Abstract
Background
Vitiligo is an acquired skin depigmentation disease. It can be misdiagnosed at an early stage and tend to relapse. Serum markers are essential to monitoring the progression of vitiligo. Exosomal miRNAs act as the communication mediator between melanocytes and immune cells. Our study aimed to use serum exosomal miRNAs as a reference for evaluating vitiligo progression.
Methods
The miRNAs were extracted from the serum exosomes of ten progressive vitiligo patients (before and after treatment) and ten healthy individuals. We profiled miRNAs expression by RNA sequencing and screened out potential miRNAs and plotted their receiver operating characteristic (ROC) curves to explore their sensitivity and specificity as prognostic biomarkers in vitiligo progression. We examined the correlation between miRNA expression and the lesion area. Different databases were used to predict gene targets of miRNAs, which were analyzed by gene ontology and Kyoto encyclopedia of genes and genomes (KEGG).
Results
Our results showed that 141 miRNAs were differentially expressed in serum exosomes of progressive vitiligo patients, and 365 miRNAs were differentially expressed in these patients after treatment compared to healthy individuals. The expression of hsa-miR-487b-3p was significantly lower in these patients compared to healthy individuals. Still, there was no difference in its levels in patients after corticosteroid treatment compared to healthy controls. ROC curve analysis (area under curve = 0.840) indicated that hsa-miR-487b-3p could serve as a biomarker for the prognosis of vitiligo progression. Its expression positively correlated with the lesion area. A total of 41 target genes of hsa-miR-487b-3p were predicted via different databases. KEGG pathways were enriched in phenylalanine metabolism, glycan degradation, and protein export.
Conclusion
Serum exosomal hsa-miR-487b-3p can be a biomarker to detect vitiligo progression. The predicted target genes of hsa-miR-487b-3p were enriched in catabolism. Thus, its in progressive vitiligo may accelerate catabolism in melanocytes and cause its impairment.
Keywords: progressive vitiligo, exosome, miRNA, biomarker
Introduction
Vitiligo is an acquired autoimmune disease that causes skin depigmentation, characterized by epidermal melanocyte destruction. The global incidence of vitiligo in a community-based study was around 0.2%, while it was about 1.8% in a hospital-centered study.1 It is currently thought to be related to autoimmunity, oxidative stress, mental stress, metabolic toxins, and genetic factors.2 The different shapes of white patches or spots on exposed areas of the body severely affect the quality of life and mental health due to the feeling of shame created by depigmentation.3 However, the exact pathogenesis of vitiligo remains unknown. A growing body of literature reported that exosomes play an important role in the autoimmune mechanism of vitiligo. The antigen-presenting cells can recognize the melanocyte-specific antigens, heat shock proteins, lipids, and miRNAs in exosomes and can play a role in the accelerated maturity of auto-reactive T cells or autoimmune attacks on melanocytes.4
Due to their immunomodulatory properties, exosomal miRNAs have been linked to autoimmune diseases.5 They have immunosuppressive effects on CD8+ T cells in the cancer microenvironment.6 In this regard, regulatory T cells (Tregs) can produce exosomes and exert immune inhibitory effects.7 Furthermore, studies exhibited that active macrophages can act as an exosomal miRNAs reservoir. These functional miRNAs can be translocated to other cells such as endothelial cells, tumor cells, monocytes, and keratinocytes via exosomes.8 B lymphocytes may influence the function of CD8+ T cells and macrophages via exosomes.8,9 Consequently, exosomes are involved in the integration of balance between immune cells. A recent study directly demonstrated that mesenchymal stem cells-derived exosomes increased Tregs and decreased the proportion of CD4+ T, Th17, and CD8+ T cells in murine.10 Therefore, the profile of exosomal miRNAs may represent the pathophysiological status and characteristics of autoimmune diseases. Hence, exosomes and miRNAs have a significant diagnostic value in autoimmune diseases because of their therapeutic properties.5 According to the literature, the down-expression of let-7b-5p may serve as a biomarker for psoriatic arthritis patients.11 In systemic lupus erythematosus patients, the up-regulation of serum exosomal miR-21 and miR-155 may be a possible biomarker.12 Peripheral blood exosomal miR-17 is up-regulated in rheumatoid arthritis patients and may contribute to the disease by interfering with Tregs maintenance.13
As one of the autoimmune diseases, melanocyte destruction in vitiligo is closely linked to the imbalance of CD8+ T, Tregs, and Th17 cells. In autoimmune diseases like vitiligo, miRNAs carried by exosomes also have important immune regulatory effects.10,14–16 Lv et al17 reported that miR-155 could inhibit the proliferation of CD8+ T cells through Tregs in vitiligo. Also, exosomes serve as a messenger between keratinocytes and melanocytes during melanogenesis. Earlier reports suggested that exosomal miR-196a-2 can affect tyrosinase (TYR) and tyrosinase-related protein-1 (TYRP-1) heterodimeric complexes, thus influencing the susceptibility of vitiligo. The miR-183-5p can regulate microphthalmia-associated transcription factor (MITF) expression in vitiligo skin depigmentation.18,19 Zhao et al20 reported that exosomal miR-200c secreted by keratinocytes could enhance melanogenesis genes, which are down-regulated in vitiligo lesions. In addition, miRNAs are involved in melanocyte survival in response to environmental stimuli. Researchers reported that in vitiligo, over-expression of miR-25 detected in serum during oxidative stress could promote the destruction of melanocytes, and miR-211 can regulate mitochondrial energy metabolism.21,22
Exosomal miRNAs have important roles in vitiligo pathogenesis and in representing pathophysiological status and disease characteristics. They can be used as potential biomarkers or therapeutic targets in this disease. Early prognosis and timely intervention after vitiligo recurrence are critical because its treatment remains a challenge in dermatological clinics. Our research intended to explore the potential biomarkers for the prognosis of vitiligo progression, further understand the role of miRNAs in progressive vitiligo and provide new insights for its treatment.
