Abstract
The doublesex/mab-3 related transcription factor (Dmrt) genes regulate sexual development in metazoans. Studies of the doublesex (dsx) gene in insects, in particular Drosophila melanogaster, reveal that alternative splicing of dsx generates sex-specific Dsx isoforms underlying sexual differentiation. Such a splicing-based mechanism underlying sex-specific Dmrt function is thought to be evolved from a transcription-based mechanism used in non-insect species, but how such transition occurs during evolution is not known. Here we identified a male-specific dsx transcript (dsxM2) through intron retention (IR), in addition to previously identified dsxM and dsxF transcripts through alternative polyadenylation (APA) with mutually exclusive exons. We found that DsxM2 had similarly masculinizing function as DsxM. We also found that the IR-based mechanism generating sex-specific dsx transcripts was conserved from flies to cockroaches. Further analysis of these dsx transcripts suggested an evolutionary pathway from sexually monomorphic to sex-specific dsx via the sequential use of IR-based and APA-based alternative splicing.
Subject terms: Evolution, Molecular biology
An ancestral male-specific doublesex isoform, dsxM2, is identified via intron retention, with a masculinizing function weaker than the modern dsxM
Introduction
Sexually dimorphic traits and behaviors are mediated by sex-specific expression of regulatory genes. Substantial studies reveal that the doublesex/mab-3 related transcription factor (Dmrt) genes have been found in all studied animal models and humans for sexual differentiation1–3. Dmrt transcription factors share a common DM (Dsx/Mab-3) domain which is a zinc-finger DNA binding motif, but show little sequence conservation in other parts1. In humans, Dmrt1 is linked to disorders of sex development and testicular germ cell tumors4,5. Dmrt1 is preferentially expressed in the male gonad and required postnatally to maintain gonadal sex in mammals and other vertebrates2,6–8. In the nematode worm Caenorhabditis elegans, mab-3 is mainly expressed in male tails and necessary for proper morphogenesis and differentiation of copulatory structures9. In the fruit fly Drosophila melanogaster, sex-specific Dsx isoforms, DsxM in males and DsxF in females, control sexual differentiation and behaviors in both sexes, and are generated through alternative polyadenylation (APA) with mutually exclusive exons10–14.
Despite conserved roles of Dmrt genes in sexual differentiation in metazoans, regulatory mechanisms underlying their sex-specific function are diversified1–3,15,16. The alternative splicing-based regulation of Dmrt genes has only been found in insects and often mediated by the female-specific expression of transformer (tra) with few exceptions10,16–19. In vertebrates and nematodes, Dmrt genes are not sex-specifically spliced, but transcribed in a predominantly male-specific manner for the development of male-specific traits2. It has been proposed that the insect-specific mechanism based on alternative splicing of dsx evolved from a more ancient mechanism based on male-specific transcription16,17, but how such transition occurs during evolution is rarely known.
The canonical insect tra-dsx pathway generates sex-specific Dsx isoforms by the selective use of distinct 3′ exons in the two sexes16,17. In D. melanogaster females, Tra and Tra-2 bind to the female-specific exon of dsx and direct female-specific splicing of dsxF, while in males the absence of Tra function permits the default splicing of dsxM including both common and male-specific exons10,20–22. DsxM and DsxF have common DM domain and two oligomerization domains, thus may bind to the same sets of target genes; however, they have been found to oppositely regulate target gene expression to establish male- or female-specific differentiation, possibly through their sex-specific C-terminus23–26.
In this study, we report that intron retention (IR) generates a male-specific dsx transcript and propose an evolutionary pathway from sexually monomorphic to sex-specific splicing of dsx. We identify another male-specific dsx transcript (dsxM2), in which the intron linking the last common exon and the female-specific exon is retained. The IR-generated DsxM2 has a masculinizing role like DsxM and is crucial for male courtship robustness in D. melanogaster. We further show that such mechanism generating sex-specific dsx transcripts is deeply conserved in insects from fruit flies to cockroaches. The male-specific intron retention depends on the presence of a weak splicing acceptor sequence inside the intron, as well as the regulation of Tra. Comparison of the structures of dsx transcripts, in light of their difference of conservation, suggests an evolutionary pathway from sexually monomorphic to sex-specific dsx via the sequential use of IR-based and APA-based alternative splicing.
Results
Identification of a novel dsx transcript dsxM2 through intron retention
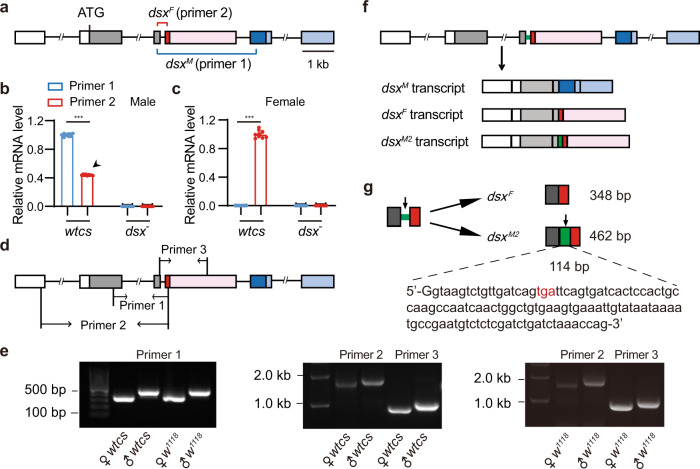
It has been determined that the dsx pre-mRNA is sex-specifically spliced to yield the male-specific dsxM and female-specific dsxF mRNAs. In an attempt to determine relative expression of dsx transcripts in the two sexes in wild-type Canton-S (wtcs) and dsx mutant (dsx683-7058/dsx1649-9625) flies, we performed quantitative PCR (qPCR) experiments using two pairs of primers targeting the male-specific or female-specific locus in dsx, respectively (Fig. 1a). We found that dsxM was indeed expressed exclusively in wtcs males, but not in wtcs females or dsx mutant flies; however, comparable levels of dsx expression were detected in both wtcs males and females, but not in dsx mutant flies, by using the primer pair targeting dsxF (Fig. 1b, c). These results suggest the presence of dsxF or a novel dsx transcript, in addition to dsxM, in male flies.
Fig. 1. Identification of a male-specific dsxM2 transcript through intron retention.
a Schematic structure of the dsx gene. Boxes: exons; white and gray boxes: common 5’-UTR and coding sequences respectively in both sexes; red and pink: coding and 3’-UTR sequences of the female-specific exon; dark and light blue: coding and 3’-UTR sequences of male-specific exons. Primers against sex-specific sequences are indicated. b, c Relative mRNA expression levels of presumed dsxMcycle at 60% humiditytranscript (b, primer 1) and dsxF transcript (c, primer 2) in wtcs and dsx mutant (dsx683-7058/dsx1649-9625) males and females. n = 9 based on three replicates for each. ***p < 0.001, Mann–Whitney U test. Note that a dsx transcript (arrowhead) is detected in males using the primer against the female-specific transcript (b). d, e Reverse transcription PCR (RT-PCR), using three pairs of primers as indicated (d), and sequencing identify the male-specific dsxM2 transcript, in addition to the female-specific dsxF transcript, in both wtcs and w1118 flies (e). For primer 1: 462bp and 348bp for male and female products, respectively; for primer 2: 1833bp and 1719bp for male and female products, respectively; for primer 3: 1069bp and 955bp for male and female products, respectively. f Illustration of the male-specific dsxM2 transcript, in addition to previously identified dsxM and dsxF transcripts. g The dsxM2 transcript differs from the dsxF transcript with the 114 bp intron (in green) retained. Red letters (tga) indicate the stop codon inside the retained intron.
