Abstract
The intestinal immune function of chickens is limited during the early growing stage. Maternal nutritional intervention has been suggested to affect the innate immunity of offspring. The present study aimed to investigate the effects of maternal stevioside supplementation on the intestinal immune function of chicken offspring. A total of 120 Jinmao yellow-feathered breeder hens were fed a basal diet or a diet supplemented with 250 mg/kg stevioside for 5 weeks. During the last week, 200 breeding eggs from each group were collected for incubation. After hatching, 80 male offspring (40 chickens from each group) were randomly selected and fed the same basal diet for 28 d. In addition, 90 well-shaped fertile eggs of non-treated breeder hens were incubated for the in ovo injection experiment. Steviol dissolved in 20% glycerol was injected at 7 d of incubation. The results showed that maternal stevioside supplementation could improve embryonic development, jejunal integrity and proliferation in the jejunal crypt (P < 0.05). Maternal stevioside supplementation could also increase the innate transcription levels of cytokines and endotoxin tolerance-related factors in the jejunum of chicken offspring (P < 0.05). At 28 d of age, the offspring following maternal stevioside supplementation exhibited higher jejunal secretory immunoglobulin A and serum interferons levels (P < 0.05). A higher abundance of Lactobacillales induced by maternal stevioside supplementation was positively correlated with intestinal immune-related factors (P < 0.05). The in ovo injection with steviol did not alter either embryonic development or intestinal immune function of hatching chickens (P > 0.05). Furthermore, maternal stevioside supplementation could induce hypo-methylation on the promoter region of suppressor of cytokine signaling 1 (SOCS1). In conclusion, maternal stevioside supplementation could improve the intestinal immune function of chicken offspring potentially via modulating the gut microbiota and down-regulating the promoter methylation level of SOCS1.
Keywords: Maternal, Stevioside, Jejunum, Transcriptome, Offspring, Immune function
1. Introduction
The intestine is one of the most important immune systems in the animal body. Intestinal immunity plays a critical role to defend against bacterial infection (Martens et al., 2018). A major problem in the poultry industry is that the development of intestinal immune function is slow in neonatal chickens (Alkie et al., 2019; Weström et al., 2020). This immature intestinal immune function could increase the susceptibility to pathogenic microorganisms in the early growing stage of chickens, which might reduce growth performance and eventually lead to heavy financial losses in intensive production (Bar-Shira and Friedman, 2006; Li et al., 2015). Thus, it is of great significance to explore strategies for improving the innate intestinal immune function of chickens. In mammals, it has been demonstrated that the manipulation of maternal nutrition could effectively enhance the intestinal innate immunity of offspring (Kelly and Coutts, 2000; Feil and Fraga, 2012). However, it remains unclear whether maternal nutrition intervention could exert such an effect on chicken offspring.
Stevioside, extracted form Stevia rebaudiana, exerts immune-regulatory effects on the intestinal cells (Alavala et al., 2019; Luo et al., 2019). Acting as an immune-modulator, stevioside has been suggested to specifically dock with a key immune receptor, toll-like receptor 4 (TLR4) (Casas-Grajales et al., 2019). Our previous study has shown that dietary supplementation with stevioside could alleviate lipopolysaccharide-induced intestinal mucosal damage in broiler chickens (Jiang et al., 2019). Additionally, stevioside treatment could increase the antibody titer of Newcastle Disease Virus (NDV) in chickens (Molina-Barrios et al., 2021). Another previous study has also demonstrated that stevioside supplementation could increase the serum concentrations of immunoglobulin G (IgG) and immunoglobulin A (IgA) in chickens (Wu et al., 2019b). If these immune-regulatory effects can be transmitted to the next generation, the intestinal immune function of chicken offspring may be enhanced. Numerous studies have proven that a changed phenotype of the parents induced by epigenetic factors—including dietary components—could be partly inherited by the offspring (Li et al., 2018; James et al., 2019; Wu et al., 2019a). It is worth mentioning that, in the colonic cells, a derivative of stevioside could affect DNA methylation, which is an important kind of epigenetic modification (Mokarram et al., 2017). However, whether stevioside could affect the intestinal epigenetic modification has not been elucidated.
Gut microbiota are also involved in modulating the intestinal immune function (Kabat et al., 2014). There is emerging evidence that the gut microbiota may affect the differentiation and function of intestinal innate immune cells (Hepworth et al., 2013; Geva-Zatorsky et al., 2017). A previous study has shown that the microbial community of the chicken embryo is partly inherited from hens, and could be altered by host genetic variation (Ding et al., 2017). Due to the fact that dietary stevioside supplementation has influence on the gut microbiota of chickens (Wu et al., 2019b; Jiang et al., 2021a), we speculate that maternal stevioside supplementation might change the microbial communities and have an impact on the intestinal immune function of chicken offspring. In addition, a change of nutrients within the egg yolk can also alter the gut microbiota of chickens (Slawinska et al., 2019; Abdel-Moneim et al., 2020; Gong et al., 2020). In the animal body, stevioside is mainly hydrolyzed into steviol by the gut microbiota, and steviol can be transferred into the liver through the blood (Renwick and Tarka, 2008). There is a possibility that steviol might enter the egg yolk through the yolk precursor synthesized by the liver, which might also change the microbial communities of neonatal chickens (Mahalak et al., 2020).
Thus, the purpose of the current study was to investigate whether maternal stevioside supplementation could improve the intestinal immune function of chicken offspring, and to explore its potential underlying molecular mechanisms.
2. Materials and methods
2.1. Animal ethics statement
All animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of Nanjing Agricultural University (SYXK (SU) 2017-0007).
2.2. Animals and experimental design
The feeding trial was performed in the experimental animal room of Tushan Breeding Farm (Changzhou, China) under a controlled environment. A total of 120 Jinmao yellow-feathered breeder hens (54 weeks of age) were randomly allocated to 2 treatment groups: (1) a control group and (2) a stevioside-supplemented group. Stevioside (Macklin Inc, Shanghai, China) with a purity of 98% was added to the basal diet at the expense of equal amount (250 mg/kg) of limestone. The dose of stevioside was selected based on the results of our previous studies showing that 250 mg/kg stevioside supplementation could affect the intestinal physiological function of chickens (Jiang et al., 2019, 2020a). The dietary treatment of breeder hens lasted for 5 weeks, and all breeder hens were subjected to artificial insemination during the last 2 weeks of the dietary treatment. During the last week of this experiment, 200 breeding eggs from each group were collected for incubation. Each group contained 8 replicates of 25 eggs. Eggs were placed in 8 trays (25 eggs per tray) and set randomly within the same incubator. One tray contained one replicate from each treatment.
After hatching, sex identification was performed by the same experienced technician. The male chickens were separated. A total of 80 one-day-old male chicken offspring (40 chicken offspring from each group) were then transferred to brood cages in a temperature- and light-controlled room with continuous light. The offspring were divided into 2 groups: (1) the offspring of breeder hens fed a basal diet (CON) and (2) the offspring of breeder hens fed a basal diet supplemented with 250 mg/kg stevioside (STE). Each group contained 8 replicates with 5 chicken offspring in each replicate. The temperature was maintained at 33 to 35 °C for one week, and it was then gradually decreased by 1 °C every 2 d to a final temperature of 26 °C. All chicks had ad libitum access to the same basal diet and water. The composition of the diet for the chick offspring is presented in Appendix Table 1. The basal diet met the nutrient requirement of Chinese Feeding Standard of Chicken (NY/T33–2004). All chicken offspring were inoculated with the NDV vaccine on 7 d and with an inactivated infectious bursal disease vaccine on 14 d. The body weight and total feed consumption were recorded to calculate growth performance of the CON and STE groups on 1 and 28 d. The growth performance of chicken offspring was analyzed using each replicate as the experimental unit (n = 8).
The in ovo injection experiment was performed in Nanjing Agricultural University. Briefly, 90 well-shaped fertile eggs of non-treated Jinmao yellow-feathered breeder hens (58 weeks of age) were collected from Tushan Breeding Farm and incubated in the same incubator. All eggs were randomly divided into 3 groups: (1) in ovo injection with 20% glycerol (Vehicle); (2) in ovo injection with 0.3 mg steviol per egg (Low-dose steviol) and (3) in ovo injection with 3.0 mg steviol per egg (High-dose steviol). Each group contained 6 replicates with 5 eggs in each replicate. At 7 d of incubation, a small hole was drilled at the blunt end. Steviol (98% purity, Shanghai Yuanye Bio-Technology, Shanghai, China) dissolved in 20% glycerol was injected into the yolk by the same operator. Each egg was injected with 100 μL of solution. After injection, all the eggs were sealed with tape and returned for continued incubation. On 18 d of incubation, all eggs were candled to identify the fertilization rate. At the hatching day, the number of dead embryos were recorded.
2.3. Sample collection
On embryonic d 11, 13, 15, 17, 19 and the hatching day, we randomly selected one normal embryo per replicate (a total of 8 embryos per group) for sampling. The embryo weight, embryo length and the length of tibia were measured to evaluate embryonic development. We further measured the weight of breast muscle and liver on embryonic d 17, 19 and the hatching day. The embryonic development of chicken offspring was analyzed using 8 embryos randomly selected (n = 8).
At the hatching day and 28 d of age, 8 chicken offspring from each group were randomly selected, respectively. The live body weight for each selected chicken was measured. The chicken offspring were then euthanized, and blood samples collected. Serum was separated and stored at −20 °C. Organs (thymus, spleen, liver, breast muscle and bursa of Fabricius) were removed and weighed. We then carefully collected the jejunum and cecum. Sections of 1 cm were cut off from the middle of the jejunum. The jejunal sections were then fixed in 4% paraformaldehyde. The remaining length of the jejunum were stored at −80 °C for further analysis. The cecal digesta of chickens at 28 d and the meconium of newly-hatched chickens were collected and frozen in liquid nitrogen for DNA extraction.
The sample collection of the in ovo injection experiment was performed on the hatching day. The mortality of the high-dose steviol group was too high. Thus, we only randomly selected 8 chickens from the Vehicle and Low-dose steviol groups for the sample collection. The hatching weight, length of tibia and the weight of breast muscle and liver were measured. After euthanasia, the jejunum was carefully collected and stored at −80 °C.
2.4. Enzyme linked immunosorbent assay (ELISA)
The levels of serum immunoglobulin, serum cytokine and jejunal secretory immunoglobulin A (sIgA) were determined using commercial ELISA kits (Shanghai Enzyme-linked Biotechnology, Shanghai, China). The serum NDV antibody titer was determined using commercial ELISA kits (IDEXX laboratories, Westbrook, USA). All experimental procedures were performed precisely according to the manufacturer’s instructions. The serum was diluted 1:5 with phosphate buffered saline (PBS) containing 1% bovine serum albumin before the test. The jejunal mucosa was homogenized in saline and then centrifuged to obtain the supernatant for measuring the jejunal sIgA levels. As indicated on the instructions of ELISA kits, the sensitivity for sIgA determination was 10 ng/mL; the sensitivity for IgA determination was 1.0 μg/mL; the sensitivity for IgG determination was 10.0 μg/mL; the sensitivity for both interferon (IFN)-α and IFN-γ determinations was 0.1 pg/mL; the intra- and inter-assay coefficients of variation were <10% and <15%, respectively, for all ELISA kits. The data were analyzed using 8 sampled chickens per group (n = 8).
2.5. Morphology analysis
After fixation in 4% paraformaldehyde for 24 h, the jejunal sections were soaked through a graded series of ethanol and xylene and embedded in paraffin. The jejunum was then sectioned at 5 μm with a Lecia RM2235 microtome (Leica Biosystems Inc., Buffalo Grove, IL). The sections were deparaffinized with xylene and rehydrated through a graded dilution of ethanol. Hematoxylin and eosin (H&E) staining and Alcian Blue-periodic acid Schiff (AB-PAS) staining were performed, respectively. The images were acquired using an Olympus microscope (Olympus Optical Co., Ltd., Beijing, China). The values of villus height (VH), crypt depth (CD), and the number of jejunal goblet cells were measured 6 times from different villus and crypts per section from each chicken using Image-Pro Plus software 6.0 (Media Cybernetics, Rockville, MD, USA). The data were analyzed using 8 sampled chickens per group (n = 8).
2.6. Immunohistochemistry
The jejunal sections for immunohistochemistry were subjected to deparaffinization, hydration, heat-induced antigen retrieval and blocking of peroxidase activity. After washing with PBS, sections were incubated with 5% bovine serum albumin for 2 h. Then, the sections were incubated overnight with proliferating cell nuclear antigen (PCNA) antibody (Abcam, Cambridge, UK) at 4 °C, and negative controls were performed by replacing the antibody with normal goat serum. The mouse IgG-SABC kit (Boster Biological Technology, Wuhan, China) was used to detect the immunoreactivity of PCNA. The immunolabeling was then visualized with 0.05% DAB in PBS. Finally, the sections were counterstained with hematoxylin. Images were acquired using an Olympus microscope (Olympus Optical Co., Ltd., Beijing, China). The ratio of PCNA positive cells in the villus and crypt of jejunum was analyzed using Image-Pro Plus software 6.0 (Media Cybernetics, Rockville, MD, USA), respectively. The data were analyzed using 8 sampled chickens per group (n = 8).
