Summary
Fiber photometry is an emerging technique for recording fluorescent sensor activity in the brain. However, significant hemoglobin absorption artifacts in fiber photometry data may be misinterpreted as sensor activity changes. Because hemoglobin exists widely in the brain, and its concentration varies temporally, such artifacts could impede the accuracy of photometry recordings. Here we present use of spectral photometry and computational methods to quantify photon absorption effects by using activity-independent fluorescence signals, which can be used to derive oxy- and deoxy-hemoglobin concentration changes. Although these changes are often temporally delayed compared with the fast-responding fluorescence spikes, we found that erroneous interpretation may occur when examining pharmacology-induced sustained changes and that sometimes hemoglobin absorption could flip the GCaMP signal polarity. We provide hemoglobin-based correction methods to restore fluorescence signals and compare our results with other commonly used approaches. We also demonstrated the utility of spectral fiber photometry for delineating regional differences in hemodynamic response functions.
Keywords: spectral fiber photometry, hemoglobin absorption, excitation, emission, photon traveling path lengths, hemodynamic response function, genetically encoded calcium indicators, isosbestic, green fluorescent sensor, red-shifted fluorescent sensor
Graphical abstract
Highlights
-
•
Hemoglobin absorbs light and creates artifacts in fiber photometry recordings in vivo
-
•
A method to derive hemoglobin concentration changes from spectral photometry
-
•
Correction of this artifact provides accurate measurements of fluorescent sensor signals
-
•
Quantification of regional differences in neurovascular transfer function
Motivation
Hemoglobin (Hb) in blood vessels is known to absorb light and affect the accuracy of optical measurements. In fiber photometry, where photons are collectively sampled from a volume of the brain tissue, changes in Hb concentration could have a significant effect on its readout. It is therefore important to quantify Hb concentration fluctuations to ensure accurate photometry-based fluorescent sensor activity measurements. Here we describe a measurement platform and analytical methods that allowquantification of oxy- and deoxy-Hb concentration changes and correction of Hb absorption artifacts in fiber photometry data. We also demonstrate the added benefit of our methods to compute hemodynamic response function using the derived Hb concentration information.
Changes in hemoglobin concentration affect the accuracy of fiber photometry measurements in vivo, even flipping the measured signal polarity under certain conditions. Zhang et al. develop an approach to derive oxy- and deoxy-hemoglobin concentration changes from spectral fiber photometry data and a pipeline to correct for these artifacts in fluorescent sensor measurements.
Introduction
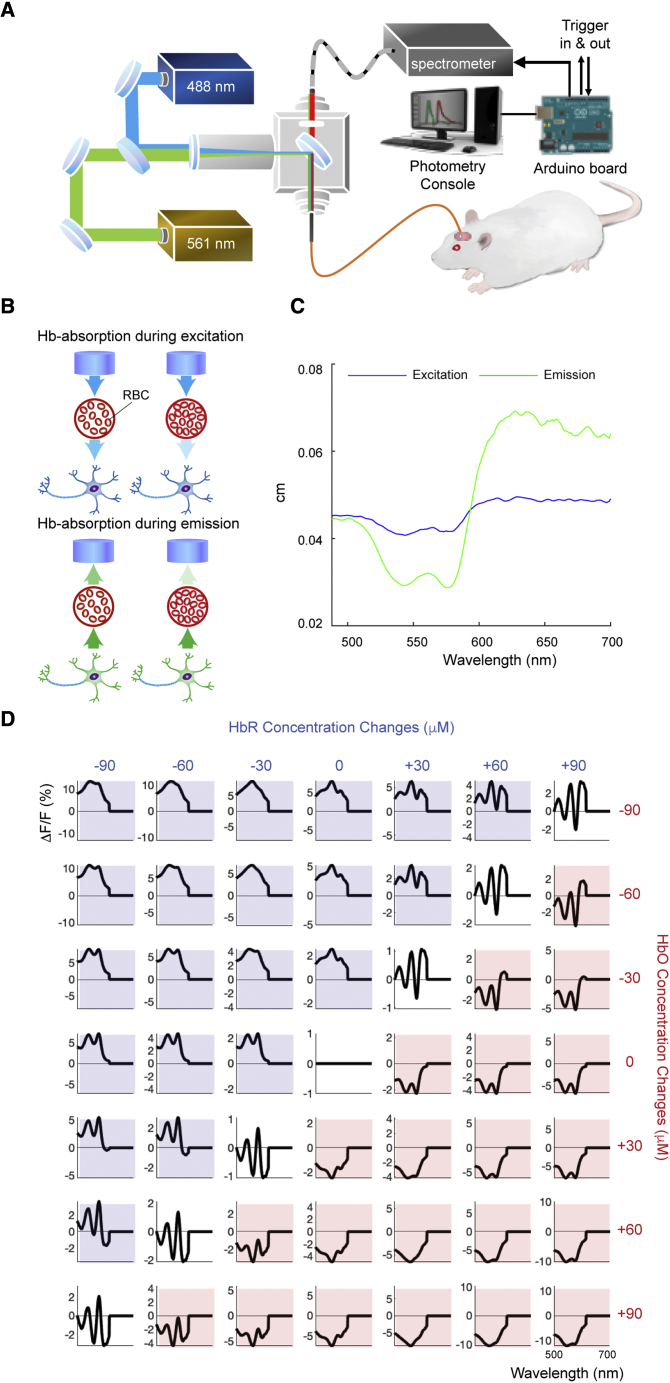
Fiber photometry relies on an implanted optical fiber to excite and detect fluorescence signals and is best known for its ability to measure ensemble neuronal or neurochemical activity in small volumes of brain tissue in animal models. The simplicity and cost effectiveness of the optical system make the technique highly accessible (Meng et al., 2018), and lightweight, miniature fiber optics make photometry compatible with a wide variety of experimental settings, such as freely moving behavioral assessment (Kim et al., 2016; Meng et al., 2018) or head-fixed brain-wide imaging (Chen et al., 2019; Pais-Roldán et al., 2020; Figure 1A). Several genetically encoded fluorescent sensors of cellular activity and neurotransmitters have become widely available (Chen et al., 2013; Dana et al., 2016; Marvin et al., 2013, 2019; Patriarchi et al., 2018; Sun et al., 2018; Tian et al., 2009; Wang et al., 2018a, 2018b), expanding the range of applications for fiber photometry. The tunable spectral specificity has also enabled new parallel sensing capabilities with wavelength multiplexing. Given its rapidly increasing usage in neuroscience (Cui et al., 2013; Luchsinger et al., 2021; Pisano et al., 2019; Sych et al., 2019), understanding the inherent confounding factors in fiber photometry data is of paramount significance.
Figure 1.
Spectrally resolved fiber photometry system and Hb absorption effects
(A) Schematic of a spectrally resolved fiber photometry system used for recording fluorescence signals.
(B) Hb in red blood cells absorbs light in two phases: excitation light from the optical fiber (left) and emission light to be recorded by the optical fiber (right). The color density of arrowheads indicates light intensity. An increase in Hb is expected to decrease the number of photons in both phases.
(C) Simulated average excitation and emission photon traveling path lengths across 488- 700-nm emission wavelengths from our database of paired excitation/emission light paths.
(D) Effect of HbO and HbR concentration on ΔF/F. Shown are simulated effects of HbO and HbR concentration changes on measurement of fluorescence signals. ΔCHb0 and ΔCHbR ranging from −90 to +90 μM were used to simulate fluorescence signal changes (ΔF/F) between 500 and 700 nm using the molar extinction coefficients of HbO and HbR (Figure S1) and simulated light paths (Figure 1C). Graphs shaded in blue indicate an overall decrease in total Hb (HbT), and graphs shaded in red indicate an overall increase in HbT.
See also Figures S1, S2, Video S1, and Table S1.
One of the most notable confounders affecting fluorescence measurements is light absorption by hemoglobin (Hb) in blood vessels (Ma et al., 2016; Prahl, 1999). Hb absorption artifacts have been studied extensively in wide-field imaging with reflective or backscattered lights (Ma et al., 2016; Valley et al., 2020) and recently in two-photon lifetime microscopy (2PLM) measuring the partial pressure of O2 (Mächler et al., 2021). Hb absorption artifacts should be present in most photometry data because the intervascular distance of capillaries is known to be less than 60 in nearly every location in the brain (Smith et al., 2019) and commonly used photometry targeted volumes are 105–106 (Pisanello et al., 2019). This absorption occurs in two phases: along the excitation path, by absorbing photons that travel from the fiber tip to the targeted fluorescent proteins, and along the emission path, by absorbing photons emitted by the fluorescent proteins as they travel back toward the optical fiber (Figure 1B). Both absorption effects add up and result in a substantial photon count reduction. Importantly, Hb absorption is nonlinear and wavelength dependent and varies as a function of dynamic oxy-Hb (HbO) and deoxy-Hb (HbR) concentration changes in vivo ( and , respectively) (Meng and Alayash, 2017; Prahl, 1999; Figures S1A and S1B; derived from Prahl, 1999). Because the power spectra of hemodynamic signals overlap significantly with most fluorescent sensors (Figure S1C), Hb absorption artifacts cannot be removed from fiber photometry data by notch filtering or regressing cerebral blood flow or cerebral blood volume (CBV) signals. To the best of our knowledge, the influence of Hb absorption on fiber photometry-derived fluorescent sensor activity has never been studied. The ability to quantify Hb concentration fluctuations in photometry data would not only enable more accurate fluorescent sensor activity measurements but also provide concurrent hemodynamic metrics, which is the foundation of several brain mapping techniques, including functional magnetic resonance imaging (fMRI) (Schulz et al., 2012), intrinsic optics (Hillman, 2007), and functional ultrasound (Rungta et al., 2017).
In this study, we propose use of spectrally resolved fiber photometry and computational methods that can independently derive HbO and HbR concentration changes from activity-independent fluorescent reporter signals. We present use of this technique to remove Hb absorption artifacts from calcium activity recording and demonstrate the added benefit to compute the transfer function between neuronal and vascular activity, which is crucial for interpreting hemodynamics-based neuroimaging data such as fMRI, functional ultrasound, and intrinsic optics. A step-by-step protocol of this method could be found in Zhang et al. (2022).
Results
Theoretical Hb absorption effect on photometry recordings
We solved the following equation using the generalized method of moments (GMM) (Hansen, 1982) with spectrometer outputs, which provides quantifiable photon counts across spectra:
| (Equation 1) |
where and represent the absorption coefficient changes at excitation wavelength and emission wavelength , respectively, at time point t compared with a reference time point t0. and are affected by and (STAR Methods, Equation 16). and represent the photon traveling path lengths (see STAR Methods for details) at the excitation and emission wavelengths, respectively. To solve Equation 1, we first performed a Monte Carlo simulation using the pipeline shown in Figure S2A to obtain and . We determined the average photon traveling path lengths for emission wavelengths from 488–700 nm in the simulated brain tissue from a total of 29 simulations (2–2.5 104 photons each; Figure 1C, detailed numbers tabulated in Table S1). These results are expected to be generalizable and can be used across labs. As an example, we illustrated spatial probability and luminance-distance profiles for the commonly used 488-nm excitation, 515-nm emission and 580-nm emission in Figures S2B–S2D (see also Video S1). These results allowed calculating the simulated effects of Hb absorption on fluorescence signal measurements, for which we observed significant nonlinear, wavelength-dependent effects (Figure 1D).
Left: Initial ballistic photon flow of a 488 nm excitation beam incident from the optical fiber. Right: Diffuse propagation of the elicited 561 nm emissions captured by the same fiber.
Ex vivo quantification of Hb absorption effects on photometry recordings
We first performed ex vivo measurements and showed that adding 1 mL of fresh venous mouse blood into a 10-mL mixture of Alexa 488 and 568 fluorescent dye solution significantly reduced both fluorescence signals measured by fiber photometry. Compared with adding the blood, adding 1 mL vehicle (0.9% saline) induced significantly less reduction in Alexa 488 and 568 signals (Figures S3A–S3C). Subsequently, we saturated the mixture with 100% oxygen to increase the HbO-HbR ratio and observed marked increases in the Alexa 568 signals but decreases in Alexa 488 signals. We did not observe significant signal changes when infusing oxygen into the solution with 1 mL saline added (Figures S3B and S3D).
