Abstract
Gain of chromosome arm 13q is one of the most prevalent DNA copy number alterations associated with colorectal adenoma-to-carcinoma progression. The oncogenic miR-17-92 cluster, located at 13q, was found to be overexpressed in colorectal cancer and in adenomas harboring 13q gain. However, to what extent overexpression of this group of microRNAs actually drives progression to cancer remains to be resolved. Therefore, we aimed to clarify the role of miR-17-92 cluster in the progression from colorectal adenoma to carcinoma.
The miR-17-92 cluster was overexpressed in human colorectal adenoma organoids without 13q gain and downstream effects on mRNA expression were investigated, along with functional consequences in vitro and in vivo.
Comparison of mRNA sequencing results of organoids overexpressing miR-17-92 and cultures transduced with control vector revealed a miR-17-92 expression signature. This signature appeared to be enriched in an independent series of colorectal cancers and adenomas with 13q gain, confirming that miR-17-92 expression is associated with malignant progression. However, tumor-associated characteristics, such as increased proliferation rate, were not observed in miR-17-92 overexpressing adenoma organoids in vitro. In addition, subcutaneous injection of these organoids in immunodeficient mice was insufficient to cause tumor outgrowth.
In conclusion, this study showed that miR-17-92 expression contributes to 13q gain-associated adenoma-to-carcinoma progression, however, this is insufficient to cause malignancy.
Keywords: miR-17-92, colorectal, adenoma-to-carcinoma progression
Introduction
Colorectal cancer (CRC) development is a multistep process that is characterized by the sequential acquisition of specific (epi)genetic alterations [1]. Mutations that activate the WNT signaling pathway are held responsible for the transformation of normal epithelial cells into an adenoma (benign lesion). About 5% of these lesions will accumulate additional genetic and epigenetic errors and progress into a carcinoma (malignant lesion) [2,3]. The majority of CRCs follow the so-called chromosomal instability (CIN) pathway, which is characterized by the accumulation of specific nonrandom DNA copy number gains, further referred to as cancer associated events (CAEs) [4]. One of these CAEs is gain of chromosome arm 13q, which occurs in 10-20% of adenomas [5,6] and 40-60% of CRCs [5,7,8]. Gain of 13q is unique to CRC, whereas other solid tumors frequently show 13q loss [9]. This implies that (onco)genes mapping to this chromosome arm may play an important role in the development of CRC. As a consequence of 13q gain, the expression of protein-coding genes (e.g. POLR1D [10], DIS3 [11] and LNX2 [12]) was shown to be upregulated in CRC. In addition, the expression of non-protein-coding transcripts derived from 13q loci, like microRNA-15a-3p [13] and the miR-17-92 microRNA cluster [14], proved to be significantly correlated with 13q gain.
The miR-17-92 miRNA cluster is transcribed from its host locus (MIR17HG) located at 13q31.3 and comprises six miRNAs: miR-17, miR-18a, miR-19a, miR-19b-1, miR-20a, and miR-92a-1. This miRNA cluster is frequently upregulated in lymphomas and solid tumors, including CRC (reviewed in [15]). Several studies showed that overexpression of miR-17-92 accelerates carcinogenesis initiated by oncogenic events, like C-Myc [16] or NOTCH1 [17] activation and deletion of Rb and p107 [18]. This indicates that the miR-17-92 cluster contributes to cancer development, but it is unclear whether overexpression of this group of microRNAs alone is sufficient to drive progression to cancer.
For a long time, functional analysis of the colorectal adenoma-to-carcinoma progression has been hampered by absence of suitable model systems. Adenoma organoid models now make it possible to grow, expand, and investigate human adenomas in vitro and to study the progression from a colorectal adenoma towards a carcinoma [19]. This is even more important as in vivo (in the patient) the natural history of adenomas is interrupted as soon as they are detected and removed during colonoscopy. Here we investigated the potential of miR-17-92 as a driver of colorectal adenoma-to-carcinoma progression in patient-derived colorectal adenoma organoids.
Material and methods
Establishment of colorectal adenoma organoid cultures
Adenoma left-over material was obtained from adenomas resected during a colonoscopy procedure either at the VU University Medical Center or MC Slotervaart, Amsterdam. Briefly, after resection, adenomas were sent to the pathology department of the respective center. After macroscopic examination of the specimen, the central part of the lesion was secured for diagnostic purposes. The left-over tissue was split for research purposes; one part was immediately snap-frozen and the other part was used for the establishment of an adenoma organoid culture.
Adenoma tissue pieces were minced using a sterile scalpel knife and washed three times in ADF+++ culture medium (see below for the composition of culture media). Next, the tissue pieces were immediately resuspended in Matrigel® Matrix basement membrane (growth factor reduced) (Corning Inc., Corning, NY, USA) and plated in pre-warmed (37°C) 24-wells culture plates (Corning Costar, New York, NY, USA). Droplets were left to solidify at 37°C during 10 minutes. Next, 500 µl of ADF+++/BCM (for exact composition of the media see ’organoid culture medium’) was applied to each well containing Matrigel® Matrix droplets. In addition, 10 µM LY27632 (Rho kinase inhibitor; Sigma-Aldrich, Saint Louis, MO, USA)) was added.
Cultures were inspected every 2-3 days for the outgrowth of organoids. Culture medium was replaced upon every inspection by fresh ADF+++/BCM. In general, organoids started to form after 2-5 days.
General organoid culturing
We used the culturing guidelines as published by Van de Wetering et al. [20] with small adaptations. Organoid cultures were inspected every 2-3 days, upon which either the culture medium was refreshed or the organoids were passaged. For passaging, the culture medium was removed, 500 µl ice-cold ADF+++ was added and the Matrigel® Matrix was broken up by pipetting. To wash away the Matrigel® Matrix, organoids were collected in a cold 50 ml tube and cold ADF+++ was added to a final volume of 15 ml. After spinning, the pellet was resuspended in 1 ml of TrypLE Express (Gibco, Waltham, MA, USA) and incubated at 37°C during 5 minutes. Then, ADF+++ was added and the tube was centrifuged again at 1050 rpm during 5 minutes. The pellet was resuspended in Matrigel® Matrix and the cells were plated in droplets in a pre-warmed 24-wells culture plate. After allowing the Matrigel® Matrix to solidify at 37°C for 10 minutes, 500 µl of ADF+++/BCM (supplemented with 10 µM LY27632) was added to each well and cells were incubated at 37°C.
Organoid culture medium
The general ADF+++ culture medium consisted of Advanced DMEM/F12 (Gibco) with 1x Pen/Strep/Glut (Gibco), 10 mM Hepes (Lonza) and 1x Primocin (InvivoGen, San Diego, CA, USA)).
For the culturing of adenoma organoids, we used ADF+++/BCM, which is ADF+++ culture medium supplemented with BCM (basal culture medium; 20% R-Spondin1 conditioned medium, 10% noggin conditioned medium, 1x B27 supplement without Vitamin A (Gibco), 1.25 mM N-Acetyl-L-cysteine (Sigma-Aldrich), 50 ng/ml human recombinant EGF (Peprotech, London, UK), 10 mM Nicotinamide (Sigma-Aldrich), 500 nM A-83-01 (Tocris Bioscience, Abington, UK), 5 µM SB202190 (Cayman Chemicals, Ann Arbor, MI, USA), 5 nM [Leu15]-Gastrin I human (Sigma-Aldrich), 10 nM Prostaglandin E2 (Sigma-Aldrich)). After each passage additional 10 µM LY27632 (Sigma-Aldrich) was added.
Quadruple mutant organoids (normal human colon organoid stem cells with introduced oncogenic mutations in KRAS, APC, TP53 and SMAD4) were kindly provided by Dr. J. Drost (Princess Máxima Center for Pediatric Oncology, Utrecht, the Netherlands) and were expanded in the recommended culture medium [21].
All conditioned media were produced in-house from the cell lines 293T-HA-RspoI-Fc and HEK293-mNoggin-Fc, as described previously [22]. These cell lines were kindly provided by Prof. C. Kuo (293T-HA-RspoI-Fc) from the Stanford University School of Medicine, Stanford, California, USA, and by Prof. H. Clevers (HEK293-mNoggin-Fc) from the Hubrecht Institute, University Medical Center Utrecht and Princess Máxima Center, Utrecht, The Netherlands.
Organoid proliferation assay
Organoids were seeded in triplicate in low-density in 5 µl of Matrigel® Matrix in 96-well plates. Afterwards, plates were incubated in the IncuCyte ZOOM Live-Cell Analysis System (Essen Bioscience, Welwyn Garden City, Hertfordshire, UK) during at least 80 hours. Four pictures per well were taken every 4 hours with 4x magnification. For each well, the best series of pictures (e.g. the series with at least 1 organoid in focus at every timepoint) was chosen.
Next, we used FIJI image software [23] to measure the area of the organoid of interest at each time point. Median areas of at least 6 organoids (two triplicate measurements) were calculated and plotted together with the standard deviation.
