Summary
Fine-scale molecular architecture is critical for nervous system and other biological functions. Methods to visualize these nanoscale structures would benefit from enhanced accessibility, throughput, and tissue compatibility. Here, we report RAIN-STORM, a rapid and scalable nanoscopic imaging optimization approach that improves three-dimensional visualization for subcellular targets in tissue at depth. RAIN-STORM uses conventional tissue samples and readily available reagents and is suitable for commercial instrumentation. To illustrate the efficacy of RAIN-STORM, we utilized the retina. We show that RAIN-STORM imaging is versatile and provide 3D nanoscopic data for over 20 synapse, neuron, glia, and vasculature targets. Sample preparation is also rapid, with a 1-day turnaround from tissue to image, and parameters are suitable for multiple tissue sources. Finally, we show that this method can be applied to clinical samples to reveal nanoscale features of human cells and synapses. RAIN-STORM thus paves the way for high-throughput studies of nanoscopic targets in tissue.
Keywords: nanoscopic imaging, STORM, super-resolution, retina, synapse, neuron
Graphical abstract
Highlights
-
•
Developed RAIN-STORM, a 3D nanoscopic imaging optimization approach
-
•
RAIN-STORM is rapid, with a 1-day turnaround from sample to image
-
•
RAIN-STORM is versatile, enabling 3D nanoscale imaging for numerous molecular targets
-
•
Tissues from humans and other species reveal nanoscale neuron and synapse features
Motivation
The precise organization of fine-scale molecular architecture is critical for the nervous system and other biological functions and would benefit from improved nanoscopic imaging methods with enhanced accessibility, throughput, and native tissue compatibility. Here, we report RAIN-STORM, a rapid and scalable nanoscopic imaging optimization approach that improves three-dimensional nanoscale target visualization for multiple subcellular and intracellular targets within tissue at depth.
Albrecht et al. develop RAIN-STORM, a nanoscopic imaging optimization method that improves the accessibility, throughput, and compatibility of current 3D nanoscopic tissue imaging. Moreover, the RAIN-STORM-optimized method is compatible with a wide range of tissue sources and a large number of molecular targets across a spectrum of biological structures.
Introduction
The advent of single-molecule localization microscopy (SMLM) techniques has greatly increased the ability to resolve the location, density, and nanoscale spatial relationships of diverse molecules using fluorescence (Klein et al., 2014). Yet, large-scale adoption of these techniques for tissue analysis remains limited, in part because most SMLM approaches are challenging to apply to thick samples where aberrations and background fluorescence can limit imaging. Furthermore, relatively little side-by-side comparative data from which to build protocols for new applications or targets are available. As a result, most analyses of the molecular architecture within tissue continue to rely on immunofluorescence microscopy and fluorescent protein reporters. These powerful tools have shown that cellular and tissue functions intimately depend on small-scale arrangements of proteins within subcellular compartments. However, the molecular composition and organization of many nanoscale biological structures are still unknown.
To help address this challenge, a number of bespoke SMLM solutions have been developed. These include in-house, specialized fluorescence microcopy and optical systems, customized analysis software, and advanced sample preparation techniques. For example, fixed fluorescently labeled cultured cells have been visualized in three-dimensional volumes acquired with a number of Z-positioning approaches, including astigmatism (Huang et al., 2008), biplane imaging (Juette et al., 2008), engineered point spread functions (PSFs) (Pavani et al., 2009), and 4pi imaging (Bewersdorf et al., 2006). Fluorescent markers in tissue samples have been visualized in 3D using adaptive optics (Mlodzianoski et al., 2018), biplane imaging (Bewersdorf et al., 2006), self-interfering PSFs (Bon et al., 2018), light-sheet approaches (Greiss et al., 2016), and ultrathin physical sample sectioning (Sigal et al., 2015). While these methods have greatly improved 3D SMLM capability in the field and led to numerous discoveries (Bowler et al., 2019; Chamma et al., 2016; Leterrier et al., 2015; Sigal et al., 2015; Suleiman et al., 2013; van den Dries et al., 2013), important challenges remain. The first is accessibility. Most 3D SMLM approaches rely on techniques, systems, and expertise that are available only to a handful of specialists. The second is simplicity. For instance, one useful approach is serial-section STORM (stochastic optical reconstruction microscopy), which requires ultrathin sectioning and sequential reconstruction (Sigal et al., 2015). However, the labor and imaging time for this technique make it best suited for deep interrogation of small sample numbers. The third is target compatibility within native tissue environments. Many current methods report imaging capabilities using only a small number of antibodies and lack a systematic comparison between relevant conditions for sample preparation and imaging (Chen et al., 2015; Ku et al., 2016; Tillberg et al., 2016; Mikhaylova et al., 2015; Monkemoller et al., 2015). As a result, only a few studies have used tissue samples for SMLM imaging (Baddeley et al., 2011; German et al., 2017; Nanguneri et al., 2012; Dani and Huang, 2010).
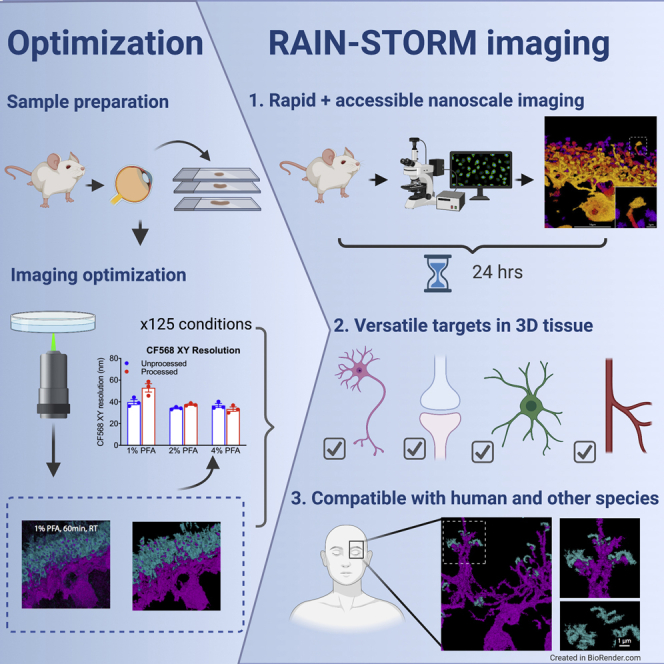
We set out to develop a nanoscopic imaging optimization method for central nervous system (CNS) tissue that would complement and combine the strengths of currently available methods for nanoscopic tissue imaging in general and for neural circuits specifically. Our criteria for this method were to improve the accessibility, throughput, and compatibility of 3D nanoscopic tissue imaging so that it can be readily applied to diverse tissue sources and multiple targets. Furthermore, we wished to understand how different conditions can be tuned to improve outcomes in a given biological specimen. Toward this goal, we present rapid imaging of tissues at the nanoscale (RAIN-STORM), an optimized method for the preparation and imaging of standard tissue samples to generate SMLM data at depth for a wide range of molecular targets using commercially available reagents and imaging systems. To achieve this goal, we first identified and then quantitatively tested 125 distinct tissue preparation, fixation, quenching, labeling, and imaging conditions. From these parameters, we identified a select set of 3D tissue imaging parameters based on resolution, antibody labeling density, and reduction of background fluorescence. We show that the resulting RAIN-STORM optimized method is rapid and versatile, enabling 3D nanoscopic sub-diffraction limited imaging with a 24-h turnaround. Furthermore, these imaging parameters are compatible with a wide range of tissue sources and a large number of molecular targets across a spectrum of biological structures. Finally, we show that the optimized set of parameters can be applied to clinically derived samples to study the nanoscale features of molecular targets in human tissue. RAIN-STORM optimization thus enables 3D nanoscopic imaging for a range of molecules, paving the way for high-throughput studies of nanoscale biological structures in intact tissue from diverse sources.
Results
To improve the accessibility of STORM imaging we set out to develop an optimization method compatible with standard tissue preparation and a turnkey SMLM imaging system. We chose to use mouse retina tissue because it has a highly defined laminar structure that provides endogenous fiduciaries for evaluating the labeling precision of multiple molecular targets (Sanes and Zipursky, 2010). For nanoscopic SMLM imaging, we utilized the Vutara 352 (Bruker, Billerica, MA). This system enables multicolor STORM imaging within a 40 × 40-μm planar region of interest and achieves single-molecule imaging of individual emitters by recording the PSF in two imaging planes simultaneously (biplane imaging; Figure S1) rather than utilization of an astigmatic lens. Thus, the system is able to refine three-dimensional information more accurately through thick samples without objective-based distortion. Single-molecule localization in 3D is based on calibration data generated a priori from fiducial imaging.
To optimize our labeling and imaging parameters, we selected the calcium buffering protein calbindin (Calb1) and the synaptic protein PSD95 (postsynaptic density protein 95, also known as Dlg4). In the outer plexiform layer (OPL) of the mouse retina, calbindin specifically and densely fills the cell bodies and neurite terminals of retina horizontal neurons (Celio, 1990; Uesugi et al., 1992), whereas PSD95 is localized at the synaptic terminals in photoreceptor neurons (Hunt et al., 1996; Koulen et al., 1998). We reasoned that optimization with these targets would enable us to evaluate the efficacy of our method across two different cell structures and protein densities. Moreover, because calbindin and PSD95 are found in other CNS regions (Celio, 1990; Hunt et al., 1996), preparation methods compatible with these targets may extend to other neural tissues.