Materials and Methods
Patients and Controls
A total of 10 patients were recruited from the dermatology department of our hospital between June and July 2022. The patients included four males and six females, ranging in age from 18 to 72 years. The area of lesions ranged from 1.6–15.4% of body surface area (BSA), with disease duration ranging from two months to 18 years. The control group consisted of 10 age and gender-matched healthy individuals. All patients and healthy controls were informed about the research and included in the study after signing the consent forms. The demographic and clinical details of patients and controls are given in Supplementary Table. Our study was conducted following the principles of the Declaration of Helsinki, 2013 edition, and approved by the Ethics Committee of Hangzhou Third People’s Hospital (No.2020KA001).
The inclusion criteria followed were as follows: 1) the patients were diagnosed by Wood’s lamp or reflectance laser confocal microscopy (RCM); 2) vitiligo disease activity (VIDA) score was ≥3. Exclusion criteria included the following: 1) segmental vitiligo patients; 2) patients who had received systemic corticosteroid therapy or phototherapy in the last three months; 3) the patients who could not return to the clinic for follow-up or take the prescribed medication; 4) patients with other autoimmune diseases; 5) patients who were pregnant or under the age of 18.
The treatment regimen for patients was oral prednisone for three months (the dose was 0.3 mg per kg/d for one month, then 5 mg every two weeks and maintained at 5 mg/d). A total of 30 serum exosome samples were collected from ten healthy controls and ten progressive vitiligo patients (before treatment and three months after systemic corticosteroid treatment).14
Exosomes Isolation, RNA Extraction and Microarray Analysis
Serum (supernatant liquid) was extracted by centrifugation of peripheral blood samples in a TGL-16B centrifuge machine (Anting Corporation, Shanghai, China), firstly at 3000 rpm (4 °C for 15 min) and then at 12,000 rpm (4 °C for 20 min), and were stored at −80 °C. Exosomes were isolated from serum using Serum Exosome Purification Kit (Norgen 57,600, ON, Canada) according to the manufacturer’s instructions. Total RNA was extracted from serum exosomes using the Exosomal RNA Isolation Kit (Norgen 58,000, ON, Canada), which was then quantified on NanoDrop ND-2000 (Thermo Fisher Scientific, Lafayette, LA, USA). RNA integrity was tested with Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). Followed by miRNA libraries establishment, cDNA libraries were constructed with TruSeq Small RNA Preparation Kit (Illumina, San Diego, CA, USA) and sequenced using Illumina HiSeq 2500 system (Lcbio, Hangzhou, China) according to the manufacturer’s standard protocols. ACGT101-miR program (LC Sciences, Houston, TX, USA) was used to dispose of raw reads and remove junk, adapter dimers, repeats, and other RNA families. Differentially expressed miRNAs were screened with fold change and P-value (Student’s t-test). A volcano and heatmap were used for visualization.14
Receiver Operating Characteristic (ROC) Curve and the Lesions Area Correlation with miRNAs
We screened significantly up-regulated or down-regulated miRNAs in progressive vitiligo patients before and after systemic corticosteroid treatment compared to healthy controls. The screened miRNAs were evaluated by ROC curve analysis. We explored the correlation between the expression of miRNAs and the area of lesions by Spearman correlation coefficient using IBM® SPSS Statistics Software (V27.0).23
Target Genes Prediction of miRNAs, GO, and KEGG Analyses