To identify the dsx transcript detected in male flies, we next performed reverse transcription PCR (RT-PCR) experiments using three different primer pairs targeting different sites of the dsxF transcript (Fig. 1d), and found that all PCR products from male flies were larger than those from females (Fig. 1e). The same results were also obtained in another broadly used wild-type w1118 flies (Fig. 1e). By sequencing these PCR products and sequence alignment, we found that the novel dsx transcript in males is quite similar to dsxF, with only 114 bp intron not being spliced out and retained between the last common exon and the female-specific exon (Fig. 1f, g). Thus, we identified a novel male-specific dsx transcript, hereafter referred to as dsxM2, in addition to the previously identified male-specific dsxM and female-specific dsxF transcripts (Supplementary Fig. 1a). Indeed, previous RNA-seq results already suggested the possibility of the 114 bp intron retention in males but not females27,28 (Supplementary Fig. 1b and Supplementary Table 1). The predicted DsxM2 protein contains the common N-terminus of Dsx proteins and a specific C-terminus with only six amino acids resulting from the early stop codon inside the retained intron (Supplementary Fig. 2).
dsxM2 functions like dsxM but has limited roles
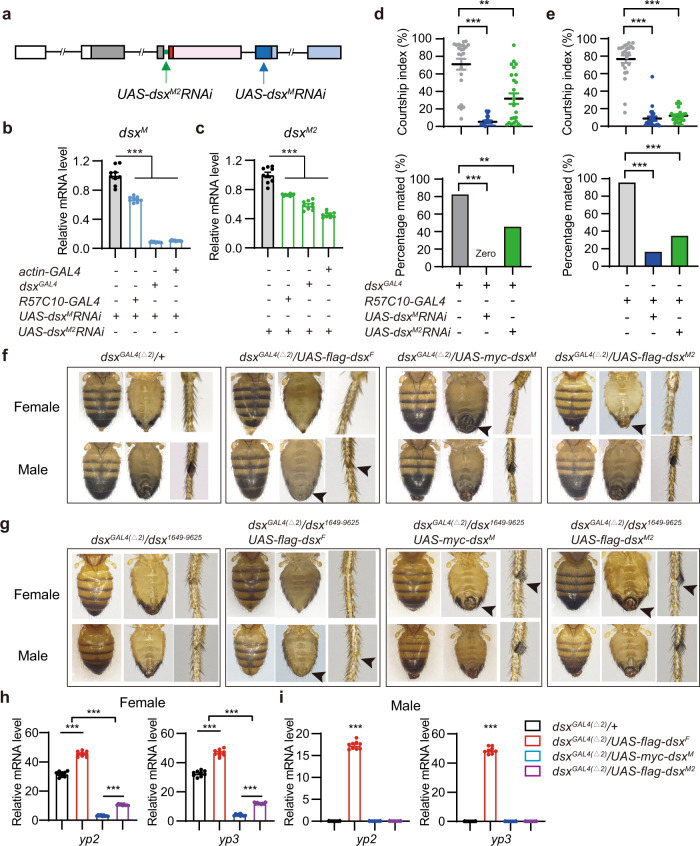
The above results identified a male-specific dsxM2 transcript, but how it is generated from intron retention and whether it plays any role in sexual development or behavior is not known. To investigate whether and where dsxM2 expresses, we tried to generate a polyclonal antibody against the eight amino acids in the C-terminus of the predicted DsxM2 protein but failed to detect any signal through immunostaining and western blot experiments. We next performed qPCR experiments to quantify the relative expression levels of dsxM and dsxM2 in various tissues. We found that dsxM2 was expressed in the fly head, thorax, and forelegs, but the level of dsxM2 mRNA was generally lower than the level of dsxM mRNA (Supplementary Fig. 3), which is also consistent with the result using the whole fly body (Fig. 1b). To investigate whether dsxM2 plays a role in sexual development and/or behavior, we generated a UAS-dsxM2RNAi construct targeting the retained intron (Fig. 2a), combined with previously used UAS-dsxMRNAi construct targeting the male-specific exon14, and validated their efficiency with three different GAL4 drivers, the actin-GAL4, the pan-neuronal R57C10-GAL429, and dsxGAL4, using qPCR (Fig. 2b, c). Both dsxM and dsxM2 RNAi lines knocked down the corresponding dsx mRNA efficiently though not in the same level (Fig. 2b, c), and did not reduce the level of the other transcript (Supplementary Fig. 4). We next knocked down dsxM or dsxM2 in all dsx-expressing cells [dsxGAL4(∆2)] and found that males with dsxM knocked down were intersexual and displayed little courtship, while males with dsxM2 knocked down had regular male appearance but reduced courtship and mating success with virgin female targets (Fig. 2d). These results suggest that dsxM2 is involved in regulating male courtship but not, if any, development of sexually dimorphic traits. To further confirm whether dsxM2 functions in the nervous system to mediate male courtship, we knocked down dsxM or dsxM2 pan-neuronally using the R57C10-GAL4 driver. Males with dsxM or dsxM2 knocked down pan-neuronally had regular male appearance but much reduced courtship levels and mating success (Fig. 2e). These results indicate a crucial role of dsxM2, like dsxM, in regulating male courtship intensity.
Fig. 2. dsxM2 has a potentially masculinizing function like dsxM.
a RNAi targeting dsxM and dsxM2 as indicated by blue and green arrows, respectively. b, c Relative mRNA expression levels of dsxM (b) and dsxM2 (c) in control and RNAi-mediated males. n = 9 based on three replicates for each. ***p < 0.001, Mann-Whitney U test. d, e Knocking down dsxM or dsxM2 in dsx-expressing cells (d) or pan-neuronally (e) impairs male courtship and mating success with females. n = 23, 24, 24, 24, 24, 24 for dsxGAL4/+, dsxGAL4/UAS-dsxMRNAi, dsxGAL4/UAS-dsxM2RNAi, R57C10-GAL4/+, R57C10-GAL4/UAS-dsxMRNAi and R57C10-GAL4/UAS-dsxM2RNAi respectively. For courtship index, **p = 0.0012 and ***p < 0.001, Kruskal-Wallis test with Dunn’s multiple comparisons test; for percentage mated, **p = 0.0087 and ***p < 0.001, Chi-square test. f, g Overexpression of DsxF feminized, while overexpression of DsxM or DsxM2 masculinized dsx-expressing cells, including external genitalia and sex comb (arrowhead), under wild-type background [f, dsxGAL4(∆2)/+] or mutant background [g, dsxGAL4(∆2)/dsx1649-9625] for dsx. h, i Relative mRNA expression levels of dsx target genes, yp2 and yp3, in females (h) and males (i). Expression levels of yp2 and yp3 were increased with DsxF overexpression and decreased with DsxM or DsxM2 overexpression. n = 9 based on three replicates for each. ***p < 0.001. Mann-Whitney U test. Error bars indicate SEM.
