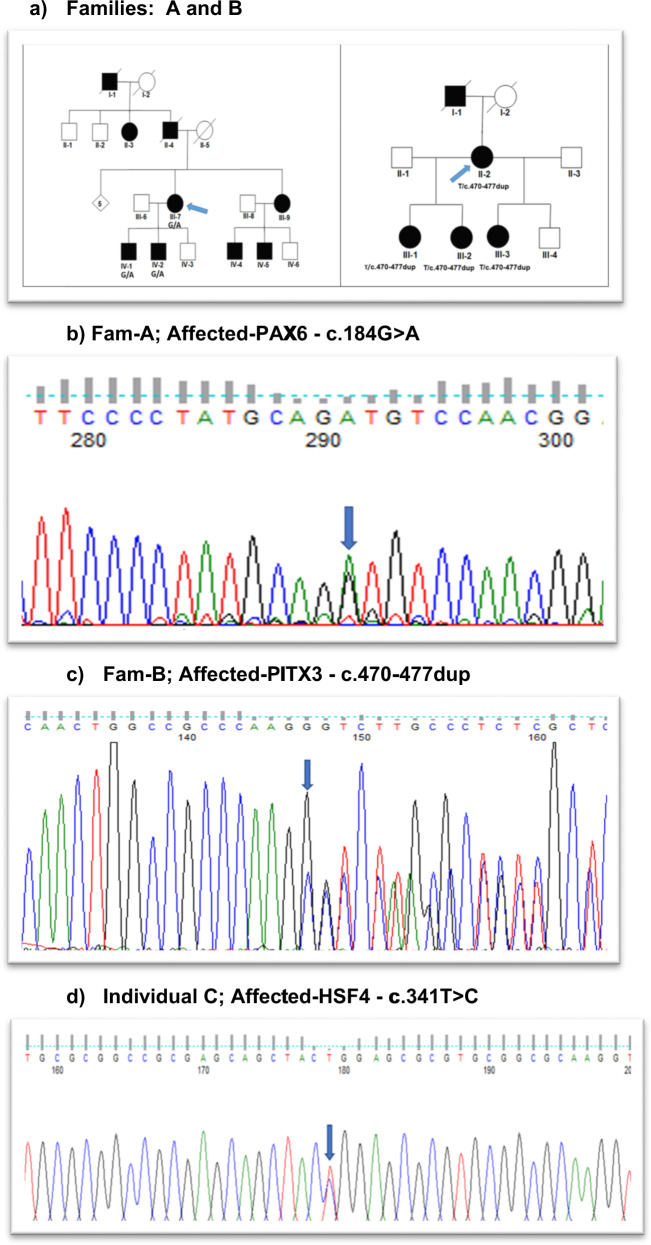
A four-generation pedigree of 22 individuals with four spouses, nine unaffected and nine affected presenting with nuclear cataract (Fig. 1a - Fam A). Individual II-7 had bilateral congenital cataract and nystagmus, Individual IV-1 had bilateral congenital cataract and surgery was performed at age 1. This individual also had congenital nystagmus. One affected individual (IV-1) was sequenced by WES. After the Phenopolis genetic variant analysis pipeline, variants were filtered by allele frequency and from a total of 3204 rare coding variants, 381 variants remained. The top scoring variant for CADD was a rare heterozygous variant NM_001258462.3: c.184 G>A; p.V62M in exon 7 of PAX6 with a score of 27.6. Direct Sanger sequencing confirmed the variant (Fig. 1b), which co-segregated in the affected family members.
Fig. 1. Pedigrees and sequence analysis.
a Family A: Abridged pedigree with nuclear cataract; Family B: Abridged pedigree with nuclear cataract. The diagonal line indicates a deceased family member. Squares and circles symbolise males and females, respectively. Open and filled symbols indicate unaffected and affected individuals, respectively. Diamond symbolises-number of unaffected siblings grouped together. The arrow indicates the family members who participated in the WES analysis. All the available members in the family were sequenced to show the segregation. b Sequence analysis of (a): PAX6–missense variant c.184 G>A in affected member of family A with nuclear cataract; (c) PITX3-a frameshift variant at c.470–477dup in an affected member of family B with nuclear cataract; (d) HSF4- missense variant c.341 T>C in an affected individual with congenital cataract.