The target genes of screened miRNAs were predicted using three databases: TargetScanHuman (http://www.targetscan.org/vert_80/), miRDB (http://www.mirdb.org/), and microRNA.org (http://www.microrna.org/). The miRNA-mRNA network was visualized in Cytoscape software (V3.8.1). All predicted gene targets were processed by gene ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG) analyses, including cellular components, molecular functions, biological processes, and enriched pathways.14
Results
Profiles of Serum Exosomal miRNAs in Progressive Vitiligo Before and After Systemic Corticosteroid Treatment
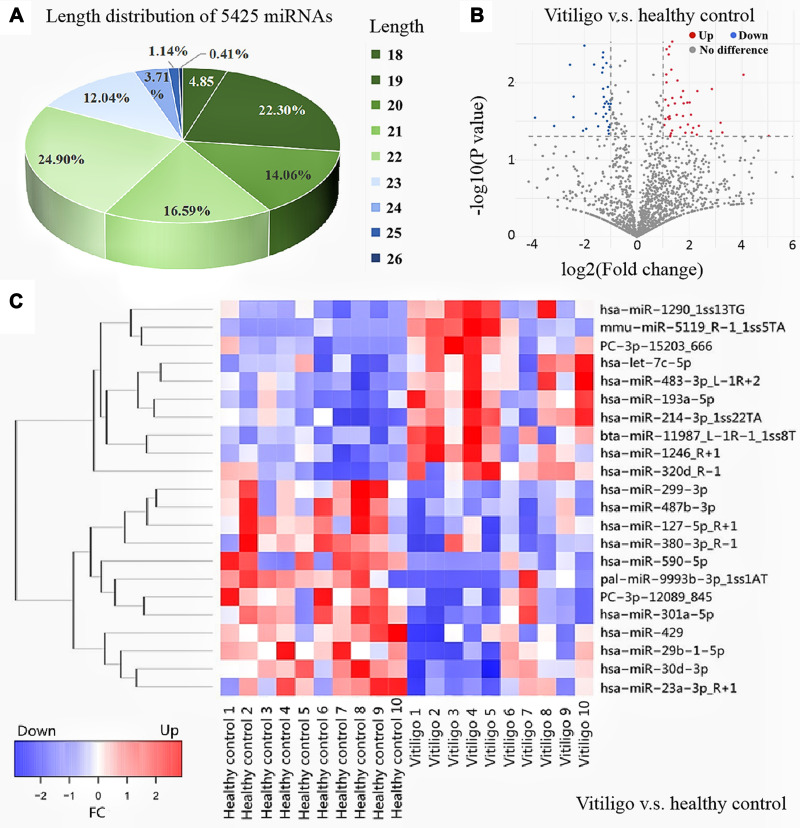
A total of 5425 miRNAs were detected in all exosomal samples. Our results showed that 141 miRNAs were differentially expressed in progressive vitiligo patients (before treatment) compared with healthy controls (P<0.05). After three months of systemic corticosteroid treatment, no new expanded lesions were captured, and the lesions remained stable as determined by RCM detection. A total of 265 differentially expressed miRNAs were detected in patients after three months of treatment compared to healthy controls. The results are summarized with a volcano, heatmap, and sector diagram in Figure 1. We screened miRNAs with P<0.05, |log2FC|>1 (FC = fold change) middle or high-intensity signals. Table 1 lists the top 15 differentially expressed miRNAs in vitiligo patients (before treatment) and healthy controls. Table 2 lists differentially expressed miRNAs in vitiligo patients (after treatment) and healthy controls.
Figure 1.
Length distribution of total miRNAs and volcano map and heat map of differential expressed miRNAs (A) Length distribution of 5425 miRNAs; (B) volcano map of differential expressed miRNAs between progressive vitiligo patients (before treatment) and healthy controls; (C) heat map of differential expressed miRNAs between vitiligo patients (before treatment) and healthy controls and healthy controls, with P<0.01, middle or high intensity signal.
Table 1.
A Total of Top 15 Differential Expressed miRNAs Between Vitiligo Patients (Before Treatment) and Healthy Controls
| NO | Name of miRNAs | Up/Down | FC | log2FC | P value |
|---|---|---|---|---|---|
| 1 | PC-5p-68634_136 | Up | 32.78 | 5.03 | 0.0492 |
| 2 | mmu-miR-5119_R-1_1ss5TA | Up | 16.84 | 4.07 | 0.0079 |
| 3 | hsa-mir-3648-1-p5 | Up | 9.62 | 3.27 | 0.0444 |
| 4 | PC-5p-120360_87 | Up | 9.16 | 3.20 | 0.0332 |
| 5 | hsa-miR-4683 | Up | 7.29 | 2.86 | 0.0122 |
| 6 | mmu-miR-5100_1ss21TC | Up | 7.13 | 2.83 | 0.0426 |
| 7 | hsa-miR-6803-3p_R-1 | Up | 5.16 | 2.37 | 0.0364 |
| 8 | mmu-miR-5106_R-4_1ss1AG | Up | 4.96 | 2.31 | 0.0129 |
| 9 | PC-5p-905_23133 | Up | 4.89 | 2.29 | 0.0474 |
| 10 | hsa-mir-3665-p5_1ss2CA | Up | 4.44 | 2.15 | 0.0387 |
| 11 | hsa-mir-8086-p3_1ss11AG | Up | 4.29 | 2.10 | 0.0445 |
| 12 | mmu-mir-1895-p3_1ss9CT | Up | 4.19 | 2.07 | 0.0261 |
| 13 | mmu-miR-5126_L-4_1ss18CT | Up | 4.04 | 2.01 | 0.0181 |
| 14 | hsa-mir-1285-1-p5_1ss12AG | Up | 3.86 | 1.95 | 0.0350 |
| 15 | ssc-mir-1285-p5_1ss20TC | Up | 3.80 | 1.93 | 0.0182 |
| 16 | hsa-miR-376c-3p | Down | 0.50 | −1.00 | 0.0189 |
| 17 | hsa-miR-429 | Down | 0.50 | −1.00 | 0.0063 |
| 18 | hsa-miR-19a-3p | Down | 0.50 | −1.01 | 0.0170 |
| 19 | hsa-miR-493-3p | Down | 0.49 | −1.02 | 0.0352 |
| 20 | hsa-miR-136-5p_R-1 | Down | 0.49 | −1.04 | 0.0381 |
| 21 | hsa-miR-136-3p | Down | 0.49 | −1.04 | 0.0207 |
| 22 | hsa-miR-410-3p | Down | 0.48 | −1.05 | 0.0225 |
| 23 | hsa-miR-542-3p | Down | 0.48 | −1.07 | 0.0461 |
| 24 | hsa-miR-758-3p_R-1 | Down | 0.48 | −1.07 | 0.0376 |
| 25 | hsa-miR-377-3p | Down | 0.47 | −1.09 | 0.0188 |
| 26 | hsa-miR-1197 | Down | 0.47 | −1.09 | 0.0418 |
| 27 | hsa-miR-381-3p | Down | 0.46 | −1.11 | 0.0114 |
| 28 | mdo-miR-200a-3p_R+2 | Down | 0.45 | −1.14 | 0.0315 |
| 29 | hsa-miR-494-3p_R+1 | Down | 0.45 | −1.16 | 0.0177 |
| 30 | hsa-miR-487b-3p | Down | 0.45 | −1.16 | 0.0056 |
Abbreviation: FC, fold change.