2.7. Western blotting
Proteins of jejunum were extracted using radioimmunoprecipitation assay buffer (Beyotime Biotechnology, Shanghai, China) containing phenylmethylsulfonyl fluoride (Beyotime Biotechnology, Shanghai, China), and total protein content was determined using a Bicinchoninic Acid Protein Assay Kit (Beyotime Biotechnology, Shanghai, China) according to the manufacturer’s protocol. Equal amounts of proteins (30 μg) were electrophoresed and then transferred on to polyvinylidene difluoride membranes (Millipore, Bedford, MA, USA). The membranes were incubated with the PCNA antibody overnight at 4 °C. Beta-actin was utilized as a loading control. After 6 washes in tris buffered saline with Tween-20 (Beyotime Biotechnology, Shanghai, China) for 5 min each, the membranes were incubated with secondary antibody for 60 min at room temperature. Finally, the blots were washed and the protein bands were detected using an enhanced chemiluminescence kit (Thermo Scientific, Wilmington, DE, USA). Signals were visualized using ChemiDoc Touch Imaging System (Bio-Rad Laboratories, CA, USA). The protein expressions were estimated by quantifying the intensities of the bands using Image J NIH software (Media Cybernetics, Rockville, MD, USA). The data of Western blotting were analyzed using 6 randomly selected samples per group (n = 6).
2.8. RNA extraction, qualification, library preparation, and sequencing
Eight samples of jejunum of chicken offspring on 1 and 28 d were randomly selected (four samples from each group) for RNA-Sequencing (RNA-Seq), respectively. The total RNA of the jejunum was extracted using the RNAiso Plus (Takara, Dalian, China). The concentration and quality of total RNA was tested by a ND-2000 micro spectrophotometer (Thermo Scientific, Wilmington, DE, USA). A total of 3 μg RNA per sample was used as the input material for the RNA sample preparations. Sequencing libraries were generated using the NEBNext Ultra RNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, MA, USA) following the manufacturer’s instructions. Briefly, first strand cDNA was synthesized using random hexamer primer and M-MuLV Reverse Transcriptase (Thermo Scientific, Wilmington, DE, USA). Second strand cDNA synthesis was subsequently performed using DNA Polymerase I and RNase H (Thermo Scientific, Wilmington, DE, USA). AMPure XP system (Beckman Coulter, Brea, CA, USA) was applied to purify the library fragments. Then, PCR was performed and the PCR products were also purified using AMPure XP system. The library quality was estimated on the Agilent Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA). After cluster generation, the library was sequenced on an Illumina Hiseq 2500 platform (Illumina, San Diego, USA) and 125 bp/150 bp paired-end reads were generated.
2.9. Data analysis of RNA-seq
After sequencing, the raw data of FASTQ reads were processed through in-house Perl scripts. Low-quality reads (including short reads and adapter sequences) were removed to obtain clean reads (clean data). The reference genome used in the present study was Gallus_gallus-5.0 (Ensembl release 94). All paired-end clean reads were aligned to the reference genome using Hisat2 (version 2.0.5). Fragments per kilobase per million were used to calculate the intensity of gene expressions. Differential expression analysis was performed by the DESeq2 R package (1.16.1). To minimize the false discovery rate, P-values were adjusted by the Benjamini and Hochberg’s approach. Genes with an adjusted P-value < 0.05 were considered to be differentially expressed genes (DEGs). Gene ontology (GO) and a Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of the DEGs were performed using the clusterProfiler R package. An adjusted P-value < 0.05 was taken as a threshold of significance. The data of RNA-Seq were analyzed using 4 randomly selected samples per group (n = 4).
2.10. Quantitative real-time PCR (qRT-PCR)
After RNA extraction, a total amount of 1 μg of RNA was reverse-transcribed into cDNA using a HiScript II 1st Strand cDNA Synthesis Kit with gDNA wiper (Vazyme Biotech, Nanjing, China). The cDNA was then diluted 1:5 using RNase free water. The qRT-PCR was carried out using the ChamQ SYBR qPCR Master Mix (Vazyme Biotech, Nanjing, China) on the QuantStudio 5 Real-Time PCR System (Thermo Scientific, Wilmington, DE, USA). Beta-actin (ACTB) was selected as the housekeeping gene. The primers were synthesized by Sangon Biotech (Sangon Biotech, Shanghai, China), and the primer sequences were presented in Appendix Table 2. The amplification of a single product was verified via a melting curve with a single peak. Relative gene expression levels were analyzed by the 2−ΔΔCt method after normalization against ACTB (Livak and Schmittgen, 2001). The data were analyzed using 8 sampled chickens per group as the experimental unit (n = 8).
2.11. Sequencing of the 16S ribosomal RNA (rRNA) gene
Ten samples of meconium and cecal digesta were randomly selected (5 samples from each group) for 16S Ribosomal RNA (rRNA) gene sequencing analysis. Generally, total genome DNA was extracted according to the protocol of a QIAampDNA Stool Mini Kit (Qiagen, CA, USA). The 16S V4 region of 16S rRNA genes was amplified by the specific primer with the barcode (515F, GTGCCAGCMGCCGCGGTAA; 806R, GGACTACHVGGGTWTCTAAT). Phusion High-Fidelity PCR Master Mix (New England Biolabs, Ipswich, MA, USA) was used to perform the PCR reactions. Sequencing libraries were generated using the Ion Plus Fragment Library Kit (Thermo Scientific, Wilmington, DE, USA). Finally, the library was sequenced on an Ion S5 XL platform (Thermo Scientific, Wilmington, DE, USA).
2.12. Data analysis of the 16S rRNA gene sequencing
High-quality clean reads were obtained after data filtration of the raw reads according to the Cutadapt (version 1.9.1) quality-controlled process. Uparse software (version v7.0.1001) was applied for the analysis of sequences. Sequences with ≥97% similarity were assigned to the same operational taxonomic units (OTUs). Representative sequence for each OTU was screened for further annotation. The taxonomy of OTU sequences was annotated from phylum to genus level. A principal co-ordinate analysis (PCoA) and unweighted pair-group method with arithmetic means (UPGMA) was calculated by Quantitative Insights Into Microbial Ecology (version 1.7.0; http://qiime.org/scripts/split_libraries_fastq.html). The analysis of linear discriminant analysis effect size (LEfSe) was performed to evaluate differentially abundant bacterium among the groups. The data of 16S rRNA gene sequencing were analyzed using 5 randomly selected samples per group (n = 5).
2.13. High-performance liquid chromatography (HPLC)
The concentrations of steviol in the egg yolk were analyzed by HPLC performed on an Agilent HPLC system (Agilent, Santa Clara, CA, USA) using the following reverse-phase conditions: a 5-μm Phenomenex Gemini C18 (4.6 mm × 250 mm) (Phenomenex, Torrance, CA, USA), a column temperature of 30 °C, diode array detector with detection wavelength of 213 nm, and mobile phases were held at water with acetic acid (A) and acetonitrile (B). The gradient conditions were set as follows: flow rate, 1 mL/min; 80% A and 20% B for 20 min, 80% B from 20 to 25 min; and re-equilibration with 80% A and 20% B for 5 min. The injection volume was 10 μL. The chromatograms of steviol were collected and integrated by software ChemStation (Agilent, Santa Clara, CA, USA). The steviol standard with a purity of 98% was purchased from Shanghai Yuanye Bio-Technology (Shanghai, China). We randomly selected 4 yolk samples for HPLC (n = 4).
2.14. DNA isolation and bisulfite modification
Genomic DNA was isolated from the jejunum using Fast Pure Cell/Tissue DNA Isolation Mini Kit (Vazyme Biotech, Nanjing, China). The concentration and quality of DNA was identified by a ND-2000 micro spectrophotometer (Thermo Scientific, Wilmington, DE, USA). Then, 1 μg of DNA was then bisulfite modified using EpiArt DNA Methylation Bisulfite Kit (Vazyme Biotech, Nanjing, China). All procedures were performed according to the manufacturer’s instructions. Modified DNA was applied as the template DNA for further PCR amplification.
2.15. Methylation-specific PCR (MSP)
The primers for MSP were designed using the online Methprimer software (http://www.urogene.org/methprimer/). The primers are presented in Appendix Table 3. The PCR reaction mixture of 50 μL contained 25 μL 2 × EpiArt HS Taq Master Mix (Vazyme Biotech, Nanjing, China), 2 μL of each primer (10 μM), 5 μL of template DNA (50 ng) and 16 μL of double-distilled H2O. The reaction program of PCR was as follows: 5 min at 95 °C, followed by 40 cycles of 95 °C for 30 s, 56 °C for 30 s, 72 °C for 30 s and finally, 5 min for extension at 72 °C. The PCR product was visualized on a 2% agarose gel under UV light using the Tanon 1600 gel image analysis system (Tanon, Shanghai, China).
2.16. Bisulfite sequencing PCR (BSP)
The methylation of the promoter region of suppressor of cytokine signaling 1 (SOCS1) was analyzed by bisulfite sequencing. The primers were designed using the Methyl Primer Express v1.0 (Thermo Scientific, Wilmington, DE, USA) and presented in Appendix Table 3. The same PCR reaction mixture as MSP was used with different primers specially for BSP. The reaction program of PCR was as follows: 10 min at 95 °C, followed by 40 cycles of 94 °C for 30 s, 55 °C for 30 s, 72 °C for 40 s, and finally, 5 min for extension at 72 °C. The PCR products were gel purified using Gel Purification Kit (Takara, Dalian, China), and then the purified DNA was cloned into the pMD18-T vector (Takara, Dalian, China). For each PCR product, 10 clones were randomly picked for final sequencing (Shanghai Personal Biotechnology, Shanghai, China). The sequencing results were analyzed using the online BiQ Analyzer software (https://biq-analyzer.bioinf.mpi-inf.mpg.de/). The data of BSP were analyzed using 3 randomly selected samples per group (n = 3).
2.17. Statistical analysis
The Shapiro–Wilk test was performed to determine the normality distribution of the data. Two-tailed Student's t-test was used to compare the results between the experimental groups using Graphpad Prism 8. G∗Power 3.1.9.7 software (http://www.gpower.hhu.de/) was applied to calculate the Cohen’s d effect size and statistical power of the data by performing a post hoc analysis. Cohen’s d effect size was analyzed according to the mean value and standard deviation of 2 groups. Statistical power was calculated based on Cohen’s d effect size, error probability (0.05) and sample size. Data were presented as the mean and standard error of the mean (SEM). Differences were considered to be statistically significant at P < 0.05.
3. Results
3.1. Embryonic development and growth performance
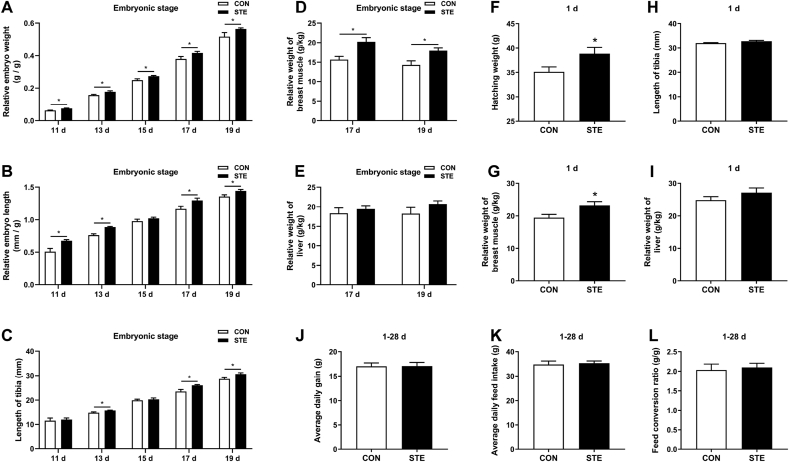
We evaluated the effects of maternal stevioside supplementation on the embryonic development and growth performance of chicken offspring, and the data are shown in Fig. 1. Maternal stevioside supplementation significantly increased the relative embryo weight, relative embryo length and length of tibia during the embryonic stage (P < 0.05, Fig. 1A–C). It also increased the relative weight of breast muscle (P < 0.05, Fig. 1 D), but did not alter the relative weight of liver at 17 and 19 d of incubation (P > 0.05, Fig. 1 E). In the newly-hatched chickens, maternal stevioside supplementation significantly increased the hatching weight and the relative weight of breast muscle (P < 0.05, Fig. 1F and G), whereas it had no effect on the length of tibia and the relative weight of liver (P > 0.05, Fig. 1H and I). In addition, maternal stevioside supplementation did not change the average daily gain, average daily feed intake and feed conversion rate of chicken offspring during 1–28 d (P > 0.05, Fig. 1J–L). No difference was found in the relative organ weight between the 2 groups on 28 d (P > 0.05, Appendix Table 4).