Molecular cloning pLV-PGK-miR-17-92-dsRED
The miR-17-92 cluster was PCR amplified from the expressing vector pcDNA3.1/V5-His-TOPO-mir17-92, which was a gift from Joshua Mendell (Addgene plasmid #21109; http://n2t.net/addgene:21109; RRID:Addgene_21109) [24]. The following primers were used; forward: 5’-GGCGCGCCGTCGACCCGGAATTCCTAAATGGACC-3’, reverse: 5’-TGCACGCGTCCGGAATTCGAAAACAAGACAAG-3’. The PCR product was cloned in the pCR 2.1-TOPO TA vector following the guidelines provided in the TOPO TA Cloning Kit for Subcloning (Thermo Fisher Scientific, Waltham, MA, USA). The miR-17-92 insert was subsequently cloned into pLV-PGK-flag2A-dsRED lentiviral vector (kindly provided by Dr. J. Drost, Princess Máxima Center for Pediatric Oncology, Utrecht, the Netherlands) using the restriction enzymes SalI and XhoI (Roche, Basel, Switzerland).
Production of lentiviral particles
The miR-17-92 expressing vector was transfected into HEK293T cells. First, cells were grown up to ∼50% confluency. On the day of the transfection, the culture medium was changed to ADF+++/BCM and the cells were subsequently transfected with 7.5 µg of DNA (packaging, envelope and pLV-vector in a 1:0.1:0.9 ratio) using 45 ug PEI (Polysciences, Warrington, PA, USA). Culture medium was refreshed 18 hours post-transfection. Medium containing lentiviral particles was harvested after an additional 24 and 48 hours, passed through a 0.45-µm filter and concentrated using PEG-8000 precipitation to increase transduction efficiency.
Lentiviral transductions
Adenoma organoid cultures were transduced with virus particles containing pLV-PGK-flag2a-dsRED as the empty vector (EV) or pLV-PGK-miR-17-92-dsRED with the miR-17-92 cluster. Organoids were first enzymatically dissociated using TrypLE Express (Gibco), after which transduction was done by spinoculation [25]. In short, cells were centrifuged for 1 hour at 900 rpm and 25°C in infection medium containing 550 µl ADF+++/BCM, 150 µl ADF+++, 0.75 µl LY27632 (Sigma-Aldrich), 0.75 µl Polybrene (8 µg/ml), and 200 µl concentrated virus supernatant. After spinoculation the cells were plated in a 24-wells plate and incubated for another 5 hours at 37°C. Subsequently, cells were harvested, washed with ADF+++ and plated in 17 µl Matrigel® Matrix droplets. Cultures were supplemented with ADF+++/BCM medium with LY27632. Culture medium was refreshed 24 hours post-transduction.
Transduced organoids were cultured under selective pressure using 1 μg/ml puromycin (Invivogen).
Quantitative reverse transcription-PCR
RNA was isolated from organoid cell pellets using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer's protocol. The quality and quantity of isolated RNA was determined using the NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific).
Expression levels of the separate miRNAs that constitute the miR-17-92 cluster (hsa-miR-17-5p (assay ID 204771), hsa-miR-17-3p (assay ID 206008), hsa-miR-18a-5p (assay ID 204207), hsa-miR-18a-3p (assay ID 204523), hsa-miR-19a-3p (assay ID 205862), hsa-miR-19b-3p (assay ID 204450), hsa-miR-20a-5p (assay ID 204292), hsa-miR-92a-3p (assay ID 204258), and hsa-miR-92a-1-5p (assay ID 204560)) were analyzed using the miRCURY LNA Universal RT microRNA PCR System (Exiqon, Vedbæk, Denmark) following the manufacturer's instructions. PCRs were run on an ABI 7500 Fast Real-time PCR system (Thermo Fisher Scientific). Relative miRNA expression levels were determined using the 2(-∆∆Ct)-method [26]. Data were normalized using the endogenous reference microRNA miR-16-5p (hsa-miR-16-5p (assay ID 205702; Exiqon).
Low-coverage whole genome sequencing
DNA from organoid cell pellets was isolated using the QIAamp DNA micro kit (QIAGEN, Venlo, The Netherlands) according to the “Genomic DNA from cultured cells” guidelines. Quality and quantity of isolated genomic dsDNA was determined using the Qubit dsDNA BR Assay Kit (Invitrogen). Up to 2000 ng of double stranded genomic DNA was fragmented by Covaris shearing to obtain fragment sizes of 160-190 bp. Samples were purified using 1.6X Agencourt AMPure XP PCR Purification beads according to manufacturer's instructions (Beckman Coulter, Brea, Ca, USA). The quality and quantity of the sheared DNA samples was inspected on a BioAnalyzer system using the DNA7500 assay kit (Agilent Technologies, Santa Clara, CA, USA). Library preparation for Illumina sequencing was performed using the KAPA HTP Library Preparation Kit (KAPA Biosystems, Wilmington, MA, USA), with an input of maximum 1 μg sheared DNA. Enrichment of the library was done by 4-8 PCR cycles. After library preparation, AMPure XP beads (Beckman Coulter) were used to remove contaminants. All DNA libraries were analyzed on a BioAnalyzer system using a DNA7500 chip to determine the molarity. Pools of twenty-nine and twenty-two uniquely indexed samples were created by equimolar pooling, in a final concentration of 10 nM, and subjected to sequencing on an Illumina HiSeq2000 machine in a single read 65bp run, according to manufacturer's instructions.
Identification of copy number aberrations was performed as described previously [27]. Briefly, low quality reads and adapter sequences were trimmed with Trimmomatic version 3 [28]. Reads were cropped to 50 bp and shorter reads were removed. Burrows-Wheeler Aligner was used to uniquely align trimmed reads to human reference genome build hg19 [29]. Read counting per bins, normalizations, corrections and filtering were done with Bioconductor package QDNAseq [30]. Wave-correction was performed after median normalization with the NoWaves R package [31]. Copy number segmentation was performed using Bioconductor package DNAcopy [32]. Gained and lost regions were identified using Bioconductor package CGHcall [33]. Copy number aberrations were considered (called) when probability was higher than 50%.
Raw DNA copy number data is made available in the European Genome-Phenome Archive (EGA; https://www.ebi.ac.uk) under the Study ID datasets: EGAS00001005947, EGAS00001005937 and EGAS00001005865.
Mutation analysis
DNA was isolated from the generated organoids as described above. Mutation analysis was performed by targeted next generation sequencing using either the TruSeq Amplicon Cancer Panel (TSACP; Illumina, San Diego, CA, USA) or a 48 gene xGen® Predesigned Gene Capture Pools (Integrated DNA Technologies, San Diego, CA, USA), as previously described ([34,35]. Both panels contain the same 48 genes (Supplementary Table 1).
Mutation calling was performed as described previously [35]. Raw DNA mutation data is made available in the European Genome-Phenome Archive (EGA; https://www.ebi.ac.uk) under the Study ID datasets: EGAS00001005947, EGAS00001005937 and EGAS00001005865.
RNA sequencing and data pre-processing
RNA was isolated from organoid cell pellets using TRIzol reagent (Invitrogen) following the manufacturer's protocol. The quality and quantity of isolated total RNA was assessed using the 2100 Bioanalyzer using a Nano chip (Agilent). Total RNA samples having a RIN>8 were subjected to library generation. Strand-specific libraries were generated using the TruSeq Stranded mRNA sample preparation kit (Illumina) according to the manufacturer's instructions. In short, polyadenylated RNA from intact total RNA was purified using oligo-dT beads. Following purification, the RNA was fragmented, random primed, and reverse transcribed using SuperScript II Reverse Transcriptase (Invitrogen) with the addition of Actinomycin D. Second strand synthesis was performed using Polymerase I and RNaseH with replacement of dTTP for dUTP. The generated cDNA fragments were 3′ end adenylated and ligated to Illumina Paired-end sequencing adapters and subsequently amplified by 12 cycles of PCR. Resulting libraries were analyzed on the 2100 Bioanalyzer using a 7500 chip (Agilent). The libraries were sequenced with 65 base single reads on a HiSeq2500 using V4 chemistry (Illumina).
RNA-seq data (pre-)processing was performed as described previously [27], using the human genome build hg19 (USCS RefSeq hg19, gencode v19 annotation). Raw RNA-seq data is made available in the European Genome-Phenome Archive (EGA; https://www.ebi.ac.uk) under the Study ID dataset: EGAS00001005949.
Differential gene expression (DGE) analysis
RNA‐seq data were subjected to differential expression analysis, using gene set enrichment analysis (GSEA) [36] and ssGSEA [37]. In this analysis EV-transduced and miR-17-92-transduced organoid cultures were compared using DESeq2 [38]. DEGs were obtained by filtering for absolute log2 fold change≥1 and adjusted p-value≤0.1. The resulting list of statistically significant overexpressed and underrepresented genes are collectively referred to as the “miR-17-92 expression signature”.