Table 2.
A Total of Top 15 Differential Expressed miRNAs Between Vitiligo Patients (After Treatment) and Healthy Controls
| NO | Name of miRNAs | Up/Down | FC | log2FC | P value |
|---|---|---|---|---|---|
| 1 | PC-5p-17405_580 | Up | 27.26 | 4.77 | 0.0043 |
| 2 | hsa-miR-561-5p | Up | 20.40 | 4.35 | 0.0404 |
| 3 | hsa-miR-4772-5p | Up | 19.09 | 4.25 | 0.0017 |
| 4 | hsa-miR-5582-3p_L+1R-1_1ss22GT | Up | 14.18 | 3.83 | 0.0143 |
| 5 | hsa-miR-29a-5p_R-1 | Up | 11.50 | 3.52 | 0.0027 |
| 6 | mmu-mir-1983-p5_1ss1GA | Up | 11.28 | 3.50 | 0.0017 |
| 7 | hsa-mir-1302-1-p5_1ss9AG | Up | 11.05 | 3.47 | 0.0049 |
| 8 | hsa-miR-190a-3p_L+1 | Up | 10.63 | 3.41 | 0.0036 |
| 9 | hsa-miR-5586-3p | Up | 9.91 | 3.31 | 0.0131 |
| 10 | hsa-miR-3176_R+2 | Up | 8.82 | 3.14 | 0.0181 |
| 11 | hsa-miR-5690_R+1 | Up | 8.70 | 3.12 | 0.0017 |
| 12 | hsa-miR-616-3p_L+1R+1 | Up | 8.35 | 3.06 | 0.0084 |
| 13 | PC-3p-92643_105 | Up | 8.10 | 3.02 | 0.0385 |
| 14 | hsa-miR-301b-5p | Up | 8.05 | 3.01 | 0.0320 |
| 15 | hsa-miR-548w_R-1 | Up | 7.92 | 2.99 | 0.0035 |
| 16 | PC-5p-5980_1907 | Down | 0.00 | −8.07 | 0.0001 |
| 17 | pal-miR-9993b-3p_1ss9GT | Down | 0.00 | −7.77 | 0.0008 |
| 18 | PC-3p-41069_236 | Down | 0.01 | −7.40 | 0.0193 |
| 19 | oga-miR-100_R+1_1ss9GT | Down | 0.01 | −6.99 | 0.0015 |
| 20 | PC-3p-22717_440 | Down | 0.01 | −6.76 | 0.0242 |
| 21 | PC-5p-40052_242 | Down | 0.02 | −5.97 | 0.0011 |
| 22 | pal-miR-9995-3p_1ss9GT | Down | 0.02 | −5.89 | 0.0008 |
| 23 | cgr-miR-15b-5p_L+2 | Down | 0.02 | −5.85 | 0.0013 |
| 24 | mmu-miR-362-5p_R-1_1ss9GT | Down | 0.04 | −4.77 | 0.0194 |
| 25 | mmu-miR-5124a_L-1R-1_1ss5CA | Down | 0.04 | −4.57 | 0.0004 |
| 26 | mmu-miR-5124a_L-1_1ss5CA | Down | 0.07 | −3.92 | 0.0004 |
| 27 | bta-miR-2478_L+2 | Down | 0.07 | −3.89 | 0.0076 |
| 28 | PC-3p-83939_113 | Down | 0.09 | −3.52 | 0.0036 |
| 29 | PC-5p-40181_241 | Down | 0.09 | −3.47 | 0.0406 |
| 30 | hsa-miR-4433b-3p_L+1_1ss9GT | Down | 0.09 | −3.46 | 0.0019 |
Abbreviation: FC, fold change.
