Abstract
Preterm infants and patients with lung disease often have excess fluid in the lungs and are frequently treated with oxygen, however long-term exposure to hyperoxia results in irreversible lung injury. Although the adverse effects of hyperoxia are mediated by reactive oxygen species, the full extent of the impact of hyperoxia on redox-dependent regulation in the lung is unclear. In this study, neonatal mice overexpressing the beta-subunit of the epithelial sodium channel (β-ENaC) encoded by Scnn1b and their wild type (WT; C57Bl6) littermates were utilized to study the pathogenesis of high fraction inspired oxygen (FiO2)-induced lung injury. Results showed that O2-induced lung injury in transgenic Scnn1b mice is attenuated following chronic O2 exposure. To test the hypothesis that reversible cysteine-redox-modifications of proteins play an important role in O2-induced lung injury, we performed proteome-wide profiling of protein S-glutathionylation (SSG) in both WT and Scnn1b overexpressing mice maintained at 21% O2 (normoxia) or FiO2 85% (hyperoxia) from birth to 11–15 days postnatal. Over 7700 unique Cys sites with SSG modifications were identified and quantified, covering more than 3000 proteins in the lung. In both mouse models, hyperoxia resulted in a significant alteration of the SSG levels of Cys sites belonging to a diverse range of proteins. In addition, substantial SSG changes were observed in the Scnn1b overexpressing mice exposed to hyperoxia, suggesting that ENaC plays a critically important role in cellular regulation. Hyperoxia-induced SSG changes were further supported by the results observed for thiol total oxidation, the overall level of reversible oxidation on protein cysteine residues. Differential analyses reveal that Scnn1b overexpression may protect against hyperoxia-induced lung injury via modulation of specific processes such as cell adhesion, blood coagulation, and proteolysis. This study provides a landscape view of protein oxidation in the lung and highlights the importance of redox regulation in O2-induced lung injury.
Keywords: Redox proteomics, Hyperoxia, Lung, S-Glutathionylation, Thiol oxidation, SCNN1B
Graphical abstract
Highlights
-
•
Scnn1b overexpression in neonatal mouse lung attenuated hyperoxia induced lung injury.
-
•
Novel molecules and biological processes identified in hyperoxia-induced lung injury.
-
•
Potential mechanisms revealed for SCNN1B protective effect under hyperoxia.
1. Introduction
Supplemental oxygen is routinely administered to premature infants and patients with respiratory failure and plays a life-saving role by enhancing airway function [1]. Unfortunately, hyperoxia is also associated with abnormal pulmonary development, chronic lung diseases such as bronchopulmonary dysplasia, and even death [2,3]. Although the precise mechanisms of hyperoxia induced lung injury remains largely unknown, it is clear that accumulation of reactive oxygen species (ROS; such as H2O2) under pro-oxidizing conditions adversely disrupts the homeostasis of antioxidant systems needed to neutralize ROS, causing oxidative stress. Preterm lungs are especially vulnerable to supplemental oxygen therapy as their cellular antioxidant systems are not fully developed [[4], [5], [6], [7]].
Glutathione (GSH) is an important component of an antioxidant defense system deficient in the preterm lungs [8]. GSH is comprised of cysteine, glutamic acid, and glycine. The reducing power of this tripeptide is attributed to the thiol group of cysteine, which catalyzes the reduction of H2O2 to water and O2 using the cytosolic enzyme glutathione peroxidase (GPx). The reduction of H2O2 requires 2 GSH molecules and releases glutathione disulfide (GSSG) as a byproduct. A representation of the biochemical reaction is shown below:
| 2GSH + H2O2→ GSSG + 2H2O | (reaction 1) |
While this reaction is generally regarded as protective against oxidative stress, the increased bioavailability of GSSG could in turn bring about functional alterations of proteins [9]. For example, GSSG can form mixed disulfides with free protein cysteine thiols as a mode of post-translational modification termed S-glutathionylation (SSG) [10]. Protein modification by SSG has been recognized as a key redox signaling paradigm that links oxidative stress and numerous pathological processes such as cancer, heart disease, and diabetes [[11], [12], [13]]. The relevance of SSG with regard to lung pathology and diseases such as asthma and idiopathic pulmonary fibrosis has also been investigated [14]; total amounts of SSG increases with lung injury and fibrosis [15], whereas decreased SSG in sputum of asthmatics was reported by Kuipers et al. [16]. We have shown that GSSG inhibits lung epithelial sodium channel (ENaC) activity, the rate limiting step of alveolar fluid clearance (AFC), thereby constraining AFC and normal lung development [17,18]. Since GSSG inhibition of ENaC activity can be restored by reducing the extracellular GSH/GSSG Eh (i.e. increase bioavailability of GSH) [17], we hypothesize that SSG modifications of the lung proteome may play an important role in the pathogenesis of, or recovery from, high oxygen induced injury.
In this study, we investigate the redox-dependent mechanisms of hyperoxia induced lung injury by quantitatively assessing protein cysteine sites bearing SSG modifications using neonatal mice overexpressing the Scnn1b gene for ENaC and their WT littermates housed under normoxic (21% O2) or chronic high (85% O2) exposures. The Scnn1b gene encodes the β-subunit of epithelial sodium channel (β-ENaC), which assembles with α- and γ-subunits to form trimeric ion channels that regulate salt and water transport across the lung [19,20]. Overexpression of Scnn1b under the Clara cell secretory promoter (CCSP) in and of itself leads to an inflammation and emphysema-like lung injury phenotype in neonatal mice [[21], [22], [23]]. Likewise, chronic exposure to high fractions of inspired oxygen (FiO2; 85%) in neonatal mice recapitulates the developmental perturbations in infants with bronchopulmonary dysplasia [24]. Given that hyperactive ENaC dysfunction and chronic hyperoxia exposure both lead to oxidative stress and severe lung injury phenotypes independently, we were surprised to observe that chronic 85% O2 exposure improves the lung architecture of transgenic Scnn1b mice when compared to their WT littermates [25]. This unexpected finding warrants further investigation into the redox proteome of Scnn1b overexpressing mice under normoxic and hyperoxic conditions, as it could lead to an improved understanding of the molecular mechanisms underpinning irreversible lung injury.
For a better understanding of the role of thiol redox modifications in hyperoxia-mediated responses, proteome-wide identification and quantification of protein modifications that are sensitive to oxygen toxicity is required. Currently, mass spectrometry (MS)-based proteomics is the method of choice for the characterization of chemical modifications and quantification of the relative changes in redox status of thousands of proteins simultaneously [[26], [27], [28], [29]]. Our lab has developed a robust resin-assisted capture (RAC) approach that can not only identify site-specific cysteine oxidation, but also quantify the occupancy of these redox modifications at the proteome level [[30], [31], [32]]. In this study, we quantitatively profiled the thiol redox proteome of both transgenic Scnn1b and WT littermates under normal and hyperoxic conditions. We found that hyperoxia induced significant changes in SSG levels of specific proteins and pathways. Moreover, our data revealed that more substantial hyperoxia-induced changes of protein-SSG occur in transgenic Scnn1b mice compared to that of WT mice. Another experiment analyzing total protein oxidation, which quantifies all forms of reversible thiol-based oxidative modifications, further corroborated our observations on protein-SSG.
2. Materials and methods
2.1. Chemicals and materials
BCA (Bicinchoninic Acid) protein assay reagents, screw cap spin columns, and tandem mass tag (TMT) reagents were purchased from Thermo Fisher Scientific (Rockford, IL). Amicon Ultra centrifugal filter units (10 kDa cutoff) were purchased from MilliporeSigma (Burlington, MA). Thiopropyl Sepharose 6B affinity resin was obtained from Amersham Biosciences (Uppsala, Sweden). Sequence grade trypsin was purchased from Promega (Madison, WI). GRX1-C14S, a mutant form of glutaredoxin used in selective reduction of protein-SSG, was purchased from Cayman Chemical (Ann Arbor, MI). All other chemicals were from Sigma-Aldrich.
2.2. Animal treatment
Animal studies were approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Utah. The investigators adhered to IACUC and the National Institutes of Health guidelines for the Care and Use of laboratory Animals guidelines, and the research was carried out in compliance with the Animal Welfare Act [33,34]. All animals were given ad libitum access to food and water and housed under a normal light/dark cycle. Ambient temperature was kept between 22.2-23.3ᴼC in the vivarium.
Transgenic Scnn1b mice were a gift from Dr. Alessandra Livraghi-Butrico (Marsico Lung Institute, Chapel Hill, NC [35]). Male and female hemizygous Scnn1b mice were bred with wild type C57Bl6 mice (Taconic Bioscience, Rensselaer, NY) to obtain litters comprising approximately 50% hemizygous transgenic and 50% homozygous wild type littermates. All newborn mice were genotyped on PN day 7 by submitting tail snips to TransnetYX, Cordova, TN and were randomly assigned to either normoxic (21% O2) or hyperoxic (85% O2) exposure treatments.
Newborn litters were paired so that lactating dams could be swapped daily between high fraction inspired oxygen (FiO2 85%) and room air (21% O2) chambers in order to avoid maternal oxygen toxicity. Newborns were continuously exposed to FiO2 85% or 21% O2 beginning on the day of birth through PN days 11–15 of alveologenesis. Pups assigned to FiO2 85% were housed in a Plexiglass chamber with continuous oxygen monitoring (Proox model 110; Biospherix, Redfield, NY). Newborn litters were not redistributed to separate males from females; in rodent studies, sex is not a biological variable in high oxygen induced lung injury [36,37].
Animals were euthanized immediately following the last day of hyperoxic/normoxic exposures using intraperitoneal injection of 100 mg/kg Ketamine (West-Ward, Eatontown, NJ) and 10 mg/kg Xylazine (MWI Animal Health, Boise, ID) mix. Lungs were removed en bloc, rinsed with PBS, and snap frozen (−80 ᴼC). Lungs were not perfused prior to mass spec analysis. The total number of observations reported (n) refers to the total number of pups evaluated from distinct litters.
2.3. BALF collection and immune cell counts
Tracheas were cannulated from non-perfused lungs and lavaged (three times) with 50 μL PBS per 1 g of body weight. Bronchoalveolar lavage fluid (BALF) was collected and snap frozen prior to mass spec analysis. Immune cells were immoblized onto glass slides by cytospin centrifugation (1000×g, 5 min). BALF cells were labeled with Wright Stain Solution (Millipore, Burlington, MA) using manufacturer protocol and analyzed under an Olympus IX71 microscope and compatible DP80 dual sensor camera attached to the side port (Olympus, Tokyo, Japan). Percent neutrophil were calculated after counting 600 cells from 3 independent Wright stain studies.
2.4. Histology and destructive index (DI) assessment
Non-perfused and uninflated lungs were fixed in 10% neutral buffered formalin, paraffin-embedded, and then sectioned (transverse 4 μcron thin sections) and mounted onto glass slides. Deparaffinization and Masson's trichrome staining of lung sections were conducted using a standard histology protocol [38] and commercially available Masson's Trichrome Stain Kit (Agilent Technologies, Santa Clara, CA). Lung destructive indexes (ratio of damaged to healthy tissue in the alveoli/duct space) were assessed as described in Refs. [39,40] from 3 independent studies using the following Image J Plug settings: Analyze > Tools > Grid; random offset; 5000 μm2 area per point.
2.5. GSH/GSSG redox potential (Eh)
Reduced and oxidized levels of glutathione (GSH and GSSG, respectively) were concurrently measured from homogenized lung tissue using a colorimetric glutathione assay (Abcam, Cambridge, MA) following manufacturer protocol. Briefly, all samples were de-proteinated by adding equal volume metaphosphoric acid (1.25 M solution; Sigma-Aldrich, St. Louis, MO) and aliquots of the de-proteinated samples were prepared. One aliquot of the lung lysate was directly assayed for GSH by measuring absorbance at 415 nm following incubation with 5,5'dithio-bis-2-(nitrobenzoic acid) (DTNB; provided in the assay kit). A separate aliquot of de-proteinated lung lysate was derivatized with 1 M solution 2-vinylpyridine solution (prior to DTNB incubation) for total GSSG measurements. After obtaining GSH and GSSG concentrations, the redox potential (Eh) was calculated in accordance with the Nernst equation where: Eh = Eo + RT/2F ln [(GSSG)/(GSH)2]; Eo is the standard potential for the redox couple, R is the gas constant, T is the absolute temperature (ᴼK), and F is Faraday's constant. Calculated Eh serves as a quantitative expression of the redox pair's affinity for electrons (i.e. oxidative stress).
2.6. Quantitative PCR
Lung tissue was homogenized in RNA Stat60 (Amsbio) and total RNA extracted using standard phenol-chloroform protocols followed by DNASE treatment and RNA-II purification (Machery-Nagel). A total of 100 ng of RNA per sample was converted into cDNA using Superscript II components (Life Technologies) per manufacturer protocol. The cDNA was treated with 2.5 U RNase H (Affymetrix) at 37 ᴼC for 20 min to remove residual RNA. Real-time qPCR reactions were performed using Quantitect SYBR Green PCR reagent (Qiagen) using 5 μL of cDNA (diluted 1:10) per manufacturer protocol and mouse primer sequences GAT GTC CGA ACA GCT ATT TAC CA (forward) and CCT TGC GAC TTCA GCC ACT (reverse) for fibronectin (FN1); CGG GAG AAA ATG ACC CAG ATT (forward) and AGG GAC AGC ACA GCC TGA ATA G (reverse) for actin (ACTA-2); and CTG GCA CAA AAG GGA CGA G (forward) and ACG TGG CCG AGA ATT TCA CC (reverse) for collagen (COL4A1). Transcripts were normalized to the housekeeping gene GAPDH and mRNA expression levels calculated using the 2−ΔΔCt method.
2.7. Sample preparation of lung BALF and tissue for proteomics
BALF samples were concentrated by speedvac (ThermoSavant, New York, NY) until dry and further processed as per manufacturer instructions for S-trap micro spin columns (Protifi, Farmingdale, NY). Briefly, all samples were re-suspended in a uniform volume of 50 μL of lysis buffer containing 5% SDS, 50 mM triethylammonium bicarbonate (TEAB) pH 8.5. The samples were reduced with dithiothreitol (DTT) and alkylated with iodoacetamide (IAA) at final concentrations of 20 mM and 40 mM, respectively. Phosphoric acid was then added to a final concentration of 2.5% to obtain a pH ≤ 1 and then 7x volume of binding buffer containing 100 mM TEAB; 90% methanol was added to the lysates. The samples were then added to S-Trap micro columns, centrifuged at 4000×g for 30 s and washed 3 times with 150 μL of binding buffer. Each sample was digested with 20 μL digestion buffer containing 50 mM TEAB and 1.5 μg of sequencing grade trypsin (Promega, Madison, WI) for 2 h at 47 °C. Peptides were eluted by 40 μL of 50 mM TEAB, 40 μL of 0.2% formic acid, and 40 μL 50% acetonitrile, where the column was centrifuged at 4000×g for 1 min for each elution buffer. The peptides were concentrated by speedvac until dry, re-suspended in 20 μL of 0.1% formic acid solution, and quantified using standard BCA protein assay.