To further investigate dsxM2 function, we generated constructs overexpressing each dsx isoform (UAS-flag-dsxF, UAS-myc-dsxM and UAS-flag-dsxM2) and validated their efficiency using qPCR experiments and immunostaining with anti-Flag and anti-Myc antibodies (Supplementary Fig. 5). We then overexpressed these dsx isoforms in all dsx-expressing cells and found that overexpressing DsxF feminized male development (genitals and sex combs), while overexpressing DsxM or DsxM2 masculinized female development (genitals) (Fig. 2f). To avoid potential interference of overexpressed and indigenous Dsx isoforms (e.g., DsxM2 and DsxF), we next overexpressed these dsx isoforms in a dsx mutant background [dsxGAL4(∆2)/dsx1649-9625] in which both sexes were intersexual, and found that expressing DsxF induced female differentiation in both sexes, while expressing DsxM or DsxM2 strongly masculinized genital and sex comb development in both sexes (Fig. 2g). These results indicate that DsxM2 has a potential masculinizing role like DsxM.
As transcription factors, Dsx proteins regulate sexual differentiation through their target genes, of which three genes encoding the female-specific Yolk Proteins (YPs) have been intensively studied under control of Dsx10,24,30. To compare the transcriptional regulation of different Dsx isoforms on target genes, we overexpressed the three Dsx isoforms driven by the dsxGAL4(∆2) and tested relative expression changes of yp2 and yp3. Compared to control females, overexpressing DsxF significantly increased yp2 and yp3 expression, while overexpressing DsxM or DsxM2 significantly reduced their expression in females (Fig. 2h). Note that yp2 and yp3 expression were more severely reduced in females expressing DsxM than those expressing DsxM2, suggesting that DsxM2 had a weaker role of transcriptional inhibition compared to DsxM (Fig. 2h). The expression of yp2 or yp3 was undetectable in control males, but significantly increased in males overexpressing DsxF, which further confirmed the role of DsxF in promoting yp2 and yp3 transcription (Fig. 2i). Taken together, the knockdown experiments indicate a crucial role of dsxM2 in the nervous system for male courtship robustness but not sexual development, while the overexpression experiments indicate that DsxM2 has a potentially masculinizing role like DsxM.
Intron retention is a common mechanism to generate sex-specific dsx isoforms
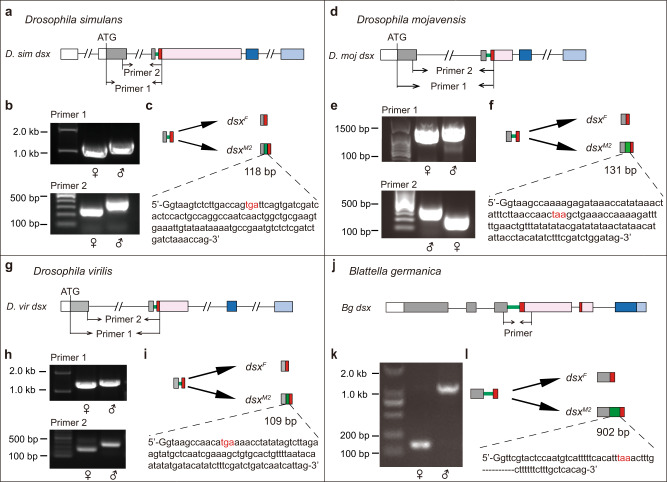
As we identified the male-specific dsxM2 transcript through intron retention, which is a potent mechanism underlying sexual differentiation, we asked if such an alternative splicing form also existed in other animal species. We tested three other Drosophila species including the closely related D. simulans and two other distant species D. mojavensis and D. virilis, by performing RT-PCR experiments using multiple primer pairs targeting sequences including the potentially retained intron and a portion of the female-specific exon, followed by sequencing. We found retention of the 118 bp intron in D. simulans males (Fig. 3a–c), 131 bp intron in D. mojavensis males (Fig. 3d–f) and 109 bp intron in D. virilis males (Fig. 3g–i), which generated dsxM2 transcripts in the same way as in D. melanogaster males. All predicted DsxM2 proteins have conserved DM domain including a zinc-finger DNA binding domain and oligomerization domains, and few amino acids in the C-terminus due to stop codons in the retained intron (Supplementary Fig. 6). Protein sequence alignment of Dsx isoforms in these Drosophila species revealed that sex-specific C-terminus of DsxF proteins were almost identical, while those of DsxM and DsxM2 proteins were less conserved (Supplementary Fig. 7), which is consistent with previous findings on the different levels of conservation of DsxM and DsxF isoforms across insect species31.
Fig. 3. Intron retention-induced sex-specific splicing is widely conserved.
Retention of the intron that separates the last common exon and the female-specific exon generates male-specific dsxM2 transcripts in D. simulans (a–c), D. mojavensis (d–f), D. virilis (g–i) and B. germanica (j–l). The dsx gene structures are illustrated in a, d, g and j. White and gray boxes indicate common 5’-UTR and coding sequences respectively in both sexes. Red and pink boxes indicate coding and 3’-UTR sequences of the female-specific exon. Dark and light blue boxes indicate coding and 3’-UTR sequences of male-specific exons. The thick green line indicates the intron spliced out in the dsxF transcript and retained in the dsxM2 transcript. Primers for RT-PCR experiments (b, e, h and k) are indicated respectively (a, d, g and j). Full sequences of the retained intron of dsxM2 are indicated (c, f and i) except for the 902bp one in B. germanica dsxM2 (l). The capital letter “G” from the upstream exon is added in front of the intron sequence to ensure that the stop codon highlighted in red is aligned within the open reading frame.