Fig. 1.
Effects of maternal stevioside supplementation on the embryonic development and growth performance of chicken offspring. (A) Relative embryo weight during embryonic stage. (B) Relative embryo length during embryonic stage. (C) Length of tibia during embryonic stage. (D) Relative weight of breast muscle during embryonic stage. (E) Relative weight of liver during embryonic stage. (F) Hatching weight. (G) Relative weight of breast muscle on 1 d. (H) Length of tibia on 1 d. (I) Relative weight of liver on 1 d. (J) Average daily gain during 1–28 d. (K) Average daily feed intake during 1–28 d. (L) Feed conversion ratio during 1–28 d. Data are presented as mean values ± SEM (n = 8). Values with an asterisk represent statistically significant differences (P < 0.05). CON, the offspring of breeder hens fed a basal diet; STE, the offspring of breeder hens fed a basal diet supplemented with 250 mg/kg stevioside. Effect size and statistical power of parameters in Fig. 1 are shown in Appendix Table 5.
3.2. Intestinal morphology and proliferation
As shown in Fig. 2, H&E staining was performed to observe the effects of maternal stevioside supplementation on the jejunal morphology of chicken offspring. AB-PAS was also conducted to determine the density of goblet cells in the jejunum of chicken offspring, and immunohistochemistry and Western blotting was also applied to evaluate the jejunal proliferation. The representative images of HE staining and AB-PAS were presented in Fig. 2A and B. At 1 d of age, maternal stevioside supplementation significantly decreased the crypt depth, and increased the villus height and villus height-to-crypt depth ratio (VCR) of the jejunum (P < 0.05, Fig. 2C). At 28 d of age, maternal stevioside supplementation also increased the jejunal villus height and VCR (P < 0.05, Fig. 2 D). However, there was no significant differences in the density of goblet cells between the 2 groups on both 1 and 28 d (P > 0.05). Moreover, the immunostainings of PCNA on the jejunum of chicken offspring were shown in Fig. 2E and H. The number of PCNA-positive cells in the crypt and villus was counted, respectively. Maternal stevioside supplementation increased the ratio of PCNA-positive cells in the jejunal crypt of newly-hatched chicken offspring (P < 0.05, Fig. 2 F). The results of Western blotting also showed a higher PCNA protein expression in the jejunum of the STE group on 1 d of age (P < 0.05, Fig. 2 G). In addition, there was no difference in the jejunal proliferation between the 2 groups on 28 d (P > 0.05, Fig. 2I and J).
Fig. 2.
Effects of maternal stevioside supplementation on the jejunal morphology and proliferation of chicken offspring. (A) The representative images of hematoxylin and eosin (H&E) staining on the jejunum. (B) The representative images of Alcian Blue-periodic acid Schiff (AB-PAS) staining on the jejunum. (C and D) Villus height, crypt depth, villus height-to-crypt depth ratio and the number of goblet cells per 100 μm jejunum in the chicken offspring on 1 and 28 d (n = 8). (E and H) The representative images of immunostaining of proliferating cell nuclear antigen (PCNA) on the jejunum of chicken offspring on 1 and 28 d. (F and I) Ratio of PCNA-positive cells in the jejunal crypt and villus on 1 and 28 d (n = 8). (G and J) Western blotting of PCNA in the jejunum on 1 and 28 d (n = 6). Data are presented as mean value ± SEM. Values with an asterisk represent statistically significant differences (P < 0.05). CON, the offspring of breeder hens fed a basal diet; STE, the offspring of breeder hens fed a basal diet supplemented with 250 mg/kg stevioside. Effect size and statistical power of parameters in Fig. 2 are shown in Appendix Table 6.
3.3. Overview of jejunal RNA-seq data
RNA-seq was performed to explore the gene expression differences in the jejunum of the chicken offspring. The RNA-seq data of the jejunum is shown in Table 1. Generally, 39358268 to 46573508 paired-end clean reads were obtained for 16 RNA-Seq libraries after removing the low-quality reads. The percentages of Q20 and Q30 for all samples were higher than 97.55% and 93.61%, respectively. The mapping rates ranged from 89.77% to 93.45% for all 16 samples. This sequencing data suggested that the RNA-Seq libraries of all 16 samples were of good quality and could be analyzed further.
Table 1.
RNA sequencing data of the jejunum.
| Sample | Raw reads number | Clean reads number | Clean bases | Q20 | Q30 | Mapping rate |
|---|---|---|---|---|---|---|
| 1 d | ||||||
| CON1 | 46150560 | 44726908 | 6.71G | 97.76% | 94.32% | 91.58% |
| CON2 | 40182190 | 39358268 | 5.90G | 97.92% | 94.69% | 91.93% |
| CON3 | 44797696 | 43702686 | 6.56G | 97.84% | 94.48% | 91.61% |
| CON4 | 45710606 | 44721016 | 6.71G | 97.77% | 94.35% | 91.27% |
| STE1 | 46210396 | 45039490 | 6.76G | 98.12% | 94.88% | 93.45% |
| STE2 | 45138352 | 44167082 | 6.63G | 98.05% | 94.77% | 93.11% |
| STE3 | 45562494 | 44456146 | 6.67G | 98.00% | 94.64% | 91.04% |
| STE4 | 45991338 | 44948946 | 6.74G | 97.90% | 94.45% | 92.49% |
| 28 d | ||||||
| CON1 | 41740430 | 40854062 | 6.13G | 97.55% | 93.61% | 93.09% |
| CON2 | 44908428 | 43993418 | 6.60G | 97.61% | 94.12% | 89.77% |
| CON3 | 45161400 | 43970600 | 6.60G | 97.63% | 94.12% | 90.82% |
| CON4 | 46573508 | 45623476 | 6.84G | 97.74% | 94.20% | 92.24% |
| STE1 | 41828024 | 40677234 | 6.10G | 98.18% | 95.12% | 92.67% |
| STE2 | 41629264 | 40292636 | 6.04G | 98.09% | 94.88% | 92.20% |
| STE3 | 44221322 | 43026038 | 6.45G | 98.05% | 94.86% | 92.37% |
| STE4 | 46044936 | 44620052 | 6.69G | 97.87% | 94.61% | 90.65% |
CON, the offspring of breeder hens fed a basal diet; STE, the offspring of breeder hens fed a basal diet supplemented with 250 mg/kg stevioside.
3.4. Differentially expressed genes
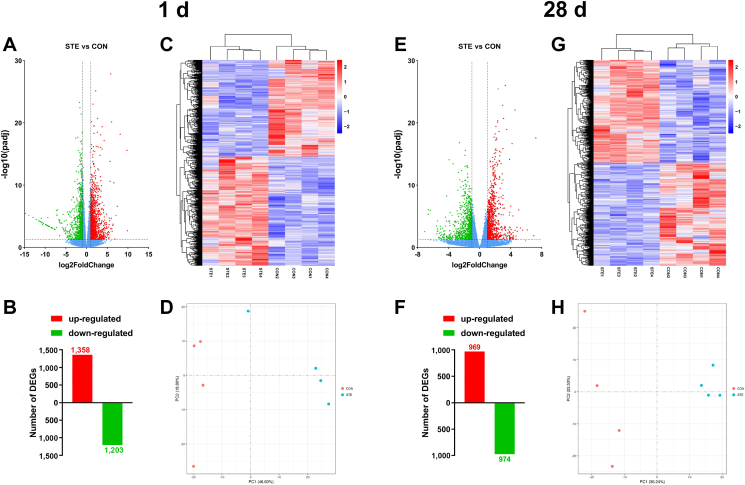
The number of differentially expressed genes is shown in Fig. 3. Compared with the CON group, a total of 2,561 and 1,943 DEGs were identified in the STE group on 1 and 28 d, respectively (Fig. 3A, B, E and F, adjusted P < 0.05). On 1 d, 1,358 genes were up-regulated and 1,203 genes were down-regulated in the STE group. On 28 d, 969 genes were up-regulated and 974 genes were down-regulated in the STE group. The cluster analysis with the DEGs, as indicated by the Heatmap plot, further suggested an obvious separation between the CON and STE groups (Fig. 3C and G). All information of DEGs were shown in Appendix Data 1 and 2. The principal component analysis also revealed that the samples between groups were scattered (Fig. 3D and H).
Fig. 3.
Analysis of the jejunal transcriptome data in the chicken offspring. (A and E) Volcano plot of differentially expressed genes (DEGs) in the jejunum of chicken offspring on 1 and 28 d. (B and F) Number of up- and down-regulated DEGs on 1 and 28 d. (C and G) Heatmap plot of DEGs between the STE and CON groups on 1 and 28 d. (D and H) Plots of Principal Component Analysis (PCA) on 1 and 28 d. CON, the offspring of breeder hens fed a basal diet; STE, the offspring of breeder hens fed a basal diet supplemented with 250 mg/kg stevioside. The data of RNA-Seq were analyzed using randomly selected 4 samples from each group.
3.5. GO and KEGG pathway analysis
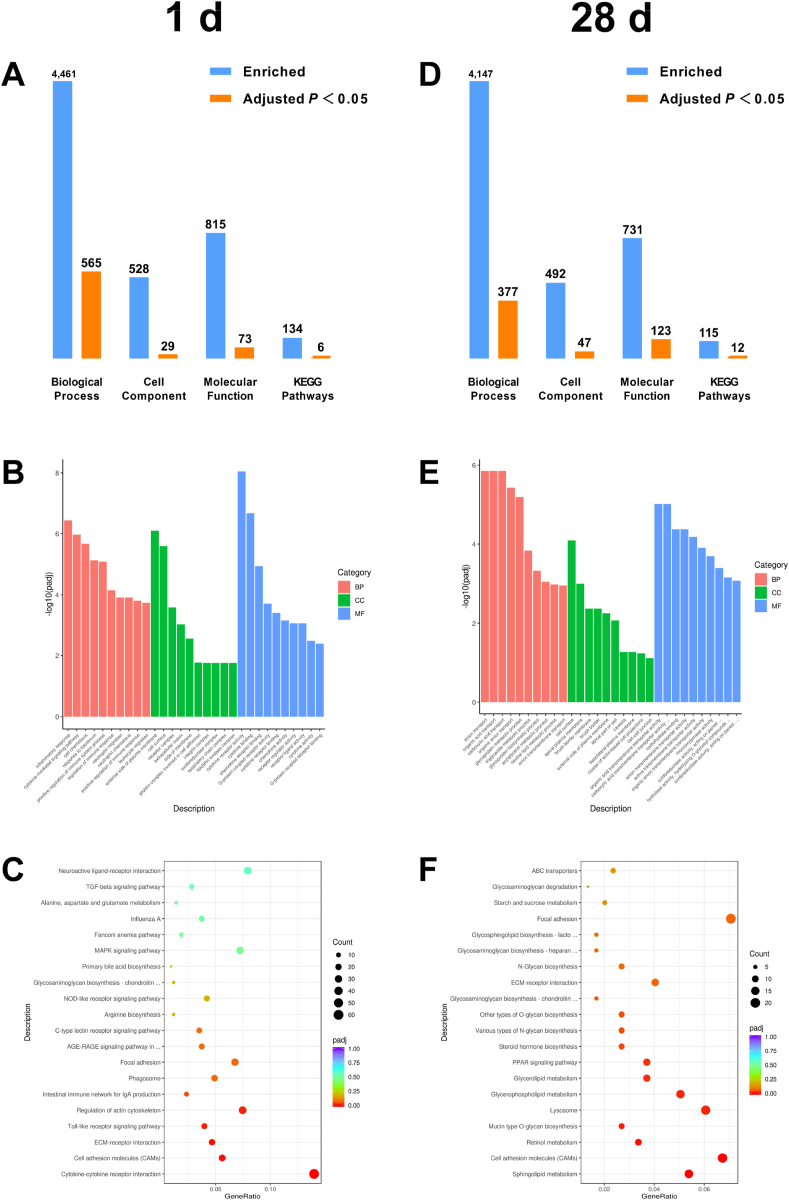
To better understand the signaling pathways that regulate the jejunal immune functions of maternal stevioside supplementation, we conducted GO and KEGG pathway analysis using the DEGs between the CON and STE groups. As shown in Fig. 4 A and D, a total of 565 and 377 Biological Process, 29 and 47 Cell Component, 73 and 123 Molecular Function, and 6 and 12 KEGG pathways were significantly enriched in the jejunum of chicken offspring on 1 and 28 d, respectively (adjusted P < 0.05). The top 10 notably enriched items of Biological Process, Cell Component and Molecular Function were presented in Fig. 4B and E. The top 20 notably enriched items of KEGG pathway analysis were shown in Fig. 4C and F. In the jejunum of chicken offspring on 1 d, the DEGs were significantly enriched into the KEGG pathway of Cytokine–cytokine receptor interaction, Cell adhesion molecules, ECM-receptor interaction, Toll-like receptor signaling pathway, Regulation of actin cytoskeleton and Intestinal immune network for IgA production. In the jejunum of chicken offspring on 28 d, the DEGs were significantly enriched into the KEGG pathway of sphingolipid metabolism, cell adhesion molecules, retinol metabolism, mucin type O-glycan biosynthesis, lysosome, glycerophospholipid metabolism, glycerolipid metabolism, PPAR signaling pathway, steroid hormone biosynthesis, various types of N-glycan biosynthesis, other types of O-glycan biosynthesis and glycosaminoglycan biosynthesis–chondroitin sulfate/dermatan sulfate. All information regarding GO and KEGG pathway analysis was shown in Appendix Data 3 to 6.