A Prognostic Biomarker for Vitiligo Progression and Its Correlation with BSA
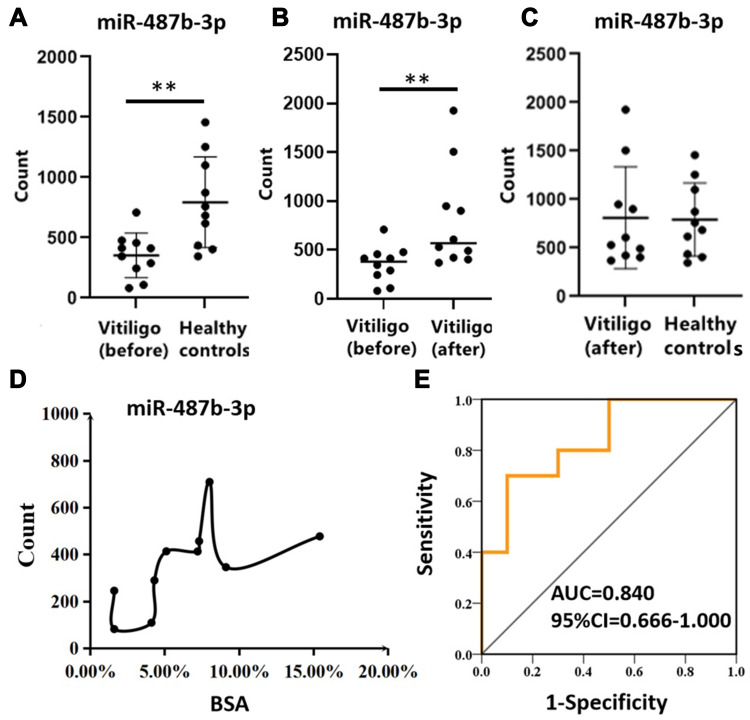
According to RNA sequencing results, hsa-miR-487b-3p expression in progressive vitiligo patients (before treatment) was significantly lower than in healthy individuals (FC = 0.45, P = 0.006). The lesions became temporarily stable after three months of systemic corticosteroid treatment according to RCM. Meanwhile, the hsa-miR-487b-3p expression was significantly up-regulated in patients after treatment (FC = 2.27, P = 0.025). There was no significant difference in hsa-miR-487b-3p expression between patients after treatment and healthy individuals (FC = 1.02, P = 0.935), as displayed in Figure 2A–C. Interestingly, hsa-miR-487b-3p expression was positively correlated with the area of lesions (P = 0.004) in progressive vitiligo, as depicted in Figure 2D. ROC curve was made to show the sensitivity and specificity of hsa-miR-487b-3p as a prognostic biomarker for vitiligo progression (Figure 2E). The hsa-miR-487b-3p was found to be an appropriate exosomal miRNA prognostic biomarker (AUC = 0.840, P = 0.001).
Figure 2.
Expression of hsa-miR-487b-3p, correlation with BSA and ROC curves. (A) differential expression of hsa-miR-487b-3p in progressive vitiligo (before treatment) and healthy controls tested by RNA sequencing; (B) differential expression of hsa-miR-487b-3p in progressive vitiligo before and after 3 months systemic corticosteroid treatment; (C) differential expression of hsa-miR-487b-3p in patients after treatment and healthy controls; (D) correlation of hsa-miR-487b-3p expression with total lesions area in progressive vitiligo patients; (E) hsa-miR-487b-3p ROC curve of progressive vitiligo before treatment vs after treatment.
Note: **P<0.05.
Abbreviations: ROC, receiver operating characteristic curve; AUC, area under ROC curve; BSA, body surface area; vitiligo (before): the progressive vitiligo patients before treatment; vitiligo (after): the progressive patients became stable after treatment.
Target Genes of Hsa-miR-487b-3p, GO, and KEGG Analyses
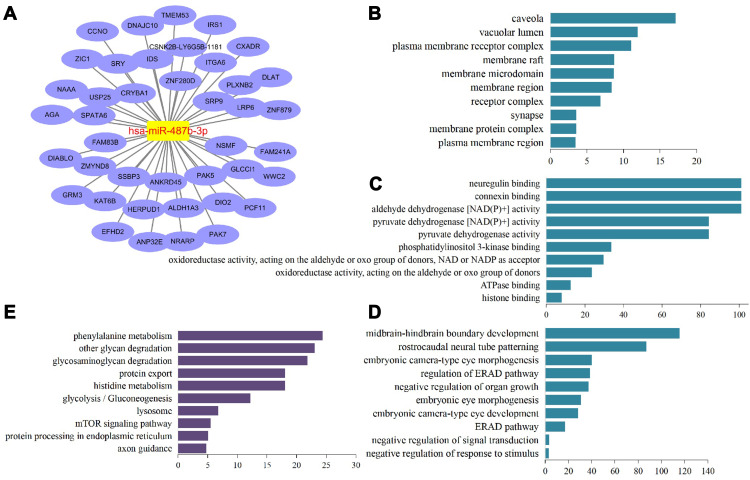
A total of 41 target genes were found after the prediction of hsa-miR-487b-3p with three databases; TargetScanHuman, miRDB, and microRNA.org (Figure 3A). The results of a comprehensive biological enrichment evaluation using GO and KEGG analyses revealed that cellular components of these target genes were mainly enriched in coveolae, vacuolar lumen, and plasma membrane receptor complex (Figure 3B). Molecular functions were primarily enriched in neuregulin binding, connexin binding, and aldehyde dehydrogenase [NAD(P)+] activity (Figure 3C). Biological processes were mainly enriched in midbrain-hindbrain boundary development, rostrocaudal neural tube patterning, embryonic camera-type eye morphogenesis, and regulation of endoplasmic reticulum (ER) associated degradation (ERAD) pathway (Figure 3D). KEGG pathways were mainly enriched in phenylalanine metabolism, other glycan degradation, glycosaminoglycan degradation, and protein export (Figure 3E). Among these genes, LRP6, ALDH1A3, PAK5, ZIC1, and GRM3 are related to melanocytes.24–28 DIABLO, also named SMAC, has been closely linked to vitiligo pathogenesis.29
Figure 3.