To further test whether intron retention could be a conserved way to generate alternative splicing of dsx, we performed RT-PCR experiments targeting the dsx gene in a hemimetabolous insect, the German cockroach Blattella germanica, as it is not only a far distant insect species apart from D. melanogaster (>300 million years), but also with the dsx gene structure being previously studied17,32. We designed a pair of primer targeting the last common exon and the first female-specific exon respectively (Fig. 3j) and obtained sex-specific fragments by PCR (Fig. 3k). Through sequencing, we revealed the existence of the male-specific dsxM2 transcript that retains the 902 bp intron linking the last common exon and the first female-specific exon (Fig. 3l), in addition to previously identified dsx transcripts17. Taken together, these results indicate that intron retention is a common mechanism to generate sex-specific dsx isoforms in distant insect species.
dsx intron retention is jointly regulated by a weak splice acceptor and transformer
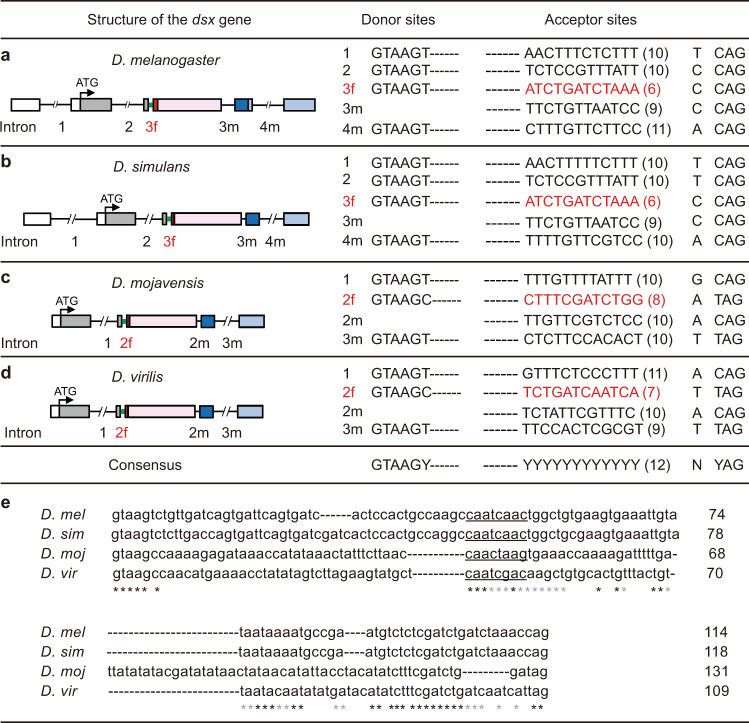
To further investigate factors affecting intron retention and generating sex-specific dsx isoforms, we compared sequences of all introns of dsx in the four Drosophila species. We did not compare the acceptor sequences in Blattella germanica, as we have insufficient knowledge about the consensus acceptor sequence in this species. We found that all introns of dsx in D. melanogaster and D. simulans start with the donor sequence GTAAGT and end with acceptor sequences (T/C)nNCAG (N indicates A, C, G or T) (Fig. 4a, b). Previous studies already suggest such consensus acceptor sequence10, where the number of pyrimidines (T/C) upstream of the last four nucleotides (NCAG) is at least 9. Indeed, the common (intron 1 and 2) and male-specific (intron 3m and 4m) acceptors have 9–11 pyrimidines upstream of NCAG. As for the retained intron (spliced out in female, 3f), the acceptor has only 6 pyrimidines upstream of NCAG (Fig. 4a, b). We next compared donor and acceptor sequences of introns of the dsx gene in D. mojavensis and D. virilis. We found similar results as in D. melanogaster and D. simulans: the common and male-specific acceptors have 9–11 pyrimidines, while the female-specific acceptors have 7 or 8 pyrimidines upstream of NCAG (Fig. 4c, d). These results indicate that the presence of a weak splice acceptor (6–8 pyrimidines) may lead to intron retention of the dsx gene in males of four Drosophila species.
Fig. 4. Retained introns share conserved sequence properties.
a–d Comparison of donor and acceptor sequences of spliced and retained introns. The structure of the dsx gene including introns (black lines), the retained intron for dsxM2 (thick green line), and exons (boxes), as well as splicing donor and acceptor sequences for each intron in D. melanogaster (a), D. simulans (b), D. mojavensis (c) and D. virilis (d). Introns were labeled with numbers followed by “f” (female-specific splicing) or “m” (male-specific splicing). respectively. As for the consensus sequences, Y indicates pyrimidine, and N indicates either A, G, T or C. The number of pyrimidines in the 12 upstream nucleotides of the acceptor sequence NYAG is indicated in the brackets for each intron. e Comparison of retained intron sequences of dsxM2 in four Drosophila species. Underlined letters indicate potential Tra binding sites. Black asterisks indicate perfect matches of amino acids among four species, and gray asterisks indicate three matches out of four species.
We next compared full sequences of retained introns in the four Drosophila species. Surprisingly, these intron sequences, despite many differences, share intensive identity in two regions: one in the 3′ end of the intron overlapping the region of the weak acceptor site that may contribute to intron retention as above mentioned, and the other in the middle of these introns containing the core sequence (CAATCAAC) of the tra binding sequence, TC(T/A)(T/A)CAATCAACA, that occurs six times in the female-specific exon (Fig. 4e). The conservation of the above core sequence, which is a potential tra binding site within the retained introns, suggests its potential role in tra-mediated alternative splicing. Indeed, we observed intron retention in females with tra knocked down in all cells (actin-GAL4/+; UAS-traRNAi/+) (Supplementary Fig. 8). Together these results suggest that the weak splice acceptor and tra regulation jointly determine the splicing modes between dsxM2 (intron retained) and dsxF (intron spliced out).
A proposed evolutionary pathway of dsx alternative splicing
The identification of the dsxM2 transcript promoted us to analyze how the two forms of alternative splicing (IR and APA) were evolved to generate the modern day dsx transcripts (Fig. 5a). Substantial studies already elucidated an evolutionary pathway from sexually monomorphic dsx to sex-specific dsx transcripts upon the evolution of tra-dsx regulation in females16,17. A simple assumption is that sex-specific dsx transcripts come from a sexually monomorphic transcript dsxM2, or dsxM, or both. Due to the nature of close relationship between dsxM2 and dsxF (whether an intron with a weak splicing acceptor is spliced out or not), and the more conservation of the female-specific exon than the male-specific exons in insect species31,33, we propose that dsxM2 could serve as an ancient monomorphic transcript in both sexes, followed by two sequential events evolving alternative splicing: the IR-based mechanism generating dsxM2 in males and dsxF in females with the evolution of Tra regulation in females, and later the APA-based mechanism generating the rapid-changing dsxM with the evolution of a stronger splicing acceptor to capture male-specific exons (Fig. 5b).
Fig. 5. An evolutionary pathway of dsx alternative splicing underlying sexual development.
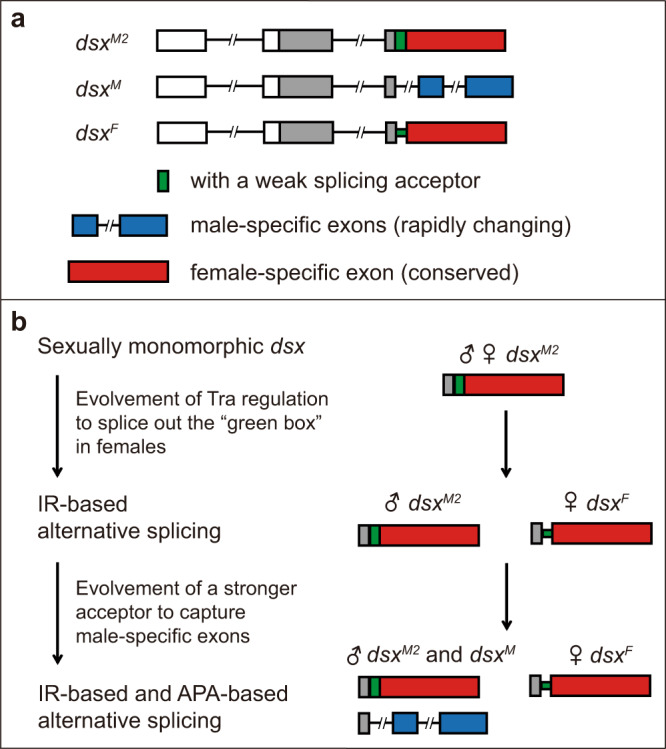
a Structures of the modern day dsx transcripts, dsxM2, dsxM, and dsxF in D. melanogaster. b A proposed evolutionary pathway from sexually monomorphic dsxM2 to sex-specific dsx transcripts sequentially via the IR-based and APA-based mechanisms.