Fig. 4.
Gene ontology (GO) and a Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis in the jejunum of chicken offspring. (A and D) The number of significantly enriched pathways based on GO and KEGG analysis in the jejunum of chicken offspring on 1 and 28 d. (B and E) The top 10 enriched items in the GO database on 1 and 28 d. (C and F) The top 20 enriched KEGG signal pathways on 1 and 28 d. The size of the dots indicates the numbers of differentially expressed genes (DEGs), and the color of the dots represents the adjusted P-value.
3.6. Jejunal gene expressions
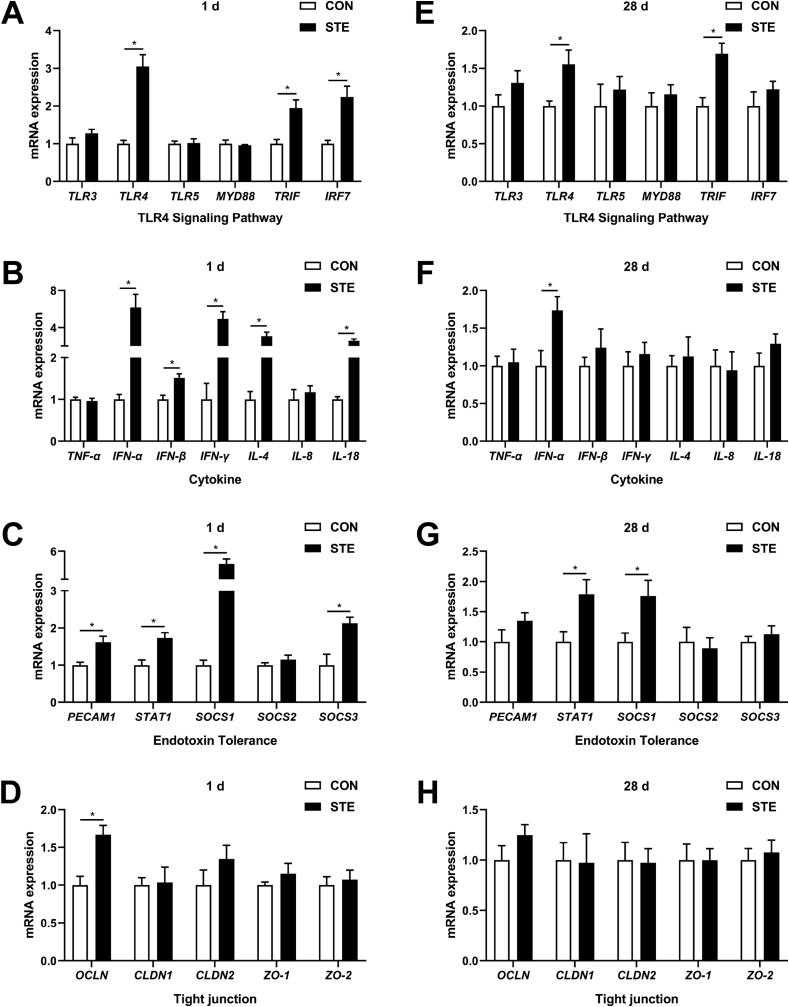
According to the data of RNA-Seq, we performed qRT-PCR to detect the transcription expressions of genes related to TLR4 signaling pathway, cytokine, endotoxin tolerance and tight junction in the jejunum of chicken offspring. At 1 d of age, maternal stevioside supplementation significantly increased the transcription expression of TLR4, toll like receptor adaptor molecule 1 (TRIF), interferon regulatory factor 7 (IRF7), IFN-α, IFN-β, IFN-γ, interleukin 4 (IL-4), interleukin 18 (IL-18), platelet and endothelial cell adhesion molecule 1 (PECAM1), signal transducer and activator of transcription 1 (STAT1), SOCS1, suppressor of cytokine signaling 3 (SOCS3) and occludin (OCLN) in the jejunum (P < 0.05, Fig. 5A–D). At 28 d of age, maternal stevioside supplementation markedly enhanced the expression of TLR4, TRIF, IFN-α, STAT1 and SOCS1 in the jejunum (P < 0.05, Fig. 5 E–H).
Fig. 5.
Effects of maternal stevioside supplementation on the gene expression in the jejunum of chicken offspring. (A and E) Relative mRNA expression of genes related with TLR4 signaling pathways on 1 and 28 d. (B and F) Relative mRNA expression of genes related with cytokine on 1 and 28 d. (C and G) Relative mRNA expression of genes related with endotoxin tolerance on 1 and 28 d. (D and H) Relative mRNA expression of genes related with tight junction on 1 and 28 d. Data are presented as mean values ± SEM (n = 8). Values with an asterisk represent statistically significant differences (P < 0.05). CON, the offspring of breeder hens fed a basal diet; STE, the offspring of breeder hens fed a basal diet supplemented with 250 mg/kg stevioside. TLR3 = toll like receptor 3; TLR4 = toll like receptor 4; TLR5 = toll like receptor 5; MYD88 = myeloid differentiation primary response gene 88; TRIF = toll like receptor adaptor molecule 1; IRF7 = interferon regulatory factor 7; TNF-α = tumor necrosis factor-alpha; IFN-α = interferon alpha; IFN-β = interferon beta; IFN-γ = interferon gamma; IL-4 = interleukin 4; IL-8 = interleukin 8; IL-18 = interleukin 18; PECAM1 = platelet and endothelial cell adhesion molecule 1; STAT1 = signal transducer and activator of transcription 1; SOCS1 = suppressor of cytokine signaling 1; SOCS2 = suppressor of cytokine signaling 2; SOCS3 = suppressor of cytokine signaling 3; OCLN = occludin; CLDN1 = claudin-1; CLDN2 = claudin-2; ZO-1 = tight junction protein 1; ZO-2 = tight junction protein 2. Effect size and statistical power of parameters in Fig. 5 are shown in Appendix Tables 7 and 8.
3.7. Serum and jejunal immune parameters
To assess the effects of maternal stevioside supplementation on the immune function, we detected the sIgA concentration within the jejunal mucosa, serum immunoglobulin and cytokine concentrations, and the serum antibody titer of NDV in the chicken offspring on 28 d (Table 2). The results showed that maternal stevioside supplementation significantly increased the sIgA concentration in the jejunal mucosa (P < 0.05). Chicken offspring with maternal stevioside supplementation also exhibited elevated serum concentrations of IgA, IFN-α and IFN-γ (P < 0.05). Furthermore, the serum antibody titer of NDV was higher in the STE group compared with that in the CON group (P < 0.05).
Table 2.
Effects of maternal stevioside supplementation on the immunoglobulin production, serum cytokine concentrations and antibody titer of chicken offspring on 28 d.
| Treatments |
||||||
|---|---|---|---|---|---|---|
| Item | CON | STE | SEM | P-value | Effect size | Statistical power |
| Jejunal mucosa | ||||||
| sIgA, ng/mg prot | 227.25 | 290.88∗ | 16.31 | 0.017 | 1.356 | 0.713 |
| Serum | ||||||
| IgA, μg/mL | 153.63 | 208.38∗ | 8.60 | <0.001 | 2.247 | 0.986 |
| IgG, μg/mL | 957.88 | 1,125.75 | 72.21 | 0.125 | 0.817 | 0.331 |
| IFN-α, pg/mL | 84.91 | 122.82∗ | 6.60 | 0.001 | 2.029 | 0.965 |
| IFN-γ, pg/mL | 167.52 | 197.34∗ | 8.98 | 0.036 | 1.162 | 0.581 |
| Serum antibody titer | ||||||
| NDV | 3.14 | 4.80∗ | 0.46 | 0.026 | 1.479 | 0.785 |
sIgA = secretory immunoglobulin A; IgA = immunoglobulin A; IgG = immunoglobulin G; IFN-α = interferon alpha; IFN-γ = interferon gamma; NDV = Newcastle Disease Virus.
CON, the offspring of breeder hens fed a basal diet; STE, the offspring of breeder hens fed a basal diet supplemented with 250 mg/kg stevioside.
Data were analyzed using each sampled chicken as the experimental unit (n = 8).
Values with an asterisk represent statistically significant differences (P < 0.05).
3.8. Description of the 16S rRNA gene sequencing data
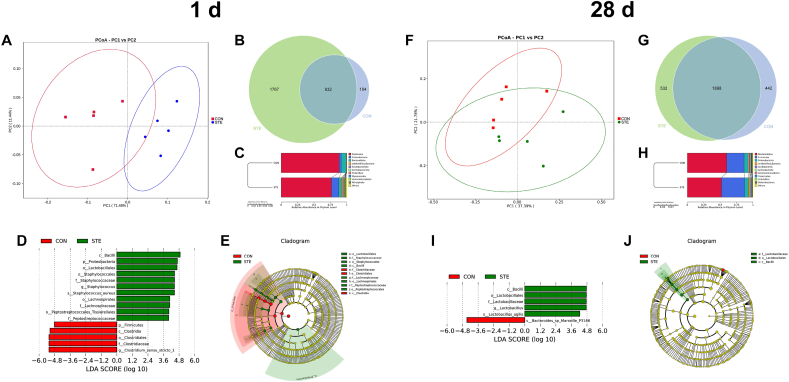
To explore the effects of maternal stevioside supplementation on the gut microbiota, we performed 16S rRNA gene sequencing on the meconium and cecal digesta of chicken offspring on 1 and 28 d, respectively. PCoA of the OTUs revealed that both meconial and cecal microbiota of the STE group was separated from the CON group (Fig. 6A and F). At 1 d of age, Venn diagram analysis revealed that 932 OTUs were overlapped between the CON and STE groups. There were 1,767 unique OTUs in the STE group, whereas 184 unique OTUs in the CON group (Fig. 6 B). At 28 d of age, Venn diagram analysis revealed that 1,898 OTUs were overlapped between the CON and STE groups. There were 532 unique OTUs in the STE group, whereas 442 unique OTUs in the CON group (Fig. 6 G). UPGMA analysis also showed that meconial and cecal microbiota of the STE group exhibited clear difference with the CON group (Fig. 6C and H).
Fig. 6.
Analysis of the gut microbiota in the chicken offspring. (A and F) Plots of Principal Co-ordinates Analysis (PCoA) on 1 and 28 d. (B and G) Venn diagram of operational taxonomic units (OTUs) on 1 and 28 d. (C and H) The results of unweighted pair-group method with arithmetic means (UPGMA) on 1 and 28 d. (D and J) Different structures of gut microbiota according to the linear discriminant analysis effect size (LEfSe) analysis on 1 and 28 d. (E and J) Cladogram plot of the biomarkers the CON and STE groups on 1 and 28 d. CON, the offspring of breeder hens fed a basal diet; STE, the offspring of breeder hens fed a basal diet supplemented with 250 mg/kg stevioside.
3.9. Structure of the meconial and cecal microbiota
The top 10 most abundant bacteria at the order level in the meconium and cecal digesta are presented in Table 3, Table 4. At 1 d of age, compared with the CON group, maternal stevioside supplementation markedly increased the relative abundance of Lactobacillales, Staphylococcales, Peptostreptococcales-Tissierellales, Lachnospirales, Oscillospirales, Burkholderiales and Rhizobiales, whereas it reduced the abundance of Clostridiales (P < 0.05). At 28 d of age, the STE group had higher relative abundance of Lactobacillales compared with the CON group (P < 0.05). No significant differences were observed in the other bacteria between the 2 groups (P > 0.05).
Table 3.