Family B- novel frameshift variant (c.470–477dup; p.A160R*) in PITX3 with nuclear cataract
A three-generation family with nuclear cataract comprising seven members, including three affected, two unaffected, and two spouses, were examined (Fig. 1a- Fam B). Affected individuals had bilateral cataract and surgery was performed in infancy. One affected individual (II-3) was sequenced by WES. Variant annotation and tiered filtering yielded 512/3164 variants and further a rare heterozygous variant NM_001040667.3: c.470_477dup (CTTGGGCG-8bp dup); p.Ala160ArgfsTer152 in exon 4 of PITX3, with a highest CADD score of 34 was found. Sanger sequencing confirmed the variant (Fig. 1c), and it co-segregated in the affected individuals.
An individual C, with congenital cataract underwent WES. Following, variant analysis and filtering, one rare variant remained with a top CADD score of 31. This variant happened to be a recurrent variant NM_001040667.3 c.341 T>C; p.L114P in exon 5 of HSF4, validated by Sanger sequencing (Fig. 1d). Pathogenicity scores for all the three variants are shown in Table 1.
Table 1.
Pathogenicity scores for PAX6, PITX3 and HSF4 Variants.
| Gene | Genomic pos./Exon | HGVSc | HGVSp | Phenotype | CADD | GERP NR | MutationTaster/verdict |
|---|---|---|---|---|---|---|---|
| PAX6 | Chr11/Ex7 | c.184 G>A | p.V62M | Nuclear | 27.6 | 5.3 | Disease causing/0.81/Pathogenic/Novel |
| PITX3 | Chr10/Ex4 | c.470–477dup | p.A160R* | Nuclear | 34 | 4.46 | Disease causing/0.81/Pathogenic/Novel |
| HSF4 | Chr16/Ex5 | c.341 T>C | p.L114P | Congenital cataract | 31 | 4.86 | Disease causing/0.81/Likely pathogenic/Recurrent |
Combined annotation dependent depletion (CADD) is score for the deleteriousness of a variant. A CADD score >20 is considered damaging; genomic evolutionary rate profiling (GERP) NR corresponds to the neutral rate conservation score of the site.
* designated to truncated protein.
Swiss modelling
The protein structure of PAX6 and HSF4 were analysed using SWISSMODEL (Fig. 2A, B)
Wt-PAX6 (https://swissmodel.expasy.org/repository/uniprot/P26367)
Mut-PAX6 (https://swissmodel.expasy.org/interactive/1hpd5p/models/)
Wt-HSF4 (https://swissmodel.expasy.org/interactive/0QmfH7/models/)
Mut-HSF4 (https://swissmodel.expasy.org/interactive/Fx4QGZ/models/)
Discussion
Transcription factor genes like PAX6 [30–32] PITX3 [33, 34] and HSF4 [26, 35] are critically important for eye development. Disease-causing variants in these cause severe ocular defects and congenital cataracts that may be part of non-lenticular diseases.
HSF4 expression is tissue specific and it has two splice forms HSF4a & HSF4b. HSF4 gene resides on chromosome 16q22.1, comprised of 15 exons and encodes a heat-shock transcriptional protein of 493 amino-acid residues with a DNA binding domain (DBD), an amino-terminal hydrophobic repeat (HR/A-B), an isoform-specific region and downstream of hydrophobic repeat (DHR) [36, 37]. Mutations in this gene cause both autosomal dominant and recessive cataracts, with higher incidence of dominant suggesting that mutations in the dna binding domain are dominant negative rather than loss of function. Apparently, all autosomal-dominant mutations in HSF4 lie within the α-helical DBD, whereas the recessive variants lie outside this highly conserved functional domain [38]. HSF4 has been shown to regulate lens fibre cell differentiation by modulating the expression of certain lens structural proteins, such as lens specific crystallins, beaded filament proteins, and fibroblast growth factors26 and play important role in the de-nucleation of lens fibre cells [39]. To date 25/27 unique pathogenic variants have been reported mostly in the Asian population (https://cat-map.wustl.edu/). Two heterozygous variants at p.L114P and R119C, both have been reported twice in Chinese [40] and Danish families [41]. The variant p.L114P has previously been described causing autosomal dominant lamellar cataract in a Chinese family and AD lamellar and sutural opacities in a Danish family suggesting further heterogeneity of the same variant in different ethnic groups perhaps due to other interacting proteins. Here we report a recurrent missense variant NM_001040667.3: c.341 T>C; L114P in the DBD region of the HSF4 protein in an isolated case of English origin with autosomal dominant bilateral congenital cataract.
