Abstract
Objectives
To investigate the role of polymorphism rs11200638 of high-temperature requirement factor A-1 (HtrA1) gene in the pathogenesis of age-related macular degeneration (AMD).
Methods
Cultured adult retinal pigment epithelial cells (ARPE-19) expressing HtrA1 gene were treated with H2O2 or lipopolysaccharides (LPS) and analysed using western blot and quantitative polymerase chain reaction to illustrate the effects of oxidative and inflammatory stress on HtrA1 gene expression. Luciferase reporter plasmid driven by HtrA1 promoter with either normal allele G or risk allele A at SNP rs11200638 was transfected to ARPE-19 cells to investigate the effect of the G/A variation on HtrA1 promoter activity. The effects of HtrA1 overexpression on ARPE-19 cells were analysed with respect to percentage of cell proliferation inhibition and cell apoptosis.
Results
HtrA1 expression was significantly increased with LPS or H2O2 stimulations (p < 0.05). In ARPE-19 cells, HtrA1 promoters (−1 to −2175 bp from translation starting point) with risk allele A or normal G at rs11200638 did not show statistically significant differences in their luciferase reporter expression (p = 0.054425173), however, both promoters showed a persistent trend of higher luciferase expressions after 100 ng/ml LPS treatment. The luciferase expression level was significantly greater in the promoter with risk A when compared to that with normal G. Overexpression of HtrA1 resulted in apoptosis of ARPE-19 cells with 53.8 ± 1.6% of proliferation inhibition (p < 0.01).
Conclusions
Risk haplotype A at rs11200638 significantly increased the responsiveness of HtrA1 promoter to inflammation and subsequently enhanced HtrA1 expression. HtrA1 overexpression induced ARPE-19 apoptosis and growth inhibition, relevant to pathogenesis of AMD.
Subject terms: Mechanisms of disease, Inflammation
Introduction
Age-related macular degeneration (AMD) is the leading cause of blindness in elder individuals, accounting for 8.7% of blindness globally [1]. Both environmental and genetic risk factors are involved in AMD pathogenesis [2, 3]. Major progress has been made in determining the genetic basis of AMD through the identification of susceptibility gene loci [4–6]. Among those reported loci, high-temperature requirement factor A-1 (HtrA1) gene has been shown to be one of the major causal genes to AMD [7–9]. Certain single nucleotide polymorphisms (SNPs) including rs11200638, rs2672598, rs1049331 and rs2293870 in the promoter region and the first exon of HtrA1 gene have been identified to be implicated in AMD [10–12].
SNP rs11200638 in HtrA1 promoter site was deemed as the AMD-associated allele primarily based on the assumption that rs11200638 upregulated HtrA1 expression. While previous studies suggested that variant rs11200638 increased the expression of HtrA1 and the susceptibility to AMD [13–15]. Yang et al. reported that rs11200638 with risk allele was correlated with elevated HtrA1 expression in lymphocytes and RPE tissue from AMD patients [14]. Subsequently, this correlation was confirmed in the retinas of patients with AMD [13, 16–19]. However, the potential role of HtrA1 encompassed by the rs11200638 haplotype in the pathogenesis of AMD remains unclear, because the previous evidences pertaining to the correlation between rs11200638 genotypes and HtrA1 expression levels were conflicting [20–23]. Some reporters claimed that variant rs11200638 increased the expression of HtrA1, but others claimed that the genotypes at rs11200638 were not correlated with HtrA1 expression. We speculated that micro-environmental risk factors were necessary to determine the role of AMD-associated allele on the pathogenesis of AMD. Variant rs11200638 may alter the HtrA1 promoter responsiveness to those risk factors, i.e., oxidative stress and inflammation, relevant to the pathogenesis of AMD.
Retinal pigment epithelium (RPE) senescence induced by oxidative stress is one of the key contributors to early AMD and drusen formation [11]. The role of inflammation in early stage has been implicated in the pathogenesis of late-stage AMD [24]. RPE cell, one of the most important cells involved in the pathogenesis of AMD, is essential for maintaining vision by keeping photoreceptors healthy [25]. There is a mutualistic relationship between the components of the photoreceptor/RPE/Bruch’s membrane/choriocapillaris complex. Aging and the accumulated effects of environmental stresses may result in death and dysfunction of all of the components in the complex and progress to AMD. HtrA1 expression and its promoter activity were investigated in cultured adult RPE (ARPE-19) cells. The effects of overexpression of HtrA1 on ARPE-19 cell growth were also studied.
Materials and methods
Cell culture
ARPE-19 cells were obtained from the American Type Culture Collection (Manassas, VA, USA) and cultured in Dulbecco’s modified Eagle’s medium (DMEM, low glucose formulation, 11330-033; Gibco, Invitrogen, CA, USA) supplemented with 10% (v/v) foetal bovine serum (FBS, 04-001-1ACS; BI, US origin), 100 U/ml penicillin and 100 μg/ml streptomycin (10378-016; Gibco).
STR (short tandom repeat) analysis
The STR analysis was performed on the cell line to genotype the cell line by iCell Bioscience Inc (Shanghai, China). DNA was extracted using the Axygen genomic DNA extraction kit (200600; Corning, USA), and the STR loci and gender gene Amelogenin were detected using the ABI 3730XL genetic analyser according to the 20-STR amplification protocol.
LPS (lipopolysaccharides) and H2O2 stimulation of ARPE-19 cells
ARPE-19 cells grown to 70–80% cell confluence in six-well cell plates were incubated overnight in fresh DMEM without FBS, and subsequently treated with various concentrations of lipopolysaccharides (LPS, 01062003; DIFCO, Livonia, MI, USA) or H2O2. For inducing oxidative stress, H2O2-conditioned media was used at final concentrations of 0.2, 2, 20 or 200 µM H2O2, respectively, in DMEM for 24 h, by dissolving 30% H2O2 stock solution (Q18755; Fisher Scientific, Fair lawn, NJ, USA) before use. For inducing inflammation, LPS was used at final concentrations of 0.1, 1, 10 or 100 ng/ml, respectively, in DMEM for 24 h. The treated cells were investigated for HtrA1 protein and mRNA expression level using western blot and quantitative polymerase chain reaction (qPCR), respectively. The doses of LPS and H2O2 that increased HtrA1 expression significantly were defined as effective doses.