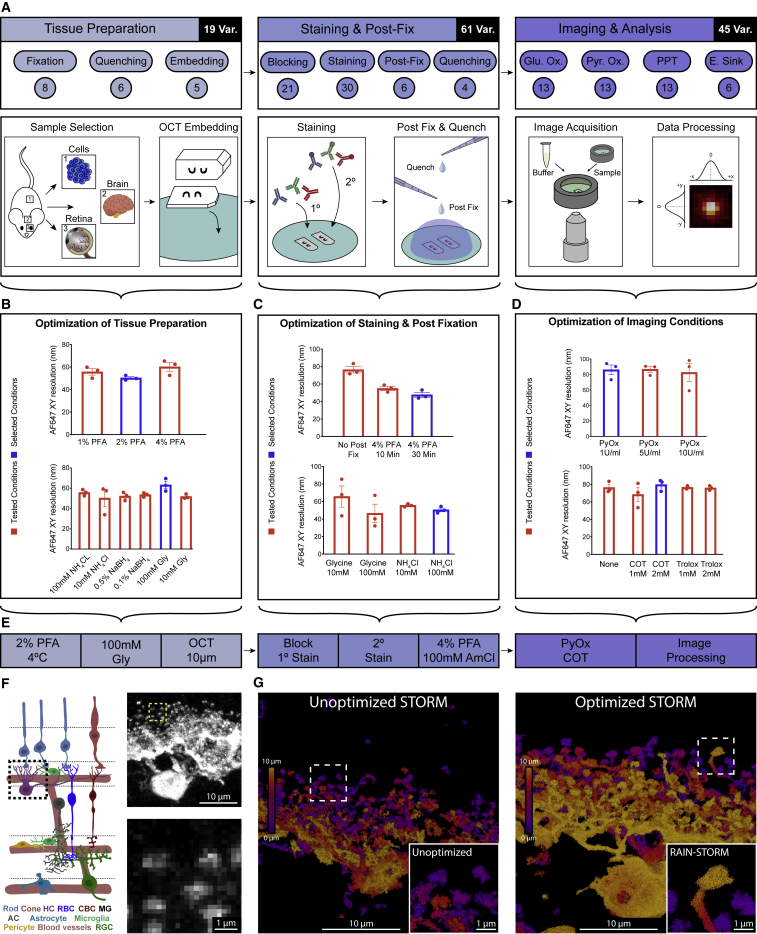
To begin RAIN-STORM optimization, we adapted a standard tissue preparation protocol for immunofluorescent labeling of mouse retina tissue that involves fixation, autofluorescence quenching, cryoprotection, embedding in freezing medium, tissue cryosectioning at 10 μm, and mounting on coated glass slides (Albrecht et al., 2018). We selected this approach because these sample preparation techniques are widely available, do not require specialized reagents or labor-intensive handling, and result in staining ready slides within hours of tissue harvest. Within this framework, we identified eight tissue preparation, staining, and imaging steps where protocol variation could significantly alter the quality and degree of nanoscale labeling and background fluorescence. We reasoned that tuning these parameters from baseline parameters previously established for 3D-STORM imaging (e.g., tissue blocking with 5% NDS, 1% BSA, 0.1% saponin in PBS, tissue fixed at 4% paraformaldehyde [PFA] for 24 h, quenched with 0.1% NaBH4 [German et al., 2017]) could markedly improve nanoscale image quality. Toward this goal, we identified a total of 125 test variations within these steps that included 19 primary fixation, primary quenching, and embedding conditions, 61 blocking, staining (Table S2), and secondary quenching conditions, and 45 imaging buffer variations (Figure 1A). We then systemically and quantitatively evaluated each variation for its effect on imaging outcomes while holding other parameters stable based on the following metrics: (1) the total number of localizations acquired, (2) epitope and morphological detail preservation, (3) the relative background fluorescence level (noise) observed as unclustered localizations or spurious antibody signal, and (4) the calculated resolution across planar (XY) and axial (XZ) image dimensions using Fourier ring correlation metrics for each channel of analysis (Nieuwenhuizen et al., 2013) (Figures 1B–1D; Table S2; Figure 2). Each of the 125 variations was tested independently in tissues from three individual animals resulting in a database of ∼375 3D-STORM images from which quantitative metrics were evaluated. Processed reconstructions were generated with the OPTICS algorithm (ordering points to identify the clustering structure [Ankerst et al., 1999]) using a particle distance of 0.16 μm and a particle count per cluster of 25. For each condition, both processed and unprocessed images were evaluated. Representative images from these conditions are presented in Figure 2, while all quantitative metrics are presented in Table S2. The best performing condition within each parameter space was selected for inclusion in the final optimized protocol.
Figure 1.
Nanoscale 3D imaging of neurons in tissue
(A) Schematic of the RAIN-STORM optimization workflow. A total of 125 condition variations were tested consisting of 19 tissue preparation parameters (fixation, autofluorescence quenching, and embedding), 61 variations in staining and post-fixation (blocking buffer formulations, primary and secondary antibody staining, post-fixation conditions, and autofluorescence quenching), and 45 different conditions in imaging buffer formulations.
(B–D) Representative AF647 quantifications for tested parameters in one analysis plane for exemplar sample conditions in tissue preparation, staining, and imaging. Red bars indicate the tested conditions, and black bars denote the chosen condition from that set. For tissue preparation, 2% PFA and 100 mM glycine performed optimally (B), whereas for staining and post-fixation 4% PFA for 30 min followed by a quenching treatment of 100 mM NH4Cl was ideal based on the resolution of the resulting imaging (C). Pyranose oxidase (1 U/mL of) and 2 mM of COT did not alter resolution but improved the number of localizations acquired (D). Data are represented as the mean ± SEM. Please see Table S2 and Figure S4 for quantitative data from all tested parameters.
(E) The best performing parameters from each test set were selected for inclusion in the final RAIN-STORM optimized protocol.
(F) Retina schematic showing the location and structure of horizontal cells (HC, boxed region) relative to other retinal cell types (RBC, rod bipolar cell; CBC, cone bipolar cell; MG, Muller glia; AC, amacrine cell; RGC, retinal ganglion cell). In confocal imaging, horizontal cell synapses are poorly resolved following calbindin staining (inset).
(G) Unoptimized STORM image of a horizontal cell labeled with calbindin. Putative synapses are indistinct with little defining morphology or contiguous structure (unoptimized inset). In optimized RAIN-STORM conditions, both horizontal cell synapses and their connecting structures are well labeled and contiguous (optimized inset), demonstrating clear structural detail across the neuronal arbor. Images are representative from N = 3 animals. All confocal images are intensity-based representations of fluorescence, whereas STORM-based images show reconstructions formed from individual localizations. STORM images are color coded by depth (purple, 0 μm, to yellow, 10 μm). Scale bars, 10 and 1 μm.
Figure 2.
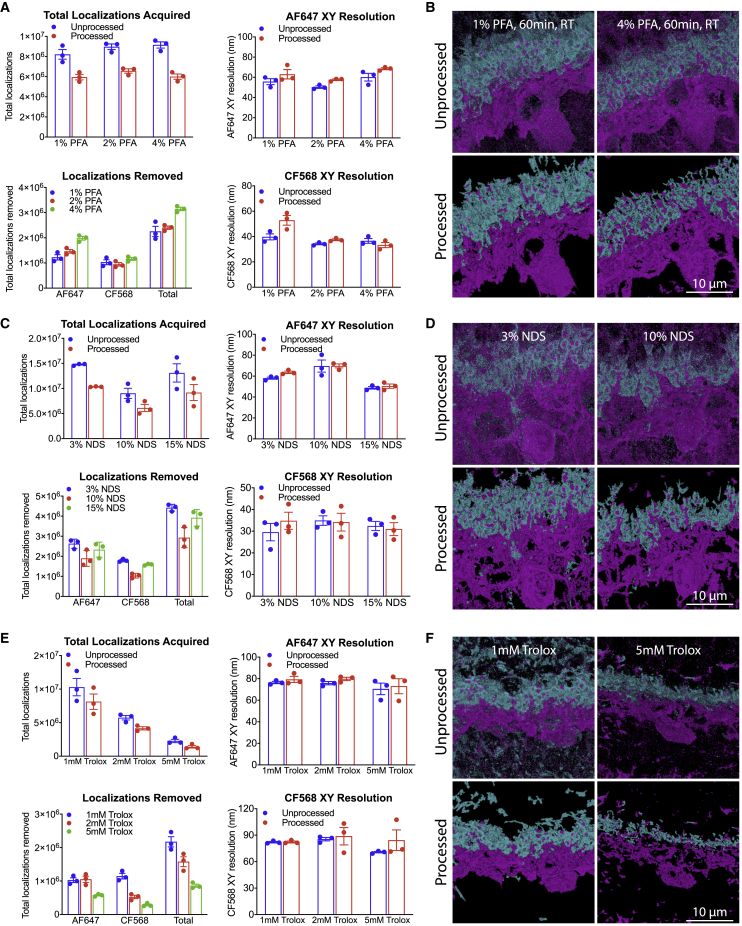
Modifying sample conditions improves visual quality and image metrics
(A and B) Representative primary data metrics (A) and corresponding unprocessed and processed images (B) are shown for variations in primary fixation concentrations. A small increase in total localizations acquired was observed with increasing concentrations of PFA, although the gain in localizations was offset by background localizations at these concentrations. Based on these parameters, 2% PFA was selected for optimized imaging.
(C and D) Representative primary data metrics (C) and corresponding unprocessed and processed images (D) are shown for variations in blocking buffer serum concentrations. In general, the best image metrics were obtained for mid-level serum concentrations (e.g., 3%–5% NDS), whereas the lower (e.g., 1% NDS) and higher (e.g., >10%) serum concentrations resulted in either increased filtered localizations or decreased resolution overall and poorer image quality.
(E and F) Representative primary data metrics (E) and corresponding unprocessed and processed images (F) are shown for variations in imaging buffer formulations using Trolox. Increasing the Trolox concentration reduced the total amount of data collected, resulting in poorer image quality relative to low Trolox concentrations (e.g., 1 mM). Images are representative of those acquired from N = 3 animals. Calbindin, magenta; PSD95, cyan. Scale bars, 10 μm. Data are represented as the mean ± SEM. Both unprocessed (blue) and processed (red, OPTICS algorithm) values are shown for each dataset. For graphs of “localizations removed,” color differences indicate a concentration change in a given reagent.
Final conditions for each step were selected through a combination of performance metrics. First, the resultant image and data had to display accurate antibody staining. Some tissue preparation conditions were noted to strongly distort staining patterns and, thus, no matter how well performing their other metrics (e.g., high resolution of poorly refined structures), they were not suitable conditions for biologically relevant imaging. Second, the number of localizations for each channel was assessed as either a gain or a loss from the basal test conditions. Gains in localizations were desirable both pre- and post-processing as these allow for better reconstructions of relevant biological structures and for more stringent post-processing if needed. Third, the number of localizations removed from the dataset was considered. Here, unclustered localizations removed during the processing steps were utilized to obtain an estimate for how much extraneous signal was introduced by a given condition compared with basal conditions.