Sample processing for redox proteomics was performed as previously described [30,32,41]. In addition, profiling of the global proteome was performed in parallel with an aliquot of proteins used in the redox experiments. A graphical representation of the modified workflow is shown in Fig. 1. First, frozen lung lobes were minced into small pieces on a glass slide placed in an aluminum tray that was chilled by dry ice to prevent the samples from thawing. The minced tissue of each sample was collected into their own 5 mL round bottom polystyrene tube (Corning) and incubated on ice for 30 min in the dark with homogenization buffer (250 mM MES; 1% SDS; 1% Triton X-100) adjusted to pH 6 to protect against artificial oxidation during homogenization, as was used in a previous study of lung tissue [42]. The pooled total thiol sample was prepared by collecting several pieces of tissue from each minced sample into a single tube. Homogenization buffer for glutathionylation or total oxidation samples included 100 mM NEM to alkylate free thiols, while this reagent was omitted in the homogenization buffer used for the total thiol sample. A tissue homogenizer (BioSpec Products) was then used to create homogenates of these samples, which were transferred to microcentrifuge tubes and spun down at top speed for 10 min at 4 °C. Supernatants were transferred to a new tube and incubated at 55 °C for 30 min at 850 rpm to promote protein denaturation and alkylation, followed by overnight acetone precipitation. The protein pellet was re-suspended with resuspension buffer (250 mM HEPES pH 7; 8 M Urea; 0.1% SDS) and measured by BCA assay. 600 μg of protein was concentrated by buffer exchange via 10K Amicon ultra filters (EMD Millipore) and the volumes were adjusted with a reducing buffer (25 mM HEPES pH 7.6; 1 M Urea) to obtain a final concentration of ∼1 μg/μL. For total oxidation analysis, samples were then reduced with DTT (20 mM final concentration) at 37 °C and 850 rpm for 30 min. For SSG analysis, de-glutathionylation of proteins was performed by adding a Glutaredoxin 1 cocktail to selectively reduce protein-SSG modifications at 37 °C and 850 rpm for 10 min (the corresponding total thiol sample was totally reduced by DTT during the SSG workflow). Following either mode of total or selective reduction, buffer exchange with 8 M urea in 25 mM HEPES pH 7 was used to remove excessive reducing reagents. Samples were then diluted with 25 mM HEPES pH 7.6 to obtain a urea concentration of 2.5 M and the resulting proteins were quantified by the BCA assay and then aliquoted for either redox proteomics or global proteomics.
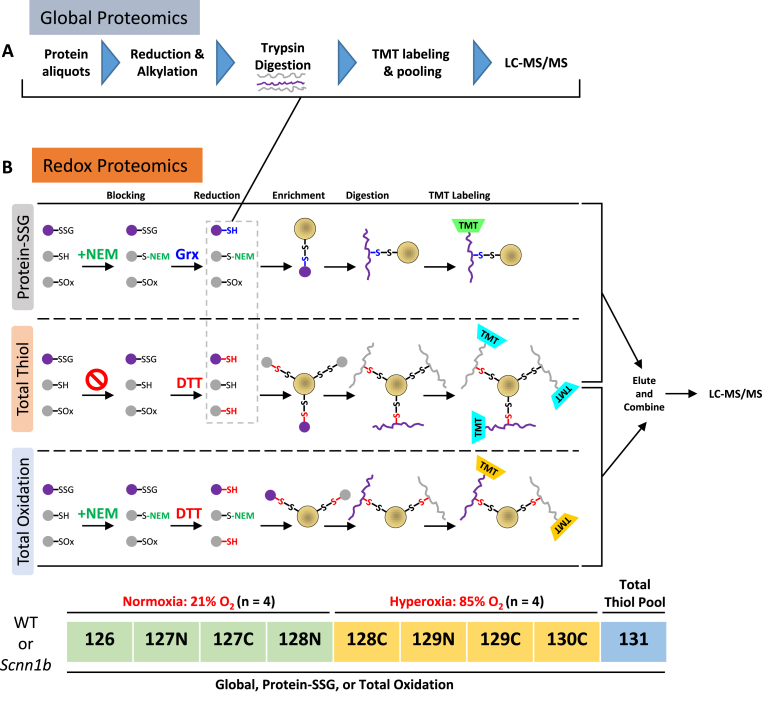
Fig. 1.
Proteomic profiling of mouse lung protein. (A) Workflow for global proteomics on protein aliquots collected after the reduction step (gray dashed box in (B)) during the redox proteomics procedures for protein-SSG and total thiol. (B) Schematics of redox proteomics. Except for S-glutathionylation, all other reversible forms of oxidation (S-nitrosylation, S-sulfenylation, disulfides, persulfides, polysulfides) are represented by “SOx”. Proteins/peptides with SSG sites are distinguished by purple-colored shapes. Protein samples from control (21% O2, n = 4) and hyperoxic-exposed (85% O2, n = 4) mice were subjected to SSG enrichment (upper panel), which consisted of NEM blocking of free thiols (denoted by green text), selective reduction of PSSG with glutaredoxin (Grx; denoted by blue text), RAC-based enrichment (yellow circle denotes resin), on-resin digestion and TMT labeling, and finally elution off the resin using DTT. A pooled sample was enriched for total thiol (middle panel), in which NEM blocking is omitted, the reduction was performed with DTT to reduce all reversible forms of oxidation (red text), and RAC is used to capture all free thiols in the sample to represent the total thiol content of the samples. The lower panel represents the total oxidation workflow, where free thiols are alkylated with NEM, all reversible forms of thiol oxidation are reduced with DTT, and nascent free thiols are enriched. All TMT-labeled samples were pooled and analyzed by LC-MS/MS. The TMT labeling scheme used for global and redox proteomic workflows for either genotype is shown below the total oxidation workflow. Note: total thiol sample prep was included in either the protein-SSG or total oxidation workflows. Two sets of experiments were performed for wild type and transgenic Scnn1b mice, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
For each sample, 120 μL of 300 μg de-glutathionylated protein or 150 μg from the total oxidation workflow were used as input for enrichment in their own columns containing 30 mg of hydrated Thiopropyl Sepharose 6B resin (Cytiva) at room temperature and shaking for 2 hr at 850 rpm. Unbound proteins were washed off the resin with 5 × 1) 25 mM HEPES pH 7.7 with 8 M urea, 2) 2 M NaCl, 3) 80% acetonitrile (ACN) with 0.1% Trifluoroacetic acid (TFA), and 4) 25 mM HEPES pH 7. Resin-bound proteins in each column were digested with 8 μg of sequencing-grade trypsin (Promega) dissolved in 25 mM HEPES pH 7.7 overnight at 37 °C and shaking at 850 rpm. Washing out of non-Cys-containing peptides was performed with 5 × 1) 25 mM HEPES pH 7.7, 2) 2 M NaCl, 3) 80% ACN with 0.1% TFA, 3x with 25 mM HEPES pH 7, and 2x with 50 mM triethylammonium bicarbonate buffer (TEAB). To label the peptides, 40 μL of 100 mM TEAB was added to each column followed by 400 μg of tandem mass tag (TMT) reagent dissolved in 70 μL anhydrous ACN and incubated for 1 hr at room temperature with shaking at 850 rpm. Unbound TMT reagent was washed out with 5 × 1) 80% ACN with 0.1% TFA, 3x with 25 mM Ammonium bicarbonate, 2x with ultra-pure water. Finally, Cys-containing peptides were eluted off the resin 2x with 20 mM DTT in 100 μL of ammonium bicarbonate after a 30 min incubation at room temperature with 850 rpm shaking and a single 10 min incubation with 80% ACN with 0.1% TFA. Samples were combined and concentrated by speedvac and subjected to standard C18 solid phase extraction (SPE) sample cleanup. Additional detailed steps regarding enrichment with Thiopropyl Sepharose 6B resin, on-resin trypsin digestion and labeling with TMT, and elution of enriched cysteine-containing peptides can be found elsewhere [[30], [31], [32],41].
To perform global proteomics analysis (Figs. 1A), 100 μg proteins were aliquoted after the selective reduction in a redox proteomic workflow. These samples were diluted with 25 mM HEPES, pH 7.7, to a final urea concentration of 1 M (∼40 μl final volume). Reduction and alkylation were performed by incubating the samples first with 5 mM DTT at 37 °C for 30 min, and then with 10 mM iodoacetamide for 1 hr in the dark. The samples were digested with 2 μg of trypsin overnight at 37 °C. The resulting peptides were labeled with 70 μL of TMT10 plex reagents (the same set used for redox proteomics) using the same labeling scheme as for the redox samples (Fig. 1B). The labeling reactions were quenched by adding 8 μl of 5% hydroxylamine, followed by shaking at 600 rpm for 30 min at room temperature. All samples were pooled together, and SPE cleanup was performed prior to LC-MS analysis.
2.8. LC-MS/MS analysis
Prior to LC-MS/MS analysis, the pooled global and redox peptides were acidified by trifluoroacetic acid and desalted by a ziptip C18 column (Agilent Technologies). Samples were then dissolved in 20 mM DTT in H2O to prevent oxidation of cysteine thiols. For BALF samples, 1.5 μg of peptide from each sample was diluted to a final concentration of 0.1 μg/μL with 0.1% formic acid. The final peptide samples were analyzed on a Q Exactive Plus (Thermo Scientific) coupled with a nanoAcquity ultra performance liquid chromatography (UPLC) system (Waters). Elution of the peptides were performed on a reverse phase C18 column (70 cm × 75 μm i.d., 3 μm particle size of Jupiter C18, Phenomenex) at 300 nL/min with buffer A (0.1% formic acid in H2O) and buffer B (0.1% formic acid in acetonitrile) using the following gradient: 0.1–8% B for 4 min, 8–12% B for 32 min, 12–30% B for 100 min, 30–45% B for 40 min, and 45–95% B for 5 min.
Mass spectra of the eluted peptides were collected using a data-dependent acquisition method that consisted of a full MS scan and subsequently up to 12 MS/MS scans for the most intense precursor ions. Full MS scans (400–2800 m/z) were performed in the Orbitrap with the following key parameters: resolution, 70 K; automatic gain control, 3E6; and maximum ion injection time: 20 ms. Precursor ions were selected by the quadrupole with an isolation window 2 m/z, and fragmentation was performed by higher-energy collisional dissociation at a normalized collision energy of 30. Scan parameters for MS/MS spectra were: resolution, 35 K; automatic gain control, 1E5; and maximum ion injection time: 100 ms.
2.9. Proteomic data processing and bioinformatics
Raw BALF data were analyzed with MaxQuant (v2.0.3.0) [43] with the match between runs option enabled for label free quantification with a matching time window of 0.7 min and alignment window of 20 min. Additional parameters include: methionine oxidation and N-terminal acetylation set as variable modifications, Cysteine carbidomethylation set as a fixed modification, parent ion tolerance of 20 ppm, and fragment ion mass tolerance of 0.5 Da. The LFQ intensities in the “proteinGroups.txt” file were log-transformed and median centered across all samples for statistical analysis using R.
Raw global and redox proteomics data were analyzed with MS-GF + against the Uniprot Mus musculus database (release 2017_04, with 16,880 entries). Addition of N-ethyl maleimide on cysteine (+125.0477), and oxidation of methionine (+15.9949) were set as dynamic modifications. Labeling of TMT reagents on peptide N terminus and lysine residues (+229.1629) was set as the static modification. A partial tryptic rule was applied for peptide identification. Similar parameters were used for global proteomics data, except that carbamidomethylation of cysteine residues (+57.0215) is also set as a dynamic modification since proteins were alkylated with iodoacetamide. Multiple criteria were applied to filter peptide spectrum matches (PSMs) to control <1% false discovery rate (FDR, calculated using a target-decoy approach) at the peptide level: 1) MSGF SpecProb < 1E-8; 2) precursor tolerance within 10 ppm; 3) PepQValue < 0.01.
Quantitative analyses of the proteomic data were performed as previously described [44,45]. Briefly, TMT reporter ion signals were first summed for PSMs corresponding to the same unique peptide. Peptides with missing values in any of the quantification channels were excluded. Raw TMT reporter ion intensities were then log2 transformed as surrogates for relative peptide abundances. Redox proteomics data was normalized against the non-cysteine containing peptides, assuming that these non-specific binding peptides have an equal distribution of peptide abundance across all channels. Non-Cys peptides and peptides only containing an NEM-blocked Cys were removed prior to subsequent analysis. To identify the Cys sites, each unique Cys-containing peptide was mapped the mouse proteome to determine the positions of the Cys residue on the protein. The unique peptide data was then aggregated to the unique Cys site level by combining the TMT reporter ion intensities (non-log2-transformed) of peptides containing the same Cys site ID. The student's t-test was used for statistical analysis, where unique Cys sites with log2 fold change of 0.3 or greater were further subjected to the Benjamini-Hochberg p value adjustment method. Cys sites with adjusted p value < 0.1 were then defined as significant. A similar workflow was used for analysis of the global proteomics data except that TMT reporter ion intensities and all subsequent analysis were performed at the protein level. Integration of data between the two genotypes was performed by merging SSG or global datasets together and only keeping Cys site or proteins that are common to both datasets. Each genotype's SSG data was mean centered, where the mean represents the average SSG intensity of all channels except total thiol. The global data was mean centered in a similar manner, except that total thiol was included in the calculation of the mean. The fold change of each replicate per genotype were then calculated, used for statistics, and used to calculate the fold change of hyperoxia response in Scnn1b overexpressing versus wild type mice. Gene ontology analysis was performed with DAVID (https://david.ncifcrf.gov/), where the background was set to all proteins identified in each analysis.
