Abstract
Background and Aims
Hepatocellular carcinoma (HCC) is a multistep process whereby abnormally proliferating cancer cells undergo extensive metabolic reprogramming. Metabolic alterations in hepatocarcinogenesis depend on the activation of specific oncogenes, thus partially explaining HCC heterogeneity. c-Myc oncogene overexpression, frequently observed in human HCCs, leads to a metabolic rewiring toward a Warburg phenotype and production of lactate, resulting in the acidification of the extracellular space, favoring the emergence of an immune-permissive tumor microenvironment. Here, we investigated whether Ldha genetic ablation interferes with metabolic reprogramming and HCC development in the mouse.
Methods
We characterized the metabolic reprogramming in tumors induced in C57BL/6J mice hydrodynamically cotransfected with c-Myc and h-Ras. Using the same experimental model, we investigated the effect of Ldha inhibition—gained through the inducible and hepatocyte-specific Ldha knockout—on cancer cell metabolic reprogramming, number and size of HCC lesions, and tumor microenvironment alterations.
Results
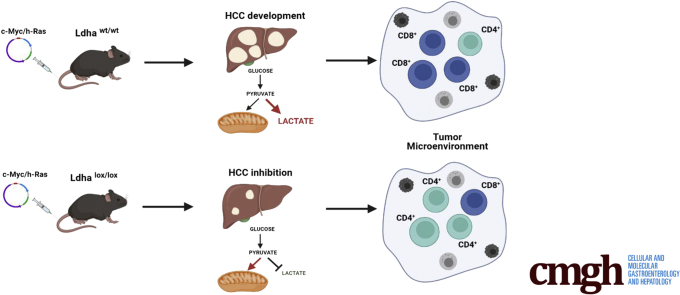
c-Myc/h-Ras–driven tumors display a striking glycolytic metabolism, suggesting a switch to a Warburg phenotype. The tumors also exhibited enhanced pentose phosphate pathway activity, the switch of glutamine to sustain glutathione synthesis instead of the tricarboxylic acid cycle, and the impairment of oxidative phosphorylation. In addition, Ldha abrogation significantly hampered tumor number and size together with an evident inhibition of the Warburg-like metabolic feature and a remarkable increase of CD4+ lymphocytes compared with Ldha wild-type livers.
Conclusions
These results demonstrate that Ldha deletion significantly impairs mouse HCC development and suggest lactate dehydrogenase as a potential target to enhance the efficacy of the current therapeutic options.
Keywords: HCC, c-Myc, Ldha, Metabolic Reprogramming, TME
Abbreviations used in this paper: Casp3, caspase 3; FACS, fluorescence-activated cell sorting; Gls, glutaminase; Gs, glutamine synthase; H&E, hematoxylin and eosin; HCC, hepatocellular carcinoma; IHC, immunohistochemistry; KO, knockout; LDH, lactate dehydrogenase; mRNA, messenger RNA; OXPHOS, oxidative phosphorylation; PPP, pentose phosphate pathway; qRT-PCR, quantitative reverse-transcription polymerase chain reaction; SDH, succinate dehydrogenase; SDL, Smart Protein Layers; TCA, tricarboxylic acid; TGCA, The Cancer Genome Atlas; TME, tumor microenvironment; WT, wild-type
Graphical abstract
Summary.
Because metabolic reprogramming characterized by a switch toward glycolysis is recognized as a hallmark of cancer, including hepatocellular carcinoma, genes playing essential roles in metabolic regulation represent important candidate targets in liver cancer therapy. The present results suggest the potential usefulness of targeting LDHA to restore normal cell metabolism and inhibit hepatocellular carcinoma development.
Hepatocellular carcinoma (HCC) is the most common type of primary liver cancer1 and the fourth leading cause of cancer-related death worldwide,2 representing a significant challenge for global health. Despite a significant increase in incidence, current therapeutic options for HCC remain unsatisfactory.3 HCC, like many other solid tumors, results from a multistep process in which malignant transformation requires the elimination of several cell-imposed barriers such as antiproliferative responses, programmed cell death, and senescence.
These cancer-associated features have been intensely investigated, leading to the identification of so-called hallmarks of cancer.4 Among the several hallmarks of cancer, metabolic reprogramming is considered a critical process, as it is necessary to enhance cell proliferation and survival. In this process, cancer cells—unlike most mammalian cells in which glucose is metabolized through glycolysis in a multistep set of reactions resulting in the production of pyruvate—preferentially rely on so-called aerobic glycolysis, in which pyruvate is redirected to generate lactate through the action of lactate dehydrogenase A (LDHA)-a process typically observed in conditions of low oxygen state.5
The consequence of lactate accumulation is the acidification of the extracellular pH in the tumor microenvironment (TME), which is believed to favor metastasis, angiogenesis, and immunosuppression by affecting several cell types within the TME, and is associated with a worse clinical prognosis.6
Moreover, the production and excretion of lactate, an otherwise energetically wasteful activity, is necessary to sustain the Warburg effect. Indeed, it ensures the continuous generation of NAD+ to serve as an electron acceptor during glucose oxidation.7
Thus, lactate should be considered a critical oncometabolite in the metabolic reprogramming of cancer and LDH as a suitable therapeutic target for cancer treatment. LDH is a tetrameric protein comprising 2 major subunits, LDHA (subunit M) and LDHB (subunit H) genes.8 Evidence to support the role of LDH in cancer progression and the need for the development of LDH inhibitors as a therapeutic option for cancer treatment stems from the finding that knockout (KO) of LDHA induced cell death in several tumor cell lines.9,10 In addition, pharmacological inhibition of mouse LDHA caused the regression of tumors in an experimental model of non-small cell lung cancer.11 In contrast, LDHB knockdown has been reported not to significantly impact tumor cell survival.9
In the present study, we sought to investigate the effect of Ldha genetic deletion on cancer cell metabolic reprogramming and its impact on mouse HCC development. For this purpose, we utilized and characterized a model in which mouse liver tumors were induced by hydrodynamic cotransfection of c-Myc and h-Ras, 2 oncogenes frequently overexpressed in human HCC.
Results
Cotransfection of c-Myc/h-Ras Leads to 100% HCC
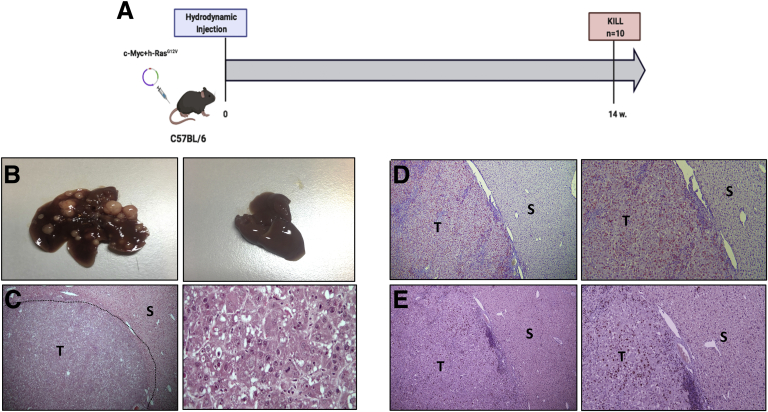
Combining the 2 oncogenes (Figure 1A) led to the formation of multifocal, widely distributed HCCs of variable size in 100% of mice at 14 weeks after in vivo liver transfection (Figure 1B). Tumoral lesions displayed an intense basophilic cytoplasm and strong nuclear atypia characterized by the presence of several nucleoli. Apoptotic bodies and mitoses were also observed (Figure 1C). All HCCs displayed robust nuclear staining of c-Myc (Figure 1D) and were characterized by a very high proliferative activity as demonstrated by uniform immunoreactivity for Ki-67 (Figure 1E).
Figure 1.
(A) Schematic representation of experimental protocol 1. (B) (Left) Macroscopic view of the liver from c-Myc/h-Ras mice 14 weeks after hydrodynamic injection. (Right) Liver from mice subjected to hydrodynamic injection without oncogene-carrying plasmids. (C) (Left) Microscopic examination of c-Myc/h-Ras tumors (H&E, 5×). (Right) Microscopic examination of c-Myc/h-Ras tumors (H&E, 40×). (D) Nuclear staining of c-Myc in HCC (T) but not in the surrounding (S) liver (left: 5×, right: 10×). (E) Serial sections showing Ki-67–positive nuclear staining in HCC (T). S indicates surrounding liver (left: 5×, right: 10×).
c-Myc/h-Ras HCCs Display Metabolic Reprogramming
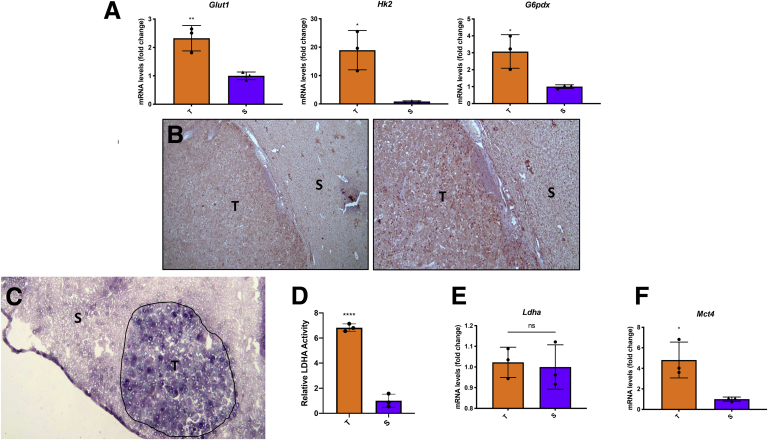
c-Myc–driven liver tumors display increased glucose uptake and enhanced glutaminolysis to fuel the tricarboxylic acid (TCA) cycle.12 Because many metabolic changes are strictly oncogene-dependent, we aimed to characterize the metabolic profile of HCCs developed by the c-Myc/h-Ras experimental model. HCCs have been shown to undergo metabolic reprogramming and exhibit a profound glycolysis dysregulation.13, 14, 15, 16 Quantitative reverse-transcription polymerase chain reaction (qRT-PCR) analysis performed on laser-microdissected tumors and peritumoral tissues revealed a strong induction of glycolysis, as shown by enhanced levels of Glut1, suggesting an increased glucose uptake by cancer cells (Figure 2A), and Hk2, a tumor-specific kinase. In addition, the isoform of the first enzyme of glycolysis, Gck, was strongly increased in the tumor compared with the surrounding tissue (Figure 2A). Moreover, G6pd, the rate-limiting enzyme of the oxidative branch of the pentose phosphate pathway (PPP), was upregulated at the messenger RNA (mRNA) level (Figure 2A) to cope with the increased demand for ribose, required for nucleic and fatty acids synthesis to sustain cell proliferation, and the need of reducing equivalent NADPH, to counteract the increased generation of reactive oxygen species and enhance tumor cell survival. Accordingly, immunohistochemistry (IHC) analysis showed that the expression of G6PD was strongly induced in tumors while being virtually negative in the peritumoral area (Figure 2B).