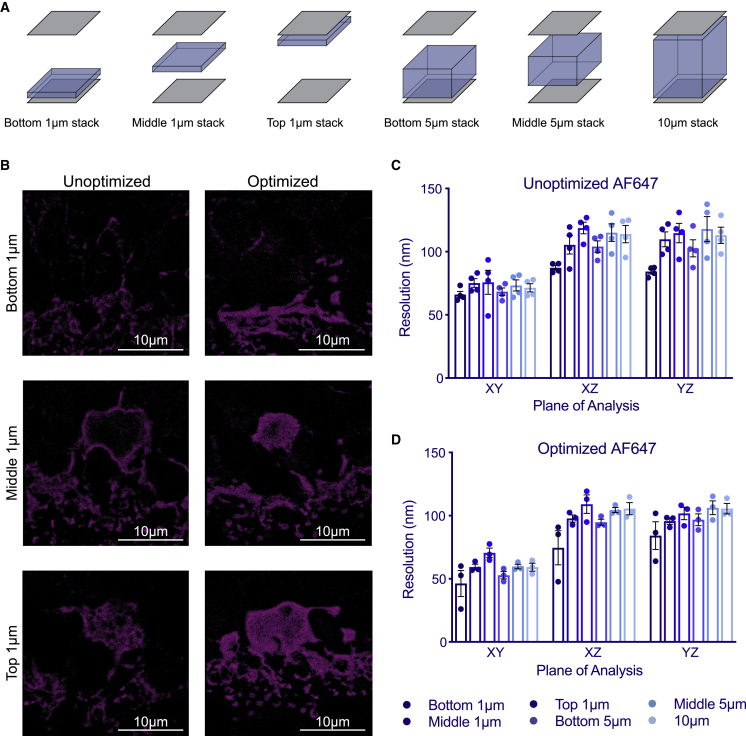
The final metric we considered was improvements in the resolution for structures labeled with either of the STORM-compatible fluorophores used in this study, AF647 and CF568, at depth over the 10-μm tissue section thickness in the XY, XZ, and XZ dimensions. We found that resolution decreased relatively modestly as a function of the distance from the objective for all conditions due to the increased material thickness and resulting diffraction artifacts (Figure 3). For example, the XY resolution of 1-μm optical slices reconstructed at a Z-position near the objective following staining for calbindin in unoptimized conditions was 61.7 nm, whereas slices reconstructed in the middle (5 μm depth) and at the farthest point from the objective (10 μm depth) within the imaging volume had XY resolutions of 69.0 and 77.2 nm, respectively. We also found that axial resolutions at depth (Rxz, Ryz) were roughly equal to each other across preparation variations (Figure 3). To account for resolution differences across the imaging volume, we acquired image stacks encompassing the entire 10-μm tissue section depth for reconstruction and report the resolution for conditions discussed below using the average planar resolution computed from the entire depth of the image stack as a conservative estimate for the resolution of the whole 3D 10-μm sample volume (denoted Rxy 10 μm average). We now highlight a selection of the tested conditions and their effects on resolution and the other metrics to demonstrate their impact on imaging outcomes.
Figure 3.
RAIN-STORM resolution as a function of sample depth
(A) Schematic of the image depths used to calculate sample resolution. The resolution was compared in: (1) single-micron slices at the bottom of the image (nearest the objective), the middle of the image stack, or the top of the image; (2) 5-μm slices from the bottom of the image to the midpoint; and (3) the entirety of the 10-μm stack.
(B) Example images of bottom, middle, and top single-micron slices in optimized and unoptimized RAIN-STORM conditions following staining with calbindin. Scale bars, 10 μm.
(C and D) Sample planar (XY dimension) and axial resolutions (XZ and YZ dimensions) in AF647 following staining for calbindin are measured in unoptimized RAIN-STORM conditions (C) and optimized conditions (D) across different imaging depths schematized in (A). Planar resolutions are improved in the optimized condition compared with the unoptimized condition across all imaging depths, whereas axial resolutions do not significantly differ. N = 3. Data are represented as the mean ± SEM.
For tissue fixation, we tested PFA, glutaraldehyde (GA), or a combination of the two in conjunction with the quenching reagents NH4Cl, glycine, or NaBH4. We found that combinations of fixatives that incorporated GA were poorly suited to high-fidelity imaging and generally resulted in a large increase in background localizations (Data S1; Table S2). For PFA-based fixation, we tested various PFA concentrations (1%, 2%, and 4%), durations (3, 60, and 120 min), and temperatures (25°C versus 4°C). From these tests, we found that autofluorescence could generally be lowered by reducing PFA concentrations and that a cold fixation (4°C) for 60 min using a 2% concentration of PFA yielded the best labeling density and imaging metrics (Table S2; Figure 2; Data S1). We also found that inclusion of a post-fixation step after antibody staining markedly improved resolution. For example, pre-clustering AF647sample resolutions without post-fixation were typically 76.3 ± 3.6 nm but the inclusion of a 4% PFA post-fixation step for 30 min improved resolution by ∼30 nm to 47.6 ± 2.9 nm (Table S2).
Antibody dilutions also significantly impacted image resolution and quality and, in some cases, gains in certain metrics came at a cost to other measures of image quality (Table S2). First, we found that targets benefitted from using increased primary antibody concentrations relative to standard histological preparations (e.g., anti-calbindin primary antibody is optimal at 1.0 μg/mL in STORM compared with 0.1 μg/mL in diffraction-limited fluorescence microscopy). Second, we also found that increasing the concentration of secondary antibodies improved the overall image quality as it resulted in denser structural filling but that this could come at a cost to image resolution. For example, using higher secondary antibody concentrations resulted in somewhat poorer resolution for AF647-labeled calbindin (Rxy 10 μm average for 5.0 μg/mL = 63.4 ± 3.3 nm, 0.5 μg/mL = 44.0 ± 1.0 nm, and 0.1 μg/mL = 38.4 ± 1.7 nm), but lower secondary concentrations reduced the amount of data collected, with commensurately less filling of the structures of interest (localizationsxy for 5.0 μg/mL = 6.07 × 106 ± 0.490 × 106, 0.5 μg/mL = 3.74 × 106 ± 0.239 × 106, and 0.1 μg/mL = 1.19 × 106 ± 0.038 × 106). In this case, imaging results suggest that the increased localization and structural filling obtained with higher secondary antibody concentrations outweighed the resolution gains of lower concentrations. Of note, resolution metrics for CF568-labeled PSD95 structures did not vary as dramatically across secondary antibody concentrations (Rxy 10 μm average for 5.0 μg/mL = 29.0 ± 1.1nm, 0.5 μg/mL = 35.5 ± 2.0nm, and 0.1 μg/mL = 29.8 ± 2.2nm) despite a similar impact on localizations (localizationsxy for 5.0 μg/mL = 6.30 × 106 ± 0.347 × 106, 0.5 μg/mL = 3.43 × 106 ± 0.254 × 106, and 0.1 μg/mL = 0.656 × 106 ± 0.079 × 106). From these variations, we thus selected 0.5 μg/mL for general use during testing of other conditions to accurately assess each condition’s impact on the sample and image quality but chose 5.0 μg/mL as the desired concentration of secondary antibodies for the final protocol.
Finally, we undertook an examination of imaging buffer conditions and tested the effects of (1) oxygen-scavenging enzymes, (2) catalase concentration, (3) the ratio of thiols (β-mercaptoethanol [BME] versus ethanolamine [MEA]), and (4) triplet-state quenching (cyclooctatetraene [COT]). Since the imaging buffer is intended to promote blinking behavior and limit photobleaching of fluorophores, we reasoned that the demands of imaging complex, 3D biological tissue structures might differ from those of more commonly imaged cell cultures. Results from these tests are summarized in Table S2. We found that the use of pyranose oxidase was preferable to glucose oxidase as it enabled longer imaging with less observable photobleaching, and we empirically determined that the addition of 2 mM COT led to an increase in the total number of localizations for both AF647 and CF568 channels (Table S2; Figure 2). In addition, we found that the total acquired localizations for each channel were affected by the relative levels of BME and MEA in pyranose oxidase-based buffers. In the case of MEA, AF647 localizations were decreased with increasing concentrations (localizationsxy for 0 mM = 6.77 × 106 ± 0.499 × 106 versus 40 mM = 4.21 × 106 ± 0.437 × 106), yet resolution remained relatively constant (Rxy 10 μm average for 0 mM = 85.3 ± 1.4 nm versus 10 mM = 77.7 ± 2.6 nm) as long as at least some amount of MEA was present (Rxy 10 μm average for 40 mM = 74.9 ± 1.7 nm). This trend was similar for the CF568 channel, although fewer localizations were acquired (localizations for 0 mM = 2.07 × 1 06 ± 0.267 × 106, 10 mM = 1.29 × 106 ± 0.326 × 106, and 40 mM = 2.27 × 106 ± 0.221 × 106). From these data, we determined that MEA is necessary for AF647 resolution gains and that CF568 performs better when MEA is at higher concentrations. Therefore, we selected a concentration of 40 mM MEA as preferred both in terms of resolution and acquired localizations across both fluorophore channels. Using a similar method, the concentration of catalase and BME were tested to arrive at final optimized concentrations (140 mM BME and 100 U/mL catalase).
Following analysis of each reagent and their variations, the final sample preparation and imaging method were chosen based on the underlying quantitative metrics using the best combination and tradeoff between localizations acquired, resolution improvements to both imaging channels, overall image quality, and antibody fidelity. Our data highlight the importance of considering factors in addition to resolution, as it is possible to have highly resolved “noise” in poorly prepared tissue that yields images of little biological relevance. For example, with post-fixation quenching, a slight decrease in resolution was accepted as the necessary tradeoff for improving overall image quality and localizations acquired. Our final sample preparation workflow is comprised of readily accessible reagents that include a moderate initial fixation (2% PFA for 60 min at 4°C), primary quenching of autofluorescence (100 mM glycine for 60 min at 4°C), primary and secondary staining using an optimized serum-based buffer (5% NDS, 0.3% Triton X-100), post-fixation of secondary antibodies (4% PFA for 30 min at 4°C), and a second round of quenching (100 mM NH4Cl for 30 min).