3. Results
3.1. The neonatal transgenic Scnn1b lung phenotype under chronic hyperoxia
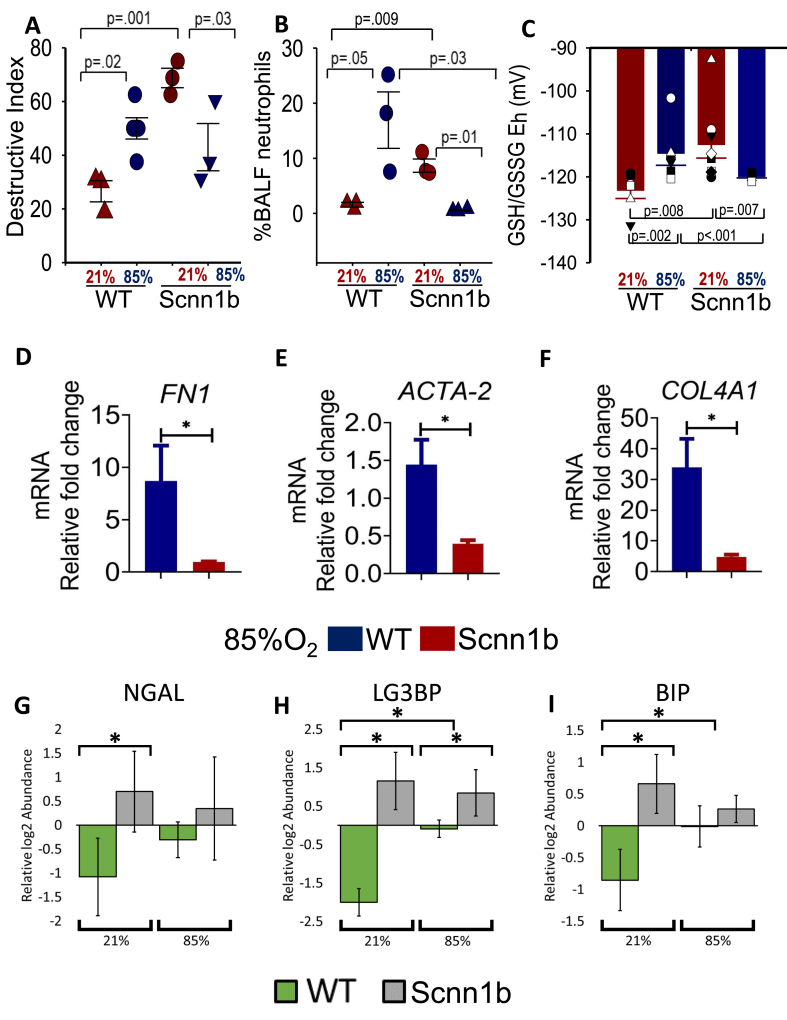
Transgenic Scnn1b and WT littermates were maintained on FiO2 85% or at room air from the day of birth up to postnatal day 11 (PN 11). At the end of exposure treatment, BALF was collected for i) neutrophil counts and ii) proteomics analysis, while lungs were excised en bloc and partitioned for iii) histological analysis, iv) redox measurements, and v) measuring the level of transcripts of genes commonly associated with epithelial lung injury in order to validate and strengthen our initial observation indicating that epithelial sodium channels play an important role in attenuating oxidative injury [25]. Alveolar septal damage and emphysematous space (i.e. destructive index; DI) were calculated from histological sections (not shown) obtained from transgenic Scnn1b and WT lungs housed as indicated in Fig. 2A. As expected, chronic FiO2 85% inhalation in WT pups and Scnn1b overexpression (at room air) can independently increase the destructive index (DI; Fig. 2A); neutrophil infiltration (Fig. 2B), and oxidative stress (Fig. 2C) compared to control WT pups housed at 21% O2. The DI of neonatal WT lung (housed at 21% O2) increases from 26.5% ± 3.9%–50.0% ± 3.2% (when housed at FiO2 85% for 11 days). Interestingly, chronic high O2 exposure significantly decreases the DI of Scnn1b overexpressing lung from 68.75% ± 3.6%–43.08% ± 8.9%, lowers neutrophil infiltration, and decreases oxidative stress (Fig. 2A–C; n = 3 pups from 3 distinct litters with standard error shown).
Fig. 2.
Scnn1b transgenic lung phenotype under chronic hyperoxia (A) Percent destructive Index (DI) calculated from histological sections of transgenic Scnn1b and WT pups housed under FiO2 85% or room air from birth to PN 11, not shown. (B) Bronchoalveolar lavage fluid (BALF) cell count of % neutrophils in WT and transgenic Scnn1b neonates housed under 85% or 21% O2 from birth to PN 11. (C) BALF GSH/GSSG redox potential (Eh) measurements obtained from transgenic Scnn1b and WT lungs maintained on chronic FiO2 85% or room air. All pairwise multiple comparisons were evaluated in 2A-C using ANOVA tests in SigmaPlot 14.0 (Systat Software Inc); p values as indicated. (D–F) Total RNA was prepared from transgenic Scnn1b and WT lungs maintained on chronic FiO2 85% and quantitative RT-qPCR was used to assess gene expression levels of various fibrotic markers (FN-1, ACTA2, and COL4A1); gene expression data are normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Data are mean ± SD, n = 3 animals from 3 litters; * = p < 0.05 using Student's t-test. (G–I) Relative log2 abundances (mean centered) of lung biomarkers (NGAL, LG3BP, and BIP) in BALF of WT or transgenic Scnn1b neonates housed under 85% or 21% O2 from birth to PN 11. BALF was analyzed from across 4 litters, where 4–6 samples were analyzed per group. Data are mean ± SD; * = p < 0.05 using Student's t-test.
Deposition of collagen and actin reorganization are key steps in the response to lung injury and inflammation. Fig. 2D–F shows that mRNA levels of genes involved in the pro-fibrotic responses namely FN1, ACTA-2, and COL4A1 are significantly lower in transgenic Scnn1b lungs vs WT lungs maintained under chronic 85% FiO2 exposure; baseline levels (i.e. room air) of hydroxyproline quantitation of collagen have recently been reported in Ref. [25]. We also collected bronchoalveolar lavage fluid (BALF) for proteomics analyses (Supplemental Data 8). Fig. 2G–I further shows that the levels of markers used to monitor lung injury and stress (NGAL [46], LG3BP [47], BIP(GRP78) [48,49]) in BALF as analyzed by MS proteomics are significantly higher for transgenic Scnn1b mice under normoxia compared to WT; however, hyperoxia further induces the levels of these markers in BALF only for the WT mice. Together, these data suggest that overexpression of Scnn1b, while itself exhibits elevated level of lung injury and inflammation, could potentially confer a protective effect against high oxygen exposure-mediated lung damage/inflammation and fibrosis. To better understand this effect and the role of oxidative modifications on hyperoxia-induced lung injury, we conducted a proteome-wide analysis of lung tissues from neonatal wild type and transgenic Scnn1b littermates housed under FiO2 85% and room air.
3.2. S-glutathionylation proteomes and global proteomes of neonatal mouse lung
To quantify the SSG proteomes in mouse lung under both normoxia and hyperoxia, a RAC-TMT based redox proteomics approach was used (Fig. 1B). For each genotype, lung samples from mice kept at either 21% or 85% O2 (n = 4) were processed for the detection of in vivo S-glutathionylation levels. In addition, a “Total Thiol” sample was created by pooling a small fraction from each of the SSG samples in the same TMT set. For the “Protein-SSG” samples, the workflow consisted of blocking protein free thiols with NEM, selective reduction of protein-SSG modifications with a glutaredoxin enzyme cocktail [50], enrichment of the nascent thiols (products of reduction of protein-SSGs) with Thiopropyl Sepharose 6B resin, on-resin trypsin digestion, and subsequent TMT labeling (Fig. 1B). For the “Total Thiol” sample, a similar processing scheme was used except that NEM blocking was omitted and the reduction was performed using DTT (Fig. 1B), thus this sample represents all free and reversibly oxidized thiols present in the sample. Quantification of total thiols reveals the overall level of protein thiols (both reduced and oxidized) present in the sample, which allows for stoichiometric estimation of average S-glutathionylation occupancy at individual Cys sites. Following TMT labeling, cysteine-containing peptides from each channel were eluted, combined, and subjected to LC-MS/MS analysis. TMT label 130N was omitted from the labeling scheme to avoid channel crosstalk from the 131N total thiol channel. In addition, we speculated that exposure to long term hyperoxia (>10 days in this study) could also result in changes in protein abundance. Therefore, we incorporated global proteome measurements into the redox proteomics workflow to detect any changes in protein abundance (Fig. 1A). Following the selective reduction step, an aliquot of 75 μg proteins from each sample (except for total thiol) were subjected to reduction, alkylation, trypsin digestion, TMT labeling, and LC-MS/MS analysis for global proteome profiling.
SSG proteome measurements identified >10,000 unique cysteine-containing peptides, which accounted for >98% of all identified peptides in both datasets, demonstrating the specificity of resin-based enrichment (Fig. S1A; Supplemental Data 1 and 2). These unique peptides cover more than 7600 Cys sites that correspond to ∼3000 proteins for each genotype. Nearly ∼6000 Cys sites and ∼2600 proteins were commonly identified from both datasets (Figs. S1B–C; Supplemental Data 1 and 2), allowing comparisons between genotypes. For global proteomics, > 3000 proteins were identified from single-shot LC-MS/MS analysis in both datasets, with 2511 proteins present in both datasets (Fig. S1D; Supplemental Data 3 and 4). The quantitative measurement of SSG and global protein abundance levels was highly reproducible among biological replicates, with median coefficients of variation (CVs) on TMT reporter ion intensities around 10% (Figs. S1E–F). Furthermore, the identified SSG proteins are distributed throughout various cellular compartments (Fig. S2A), indicating a broad coverage of the redox proteome by our workflow.
3.3. Hyperoxia-induced changes in global protein expression
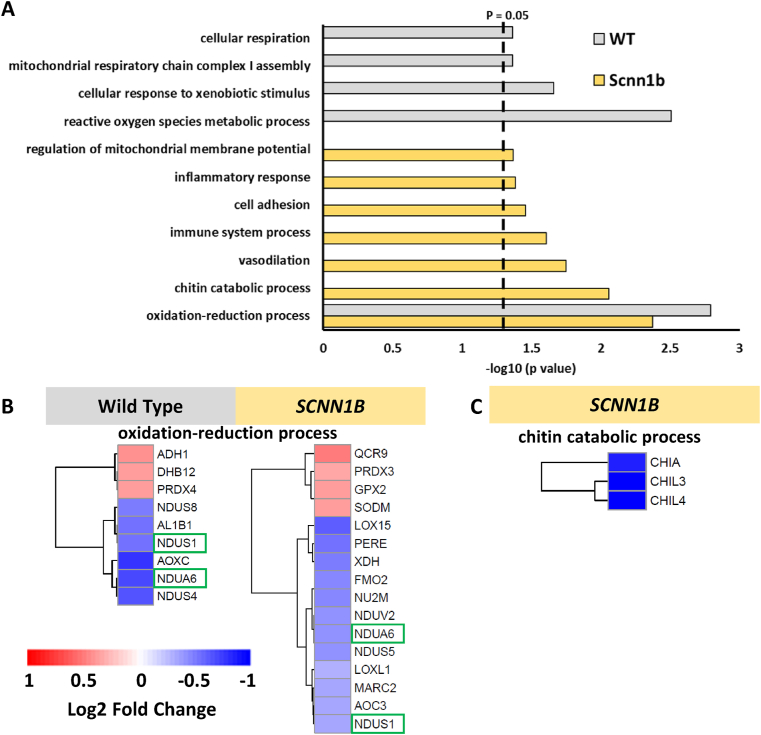
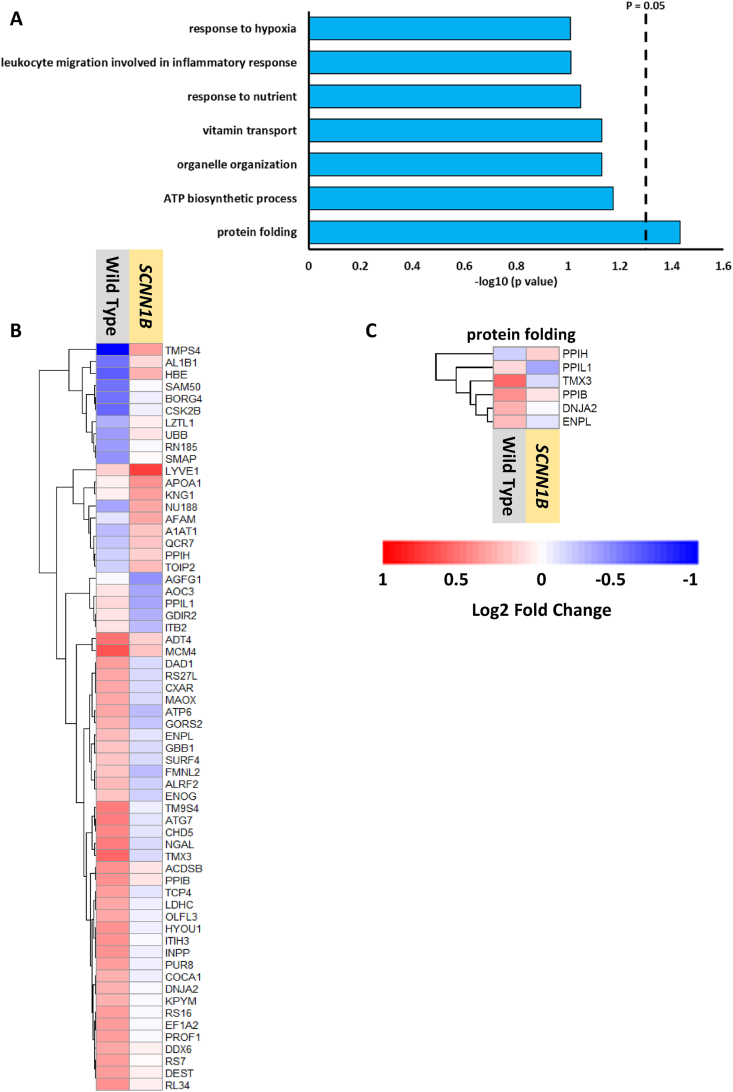
To identify proteins with significant changes in their abundance following hyperoxia exposure in each genotype, we used a threshold of a log2 fold change that is greater than ±0.3 (upregulated or downregulated) followed by a cutoff of an adjusted p value that is less than 0.1. Volcano plots illustrate the distribution of proteins that meet these criteria, where hyperoxia induces changes in the abundance of 32 proteins (15 increase; 17 decreased) in wild type mice, while transgenic Scnn1b mice exhibited 95 significantly changing proteins (35 increased; 60 decreased) (Figs. S3A–B). Gene ontology analysis shows that “oxidation-reduction process” is the most significant and only pathway common to both genotypes (Fig. 3A). NADH dehydrogenase 1 alpha subcomplex subunit 6 (NDUA6) and NADH-ubiquinone oxidoreductase 75 kDa subunit (NDUS1) are observed in both genotypes for this pathway, where each exhibited reduced abundance under hyperoxic conditions (green boxes; Fig. 3B). Other proteins related to this process and observed exclusively for either genotype include NADH dehydrogenase iron-sulfur protein 4/8 (NDUS4/NDUS8) in wild type or NADH-ubiquinone oxidoreductase chain 2 (NU2M), NADH dehydrogenase flavoprotein 2 (NDUV2), or NADH dehydrogenase iron-sulfur protein 5 (NDUS5) in Scnn1b overexpressing lung. This abundance of downregulated mitochondrial complex I proteins in either genotype emphasizes this pathway as potentially sensitive to hyperoxia. Other pathways such as “mitochondrial respiratory chain complex I assembly” and “cellular respiration” in wild type mice indicate a perturbation to bioenergetic processes following hyperoxia treatment. Downregulated expression of these proteins could lead to mitochondrial dysfunction, especially NDUS1, NU2M, or NDUV2, which are core subunits for function and structure of complex I in the mitochondria [51,52]. A similar finding on reduced expression of complex I mitochondrial proteins following 85% oxygen was previously reported [53] and hyperoxia-induced mitochondrial damage is also proposed to cause simplification of lung structure, or bronchopulmonary dysplasia (BPD) in infant lungs [54].
