Summary
With mass and flow cytometry, millions of single-cell profiles with dozens of parameters can be measured to comprehensively characterize complex tumor ecosystems. Here, we present scQUEST, an open-source Python library for cell type identification and quantification of tumor ecosystem heterogeneity in patient cohorts. We provide a step-by-step protocol on the application of scQUEST on our previously generated human breast cancer single-cell atlas using mass cytometry and discuss how it can be adapted and extended for other datasets and analyses.
For complete details on the use and execution of this protocol, please refer to Wagner et al. (2019).
Subject areas: Bioinformatics, Single Cell, Flow Cytometry/Mass Cytometry, Cancer
Graphical abstract

Highlights
-
•
Tumor ecosystems exhibit a strong degree of phenotypic heterogeneity
-
•
scQUEST facilitates the analysis of large cytometry datasets with millions of cells
-
•
scQUEST implements scores to quantify tumor heterogeneity based on phenotypic profiles
-
•
All scores are easily customizable allowing users to adapt them to their needs
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
With mass and flow cytometry, millions of single-cell profiles with dozens of parameters can be measured to comprehensively characterize complex tumor ecosystems. Here, we present scQUEST, an open-source Python library for cell type identification and quantification of tumor ecosystem heterogeneity in patient cohorts. We provide a step-by-step protocol on the application of scQUEST on our previously generated human breast cancer single-cell atlas using mass cytometry and discuss how it can be adapted and extended for other datasets and analyses.
Before you begin
Cancer is a tissue disease in which tumor cells and cells of the microenvironment form an ecosystem that drives tumor progression and therapy response. Current single-cell technologies generate highly multiplexed profiles for millions of cells and up to hundreds of patients, thus allowing unprecedented insights into the complexity of tumor ecosystem biology. However, new computational challenges arise from large datasets with millions of cells. These datasets can be prohibitive in terms of computational resources for available clustering algorithms that partition high-dimensional data into populations, thus hindering cell type identification and annotation. Another challenge is the quantification of different aspects of tumor heterogeneity. Although several computational scores that quantify heterogeneity from single-cell genomics, transcriptomics or proteomics measurements have been proposed in recent years (Kashyap et al., 2022; Martinelli and Rapsomaniki, 2022), data-driven approaches that directly learn tumor heterogeneity patterns from those measurements are largely missing.
The following protocol has been designed for mass cytometry data but can also be directly applied to flow cytometry data. Several common mass cytometry data preprocessing steps should be performed before starting this protocol, including data normalization, deconvolution of cellular barcodes, and signal compensation (Chevrier et al., 2018). Deconvolution and signal compensation are also important steps for flow cytometry data.
Data preprocessing
Timing: ∼1 day
-
1.
Normalization: During long mass cytometry data acquisitions, changes in instrument performance can introduce signal variation over time (Finck et al., 2013). To address this, metal-containing polystyrene beads are measured together with the biological samples and the bead-derived signature is then used to correct (normalize) occurring signal fluctuations post-acquisition (Chevrier et al., 2018).
-
2.
Deconvolution of sample barcodes: To minimize inter-sample antibody staining variation, sample multiplexing can be achieved through mass-tag barcoding or fluorescent cell barcoding (Krutzik and Nolan, 2006; Zunder et al., 2015). Here, unique combinations of barcoding reagents are used to stain individual samples, which can then be pooled for staining with an antibody master mix. Computational deconvolution of barcodes allows the direct comparison of signal intensities for dozens of samples (Chevrier et al., 2018).
-
3.
Signal compensation: Current mass cytometry and flow cytometry applications allow the simultaneous assessment of 40 and more antibody targets per cell. As reporters, mass cytometry employs heavy metal isotopes as antibody tags and flow cytometry uses fluorescent dyes. Both approaches suffer from signal crosstalk between detection channels and require signal compensation prior to data analysis. Channel crosstalk is more pronounced in flow cytometry due to the broad emission spectra of fluorochromes that lead to spillover effects, than in mass cytometry, where it is caused by isotopic impurities, oxidation, and instrument properties. In both approaches, single-stained controls are used to generate a compensation matrix, which can be applied to the data post-acquisition for signal compensation, e.g., using commercial software like FlowJoTM (BD Life Sciences) and Cytobank (Kotecha et al., 2010), or the R package CATALYST (Chevrier et al., 2018).
Key resources table
Step-by-step method details
Install scQUEST and download dataset
Timing: 15–30 min
scQUEST is a Python package that runs on all operating systems (Windows, Linux, and MAC OSX) with Python (ideally version 3.8) installed. To simplify the installation process, we recommend using Miniconda (key resources table), as described below. The minimal hardware requirements to complete this protocol largely depend on the dataset used. As an example, to successfully complete this protocol using the supplied mass cytometry dataset of ∼13.5 million cells, a system with 16 GB of RAM is recommended. A detailed Jupyter Notebook tutorial with step-by-step instructions to reproduce all results and figures of this protocol is supplied (key resources table). Although prior knowledge of popular Python libraries (e.g., numpy, matplotlib, pandas) is a plus, it is still possible to follow all steps of the protocol using the supplied code in the Jupyter Notebook. Furthermore, additional detailed tutorials demonstrate how users can customize the analysis to fit their dataset or question or interest (key resources table). scQUEST runs on top of AnnData, a Python package for handling annotated single-cell datasets. For users without prior experience with AnnData, we strongly recommend the corresponding paper (Virshup et al., 2021) (key resources table) to familiarize themselves with the structure of the AnnData object prior to executing this protocol. To simplify the use of this protocol, a pre-uploaded version of the mass cytometry dataset measured with the tumor epithelial cell-centric antibody panel from Wagner et al. (2019) is included in scQUEST as an AnnData object. The dataset measured with the immune cell-centric antibody panel from Wagner et al. (2019) is not included here and can be found in the original publication (key resources table). For users interested in using scQUEST with their own mass or flow cytometry data, we also provide a simple tutorial that illustrates how to load and process .fcs files, and construct an AnnData object (key resources table). We note that in our original paper Wagner et al. (2019), our analyses focused on patient samples for which both a tumor epithelial cell-centric and an immune cell-centric measurement was available, resulting in 144 breast tumor and 50 non-tumor tissue samples. Since the original tumor epithelial cell centric-only measurements involved 23 additional tumor samples and nine additional non-tumor samples, we included these in the dataset provided with this protocol (AnnData object), resulting in the final dataset of 226 samples from 163 patients. We next outline all major steps and explain how to tailor the protocol to different datasets or analyses.