Western blot analysis
The ARPE-19 cells were lysed in lysis buffer containing 1% Triton X-100 (0694; Amersco, OH, USA), 1% deoxycholate (0613; Amersco), and proteinase inhibitor cocktail (04693132001; Roche, Rotkreuz, Switzerland). Total protein concentrations were measured using a modified Lowry protein assay kit (500-0001; BioRad, CA, USA). Equal amounts of denatured cellular protein (25 μg) from each sample were applied to 10% SDS-polyacrylamide gels, and resolved proteins were transferred to nitrocellulose membrane for sequential probing with specific antibodies, i.e., rabbit polyclonal anti-htrA1 antibody (ab38610; Abcam, MA, USA), anti-superoxide dismutase 1 (SOD1) antibody (2582-1; Epitomics, CA, USA) or monoclonal antibody against interleukin-6 (IL-6) (1457-1; Epitomics, CA, USA), respectively, and the secondary antibody against rabbit IgG conjugated with horseradish peroxidase (611035215; Jackson ImmunoResearch, PA, USA). PBS-treated cells were used as negative control. In order to avoid the false-positive caused by non-specific adsorption, the sample without cells was used as a negative control in the experiment. The blots were developed with BeyoECL plus enhanced chemiluminescence detection kit (P0018S; Beyotime, Shanghai, China). Band density was analysed using Quantity One software (Bio-Rad, CA, USA) and the grey value (area under the optical density distribution curve) was used to determine the relative protein levels.
Preparation of RNA
Total RNA was isolated from the cultured ARPE-19 cells with TRIzol Reagent (15596026, Invitrogen, CA, USA) following the manufacturer’s instructions. The RNA pellets were washed with 75% ethanol and dissolved in RNase-free water. The residual DNA was digested with DNase I. Concentration and purity (i.e., OD260/OD280) of RNA samples were monitored with a SmartSpec Plus spectrophotometer (Bio-Rad, CA, USA). RNA integrity was evaluated by examining the 28S and 18S rRNA bands after separation in a 1% agarose gel.
QPCR analysis
Total RNA was reverse-transcribed to complementary DNA (cDNA) using RevertAid™ first-strand cDNA synthesis kit (K1622, Fermentas, Vilnius, Lithuania). In brief, a 12 µl mixture containing 1 µg RNA and 1 µl oligo (dT)18 (0.5 µg/µl) was incubated at 65 °C for 5 min, followed by adding 4 µl reaction buffer, 1 µl RNase inhibitor (20U/µl), 2 µl dNTP Mix (10 mM), and 1µl M-MuLv Reverse Transcriptase (200 U/µl). Then, the total reverse-transcription system above was incubated sequentially at 42 °C for 60 min and 70 °C for 5 min.
QPCR was carried out on iCycler-iQ5® (Bio-Rad, CA, USA) as described [26]. In brief, a 20 µl reaction mixture, containing 1× SoFast EVAGreen supermix (1725202, Bio-Rad), 2 µl of cDNA, and 330 nM of each of the forward and reverse primers, was cycled sequentially, 30 s at 95 °C for one cycle, 5 s at 95 °C and 10 s at optimum annealing temperature (Table 1 of Supplementary materials) for 40 cycles. Primers were synthesised (HPLC purification grade) at Beijing Genomics Institute, China (Supplementary Table 1). Assays for each cDNA sample were performed in triplicate (maximum ∆Ct of replicate samples <0.5). Relative expression calculations, corrected with PCR efficiency and normalised versus the reference gene GAPDH (NM002046.3; GenBank), were performed with IQ 5 software (Bio-Rad). The results were expressed as the ratio of specific mRNA/GAPDH mRNA. The experiment was repeated three times for consistency.
Luciferase reporter assays
A DNA fragment containing −1 to −2175 bp from the HtrA1 translation site including protective haplotype G allele at rs11200638 (wild type, WT) was amplified from genomic DNA of an individual using nested-PCR with the following primers: inner forward, CGACGCGTCGCTGGAGGGGAGGAGGGGT, inner reverse, CCCAAGCTTGGCGACTCTGGCGGCGGC; outer forward, GGGCACGAGGATGGAAGAGG, outer reverse, AGCAGCAGCGGGAGAAGAGC. The construct was subcloned into the Mlu I- Hind III site of the pGL3-Basic vector (E1751, Promega, WI, USA), and then verified using complete bidirectional DNA sequencing. The risk haplotype construct carrying risk A allele at rs11200638 (mutant type, MT) was generated using site-directed mutagenesis kit (210518, Agilent Technologies, CA, USA), with the following primers: forward, GGACGCTGCCTTCGTCCAGCCGCAGAGGCCCCG, and reverse, GGGGCCTCTGCGGCTGGACGAAGGCAGCGTCCGC. The pGL3-Basic vector without insert (negative) was transfected into ARPE-19 cells as a negative control. A positive control plasmid (pGL3-control Vector, E1741, Promega, WI, USA) containing an SV40 enhancer and promoter driving luciferase reporter was obtained from Promega.
ARPE-19 cells were split into 96-well plates and cultured overnight, then co-transfected with 1 ng of the transfection control Renilla luciferase plasmid pTK-RL (Promega) and 200 ng of WT or MT plasmid. Transfections were performed using the Lipofectamine® LTX&PLUS™ Reagent (15338100; Invitrogen, CA, USA). Each type of plasmid was transfected in three wells simultaneously. Twenty-four hours after transfection, cells were washed twice with PBS and then harvested. Luciferase activities were measured using the Luciferase Assay Kit (E1910; Promega, WI, USA). Fold induction was derived relatively to the normalised reporter activity.
To investigate the effect of inflammatory and oxidative stress on the HtrA1 reporter activity, after pre-treatment with 200 μM H2O2 or 100 ng/ml LPS for 6 h to induce cellular stress, ARPE-19 cells were then transiently transfected with a luciferase reporter plasmid driven by the HtrA1 promoter, with WT or MT at SNP rs11200638. Six hours after transfection, ARPE-19 cells were further treated with 200 μM H2O2 or 100 ng/ml LPS for 24 h. Luciferase activities were finally measured as described above.
Cloning of human HtrA1
A 1443-bp open reading frame of the HtrA1 gene (NM 002775.4; GenBank) was cloned into an empty OmicsLink™ expression clone pReceiver-M06 (GeneCopoeia, Guangzhou, China) to obtain HtrA1 expression clone of EX-M0558-M06. The information pertaining to the vector and restriction enzyme for EX-M0558-M06 was presented in Fig. 4a. The construct was verified by direct sequencing. Sub-confluent ARPE-19 cells in 96-well plate were transfected with 3 μg constructed plasmid per well in transfection reagent (Lipofectamine® LTX&PLUS™ Reagent). Twenty-four hours after transfection, expression of the recombinant human HtrA1 protein was validated by western blot.
Fig. 4. Overexpression of HtrA1 in cultured ARPE-19 cells.