Discussion
Previous studies have shown that alternative splicing of dsx generates sex-specific isoforms (DsxM and DsxF) underlying sexual differentiation in D. melanogaster and many other insects1,2,10,16. Such splicing mechanism involves the selective use of sex-specific 3′ exons, which is under regulation of Tra and Tra-210,20–22. Our results identified the male-specific DsxM2 isoform through intron retention, which is a common mechanism generating sex-specific Dsx isoforms in insects across 300 million years.
Recent studies on the alternative splicing of dsx using a variety of insect species revealed that the tra-mediated female-specific splicing of dsx could be evolved from sexually monomorphic isoforms with male-biased expression16,17. Indeed, vertebrates, nematodes and crustaceans use male-biased transcription of Dmrt genes to direct male-specific function1,2,34. In this regard, the evolution of dsxF may come from dsxM and/or dsxM2 under Tra regulation, possibly through the functional gain of Tra binding sites in the female-specific exon of dsx. Indeed, knocking down Tra function in females induced intron retention-based splicing of dsxM2 instead of dsxF. The coding sequence of the female-specific exon, which is also included as 3′-UTR of dsxM2, is more conserved than the male-specific exon of dsxM that rapidly changes not only across insect orders but also across closely related genera within each insect order31,33, excluding the possibility of dsxM as the origin of sexually monomorphic dsx transcript. The identification of the intron retention-based dsxM2 transcript fits well into a position as the ancestral isoform of dsx expressed in both sexes before the evolution of dsxM and dsxF. The conserved presence of a weak splicing acceptor upstream of the female-specific exon provides the selection basis for later recruitment of Tra regulation to generate the female-specific dsxF from dsxM2 in females, as well as the evolution of a stronger splicing acceptor to capture male-specific exons and generate dsxM in males. Such an evolutionary pathway of generating sex-specific Dsx isoforms could fill the gap between the splicing-based mechanism in insects and transcription-based mechanism in vertebrates and nematodes.
The DsxM2 isoform contains all common Dsx amino acids and six specific C-terminal residues, thus could act like other Dmrt transcription factors. Our knockdown experiments reveal that DsxM2 still functions in the nervous system to promote courtship robustness in D. melanogaster, but not in the development of male-specific traits such as genitals and sex combs. In addition, the overexpression experiments indicate that DsxM2 has a potential masculinizing role just like DsxM. However, a direct comparison of DsxM and DsxM2 function could not be faithfully achieved as knockdown and overexpression levels of dsxM and dsxM2 may be different. Another caveat of these results is the lack of direct evidence of DsxM2 expression in males. It is also possible that DsxM2 only serves as an ancient Dsx isoform whose function has largely been replaced during evolution (e.g., by DsxM), and now has limited expression and function. Future studies could generate better reagents to testify DsxM2 expression in a temporal and spatial manner in D. melanogaster and other insect species to better understand its function as a possible origin of sex-specific Dsx isoforms underlying sexual development and sexual dimorphism.
Methods
Fly stocks
Flies were raised at 22 °C or 25 °C at 12 hr light/12 hr dark cycle at 60% humidity. Canton-S (wtcs) and w1118 were used as wild-type strains. dsx mutant lines used in Fig. 1b and c include dsx683-7058 and dsx1649-9625, which were used as previously13. Drosophila mojavensis, Drosophila simulans and Drosophila virilis13, R57C10-GAL4 (attP2, BDSC_39171), actin-GAL4 (BDSC_25374), UAS-dsxMRNAi (attP2)14, UAS-flag-dsxF (attP40) and UAS-myc-dsxM (attP40)35, dsxGAL412 and dsxGAL4(Δ2)36 were used as described previously. UAS-dsxM2RNAi (attP2) and UAS-flag-dsxM2 (attP40) were generated in this study and described below in details. Detailed information about fly stocks and other materials used in this study is listed as Table 1.
Table 1.
Detailed information for key resources and fly stocks.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Mouse monoclonal anti-Flag | Sigma–Aldrich | Cat# F1804 | IHC (1:500) |
| Antibody | Mouse monoclonal anti-Myc | MBL | M047-3 | IHC (1:200) |
| Antibody | Donkey polyclonal anti-Mouse, Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-21202, RRID: AB_141607 | IHC (1:500) |
| Antibody | Donkey polyclonal anti-Mouse, Alexa Fluor 555 | Thermo Fisher Scientific | Cat# A-31570, RRID: AB_2536180 | IHC (1:500) |
| Plasmid | pVALIUM20 | Tsinghua University | ||
| Plasmid | pJFRC2-10×UAS-IVS-mCD8::GFP | Addgene | #26214 | |
| Chemical compound drug | Normal Goat Serum (NGS) | Jackson ImmunoResearch Laboratories | Code# 005-000-121 RRID: AB_2336990 | 3% NGS in 1×PBS |
| Chemical compound drug | Paraformaldehyde (PFA) | Sigma–Aldrich | CAS# 30525-89-4 | 4% PFA in 1×PBS |
| Chemical compound drug | TRIzol™ reagent | Invitrogen | Cat# 15596026 | |
| Chemical compound drug | SuperScript™ IV | Invitrogen | Cat# 18091050 | |
| Chemical compound drug | DNA Polymerase High Fidelity | Transgen | Cat# AS131-21 | |
| Chemical compound drug | EvaGreen Dye | Biotium | Cat# 31000 | |
| Genetic reagent (D. melanogaster) | UAS-dsxMRNAi | 14 | ||
| Genetic reagent (D. melanogaster) | dsxGAL4 | 12 | ||
| Genetic reagent (D. melanogaster) | dsxGAL4(Δ2) | 36 | ||
| Genetic reagent (D. melanogaster) | R57C10-GAL4 | Bloomington Drosophila Stock Center | BDSC_39171 | |
| Genetic reagent (D. melanogaster) | actin-GAL4 | Bloomington Drosophila Stock Center | BDSC_25374 | |
| Genetic reagent (D. melanogaster) | UAS-traRNAi | Bloomington Drosophila Stock Center | BDSC_28512 | |
| Genetic reagent (D. simulans) | Drosophila simulans | 13 | ||
| Genetic reagent (D. mojavensis) | Drosophila mojavensis | 13 | ||
| Genetic reagent (D. virilis) | Drosophila virilis | 13 | ||
| Genetic reagent (D. melanogaster) | UAS-dsxM2RNAi | This study | Described below | |
| Genetic reagent (D. melanogaster) | UAS-flag-dsxM2 | This study | Described below | |
| Genetic reagent (D. melanogaster) | UAS-flag-dsxF | 35 | Described below | |
| Genetic reagent (D. melanogaster) | UAS-myc-dsxM | 35 | Described below | |
| Software,algorithm | LaserGene | DNAStar | http://www.dnastar.com/ | |
| Software,algorithm | Clustal Omega | EMBL-EBI | https://www.ebi.ac.uk/Tools/msa/clustalo | |
| Software,algorithm | ImageJ | ImageJ National Institutes of Health | https://imagej.nih.gov/ij/ | |
| Software,algorithm | Prism 8 | GraphPad | https://www.graphpad.com/ | |
| Software,algorithm | Integrative Genomics Viewer | 37 | https://www.igv.org/ |
dsx sequences and multiple sequence alignment
We downloaded dsx gene sequence of D. melanogaster from FlyBase (http://flybase.org/). We used NCBI (https://www.ncbi.nlm.nih.gov/) to find the location and gene sequences of dsx in following species: D. simulans (Gene ID: 6727147), D. mojavensis (Gene ID: 6574377), D. virilis (Gene ID: 6633147) and B. germanica17,32. Gene sequence or amino acid sequences comparisons were performed using Clustal Omega and LaserGene software.