PAX6 homozygous variants typically give rise to aniridia or anophthalmia; but missense variants (nearly 12%) may result in milder phenotypes including isolated foveal hypoplasia, Peter’s anomaly, partial aniridia, optic nerve defect and bilateral cataract [42, 43]. To date, nearly 500 mutations have been reported in Human PAX6 database (http://lsdb.hgu.mrc.ac.uk/home.php?select_db=PAX6) displaying (>80%) of heterozygous null mutation [44]. Most of the missense variants have been found in the PD region, except one affecting only the C-terminal PST domain, associated with lamellar cataracts and later-onset corneal dystrophy [45]. In the lens, PAX6 plays a key role in the regulation of lens-specific crystallins [46, 47] and over-expression of the PAX6 (5a) isoform dramatically alters lens fibre cell shape and organisation in the lenses of transgenic mice [48]. It is a key transcription factor in lens development as it regulates the expression of crystallins and other structural proteins in the lens [49]. A list of PAX6 disease causing variants are shown in Table 2 causing milder ocular phenotypes and congenital cataract [50–55]. We have identified a novel missense pathogenic variant in exon 6 (c.184 G>A; p.V62M) encompassing the paired domain region of PAX6 gene, causing nuclear cataract.
Table 2.
List of PAX6 mutations causing cataract including spectrum of other eye anomalies.
| S.no | Origin | HGVSc | HGVSp | Phenotype | Reference |
|---|---|---|---|---|---|
| 1. | America |
c.388 C>T c.1058 C>G |
p.Arg130*, p.Ser353* compound heterozygous |
Aniridia, nystagmus, foveal hypoplasia, congenital lamellar cataract, late onset corneal dystrophy | Glaser et al. 1994 |
| 2. | France | c.137 T>C | p. Leu46Pro | Bilateral microphthalmia, nystagmus and cataract | Dansault et al. 2007 |
| 3. | France | c.143delG | P. Val48fsX53 | bilateral aniridia associated with congenital cataract, foveal hypolasia, and nystagmus | Dansault et al. 2007 |
| 4. | America | c.112 C>T | p.Arg38Trp | Microcornea, cataract | Solomon et al. 2009 |
| 5. | France | c.718 C>T | p.Arg240*Ter | Aniridia, congenital cataract | Brémond-Gignac et al. 2010 |
| 6. | China | c.113_129del17 | p.Arg38ProfsX12 | Aniridia, congenital cataract | Cai F et al. 2010 |
| 7. | UK | c.227 C>G | p.Pro76Arg | Nystagmus, foveal hypoplasia and presenile cataract | Thomas et al. 2014 |
| 8. | South Africa | c.622 G>A | p.Arg208Thr | Coloboma, nystagmus, variable cataract | Goolam et al. 2018 |
| 9. | UK | c.184 G>A | p.Val62Met | Congenital cataract, congenital nystagmus | Present study 2021 |
* designated to truncated protein.
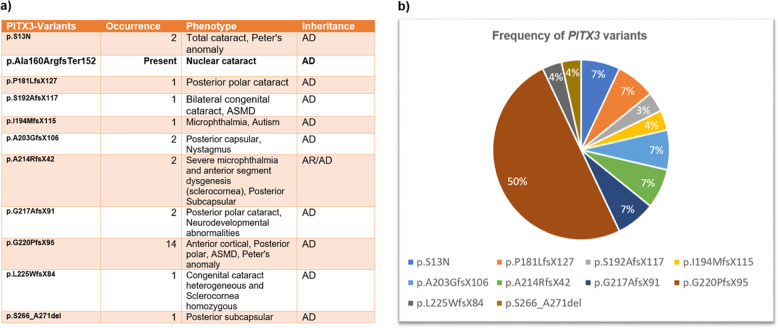
PITX3 gene comprises of four exons and encodes a protein of 303 amino acid residues located at chromosome 10q25. To date, 28 variants have been identified mostly with an autosomal dominant inheritance. Two homozygous variants have been observed in recessive families with severe ocular abnormalities; one at c.650delG; p.G217AfsX91 causing microphthalmia and central nervous system defects [56] and second at p.A214RfsX42 causing severe microphthalmia, anterior segment dysgenesis and sclerocornea [57]. Heterozygous 650delG variant has been reported with only progressive posterior polar cataract in autosomal dominant families [58]. Half of the PITX3 variants, are represented by a single hot spot in exon 4 at c.640_656dup17bp; p.G220PfsX95 in the c-terminal region of the gene that causes mainly AD posterior polar cataracts and anterior segment dysgenesis in several families around the globe [58–64] (Fig. 3). The PITX3 variants, a recurrent 17-bp duplication at c.640_656dup17bp, the p.G220PfsX95 and lastly a 1-bp deletion at c.573delC; p.S192AfsX117 have been shown to reduce PITX3 DNA-binding and transactivation activity due to altered protein length [34, 64]. In a mouse model removal of the C-terminal domain which contains the highly conserved otp, aristaless, and rax (OAR) domain leads to micro-ophthalmia and aphakia in mice [65].