The top 10 most abundant bacteria at the order level in the meconium of chicken offspring on 1 d (%).
| Treatments |
||||||
|---|---|---|---|---|---|---|
| Item | CON | STE | SEM | P-value | Effect size | Statistical power |
| Clostridiales | 71.58 | 39.48∗ | 4.42 | 0.001 | 3.119 | 0.990 |
| Lactobacillales | 10.16 | 18.36∗ | 2.10 | 0.033 | 1.624 | 0.616 |
| Bacteroidales | 7.21 | 3.36 | 1.17 | 0.071 | 1.317 | 0.449 |
| Enterobacterales | 2.82 | 7.82 | 2.83 | 0.284 | 0.726 | 0.174 |
| Staphylococcales | 2.02 | 7.88∗ | 0.46 | <0.001 | 5.282 | 1.000 |
| Peptostreptococcales-Tissierellales | 1.68 | 3.62∗ | 0.39 | 0.008 | 2.213 | 0.864 |
| Lachnospirales | 0.95 | 3.51∗ | 0.63 | 0.021 | 1.805 | 0.706 |
| Oscillospirales | 0.54 | 1.46∗ | 0.15 | 0.003 | 2.701 | 0.961 |
| Burkholderiales | 0.16 | 1.30∗ | 0.14 | <0.001 | 3.456 | 0.997 |
| Rhizobiales | 0.05 | 0.85∗ | 0.19 | 0.042 | 1.533 | 0.567 |
CON, the offspring of breeder hens fed a basal diet; STE, the offspring of breeder hens fed a basal diet supplemented with 250 mg/kg stevioside.
Data were analyzed using randomly selected sampled chicken as the experimental unit (n = 5).
Values with an asterisk represent statistically significant differences (P < 0.05).
Table 4.
The top 10 most abundant bacteria at the order level in the cecal digesta of chicken offspring on 28 d (%).
| Treatments |
||||||
|---|---|---|---|---|---|---|
| Item | CON | STE | SEM | P-value | Effect size | Statistical power |
| Bacteroidales | 58.63 | 50.31 | 4.35 | 0.248 | 0.789 | 0.196 |
| Clostridiales | 16.72 | 14.42 | 1.18 | 0.205 | 0.870 | 0.229 |
| Selenomonadales | 5.23 | 4.69 | 1.07 | 0.731 | 0.225 | 0.061 |
| Unidentified_Gammaproteobacteria | 1.78 | 1.55 | 0.43 | 0.720 | 0.235 | 0.063 |
| Lactobacillales | 1.77 | 15.49∗ | 2.89 | 0.025 | 1.734 | 0.672 |
| Campylobacterales | 1.50 | 1.90 | 0.41 | 0.520 | 0.427 | 0.092 |
| Cytophagales | 0.97 | 0.68 | 0.20 | 0.350 | 0.627 | 0.142 |
| Pseudomonadales | 0.65 | 0.75 | 0.43 | 0.878 | 0.100 | 0.052 |
| Aeromonadales | 0.39 | 0.48 | 0.25 | 0.828 | 0.142 | 0.055 |
| Enterobacteriales | 0.23 | 0.81 | 0.15 | 0.061 | 1.381 | 0.484 |
CON, the offspring of breeder hens fed a basal diet; STE, the offspring of breeder hens fed a basal diet supplemented with 250 mg/kg stevioside.
Data were analyzed using each randomly selected sampled chicken as the experimental unit (n = 5).
Values with an asterisk represent statistically significant differences (P < 0.05).
Additionally, the LEfSe analysis was also conducted to detect distinctive bacterium between the CON and STE groups. In the newly-hatched chicken offspring, the STE group exhibited a higher relative abundance of Bacilli, Proteobacteria, Lactobacillales, Staphylococcaceae, Staphylococcus, Staphylococcus_aureus, Lachnospiraceae, Peptostreptococcales-Tissierellales and Peptostreptococcaceae; whereas it had a reduced abundance of Firmicutes, Clostridia, Clostridiales, Clostridiaceae and Clostridium_sensu_stricto_1. In the chicken offspring on 28 d, maternal stevioside supplementation increased the relative abundance of Bacilli, Lactobacillales, Lactobacillaceae, Lactobacillus and Lactobacillus_agilis; whereas the abundance of Bacteroides_sp_Marseille_P3166 was higher in the CON group.
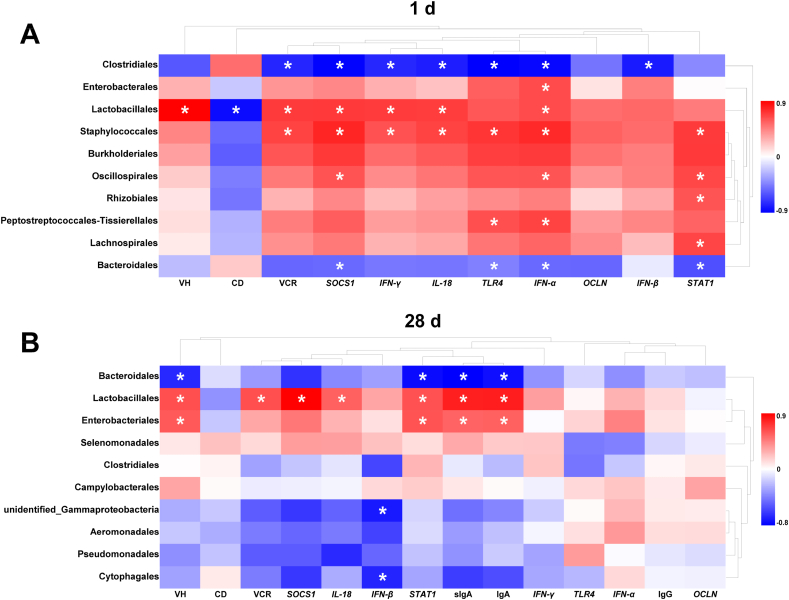
3.10. Correlation between gut microbiota (at order level) and intestinal immune function-related parameters
Pearson correlation analysis was performed to explore the potential relationship between gut microbiota (at the order level) and intestinal immune function-related parameters, including the intestinal morphology, immunoglobulin concentrations, and gene expression involving the TLR4 signaling pathway, cytokine, endotoxin tolerance and tight junction (Fig. 7). In the newly-hatched chicken offspring, Clostridiales and Bacteroidales showed a significant negative correlation with the expression of jejunal SOCS1, TLR4 and IFN-α; Clostridiales was also negatively correlated with VCR and the expression of IFN-γ, IL-18 and IFN-β. It was worth noting that Lactobacillales was negatively correlated with CD, but positively correlated with VH. Both Lactobacillales and Staphylococcales showed a significantly positive correlation with VCR and the expression of jejunal SOCS1, IFN-γ, IL-18 and IFN-α. Oscillospirales was positively correlated with SOCS1 and IFN-α expression, and Peptostreptococcales-Tissierellales had a positive correlation with TLR4 and IFN-α expression. Staphylococcales, Oscillospirales, Rhizobiales and Lachnospirales were positively correlated with the expression of STAT1. In chicken offspring at 28 d of age, Bacteroidales was negatively correlated with VH, STAT1 expression, sIgA and IgA concentrations. Additionally, both Unidentified_Gammaproteobacteria and Cytophagales were negatively correlated with IFN-β expression; whereas Lactobacillales showed a significantly positive correlation with VCR, SOCS1 and IL-18 expression. Finally, both Lactobacillales and Enterobacteriales showed a significantly positive correlation with VH, STAT1 expression, sIgA and IgA concentrations.
Fig. 7.
Heatmap of the correlation analysis between gut microbiota (at order level) and intestinal immune-related factors on (A and B) 1 and 28 d. Positive and negative correlations are shown by the red and blue matrices, respectively. The color intensity shows the Pearson correlation coefficient (r) value in each matrix. The asterisk represents a significant correlation between the 2 parameters (P < 0.05). VH = villus height; CD = crypt depth; VCR = villus height-to-crypt depth ratio; SOCS1 = suppressor of cytokine signaling 1; IFN-γ = interferon gamma; IL-8 = interleukin 8; TLR4 = toll like receptor 4; IFN-α = interferon alpha; OCLN = occludin; IFN-β = interferon beta; STAT1 = signal transducer and activator of transcription 1; sIgA = secretory immunoglobulin A; IgA = immunoglobulin A; IgG = immunoglobulin G.
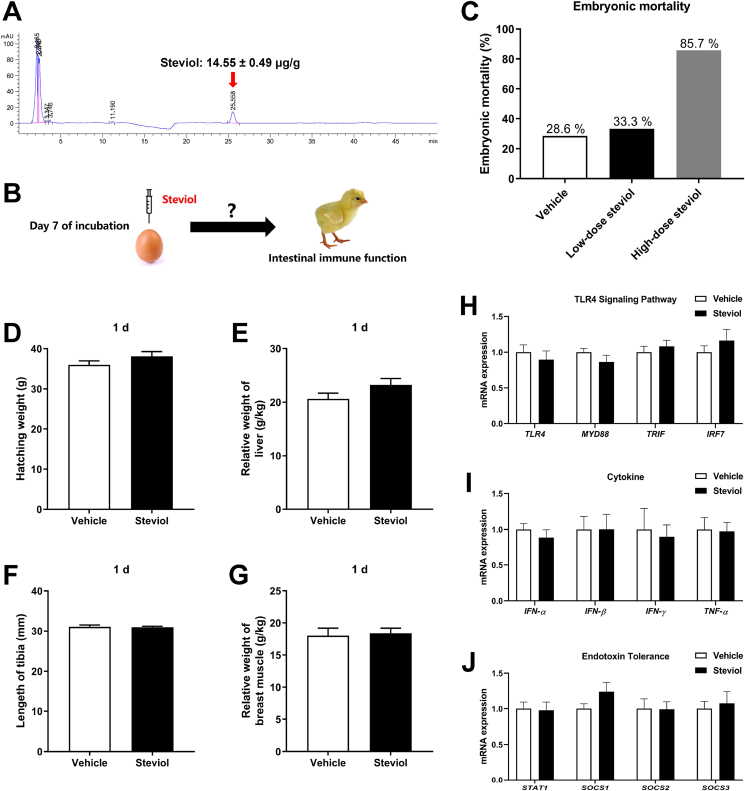
3.11. In ovo injection with steviol
With HPLC analysis, it was determined that steviol could deposit in the egg yolk through maternal stevioside supplementation. The amount of steviol deposited in the egg yolk was 14.55 ± 0.49 μg steviol per g egg yolk (about 0.29 mg/egg) (Fig. 8 A). To investigate whether the altered intestinal immune function induce by maternal stevioside supplementation was caused by steviol deposited in the yolk during the embryonic development, we performed an in ovo injection experiment (Fig. 8 B). The results showed that the in ovo injection with low-dose steviol (0.3 mg/egg) had similar embryonic mortality with the Vehicle group, but the injection with high-dose steviol (3 mg/egg) resulted in severe embryonic death (Fig. 8C). Hence, we performed the following experiment with the Vehicle group and the low-dose steviol group (Steviol). The results showed that the in ovo injection with steviol had no effect on the embryonic development as revealed by similar hatching weight, length of tibia and relative weight of liver and breast muscle (P > 0.05, Fig. 8D–G). Moreover, the in ovo injection with steviol did not alter the transcription expression levels of genes related with TLR4 signaling pathway, cytokine and endotoxin tolerance (P > 0.05, Fig. 8H–J).
Fig. 8.
Effects of in ovo injection with steviol on embryonic development and intestinal innate immune function. (A) Concentration of steviol deposited in the egg yolk after stevioside supplementation reveled by high-performance liquid chromatography (HPLC). The amount of steviol was shown to be 14.55 ± 0.49 μg steviol per g egg yolk. (B) The scheme of in ovo injection experiment. (C) The embryonic mortality after in ovo injection with steviol. (D) Hatching weight. (E) Relative weight of liver. (F) Length of tibia. (G) Relative weight of breast muscle. (H) Relative mRNA expression of genes related with TLR4 signaling pathways in the jejunum. (I) Relative mRNA expression of genes related with cytokine in the jejunum. (J) Relative mRNA expression of genes related with endotoxin tolerance in the jejunum. The data apart from the embryonic mortality are presented as mean value ± SEM (n = 8). TLR4 = toll like receptor 4; MYD88 = myeloid differentiation primary response gene 88; TRIF = toll like receptor adaptor molecule 1; IRF7 = interferon regulatory factor 7; IFN-α = interferon alpha; IFN-β = interferon beta; IFN-γ = interferon gamma; TNF-α = tumor necrosis factor-alpha; STAT1 = signal transducer and activator of transcription 1; SOCS1 = suppressor of cytokine signaling 1; SOCS2 = suppressor of cytokine signaling 2; SOCS3 = suppressor of cytokine signaling 3. Effect size and statistical power of parameters in Fig. 8 are shown in Appendix Table 9.