Fig. 3.
Hyperoxia impacts the abundance of proteins belonging to specific biological processes. (A) Bar plot representing biological processes recovered from gene ontology analysis of proteins with significant changes in abundance following hyperoxia in either genotype. The dashed line denotes the cutoff for category significance (p < 0.05) on the -log10 transformed x axis. (B) Heat map representing the log2 fold change values of proteins in wild type or transgenic Scnn1b mice that belong to the most significant category in both datasets, “oxidation-reduction process”. Protein names with green boxes denote that they are shared between both genotypes. (C) Heat map representing the log2 fold change values of proteins belonging to the “chitin catabolic process” category in the transgenic Scnn1b dataset. The heat map scale in panel (B) also applies to the heat map in (C). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
In contrast, Scnn1b overexpressing lung show significant changes in protein expression for a wider range of processes, including immune and inflammatory signaling. Interestingly, transgenic Scnn1b mice maintained under chronic hyperoxia show perturbed chitin catabolism based on recovery of “chitin catabolic process” (Fig. 3A). Chitinase-like protein 3/4 (CHIL3; CHIL4) and the acidic mammalian chitinase (CHIA) are significantly downregulated in Scnn1b overexpressing lungs (Fig. 3C). These proteins bind and degrade chitin and other forms of saccharides that are found on pathogens, but also extends to chitin polymers present in airways of lungs with fibrotic diseases [55]. Different cell types found within the lung, such as macrophages and neutrophils, can secrete chitinases [56,57]. In our study, we observed a significant reduction in chitinase content in transgenic Scnn1b lungs maintained under chronic hyperoxia (Fig. 3C), which may result from the marked reduction of neutrophil infiltration in these mice (Fig. 2B). A previous study found that deficiency of chitinase CHIA, leads to the accumulation of chitin polymers and a fibrotic lung phenotype [58]. Other members of the chitinase family were identified in our dataset (CH3L1: Chitinase-3-like protein 1 and CHID1: Chitinase domain-containing protein 1; Supplemental Data 4), however they did not exhibit significant changes in expression. Together, the global proteomics data revealed that hyperoxia induces more diverse changes in protein expression in transgenic Scnn1b mice than wild type mice.
3.4. Hyperoxia-induced changes in protein-SSG modifications
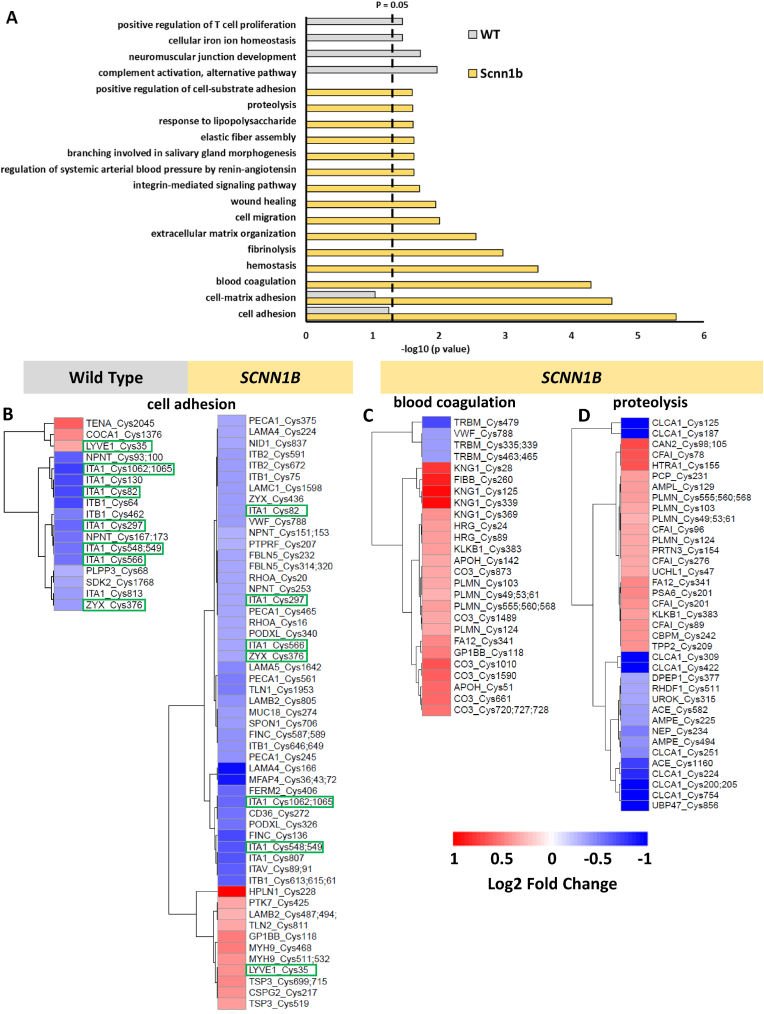
To find proteins and pathways that are responsive to hyperoxia via SSG regulation, we first identified differentially modified Cys sites. The impact of hyperoxia on the S-glutathionylation proteome is visualized by plotting the extent of change (log2 fold change) of all identified S-glutathionylated unique Cys sites (Figs. S3C–D). These plots show that the S-glutathionylation proteome was altered by hyperoxia in both genotypes, however the perturbation is greater in transgenic Scnn1b mice. The distribution of proteins with altered Cys site S-glutathionylation were found across all major subcellular compartments (Figs. S2B–C), suggesting that long-term exposure to hyperoxia caused a systematic change in the SSG proteome across multiple organelles. Cys sites with significant changes in SSG levels were identified on the basis that they met the same criteria as defined for the global data (log2 fold change > ±0.3 and adjusted p value < 0.1). In wild type mice, 85 proteins yielding 110 unique Cys sites showed a significant increase (25) or decrease (85) in S-glutathionylation, respectively (Fig. S3C). Transgenic Scnn1b mice exhibited a greater abundance of proteins (269) with 425 significantly altered S-glutathionylated Cys sites (190 increased and 235 decreased) (Fig. S3D). Notably, while S-glutathionylation of Cys98 of lung β-ENaC subunit protein was detected in wild type and Scnn1b overexpressing lung, there was no significant change in SSG at this site following hyperoxia for either genotype (Supplemental Data 1 and 2). Given the rapid and continuous oxidation of thiols that can occur in vivo, it remains unclear whether glutathionylation of β-ENaC subunit directly mediates channel function, or if intermediate redox-sensitive modulators (yet to be identified) alters effective ion transport in the lung under oxidative stress. Consequently, we examined what other proteins exhibit significant changes in SSG levels to uncover alternative processes that may contribute to the protective effect of Scnn1b overexpression during hyperoxia. Gene ontology of these proteins with significant changes in SSG modifications showed that a variety of biological processes are distinctly over-represented in either wild type or transgenic Scnn1b mice under hyperoxia (Fig. 4A). Their enrichment not only highlights what biological processes are perturbed by hyperoxia, but more importantly provides a list of novel candidate proteins that may mediate the effect of hyperoxia via SSG regulation.
Fig. 4.
Hyperoxia impacts glutathionylation of proteins associated with multiple biological processes in mouse lung. (A) Bar plot representing the biological processes recovered from gene ontology analysis of proteins with significant changes in Cys site glutathionylation following hyperoxia in either genotype. The dashed line denotes the cutoff for category significance (p < 0.05) on the -log10 transformed x axis. (B) Heat map representing the log2 fold change values of Cys site glutathionylation in wild type or transgenic Scnn1b mice that belong to the “cell adhesion” category. Cys sites with green boxes denote that they are shared between both genotypes. (C) Heat map representing the log2 fold change values of Cys sites belonging to the “blood coagulation” category in the transgenic Scnn1b dataset. (D) Heat map representing the log2 fold change values of Cys sites belonging to the “proteolysis” category in the transgenic Scnn1b dataset. The heat map scale below panel C–D also applies to the heat map in (B). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
“Cell adhesion” was identified as the most over-represented category in transgenic Scnn1b mice, which is in agreement with the assertion that cell adhesion molecules/proteins are a major part of fibrotic lung pathologies [59]. Strong representation of cell-adhesion in transgenic Scnn1b mice is supported by the presence of many perturbed Cys sites in the dataset, while wild type mice have fewer Cys sites (Fig. 4B), resulting in less significant representation (p value = 0.0915; Fig. 4A). Nonetheless, either genotype contains significantly altered cell adhesion-related glutathionylation Cys sites that are predominantly downregulated (Fig. 4B). In particular, five downregulated integrin alpha-1 (ITA1) glutathionylation Cys sites were observed in both wild type and transgenic Scnn1b datasets (green boxes; Fig. 4B), suggesting that this protein is sensitive to redox regulation under hyperoxia. A previous study found glutathionylation of neutrophil ITA4 was necessary for its interaction with vascular adhesion molecule 1 (VCAM-1) on endothelial cells [60], which may be relevant to the observation of reduced neutrophil cell count in lung tissue that overexpresses Scnn1b under hyperoxic conditions (Fig. 1B). In contrast to “cell adhesion”, a group of high-ranking biological processes related to blood clotting (Fig. 4A; “blood coagulation”, “hemostasis”, “fibrinolysis”) have proteins with Cys sites that exhibited predominantly upregulated glutathionylation following hyperoxia in transgenic Scnn1b mice (as represented by “blood coagulation” in Fig. 4C). This suggests that blood clotting may be a hyperoxia-sensitive process and a contributor to the protective effect of Scnn1b overexpression. These upregulated Cys sites may play a potentially important redox-dependent role since the corresponding proteins belong to processes that are involved in fibrotic lung pathology [61].
A more in-depth view of the “proteolysis” category revealed a variety of proteins that have Cys sites with altered SSG levels (Fig. 4D). Notably, the Calcium-activated chloride channel regulator 1 (CLCA1) has 7 Cys sites that exhibited a strong and consistent downregulation of glutathionylation following hyperoxia. CLCA1 functions as a protease to regulate mucus production in the colon [62,63] as well as the lung [64] and has additional putative roles in cancer and inflammatory and immune responses [65]. Among the CLCA1 Cys sites we identified, 2 are highly conserved and are located within the metalloprotease domain of the catalytic region (positions 125 and 187) that is used for its self-cleavage and activity [66]. Cys sites 754 and 856 recovered in our dataset correspond to the C-terminal region, which may participate in intermolecular disulfide bridges that form C-terminal homodimers and lead to large oligomers, as was shown in a previous report [62]. The von Willebrand factor (VWF) type A (VWA) domain is another region of CLCA1 that is important for interaction with and activation of the calcium-activated chloride channel TMEM16A [67]. This VWA domain contains 3 highly conserved Cys sites (positions 309, 387, and 422), where sites 387 and 422 form a disulfide to promote folding and stability, while site 309 may contribute to domain folding [68]. We observed sites 309 and 422 in our datasets, suggesting that glutathionylation could be a potential regulatory mechanism of CLCA1's VWA domain and may be important for modulating TMEM16A activity. Interestingly, the VWA domain is found within many proteins associated with cell adhesion, including integrins [69,70], meaning that glutathionylation of this domain could be a common regulatory mechanism of interaction with other binding partners in lung physiology.
Since we observed that hyperoxia induces both increases and decreases in SSG levels, we next quantified the redox occupancy to see whether hyperoxia leads to an overall shift of S-glutathionylation. Under normal conditions, the median SSG occupancy was 3.24% in wild type (Fig. S4A), consistent with our observations on basal SSG occupancy levels in cultured cells and other mouse tissues such as heart and muscle [41,71]. No apparent shift of the SSG occupancy distribution was observed under hyperoxia (Fig. S4C), suggesting that hyperoxia did not cause an overall change in redox status. Likewise, similar SSG occupancy distributions were seen in both control and hyperoxia exposed Scnn1b overexpressing lung (Fig. S4B,D).
3.5. Hyperoxia-induced changes in protein total oxidation
To further corroborate our findings on protein-SSG, we performed an independent experiment to measure protein thiol total oxidation, which includes all forms of reversible oxidation and thus represents the overall redox status in transgenic Scnn1b mice. A similar workflow was used, where free thiols are initially blocked with NEM (except for the total thiol sample) and DTT was used as the reductant to reduce all reversible protein oxidation, in contrast to the selective reduction of protein-SSG by glutaredoxin utilized in SSG workflow (Fig. 1B; compare “Total Oxidation” to “Protein-SSG”). We identified a total of 10,836 unique peptides, 10,629 of which were cysteine-containing peptides (Fig. S1A) and amounts to more than 7700 Cys sites (Supplemental Data 5). Like PSSG and global protein abundance, the quantitative measurement of total oxidation levels was highly reproducible among biological replicates, with median coefficients of variation being less than 10% (Fig. S1G). Consistent with the SSG data, we observed that hyperoxia causes a similar bidirectional change of protein total oxidation (Fig. S3E). Therefore, we used the same cutoff for significance (log2 fold change > ±0.3 and adjusted p value < 0.1) to identify 428 proteins with 764 Cys sites exhibiting a significant increase (517) or decrease (247) in total oxidation upon hyperoxia exposure.