In vivo quantification of Hb absorption effects on photometry recordings
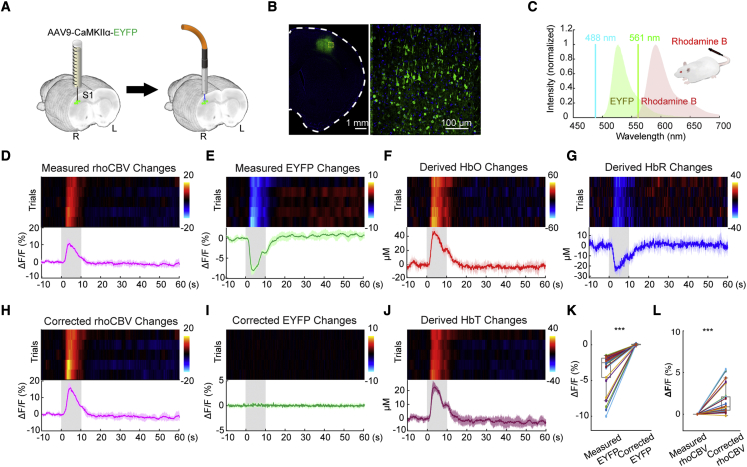
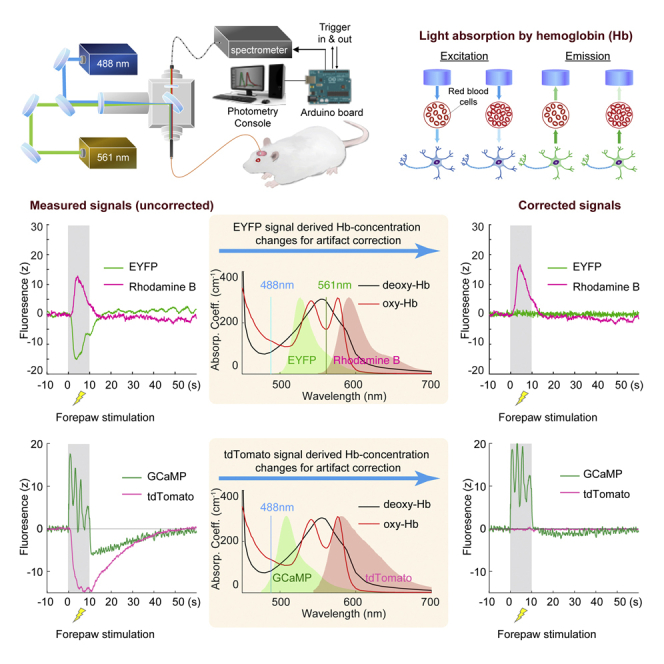
We performed an in vivo experiment to measure the Hb absorption of activity-independent fluorescence signals in fiber photometry. We selected a commonly used fluorophore, enhanced yellow fluorescent protein (EYFP), whose fluorescence signal is activity independent and has an emission spectrum peaking at 526 nm. We virally expressed EYFP using an adeno-associated virus (AAV) in the primary somatosensory cortex of the forelimb region (S1FL) under the Ca2+/calmodulin-dependent protein kinase IIα (CaMKIIα) promoter and then implanted an optical fiber into this target area (Figures 2A and 2B). On the day of photometry recording, we intravenously administered Rhodamine B, a long-circulating red fluorescent dye, to enable steady-state CBV measurements (Unekawa et al., 2015; Figure 2C) and cross-validate our proposed method for calculating total Hb concentration changes. The resulting EYFP and Rhodamine B spectra overlapped significantly with the Hb absorption spectra (Figures S1A and S1B), suggesting a significant effect of Hb absorption on the data from these fluorescence signals. Upon electrical stimulation of the contralateral forepaw (9 Hz, 3 mA, 0.5-ms pulse width for 10 s), we observed robust positive Rhodamine-derived CBV (rhoCBV) changes, as expected (Figure 2D). Intriguingly, the EYFP signal concurrently recorded from the same fiber showed robust negative changes (Figure 2E), which indicated significant Hb absorption of the EYFP signal in vivo. Although a pH-dependent effect on the EYFP signal cannot be completely excluded, a previous study has shown that, at pH 6.5–9, where typical pH homeostasis of a hippocampal neuron (∼7.03–7.46) is located (Ruffin et al., 2014), EYFP excited by 470 nm was not significantly affected by the pH changes (Nakabayashi et al., 2012). Using well-documented molar extinction coefficients (Figures S1A and S1B), photon-traveling path lengths derived from a Monte Carlo simulation (Figure 1C) and spectral fiber photometry datapoints across 502- to 544-nm wavelengths over time (see Equations 17, 18, 19, 20, and 21 in STAR Methods), we derived and using GMM. This is feasible and robust because we have access to a range of spectral datapoints for every single time point and therefore could use numerous empirical data to solve two unknowns ( and ; see also Figures S4A and S4B for the effect of input spectral datapoints). The derived concentration changes (Figures 2F and 2G) were within physiological ranges and comparable with those reported in the literature (Ma et al., 2016), and the summation of the two (i.e., total Hb concentration changes, termed ) informs CBV changes (Ma et al., 2016; Figure 2J). The calculated highly resembled the rhoCBV changes measured from a distinct and less affected red spectrum (Figures S4C–S4E). With and obtained, we then calculated and and entered these parameters into Equation 1 to correct the rhoCBV changes (Figure 2H) and EYFP signals (Figure 2I). Figure 2I shows a flattened EYFP trace after the proposed correction, demonstrating the validity of our approach. Although the influence of Hb absorption on EYFP signals varied by trials, our proposed method accurately corrected EYFP signals in all trials (Figure 2K). Despite the smaller influence on the rhoCBV signal measured from the red spectrum, the absorption effect remained statistically significant (Figure 2L). In addition to the fluorescence signal changes induced by forepaw sensory stimulations, we also replicated these findings using a 5% CO2 hypercapnic challenge paradigm and successfully restored the contaminated EYFP signal changes (Figure S5).
Figure 2.
In vivo quantification of HbO and HbR changes using EYFP emission in fiber photometry and correction of EYFP signals for Hb absorption
(A) Preparation for fiber photometry measurement of EYFP signals in S1FL.
(B) Confocal image showing EYFP expression.
(C) Simultaneous dual spectral photometry recording of EYFP signals and hemodynamic responses (CBV) was achieved by intravenously injecting a long-circulating red fluorescent dye, Rhodamine B.
(D–J) Peri-stimulus heatmaps showing repeated trials and average response time courses of the changes in measured rhoCBV, measured EYFP, derived HbO, derived HbR, corrected rhoCBV, corrected EYFP, and derived HbT, respectively. The gray-shaded segment indicates the forepaw stimulation period. Hb absorption induces pseudo-negative changes in measured EYFP signals during stimulation.
(K and L) Correction of the contaminated EYFP and rhoCBV signals, respectively, in 5 hemispheres and 30 trials (∗∗∗p < 0.001, paired t test). Measured and corrected EYFP signals are compared. Measured rhoCBV was normalized to 0 to show the difference before and after correction.
All color bars use the same unit as the y axis of the time course. Error bars represent standard deviation.
See also Figures S3–S5.
Effect of Hb absorption on acute GCaMP signal measurements using fiber photometry
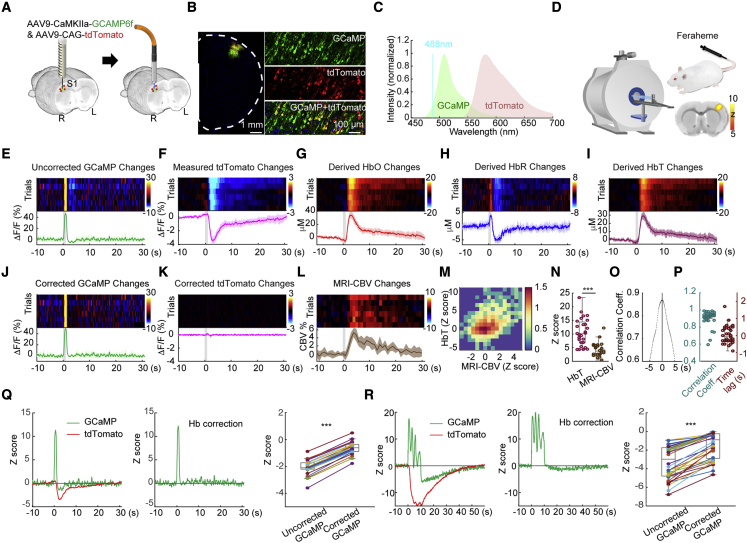
We virally expressed GCaMP6f (hereafter called GCaMP), one of the most widely used genetically encoded calcium indicators (Chen et al., 2013), using an AAV under the CaMKIIα promoter in the S1FL. We also expressed tandem dimer tomato (tdTomato), an activity-independent red fluorescent protein, under the CAG promoter (Figures 3A and 3B) and then implanted an optical fiber into this target area. Our rationale for choosing tdTomato was similar to that for EYFP (Figure 2): it does not encode activity changes, and, thus, its photon counts can be used to derive and . Unlike in the experiment described in Figure 2, Rhodamine B could no longer be used for CBV cross-validation because the red spectrum was occupied by tdTomato (Figure 3C). Instead, we performed this GCaMP-tdTomato experiment in an MRI scanner, where CBV changes could be measured by intravenously administering an iron oxide contrast agent, Feraheme23 (Figure 3D). This allowed CBV to be validated using an established and completely independent modality. After a short, 1-s electrical stimulation of the contralateral forepaw (9 Hz, 3 mA, 0.5-ms pulse width), we observed a rapid increase in GCaMP signal followed by an undershoot (Figures 3E–3Q). Such an undershoot is absent in cultured neurons where Hb is not present and rarely observed in two-photon imaging data, where neurons and blood vessels are mostly segregated in space (Chen et al., 2013; Mittmann et al., 2011). The tdTomato signal decreased significantly upon forepaw stimulation and showed a much longer delay in its kinetics, but its peak temporally matched that of the GCaMP undershoot (Figure 3F). Using methods similar to that described in Figure 2, but with spectral photometry datapoints across 575- 699-nm wavelengths over time, we derived (t) and (t) (Figures 3G and 3H). Interestingly, the derived ΔCHbR data exhibited a rapid initial increase (Figure 3H) before a more prolonged reduction, a phenomenon well documented in the optical literature (Devor et al., 2003) and termed “initial dip” in blood-oxygen-level-dependent fMRI (Hu and Yacoub, 2012). This initial increase in HbR was not detectable in the raw tdTomato data, indicating that our method is capable of deriving parameters with temporal kinetics distinct from the detected raw fluorescence activity. Figure 3J shows the corrected GCaMP signals, where the after-stimulus undershoot no longer exists. Accordingly, Figure 3K shows a flattened tdTomato trace after the correction. The derived ΔCHbT(t) (Figure 3I) highly resembled the CBV changes measured by fMRI (Figure 3L), and these two time-courses correlated significantly (Figure 3M). The derived ΔCHbT showed higher sensitivity than MRI-CBV (Figure 3N). We observed no significant time lag between the ΔCHbT and CBV-fMRI time courses (Figures 3O and 3P).
Figure 3.
In vivo quantification of HbO and HbR changes using tdTomato emission in fiber photometry and correction of GCaMP signals for Hb absorption
(A) Preparation for fiber photometry measurement of GCaMP and tdTomato signals in S1FL.
(B) Confocal images showing GCaMP and tdTomato expression.
(C) GCaMP and tdTomato emission spectra from 488-nm excitation.
(D) Concurrent CBV measurement was achieved by MRI and intravenous injection of long-circulating iron oxide nanoparticles (Feraheme). The inset shows a map of group-level fMRI activation induced by forepaw stimulation (corrected p < 0.05, n = 5 hemispheres).
(E–L) Peri-stimulus heatmaps showing repeated trials and average response time courses of the changes in measured GCaMP, measured tdTomato, derived HbO, derived HbR, derived HbT, corrected GCaMP, corrected tdTomato, and measured MRI-CBV, respectively. The gray-shaded segment indicates the forepaw stimulation period. All color bars use the same unit as the y axis of the time course.
(M) Two-dimensional histogram summarizing the changes in HbT and MRI-CBV in 5 hemispheres and 27 trials. The color bar indicates counts on the log scale.
(N) HbT derived from photometry exhibited higher sensitivity than MRI-CBV (∗∗∗p < 0.001, paired t-test).
(O) Cross-correlation at the trial level between the Z scores of the derived HbT and the Z scores of the MRI-CBV over the 10-s peri-stimulus period.
(P) Scatterplots showing the cross-correlation coefficients (left) and time lag (right) of all trials.
(Q and R) Correction of pseudo-undershoot artifacts in GCaMP time courses of (Q) short (1 s), and (R) long (10 s) stimulations, respectively (∗∗∗p < 0.001, paired t-test).
All error bars represent standard deviation.
See also Figures S6, S9, and S10.
Figures 3Q and 3R demonstrate the corrections of pseudo-undershoot artifacts in GCaMP time courses of short (1 s) and long (10 s) stimulations, respectively. These artifacts after short bursts of activation were also apparent when optogenetic stimulation was applied in the brain (Figures S6A–S6D). We also observed a significant correction effect on GCaMP response amplitudes (Figure S6E). Without proper correction of Hb absorption artifacts, the accuracy of quantified fluorescence signals is compromised, leading to misinterpretation of fiber photometry data. This problem could become exacerbated in the absence of time-locked stimulations because the desired activity could be superimposed on physiological hemodynamic signal changes.