Figure 2.
(A) qRT-PCR analysis of Glut1, Hk2, and G6pdx in tumors compared with the surrounding areas. (B) Microphotographs showing G6pd immunostaining in HCC compared with peritumoral areas (G6pd staining: 5× [left], 10× [right]. (C) LDHA activity in tumors (T) and surrounding areas (S). (D) Relative LDHA activity. Measurement of LDHA activity was performed using the ImageJ software. (E) qRT-PCR analysis of Ldha. (F) qRT-PCR analysis of Mct4. qRT-PCR analysis was performed on dissected tumors or surrounding livers. Gene expression is reported as a fold change of tumor mRNA relative to surrounding livers. Relative quantification analysis for each gene was calculated by the 2ΔΔCt method. The histogram represents the means ± SD of 3 tumors and 3 surrounding livers (t test). ∗P < .05; ∗∗P < .01; ∗∗∗∗P < .0001.
In agreement with the increased glucose uptake, LDHA activity was significantly enhanced in tumoral tissue compared with the surrounding liver (Figure 2C and D), even without a significant change in mRNA levels (Figure 2E). In accord with the enhanced LDHA activity, Mct4 gene expression was also enhanced in HCCs vs peritumoral tissues. (Figure 2F)
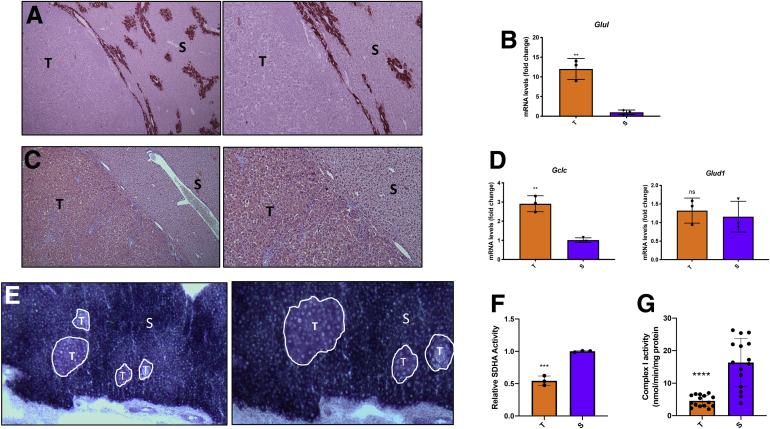
Together with increased glucose uptake, tumor cells are often characterized by changes in glutamine metabolism, and c-Myc has been shown to promote glutaminolysis to fuel the TCA cycle.12 Therefore, we wished to determine the fate of glutamine in the tumors generated by the c-Myc/h-Ras model. The results showed that while glutamine synthase (GS), the enzyme that catalyzes the conversion of glutamate into glutamine, was present in the hepatocytes surrounding the terminal hepatic venules as expected,17,18 it was completely absent within the tumors (Figure 3A).
Figure 3.
(A) Microphotographs showing the absence of GS immunostaining in HCC. Surrounding liver (S) shows intense staining of the hepatocytes encompassing the hepatic terminal venules (GS: 5× [left], 10× [right]). (B) qRT-PCR analysis of Glul mRNA levels in tumors (T) compared with the surrounding areas (S). (C) GLS-positive tumors (T) compared with the peritumoral areas (S). (left: 5×, right: 10×). (D) qRT-PCR analysis of Gclc and Glud1 mRNA levels in tumors compared with the surrounding areas. (E) Histochemistry of SDHA showing a remarkable decrease of enzyme activity in c-Myc/h-Ras tumors (outlined areas) compared with the surrounding tissue (left: 5×, right: 10×). (F) Relative SDHA activity. Measurement of SDHA activity was performed using the ImageJ software. (G) complex I activity. The activity of complex 1 was calculated as NADH consumption assessing the difference between the trace slopes with and without complex I inhibitor rotenone (10 μM). Values were then normalized for protein amount. Gene expression is reported as a fold-change of tumor mRNA relative to surrounding livers. Relative quantification analysis for each gene was calculated by the 2ΔΔCt method. The histogram represents means ± SD of 3 tumors and 3 surrounding livers (t test). ∗∗P < .01; ∗∗∗∗P < .0001. ns, not significant.
Further analysis showed that the lack of GS protein was the consequence of a severe downregulation of transcription of Glul, the gene coding for GS, in tumoral hepatocytes compared with the surrounding tissue. (Figure 3B).
On the other hand, glutaminolysis was profoundly increased in tumors compared with surrounding livers, as shown by the intense immunostaining for glutaminase (GLS), the enzyme responsible for converting glutamine into glutamate, unveiling that the combination of c-Myc/h-Ras oncogenes promotes glutamine catabolism (Figure 3C).
Interestingly, unlike c-Myc tumors, those generated by c-Myc/h-Ras showed enhanced mRNA levels of Gclc, which is involved in the conversion of glutamate into glutathione, but no change in the expression of Glud1, the enzyme responsible for the conversion of glutamate into α-ketoglutarate, compared with the surrounding liver (Figure 3D).
The results suggest that the excess of catabolized glutamine is not utilized as a fuel for the TCA cycle, but rather is used to convert glutamate into glutathione in order to protect cancer cells from oxidative damage usually occurring in rapidly growing tumors.
Although increased glycolysis in HCC has often been associated with a decreased oxidative phosphorylation (OXPHOS),16,19 our understanding of the modifications of TCA and OXPHOS occurring in HCC and Myc-driven tumors remains controversial.16,20, 21, 22 Hence, we measured the activity of succinate dehydrogenase A (SDHA), the major catalytic subunit of succinate-ubiquinone oxidoreductase, which converts succinate into fumarate. Importantly, SDHA not only is part of the TCA cycle, but also is a complex of the mitochondrial respiratory chain, specifically complex II.23,24 Histochemical analysis performed on cryostat sections showed that SDHA activity was severely impaired in c-Myc/h-Ras tumors compared with the peritumoral area (Figure 3E and F).
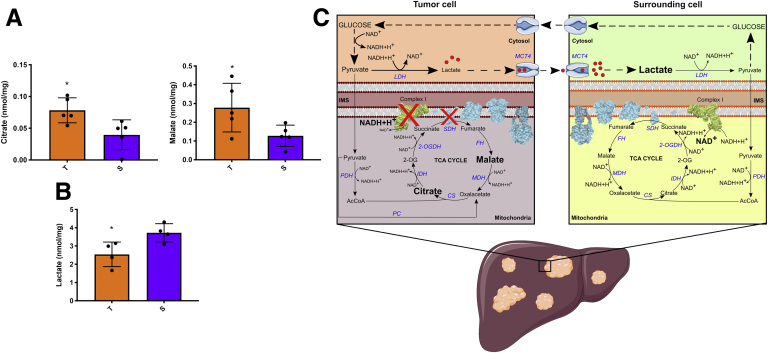
In support of the metabolic switch from OXPHOS to glycolysis in HCC, we measured complex I activity and TCA cycle intermediate levels in c-Myc/h-Ras tumors and the corresponding adjacent nontumoral livers. As indicated in Figure 3G, complex I activity was significantly lower in tumor areas than surrounding tissues. The subsequent increase in NADH/NAD+ ratio inhibited the activities of the TCA cycle dehydrogenases, leading to citrate and malate accumulation in tumor areas compared with surrounding tissues (Figure 4A). These findings, together with the inhibition of complex II (Figure 3E and F), point to a substantial impairment of OXPHOS in tumor tissues associated with a slowing of the TCA cycle flux. Interestingly, lactate levels were lower in tumors compared with surrounding tissues (Figure 4B), although LDH activity was higher to support NADH oxidation required to sustain the increased glycolytic flux (Figure 2C and D). It is conceivable that lactate is rapidly produced and secreted by cancer cells to prevent intracellular acidification.25 Therefore, extracellular lactate can be used by surrounding cells to convert it back to pyruvate and produce glucose (Figure 4C). This rapid secretion, facilitated by the upregulation of the lactate transporter (Figure 2E), prevents lactate from feedback inhibiting LDH and other glycolytic enzymes.
Figure 4.