Fig. 3. Frequency and phenotypic presentation of PITX3.
a Spectrum of PITX3 variants showing cataract phenotypes and anterior segment dysgenesis; (b) Frequency pie chart of PITX3 variants to date.
So far, only one PITX3 missense variant p.S13N has been reported twice causing total cataract in one isolated case [21] and Peter’s anomaly in another [62]. Interestingly nearly all the frameshift variants truncate the protein in the COOH region display mostly isolated posterior polar cataract phenotypes and other ocular defects (Fig. 3). Here we report a novel frameshift variant at c.470_477dup (CTTGGGCG-8bp dup); p. Ala160ArgfsTer152 in exon 4 of PITX3 results in a premature termination of the protein at amino acid residue A160, responsible for an isolated nuclear cataract in an English Pedigree. This is the first report of nuclear cataract caused by mutation in PITX3. PITX3 expression is seen in the developing lens, skeletal muscle, and dopaminergic neurons of the substantia nigra in the brain and the PITX3 polymorphisms have been shown to be associated with Parkinson disease [66] dementia [67] and with neurological abnormalities [68, 69].
Conclusions
We report two novel and one recurrent cataract-causing variant in three transcription factor genes important in lens development. Both the identified novel variants that cause autosomal dominant cataract provide further evidence of phenotypic heterogeneity. These variants and their discovery provide further evidence of the importance of combining clinical characterisation with NGS in order to understand the biological basis for the phenotypic variation often associated with familial cataract. The clinical and genetic heterogeneity now reported in congenital cataract has begun to rival the vast variability documented in inherited and isolated eye disease, making ophthalmic genetics the most heterogeneous in clinical medicine. Our study extends the mutation spectrum associated with the transcriptional factors essential to lens development to the benefit of patients through the improved genetic counselling this knowledge delivers.
Summary
What was known before
Congenital cataract is clinically and genotypically heterogeneous disease. Mutations in transcription factors are known to cause congenital cataracts and other eye anomalies.
What this study adds
We have identified novel heterozygous mutations in PAX6 and PITX3 in two English families with autosomal-dominant congenital nuclear cataract.
Acknowledgements
We thank the family members for their cooperation in this study. For this work VB and MM were supported by grant from the National Institute for Health Research Biomedical Research Centre at Moorfields Eye Hospital NHS Foundation Trust and UCL Institute of Ophthalmology (BRC-D-CON 546795).
Author contributions
VB conceived, analysed the data, wrote and provided critical revision of the paper. AI provided clinical information, AM, RQ and MM provided critical revision of the paper. NP contributed as data curator.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Vanita Berry, Email: v.berry@ucl.ac.uk.
Michel Michaelides, Email: michel.michaelides@ucl.ac.uk.
References
- 1.Traboulsi EI, Vanderveen D, Morrison D, Drews-Botsch CD, Lambert SR. Associated systemic and ocular disorders in patients with congenital unilateral cataracts: the Infant Aphakia Treatment Study experience. Eye (Lond) 2016;30:1170–4. doi: 10.1038/eye.2016.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Messina-Baas O, Cuevas-Covarrubias SA. Inherited congenital cataract: a guide to suspect the genetic etiology in the cataract genesis. Mol Syndromol. 2017;8:58–78. doi: 10.1159/000455752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ionides A, Francis P, Berry V, Mackay D, Bhattacharya S, Shiels A, et al. Clinical and genetic heterogeneity in autosomal dominant cataract. Br J Ophthalmol. 1999;83:802–8. doi: 10.1136/bjo.83.7.