Effect of Hb absorption on sustained GCaMP signal measurement using fiber photometry
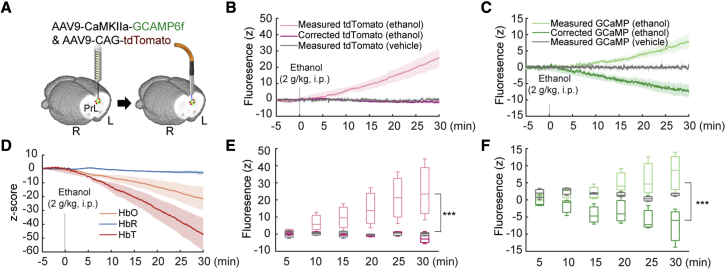
To demonstrate the effect of Hb absorption on GCaMP signals over an extended period of time, we leveraged an acute ethanol challenge protocol (Broadwater et al., 2018; Vaagenes et al., 2015) that has been shown to decrease neuronal activity (Lotfullina and Khazipov, 2018) as well as cerebral perfusion (Broadwater et al., 2018; Kelly et al., 1997; Sano et al., 1993). We virally expressed GCaMP using an AAV under the CaMKIIα promoter and tdTomato under the CAG promoter in the prelimbic cortex (PrL) (Figure 4A) and then implanted an optical fiber in this target area. On the day of the experiment, ethanol (2 g/kg, 20% [v/v], intraperitoneally [i.p.]) was given 5 min after onset of fiber photometry recording. After ethanol administration, we observed robust increases in the raw, measured tdTomato (Figure 4B) and GCaMP (Figure 4C) signals. These changes were ethanol specific because we did not observe any changes in a separate session where the same group of subjects received a vehicle injection (an equal volume of 0.9% saline, i.p.) (Figures 4B and 4C). Next, we derived , , and (Figure 4D) from the ethanol data using correction methods identical to that described in Figure 3 and found robust decreases in and , in agreement with studies showing reduced Hb affinity for oxygen (Van De Borne et al., 1997) and vasoconstriction (Kudo et al., 2015) following acute alcohol administration. Following our proposed correction, the tdTomato trace flattened as expected (Figures 4B and 4E). Importantly, the trend of the GCaMP signal shifted from unexpected positive changes to the expected negative changes (Lotfullina and Khazipov, 2018; Figures 4C and 4F). In another two proof-of-concept experiments, we also noted the influence of Hb absorption on anesthetics- and pharmacology-induced fluorescence signal changes (Figure S7). These data highlight the importance of correcting for Hb absorption in fiber photometry studies because misinterpretation may otherwise occur.
Figure 4.
Hb absorption effects on sustained GCaMP signal measurement using fiber photometry
(A) Preparation for fiber photometry measurement of GCaMP and tdTomato signals in the PrL region.
(B) Measured and corrected tdTomato time courses before and after ethanol administration. The same volume of saline was administered in a separate session to generate vehicle control data within the subject.
(C) Measured and corrected GCaMP time courses before and after ethanol administration. The same volume of saline was administered in a separate session to generate vehicle control data within the subject. Data were collected concurrently with those shown in (B).
(D) Derived HbO, HbR, and HbT time courses before and after ethanol administration. Higher HbT Z scores were attributed to the lower baseline variation after addition of HbO and HbR.
(E) Measured and corrected tdTomato signals were sampled and displayed at 5 min. A significant effect was found between the three groups, and a post hoc test showed a significant difference between the measured and corrected signals. ∗∗∗p < 0.001, two-way ANOVA (n = 3 subjects). Error bars represent standard deviation.
(F) Measured and corrected GCaMP signals were sampled and displayed at 5-min intervals. A significant effect was found between the three groups, and a post hoc test showed a significant difference between the measured and corrected signals. ∗∗∗p < 0.001, two-way ANOVA (n = 3 subjects). Error bars represent standard deviation.
See also Figure S7.
Correction of Hb absorption artifacts in other fiber photometry settings
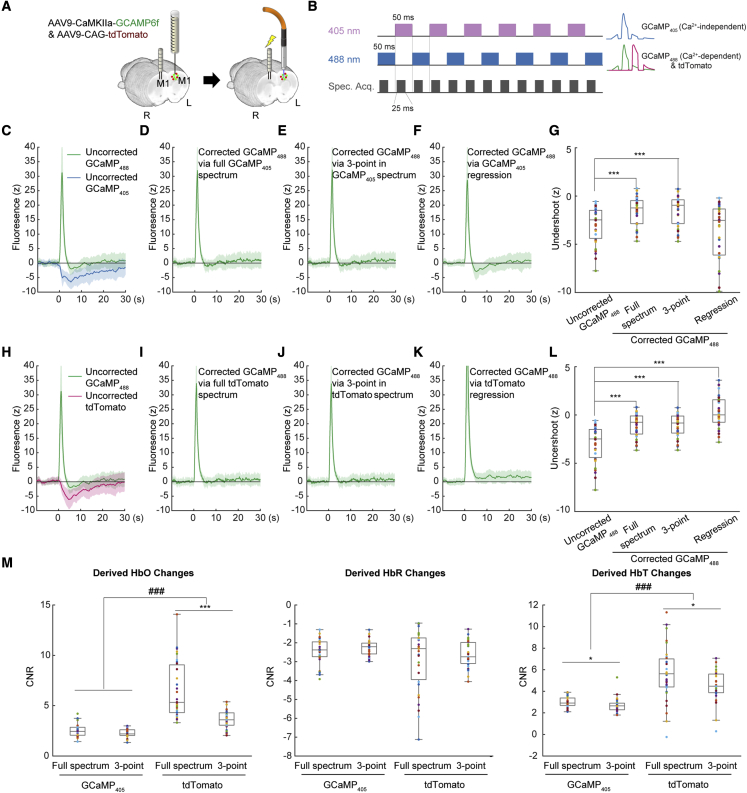
To expand the utility of our method, we modeled and compared the Hb absorption correction techniques among three settings. The first method involves using a spectrometer with a sequence that interleaves between 405 and 488 nm excitation. The GCaMP isosbestic point is around 405 nm (Tian et al., 2009; Dana et al., 2019), where excitation light delivered at this range would evoke GCaMP emission photons irrelevant to the changes in calcium concentration. The GCaMP emission signals from the 405-nm excitation is abbreviated as GCaMP405, in contrast to GCaMP488 excited by the 488 nm laser. The fact that apparent Hb absorption exists at the GCaMP405 emission spectrum (Figure S1A) makes this an appealing alternative approach when another activity-independent fluorescent protein is not expressed. In essence, this provides reference fluorescence signals similar to that of EYFP (Figure 2) or tdTomato (Figure 3) and allows and to be calculated for Hb absorption correction. The second method involves using GCaMP405 data from a spectrometer but only selecting three spectral datapoints to mimic the settings using band-pass filters with a limited number of photodetectors (i.e., without using the full GCaMP405 spectrum for correction). The third method involves regressing out the activity-independent GCaMP405 fluorescence signal intensity as in Kim et al. (2016). To provide data for these evaluations, we virally expressed GCaMP6f using an AAV under the CaMKIIα promoter and tdTomato under the CAG promoter in the primary motor cortex (M1) of the left hemisphere and implanted a microelectrode into the M1 of the right hemisphere, allowing electrical stimulation to be delivered (60 Hz, 300 μA, 0.5-ms pulse width for 2 s); thus, time-locked GCaMP signal changes can be induced in the left M1 (Figure 5A). The interleaved excitation and acquisition sequences are shown in Figure 5B. The raw responses of GCaMP405 and GCaMP488 to stimulation are shown in Figure 5C. Similar to Figures 3Q and 3R, we observed a rapid increase in GCaMP488 signal, followed by a pseudo-undershoot (Chen et al., 2013; Mittmann et al., 2011). We then compared the three settings using a single set of in vivo data, thus avoiding subject and trial-by-trial variability confounds. With respect to the three settings, we present (1) corrected GCaMP488 using and derived from the full GCaMP405 spectrum in Figure 5D, (2) corrected GCaMP488 using and derived from three spectral datapoints from the GCaMP405 spectrum in Figure 5E, and (3) corrected GCaMP488 by regressing out the GCaMP405 signal intensity in Figure 5F. Comparisons of the post-stimulus pseudo-undershoot artifacts suggested successful restoration of the GCaMP488 signals (i.e., signals rapidly return to zero) via both Hb absorption-based correction methods but not via the regression-based method (Figure 5G). Because tdTomato signals are available from 488-nm excitation (Figure 5B), we performed a similar set of analysis using tdTomato instead of GCaMP405 emission signals (Figures 5H–5K) and observed successful restoration of GCaMP488 signals using both Hb absorption-based correction methods but sustained elevated signals using the regression-based method (Figures 5H–5L). No significant difference was observed between the full spectrum versus the three-point correction methods (Figures 5G and 5L). As expected, the calculated HbT from the GCaMP405 or tdTomato spectrum exhibited a lower CNR when using only three spectral datapoints compared with the full spectrum (Figure 5M).
Figure 5.
Correction of Hb absorption artifacts in other fiber photometry settings
(A) Preparation for fiber photometry measurement of GCaMP and tdTomato signals in the left M1, with the right M1 receiving local micro-electrical stimulation.
(B) Interleaved acquisition paradigm with 405- and 488-nm excitation lasers. Ca2+-independent GCaMP signals (GCaMP405) were excited by a 405-nm laser, whereas Ca2+-dependent GCaMP signals (GCaMP488) and tdTomato signals were excited by a 488-nm laser. This interleaved paradigm was used because GCaMP405 and GCaMP488 share nearly identical emission spectra and cannot be spectrally unmixed. Data were acquired at 50% duty cycle every 50 ms using a box paradigm to avoid spectrometer frame loss during trigger mode. The final sampling rate is 10 Hz for all signals.
(C) Measured GCaMP488 and GCaMP405 signals without any correction.
(D) Corrected GCaMP488 using HbO and HbR dynamics derived from the full GCaMP405 spectrum.
(E) Corrected GCaMP488 using HbO and HbR dynamics from 3 datapoints in the GCaMP405 spectrum (at 502, 523, and 544 nm).
(F) Corrected GCaMP488 by regressing out the GCaMP405 signal dynamics.
(G) Comparisons of the post-stimulus undershoot artifacts among uncorrected GCaMP488 and corrected GCaMP488 signals from three different settings, as shown, respectively, in (D)–(F). ∗∗∗p < 0.001, paired t-test (4 subjects, 31 trials).
(H) Measured GCaMP488 and tdTomato signals without any correction.
(I) Corrected GCaMP488 using HbO and HbR dynamics derived from full the tdTomato spectrum.
(J) Corrected GCaMP488 using HbO and HbR dynamics from 3 datapoints in the tdTomato spectrum (at 578, 594, and 610 nm).
(K) Corrected GCaMP488 by regressing out the tdTomato signal dynamics.
(L) Comparisons of the post-stimulus undershoot artifacts among uncorrected GCaMP488 and corrected GCaMP488 signals from three different settings, as shown, respectively, in (I)–(K). ∗∗∗p < 0.001, paired t-test (4 subjects, 31 trials).
(M) Sensitivity of HbO, HbR, and HbT derived from full or 3-point emission spectra. A comparison was made between these two conditions and between use of GCaMP405 and tdTomato signals. ∗∗∗p < 0.001, paired t-test (4 subjects, 31 trials); ###p < 0.001, two-way ANOVA (4 subjects, 31 trials).
All error bars represent standard deviation.
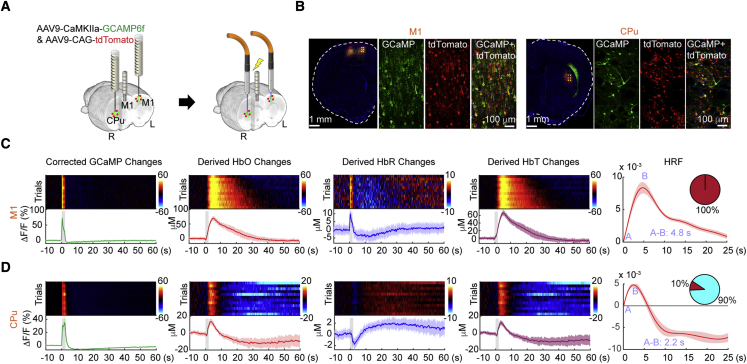
Brain-regional differences in hemodynamic response function
Because our proposed technique allows simultaneous measurements of neuronal activity (e.g., GCaMP) and vascular responses (e.g., ) with minimized Hb absorption artifacts, it offers a significant added benefit for studying the relationship between the two. This relationship, often termed the hemodynamic response function (HRF), governs data interpretation of many noninvasive neuroimaging technologies such as fMRI, intrinsic optics, and functional ultrasound (Taylor et al., 2018). Although use of a canonical HRF to model activity throughout the brain remains the most common practice in the field, accumulating evidence shows that HRF is region dependent (Devonshire et al., 2012; Taylor et al., 2018) and that a thorough understanding of HRF affected by regional cell types, neurochemicals, and subject conditions is crucial to accurately interpret most neuroimaging data (Lecrux and Hamel, 2016). To demonstrate the utility of our proposed technique for computing region- and sensor-dependent HRFs (Chao et al., 2022), we first described the method to compute HRF (Figure S8A), similar to that in Chao et al. (2022), and then validated the results on a dataset not used in HRF computation (Figure S8B). Next, we virally expressed GCaMP6f using an AAV under the CaMKIIα promoter and tdTomato under the CAG promoter in the M1 of the left hemisphere and the caudate putamen (CPu; also known as the dorsal striatum) of the right hemisphere (Figures 6A and 6B). We implanted a microelectrode into the M1 of the right hemisphere (Figure 6A) so that transcallosal and corticostriatal stimulation could be achieved simultaneously. Upon electrical stimulation (60 Hz, 300 μA, 0.5-ms pulse width for 2 s), we observed robust GCaMP changes in the left M1 and right CPu (Figures 6C and 6D), which are both monosynaptically connected to the stimulation target. After calculation of Hb concentration changes, Hb absorption correction, and computation of HRFs, we observed that the M1 and CPu exhibited distinct HRF profiles (Figures 6C and 6D). Interestingly, we also observed distinct HbR polarities between these two regions.
Figure 6.
Regional differences in brain HRF
(A) Preparation for fiber photometry measurement of GCaMP and tdTomato signals in the left M1 and right CPu. Both have a direct anatomical connection with the right M1 receiving local micro-electrical stimulation.