(A,B) Metabolomic analysis of citrate, malate, and lactate in tumors vs surrounding tissue. Values are expressed as nmol/mg of protein. The histogram represents means ± SD of 5 tumors and 5 surrounding livers (t test) ∗P < .05. (C) Schematic representation of the metabolic changes displayed by tumor and surrounding tissues in an HCC mouse model. Acquisition of KRAS and c-Myc mutations induces the downregulation of mitochondrial complex I and complex II (ie, SDH) activities. That inhibition reduces mitochondrial NAD+ conversion from NADH, leading to inhibition of the TCA cycle dehydrogenases with the accumulation of citrate and malate. Pyruvate to lactate conversion through LDH sustains reoxidation of NADH, which supports the highly glycolytic flux of cancer cells. Lactate is rapidly secreted by tumor areas to prevent feedback inhibition of LDH. Surrounding healthy cells can then uptake it and convert it back to pyruvate, which could be channeled into glucose synthesis through gluconeogenesis. 2-OG, 2-oxoglutarate; 2-OGDH, 2-oxoglutarate dehydrogenase; AcCoA, acetyl-CoA; CS, citrate synthase; FH, fumarate hydratase, IDH, isocitrate dehydrogenase, IMS, intermembrane space; MCT4, monocarboxylate transporter 4; MDH, malate dehydrogenase; PC, pyruvate carboxylase; PDH, pyruvate dehydrogenase.
Taken together, these results demonstrate that the c-Myc/h-Ras oncogene model leads to a metabolic reprogramming characterized by increased glucose uptake and lactate production, enhanced PPP, and impairment of OXPHOS.
A Similar Metabolic Reprogramming and Enhanced LDHA Occurs in Human HCC
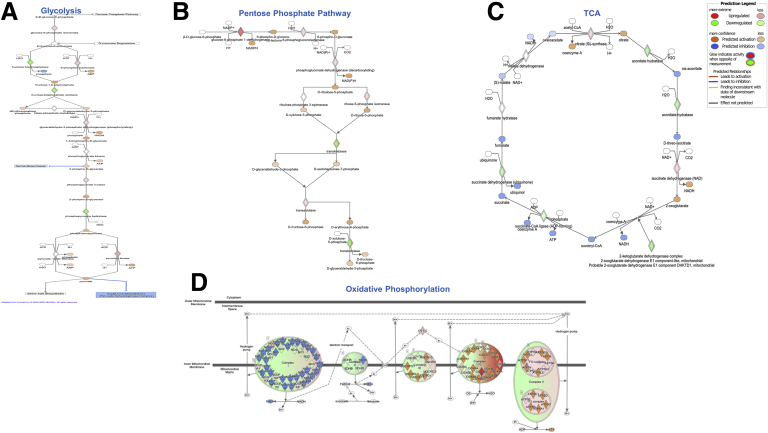
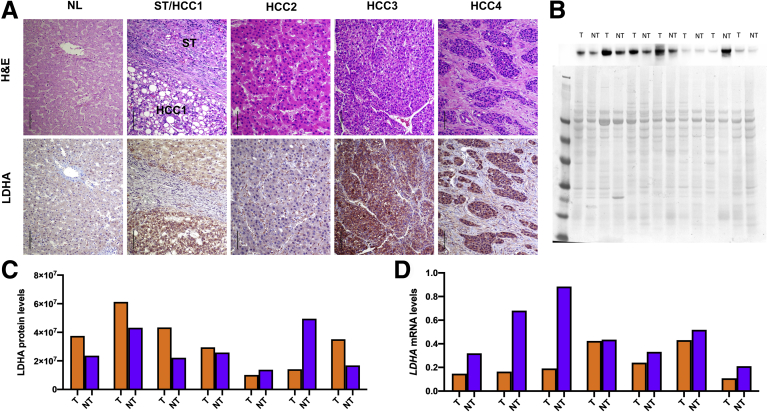
To investigate whether these findings could have a translational value, we interrogated the dataset from The Cancer Genome Atlas (TCGA) to determine the pattern of metabolic changes occurring in c-Myc overexpressing human HCCs. As shown in Figure 5A–D, human HCCs were characterized by a metabolic switch involving glycolysis, PPP, TCA, and OXPHOS strikingly similar to our mouse model. In addition, similar to what was observed in mouse HCC, when we evaluated by IHC the LDHA protein levels in a vast collection of human HCC specimens (n = 228), we found a robust cytoplasmic, membranous, or nuclear LDHA staining in most tumors (n = 148 of 228, 64.9%) (Figure 6A). Notably, upregulation of LDHA compared with nontumoral surrounding liver tissues was observed in the vast majority of poorly-differentiated tumors (n = 52 of 68, 76.5%) and, at a lower frequency, in moderately differentiated (n = 38 of 62, 61.3%) and well-differentiated tumors (n = 58 of 98, 59.2%) (Figure 6A), in accordance with a recent study.26
Figure 5.
Ingenuity Pathways Analysis of (A):glycolysis, (B) PPP, (C) TCA cycle and (D) OXPHOS in human HCCs.
Figure 6.
(A) Representative staining patterns in human HCC specimens for LDHA. In normal liver (NL), a faint or absent staining for LDHA was detected. Surrounding nontumorous liver tissues (ST) exhibited weak to moderate cytoplasmic LDHA immunoreactivity, which was generally lower than in corresponding HCC lesions. In this case (HCC1), a steatotic HCC shows a more pronounced LDHA immunolabeling than the neighboring ST. HCC2 depicts instead a well-differentiated HCC displaying weak immunoreactivity for LDHA. Tumors with moderate (HCC3) and poor (HCC4) differentiation were instead most frequently characterized by robust LDHA immunoreactivity. Strong immunolabeling for LDHA was often observed not only in the cytoplasm but also in the membrane (HCC3) and the nucleus (HCC4). Original magnification = 200×. scale bar = 100 μM. (B) Representative Western blot analysis results for LDHA protein levels in human HCC. The blot of a series of 7 tumors (T) and corresponding nontumorous surrounding livers (NT), whose quantifications are shown in panel B, is depicted here. Specifically, tumors 1–4 and 7 displayed higher LDHA levels than corresponding nontumorous livers, whereas tumors 5 and 6 exhibited lower levels than respective non-neoplastic livers. The lower panel shows equal loading in the various samples as assessed by reversible Ponceau red staining. (C) Graph showing the results of Western Blot analysis of the levels of LDHA protein in human HCCs and their respective peritumoral counterparts. (D) qRT-PCR analysis of LDHA. qRT-PCR analysis was performed on the same samples shown in B. Relative quantification analysis for each gene was calculated by the 2ΔΔCt method. M, marker
Western blot analysis confirmed the presence of high LDHA protein levels in most human HCCs analyzed (Figure 6B and C). On the other hand, when we determined in the same samples the LDHA mRNA levels, no significant differences were detected between human HCCs and their respective peritumoral counterparts, in accord with the results of the mouse study (Figure 2E). Thus, the present data suggest that posttranscriptional mechanisms are responsible for enhanced LDHA protein levels in HCC (Figure 6D).
Ldha KO Strongly Impairs HCC Development
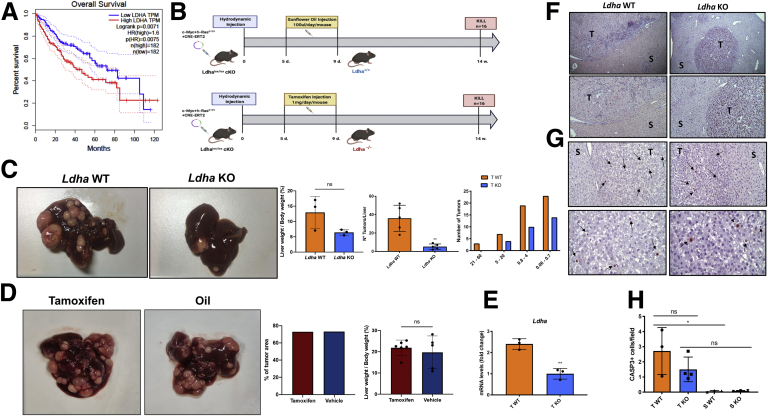
Once we characterized the mouse c-Myc/h-Ras model, we then wished to investigate whether the knocking out of Ldha could impact HCC progression. Notably, TCGA data showed improved overall survival in patients with human HCCs displaying low levels of LDH (Figure 7A). Using the hepato-specific Ldha KO mouse model represented in Figure 7B, we found a strong tumor reduction in terms of relative liver weight, number, and size (Figure 7C). This effect was not due to some unknown tamoxifen effect because no difference in number and % of tumor area was found in tamoxifen-treated c-Myc/h-Ras mice compared with animals treated with vehicle (Figure 7D). Using laser microdissection technology, we extracted mRNA from tumors and surrounding livers of wild-type (WT) and KO mice. To validate our KO procedures, we evaluated the levels of Ldha transcripts in WT and KO tumors. As shown in Figure 7E, the expression of Ldha in KO tumors was significantly lower (∼70%) than that observed in WT HCCs.
Figure 7.
