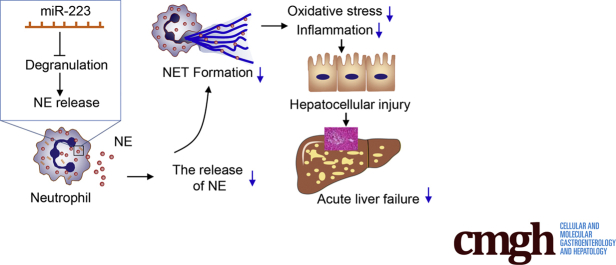
Abstract
Background & Aims
Marked enhancement of neutrophil infiltration in the liver is a hallmark of acute liver failure (ALF), a severe life-threatening disease with varying etiologies. However, the mechanisms and pathophysiological role corresponding to hepatic neutrophil infiltration during ALF development remain poorly characterized.
Methods
Experimental ALF was induced in 10-week-old male microRNA-223 (miR-223) knockout (KO) mice, neutrophil elastase (NE) KO mice, and wild-type controls by intraperitoneal injection of galactosamine hydrochloride and lipopolysaccharide. Age-matched mice were injected with phosphate-buffered saline and served as vehicle controls.
Results
Mouse liver with ALF showed evident formation of neutrophil extracellular traps (NETs), which were enhanced markedly in miR-223 KO mice. The blockade of NETs by pharmacologic inhibitor GSK484 significantly attenuated neutrophil infiltration and massive necrosis in mouse liver with ALF. ALF-related hepatocellular damage and mortality in miR-223 KO mice were aggravated significantly and accompanied by potentiated neutrophil infiltration in the liver when compared with wild-type controls. Transcriptomic analyses showed that miR-223 deficiency in bone marrow predominantly caused the enrichment of pathways involved in neutrophil degranulation. Likewise, ALF-induced hepatic NE enrichment was potentiated in miR-223 KO mice. Genetic ablation of NE blunted the formation of NETs in parallel with significant attenuation of ALF in mice. Pharmaceutically, pretreatment with the NE inhibitor sivelestat protected mice against ALF.
Conclusions
The present study showed the miR-223/NE axis as a key modulator of NETs, thereby exacerbating oxidative stress and neutrophilic inflammation to potentiate hepatocellular damage and liver necrosis in ALF development, and offering potential targets against ALF.
Keywords: Hepatocellular Injury, NET Inhibitors, Neutrophil Degranulation
Abbreviations used in this paper: ALF, acute liver failure; ALT, alanine aminotransferase; AST, aspartate aminotransferase; DAPI, 4′,6-diamidino-2-phenylindole dihydrochloride; D-GalN, D-(+)-galactosamine; GSEA, gene set enrichment analysis; H3Cit, citrullinated histone H3; KO, knockout; LPS, lipopolysaccharide; miR-223, microRNA-223; MPO, myeloperoxidase; NE, neutrophil elastase; NET, neutrophil extracellular trap; NETosis, neutrophil extracellular trap formation; PAD4, protein arginine deiminase 4; PBS, phosphate-buffered saline; PCR, polymerase chain reaction; RNA-seq, RNA sequencing; TLR4, Toll-like receptor 4; TNF-α, tumor necrosis factor α; WT, wild type
Graphical abstract
Summary.
This work highlights the pathogenic role of neutrophil extracellular traps in the development of acute liver failure. The data support neutrophil extracellular traps and their modulators microRNA-223 and neutrophil elastase as potential therapeutic targets of acute liver failure.
Acute liver failure (ALF) represents a life-threatening outcome resulting from a variety of causes, among which viral infection (hepatitis A, B, and E) and drug-induced liver injury are predominant.1 Clinically, ALF is closely associated with coagulopathy, encephalopathy, and multiple organ failure, with death occurring in up to half of all cases. Although survival of patients with ALF has improved substantially in recent years with advances in critical care management and availability of emergency liver transplantation, the evidence to guide supportive care is rather limited, largely owing to the severity and heterogeneity of the condition.1 Most importantly, the key mechanisms underlying the initiation and progression of ALF have not been fully elucidated, ensuring that prevention and therapy remain a challenge.
Enhanced neutrophil infiltration coupled with activation of neutrophilic inflammation in the liver has been established as a hallmark of early stage acute liver injury and ALF in both human beings and rodent models.2,3 After extravasation into the liver parenchyma, neutrophils crosstalk with hepatocytes to potentiate the production and release of cytotoxic reactive oxygen species to induce cellular damage in local and adjacent hepatocytes and subsequent massive liver necrosis.4 Apart from neutrophils, the liver is enriched selectively with key components of the innate immune system, especially hepatic macrophages (including circulating macrophages and resident Kupffer cells) and natural killer cells. Infiltrated neutrophils interact with other innate immune cells through various mediators such as proinflammatory cytokines, chemokines, and a vast array of neutrophil granule proteins (including neutrophil elastase [NE]) to aggravate sterile inflammation and liver injury.5 Antibody-mediated neutrophil depletion has been shown to protect against hepatocellular damage and acute liver inflammation resulting from acetaminophen overdose in mice,6 highlighting the detrimental role of neutrophils in the initiation of acute liver injury and ALF. Hence, it is of paramount importance to identify the mechanisms and the key modulators contributing to ALF-induced robust infiltration of neutrophils in the liver, and potential target(s) for novel therapeutic approaches to ALF.
In response to strong activation signals, neutrophils release decondensed chromatin filaments and granular proteins into the extracellular space, forming sticky web-like structures called neutrophil extracellular traps (NETs).4 NETs represent the consequence of a unique form of cell death of neutrophils that is characterized morphologically by the loss of intracellular membranes before the integrity of the plasma membrane is compromised. Functionally, NETs have been well documented as one way by which neutrophils catch and kill microorganisms.7 In addition to their role as a bactericidal and antifungal armamentarium, there is an increasing body of evidence that they are a pivotal player in modulating the innate immune response under various conditions related to sterile inflammation, including autoimmunity-mediated diabetes, cancer metastasis, and inappropriate thrombosis.8,9 In the liver, excessive formation of NETs exacerbates tissue damage and the transition from nonalcoholic steatohepatitis to hepatocellular carcinoma in rodent models.10 Likewise, abundant NETs represent a typical feature of hepatic metastatic lesions, helping to attract cancer cells to form distant metastases in the liver.11 Specifically, NETs are detectable in liver tissues from ALF patients. Increased plasma levels of NET marker were associated with poor outcome in patients with ALF.12 Nonetheless, the potential role of NETs in the onset and/or progression of ALF and the regulatory mechanisms underlying the formation of NETs in ALF remain elusive.
Of the microRNAs that are endogenous, noncoding, and single-stranded of approximately 20–30 nucleotides in length, microRNA-223 (miR-223) is typical, with a predominantly enriched abundance in neutrophils.13,14 miR-223 has been shown as a key player in the maintenance of innate immune homeostasis and regulation of myeloid differentiation and granulocyte function.15 Under various disease conditions, miR-223 exerts modulatory effects on the apoptosis of macrophages in patients with tuberculosis infection16 and the activation of macrophages in obesity-associated adipose tissue inflammation.17 In the liver, miR-223 deficiency protects against hepatocyte apoptosis and liver injury by targeting the insulin-like growth factor 1 receptor.18 Specifically, miR-223 functions as a master regulator of neutrophilic inflammation, thereby modulating acetaminophen-induced acute liver injury,19 alcoholic hepatitis,20 and nonalcoholic steatohepatitis.21 Nonetheless, its role in the regulation of the inflammatory response in neutrophils during ALF remains largely unknown.
In the present study, our results showed that neutrophil-derived miR-223 and NE played a distinct pathophysiological role in the development of experimental ALF. Mechanistically, miR-223 regulated NE, thereby modulating the formation of NETs to promote neutrophilic inflammation and hepatocellular injury and exacerbate ALF in mice.
Results
MiR-223 Deficiency Potentiates NET Formation in Mouse Liver With D-(+)-Galactosamine/Lipopolysaccharide-Induced ALF
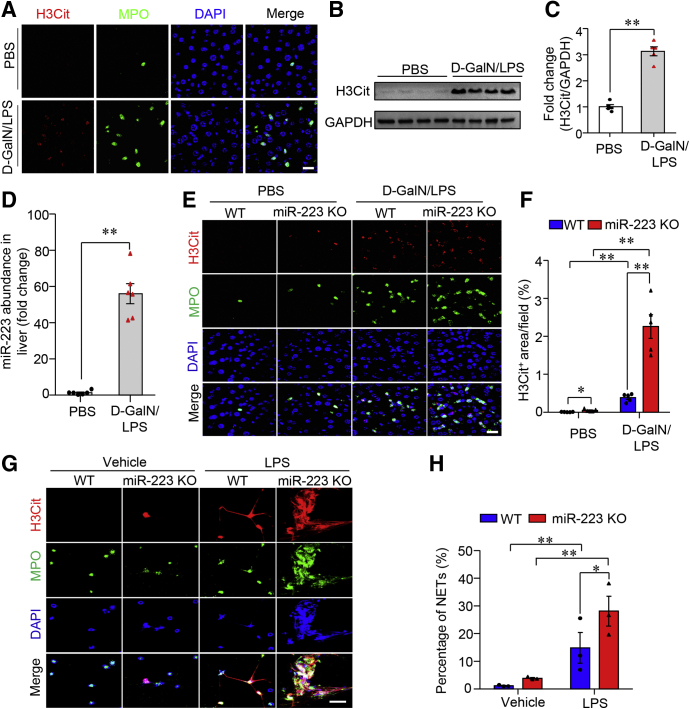
In response to various stimuli (such as viruses, bacteria, cytokine-mediated sterile inflammation), neutrophils that have infiltrated tissue form NETs that are large, extracellular, web-like structures composed of cytosolic and granule proteins assembled on a scaffold of decondensed chromatin.7 In the regard that marked enhancement in hepatic infiltration of neutrophils is a hallmark of acute liver injury,2,3 we first tested whether infiltrated neutrophils had undergone NET formation (also known as NETosis) in mouse liver with D-(+)-galactosamine (D-GalN)/lipopolysaccharide (LPS)–induced ALF. NETs were detected in paraffin-embedded sections by identifying the colocalization of myeloperoxidase (MPO), citrullinated histone 3 (H3Cit), and 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI). We detected evident formation of NETs in mouse liver with D-GalN/LPS–induced ALF, whereas neutrophil morphology remained intact in the phosphate-buffered saline (PBS) control group (Figure 1A). Western blot analysis showed that the contents of H3Cit, a specific marker of NET formation,22 were increased substantially in the liver of mice with D-GalN/LPS-induced ALF (Figure 1B and C). Given that miR-223 is one of the most abundant microRNAs in neutrophils,13,14 our results showed that the hepatic abundance of miR-223 was dramatically enriched approximately 40-fold in WT mice with D-GalN/LPS–induced ALF when compared with that in the control group (Figure 1D). Intriguingly, ALF-induced NETosis in the liver was augmented significantly in mice lacking miR-223 (Figure 1E and F). Consistently, the data from primary mouse neutrophils stimulated with LPS under ex vivo conditions showed markedly enhanced NETosis in cells lacking miR-223 (Figure 1G and H). Previous study showed the presence of NETs in liver tissues from mouse model of LPS-induced sepsis.23 We next detected NETs in mouse liver 4 hours after LPS treatment alone, which showed mild hepatocellular injury. Notably, NETs in mouse liver with a single LPS treatment were dramatically less in comparison with that in mice with D-GalN/LPS combination strategy-induced ALF (Figure 2), supporting that ALF-induced NET formation was a response to combined stimulation by GalN and LPS, not LPS alone. Collectively, these data indicate that the presence of miR-223 significantly suppressed NETosis in liver tissues under condition of D-GalN/LPS-induced ALF in mice.
Figure 1.