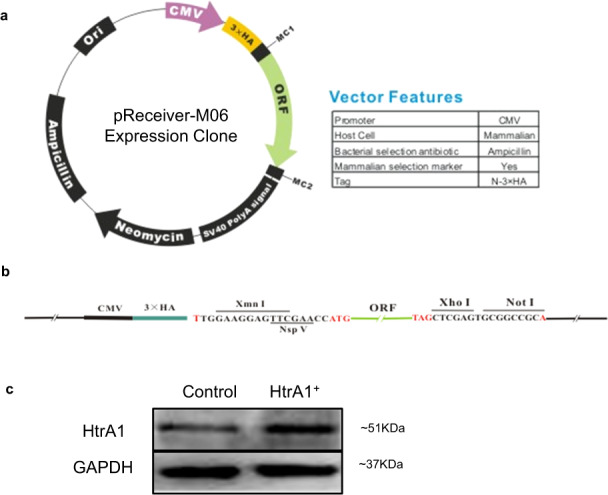
a, b Vector and restriction enzyme information for EX-M0558-M06. c Western blot analysis of HtrA1 expression in ARPE-19 cells after transfected with the expression vector hosting HtrA1 cDNA. HtrA1+, ARPE-19 cells transfected with HtrA1 construct EX-M0558-M06; Control, empty vector control.
Cell proliferation assay
Cell proliferation was analysed using Cell Counting Assay Kit-8 (CCK-8) (DJDB4000X, Dojindo Molecular Technologies, Kumamoto, Japan). Briefly, 100 μl of ARPE-19 cell suspension (5 × 103 cells/well) was dispensed into a 96-well plate and pre-incubated overnight, then transfected with 3 μg HtrA1 construct EX-M0558-M06 per well in transfection reagent (Lipofectamine® LTX&PLUS™ Reagent). Twenty-four hours after transfection, 10 μl CCK-8 solutions were added to each well. After an additional incubation for 1 h, the absorbance at 450 nm was measured using a microplate reader. The inhibition percentage of cell proliferation was recorded using transfection reagent treated cells at 100%.
Cell apoptosis detection
Cell apoptosis was analysed using the Annexin V-FITC/propidium iodide (PI) apoptosis assay kit (KGA108; KeyGEN Biotech, Nanjing, China). Briefly, 24 h after transfection with HtrA1 construct EX-M0558-M06, ARPE-19 cells were harvested and washed twice with cold PBS (pH 7.4) and stained with 3 μl of Annexin V and 3 μl of PI in 300 μl of binding buffer for 10 min in the dark. The number of apoptotic cells was analysed using Becton Dickinson FACScan Flow Cytometer (Becton Dickinson, MD, USA).
Statistical analysis
Each sample was triplicated. Every experiment was repeated at least three times for consistency. The experimental data were expressed as means () ± standard deviation (SD), and analysed with software SPSS Statistics 17.0 (SPSS Inc., IL, USA) for group-wise comparisons and statistical analyses. T-test was used for comparison between two groups. Differences were considered statistically significant when P < 0.05.
Results
STR analysis
The DNA genotype of the cell line was found to completely match in the cell line retrieval, which showed that the cells named ARPE-19 and ARPE-19/HPV-16 in DSMZ database. The cell numbers were related to CRL-2302 and CRL-2502, respectively. No multiple alleles were found in this cell line. The results of the genotype were listed in Supplementary materials (including Supplementary Table 2).
H2O2-mediated induction of HtrA1 expression in ARPE-19 cells
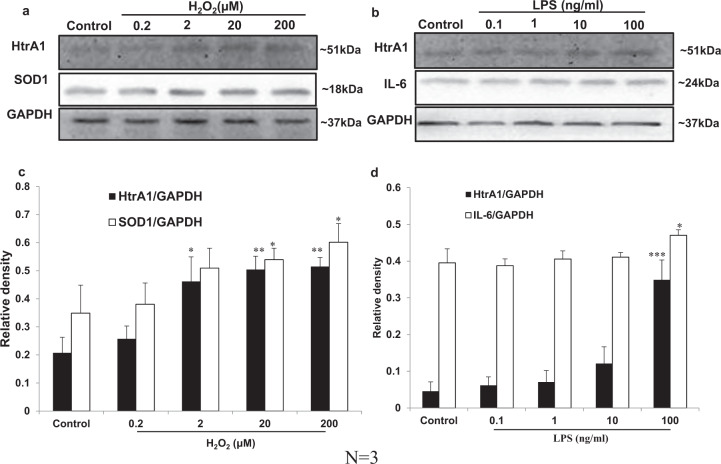
To investigate the involvement of oxidative stress in HtrA1 expression, ARPE-19 cells were treated with diverse concentrations of H2O2 for 24 h. The effects of oxidative stress were evidenced by the increase in SOD-1 protein level (Fig. 1a, c). Western blot result showed that H2O2 at concentrations of 2, 20 and 200 μM induced 2.23 ± 0.42 (n = 3, p < 0.05), 2.43 ± 0.23 (n = 3, p < 0.01), and 2.48 ± 0.15-fold (n = 3, p < 0.01) increases in HtrA1 protein level, respectively (Fig. 1a, c).
Fig. 1. Western blot analysis of HtrA1 expression in ARPE-19 cells treated with H2O2 or LPS.
Serum-starved ARPE-19 cells were plated on dishes in medium supplemented with varying concentrations of H2O2 (a) or LPS (b) for 24 h and then lysed. The lysates were analysed by western blot. The control was performed with vehicle. Blots were developed with HRP-conjugated secondary antibodies and an enhanced chemiluminescence detection kit. c, d Densitometry of three independent experiments. The western blot gels were cropped according to the molecular size of protein. Band density (grey value) was analysed using Quantity One software and used to determine the relative protein level. Results were expressed as means ± SD (n = 3). *p < 0.05, **p < 0.01, ***p < 0.001, compared with control.
Similarly, qPCR data showed a 2.94 ± 0.06-fold (n = 3, p < 0.001) significant increase in HtrA1 mRNA expression level with the minimum effective dose of 200 μM H2O2 (Fig. 2a). These results suggest that oxidative stress simulated by H2O2 may enhance HtrA1 expression at both the mRNA and protein levels. Due to the volatility of H2O2, the maximum effective dose of 200 μM H2O2 was used in subsequent experiments to obtain relative certain oxidative stress induction for evaluation of the HtrA1 promoter activity.
Fig. 2. Changes in the relative expression of HtrA1 mRNA expression in ARPE-19 cells treated with H2O2 or LPS.