Regularized logarithmic transformation of expression values was performed, Euclidian distance between samples was calculated and the RNA expression data were visualized using the multidimensional scaling algorithm.
In vivo transplantation of organoids
Organoids transduced with either miR-17-92 or EV were injected subcutaneously in NOD-Scid IL2Rgnull mice (The Jackson Laboratory (Jax), Ellsworth, ME, USA) at the Preclinical Intervention Unit of the Mouse Cancer Clinic of the Netherlands Cancer Institute (Amsterdam, the Netherlands). As a positive control, we used quadruple mutant organoids [21], which we also transduced with the EV and miR-17-92 cluster vectors.
For each transduction (empty vector or miR-17-92 insert), 5 mice were injected with 4×10e5 cells in a volume of 100 µl (50 µl organoids in ADF/2xBCM or quadruple mutant organoid medium, and 50 µl Matrigel® Matrix). Body weight and tumor outgrowth were monitored twice a week, up to a maximum of 120 days. Mice were sacrificed if tumors grew bigger than 1500mm3. Tumors (if present), injection sites, liver and spleen were secured for downstream analysis.
Results
Establishment and characterization of adenoma organoid cultures
In total, 54 adenoma tissue pieces were collected and used to set up organoid cultures. Successful establishment of good growing organoid cultures was achieved for 39 of the adenoma pieces, a success rate of 72%. Reasons for failure were bacterial contamination (7 cases), no outgrowth of organoids from the tissue (1 case), lack of organoid proliferation (4 cases) and poor growth in terms of organoid size (3 cases).
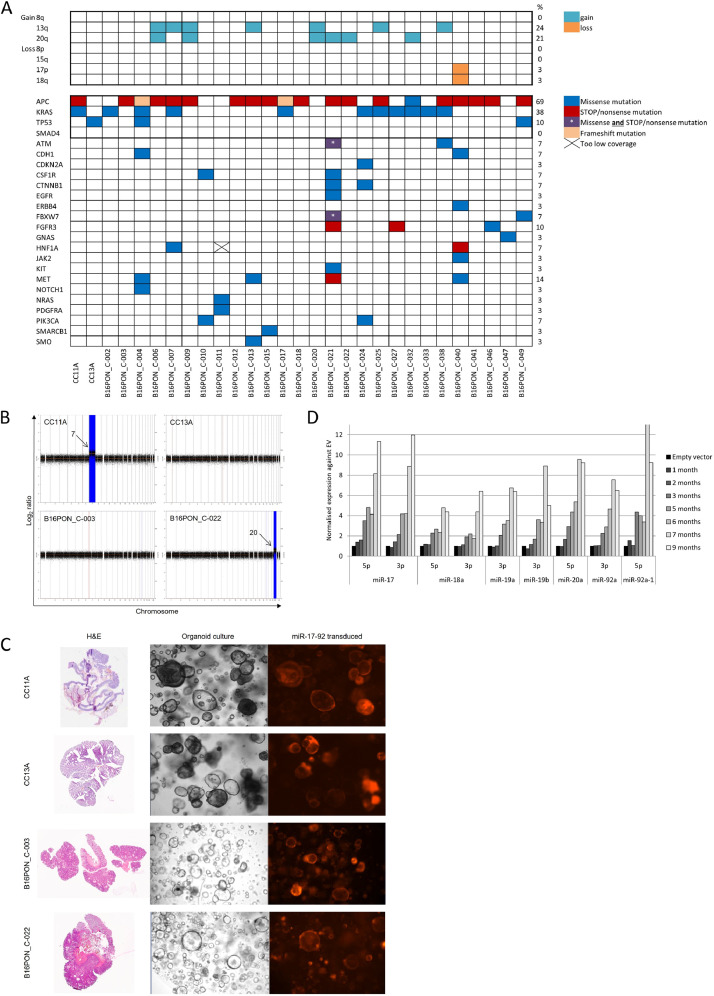
A subset of organoid cultures (n=29) were genetically characterized for cancer-related mutations and whole genome DNA copy number alterations (Fig. 1a and Supp Table 1). As expected, the majority (69%) of the adenoma-derived organoids harbored a mutation in the APC gene. In addition, genetic alterations in other well-known drivers of colorectal cancer progression were identified, like KRAS (38%) and TP53 (10%). Mutations in other cancer-related genes varied between 3% and 10% of the samples (Fig. 1a), with the exception of the mutation rate of MET (14%). Regarding DNA copy number profiles, we looked at the presence of the previously identified cancer-associated events (CAEs), namely 8q gain, 13q gain, 20q gain, 8p loss, 15q loss, 17p loss and 18q loss [4,39], as markers of high risk of progression to cancer. The most frequently detected CAEs are gains of chromosome arms 13q (24%, 7/29) and 20q (21%, 6/29) (Fig. 1a). Taken together, we created a very rich source of adenoma organoid cultures with diverse genetic backgrounds that are representative of colorectal adenomas.
Fig. 1.
The colorectal adenoma organoid biobank is genetically diverse and several organoids were successfully transduced with miR-17-92A) Adenoma organoid cultures (bottom) were characterized for the presence of specific DNA copy number changes - cancer associated events (CAEs; top) and mutations in cancer associated genes (left). Colors in the matrix indicate the type of mutation detected. The column on right of the matrix shows the percentage of samples that harbors a mutation in that specific gene (e.g. 69% of the adenoma organoids has a mutation in APC). B) DNA copy number profile of the adenoma organoid cultures used for the transduction of miR-17-92. B16PON_C-003 and CC13A do not have any aberrations. CC11A has a gain of both arms of chromosome 7 (not cancer associated) and B16PON_C-022 has a cancer associated gain of chromosome 20q. C) H&E stainings of the adenoma tissue used to establish adenoma cultures selected for miR-17-92 transduction (left). Second and third columns show the corresponding organoid culture and miR-17-92 transduced culture, respectively. Pictures of organoid cultures were made using 5x magnification. D) mRNA expression of the individual members of the miR-17-92 cluster in the miR-17-92 transduction of organoid CC11A over time. Expression values are normalized to EV transduced cultures at the corresponding time point. Data is representative for all organoid cultures, however the maximum miRNA expression varies between the organoid cultures.
Overexpression of miR-17-92 leads to a CRC-like expression profile
In the present study, we were interested in the potential role of the miR-17-92 cluster as driver of the 13q amplicon during colorectal adenoma-to-carcinoma progression. If so, overexpression of the miR-17-92 cluster will mimic gain of 13q and may drive progression to carcinoma. To investigate this, four low-risk adenoma organoid cultures with mutational backgrounds representative for the most common genetic alterations, but all without a 13q gain, were selected for overexpression of the miR-17-92 cluster. These organoids either carried an APC mutation (B16PON_C-003), APC and KRAS mutations (CC11A), a TP53 mutation (CC13A), or KRAS mutation and gain of chromosome 20q (B16PON_C-022) (Supp Table 1). Gain of chromosome arm 13q is often observed in combination with 20q gain (another frequently observed CAE), indicating that these two DNA copy alterations may interact with each other in promoting the progression from adenoma to carcinoma (Fig. 1a/b, Table I). The four cultures were transduced with a vector expressing the miR-17-92 cluster or an empty vector (EV) as a control (Supp Fig. 1). Following puromycin selection, both the expression of the dsRED tag (Fig. 1c, red fluorescent signal) and the mRNA expression of the miR-17-92 cluster were monitored in the organoids. A clear gradual increase over time in mRNA expression of all individual miRNAs that are part of the miR-17-92 cluster was detected (Fig. 1d). This confirms that the transduction of the miRNA cluster into the adenoma organoids was successful.
Table 1.