ALF-induced NETosis in the liver is promoted by miR-223 deficiency. (A) The formation of NETs was characterized and identified by immunofluorescence of H3Cit (red), MPO, and DAPI (blue) in liver sections from 10-week-old, male C57 BL/6J mice treated with either an intraperitoneal injection of D-GalN (0.75 mg g-1 body weight) and LPS (2.5 μg g-1 body weight) or an equal volume of PBS as vehicle. Scale bar: 5 μm. (B) Immunoblotting analysis of H3Cit in liver lysate from ALF mice or PBS-treated controls and (C) densitometric analysis of H3Cit protein abundance relative to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as loading control. (D) The abundance of miR-223 in the liver of C57 BL/6J mice after treatment with either D-GalN/LPS or PBS was determined by quantitative real-time PCR. (E) NETs were detected by immunofluorescence of H3Cit, MPO, and DAPI in liver section from 10-week-old, male miR-223 KO mice and WT control mice treated with intraperitoneal injection of D-GalN (0.75 mg g-1 body weight) and LPS (2.5 μg g-1 body weight) or an equal volume of PBS as vehicle (Scale bar: 5 μm) and (F) semiquantified by the measurement of H3Cit-positive area per field. (G) Immunofluorescence of H3Cit (red), MPO, and DAPI (blue) in primary neutrophils isolated from miR-223 KO mice and WT controls with ex vivo stimulation by LPS (200 μg mL-1) to identify the formation of NETs (Scale bar: 10 μm) and (H) semiquantification under 40× microscopic fields. Ex vivo data were analyzed according to 3 independent experiments. Data in animal studies are expressed as means ± SEM. n = 5–6. ∗P < .05, ∗∗P < .01.
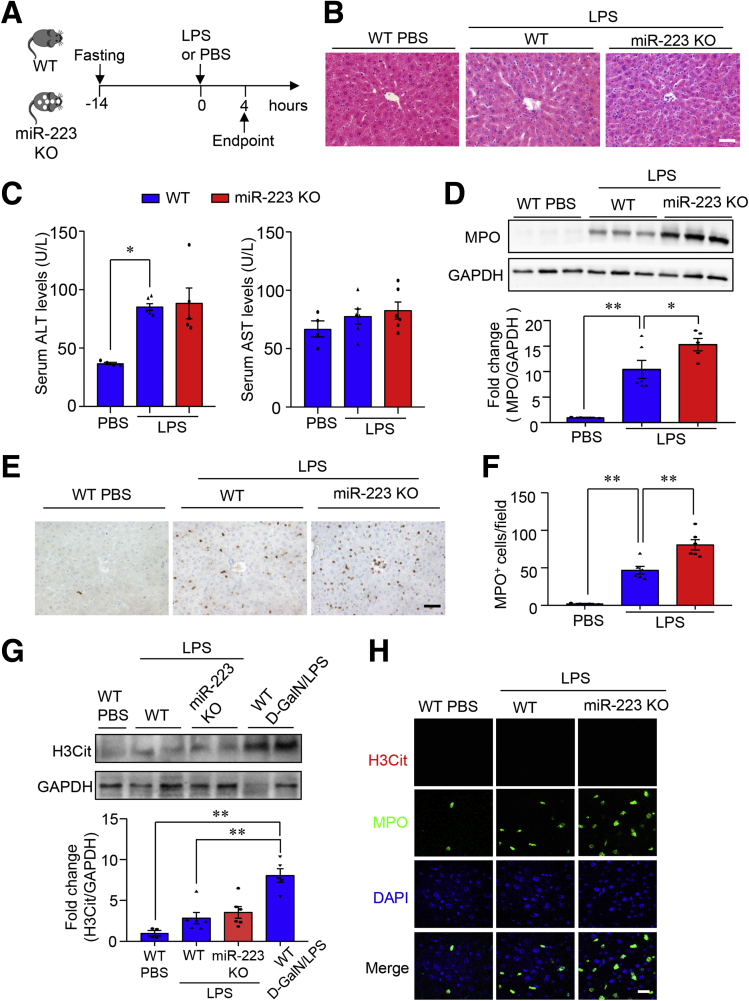
Figure 2.
LPS treatment induces slight hepatocellular injury and mild NET formation in liver in miR-223 KO mice and WT controls. (A) Ten-week-old male miR-223 KO mice with C57 BL/6J genetic background and WT control mice were injected intraperitoneally with LPS (2.5 μg g-1 body weight) dissolved in PBS as vehicle to induce ALF. (B) Representative images of H&E-stained liver sections. Scale bar: 10 μm. (C) Hepatocellular damage as determined by measured serum activity of ALT (left) and AST (right). (D) Hepatic influx of neutrophils was determined by immunoblotting analysis of MPO in liver lysate. (E) Immunohistochemistry of MPO in liver tissues (scale bar: 10 μm) and (F) semiquantification of MPO-positive cells. (G) Immunoblotting analysis of H3Cit in liver lysate from miR-223 KO mice and WT controls with treatment of either LPS or PBS (upper), and densitometric analysis of H3Cit protein abundance relative to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as loading control (lower). Liver samples from WT mice with D-GalN/LPS–induced ALF serve as positive controls. (H) NETs were detected by immunofluorescence of H3Cit, MPO, and DAPI in liver sections from miR-223 KO mice and WT controls with treatment of either LPS or PBS. Scale bar: 5 μm. Data are expressed as means ± SEM. n = 5–6. ∗P < .05, ∗∗P < .01.
MiR-223 Deficiency Sensitize Mice to D-GalN/LPS–Induced ALF
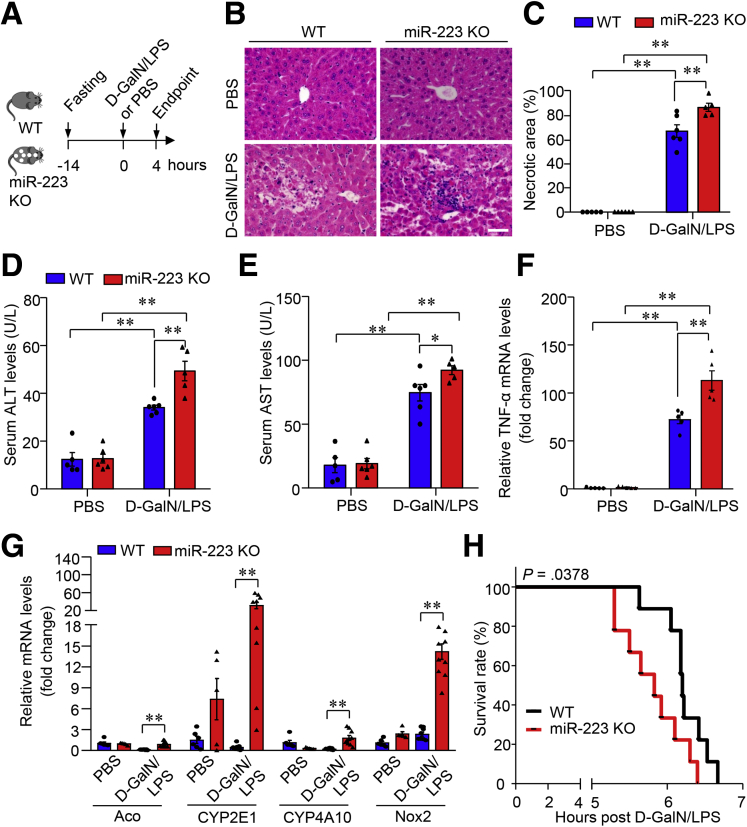
We next investigated whether miR-223 contributes to ALF pathogenesis in mice by phenotypical comparison of miR-223 knockout (miR-223 KO) and wild-type (WT) controls (Figure 3A). MiR-223 KO mice showed significantly aggravated hepatocellular injury compared with WT controls, evidenced by the markedly increased massive necrotic area in H&E-stained liver sections (Figure 3B and C) and increased serum levels of alanine aminotransferase (ALT) (Figure 3D) and aspartate aminotransferase (AST) (Figure 3E), the 2 most commonly used biomarkers for liver injury and hepatocyte damage. The ALF-induced marked increase in transcriptional abundance of proinflammatory tumor necrosis factor α (TNF-α) and pro-oxidative genes (Aco, CYP2E1, CYP4A10, and Nox2) was boosted substantially in liver tissues from miR-223 KO mice relative to that in WT controls (Figure 3F and G). Likewise, survival of miR-223 KO mice was significantly lower than that of WT mice in response to treatment with a lethal dose of D-GalN/LPS (Figure 3H). Taken together, miR-223 deficiency significantly enhanced susceptibility to D-GalN/LPS-induced ALF.
Figure 3.
miR-223 deficiency promotes liver injury and mortality rates in mice with D-GalN/LPS–induced ALF. (A) Ten-week-old male miR-223 KO mice with C57 BL/6J genetic background and WT control mice were injected intraperitoneally with D-GalN (0.75 mg g-1 body weight) and LPS (2.5 μg g-1 body weight) dissolved in PBS as vehicle to induce ALF. (B and C). Representative images of H&E-stained liver sections and quantification data of necrotic area based on H&E staining images of the liver. Scale bar: 10 μm. Hepatocellular damage as determined by measured serum activity of (D) ALT and (E) AST. mRNA expression level of (F) TNF-α and (G) pro-oxidative genes in liver tissue determined by quantitative PCR. Data are expressed as means ± SEM. n = 5–6. ∗P < .05, ∗∗P < .01. (H) Survival rate in miR-223 KO mice and WT controls after intraperitoneal injection of D-GalN (0.75 mg g-1 body weight) and LPS (0.01 μg g-1 body weight). n = 9 per group.
GSK484-Mediated Blockade of NETosis Protects Against ALF With Blunted Response in miR-223 KO Mice
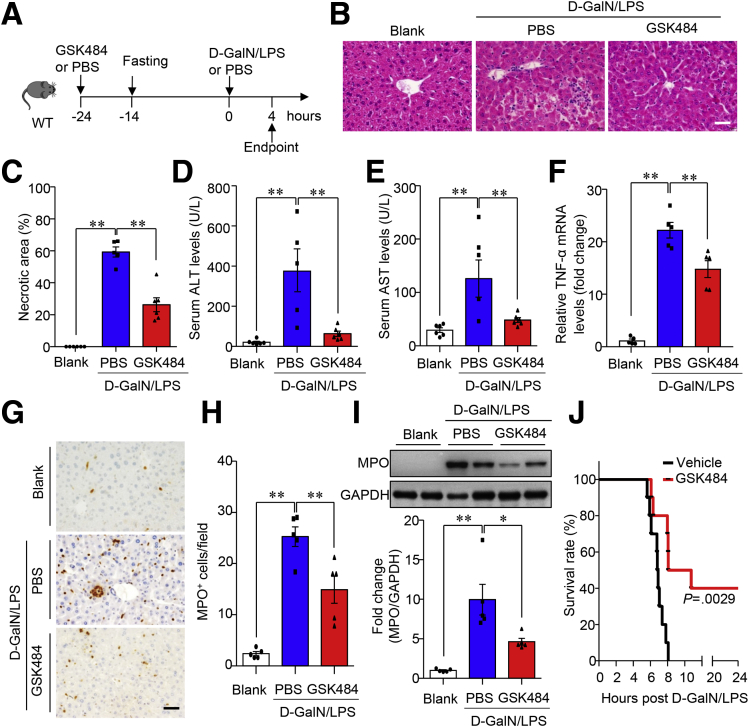
To determine whether ALF-associated NETs in the liver function as a key player in modulating disease development, we used a selective and potent inhibitor, GSK484, to block protein arginine deiminase 4 (PAD4), which is critically required for the formation of NET.24 GSK484 was administered to mice 24 hours before D-GalN/LPS injection via intraperitoneal injection at a dose of 20 mg kg-1 body weight (Figure 4A). GSK484 treatment substantially inhibited NET formation in both primary mouse neutrophils with ex vivo stimulation with LPS (Figure 5A and B) and liver tissues from mice with D-GalN/LPS treatment (Figure 5C and D). Therapeutically, D-GalN/LPS–induced massive liver necrosis was reduced significantly in mice with GSK484 pretreatment when compared with those in the vehicle control group, as shown on H&E staining of liver sections (Figure 4B) and semiquantification of the necrotic area (Figure 4C). The improvement in histologic lesions consequent to GSK484 pretreatment was accompanied by notably lowered serum levels of ALT (Figure 4D) and AST (Figure 4E). Meanwhile, ALF-evoked increase in messenger RNA levels of TNF-α in the liver were down-regulated significantly in mice with GSK484 pretreatment relative to vehicle controls (Figure 4F). ALF-induced hepatic infiltration of neutrophils also markedly was blunted in the GSK484 treatment group, as evidenced by the detection and measurement of neutrophil marker MPO by either immunohistochemistry (Figure 4G and H) or immunoblotting analysis (Figure 4I). In addition, the high mortality associated with a lethal dose of D-GalN/LPS in the vehicle control group evidently was reversed in mice with GSK484 pretreatment (Figure 4J). These data imply that pharmacologic inhibition of PAD4 blunts ALF-induced NETosis in the liver, thereby exerting protective effects against ALF and its associated mortality.