We also used public RNA-seq data27 (GSM694258 and GSM694259 for D. melanogaster females, GSM694260 and GSM694261 for D. melanogaster males, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE28078) to analyze splicing events of dsx (Supplementary Fig. 1b) using the Integrative Genomics Viewer (IGV)37.
Generation of UAS-dsxM2RNAi line
To generate the UAS-dsxM2RNAi construct, the UAS-dsxFRNAi plasmid (based on the pVALIUM20)14 was digested to be linearized with NheI and EcoRI. We selected a 21 nucleotides sequence (tggctgtgaagtgaaattgta) targeting the 114 bp intron, which was synthesized and generated into hairpin by annealing. The resulting DNA fragment and linearized vector were ligated. The resulting constructs were injected into embryos of attP2 site with PhiC31-mediated transgenesis. The correct insertion was screened by vermillion (vermillion positive eye color) and verified by PCR and followed by DNA sequencing. Oligo primers are as following:
Oligo forward:
5′-ctagcagtTGGCTGTGAAGTGAAATTGTAtagttatattcaagcataTACAATTTCACTTCACAGCCAgcg-3′
Oligo reverse:
5′-aattcgcTCGCACTGTAGCCCAGATCTAtatgcttgaatataactaTACAATTTCACTTCACAGCCAactg-3′.
Generation of UAS-flag-dsxF, UAS-myc-dsxM and UAS-flag-dsxM2 lines
UAS-flag-dsxF and UAS-myc-dsxM lines were generated previously in our lab35. The UAS-flag-dsxM2 line was generated in this study. In brief, the 3×Flag tags (MDYKDHDG-DYKDHDI-DYKDDDDKL) were added to the N terminus of DsxF and DsxM2 protein separately. We simultaneously amplified the flag sequence and the full-length CDS fragment of dsxF and dsxM2 from wild-type cDNAs using the following overhang primers and then cloned into pJFRC2-10XUAS-IVS-mCD8::GFP plasmid (Addgene #26214) to replace the mCD8::GFP sequence.
dsxF-forward: CCGAGATCTATGGACTACAAAGACCATGACGGTGATTATAAAGATCATGACATCGATTACAAGGATGACGATGACAAGCTTGTTTCGGAGGAGAACTGG
dsxF-reverse: CCATCTAGATCATCCACATTGCCGCGTTG.
dsxM2-forward: CCGAGATCTATGGACTACAAAGACCATGACGGTGATTATAAAGATCATGACATCGATTACAAGGATGACGATGACAAGCTTGTTTCGGAGGAGAACTGG
dsxM2- reverse: CCATCTAGATCACTGATCAACAGACTTACC.
For the UAS-myc-dsxM transgenic line, the 6×Myc tag (MEQKLISEEDL) was added to the N terminus of DsxM. We first cloned the 6×myc sequence into pJFRC2-10XUAS-IVS-mCD8::GFP between the BglII and the XhoI sites, and then amplified the full-length CDS of dsxM and cloned into C terminus of 6×Myc to replace the mCD8::GFP sequences.
myc-F: CCGAGATCTTCCCATCGACTTAAAGCTATG
myc-R: CCACTCGAGACTAGTCTCAAGAGGCCTTGAGTTC
dsxM-F: CCGACTAGTGTTTCGGAGGAGAACTGG
dsxM-R: CCATCTAGACTACGTGGCAGCCGTGGAG
The constructed plasmids were injected and integrated into the attP40 sites on the second chromosome through phiC31 integrase mediated transgenesis. The correct strains were screened by mini-white (orange eyes) and verified by PCR and followed by DNA sequencing.
RT-PCR analysis
To acquire cDNA template, Canton-S, w1118 and other species including D. simulans, D. mojavensis, D. virilis and Blattella germanica were used as wild-type strains for RT-PCR. We obtained total RNA from approximate thirty adult females and/or males using a commercial TRizol™ reagent (15596026, Invitrogen, USA) and purified RNA with DNA-free™ kit (AM1906, AMbion) according to manufacturer’s protocol. Purified RNA was finally resuspended in 60μL of DEPC-treated water. First strand cDNA was synthesized for each RNA sample using SuperScript™ IV reverse transcriptase (18091050, Invitrogen). dsx-specific primers are listed in Table 2 to detect different dsx transcripts.
Table 2.
Primers used for RT-PCR experiments.