To illustrate the improvement achieved through RAIN-STORM optimization we reconstructed retinal horizontal cells in 3D. We found a marked increase in the resolution of nanoscopic horizontal cell features (Figure 1G). For example, in optimized RAIN-STORM imaging, we were able to observe specific and contiguous well-labeled neurites arising from the cell body, which terminated in a distinct postsynaptic invagination (Figure 1G; Videos S1 and S2). These synaptic terminals were easily resolved and structurally separated from their neighbors, demonstrating markedly improved spatial resolution (Rxy 10 μm average = 44.0 ± 1.0 nm for calbindin labeled with AF647, compared with ∼70–90 nm for unoptimized STORM conditions and ∼200–250 nm resolution for excellent confocal microscopy). These data suggest RAIN-STORM is well suited for 3D super-resolution imaging of molecular targets within tissue volumes. We also note that lower resolutions were achieved when focusing on a sub-volume of interest, with single plane XY resolution as low as 33.3 ± 1.1 and 21.7 ± 0.9 nm for AF647 and CF568, respectively.
Tissue was prepared using a standard preparatory method (see supplemental Table 1, materials and methods) to image calbindin-labeled horizontal cells across 10 μm. The image is color coded by depth, with colors indicating the axial position throughout the stack (purple 0 μm to yellow 10 μm). Synapses appear jagged and incomplete, with little structural detail connecting them back to the cell body.
Tissue was prepared using our optimized RAIN-STORM protocol, and calbindin-labeled horizontal cells were imaged across 10 μm. The image is color coded by depth, with colors indicating the axial position throughout the stack (purple 0 μm to yellow 10 μm). Fine neurite structural details are observed and individual synapses are clearly visible.
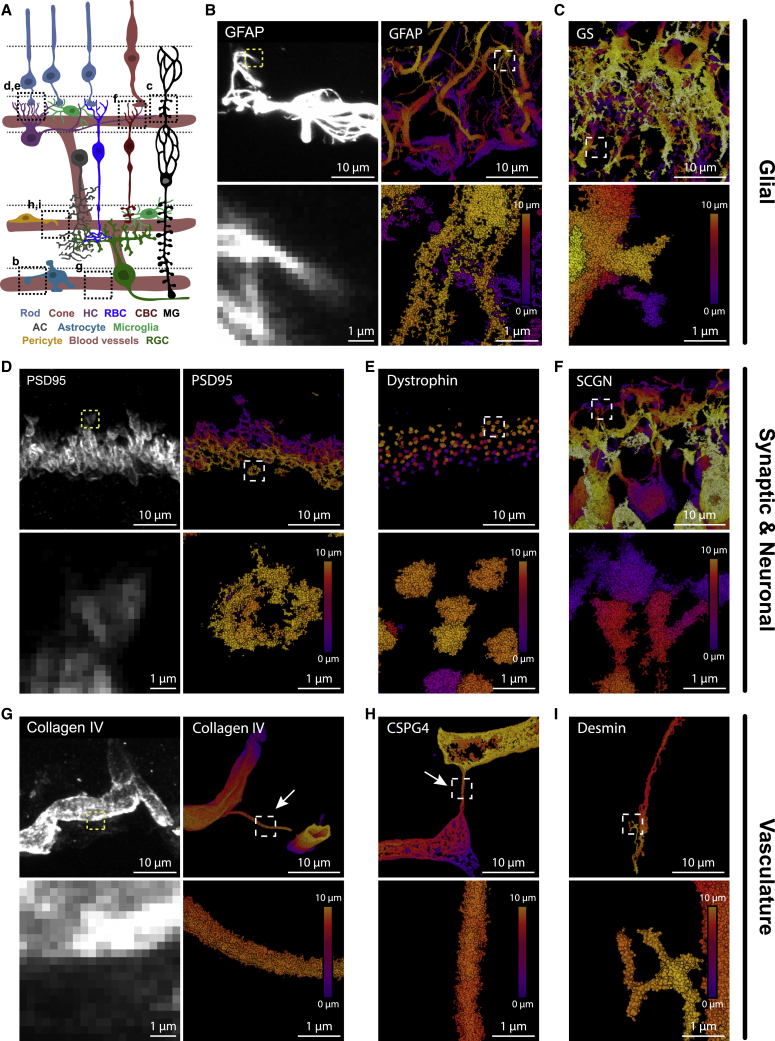
RAIN-STORM optimized parameters also readily extend to a diverse array of molecular targets that span diverse structures and cell types and were robust in other commercial STORM imaging systems. To assess this, we took advantage of the wide spectrum of specific antibody markers available in the retina (Sanes and Zipursky, 2010) and tested the robustness of optimized methods across 21 validated molecular targets (Table S1; Figures 4, and S3). These included diverse cell structures, types, and molecular targets, such as synapse proteins (Bassoon, Bsn; RIBEYE, Ctbp2; Connexin 43, Gja1; dystrophin, Dmd; Piccolo, Pclo; PSD95, Dlg4), vasculature markers (CD31, Pcam1; collagen IV, Col4A1; desmin, Des; CSPG4, Cspg4), glial proteins (Iba1, Aif1; GFAP, Gfap; GS, Glul), excitatory interneuron markers (PKCα, Prkca; SCGN, Scgn), presynaptic photoreceptor targets (CAR, Arr3), and intracellular proteins and structures (Tau, Mapt; Tomm20, mitochondria) (Figures 4 and S3). We found that approximately 90% of the commercially available antibodies tested were compatible with optimized methods and could be used to successfully obtain SMLM images of individual targets at depth within intact tissue slices (Figure 4; Videos S2, S3, and S4). Furthermore, in all cases, labeling of the proper cell type or structure at the proper location was observed relative to the well-documented confocal staining patterns for these targets (Figures 4A and S3). In addition, our results resolve novel nanoscale structural features and molecular distributions. For example, we observed that astrocyte endfeet form fine (∼100–200 nm) filamentous structures and what appears to be a mesh of fibers interdigitating the ganglion cell layer (Figure 4B; Video S4). Alongside these, we show that other glial cell types, such as Müller glia, display many fine protrusions thought to encapsulate nearby vessels and synapses (Figure 4C) (Bringmann et al., 2013; Vecino et al., 2016). To determine whether RAIN-STORM optimized conditions could be readily extended to other commercial STORM systems, we tested our method on the Nikon N-STORM system. We found that the RAIN-STORM imaging protocol was robust in this system and resolved nanoscale features of both synaptic and neural targets in mouse retina cryosections from both 2D- and 3D-STORM (astigmatism) acquisitions with good reconstruction metrics in both cases (Figure S4). Notably, the z-scanning procedure that we used for RAIN-STORM reconstructions improved the achievable z section depth in the N-STORM system by at least 10-fold over that using astigmatism from a single focal plane, a method previously reported to reconstruct a z section depth of <1 μm (Robichaux et al., 2019) (Figure S4).
Figure 4.
RAIN-STORM delivers robust imaging for a diverse array of molecular targets
(A) Retina schematic showing the diverse cell types and structural features of interest for this figure. Lettered inset boxes indicate the cell type or structure of interest displayed in the corresponding confocal and STORM images. RBC, rod bipolar cell; CBC, cone bipolar cell; MG, Muller glia; AC, amacrine cell; RGC, retinal ganglion cell; HC, horizontal cell.
(B and C) RAIN-STORM imaging of glial proteins. Using GFAP (B), a marker for astrocytes, RAIN-STORM (right column) offers markedly improved detail of nanoscale structures over confocal microscopy (left column). These include visualization of small filament-like protrusions from the astrocytes. RAIN-STORM imaging of glutamine synthetase (GS) (C), a marker for Müller glia, also shows the small protrusions arising from these cells.
(D–F) RAIN-STORM imaging of diverse synaptic and neuronal proteins. Following staining with PSD95 (D), a marker for photoreceptor synaptic terminals, individual synapse terminals can be observed more clearly using RAIN-STORM (right column) relative to confocal (left column). For example, the interior space of the magnified terminal is now visible. RAIN-STORM imaging of dystrophin (E), a synaptic structural protein in the outer plexiform layer shows that individual puncta for each synapse can be clearly observed and isolated. RAIN-STORM imaging of SCGN (F), a marker for excitatory cone bipolar cells, shows individual postsynaptic invaginating terminals.
(G–I) RAIN-STORM imaging of diverse vasculature associated proteins. Following staining with collagen IV (G), a marker for select blood vessels, clear structural detail can be observed in RAIN-STORM images (right column) relative to diffraction-limited confocal images (left column). These include defined vessel walls, interpericyte tunneling nanotubes (IP-TNTs, boxed region), and the hollow interior region of the microvasculature. RAIN-STORM imaging of CSPG4 (H), a marker for vascular pericytes, also shows IP-TNTs connecting two blood vessels. RAIN-STORM imaging of desmin (I) a filament protein that marks subsets of pericytes and vascular-associated smooth muscle cells shows fine protrusions arising from these cells (boxed region). Images are representative from N = 3 animals. All confocal images are intensity-based representations of fluorescence, whereas STORM-based images show reconstructions formed from individual localizations. STORM images are color-coded by depth (purple, 0 μm, to yellow, 10 μm). Scale bars, 10 and 1 μm.
Tissue was prepared using our optimized RAIN-STORM protocol, and collagen IV-labeled vessels were imaged across 10 μm. The image is color coded by depth, with colors indicating the axial position throughout the stack (purple 0 μm to yellow 10 μm). The structure and morphology of hollow vessels are well preserved, and cross-vessel nanotube connections are apparent.
Tissue was prepared using our optimized RAIN-STORM protocol, and GFAP-labeled astrocytes were imaged across 10 μm. The image is color coded by depth, with colors indicating the axial position throughout the stack (purple 0 μm to yellow 10 μm). The structure and morphology of astrocyte filamentous fibers were observed, which appear to form a highly branched network.
RAIN-STORM optimized imaging was also robust across a variety of synaptic and neuronal targets (Figures 4D–4F and S3). For example, with RAIN-STORM, we resolved the morphology of individual PSD95-labeled synapses and showed that the sub-synaptic space within rod terminals could be visualized (Figure 4D). We also resolved individual dystrophin puncta in the outer retina synapse layer, which are thought to interact with actin filaments to enable contact formation between photoreceptors and ON-bipolar cells (Schmitz and Drenckhahn, 1997) (Figure 4E). In parallel, we tested a range of neural markers, including the cone bipolar cell marker secretagogin (SCGN) (Puthussery et al., 2010) (Figure 4F). Here, the fine arbors and neurites of bipolar cells are seen as distinct and separate from their neighbors, enabling the study of how individual neurite tips may be organized with individual presynaptic terminals.