-
1.Installation:
-
a.If you are new to Python, install the latest version of Miniconda (key resources table) according to your system specifications.
-
b.Once Miniconda is installed, open up your system terminal/console, create and activate a new virtual environment by typing:> conda create -y -n scquest python=3.8> conda activate scquest
-
c.Install scQUEST:> pip installai4scr-scQUEST
-
d.Install Jupyter Notebook:> pip install jupyterlabDownload the supplied Jupyter Notebook tutorial (key resources table).> curl -o scQUEST_tutorial.ipynbhttps://raw.githubusercontent.com/AI4SCR/scQUEST/master/tutorials/scQUEST_tutorial.ipynb
-
e.In your terminal/console, fire up a Jupyter Notebook by typing:> jupyter notebook scQUEST_tutorial.ipynbAlternatively, you can run the notebook on a remote host (e.g., an HPC environment). To do that, start the server on the remote:> jupyter notebook scQUEST_tutorial.ipynb --no-browser --port=8080and setup an SSH tunnel. On your local machine run:> ssh -L 8080:localhost:8080 <REMOTE_USER>@<REMOTE_HOST>Note: scQUEST automatically installs all dependencies, such as pandas, PyTorch, sklearn, and AnnData.
-
a.
-
2.Explore example dataset:
-
a.Load pre-uploaded mass cytometry dataset in an AnnData object by simply typing:> ad = scq.dataset.breastCancerAtlasRaw()
-
b.Explore the AnnData object:> ad
-
c.Notice that ad contains 13,384,828 single-cell measurements of 68 channels (stored in .X) with channel annotations (stored in .var). Cell-level annotations (stored in .obs) include various patient metadata, such as tissue type of origin or patient ID (patient_number).
-
a.
Cell type assignment
Timing: 1–2 h
Identification of cell populations of interest is a common task in cytometry-based single-cell analyses, performed traditionally through gating, or, more recently, using clustering algorithms (e.g., PhenoGraph (Levine et al., 2015) or FlowSOM (Van Gassen et al., 2015)). An emerging challenge is that often, the acquired single-cell measurements are in the order of millions of cells, which prohibits analyzing the whole dataset at once. A common solution is subsampling, which however implies that a large part of data will be discarded. In our previous work (Wagner et al., 2019), we were interested in detecting cells of an epithelial phenotype among our ∼13.5 million single-cells dataset stained with 37 different antibodies, and designed an alternative approach to overcome this challenge. The main idea behind our approach was to cluster and annotate a smaller, representative subset of the whole dataset, acquired by a custom down-sampling approach (Figure 1A, Tutorial: Downsampling and clustering, key resources table). This smaller dataset was clustered using PhenoGraph (Levine et al., 2015), resulting in 42 clusters, which were in turn annotated as epithelial based on the expression of one or more of the following epithelial markers: Epithelial cell adhesion molecule (EpCAM), E-Cadherin, cytokeratin 5 (K5), K7, K8, K14, K18, and/or a pan-cytokeratin marker. All clusters negative for all the above markers were labeled as non-epithelial (Figure 1B). The single-cell proteomic measurements with their class assignments were used to train a neural network classifier (Figure 1C), that, once trained, was used to classify the remaining data into epithelial or non-epithelial (Figure 1D). Here, we show how to perform these steps using the same annotated dataset, which is also supplied with scQUEST.
-
3.Load and explore the annotated dataset, ad_anno:> ad_anno = scq.dataset.breastCancerAtlas()
-
a.Notice how ad_anno consists of 687,161 single-cell measurements that have been previously clustered in 42 clusters (cluster label found in .obs['cluster']).
-
b.The 42 clusters have in turn been annotated as epithelial or non-epithelial based on marker expression. We see that in total, there are 484,279 non-epithelial and 202,882 epithelial cells (information included in .obs[celltype_class]).
 CRITICAL: When working with your own data, you can use your algorithm of choice to cluster and annotate the single-cell measurements. It is critical, however, that the smaller, subsampled dataset that you will annotate is representative of the whole population of cells you want to analyze. See also troubleshooting for more information on how to evaluate the quality of your annotated dataset.
CRITICAL: When working with your own data, you can use your algorithm of choice to cluster and annotate the single-cell measurements. It is critical, however, that the smaller, subsampled dataset that you will annotate is representative of the whole population of cells you want to analyze. See also troubleshooting for more information on how to evaluate the quality of your annotated dataset.
-
a.
-
4.Prepare the dataset for classification:
-
a.First, subset the dataset such that it only contains the features/markers relevant for the classification (typically, the ones used for annotating the data).