Structure of miRNA-mRNA network and biological enrichment (A) network of hsa-miR-487b-3p and target mRNAs (genes); (B) cellular component; (C) molecular function; (D) biological process; (E) KEGG pathway.
Discussion
Vitiligo is a depigmented autoimmune disease characterized by the death of epidermal melanocytes. Treatment of vitiligo is still a challenge in dermatological clinics. Thus, early prognosis and timely intervention after vitiligo recurrence are pivotal.1 Various miRNAs are important in vitiligo pathogenesis as they regulate autoimmune tolerance, melanogenesis, melanocyte survival, and cell metabolism under oxidative stress.16–18 Mansuri et al30 studied the miRNA signatures in the whole blood and peripheral blood mononuclear cells of vitiligo patients. Their research showed that miR-1, miR-184, miR-211, miR-328, miR-383, and miR-577 could be the potential markers of vitiligo. By analyzing their target genes, the researchers demonstrated that miRNAs are involved in many aspects of vitiligo pathogenesis, such as autoimmune regulation, skin pigmentation, and cellular apoptosis in response to the oxidative stimulus. In addition, Utpreksha et al31 reported a group of six miRNAs (miR-185, miR-202-3p, miR-525-5p, miR-326, miR-518a-5p, and miR-518c) down-regulated TRP1 expression in keratinocyte which inhibited the crosstalk in melanosomes transfer from melanocytes to keratinocytes, causing skin pigmentation in vitiligo. All of these miRNAs may regulate target genes associated with vitiligo pathogenesis. These genes are classified into the following categories: 1) anti-oxidative, damage repair, and melanocyte survival-related genes (PRDX3, G6PD, TMX4, HSP70, SERP1, SIRT1, and TRPM1); 2) melanogenesis-related genes (TYR, TYRP1, TYRP2, and MITF); 3) vesicular trafficking genes (VAMP1, RAB25, and RAB27a).30,31 Since miRNAs play important roles in all aspects of vitiligo pathogenesis, further research is particularly important.
No consistent and effective biomarkers for the prognosis of vitiligo progression have been discovered until now. Our study extracted serum exosomal miRNAs from ten non-segmental progressive vitiligo patients and ten healthy individuals before and after systemic corticosteroid treatment. We found that 141 miRNAs were differentially expressed in progressive vitiligo (before treatment) compared to healthy individuals (Figure 1). The profile of miRNAs expression changed after systemic corticosteroid treatment. The hsa-miR-487b-3p was down-regulated in progressive vitiligo but was significantly up-regulated three months after treatment (Figure 2A), without significant difference compared to healthy individuals (Figure 2C). According to ROC analysis (Figure 2D), serum exosomal hsa-miR-487b-3p could be a marker for the prognosis of vitiligo progression (Figure 2E). In other words, it could serve as a biomarker for vitiligo recurrence surveillance. In addition, our results showed that hsa-miR-487b-3p was positively correlated with the total lesion area in progressive vitiligo patients. The higher the area, the more hsa-miR-487b-3p is expressed in serum exosomes. However, it remains unknown what roles this miRNA plays in the pathogenesis of vitiligo. So far, there has been little research on hsa-miR-487b-3p. It was reported to be down-regulated in human osteosarcoma cells and ossification of the posterior longitudinal ligament.32,33 It may increase in U251 glioma cells after treatment with glial cell line-derived neurotrophic factor.34 There has been no reported research on it regarding vitiligo. More specific mechanisms will need to be investigated in the future.
Target genes are important for understanding miRNA functions because a single miRNA could bind to multiple target genes and inhibit their expression.35 Thus, we predicted the target genes of hsa-miR-487b-3p and analyzed them using GO and KEGG enrichment analyses. These genes were mostly involved in the negative regulation of organ growth, such as glycan degradation, protein export, phenylalanine metabolism, and ER-associated degradation (Figure 3). In the case of progressive vitiligo, the low expression level of hsa-miR-487b-3p was unable to effectively inhibit these genes, resulting in abnormal substance metabolism of melanocytes. The imbalance of melanocyte metabolism could further induce autoimmune attacks and accelerate the progression of vitiligo. Among these genes, LRP6, ALDH1A3, PAK5, ZIC1, and GRM3 were related to the physiological functions of melanocytes.24–28 However, their more specific mechanisms in vitiligo pathogenesis have not been studied. DIABLO, also known as Smac, has been closely linked to vitiligo pathogenesis. A high level of DIABLO protein was detected in keratinocytes from perilesional vitiligo skin. It was found to play an important role in keratinocyte damage via the p38/MAPK pathway in vitiligo patients.29 However, there is no functional evidence that hsa-miR-487b-3p affects these genes and pathways in vitiligo pathogenesis, which needs further exploration.
Conclusion
In general, the serum exosomal miRNAs profile of patients with progressive vitiligo differed significantly from those of healthy individuals. The hsa-miR-487b-3p could be an effective serum biomarker for diagnosing vitiligo progression. Its predicted target genes were enriched in catabolism. Down-regulation of hsa-miR-487b-3p in progressive vitiligo may decrease the inhibition of these genes because miRNA can bind to target genes and inhibit its expression. Accordingly, we speculated that negative regulation of cell growth, such as glycan degradation, protein export, phenylalanine metabolism, and ER-associated degradation, could accelerate in melanocytes, leading to their impairment, which could be one of the mechanisms in vitiligo progression. However, more specific studies are needed.