Figure 4.
GSK484-mediated blockade of NET formation attenuates ALF in mice. (A) Work flow of pretreatment with the PAD4 inhibitor GSK484 (20 mg kg-1 body weight) in 10-week-old male C57 BL/6J mice with D-GalN/LPS–induced ALF (D-GalN 0.75 mg g-1 body weight, LPS 2.5 μg g-1 body weight). PBS-treated mice served as controls. n = 5–6. (B) Representative images of H&E-stained liver tissue (original magnification, 400×) and the (C) quantification of necrotic area in H&E staining. Scale bar: 10 μm. Serum activity of (D) ALT and (E) AST. (F) mRNA expression level of TNF-α in the liver tissue determined by quantitative PCR. (G, H). Hepatic infiltration of neutrophils was determined by (G) immunohistochemistry of MPO in liver sections as shown in representative images (original magnification, 400×) and (H) semiquantification of the numbers of MPO+ cells under 400× microscopic fields. Scale bar: 10 μm. (I) Immunoblotting analysis of MPO in liver lysate and density ratio of MPO relative to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as loading control. (J) Survival in 10-week-old male C57 BL/6J mice after intraperitoneal injection of D-GalN (0.75 mg g-1 body weight) and LPS (0.01 μg g-1 body weight) in the presence or absence of GSK484 pretreatment. n = 10 per group. ∗P < .05, ∗∗P < .01.
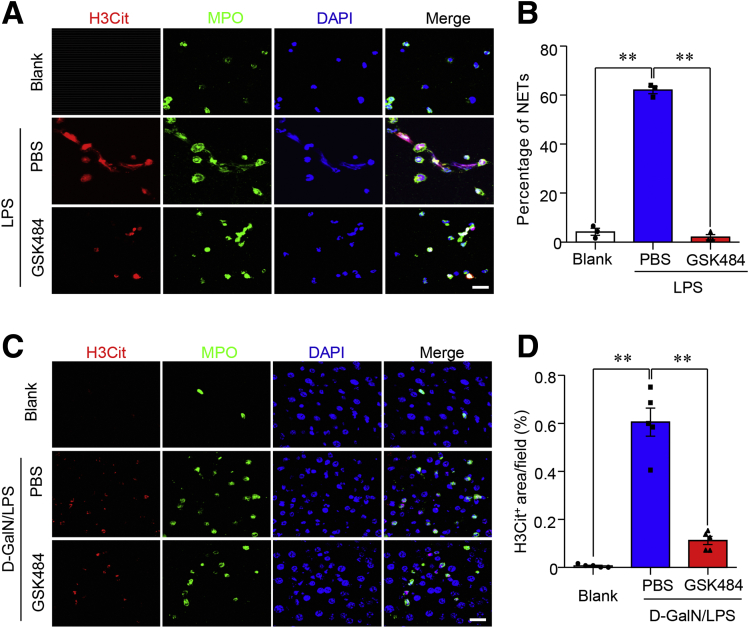
Figure 5.
Pretreatment with GSK484 markedly diminished NET formation ex vivo and in vivo. (A) Immunofluorescence of H3Cit (red), MPO (green), and DAPI (blue) in primary neutrophils isolated from C57 BL/6J mice with ex vivo stimulation by LPS (200 μg mL-1), GSK484 (10 μmol/L), and PBS to detect the formation of NETs. Scale bar: 5 μm. (B) The quantification of cells that have undergone NET formation. (C) Representative images of immunofluorescence of H3Cit (red), MPO (green), and DAPI (blue) in liver sections from 10-week-old male C57 BL/6J mice with pretreatment of GSK484 (20 mg kg-1) and D-GalN/LPS–induced ALF according to the procedures as indicated in Figure 4. Scale bar: 5 μm. (D) The formation of NETs in panel C was quantified as the percentage of the H3Cit-positive area per field. ∗∗P < .01.
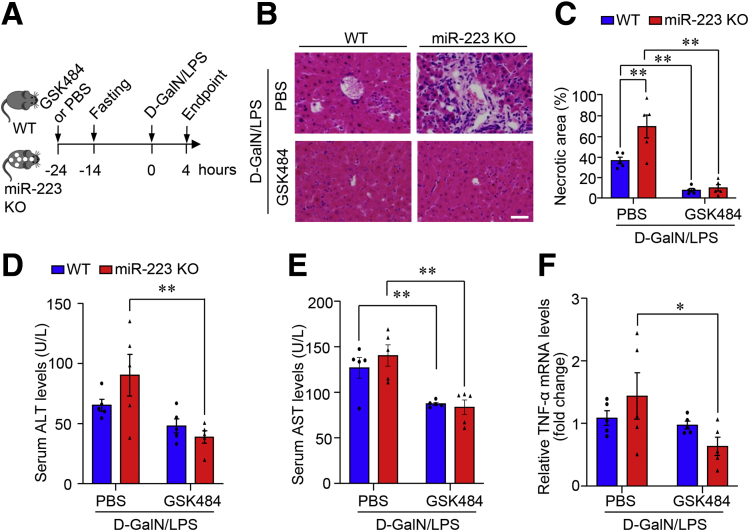
To delineate the link between the pathophysiological role of miR-223 and NETosis during ALF development, the response to GSK484 treatment in miR-223 KO mice was compared with that in WT controls. GSK484-mediated inhibition of NETosis exerted beneficial effects in both miR-223 KO mice and WT controls, evidenced by comparable end point levels in liver histology and serum activity of ALT and AST (Figure 6). Nonetheless the magnitude of beneficial effects of GSK484 pretreatment was higher in miR-223 KO mice relative to WT controls, majorly owing to the increased severity of hepatocellular injury in miR-223 KO mice in basal condition of vehicle treatment (Figure 6). These data suggest that miR-223 deficiency potentiated hepatocellular damage and liver inflammation in mice with ALF, at least in part through modulating NETosis in the liver.
Figure 6.
GSK484-mediated blockade of NET formation protected against ALF in miR-223 KO mice and WT controls. (A) Work flow of pretreatment with PAD4 inhibitor GSK484 in 10-week-old male miR-223 KO mice and WT control mice with D-GalN/LPS–induced ALF. PBS-treated mice served as controls. n = 5–6. (B) Representative images of H&E-stained liver tissue (original magnification, 400×) and the (C) quantification of necrotic area in H&E staining. Scale bar: 10 μm. Serum activity of (D) ALT and (E) AST. (F) mRNA expression level of TNF-α in liver tissue determined by quantitative PCR. n = 5 per group. ∗P < .05, ∗∗P < .01.
MiR-223 Deficiency Promotes Neutrophil Infiltration in Association With NE Enrichment in Mouse Liver With ALF
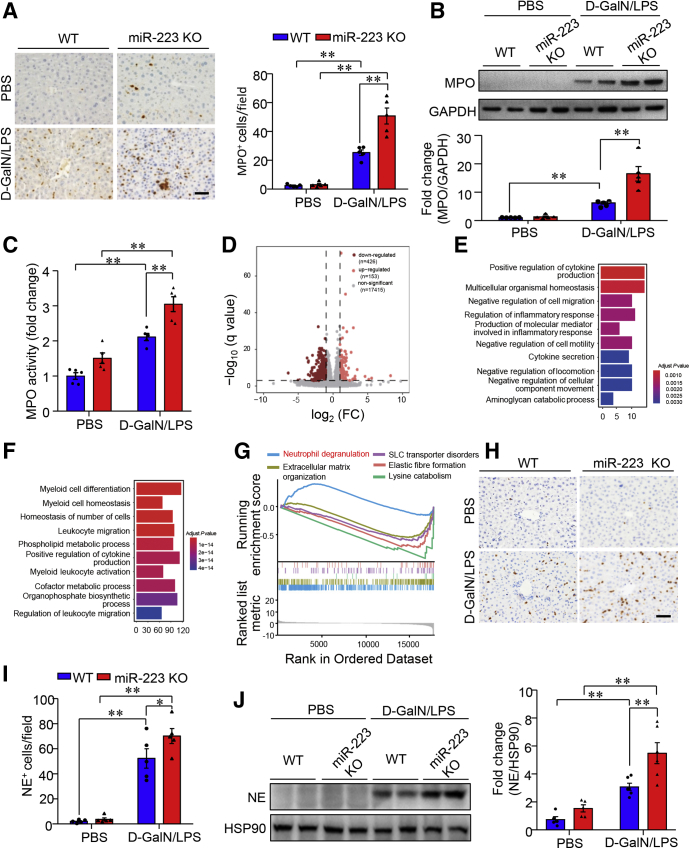
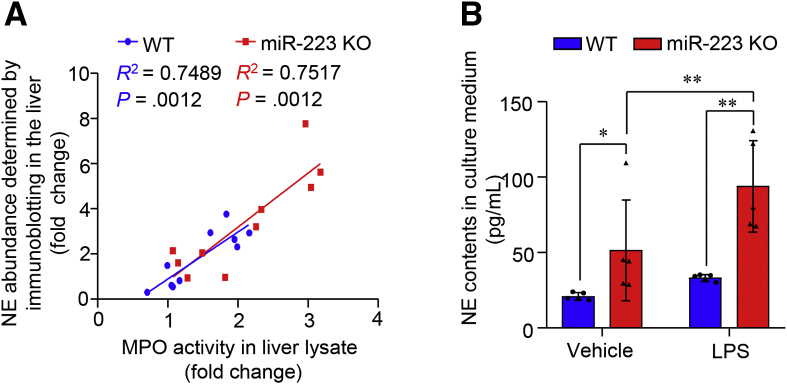
MiR-223 has been shown to be a key player in the regulation of biological and immunologic homeostasis of neutrophils.25 We next explored the regulatory effect of miR-223 on neutrophil infiltration in the liver and the expression of neutrophil elastase, one of the prominent neutrophil serine proteases with selective enrichment in azurophil granules in neutrophils. To this end, hepatic infiltration of neutrophils was detected and semiquantified by immunostaining of MPO, a specific marker of neutrophils.26 In WT mice, D-GalN/LPS treatment invoked markedly increased infiltration of MPO-positive cells into the liver. Notably, miR-223 KO mice showed a 2-fold increase in hepatic infiltration of MPO-positive cells compared with WT controls (Figure 7A). Furthermore, protein levels (Figure 7B) and enzymatic activity of MPO (Figure 7C) were increased significantly in miR-223 KO liver when compared with WT controls in response to ALF induction, confirming that ALF-associated hepatic infiltration of neutrophils was enhanced significantly in miR-223 KO mice relative to WT mice. To gain insight into the potential mechanisms underlying the sensitized response to ALF in miR-223 KO mice and the downstream target(s) of miR-223, we compared the transcriptome in bone marrow cells by RNA sequencing (RNA-seq) and identified 153 up-regulated genes and 426 down-regulated genes in miR-223 KO mice vs. WT controls (Figure 7D). Gene Ontology pathway enrichment analysis showed that genes with differentiated expression between miR-223 KO and WT mice were involved in innate immune responses, such as cytokine production and myeloid differentiation (Figure 7E and F). Furthermore, the gene set enrichment analysis (GSEA) using the Reactome pathway database highlights the activation of neutrophil degranulation as the top ranked pathway linked to differentially regulated genes in miR-223 KO bone marrow (Figure 7G). Given that NE functions as one of the prominent granule proteins stored in the azurophil granules of neutrophils, of which the release depends heavily on neutrophil degranulation,8 we compared NE contents in the liver of miR-223 KO and WT control mice under ALF conditions. The data of immunostaining and immunoblotting analysis consistently showed that D-GalN/LPS–induced NE enrichment in the liver was boosted significantly in miR-223 KO mice relative to WT controls (Figure 7H–J). In addition, hepatic NE content was correlated positively with MPO activities in liver tissues from miR-223 KO mice and WT control mice with or without D-GalN/LPS–induced ALF (Figure 8A). Furthermore, NE release into culture medium in response to ex vivo stimulation with LPS was significantly higher in primary neutrophils isolated from miR-223 KO mice compared with WT controls (Figure 8B). Taken together, these data suggest that enhanced hepatic infiltration of neutrophils and increased neutrophilic NE release contribute to increased NE content in the liver in miR-223 KO mice after ALF induction.