-
b.Create a binary integer label with 0: non-epithelial and 1: epithelial:> ad_anno.obs['is_epithelial'] = (ad_anno.obs.celltype_class == 'epithelial').astype(int)
-
c.Apply the inverse hyperbolic sine (arcsinh) transformation and save the transformed data in a new layer of the AnnData object:> X = ad_anno.X.copy()> cofactor = 5> np.divide(X, cofactor, out=X)> np.arcsinh(X, out=X)> ad_anno.layers['arcsinh'] = X
-
d.Normalize the data from 0 to 1 using a min-max scaler and save the results in a new layer of the AnnData object:> minMax = MinMaxScaler()> X = minMax.fit_transform(X)> ad_anno.layers['arcsinh_norm'] = XNote: Before training the classifier, you can visualize the data distribution per channel for epithelial and non-epithelial cells (Figure 2). Notable differences in abundance for multiple marker channels are observed, among which are the epithelial markers (e.g., EpCAM, E-Cadherin), as expected.
-
a.
-
5.Train the neural network classifier:
-
a.Initialize the model by setting the dimensions of the input layer equal to the number of markers in the training data. Also set a seed for reproducibility:> clf = scq.Classifier(n_in=ad_anno.shape[1], seed=1)
-
b.Train the classifier:> clf.fit(ad_anno, layer='arcsinh_norm', target='is_epithelial', max_epochs=20, seed=1)Note: By default, the model architecture consists of 1 hidden layer of 20 neurons with a ReLU activation function and one output layer of two neurons. The dataset is split in a stratified fashion into training (90%) and test (10%) sets. Of the training set, 10% are used to validate the model. The classifier is trained using the Adam optimizer (Kingma and Ba, 2017) with a learning rate of 0.001 and a batch size of 256. Its performance is evaluated using a standard cross-entropy loss function. Training is finalized after 20 epochs; we further use an early stopping criterion by terminating training when the model’s performance fails to improve for 10 consecutive runs. In our online documentation (key resources table), we provide an example on how to create custom model architectures.
-
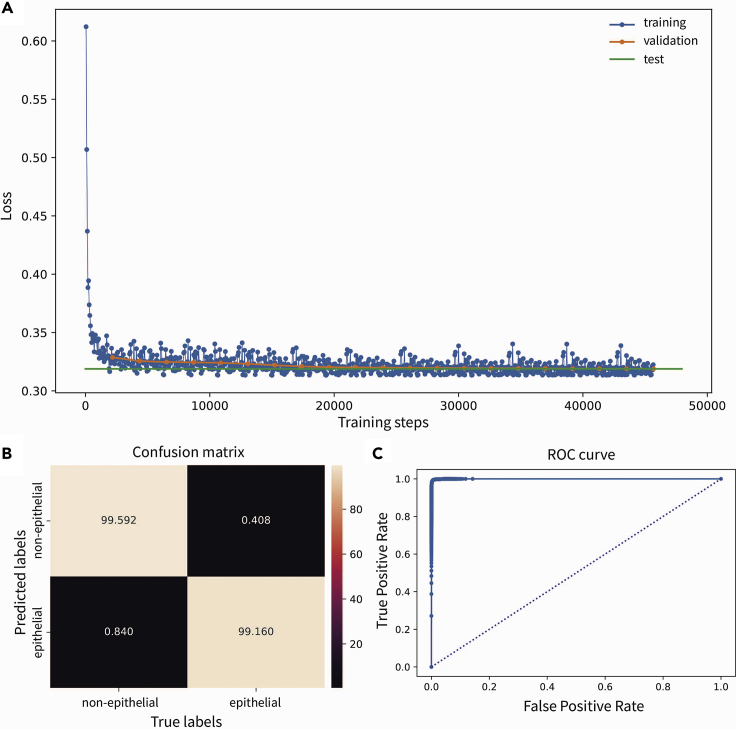
c.Examine model performance by assessing the performance of the classifier in the test data. Some additional ways to evaluate the model is by observing the evolution of the loss, the percentage of accurately predicted epithelial or non-epithelial cells in the form of a confusion matrix and the ROC curve (Figure 3).
 CRITICAL: If the model fails during training, a number of issues may be the reason. Check troubleshooting for possible solutions. It is important to make sure that your model is not overfitting the data. Overfitting occurs when the model fits precisely the training data, but cannot generalize to unseen test data. To learn how to detect and avoid overfitting, check troubleshooting.
CRITICAL: If the model fails during training, a number of issues may be the reason. Check troubleshooting for possible solutions. It is important to make sure that your model is not overfitting the data. Overfitting occurs when the model fits precisely the training data, but cannot generalize to unseen test data. To learn how to detect and avoid overfitting, check troubleshooting.
-
a.
-
6.Once you are confident about the model performance, you can use the trained classifier to predict all epithelial cells across in the ∼13.5 million cell measurements:
-
a.Again, prepare the whole dataset for classification by applying an arcsinh transformation and scaling the data, as in step 4c:> ad_pred = ad[:, ad.var.used_in_clf]> X = ad_pred.X.copy()> np.divide(X, cofactor, out=X)> np.arcsinh(X, out=X)> ad_pred.layers['arcsinh'] = X> X = minMax.transform(X)> ad_pred.layers['arcsinh_norm'] = X
 CRITICAL: When scaling the unseen data, it is important to apply the min-max scaler that was fitted on the training data without re-fitting it on the new data (use transform instead of fit_transform).