Overview of the mutations detected in the adenoma organoid cultures selected for miR-17-92 transduction.
| Organoid | Mutated gene | Genomic description | Protein description | Mutation effect |
|---|---|---|---|---|
| B16PON_C-003 | APC | c.637C>T | p.R213* | STOP |
| CC13A | TP53 | c.473G>A | p.R158H | missense |
| CC11A | APC | c.4348C>T | p.R1450* | STOP |
| KRAS | c.34G>A | p.G12S | missense | |
| B16PON_C-022 | APC | c.2626C>T | p.R876* | STOP |
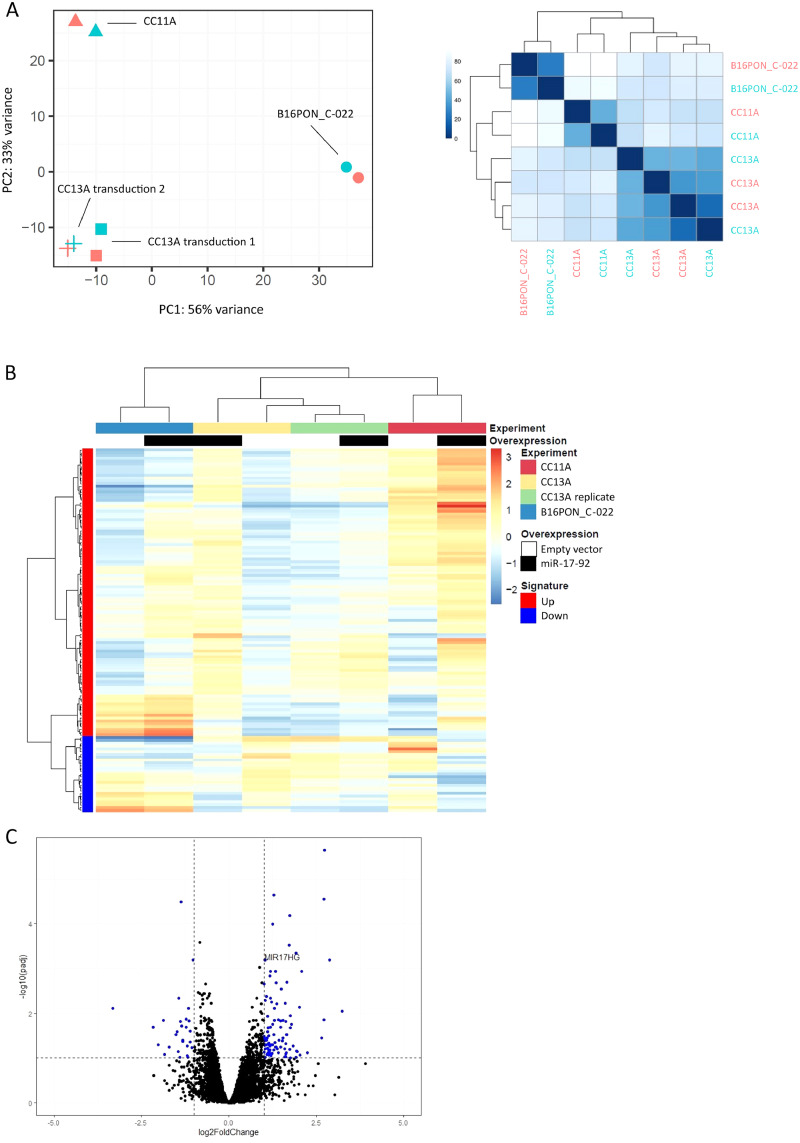
To study the downstream effect of miR-17-92 overexpression in the transcriptome, RNA was isolated from all four miR-17-92 transduced adenoma organoid cultures and matched EV transduced cultures (same passages). For the CC11A and CC13A cultures, two independent rounds of transduction were included to have duplicates meant to investigate the robustness of the experimental system. RNAseq results showed that the expression of the MIR17HG locus (positive control) was not higher in the B16PON_C-003 organoid culture transduced with the miR-17-92 cluster as compared to the EV transduced culture, and therefore B16PON_C-003 organoid results were excluded from further analysis. For the same reason, the first transduction of CC11A was excluded (Supp Fig. 2). Principal component analysis (PCA) of the 8 samples left, showed that variation in the genetic background of the adenoma organoids is more pronounced than the variation between the EV and miR-17-92 transductions within the same organoid background (Fig. 2a). Furthermore, the two independent CC13A transductions cluster close together, pointing out that the two independent transductions led to similar expression profiles.
Fig. 2.
Overexpression of miR-17-92 results in changes in mRNA expression.A) Principle component analysis of the four transductions that passed quality control (left). EV (pink) and miR-17-92 (green) transductions cluster together per genetic background. The similarity plot (right) shows the percentage of similarity between the EV (pink labels) and the miR-17-92 transduced (green labels) organoid cultures. B) Hierarchical clustering of the mRNAs that compose the miR-17-92 expression signature (left column, red is overexpressed, blue is underrepresented;|FC|>2, adjusted p-value p<0.1) in the four transduced organoid cultures (top row) with either EV (white in second row) or miR-17-92 overexpression (black in second row). Euclidian distance between samples is displayed. C) mRNA expression of MIR17HG, the host gene for the miR-17-92 cluster, is one of the most overrepresented mRNAs in miR-17-92 transduced adenoma organoids cultures.
Next, we extracted differentially expressed genes (|FC|≥2 and adj. p-value≤0.1) between the four EV and miR-17-92 transduced organoid cultures. The resulting 103 overexpressed and 27 underrepresented genes were collectively considered to reflect the expression of the miR-17-92 cluster and were therefore referred to as miR-17-92 expression signature (Table 2). Hierarchical clustering of the transduced organoids using the mRNA expression of the genes included in the miR-17-92 expression signature again showed that the samples cluster mainly based on genetic background, although one observes differences in expression between each miR-17-92 overexpressing culture in comparison to the matched EV control (Fig. 2b). Amongst the most overrepresented mRNAs in miR-17-92 transduced adenoma organoids, we found MIR17HG, the host gene for the miR-17-92 cluster of microRNAs (Fig. 2c). In addition, several experimentally validated targets of the miR-17-92 cluster of microRNAs were significantly downregulated in the miR-17-92 transduced organoid cultures as compared to the EV transduced organoids (i.e. PRKACB, MCOLN2, CYBRD1, ITPR1, PTPRG, NBEA) (miRTarBase release 7.0 [40]).
Table 2.
The miR-17-92 expression signature consists of 27 underrepresented and 103 overrepresented mRNAs.
| Underrepresented | Overrepresented | |||
|---|---|---|---|---|
| AASS | ABCC2 | EEF1A2 | LOC100190940 | SLC51A |
| ABCC4 | ADAM8 | EFEMP2 | MFAP2 | SLC51B |
| BICC1 | ADCY4 | EFNA2 | MIR17HG | SLC5A9 |
| CELF2 | ADM2 | ERICH4 | MLLT4-AS1 | SLC6A12 |
| CXCL8 | ADRA2C | EVA1B | MLXIPL | SLC6A4 |
| CYBRD1 | ALOX15B | F10 | MOGAT3 | SMIM24 |
| DOCK4 | ANG | F13A1 | MST1 | SPAG4 |
| DSC3 | ANGPTL4 | FADS6 | MYO1F | SPN |
| IFI6 | ASNS | FAM3B | NOTCH3 | STC2 |
| ITPR1 | ASPDH | FAXDC2 | NOTUM | STKLD1 |
| KBTBD8 | AXL | FBXL16 | NR1H4 | STRA6 |
| LOC101928100 | BMF | FBXO2 | PCBP3 | TFF3 |
| MAOB | CACNA1F | FGD5 | PDE2A | TGM2 |
| MCOLN2 | CACNA2D4 | G0S2 | PDLIM4 | TM6SF2 |
| MT1E | CACNG4 | GAL3ST1 | PDZK1 | TMEM178A |
| NBEA | CDH16 | GIPR | PHGDH | TMEM63C |
| NR2F1 | CDHR2 | GMFG | PLXND1 | TMEM92 |
| ODAM | COL27A1 | GS1-259H13.2 | POU6F1 | TRIB3 |
| OXCT1 | COL5A3 | HAPLN4 | PPFIA4 | UNC93A |
| PDE10A | COL7A1 | IL6R | RASA4CP | UPK3A |
| PRKACB | CYP27A1 | KIF12 | SARDH | VAV2 |
| PTGER2 | CYP2B6 | KIRREL2 | SH3D21 | WFDC2 |
| PTPRG | CYP4F2 | LAMA1 | SHC2 | |
| REG4 | DDIT3 | LCN12 | SLC22A17 | |
| RHOBTB1 | DDIT4 | LINC00483 | SLC29A4 | |
| SLC6A14 | DNM1 | LINC00668 | SLC39A5 | |
| SOSTDC1 | EBF4 | LINC01207 | SLC3A1 | |
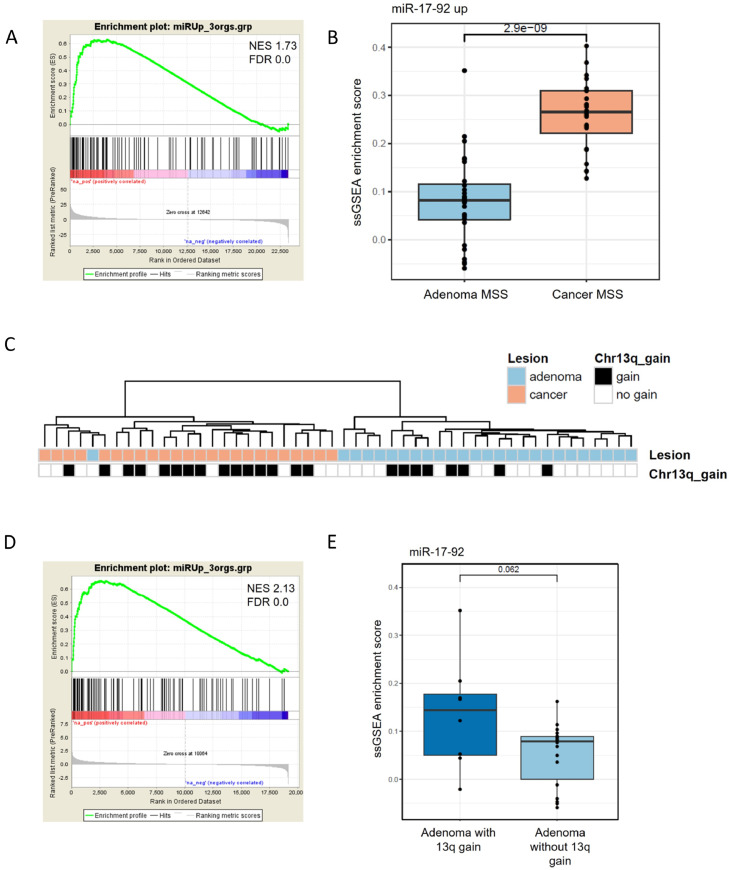
To investigate the potential role of miR-17-92 in promoting progression from a colorectal adenoma towards a malignant phenotype, we set out to compare the expression of the 130 gene set (miR-17-92 expression signature) between colorectal adenomas and cancers. For this, previously published mRNA sequencing data on independent tissues of colorectal adenomas and cancers [27] were used. GSEA analysis of the mRNA expression data of 26 microsatellite stable (MSS) colorectal adenomas and 24 MSS colorectal cancers showed that the miR-17-92 expression signature was enriched in colorectal cancers compared to adenomas. (Fig. 3a). Calculation of the single sample GSEA (ssGSEA) scores for these samples showed a significant difference in the enrichment of the miR-17-92 expression signature in the cancer samples (p-value=2.9×10e-9) (Fig. 3b). This is also reflected in the hierarchical clustering of the 50 samples, where the miR-17-92 expression signature almost perfectly separates the adenomas from the cancers (Fig. 3c), irrespective of the presence or absence of chromosome arm 13q gain.