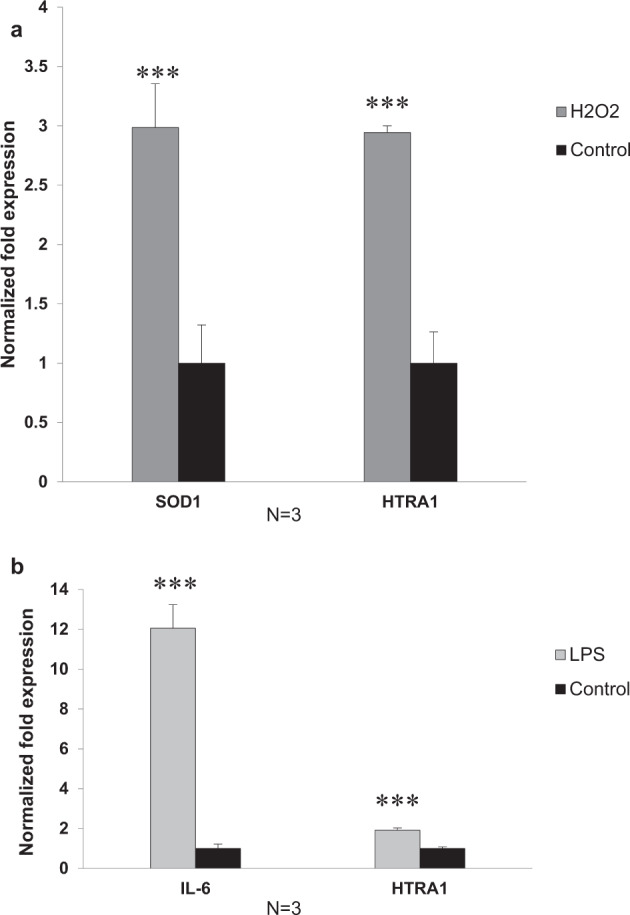
Serum-starved ARPE-19 cells were plated on dishes in medium supplemented with 2 μM H2O2 (a) or 100 ng/ml LPS (b) for 24 h. The control was performed with vehicle. Total RNA extracted from those ARPE-19 cells was subjected to qPCR analysis and normalised with that of the reference gene GAPDH as described in “Methods”. Results were expressed as means ± SD (n = 3). ***p < 0.001 vs. control.
LPS-mediated induction of HtrA1 expression in ARPE-19 cells
To determine the effects of inflammation on HtrA1 expression, ARPE-19 cells were stimulated with different concentrations of LPS for 24 h. The effects of inflammation were evidenced by the increase in IL-6 protein level (Fig. 1b, d). Western blot results showed that HtrA1 protein expression increased at doses of 1, 10 and 100 ng/ml and induced a 1.55 ± 0.69 (n = 3, p > 0.05), 2.65 ± 1.00 (n = 3, p > 0.05), and 7.66 ± 1.18-fold (n = 3, p < 0.001) increases in HtrA1 protein level, respectively (Fig. 1b, d). HtrA1 protein expression after using the maximum dose of 100 ng/ml of LPS was statistically significantly increased when compared to the control (Fig. 1d). Hence, the effective dose of 100 ng/ml LPS was used in subsequent experiments to induce inflammatory stress for evaluation of the HtrA1 promoter activity.
Consistent with the results obtained from the western blot, qPCR data showed a 1.92 ± 0.11-fold (n = 3, p < 0.001) increase in HtrA1 mRNA expression (Fig. 2b) with the effective dose of 100 ng/ml LPS induction. These findings suggest that inflammation stress stimulated by LPS may induce HtrA1 over-expression at both the mRNA and protein levels.
Effect of rs11200638 on HtrA1 expression
The SNP rs11200638, located in the promoter region of HtrA1 gene, is highly associated with development of AMD. To evaluate the effects of SNP rs11200638 on HtrA1 promoter activity, as in previous reports [13, 14], ARPE-19 cells were transiently transfected with a luciferase reporter plasmid driven by the HtrA1 promoter, carrying either allele G (WT) or the risk allele A (MT) genotype at SNP rs11200638. In ARPE-19 cells, WT HtrA1 promoter and MT did not show statistically significant differences in their luciferase reporter expression (P = 0.054425173, Fig. 3a).
Fig. 3. Luciferase activities in cultured ARPE-19 cells transfected with HtrA1 promoter reporter constructs after treated with LPS or H2O2.
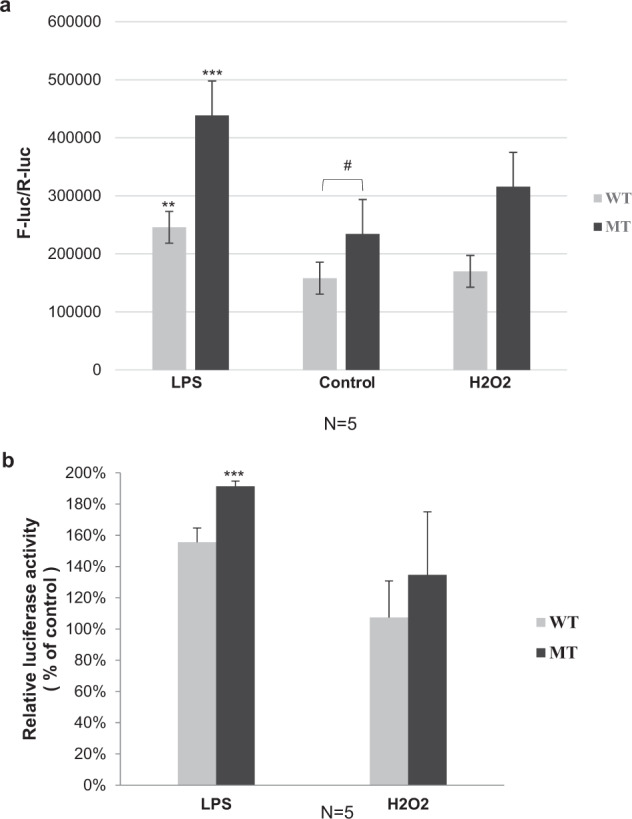
Effects of the rs11200638 variants on HtrA1 expression in ARPE-19 cells. After pre-treatment with 100 ng/ml LPS or 200 μM H2O2 for 6 h to induce cellular stress, ARPE-19 cells were transiently transfected with WT or MT HtrA1 promoter, another 6 h after transfection, ARPE-19 cells were further treated with 100 ng/ml LPS or 200 μM H2O2 for 24 h. The control was performed with vehicle. Normalised luciferase activity was measured in five independent experiments and shown in (a). Results were expressed as means ± SD. **p < 0.01 vs. WT control treated with LPS; ***p < 0.001 vs. MT control treated with LPS. #P = 0.054425173. The relative luciferase activity (% of control) was shown in (b). ***p < 0.001 vs. LPS-treated control.