CRITICAL: When scaling the unseen data, it is important to apply the min-max scaler that was fitted on the training data without re-fitting it on the new data (use transform instead of fit_transform). -
b.Feed the data to the classifier and save the results:> clf.predict(ad_pred, layer='arcsinh_norm')> ad.obs['is_epithelial'] = ad_pred.obs.clf_is_epithelial.values
-
c.Examining the final classification results, you can see that the model classified the ∼13.5 million cells into ∼3.97 million epithelial cells and ∼9.4 million non-epithelial cells. The classification results can be further examined, for example by generating a dimensionality reduction uniform manifold approximation and projection (UMAP) plot (McInnes et al., 2018). The UMAP plot shows the expected separation of epithelial and non-epithelial cells and at the same time, annotated and predicted cells of each respective class are well-mixed (Figure 4).
-
a.
Figure 1.
Workflow for cell type assignment from single-cell mass cytometry data
(A) Random subsampling step for large datasets.
(B) Clustering of the data, cluster annotation by class based on the respective marker expression profiles, and labeling of each cell according to class.
(C) Splitting the annotated data and using the training set to train a neural network to achieve an accurate class prediction for each cell. The test set is used to evaluate the model performance.
(D) Using the trained neural network to assign the remaining cells to their respective class based on the marker expression profile. Some figure elements have been adapted from Wagner et al. (2019).
Figure 2.
Distribution of marker expression per channel for epithelial (orange) and non-epithelial cells (blue) in the annotated training dataset
Figure 3.
Assessing the performance of the classifier
(A) Evolution of the loss during model training, with training loss indicated in blue, validation loss indicated in orange, and test loss indicated in green.
(B and C) Confusion matrix and (C) receiver operator characteristic (ROC) curve, indicating very high accuracy in predicting epithelial and non-epithelial cells.
Figure 4.
UMAP projection of all single-cell measurements, marked with blue hues for non-epithelial cells (dark blue: annotated, light blue: predicted) and orange hues for epithelial cells (dark orange: annotated, light orange: predicted)
Phenotypic abnormality score
Timing: 1–2 h
Tumor cell heterogeneity is believed to be a source of cancer aggressiveness and an obstacle for complete elimination of tumor cells during therapy (Ramos and Bentires-Alj, 2015). Molecular phenotypic deviation of breast tumor cells from normal mammary epithelial cells is routinely assessed in the clinic and has prognostic value, e.g., protein levels of estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor 2 (HER2), and proliferation marker Ki-67 (Amin et al., 2017). In our previous work, we showed how a type of unsupervised artificial neural network called an autoencoder (Goodfellow et al., 2016; Hinton and Salakhutdinov, 2006) could be used to comprehensively describe the phenotypic abnormality of breast tumor cells in relation to non-cancerous mammary epithelial cells. Briefly, we first trained a “reference” autoencoder with epithelial cells derived from juxta-tumoral non-cancerous mammary gland tissue so it could reconstruct their proteomic phenotypes (Figure 5A). We then fed the trained autoencoder with a “query” dataset with tumor-derived epithelial cells to determine how much they had deviated from the reference phenotypes. The autoencoder calculated a mean squared error (MSE) of the reconstruction for each tumor cell, which represented the phenotypic abnormality (dissimilarity) of that cell to non-cancerous tissue-derived cells (Figure 5B). A tumor-based phenotypic abnormality score was computed as the median score of all epithelial cells of a sample (Figure 5C), and a tissue-based phenotypic abnormality score was computed as the median score of all cells of a tissue type (Figure 5D). In our previous work, we observed a correlation of tumor phenotypic abnormality with features of abnormal growth conditions within the tumor ecosystem, such as a high number of cells positive for the proliferation marker Ki-67 and cells positive for the hypoxia marker carbonic anhydrase 9 (Wagner et al., 2019).
Figure 5.
Steps to compute the phenotypic abnormality scores per cell, tumor, and tissue type
(A) Epithelial cells from juxta-tumoral tissue are used to train an autoencoder, which computes a reconstruction error for each cell.
(B) Tumor-derived epithelial cells are fed to the trained autoencoder and a reconstruction error is computed for each cell.
(C and D) From this, the median reconstruction error can be calculated per sample (C) and per tissue type (D). Some figure elements have been adapted from Wagner et al. (2019).
Here, we explain how to follow the same approach using as example the epithelial measurements of the supplied data.
-
7.Prepare dataset:
-
a.Select a list of patient samples (patients) that will serve as reference (optional).
-
b.Similarly, select a list of markers (markers) as features of the input data.
-
c.Subset the whole ad dataset to include only epithelial cells (ad.obs.is_epithelial == 1) from juxta-tumoral tissue (ad.obs.tissue_type == 'N') of the selected patient samples (ad.obs.patient_number.isin(patients)) and markers (ad.var.used_in_abnormality):> ad_train = ad[(ad.obs.patient_number.isin(patients)) & (ad.obs.tissue_type == 'N') & (ad.obs.is_epithelial == 1), ad.var.used_in_abnormality]
-
d.Preprocess the dataset by applying the arcsinh transformation and min-max normalization, as in step 4c.
-
a.
-
8.Train the abnormality autoencoder:
-
a.Initialize the model by setting the dimensions of the input/output layer equal to the number of markers in the training data:> Abn = scq.Abnormality(n_in=ad_train.shape[1])
-
b.Fit the model to the training data:> Abn.fit(ad_train, layer='arcsinh_norm', max_epochs=20)
-
c.As in step 5c, the model performance can be evaluated in terms of the evolution of the loss.
-
a.