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shiels A, Hejtmancik JF. Biology of inherited cataracts and opportunities for treatment. Annu Rev Vis Sci. 2019;5:123–49. doi: 10.1146/annurev-vision-091517-034346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reis LM, Semina EV. Genetic landscape of isolated pediatric cataracts: extreme heterogeneity and variable inheritance patterns within genes. Hum Genet. 2019;138:847–63. doi: 10.1007/s00439-018-1932-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berry V, Georgiou M, Fujinami K, Quinlan R, Moore A, Michaelides M. Inherited cataracts: molecular genetics, clinical features, disease mechanisms and novel therapeutic approaches. Br J Ophthalmol. 2020;104:1331–7. doi: 10.1136/bjophthalmol-2019-315282. [DOI] [PubMed] [Google Scholar]
- 7.Sinn R, Wittbrodt J. An eye on eye development. Mech Dev. 2013;130:347–58. doi: 10.1016/j.mod.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 8.Cvekl A, Ashery-Padan R. The cellular and molecular mechanisms of vertebrate lens development. Development. 2014;141:4432–47. doi: 10.1242/dev.107953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harding P, Moosajee M. The molecular basis of human anophthalmia and microphthalmia. J Dev Biol. 2019;7. 10.3390/jdb7030016. [DOI] [PMC free article] [PubMed]
- 10.Bakrania P, Robinson DO, Bunyan DJ, Salt A, Martin A, Crolla JA, et al. SOX2 anophthalmia syndrome: 12 new cases demonstrating broader phenotype and high frequency of large gene deletions. Br J Ophthalmol. 2007;91:1471–6. doi: 10.1136/bjo.2007.117929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ton CC, Hirvonen H, Miwa H, Weil MM, Monaghan P, Jordan T, et al. Positional cloning and characterization of a paired box- and homeobox-containing gene from the aniridia region. Cell. 1991;67:1059–74. doi: 10.1016/0092-8674(91)90284-6. [DOI] [PubMed] [Google Scholar]
- 12.Terzić J, Saraga-Babić M. Expression pattern of PAX3 and PAX6 genes during human embryogenesis. Int J Dev Biol. 1999;43:501–8. [PubMed] [Google Scholar]
- 13.Ogino H, Yasuda K. Sequential activation of transcription factors in lens induction. Dev Growth Differ. 2000;42:437–48. doi: 10.1046/j.1440-169x.2000.00532.x. [DOI] [PubMed] [Google Scholar]
- 14.Kamachi Y, Uchikawa M, Tanouchi A, Sekido R. Kondoh H. Pax6 and SOX2 form a co-DNA-binding partner complex that regulates initiation of lens development. Genes Dev. 2001;15:1272–86. doi: 10.1101/gad.887101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanson I, Churchill A, Love J, Axton R, Moore T, Clarke M, et al. Missense mutations in the most ancient residues of the PAX6 paired domain underlie a spectrum of human congenital eye malformations. Hum Mol Genet. 1999;8:165–72. doi: 10.1093/hmg/8.2.165. [DOI] [PubMed] [Google Scholar]
- 16.Simpson TI, Price DJ. Pax6; a pleiotropic player in development. Bioessays. 2002;24:1041–51. doi: 10.1002/bies.10174. [DOI] [PubMed] [Google Scholar]
- 17.Wen JH, Chen YY, Song SJ, Ding J, Gao Y, Hu QK, et al. Paired box 6 (PAX6) regulates glucose metabolism via proinsulin processing mediated by prohormone convertase 1/3 (PC1/3) Diabetologia. 2009;52:504–13. doi: 10.1007/s00125-008-1210-x. [DOI] [PubMed] [Google Scholar]
- 18.Sun J, Rockowitz S, Xie Q, Ashery-Padan R, Zheng D, Cvekl A. Identification of in vivo DNA-binding mechanisms of Pax6 and reconstruction of Pax6-dependent gene regulatory networks during forebrain and lens development. Nucleic Acids Res. 2015;43:6827–46. doi: 10.1093/nar/gkv589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh S, Chao LY, Mishra R, Davies J, Saunders GF. Missense mutation at the C-terminus of PAX6 negatively modulates homeodomain function. Hum Mol Genet. 2001;10:911–8. doi: 10.1093/hmg/10.9.911. [DOI] [PubMed] [Google Scholar]
- 20.Epstein JA, Glaser T, Cai J, Jepeal L, Walton DS, Maas RL. Two independent and interactive DNA-binding subdomains of the Pax6 paired domain are regulated by alternative splicing. Genes Dev. 1994;8:2022–34. doi: 10.1101/gad.8.17.2022. [DOI] [PubMed] [Google Scholar]