(A) Kaplan-Meier survival curve for overall survival of patients with low and high expression of LDHA. mRNA data were obtained from TCGA Liver Hepatocellular Carcinoma dataset. Based on LDHA expression, patients are classified as LDHA high expression and LDHA low expression. The red curve indicates a statistically significant reduction in overall survival with higher LDHA mRNA expression (P < .0071). (B) Schematic representation of experimental protocol 2. (C) Macroscopic view of livers from Ldha WT (left) or KO mice (right), liver-weight-to-body-weight ratio of WT, percentage of the tumor area, and the average of the size of Ldha WT or KO tumors (t test). ∗∗P < .01. Number and tumor size were evaluated in H&E-stained sections as described in the Materials and Methods. The histogram represents the means ± SD of 5 tumors per group (t test). (D) Macroscopic view of livers from Ldha WT treated with tamoxifen or oil, percentage of the tumor area, and liver-weight-to-body-weight ratio. The histograms represent means ± SD of 6 samples/group (t test). ns, not significant. (E) qRT-PCR analysis of Ldha mRNA levels in laser-microdissected WT HCCs compared with Ldha KO tumors. Gene expression is reported as a fold-change of mRNA from WT tumors relative to those from Ldha KO livers. Relative quantification analysis for each gene was calculated by the 2ΔΔCt method. The histogram represents the means ± SD of 3 tumors from 3 mice per group (t test). ∗∗P < .01. (F) Representative microphotographs of immunohistochemical staining for Ki-67 in the tumors (T) or surrounding livers (S) of Ldha WT or Ldha KO mice (top: 5×; bottom: 10×; sections counterstained with hematoxylin). (G) Representative microphotographs showing immunostaining for the cleaved form of CASP3 in the tumors (T) or surrounding liver (S) of Ldha WT and KO mice (CASP3 counterstained with Hematoxylin, Top 20×, Bottom 40×). Arrows indicate single CASP3-positive cells. (H) Apoptotic index (AI). AI was calculated as the number of Casp3-positive cells per field at 40× magnification. From 20 to 40 fields per liver were scored. Results were expressed as means ± SD of 3–4 mice per group (1-way analysis of variance). ∗P < .05.
Next, we investigated whether the reduced number and size of Ldha KO HCCs could result from (1) a reduced proliferation rate or (2) an increased death of cancer cells by apoptosis. However, immunostaining for Ki-67 and the cleaved form of caspase 3 (Casp3) showed that the proliferative and the apoptotic indices in both WT and Ldha KO tumors were much higher than their corresponding peritumoral tissues (2.7 and 1.5 vs 0.2 and 0.3, respectively) and were not significantly different between the 2 groups (Figure 7F–H).
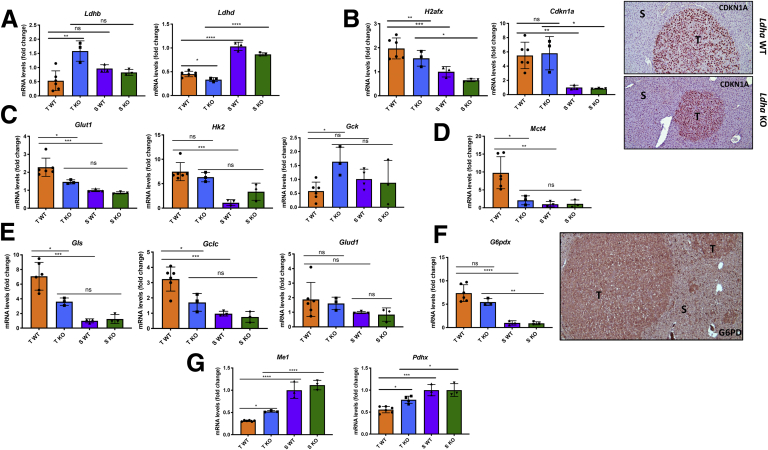
To determine whether upregulation of the other isoforms of the tetrameric LDH could compensate for Ldha loss, we assessed mRNA expression levels of Ldhb, Ldhc, and Ldhd. qRT-PCR analysis (Figure 8A) showed that Ldhb expression was increased in the Ldha KO tumors compared with WT tumors. However, no significant differences were observed in the expression of Ldhb in tumors vs surrounding peritumoral livers. As to the other 2 isoforms, the results showed that Ldhd expression was lower in both tumor groups than in peritumoral tissues, while Ldhc was not detectable.
Figure 8.
(A) qRT-PCR analysis of Ldhb and Ldhd mRNA levels in laser-microdissected tumors (T) and nontumorous surrounding livers (S) from Ldha WT and Ldha KO mice. (B) qRT-PCR analysis of Cdkn1a and H2afx mRNA levels in laser-microdissected tumors (T) and surrounding livers (S) from Ldha WT and Ldha KO mice (left); microphotographs showing CDKN1A immunostaining in tumors and surrounding liver of WT and Ldha KO mice (right). (C) qRT-PCR analysis of Glut1, Hk2, and Gck mRNA levels in microdissected tumors (T) and nontumorous surrounding livers (S) from Ldha WT and KO mice. (D) qRT-PCR analysis of Mct4 mRNA levels in microdissected tumors (T) and surrounding livers (S) from Ldha WT and KO mice. (E) qRT-PCR analysis of Gls, Glud1, and Gclc mRNA levels in microdissected tumors (T) and surrounding livers (S) from Ldha WT and KO mice. (F) qRT-PCR analysis of the expression of G6pdx and microphotograph showing G6PD immunostaining in Ldha KO tumors (T) compared with peritumoral areas (S) (10×). (G) qRT-PCR analysis of Me1 and Pdhx mRNA levels in laser-microdissected tumors (T) and surrounding livers (S) from Ldha WT and KO mice. Gene expression is reported as a fold change of mRNA relative to those from Ldha WT surrounding livers. Relative quantification analysis for each gene was calculated by the 2ΔΔCt method. The histogram represents means ± SD of 6 WT tumors, 3 KO tumors, and 3 WT and KO surrounding livers (1-way analysis of variance). ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. ns, not significant.
To further probe into the mechanisms responsible for the impairment of tumor development observed in Ldha KO mice, we assessed the expression of Cdkn1a and H2afx, 2 genes associated with senescence and DNA damage, respectively. However, the results showed that mRNA and protein levels of CDKN1A were significantly upregulated in both WT and KO tumors compared with the surrounding tissue, whereas no difference was detected between WT and KO tumors. The expression of H2afx was overall upregulated in tumors compared with the nontumoral area, but this induction was slightly lower in KO vs WT tumors (Figure 8B).
Ldha KO Impairs Metabolic Reprogramming
Because neither inhibition of proliferation nor increased apoptosis or senescence seem to be involved in the arrest of the hepatocarcinogenic process, we sought to investigate whether LDH loss could affect the metabolic reprogramming operating in c-Myc/h-Ras tumors.
To this aim, we examined by qRT-PCR and IHC the expression of several components implicated in glycolysis, PPP, and OXPHOS. As shown in Figure 8C, the expression of Glut1 was significantly downregulated in KO tumors compared with WT ones, and was not different from that observed in the surrounding livers, highlighting a lesser glucose uptake by cancer cells in the absence of Ldha. Notably, Ldha KO HCC also displayed decreased expression, albeit not significant, of the protumoral isoform Hk2 and an enhanced expression of Gck compared with WT HCC.
According to the reduced glucose uptake by Ldha KO tumors, the expression of the lactate transporter Mct4 was profoundly downregulated when compared with WT HCCs and did not show significant changes in mRNA levels compared with the surrounding livers (Figure 8D).
Ldha loss also affected glutamine metabolism. Indeed, significant inhibition of Gls expression compared with WT tumors, indicating a lesser glutamine utilization, was observed in Ldha KO tumors (Figure 8E). Notably, Gclc expression was significantly enhanced in WT tumors compared with the surrounding. Still, it was downregulated in KO tumors, indicating that this pathway is deeply affected by Ldha KO (Figure 8E). On the other hand, Glud1 expression was not affected by Ldha deletion (Figure 8E).
Next, we examined the activation of the PPP pathway by measuring the expression of the rate-limiting enzyme G6PD. Accordingly to IHC, the expression of G6PD was significantly enhanced in KO HCCs compared with surrounding livers (Figure 8F). The levels of G6pd mRNA were also elevated compared with peritumoral livers, although slightly lower than those observed in WT animals (Figure 8F). These results suggest that loss of Ldha only minimally affects PPP activation.
Interestingly, malic enzyme, a cytosolic protein that catalyzes the interconversion of malate into pyruvate while concomitantly generating NADPH from NADP, and pyruvate dehydrogenase, responsible for converting pyruvate into acetyl-CoA, was downregulated in c-Myc/h-Ras–driven tumors compared with the peritumoral areas. On the opposite, KO of Ldha led to a slight increase in the expression of Me-1 and Pdhx expression in the tumors compared with WT HCCs, suggesting an impairment of the TCA cycle (Figure 8G).
Taken together, these results suggest that hepatocyte-specific Ldha KO in a c-Myc/h-Ras tumor model significantly impairs the development and progression of hepatic tumors by inhibiting the metabolic switch toward Warburg metabolism.
Ldha KO Affects the TME
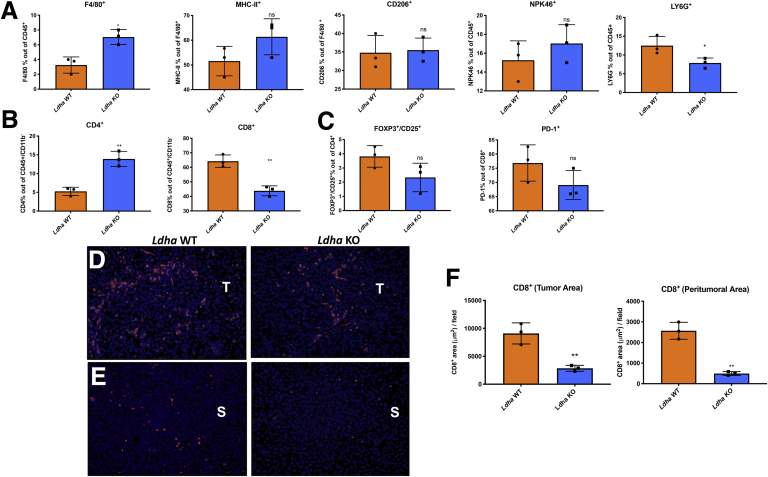
Extrusion of lactate through its transporter Mct4 can influence the extracellular environment. Therefore, the next step was to investigate if the reduction of lactate, due to Ldha KO, could affect cell components of the TME. Fluorescence-activated cell sorting (FACS) analysis unveiled a decrease of neutrophils as shown by the percentage of Ly6g-positive cells in Ldha KO tumors (Figure 9A), which was associated with an increased F4/80+ macrophage population. However, no change of polarization from M1 to M2 was observed, as the expression of major histocompatibility complex class II, a typical M1 marker, and CD206, whose upregulation is a classical M2 feature, was not modified compared with WT HCCs. In the same line, no differences were detected in the number of natural killer cells, as suggested by the expression of the NKp46 marker (Figure 9A).