-
9.Use the trained abnormality autoencoder to reconstruct all epithelial cell measurements:
-
a.Select markers and preprocess the dataad_pred = ad[ad.obs.is_epithelial == 1, ad.var.used_in_abnormality]X = ad_pred.X.copy()np.divide(X, cofactor, out=X)np.arcsinh(X, out=X)X = minMax.transform(X)ad_pred.layers['arcsinh_norm'] = X
-
b.Reconstruct the measurements and estimate the mean squared error (MSE):Abn.predict(ad_pred, layer='arcsinh_norm')mse = (ad_pred.layers['abnormality'] ∗∗ 2).mean(axis=1)ad_pred.obs['abnormality'] = mse
-
c.Examine the results across all juxta-tumoral tissue, mammoplasty, and tumor-derived epithelial cells by visualizing and comparing the min-max-normalized protein levels of the input data (Figure 6A) with the reconstructed data (Figure 6B) and the resulting reconstruction errors, called residuals (Figure 6C). The autoencoder nicely reconstructs the protein expression patterns observed in cells derived from juxta-tumoral tissue and mammoplasty samples, resulting in close-to-zero reconstruction errors. For tumor-derived epithelial cells, the reconstruction errors are much more pronounced (Figure 6C – rows labeled with green).Note: The reconstruction errors and phenotypic abnormality score are not zero for juxta-tumoral-derived, non-cancerous epithelial cells of the training set because data reconstruction by the autoencoder is by definition lossy.
 CRITICAL: Before drawing biologically relevant conclusions from the computed score, make sure your data are not confounded by batch effects (troubleshooting).
CRITICAL: Before drawing biologically relevant conclusions from the computed score, make sure your data are not confounded by batch effects (troubleshooting).
-
a.
Figure 6.
Evaluation of the performance of the abnormality autoencoder
(A) Normalized protein expression levels per epithelial cell and marker of the input data.
(B) Normalized protein expression levels per epithelial cell and marker after reconstruction.
(C) Reconstruction error (residuals) per cell and marker, ordered by tissue type mammoplasty (blue), juxta-tumoral (orange) and tumor (green). The order of cells is identical across (A), (B), and (C).
(D–G) Phenotypic abnormality scores by (D) tissue type, (E) tumor grade, (F) estrogen receptor status, and (G) clinical subtype. M = mammoplasty, JT = juxta-tumoral, T = tumor, G = grade, ER = estrogen receptor.
Sample individuality score
Timing: 30 min
Tumor ecosystems may represent unique compositions of tumor cell molecular phenotypes. To quantify and compare the individuality of tumors, in Wagner et al. (2019) we applied a graph-based approach using the epithelial single-cell data from all samples. First, a k-nearest neighbor graph was constructed that placed phenotypically similar epithelial cells close to one another in the high-dimensional space (Figure 7A). Then, for each cell, the respective sample origin labels of their k nearest neighboring cells were determined, and a probability distribution was computed based on its neighborhood proportion (Figure 7B). For each sample, the median value of the probability distribution of all single cells belonging to that sample was computed, resulting in a sample x sample individuality matrix that quantified whether cells were more similar to cells of the same or to cells of other samples. Last, a sample individuality score was obtained using the diagonal of this matrix (Figure 7C). This analysis revealed that tumors had higher individuality scores than non-cancerous tissue. We further observed higher individuality scores for grade 3 aggressive breast cancers compared with grade 1 and grade 2 less aggressive cancers. Here, we show how to apply this analysis using the single-cell epithelial measurements across all samples.
Figure 7.
Workflow for computing the sample individuality score
(A) A k-nearest neighbor graph is constructed using all epithelial cells.
(B) For each cell, the neighborhood proportion is computed for a chosen k.
(C) The median neighborhood proportions for all cells of a sample are determined. Scores on the diagonal represent the sample individuality score. Some figure elements have been adapted from Wagner et al. (2019).
To compute the sample-level individuality score, follow the steps below:
-
10.Prepare the dataset:
-
a.Subset the ad dataset to include only epithelial cells and selected markers (here, the same markers as in step 7b):> ad_indiv = ad[ad.obs.is_epithelial == 1, ad.var.used_in_abnormality]
-
b.Preprocess the dataset by applying the arcsinh transformation, as in step 4c.
-
c.Subsample 200 cells per sample (for samples that have less cells, use them all):ad_indiv.obs['sample_id'] = ad_indiv.obs.groupby(['tissue_type', 'breast', 'patient_number']).ngroup()tmp = ad_indiv.obs.groupby(['sample_id']).indicesn_cells = 200indices = []for key, item in tmp.items():size = min(len(item), n_cells)idx = np.random.choice(range(len(item)), size, replace=False)indices.extend(item[idx])indices = np.array(indices)ad_indiv = ad_indiv[indices]
-
a.
-
11.
Initialize the model and compute the individuality score:
> Indiv = scq.Individuality()
> Indiv.predict(ad_indiv, ad_indiv.obs.sample_id, layer='arcsinh')
-
12.Assess the results:
-
a.The observation-level scores, i.e., one vector per single cell, indicating a cell’s similarity to all other samples, are saved in the .obsm attribute of the AnnData object, as a matrix of size n_cells x n_samples:> ad_indiv.obsm['individuality']
-
b.The aggregated sample-level scores (one vector per sample, indicating a sample’s similarity to all other samples) are saved in the .uns attribute of the AnnData object, as a matrix of size n_samples x n_samples:> ad_indiv.uns['individuality_agg']
-
c.To assess a sample’s individuality, we will use the diagonal of that matrix:>dat=pd.DataFrame(np.diag(ad_indiv.uns['individuality_agg']), index=dat.index, columns=['individuality'])
-
d.Finally, assess the individuality score with respect to different patient clinical data (Figure 8).