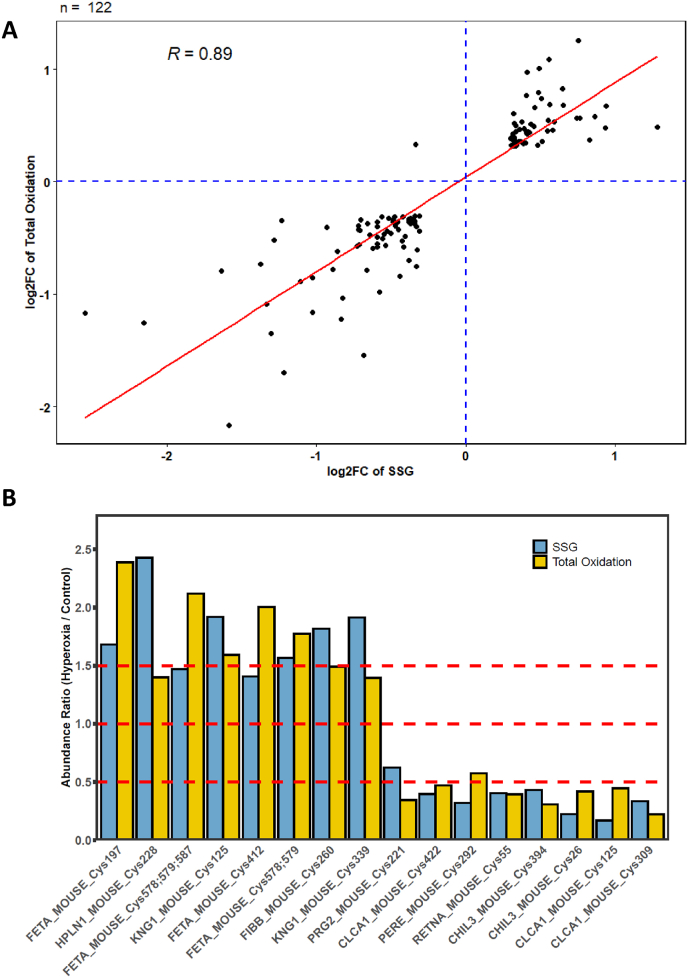
Proteome-wide estimations of stoichiometry also showed a minimum shift in total Cys site oxidation occupancy, with the median value increasing from ∼31.5% under normal air to ∼32.6% under hyperoxia (Figs. S4E–F). We note that the median total oxidation occupancy in the lung was much higher than a typical 10–15% occupancy observed in cultured mammalian cells and other tissue types. Indeed, the higher median occupancy observed in the total oxidation of the lung tissue obtained from Scnn1b overexpressing mice is comparable to what is found in the rat lung [42], suggesting the lung as a highly oxidized environment. These findings suggest that SSG may not be a primary oxidative modification in the lung under normal or hyperoxic conditions, as it only makes up a small fraction of total site occupancy. Next, we explored the correlation between Cys site SSG and total oxidation. To this end, we plotted the extent of changes of significant Cys sites commonly identified in both SSG and total oxidation datasets (122 sites total) (Fig. 5A). We observed a strong correlation between the two datasets, supporting the confidence level in quantifying Cys SSG and total oxidation in two separate experiments. The data also highlighted proteins whose Cys site oxidation levels show consistent perturbation by hyperoxia at the SGG or total oxidation level, suggesting that SSG is a good surrogate for protein cysteine oxidation levels. Prominent examples such as Alpha-fetoprotein (FETA) and Kininogen-1 (KNG1) showed increases greater than 1.5X, while others such as chitinase-like protein 3 (CHIL3) and Calcium-activated chloride channel regulator 1 (CLCA1) show a decrease in both SSG and total oxidation levels (Fig. 5B).
Fig. 5.
Total thiol oxidation of Cys sites in Scnn1b overexpressing mice following hyperoxia. (A) Correlation of log2 fold change values for Cys sites found in both SSG and total thiol oxidation datasets. (B) Representative Cys sites found in both datasets showing an increase or decrease in thiol oxidation following exposure to hyperoxia. Cys sites are plotted as raw, non-log-transformed fold changes. Dashed lines denote fold change thresholds of 0.5 (decrease), 1 (no change), and 1.5 (increase), as plotted on y axis.
3.6. Differential regulation of the global proteome contributes to the protective role of SCNN1B
The redox and global proteomics data showed that hyperoxia alters the abundance or redox status of specific proteins under hyperoxia in both the wild type and transgenic Scnn1b mice. Next, we aimed to identify proteins that could contribute to the protective role of β-ENaC against hyperoxia-induced lung injury. Fig. 2 shows that overexpression of Scnn1b contributes to lung injury at room air, but dampens lung destruction, neutrophil migration, and redox status at FiO2 85%. The specific protein(s) that may play a protective role against oxidative conditions in these Scnn1b transgenic mice remain unclear. To compare the effect of hyperoxia on the two genotypes directly and quantitatively, we integrated the hyperoxia induced fold-change data for both genotypes. We used the same cutoff (log2 fold change > ±0.3 and adjusted p value < 0.1) to identify any significant differences in hyperoxia induced perturbation at the SSG and protein abundance levels between the two genotypes (Supplemental Data 6–7). At the SSG level, only 1 Cys site was identified as significant: Cys 734 on ATP-dependent RNA helicase A (DHX9) (Supplemental Data 6). Similar analysis of total protein abundance revealed 62 differentially regulated proteins between wild type and transgenic Scnn1b mice (Fig. S3F, Supplemental Data 7). These proteins were involved in diverse biological processes including protein folding, ATP synthesis, and organelle organization (Fig. 6A). Many of these proteins show increased abundance in the wild type (Fig. 6B) mice, including the proteins belonging to the single most significantly enriched category, protein folding (Fig. 6C), a process tightly associated with oxidative stress. In line with our previous observation of oxidative stress as a prominent theme in wild type mice (Fig. 3A), upregulation of protein folding might be a compensatory response to hyperoxia, where protein folding is needed to prevent accumulation of aberrantly oxidized and misfolded proteins [72]. This may be further corroborated by the observation of the next highest ranking category “ATP biosynthetic process”, as ATP is a necessary cofactor for the protein folding/chaperone function of some proteins, such as heat shock protein 90B1 (ENPL; endoplasmin) [73].
Fig. 6.
Overexpressing Scnn1b results in differential regulation of hyperoxia-induced protein abundance changes. (A) Gene ontology analysis showing biological processes enriched from proteins that exhibit significant changes in abundance following exposure to hyperoxia and are significantly different between transgenic Scnn1b and wild type mice. The dashed line denotes the cutoff for category significance (p < 0.05) on the -log10 transformed x axis. (B) Heat map representing the log2 fold changes (hyperoxia versus control) of each significant protein identified in the analysis for each genotype (Supplemental Data 7). (C) Log2 fold changes of proteins associated with the “protein folding” category for each genotype.
4. Discussion
The current study provides quantitative profiling of the proteome and thiol redox proteome of the mouse lung under both normoxia and hyperoxia conditions, through utilization of a wild type and a genetically altered mouse model overexpressing Scnn1b, the gene that codes for β-ENaC. The lung is a highly complex tissue comprised of over 40 cell types [74]—constantly exposed to both highly oxidative conditions and environmental toxins, and thus must maintain redox homeostasis for proper function. In addition to the many exogenous sources that promote ROS generation, lung cells also generate ROS either as byproducts of aerobic metabolism or by various highly controlled biological pathways in immune defense and other cellular processes [75,76]. The role of ROS as signaling molecules has been firmly established, and specific modifications of thiols on redox sensitive proteins represents a central mechanism by which ROS mediates downstream cellular activities. This study not only created a comprehensive atlas of the lung redox proteome with site-specific occupancy estimates, but also evaluated the overall impact of hyperoxia on the redox proteome.
4.1. Hyperoxia is not a strong inducer of protein S-glutathionylation
Comparing the SSG proteomes under normoxia and hyperoxia conditions for both genotypes (WT and hemizygous Scnn1b mice) revealed that hyperoxia does lead to increasing or decreasing changes in Cys site glutathionylation (Figs. S3C–D). However, this does not induce a significant overall increase in redox occupancy (Fig. S4), most likely due to a newly established redox homeostatic baseline under hyperoxia conditions. Instead, non-stochastic redox regulation of specific proteins was observed under hyperoxia exposure. We found that the basal level of SSG modifications is low, with a median level no greater than 4.2% in both wild type and transgenic Scnn1b mice under normoxia conditions (Figs. S4A–B). This data aligns well with the overall SSG occupancy observed in other tissue types [41], suggesting that a basal level of ROS production may mediate formation of redox modifications to maintain lung homeostasis without any apparent oxidative stress. At the most, we observed that roughly 5% of all quantified Cys sites exhibited significant differential SSG modification between normoxia and hyperoxia for each genotype. Given this outcome, it is not surprising that our bio-molecular attempts at measuring oxidized Cys sites using fluorescein-5-maleimide (F5M) labeling and SDS-PAGE analysis (as described in Ref. [77]) could not delineate significant differences in redox sensitive Cys modifications in WT and Scnn1b overexpressing mouse lung protein. Normalized F5M for WT and transgenic Scnn1b pups maintained on chronic 85% O2 were 0.99 ± 0.08 and 1.05 ± 0.15, respectively, from 6 independent observations/litters (data not shown). Hyperoxia-induced changes of the SSG proteome were small compared to the overall shift in SSG levels observed in other biological systems such as macrophages in response to nanomaterial treatment [78] and mouse skeletal muscle under fatiguing contractions [41].
The notion that hyperoxia does not result in drastic perturbation of the overall redox status is also supported by only moderate changes observed in abundance changes for proteins involved in ROS clearance. For example, we found the protein abundances of SODM (superoxide dismutase Mn, mitochondrial) and PRDXs 3 and 4 (Thioredoxin-dependent peroxide reductase), which are important intracellular ROS scavenging enzymes, only exhibited moderate, but still significant increases in wild type or transgenic Scnn1b mice under hyperoxia (Fig. 3B). A similar observation was made in mouse lung under 75% O2 using immunoblot [79]. Other forms of SOD such as SODC and SODE, as well as catalase, do not show significant increases in protein abundance under hyperoxia. As multiple non-enzymatic and enzymatic antioxidant defense systems exist in the lung, it is likely that a new redox balance and baseline are established in response to long term hyperoxia under our experimental conditions. This is in contrast to short durations of hyperoxia, in which significant increases in mitochondrial ROS are observed [80].
Further evidence supporting the overall small impact of hyperoxia on redox homeostasis of the lung is provided by examining proteome-wide total oxidation, where the overall shift of the median occupancy from normal to hyperoxic conditions was roughly ∼1% (Figs. S4E–F). Interestingly, we observed a high correlation between our SSG and total oxidation data, which suggests that the biological change and response is consistent between this specific modification, versus overall Cys site oxidation (Fig. 5). These findings also support the notion that multiple forms of thiol modifications could coexist on the same cysteine residues [[81], [82], [83]], where SSG only comprises a small fraction of total oxidation sites.
It is also noteworthy that basal oxidation occupancy in the lung is relatively high (∼31.5% as the median; Fig. S4E). While previously we observed ∼22% oxidation in rat lung [42], using a similar experimental approach, 5–20% total oxidation in cyanobacteria [84] and 11.9% total oxidation in mouse macrophages were observed [32]. Using a thiol trapping technique, (OxICAT, reviewed in Ref. [85], Leichert et al. report that >20% of the E. coli proteome is reversibly oxidized under aerobic growth conditions. Moreover, Xiao et al., recently reported that the lung has the highest average oxidation level compared to other types of murine tissue studied [86]. Theoretical estimates by D.P. Jones indicate that 5% of the global thiol content could be oxidized per minute [87]. Given these published reports, and since the lungs are constantly exposed to high levels of oxygen [88], it is plausible that a large proportion of the lung proteome is indeed oxidized (and that the high basal measurements are not in large part due to artifacts in tissue storage/preparation).
4.2. SSG changes points to potential regulatory mechanisms at the molecular level
We and others, have shown that Scnn1b overexpression attenuates normal lung development, enlarges airways, and increases susceptibility to spontaneous infection during the early neonatal period [22,25,35]. Interestingly, exposing transgenic Scnn1b pups to chronic FiO2 85% facilitated normal alveologenesis [25] and accordingly decreased the % DI and neutrophil infiltration in the present study (as shown in Fig. 2 compared to Scnn1b overexpressing pups maintained at room air and hyperoxic WT pups). Subsequently, proteomics of lung BALF revealed that the protein abundances of markers associated with lung injury or stress in transgenic Scnn1b pups maintained under chronic hyperoxia were not elevated compared to the normoxia group (Fig. 2G–I). Given these observed differences in Scnn1b over-expressing and WT lung phenotypes at 21% and 85% O2, we were surprised to observe no significant change in ENaC glutathionylation in response to hyperoxia for either genotype in our current data, and as such, we evaluated alternative post-translational mechanisms that may be involved. Moreover, prior research has shown increased levels of GSSG (whether administered by insufflation or induced by hyperoxia) leads to attenuated ENaC activity [17,25]. This observation also warranted additional research into translational and post-translational regulation using a mass spec approach, especially since high oxygen tensions are known to increase ENaC expression in fetal distal lung epithelial cells [89,90]. The perturbed biological processes identified in Fig. 4 (and discussed below) may provide insight into the underlying mechanisms behind the protective effect of ENaC overexpression during hyperoxia.
Depending on the specific protein being modified, the reversible nature of SSG modifications could have a direct regulatory role on protein function. For instance, formation of protein-SSG on the active site of glyceraldehyde-3-phosphate dehydrogenase (G3P) protects the protein from irreversible oxidation, resulting in complete inactivation of enzyme under oxidative stress [91]. In addition to some of the molecular mechanisms that were highlighted for proteins with significantly altered SSG status (ITA1, CLCA1; see Results section), the data in this study is rich in many other potentially biologically significant sites. Our study identified 2 significantly altered glutathionylation sites in Scnn1b overexpressing mice that are found at or near annotated active sites (Supplemental Data 2), suggesting that SSG-modifications on these Cys sites provides a direct link between redox regulation and its biological consequences. Mitochondrial thiosulfate sulfurtransferase (THTR; TST) was identified in our dataset as a protein with significant downregulation of SSG at the active site Cys248 in mice overexpressing Scnn1b in the lungs. (Supplemental Data 2). This enzyme was described to have a role cyanide detoxification, likely through its catalytic Cys248 site, which uses sulfur-containing donors such as thiosulfate to form persulfides at the active site, which are then transferred to an acceptor, such as cyanide or even glutathione [92,93]. Persulfidated glutathione is capable of serving as the sulfur donor for THTR as well, and is more catalytically efficient in this way [92], leading to regeneration of reduced glutathione. Downregulated SSG at the catalytic site in transgenic Scnn1b mice might be indicative of a more active THTR enzymatic state, however more work is needed to better understand the implications of this mechanism.