Finally, we expanded our target analysis to the retina vasculature. Using antibodies to CSPG4, a pericyte marker (Ivanova et al., 2021), and collagen IV, a structural component of the vasculature (Sarthy, 1993; Ishizaki et al., 1993), we observed vascular associated interpericyte tunneling nanotubes (IP-TNTs) (Figure 4G; Video S3). These ∼500-nm diameter structures enable pericyte-driven neurovascular coupling (Alarcon-Martinez et al., 2020) but their molecular composition is largely unknown (Video S3). Our imaging results suggest that collagen IV may be a primary structural component of IP-TNTs (Figure 4H). We also stained for desmin, a filament protein that labels select pericytes and vascular-associated smooth muscle cells (Mendes-Jorge et al., 2012), and observed small filamentous structures arising from these cells (Figure 4I). These results suggest that RAIN-STORM optimization is effective for imaging many diverse targets and can be used to discover unknown nanoscopic cellular structures with high fidelity.
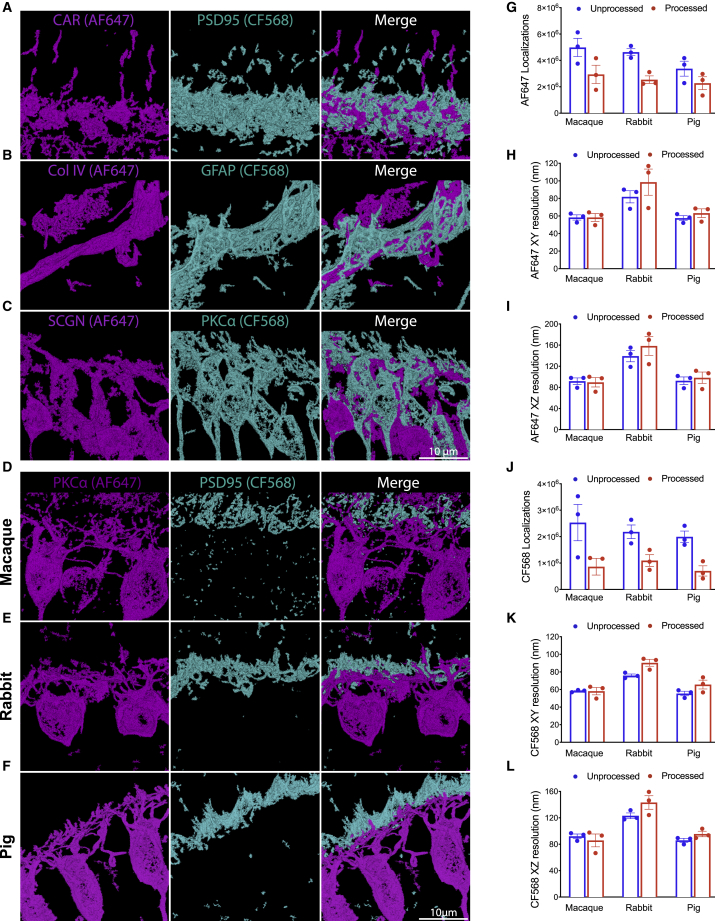
RAIN-STORM is also useful for multicolor imaging, is robust across multiple intracellular targets and genetic models, and is useful for tissue from diverse species (Figure 5). For multicolor imaging, we used our optimized method in multiple target combinations. These included a neuron subtype marker together with synaptic antibodies, staining for two distinct postsynaptic neuron types, and co-staining for vascular markers and astrocytes (Figures 5A–5C). Using these combinations, we were able to resolve contact sites, overlapping and non-overlapping cellular structures, and fine cellular interactions. For example, astrocytes and blood vessel co-staining revealed fine astrocytic filaments enshrouding neighboring vessels (Figure 5B). RAIN-STORM optimized imaging was also compatible with tissue from diverse species, revealing both conserved and unique nanoscopic features in mouse, rabbit, macaque, and pig tissue (Figures 5D–5L). For instance, co-staining for rod bipolar cell dendrites and the presynaptic terminals they invaginate showed that, despite similar functions and molecular identities, rod bipolar cells have incredibly diverse dendritic structures across species (Figures 5D–5F). Rabbit rod bipolar cell bodies were rounder and less elongated relative to pig, macaque, and mouse, with arbors that branched closer to the cell body. Notably, PSD95 labeling was confined to the outer layer of rod bipolar dendrites in all samples, suggesting that photoreceptor terminal staining is consistent across species.
Figure 5.
RAIN-STORM enables dual-channel super resolution imaging among diverse species
(A–C) RAIN-STORM imaging of diverse sets of two independent molecular targets, including (A) cones (CAR, magenta, AF647) and synaptic terminals (PSD95, cyan, CF568), (B) vasculature (collagen IVa, magenta, AF647) and astrocyte (GFAP, cyan, CF568) interactions, and (C) cone bipolar cells (SCGN, magenta, AF647) and rod bipolar cells (PKCα, cyan, CF568).
(D–F) RAIN-STORM imaging can be applied to tissue derived from diverse species. Retinas from (D) macaque (N = 2 animals), (E) rabbit (N = 2 animals), and (F) pig (N = 3 animals) were labeled with antibodies to rod bipolar cells (PKCα, magenta, AF647) and rod photoreceptor terminals (PSD95, cyan, CF568). Representative RAIN-STORM images from these species are shown.
(G–I) The number of AF647 localizations (G) and resolution for the XY (H) and XZ imaging planes (I) for PKCα AF647 images in (D–F). Pre- (blue bars) and post-processing (red bars) values for each dataset are shown.
(J–L) The number of AF647 localizations (J) and resolution for the XY (K) and XZ imaging planes (L) for PSD95 CF568 images in (D–F). Pre- (blue bars) and post-processing (red bars) values for each dataset are shown. These data show comparable localization and image resolution to that obtained using mouse tissue for these antibody combinations. Scale bars, 10 and 1 μm. Data are represented as the mean ± SEM.
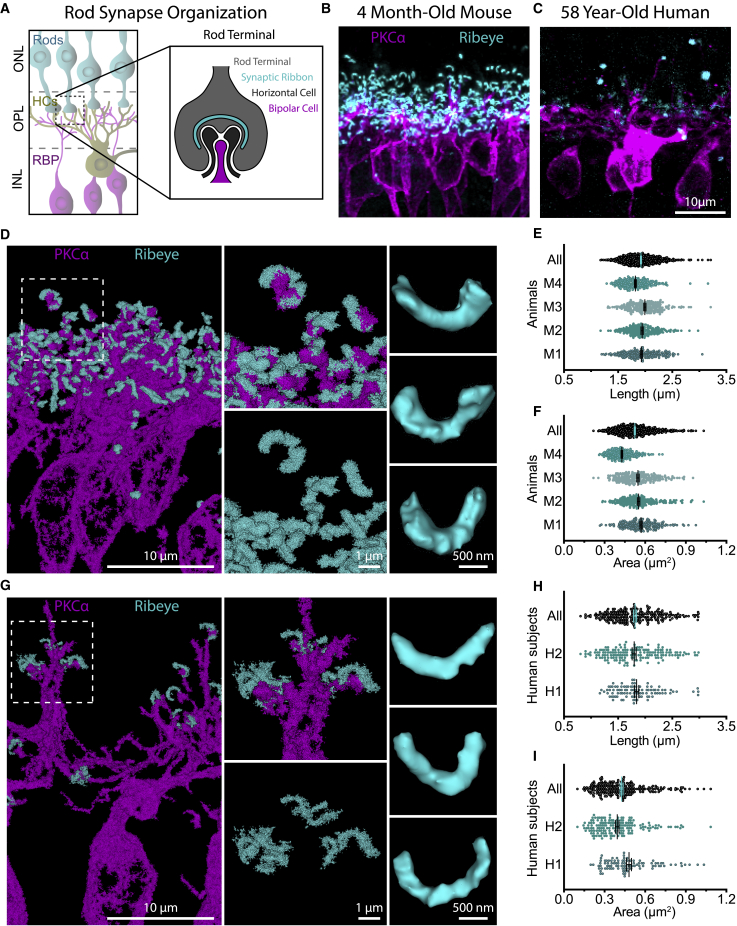
We next used RAIN-STORM to visualize and quantify individual ribbon synapses in the retina OPL (Figure 6A). This region has two advantages. First, the architecture and composition of outer retina synapses have been well characterized, allowing comparisons between our method and metrics reported using other approaches. Second, because each photoreceptor forms connections at one distal location, the relationship between the structure of both pre- and postsynaptic neurons relative to their connectivity can be directly examined. To resolve both murine ribbon synapses and their postsynaptic partners, we applied our optimized method using antibodies against the synapse scaffolding protein RIBEYE (Moser et al., 2020) together with the postsynaptic bipolar marker PKCα (Figures 6A–6C) and used CF568- and AF647-conjugated secondary antibodies to serially visualize both targets. We reconstructed individual RIBEYE labeled ribbons and assessed their 2D-projected length and 2D-projected area. RIBEYE showed a rich variety of ribbon morphologies, including many that appeared in a horseshoe shape and others that appear flatter, consistent with the more elongated contacts of basally located rods (Li et al., 2016). Measurements taken of individual ribbons (n = 844) across four adult mice showed an average ribbon length of 1.92 ± 0.30 μm and a 2D projected area of 0.52 ± 0.13 μm2 (Figures 6D–6F, Video S5). Notably, these values are comparable with those obtained using structured illumination microscopy (1.25 ± 0.05 to 1.95 ± 0.07 μm in length) (Dembla et al., 2020), validating the specificity and accuracy of our optimized methods for measuring cellular structures. This comparison also demonstrates that our tissue preparation and imaging optimization do not distort the structures being measured.
Figure 6.
RAIN-STORM resolves structural and molecular features of synapses
(A) Schematic of retina outer plexiform synapse organization. Rod photoreceptor terminals (cyan) are presynaptic to invaginating postsynaptic horizontal cells (gray) and bipolar cells (magenta).
(B and C) Diffraction-limited imaging of murine (B) and human (C) retina outer plexiform synapses. Presynaptic photoreceptor terminals are labeled with RIBEYE (cyan), whereas postsynaptic bipolar cells are labeled with PKCα (magenta).