(B) Confocal images showing expression of GCaMP and tdTomato in the M1 and CPu.
(C and D) Peri-stimulus heatmaps and response time courses of the changes in corrected GCaMP, derived HbO, derived HbR, and derived HbT and modeled HRFs in the M1 and CPu, respectively. Pie charts in the insets indicate the ratio between positive (red) and negative (blue) HRF polarities. All pre-stimulus heatmap color bars use the same unit as the time course y axis. Error bars represent standard deviation (4 subjects, 12 trials).
See also Figure S8.
Discussion
Techniques measuring fluorescence signals in vivo, such as fiber photometry (Liang et al., 2017; Schlegel et al., 2018; Chao et al., 2022), one-photon microscopy (Bollimunta et al., 2021), two-photon microscopy (Baraghis et al., 2011), or macroscopic imaging through a thinned skull or implanted window (Cramer et al., 2021), may suffer from Hb absorption. Unlike many imaging techniques that provide spatial information, fiber photometry does not resolve fluorescence signals emitted from individual cells, and its analysis cannot spatially circumvent the blood vessels. Instead, fiber photometry uses an optical fiber to collect all photons from the targeted volume (Figures S2B–S2D). With the conventional fiber photometry setting, the targeted volume has been reported to be 105–106 μm3 with a recording depth approximately 200 μm from the fiber tip (Pisanello et al., 2019). Our simulated detection volume is well within this range (Figure S9A). With the average blood volume fraction in the brain and the Hb concentration documented in the literature (van Zijl et al., 1998), about 9,860 μM Hb would appear in the photometry-detectable brain volume and affect the accuracy of fluorescent sensor measurement by absorbing excitation and emission photons. Under normal physiological conditions, , , and could change up to 40 μM (Hillman, 2014), and CBV could change up to ∼20% (Uludağ and Blinder, 2018; Hua et al., 2019) in response to neuronal activity, making such correction a time dependent problem. Our derived changes in , , and (Figures 2F, 2G, 2J, and 3G–3I) and measured rhoCBV (Figure 2H) and MRI-CBV (Figure 3L) confirm that the amplitudes of those changes are possible in vivo. Our data indicated that roughly 20% of the CBV increase would contribute to a 4.08% decrease in green fluorescence signal changes (Figure S9B; 95% confidence interval [CI], 1.33%–6.83%) and 2.20% decrease in red fluorescence signal changes (Figure S9C; 95% CI, 0.57%–3.84%). This level of fluorescence signal changes (ΔF/F) on green and red sensors is commonly reported in the photometry literature (Chen et al., 2013; Sun et al., 2018) and could be interpreted as changes in specific sensor activity when caution was not exercised. Conventionally, in brain regions where CBV increases after local neuronal activation, pseudo-negative fluorescent sensor activity would be expected. However, in cases where CBV decreases with neuronal inhibition (e.g., Figure 4), or on occasion with activation (Shih et al., 2009, 2011, 2014; Schridde et al., 2008; Mishra et al., 2011), a pseudo-positive fluorescence signal would occur. Although several techniques are readily available to measure CBV (Kwong et al., 1992; Mintun et al., 1984; Sakai et al., 1985; Siegel et al., 2003; Figures 2D–2H and 3L), correction of Hb absorption cannot be achieved simply by regressing the CBV signal measured concurrently (Figures S9D and S9E). This is mainly due to the fact that HbO and HbR absorption are wavelength dependent, and their respective concentrations change over time (Meng and Alayash, 2017; Prahl, 1999; Figures S1A and S1B). Our simulation suggested that the accuracy of the fluorescence signal could be compromised even without a net change in CBV (see Figure 1D for plots without a shaded background). This makes it crucial to quantify and so that the true changes in the fluorescence signal can be restored.
At present, two major fluorescence-measurement strategies are used in fiber photometry. One is non-spectrally resolved using a narrow band-pass filter paired with a photodetector (Gunaydin et al., 2014; Kim et al., 2016; Lazarjan et al., 2021). The other is spectrally resolved using a photomultiplier tube array (Cui et al., 2013, 2014) or a spectrometer (Meng et al., 2018; Sun et al., 2018; Chao et al., 2022). The non-spectrally resolved method has the advantage of recording fluorescence signals at a faster speed and can be used with modulated excitation lights and a lock-in amplifier (Gunaydin et al., 2014). In contrast, the spectrally resolved method provides wavelength-dependent information for the fluorescence, where the detected photons can be matched with the specific spectral profile of the sensor to improve specificity or perform spectral unmixing for multiplexing (Meng et al., 2018). If a single non-spectral photodetector is used and and cannot be obtained, the Hb absorption spectra (Figures S1A and S1B) and our simulation results (Figure 1D) indicated that far-red sensors are less prone to Hb absorption artifacts. This highlights the importance of continually developing fluorescent sensors in the red-shifted and far-red range (Patriarchi et al., 2020; Shcherbakova, 2021).
Although we did not obtain photodetector-based data in this work, we mimicked the photodetector-based setting by sampling data only from three specific spectral datapoints to enter our Hb absorption correction pipeline. These modeling data support the utility of our proposed technique in the photodetector setting (Figures 5E and 5J). The detector in the spectrometer used in this study is a high-end back-thinned charge-coupled device (CCD) array (90% quantum efficiency) with TE cooling (40°C–50°C below ambient) to reduce the dark current (Ocean Insight, 2022). For a rough comparison of the reported detector sensitivity, the most common commercial photodiode detectors used in fiber photometry have a sensitivity (reported as responsivity) of 0.38 A/W at 550 nm (Doric, 2021), which is similar to 0.40 A/W at 550 nm of the system used in this study (calculated based on 90% quantum efficiency). We do not have access to ground-truth GCaMP signals (i.e., the “real” GCaMP signal) using fiber photometry to make an unequivocal comparison. However, GCaMP measurements from cultured neurons without Hb or using two-photon imaging to circumvent blood vessels have collectively demonstrated the absence of post-stimulus undershoot (Chen et al., 2013; Mittmann et al., 2011). Leveraging these observations in the literature, together with the results we obtained in Figures 2E and 3F, we believe in the utility of our proposed Hb absorption correction method. We recognize that the post-stimulus undershoot artifacts may not significantly compromise all fiber photometry studies, especially for those studying brief time-locked stimulations/tasks. We consider the post-stimulus undershoot artifact to be a useful marker to benchmark methods dealing with Hb absorption. We suggest caution when studying spontaneous ongoing neuronal/neurochemical activity dynamics or slower brain-state changes using fiber photometry because the HbO and HbR concentration changes may cause erroneous readings of the desired sensor activity (Figures 4 and S7). Our results also suggest that regression-based models may be suboptimal to restore the GCaMP signal changes in fiber photometry (Figures 5F–5K) and highlighted the benefits of CNR when using a spectrometer system to derive HbO and HbR changes (Figure 5M).
Fluorescence signals generated from GCaMP isosbestic excitation can be used as the activity-independent fluorescent reference signal in the analytical method proposed in this study and derive and to restore the functional GCaMP signals (Figure 5). Potential limitations of this approach are (1) trade-off in temporal resolution because interleaved excitation is required, (2) relatively low CNR (Figure 5M; approximately one-tenth of the emission signal was obtained compared with 488-nm excitation), (3) higher susceptibility to pH-related confounding factors (Shaner et al., 2005; Barnett et al., 2017), and (4) although GCaMP has an isosbestic point at an amenable wavelength for this method, other functional fluorescent sensors may perform optimally at a different wavelength (Akerboom et al., 2012; Sun et al., 2020).
The opportunity to examine the interactions between cell types and/or neurotransmitters has become increasingly popular in fiber photometry. When the green and the red spectrum are used to encode fluorescent sensor activities, it may be possible to use the interleaved sequence, as shown in Figure 5, or spectral signals from the unmixing residuals (i.e., the remaining autofluorescence) to achieve hemodynamic correction (Figure S10). In our hands, the residual method suffers significantly from CNR issues because of very low residual photon counts. However, this approach may demonstrate its utility with the advent of a more sensitive spectrometer system or an improved unmixing algorithm. Considering the increasing need for multiplexing, a valuable next step should consider establishing an experimental pipeline to achieve motion and Hb absorption artifact-free measurement of green and a red fluorescent sensor (e.g., GCaMP and jRGECO) in freely moving animals. This may be achieved by incorporating a far-red activity-independent fluorescent sensor for motion correction first (because Hb absorption is negligible in this range; Figure S1), followed by Hb absorption correction using data from isosbestic GCaMP excitation, as shown in Figure 5.
Given the irreplaceable roles of noninvasive hemodynamics-based neuroimaging techniques (such as fMRI, near-infrared, intrinsic optics, functional ultrasound, positron emission tomography, etc.) for studying brain function, identifying brain region-dependent HRF is crucial for accurate interpretation of hemodynamics-based neuroimaging data. Current standard fMRI analysis uses one HRF shape throughout the brain. Increasingly, literature has pointed to potential problems with this analysis pipeline because HRF is brain region dependent (Devonshire et al., 2012; Ekstrom, 2021; Jin et al., 2020; Sloan et al., 2010). The current study and other pioneering studies (Albers et al., 2017; He et al., 2018; Liang et al., 2017; Schlegel et al., 2018; Schmid et al., 2016; Schulz et al., 2012; Schwalm et al., 2017; Wang et al., 2018a, 2018b; Zhang et al., 2022; Chao et al., 2022) have demonstrated that fiber photometry could be seamlessly coupled with fMRI and provide ground-truth cellular activity to help unravel the relationship between neuronal and hemodynamic responses. These studies have bridged a crucial knowledge gap in interpretation of fMRI data. The method proposed in this study expands the scope of this research area because it generates hemodynamic signals “free of charge” with better sensitivity than fMRI (Figure 3N), and the results can be readily compared with the concurrently recorded fluorescent sensor activity. With spectral photometry recording becoming increasingly popular, the proposed method opens up a unique opportunity to compute HRF in a cell-type-dependent, stimulation-circuit-dependent, and neurotransmission-dependent manner. We expect that this information can be obtained by performing a meta-analysis on fiber photometry data shared through an open-science platform and contribute significantly to a more accurate interpretation of hemodynamics-based neuroimaging results.
We demonstrated that significant Hb absorption artifacts in fiber photometry data may cause inaccurate measurements and, therefore, erroneous interpretation. We proposed a method that addresses this issue by quantifying dynamic HbO and HbR absorption changes using activity-independent fluorescence spectra and demonstrated that these additional quantitative measurements were efficient in correcting the artifact and retrieve the desired, unperturbed fluorescence signals in fiber photometry. This approach simultaneously measures HbO, HbR, and the activity of a selected fluorescent sensor, providing an additional experimental capability to investigate the influence of neuronal or neurochemical activity on cerebral hemodynamics. Through a proof-of-principle study, we provide direct evidence that HRF is brain region dependent, suggesting that analysis of hemodynamics-based neuroimaging data should not apply a single HRF across all brain regions.
Limitations of the study
Lack of access to ground-truth GCaMP signals is a limitation when making unequivocal comparisons between the “corrected” GCaMP and the “real” GCaMP signals. Second, post-stimulus undershoot may not be significant in all fiber photometry studies. Finally, we recognize that applying this method in a non-spectrally resolved fiber photometry system might be challenging because multiple band-pass filters, each combined with a photodetector, are required.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| AAV5-CaMKIIα-EYFP | UNC Vector Core | N/A |
| AAV9-CaMKIIα-GCaMP6f-WPRE-SV40 | U.Penn Vector Core | AV-9-PV3435 |
| AAV9-CAG-tdTomato | UNC Vector Core | N/A |
| AAV9-CaMKIIα-ChR2-EYFP | UNC Vector Core | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Rhodamine B, 70,000 MW, Neutral | Fisher scientific | D1841 |
| Phosphate Buffered Saline (10 x) | Sigma-Aldrich | P4417 |
| Lidocaine HCl jelly (2%, 5mL) | UNC SSC Pharmacy | N/A |
| Meloxicam oral suspension | Henry Schein | N/A |
| Software and algorithms | ||
| MATLAB | Mathworks | R2019b |
| RStudio | RStudio Inc. | Version 1.0.136 |
| Oceanview | Ocean Insight | N/A |
| Correction for Hb-absorption artifact in fiber-photometry | In this paper | https://github.com/CAMRIatUNC/HemoCorrection (https://doi.org/10.5281/zenodo.6599234) |
| Computation of hemodynamic response function | Chao et al., 2022 | https://github.com/CAMRIatUNC/Photometry-tools (https://doi.org/10.5281/zenodo.6599442) |
| Other | ||
| 488 nm laser and remote | Coherent | OBIS 488 LS (60 mW) |
| 561 nm laser and remote | Coherent | OBIS 561 LS (50 mW) |
| Spectrometer | Ocean Insight | QEPro-FL |
| Optic patch cable | Thorlabs | FT200EMT (200 mm, 0.39 NA) |
| Uncleaved fiber optic cannula | Thorlabs | CFMC12U-20 (200 mm, 0.39 NA) |
| Bipolar Tungsten electrode | P1 Technologies | MS303T/2C-B/SP ELEC .008/.2MM TW TUNGSTEN (MRI) |
| Dual-edge dichroic mirror | Chroma | ZT488/561rpc |
| Dual-band emission filter | Chroma | ZET488/561m |
| Smart control panel | Med Associate inc. | DIG-716P2 |
Resource availability
Lead contact
Further information and requests for data and resources should be directed to and will be fulfilled by the lead contact, Yen-Yu Ian Shih (shihy@unc.edu).