- 21.Semina EV, Ferrell RE, Mintz-Hittner HA, Bitoun P, Alward WL, Reiter RS, et al. A novel homeobox gene PITX3 is mutated in families with autosomal-dominant cataracts and ASMD. Nat Genet. 1998;19:167–70. doi: 10.1038/527. [DOI] [PubMed] [Google Scholar]
- 22.Gage PJ, Suh H, Camper SA. The bicoid-related Pitx gene family in development. Mamm Genome. 1999;10:197–200. doi: 10.1007/s003359900970. [DOI] [PubMed] [Google Scholar]
- 23.Amendt BA, Sutherland LB, Semina EV, Russo AF. The molecular basis of Rieger syndrome. Analysis of Pitx2 homeodomain protein activities. J Biol Chem. 1998;273:20066–72. doi: 10.1074/jbc.273.32.20066. [DOI] [PubMed] [Google Scholar]
- 24.Semina EV, Reiter RS, Murray JC. Isolation of a new homeobox gene belonging to the Pitx/Rieg family: expression during lens development and mapping to the aphakia region on mouse chromosome 19. Hum Mol Genet. 1997;6:2109–16. doi: 10.1093/hmg/6.12.2109. [DOI] [PubMed] [Google Scholar]
- 25.Fujimoto M, Izu H, Seki K, Fukuda K, Nishida T, Yamada S-I, et al. HSF4 is required for normal cell growth and differentiation during mouse lens development. EMBO J. 2004;23:4297–306. doi: 10.1038/sj.emboj.7600435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Somasundaram T, Bhat SP. Developmentally dictated expression of heat shock factors: exclusive expression of HSF4 in the postnatal lens and its specific interaction with αB-crystallin heat shock promoter*. J Biol Chem. 2004;279:44497–503. doi: 10.1074/jbc.M405813200. [DOI] [PubMed] [Google Scholar]
- 27.Berry V, Ionides A, Pontikos N, Georgiou M, Yu J, Ocaka LA, et al. The genetic landscape of crystallins in congenital cataract. Orphanet J Rare Dis. 2020;15:333. doi: 10.1186/s13023-020-01613-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pontikos N, Yu J, Moghul I, Withington L, Blanco-Kelly F, Vulliamy T, et al. Phenopolis: an open platform for harmonization and analysis of genetic and phenotypic data. Bioinformatics. 2017;33:2421–3. doi: 10.1093/bioinformatics/btx147. [DOI] [PubMed] [Google Scholar]
- 29.Glusman G, Caballero J, Mauldin DE, Hood L, Roach JC. Kaviar: an accessible system for testing SNV novelty. Bioinformatics. 2011;27:3216–7. doi: 10.1093/bioinformatics/btr540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cvekl A, Callaerts P. PAX6: 25th anniversary and more to learn. Exp Eye Res. 2017;156:10–21. doi: 10.1016/j.exer.2016.04.017. [DOI] [PubMed] [Google Scholar]
- 31.Graw J. Eye development. Curr Top Dev Biol. 2010;90:343–86. doi: 10.1016/S0070-2153(10)90010-0. [DOI] [PubMed] [Google Scholar]
- 32.Aryal S, Viet J, Weatherbee BAT, Siddam AD, Hernandez FG, Gautier-Courteille C, et al. The cataract-linked RNA-binding protein Celf1 post-transcriptionally controls the spatiotemporal expression of the key homeodomain transcription factors Pax6 and Prox1 in lens development. Hum Genet. 2020;139:1541–54. doi: 10.1007/s00439-020-02195-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahmad N, Aslam M, Muenster D, Horsch M, Khan MA, Carlsson P, et al. Pitx3 directly regulates Foxe3 during early lens development. Int J Dev Biol. 2013;57:741–51. doi: 10.1387/ijdb.130193jg. [DOI] [PubMed] [Google Scholar]
- 34.Verdin H, Sorokina EA, Meire F, Casteels I, de Ravel T, Semina EV, et al. Novel and recurrent PITX3 mutations in Belgian families with autosomal dominant congenital cataract and anterior segment dysgenesis have similar phenotypic and functional characteristics. Orphanet J Rare Dis. 2014;9:1–11. doi: 10.1186/1750-1172-9-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anand D, Agrawal SA, Slavotinek A, Lachke SA. Mutation update of transcription factor genes FOXE3, HSF4, MAF, and PITX3 causing cataracts and other developmental ocular defects. Hum Mutat. 2018;39:471–94. doi: 10.1002/humu.23395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakai A, Tanabe M, Kawazoe Y, Inazawa J, Morimoto RI, Nagata K. HSF4 a new member of the human heat shock factor family which lacks properties of a transcriptional activator. Mol Cell Biol. 1997;17:469–81. doi: 10.1128/MCB.17.1.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanabe M, Sasai N, Nagata K, Liu XD, Liu PC, Thiele DJ, et al. The mammalian HSF4 gene generates both an activator and a repressor of heat shock genes by alternative splicing. J Biol Chem. 1999;274:27845–56. doi: 10.1074/jbc.274.39.27845. [DOI] [PubMed] [Google Scholar]