 CRITICAL: Ensure your data are not confounded by batch effects (troubleshooting).
CRITICAL: Ensure your data are not confounded by batch effects (troubleshooting).
-
a.
Figure 8.
Association of individality score with clinical metadata
Samples individuality scores by (A) tissue type, (B) tumor grade, (C) estrogen receptor status, and (D) clinical subtype. M = mammoplasty, JT = juxta-tumoral, T = tumor, G = grade, ER = estrogen receptor.
Expected outcomes
scQUEST enables a robust discrimination of cell populations in large datasets as well as quantification of different aspects of tumor heterogeneity for inter-patient comparisons and comparisons with a reference tissue. All computations are done seamlessly, and the outcomes are integrated in different components of the AnnData object to facilitate downstream analyses.
Cell type assignment
The trained classifier model (clf) can be saved and reused for cell type assignment in future datasets:
torch.save(clf.model.state_dict(), PATH)
clf = scq.Classifier(n_in=ad_anno.shape[1])
clf.model.load_state_dict(torch.load(PATH))
The predicted labels are automatically saved as cell-level annotations in the .obs attribute of the AnnData object.
Phenotypic abnormality score
The autoencoder model can be saved and reused in the future. The single-cell level abnormality values can be stored in the .obs attribute of the AnnData object.
Tumor individuality score
The single-cell level and sample-level individuality scores are automatically saved in the .obsm and .uns attributes of the AnnData object, as explained in steps 4–12.
Data export
The data can be exported to different commonly used data formats (e.g., .csv, .xls) to enable data analysis with other tools. For example, the epithelial cell data as classified by the model can be exported together with the associated metadata:
mask=ad.obs['is_epithelial']==1
temp=pd.DataFrame(data=ad.X[mask], columns = ad.var['desc'])
temp.to_csv('all_epithelial_cells.csv')
ad.obs[mask].to_csv('epithelial_metadata.csv')
All resulting plots in this protocol can simply be exported using the matplotlib savefig function:
> plt.savefig('figure_name.pdf', dpi=300)
Limitations
The performance of the different components of scQUEST depends on several factors, summarized by step below:
Cell type assignment
The most crucial aspect for accurate cell type identification is the quality and size of the annotated dataset used for training, which in turn largely depends on the use of cell type-stratifying markers. In this example, we used epithelial lineage markers to separate epithelial cells from cells of other lineages (e.g., mesenchymal and immune cells). Another important aspect is to titrate antibodies for data generation so that they yield an optimal signal-to-noise ratio. Low signal-to-noise ratios in marker channels used for training the neural network may result in poorer separation results for the cell populations of interest. Concerning the dataset size, the more data available to train a neural network for cell type identification the better, with the minimum amount depending largely on the difficulty of the task. Typically, for training datasets, a few thousand cells should suffice. The results of data clustering and applying the neural network to detect cell types of interest may be compared with other approaches that do not use annotated cell populations as input for a neural network but rather pre-specify marker profiles for cell type detection (Geuenich et al., 2021). Finally, although the same model can be reused to classify cells from additional, unseen datasets measured with the same panel, it is crucial that the effect of different experimental batches is minimal, or corrected for (see also troubleshooting).
Phenotypic abnormality score
It is important to select a proper reference to serve as the basis to train the autoencoder, and one (or more) query datasets that will be used to compute the abnormality score. Ideally, these datasets deviate considerably in phenotypic space. For calculating a sample abnormality score for tumors, ideally, the corresponding normal tissue-derived cells are used as a reference. It is crucial that the reference and query datasets are free of batch effects (see also troubleshooting), so that the estimated phenotypic deviation is attributed to biological and not technical variability. The abnormality score indicates how similar (low score) or dissimilar (high score) a tumor cell is compared with the median of the reference. However, two tumor cells with the same abnormality score may be phenotypically very distant, as the score does not suggest a trajectory (i.e., it is not directional). For single-cell trajectory reconstruction, other tools can be explored, as reviewed in (Cannoodt et al., 2016). As mentioned in step 2, the more data available to train the autoencoder the better.
Tumor individuality score
The individuality score captures the uniqueness of the phenotypic pattern of a sample within a cohort, and as such, is dependent on the choice of markers to describe the phenotypic pattern and the variability of the cohort itself. Therefore, care should be taken when choosing markers to assess cell phenotypes by mass cytometry or flow cytometry and when assembling a patient cohort. As for phenotypic abnormality, the k-nearest neighbor graph and individuality scores do not suggest a trajectory of disease evolution, but rather describe how rare or ubiquitous a patient-level tumor profile is within the cohort. Finally, the sample individuality score is highly affected by the number of cells per sample, with estimates of smaller samples being less robust. For imbalanced datasets, we recommend re-sampling an equal number of cells per sample to circumvent this issue.
Due to the modular structure of scQUEST, individual components of the package can be used for alternative applications beyond the dataset presented here. For example, the phenotypic abnormality score approach could be applied to cells before and after drug perturbations or genetic alterations. Alternatively, cells at different timepoints during in vitro experiments, in disease models, or during patient disease progression can be compared.
Troubleshooting
Problem 1
The model fails to accurately classify the training dataset (step 2).