(D) Dual-color RAIN-STORM imaging of murine rod bipolar cells (PKCα) and ribbon synapses (RIBEYE) allows individual quantification of ribbons and shows a rich variety of morphologies.
(E and F) Individual outer retina synapses (n = 844) were reconstructed from adult mice (N = 4), and the largest 2D projected length (E) (1.92 ± 0.30 μm) and total area (F) (0.52 ± 0.13 μm2) were quantified for each synapse.
(G) RAIN-STORM imaging of human rod bipolar cells (PKCα, magenta) and ribbon synapses (RIBEYE, cyan) resolves interactions between pre- and postsynaptic neurons.
(H and I) Individual outer retina synapses (n = 263) were reconstructed from human adult donors aged 43–58 years (N = 2), and the largest 2D projected length (H) (1.81 ± 0.03 μm) and total area (I) (0.43 ± 0.01 μm2) were quantified for each synapse. Scale bars, 10 μm, 1 μm, and 500 nm. Data are represented as the mean ± SEM.
Tissue was prepared using our optimized RAIN-STORM protocol, and mouse bipolar neurons in the outer plexiform layer were labeled with PKCα (magenta) and presynaptic ribbons were labeled with RIBEYE (cyan). The image is colored by channel but covers a full 10 μm depth through the tissue slice. Individual neurite tips can clearly be observed invaginating individual presynaptic ribbons.
Finally, we found that our RAIN-STORM optimized protocol is compatible with human tissue. Ribbon synapses are also present in the human outer retina (Moser et al., 2020), but their relative size and organization have not been well mapped. Eyes from two adult human donors (a 43-year-old female and a 58-year-old male) were processed for RAIN-STORM with similar parameters to those in mice and stained with antibodies against RIBEYE and PKCα. As above, images were acquired using sequential imaging for each of the two channels. Reconstruction and quantification of human RIBEYE-labeled synapses showed an average length of 1.81 ± 0.03 μm. Human ribbons also displayed a similar variety in shape and morphology as those found in mice (Figures 6G–6I, Video S6). Of note, we did observe fewer synapses in the human samples relative to the mouse, although this may be due to the inherent delay in human postmortem sample collection rather than a biologically relevant difference.
Human retina was prepared using our optimized RAIN-STORM protocol, and human bipolar neurons in the outer plexiform layer were labeled with PKCα (magenta) and presynaptic ribbons were labeled with RIBEYE (cyan). The image is colored by channel but covers a full 10 μm depth through the tissue slice. As in mouse samples, individual human bipolar neurite tips are directly apposed to invaginating individual presynaptic ribbons. While fewer synapses were observed in the human samples relative to the mouse, this decrease is likely due to the inherent delay in human postmortem sample collection rather than a biologically relevant difference.
Discussion
In this paper, we introduce RAIN-STORM, a rapid and scalable nanoscopic imaging optimization approach that enables 3D target visualization for multiple subcellular and intracellular fluorescent targets within the tissue at depth. We took advantage of the well organized but structurally complex retina circuit to demonstrate that RAIN-STORM optimization can resolve nanoscale features for a wide range of cell types and structures. This enabled us to validate known nanoscale structures as well as map novel cellular features of neurons, glia, and vasculature. In addition, we visualized and quantified hundreds of single human and mouse synapses across multiple individuals. The acquisition of this dataset was facilitated by the high-throughput nature of RAIN-STORM methods, and we show that this approach is practical for analyzing specimens from multiple samples and species. Finally, because RAIN-STORM was developed to be compatible with a commercial imaging system and standard tissue processing, this approach is open to a range of researchers, applications, and clinical samples.
How does RAIN-STORM optimization compare with other efforts aimed at 3D nanoscopic fluorescent imaging of CNS tissue? Dani et al. (2010), achieved 2 to 3 μm imaging depth in brain tissue sections using astigmatism and was able to localize a number of pre- and postsynaptic proteins with excellent resolution using photoswitchable dye pairs (14 nm in a single XY plane and 35 nm in the z direction). Similarly, approaches suitable for synapse imaging at 20–30 nm lateral and 50–70 nm axial resolution at a depth of 0.6–4 μm have also been reported (Herrmannsdorfer et al., 2017). However, the former approach used photoswitchable dye pairs, increasing the level of technical difficulty for new users. In addition, in both cases, the restricted imaging depth precluded examination of whole-cell structures we demonstrate here. Thus, RAIN-STORM methods are complementary to these approaches but offer the advantage of a large set of comparative quantitative data for different imaging parameters and a step-by-step guide to adopting user-friendly sample preparations to acquire nanoscopic data for a host of targets at depths not reported previously.
With these advances, optimized 3D tissue RAIN-STORM imaging has the capacity to be as routine as confocal imaging in many laboratories. This contrasts with many other 3D-STORM imaging approaches that, while excellent (Xu et al., 2015; Huang et al., 2008; Nehme et al., 2020; Punge et al., 2008), rely on custom-built microscopes and require optics expertise and resources that are unavailable to most researchers. Furthermore, unlike serial array tomography electron microscopy (Micheva and Smith, 2007), serial section SMLM (Nanguneri et al., 2012), or related approaches (Li et al., 2016), RAIN-STORM does not require ultrathin sectioning, successive section imaging, alignment, or reconstruction. This further simplifies the application of our method and enables comparative data to be generated quickly both for optimization and application. For instance, in our study of human and mouse outer retina synapses, each dataset consisted of hundreds of synapses and was acquired with a 24-h turnaround from sample collection to image-ready tissue, followed by approximately 8 h of imaging time per species. RAIN-STORM optimization is thus uniquely suitable for large sample numbers and the acquisition and quantitative analysis of extensive datasets.
While improvements in accessibility, speed, and target compatibility were major motivations for this work, several parameters were uncovered that could be useful in other nanoscopic imaging applications. Toward this end, we report quantitative imaging metrics for all 125 tested parameters as a resource for the community and validated our approach using two independent commercial STORM systems. For some applications, we anticipate that the final optimized method we provide will be immediately useful, whereas for others additional tissue- or target-specific testing using the steps we outline may improve outcomes. Among the parameters we tested, some unexpected advantages of various conditions were uncovered. For example, including a post-fixation step after primary and secondary antibody staining markedly improved resolution by ∼10–40 nm. We also found that the ratio of BME to MEA could be used to tune the relative frequency of localizations between the AF647 and CF568 imaging channels. This enabled us to arrive at concentrations that provide a balanced blinking behavior between both channels such that the acquired localizations in one channel are not heavily favored at a cost to the other. Taken together with our other results, these data demonstrate the importance of titrating concentrations for optimal image quality (Figure S2).
Results of this study also shed light on the importance of considering parameters in addition to resolution when selecting conditions for nanoscopic imaging. In short, resolution, although important, is not a sole indicator of performance for a given condition. It is possible to generate images with highly resolved noise that yield good resolution metrics but offer little biological insight. These artifacts could be the result of antibodies binding non-specifically within the tissue or autofluorescent signal, both of which can contribute to the signal observed during acquisition. To account for this, we prioritized the preservation of biological structures and tuning data acquisition and post-processing to remove small localization clusters (noise or “background”) that were not associated with major structures. Our effort was greatly aided by the well-mapped structure of the retina, which provided a backdrop against which we could judge the reliability of imaging parameters to select optimal conditions for inclusion in our final method.
RAIN-STORM optimization also offers opportunities for the discovery of novel nanoscopic structures or molecular components. We found that a diverse array of targets could be readily visualized across a range of cell types and subcellular structures, with ∼90% of tested commercial antibodies showing good labeling and resolution. This broad target compatibility enabled us to examine novel aspects of neural, glial, and vasculature architecture. For example, we visualized IP-TNTs and showed that they appeared to contain a solid collagen IV core, which may provide stability to these small structures as they engage in neurovascular coupling (Alarcon-Martinez et al., 2020). In addition, we documented ∼100–200-nm filaments arising from astrocytes in the ganglion cell layer. Various roles for retinal astrocytes have been described, but these data suggest that they may also physically interact with and perhaps modulate adult retinal neurons (Arizono et al., 2020; O'Sullivan et al., 2017; Tao and Zhang, 2016).
Finally, as with other SMLM methods, the relative density of a given protein and the specificity of available antibodies are key factors in determining good targets for RAIN-STORM optimization. Because single molecules are visualized with this approach, the labeling density of a given target can report on the location and area of individual substructures. In addition, the relative density of a given protein and the availability of high-quality antibodies can make some targets more useful than others. For example, PKCα densely and specifically labels rod bipolar cells and reliably reveals the entirety of the bipolar cellular architecture at the nanoscale, whereas Iba1, a microglia marker, appears less dense. Notably, RAIN-STORM performed consistently well across molecular targets and species, suggesting that these parameters and their variations should provide a good baseline for imaging additional targets and tissues not tested here.
In sum, RAIN-STORM enables precise and data-driven imaging resulting in improved 3D nanoscopic tissue visualization approaches that reveal important aspects of neural circuit organization. In addition, because the optimized RAIN-STORM methods presented here are readily compatible with a wide array of molecular targets and tissue sources, this protocol may prove useful in a number of other applications. These include studies of human clinical specimens, which will enable direct comparisons between human disease and animal models at the nanoscale.