| Usage | Primer names | Sequences (5′–3′) |
|---|---|---|
| Fig. 1d, e | Primer 1 | Forward: CAATCGCTGGAGGGGTCCTG |
| Reverse: TCATCCACATTGCCGCGTTG | ||
| Fig. 1d, e | Primer 2 | Forward: GCAAAGCACACCTCGCGGAG |
| Reverse: TCATCCACATTGCCGCGTTG | ||
| Fig. 1d, e | Primer 3 | Forward: TTCCGCTATCCTTGGGAGCT |
| Reverse: TTGGCTTGTATGCCTATTCG | ||
| Fig. 3a, b | Primer 1 | Forward: GAATCATGGTTTCGGAGGAGAACTG |
| Reverse: CTACGTGGCAGCCGTGGAGCTCACC | ||
| Fig. 3a, b | Primer 2 | Forward: GAATCATGGTTTCGGAGGAGAACTG |
| Reverse: TCATCCACATTGCCGCGTTGTGTTGC | ||
| Fig. 3d, e | Primer 1 | Forward: ATGGTTTCGGAGGAGAACTGG |
| Reverse: TCATCCACATTGCCGCGTTGT | ||
| Fig. 3d, e | Primer 2 | Forward: ACGCAAGAATGTGCCACTCG |
| Reverse: TCATCCACATTGCCGCGTTGT | ||
| Fig. 3g, h | Primer 1 | Forward: ATGGTTTCAGAGGAGAATTGGAACA |
| Reverse: TCATCCACATTGCCGCGTTGTGTTG | ||
| Fig. 3g, h | Primer 2 | Forward: AGCACACGCAAGAATGTGCCACTGG |
| Reverse: TCATCCACATTGCCGCGTTGTGTTG | ||
| Fig. 3j, k | Primer | Forward: AGAACGGCAGCGAGACAGGC |
| Reverse: TGTGGAGACGGGCGATGAGG | ||
| Supplementary Fig. 8b, c | Primer 1 | Forward: ATGGTTTCGGAGGAGAACTG |
| Reverse: TCATCCACATTGCCGCGTTG | ||
| Supplementary Fig. 8b, c | Primer 2 | Forward: CAATCGCTGGAGGGGTCCTG |
| Reverse: TCATCCACATTGCCGCGTTG |
qPCR
Total RNA extraction and first strand cDNA template were acquired as described above, except for the data in Supplementary Fig. 3, which used approximately 100 flies to obtain different tissues (head, thorax, abdomen, forelegs, and wings). qPCR was performed using a LightCycler® 96 SW 1.1 system (Roche). We used EvaGreen Dye (31000, Biotium, USA) and High Fidelity PCR SuperMix (AS131–21, TransGen, Beijing) to conduct qPCR. actin was amplified as an internal control for normalization. The primers for qPCR used in this study are listed in Table 3.
Table 3.
Primers used for qPCR experiments.
| Sequences (5′–3′) | |
|---|---|
| actin | Forward: CAGGCGGTGCTTTCTCTCTA |
| Reverse: AGCTGTAACCGCGCTCAGTA | |
| dsxM | Forward: GAAGAGGCTTCCCGGCGAAT |
| Reverse: GGACAAATCTGTGTGAGCGG | |
| dsxF | Forward: TTCCGCTATCCTTGGGAGCT |
| Reverse: CATCCACATTGCCGCGTTGT | |
| dsxM2 | Forward: GCCGATCTCAGTTTCCGTCA |
| Reverse: TCACTGATCAACAGACTTAC | |
| yp2 | Forward: TGGGTCAATCCACGTGAAGT |
| Reverse: ACAATGTAGCCCCTGATCTG | |
| yp3 | Forward: GAAGCCGACCAAGTGGCTGA |
| Reverse: TCCAGACGGGCACATTGCTC |
Male courtship assay
Newly enclosed males were collected and group housed for 4–7 days at 25°C and 60% humidity with a 12 hr: 12 hr light/dark cycle. Virgin Canton-S females were group-housed and aged under similar conditions. To measure courtship, a male of each genotype and a Canton-S virgin female were loaded individually into two-layer chambers (diameter: 1 cm; height: 3 mm per layer) which were separated by a plastic transparent barrier until courtship test for 30 min. Courtship index (CI), which is the percentage of observation time a fly displayed any courtship step in 10 min, and percentages mated were counted based on successful copulation in 30 min.
Tissue dissection, staining, and imaging
Adult flies were reared at 25°C and aged for 4–7 days old, and were dissected in Schneider’s insect medium (Thermo Fisher Scientific, Waltham, MA) and fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) for 20–30 min at room temperature (RT). Tissues were washed at least 4 times for 15–20 min with PAT3 (0.5% Triton X-100, 0.5% bovine serum albumin in PBS), and then blocked in 3% normal goat serum (NGS) for 60 min at RT. In Supplementary Fig. 5e and f, samples were incubated with anti-Flag mouse (Sigma, F1804, 1:500) antibody or anti-Myc mouse (MBL, M047-3, 1:200) antibody diluted in 3% NGS for overnight at 4 °C, then washed four times in PAT3, and incubated in secondary antibodies anti-mouse IgG conjugated to Alexa 488 (Invitrogen, A21202, 1:500) diluted in 3% NGS for 1–2 days at 4 °C. Tissues were then washed thoroughly in PAT3 and mounted for confocal imaging.
For visualizing morphological appearances of flies (Fig. 2f, g and Supplementary Fig. 8a), 4–7 days old flies were frozen at -80°C for 30 min. Fly forelegs and abdomens were dissected and imaged by a Nikon Shuttle pix P400RV stereoscopic microscope.
Statistics and reproducibility
All statistical analyses were performed using the Prism 8 (GraphPad software). Experimental flies and genetic controls were tested at the same condition, and data are collected from at least two independent experiments and are reproducible. The Mann-Whitney U test was used for pairwise comparisons. For comparing mating success, Chi-square tests were performed to compare two different groups at 30 min time point.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
We thank the Bloomington Drosophila Stock Center and the Tsinghua Fly Center for fly stocks, and Dr. Sheng Li and Nan Chen (South China Normal University) for generously providing cDNA of Blattella germanica. This work was supported by grants from National Key R&D Program of China (2019YFA0802400), the National Natural Science Foundation of China (31970943 and 31700905), and the Jiangsu Innovation and Entrepreneurship Team Program.
Author contributions
C.H., Q.P. and Y.P. designed research; C.H., Q.P., X.S. and L.X. generated transgenic flies; C.H., and X.S. performed behavioral experiments; C.H. and X. Ji. performed immunostaining and qPCR experiments; C.H., Q.P. and Y.P. analyzed data; and Y.P. wrote the manuscript with input from all authors.
Peer review
Peer review information
Communications Biology thanks Maroun Bou Sleiman and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Luke R. Grinham. Peer reviewer reports are available.
Data availability
All data generated or analyzed during this study are included in the manuscript and its supplementary information files. Source data underlying figures are presented in Supplementary Data 1. Uncropped gel images are provided in Supplementary Fig. 9. All other relevant data supporting the findings of this study are available from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Caihong Han, Qionglin Peng.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-022-03664-7.