Figure 7.
Deletion of miR-223 augments hepatic recruitment of neutrophils and NE contents in mouse liver with ALF. Ten-week-old male miR-223 KO mice and WT controls were subjected to intraperitoneal injection of D-GalN/LPS to induce ALF (D-GalN 0.75 mg g-1 body weight, LPS 2.5 μg g-1 body weight). (A) Neutrophil infiltration in the liver was determined by immunohistochemistry of MPO (left, representative images) and semiquantification of MPO+ cells/per 40× microscopic field (right). Scale bar: 10 μm. The levels of MPO (B) protein contents and (C) enzymatic activities in liver lysates. (D) Volcano plot of RNA-seq data using bone marrow cells from miR-223 KO and WT control mice. n = 4 per group. Gene Ontology enrichment analyses of the target genes with marked (E) up-regulation and (F) down-regulation in miR-223 KO mice relative to WT controls. (G) Top 5 ranked Reactome pathways enriched in miR-223 KO bone marrow cells vs WT controls. (H–J) The abundance of NE in the liver from miR-223 KO and WT mice treated with either D-GalN/LPS or PBS were measured by (H) immunohistochemistry (representative images with original magnification, 400×; scale bar: 10 μm) with (I) semiquantification of NE+ cells per 40× microscopic field, and (J) immunoblotting analysis. Data are expressed as means ± SEM. n = 5 in functional studies. ∗P < .05, ∗∗P < .01. HSP90 heat shock protein 90.
Figure 8.
NE production is increased in primary mouse neutrophils lacking miR-223. (A) Correlation analysis of MPO activities in liver lysate shown in Figure 7C and NE contents measured by immunoblotting analysis shown in Figure 7J. (B) NE content in culture medium of equal amounts of primary neutrophils (1 × 105 cells per well) isolated from miR-223 KO mice and age-matched, WT control mice after ex vivo stimulation with LPS (200 μg/mL) or PBS as vehicle was determined by enzyme-linked immunosorbent assay at 3 hours after LPS treatment. ∗P < .05, ∗∗P < .01.
NE Plays a Key Role in ALF-Induced NETosis
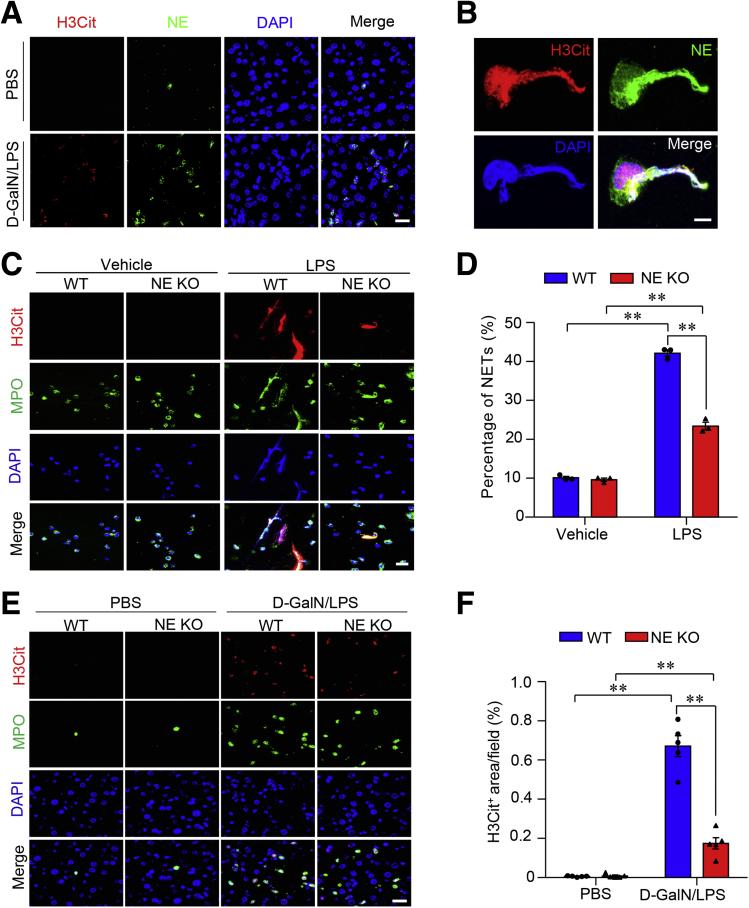
Because NE content was enriched significantly in miR-223–deficient primary mouse neutrophils and mouse liver with ALF, we next tested whether NE represented the component within ALF-induced NETs. Morphologic examination showed the colocalization of NE signals with H3Cit and DAPI in NET structure in mouse liver with D-GalN/LPS–induced ALF (Figure 9A). This finding was consolidated by immunofluorescence detection of NE in primary mouse neutrophils stimulated with LPS (Figure 9B). In contrast, primary neutrophils isolated from NE KO mice (Figure 10) showed notably impaired capacity for NETosis in response to ex vivo stimulation by LPS, compared with WT controls (Figure 9C and D). Consistently, NE KO mice with D-GalN/LPS–induced ALF displayed significantly suppressed NETosis in liver tissues relative to that in WT control mice (Figure 9E and F), suggesting NE as a key driving factor that is required for NETosis.
Figure 9.
ALF results in the formation of NETs in the liver under regulation of NE. (A) Immunofluorescence of H3Cit (red), NE (green), and DAPI (blue) in liver sections showing NE as a component of NETs in mouse liver with ALF (scale bar: 5 μm). (B) The formation of NETs was detected by immunofluorescence of H3Cit (red), NE (green), and DAPI (blue) in mouse primary neutrophils with ex vivo stimulation by LPS (200 μg mL-1) for 3 hours (scale bar: 1 μm). Immunofluorescence of H3Cit (red), MPO (green), and DAPI (blue) in primary neutrophils isolated from NE KO mice and WT control mice with ex vivo stimulation by LPS (C, scale bar: 5 μm) to identify the formation of NETs and (D) semiquantification under 40× microscopic fields. Representative images of immunofluorescence of H3Cit, MPO, and DAPI in liver sections from 10-week-old male NE KO mice and WT control mice treatment with (E, scale bar: 5 μm) D-GalN/LPS or PBS as vehicle and (F) semiquantification of the number of NETs under 40× microscopic fields, indicating the impaired capacity for NET formation in NE KO mice under ALF conditions. Ex vivo data were analyzed according to 3 independent experiments. Data in animal studies are expressed as means ± SEM. n = 5. ∗∗P < .01.
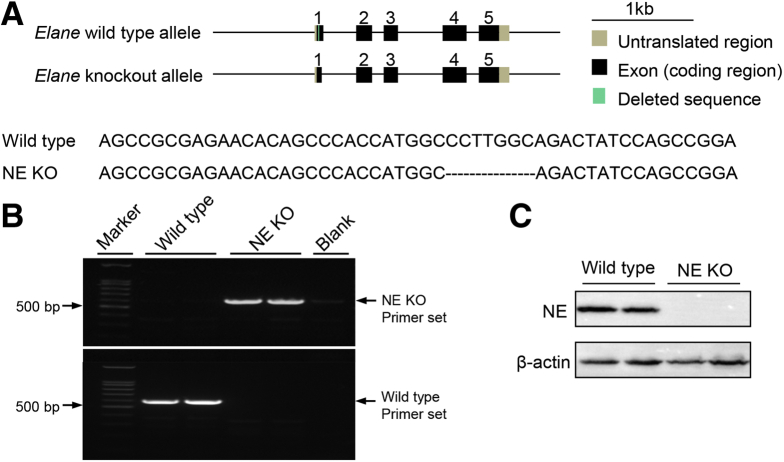
Figure 10.
Schematic diagram for the genetic modification to generate NE KO mice. (A) Schematic diagram of the genetic modification strategy to generate NE KO mice. (B) Representative gel images of PCR product in genotyping analysis to detect genetic modification of NE using primers (forward: gaacacagcccaccatggcag; reverse: gcatggacattgagggctaagagg). (C) Immunoblotting analysis of NE in primary bone marrow cells isolated from 10-week-old male NE KO mice and age-matched WT controls.
Loss of NE Protected Mice Against D-GalN/LPS–Induced ALF
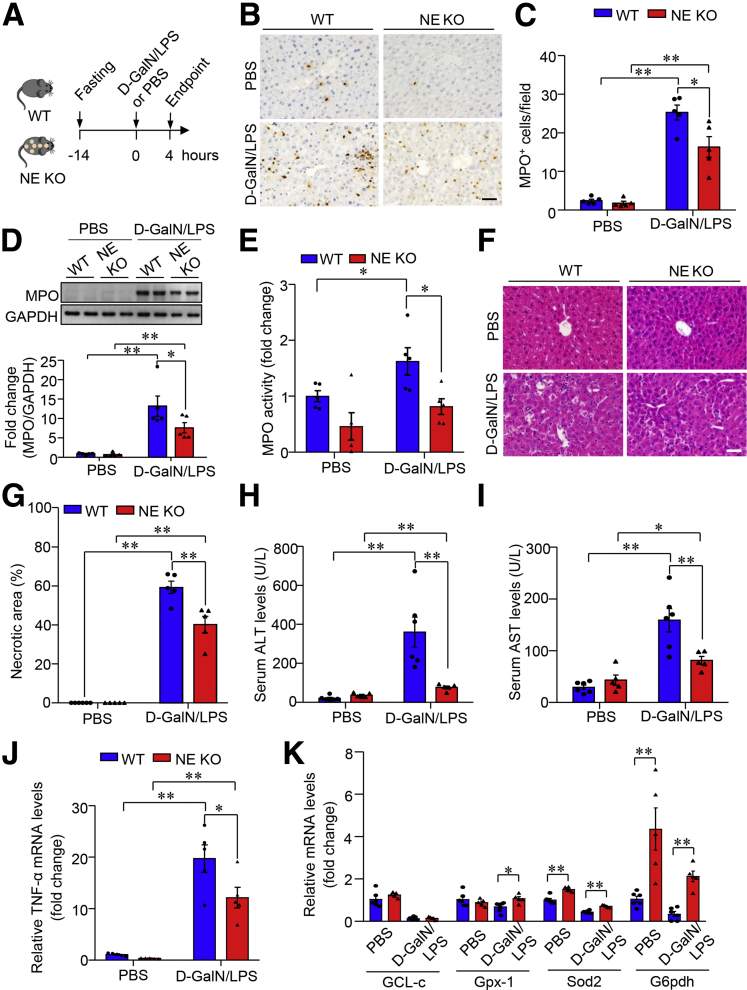
Markedly enhanced NE-positive cells in mouse liver that lacked miR-223 prompted us to investigate whether NE acts as a key player in the regulation of hepatic infiltration of neutrophils and ALF development (Figure 11A). Intriguingly, D-GalN/LPS stimulation–induced enhancement of neutrophil infiltration in the liver notably was blunted in NE KO mice, evidenced by reduced MPO contents as determined by immunostaining (Figure 11B and C) and immunoblotting analysis (Figure 11D) together with marked decreased MPO activity in the liver (Figure 11E), indicating that NE is required during neutrophil entry into the liver in the presence of ALF. Pathophysiologically, the D-GalN/LPS treatment–induced necrotic area in the liver of NE KO mice was reduced by approximately 20% relative to that in WT controls, as shown by H&E staining of liver sections (Figure 11F and G). Compared with WT controls, NE KO mice showed marked suppression of ALF-increased serum levels of ALT and AST by 70% and 50%, respectively (Figure 11H and I). Furthermore, NE deficiency caused marked suppression in the transcriptional abundance of TNF-α (Figure 11J), but up-regulation of anti-oxidative genes in the liver (Figure 11K) in response to D-GalN/LPS stimulation. Collectively, these data show that NE mediates ALF-related neutrophil infiltration in the liver, thereby potentiating liver necrosis and hepatocellular injury in mice.