Limitations of the study
In this study we focused on the retina for optimization, so ideal imaging conditions for other types of tissue may require additional parameter testing. However, the methodology we present can be used for protocol optimization for different applications.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Calbindin D-28K | Swant | Cat# CB38a, RRID: AB_10000340 |
| CD31 | BD Biosciences | Cat# BDB5500274, RRID: AB_393571 |
| Cone arrestin | EMD Millipore | Cat# AB15282, RRID: AB_11210270 |
| Collagen IV | EMD Millipore | Cat# AB769, RRID: AB_92262 |
| Connexin 43 | Sigma-Aldrich | Cat#C6219, RRID: AB_476857 |
| Desmin | Thermo Fisher Scientific | Cat# MA513259, RRID: AB_11000611 |
| Dystrophin | Abcam | Cat# ab15277, RRID: AB_301813 |
| Glutamine synthetase (GS) | BD Biosciences | Cat# 610517, RRID: AB_397879 |
| GFAP | Sigma-Aldrich | Cat# G3893, RRID: AB_477010 |
| Iba1 | Abcam | Cat# ab5076, RRID: AB_2224402 |
| Islet1 | R&D System | Cat# AF1837, RRID: AB_2126324 |
| NG2 | Abcam | Cat# ab129051, RRID: AB_2877152 |
| PKCα | Abcam | Cat# ab31, RRID: AB_303507 |
| PSD95 | Abcam | Cat# ab12093, RRID: AB_298846 |
| RIBEYE | Synaptic Systems | Cat#192103, RRID: AB_2086775 |
| Secretagogin (SCGN) | BioVendor | Cat# RD181120100, RRID: AB_2034060 |
| Tau | Proteintech | Cat#66499-1-Ig, RRID: AB_2881863 |
| α-Tubulin | Sigma-Aldrich | Cat#T5168-.2ML, RRID: AB_477579 |
| Tomm20 | Abcam | Cat# ab78547, RRID: AB_2043078 |
| Tyrosine hydroxylase (TH) | EMD Millipore | Cat# AB1542, RRID: AB_90755 |
| Vesicular glutamate transporter 1 (VGlut1) | Abcam | Cat# ab77822, RRID: AB_2187677 |
| Vesicular glutamate transporter 3 (VGlut3) | Millipore | Cat#AB5421, RRID: AB_2187832 |
| Biological samples | ||
| Macaque eyes | Baylor College of Medicine Center for Comparative Medicine veterinary | N/A |
| Rabbit eyes | Baylor College of Medicine Center for Comparative Medicine veterinary | N/A |
| Pig eyes | Baylor College of Medicine Center for Comparative Medicine veterinary | N/A |
| Human eyes | Lions Eye Bank of Texas | |
| Chemicals, peptides, and recombinant proteins | ||
| Sodium hydroxide | Fisher Scientific | 1310-73-2 |
| Ethanol | Decon Labs | 2716 |
| Gelatin A | Sigma | G2500 |
| Chromium (III) potassium sulfate | Sigma | 243361 |
| Glutaraldehyde | Electron Microscopy Sciences | 16019 |
| Paraformaldehyde | Electron Microscopy Sciences | 15710 |
| Ammonium chloride | Sigma | A9434 |
| Glycine | Sigma | G8898 |
| Sodium borohydride | Sigma | 452882 |
| OCT compound | Sakura | 4583 |
| Normal donkey serum | Jackson ImmunoResearch | 017-000-121 |
| Triton-X | EMD Millipore | TX1568-1 |
| Saponin | Sigma | 47036-50G-F |
| Pyranose oxidase | Sigma | P4234-250UN |
| Glucose oxidase | Sigma | G2133-50KU |
| Protocatechuate 3,4-dioxygenase | Sigma | P8279-25UN |
| Catalase | Sigma | C1345-10G |
| Cysteamine hydrochloride | Chem-Impex International | 02839 |
| 2-mercaptoethanol | Sigma | M6250-250ML |
| Protocatechuic acid | Sigma | 37580-25G-F |
| Cyclooctatetraene | Sigma | 138924-1G |
| Trolox | Sigma | 238813-1G |
| Experimental models: Organisms/strains | ||
| C57BL/6J mice | Jackson Laboratory | 000664 |
| Software and algorithms | ||
| Vutara Software | Bruker | https://www.bruker.com/en/products-and-solutions/fluorescence-microscopy/super-resolution-microscopes/srx-software.html |
| GraphPad Prism Version 8.4.3 (471) | GraphPad software | https://www.graphpad.com |
| Other | ||
| #1.5H 22mm round glass coverslips | Neuvitro | GG-25-1.5H |
| Cell chamber for microscopy | Thermo Fisher Scientific | A7816 |
Resource availability
Lead contact
Information and requests for resources and reagents can be directed to the lead contact, Melanie Samuel (msamuel@bcm.edu).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
For mouse tissue, retinas were collected from four 6 to 8-week-old C57BL/6J animals. Experiments were carried out in male and female mice in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the NIH under protocols approved by the BCM Institutional Animal Care and Use Committee. Macaque (N = 2), rabbit (N = 2), and pig (N = 3) retinal tissue were obtained via the Baylor College of Medicine Center for Comparative Medicine from unrelated surgical procedures. Human donor eyes (N = 2) were obtained in collaboration with the Lions Eye Bank of Texas at Baylor College of Medicine. Informed consent was acquired from all patients and/or participating family members in accordance with EBAA and FDA regulatory standards. Subjects were a 43-year-old female and a 58-year-old male and had no documented history of eye disease.
Method details
Coverslip preparation
All samples were mounted and prepared on #1.5H 22mm round glass coverslips. To prepare the coverslips, small batches were placed in beakers and submerged in 5M sodium hydroxide and sonicated for 30 min. Coverslips were subsequently rinsed with deionized water and sonicated for 30 min, three times, using fresh deionized water each time. Coverslips were then sonicated in serial dilutions of ethanol (70%, 90%, 100%) for 30 min each. Once the last wash was complete, slides were removed from the solution and allowed to dry. Coverslips were then dipped in a solution of 0.5% (m/v) Gelatin A and 7mM Chromium Potassium Sulfate in PBS. Excess solution was allowed to drain prior to placing the coverslips upright to air dry. Coverslips were stored for up to one month at room temperature once fully dry.
Tissue preparation
Details for tissue preparation and staining methods are provided for the optimized RAIN-STORM protocol. Tested variations on this protocol are listed in Table S2. Briefly, tested parameters were analyzed in parallel and considered as independent variables from one another. Tested variations for each parameter included both 25 and 4°C fixations, PFA concentration gradients, and fixation duration. Glutaraldehyde preparations were also tested alone (0.3% GA) or in combination with PFA (2% PFA, 0.3% GA) prepared in phosphate buffered saline (PBS). Quenching conditions included ammonium chloride (10mM, 100mM), glycine (10mM, 100mM) or sodium borohydride (0.1%, 0.5% w/v). For the final selected tissue preparation condition, mouse eyes were enucleated and placed in 2% PFA for 1 h at 4°C, and then subsequently rinsed in 100mM glycine solution for 1 h at 4°C. Samples were then washed in PBS for 30 min and stored in PBS until processed for the next steps.
Following fixation, eyecups were dissected, and the cornea and lens were removed from the optic cup. Samples were then allowed to fully equilibrate in 30% sucrose at 25°C until the tissue sank (∼45–60 min). Tissue was serially washed by hand in Optimal Cutting Temperature (OCT) compound to remove excess sucrose and subsequently placed into molds filled with OCT compound (Sakura, Torrance, CA). Embedded tissue was then frozen by submersion into methyl butane chilled on dry ice. Human eyes were prepared using our final selected fixation condition of 2% PFA at 4°C but were fixed for 4 h given the increase in tissue thickness and size of the optic cup. All other conditions were identical to those detailed above. All frozen blocks were then stored at −80°C until ready for use. For analysis and quantification, tissue was sectioned at 10μm and mounted on prepared coverslips.
Antibody staining
For quantification and final images, slides were incubated in the optimized blocking solution (5% normal donkey serum and 0.3% Triton X-100 in PBS) for 1 h and then with primary antibodies diluted in blocking solution for a minimum of 12 h at 4°C. Other tested conditions include variations in concentrations of the blocking buffer components (serum, Triton-X100, or Saponin). Components of the blocking buffer were tested on an individual basis, keeping all other components of the buffer at baseline conditions (NDS 3%, Triton-X100 0.3%) and included: Triton-X100 (0.1%, 0.3%, 0.5%, 1.0%, 2.0%) or Saponin (0.1%, 0.3%, 0.5%, 1.0%, 2.0%), and Normal Donkey Serum (1%, 3%, 5%, 10%, 15%), made to the final concentration using PBS.
After blocking and staining, samples were washed with PBS three times for 20 min and then incubated with commercial dye-conjugated secondary antibodies diluted in the blocking solution being tested for 1 h at room temperature. Slides were next washed with PBS three times for 20 min prior to applying a 4% PFA solution as a postfix for 30 min. Slides were washed with PBS three times for 20 min each, 100mM NH4Cl was applied for 30min, and slides were again washed with PBS three times for 20 min each. Samples were stored in PBS until imaging.
Imaging buffer
Imaging buffer for use in STORM was prepared using stock solutions of one of three oxygen scavenging enzymes: pyranose oxidase, glucose oxidase, or protocatechuate 3,4-dioxygenase. Each oxygen-scavenging enzyme was used to name the associated buffer formulation and any of its derived variations. For Nikon N-STORM experiments, 10 mM of sodium sulfate was used in place of oxygen-scavenging enzymes, based on a previous report (Hartwich et al., 2018). Tested imaging buffers also contained bovine-derived catalase, cysteamine hydrochloride, and 2-mercaptoethanol. When PCD was used, its substrate protocatechuic acid was also included.
One of the four components of a given buffer formulation was varied in concentration while the others remained constant. This testing of each component was carried out for a single type of enzyme (e.g., pyranose oxidase, glucose oxidase, or PCD), and repeated for each enzymatic type, yielding three different buffer types, each with four components tested in a cross-comparative manner. Once this was performed for each buffer-enzyme combination, the final selected buffer was tested with either cyclooctatetraene or Trolox in varying concentrations (1, 2, or 5mM) as an additive known to affect photostability of the fluorophores and blinking of the fluorophores (Olivier et al., 2013; Rasnik et al., 2006). Stock solution aliquots were kept frozen at −80°C until just prior to use. Aliquots were thawed at room temperature and added to a freshly prepared solution of 30% (m/v) glucose in PBS. Once all components for a given buffer were added together to the base solution, the imaging buffer was allowed to equilibrate for 20 min and subsequently used.
Imaging and image processing
Image acquisition was performed on a Bruker Vutara 352 (Bruker, Billerica, MA) using a water objective (UPLSAPO60XW). Stained samples were mounted in a collared well, and 1mL of imaging buffer was added on top of the sample, submersing it in an open well configuration. All images for quantification purposes were acquired in the following standard manner unless otherwise stated: the sample was interrogated at 200nm axial steps using a framerate of 67 Hz for AF647 and 40Hz for CF568 across the full 10μm thickness of tissue. Thus, in a 10μm volume, 51 “steps” were imaged and 250 frames were acquired per step. The entire 51 steps were imaged three sequential times in this manner, yielding a total of 38,250 frames (51∗250∗3) for a single channel per sample. Between each cycle, a timed 600-s pause was included to allow for the imaging buffer to re-equilibrate and recover. 3D nanoscopic imaging volumes were thus 40μm by 40μm by 10μm (16,000 μm3).