The active site in the catalytic subunit of the calcium regulated thiol protease Calpain-2 (CAN2) contains a Cys site at position 105, which was also identified in our data as having significantly upregulated glutathionylation in Scnn1b overexpressing mice following hyperoxia, along with Cys 98 on the same peptide of origin (Fig. 4D). While we are unable to resolve whether one or both sites underwent a significant change in SSG, a modification on the active site Cys105 and/or in close proximity at Cys98, which is near a Calcium ion binding site [94], is suggestive of modulation of Calpain function. Studies have shown that inhibition of Calpain by calpeptin leads to a reduction in bleomycin-induced pulmonary lung fibrosis, which was also evidenced by a marked reduction in transforming growth factor β1 that contributes to lung fibrosis by inducing synthesis of matrix proteins [95,96]. Glutathionylation may have a similar effect on Calpain as calpeptin, giving rise to an anti-fibrotic response in the lung that may also have benefits that extend beyond the lung, based on Calpain's ubiquitous expression and involvement in other pathologies [97]. Together, our data uncovered numerous glutathionylation sites that may exert a variety of modulatory effects on proteins and related biological processes that are responsible for the protective role of Scnn1b overexpression under a hyperoxic environment.
4.3. Several biological processes are involved in the protective effect of SCNN1B against hyperoxia
Individual SSG proteome-wide analysis between normoxia and hyperoxia for each genotype revealed the differential effects of high O2 concentrations, leading us to uncover protein signatures and mechanisms that contribute to the protective role of β-ENaC in the lungs. In wild type mice, hyperoxia appears to induce an immunoregulatory response, based on the recovery of the top ranking category “complement activation, alternative pathway” (Fig. 4A). In contrast, the most significantly enriched biological process in transgenic Scnn1b mice was cell adhesion, suggesting that regulation of adhesion molecules is a critical factor determining the outcome of hyperoxic lung injury. This is consistent with other findings that adhesion molecules contribute to the protective effect of nitric oxide inhalation against hyperoxic lung injury [98]. Intriguingly, comparison of cell adhesion Cys sites found in both wild type and transgenic Scnn1b mice both show the same trends of hyperoxia-induced modulation of glutathionylation (green boxes; Fig. 4B), suggesting that this process could be part of a common pathway used in response to high oxygen exposure. In transgenic Scnn1b mice, this response pathway is likely more complicated due to the altered physiological state of the fibrotic lung, hence the enrichment of other pathways like blood coagulation and proteolysis (Fig. 4C–D), which may help to confer protection against hyperoxia. Indeed, proteins belonging to these categories exhibited significant changes in Cys site SSG levels that may be important. A previous report demonstrated the modulatory effects of shifts in the GSH/GSSG equilibrium, where increased levels of GSSG prolongs clotting time and thiol oxidation was speculated as the mechanism of inhibition [99]. For example, thrombomodulin (TRBM), an inhibitor of the pro-fibrotic role of the enzyme thrombin [100], shows several downregulated SSG Cys sites that may lead to its activation and therefore prevent thrombin-mediated deposition of fibrin in the lung. Coincidentally, fibrinogen (FIBB) has a strongly upregulated SSG site, which may impair its ability to be cleaved by thrombin and polymerize into a blood clot or a matrix deposition in the lung. Glutathionylation of multiple Cys sites on plasminogen (PLMN) could prevent it from proper function, which includes binding and dissolving fibrin-containing clots [101]. Collectively, these results suggest that up- or down-regulation of SSG on different proteolysis proteins may be part of a combinatorial mechanism where coagulation and protein degradation are selectively modulated in response to hyperoxia to confer protection against high oxygen levels in transgenic Scnn1b mice.
Direct comparison of the hyperoxia induced protein abundance changes between the wild type and transgenic Scnn1b mice (Supplemental Data 7) revealed fewer significant biological processes compared to the genotype specific SSG analysis. Nonetheless, we found that protein folding is the most significant biological process that is altered between wild type and transgenic Scnn1b mice in terms of significant changes in protein expression (Fig. 6A). Together with the redox proteomics data, these datasets provide novel candidate proteins and biological processes to further study the mechanisms behind β-ENaC as a protective molecule against lung hyperoxia.
5. Conclusion
Our data presents a unique landscape of the lung thiol redox proteome, which together with the global proteome indicates that hyperoxia does not elicit a drastic perturbation in redox homeostasis. In addition, we found Scnn1b overexpression may protect against hyperoxia-induced lung injury via modulation of specific processes such as cell adhesion, blood coagulation, and proteolysis. Novel findings including regulation of SSG on specific proteins deepen our understanding of the landscape of redox regulation of the lung in response to high O2. Together with the quantification of protein total oxidation, our current study underlines the importance of redox regulation in lung.
Data availability
All mass spectrometry raw data and processed proteomics data are available upon request.
Declaration of competing interest
The authors have no conflicts of interest to declare.
Acknowledgments
Portions of this work was supported by R01HL137033 awarded to MNH. The proteomic experimental work described herein was performed in the Environmental Molecular Sciences Laboratory, Pacific Northwest National Laboratory, a national scientific user facility sponsored by the Department of Energy under Contract DE-AC05-76RL0 1830.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2022.102405.
Contributor Information
Wei-Jun Qian, Email: Weijun.Qian@pnnl.gov.
My N. Helms, Email: my.helms@hsc.utah.edu.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Dataset metrics. (A) Resin assisted capture identifies more than 10,000 peptides in wild type and transgenic Scnn1b mice exposed to control or hyperoxic conditions in SSG or total oxidation workflows, where more than 98% of these peptides contain cysteine. (B) The more than 10,000 SSG-modified peptides correspond to >7600 Cys sites for each genotype, with nearly 6000 Cys sites being common to both wild type and transgenic Scnn1b mice. (C) More than 3000 proteins contain a SSG-modified Cys site in each genotype, with almost 2600 of these proteins found in both wild type and transgenic Scnn1b mice. (D) Global proteomics identifies more than 3000 proteins in both wild type and transgenic Scnn1b mice, with ∼2500 present in both samples. (E-F) Coefficients of variation of TMT reporter ion intensities for SSG-modified Cys sites (E), proteins in global (F), or total oxidation of Cys sites (G) identified in this study for both genotypes, where the dashed line represents the 10% boundary as a reference.
Subcellular distribution of PSSG-modified proteins. (A) Scatter plot illustrating how many glutathionylated proteins belong to a specific compartment in cells obtained from transgenic Scnn1b mice. (B–C) Box and whisker plots illustrating the fold change differences in Cys site SSG levels for proteins that reside in a specific subcellular compartment in wild type (B) or Scnn1b overexpressing (C) mice. The red and blue dashed lines denote the log2fold change cutoffs of 0.3 and −0.3, respectively, which are plotted on a non-log-transformed y axis.
Visualization of the impact of hyperoxia on global and redox proteomes. (A-F) For each panel, the plot on the left represents all features identified (protein or Cys site; as specified in the following legend descriptions), while the plot on the right illustrates all features that met the significance cutoff of log2 fold change > 0.3 and p adjusted < 0.1 (red = upregulated; blue = downregulated). (A-B) Volcano plots representing the log2 fold changes in protein expression under hyperoxic versus control conditions at the protein level for wild type (A) and transgenic Scnn1b mice (B). (C-D) Volcano plots representing the log2 fold changes in SSG modifications at the Cys site level under hyperoxic versus control conditions for wild type (C) and transgenic Scnn1b mice (D). (E) Volcano plots representing the log2 fold changes in total oxidation under hyperoxic versus control conditions at the Cys site level for transgenic Scnn1b mice. (F) Volcano plots representing the integrated global abundance log2 fold change of proteins found in both wild type and transgenic Scnn1b mice. The values in this plot represent how much more change was observed in response to hyperoxia in transgenic Scnn1b mice versus wild type mice for a given protein (red = upregulated in transgenic Scnn1b/downregulated in wild type mice; blue = downregulated in transgenic Scnn1b/upregulated in wild type mice).
Occupancy distribution of Cys sites in each genotype. (A-F) Data was transformed to plot on log10 scale and the red vertical line denotes the median occupancy in the dataset. (A,C) The distribution of SSG occupancy on Cys sites in wild type mice under control and hyperoxic conditions with minimal change between conditions. (B,D) The distribution of SSG occupancy on Cys sites in transgenic Scnn1b mice under control and hyperoxic conditions with minimal change between conditions. (E,F) The distribution of total oxidation occupancy on Cys sites in Scnn1b overexpressing lungs under control and hyperoxic conditions with minimal change between conditions.
References
- 1.Bhandari V. Hyperoxia-derived lung damage in preterm infants. Semin. Fetal Neonatal Med. 2010;15(4):223–229. doi: 10.1016/j.siny.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bourbon J., Boucherat O., Chailley-Heu B., Delacourt C. Control mechanisms of lung alveolar development and their disorders in bronchopulmonary dysplasia. Pediatr. Res. 2005;57(5 Pt 2):38R–46R. doi: 10.1203/01.PDR.0000159630.35883.BE. [DOI] [PubMed] [Google Scholar]
- 3.Min J.H., Codipilly C.N., Nasim S., Miller E.J., Ahmed M.N. Synergistic protection against hyperoxia-induced lung injury by neutrophils blockade and EC-SOD overexpression. Respir. Res. 2012;13(1):58. doi: 10.1186/1465-9921-13-58. 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cannavo L., Perrone S., Viola V., Marseglia L., Di Rosa G., Gitto E. Oxidative stress and respiratory diseases in preterm newborns. Int. J. Mol. Sci. 2021;22(22) doi: 10.3390/ijms222212504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robbins M.E., Cho H.Y., Hansen J.M., Luchsinger J.R., Locy M.L., Velten M., Kleeberger S.R., Rogers L.K., Tipple T.E. Glutathione reductase deficiency alters lung development and hyperoxic responses in neonatal mice. Redox Biol. 2021;38 doi: 10.1016/j.redox.2020.101797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rogers S., Witz G., Anwar M., Hiatt M., Hegyi T. Antioxidant capacity and oxygen radical diseases in the preterm newborn. Arch. Pediatr. Adolesc. Med. 2000;154(6):544–548. doi: 10.1001/archpedi.154.6.544. [DOI] [PubMed] [Google Scholar]
- 7.Torres-Cuevas I., Parra-Llorca A., Sanchez-Illana A., Nunez-Ramiro A., Kuligowski J., Chafer-Pericas C., Cernada M., Escobar J., Vento M. Oxygen and oxidative stress in the perinatal period. Redox Biol. 2017;12:674–681. doi: 10.1016/j.redox.2017.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jain A., Mehta T., Auld P.A., Rodrigues J., Ward R.F., Schwartz M.K., Mårtensson J. Glutathione metabolism in newborns: evidence for glutathione deficiency in plasma, bronchoalveolar lavage fluid, and lymphocytes in prematures. Pediatr. Pulmonol. 1995;20(3):160–166. doi: 10.1002/ppul.1950200306. [DOI] [PubMed] [Google Scholar]
- 9.Yang J., Carroll K.S., Liebler D.C. The expanding landscape of the thiol redox proteome. Mol. Cell. Proteomics. 2016;15(1):1–11. doi: 10.1074/mcp.O115.056051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mieyal J.J., Chock P.B. Posttranslational modification of cysteine in redox signaling and oxidative stress: focus on s-glutathionylation. Antioxidants Redox Signal. 2012;16(6):471–475. doi: 10.1089/ars.2011.4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grek C.L., Zhang J., Manevich Y., Townsend D.M., Tew K.D. Causes and consequences of cysteine S-glutathionylation. J. Biol. Chem. 2013;288(37):26497–26504. doi: 10.1074/jbc.R113.461368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J., Ye Z.W., Singh S., Townsend D.M., Tew K.D. An evolving understanding of the S-glutathionylation cycle in pathways of redox regulation. Free Radic. Biol. Med. 2018;120:204–216. doi: 10.1016/j.freeradbiomed.2018.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ullevig S.L., Kim H.S., Short J.D., Tavakoli S., Weintraub S.T., Downs K., Asmis R. Protein S-glutathionylation mediates macrophage responses to metabolic cues from the extracellular environment. Antioxidants Redox Signal. 2016;25(15):836–851. doi: 10.1089/ars.2015.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chia S.B., Elko E.A., Aboushousha R., Manuel A.M., van de Wetering C., Druso J.E., van der Velden J., Seward D.J., Anathy V., Irvin C.G., Lam Y.W., van der Vliet A., Janssen-Heininger Y.M.W. Dysregulation of the glutaredoxin/S-glutathionylation redox axis in lung diseases. Am. J. Physiol. Cell Physiol. 2020;318(2):C304–C327. doi: 10.1152/ajpcell.00410.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aesif S.W., Anathy V., Havermans M., Guala A.S., Ckless K., Taatjes D.J., Janssen-Heininger Y.M.W. In situ analysis of protein S-glutathionylation in lung tissue using glutaredoxin-1-catalyzed cysteine derivatization. Am. J. Pathol. 2009;175(1):36–45. doi: 10.2353/ajpath.2009.080736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuipers I., Louis R., Manise M., Dentener M.A., Irvin C.G., Janssen-Heininger Y.M.W., Brightling C.E., Wouters E.F.M., Reynaert N.L. Increased glutaredoxin-1 and decreased protein<em> S</em>-glutathionylation in sputum of asthmatics. Eur. Respir. J. 2013;41(2):469. doi: 10.1183/09031936.00115212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Downs C.A., Kreiner L., Zhao X.M., Trac P., Johnson N.M., Hansen J.M., Brown L.A., Helms M.N. Oxidized glutathione (GSSG) inhibits epithelial sodium channel activity in primary alveolar epithelial cells. Am. J. Physiol. Lung Cell Mol. Physiol. 2015;308(9):L943–L952. doi: 10.1152/ajplung.00213.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grant G.J., Coca C., Zhao X.M., Helms M.N. Oxidized glutathione increases delta-subunit expressing epithelial sodium channel activity in Xenopus laevis oocytes. Emed Res. 2020;2 [PMC free article] [PubMed] [Google Scholar]