Fig. 3.
The miR-17-92 expression signature is enriched in adenomas with 13q gain and in cancers versus adenomas.A) GSEA using the miR-17-92 expression signature in 26 adenoma tissue samples and 24 colorectal cancers (NES 1.73, FDR q value 0.0). B) ssGSEA enrichment score for the same samples (p-value 2.9x10-9). C) Hierarchical clustering of the differential mRNAs that compose the miR-17-92 expression signature in 26 adenoma tissue samples (blue in first row) and 24 colorectal cancers (pink in first row). The samples cluster independent of the presence of a 13q gain (black in second row). D) GSEA using the miR-17-92 expression signature in 18 adenoma tissue samples without 13q gain and 8 adenomas with 13q gain (NES 2.13, FDR q value 0.0). E) ssGSEA enrichment score for the same samples (p-value 0.062).
Next, we investigated whether the miR-17-92 expression signature was able to distinguish adenomas with an overexpression of the miR-17-92 cluster, due to a gain of chromosome arm 13q, from those that do not have the amplification. The independent validation series comprised 18 adenomas without a 13q gain and 8 adenomas with this specific gain of the chromosome arm. GSEA analysis showed that the miR-17-92 signature was significantly enriched in the adenomas that have a 13q gain (Fig. 3d). When the ssGSEA scores were calculated for these adenomas using the miR-17-92 expression signature followed by comparison of samples with and without 13q gain, the same trend was observed (Fig. 3e). However, this did not reach statistical significance (p-value=0.062).
miR-17-92 overexpression has no effect on organoid proliferation
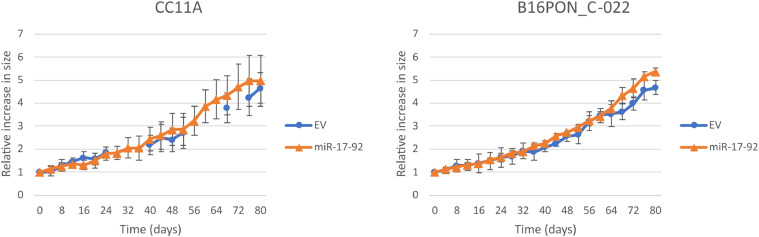
Next, we investigated whether the overexpression of the miR-17-92 cluster induced cancer-specific characteristics in the adenoma organoids. A well-known trait of tumor cells is the increase in cell proliferation, so therefore the growth rate of the organoid cultures transduced with either EV or miR-17-92 was examined by measuring the area of intact organoids over time. In Fig. 4 representative growth curves of two different organoid cultures are depicted. There was no significant difference in growth rate between the EV and miR-17-92 transduced cultures for any of the genetic backgrounds. In addition, CC11A, the organoid culture without CAEs (Fig. 1b) did not significantly grow faster nor slower than the culture with a 20q gain (B16PON_C-022).
Fig. 4.
Overexpressing miR-17-92 in adenoma organoids does not alter growth rate in 3D cultures.EV (dots) and miR-17-92 (triangle) transduced organoids were monitored for proliferation for 80 hours. Every four hours, the culture area (referred to as organoid size) occupied by the organoid was measured. The graphs display the relative increase in size as compared to the start of the measurement. At least four organoids per culture were followed and mean sizes and standard deviations are shown.
Overexpression of the miR-17-92 cluster is not sufficient to grow tumors in mice
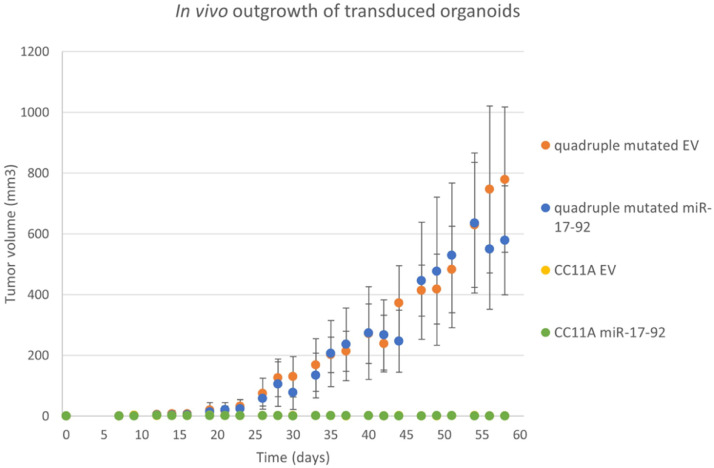
Due to their non-invasive, benign nature, adenomas are not able to form tumors in mice. To investigate if overexpression of the miR-17-92 cluster is sufficient to provide an adenoma organoid with the ability to grow out in vivo, we injected CC11A organoids transduced with either EV or miR-17-92 expression constructs s.c. in immunodeficient mice. The same constructs were transduced in human normal colon epithelium derived organoids where the main mutations described in the Vogelstein progression model, namely APC, KRAS, TP53 and SMAD4, were introduced. These engineered organoids (hereafter called quadruple mutated organoids) are able to form invasive tumors in mice when injected s.c. [21] and therefore function as our positive control. There was no difference in in vitro growth rate between the CC11A organoids and the quadruple mutated organoid (Supp Fig. 3).
Twenty-one days after s.c. injection, the transduced quadruple mutated organoids (both EV and miR-17-92 transduced) showed outgrowth of the injected cells. This was not the case for the EV or miR-17-92 transduced CC11A organoids (Fig. 5). By day 26 post-injection, the tumor volume of both CC11A and the quadruple mutated organoids that were transduced with miR-17-92 transduced was significantly different (p<0.05, Student's t-test), as was the difference in outgrowth between the CC11A EV transduced and the quadruple mutated EV organoids. We also did not observe a significant difference in tumor growth between the EV and miR-17-92 transduced quadruple mutated organoid cultures. From day sixty post-injection onwards, tumors with sizes >1500 mm3 were harvested from the quadruple mutated organoid groups (both EV and miR-17-92). In mice inoculated with the CC11A manipulated organoids, bumps of <7 mm3 (miR-17-92) or <5 mm3 (EV) at the inoculation site were measured. For both EV and miR-17-92 transduced CC11A organoids, the sizes of these bulges did not increase during the next sixty days. After termination of the in vivo experiment, we inspected microscopically the tissue surrounding the organoid injection site and could not find any evidence for organoid outgrowth.
Fig. 5.
Overexpressing miR-17-92 in adenoma organoids does not induce malignant tumorigenicity.EV (yellow) and miR-17-92 transduced (green) CC11A organoids were subcutaneously injected in immunodeficient mice. As a positive control, quadruple mutated organoids were also transduced with both constructs and inoculated in mice (blue and orange). Median tumor volumes are displayed, together with the standard deviation.Tumor outgrowth was measured twice a week over a period of 60 days.