- 38.Nakai A. Molecular basis of HSF regulation. Nat Struct Mol Biol. 2016;23:93–5. doi: 10.1038/nsmb.3165. [DOI] [PubMed] [Google Scholar]
- 39.Cui X, Wang L, Zhang J, Du R, Liao S, Li D, et al. HSF4 regulates DLAD expression and promotes lens de-nucleation. Biochim Biophys Acta. 2013;1832:1167–72. doi: 10.1016/j.bbadis.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 40.Bu L, Jin Y, Shi Y, Chu R, Ban A, Eiberg H, et al. Mutant DNA-binding domain of HSF4 is associated with autosomal dominant lamellar and Marner cataract. Nat Genet. 2002;31:276–8. doi: 10.1038/ng921. [DOI] [PubMed] [Google Scholar]
- 41.Hansen L, Mikkelsen A, Nürnberg P, Nürnberg G, Anjum I, Eiberg H, et al. Comprehensive mutational screening in a cohort of Danish families with hereditary congenital cataract. Invest Ophthalmol Vis Sci. 2009;50:3291–303. doi: 10.1167/iovs.08-3149. [DOI] [PubMed] [Google Scholar]
- 42.Hever AM, Williamson KA, van Heyningen V. Developmental malformations of the eye: the role of PAX6, SOX2 and OTX2. Clin Genet. 2006;69:459–70. doi: 10.1111/j.1399-0004.2006.00619.x. [DOI] [PubMed] [Google Scholar]
- 43.Davis LK, Meyer KJ, Rudd DS, Librant AL, Epping EA, Sheffield VC, et al. Pax6 3’ deletion results in aniridia, autism and mental retardation. Hum Genet. 2008;123:371–8. doi: 10.1007/s00439-008-0484-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lima Cunha D, Arno G, Corton M, Moosajee M. The spectrum of PAX6 mutations and genotype-phenotype correlations in the eye. Genes. 2019;10. 10.3390/genes10121050. [DOI] [PMC free article] [PubMed]
- 45.Glaser T, Jepeal L, Edwards JG, Young SR, Favor J, Maas RL. PAX6 gene dosage effect in a family with congenital cataracts, aniridia, anophthalmia and central nervous system defects. Nat Genet. 1994;7:463–71. doi: 10.1038/ng0894-463. [DOI] [PubMed] [Google Scholar]
- 46.Cvekl A, Piatigorsky J. Lens development and crystallin gene expression: many roles for Pax-6. Bioessays. 1996;18:621–30. doi: 10.1002/bies.950180805. [DOI] [PubMed] [Google Scholar]
- 47.Walther C, Gruss P. Pax-6, a murine paired box gene, is expressed in the developing CNS. Development. 1991;113:1435–49. doi: 10.1242/dev.113.4.1435. [DOI] [PubMed] [Google Scholar]
- 48.Duncan MK, Kozmik Z, Cveklova K. Overexpression of PAX6 (5a) in lens fiber cells results in cataract and upregulation of (alpha) 5 (beta) 1 integrin expression. J Cell. 2000. https://jcs.biologists.org/content/113/18/3173.short. [DOI] [PubMed]
- 49.Cvekl A, Zhang X. Signaling and gene regulatory networks in mammalian lens development. Trends Genet. 2017;33:677–702. doi: 10.1016/j.tig.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dansault A, David G, Schwartz C, Jaliffa C, Vieira V, de la Houssaye G, et al. Three new PAX6 mutations including one causing an unusual ophthalmic phenotype associated with neurodevelopmental abnormalities. Mol Vis. 2007;13:511–23. [PMC free article] [PubMed] [Google Scholar]
- 51.Solomon BD, Pineda-Alvarez DE, Balog JZ, Hadley D, Gropman AL, Nandagopal R, et al. Compound heterozygosity for mutations in PAX6 in a patient with complex brain anomaly, neonatal diabetes mellitus, and microophthalmia. Am J Med Genet A. 2009;149A:2543–6. doi: 10.1002/ajmg.a.33081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brémond-Gignac D, Bitoun P, Reis LM, Copin H, Murray JC, Semina EV. Identification of dominant FOXE3 and PAX6 mutations in patients with congenital cataract and aniridia. Mol Vis. 2010;16:1705. [PMC free article] [PubMed] [Google Scholar]
- 53.Cai F, Zhu J, Chen W, Ke T, Wang F, Tu X. A novel PAX6 mutation in a large Chinese family with aniridia and congenital cataract. Molecular. 2010. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2901194/. [PMC free article] [PubMed]