Effect of inflammatory and oxidative stress on HtrA1 expression
To evaluate the effect of inflammatory and oxidative stress on the promoter activity, ARPE-19 cells were transfected with either WT or MT reporter plasmid encompassed with SNP rs11200638, then further treated with identified most effective dose of 200 μM H2O2 or 100 ng/ml LPS, luciferase activities were then measured as described above. Both MT and WT showed a persistent trend of higher relative luciferase expressions compared with their respective control when treated with LPS or H2O2 (P < 0.001 and P < 0.01, respectively) (Fig. 3a). When treated with 100 ng/ml LPS, the fold increase of luciferase expression in MT group was significantly greater than that in the WT (P < 0.001) (Fig. 3b). These results suggest that the risk haplotype (AA) of SNP rs11200638 increased the responsiveness of HtrA1 promoter to inflammation induction.
Effects of HtrA1 overexpression on the growth of ARPE-19 cells
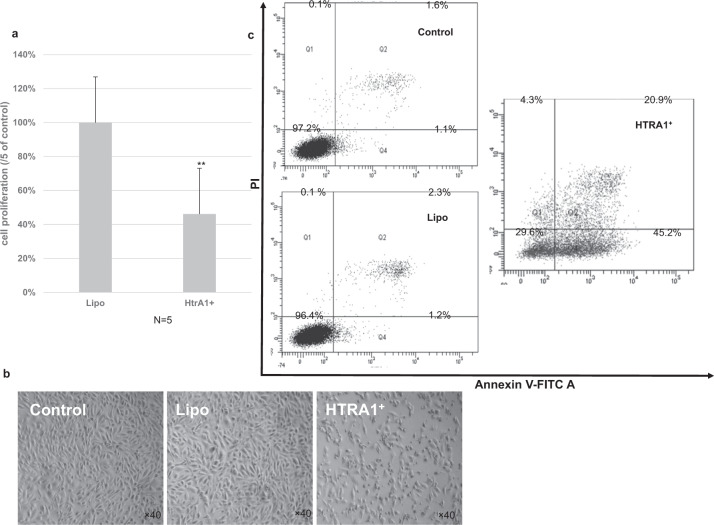
RPE cell is a crucial component in the pathogenesis of AMD [25, 27]. Both local inflammation and oxidative stress are related to RPE apoptosis in AMD [12, 14, 28]. To investigate the effects of HtrA1 overexpression on the growth of RPE, ARPE-19 cells were transfected with an expression vector containing HtrA1 cDNA (Fig. 4a, b). Western blot analysis revealed an overexpression of HtrA1 in the transfected ARPE-19 cells (Fig. 4c). Overexpression of HtrA1 resulted in significant apoptotic changes in ARPE-19 cells and 53.8 ± 1.6% (n = 5) of proliferation inhibition in ARPE-19 cells using CCK-8 assay (P < 0.01) (Fig. 5a, b).
Fig. 5. Effects of HtrA1 overexpression on the growth of ARPE-19 cells.
a Cell proliferation of ARPE-19 cells transfected with HtrA1 construct EX-M0558-M06 analysed using CCK-8, and the result was recorded using lipo-treated cells at 100%. b Cell morphology of ARPE-19 cells after transfected with the HtrA1 construct EX-M0558-M06. The magnification is ×40. c Flow cytometric evaluation of ARPE-19 apoptosis. Histograms derived from flow cytometry comparing apoptotic cells between vehicle and HtrA1 transfected cells. Twenty-four hours after transfection, the induction of apoptosis was determined using flow cytometric analysis of Annexin V-FITC and PI-stained ARPE-19 cells. Cells in the lower right quadrant indicate Annexin positive, early apoptotic cells, and cells in the upper right quadrant indicate Annexin positive/PI positive, late apoptotic cells. Results were expressed as means ± SD. **p < 0.01 vs. Lipo-treated control. Control, non-transfected ARPE-19 cells. Lipo, performed with Lipofectamine® LTX&PLUS™ reagent. HtrA1+, transfected with HtrA1 construct EX-M0558-M06.
To elucidate whether the growth-inhibitory effect was related to the induction of apoptosis, flow cytometry analysis was performed on the Annexin V-FITC/PI stained ARPE-19 cells. It was determined that the overexpression of HtrA1 induced apoptotic process in ARPE-19 cells, with a higher percentage of early apoptotic cells in the transfected cells compared to the control (Fig. 5c, 45.2% vs 1.1%).
Discussion
Various aetiological factors including advanced age, micro-environmental risk factors and inherited susceptibility gene(s) have been identified to be involved in the pathogenesis of AMD. Oxidative stress, an important micro-environmental risk factor, results in significant structural and functional changes in RPE, which could result in RPE senescence and drusen formation in early AMD [11, 24, 29]. The previous study suggested that the oxidative damage and immune response contributing to AMD [30]. Kauppinen et al. also demonstrated that oxidative stress can activate NLRP3 inflammasomes in RPE cells which occupy centre stage in the pathogenesis of AMD [31].
HtrA1 protein is involved in many diseases, including AMD. However, the mechanisms underlying regulation of HtrA1 expression remain to be determined. Supanji et al. suggested that oxidative stress induced by H2O2 increased HtrA1 expression during premature cell senescence [32]. Hou et al. have demonstrated that toll-like receptor-4 (TLR4) activation by LPS significantly induced HtrA1 expression in fibroblasts and macrophages [33]. In our study, diverse doses of H2O2 and LPS were independently applied in cultured ARPE-19 cells to mimic oxidative stress and inflammation, which were evidenced by the significant increases in SOD-1 and IL-6 protein level, respectively (p < 0.05). The HtrA1 expressions at both protein and mRNA increased with the identified concentrations of H2O2 and LPS inductions. The concomitant augmentation of SOD-1 and IL-6 with HtrA1 expression suggests a potential modelling of HtrA1 in the pathogenesis of AMD at a molecular basis under the oxidative stress and inflammation. However, results from other independent groups showed that the genotypes at rs11200638 were not correlated with HtrA1 expression at either mRNA or protein level either in the retina and white blood cells [21, 23]. Wang et al. reported that rs11200638 genotype has no effects on HtrA1 expression after examining 24 retina-RPE-choroids samples [21]. Furthermore, they studied AMD-associated variants at chromosome 10q26 locus and the expression of HtrA1 in 82 retina-RPE-choroids samples and suggested that there was no significant change of HtrA1 mRNA level [34]. In the current study, we also demonstrated that upregulation of HtrA1 expressions induced by H2O2 and LPS were in a dose-dependent manner. HtrA1 had been identified as one of the most important genes for the risk of advanced AMD [35]. Previous studies suggested that overexpression of HtrA1 may alter the integrity of Bruch’s membrane, favour neovascularization, and promote the invasion of choroid capillaries across the extracellular matrix, as what happened in wet AMD or advanced AMD [22, 36], suggesting that a certain threshold of oxidative and inflammative damage may be required to induce significant changes in HtrA1 expression. We speculated that the upregulation process on HtrA1 gene expression in vivo may not occur until the late stage of the disease, and therefore may explain the above incongruous results on HtrA1 expression.