Figure 11.
Lack of NE alleviates D-GalN/LPS–induced hepatic infiltration of neutrophils and ALF in mice. (A) Ten-week-old male NE KO mice and WT controls were injected intraperitoneally with D-GalN (0.75 mg g-1 body weight) and LPS (2.5 μg g-1 body weight) dissolved in PBS as vehicle to induce ALF. (B) Hepatic infiltration of neutrophils determined by immunohistochemistry of MPO (representative images, scale bar: 10 μm) and (C) semiquantification of MPO+ cells in each 40× microscopic field. (D) Immunoblotting analysis and (E) enzymatic activities of MPO in liver lysate from NE KO mice and WT controls. (F) Representative images of H&E-stained liver sections with original magnification 400×. Scale bar: 10 μm. (G) Quantification of necrotic area in liver sections according to H&E staining as shown in panel F. Serum activity of (H) ALT and (I) AST. mRNA expression level of (J) TNF-α and (K) anti-oxidative genes in the liver tissue determined by quantitative PCR. Data are expressed as means ± SEM. n = 5–6. ∗P < .05, ∗∗P < .01. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
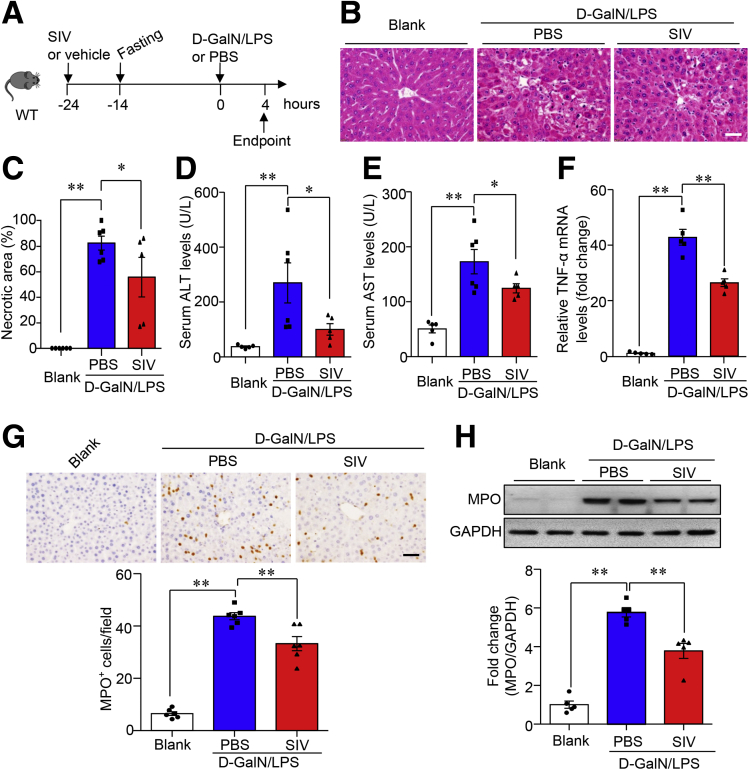
Pharmacologic Inhibition of NE With Sivelestat Alleviated ALF in Mice
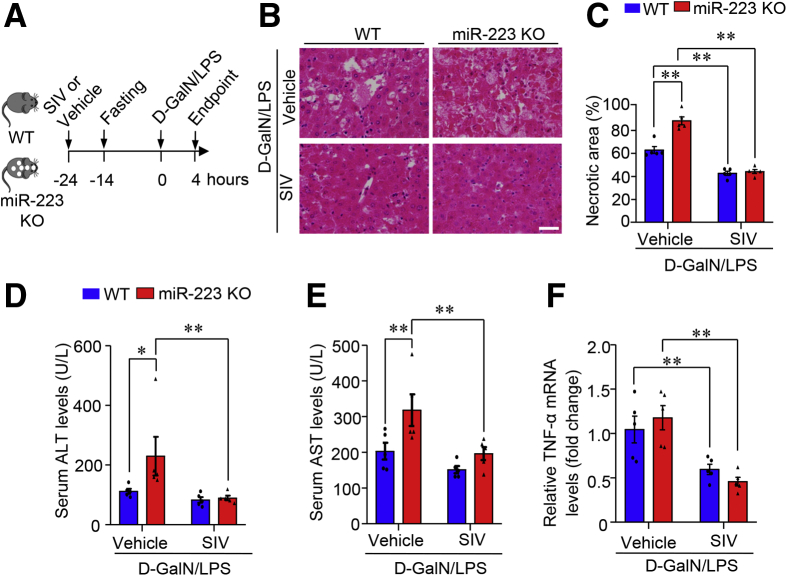
Given that D-GalN/LPS–induced hepatocellular damage and histologic lesions in the liver were attenuated markedly in mice with NE deficiency but potentiated in miR-223 KO mice with NE enrichment in the liver, we next tested the effect of pharmacologic blockade of NE on ALF in mice given sivelestat (also known as ONO-5046), a selective, reversible, and competitive inhibitor of NE.27 Sivelestat pretreatment was administered 24 hours before that of D-GalN/LPS (Figure 12A). H&E staining showed that D-GalN/LPS treatment induced massive liver necrosis in the vehicle control group, whereas liver histologic lesions in mice with sivelestat pretreatment were reduced significantly (Figure 12B and C). Likewise, the amplitude of D-GalN/LPS–induced increase of serum levels of ALT (Figure 12D) and AST (Figure 12E) was substantially lower in the sivelestat pretreatment group. The level of ALF-evoked up-regulation of TNF-α also was attenuated in mice with sivelestat pretreatment in response to D-GalN/LPS (Figure 12F). Notably, the preventive benefit of sivelestat-mediated pharmacologic inhibition of NE in experimental ALF was accompanied by a marked reduction in neutrophil infiltration of the liver, evidenced by the hepatic abundance of neutrophil marker MPO on immunohistochemistry (Figure 12G) and Western blot analysis (Figure 12H). Because miR-223 KO mice showed significantly increased susceptibility to D-GalN/LPS–induced ALF (Figure 3) and ALF-associated enrichment of NE content in the liver (Figure 7H–J), sivelestat pretreatment was used to inhibit NE in miR-223 KO mice and WT controls (Figure 13A). Sivelestat treatment protected against ALF in both miR-223 KO mice and WT controls (Figure 13B–F). Hence, NE blockade protected against ALF in mice, at least partially, through suppression of hepatic infiltration of neutrophils.
Figure 12.
Sivelestat-mediated NE blockade protects against ALF in mice. (A) Ten-week-old male C57 BL/6J mice were pretreated with sivelestat (SIV, 150 mg kg-1 body weight) via a single intraperitoneal injection 24 hours before ALF induction with intraperitoneal injection of D-GalN (0.75 mg g-1 body weight) and LPS (2.5 μg g-1 body weight). (B) Histologic lesions in the liver were assessed by H&E staining as shown in representative images (original magnification, 400×). Scale bar: 10 μm. (C) Quantification of necrotic area in H&E-stained liver sections. Serum activity of (D) ALT and (E) AST was measured to evaluate hepatocellular damage. (F) mRNA expression level of TNF-α in the liver tissue determined by quantitative PCR. (G) Representative images of immunohistochemistry of MPO in liver section (upper; scale bar: 10 μm) and semiquantification of MPO+ cells in each 40× microscopic field (lower). (H) Immunoblotting analysis of MPO in liver lysate (upper) and densitometric analysis of MPO protein abundance relative to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as loading control (lower). Data are expressed as means ± SEM. n = 5–6. ∗P < .05, ∗∗P < .01.
Figure 13.
Pharmacologic inhibition of NE protects against ALF in miR-223 KO mice and WT control mice. (A) The pretreatment with sivelestat (SIV) was conducted in 10-week-old male miR-223 KO mice and WT control mice via a single intraperitoneal injection of D-GalN (0.75 mg g-1 body weight) and LPS (2.5 μg g-1 body weight) 24 hours before ALF induction. (B) Histologic lesions were assessed by H&E staining in liver sections as shown in representative images (original magnification, 400×) Scale bar: 10 μm. (C) Quantification data of necrotic area in H&E-stained liver sections. Serum levels of (D) ALT and (E) AST was measured to evaluate hepatocellular damage. (F) TNF-α mRNA levels in liver tissue were determined by quantitative PCR. Data are expressed as means ± SEM. n = 5–6. ∗P < .05, ∗∗P < .01.
Discussion
Although augmented infiltration of neutrophils has been widely recognized as a hallmark of ALF in patients and rodent models, the mechanisms responsible for regulation of neutrophil entry into the liver and their pathophysiological role during ALF development remain poorly characterized. We provide evidence that miR-223 and NE are versatile regulators with distinct roles in modulating neutrophilic inflammation and hepatocellular injury during the pathogenesis of ALF. MiR-223 deficiency promotes NE production in neutrophils, thereby potentiating the formation of NETs to drive necroinflammation in the liver and ALF progression. Therapeutically, blockade of NE or NETs with pharmacologic inhibitors significantly attenuated hepatic infiltration of neutrophils and massive necrosis in the liver of mice with ALF.
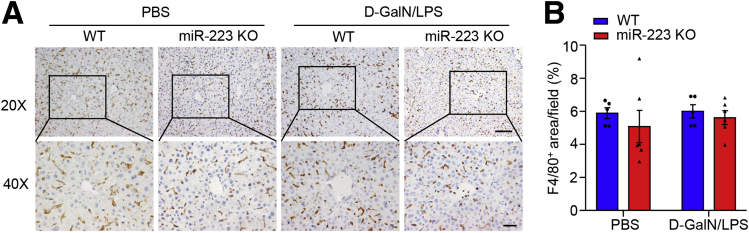
MiR-223 plays important roles in the development of liver disease by modulating the functions of neutrophils,19,20 macrophages,17 and hepatocytes.21 Our data show neutrophils as the key mediators for the beneficial effects of miR-223 on ALF in mice. On the one hand, ALF-induced massive migration of neutrophils into the liver was enhanced significantly in mice that lacked miR-223 relative to their wild-type controls (Figure 7A–C). On the other hand, the formation of NETs driving the development of necroinflammation in the liver and hepatocellular injury was boosted significantly in the absence of miR-223. These findings coincide with the predominant enrichment of miR-223 in neutrophils.28 In addition, miR-223 acts as a crucial regulator of macrophage polarization to inhibit its proinflammatory activation and protect against the diet-induced adipose tissue inflammatory response and systemic insulin resistance in mice,17 although its abundance in macrophages is approximately 10-fold lower than in neutrophils. Nonetheless, macrophages may not be the prime target of miR-223 to control liver damage in the initial stage of ALF. Acute stimuli-induced entry of circulating macrophages into tissues predominantly occurred 7 hours after proinflammatory stimulation, as shown in glycogen-induced peritonitis, a well-stablished in vivo model of chemotaxis.29 Meanwhile, hepatic infiltration of macrophages showed no marked increase 4 hours (the time point for tissue harvesting in the present study) after injection of D-GalN/LPS in both miR-223 KO mice and WT controls, as measured by immunohistochemistry of macrophage marker F4/80 in the liver (Figure 14). In hepatocytes, miR-223 abundance is enhanced in mice fed a high-fat diet.21 Continuous infusion of miR-223 mimics modulates hepatocyte apoptosis.18,30 In light of recent data showing miR-223 as a key mediator of intercellular communications through the transfer of extracellular vesicle cargo,31 it is plausible that miR-223 released from neutrophils might exert direct protective effects on hepatocyte death, thereby contributing to the attenuation of massive liver necrosis in ALF. Our data expand the current understanding of the regulation of neutrophil migration into the liver under conditions of acute severe stimuli to the liver.
Figure 14.
Hepatic infiltration of macrophages showed no marked increase at 4 hours after injection of D-GalN/LPS. (A) The infiltration of hepatic macrophages was determined by immunohistochemistry of F4/80 in liver tissues from 10-week-old male C57 BL/6J mice with peritoneal injection of D-GalN/LPS or PBS (as vehicle), as shown in representative images. Upper: original magnification, 200×; scale bar: 50 μm; lower: original magnification, 400×; scale bar: 20 μm. (B) F4/80-positive areas were quantified as the percentage in 400× microscopic fields.