Discussion
The oncogenic miR-17-92 cluster is located on chromosome arm 13q, and gain of this region is one of the genetic events associated with the progression from colorectal adenoma to CRC. In this study, we investigated the potential role of the miR-17-92 cluster as driver of 13q amplicon in the colorectal adenoma-to-carcinoma progression. For this purpose, we established patient-derived colorectal adenoma organoid cultures and overexpressed the miR-17-92 cluster. The comparison of mRNA expression data between cultures transduced with miR-17-92 and empty vector led to the identification of a miR-17-92 expression signature, which showed to be enriched in an independent series of CRC tissues, as well as in adenoma tissues with 13q gain. This confirms that miR-17-92 expression is associated with malignant progression and supports a role as a driver of 13q gain. However, overexpression of the miRNA cluster did not have an effect on the proliferation rate of adenoma organoids in vitro, nor did it cause tumor outgrowth upon subcutaneous injection in immunodeficient mice, showing that overexpression of miR-17-92 alone is not sufficient to transform a colorectal adenoma into carcinoma.
Colorectal adenoma cells do not easily grow in vitro under standard cell line culture conditions. Therefore, it was for a long time challenging to culture adenoma-derived cell lines. With the recent advances made in organoid technology [19], we were able to culture patient-derived colorectal adenoma cells as organoids in vitro. With a success rate of 72%, we assembled a panel of 39 adenoma organoid cultures. This resource now makes it possible to investigate step-by-step the events that drive colorectal adenoma-to-carcinoma progression.
The oncogenic miR-17-92 cluster has previously been described to be involved in CRC development [14,15]. To investigate this further, we transduced adenoma organoid cultures with the miR-17-92 cluster, or an empty vector as control, and obtained a miR-17-92 expression signature from RNA sequencing data. Interestingly, despite the fact that this signature was acquired by overexpression of the miR-17-92 cluster in pure epithelial cell cultures (the organoids), the signature was also able to discriminate carcinoma from adenoma tissue samples, which next to the epithelium also contain other cell types in the stroma component. This observation shows that adenoma cell cultures are a valuable addition to the armamentarium for in vitro investigation of carcinogenesis and that the results derived from research involving adenoma organoids are translatable to tumors found in patients.
The overexpression of the miR-17-92 cluster in the adenoma organoid cultures was verified by inspecting the expression of all individual members of the miRNA cluster using RT-qPCR. We could confirm that the transduction of the cluster was successful, however, the established increase in miRNA expression varied among the cluster members. This is similar to what has been seen in clinical samples when comparing normal colorectal mucosa and colorectal cancer [14,41,42] or colorectal tumors with 13q gain versus those without [14]. Specifically, the higher expression of miR-92a, also seen in the transduced organoids, is of interest as it has been proposed as a potential biomarker in the early diagnosis of colorectal cancer [43,44], even in stool samples [45]. In contrast, the expression of miR-18a was less elevated than other members of the miR-17-92 cluster in the transduced organoids. This might be explained by the antiproliferative effect of miR-18a shown in experiments using colon [41], stomach [46], and bladder cancer cell lines [47] and in pancreatic progenitor cells [48].
Not only RT-qPCR results showed that the transduction of the miR-17-92 cluster was successful in the organoid cultures, also the overexpression of its host gene MIR17HG, identified by RNA sequencing, confirms this. Next to MIR17HG, multiple differentially expressed mRNAs were identified when comparing EV and miR-17-92 transduced organoids. These were collectively called the miR-17-92 expression signature, which was enriched in colorectal cancers when compared to adenomas. This is in line with literature on the oncogenic role of miR-17-92 (reviewed in [15]). The miR-17-92 expression signature was also significantly enriched in adenomas with a 13q gain, indicating that artificial overexpression of the miR-17-92 cluster in adenoma organoids causes changes in mRNA expression that closely resemble the situation in adenomas with endogenous amplification of the miR cluster locus. Upregulation of miR-17-92 showed to block differentiation [49], accelerate cell proliferation [50], evade apoptosis [51,52], trigger angiogenesis [53], and promote metastasis [54]. Indeed, looking at the genes in the miR-17-92 expression signature obtained in the present study, we identified genes that play roles in these oncogenic processes, such as PRKACB [55] and DOCK4 [56]. In addition, several of the genes in the miR-17-92 expression signature (PRKACB, MCOLN2, CYBRD1, ITPR1, PTPRG and NBEA) are confirmed targets of multiple members of the miR-17-92 cluster according to miRTarBase [40]. We observed downregulation of ITPR1 expression, a gene with a pro-apoptotic function [57]. ITPR1 induces calcium release from the endoplasmic reticulum, which leads to the subsequent activation of downstream apoptosis pathways [58]. Furthermore, mRNA expression of the tyrosine phosphatase PTPRG was significantly downregulated in the signature. PTPRG is a candidate tumor suppressor and loss of function by deletion [59,60] or mutation [61,62] was associated with various types of cancer. PTPRG antagonizes the activity of protein tyrosine kinases, thereby inhibiting the activity of crucial signaling pathways that control cell cycle progression, proliferation, invasion, and angiogenesis. Taken together, the miR-17-92 expression signature is composed of multiple genes that are confirmed to be differentially expressed upon miR-17-92 overexpression and that play a role in malignant processes known to be influenced by the miR-17-92 cluster.
It was shown previously that numerical changes in chromosomes or chromosome arms, in addition to driver mutations, might be required for full transformation of colorectal epithelial cells, specifically concerning invasive and metastatic potential [63]. This feeds the hypothesis that other genetic alterations, on 13q or elsewhere in the genome, might be required to drive progression to cancer. In line with this, several studies showed that full-blown tumors were not detected upon miR-17-92 overexpression in mouse lymphocytes [64] and that overexpression of miR-17-92 on top of other oncogenic events accelerated the development of cancer [16], [17], [18]. Gain of 13q often co-occurs with 20q gain, which suggests that genes located in these two genomic locations may interact and play a role in the progression from adenoma to cancer. We previously showed that AURKA and TPX2 (mapping at the gained region of 20q) are drivers of that amplicon [65]. In addition, an interesting novel candidate driver of tumor progression located on 20q is POFUT1, a gene recently linked to increased Notch signaling [66]. In light of the hypothesis that miR-17-92 accelerates carcinogenesis initiated by NOTCH1 [17], the identification of POFUT1 could reinforce the theory that genes located in 13q and 20q interact in the same pathways, and contribute to the progression to cancer. Since one of the organoid cultures we transduced with the miR-17-92 expression vector (B16PON_C-022) harbors a 20q gain, we have a model in hands to further investigate the interaction of miR-17-92 and genes located on chromosome arm 20q.
Overexpression of miR-17-92, as a candidate driver of adenoma-to-carcinoma progression on the chromosome 13q, in itself did not show to be sufficient for adenoma organoids to acquire hallmarks of cancer, like accelerated proliferation rates in vitro and tumor outgrowth in vivo. These results demonstrate that, while the induced overexpression of a potential driver located on chromosome 13q may transform a low-risk adenoma into an adenoma with higher risk of progression, overexpression of the miR-17-92 cluster alone is not sufficient to fully force malignant transformation. Overexpression of the miR-17-92 cluster in chromosomal instable quadruple mutant organoids (our positive control for tumorigenesis in vivo) did not result in accelerated tumor outgrowth in mice, nor invasion in surrounding tissues, compared to its respective EV-control. Apparently, the quadruple mutations themselves are sufficient to drive a malignant phenotype. However, only a minority of human colorectal cancers harbor all four mutations, suggesting that in other cancers the effect of one of the mutations may be substituted by 13q gain.
Taken together, we established a large biobank of colorectal adenoma organoids that provides a valuable source of adenoma cultures. The adenoma organoids can be expanded and genetically manipulated in order to investigate colorectal adenoma-to-carcinoma progression. Overexpressing the miR-17-92 cluster in several adenoma organoids led to the identification of a miR-17-92 expression signature. This signature proved to be enriched in both adenomas with 13q gain and in colorectal cancers, supporting its role as a driver of 13q gain. However, the results of both in vitro and in vivo experiments lead to the conclusion that overexpression of miR-17-92 alone is not sufficient to transform colorectal adenoma cells into carcinoma cells. We hypothesize that additional genetic alterations, like chromosomal gains and losses of CAEs, as well as other driver mutations might be required for full transformation of colorectal adenoma cells.
CRediT authorship contribution statement
Sanne R. Martens-de Kemp: Conceptualization, Investigation, Visualization, Writing – original draft. Malgorzata A. Komor: Software, Formal analysis, Data curation, Visualization. Rosa Hegi: Formal analysis. Anne S. Bolijn: Investigation. Marianne Tijssen: Investigation. Florence L.M. de Groen: Investigation. Annekatrien Depla: Resources. Monique van Leerdam: Resources. Gerrit A. Meijer: Writing – review & editing. Remond J.A. Fijneman: Conceptualization, Writing – review & editing. Beatriz Carvalho: Conceptualization, Writing – review & editing.