Materials availability
This study did not generate new materials or new reagents.
Experimental models and subject details
This study employed a total of 26 male Sprague Dawley (SD) rats weighing between 300–600g. All procedures were carried out in accordance with the National Institutes of Health Guidelines for Animal Research (Guide for the Care and Use of Laboratory Animals) and approved by the University of North Carolina (UNC) Institutional Animal Care and Use Committee. These rats were separated into 6 cohorts. In the first cohort (n = 3), EYFP was expressed in the bilateral S1FL using AAV5-CaMKIIα-EYFP (titer 4.2 1012 molecules/mL, UNC Vector Core) to demonstrate the impact of Hb concentration changes on activity-independent fluorescence signals and evaluate the proposed correction method. In the second cohort (n = 4), GCaMP and tdTomato were co-expressed in the bilateral S1FL using a mixture of AAV9-CaMKIIα-GCaMP6f-WPRE-SV40 (titer 1 1013 vg/mL, Penn Vector Core) and AAV9-CAG-tdTomato (titer 3.8 1012 molecules/mL, UNC Vector Core) in a 5:1 ratio to demonstrate the impact of Hb concentration on a commonly used fluorescent sensor and evaluate the proposed correction method. In the third cohort (n = 3), GCaMP and tdTomato were co-expressed in the left PrL using a viral mixture identical to the second cohort. The fourth cohort (n = 4) received a twisted tungsten electrode implanted into the M1, and GCaMP and tdTomato were co-expressed in the contralateral M1 using a viral mixture identical to the second cohort. The fifth cohort (n = 5) was used in a supplementary study of isoflurane challenge and only GCaMP was expressed in the left M1. The sixth cohort (n = 4) received a twisted tungsten electrode implanted into the M1, and GCaMP and tdTomato were co-expressed in the contralateral M1 and ipsilateral striatum using a viral mixture identical to the second cohort. The seventh cohort (n = 3) was used in a supplementary study and had channelrhodopsin2 (ChR2) expressed in the ventroposterior thalamus (VP) using AAV9-CaMKIIα-ChR2-EYFP (titer 3.6 1012 molecules/mL, UNC Vector Core) and GCaMP expressed in the ipsilateral S1 using AAV9-CaMKII-GCaMP6f-WPRE-SV40 (titer 1 1013 vg/mL, Penn Vector Core). All rats in this study were housed under environmentally controlled conditions (12 h normal light/dark cycles, lights on at 7a.m.; 20–23°C and 40–60% relative humidity), with ad libitum access to food and water.
Method details
Monte Carlo simulation for photon transport in fiber-photometry recording
The Monte Carlo simulation of photon transport during fiber-photometry recording was achieved in two steps: 1) excitation light path simulation and 2) emission light path simulation. The goal was to first build a database composed of a large number of possible excitation light paths. After which, an emission starting point can be randomly selected from the excitation light path database for emission light path simulation.
For the excitation light path simulation, we built the database by repeatedly launching excitation photon packets that started with a unity weight = 1. The photon packets deposited weight due to absorption at each step of traveling through the simulated brain, and the photon packet’s current locations and remaining weights were logged along the way. Each excitation photon packet repeatedly underwent the following steps (Figure S3, left panel), until it was either terminated or transmitted out of the brain.
Step 1: Launching a photon packet (Figure S3, left panel a and b):
In our model, we launched our photon packets from an optic fiber tip with a diameter of 200 μm; the range of possible launch directions, or so-called acceptance angles, would be limited by the fiber numerical aperture (NA) according to Snell’s law. For applications, the NA is most commonly expressed as:
| (Equation 2) |
where is the maximum ½ acceptance angle of the fiber, and is the index of refraction of the material outside of the fiber. With this in mind, we simply set the initial launch position randomly within the surface of fiber tip, along with a random initial launch direction that was within the range of acceptance angles. This can be easily done by using three Cartesian coordinates and three direction cosines to determine the initial position and direction, respectively.
For initial position:
| (Equation 3) |
For initial direction:
| (Equation 4) |
where is the radius of the optic fiber, and are the independent random numbers between 0 and 1.
Step 2: Step size selection and photon packet movement (Figure S3, left panel 1-2 and right panel 5–6):
The step size is the distance that the photon packet travels between interaction sites. In our study, we selected a step size using the following function derived from the inverse distribution method and the Beer–Lambert law:
| (Equation 5) |
where is a random number between 0 and 1, and is the total interaction coefficient (the sum of the absorption coefficient () and scattering coefficient () of brain tissue). The and for distinct wavelengths of a traveling photon were derived from the optical absorption and scattering properties provided by an online database from Oregon Medical Laser Center, Oregon Health & Science University (https://omlc.org/software/mc/mcxyz/index.html). We assumed 3% blood with 75% average oxygen saturation in our simulated brain.
Once the step size was selected, the photon packet traveled for distance in the direction defined by direction cosines, and the coordinates were updated as follows:
| (Equation 6) |
Step 3: Absorption and scattering (Figure S3, left panel 3–4 and right panel 7–8):
At each interaction site, photon weight was partially absorbed by brain tissue, and the decreased fraction of the weight can be defined as:
| (Equation 7) |
The weight of the photon packet was then updated as follows:
| (Equation 8) |
Following absorption, the photon packet was scattered. The scattering direction was defined by both scattering angle and polar angle . The scattering angle was determined using the Henyey-Greenstein phase function (Henyey and Greenstein, 1941):
| (Equation 9) |
where refers to scattering anisotropy, which has a value between −1 and 1. When is 0, it generally indicates that the scattering is isotropic. As approaches 1 or -1, the scattering is confined to forwards and backwards, respectively.
The polar angle was assumed to be uniformly distributed between 0 and 2π, as below:
| (Equation 10) |
With these angles and the original direction cosines, we were able to calculate the new set of propagation direction cosines, which can be represented in the global coordinate system as follows:
| (Equation 11) |
Step 4: Photon termination
Steps 2 and 3 were repeated until the weight of the photon packet reached 10−4 or less, then the simulation for this particular photon packet was terminated. The excitation light path and weight of each step were saved to the database, after which a new photon packet simulation was started from step 1.
The emission light path simulation was similar to the excitation light path simulation. Specifically, we need to first determine the initial position, the initial direction, and the initial weight before launching an emission photon packet (Figure S3, right panel C-D). The initial coordinates of each emission photon packet were randomly selected from the excitation light path database. All coordinates in the database had an equal probability to be selected regardless of excitation light path. If the selected coordinates were within an area containing fluorophore, its initial coordinates were determined first, and then the corresponding excitation light path history (from the start to the point where the emission occurred) was recorded. In this study, we assumed the fluorophore existed equally within a 1 mm diameter sphere of viral expression area centered 0.3 mm below the fiber tip.
The initial direction of an emission photon was assumed to have isotropic probability in all directions; therefore, the direction cosines can be derived using the following:
| (Equation 12) |
Lastly, the initial weights were inherited from the remaining weights of the excitation photon packet at the initial coordinates.
Steps 2 and 3 were repeated until the weight of the photon packet reached 10−4 or less, and then the simulation of this photon packet was terminated (step 4). In emission step 4, only when the photon packet reached the fiber tip at smaller than ½ acceptance angle and a weight beyond 10−4, the paired excitation/emission light path and the weight of each step were saved to the database. In order to improve signal-to-noise ratio, we collected 2–2.5 104 pairs of excitation and emission light paths.
The final weight () when the emission photon reaching the optical fiber tip represents the survival rate of the current combined excitation/emission light path. Therefore, the total average pathlength of excitation and emission lights are then calculated by summing each survived photon’s multiplied by its paired excitation and emission pathlength respectively:
| (Equation 13) |
Since the is affected during both excitation and emission phase, the emission measured at different wavelengths have not only distinct average emission pathlength (, but also distinct average excitation pathlength () even when these emission wavelengths are excited by the same excitation wavelength. In this study, only the 488 nm excitation wavelength was used and simulated.
To plot the 488 nm excitation light illuminating distribution (Figures S3A S3B), all excitation light paths’ coordinates generated from the excitation light path simulation, regardless to the emission light detection, were first compressed into 2D coordinates, and then used for the 2D histogram. To plot the 515 or 580 nm emission light receptive probability map (Figures S3B-S3C), the initial emission 3D coordinate of each successfully detected 515 or 580 emission photons were compressed into 2D coordinates and shown in a 2D histogram.
Correction of Hb-absorption
Given the different absorption of HbO and HbR (Figure S1), their dynamic concentration changes must be quantified in order to properly correct fluorescent sensor signals. To address this, we proposed the following methods to derive changes in the HbO and HbR concentration ( and ) using spectral datapoints.
In a simple, non-absorbing and non-scattering medium, the measured fluorescence signal F at any time t is linearly related to the fluorescent protein concentration :
| (Equation 14) |
where and represent the fluorescence signal and the fluorescent protein concentration at the reference time point 0, respectively. When the absorption of the medium is considered, and F is measured at multiple wavelengths , Equation (14) can be further modified according to an established model (Ma et al., 2016):
Then after natural logarithmic transformation:
| (Equation 15) |
where represents the change of the absorption coefficient at excitation wavelength at the time point t compared to the time point 0; represents the change of the absorption coefficient at emission wavelength at the time point t compared to the time point 0; and represent the photon traveling pathlengths at the given wavelength, respectively. Next, HbO and HbR concentration changes ( and ) as a function of time need to be incorporated. When using 488 nm excitation, the absorption term in Equation (15) that contains could be expressed as:
| (Equation 16) |
where and are well-established molar extinction coefficients at different wavelengths of HbO and HbR, respectively (Prahl, 1999).
At the time point t, the measured fluorescence ratio after natural logarithmic transformation, ranged from to could be put together in a vector form :
| (Equation 17) |
Additionally, two vectors A and B could be used to represent the following:
| (Equation 18) |
| (Equation 19) |
With any activity-independent fluorescence signals, the last component should be 0. Practically, we designate an error value σ. Taken together, Equation (15) could then be simplified and reorganized as Equation (20). To solve and at any time point t, we used generalized method of moment (GMM) (Hansen, 1982) to minimize L in Equation (21). By performing the GMM computation for each time point, we can obtain time-courses of , and .
| (Equation 20) |
| (Equation 21) |
In this case, we have numerous spectral datapoints that can be used to resolve two unknowns that are of interest – and . After which, and can be calculated using Equation (16) and can be subsequently calculated using Equation (15) to correct for HbO and HbR absorptions.
Stereotactic surgery
For all surgical procedures, rats were initially anesthetized with 5% isoflurane, and maintained with a constant flow of 2–3% isoflurane mixed with medical air. Rectal temperature was continuously monitored and maintained within 37 ± 0.5°C using a feedback-controlled heating pad (Harvard Apparatus, Model 557,020, Holliston, MA). Rats were head-fixed to a stereotactic frame (Kopf Instruments, Model 962, Tujunga, CA). The skin was opened to expose the skull surface, and burr holes were prepared according to experimental coordinates. All coordinates used for microinjection are listed as follows. S1: AP = +0.5 mm and ML = +3.7 mm, DV = 1.3 mm, M1: AP = +1.8 mm and ML = −2.5 mm, DV = 2.0 mm, Striatum: AP = 0.5 mm and ML = 4.0 mm, DV = 5.2 mm, VP: AP = −2.8 mm and ML = +2.5 mm, DV = 6.5 mm, PrL: AP = 2.2 mm and ML = 0.9 mm, DV = 4.1 mm. Microinjections were performed at a flow rate of 0.1 μL/min for 1 μL, and an additional 10 min was given for virus diffusion prior to slow retraction of the microsyringe needle. The burr holes were then sealed with bone wax (Fisher Scientific, Pittsburgh, PA), and the wound was sutured. A month after virus microinjection, the skin was reopened to expose the skull, then bone wax was removed and optical fibers (200 μm in diameter; NA: 0.39) were chronically implanted to the coordinates 0.2–0.5 mm above the virus injection sites. Four MR-compatible miniature brass screws (Item #94070A031, McMaster Carr, Atlanta, GA) were anchored to the skull, then the surface of the skull was covered with dental cement to seal implanted components and the wound was sutured to further protect the surgical site.
At the end of each surgical procedure, lidocaine jelly (#866096, Henry Schein Inc., Melville, NY) was applied around the surgical wound to relieve pain and prevent the rat from scratching the wound. Meloxicam (#6451720670, Henry Schein Inc., Melville, NY) was also given by oral administration for further pain relief. Rats were allowed at least 1 week to recover from surgical procedures before any further experiments.