- 19.Hanukoglu I., Hanukoglu A. Epithelial sodium channel (ENaC) family: phylogeny, structure-function, tissue distribution, and associated inherited diseases. Gene. 2016;579(2):95–132. doi: 10.1016/j.gene.2015.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goodson P., Kumar A., Jain L., Kundu K., Murthy N., Koval M., Helms M.N. Nadph oxidase regulates alveolar epithelial sodium channel activity and lung fluid balance in vivo via O2- signaling. Am. J. Physiol. Lung Cell Mol. Physiol. 2012;302(4):L410–L419. doi: 10.1152/ajplung.00260.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gehrig S., Duerr J., Weitnauer M., Wagner C.J., Graeber S.Y., Schatterny J., Hirtz S., Belaaouaj A., Dalpke A.H., Schultz C., Mall M.A. Lack of neutrophil elastase reduces inflammation, mucus hypersecretion, and emphysema, but not mucus obstruction, in mice with cystic fibrosis-like lung disease. Am. J. Respir. Crit. Care Med. 2014;189(9):1082–1092. doi: 10.1164/rccm.201311-1932OC. [DOI] [PubMed] [Google Scholar]
- 22.Mall M., Grubb B.R., Harkema J.R., O'Neal W.K., Boucher R.C. Increased airway epithelial Na+ absorption produces cystic fibrosis-like lung disease in mice. Nat. Med. 2004;10(5):487–493. doi: 10.1038/nm1028. [DOI] [PubMed] [Google Scholar]
- 23.Trojanek J.B., Cobos-Correa A., Diemer S., Kormann M., Schubert S.C., Zhou-Suckow Z., Agrawal R., Duerr J., Wagner C.J., Schatterny J., Hirtz S., Sommerburg O., Hartl D., Schultz C., Mall M.A. Airway mucus obstruction triggers macrophage activation and matrix metalloproteinase 12-dependent emphysema. Am. J. Respir. Cell Mol. Biol. 2014;51(5):709–720. doi: 10.1165/rcmb.2013-0407OC. [DOI] [PubMed] [Google Scholar]
- 24.Nardiello C., Mizikova I., Silva D.M., Ruiz-Camp J., Mayer K., Vadasz I., Herold S., Seeger W., Morty R.E. Standardisation of oxygen exposure in the development of mouse models for bronchopulmonary dysplasia. Dis Model Mech. 2017;10(2):185–196. doi: 10.1242/dmm.027086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grant G.J., Mimche P.N., Paine R., 3rd, Liou T.G., Qian W.J., Helms M.N. Enhanced epithelial sodium channel activity in neonatal Scnn1b mouse lung attenuates high oxygen induced lung injury. Am. J. Physiol. Lung Cell Mol. Physiol. 2021;321:L29–L41. doi: 10.1152/ajplung.00538.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duan J.C., Gaffrey M.J., Qian W.J. Quantitative proteomic characterization of redox-dependent post-translational modifications on protein cysteines. Mol. Biosyst. 2017;13(5):816–829. doi: 10.1039/c6mb00861e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang T., Gaffrey M.J., Qian W.-J., Thrall B.D. In: Interaction of Nanomaterials with the Immune System. Bonner J.C., Brown J.M., editors. Springer International Publishing; Cham: 2020. Oxidative stress and redox modifications in nanomaterial–cellular interactions; pp. 127–148. [Google Scholar]
- 28.McDonagh B. Detection of ROS induced proteomic signatures by mass spectrometry. Front. Physiol. 2017;8:470. doi: 10.3389/fphys.2017.00470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang T., Zhu M., Zhu N., Strul J.M., Dufresne C.P., Schneider J.D., Harmon A.C., Chen S. Identification of thioredoxin targets in guard cell enriched epidermal peels using cysTMT proteomics. J. Proteonomics. 2016;133:48–53. doi: 10.1016/j.jprot.2015.12.008. [DOI] [PubMed] [Google Scholar]
- 30.Guo J., Gaffrey M.J., Su D., Liu T., Camp D.G., Smith R.D., Qian W.-J. Resin-assisted enrichment of thiols as a general strategy for proteomic profiling of cysteine-based reversible modifications. Nat. Protoc. 2014;9(1):64–75. doi: 10.1038/nprot.2013.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gaffrey M.J., Day N.J., Li X., Qian W.J. Resin-assisted capture coupled with isobaric tandem mass tag labeling for multiplexed quantification of protein thiol oxidation. JoVE. 2021;172 doi: 10.3791/62671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duan J., Zhang T., Gaffrey M.J., Weitz K.K., Moore R.J., Li X., Xian M., Thrall B.D., Qian W.J. Stochiometric quantification of the thiol redox proteome of macrophages reveals subcellular compartmentalization and susceptibility to oxidative perturbations. Redox Biol. 2020;36 doi: 10.1016/j.redox.2020.101649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Percie du Sert N., Hurst V., Ahluwalia A., Alam S., Avey M.T., Baker M., Browne W.J., Clark A., Cuthill I.C., Dirnagl U., Emerson M., Garner P., Holgate S.T., Howells D.W., Karp N.A., Lazic S.E., Lidster K., MacCallum C.J., Macleod M., Pearl E.J., Petersen O.H., Rawle F., Reynolds P., Rooney K., Sena E.S., Silberberg S.D., Steckler T., Wurbel H. The ARRIVE guidelines 2.0: updated guidelines for reporting animal research. BMC Vet. Res. 2020;16(1):242. doi: 10.1186/s12917-020-02451-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.I.f.L.A.R.U.S. National Research Council (US) 8 ed. The National Academies Press; Washington, D.C.: 2011. Committee for the Update of the Guide for the Care and Use of Laboratory Animals., and National Academies Press (U.S.), Guide for the Care and Use of Laboratory Animals. [Google Scholar]
- 35.Livraghi-Butrico A., Grubb B.R., Kelly E.J., Wilkinson K.J., Yang H., Geiser M., Randell S.H., Boucher R.C., O'Neal W.K. Genetically determined heterogeneity of lung disease in a mouse model of airway mucus obstruction. Physiol. Genom. 2012;44(8):470–484. doi: 10.1152/physiolgenomics.00185.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bouch S., O'Reilly M., Harding R., Sozo F. Neonatal exposure to mild hyperoxia causes persistent increases in oxidative stress and immune cells in the lungs of mice without altering lung structure. Am. J. Physiol. Lung Cell Mol. Physiol. 2015;309(5):L488–L496. doi: 10.1152/ajplung.00359.2014. [DOI] [PubMed] [Google Scholar]
- 37.Mankouski A., Kantores C., Wong M.J., Ivanovska J., Jain A., Benner E.J., Mason S.N., Tanswell A.K., Auten R.L., Jankov R.P. Intermittent hypoxia during recovery from neonatal hyperoxic lung injury causes long-term impairment of alveolar development: a new rat model of BPD. Am. J. Physiol. Lung Cell Mol. Physiol. 2017;312(2):L208–L216. doi: 10.1152/ajplung.00463.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou X., Moore B.B. Lung section staining and microscopy. Bio Protoc. 2017;7(10) doi: 10.21769/BioProtoc.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eidelman D.H., Ghezzo H., Kim W.D., Cosio M.G. The destructive index and early lung destruction in smokers. Am. Rev. Respir. Dis. 1991;144(1):156–159. doi: 10.1164/ajrccm/144.1.156. [DOI] [PubMed] [Google Scholar]
- 40.Saetta M., Shiner R.J., Angus G.E., Kim W.D., Wang N.S., King M., Ghezzo H., Cosio M.G. Destructive index: a measurement of lung parenchymal destruction in smokers. Am. Rev. Respir. Dis. 1985;131(5):764–769. doi: 10.1164/arrd.1985.131.5.764. [DOI] [PubMed] [Google Scholar]
- 41.Kramer P.A., Duan J., Gaffrey M.J., Shukla A.K., Wang L., Bammler T.K., Qian W.J., Marcinek D.J. Fatiguing contractions increase protein S-glutathionylation occupancy in mouse skeletal muscle. Redox Biol. 2018;17:367–376. doi: 10.1016/j.redox.2018.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang J., Zhang T., Johnston C.J., Kim S.Y., Gaffrey M.J., Chalupa D., Feng G., Qian W.J., McGraw M.D., Ansong C. Protein thiol oxidation in the rat lung following e-cigarette exposure. Redox Biol. 2020;37 doi: 10.1016/j.redox.2020.101758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26(12):1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 44.Chiao Y.A., Zhang H., Sweetwyne M., Whitson J., Ting Y.S., Basisty N., Pino L.K., Quarles E., Nguyen N.H., Campbell M.D., Zhang T., Gaffrey M.J., Merrihew G., Wang L., Yue Y., Duan D., Granzier H.L., Szeto H.H., Qian W.J., Marcinek D., MacCoss M.J., Rabinovitch P. Late-life restoration of mitochondrial function reverses cardiac dysfunction in old mice. Elife. 2020 Jul 10;9 doi: 10.7554/eLife.55513. PMID: 32648542; PMCID: PMC7377906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang T., Chhajed S., Schneider J.D., Feng G., Song W.-Y., Chen S. Proteomic characterization of MPK4 signaling network and putative substrates. Plant Mol. Biol. 2019;101(3):325–339. doi: 10.1007/s11103-019-00908-9. [DOI] [PubMed] [Google Scholar]
- 46.Xiao R., Chen R. Neutrophil gelatinase-associated lipocalin as a potential novel biomarker for ventilator-associated lung injury. Mol. Med. Rep. 2017;15(6):3535–3540. doi: 10.3892/mmr.2017.6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seijo L.M., Peled N., Ajona D., Boeri M., Field J.K., Sozzi G., Pio R., Zulueta J.J., Spira A., Massion P.P., Mazzone P.J., Montuenga L.M. Biomarkers in lung cancer screening: achievements, promises, and challenges. J. Thorac. Oncol. 2019;14(3):343–357. doi: 10.1016/j.jtho.2018.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aksoy M.O., Kim V., Cornwell W.D., Rogers T.J., Kosmider B., Bahmed K., Barrero C., Merali S., Shetty N., Kelsen S.G. Secretion of the endoplasmic reticulum stress protein, GRP78, into the BALF is increased in cigarette smokers. Respir. Res. 2017;18(1):78. doi: 10.1186/s12931-017-0561-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang F., Li X., Zhao N., Duan L., Chen Y. Circulating GRP78 acts as a biomarker in the early diagnosis of lung cancer. Int. J. Clin. Exp. Pathol. 2018;11(11):5223–5231. [PMC free article] [PubMed] [Google Scholar]
- 50.Su D., Gaffrey M.J., Guo J., Hatchell K.E., Chu R.K., Clauss T.R., Aldrich J.T., Wu S., Purvine S., Camp D.G., Smith R.D., Thrall B.D., Qian W.J. Proteomic identification and quantification of S-glutathionylation in mouse macrophages using resin-assisted enrichment and isobaric labeling. Free Radic. Biol. Med. 2014;67:460–470. doi: 10.1016/j.freeradbiomed.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stroud D.A., Surgenor E.E., Formosa L.E., Reljic B., Frazier A.E., Dibley M.G., Osellame L.D., Stait T., Beilharz T.H., Thorburn D.R., Salim A., Ryan M.T. Accessory subunits are integral for assembly and function of human mitochondrial complex I. Nature. 2016;538(7623):123–126. doi: 10.1038/nature19754. [DOI] [PubMed] [Google Scholar]
- 52.Mimaki M., Wang X., McKenzie M., Thorburn D.R., Ryan M.T. Understanding mitochondrial complex I assembly in health and disease. Biochim. Biophys. Acta Bioenerg. 2012;1817(6):851–862. doi: 10.1016/j.bbabio.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 53.Ramani M., Miller K., Brown J., Kumar R., Kadasamy J., McMahon L., Ballinger S., Ambalavanan N. Early life supraphysiological levels of oxygen exposure permanently impairs hippocampal mitochondrial function. Sci. Rep. 2019;9(1) doi: 10.1038/s41598-019-49532-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xuefei Y., Xinyi Z., Qing C., Dan Z., Ziyun L., Hejuan Z., Xindong X., Jianhua F. Effects of hyperoxia on mitochondrial homeostasis: are mitochondria the hub for bronchopulmonary dysplasia? Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.642717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van Dyken S.J., Locksley R.M. Chitins and chitinase activity in airway diseases. J. Allergy Clin. Immunol. 2018;142(2):364–369. doi: 10.1016/j.jaci.2018.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cassatella M.A., Östberg N.K., Tamassia N., Soehnlein O. Biological roles of neutrophil-derived granule proteins and cytokines. Trends Immunol. 2019;40(7):648–664. doi: 10.1016/j.it.2019.05.003. [DOI] [PubMed] [Google Scholar]
- 57.Przysucha N., Górska K., Krenke R. Chitinases and chitinase-like proteins in obstructive lung diseases - current concepts and potential applications. Int. J. Chronic Obstr. Pulm. Dis. 2020;15:885–899. doi: 10.2147/COPD.S236640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Van Dyken S.J., Liang H.-E., Naikawadi R.P., Woodruff P.G., Wolters P.J., Erle D.J., Locksley R.M. Spontaneous chitin accumulation in airways and age-related fibrotic lung disease. Cell. 2017;169(3):497–509. doi: 10.1016/j.cell.2017.03.044. e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paine R., 3rd, Ward P.A. Cell adhesion molecules and pulmonary fibrosis. Am. J. Med. 1999;107(3):268–279. doi: 10.1016/s0002-9343(99)00226-0. [DOI] [PubMed] [Google Scholar]
- 60.You Y., Chen J., Zhu F., Xu Q., Han L., Gao X., Zhang X., Luo H.R., Miao J., Sun X., Ren H., Du Y., Guo L., Wang X., Wang Y., Chen S., Huang N., Li J. Glutaredoxin 1 up-regulates deglutathionylation of α4 integrin and thereby restricts neutrophil mobilization from bone marrow. J. Biol. Chem. 2019;294(8):2616–2627. doi: 10.1074/jbc.RA118.006096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chambers R.C., Scotton C.J. Coagulation cascade proteinases in lung injury and fibrosis. Proc. Am. Thorac. Soc. 2012;9(3):96–101. doi: 10.1513/pats.201201-006AW. [DOI] [PubMed] [Google Scholar]