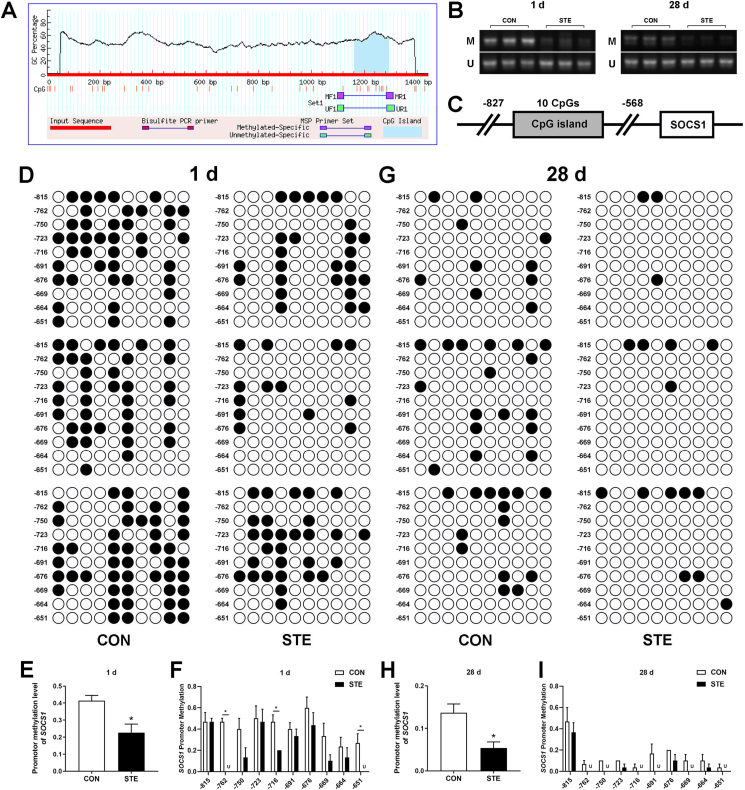
3.12. SOCS1 methylation status
As maternal stevioside supplementation had an intergenerational regulatory effect on the TLR4-IFNγ-SOCS1 pathway in the jejunum of chicken offspring, we investigated the promoter methylation of SOCS1 to explore the potential mechanism of improved intestinal immune function induced by maternal stevioside supplementation. Initially, MSP was performed to qualitatively evaluate the effect of maternal stevioside supplementation on the methylation of SOCS1 promoter (Fig. 9 A). The STE group exhibited an obvious down-regulation on the methylation level of SOCS1 (Fig. 9 B). We then performed the BSP to quantitatively evaluate the promoter methylation level of SOCS1 in the jejunum of chicken offspring. Methylation site mapping of cytosine-guanine (CpG) islands allowed analysis of the methylation levels of 10 CpGs in the jejunum (Fig. 9C). An overview of the methylation status of each CpG site was shown in Fig. 9 D and G. Maternal stevioside supplementation significantly reduced the overall methylation level of SOCS1 promoter in the jejunum on both 1 and 28 d (P < 0.05, Fig. 9 E and H). Of all the 10 CpGs analyzed, maternal stevioside supplementation specifically decreased the methylation level of 3 CpGs (−762, −716 and −651) in the jejunum of newly-hatched chicken offspring (P < 0.05, Fig. 9 F). At 28 d of age, maternal supplementation with stevioside had no significant effect on the specific CpGs compared with the CON group (P > 0.05), but led to the unmethylation of 6 CpGs (−762, −750, −716, −691, −669 and −651) in the jejunum (Fig. 9 I).
Fig. 9.
Effects of maternal stevioside supplementation on the methylation levels of suppressor of cytokine signaling 1 (SOCS1) promoter cytosine-guanine (CpG) island. (A) Primer sequences of SOCS1 for methylation-specific PCR (MSP). (B) SOCS1 methylation revealed by MSP in the jejunum of chicken offspring on 1 and 28 d. M, methylated; U, unmethylated. (C) Map of SOCS1 promoter CpG island. Numbers above the line are nucleotides relative to the transcription start site. (D and G) Bisulphite sequencing of SOCS1 promotor in the jejunum on 1 and 28 d. A black circle represents a methylated cytosine, and a blank circle represents an unmethylated cytosine. (E and H) The overall methylation level of the SOCS1 promoter CpG island on 1 and 28 d. (F and I) The methylation level of each CpG site in the SOCS1 promoter CpG island on 1 and 28 d. U means unmethylation in the specific CpG site. Data are presented as mean value ± SEM (n = 3). Values with an asterisk represent statistically significant differences (P < 0.05).
4. Discussion
Stevioside supplementation can improve the growth rate and reproductive performance of chickens (Jiang et al., 2020a, 2020b). Herein, we found that maternal stevioside supplementation could intergenerationally promote the embryonic development of chicken offspring. After 5 weeks’ supplementation with stevioside, steviol was deposited in egg yolk (about 0.29 mg/egg). However, in ovo injection with steviol did not alter the embryonic development of hatching chickens; an opposite effect to the results of maternal stevioside supplementation. Similarly, a previous study has also indicated that in ovo injection with steviol has no influence on embryonic development (Geuns et al., 2003). In addition, the unchanged growth performance and organ weight on 28 d suggested that maternal stevioside supplementation induced a potential growth promoting effect on chicken offspring which might only exist in the embryonic stage. Maternal nutrition could significantly alter the epigenome of the offspring (Dunislawska et al., 2021). Developmental changes of chicken offspring have been suggested to partly result from the intergenerational epigenetic inheritance induced by maternal factors (Hu et al., 2018; Jiang et al., 2020b). Moreover, other maternal effects including maternal hormone levels and mitochondrial inheritance could also affect embryonic development (Darras, 2019; Ruebel and Latham, 2020). Promotion of embryonic development may be the consequence of these maternal effects or maternal stevioside supplementation-induced changes on the other nutritional components in the eggs. Future molecular studies are required to identify the exact mechanisms.
The turnover of intestinal epithelium is strongly related with intestinal immune function, which is important for the protection against pathogenic invasions (Bussières-Marmen et al., 2018). Intestinal integrity can represent the extent of epithelial turnover. The VH is associated with the ability of digesting and absorbing nutrients. Higher VCR is generally believed to be an indicator of better intestinal epithelial turnover. In the current study, maternal stevioside supplementation could improve the intestinal turnover of chicken offspring as reflected by increased VH and VCR on both d 1 and d 28. The maintenance of intestinal integrity also requires the involvement of tight junction proteins (Singh et al., 2020). OCLN is one of the most important tight junction proteins, and its alteration is closely relevant to intestinal permeability and integrity (Al-Sadi et al., 2011). Increased transcription level of OCLN in the STE group provided evidence for a better intestinal integrity after maternal stevioside supplementation. In addition, intestinal proliferation is essential to the maintenance of homeostatic epithelial turnover (Bankaitis et al., 2018). It can be detected by the immunostaining of PCNA in the intestine. In this study, increased intestinal proliferation was found in the crypt, further suggesting an enhanced integrity of intestinal mucosa in the STE group. Interestingly, this effect only occurred in the neonatal chickens, whereas the intestinal proliferation in chickens at 28 d of age remained unchanged. The innate intestinal proliferation may be improved via maternal stevioside supplementation in the embryonic stage. Whether this effect was inherited through epigenetic modifications or not remains unclear. With the days increased, the intestinal epithelial cells could gradually reach a balance between the apoptosis and proliferation, causing similar cell proliferation between the 2 groups. Therefore, maternal stevioside supplementation might enhance the innate intestinal proliferation, which may in turn improve the intestinal integrity of chicken offspring.
The jejunal transcriptome data revealed that a number of immune response-related pathways were activated. Among them, Toll-like receptor signaling pathway is the main immune pathway in the intestine (Kawasaki and Kawai, 2014). On the intestinal epithelial cells, the ligation of TLRs by bacterial products can promote epithelial proliferation (Abreu, 2010). Increased transcription of TLR4 induced by maternal stevioside supplementation could be one of the mechanisms for enhanced intestinal proliferation. Additionally, there are 2 distinct downstream signaling pathways for TLR4: MyD88-dependent and TRIF-dependent pathways (Häcker et al., 2011). The MyD88-dependent pathway is responsible of regulating the NF-κB phosphorylation, which could result in a pro-inflammatory response (Neves et al., 2010); whereas the TRIF-dependent pathway is involved in mediating the activation of TNF receptor-associated factor 3, which could inhibit the MyD88-dependent pathway and improve intestinal immune function to defend against pathogens (Kawasaki and Kawai, 2014; Li et al., 2018). Our results showed that maternal stevioside supplementation could specifically activate the TRIF-dependent (rather than MyD88-dependent pathway) in the jejunum of chicken offspring. It is well known that the activation of TLR4–TRIF signaling would drive the production of interferons (Perkins et al., 2018). As a result, the transcription levels of various interferons in the neonatal chickens were increased. The immune system in neonatal animals is immature, which makes them highly susceptible to pathogenic infections (Liu et al., 2009). Neonatal deficiency of innate immunity involves low production of cytokines, including interferons (Maródi, 2006). Both IFN-α and IFN-β belong to type І interferons, and they are crucial to the regulation of innate immune responses (Ivashkiv and Donlin, 2014). IFN-γ is a vital mediator for the immunity of chickens with antimicrobial properties, and it also acts as a biomarker for the activation of the innate immune system (Liu et al., 2009; Yang et al., 2020). In the early response to microbial infection, interferons may upregulate the transcription of antiviral genes to interrupt the viral replication, providing sufficient time for the immune response to develop (Samuel, 2001). IL-18, known as IFN-γ-inducing factor, plays a pivotal role in activating immune responses (Hung et al., 2010). Under normal circumstances, the basal transcription levels of cytokines in chickens begin to continuously increase at the late embryonic stage until week 2 post–hatch (Maródi, 2006). In the present study, significantly higher neonatal cytokine responses suggested a stronger innate immune function in the intestine of the chicken offspring following maternal stevioside supplementation. Moreover, improved innate immunity might further promote sustainable immune modifications, which would result in reinforced intestinal immune function in the juvenile period, as shown by increased sIgA concentrations in the jejunal mucosa on 28 d. The secretion of sIgA is required for the maintenance of intestinal immune homeostasis (Cerutti and Rescigno, 2008). In the intestinal epithelial cells, IL-4 and IFN-γ can synergistically contribute to the transfer of sIgA in mucosal secretions (Le Bourgot et al., 2017). Enhanced basal expression levels of IL-4 and IFN-γ might also explain the elevated jejunal sIgA concentrations on 28 d. Furthermore, the antibody titer reflects the humoral immunity against a specific antigen. Thus, the serum IFN-γ levels could indicate the degree of immune response against NDV vaccine (Yang et al., 2019). Consistently, the variation in the tendency of serum IFN-γ and NDV antibody titer levels was similar in the present study. It has been suggested that IL-18 exerts adjuvant effect on the NDV vaccine (Hung et al., 2010). The increased serum antibody titer of NDV might be attributed to enhanced basal expression of IL-18 induced by maternal stevioside supplementation (Wang et al., 2015). Collectively, our results suggested that maternal stevioside supplementation could increase the levels of interferons to enhance the immune response of chicken offspring.
Increased production of interferons would subsequently activate the IFNγ-SOCS1 pathway, and this could cause an endotoxin tolerance-like immune response in the intestine (Baker et al., 2009; Chinen et al., 2011). SOCS1 has been found to regulate a variety of immune responses, including antiviral interferon response, T-lymphocyte cell differentiation and the inhibition of TLR4-MyD88-NF-κB signaling pathway (Croker et al., 2003; Hashimoto et al., 2011; Strebovsky et al., 2011). In innate immune cells, SOCS1 is essential for the regulation of the immune response (Alice et al., 2018). The lack of SOCS1 would thus down-regulate the cytokine production, disturb the immune response and consequently induce intestinal inflammation—suggesting a critical role for SOCS1 in the maintenance of intestinal mucosal homeostasis (Chinen et al., 2011; Pathak et al., 2015). In the present study, increased transcription level of SOCS1 induced by maternal stevioside supplementation was beneficial for both intestinal immune function and endotoxin tolerance. Intestinal immune function could also promote the turnover of intestinal epithelium, resulting in a better intestinal morphology (Cliffe et al., 2005; Garrett et al., 2010). Furthermore, a previous study has shown that the up-regulation of SOCS1 is accompanied by increased IgA concentration (Kanti Ghosh et al., 2016). This phenomenon was also found in the 28-day-old chicken offspring following maternal stevioside supplementation. Thus, our data demonstrates that maternal stevioside supplementation may up-regulate the expression of SOCS1 to improve intestinal immune function.