AMD-associated variant rs11200638 locates in HtrA1 promoter site and influences the transcriptional regulation of HtrA1 gene [14]. With the hypothesis that the variant rs11200638 alters the responsiveness of HtrA1 promoter and subsequently enhances the HtrA1 expression under oxidative stress and inflammation, the luciferase reporter plasmids driven by HtrA1 short promoter (−1 to −2175 bp from translation site), encompassed with either protective allele G (WT) or risk allele A (MT) at rs11200638, were transfected into ARPE-19 cells. The identified maximum effective doses of H2O2 and LPS were used to induce AMD-like oxidative and inflammative damage in cells. Before the introduction of H2O2 or LPS, we measured the luciferase expression and found that there were no statistically significant differences in luciferase reporter either with WT or MT genotype. After ARPE-19 cells were treated with LPS, reporter plasmid with both MT and WT HtrA1 promoter exhibited higher luciferase expressions compared with their respective controls. And samples with MT exhibited a significant higher fold increase in luciferase expression when compared to that with WT samples (p < 0.001), indicating that inflammatory damage involved in variant rs11200638 upregulating HtrA1 promoter responsiveness. Risk haplotype A had a more significant effect on altering HtrA1 promoter activity, as demonstrated by a greater fold increase in luciferase expression in MT group when compared to that in WT group.
Variant rs11200638 locates in a conserved CpG island, which extended 2.1 kb upstream from the transcription starting site within promoter region of HtrA1 gene. AMD-risk allele A may disrupt the CpG island [14] by removing the inhibition effect on methylated CpG island, increasing the responsiveness of HtrA1 gene to the micro-environmental factors. However, the mechanism of over-expression of HtrA1 gene in RPE cells remains unclear and requires investigation, it has been described that LPS can induce HtrA1 expression in human synovial fibroblasts and monocyte/macrophages via the classical NF-κB pathway [33]. And, TLR4 activation from LPS results in preferential binding of NF-κB p65 to the HtrA1 promoter, which subsequently induces the upregulation of HtrA1 transcription [33].
We noticed that HtrA1 expression and its promoter potency did not respond similarly to the oxidative stress induction. That is, certain doses of H2O2 stimulations significantly increased the HtrA1 expressions at protein and mRNA level(p < 0.05), but did not present corresponding effects on its promoter potency, neither with MT nor MT (p > 0.05). We speculated that the short promoter (−1 to −2175bp from translation starting point) constructs in the study did not harbour those necessary response elements for transcription factor(s) elicited by oxidative stress; or that the AMD risk, conferred by this region is potentially driven by multiple variants, such as LOC387715 insertion/deletion(ins/del) and rs2672598, which were not encompassed or investigated with our promoter constructs [18, 37].
In our study, we only focused on the role of single variant rs11200638 in the promoter region of HtrA1 gene. Previous studies [18, 21, 23, 38] suggested AMD risk conferred by HtrA1 gene may be driven by multiple alleles in risk haplotype with binary model. The intergenic region between LOC387715 (ins/del) and the SNP rs11200638, rs3793917, and rs11200638 in the HtrA1 gene were all reported the likely sites to be significantly associated with AMD [18, 39]. Further studies may combine other AMD-associated variants at chromosome 10q26 region to refine our understanding of their role on HtrA1 gene.
HtrA1 gene is thought to control extra-cellular matrix degradation and RPE cell growth or survival [11, 40–42] and enhance cell senescence through p38 MAPK pathway [32]. Excessive HtrA1 accumulation results in drusen formation which may compromise Bruch’s membrane integrity, favouring the invasion of choroid capillaries across the extra-cellular matrix, as occurs in wet AMD [11, 14]. We found that the overexpression of HtrA1 significantly inhibited the growth of ARPE-19 cells. Apoptotic processes were involved in the growth inhibition from enhanced HtrA1 expression on RPE cells. This functional property of HtrA1 could explain the enhanced expression of this protein in dry and wet AMD lesions, both preceded by early and intermediate stages, which are characterised by RPE pigmentary abnormalities and drusen. The primary clinical characteristic of advanced dry AMD is the appearance of RPE cell atrophy, which is the consequence of stressed RPE dying and defects. The loss of RPE could lead to the gradual degeneration of nearby photoreceptors and progressive visual impairment [43]. We also demonstrated that HtrA1 protein increased significantly in cultured ARPE-19 cell nuclei when it was treated with the effective dose of 100 ng/ml LPS or 200 μM H2O2 (data not shown). Whether the accumulation of HtrA1 protein in nuclei increased its role in AMD pathogenesis remains unclear, further studies are needed to address it.
In our study, diverse doses of H2O2 and LPS were tested for the induction of oxidative and inflammatory reactions. The identified maximum effective doses of H2O2 and LPS were finally applied to treat ARPE-19 cells to imitate the AMD-like damage. We found variant rs11200638 upregulated the responsiveness of HtrA1 promoter after inflammation induction. However, the involvement of AMD-risk factors to AMD pathogenesis may be more complex in vivo.
So, it suggests that oxidative stress and inflammation increased HtrA1 expression. Variant rs11200638 enhances the responsiveness of HtrA1 promoter with LPS induction. The increased HtrA1 expression induces RPE cell apoptosis and growth inhibition.
Summary
What was known before
Both environmental and genetic risk factors are involved in AMD pathogenesis.
High-temperature requirement factor A-1 (HtrA1) gene has been shown to be one of the major causal genes to AMD.
SNP rs11200638 in HtrA1 promoter site was deemed as the AMD-associated allele primarily based on the assumption that SNP rs11200638 up-regulated HtrA1 expression.
What this study adds
In the present study, diverse doses of H2O2 and LPS were tested for the induction of oxidative and inflammatory reactions.
We found variant rs11200638 upregulated the responsiveness of HtrA1 promoter after inflammative induction.
The increased HtrA1 expression induces RPE cell apoptosis and growth inhibition. However, the involvement of AMD-risk factors to AMD pathogenesis may be more complex in vivo eyes.
Supplementary information
Supplemental Table 1 - Primers of the specific genes for QPCR analysis
Supplemental Table 2 - Results of the genotype of STR and Amelogenin analysis for the cells
Acknowledgments
Author contributions
FH, XL, SC, and XL conceived and designed the presented study. FH, XL, SC, LL, TZ and MY performed the data collection. FH, XL, SC, and XL performed the analysis and wrote the manuscript. XW and NF provided critical review of the manuscript.