- 62.Nyström E.E.L., Arike L., Ehrencrona E., Hansson G.C., Johansson M.E.V. Calcium-activated chloride channel regulator 1 (CLCA1) forms non-covalent oligomers in colonic mucus and has mucin 2-processing properties. J. Biol. Chem. 2019;294(45):17075–17089. doi: 10.1074/jbc.RA119.009940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nyström E.E.L., Birchenough G.M.H., van der Post S., Arike L., Gruber A.D., Hansson G.C., Johansson M.E.V. Calcium-activated chloride channel regulator 1 (CLCA1) controls mucus expansion in colon by proteolytic activity. EBioMedicine. 2018;33:134–143. doi: 10.1016/j.ebiom.2018.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Centeio R., Ousingsawat J., Schreiber R., Kunzelmann K. CLCA1 regulates airway mucus production and ion secretion through TMEM16A. Int. J. Mol. Sci. 2021;22(10) doi: 10.3390/ijms22105133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu C.-L., Shi G.-P. Calcium-activated chloride channel regulator 1 (CLCA1): more than a regulator of chloride transport and mucus production. World Allergy Organization Journal. 2019;12(11) doi: 10.1016/j.waojou.2019.100077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yurtsever Z., Sala-Rabanal M., Randolph D.T., Scheaffer S.M., Roswit W.T., Alevy Y.G., Patel A.C., Heier R.F., Romero A.G., Nichols C.G., Holtzman M.J., Brett T.J. Self-cleavage of human CLCA1 protein by a novel internal metalloprotease domain controls calcium-activated chloride channel activation. J. Biol. Chem. 2012;287(50):42138–42149. doi: 10.1074/jbc.M112.410282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sala-Rabanal M., Yurtsever Z., Berry K.N., Nichols C.G., Brett T.J. Modulation of TMEM16A channel activity by the von Willebrand factor type A (VWA) domain of the calcium-activated chloride channel regulator 1 (CLCA1) J. Biol. Chem. 2017;292(22):9164–9174. doi: 10.1074/jbc.M117.788232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Berry K.N., Brett T.J. Structural and biophysical analysis of the CLCA1 VWA domain suggests mode of TMEM16A engagement. Cell Rep. 2020;30(4):1141–1151. doi: 10.1016/j.celrep.2019.12.059. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Whittaker C.A., Hynes R.O. Distribution and evolution of von Willebrand/integrin A domains: widely dispersed domains with roles in cell adhesion and elsewhere. Mol. Biol. Cell. 2002;13(10):3369–3387. doi: 10.1091/mbc.E02-05-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Springer T.A. Complement and the multifaceted functions of VWA and integrin I domains. Structure. 2006;14(11):1611–1616. doi: 10.1016/j.str.2006.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Campbell M.D., Duan J., Samuelson A.T., Gaffrey M.J., Merrihew G.E., Egertson J.D., Wang L., Bammler T.K., Moore R.J., White C.C., Kavanagh T.J., Voss J.G., Szeto H.H., Rabinovitch P.S., MacCoss M.J., Qian W.J., Marcinek D.J. Improving mitochondrial function with SS-31 reverses age-related redox stress and improves exercise tolerance in aged mice. Free Radic. Biol. Med. 2019;134:268–281. doi: 10.1016/j.freeradbiomed.2018.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Reichmann D., Voth W., Jakob U. Maintaining a healthy proteome during oxidative stress. Mol. Cell. 2018;69(2):203–213. doi: 10.1016/j.molcel.2017.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hoter A., El-Sabban M.E., Naim H.Y. The HSP90 family: structure, regulation, function, and implications in health and disease. Int. J. Mol. Sci. 2018;19(9):2560. doi: 10.3390/ijms19092560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tata P.R., Rajagopal J. Plasticity in the lung: making and breaking cell identity. Development. 2017;144(5):755–766. doi: 10.1242/dev.143784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.van der Vliet A., Janssen-Heininger Y.M.W., Anathy V. Oxidative stress in chronic lung disease: from mitochondrial dysfunction to dysregulated redox signaling. Mol. Aspect. Med. 2018;63:59–69. doi: 10.1016/j.mam.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rosanna D.P., Salvatore C. Reactive oxygen species, inflammation, and lung diseases. Curr. Pharmaceut. Des. 2012;18(26):3889–3900. doi: 10.2174/138161212802083716. [DOI] [PubMed] [Google Scholar]
- 77.Nishi T., Shimizu N., Hiramoto M., Sato I., Yamaguchi Y., Hasegawa M., Aizawa S., Tanaka H., Kataoka K., Watanabe H., Handa H. Spatial redox regulation of a critical cysteine residue of NF-kappa B in vivo. J. Biol. Chem. 2002;277(46):44548–44556. doi: 10.1074/jbc.M202970200. [DOI] [PubMed] [Google Scholar]
- 78.Duan J., Kodali V.K., Gaffrey M.J., Guo J., Chu R.K., Camp D.G., Smith R.D., Thrall B.D., Qian W.J. Quantitative profiling of protein S-glutathionylation reveals redox-dependent regulation of macrophage function during nanoparticle-induced oxidative stress. ACS Nano. 2016;10(1):524–538. doi: 10.1021/acsnano.5b05524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kindermann A., Binder L., Baier J., Gündel B., Simm A., Haase R., Bartling B. Severe but not moderate hyperoxia of newborn mice causes an emphysematous lung phenotype in adulthood without persisting oxidative stress and inflammation. BMC Pulm. Med. 2019;19(1):245. doi: 10.1186/s12890-019-0993-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Farrow K.N., Lee K.J., Perez M., Schriewer J.M., Wedgwood S., Lakshminrusimha S., Smith C.L., Steinhorn R.H., Schumacker P.T. Brief hyperoxia increases mitochondrial oxidation and increases phosphodiesterase 5 activity in fetal pulmonary artery smooth muscle cells. Antioxidants Redox Signal. 2012;17(3):460–470. doi: 10.1089/ars.2011.4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gould N.S., Evans P., Martinez-Acedo P., Marino S.M., Gladyshev V.N., Carroll K.S., Ischiropoulos H. Site-specific proteomic mapping identifies selectively modified regulatory cysteine residues in functionally distinct protein networks. Chem. Biol. 2015;22(7):965–975. doi: 10.1016/j.chembiol.2015.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ma T., Yoo M.J., Zhang T., Liu L., Koh J., Song W.Y., Harmon A.C., Sha W., Chen S. Characterization of thiol-based redox modifications of Brassica napusSNF1-related protein kinase 2.6-2C. FEBS Open Bio. 2018;8(4):628–645. doi: 10.1002/2211-5463.12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhu M., Zhang T., Ji W., Silva-Sanchez C., Song W.Y., Assmann S.M., Harmon A.C., Chen S. Redox regulation of a guard cell SNF1-related protein kinase in Brassica napus, an oilseed crop. Biochem. J. 2017;474(15):2585–2599. doi: 10.1042/BCJ20170070. [DOI] [PubMed] [Google Scholar]
- 84.Guo J., Nguyen A.Y., Dai Z., Su D., Gaffrey M.J., Moore R.J., Jacobs J.M., Monroe M.E., Smith R.D., Koppenaal D.W., Pakrasi H.B., Qian W.J. Proteome-wide light/dark modulation of thiol oxidation in cyanobacteria revealed by quantitative site-specific redox proteomics. Mol. Cell. Proteomics. 2014;13(12):3270–3285. doi: 10.1074/mcp.M114.041160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Leichert L.I., Gehrke F., Gudiseva H.V., Blackwell T., Ilbert M., Walker A.K., Strahler J.R., Andrews P.C., Jakob U. Quantifying changes in the thiol redox proteome upon oxidative stress in vivo. Proc. Natl. Acad. Sci. U. S. A. 2008;105(24):8197–8202. doi: 10.1073/pnas.0707723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xiao H., Jedrychowski M.P., Schweppe D.K., Huttlin E.L., Yu Q., Heppner D.E., Li J., Long J., Mills E.L., Szpyt J., He Z., Du G., Garrity R., Reddy A., Vaites L.P., Paulo J.A., Zhang T., Gray N.S., Gygi S.P., Chouchani E.T. A quantitative tissue-specific landscape of protein redox regulation during aging. Cell. 2020;180(5):968–983 e24. doi: 10.1016/j.cell.2020.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jones D.P. Radical-free biology of oxidative stress. Am. J. Physiol. Cell Physiol. 2008;295(4):C849–C868. doi: 10.1152/ajpcell.00283.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rogers L.K., Cismowski M.J. Oxidative stress in the lung - the essential paradox. Curr Opin Toxicol. 2018;7:37–43. doi: 10.1016/j.cotox.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Otulakowski G., Rafii B., Harris M., O'Brodovich H. Oxygen and glucocorticoids modulate alphaENaC mRNA translation in fetal distal lung epithelium. Am. J. Respir. Cell Mol. Biol. 2006;34(2):204–212. doi: 10.1165/rcmb.2005-0273OC. [DOI] [PubMed] [Google Scholar]
- 90.Rafli B., Tanswell A.K., Pitkänen O., O'Brodovich H. In: Current Topics in Membranes. Benos D.J., editor. Academic Press; 1999. Chapter 14 induction of epithelial sodium channel (ENaC) expression and sodium transport in distal lung epithelia by oxygen; pp. 239–254. [Google Scholar]
- 91.Muronetz V.I., Melnikova A.K., Saso L., Schmalhausen E.V. Influence of oxidative stress on catalytic and non-glycolytic functions of glyceraldehyde-3-phosphate dehydrogenase. Curr. Med. Chem. 2020;27(13):2040–2058. doi: 10.2174/0929867325666180530101057. [DOI] [PubMed] [Google Scholar]
- 92.Libiad M., Yadav P.K., Vitvitsky V., Martinov M., Banerjee R. Organization of the human mitochondrial hydrogen sulfide oxidation pathway. J. Biol. Chem. 2014;289(45):30901–30910. doi: 10.1074/jbc.M114.602664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kruithof P.D., Lunev S., Aguilar Lozano S.P., de Assis Batista F., Al-dahmani Z.M., Joles J.A., Dolga A.M., Groves M.R., van Goor H. Unraveling the role of thiosulfate sulfurtransferase in metabolic diseases. Biochim. Biophys. Acta (BBA) - Mol. Basis Dis. 2020;1866(6) doi: 10.1016/j.bbadis.2020.165716. [DOI] [PubMed] [Google Scholar]
- 94.Moldoveanu T., Hosfield C.M., Lim D., Elce J.S., Jia Z., Davies P.L. A Ca2+ switch aligns the active site of calpain. Cell. 2002;108(5):649–660. doi: 10.1016/s0092-8674(02)00659-1. [DOI] [PubMed] [Google Scholar]
- 95.Liu Y., Liu B., Zhang G.-Q., Zou J.-F., Zou M.-L., Cheng Z.-S. Calpain inhibition attenuates bleomycin-induced pulmonary fibrosis via switching the development of epithelial-mesenchymal transition. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2018;391(7):695–704. doi: 10.1007/s00210-018-1499-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tabata C., Tabata R., Nakano T. The calpain inhibitor calpeptin prevents bleomycin-induced pulmonary fibrosis in mice. Clin. Exp. Immunol. 2010;162(3):560–567. doi: 10.1111/j.1365-2249.2010.04257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Potz B.A., Abid M.R., Sellke F.W. Role of calpain in pathogenesis of human disease processes. J. Nat. Sci. 2016;2(9):e218. [PMC free article] [PubMed] [Google Scholar]
- 98.Wright C.J., Agboke F., Chen F., La P., Yang G., Dennery P.A. NO inhibits hyperoxia-induced NF-kappaB activation in neonatal pulmonary microvascular endothelial cells. Pediatr. Res. 2010;68(6):484–489. doi: 10.1203/PDR.0b013e3181f917b0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bayele H.K., Murdock P.J., Perry D.J., Pasi K.J. Simple shifts in redox/thiol balance that perturb blood coagulation. FEBS (Fed. Eur. Biochem. Soc.) Lett. 2002;510(1):67–70. doi: 10.1016/s0014-5793(01)03209-4. [DOI] [PubMed] [Google Scholar]
- 100.Bajzar L., Nesheim M., Morser J., Tracy P.B. Both cellular and soluble forms of thrombomodulin inhibit fibrinolysis by potentiating the activation of thrombin-activable fibrinolysis inhibitor. J. Biol. Chem. 1998;273(5):2792–2798. doi: 10.1074/jbc.273.5.2792. [DOI] [PubMed] [Google Scholar]
- 101.Chapin J.C., Hajjar K.A. Fibrinolysis and the control of blood coagulation. Blood Rev. 2015;29(1):17–24. doi: 10.1016/j.blre.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dataset metrics. (A) Resin assisted capture identifies more than 10,000 peptides in wild type and transgenic Scnn1b mice exposed to control or hyperoxic conditions in SSG or total oxidation workflows, where more than 98% of these peptides contain cysteine. (B) The more than 10,000 SSG-modified peptides correspond to >7600 Cys sites for each genotype, with nearly 6000 Cys sites being common to both wild type and transgenic Scnn1b mice. (C) More than 3000 proteins contain a SSG-modified Cys site in each genotype, with almost 2600 of these proteins found in both wild type and transgenic Scnn1b mice. (D) Global proteomics identifies more than 3000 proteins in both wild type and transgenic Scnn1b mice, with ∼2500 present in both samples. (E-F) Coefficients of variation of TMT reporter ion intensities for SSG-modified Cys sites (E), proteins in global (F), or total oxidation of Cys sites (G) identified in this study for both genotypes, where the dashed line represents the 10% boundary as a reference.
Subcellular distribution of PSSG-modified proteins. (A) Scatter plot illustrating how many glutathionylated proteins belong to a specific compartment in cells obtained from transgenic Scnn1b mice. (B–C) Box and whisker plots illustrating the fold change differences in Cys site SSG levels for proteins that reside in a specific subcellular compartment in wild type (B) or Scnn1b overexpressing (C) mice. The red and blue dashed lines denote the log2fold change cutoffs of 0.3 and −0.3, respectively, which are plotted on a non-log-transformed y axis.
Visualization of the impact of hyperoxia on global and redox proteomes. (A-F) For each panel, the plot on the left represents all features identified (protein or Cys site; as specified in the following legend descriptions), while the plot on the right illustrates all features that met the significance cutoff of log2 fold change > 0.3 and p adjusted < 0.1 (red = upregulated; blue = downregulated). (A-B) Volcano plots representing the log2 fold changes in protein expression under hyperoxic versus control conditions at the protein level for wild type (A) and transgenic Scnn1b mice (B). (C-D) Volcano plots representing the log2 fold changes in SSG modifications at the Cys site level under hyperoxic versus control conditions for wild type (C) and transgenic Scnn1b mice (D). (E) Volcano plots representing the log2 fold changes in total oxidation under hyperoxic versus control conditions at the Cys site level for transgenic Scnn1b mice. (F) Volcano plots representing the integrated global abundance log2 fold change of proteins found in both wild type and transgenic Scnn1b mice. The values in this plot represent how much more change was observed in response to hyperoxia in transgenic Scnn1b mice versus wild type mice for a given protein (red = upregulated in transgenic Scnn1b/downregulated in wild type mice; blue = downregulated in transgenic Scnn1b/upregulated in wild type mice).
Occupancy distribution of Cys sites in each genotype. (A-F) Data was transformed to plot on log10 scale and the red vertical line denotes the median occupancy in the dataset. (A,C) The distribution of SSG occupancy on Cys sites in wild type mice under control and hyperoxic conditions with minimal change between conditions. (B,D) The distribution of SSG occupancy on Cys sites in transgenic Scnn1b mice under control and hyperoxic conditions with minimal change between conditions. (E,F) The distribution of total oxidation occupancy on Cys sites in Scnn1b overexpressing lungs under control and hyperoxic conditions with minimal change between conditions.
Data Availability Statement
All mass spectrometry raw data and processed proteomics data are available upon request.