Acknowledgments
This research was supported by Dutch Cancer Society (KWF Kankerbestrijding), project numbers 2013‐6025 and 2014-6813. The authors would like to thank the Genomics Core Facility (Netherlands Cancer Institute) for performing the low-coverage whole genome sequencing of the organoid cultures and the mRNA sequencing of the transduced organoids, the Bioimaging Facility (Netherlands Cancer Institute) for the imaging capture of the organoid cultures and the Preclinical Intervention Unit of the Mouse Cancer Clinic of the Netherlands Cancer Institute (Amsterdam, the Netherlands) for performing the mouse experiments. This work was performed within the frame of the COST Action [CA17118], supported by COST (European Cooperation in Science and Technology).
Footnotes
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neo.2022.100820.
Appendix. Supplementary materials
References
- 1.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 2.Church JM. Clinical significance of small colorectal polyps. Dis Colon Rectum. 2004;47:481–485. doi: 10.1007/s10350-003-0078-6. [DOI] [PubMed] [Google Scholar]
- 3.Heitman SJ, Ronksley PE, Hilsden RJ, Manns BJ, Rostom A, Hemmelgarn BR. Prevalence of adenomas and colorectal cancer in average risk individuals: a systematic review and meta-analysis. Clin Gastroenterol Hepatol. 2009;7:1272–1278. doi: 10.1016/j.cgh.2009.05.032. [DOI] [PubMed] [Google Scholar]
- 4.Hermsen M, Postma C, Baak J, Weiss M, Rapallo A, Sciutto A, Roemen G, Arends JW, Williams R, Giaretti W, et al. Colorectal adenoma to carcinoma progression follows multiple pathways of chromosomal instability. Gastroenterology. 2002;123:1109–1119. doi: 10.1053/gast.2002.36051. [DOI] [PubMed] [Google Scholar]
- 5.Carvalho B, Postma C, Mongera S, Hopmans E, Diskin S, van de Wiel MA, van Criekinge W, Thas O, Matthai A, Cuesta MA, et al. Multiple putative oncogenes at the chromosome 20q amplicon contribute to colorectal adenoma to carcinoma progression. Gut. 2009;58:79–89. doi: 10.1136/gut.2007.143065. [DOI] [PubMed] [Google Scholar]
- 6.Voorham QJ, Carvalho B, Spiertz AJ, van Grieken NC, Mongera S, Rondagh EJ, van de Wiel MA, Jordanova ES, Ylstra B, Kliment M, et al. Chromosome 5q loss in colorectal flat adenomas. Clin Cancer Res. 2012;18:4560–4569. doi: 10.1158/1078-0432.CCR-11-2385. [DOI] [PubMed] [Google Scholar]
- 7.Douglas EJ, Fiegler H, Rowan A, Halford S, Bicknell DC, Bodmer W, Tomlinson IP, Carter NP. Array comparative genomic hybridization analysis of colorectal cancer cell lines and primary carcinomas. Cancer Res. 2004;64:4817–4825. doi: 10.1158/0008-5472.CAN-04-0328. [DOI] [PubMed] [Google Scholar]
- 8.Cancer Genome Atlas N Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taylor AM, Shih J, Ha G, Gao GF, Zhang X, Berger AC, Schumacher SE, Wang C, Hu H, Liu J, et al. Genomic and Functional Approaches to Understanding Cancer Aneuploidy. Cancer Cell. 2018;33:676–689. doi: 10.1016/j.ccell.2018.03.007. e673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang M, Niu W, Hu R, Wang Y, Liu Y, Liu L, Zhong J, Zhang C, You H, Zhang J, et al. POLR1D promotes colorectal cancer progression and predicts poor prognosis of patients. Mol Carcinog. 2019;58:735–748. doi: 10.1002/mc.22966. [DOI] [PubMed] [Google Scholar]
- 11.de Groen FL, Krijgsman O, Tijssen M, Vriend LE, Ylstra B, Hooijberg E, Meijer GA, Steenbergen RD, Carvalho B. Gene-dosage dependent overexpression at the 13q amplicon identifies DIS3 as candidate oncogene in colorectal cancer progression. Genes Chromosomes Cancer. 2014;53:339–348. doi: 10.1002/gcc.22144. [DOI] [PubMed] [Google Scholar]
- 12.Camps J, Pitt JJ, Emons G, Hummon AB, Case CM, Grade M, Jones TL, Nguyen QT, Ghadimi BM, Beissbarth T, et al. Genetic amplification of the NOTCH modulator LNX2 upregulates the WNT/beta-catenin pathway in colorectal cancer. Cancer Res. 2013;73:2003–2013. doi: 10.1158/0008-5472.CAN-12-3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Groen FL, Timmer LM, Menezes RX, Diosdado B, Hooijberg E, Meijer GA, Steenbergen RD, Carvalho B. Oncogenic role of miR-15a-3p in 13q Amplicon-Driven colorectal adenoma-to-carcinoma progression. PLoS One. 2015;10 doi: 10.1371/journal.pone.0132495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diosdado B, van de Wiel MA, Terhaar Sive Droste JS, Mongera S, Postma C, Meijerink WJ, Carvalho B, Meijer GA. MiR-17-92 cluster is associated with 13q gain and c-myc expression during colorectal adenoma to adenocarcinoma progression. Br J Cancer. 2009;101:707–714. doi: 10.1038/sj.bjc.6605037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mogilyansky E, Rigoutsos I. The miR-17/92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013;20:1603–1614. doi: 10.1038/cdd.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mavrakis KJ, Wolfe AL, Oricchio E, Palomero T, de Keersmaecker K, McJunkin K, Zuber J, James T, Khan AA, Leslie CS, et al. Genome-wide RNA-mediated interference screen identifies miR-19 targets in Notch-induced T-cell acute lymphoblastic leukaemia. Nat Cell Biol. 2010;12:372–379. doi: 10.1038/ncb2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conkrite K, Sundby M, Mukai S, Thomson JM, Mu D, Hammond SM, MacPherson D. miR-17∼92 cooperates with RB pathway mutations to promote retinoblastoma. Genes Dev. 2011;25:1734–1745. doi: 10.1101/gad.17027411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S, Van Houdt WJ, Pronk A, Van Gorp J, Siersema PD, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 20.van de Wetering M, Francies HE, Francis JM, Bounova G, Iorio F, Pronk A, van Houdt W, van Gorp J, Taylor-Weiner A, Kester L, et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161:933–945. doi: 10.1016/j.cell.2015.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drost J, van Jaarsveld RH, Ponsioen B, Zimberlin C, van Boxtel R, Buijs A, Sachs N, Overmeer RM, Offerhaus GJ, Begthel H, et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature. 2015;521:43–47. doi: 10.1038/nature14415. [DOI] [PubMed] [Google Scholar]
- 22.Jung P, Sato T, Merlos-Suarez A, Barriga FM, Iglesias M, Rossell D, Auer H, Gallardo M, Blasco MA, Sancho E, et al. Isolation and in vitro expansion of human colonic stem cells. Nat Med. 2011;17:1225–1227. doi: 10.1038/nm.2470. [DOI] [PubMed] [Google Scholar]
- 23.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 25.Koo BK, Stange DE, Sato T, Karthaus W, Farin HF, Huch M, van Es JH, Clevers H. Controlled gene expression in primary Lgr5 organoid cultures. Nat Methods. 2011;9:81–83. doi: 10.1038/nmeth.1802. [DOI] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Komor MA, Bosch LJ, Bounova G, Bolijn AS, Delis-van Diemen PM, Rausch C, Hoogstrate Y, Stubbs AP, de Jong M, Jenster G, et al. Consensus molecular subtype classification of colorectal adenomas. J Pathol. 2018;246:266–276. doi: 10.1002/path.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scheinin I, Sie D, Bengtsson H, van de Wiel MA, Olshen AB, van Thuijl HF, van Essen HF, Eijk PP, Rustenburg F, Meijer GA, et al. DNA copy number analysis of fresh and formalin-fixed specimens by shallow whole-genome sequencing with identification and exclusion of problematic regions in the genome assembly. Genome Res. 2014;24:2022–2032. doi: 10.1101/gr.175141.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van de Wiel MA, Brosens R, Eilers PH, Kumps C, Meijer GA, Menten B, Sistermans E, Speleman F, Timmerman ME, Ylstra B. Smoothing waves in array CGH tumor profiles. Bioinformatics. 2009;25:1099–1104. doi: 10.1093/bioinformatics/btp132. [DOI] [PubMed] [Google Scholar]
- 32.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 33.van de Wiel MA, Kim KI, Vosse SJ, van Wieringen WN, Wilting SM, Ylstra B. CGHcall: calling aberrations for array CGH tumor profiles. Bioinformatics. 2007;23:892–894. doi: 10.1093/bioinformatics/btm030. [DOI] [PubMed] [Google Scholar]