Experimental setup
The spectrally resolved fiber-photometry system in this study replicates an established system described previously (Meng et al., 2018). Laser beams from a 488 nm 60 mW continuous wave (CW) laser (OBIS 488 LS-60, Coherent, Santa Clara, CA) and a 561 nm 50 mW CW laser (OBIS 561 LS-50, Coherent, Inc.) are aligned and combined by broadband dielectric mirrors (BB1-E02, Thorlabs, newton, NJ) and a long-pass dichroic mirror (ZT488rdc, Chroma Technology Corp), then launched into a fluorescence cube (DFM1, Thorlabs, newton, NJ). Extra neutral density filters (NEK01, Thorlabs, newton, NJ) are placed between the combined laser beam and the fluorescence cube to adjust the final laser power. The fluorescence cube contains a dichroic mirror (ZT488/561rpc, Chroma Technology Corp) to reflect and launch the combined laser beam through an achromatic fiber port (PAFA-X-4-A, Thorlabs, newton, NJ) into the core of a 105/125 mm core/cladding multi-mode optical fiber patch cable (FG105UCA, Thorlabs, newton, NJ). The distal end of the patch cable is connected to an implantable optical fiber probe for both excitation laser delivery and emission fluorescence collection. The emission fluorescence collected from the fiber travels back along the patch cable into the fluorescence cube, passes through the dichroic mirror and an emission filter (ZET488/561 m, Chroma Technology Corp, Bellows Falls, VT), then launches through an aspheric fiber port (PAF-SMA-11-A, Thorlabs, newton, NJ) into the core of an AR-coated 200/230 mm core/cladding multi-mode patch cable (M200L02S-A, Thorlabs, newton, NJ). The AR-coated multi-mode patch cable is connected to a spectrometer (QE Pro-FL, Ocean Optics, Largo, FL) for spectral data acquisition, which can be operated by a UI software OceanView (Ocean Optics, Largo, FL). In order to achieve concurrent recording during fMRI, trigger mode is used in OceanView, where the photometry system is synchronized with MRI using an Arduino micro-controller board.
The stimulation system uses a DAQ board (1208Hs-2AO, Measurement Computing Corp., Norton, MA) to send out stimulus triggers according to the stimulus paradigm set in a homemade software program. During fMRI experiments, the DAQ synchronizes stimulation pulses via triggers from the MRI system. The stimulation pulses were driven by a constant current stimulus isolator (A385RC, World Precision Instruments, Sarasota, FL) for forepaw and micro-electrical stimulation experiments; and by a 473 nm blue diode laser (Shang Laser and Optics Century, BL473T8-200FC + ADR-800A, Shanghai, China) for optogenetic experiments.
Animal subject preparation and physiology management
The rats were initially anesthetized with 4% isoflurane (Vaporizer #911103, VetEquip Inc., Livermore, CA, USA) mixed with medical air and endotracheally intubated using a 14G x 2"(>400 g) or 16G x 2"(<400 g) i.v. catheter (Surflash Polyurethane Catheter, TERUMO, Somerset, NJ, USA). Respiration was maintained by a ventilator (SAR-830 or MRI-1, CWE Inc, Ardmore, PA, USA) set at 60 breaths/min and an inspiration time ratio of 40%. A rectal probe was used to monitor core body temperature (OAKTON Temp9500, Cole-Parmer, Vernon Hills, IL, USA) and a capnometer was used to monitor heart rate, peripheral blood oxygen saturation, and end-tidal CO2 (SURGIVET V90041LF, Smith Medical, Dublin, OH, USA). Body temperature was maintained at 37 ± 0.5 °C using a circulating water blanket connected to a temperature adjustable water bath (Haake S13, Thermo Fisher Scientific, Waltham, MA, USA). Ventilation tidal volume was adjusted to keep the heart rate at 300 ± 50 beats per minute, peripheral blood oxygen saturation above 90%, and end-tidal CO2 between 2.8 and 3.2%. End-tidal CO2 values from this capnometer system were previously calibrated against invasive sampling of arterial blood gas, reflecting a partial pressure of carbon dioxide (pCO2) level of 30–40 mm Hg (Shih et al., 2012, 2013). For studies using Rhodamine B for CBV measurements, a bolus dose of 40 mg/kg (Sigma–Aldrich, St. Louis, MO) was injected via tail vein.
Concurrent fMRI scan with fiber-photometry recording
All fMRI data in this study were collected on a Bruker BioSpec 9.4-Tesla, 30 cm bore system with 6.0.1 on an AVANCE II console (Bruker BioSpin Corp., Billerica, MA). An RRI BFG 150/90 gradient insert (Resonance Research, Inc, Billerica, MA) paired with a Copley C700 gradient amplifier (Copley Controls Corp., Canton, MA) was used. A homemade single-loop surface coil with an internal diameter of 1.6 cm was used as a radio-frequency transceiver. Isoflurane concentrations were adjusted to 2% and animals were secured in to a custom-built, MR-compatible rat cradle. Animal physiology was monitored and maintained as described in the previous paragraph.
Upon stabilizing the animals, a pair of needle electrodes was inserted under the skin of forepaw for stimulation, or electrode/optical fiber patch cables were connected according to experimental design. Before connecting the fiber-photometry patch cable, all light in the room was turned off, the final output power of the 488 nm laser and 561 nm laser were 51.3 ± 18.6 μW (mean ± SD, n = 26), which were adjusted to balance spectral amplitudes (Meng et al., 2018). The 100 μW power is the maximum we suggested when the virus expression is less robust. This power range is consistent with published fiber-photometry studies (Gunaydin et al., 2014; Jones-Tabah et al., 2021, p.; Liang et al., 2017; Zhang et al., 2021). A background spectrum was measured as a reference by pointing the fiber tip to a nonreflective background in the dark room. This background spectrum was then automatically subtracted by OceanView during photometry recording.
Following setup processes, the cradle was pushed into MRI bore, and a bolus of dexmedetomidine (0.025 mg/kg; Dexdormitor, Orion, Espoo, Finland) cocktailed with paralytic agent rocuronium bromide (4.5 mg/kg; Sigma–Aldrich, St. Louis, MO) was injected into the tail vein (Chao et al., 2018). Fifteen minutes after bolus injection, a continuous intravenous infusion of dexmedetomidine (0.05 mg/kg/h) and rocuronium bromide (9 mg/kg/h) cocktail was initiated and the isoflurane concentration was adjusted to 0.5–1% for the entire scanning period.
Magnetic field homogeneity was optimized first by global shim and followed by local first- and second-order shims according to B0 map. Anatomical images for referencing were acquired using a rapid acquisition with relaxation enhancement (RARE) sequence (12 coronal slices, thickness = 1 mm, repetition time (TR) = 2500 ms, echo time (TE) = 33 ms, matrix size = 256 × 256, field-of-view (FOV) = 25.6 × 25.6 mm2, in plane resolution 0.1 × 0.1 mm, average = 8, RARE factor = 8). The center of the fifth slice from the anterior direction was aligned with the anterior commissure. Cerebral blood volume (CBV) fMRI scans were acquired using a multi-slice single-shot gradient echo echo-planar imaging sequence (GE-EPI) (slice thickness = 1 mm, TR = 1000 ms, TE = 8.1 ms, matrix size = 80 × 80, FOV = 25.6 × 25.6 mm2, in plane resolution 0.32 × 0.32 mm, bandwidth = 250 kHz), with the same image slice geometry imported from the previously acquired T2-weighted anatomical image. A session of GE-EPI scans with 300 repetitions was taken, and at about the 100th scan, Feraheme (30 mg Fe/kg, i.v.) was administered for CBV percentage change calculations.
CBV fMRI data processing and statistical analyses
All fMRI data were analyzed using the Analysis of Functional Neuroimages (AFNI) (Cox, 1996) general linear model (GLM) framework (Worsley and Friston, 1995). All EPI images were skull-stripped, and slice-timing was corrected. Then automatic co-registration was applied to realign time-courses within subjects to correct subtle drift of EPI images. Finally all EPI images were aligned to a T2-weighted rat brain template (Valdés-Hernández et al., 2011) to generate normalized fMRI images to allow for group-level comparisons. Gaussian smooth (FWHM = 0.5 mm) was performed before feeding into the GLM fitting. To test the significant consistency of the stimulus-evoked responses at the group-level, we employed a parametric one-sample t test implemented in AFNI. The false discovery rate correction was used to adjust for multiple comparisons of fMRI maps (p < 0.05). A 3x3 voxel region of interest (ROI) was placed at the fiber tip in the S1 to extract fMRI time-course data. To account for Feraheme kinetics during the course of the experiments, the following equations were used to calculate the , , and (Decot et al., 2017).
| (Equation 22) |
| (Equation 23) |
| (Equation 24) |
where and represents MR signal intensity after and before Feraheme injection, respectively, and are the MR signal intensities during the stimulation.
Fiber-photometry spectral unmixing
Mixed spectra acquired by fiber-photometry were analyzed using a spectral linear unmixing algorithm (Meng et al., 2018). Briefly, at any time point n, the mixed spectrum was modeled as:
| (Equation 25) |
where and are the normalized reference emission spectra of the two fluorescence signal sources. Coeff1 and Coeff2 are the unknown regression coefficients corresponding to the and respectively. is the unknown constant, and is random error. , , and at each time point were estimated using the lm() function in the RStudio package (RStudio Inc. V1.0.136, Boston, MA).
Modeling the hemodynamic response function
The relationship between neuronal activity and hemodynamic response can be expressed as:
| (Equation 26) |
where the represents hemoglobin fluctuation time-course, the represents neuronal activity time-course, the represents an impulse hemodynamic response function with sampling points, is a constant for baseline offset and is for linear drift over time. Assuming ,this equation can be expressed as:
| (Equation 27) |
Therefore, the , and (slope of ) can be solved using Ordinary Least Squares solution with known and .
To avoid physiological noise contamination, were low-pass filtered with a cutoff frequency at 0.5 Hz before the HRF estimation.
Histology
At the end of the experiments, the rats were euthanized with a mixture of 1–2 mL of sodium pentobarbital and phenytoin sodium (Euthasol, Virbac AH, Inc., Westlake, TX), and transcardially perfused with saline followed by 10% formalin. The brains were removed and stored in 10% formalin overnight, then transferred to a 30% sucrose solution (in 0.1 M phosphate buffer) for 2 to 3 days, until the brains sunk to the bottom of storage bottles. These brains were cut into serial coronal sections (40 μm) using a cryotome (#HM450, Thermo Fisher Scientific, Waltham, MA) and mounted on glass slides. The Fluoro-Gel II Mounting Medium (#17985-50, Electron Microscopy Sciences, Hatfield, PA) was covered on the brain slides to provide DAPI stain and for fluorescence imaging. The slides were imaged using a Zeiss LSM780 confocal microscope.
Quantification and statistical analysis
The fiber-photometry data a nalysis was carried out using customized codes in MATLAB (Mathworks, R2019b). A customized program was written in R to fit Hb changes and called by MATLAB internally. Linear regression (in Figure 5), two-way ANOVA, and paired t test were carried out using MATLAB (Mathworks, R2019b). Statistical data including means, sample numbers n, what n represents and significance values were indicated in the main body or figure legends. One hemisphere in cohort 1 and three hemispheres in cohort 2 were excluded due to the lack of response to forepaw stimulation. All error bars in the figures are SD.
Acknowledgments
We thank Drs. Elizabeth Hillman, Anna Devor, Lindsay Walton, Dom Cerri, and UNC CAMRI members for their helpful discussions and critiques. This work is supported in part by NIH grants (RF1MH117053, RF1NS086085, R01MH126518, R01MH111429, R01NS091236, P60AA011605, U01AA020023, P50HD103573, S10MH124745, and S10OD026796 to Y.Y.I.S.), the Intramural Research Program of the NIH/NIEHS (1ZIAES103310 to G.C.), the Burroughs Wellcome Fund (CASI 5113244 to N.C.P.), and the Arnold and Mabel Beckman Foundation (2021BYI to N.C.P.).
Author contributions
W.-T.Z., T.-H.H.C., G.C., and Y.-Y.I.S. conceived the project and designed the experiments. W.-T.Z., T.-H.H.C., T.-W.W., E.A.O., J.Z., and R.N. implemented the methods. W.-T.Z., T.-H.H.C., Y.Y., S.-H.L., and N.C.P. analyzed data. W.-T.Z., T.-H.H.C., and Y.-Y.I.S. wrote the manuscript with input from all authors. H.Z., G.C., and Y.-Y.I.S. supervised the study and provided funding support.
Declaration of interests
The authors declare no competing interests.
Published: June 29, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.crmeth.2022.100243.
Contributor Information
Guohong Cui, Email: cuig@mail.nih.gov.
Yen-Yu Ian Shih, Email: shihy@unc.edu.
Supporting citations
The following references appear in the supplemental information: Bekar et al. (2012).
Supplemental information
Data and code availability
All original code has been deposited at Github and is publicly available as of the date of publication. DOIs are listed in the Key Resources Table.