Potential solution
Step 2 may fail if the quality of your annotated dataset is suboptimal. To correct this issue, you first need to evaluate the clustering and ensure that the identified clusters represent distinct cell phenotypes or subpopulations. Maximize the number of cells to cluster according to your computational resources to increase robustness. Decide which markers you want to use for the clustering step, as this will strongly affect the result. An ideal clustering groups cells together with a very similar marker profile, i.e., low signal variance in each marker channel. Ideally, marker channels are chosen for clustering that are expressed by cells of one class and not by cells of another class and vice versa. You may want to exclude markers from the clustering with very low signal-to-noise ratios and markers with signal spill into other channels, which may create possible false-positive cells. Examine the robustness of the clustering with respect to the inherent stochasticity of the algorithm (e.g., different random initializations) or different parameter values (e.g., k number of neighbors in PhenoGraph), by exploiting metrics such as the Adjusted Rand Index (Hubert and Arabie, 1985), the Silhouette coefficient (Rousseeuw, 1987) or the adjusted mutual information score (Vinh et al., 2010) from repetitive clustering runs. When working with imbalanced data, we recommend subsampling strategies that balance out large discrepancies in sample sizes (see our Downsamplng and clustering tutorial, key resources table).
Problem 2
The model fails to accurately classify the test or unseen data (step 2).
Potential solution
Even if the model achieved high accuracy in the training set, it may fail in accurately classifying the test set. One common pitfall may be overfitting. To detect overfitting, make sure the training loss is not getting progressively lower while at the same time the test loss is getting higher. Using a k-fold cross validation scheme will allow you to evaluate the model performance. If you have issues with overfitting, you can try reducing the complexity of the model, training with more data and exploiting techniques such as regularization, dropout and early stopping. Another common pitfall may be batch effects, addressed in problem 3 below.
Problem 3
The abnormality or individuality scores are not capturing patterns relevant to disease variability (steps 3 and 4).
Potential solution
The first factor you need to assess is whether your data is confounded by batch effects. Ideally, all samples to be compared should be measured in the same acquisition, so that systematic biases introduced by antibody staining variability, sample handling or machine calibration are absent. If this is not the case, then both the abnormality and individuality scores may highlight samples as abnormal or unique that differ in experimental setup instead of differing in biological means. To address this issue, batch effects first need to be detected and then corrected for, e.g., by comparing different available computational approaches (Haghverdi et al., 2018; Leek et al., 2010).
Problem 4
The Classifier model always converges to the same solution, which makes it hard to test its robustness.
Potential solution
In the current tutorial, we are using a fixed seed both in the initialization and fitting of the Classifier module for reproducibility reasons. To test how different stochastic initializations of the model affect the results, change the value of the seed, and compare the outcomes. The same process can be followed for the Abnormality module as well.
Problem 5
My dataset is highly unbalanced, resulting in low prediction accuracy of the underrepresented class.
Potential solution
This is a common problem in real world cytometry datasets, which can be circumvented by using class weights, as we show in step 2: Train the neural network classifier of our scQUEST AML tutorial (key resources table).
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Maria Anna Rapsomaniki (aap@zurich.ibm.com).
Materials availability
This study did not generate new unique reagents.
Acknowledgments
This work has been supported by the Swiss National Science Foundation Sinergia grant CRSII5_202297. J.W. is supported by a European Molecular Biology Organization (EMBO) Postdoctoral Fellowship ALTF 599-2021.
Author contributions
Conceptualization, J.W., B.B., and M.A.R.; methodology, A.L.M. and M.A.R.; software, A.L.M.; investigation, A.L.M.; visualization and data interpretation, J.W., A.L.M., and M.A.R.; writing, J.W. and M.A.R. with input from B.B.; supervision, M.A.R.; funding acquisition, J.W. and M.A.R.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Adriano Luca Martinelli, Email: art@zurich.ibm.com.
Maria Anna Rapsomaniki, Email: aap@zurich.ibm.com.
Data and code availability
The code generated during this study is available at https://github.com/AI4SCR/scQUEST. Original data used in this protocol were generated as described in Wagner et al. (2019) and are deposited in Mendeley data: https://doi.org/10.17632/gb83sywsjc.1. The single-cell measurements of the tumor epithelial cell-centric panel (raw and pre-annotated) are also pre-uploaded in scQUEST as AnnData objects. The AnnData object contains 23 additional tumors and 9 additional non-tumor samples which were not previously included in the Wagner et al. dataset deposited on Mendeley data.