Acknowledgment
This work was supported by [National Natural Science Foundation of China] under grant [number 81872517], [Hangzhou medical key discipline construction project] under grant [number (2021)21-3], and [Science and Technology Special Project of Biomedicine and Health Industry in Hangzhou (the 3rd batch in 2021)] under grant [number 2021WJCY156].
Disclosure
The authors declare that they have no conflicts of interest.
References
- 1.Ezzedine K, Eleftheriadou V, Whitton M, et al. Vitiligo. Lancet. 2015;386:74–84. doi: 10.1016/S0140-6736(14)60763-7 [DOI] [PubMed] [Google Scholar]
- 2.Bergqvist C, Ezzedine K. Vitiligo: a Review. Dermatology. 2020;236:571–592. doi: 10.1159/000506103 [DOI] [PubMed] [Google Scholar]
- 3.Elbuluk N, Ezzedine K. Quality of life, burden of disease, co-morbidities, and systemic effects in vitiligo patients. Dermatol Clin. 2017;35:117–128. doi: 10.1016/j.det.2016.11.002 [DOI] [PubMed] [Google Scholar]
- 4.Xie B, Song X. The impaired UPR-PMEL and TRP channels-autophagy axes in apoptotic melanocytes in vitiligo. Pigment Cell Melanoma Res. 2021;35. doi: 10.1111/pcmr.13006 [DOI] [PubMed] [Google Scholar]
- 5.Mirzaei R, Zamani F, Hajibaba M, et al. The pathogenic, therapeutic and diagnostic role of exosomal microRNA in the autoimmune diseases. J Neuroimmunol. 2021;358:577640. doi: 10.1016/j.jneuroim.2021.577640 [DOI] [PubMed] [Google Scholar]
- 6.Wang X, Shen H, He Q, et al. Exosomes derived from exhausted CD8+ T cells impaired the anticancer function of normal CD8+ T cells. J Med Genet. 2019;56:29–31. doi: 10.1136/jmedgenet-2018-105439 [DOI] [PubMed] [Google Scholar]
- 7.Okoye IS, Coomes SM, Pelly VS. MicroRNA-containing T-regulatory-cell-derived exosomes suppress pathogenic T helper 1 cells. Immunity. 2014;41(1):89–103. doi: 10.1016/j.immuni.2014.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernandez-Messina L, Gutierrez-Vazquez C, Rivas-García E, et al. Immunomodulatory role of microRNAs transferred by extracellular vesicles. Biol Cell. 2015;107(3):61–77. doi: 10.1111/boc.201400081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Almanza G, Anufreichik V, Rodvold JJ, et al. Synthesis and delivery of short, noncoding RNA by B lymphocytes. Proc Natl Acad Sci USA. 2013;110(50):20182–20187. doi: 10.1073/pnas.1311145110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu M, Wang J, Liu M, et al. Study of immunomodulatory function of exosomes derived from human umbilical cord mesenchymal stem cells. Zhonghua Yi Xue Za Zhi. 2015;95:2630–2633. [PubMed] [Google Scholar]
- 11.Pasquali L, Svedbom A, Srivastava A, et al. Circulating microRNAs in extracellular vesicles as potential biomarkers for psoriatic arthritis in patients with psoriasis. J Eur Acad Dermatol Venereol. 2020;34:1248–1256. doi: 10.1111/jdv.16203 [DOI] [PubMed] [Google Scholar]
- 12.Ermakov EA, Kabirova EM, Sizikov AE, et al. IgGs-Abzymes from the sera of patients with systemic lupus erythematosus hydrolyzed miRNAs. J Inflamm Res. 2020;13:681–699. doi: 10.2147/JIR.S258558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang L, Wang C, Jia X, et al. Circulating exosomal miR-17 inhibits the induction of regulatory T cells via suppressing TGFBR II expression in rheumatoid arthritis. Cell Physiol Biochem. 2018;50(5):1754–1763. doi: 10.1159/000494793 [DOI] [PubMed] [Google Scholar]
- 14.Shang Z, Li H. Altered expression of four miRNA (miR-1238-3p, miR-202-3p, miR-630 and miR-766-3p) and their potential targets in peripheral blood from vitiligo patients. J Dermatol. 2017;44:1138–1144. doi: 10.1111/1346-8138.13886 [DOI] [PubMed] [Google Scholar]
- 15.Wang Y, Wang K, Dang N, et al. Downregulation of miR-3940-5p promotes T-cell activity by targeting the cytokine receptor IL-2R gamma on human cutaneous T-cell lines. Immunobiology. 2016;221:1378–1381. doi: 10.1016/j.imbio.2016.07.008 [DOI] [PubMed] [Google Scholar]
- 16.Wong PM, Yang L, Yang L, et al. New insight into the role of exosomes in vitiligo. Autoimmun Rev. 2020;19:102664. doi: 10.1016/j.autrev.2020.102664 [DOI] [PubMed] [Google Scholar]
- 17.Lv M, Li Z, Liu J, et al. MicroRNA‑155 inhibits the proliferation of CD8+ T cells via upregulating regulatory T cells in vitiligo. Mol Med Rep. 2019;20:3617–3624. doi: 10.3892/mmr.2019.10607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cui TT, Yi XL, Zhang WG, et al. miR-196a-2 rs11614913 polymorphism is associated with vitiligo by affecting heterodimeric molecular complexes of Tyr and Tyrp1. Arch Dermatol Res. 2015;307:683–692. doi: 10.1007/s00403-015-1563-1 [DOI] [PubMed] [Google Scholar]