Data reported in this paper will be shared by the lead contact upon request.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Akerboom J., Chen T.-W., Wardill T.J., Tian L., Marvin J.S., Mutlu S., Calderón N.C., Esposti F., Borghuis B.G., Sun X.R., et al. Optimization of a GCaMP calcium indicator for neural activity imaging. J. Neurosci. 2012;32:13819–13840. doi: 10.1523/jneurosci.2601-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albers F., Wachsmuth L., van Alst T.M., Faber C. Multimodal functional neuroimaging by simultaneous BOLD fMRI and fiber-optic calcium recordings and optogenetic control. Mol. Imaging Biol. 2017:171–182. doi: 10.1007/s11307-017-1130-6. [DOI] [PubMed] [Google Scholar]
- Baraghis E., Devor A., Fang Q., Srinivasan V.J., Wu W., Lesage F., Ayata C., Kasischke K.A., Boas D.A., Sakadzic S. Two-photon microscopy of cortical NADH fluorescence intensity changes: correcting contamination from the hemodynamic response. J. Biomed. Opt. 2011;16:106003. doi: 10.1117/1.3633339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett L.M., Hughes T.E., Drobizhev M. Deciphering the molecular mechanism responsible for GCaMP6m's Ca2+-dependent change in fluorescence. PLoS One. 2017;12:e0170934. doi: 10.1371/journal.pone.0170934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekar L.K., Wei H.S., Nedergaard M. The locus coeruleus-norepinephrine network optimizes coupling of cerebral blood volume with oxygen demand. J. Cereb. Blood Flow Metab. 2012;32:2135–2145. doi: 10.1038/jcbfm.2012.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollimunta A., Santacruz S.R., Eaton R.W., Xu P.S., Morrison J.H., Moxon K.A., Carmena J.M., Nassi J.J. Head-mounted microendoscopic calcium imaging in dorsal premotor cortex of behaving rhesus macaque. Cell Rep. 2021;35:109239. doi: 10.1016/j.celrep.2021.109239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadwater M.A., Lee S.-H., Yu Y., Zhu H., Crews F.T., Robinson D.L., Shih Y.-Y.I. Adolescent alcohol exposure decreases frontostriatal resting-state functional connectivity in adulthood. Addict. Biol. 2018;23:810–823. doi: 10.1111/adb.12530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao T.-H.H., Chen J.-H., Yen C.-T. Plasticity changes in forebrain activity and functional connectivity during neuropathic pain development in rats with sciatic spared nerve injury. Mol. Brain. 2018;11:55. doi: 10.1186/s13041-018-0398-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao T.-H.H., Zhang W.-T., Hsu L.-M., Cerri D.H., Wang T.W.W., Shih Y.-Y.I. Computing hemodynamic response functions from concurrent spectral fiber-photometry and fMRI data. Neurophotonics. 2022;9:032205. doi: 10.1117/1.nph.9.3.032205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T.W., Wardill T.J., Sun Y., Pulver S.R., Renninger S.L., Baohan A., Schreiter E.R., Kerr R.A., Orger M.B., Jayaraman V., et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Sobczak F., Chen Y., Jiang Y., Qian C., Lu Z., Ayata C., Logothetis N.K., Yu X. Mapping optogenetically-driven single-vessel fMRI with concurrent neuronal calcium recordings in the rat hippocampus. Nat. Commun. 2019;10:5239. doi: 10.1038/s41467-019-12850-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox R.W. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput. Biomed. Res. Int. J. 1996;29:162–173. doi: 10.1006/cbmr.1996.0014. [DOI] [PubMed] [Google Scholar]
- Cramer S.W., Carter R.E., Aronson J.D., Kodandaramaiah S.B., Ebner T.J., Chen C.C. Through the looking glass: a review of cranial window technology for optical access to the brain. J. Neurosci. Methods. 2021;354:109100. doi: 10.1016/j.jneumeth.2021.109100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui G., Jun S.B., Jin X., Luo G., Pham M.D., Lovinger D.M., Vogel S.S., Costa R.M. Deep brain optical measurements of cell type-specific neural activity in behaving mice. Nat. Protoc. 2014;9:1213–1228. doi: 10.1038/nprot.2014.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui G., Jun S.B., Jin X., Pham M.D., Vogel S.S., Lovinger D.M., Costa R.M. Concurrent activation of striatal direct and indirect pathways during action initiation. Nature. 2013;494:238–242. doi: 10.1038/nature11846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dana H., Mohar B., Sun Y., Narayan S., Gordus A., Hasseman J.P., Tsegaye G., Holt G.T., Hu A., Walpita D., et al. Sensitive red protein calcium indicators for imaging neural activity. Elife. 2016;5:e12727. doi: 10.7554/eLife.12727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dana H., Sun Y., Mohar B., Hulse B.K., Kerlin A.M., Hasseman J.P., Tsegaye G., Tsang A., Wong A., Patel R., et al. High-performance calcium sensors for imaging activity in neuronal populations and microcompartments. Nat. Methods. 2019;16:649–657. doi: 10.1038/s41592-019-0435-6. [DOI] [PubMed] [Google Scholar]
- Decot H.K., Namboodiri V.M.K., Gao W., McHenry J.A., Jennings J.H., Lee S.-H., Kantak P.A., Jill Kao Y.-C., Das M., Witten I.B., et al. Coordination of brain-wide activity dynamics by dopaminergic neurons. Neuropsychopharmacology. 2017;42:615–627. doi: 10.1038/npp.2016.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devonshire I.M., Papadakis N.G., Port M., Berwick J., Kennerley A.J., Mayhew J.E., Overton P.G. Neurovascular coupling is brain region-dependent. Neuroimage. 2012;59:1997–2006. doi: 10.1016/j.neuroimage.2011.09.050. [DOI] [PubMed] [Google Scholar]
- Devor A., Dunn A.K., Andermann M.L., Ulbert I., Boas D.A., Dale A.M. Coupling of total hemoglobin concentration, oxygenation, and neural activity in rat somatosensory cortex. Neuron. 2003;39:353–359. doi: 10.1016/s0896-6273(03)00403-3. [DOI] [PubMed] [Google Scholar]
- Doric . Doric; 2021. Integrated Fluorescence Mini Cubes (Gen2) User Manual. [Google Scholar]
- Ekstrom A.D. Regional variation in neurovascular coupling and why we still lack a Rosetta Stone. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2021;376:20190634. doi: 10.1098/rstb.2019.0634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunaydin L.A., Grosenick L., Finkelstein J.C., Kauvar I.V., Fenno L.E., Adhikari A., Lammel S., Mirzabekov J.J., Airan R.D., Zalocusky K.A., et al. Natural neural projection dynamics underlying social behavior. Cell. 2014;157:1535–1551. doi: 10.1016/j.cell.2014.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen L.P. Large sample properties of generalized method of moments estimators. Econometrica. 1982;50:1029. doi: 10.2307/1912775. [DOI] [Google Scholar]
- He Y., Wang M., Chen X., Pohmann R., Polimeni J.R., Scheffler K., Rosen B.R., Kleinfeld D., Yu X. Ultra-slow single-vessel BOLD and CBV-based fMRI spatiotemporal dynamics and their correlation with neuronal intracellular calcium signals. Neuron. 2018;97:925–939.e5. doi: 10.1016/j.neuron.2018.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henyey L.C., Greenstein J.L. Diffuse radiation in the galaxy. Astrophys. J. 1941;93:70. doi: 10.1086/144246. [DOI] [Google Scholar]
- Hillman E.M. Coupling mechanism and significance of the BOLD signal: a status report. Annu. Rev. Neurosci. 2014;37:161–181. doi: 10.1146/annurev-neuro-071013-014111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillman E.M.C. Optical brain imaging in vivo: techniques and applications from animal to man. J. Biomed. Opt. 2007;12:051402. doi: 10.1117/1.2789693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Yacoub E. The story of the initial dip in fMRI. Neuroimage. 2012;62:1103–1108. doi: 10.1016/j.neuroimage.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua J., Liu P., Kim T., Donahue M., Rane S., Chen J.J., Qin Q., Kim S.-G. MRI techniques to measure arterial and venous cerebral blood volume. Neuroimage. 2019;187:17–31. doi: 10.1016/j.neuroimage.2018.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin M., Wang L., Wang H., Han X., Diao Z., Guo W., Yang Z., Ding H., Wang Z., Zhang P., et al. Disturbed neurovascular coupling in hemodialysis patients. PeerJ. 2020;8:e8989. doi: 10.7717/peerj.8989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Tabah J., Mohammad H., Clarke P.B.S., Hébert T.E. In vivo detection of GPCR-dependent signaling using fiber photometry and FRET-based biosensors. Methods. 2021:422–430. doi: 10.1016/j.ymeth.2021.05.002. [DOI] [PubMed] [Google Scholar]
- Kelly D.F., Lee S.M., Pinanong P.A., Hovda D.A. Paradoxical effects of acute ethanolism in experimental brain injury. J. Neurosurg. 1997;86:876–882. doi: 10.3171/jns.1997.86.5.0876. [DOI] [PubMed] [Google Scholar]
- Kim C.K., Yang S.J., Pichamoorthy N., Young N.P., Kauvar I., Jennings J.H., Lerner T.N., Berndt A., Lee S.Y., Ramakrishnan C., et al. Simultaneous fast measurement of circuit dynamics at multiple sites across the mammalian brain. Nat. Methods. 2016;13:325–328. doi: 10.1038/nmeth.3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo R., Yuui K., Kasuda S., Hatake K. [Effect of alcohol on vascular function] Nihon Arukoru Yakubutsu Igakkai Zasshi. 2015;50:123–134. [PubMed] [Google Scholar]
- Kwong K.K., Belliveau J.W., Chesler D.A., Goldberg I.E., Weisskoff R.M., Poncelet B.P., Kennedy D.N., Hoppel B.E., Cohen M.S., Turner R., et al. Dynamic magnetic resonance imaging of human brain activity during primary sensory stimulation. Proc. Natl. Acad. Sci. U S A. 1992;89:5675–5679. doi: 10.1073/pnas.89.12.5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarjan V.K., Gashti A.B., Feshki M., Garnier A., Gosselin B. Miniature fiber-spectrophotometer for real-time biomarkers detection. IEEE Sens. J. 2021;21:14822–14837. doi: 10.1109/JSEN.2021.3072578. [DOI] [Google Scholar]
- Lecrux C., Hamel E. Neuronal networks and mediators of cortical neurovascular coupling responses in normal and altered brain states. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2016;371:20150350. doi: 10.1098/rstb.2015.0350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z., Ma Y., Watson G.D.R., Zhang N. Simultaneous GCaMP6-based fiber photometry and fMRI in rats. J. Neurosci. Methods. 2017;289:31–38. doi: 10.1016/j.jneumeth.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotfullina N., Khazipov R. Ethanol and the developing brain: inhibition of neuronal activity and neuroapoptosis. Neuroscientist. 2018;24:130–141. doi: 10.1177/1073858417712667. [DOI] [PubMed] [Google Scholar]
- Luchsinger J.R., Fetterly T.L., Williford K.M., Salimando G.J., Doyle M.A., Maldonado J., Simerly R.B., Winder D.G., Centanni S.W. Delineation of an insula-BNST circuit engaged by struggling behavior that regulates avoidance in mice. Nat. Commun. 2021;12:3561. doi: 10.1038/s41467-021-23674-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Shaik M.A., Kim S.H., Kozberg M.G., Thibodeaux D.N., Zhao H.T., Yu H., Hillman E.M.C. Wide-field optical mapping of neural activity and brain haemodynamics: considerations and novel approaches. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2016;371:20150360. doi: 10.1098/rstb.2015.0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mächler P., Fomin-Thunemann N., Thunemann M., Sætra M.J., Desjardins M., Kılıç K., Şencan I., Li B., Saisan P., Cheng Q., et al. Microscopic quantification of oxygen consumption across cortical layers. bioRxiv. 2021 doi: 10.1101/2021.10.13.464176. Preprint at. [DOI] [Google Scholar]
- Marvin J.S., Borghuis B.G., Tian L., Cichon J., Harnett M.T., Akerboom J., Gordus A., Renninger S.L., Chen T.-W., Bargmann C.I., et al. An optimized fluorescent probe for visualizing glutamate neurotransmission. Nat. Methods. 2013;10:162–170. doi: 10.1038/nmeth.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvin J.S., Shimoda Y., Magloire V., Leite M., Kawashima T., Jensen T.P., Kolb I., Knott E.L., Novak O., Podgorski K., et al. A genetically encoded fluorescent sensor for in vivo imaging of GABA. Nat. Methods. 2019;16:763–770. doi: 10.1038/s41592-019-0471-2. [DOI] [PubMed] [Google Scholar]
- Meng C., Zhou J., Papaneri A., Peddada T., Xu K., Cui G. Spectrally resolved fiber photometry for multi-component analysis of brain circuits. Neuron. 2018;98:707–717.e4. doi: 10.1016/j.neuron.2018.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F., Alayash A.I. Determination of extinction coefficients of human hemoglobin in various redox states. Anal. Biochem. 2017;521:11–19. doi: 10.1016/j.ab.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintun M.A., Raichle M.E., Martin W.R., Herscovitch P. Brain oxygen utilization measured with O-15 radiotracers and positron emission tomography. J. Nucl. Med. 1984;25:177–187. [PubMed] [Google Scholar]
- Mishra A.M., Ellens D.J., Schridde U., Motelow J.E., Purcaro M.J., DeSalvo M.N., Enev M., Sanganahalli B.G., Hyder F., Blumenfeld H. Where fMRI and electrophysiology agree to disagree: corticothalamic and striatal activity patterns in the WAG/Rij rat. J. Neurosci. 2011;31:15053–15064. doi: 10.1523/jneurosci.0101-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittmann W., Wallace D.J., Czubayko U., Herb J.T., Schaefer A.T., Looger L.L., Denk W., Kerr J.N.D. Two-photon calcium imaging of evoked activity from L5 somatosensory neurons in vivo. Nat. Neurosci. 2011;14:1089–1093. doi: 10.1038/nn.2879. [DOI] [PubMed] [Google Scholar]