References
- Amin M.B., Greene F.L., Edge S.B., Compton C.C., Gershenwald J.E., Brookland R.K., Meyer L., Gress D.M., Byrd D.R., Winchester D.P. The Eighth Edition AJCC Cancer Staging Manual: continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J. Clin. 2017;67:93–99. doi: 10.3322/caac.21388. [DOI] [PubMed] [Google Scholar]
- Cannoodt R., Saelens W., Saeys Y. Computational methods for trajectory inference from single-cell transcriptomics. Eur. J. Immunol. 2016;46:2496–2506. doi: 10.1002/eji.201646347. [DOI] [PubMed] [Google Scholar]
- Chevrier S., Crowell H.L., Zanotelli V.R.T., Engler S., Robinson M.D., Bodenmiller B. Compensation of signal spillover in suspension and imaging mass cytometry. Cell Syst. 2018;6:612–620.e5. doi: 10.1016/j.cels.2018.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finck R., Simonds E.F., Jager A., Krishnaswamy S., Sachs K., Fantl W., Pe’er D., Nolan G.P., Bendall S.C. Normalization of mass cytometry data with bead standards. Cytometry A. 2013;83:483–494. doi: 10.1002/cyto.a.22271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuenich M.J., Hou J., Lee S., Ayub S., Jackson H.W., Campbell K.R. Automated assignment of cell identity from single-cell multiplexed imaging and proteomic data. Cell Syst. 2021;12:1173–1186.e5. doi: 10.1016/j.cels.2021.08.012. [DOI] [PubMed] [Google Scholar]
- Goodfellow I., Bengio Y., Courville A. MIT press; 2016. Deep Learning. [Google Scholar]
- Haghverdi L., Lun A.T.L., Morgan M.D., Marioni J.C. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat. Biotechnol. 2018;36:421–427. doi: 10.1038/nbt.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinton G.E., Salakhutdinov R.R. Reducing the dimensionality of data with neural networks. Science. 2006;313:504–507. doi: 10.1126/science.1127647. [DOI] [PubMed] [Google Scholar]
- Hubert L., Arabie P. Comparing partitions. J. Classif. 1985;2:193–218. doi: 10.1007/BF01908075. [DOI] [Google Scholar]
- Kashyap A., Rapsomaniki M.A., Barros V., Fomitcheva-Khartchenko A., Martinelli A.L., Rodriguez A.F., Gabrani M., Rosen-Zvi M., Kaigala G. Quantification of tumor heterogeneity: from data acquisition to metric generation. Trends Biotechnol. 2022;40:647–676. doi: 10.1016/j.tibtech.2021.11.006. [DOI] [PubMed] [Google Scholar]
- Kingma D.P., Ba J. Adam: a method for stochastic optimization. arXiv. 2017 doi: 10.48550/ARXIV.1412.6980. Preprint at. [DOI] [Google Scholar]
- Kotecha N., Krutzik P.O., Irish J.M. Web-based analysis and publication of flow cytometry experiments. Curr. Protoc. Cytom. 2010;Chapter 10:Unit10.17. doi: 10.1002/0471142956.cy1017s53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krutzik P.O., Nolan G.P. Fluorescent cell barcoding in flow cytometry allows high-throughput drug screening and signaling profiling. Nat. Methods. 2006;3:361–368. doi: 10.1038/nmeth872. [DOI] [PubMed] [Google Scholar]
- Leek J.T., Scharpf R.B., Bravo H.C., Simcha D., Langmead B., Johnson W.E., Geman D., Baggerly K., Irizarry R.A. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat. Rev. Genet. 2010;11:733–739. doi: 10.1038/nrg2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine J.H., Simonds E.F., Bendall S.C., Davis K.L., Amir E.a.D., Tadmor M.D., Litvin O., Fienberg H.G., Jager A., Zunder E.R., et al. Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell. 2015;162:184–197. doi: 10.1016/j.cell.2015.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli A.L., Rapsomaniki M.A. ATHENA: analysis of tumor heterogeneity from spatial omics measurements. Bioinformatics. 2022;38:3151–3153. doi: 10.1093/bioinformatics/btac303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes L., Healy J., Melville J. Umap: Uniform manifold approximation and projection for dimension reduction. ArXiv. 2018 [Google Scholar]
- Ramos P., Bentires-Alj M. Mechanism-based cancer therapy: resistance to therapy, therapy for resistance. Oncogene. 2015;34:3617–3626. doi: 10.1038/onc.2014.314. [DOI] [PubMed] [Google Scholar]
- Rousseeuw P.J. Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 1987;20:53–65. doi: 10.1016/0377-0427(87)90125-7. [DOI] [Google Scholar]
- Van Gassen S., Callebaut B., Van Helden M.J., Lambrecht B.N., Demeester P., Dhaene T., Saeys Y. FlowSOM: using self-organizing maps for visualization and interpretation of cytometry data. Cytometry A. 2015;87:636–645. doi: 10.1002/cyto.a.22625. [DOI] [PubMed] [Google Scholar]
- Vinh N.X., Epps J., Bailey J. Information theoretic measures for clusterings comparison: variants, properties, normalization and correction for chance. J. Mach. Learn. Res. 2010;11:2837–2854. [Google Scholar]
- Virshup I., Rybakov S., Theis F.J., Angerer P., Wolf F.A. anndata: annotated data. bioRxiv. 2021 doi: 10.1101/2021.12.16.473007. Preprint at. [DOI] [Google Scholar]
- Wagner J., Rapsomaniki M.A., Chevrier S., Anzeneder T., Langwieder C., Dykgers A., Rees M., Ramaswamy A., Muenst S., Soysal S.D., et al. A single-cell atlas of the tumor and immune ecosystem of human breast cancer. Cell. 2019;177:1330–1345.e18. doi: 10.1016/j.cell.2019.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zunder E.R., Finck R., Behbehani G.K., Amir E.-A.D., Krishnaswamy S., Gonzalez V.D., Lorang C.G., Bjornson Z., Spitzer M.H., Bodenmiller B., et al. Palladium-based mass tag cell barcoding with a doublet-filtering scheme and single-cell deconvolution algorithm. Nat. Protoc. 2015;10:316–333. doi: 10.1038/nprot.2015.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The code generated during this study is available at https://github.com/AI4SCR/scQUEST. Original data used in this protocol were generated as described in Wagner et al. (2019) and are deposited in Mendeley data: https://doi.org/10.17632/gb83sywsjc.1. The single-cell measurements of the tumor epithelial cell-centric panel (raw and pre-annotated) are also pre-uploaded in scQUEST as AnnData objects. The AnnData object contains 23 additional tumors and 9 additional non-tumor samples which were not previously included in the Wagner et al. dataset deposited on Mendeley data.