Another important finding in the present study was the presence of NETs in liver tissue with ALF, which distinctly is regulated by miR-223 and NE to potently drive ALF development. Although the formation of NETs has been documented in the liver with nonalcoholic steatohepatitis10 and distant metastases,11 the regulatory mechanisms underlying the formation of NETs in ALF remain largely unclear. Our data showed that miR-223 and NE play opposing roles in modulating NET formation, evidenced by notably enhanced NET formation in neutrophils lacking miR-223 but marked inhibition in those without NE. Thus, miR-223 and NE function as pivotal regulators in the formation of NET in the liver with ALF. NET formation is reported to be triggered by activated platelets via Toll-like receptor 4 (TLR4).32,33 In mouse liver, TLR4 stimulates the production of proinflammatory cytokines by activating the MyD88 pathway, thereby acting as one of the key mediators of cell death induced by extracellular histones, the key component of NETs.34 It is likely that TLR4 signaling is actively involved in the regulation of NET formation by miR-223, especially in experimental ALF induced by hepatotoxic D-GalN in combination with LPS (well-established ligand for TLR4). The present study used a D-GalN/LPS–induced ALF mouse model, and consistent with the etiological features of ALF in human beings, showed the frequent co-existence of hepatotoxic stimuli with infection.1 Functionally, our data showed that GSK484-mediated blockade of NET resulted in markedly decreased neutrophil infiltration in the liver, suggesting that ALF-evoked NETs might exert a chemoattractant effect to potentiate hepatic infiltration of neutrophils via a positive feedback loop. This is in accordance with findings in other disease conditions closely associated with sterile neutrophilic inflammation.35
Serine proteases (myeloperoxidase, cathepsin G, lactotransferrin, leukocyte proteinase 3) have been documented to be key components coupled with extracellular strands of decondensed DNA, histones, and antibacterial peptide LL37 to form NETs in response to infectious or sterile stimulation of neutrophils.4 Nonetheless, the contribution of NE, one of the key serine proteases prominently abundant in azurophil granules in neutrophils, to the formation of NET in the necrotic liver is less well understood. Our data in this study showed that the capacity for NET formation was blunted substantially in neutrophils lacking NE in vitro and in vivo, confirming that NE is required for NETosis. Furthermore, D-GalN/LPS–invoked neutrophil recruitment in the liver was inhibited markedly in the presence of genetic ablation or pharmacologic inhibition of NE. The exact mechanism by which NE promotes neutrophil infiltration and NET formation during ALF remains partially unclear. It is worth noting that NE absence resulted in a substantial decrease, but not complete blockade, in LPS- or ALF-induced NET formation, indicating that NE acts as a key player in the complex scenario of NET regulation within the context of ALF progression. Given that NETosis is modulated predominantly by oxidant-dependent mechanisms and that ALF is characterized by robust generation of reactive oxygen species derived from damaged hepatocytes and recruited neutrophils,4,36 NE may coordinate modulators of oxidative stress to regulate NET formation, thereby mediating liver inflammation and damage in ALF.
Therapeutically, our data suggest that miR-223, NE, and NET are promising targets for ALF treatment. First, the delivery of miR-223 mimetics may represent a novel therapeutic intervention for ALF. Currently, the first-in-class miRNA oligonucleotide therapeutic (miravirsen) has shown efficacy for hepatitis C in clinical trials.37 In mice with experimental colitis, synthetic murine miR-223 mimics delivered by nanoparticle lipid emulsion resulted in a marked increase in miR-223 abundance in the colon and significant alleviation of colitis.38 Likewise, the injection of pre–miR-223 lentivirus attenuated chronic-plus-binge ethanol-induced liver injury.20 Nonetheless hepatic miR-223 promoted hepatocyte apoptosis.18 Advances in the development of neutrophil-specific delivery may hold great promise to yield optimal therapeutic outcomes for miR-223–based treatment of ALF. Second, small molecules targeting NE for ALF therapy warrant further investigation. Current attempts to characterize exogenous, synthetic, or recombinant inhibitors have yielded inhibitors against NE for at least 5 generations, among which the newest NE inhibitors are small molecules with pre-adaptive pharmacophores.39 In addition, several endogenous inhibitors against NE have been characterized including α1-protease inhibitors (irreversible inhibitors) or α1-antitrypsin, elafin, monocyte-neutrophil elastase inhibitor (serpin B1), and α-2-macroglobulin.8 Functionally, recombinant elafin/trappin-2–mediated inhibition of NE and proteinase 3 attenuates neutrophil-modulated tissue damage and ischemia-induced vascular injury.40 Third, a vast array of modulators with potent power to regulate NET formation, NET-resident components, and NET-evoked inflammation evidently stand out as promising therapeutic targets in ALF, in light of the evidence showing significant attenuation of tissue damage consequent to the prevention of NET formation through pharmacologic inhibition or global genetic abrogation of PAD4 under various inflammation-related conditions, such as acute kidney injury, rhabdomyolysis, cancer, or a combination of hemorrhage and sepsis.41 Of note, our results showed that post-treatment GSK484 or sivelestat is insufficient to protect against ALF development (data not shown), highlighting the need to explore novel more potent and fast-acting pharmacologic agents that target NE or NETs for treatment of ALF. Collectively, the key mediators identified during comprehensive pathophysiological exploration in this study may pave the way for the development of novel therapeutic approaches to ALF.
In summary, the present study showed the miR-223/NE axis as a key modulator of hepatic infiltration of neutrophils that trigger necroinflammation in the liver largely dependent on the formation of NETs, thereby exacerbating neutrophilic inflammation and oxidative stress to potentiate hepatocellular damage and liver necrosis in ALF development. These findings provide further evidence that therapeutic interventions that modulate miR-223/NE and NETs may represent a promising strategy for the optimal management of ALF.
Materials and Methods
Animal Studies
MiR-223 KO mice, NE KO mice, and WT controls on a C57BL/6J background were used in this study. NE KO mice were kindly provided by Professor Aimin Xu (State Key Laboratory of Pharmaceutical Biotechnology, The University of Hong Kong). miR-223 KO mice were provided by Professor Rongxin Zhang’s research group (Guangdong Province Key Laboratory for Biotechnology Drug Candidates, School of Life Sciences and Biopharmaceutics, Guangdong Pharmaceutical University), in which the genetic modification was performed as previously reported.42 Genotype was determined by polymerase chain reaction (PCR) analysis of genomic DNA. All experimental animals routinely were maintained under 12-hour light-dark cycles with free access to food and water. All experimental procedures for animal studies were approved by the Ethics Committee on the Use of Live Animals for Teaching and Research of Guangdong Pharmaceutical University (approval: gdpulac2019064) and were performed in accordance with the Guide for the Care and Use of Laboratory Animals.
Induction of Experimental ALF in Mice
Experimental ALF was induced in 10-week-old male mice with the indicated genotypes by intraperitoneal injection of galactosamine hydrochloride (D-GalN, 0.75 mg g-1 body weight, G1639; Sigma-Aldrich, Saint Louis, MO) and LPS (2.5 μg g-1 body weight or 0.01 μg g-1 body weight as indicated, from Escherichia coli serotype 026: B6, L-8274; Sigma-Aldrich, Saint Louis, MO) dissolved in PBS, after overnight fasting.43 Age-matched mice were injected with PBS and served as vehicle controls. Blood and liver samples were collected 4 hours after D-GalN/LPS administration.
Treatment With Pharmacologic Inhibitors Against Either NE or NETs in Mice
Ten-week-old male C57BL/6J mice received a peritoneal injection of either Sivelestat (pharmacologic inhibitor against NE, 150 mg kg-1 body weight, dissolved in 5% dimethyl sulfoxide, 40% polyethylene glycol 300, 5% Tween 80, 50% double-distilled H2O, 127373-66-4; Selleck, Houston, TX)44 or GSK484 (pharmacologic inhibitor against NETs, 20 mg kg-1 body weight, dissolved in PBS, SML1658; Sigma-Aldrich) 24 hours before ALF induction.
Visualization and Detection of NETs
To detect NETs in liver tissue, deparaffined liver sections were subjected to antigen retrieval with Tris-EDTA buffer (pH = 9.0) in a microwave oven for 10 minutes. After the blocking of nonspecific binding with 5% bovine serum albumin/phosphate-buffered saline with Tween 20 for 60 minutes at room temperature, sections were incubated with primary antibodies against H3Cit and MPO overnight at 4°C, followed by incubation with corresponding secondary antibodies conjugated with fluorescence dyes (anti-rabbit Alexa Fluor 568 and anti-goat Alexa Fluor 488). After counterstaining with DAPI, immunofluorescence images were captured.
For ex vivo detection of NETs, mouse primary neutrophils were isolated as described previously.5 To induce NETs, isolated mouse primary neutrophils were stimulated with LPS for 3 hours, followed by incubation with primary antibodies against H3Cit and MPO, followed by secondary antibodies including anti-rabbit Alexa Fluor 568 and anti-goat Alexa Fluor 488. Immunofluorescent images were visualized and captured using a color digital camera (DP80; Olympus, Tokyo, Japan) or confocal microscopy (FV3000; Olympus). NETs in liver sections were quantified as the percentage H3Cit-positive signal in each microscopic field using National Institutes of Health ImageJ software (Bethesda, MD).11
Biochemical Assays of Blood Samples
Mice were anesthetized and blood samples were collected via the retro-orbital venous plexus at the indicated time points. Serum activities of ALT and AST were measured using commercially available kits (C009-2-1 and C010-2-1 for ALT and AST, respectively; Nanjing Jiancheng Bioengineering Institute, Nanjing, China).
RNA Isolation and Real-Time Quantitative PCR
Total RNA in tissues and primary cells was extracted with TRIzol (15596018; Thermo Fisher Scientific, Waltham, MA) following the manufacturer’s instructions. Purified total RNAs were reverse-transcribed into complementary DNA using the PrimeScript RT reagent kit (RR047A; Takara, Kyoto, Japan). Reverse-transcription PCR was performed using TB Green Premix Ex Taq II (RR820A; Takara, Kyoto, Japan) with a primer set (sequences are listed in Table 1) on a real-time PCR detection system (LightCycler 480 Instrument II; Roche,Basel, Switzerland). To measure the abundance of miR-223, total RNA was isolated from liver tissues using TRIzol reagents. The mature miR-223 strand complementary DNA was synthesized using a Mir-X miRNA First-Strand Synthesis Kit (638313; Takara, Kyoto, Japan), according to the manufacturer's instructions. Reverse-transcription PCR analysis for miR-223 abundance was performed using TB Green Premix Ex Taq II with U6 as a housekeeping control. Relative expression of target genes was analyzed using the 2ˆ(–delta delta threshold cycle) method and normalized against the expression of housekeeping control β-actin for TNF-α and U6 for miR-223, respectively.
Table 1.
Sequences of Primers Used in Reverse-Transcription PCR in This Study
| Gene | Forward (5' to 3') | Reverse (5' to 3') |
|---|---|---|
| Mouse TNF-α | CCCTCACACTCAGATCATCTTCT | GCTACGACGTGGGCTACAG |
| Mouse Aco | TGGTATGGTGTCGTACTTGAATGAC | AATTTCTACCAATCTGGCTGCAC |
| Mouse CYP2E1 | TGTGACTTTGGCCGACCTGTTC | CAACACACACGCGCTTTCCTGC |
| Mouse CYP4A10 | CAACTTGCCCATGATCACACA | CATCCTGCAGCTGATCCTTTC |
| Mouse Nox2 | GAAAACTCCTTGGGTCAGCACT | ATTTCGACACACTGGCAGCA |
| Mouse GCL-c | GTTATGGCTTTGAGTGCTGCAT | ATCACTCCCCAGCGACAATC |
| Mouse GPX-1 | CCAGGAGAATGGCAAGAATGA | TCTCACCATTCACTTCGCACTT |
| Mouse Sod-2 | TCCCAGACCTGCCTTACGACTAT | GGTGGCGTTGAGATTGTTCA |
| Mouse G6pdh | CTGGAACCGCATCATCGTGGAG | CCTGATGATCCCAAATTCATCAAAATAG |
| Mouse β-actin | TACCACCATGTACCCAGGCA | CTCAGGAGGAGCAATGATCTTGAT |
Histology, Immunohistochemistry, and Immunofiuorescence
Paraffin-embedded liver tissue was sectioned for H&E staining to evaluate histologic lesions and to semiquantify the necrotic area. Immunohistochemistry detection of various target proteins was performed in paraffin-embedded liver sections after antigen retrieval with Tris-EDTA buffer (pH = 9.0). Nonspecific binding was blocked with 10% bovine serum albumin/phosphate-buffered saline with Tween 20 for 60 minutes at room temperature. Tissue was incubated with primary antibodies against mouse myeloperoxidase and mouse NE overnight at 4°C, and then with a horseradish peroxidase–conjugated secondary antibody against corresponding host IgG. For immunofiuorescence staining, deparaffinized tissue sections were incubated with primary antibodies against mouse H3Cit, mouse NE, followed by the incubation with secondary antibodies conjugated with various fluorescence dyes (including anti-rabbit Alexa Fluor 568 and anti-rat Alexa Fluor 488). Sections were counterstained with DAPI. Details of antibodies used in this study are listed in Table 2. All slides were examined under an Olympus biological microscope BX53 and images were captured using a color digital camera (DP80; Olympus) or confocal microscopy (FV3000; Olympus).