- Nakabayashi T., Oshita S., Sumikawa R., Sun F., Kinjo M., Ohta N. pH dependence of the fluorescence lifetime of enhanced yellow fluorescent protein in solution and cells. J. Photochem. Photobiol. Chem. 2012;235:65–71. doi: 10.1016/j.jphotochem.2012.02.016. [DOI] [Google Scholar]
- Ocean Insight . Ocean Insight; 2022. QE Pro-FL Product Details.https://www.oceaninsight.com/products/spectrometers/high-sensitivity/qepro-fl/?qty=1 [Google Scholar]
- Pais-Roldán P., Takahashi K., Sobczak F., Chen Y., Zhao X., Zeng H., Jiang Y., Yu X. Indexing brain state-dependent pupil dynamics with simultaneous fMRI and optical fiber calcium recording. Proc. Natl. Acad. Sci. U S A. 2020;117:6875–6882. doi: 10.1073/pnas.1909937117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patriarchi T., Cho J.R., Merten K., Howe M.W., Marley A., Xiong W.-H., Folk R.W., Broussard G.J., Liang R., Jang M.J., et al. Ultrafast neuronal imaging of dopamine dynamics with designed genetically encoded sensors. Science. 2018;360:eaat4422. doi: 10.1126/science.aat4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patriarchi T., Mohebi A., Sun J., Marley A., Liang R., Dong C., Puhger K., Mizuno G.O., Davis C.M., Wiltgen B., et al. An expanded palette of dopamine sensors for multiplex imaging in vivo. Nat. Methods. 2020;17:1147–1155. doi: 10.1038/s41592-020-0936-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisanello M., Pisano F., Hyun M., Maglie E., Balena A., De Vittorio M., Sabatini B.L., Pisanello F. The three-dimensional signal collection field for fiber photometry in brain tissue. Front. Neurosci. 2019;13:82. doi: 10.3389/fnins.2019.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisano F., Pisanello M., Lee S.J., Lee J., Maglie E., Balena A., Sileo L., Spagnolo B., Bianco M., Hyun M., et al. Depth-resolved fiber photometry with a single tapered optical fiber implant. Nat. Methods. 2019;16:1185–1192. doi: 10.1038/s41592-019-0581-x. [DOI] [PubMed] [Google Scholar]
- Prahl S. Optical absorption of hemoglobin. 1999. http://omlc.org/spectra/hemoglobin
- Ruffin V.A., Salameh A.I., Boron W.F., Parker M.D. Intracellular pH regulation by acid-base transporters in mammalian neurons. Front. Physiol. 2014;5:43. doi: 10.3389/fphys.2014.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rungta R.L., Osmanski B.-F., Boido D., Tanter M., Charpak S. Light controls cerebral blood flow in naive animals. Nat. Commun. 2017;8:14191. doi: 10.1038/ncomms14191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai F., Nakazawa K., Tazaki Y., Ishii K., Hino H., Igarashi H., Kanda T. Regional cerebral blood volume and hematocrit measured in normal human volunteers by single-photon emission computed tomography. J. Cereb. Blood Flow Metab. 1985;5:207–213. doi: 10.1038/jcbfm.1985.27. [DOI] [PubMed] [Google Scholar]
- Sano M., Wendt P.E., Wirsén A., Stenberg G., Risberg J., Ingvar D.H., Wirsén A. Acute effects of alcohol on regional cerebral blood flow in man. J. Stud. Alcohol. 1993;54:369–376. doi: 10.15288/jsa.1993.54.369. [DOI] [PubMed] [Google Scholar]
- Schlegel F., Sych Y., Schroeter A., Stobart J., Weber B., Helmchen F., Rudin M. Fiber-optic implant for simultaneous fluorescence-based calcium recordings and BOLD fMRI in mice. Nat. Protoc. 2018;13:840–855. doi: 10.1038/nprot.2018.003. [DOI] [PubMed] [Google Scholar]
- Schmid F., Wachsmuth L., Schwalm M., Prouvot P.H., Jubal E.R., Fois C., Pramanik G., Zimmer C., Faber C., Stroh A. Assessing sensory versus optogenetic network activation by combining (o)fMRI with optical Ca2+ recordings. J. Cereb. Blood Flow Metab. 2016;36:1885–1900. doi: 10.1177/0271678x15619428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schridde U., Khubchandani M., Motelow J.E., Sanganahalli B.G., Hyder F., Blumenfeld H. Negative BOLD with large increases in neuronal activity. Cereb. Cortex. 2008;18:1814–1827. doi: 10.1093/cercor/bhm208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz K., Sydekum E., Krueppel R., Engelbrecht C.J., Schlegel F., Schröter A., Rudin M., Helmchen F. Simultaneous BOLD fMRI and fiber-optic calcium recording in rat neocortex. Nat. Methods. 2012;9:597–602. doi: 10.1038/nmeth.2013. [DOI] [PubMed] [Google Scholar]
- Schwalm M., Schmid F., Wachsmuth L., Backhaus H., Kronfeld A., Aedo Jury F., Prouvot P.H., Fois C., Albers F., van Alst T., et al. Cortex-wide BOLD fMRI activity reflects locally-recorded slow oscillation-associated calcium waves. Elife. 2017;6:e27602. doi: 10.7554/eLife.27602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner N.C., Steinbach P.A., Tsien R.Y. A guide to choosing fluorescent proteins. Nat. Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- Shcherbakova D.M. Near-infrared and far-red genetically encoded indicators of neuronal activity. J. Neurosci. Methods. 2021;362:109314. doi: 10.1016/j.jneumeth.2021.109314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih Y.Y.I., Chen C.C.V., Shyu B.C., Lin Z.J., Chiang Y.C., Jaw F.S., Chen Y.Y., Chang C. A new scenario for negative functional magnetic resonance imaging signals: endogenous neurotransmission. J. Neurosci. 2009;29:3036–3044. doi: 10.1523/JNEUROSCI.3447-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih Y.Y.I., Li G., Muir E.R., De La Garza B.H., Kiel J.W., Duong T.Q. Pharmacological MRI of the choroid and retina: blood flow and BOLD responses during nitroprusside infusion. Magn. Reson. Med. 2012;68:1273–1278. doi: 10.1002/mrm.24112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih Y.-Y.I., Huang S., Chen Y.-Y., Lai H.-Y., Kao Y.-C.J., Du F., Hui E.S., Duong T.Q. Imaging neurovascular function and functional recovery after stroke in the rat striatum using forepaw stimulation. J. Cereb. Blood Flow Metab. 2014;34:1483–1492. doi: 10.1038/jcbfm.2014.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih Y.-Y.I., Wang L., De La Garza B.H., Li G., Cull G., Kiel J.W., Duong T.Q. Quantitative retinal and choroidal blood flow during light, dark adaptation and flicker light stimulation in rats using fluorescent microspheres. Curr. Eye Res. 2013;38:292–298. doi: 10.3109/02713683.2012.756526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih Y.-Y.I., Wey H.-Y., De La Garza B.H., Duong T.Q. Striatal and cortical BOLD, blood flow, blood volume, oxygen consumption, and glucose consumption changes in noxious forepaw electrical stimulation. J. Cereb. Blood Flow Metab. 2011;31:832–841. doi: 10.1038/jcbfm.2010.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel A.M., Culver J.P., Mandeville J.B., Boas D.A. Temporal comparison of functional brain imaging with diffuse optical tomography and fMRI during rat forepaw stimulation. Phys. Med. Biol. 2003;48:1391–1403. doi: 10.1088/0031-9155/48/10/311. [DOI] [PubMed] [Google Scholar]
- Sloan H.L., Austin V.C., Blamire A.M., Schnupp J.W.H., Lowe A.S., Allers K.A., Matthews P.M., Sibson N.R. Regional differences in neurovascular coupling in rat brain as determined by fMRI and electrophysiology. Neuroimage. 2010;53:399–411. doi: 10.1016/j.neuroimage.2010.07.014. [DOI] [PubMed] [Google Scholar]
- Smith A.F., Doyeux V., Berg M., Peyrounette M., Haft-Javaherian M., Larue A.-E., Slater J.H., Lauwers F., Blinder P., Tsai P., et al. Brain capillary networks across species: a few simple organizational requirements are sufficient to reproduce both structure and function. Front. Physiol. 2019;10:233. doi: 10.3389/fphys.2019.00233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F., Zeng J., Jing M., Zhou J., Feng J., Owen S.F., Luo Y., Li F., Wang H., Yamaguchi T., et al. A genetically encoded fluorescent sensor enables rapid and specific detection of dopamine in flies, fish, and mice. Cell. 2018;174:481–496.e19. doi: 10.1016/j.cell.2018.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F., Zhou J., Dai B., Qian T., Zeng J., Li X., Zhuo Y., Zhang Y., Wang Y., Qian C., et al. Next-generation GRAB sensors for monitoring dopaminergic activity in vivo. Nat. Methods. 2020;17:1156–1166. doi: 10.1038/s41592-020-00981-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sych Y., Chernysheva M., Sumanovski L.T., Helmchen F. High-density multi-fiber photometry for studying large-scale brain circuit dynamics. Nat. Methods. 2019;16:553–560. doi: 10.1038/s41592-019-0400-4. [DOI] [PubMed] [Google Scholar]
- Taylor A.J., Kim J.H., Ress D. Characterization of the hemodynamic response function across the majority of human cerebral cortex. Neuroimage. 2018;173:322–331. doi: 10.1016/j.neuroimage.2018.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L., Hires S.A., Mao T., Huber D., Chiappe M.E., Chalasani S.H., Petreanu L., Akerboom J., McKinney S.A., Schreiter E.R., et al. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat. Methods. 2009;6:875–881. doi: 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uludağ K., Blinder P. Linking brain vascular physiology to hemodynamic response in ultra-high field MRI. Neuroimage. 2018;168:279–295. doi: 10.1016/j.neuroimage.2017.02.063. [DOI] [PubMed] [Google Scholar]
- Unekawa M., Tomita Y., Toriumi H., Osada T., Masamoto K., Kawaguchi H., Itoh Y., Kanno I., Suzuki N. Hyperperfusion counteracted by transient rapid vasoconstriction followed by long-lasting oligemia induced by cortical spreading depression in anesthetized mice. J. Cereb. Blood Flow Metab. 2015;35:689–698. doi: 10.1038/jcbfm.2014.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaagenes I.C., Tsai S.-Y., Ton S.T., Husak V.A., McGuire S.O., O’Brien T.E., Kartje G.L. Binge ethanol prior to traumatic brain injury worsens sensorimotor functional recovery in rats. PLoS One. 2015;10:e0120356. doi: 10.1371/journal.pone.0120356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdés-Hernández P.A., Sumiyoshi A., Nonaka H., Haga R., Aubert-Vásquez E., Ogawa T., Iturria-Medina Y., Riera J.J., Kawashima R. An in vivo MRI template set for morphometry, tissue segmentation, and fMRI localization in rats. Front. Neuroinf. 2011;5:26. doi: 10.3389/fninf.2011.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valley M.T., Moore M.G., Zhuang J., Mesa N., Castelli D., Sullivan D., Reimers M., Waters J. Separation of hemodynamic signals from GCaMP fluorescence measured with wide-field imaging. J. Neurophysiol. 2020;123:356–366. doi: 10.1152/jn.00304.2019. [DOI] [PubMed] [Google Scholar]
- Van De Borne P., Mark A.L., Montano N., Mion D., Somers V.K. Effects of alcohol on sympathetic activity, hemodynamics, and chemoreflex sensitivity. Hypertension. 1997;29:1278–1283. doi: 10.1161/01.hyp.29.6.1278. [DOI] [PubMed] [Google Scholar]
- van Zijl P.C., Eleff S.M., Ulatowski J.A., Oja J.M., Uluǧ A.M., Traystman R.J., Kauppinen R.A. Quantitative assessment of blood flow, blood volume and blood oxygenation effects in functional magnetic resonance imaging. Nat. Med. 1998;4:159–167. doi: 10.1038/nm0298-159. [DOI] [PubMed] [Google Scholar]
- Wang H., Jing M., Li Y. Lighting up the brain: genetically encoded fluorescent sensors for imaging neurotransmitters and neuromodulators. Curr. Opin. Neurobiol. 2018;50:171–178. doi: 10.1016/j.conb.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., He Y., Sejnowski T.J., Yu X. Brain-state dependent astrocytic Ca(2+) signals are coupled to both positive and negative BOLD-fMRI signals. Proc. Natl. Acad. Sci. U S A. 2018;115:E1647–E1656. doi: 10.1073/pnas.1711692115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worsley K.J., Friston K.J. Analysis of fMRI time-series revisited--again. Neuroimage. 1995;2:173–181. doi: 10.1006/nimg.1995.1023. [DOI] [PubMed] [Google Scholar]
- Zhang W.-T., Chao T.-H.H., Cui G., Shih Y.-Y.I. Simultaneous recording of neuronal and vascular activity in the rodent brain using fiber-photometry. STAR Protoc. 2022;3:101497. doi: 10.1016/j.xpro.2022.101497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Cramer S.R., Ma Z., Turner K.L., Gheres K.W., Liu Y., Drew P.J., Zhang N. Brain-wide ongoing activity is responsible for significant cross-trial BOLD variability. Cereb. Cortex. 2022 doi: 10.1093/cercor/bhac016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.-F., Vargas Cifuentes L., Wright K.N., Bhattarai J.P., Mohrhardt J., Fleck D., Janke E., Jiang C., Cranfill S.L., Goldstein N., et al. Ventral striatal islands of Calleja neurons control grooming in mice. Nat. Neurosci. 2021;24:1699–1710. doi: 10.1038/s41593-021-00952-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Left: Initial ballistic photon flow of a 488 nm excitation beam incident from the optical fiber. Right: Diffuse propagation of the elicited 561 nm emissions captured by the same fiber.
Data Availability Statement
All original code has been deposited at Github and is publicly available as of the date of publication. DOIs are listed in the Key Resources Table.
Data reported in this paper will be shared by the lead contact upon request.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.