Funding
This work was supported by grants from the National Basic Research Program (973 Program, 2011CB510201), the National Natural Science Foundation of China (NNSF 81770924 and 82070963), Guangdong Basic and Applied Basic Research Foundation (2019A1515011234), and the Science and Technology Project of the Health Planning Committee of Sichuan (16PJ226). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Fen He, Xiaohong Li, Suping Cai.
Supplementary information
The online version contains supplementary material available at 10.1038/s41433-021-01706-8.
References
- 1.Jonas JB, Cheung CM, Panda-Jonas S. Updates on the epidemiology of age-related macular degeneration. Asia Pac J Ophthalmol. 2017;6:493–7. doi: 10.22608/APO.2017251. [DOI] [PubMed] [Google Scholar]
- 2.Ratnapriya R, Chew EY. Age-related macular degeneration-clinical review and genetics update. Clin Genet. 2013;84:160–6. doi: 10.1111/cge.12206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwartz SG, Hampton BM, Kovach JL, Brantley MA., Jr Genetics and age-related macular degeneration: a practical review for the clinician. Clin Ophthalmol. 2016;10:1229–35. doi: 10.2147/OPTH.S109723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jabbarpoor Bonyadi MH, Yaseri M, Bonyadi M, Soheilian M, Karimi S. Association of combined complement factor H Y402H and ARMS/LOC387715 A69S polymorphisms with age-related macular degeneration: a meta-analysis. Curr Eye Res. 2016;41:1519–25. doi: 10.3109/02713683.2016.1158274. [DOI] [PubMed] [Google Scholar]
- 5.Wu M, Guo Y, Ma Y, Zheng Z, Wang Q, Zhou X. Association of two polymorphisms, rs1061170 and rs1410996, in complement factor H with age-related macular degeneration in an asian population: a meta-analysis. Ophthalmic Res. 2016;55:135–44. doi: 10.1159/000442257. [DOI] [PubMed] [Google Scholar]
- 6.Ryu E, Fridley BL, Tosakulwong N, Bailey KR, Edwards AO. Genome-wide association analyses of genetic, phenotypic, and environmental risks in the age-related eye disease study. Mol Vis. 2010;16:2811–21. [PMC free article] [PubMed] [Google Scholar]
- 7.Tang NP, Zhou B, Wang B, Yu RB. HTRA1 promoter polymorphism and risk of age-related macular degeneration: a meta-analysis. Ann Epidemiol. 2009;19:740–5. doi: 10.1016/j.annepidem.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 8.Soysal Y, Inan U, Küsbeci T, Imirzalioğlu N. Age-related macular degeneration and association of CFH Y402H and LOC387715 A69S polymorphisms in a Turkish population. DNA Cell Biol. 2012;31:323–30. doi: 10.1089/dna.2011.1214. [DOI] [PubMed] [Google Scholar]
- 9.Askari M, Nikpoor AR, Gorjipour F, Mazidi M, Sanati MH, Aryan H, et al. Association of Htra1 gene polymorphisms with the risk of developing AMD in Iranian population. Rep. Biochem Mol Biol. 2015;4:43–49. [PMC free article] [PubMed] [Google Scholar]
- 10.Xu Y, Guan N, Xu J, Yang X, Ma K, Zhou H, et al. Association of CFH, LOC387715, and HTRA1 polymorphisms with exudative age-related macular degeneration in a northern Chinese population. Mol Vis. 2008;14:1373–81. [PMC free article] [PubMed] [Google Scholar]
- 11.Deangelis MM, Ji F, Adams S, Morrison MA, Harring AJ, Sweeney MO, et al. Alleles in the HtrA serine peptidase 1 gene alter the risk of neovascular age-related macular degeneration. Ophthalmology. 2008;115:1209–15. doi: 10.1016/j.ophtha.2007.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tam PO, Ng TK, Liu DT, Chan WM, Chiang SW, Chen LJ, et al. HTRA1 variants in exudative age-related macular degeneration and interactions with smoking and CFH. Investig Ophthalmol Vis Sci. 2008;49:2357–65. doi: 10.1167/iovs.07-1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeWan A, Liu M, Hartman S, Zhang SS, Liu DT, Zhao C, et al. HTRA1 promoter polymorphism in wet age-related macular degeneration. Science. 2006;314:989–92. doi: 10.1126/science.1133807. [DOI] [PubMed] [Google Scholar]
- 14.Yang Z, Camp NJ, Sun H, Tong Z, Gibbs D, Cameron DJ, et al. A variant of the HTRA1 gene increases susceptibility to age-related macular degeneration. Science. 2006;314:992–3. doi: 10.1126/science.1133811. [DOI] [PubMed] [Google Scholar]
- 15.Zhou YL, Chen CL, Wang YX, Tong Y, Fang XL, Li L, et al. Association between polymorphism rs11200638 in the HTRA1 gene and the response to anti-VEGF treatment of exudative AMD: a meta-analysis. BMC Ophthalmol. 2017;17:97. doi: 10.1186/s12886-017-0487-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan CC, Shen D, Zhou M, Ross RJ, Ding X, Zhang K, et al. Human HTRA1 in the archived eyes with age-related macular degeneration. Trans Am Ophthalmol Soc. 2007;105:92–97. [PMC free article] [PubMed] [Google Scholar]
- 17.Tuo J, Ross RJ, Reed GF, Yan Q, Wang JJ, Bojanowski CM, et al. The HTRA1 promoter polymorphism, smoking, and age-related macular degeneration in multiple case-control samples. Ophthalmology. 2008;115:1891–8. doi: 10.1016/j.ophtha.2008.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Z, Tong Z, Chen Y, Zeng J, Lu F, Sun X, et al. Genetic and functional dissection of HTRA1 and LOC387715 in age related macular degeneration. PLoS Genet. 2010;6:e1000836. doi: 10.1371/journal.pgen.1000836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gibbs D, Yang Z, Constantine R, Ma X, Camp NJ, Yang X, et al. Further mapping of 10q26 supports strong association of HTRA1 polymorphisms with age related macular degeneration. Vis Res. 2008;48:685–9. doi: 10.1016/j.visres.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 20.Lee SJ, Kim NR, Chin HS. LOC387715/HTRA1 polymorphisms, smoking and combined effects on exudative age-related macular degeneration in a Korean population. Clin Exp Ophthalmol. 2010;38:698–704. doi: 10.1111/j.1442-9071.2010.02316.x. [DOI] [PubMed] [Google Scholar]
- 21.Wang G, Scott WK, Haines JL, Pericak-Vance MA. Genotype at polymorphism rs11200638 and HTRA1 expression level. Arch Ophthalmol. 2010;128:1491–3. doi: 10.1001/archophthalmol.2010.256. [DOI] [PubMed] [Google Scholar]