There is a close interaction between intestinal immune function and gut microbiota. The differentiation of intestinal innate immune cells requires the involvement of gut microbiota (Geva-Zatorsky et al., 2017). The present study reveals that the microbiota of neonatal chickens could be markedly affected by maternal stevioside supplementation, which may subsequently modulate the gut microbiota of juvenile chicken offspring. Of all changed bacteria, the abundance of Lactobacillales significantly increased on both 1 and 28 d. Acting as a probiotic for animals, Lactobacillales promotes gut health and has the potential for regulating intestinal immune function and maintaining microbial homeostasis (Valeriano et al., 2017). Higher abundance of Lactobacillales is beneficial for the inhibition of intestinal inflammation (Jiang et al., 2021b). Moreover, Lactic acid bacteria are positively correlated with many immune regulators. The production of antiviral interferons has been shown to be promoted by Lactic acid bacterial strains (Kawashima et al., 2018; Gutierrez-Merino et al., 2020). In Caco-2 cells, Lactobacillus can induce endotoxin tolerance through up-regulation of SOCS1 (Chiu et al., 2013). Lactobacillus spp. is also able to augment sIgA production (Wang et al., 2016; Cheng et al., 2020). In the present study, we consistently found a positive correlation between Lactobacillales abundance and immune-related factors. Increased Lactobacillales abundance resulting from maternal stevioside supplementation might contribute to an improved intestinal immune function. In addition, a previous study suggests a coordinated regulation between the gut microbiota and intestinal integrity (Kayama et al., 2020). In agreement, the results of the present correlation analysis have shown that maternal stevioside supplementation-induced modulation of gut microbiota is closely associated with improved intestinal integrity, which might enhance the intestinal immune function.
Although steviol deposited in the egg yolk may affect the phenotype of chicken offspring, our data suggested that in ovo injection with steviol had no influence on either embryonic development or intestinal immune function. Epigenetic modification is the most likely mechanism for the intergenerational effect of maternal nutritional intervention. The status of epigenetic modification is believed to be inherited through the germ cells, and this can lead to altered innate immune responses in chickens (Gou et al., 2012; Janke et al., 2015). DNA methylation is a core process of epigenetic modification. The transcription pattern of the gene can be stabilized by the methylation on its promotor region. The methyl-CpG-binding domain (MBD) is responsible for recognizing and binding with methylated sequences. MBD then competitively inhibits the combination of transcription factors, which in turn modulates the transcription expression of the gene. As a consequence, hyper- and hypo-methylation can generally down- and up-regulate the gene expression, respectively (Jones, 2012; Zou et al., 2012). Based on the current data, we considered SOCS1 as the core regulator for improved intestinal immune function. The methylation level of the promoter CpG island of SOCS1 was analyzed. At both growing stages of 1 and 28 d, maternal stevioside supplementation induced hypo-methylation on the promoter CpG island of SOCS1 in the jejunum of chicken offspring. One important characteristic of DNA methylation is high stability, and its inheritable effect on gene expression can last for a long time (Bonasio et al., 2010). Additionally, in female animals, epigenetic modifications can be transmitted to the offspring through the oocyte, and induce the alteration of phenotypic traits (Li and Albertini, 2013). In this study, with the development of follicles, deposited steviol might affect DNA methylation in the oocyte. Due to the fact that the whole egg is developed from the oocyte, altered DNA methylation could be carried to chicken offspring. Hence, for the first time, we found that maternal stevioside supplementation could regulate the intestinal DNA methylation. Increased transcription level of SOCS1 in the jejunum of chicken offspring might be attributed to the hypo-methylation on its promoter CpG island. It would also be interesting to investigate whether this improved intestinal immune function in the chicken offspring could be inherited to the next generation via non-DNA sequence-based inheritance.
5. Conclusion
In conclusion, maternal stevioside supplementation can improve the intestinal immune function of chicken offspring. This intergenerational effect is highly related with the modulation of gut microbiota and down-regulation of SOCS1 promoter CpG island in the jejunum. Our findings have provided a novel strategy for improving the intestinal immunity of chickens.
Author contributions
Jiang, Jingle: Conceptualization, Methodology, Investigation, Writing—original draft, Visualization. Lina Qi: Investigation, Writing—review & editing. Quanwei Wei: Resources. Fangxiong Shi: Writing - review & editing, Supervision, Project administration.
Declaration of competing interest
We declare that we have no financial and personal relationships with other people or organizations that can inappropriately influence our work, and there is no professional or other personal interest of any nature or kind in any product, service and/or company that could be construed as influencing the content of this paper.
Acknowledgements
This study was supported by Jiangsu Agriculture Science and Technology Innovation Fund (JASTIF, CX(18)2002).
Footnotes
Peer review under responsibility of Chinese Association of Animal Science and Veterinary Medicine.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.aninu.2022.06.002.
Appendix ASupplementary data
The following are the Supplementary data to this article:
References
- Abdel-Moneim A.E., Elbaz A.M., Khidr R.E., Badri F.B. Effect of in ovo inoculation of Bifidobacterium spp. on growth performance, thyroid activity, ileum histomorphometry, and microbial enumeration of broilers. Probiotics Antimicrob Proteins. 2020;12:873–882. doi: 10.1007/s12602-019-09613-x. [DOI] [PubMed] [Google Scholar]
- Abreu M.T. Toll-like receptor signalling in the intestinal epithelium: how bacterial recognition shapes intestinal function. Nat Rev Immunol. 2010;10:131–144. doi: 10.1038/nri2707. [DOI] [PubMed] [Google Scholar]
- Al-Sadi R., Khatib K., Guo S., Ye D., Youssef M., Ma T. Occludin regulates macromolecule flux across the intestinal epithelial tight junction barrier. Am J Physiol Gastrointest Liver Physiol. 2011;300:G1054–G1064. doi: 10.1152/ajpgi.00055.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alavala S., Sangaraju R., Nalban N., Sahu B.D., Jerald M.K., Kilari E.K., et al. Stevioside, a diterpenoid glycoside, shows anti-inflammatory property against Dextran Sulphate Sodium-induced ulcerative colitis in mice. Eur J Pharmacol. 2019;855:192–201. doi: 10.1016/j.ejphar.2019.05.015. [DOI] [PubMed] [Google Scholar]
- Alice A.F., Kramer G., Bambina S., Baird J.R., Bahjat K.S., Gough M.J., et al. Amplifying IFN-γ signaling in dendritic cells by CD11c-specific loss of SOCS1 increases innate immunity to infection while decreasing adaptive immunity. J Immunol. 2018;200:177–185. doi: 10.4049/jimmunol.1700909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkie T.N., Yitbarek A., Hodgins D.C., Kulkarni R.R., Taha-Abdelaziz K., Sharif S. Development of innate immunity in chicken embryos and newly hatched chicks: a disease control perspective. Avian Pathol. 2019;48:288–310. doi: 10.1080/03079457.2019.1607966. [DOI] [PubMed] [Google Scholar]
- Baker B.J., Akhtar L.N., Benveniste E.N. SOCS1 and SOCS3 in the control of CNS immunity. Trends Immunol. 2009;30:392–400. doi: 10.1016/j.it.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankaitis E.D., Ha A., Kuo C.J., Magness S.T. Reserve stem cells in intestinal homeostasis and injury. Gastroenterology. 2018;155:1348–1361. doi: 10.1053/j.gastro.2018.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Shira E., Friedman A. Development and adaptations of innate immunity in the gastrointestinal tract of the newly hatched chick. Dev Comp Immunol. 2006;30:930–941. doi: 10.1016/j.dci.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Bonasio R., Tu S., Reinberg D. Molecular signals of epigenetic states. Science. 2010;330:612–616. doi: 10.1126/science.1191078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussières-Marmen S., Vinette V., Gungabeesoon J., Aubry I., Pérez-Quintero L.A., Tremblay M.L. Loss of T-cell protein tyrosine phosphatase in the intestinal epithelium promotes local inflammation by increasing colonic stem cell proliferation. Cell Mol Immunol. 2018;15:367–376. doi: 10.1038/cmi.2016.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas-Grajales S., Ramos-Tovar E., Chávez-Estrada E., Alvarez-Suarez D., Hernández-Aquino E., Reyes-Gordillo K., et al. Antioxidant and immunomodulatory activity induced by stevioside in liver damage: in vivo, in vitro and in silico assays. Life Sci. 2019;224:187–196. doi: 10.1016/j.lfs.2019.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti A., Rescigno M. The biology of intestinal immunoglobulin A responses. Immunity. 2008;28:740–750. doi: 10.1016/j.immuni.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Tang S., Huang Y., Liang F., Fang Y., Pan S., et al. Lactobacillus casei-fermented blueberry pomace augments sIgA production in high-fat diet mice by improving intestinal microbiota. Food Funct. 2020;11:6552–6564. doi: 10.1039/d0fo01119c. [DOI] [PubMed] [Google Scholar]
- Chinen T., Komai K., Muto G., Morita R., Inoue N., Yoshida H., et al. Prostaglandin E2 and SOCS1 have a role in intestinal immune tolerance. Nat Commun. 2011;2:190. doi: 10.1038/ncomms1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu Y.H., Lu Y.C., Ou C.C., Lin S.L., Tsai C.C., Huang C.T., et al. Lactobacillus plantarum MYL26 induces endotoxin tolerance phenotype in Caco-2 cells. BMC Microbiol. 2013;13:190. doi: 10.1186/1471-2180-13-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliffe L.J., Humphreys N.E., Lane T.E., Potten C.S., Booth C., Grencis R.K. Accelerated intestinal epithelial cell turnover: a new mechanism of parasite expulsion. Science. 2005;308:1463–1465. doi: 10.1126/science.1108661. [DOI] [PubMed] [Google Scholar]
- Croker B.A., Krebs D.L., Zhang J.G., Wormald S., Willson T.A., Stanley E.G., et al. SOCS3 negatively regulates IL-6 signaling in vivo. Nat Immunol. 2003;4:540–545. doi: 10.1038/ni931. [DOI] [PubMed] [Google Scholar]
- Darras V.M. The role of maternal thyroid hormones in avian embryonic development. Front Endocrinol. 2019;10:66. doi: 10.3389/fendo.2019.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J., Dai R., Yang L., He C., Xu K., Liu S., et al. Inheritance and establishment of gut microbiota in chickens. Front Microbiol. 2017;8:1967. doi: 10.3389/fmicb.2017.01967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunislawska A., Pietrzak E., Wishna Kadawarage R., Beldowska A., Siwek M. Pre-hatching and post-hatching environmental factors related to epigenetic mechanisms in poultry. J Anim Sci. 2021 doi: 10.1093/jas/skab370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil R., Fraga M.F. Epigenetics and the environment: emerging patterns and implications. Nat Rev Genet. 2012;13:97–109. doi: 10.1038/nrg3142. [DOI] [PubMed] [Google Scholar]
- Garrett W.S., Gordon J.I., Glimcher L.H. Homeostasis and inflammation in the intestine. Cell. 2010;140:859–870. doi: 10.1016/j.cell.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuns J.M., Bruggeman V., Buyse J.G. Effect of stevioside and steviol on the developing broiler embryos. J Agric Food Chem. 2003;51:5162–5167. doi: 10.1021/jf020931p. [DOI] [PubMed] [Google Scholar]
- Geva-Zatorsky N., Sefik E., Kua L., Pasman L., Tan T.G., Ortiz-Lopez A., et al. Mining the human gut microbiota for immunomodulatory organisms. Cell. 2017;168:928–943. doi: 10.1016/j.cell.2017.01.022. e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong H.Z., Lang W.Y., Lan H.N., Fan Y.Y., Wang T.P., Chu Q.R., et al. Effects of laying breeder hens dietary β-carotene, curcumin, allicin, and sodium butyrate supplementation on the jejunal microbiota and immune response of their offspring chicks. Poultry Sci. 2020;99:3807–3816. doi: 10.1016/j.psj.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou Z., Liu R., Zhao G., Zheng M., Li P., Wang H., et al. Epigenetic modification of TLRs in leukocytes is associated with increased susceptibility to Salmonella enteritidis in chickens. PLoS One. 2012;7 doi: 10.1371/journal.pone.0033627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Merino J., Isla B., Combes T., Martinez-Estrada F., Maluquer De Motes C. Beneficial bacteria activate type-I interferon production via the intracellular cytosolic sensors STING and MAVS. Gut Microb. 2020;11:771–788. doi: 10.1080/19490976.2019.1707015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häcker H., Tseng P.H., Karin M. Expanding TRAF function: TRAF3 as a tri-faced immune regulator. Nat Rev Immunol. 2011;11:457–468. doi: 10.1038/nri2998. [DOI] [PubMed] [Google Scholar]