Figure 9.
(A) FACS analysis showing the percentage of F4/80+macrophage population out of CD45+alive cells, major histocompatibility complex class II–positive and CD206+population out of F4/80+cells, Npk46+ population out of CD45+cells, and Ly6g+population out of CD45+in Ldha WT and KO tumors. ∗P < .05. (B) FACS analysis illustrating the % of T helper CD4+ and cytotoxic CD8+ cells performed on CD45+/CD11b– lymphocytes. (C) The % of Foxp3+/CD25+ T regulatory population out of CD4+ cells, and PD-1+ cells out of CD8+ population in Ldha WT and KO tumors. Results are expressed as means ± SD of 3 livers per group (t test). ∗P < .05; ∗∗P < .01. (D, E) Immunofluorescence staining showing CD8+ cells in Ldha WT and KO tumors and in peritumoral area: blue is DAPI, red is CD8+. (F) Quantification of immunofluorescence results. ns, not significant.
We also analyzed the lymphocyte cell population. As shown in Figure 9B, lymphocytes were the most affected immune population by Ldha KO in cancer cells. In particular, deprivation of lactate in the extracellular TME was associated with reducing CD8+ T cells with a concomitant increase of CD4+ T lymphocytes. Nevertheless, the percentage of Foxp3+/CD25+ T regulatory cells was not modified (Figure 9C). Loss of CD8+ lymphocytes in Ldha KD mice was further confirmed by immunofluorescence analysis, in both tumoral and peritumoral areas (Figure 9D and E), although the percentage of PD-1–positive cells suggests that the activation/exhaustion rate among the 2 groups remained unaltered (Figure 9C).
Discussion
The most relevant findings are that (1) genetic deletion of Ldha strongly impairs tumor development induced by the combined transfection of 2 oncogenes—c-Myc/h-Ras—often overexpressed in human HCC; (2) the inhibitory effect is associated with a significant reversion of the oncogene-triggered metabolic reprogramming occurring in WT hepatocytes; and (3) the effect of Ldha deletion on hepatocyte metabolism is accompanied by changes in TME as shown by altered immune cell number, in particular a decrease of CD8+ and an increase of CD4+ lymphocytes.
Although the possibility that LDHA could represent a suitable therapeutic target in cancer has been proposed long ago, the use of pharmacological inhibitors did not provide an unequivocal answer as to whether LDHA plays a critical role in cancer progression. Indeed, on the one hand, while many chemicals generated against Ldha were quite effective in in vitro assays, they were ineffective in vivo due to poor pharmacodynamic properties.27,28 On the other hand, LDHA inhibitors that were effective in vivo often showed off-target effects, thus implicating that their antitumorigenic activity may not be due entirely to LDHA inhibition.11,29 Therefore, whether LDHA represents a suitable target for tumorigenesis, in particular for HCC, remains elusive.
In the present work, we made use of a mouse model in which HCCs were induced by combined transfection of hepatocytes with 2 oncogenes frequently overexpressed in human liver tumors, namely c-Myc and h-Ras. In this animal model, genetic deletion of Ldha caused a robust impairment of both the number and size of HCCs, in the absence of overt toxicity or mortality. Interestingly, the inhibitory effect, although highly significant, was not complete. Indeed, tumors were still present in Ldha KO livers, although their number and average size were strongly reduced. While the reason for the incomplete inhibition of tumors in Ldha KO mice is unclear, it is possible to hypothesize that the enhanced expression of Ldhb, found in Ldha KO, compensates for the Ldha loss. Irrespectively of the reason(s), the results clearly demonstrated the relevance of Ldha in sustaining the growth of HCCs, at least in mice.
The second significant finding of this work is the reversion of metabolic reprogramming taking place in Ldha KO tumors. As already reported, oncogene activation represents a constitutive drive for cancer cells to proliferate, a condition generating a dramatic metabolic change. This process generally consists of rewiring from an oxidative to glycolytic metabolism. Our data are in full accordance with this statement, as the increased NADH/NAD+ ratio due to complex I inhibition reduces the activity of TCA cycle dehydrogenases such as isocitrate and malate dehydrogenases, thus suppressing the mitochondrial oxidative metabolism. The higher glycolytic flux is then supported by pyruvate being converted to lactate via LDH, which responds to the metabolic need for oxidation of NADH to NAD+ typical of highly glycolytic cells undergoing uncontrolled proliferation. On this basis, several compounds targeting glucose metabolism have been developed and used to inhibit glucose import, glucose phosphorylation by HK2, conversion of pyruvate into lactate, and export of lactate out of the cell.11,30, 31, 32
Unfortunately, the heterogeneity of tumor metabolism and the cellular complexity of the TME represent severe limitations to the therapeutic efficacy of the metabolism inhibitors. Thus, inhibitors of the lactate transporter MCT1 or activators of PKM2 display variable responses in different models.33,34 Similarly, the effectiveness of Gls inhibition is also variable, depending not only on the nature of the tumor but also on the presence of different oncogenes, as clearly demonstrated by Yuneva et al.12 Indeed, they showed that MET oncogene–driven tumors were less glycolytic than those driven by Myc.12
The notion that Myc overexpression is highly frequent in human HCCs35 confers further translational value to our data, showing that loss of Ldha has a profound impact on the metabolism of tumor cells associated with potent inhibition of tumor growth. Interestingly, the only exception to the metabolic switch induced by Ldha loss was the PPP. Indeed, the levels of G6PD mRNA and protein did not show significant differences between WT and KO tumors. In this context, it is essential to note that NAD+ generated during the LDHA-induced conversion of pyruvate into lactate is required for the glycolytic step catalyzed by GAPDH.7 Therefore, we can speculate that Ldha loss has a profound impact mainly on the second phase of the glycolytic pathway, leaving G6pd activity unaffected. In turn, the high levels of G6PD in Ldha KO tumors—compared with surrounding tissue—may justify the enhanced proliferative activity of these tumors.
An additional finding of the present work was the significant change in the number of immune cells in KO livers. In particular, the selective loss of CD8+ with a concomitant increase of CD4+ lymphocytes observed in the extracellular TME of Ldha KO livers agrees with the inhibitory effect of lactate deprivation on tumor invasion and growth. Notably, these findings are in accordance with Ma et al,36 who demonstrated that the selective loss of CD4+ T lymphocytes promotes cancer development in a nutritional mouse model of hepatocarcinogenesis, and also with Serganova et al,37 who showed that Ldha ablation regulates TME by modulating Hif-1α and promoting the enhancement of CD4+ infiltrate population. These results further support the notion that lactate should not be thought of only as a cellular metabolic waste of tumor cells, but also as a key molecule involved in carcinogenesis as well as in tumor immune evasion.
In conclusion, we believe that our results on the effect of Ldha loss on HCC development may be potentially useful in designing therapeutic strategies based on targeting lactate metabolism in combination with immunotherapy. Nevertheless, additional studies are mandatory to elucidate the exact molecular mechanisms responsible for the impaired tumor formation caused by Ldha KO and its possible impact on TME.
Materials and Methods
Animals
Ldhalox/lox cKO male mice in a C57BL/6 background were kindly provided by Prof. David Scadden (Harvard Stem Cell Institute, Cambridge, MA) to Prof. Mazzone (VIB-KU Leuven, Leuven, Belgium), while C57BL/6 WT mice were purchased from Charles River Laboratories (Brussels, Belgium). Animals were fed a standard rodent diet and maintained at 25°C temperature and 12-hour light-dark daily cycle, with food and water ad libitum. Housing and all experimental animal procedures were approved by the Institutional Animal Care and Research Advisory Committee of KU Leuven.
Experimental Protocols
For the experimental protocol 1, 6- to 8-week-old C57BL/6 male mice were hydrodynamically cotransfected with 0.1 μg of PB_h-RasG12V and PB_c-Myc, in conjunction with a plasmid encoding a hyperactive PB transposase.38,39
As to the experimental protocol 2, Ldhalox/lox cKO male mice in a C57BL/6 background were hydrodynamically cotransfected with 0.1 μg of PB_h-RasG12V, PB_c-Myc, and PB_CRE_ERT2, in conjunction with a plasmid encoding a hyperactive PB transposase. Five days after hydrodynamic injection, mice were subjected to (1) intraperitoneal tamoxifen (Sigma-Aldrich, St Louis, MO; T5648) injection (1 mg/d/mouse) or (2) intraperitoneal corn oil (Santa Cruz Biotechnology, Santa Cruz, CA; sc-215936) injection (100 μL/mouse/d), resulting in Ldha+/+ and Ldha–/– mice, respectively. In all the experimental protocols, the liver-specific promoter minimal transthyretin was used to drive gene expression in all PB constructs.38 Tumor-bearing mice were sacrificed 14 weeks after hydrodynamic injection.
Evaluation of Tumor Burden
To evaluate tumor burden, histological sections from mouse liver samples were stained with H&E. For each section, neoplastic areas were identified at high microscope magnification, acquired with a slide scanner, and the total neoplastic area was calculated with the software ImageJ 1.50i (National Institutes of Health, Bethesda, MD). The average neoplastic area/group was calculated as the mean of the sum of each lesion. Data were expressed in mm2 ± SD.