References
- 1.Kopp A. Dmrt genes in the development and evolution of sexual dimorphism. Trends Genet. 2012;28:175–184. doi: 10.1016/j.tig.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matson CK, Zarkower D. Sex and the singular DM domain: insights into sexual regulation, evolution and plasticity. Nat. Rev. Genet. 2012;13:163–174. doi: 10.1038/nrg3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Williams TM, Carroll SB. Genetic and molecular insights into the development and evolution of sexual dimorphism. Nat. Rev. Genet. 2009;10:797–804. doi: 10.1038/nrg2687. [DOI] [PubMed] [Google Scholar]
- 4.Raymond CS, et al. Evidence for evolutionary conservation of sex-determining genes. Nature. 1998;391:691–695. doi: 10.1038/35618. [DOI] [PubMed] [Google Scholar]
- 5.Veitia R, et al. Deletions of distal 9p associated with 46,XY male to female sex reversal: definition of the breakpoints at 9p23.3-p24.1. Genomics. 1997;41:271–274. doi: 10.1006/geno.1997.4648. [DOI] [PubMed] [Google Scholar]
- 6.Smith CA, McClive PJ, Western PS, Reed KJ, Sinclair AH. Conservation of a sex-determining gene. Nature. 1999;402:601–602. doi: 10.1038/45130. [DOI] [PubMed] [Google Scholar]
- 7.Kim S, Bardwell VJ, Zarkower D. Cell type-autonomous and non-autonomous requirements for Dmrt1 in postnatal testis differentiation. Dev. Biol. 2007;307:314–327. doi: 10.1016/j.ydbio.2007.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matson CK, et al. DMRT1 prevents female reprogramming in the postnatal mammalian testis. Nature. 2011;476:101–104. doi: 10.1038/nature10239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lints R, Emmons SW. Regulation of sex-specific differentiation and mating behavior in C. elegans by a new member of the DM domain transcription factor family. Genes Dev. 2002;16:2390–2402. doi: 10.1101/gad.1012602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burtis KC, Baker BS. Drosophila doublesex gene controls somatic sexual differentiation by producing alternatively spliced mRNAs encoding related sex-specific polypeptides. Cell. 1989;56:997–1010. doi: 10.1016/0092-8674(89)90633-8. [DOI] [PubMed] [Google Scholar]
- 11.Robinett CC, Vaughan AG, Knapp JM, Baker BS. Sex and the single cell. II. There is a time and place for sex. PLoS Biol. 2010;8:e1000365. doi: 10.1371/journal.pbio.1000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rideout EJ, Dornan AJ, Neville MC, Eadie S, Goodwin SF. Control of sexual differentiation and behavior by the doublesex gene in Drosophila melanogaster. Nat. Neurosci. 2010;13:458–466. doi: 10.1038/nn.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan Y, Baker BS. Genetic identification and separation of innate and experience-dependent courtship behaviors in Drosophila. Cell. 2014;156:236–248. doi: 10.1016/j.cell.2013.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peng Q, et al. The sex determination gene doublesex is required during adulthood to maintain sexual orientation. J. Genet Genomics. 2022;49:165–168. doi: 10.1016/j.jgg.2021.08.006. [DOI] [PubMed] [Google Scholar]
- 15.Zarkower D. Establishing sexual dimorphism: conservation amidst diversity? Nat. Rev. Genet. 2001;2:175–185. doi: 10.1038/35056032. [DOI] [PubMed] [Google Scholar]
- 16.Hopkins BR, Kopp A. Evolution of sexual development and sexual dimorphism in insects. Curr. Opin. Genet Dev. 2021;69:129–139. doi: 10.1016/j.gde.2021.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wexler J, et al. Hemimetabolous insects elucidate the origin of sexual development via alternative splicing. eLife. 2019;8:e47490. doi: 10.7554/eLife.47490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhuo JC, et al. A feminizing switch in a hemimetabolous insect. Sci. Adv. 2021;7:eabf9237. doi: 10.1126/sciadv.abf9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiuchi T, et al. A single female-specific piRNA is the primary determiner of sex in the silkworm. Nature. 2014;509:633–636. doi: 10.1038/nature13315. [DOI] [PubMed] [Google Scholar]
- 20.Nagoshi RN, McKeown M, Burtis KC, Belote JM, Baker BS. The control of alternative splicing at genes regulating sexual differentiation in D. melanogaster. Cell. 1988;53:229–236. doi: 10.1016/0092-8674(88)90384-4. [DOI] [PubMed] [Google Scholar]
- 21.Goralski TJ, Edstrom JE, Baker BS. The sex determination locus transformer-2 of Drosophila encodes a polypeptide with similarity to RNA binding proteins. Cell. 1989;56:1011–1018. doi: 10.1016/0092-8674(89)90634-X. [DOI] [PubMed] [Google Scholar]
- 22.McKeown M, Belote JM, Baker BS. A molecular analysis of transformer, a gene in Drosophila melanogaster that controls female sexual differentiation. Cell. 1987;48:489–499. doi: 10.1016/0092-8674(87)90199-1. [DOI] [PubMed] [Google Scholar]
- 23.Williams TM, et al. The regulation and evolution of a genetic switch controlling sexually dimorphic traits in Drosophila. Cell. 2008;134:610–623. doi: 10.1016/j.cell.2008.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burtis KC, Coschigano KT, Baker BS, Wensink PC. The doublesex proteins of Drosophila melanogaster bind directly to a sex-specific yolk protein gene enhancer. EMBO J. 1991;10:2577–2582. doi: 10.1002/j.1460-2075.1991.tb07798.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coschigano KT, Wensink PC. Sex-specific transcriptional regulation by the male and female doublesex proteins of Drosophila. Genes Dev. 1993;7:42–54. doi: 10.1101/gad.7.1.42. [DOI] [PubMed] [Google Scholar]
- 26.Sanders LE, Arbeitman MN. Doublesex establishes sexual dimorphism in the Drosophila central nervous system in an isoform-dependent manner by directing cell number. Dev. Biol. 2008;320:378–390. doi: 10.1016/j.ydbio.2008.05.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Graveley BR, et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471:473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown JB, et al. Diversity and dynamics of the Drosophila transcriptome. Nature. 2014;512:393–399. doi: 10.1038/nature12962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jenett A, et al. A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2012;2:991–1001. doi: 10.1016/j.celrep.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Belote JM, Handler AM, Wolfner MF, Livak KJ, Baker BS. Sex-specific regulation of yolk protein gene expression in Drosophila. Cell. 1985;40:339–348. doi: 10.1016/0092-8674(85)90148-5. [DOI] [PubMed] [Google Scholar]
- 31.Baral, S., Arumugam, G., Deshmukh, R. & Kunte, K. Genetic architecture and sex-specific selection govern modular, male-biased evolution of doublesex. Sci. Adv.5, eaau3753 (2019). [DOI] [PMC free article] [PubMed]
- 32.Li S, et al. The genomic and functional landscapes of developmental plasticity in the American cockroach. Nat. Commun. 2018;9:1008. doi: 10.1038/s41467-018-03281-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hughes AL. Runaway evolution of the male-specific exon of the doublesex gene in Diptera. Gene. 2011;472:1–6. doi: 10.1016/j.gene.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kato Y, Kobayashi K, Watanabe H, Iguchi T. Environmental sex determination in the branchiopod crustacean Daphnia magna: deep conservation of a Doublesex gene in the sex-determining pathway. PLoS Genet. 2011;7:e1001345. doi: 10.1371/journal.pgen.1001345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peng Q, et al. The sex determination gene doublesex regulates expression and secretion of the basement membrane protein Collagen IV. J Genet. Genomics. 2022;49:636–644. doi: 10.1016/j.jgg.2021.12.010. [DOI] [PubMed] [Google Scholar]
- 36.Pan Y, Robinett CC, Baker BS. Turning males on: activation of male courtship behavior in Drosophila melanogaster. PLoS One. 2011;6:e21144. doi: 10.1371/journal.pone.0021144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
All data generated or analyzed during this study are included in the manuscript and its supplementary information files. Source data underlying figures are presented in Supplementary Data 1. Uncropped gel images are provided in Supplementary Fig. 9. All other relevant data supporting the findings of this study are available from the corresponding author upon reasonable request.