Table 2.
Key Resources
| Antibodies and reagents | Source | Identifier |
|---|---|---|
| Goat anti-myeloperoxidase | R&D Systems (Minneapolis, MN) | Cat# AF3667; RRID: AB_2250866 (0.5 μg/mL in WB; 5 μg/mL in IHC; 10 μg/mL in IF) |
| Rabbit anti–histone H3 | Abcam (Cambridge, UK) | Cat# ab5103; RRID: AB_304752 (1 μg/mL in WB; 10 μg/mL in IF) |
| Rat anti–neutrophil elastase | R&D Systems (Minneapolis, MN) | Cat# MAB4517 (0.5 μg/mL in WB; 5 μg/mL in IHC; 10 μg/mL in IF) |
| Rabbit anti-HSP90 | Cell Signaling Technology (Boston, MA) | Cat# 4877S; RRID: AB_2233307 (1:1000 dilution in WB) |
| Rabbit anti-GAPDH | Cell Signaling Technology (Boston, MA) | Cat# 2118S; RRID: AB_561053 (1:1000 dilution in WB) |
| Rabbit anti-F4/80 | Cell Signaling Technology (Boston, MA) | Cat# 70076S; RRID: AB_2799771 (1:300 dilution in IHC) |
| Donkey anti-goat secondary Ab | R&D Systems (Minneapolis, MN) | Cat# HAF109; RRID: AB_357236 (1:1000 dilution in WB) |
| Goat anti-rabbit secondary Ab | Cell Signaling Technology (Boston, MA) | Cat# 7074S; RRID: AB_2099233 (1:2000 dilution in WB) |
| Goat anti-rat secondary Ab | Cell Signaling Technology (Boston, MA) | Cat# 7077S; RRID: AB_10694715 (1:2000 dilution in WB; 1:500 dilution in IHC) |
| Rabbit anti-goat secondary Ab | Thermo Fisher Scientific (Waltham, MA) | Cat# 31402; RRID: AB_228395 (1.5 μg/mL in IHC) |
| Donkey anti-rabbit Alexa Fluor 568 | Thermo Fisher Scientific (Waltham, MA) | Cat# A10042; RRID: AB_2534017 (5 μg/mL in IF) |
| Chicken anti-rat Alexa Fluor 488 | Thermo Fisher Scientific (Waltham, MA) | Cat# A21470; RRID: AB_2535873 (5 μg/mL in IF) |
| Chicken anti-goat Alexa Fluor 488 | Thermo Fisher Scientific (Waltham, MA) | Cat# A21467; RRID: AB_2535870 (5 μg/mL in IF) |
| DAPI | Thermo Fisher Scientific (Waltham, MA) | Cat# D1306; RRID: AB_2629482 |
Ab, antibody; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HSP90, heat shock protein 90; IF, immunofluorescence; IHC, immunohistochemistry; RRID, Research Resource Identifier; WB, Western blot analysis.
Immunoblotting Analysis
Proteins extracted from liver tissue with RIPA buffer in the presence of protease inhibitor cocktail (11873580001; Roche, Basel, Switzerland) were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis, then transferred to polyvinylidene difluoride membranes. The membranes then were probed with primary antibodies against MPO, H3Cit, or glyceraldehyde-3-phosphate dehydrogenase, followed by incubation with corresponding secondary antibodies conjugated with horseradish peroxidase. Protein bands were visualized with clarity Western ECL substrate (170-5061; Bio-Rad, Hercules, CA). Intensity of the bands was quantified using National Institutes of Health ImageJ software.
MPO Activity Measurement in Mouse Liver
MPO activities in liver tissues were measured using a commercially available assay kit (600620; Cayman Chemical, Ann Arbor, MI), following the manufacturer’s instructions. The values of MPO activity were normalized with total protein amount in liver lysate, as measured by the Pierce BCA protein assay kit (23227; Thermo Scientific).
RNA-seq and Data Analysis
Total RNA from 8-week-old, male miR-223 KO mice and age-matched male WT controls was purified using TRIzol reagents (15596018; Thermo Fisher Scientific, Waltham, MA). Library preparation, cluster generation, and sequencing were performed by BGI (Wuhan, China). In brief, samples of RNA-seq libraries were sequenced on the DNBSEQ platform. Raw reads were filtered to obtain clean reads, which were mapped further to the mouse reference genome (GCF_000001635.26_GRCm38.p6). Normalization and identification of differentially expressed genes was performed using R package DESeq2. The criteria of log2|fold change|>1 and a q value less than 0.001 was used to identify differentially expressed genes. Gene Ontology enrichment was performed on significantly differentially expressed genes using the R package clusterProfiler. GSEA was performed to identify Reactome pathways enriched in miR-223 KO bone marrow cells vs WT cells. The R package msigdbr was used to extract the Reactome gene sets of mouse species from the Molecular Signatures Database, and the GSEA plot was drawn using R package gseaplot2. The R package (version 4.0.2) was used for bioinformatic analysis.
Statistical Analysis
Two-sided tests were used for all statistical analyses. Quantitative data are presented as means ± SEM. Statistical analyses were performed using the Statistical Package for Social Sciences version 16.0 (SPSS, Chicago, IL). The normality of data was assessed by the Kolmogorov–Smirnov test. Statistical differences between 2 groups were analyzed by an unpaired 2-tailed Student t test or the Mann–Whitney test for comparison of variables with or without normal distribution, respectively. Differences among multiple groups were assessed by 1- or 2-way analysis of variance. In all statistical comparisons, P < .05 was used to indicate a significant difference.
All authors had access to the study data and reviewed and approved the final manuscript.
CRediT Authorship Contributions
Dewei Ye (Writing – review & editing, Writing – original draft, Supervision, Funding acquisition, Conceptualization)
Jianyu Yao (Formal analysis, Data curation)
Wenfa Du (Methodology, Formal analysis, Data curation)
Cuishan Chen (Methodology, Formal analysis, Data curatio)
Yong Yang (Methodology, Formal analysis, Data curation)
Kaixuan Yan (Software, Formal analysis, Data curation)
Jufei Li (Methodology, Investigation)
Ying Xu (Methodology, Formal analysis)
Shufei Zang (Methodology)
Yuying Zhang (Formal analysis)
Xianglu Rong (Methodology)
Rongxin Zhang (Resources)
Aimin Xu (Writing – review & editing, Investigation, Conceptualization)
Jiao Guo (Writing – review & editing, Funding acquisition, Conceptualization)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This work was financially supported by National Natural Science Foundation of China grants 81922074, 81570701, and 81802551; Key Laboratory of Model Animal Phenotyping and Basic Research in Metabolic Diseases (2018KSYS003), and major basic and applied basic research projects in Guangdong Province (2019B030302005).
Contributor Information
Dewei Ye, Email: deweiye@gdpu.edu.cn.
Aimin Xu, Email: amxu@hku.hk.
Jiao Guo, Email: gyguoyz@163.com.
References
- 1.Bernal W., Wendon J. Acute liver failure. N Engl J Med. 2013;369:2525–2534. doi: 10.1056/NEJMra1208937. [DOI] [PubMed] [Google Scholar]
- 2.Triantafyllou E., Pop O.T., Possamai L.A., Wilhelm A., Liaskou E., Singanayagam A., Bernsmeier C., Khamri W., Petts G., Dargue R., Davies S.P., Tickle J., Yuksel M., Patel V.C., Abeles R.D., Stamataki Z., Curbishley S.M., Ma Y., Wilson I.D., Coen M., Woollard K.J., Quaglia A., Wendon J., Thursz M.R., Adams D.H., Weston C.J., Antoniades C.G. MerTK expressing hepatic macrophages promote the resolution of inflammation in acute liver failure. Gut. 2018;67:333–347. doi: 10.1136/gutjnl-2016-313615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Graubardt N., Vugman M., Mouhadeb O., Caliari G., Pasmanik-Chor M., Reuveni D., Zigmond E., Brazowski E., David E., Chappell-Maor L., Jung S., Varol C. Ly6C(hi) monocytes and their macrophage descendants regulate neutrophil function and clearance in acetaminophen-induced liver injury. Front Immunol. 2017;8:626. doi: 10.3389/fimmu.2017.00626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Honda M., Kubes P. Neutrophils and neutrophil extracellular traps in the liver and gastrointestinal system. Nat Rev Gastroenterol Hepatol. 2018;15:206–221. doi: 10.1038/nrgastro.2017.183. [DOI] [PubMed] [Google Scholar]
- 5.Ye D., Yang K., Zang S., Lin Z., Chau H.T., Wang Y., Zhang J., Shi J., Xu A., Lin S., Wang Y. Lipocalin-2 mediates non-alcoholic steatohepatitis by promoting neutrophil-macrophage crosstalk via the induction of CXCR2. J Hepatol. 2016;65:988–997. doi: 10.1016/j.jhep.2016.05.041. [DOI] [PubMed] [Google Scholar]
- 6.Liu Z.X., Han D., Gunawan B., Kaplowitz N. Neutrophil depletion protects against murine acetaminophen hepatotoxicity. Hepatology. 2006;43:1220–1230. doi: 10.1002/hep.21175. [DOI] [PubMed] [Google Scholar]
- 7.Papayannopoulos V. Neutrophil extracellular traps in immunity and disease. Nat Rev Immunol. 2018;18:134–147. doi: 10.1038/nri.2017.105. [DOI] [PubMed] [Google Scholar]
- 8.Wu L., Gao X., Guo Q., Li J., Yao J., Yan K., Xu Y., Jiang X., Ye D., Guo J. The role of neutrophils in innate immunity-driven nonalcoholic steatohepatitis: lessons learned and future promise. Hepatol Int. 2020;14:652–666. doi: 10.1007/s12072-020-10081-7. [DOI] [PubMed] [Google Scholar]
- 9.Wang Y., Xiao Y., Zhong L., Ye D., Zhang J., Tu Y., Bornstein S.R., Zhou Z., Lam K.S., Xu A. Increased neutrophil elastase and proteinase 3 and augmented NETosis are closely associated with beta-cell autoimmunity in patients with type 1 diabetes. Diabetes. 2014;63:4239–4248. doi: 10.2337/db14-0480. [DOI] [PubMed] [Google Scholar]
- 10.van der Windt D.J., Sud V., Zhang H., Varley P.R., Goswami J., Yazdani H.O., Tohme S., Loughran P., O'Doherty R.M., Minervini M.I., Huang H., Simmons R.L., Tsung A. Neutrophil extracellular traps promote inflammation and development of hepatocellular carcinoma in nonalcoholic steatohepatitis. Hepatology. 2018;68:1347–1360. doi: 10.1002/hep.29914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang L., Liu Q., Zhang X., Liu X., Zhou B., Chen J., Huang D., Li J., Li H., Chen F., Liu J., Xing Y., Chen X., Su S., Song E. DNA of neutrophil extracellular traps promotes cancer metastasis via CCDC25. Nature. 2020;583:133–138. doi: 10.1038/s41586-020-2394-6. [DOI] [PubMed] [Google Scholar]
- 12.von Meijenfeldt F.A., Stravitz R.T., Zhang J., Adelmeijer J., Zen Y., Durkalski V., Lee W.M., Lisman T. Generation of neutrophil extracellular traps in patients with acute liver failure is associated with poor outcome. Hepatology. 2022;75:623–633. doi: 10.1002/hep.32174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gantier M.P. The not-so-neutral role of microRNAs in neutrophil biology. J Leukoc Biol. 2013;94:575–583. doi: 10.1189/jlb.1012539. [DOI] [PubMed] [Google Scholar]