Histology and Immunohistochemistry
Mouse Study
Four-micron-thick paraffin-embedded sections were collected and stained for H&E and were incubated overnight with the following antibodies: anti-c-Myc (Abcam, Cambridge, United Kingdom; ab32072), anti-KI-67 (Abcam, ab15580), anti-G6PD (Abcam, ab87230), anti-GLUL (Sigma-Aldrich G2781), anti-GLS (Abcam, ab 262716), and anti-CASP3 (Cell Signaling Technology, Danvers, MA; 9664L), and anti-CDKN1A (Abcam, ab107099). The secondary antibodies anti-rabbit Dako EnVision+ System Labelled Polymer-HRP (Dako, Carpinteria, CA) or goat anti-rabbit Biotinylated IgG Antibody (H+L) (Vector Laboratories, Newark, CA; BA-1000) were used. Peroxidase binding sites were detected by Vector NovaRED Peroxidase (HRP) Substrate Kit (Vector Laboratories).
Human Study
Frozen and formalin-fixed, paraffin-embedded human HCC and corresponding surrounding nontumorous liver tissues were collected at the University of Regensburg (Regensburg, Germany). The local Ethical Committee of the Medical University of Regensburg (approval code: 17-1015-101) provided the Institutional Review Board approval in compliance with the Helsinki Declaration. All individuals signed the written informed consent. Four-micron-thick paraffin-embedded sections were collected and stained for H&E, and were incubated overnight with the LDHA antibody (Cell Signaling Technology; 3582S). The HRP-Polymer anti-rabbit (Zytomed Systems, Berlin, Germany; ZUC032-100) was used as the secondary antibody.
Immunofluorescence Staining
For serial sections cut at 7-μm thickness, tissue samples were fixed in 2% paraformaldehyde overnight at 4°C, dehydrated, and embedded in paraffin. Paraffin slides were first rehydrated to proceed with antigen retrieval in citrate solution (Dako). Subsequently, the sections were blocked with the appropriate serum (Dako) and incubated overnight with the rabbit anti-CD8 antibody (D4W2Z; Cell Signaling Technology) at 1:200 dilution. Next, the secondary goat anti-rabbit antibody (Jackson Immunoresearch, West Grove, PA) was used at 1:300 dilution. Whenever sections were stained in fluorescence, ProLong Gold mounting medium with DAPI (Invitrogen, San Diego, CA) was used. Microscopic analysis was done with an Olympus BX41 microscope (Olympus, Tokyo, Japan) and CellSense imaging software V2.3 (Olympus Lifescience, Tokyo, Japan).
LDH Histochemical Assay
Eight-micron-thick frozen sections were briefly fixed on slides containing 4% formalin for 5 minutes and then incubated at 37°C in a medium containing 50 mM Tris (Merck, Darmstadt, Germany) pH 7.4, 750 μM NADP (Merck), 80 μM phenazine methosulfate (Merck), 600 μM nitro blue tetrazolium chloride (Merck), and 30 mM lactate (Merck). Successively, liver sections were rinsed in physiological saline and mounted with an aqueous mounting medium.40
Complex II/SDH Histochemical Assay
Eight-micron-thick frozen sections were incubated at 37°C in an incubation medium containing 0.2 M phosphate buffer, sodium succinate solution, nitro blue tetrazolium solution (Sigma-Aldrich), and distilled water. Successively, liver sections were rinsed in physiological saline, fixed in 10% formalin-saline solution, rinsed in 15% alcohol, and finally mounted with an aqueous mounting medium. As succinate was oxidized to fumarate, the reduced form of NADH was produced, and the reaction was visualized concomitantly with the electron acceptor reacting with the purple salt nitro blue tetrazolium.
Laser-Capture Microdissection
Sixteen-μm-thick serial frozen sections of mouse HCCs and non-neoplastic surrounding livers were attached to 2-μm RNase free PEN-membrane slides (Leica, Wetzlar, Germany). Microdissection (LMD6000; Leica) was preceded by H&E staining on serial sections.
Analysis of mRNA Expression Levels
The PicoPure RNA Isolation Kit (Applied Biosystems, Waltham, MA; KIT0204) was used for the extraction of total RNA from microdissected HCCs and corresponding surrounding livers. Total RNA extraction was then performed using the RNeasy FFPE Kit (Qiagen, Hilden, Germany; 73504) to extract the RNA from 10-μm-thick sections of formalin-fixed paraffin-embedded samples. RNA was quantified by a Nanodrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA), and its integrity was evaluated by an Agilent Bioanalyzer 2100 (Agilent, Santa Clara, CA). As to microdissected cryo samples, only RNAs with a RIN equal to or higher than 7 were used in the study. However, all formalin-fixed paraffin-embedded RNA samples displayed a RIN lower than 7. Nevertheless, because qRT-PCR analyses showed an almost complete similarity in the levels of gene expression changes compared with those measured in the corresponding cryo samples, formalin-fixed paraffin-embedded samples were also used. All procedures were performed according to the manufacturer’s protocol.
Quantitative Real-Time PCR
To investigate by qRT-PCR mRNA expression levels of Glut1 (Mm00441480 _m1), Hk2 (Mm00443385 _m1), Gck (Mm00439129_m1), G6pdx (Mm00656735 _g1), Mct4 (Mm00446102_m1), Glul (Mm00725701_s1), Glud1(Mm00492353_m1), Gls (Mm01257297 _m1), Gclc (Mm00802655_m1), Nqo1 (Mm01253561_m1), Ldha (Mm001612132_g1), Ldhb (Mm00493146_m1), Ldhc (Mm00496648_m1), Ldhd (Mm00459144_m1), Pdhx (Mm00558275_m1), Me1 (Mm01253561_m1), and LDHA (Hs01378790_g1), total RNA was retro-transcribed to complementary DNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Milan, Italy; Life Technologies, Milan, Italy). Once retrotranscribed, the reverse transcription product was used to assess mRNA gene expression analysis performed by qRT-PCR with the TaqMan 2× Universal PCR Master Mix, No AmpErase UNG (Thermo Fisher Scientific). The probe mix was from Thermo Fisher Scientific. The endogenous control β-actin (4352341E) was used to normalize gene expression levels.
Measurement of Enzymatic Activity of Mitochondrial Complex I
Liver samples obtained from tumor and surrounding tissues were homogenized in a buffer composed of 25 mM potassium phosphate pH 7.4, protease and phosphatase inhibitors. Complex I activity was assessed through spectrophotometric recordings following the oxidation of NADH (ε = 6.22 mM–1 cm–1) at 340 nm. Samples (5 μg of proteins per trace) were preincubated for 10 minutes at 37ºC in the presence of 2 μM alamethicin, 3 mg/mL fatty acid–free bovine serum albumin, 500 μM sodium azide, and 6.75 μM coenzyme Q10. The reaction was initiated after the addition of 30 μM NADH. Complex I activity was calculated as NADH consumption assessing the difference between the trace slopes with and without complex I inhibitor rotenone (10 μM). Values were then normalized for protein amount.
Protein Extraction, Western Blot Analysis, and Smart Protein Layers Approach
Human HCC tumors and corresponding nontumoral tissues were homogenized in 100–250 μL T-PER lysis buffer (78510; Thermo Fisher Scientific) containing 1× Halt Protease and Phosphatase Inhibitor Cocktail 1:100 (78443; Thermo Fisher Scientific) in the Bullet Blender Storm 24 (Next Advance, Troy, NY) for 3 minutes at speed 8, placed in the rotator mixer for 30–60 minutes at +4°C, and sonicated. Protein concentrations were determined using the colorimetric Bio-Rad Protein Assay Dye Reagent Concentrate (500-0006; Bio-Rad, Hercules, CA) and bovine serum albumin standards.
For accurate target protein quantification of human HCC samples, we performed Western Blot analysis using the Smart Protein Layers (SPL) Kit Red from NH DyeAgnostics (PR913; Halle, Germany) according to the manufacturer’s instruction. The SPL technology is based on 2 components: (1) red-fluorescent Smart Label (SMA label) reagent for labeling and visualization of total protein on gels and blots and (2) bifluorescent sample-specific standard Smartalyzer L (SMA basic L) for normalization, standardization, and quantification of total protein per sample. Briefly, before gel electrophoresis, every protein sample was labelled with 2 μL SMA basic L and 1 μL Smart label working solution. A total of 10 μg protein per sample and 5 μL Biotinylated Protein Ladder (SeeBlue Plus 2; Thermo Fisher Scientific) were applied to the gels. After conventional gel electrophoresis, the gel fluorescences of SMA basic L (signal: band at 80 kDa; channel: Alexa Fluor 488; gel loading control = GLO) and SMA label (signal: whole lane; channel: Cy5; gel total protein = GTO) were imaged. Following blotting, the blot fluorescence of the SMA label (signal: whole lane; channel: Cy5; blot total protein = BTO) and the blot chemiluminescent signal of the LDHA (Cell Signaling Technology; 3582S) target protein (signal: band; chemiluminescence; band target protein = BTA) were detected. Finally, normalization of gel load (based on SMA basic; GLO), normalization of Smart Label (based on SMA label), and normalization of target protein signal (based on total protein) were performed using the SPL normalization template following the manufacturer’s protocol. Results were expressed as SPL normalized target volume. Visualization and quantification were performed with the Chemidoc MP Imaging System and Image Lab software (version 6.0.1; both from Bio-Rad), respectively.