- 54.Goolam S, Carstens N, Ross M, Bentley D, Lopes M, Peden J, et al. Familial congenital cataract, coloboma, and nystagmus phenotype with variable expression caused by mutation in PAX6 in a South African family. Mol Vis. 2018;24:407. [PMC free article] [PubMed] [Google Scholar]
- 55.Thomas S, Thomas MG, Andrews C, Chan W-M, Proudlock FA, McLean RJ, et al. Autosomal-dominant nystagmus, foveal hypoplasia and presenile cataract associated with a novel PAX6 mutation. Eur J Hum Genet. 2014;22:344–9. doi: 10.1038/ejhg.2013.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bidinost C, Matsumoto M, Chung D, Salem N, Zhang K, Stockton DW, et al. Heterozygous and homozygous mutations in PITX3 in a large Lebanese family with posterior polar cataracts and neurodevelopmental abnormalities. Invest Ophthalmol Vis Sci. 2006;47:1274–80. doi: 10.1167/iovs.05-1095. [DOI] [PubMed] [Google Scholar]
- 57.Aldahmesh MA, Khan AO, Mohamed J, Alkuraya FS. Novel recessive BFSP2 and PITX3 mutations: insights into mutational mechanisms from consanguineous populations. Genet Med. 2011;13:978–81. doi: 10.1097/GIM.0b013e31822623d5. [DOI] [PubMed] [Google Scholar]
- 58.Berry V, Yang Z, Addison PKF, Francis PJ, Ionides A, Karan G, et al. Recurrent 17 bp duplication in PITX3 is primarily associated with posterior polar cataract (CPP4) J Med Genet. 2004;41:e109. doi: 10.1136/jmg.2004.020289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Finzi S, Li Y, Mitchell TN, Farr A, Maumenee IH, Sallum JMF, et al. Posterior polar cataract: genetic analysis of a large family. Ophthalmic Genet. 2005;26:125–30. doi: 10.1080/13816810500229124. [DOI] [PubMed] [Google Scholar]
- 60.Burdon KP, McKay JD, Wirth MG, Russell-Eggit IM, Bhatti S, Ruddle JB, et al. The PITX3 gene in posterior polar congenital cataract in Australia. Mol Vis. 2006;12:367–71. [PubMed] [Google Scholar]
- 61.Summers KM, Withers SJ, Gole GA, Piras S, Taylor PJ. Anterior segment mesenchymal dysgenesis in a large Australian family is associated with the recurrent 17 bp duplication in PITX3. Mol Vis. 2008;14:2010. [PMC free article] [PubMed] [Google Scholar]
- 62.Zazo Seco C, Plaisancié J, Lupasco T, Michot C, Pechmeja J, Delanne J, et al. Identification of PITX3 mutations in individuals with various ocular developmental defects. Ophthalmic Genet. 2018;39:314–20. doi: 10.1080/13816810.2018.1430243. [DOI] [PubMed] [Google Scholar]
- 63.Zhang XH, Da Wang J, Jia HY, Zhang JS, Li Y, Xiong Y, et al. Mutation profiles of congenital cataract genes in 21 northern Chinese families. Mol Vis. 2018;24:471. [PMC free article] [PubMed] [Google Scholar]
- 64.Sakazume S, Sorokina E, Iwamoto Y, Semina EV. Functional analysis of human mutations in homeodomain transcription factor PITX3. BMC Mol Biol. 2007;8:84. doi: 10.1186/1471-2199-8-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wada K, Matsushima Y, Tada T, Hasegawa S, Obara Y, Yoshizawa Y, et al. Expression of truncated PITX3 in the developing lens leads to microphthalmia and aphakia in mice. PLoS ONE. 2014;9:e111432. doi: 10.1371/journal.pone.0111432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Le W, Nguyen D, Lin XW, Rawal P, Huang M, Ding Y, et al. Transcription factor PITX3 gene in Parkinson’s disease. Neurobiol Aging. 2011;32:750–3. doi: 10.1016/j.neurobiolaging.2009.03.015. [DOI] [PubMed] [Google Scholar]
- 67.Bäckström D, Domellöf ME, Granåsen G, Linder J, Mayans S, Elgh E, et al. PITX3 genotype and risk of dementia in Parkinson’s disease: a population-based study. J Neurol Sci. 2017;381:278–84. doi: 10.1016/j.jns.2017.08.3259. [DOI] [PubMed] [Google Scholar]
- 68.Derwińska K, Mierzewska H, Goszczańska A, Szczepanik E, Xia Z, Kuśmierska K, et al. Clinical improvement of the aggressive neurobehavioral phenotype in a patient with a deletion of PITX3 and the absence of L-DOPA in the cerebrospinal fluid. Am J Med Genet B Neuropsychiatr Genet. 2012;159:236–42. doi: 10.1002/ajmg.b.32020. [DOI] [PubMed] [Google Scholar]
- 69.Graw J. From eyeless to neurological diseases. Exp Eye Res. 2017;156:5–9. doi: 10.1016/j.exer.2015.11.006. [DOI] [PubMed] [Google Scholar]