- 19.Al Robaee AA, Alzolibani AA, Rasheed Z. MicroRNA-183-5p regulates MITF expression in vitiligo skin depigmentation. Nucleosides Nucleotides Nucleic Acids. 2022;1–21. doi: 10.1080/15257770.2022.2066126 [DOI] [PubMed] [Google Scholar]
- 20.Zhao C, Wang D, Wang X, et al. Down-regulation of exosomal miR-200c derived from keratinocytes in vitiligo lesions suppresses melanogenesis. J Cell Mol Med. 2020;24:12164–12175. doi: 10.1111/jcmm.15864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi Q, Zhang W, Guo S, et al. Oxidative stress-induced overexpression of miR-25: the mechanism underlying the degeneration of melanocytes in vitiligo. Cell Death Differ. 2016;23:496–508. doi: 10.1038/cdd.2015.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spiegelman VS, Elcheva IA. Metabo-miR: miR-211 regulates mitochondrial energy metabolism in Vitiligo. J Invest Dermatol. 2017;137:1828–1830. doi: 10.1016/j.jid.2017.06.012 [DOI] [PubMed] [Google Scholar]
- 23.Zhang Z, Yang X, Liu O, et al. Differentially expressed microRNAs in peripheral blood mononuclear cells of non-segmental vitiligo and their clinical significance. J Clin Lab Anal. 2021;35:e23648. doi: 10.1002/jcla.23648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, Helke KL, Coelho SG, et al. Essential role of the molecular chaperone gp96 in regulating melanogenesis. Pigment Cell Melanoma Res. 2014;27:82–89. doi: 10.1111/pcmr.12165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pérez-Alea M, McGrail K, Sánchez-Redondo S, et al. ALDH1A3 is epigenetically regulated during melanocyte transformation and is a target for melanoma treatment. Oncogene. 2017;36:5695–5708. doi: 10.1038/onc.2017.160 [DOI] [PubMed] [Google Scholar]
- 26.LaPak KM, Vroom DC, Garg AA, et al. Melanoma-associated mutants within the serine-rich domain of PAK5 direct kinase activity to mitogenic pathways. Oncotarget. 2018;9:25386–25401. doi: 10.18632/oncotarget.25356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Q, Kumar S, Mitsios N, et al. Investigation of downstream target genes of PAX3c, PAX3e and PAX3g isoforms in melanocytes by microarray analysis. Int J Cancer. 2007;120:1223–1231. doi: 10.1002/ijc.22316 [DOI] [PubMed] [Google Scholar]
- 28.Neto A, Ceol CJ. Melanoma-associated GRM3 variants dysregulate melanosome trafficking and cAMP signaling. Pigment Cell Melanoma Res. 2018;31:115–119. doi: 10.1111/pcmr.12610 [DOI] [PubMed] [Google Scholar]
- 29.Becatti M, Prignano F, Fiorillo C, et al. The involvement of Smac/DIABLO, p53, NF-kB, and MAPK pathways in apoptosis of keratinocytes from perilesional vitiligo skin: protective effects of curcumin and capsaicin. Antioxid Redox Signal. 2010;13:1309–1321. [DOI] [PubMed] [Google Scholar]
- 30.Mansuri MS, Singh M, Begum R, et al. miRNA signatures and transcriptional regulation of their target genes in vitiligo. J Dermatol Sci. 2016;84:50–58. doi: 10.1016/j.jdermsci.2016.07.003 [DOI] [PubMed] [Google Scholar]
- 31.Vaish U, Kumar AA, Varshney S, et al. Micro RNAs upregulated in Vitiligo skin play an important role in its aetiopathogenesis by altering TRP1 expression and keratinocyte-melanocytes cross-talk. Sci Rep. 2019;9:10079. doi: 10.1038/s41598-019-46529-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shu X, Liu W, Liu H, et al. Analysis of microRNA expression in CD133 positive cancer stem-like cells of human osteosarcoma cell line MG-63. PeerJ. 2021;9:e12115. doi: 10.7717/peerj.12115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang BL, Dong FL, Guo TW, et al. MiRNAs mediate GDNF-induced proliferation and migration of glioma cells. Cell Physiol Biochem. 2017;44:1923–1938. doi: 10.1159/000485883 [DOI] [PubMed] [Google Scholar]
- 34.Yayama T, Mori K, Okumura N, et al. Wnt signaling pathway correlates with ossification of the spinal ligament: a microRNA array and immunohistochemical study. J Orthop Sci. 2018;23:26–31. doi: 10.1016/j.jos.2017.09.024 [DOI] [PubMed] [Google Scholar]
- 35.Hausser J, Zavolan M. Identification and consequences of miRNA-target interactions beyond repression of gene expression. Nat Rev Genet. 2014;15(9):599–612. doi: 10.1038/nrg3765 [DOI] [PubMed] [Google Scholar]