- Hashimoto M., Hiwatashi K., Ichiyama K., Morita R., Sekiya T., Kimura A., et al. SOCS1 regulates type I/type II NKT cell balance by regulating IFNgamma signaling. Int Immunol. 2011;23:165–176. doi: 10.1093/intimm/dxq469. [DOI] [PubMed] [Google Scholar]
- Hepworth M.R., Monticelli L.A., Fung T.C., Ziegler C.G., Grunberg S., Sinha R., et al. Innate lymphoid cells regulate CD4+ T-cell responses to intestinal commensal bacteria. Nature. 2013;498:113–117. doi: 10.1038/nature12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Sun Q., Hu Y., Hou Z., Zong Y., Omer N.A., et al. Corticosterone-induced lipogenesis activation and lipophagy inhibition in chicken liver are alleviated by maternal betaine supplementation. J Nutr. 2018;148:316–325. doi: 10.1093/jn/nxx073. [DOI] [PubMed] [Google Scholar]
- Hung L.H., Li H.P., Lien Y.Y., Wu M.L., Chaung H.C. Adjuvant effects of chicken interleukin-18 in avian Newcastle disease vaccine. Vaccine. 2010;28:1148–1155. doi: 10.1016/j.vaccine.2009.11.042. [DOI] [PubMed] [Google Scholar]
- Ivashkiv L.B., Donlin L.T. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P.T., Jawla O., Mohammed N.I., Ceesay K., Akemokwe F.M., Sonko B., et al. A novel nutritional supplement to reduce plasma homocysteine in nonpregnant women: a randomised controlled trial in the Gambia. PLoS Med. 2019;16 doi: 10.1371/journal.pmed.1002870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke R., Dodson A.E., Rine J. Metabolism and epigenetics. Annu Rev Cell Dev Biol. 2015;31:473–496. doi: 10.1146/annurev-cellbio-100814-125544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J., Liu S., Jamal T., Ding T., Qi L., Lv Z., et al. Effects of dietary sweeteners supplementation on growth performance, serum biochemicals, and jejunal physiological functions of broiler chickens. Poultry Sci. 2020;99:3948–3958. doi: 10.1016/j.psj.2020.03.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J., Qi L., Dai H., Hu C., Lv Z., Wei Q., et al. Dietary stevioside supplementation improves laying performance and eggshell quality through increasing estrogen synthesis, calcium level and antioxidant capacity of reproductive organs in aged breeder hens. Anim Feed Sci Technol. 2020;269 [Google Scholar]
- Jiang J., Qi L., Lv Z., Jin S., Wei X., Shi F. Dietary stevioside supplementation alleviates lipopolysaccharide-induced intestinal mucosal damage through anti-inflammatory and antioxidant effects in broiler chickens. Antioxidants. 2019;8:575. doi: 10.3390/antiox8120575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J., Qi L., Lv Z., Wei Q., Shi F. Dietary stevioside supplementation increases feed intake by altering the hypothalamic transcriptome profile and gut microbiota in broiler chickens. J Sci Food Agric. 2021;101:2156–2167. doi: 10.1002/jsfa.10838. [DOI] [PubMed] [Google Scholar]
- Jiang J., Qi L., Wei Q., Shi F. Maternal stevioside supplementation ameliorates intestinal mucosal damage and modulates gut microbiota in chicken offspring challenged with lipopolysaccharide. Food Funct. 2021;12:6014–6028. doi: 10.1039/d0fo02871a. [DOI] [PubMed] [Google Scholar]
- Jones P.A. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- Kabat A.M., Srinivasan N., Maloy K.J. Modulation of immune development and function by intestinal microbiota. Trends Immunol. 2014;35:507–517. doi: 10.1016/j.it.2014.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanti Ghosh A., Sinha D., Mukherjee S., Biswas R., Biswas T. IL-15 temporally reorients IL-10 biased B-1a cells toward IL-12 expression. Cell Mol Immunol. 2016;13:229–239. doi: 10.1038/cmi.2015.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki T., Kawai T. Toll-like receptor signaling pathways. Front Immunol. 2014;5:461. doi: 10.3389/fimmu.2014.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima T., Ikari N., Watanabe Y., Kubota Y., Yoshio S., Kanto T., et al. Double-stranded RNA derived from lactic acid bacteria augments Th1 immunity via interferon-β from human dendritic cells. Front Immunol. 2018;9:27. doi: 10.3389/fimmu.2018.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayama H., Okumura R., Takeda K. Interaction between the microbiota, epithelia, and immune cells in the Intestine. Annu Rev Immunol. 2020;38:23–48. doi: 10.1146/annurev-immunol-070119-115104. [DOI] [PubMed] [Google Scholar]
- Kelly D., Coutts A.G. Early nutrition and the development of immune function in the neonate. Proc Nutr Soc. 2000;59:177–185. doi: 10.1017/s0029665100000197. [DOI] [PubMed] [Google Scholar]
- Le Bourgot C., Le Normand L., Formal M., Respondek F., Blat S., Apper E., et al. Maternal short-chain fructo-oligosaccharide supplementation increases intestinal cytokine secretion, goblet cell number, butyrate concentration and Lawsonia intracellularis humoral vaccine response in weaned pigs. Br J Nutr. 2017;117:83–92. doi: 10.1017/S0007114516004268. [DOI] [PubMed] [Google Scholar]
- Li C., Guo S., Gao J., Guo Y., Du E., Lv Z., et al. Maternal high-zinc diet attenuates intestinal inflammation by reducing DNA methylation and elevating H3K9 acetylation in the A20 promoter of offspring chicks. J Nutr Biochem. 2015;26:173–183. doi: 10.1016/j.jnutbio.2014.10.005. [DOI] [PubMed] [Google Scholar]
- Li R., Albertini D.F. The road to maturation: somatic cell interaction and self-organization of the mammalian oocyte. Nat Rev Mol Cell Biol. 2013;14:141–152. doi: 10.1038/nrm3531. [DOI] [PubMed] [Google Scholar]
- Li Y., Lei X., Guo W., Wu S., Duan Y., Yang X., et al. Transgenerational endotoxin tolerance-like effect caused by paternal dietary Astragalus polysaccharides in broilers' jejunum. Int J Biol Macromol. 2018;111:769–779. doi: 10.1016/j.ijbiomac.2018.01.095. [DOI] [PubMed] [Google Scholar]
- Liu T., Nerren J., Liu M., Martens R., Cohen N. Basal and stimulus-induced cytokine expression is selectively impaired in peripheral blood mononuclear cells of newborn foals. Vaccine. 2009;27:674–683. doi: 10.1016/j.vaccine.2008.11.040. [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Luo X.C., Chen Z.H., Xue J.B., Zhao D.X., Lu C., Li Y.H., et al. Infection by the parasitic helminth Trichinella spiralis activates a Tas2r-mediated signaling pathway in intestinal tuft cells. Proc Natl Acad Sci U S A. 2019;116:5564–5569. doi: 10.1073/pnas.1812901116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahalak K.K., Firrman J., Tomasula P.M., Nuñez A., Lee J.J., Bittinger K., et al. Impact of steviol glycosides and erythritol on the human and Cebus apella gut microbiome. J Agric Food Chem. 2020;68:13093–13101. doi: 10.1021/acs.jafc.9b06181. [DOI] [PubMed] [Google Scholar]
- Maródi L. Innate cellular immune responses in newborns. Clin Immunol. 2006;118:137–144. doi: 10.1016/j.clim.2005.10.012. [DOI] [PubMed] [Google Scholar]
- Martens E.C., Neumann M., Desai M.S. Interactions of commensal and pathogenic microorganisms with the intestinal mucosal barrier. Nat Rev Microbiol. 2018;16:457–470. doi: 10.1038/s41579-018-0036-x. [DOI] [PubMed] [Google Scholar]
- Mokarram P., Mohammadi Z., Khazayel S., Dayong Z. Induction of epigenetic alteration by CPUK02, an ent- kaurenoid derivative of stevioside. Avicenna J Med Biotechnol (AJMB) 2017;9:13–18. [PMC free article] [PubMed] [Google Scholar]
- Molina-Barrios R.M., Avilés-Trejo C.R., Puentes-Mercado M.E., Cedillo-Cobián J.R., Hernández-Chavez J.F. Effect of dietary stevia-based sweetener on body weight and humoral immune response of broiler chickens. Vet World. 2021;14:913–917. doi: 10.14202/vetworld.2021.913-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves P., Lampropoulou V., Calderon-Gomez E., Roch T., Stervbo U., Shen P., et al. Signaling via the MyD88 adaptor protein in B cells suppresses protective immunity during Salmonella typhimurium infection. Immunity. 2010;33:777–790. doi: 10.1016/j.immuni.2010.10.016. [DOI] [PubMed] [Google Scholar]
- Pathak S., Grillo A.R., Scarpa M., Brun P., D'Incà R., Nai L., et al. MiR-155 modulates the inflammatory phenotype of intestinal myofibroblasts by targeting SOCS1 in ulcerative colitis. Exp Mol Med. 2015;47:e164. doi: 10.1038/emm.2015.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins D.J., Richard K., Hansen A.M., Lai W., Nallar S., Koller B., et al. Autocrine-paracrine prostaglandin E(2) signaling restricts TLR4 internalization and TRIF signaling. Nat Immunol. 2018;19:1309–1318. doi: 10.1038/s41590-018-0243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renwick A.G., Tarka S.M. Microbial hydrolysis of steviol glycosides. Food Chem Toxicol. 2008;46(Suppl 7):S70–S74. doi: 10.1016/j.fct.2008.05.008. [DOI] [PubMed] [Google Scholar]
- Ruebel M.L., Latham K.E. Listening to mother: long-term maternal effects in mammalian development. Mol Reprod Dev. 2020;87:399–408. doi: 10.1002/mrd.23336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel C.E. Antiviral actions of interferons. Clin Microbiol Rev. 2001;14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh V., Gowda C.P., Singh V., Ganapathy A.S., Karamchandani D.M., Eshelman M.A., et al. The mRNA-binding protein IGF2BP1 maintains intestinal barrier function by up-regulating occludin expression. J Biol Chem. 2020;295:8602–8612. doi: 10.1074/jbc.AC120.013646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slawinska A., Dunislawska A., Plowiec A., Radomska M., Lachmanska J., Siwek M., et al. Modulation of microbial communities and mucosal gene expression in chicken intestines after galactooligosaccharides delivery in ovo. PLoS One. 2019;14 doi: 10.1371/journal.pone.0212318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strebovsky J., Walker P., Lang R., Dalpke A.H. Suppressor of cytokine signaling 1 (SOCS1) limits NFkappaB signaling by decreasing p65 stability within the cell nucleus. Faseb J. 2011;25:863–874. doi: 10.1096/fj.10-170597. [DOI] [PubMed] [Google Scholar]
- Valeriano V.D., Balolong M.P., Kang D.K. Probiotic roles of Lactobacillus sp. in swine: insights from gut microbiota. J Appl Microbiol. 2017;122:554–567. doi: 10.1111/jam.13364. [DOI] [PubMed] [Google Scholar]
- Wang C., Li X., Zhang C., Wu T., Li Y., Cheng X. A eukaryotic expression plasmid carrying chicken interleukin-18 enhances the response to newcastle disease virus vaccine. Clin Vaccine Immunol. 2015;22:56–64. doi: 10.1128/CVI.00636-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Li Y., Xiao H., Shi Y., Le G.W., Sun J. Isolation of lactobacillus reuteri from Peyer's patches and their effects on sIgA production and gut microbiota diversity. Mol Nutr Food Res. 2016;60:2020–2030. doi: 10.1002/mnfr.201501065. [DOI] [PubMed] [Google Scholar]
- Weström B., Arévalo Sureda E., Pierzynowska K., Pierzynowski S.G., Pérez-Cano F.J. The immature gut barrier and its importance in establishing immunity in newborn mammals. Front Immunol. 2020;11:1153. doi: 10.3389/fimmu.2020.01153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Guo W., Li X., Liu Y., Li Y., Lei X., et al. Paternal chronic folate supplementation induced the transgenerational inheritance of acquired developmental and metabolic changes in chickens. Proc R Soc B-Biol Sci. 2019;286 doi: 10.1098/rspb.2019.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Yang P., Sifa D., Wen Z. Effect of dietary stevioside supplementation on growth performance, nutrient digestibility, serum parameters, and intestinal microflora in broilers. Food Funct. 2019;10:2340–2346. doi: 10.1039/c8fo01883a. [DOI] [PubMed] [Google Scholar]
- Yang H., Zhang P., Xu X., Chen X., Liu Q., Jiang C. The enhanced immunological activity of Paulownia tomentosa flower polysaccharide on Newcastle disease vaccine in chicken. Biosci Rep. 2019;39 doi: 10.1042/BSR20190224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Arslan M., Liu X., Song H., Du M., Li Y., et al. IFN-γ establishes interferon-stimulated gene-mediated antiviral state against Newcastle disease virus in chicken fibroblasts. Acta Biochim Biophys Sin. 2020;52:268–280. doi: 10.1093/abbs/gmz158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou X., Ma W., Solov'yov I.A., Chipot C., Schulten K. Recognition of methylated DNA through methyl-CpG binding domain proteins. Nucleic Acids Res. 2012;40:2747–2758. doi: 10.1093/nar/gkr1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.