- 22.Kanda A, Stambolian D, Chen W, Curcio CA, Abecasis GR, Swaroop A. Age-related macular degeneration-associated variants at chromosome 10q26 do not significantly alter ARMS2 and HTRA1 transcript levels in the human retina. Mol Vis. 2010;16:1317–23. [PMC free article] [PubMed] [Google Scholar]
- 23.Friedrich U, Myers CA, Fritsche LG, Milenkovich A, Wolf A, Corbo JC, et al. Risk- and non risk- associated variants at the 10q26 AMD locus influence ARMS2 mRNA expression but exclude pathogenic effects due to protein deficiency. Hum Mol Genet. 2011;20:1387–99. doi: 10.1093/hmg/ddr020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ardeljan CP, Ardeljan D, Abu-Asab M, Chan CC. Inflammation and cell death in age-related macular degeneration: an immunopathological and ultrastructural model. J Clin Med. 2014;3:1542–60. doi: 10.3390/jcm3041542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Datta S, Cano M, Ebrahimi K, Wang L, Handa JT. The impact of oxidative stress and inflammation on RPE degeneration in non-neovascular AMD. Prog Retin Eye Res. 2017;60:201–18. doi: 10.1016/j.preteyeres.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li X, He F, Gabelt BT, Wang Y, Cai S, Cao J, et al. Effects of latanoprost and bimatoprost on the expression of molecules relevant to ocular inflow and outflow pathways. PLoS ONE. 2016;11:e0151644. doi: 10.1371/journal.pone.0151644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhutto I, Lutty G. Understanding age-related macular degeneration (AMD): relationships between the photoreceptor/retinal pigment epithelium/Bruch’s membrane/choriocapillaris complex. Mol Asp Med. 2012;33:295–317. doi: 10.1016/j.mam.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cameron DJ, Yang Z, Gibbs D, Chen H, Kaminoh Y, Jorgensen A, et al. HTRA1 variant confers similar risks to geographic atrophy and neovascular age-related macular degeneration. Cell Cycle. 2007;6:1122–5. doi: 10.4161/cc.6.9.4157. [DOI] [PubMed] [Google Scholar]
- 29.Beatty S, Koh H, Phil M, Boulton M. The role of oxidative stress in the pathogenesis of age-related macular degeneration. Surv Ophthalmol. 2000;45:115–34. doi: 10.1016/S0039-6257(00)00140-5. [DOI] [PubMed] [Google Scholar]
- 30.Hollyfield JG, Bonilha V, Rayborn ME, Yang X, Shadrach KG, Lu L, et al. Oxidative damage-induced inflammation initiates age-related macular degeneration. Nat Med. 2008;14:194–8. doi: 10.1038/nm1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kauppinen A, Niskanen H, Suuronen T, Kinnunen K, Salminen A, Kaarniranta K. Oxidative stress activates NLRP3 inflammasomes in ARPE-19 cells−implications for age-related macular degeneration (AMD) Immunol Lett. 2012;147:29–33. doi: 10.1016/j.imlet.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 32.Supanji, Shimomachi M, Hasan MZ, Kawaichi M, Oka C. HtrA1 is induced by oxidative stress and enhances cell senescence through p38 MAPK pathway. Exp Eye Res. 2013;112:79–92. doi: 10.1016/j.exer.2013.04.013. [DOI] [PubMed] [Google Scholar]
- 33.Hou Y, Lin H, Zhu L, Liu Z, Hu F, Shi J, et al. Lipopolysaccharide increases the incidence of collagen-induced arthritis in mice through induction of protease HTRA-1 expression. Arthritis Rheum. 2013;65:2835–46. doi: 10.1002/art.38124. [DOI] [PubMed] [Google Scholar]
- 34.Wang G, Dubovy SR, Kovach JL, Schwartz SG, Agarwal A, Scott WK, et al. Variants at chromosome 10q26 locus and the expression of HTRA1 in the retina. Exp Eye Res. 2013;112:102–5. doi: 10.1016/j.exer.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rajendran A, Dhoble P, Sundaresan P, Saravanan V, Vashist P, Nitsch D, et al. Genetic risk factors for late age-related macular degeneration in India. Br J Ophthalmol. 2018;102:1213–7. doi: 10.1136/bjophthalmol-2017-311384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sobrin L, Reynolds R, Yu Y, Fagerness J, Leveziel N, Bernstein PS, et al. ARMS2/HTRA1 locus can confer differential susceptibility to the advanced subtypes of age-related macular degeneration. Am J Ophthalmol. 2011;151:345. doi: 10.1016/j.ajo.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ng TK, Liang XY, Lai TY, Ma L, Tam PO, Wang JX, et al. HTRA1 promoter variantdifferentiates polypoidal choroidal vasculopathy from exudative age-related macular degeneration. Sci Rep. 2016;6:28639. doi: 10.1038/srep28639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kanda A, Chen W, Othman M, Branham KE, Brooks M, Khanna R, et al. A variant of mitochondrial protein LOC387715/ARMS2, not HTRA1, is strongly associated with age-related macular degeneration. Proc Natl Acad Sci USA. 2007;104:16227–32. doi: 10.1073/pnas.0703933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Richardson AJ, Islam FM, Aung KZ, Guymer RH, Baird PN. An intergenic region between the tagSNP rs3793917 and rs11200638 in the HTRA1 gene indicates association with age. Investig Ophthalmol Vis Sci. 2010;51:4932–6. doi: 10.1167/iovs.09-5114. [DOI] [PubMed] [Google Scholar]
- 40.Zurawa-Janicka D, Skorko-Glonek J, Lipinska B. HtrA proteins as targets in therapy of cancer and other diseases. Expert Opin Ther Targets. 2010;14:665–79. doi: 10.1517/14728222.2010.487867. [DOI] [PubMed] [Google Scholar]
- 41.Jones A, Kumar S, Zhang N, Tong Z, Yang JH, Watt C, et al. Increased expression of multifunctional serine protease, HTRA1, in retinal pigment epithelium induces polypoidal choroidal vasculopathy in mice. Proc Natl Acad Sci USA. 2011;108:14578–83. doi: 10.1073/pnas.1102853108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vierkotten S, Muether PS, Fauser S. Overexpression of HTRA1 leads to ultrastructural changes in the elastic layer of Bruch’s membrane via cleavage of extracellular matrix components. PLoS ONE. 2011;6:e22959. doi: 10.1371/journal.pone.0022959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Giansanti V, Rodriguez G, Savoldelli M, Gioia R, Forlino A, Mazzini G, et al. Characterization of stress response in human retinal epithelial cells. J Cell Mol Med. 2013;17:103–15. doi: 10.1111/j.1582-4934.2012.01652.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1 - Primers of the specific genes for QPCR analysis
Supplemental Table 2 - Results of the genotype of STR and Amelogenin analysis for the cells