- 34.Sie D, Snijders PJ, Meijer GA, Doeleman MW, van Moorsel MI, van Essen HF, Eijk PP, Grunberg K, van Grieken NC, Thunnissen E, et al. Performance of amplicon-based next generation DNA sequencing for diagnostic gene mutation profiling in oncopathology. Cell Oncol (Dordr) 2014;37:353–361. doi: 10.1007/s13402-014-0196-2. [DOI] [PubMed] [Google Scholar]
- 35.van Lanschot MCJ, Carvalho B, Rausch C, Snaebjornsson P, van Engeland M, Kuipers EJ, Stoker J, Tutein Nolthenius CJ, Dekker E, Meijer GA. Molecular profiling of longitudinally observed small colorectal polyps: a cohort study. EBioMedicine. 2019;39:292–300. doi: 10.1016/j.ebiom.2018.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, Schinzel AC, Sandy P, Meylan E, Scholl C, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meijer GA, Hermsen MA, Baak JP, van Diest PJ, Meuwissen SG, Belien JA, Hoovers JM, Joenje H, Snijders PJ, Walboomers JM. Progression from colorectal adenoma to carcinoma is associated with non-random chromosomal gains as detected by comparative genomic hybridisation. J Clin Pathol. 1998;51:901–909. doi: 10.1136/jcp.51.12.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chou CH, Shrestha S, Yang CD, Chang NW, Lin YL, Liao KW, Huang WC, Sun TH, Tu SJ, Lee WH, et al. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018;46:D296–D302. doi: 10.1093/nar/gkx1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Humphreys KJ, McKinnon RA, Michael MZ. miR-18a inhibits CDC42 and plays a tumour suppressor role in colorectal cancer cells. PLoS One. 2014;9 doi: 10.1371/journal.pone.0112288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu G, Tang JQ, Tian ML, Li H, Wang X, Wu T, Zhu J, Huang SJ, Wan YL. Prognostic values of the miR-17-92 cluster and its paralogs in colon cancer. J Surg Oncol. 2012;106:232–237. doi: 10.1002/jso.22138. [DOI] [PubMed] [Google Scholar]
- 43.Ng EK, Chong WW, Jin H, Lam EK, Shin VY, Yu J, Poon TC, Ng SS, Sung JJ. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut. 2009;58:1375–1381. doi: 10.1136/gut.2008.167817. [DOI] [PubMed] [Google Scholar]
- 44.Huang Z, Huang D, Ni S, Peng Z, Sheng W, Du X. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer. 2010;127:118–126. doi: 10.1002/ijc.25007. [DOI] [PubMed] [Google Scholar]
- 45.Wu CW, Ng SS, Dong YJ, Ng SC, Leung WW, Lee CW, Wong YN, Chan FK, Yu J, Sung JJ. Detection of miR-92a and miR-21 in stool samples as potential screening biomarkers for colorectal cancer and polyps. Gut. 2012;61:739–745. doi: 10.1136/gut.2011.239236. [DOI] [PubMed] [Google Scholar]
- 46.Zheng Y, Li S, Ding Y, Wang Q, Luo H, Shi Q, Hao Z, Xiao G, Tong S. The role of miR-18a in gastric cancer angiogenesis. Hepatogastroenterology. 2013;60:1809–1813. [PubMed] [Google Scholar]
- 47.Tao J, Wu D, Li P, Xu B, Lu Q, Zhang W. microRNA-18a, a member of the oncogenic miR-17-92 cluster, targets Dicer and suppresses cell proliferation in bladder cancer T24 cells. Mol Med Rep. 2012;5:167–172. doi: 10.3892/mmr.2011.591. [DOI] [PubMed] [Google Scholar]
- 48.Li X, Zhang Z, Li Y, Zhao Y, Zhai W, Yang L, Kong D, Wu C, Chen Z, Teng CB. miR-18a counteracts AKT and ERK activation to inhibit the proliferation of pancreatic progenitor cells. Sci Rep. 2017;7:45002. doi: 10.1038/srep45002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu Y, Thomson JM, Wong HY, Hammond SM, Hogan BL. Transgenic over-expression of the microRNA miR-17-92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Dev Biol. 2007;310:442–453. doi: 10.1016/j.ydbio.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, Yatabe Y, Kawahara K, Sekido Y, Takahashi T. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 51.Li Y, Choi PS, Casey SC, Dill DL, Felsher DW. MYC through miR-17-92 suppresses specific target genes to maintain survival, autonomous proliferation, and a neoplastic state. Cancer Cell. 2014;26:262–272. doi: 10.1016/j.ccr.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dews M, Homayouni A, Yu D, Murphy D, Sevignani C, Wentzel E, Furth EE, Lee WM, Enders GH, Mendell JT, et al. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet. 2006;38:1060–1065. doi: 10.1038/ng1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu Q, Yang Z, An Y, Hu H, Yin J, Zhang P, Nie Y, Wu K, Shi Y, Fan D. MiR-19a/b modulate the metastasis of gastric cancer cells by targeting the tumour suppressor MXD1. Cell Death Dis. 2014;5:e1144. doi: 10.1038/cddis.2014.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen Y, Gao Y, Tian Y, Tian DL. PRKACB is downregulated in non-small cell lung cancer and exogenous PRKACB inhibits proliferation and invasion of LTEP-A2 cells. Oncol Lett. 2013;5:1803–1808. doi: 10.3892/ol.2013.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yajnik V, Paulding C, Sordella R, McClatchey AI, Saito M, Wahrer DC, Reynolds P, Bell DW, Lake R, van den Heuvel S, et al. DOCK4, a GTPase activator, is disrupted during tumorigenesis. Cell. 2003;112:673–684. doi: 10.1016/s0092-8674(03)00155-7. [DOI] [PubMed] [Google Scholar]
- 57.Khan AA, Soloski MJ, Sharp AH, Schilling G, Sabatini DM, Li SH, Ross CA, Snyder SH. Lymphocyte apoptosis: mediation by increased type 3 inositol 1,4,5-trisphosphate receptor. Science. 1996;273:503–507. doi: 10.1126/science.273.5274.503. [DOI] [PubMed] [Google Scholar]
- 58.Gerber S, Alzayady KJ, Burglen L, Bremond-Gignac D, Marchesin V, Roche O, Rio M, Funalot B, Calmon R, Durr A, et al. Recessive and dominant De Novo ITPR1 mutations cause gillespie syndrome. Am J Hum Genet. 2016;98:971–980. doi: 10.1016/j.ajhg.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Druck T, Kastury K, Hadaczek P, Podolski J, Toloczko A, Sikorski A, Ohta M, LaForgia S, Lasota J, McCue P, et al. Loss of heterozygosity at the familial RCC t(3;8) locus in most clear cell renal carcinomas. Cancer Res. 1995;55:5348–5353. [PubMed] [Google Scholar]
- 60.LaForgia S, Morse B, Levy J, Barnea G, Cannizzaro LA, Li F, Nowell PC, Boghosian-Sell L, Glick J, Weston A, et al. Receptor protein-tyrosine phosphatase gamma is a candidate tumor suppressor gene at human chromosome region 3p21. Proc Natl Acad Sci USA. 1991;88:5036–5040. doi: 10.1073/pnas.88.11.5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang Z, Shen D, Parsons DW, Bardelli A, Sager J, Szabo S, Ptak J, Silliman N, Peters BA, van der Heijden MS, et al. Mutational analysis of the tyrosine phosphatome in colorectal cancers. Science. 2004;304:1164–1166. doi: 10.1126/science.1096096. [DOI] [PubMed] [Google Scholar]
- 62.Laczmanska I, Sasiadek MM. Tyrosine phosphatases as a superfamily of tumor suppressors in colorectal cancer. Acta Biochim Pol. 2011;58:467–470. [PubMed] [Google Scholar]
- 63.Matano M, Date S, Shimokawa M, Takano A, Fujii M, Ohta Y, Watanabe T, Kanai T, Sato T. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat Med. 2015;21:256–262. doi: 10.1038/nm.3802. [DOI] [PubMed] [Google Scholar]
- 64.Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, Henderson JM, Kutok JL, Rajewsky K. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9:405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sillars-Hardebol AH, Carvalho B, Tijssen M, Belien JA, de Wit M, Delis-van Diemen PM, Ponten F, van de Wiel MA, Fijneman RJ, Meijer GA. TPX2 and AURKA promote 20q amplicon-driven colorectal adenoma to carcinoma progression. Gut. 2012;61:1568–1575. doi: 10.1136/gutjnl-2011-301153. [DOI] [PubMed] [Google Scholar]
- 66.Komor MA, de Wit M, van den Berg J, Martens de Kemp SR, Delis-van Diemen PM, Bolijn AS, Tijssen M, Schelfhorst T, Piersma SR, Chiasserini D, et al. Molecular characterization of colorectal adenomas reveals POFUT1 as a candidate driver of tumor progression. Int J Cancer. 2020;146:1979–1992. doi: 10.1002/ijc.32627. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.