Mass Spectrometry Analysis
Metabolite quantification was performed using liquid chromatography tandem mass spectrometry. Tumor and surrounding tissue specimens were weighted and sonicated in a solution containing 80% methanol to extract polar metabolites. Samples were centrifuged at 13,000 rpm for 10 minutes and the supernatants were transferred to a clean tube and dried using Speedvac (Thermo Fisher Scientific). Dried samples were reconstituted and frozen at –80ºC. Liquid chromatography tandem mass spectrometry analysis was carried out with a Quattro Premier mass spectrometer interfaced with an Acquity UPLC system (Waters, Milford, MA). The multiple reaction monitoring transitions selected in the negative ion mode were m/z 191.16 > 110.97 for citrate, m/z 132.98 > 115.04 for malate, and m/z 88.99 > 43.3 for lactate. Separation was achieved with a CSH Phenyl-Hexyl column (2.1 × 100 mm, 1.7-μm particle size; Waters), with a flow rate set at 0.3 mL/min. Calibration curves were established using standards, processed in the same conditions as samples, at 5 increasing concentrations. The best fit was determined using regression analysis based on the peak area of the analyses.
Case Selection from the TCGA Dataset
Subset of cases with copy number alteration in the MYC gene such as deep deletion, shallow deletion, diploid, gain, and amplifications were identified using the Liver HCC (TCGA, PanCancer Atlas) dataset in the cBioPortal. Htseq-Counts for the top 20 samples having high MYC amplification and 20 random samples of “Solid Tissue Normal” were downloaded using the Xena browser.
Ingenuity Pathway Analysis
For identifying genes associated with the canonical pathways, disease and functions, and gene networks, Ingenuity Pathway Analysis (Redwood City CA) was used. Core analysis was performed on the dataset with log2foldchange cutoff –1.6 and 1.6, and species selected to mammal only.
FACS Staining and Flow Cytometric Analysis
HCC-bearing mice were sacrificed by cervical dislocation and tumors were harvested. Livers were minced in RPMI medium supplemented with 40 U/mL of Dnase I (Sigma-Aldrich; D4527), 0.1% collagenase type I (Thermo Fisher Scientific; 17100-017), and 0.2% dispase type I (Thermo Fisher Scientific; 17105041), and incubated in the same solution for 30 minutes at 37°C. The tumor single-cell suspension was stained for the pan-leucocyte marker CD45 (BioLegend, San Diego, CA; 103116), the natural killer marker NKp46 (BioLegend 137612), the pan-macrophage marker F4/80 (BioLegend; 123128), the M2 macrophage marker CD206 (Bio-Rad; MCA2235A647T), the M1 macrophage marker major histocompatibility complex class II (Invitrogen; 11-5321-82), the pan-myeloid marker CD11b (eBioscience, San Diego, CA; 48-0112-82), the cytotoxic lymphocytes marker CD8a (eBioscience; 53-0081-82), the T helper lymphocytes marker CD4 (BioLegend; 100540), the activation T cell marker CD279 (PD-1) (BioLegend; 135206), and the combination of the T regulatory lymphocytes markers CD25 (eBioscience; 25-0251-82)/Foxp3 (eBioscience; 17-5773-82). The Fixable Viability Dye eFluor 506 (Invitrogen; 65-0866-14) was used to detect viable cells. After beads compensation with UltraComp eBeads Compensation Beads (Invitrogen; 01-2222-42) to set voltages and gating parameters for obtaining an accurate fluorescence signal, data were collected with both FACS Canto (BD Biosciences, Franklin Lakes, NJ) and FACS Verse (BD Biosciences).
Statistical Analysis
Data are expressed as mean ± SEM. Analysis of significance was done by Student’s t test and by 1-way analysis of variance (Tukey’s post hoc test), using the GraphPad Software 7.0 (GraphPad Software, San Diego, California).
CRediT Authorship Contributions
Marina Serra, PhD (Conceptualization: Equal; Investigation: Lead; Methodology: Lead; Supervision: Equal; Writing – original draft: Equal; Writing – review & editing: Equal)
Mario Di Matteo, PhD (Conceptualization: Equal; Investigation: Equal; Methodology: Equal; Writing – original draft: Supporting)
Jens Serneels (Methodology: Supporting)
Rajesh Pal, PhD (Formal analysis: Supporting)
Sarah Trusso Cafarello (Methodology: Supporting)
Martina Lanza (Methodology: Supporting)
Carlos Sanchez Martin, PhD (Methodology: Equal)
Matthias Evert, MD (Formal analysis: Equal; Investigation: Equal)
Alessandra Castegna, PhD (Investigation: Equal; Methodology: Lead; Validation: Equal)
Diego Franceso Calvisi, MD, PhD (Formal analysis: Equal; Methodology: Equal)
Massimiliano Mazzone, PhD (Conceptualization: Lead; Funding acquisition: Lead; Investigation: Lead; Supervision: Equal; Writing – original draft: Equal; Writing – review & editing: Equal)
Amedeo Columbano, PhD (Conceptualization: Lead; Funding acquisition: Lead; Investigation: Lead; Supervision: Lead; Writing – original draft: Lead; Writing – review & editing: Equal)
Footnotes
Conflicts of Interest The authors disclose no conflicts.
Funding This work was supported by Associazione Italiana Ricerca sul Cancro (Grant IG-20176 to Amedeo Columbano), Fondo Coesione e Sviluppo Regione Autonoma della Sardegna 2017 (to Amedeo Columbano), and an ERC Consolidator grant (ImmunoFit) (to Massimiliano Mazzone).
References
- 1.Forner A., Llovet J.M., Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245–1255. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- 2.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Llovet J.M., Ricci S., Mazzaferro V., Hilgard P., Gane E., Blanc J.-F., de Oliveira A.C., Santoro A., Raoul J.-L., Forner A., Schwartz M., Porta C., Zeuzem S., Bolondi L., Greten T.F., Galle P.R., Seitz J.-F., Borbath I., Häussinger D., Giannaris T., Shan M., Moscovici M., Voliotis D., Bruix J. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 4.Hanahan D., Weinberg R.A. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 5.Vander Heiden M.G., Cantley L.C., Thompson C.B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Consoli A., Nurjhan N., Reilly J.J.J., Bier D.M., Gerich J.E. Contribution of liver and skeletal muscle to alanine and lactate metabolism in humans. Am J Physiol. 1990;259:E677–E684. doi: 10.1152/ajpendo.1990.259.5.E677. [DOI] [PubMed] [Google Scholar]
- 7.Goetzman E.S., Prochownik E.V. The role for Myc in coordinating glycolysis, oxidative phosphorylation, glutaminolysis, and fatty acid metabolism in normal and neoplastic tissues. Front Endocrinol (Lausanne) 2018;9:129. doi: 10.3389/fendo.2018.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rai G., Urban D.J., Mott B.T., Hu X., Yang S.-M., Benavides G.A., Johnson M.S., Squadrito G.L., Brimacombe K.R., Lee T.D., Cheff D.M., Zhu H., Henderson M.J., Pohida K., Sulikowski G.A., Dranow D.M., Kabir M., Shah P., Padilha E., Tao D., Fang Y., Christov P.P., Kim K., Jana S., Muttil P., Anderson T., Kunda N.K., Hathaway H.J., Kusewitt D.F., Oshima N., Cherukuri M., Davies D.R., Norenberg J.P., Sklar L.A., Moore W.J., Dang C.V., Stott G.M., Neckers L., Flint A.J., Darley-Usmar V.M., Simeonov A., Waterson A.G., Jadhav A., Hall M.D., Maloney D.J. Pyrazole-based lactate dehydrogenase inhibitors with optimized cell activity and pharmacokinetic properties. J Med Chem. 2020;63:10984–11011. doi: 10.1021/acs.jmedchem.0c00916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allison S.J., Knight J.R.P., Granchi C., Rani R., Minutolo F., Milner J., Phillips R.M. Identification of LDH-A as a therapeutic target for cancer cell killing via (i) p53/NAD(H)-dependent and (ii) p53-independent pathways. Oncogenesis. 2014;3:e102. doi: 10.1038/oncsis.2014.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheng S.L., Liu J.J., Dai Y.H., Sun X.G., Xiong X.P., Huang G. Knockdown of lactate dehydrogenase A suppresses tumor growth and metastasis of human hepatocellular carcinoma. FEBS J. 2012;279:3898–3910. doi: 10.1111/j.1742-4658.2012.08748.x. [DOI] [PubMed] [Google Scholar]