- 14.Landgraf P., Rusu M., Sheridan R., Sewer A., Iovino N., Aravin A., Pfeffer S., Rice A., Kamphorst A.O., Landthaler M., Lin C., Socci N.D., Hermida L., Fulci V., Chiaretti S., Foa R., Schliwka J., Fuchs U., Novosel A., Muller R.U., Schermer B., Bissels U., Inman J., Phan Q., Chien M., Weir D.B., Choksi R., De Vita G., Frezzetti D., Trompeter H.I., Hornung V., Teng G., Hartmann G., Palkovits M., Di Lauro R., Wernet P., Macino G., Rogler C.E., Nagle J.W., Ju J., Papavasiliou F.N., Benzing T., Lichter P., Tam W., Brownstein M.J., Bosio A., Borkhardt A., Russo J.J., Sander C., Zavolan M., Tuschl T. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aziz F. The emerging role of miR-223 as novel potential diagnostic and therapeutic target for inflammatory disorders. Cell Immunol. 2016;303:1–6. doi: 10.1016/j.cellimm.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 16.Xi X., Zhang C., Han W., Zhao H., Zhang H., Jiao J. MicroRNA-223 Is upregulated in active tuberculosis patients and inhibits apoptosis of macrophages by targeting FOXO3. Genet Test Mol Biomarkers. 2015;19:650–656. doi: 10.1089/gtmb.2015.0090. [DOI] [PubMed] [Google Scholar]
- 17.Zhuang G., Meng C., Guo X., Cheruku P.S., Shi L., Xu H., Li H., Wang G., Evans A.R., Safe S., Wu C., Zhou B. A novel regulator of macrophage activation: miR-223 in obesity-associated adipose tissue inflammation. Circulation. 2012;125:2892–2903. doi: 10.1161/CIRCULATIONAHA.111.087817. [DOI] [PubMed] [Google Scholar]
- 18.Qadir X.V., Chen W., Han C., Song K., Zhang J., Wu T. miR-223 Deficiency protects against fas-induced hepatocyte apoptosis and liver injury through targeting insulin-like growth factor 1 receptor. Am J Pathol. 2015;185:3141–3151. doi: 10.1016/j.ajpath.2015.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He Y., Feng D., Li M., Gao Y., Ramirez T., Cao H., Kim S.J., Yang Y., Cai Y., Ju C., Wang H., Li J., Gao B. Hepatic mitochondrial DNA/Toll-like receptor 9/microRNA-223 forms a negative feedback loop to limit neutrophil overactivation and acetaminophen hepatotoxicity in mice. Hepatology. 2017;66:220–234. doi: 10.1002/hep.29153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li M., He Y., Zhou Z., Ramirez T., Gao Y., Gao Y., Ross R.A., Cao H., Cai Y., Xu M., Feng D., Zhang P., Liangpunsakul S., Gao B. MicroRNA-223 ameliorates alcoholic liver injury by inhibiting the IL-6-p47(phox)-oxidative stress pathway in neutrophils. Gut. 2017;66:705–715. doi: 10.1136/gutjnl-2016-311861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He Y., Rodrigues R.M., Wang X., Seo W., Ma J., Hwang S., Fu Y., Trojnar E., Matyas C., Zhao S., Ren R., Feng D., Pacher P., Kunos G., Gao B. Neutrophil-to-hepatocyte communication via LDLR-dependent miR-223-enriched extracellular vesicle transfer ameliorates nonalcoholic steatohepatitis. J Clin Invest. 2021;131 doi: 10.1172/JCI141513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leshner M., Wang S., Lewis C., Zheng H., Chen X.A., Santy L., Wang Y. PAD4 mediated histone hypercitrullination induces heterochromatin decondensation and chromatin unfolding to form neutrophil extracellular trap-like structures. Front Immunol. 2012;3:307. doi: 10.3389/fimmu.2012.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sakurai K., Miyashita T., Okazaki M., Yamaguchi T., Ohbatake Y., Nakanuma S., Okamoto K., Sakai S., Kinoshita J., Makino I., Nakamura K., Hayashi H., Oyama K., Tajima H., Takamura H., Ninomiya I., Fushida S., Harada K., Harmon J.W., Ohta T. Role for neutrophil extracellular traps (NETs) and platelet aggregation in early sepsis-induced hepatic dysfunction. In Vivo. 2017;31:1051–1058. doi: 10.21873/invivo.11169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewis H.D., Liddle J., Coote J.E., Atkinson S.J., Barker M.D., Bax B.D., Bicker K.L., Bingham R.P., Campbell M., Chen Y.H., Chung C.W., Craggs P.D., Davis R.P., Eberhard D., Joberty G., Lind K.E., Locke K., Maller C., Martinod K., Patten C., Polyakova O., Rise C.E., Rudiger M., Sheppard R.J., Slade D.J., Thomas P., Thorpe J., Yao G., Drewes G., Wagner D.D., Thompson P.R., Prinjha R.K., Wilson D.M. Inhibition of PAD4 activity is sufficient to disrupt mouse and human NET formation. Nat Chem Biol. 2015;11:189–191. doi: 10.1038/nchembio.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roffel M.P., Bracke K.R., Heijink I.H., Maes T. miR-223: a key regulator in the innate immune response in asthma and COPD. Front Med (Lausanne) 2020;7:196. doi: 10.3389/fmed.2020.00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bradley P.P., Priebat D.A., Christensen R.D., Rothstein G. Measurement of cutaneous inflammation: estimation of neutrophil content with an enzyme marker. J Invest Dermatol. 1982;78:206–209. doi: 10.1111/1523-1747.ep12506462. [DOI] [PubMed] [Google Scholar]
- 27.Kawabata K., Suzuki M., Sugitani M., Imaki K., Toda M., Miyamoto T. ONO-5046, a novel inhibitor of human neutrophil elastase. Biochem Biophys Res Commun. 1991;177:814–820. doi: 10.1016/0006-291x(91)91862-7. [DOI] [PubMed] [Google Scholar]
- 28.Johnnidis J.B., Harris M.H., Wheeler R.T., Stehling-Sun S., Lam M.H., Kirak O., Brummelkamp T.R., Fleming M.D., Camargo F.D. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451:1125–1129. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- 29.Bozic C.R., Kolakowski L.F., Jr., Gerard N.P., Garcia-Rodriguez C., von Uexkull-Guldenband C., Conklyn M.J., Breslow R., Showell H.J., Gerard C. Expression and biologic characterization of the murine chemokine KC. J Immunol. 1995;154:6048–6057. [PubMed] [Google Scholar]
- 30.Wan L., Yuan X., Liu M., Xue B. miRNA-223-3p regulates NLRP3 to promote apoptosis and inhibit proliferation of hep3B cells. Exp Ther Med. 2018;15:2429–2435. doi: 10.3892/etm.2017.5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Calvente C.J., Tameda M., Johnson C.D., Del Pilar H., Lin Y.C., Adronikou N., De Mollerat Du Jeu X., Llorente C., Boyer J., Feldstein A.E. Neutrophils contribute to spontaneous resolution of liver inflammation and fibrosis via microRNA-223. J Clin Invest. 2019;129:4091–4109. doi: 10.1172/JCI122258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clark S.R., Ma A.C., Tavener S.A., McDonald B., Goodarzi Z., Kelly M.M., Patel K.D., Chakrabarti S., McAvoy E., Sinclair G.D., Keys E.M., Allen-Vercoe E., Devinney R., Doig C.J., Green F.H., Kubes P. Platelet TLR4 activates neutrophil extracellular traps to ensnare bacteria in septic blood. Nat Med. 2007;13:463–469. doi: 10.1038/nm1565. [DOI] [PubMed] [Google Scholar]
- 33.Sreeramkumar V., Adrover J.M., Ballesteros I., Cuartero M.I., Rossaint J., Bilbao I., Nacher M., Pitaval C., Radovanovic I., Fukui Y., McEver R.P., Filippi M.D., Lizasoain I., Ruiz-Cabello J., Zarbock A., Moro M.A., Hidalgo A. Neutrophils scan for activated platelets to initiate inflammation. Science. 2014;346:1234–1238. doi: 10.1126/science.1256478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu J., Zhang X., Monestier M., Esmon N.L., Esmon C.T. Extracellular histones are mediators of death through TLR2 and TLR4 in mouse fatal liver injury. J Immunol. 2011;187:2626–2631. doi: 10.4049/jimmunol.1003930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nemeth T., Mocsai A. Feedback amplification of neutrophil function. Trends Immunol. 2016;37:412–424. doi: 10.1016/j.it.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 36.Wu Z., Han M., Chen T., Yan W., Ning Q. Acute liver failure: mechanisms of immune-mediated liver injury. Liver Int. 2010;30:782–794. doi: 10.1111/j.1478-3231.2010.02262.x. [DOI] [PubMed] [Google Scholar]
- 37.Janssen H.L., Reesink H.W., Lawitz E.J., Zeuzem S., Rodriguez-Torres M., Patel K., van der Meer A.J., Patick A.K., Chen A., Zhou Y., Persson R., King B.D., Kauppinen S., Levin A.A., Hodges M.R. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368:1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 38.Neudecker V., Haneklaus M., Jensen O., Khailova L., Masterson J.C., Tye H., Biette K., Jedlicka P., Brodsky K.S., Gerich M.E., Mack M., Robertson A.A.B., Cooper M.A., Furuta G.T., Dinarello C.A., O'Neill L.A., Eltzschig H.K., Masters S.L., McNamee E.N. Myeloid-derived miR-223 regulates intestinal inflammation via repression of the NLRP3 inflammasome. J Exp Med. 2017;214:1737–1752. doi: 10.1084/jem.20160462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bronze-da-Rocha E., Santos-Silva A. Neutrophil elastase inhibitors and chronic kidney disease. Int J Biol Sci. 2018;14:1343–1360. doi: 10.7150/ijbs.26111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zani M.L., Tanga A., Saidi A., Serrano H., Dallet-Choisy S., Baranger K., Moreau T. SLPI and trappin-2 as therapeutic agents to target airway serine proteases in inflammatory lung diseases: current and future directions. Biochem Soc Trans. 2011;39:1441–1446. doi: 10.1042/BST0391441. [DOI] [PubMed] [Google Scholar]
- 41.Daniel C., Leppkes M., Munoz L.E., Schley G., Schett G., Herrmann M. Extracellular DNA traps in inflammation, injury and healing. Nat Rev Nephrol. 2019;15:559–575. doi: 10.1038/s41581-019-0163-2. [DOI] [PubMed] [Google Scholar]
- 42.Li Y., Zhou D., Ren Y., Zhang Z., Guo X., Ma M., Xue Z., Lv J., Liu H., Xi Q., Jia L., Zhang L., Liu Y., Zhang Q., Yan J., Da Y., Gao F., Yue J., Yao Z., Zhang R. Mir223 restrains autophagy and promotes CNS inflammation by targeting ATG16L1. Autophagy. 2019;15:478–492. doi: 10.1080/15548627.2018.1522467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gehrke N., Hovelmeyer N., Waisman A., Straub B.K., Weinmann-Menke J., Worns M.A., Galle P.R., Schattenberg J.M. Hepatocyte-specific deletion of IL1-RI attenuates liver injury by blocking IL-1 driven autoinflammation. J Hepatol. 2018;68:986–995. doi: 10.1016/j.jhep.2018.01.008. [DOI] [PubMed] [Google Scholar]
- 44.Raevens S., Van Campenhout S., Debacker P.J., Lefere S., Verhelst X., Geerts A., Van Vlierberghe H., Colle I., Devisscher L. Combination of sivelestat and N-acetylcysteine alleviates the inflammatory response and exceeds standard treatment for acetaminophen-induced liver injury. J Leukoc Biol. 2020;107:341–355. doi: 10.1002/JLB.5A1119-279R. [DOI] [PubMed] [Google Scholar]