- 11.Le A., Cooper C.R., Gouw A.M., Dinavahi R., Maitra A., Deck L.M., Royer R.E., Vander Jagt D.L., Semenza G.L., Dang C.V. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proc Natl Acad Sci U S A. 2010;107:2037–2042. doi: 10.1073/pnas.0914433107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuneva M.O., Fan T.W.M., Allen T.D., Higashi R.M., Ferraris D.V., Tsukamoto T., Matés J.M., Alonso F.J., Wang C., Seo Y., Chen X., Bishop J.M. The metabolic profile of tumors depends on both the responsible genetic lesion and tissue type. Cell Metab. 2012;15:157–170. doi: 10.1016/j.cmet.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amann T., Maegdefrau U., Hartmann A., Agaimy A., Marienhagen J., Weiss T.S., Stoeltzing O., Warnecke C., Schölmerich J., Oefner P.J., Kreutz M., Bosserhoff A.K., Hellerbrand C. GLUT1 expression is increased in hepatocellular carcinoma and promotes tumorigenesis. Am J Pathol. 2009;174:1544–1552. doi: 10.2353/ajpath.2009.080596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen H.-L., OuYang H.-Y., Le Y., Jiang P., Tang H., Yu Z.-S., He M.-K., Tang Y.-Q., Shi M. Aberrant MCT4 and GLUT1 expression is correlated with early recurrence and poor prognosis of hepatocellular carcinoma after hepatectomy. Cancer Med. 2018;7:5339–5350. doi: 10.1002/cam4.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nwosu Z.C., Megger D.A., Hammad S., Sitek B., Roessler S., Ebert M.P., Meyer C., Dooley S. Identification of the consistently altered metabolic targets in human hepatocellular carcinoma. Cell Mol Gastroenterol Hepatol. 2017;4:303–323.e1. doi: 10.1016/j.jcmgh.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kowalik M.A., Guzzo G., Morandi A., Perra A., Menegon S., Masgras I., Trevisan E., Angioni M.M., Fornari F., Quagliata L., Ledda-Columbano G.M., Gramantieri L., Terracciano L., Giordano S., Chiarugi P., Rasola A., Columbano A. Metabolic reprogramming identifies the most aggressive lesions at early phases of hepatic carcinogenesis. Oncotarget. 2016;7:32375–32393. doi: 10.18632/oncotarget.8632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaasbeek Janzen J.W., Lamers W.H., Moorman A.F., de Graaf A., Los J.A., Charles R. Immunohistochemical localization of carbamoyl-phosphate synthetase (ammonia) in adult rat liver; evidence for a heterogeneous distribution. J Histochem Cytochem. 1984;32:557–564. doi: 10.1177/32.6.6373912. [DOI] [PubMed] [Google Scholar]
- 18.Gebhardt R., Mecke D. Heterogeneous distribution of glutamine synthetase among rat liver parenchymal cells in situ and in primary culture. EMBO J. 1983;2:567–570. doi: 10.1002/j.1460-2075.1983.tb01464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee Y.-K., Lim J.J., Jeoun U.-W., Min S., Lee E.-B., Kwon S.M., Lee C., Yoon G. Lactate-mediated mitoribosomal defects impair mitochondrial oxidative phosphorylation and promote hepatoma cell invasiveness. J Biol Chem. 2017;292:20208–20217. doi: 10.1074/jbc.M117.809012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graves J.A., Wang Y., Sims-Lucas S., Cherok E., Rothermund K., Branca M.F., Elster J., Beer-Stolz D., Van Houten B., Vockley J., Prochownik E.V. Mitochondrial structure, function and dynamics are temporally controlled by c-Myc. PLoS One. 2012;7 doi: 10.1371/journal.pone.0037699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li F., Wang Y., Zeller K.I., Potter J.J., Wonsey D.R., O’Donnell K.A., Kim J.-W., Yustein J.T., Lee L.A., Dang C.V. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol Cell Biol. 2005;25:6225–6234. doi: 10.1128/MCB.25.14.6225-6234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang H., Lu J., Dolezal J., Kulkarni S., Zhang W., Chen A., Gorka J., Mandel J.A., Prochownik E.V. Inhibition of hepatocellular carcinoma by metabolic normalization. PLoS One. 2019;14 doi: 10.1371/journal.pone.0218186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iverson T.M. Catalytic mechanisms of complex II enzymes: a structural perspective. Biochim Biophys Acta2013. 1827:648–657. doi: 10.1016/j.bbabio.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cecchini G. Function and structure of complex II of the respiratory chain. Annu Rev Biochem. 2003;72:77–109. doi: 10.1146/annurev.biochem.72.121801.161700. [DOI] [PubMed] [Google Scholar]
- 25.Brown T.P., Ganapathy V. Lactate/GPR81 signaling and proton motive force in cancer: Role in angiogenesis, immune escape, nutrition, and Warburg phenomenon. Pharmacol Ther. 2020;206 doi: 10.1016/j.pharmthera.2019.107451. [DOI] [PubMed] [Google Scholar]
- 26.Guo Y., Li X., Sun X., Wang J., Yang X., Zhou X., Liu X., Liu W., Yuan J., Yao L., Li X., Shen L. Combined aberrant expression of NDRG2 and LDHA predicts hepatocellular carcinoma prognosis and mediates the anti-tumor effect of gemcitabine. Int J Biol Sci. 2019;15:1771–1786. doi: 10.7150/ijbs.35094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wolpaw A.J., Dang C.V. Exploiting metabolic vulnerabilities of cancer with precision and accuracy. Trends Cell Biol. 2018;28:201–212. doi: 10.1016/j.tcb.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cui W., Lv W., Qu Y., Ma R., Wang Y.-W., Xu Y.-J., Wu D., Chen X. Discovery of 2-((3-cyanopyridin-2-yl)thio)acetamides as human lactate dehydrogenase A inhibitors to reduce the growth of MG-63 osteosarcoma cells: virtual screening and biological validation. Bioorg Med Chem Lett. 2016;26:3984–3987. doi: 10.1016/j.bmcl.2016.06.083. [DOI] [PubMed] [Google Scholar]
- 29.Chen C.-Y., Feng Y., Chen J.-Y., Deng H. Identification of a potent inhibitor targeting human lactate dehydrogenase A and its metabolic modulation for cancer cell line. Bioorg Med Chem Lett. 2016;26:72–75. doi: 10.1016/j.bmcl.2015.11.025. [DOI] [PubMed] [Google Scholar]
- 30.Doherty J.R., Yang C., Scott K.E.N., Cameron M.D., Fallahi M., Li W., Hall M.A., Amelio A.L., Mishra J.K., Li F., Tortosa M., Genau H.M., Rounbehler R.J., Lu Y., Dang C.V., Kumar K.G., Butler A.A., Bannister T.D., Hooper A.T., Unsal-Kacmaz K., Roush W.R., Cleveland J.L. Blocking lactate export by inhibiting the Myc target MCT1 disables glycolysis and glutathione synthesis. Cancer Res. 2014;74:908–920. doi: 10.1158/0008-5472.CAN-13-2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan D.A., Sutphin P.D., Nguyen P., Turcotte S., Lai E.W., Banh A., Reynolds G.E., Chi J.-T., Wu J., Solow-Cordero D.E., Bonnet M., JU Flanagan, Bouley D.M., Graves E.E., Denny W.A., Hay M.P., Giaccia A.J. Targeting GLUT1 and the Warburg effect in renal cell carcinoma by chemical synthetic lethality. Sci Transl Med. 2011;3 doi: 10.1126/scitranslmed.3002394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schulze A., Harris A.L. How cancer metabolism is tuned for proliferation and vulnerable to disruption. Nature. 2012;491:364–373. doi: 10.1038/nature11706. [DOI] [PubMed] [Google Scholar]
- 33.Polański R., Hodgkinson C.L., Fusi A., Nonaka D., Priest L., Kelly P., Trapani F., Bishop P.W., White A., Critchlow S.E., Smith P.D., Blackhall F., Dive C., Morrow C.J. Activity of the monocarboxylate transporter 1 inhibitor AZD3965 in small cell lung cancer. Clin cancer Res an Off J Am Assoc Cancer Res. 2014;20:926–937. doi: 10.1158/1078-0432.CCR-13-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anastasiou D., Yu Y., Israelsen W.J., Jiang J.-K., Boxer M.B., Hong B.S., Tempel W., Dimov S., Shen M., Jha A., Yang H., Mattaini K.R., Metallo C.M., Fiske B.P., Courtney K.D., Malstrom S., Khan T.M., Kung C., Skoumbourdis A.P., Veith H., Southall N., Walsh M.J., Brimacombe K.R., Leister W., Lunt S.Y., Johnson Z.R., Yen K.E., Kunii K., Davidson S.M., Christofk H.R., Austin C.P., Inglese J., Harris M.H., Asara J.M., Stephanopoulos G., Salituro F.G., Jin S., Dang L., Auld D.S., Park H.-W., Cantley L.C., Thomas C.J., Vander Heiden M.G. Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis. Nat Chem Biol. 2012;8:839–847. doi: 10.1038/nchembio.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vita M., Henriksson M. The Myc oncoprotein as a therapeutic target for human cancer. Semin Cancer Biol. 2006;16:318–330. doi: 10.1016/j.semcancer.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 36.Ma C., Kesarwala A.H., Eggert T., Medina-Echeverz J., Kleiner D.E., Jin P., Stroncek D.F., Terabe M., Kapoor V., ElGindi M., Han M., Thornton A.M., Zhang H., Egger M., Luo J., Felsher D.W., McVicar D.W., Weber A., Heikenwalder M., Greten T.F. NAFLD causes selective CD4(+) T lymphocyte loss and promotes hepatocarcinogenesis. Nature. 2016;531:253–257. doi: 10.1038/nature16969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Serganova I., Cohen I.J., Vemuri K., Shindo M., Maeda M., Mane M., Moroz E., Khanin R., Satagopan J., Koutcher J.A., Blasberg R. LDH-A regulates the tumor microenvironment via HIF-signaling and modulates the immune response. PLoS One. 2018;13 doi: 10.1371/journal.pone.0203965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tipanee J., Di Matteo M., Tulalamba W., Samara-Kuko E., Keirsse J., Van Ginderachter J.A., Chuah M.K., VandenDriessche T. Validation of miR-20a as a tumor suppressor gene in liver carcinoma using hepatocyte-specific hyperactive piggyBac transposons. Mol Ther Nucleic Acids. 2020;19:1309–1329. doi: 10.1016/j.omtn.2020.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Di Matteo M., Samara-Kuko E., Ward N.J., Waddington S.N., McVey J.H., Chuah M.K.L., VandenDriessche T. Hyperactive piggyBac transposons for sustained and robust liver-targeted gene therapy. Mol Ther. 2014;22:1614–1624. doi: 10.1038/mt.2014.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jelinek D., Flores A., Uebelhoer M., Pasque V., Plath K., Iruela-Arispe M.L., Christofk H.R., Lowry W.E., Coller H.A. Mapping metabolism: monitoring lactate dehydrogenase activity directly in tissue. J Vis Exp. 2018;136 doi: 10.3791/57760. [DOI] [PMC free article] [PubMed] [Google Scholar]