For the 640, 561 and 405nm excitation lasers, laser powers of ∼98–110mW (6125–6875 W/cm2), ∼90–100mW (5625–6250 W/cm2), and ∼2-6mW (125–375 W/cm2), were measured at the sample using a 40μm-by-40μm field of view. Data were then clustered to determine associated and contiguous structures within the image using the Ordering Points to Identify the Clustering Structure (OPTICS) algorithm (Ankerst et al., 1999) encoded with the Vutara software. To analyze images, a general particle distance of 0.16 μm and a particle count per cluster of 25 was used for all channels on all images. The particle distance refers to the maximum allowed distance that a particle can be from another particle in order to be included in a given cluster while the particle count reflects the minimum required number of particles required to form a cluster. These general parameters were selected as the most inclusive (least stringent) for filtering out data with the goal of preventing the removal of “real” data from structures that are imaged. Of note, depending on the application, stricter clustering can be performed, but this may not be suitable for all biological targets of interest. Any localizations that failed to fall into clusters as determined by these parameters was removed from the image and considered unstructured data. Once filtering and clustering were complete, an FRC analysis was performed (Banterle et al., 2013; Saxton and Baumeister, 1982) to determine the global aggregate resolution of the sample in the XY and XZ dimensions for each target imaged using three repeat measurements for curve smoothing.
For Nikon N-STORM experiments, image acquisition was performed on an N-STORM 5.0 (Nikon Instruments Inc.) with a CFI SR HP Apo TIRF 100x oil objective (NA 1.49). 10 μm retinal cryosections were prepared and immunostained as described above and then attached onto empty glass-bottom dishes (MatTek 35 mm dish, No. 1.5 coverslip). Sections were covered with 1 mL of imaging buffer and an additional No. 1.5 coverslip. For 2D-STORM on the N-STORM, acquisitions were performed at a single focal plane, while for 3D-STORM acquisitions, a cylindrical lens was added to the light path for astigmatism Z-position fitting based on a fluorescent bead calibration. For both modes, 40,000 frames were acquired from 561 to 647 nm laser channels sequentially at ∼33 frames per second for a 40 μm2 imaging area. STORM analysis details for these experiments are described in the legend for Figure S4.
Quantification and statistical analysis
Image resolution quantification
Image resolution was measured using a Fourier Ring Correlation analysis within Vutara software. Statistical analysis was performed using the GraphPad Prism software. For image resolution quantifications, three images were used per condition tested, and three repeat measurements per image were taken for curve smoothing. Data is reported as the mean ± SEM.
Acknowledgments
We thank Courtney Burger, Mrinalini Hoon, Francóis St. Pierre, Amber Stratman, and members of our laboratory for scientific discussions. This work was supported by the National Institutes of Health (NIH) (R00AG044444, DP2EY02798, and R01EY030458 to M.A.S., P20-GM144230 to M.A.R., and DP2MH125812 to C.M.S.), the BrightFocus Foundation and the Ted Nash Long Life Foundation (to M.A.S.), and the Knights Templar Eye Foundation (to M.A.R.). N.E.A. was supported by the NIH and the National Institute of General Medical Sciences under award no. T32GM088129.
Author contributions
M.A.S. and N.E.A. conceived and designed the experiments. N.E.A. and V.A. performed RAIN-STORM imaging and analysis. D.J. performed the confocal imaging and synapse quantifications. R.H. and C.M.S. helped develop and optimize the imaging protocol. M.A.R. performed N-STORM imaging and analyzed the data. N.E.A., D.J., and V.A. designed and made the figures. M.A.S., D.J., and N.E.A. wrote the paper with input from C.M.S. and M.A.R.
Declaration of interests
R.H. is an employee of Bruker Nano Surfaces.
Published: July 11, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.crmeth.2022.100253.
Supporting citation
The following reference appears in the supplemental information: Postma and Goedhart, 2019.
Supplemental information
Data and code availability
-
•
All data reported in this paper will be shared by the Lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the Lead contact upon request.
References
- Alarcon-Martinez L., Villafranca-Baughman D., Quintero H., Kacerovsky J.B., Dotigny F., Murai K.K., Prat A., Drapeau P., Di Polo A. Interpericyte tunnelling nanotubes regulate neurovascular coupling. Nature. 2020;585:91–95. doi: 10.1038/s41586-020-2589-x. [DOI] [PubMed] [Google Scholar]
- Albrecht N.E., Alevy J., Jiang D., Burger C.A., Liu B.I., Li F., Wang J., Kim S.Y., Hsu C.W., Kalaga S., et al. Rapid and integrative discovery of retina regulatory molecules. Cell Rep. 2018;24:2506–2519. doi: 10.1016/j.celrep.2018.07.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ankerst M., Breunig M., Kriegel H., Sander J. In: In ACM Sigmod Record. Davidson S., Faloutsos C., editors. ACM; 1999. OPTICS: ordering points to identify the clustering structure; pp. 49–60. [Google Scholar]
- Arizono M., Inavalli V.V.G.K., Panatier A., Pfeiffer T., Angibaud J., Levet F., Veer M.J.T.T., Stobart J., Bellocchio L., Mikoshiba K., et al. Author Correction: structural basis of astrocytic Ca2+ signals at tripartite synapses. Nat. Commun. 2020;11:2541. doi: 10.1038/s41467-020-16453-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baddeley D., Crossman D., Rossberger S., Cheyne J.E., Montgomery J.M., Jayasinghe I.D., Cremer C., Cannell M.B., Soeller C. 4D super-resolution microscopy with conventional fluorophores and single wavelength excitation in optically thick cells and tissues. PLoS One. 2011;6:e20645. doi: 10.1371/journal.pone.0020645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banterle N., Bui K.H., Lemke E.A., Beck M. Fourier ring correlation as a resolution criterion for super-resolution microscopy. J. Struct. Biol. 2013;183:363–367. doi: 10.1016/j.jsb.2013.05.004. [DOI] [PubMed] [Google Scholar]
- Bewersdorf J., Schmidt R., Hell S.W. Comparison of I5M and 4Pi-microscopy. J. Microsc. 2006;222:105–117. doi: 10.1111/j.1365-2818.2006.01578.x. [DOI] [PubMed] [Google Scholar]
- Bon P., Linarès-Loyez J., Feyeux M., Alessandri K., Lounis B., Nassoy P., Cognet L. Self-interference 3D super-resolution microscopy for deep tissue investigations. Nat. Methods. 2018;15:449–454. doi: 10.1038/s41592-018-0005-3. [DOI] [PubMed] [Google Scholar]
- Bowler M., Kong D., Sun S., Nanjundappa R., Evans L., Farmer V., Holland A., Mahjoub M.R., Sui H., Loncarek J. High-resolution characterization of centriole distal appendage morphology and dynamics by correlative STORM and electron microscopy. Nat. Commun. 2019;10:993. doi: 10.1038/s41467-018-08216-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringmann A., Grosche A., Pannicke T., Reichenbach A. GABA and glutamate uptake and metabolism in retinal glial (müller) cells. Front. Endocrinol. 2013;4:48. doi: 10.3389/fendo.2013.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celio M.R. Calbindin D-28k and parvalbumin in the rat nervous system. Neuroscience. 1990;35:375–475. doi: 10.1016/0306-4522(90)90091-h. [DOI] [PubMed] [Google Scholar]
- Chamma I., Letellier M., Butler C., Tessier B., Lim K.H., Gauthereau I., Choquet D., Sibarita J.B., Park S., Sainlos M., Thoumine O. Mapping the dynamics and nanoscale organization of synaptic adhesion proteins using monomeric streptavidin. Nat. Commun. 2016;7:10773. doi: 10.1038/ncomms10773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Tillberg P.W., Boyden E.S. Expansion microscopy. Science. 2015;347:543–548. doi: 10.1126/science.1260088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dani A., Huang B. New resolving power for light microscopy: applications to neurobiology. Curr. Opin. Neurobiol. 2010;20:648–652. doi: 10.1016/j.conb.2010.07.006. [DOI] [PubMed] [Google Scholar]
- Dani A., Huang B., Bergan J., Dulac C., Zhuang X. Super-resolution imaging of chemical synapses in the brain. Neuron. 2010;68:843–856. doi: 10.1016/j.neuron.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembla E., Dembla M., Maxeiner S., Schmitz F. Synaptic ribbons foster active zone stability and illumination-dependent active zone enrichment of RIM2 and Cav1.4 in photoreceptor synapses. Sci. Rep. 2020;10:5957. doi: 10.1038/s41598-020-62734-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- German C.L., Gudheti M.V., Fleckenstein A.E., Jorgensen E.M. Brain slice staining and preparation for three-dimensional super-resolution microscopy. Methods Mol. Biol. 2017;1663:153–162. doi: 10.1007/978-1-4939-7265-4_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiss F., Deligiannaki M., Jung C., Gaul U., Braun D. Single-molecule imaging in living Drosophila embryos with reflected light-sheet microscopy. Biophys. J. 2016;110:939–946. doi: 10.1016/j.bpj.2015.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwich T.M.P., Hin Chung K.K., Schroeder L., Bewersdorf J., Soeller C., Baddeley D. A stable, high refractive index, switching buffer for super-resolution imaging. bioRxiv. 2018 doi: 10.1101/465492. Preprint at. [DOI] [Google Scholar]
- Herrmannsdorfer F., Flottmann B., Nanguneri S., Venkataramani V., Horstmann H., Kuner T., Heilemann M. 3D d STORM imaging of fixed brain tissue. Methods Mol. Biol. 2017;1538:169–184. doi: 10.1007/978-1-4939-6688-2_13. [DOI] [PubMed] [Google Scholar]
- Huang B., Wang W., Bates M., Zhuang X. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science. 2008;319:810–813. doi: 10.1126/science.1153529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt C.A., Schenker L.J., Kennedy M.B. PSD-95 is associated with the postsynaptic density and not with the presynaptic membrane at forebrain synapses. J. Neurosci. 1996;16:1380–1388. doi: 10.1523/jneurosci.16-04-01380.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki M., Westerhausen-Larson A., Kino J., Hayashi T., Hayashi T., Kao W., WesterhAusen-LArson A. Distribution of collagen IV in human ocular tissues. Invest. Ophthalmol. Vis. Sci. 1993;34:2680–2689. [PubMed] [Google Scholar]
- Ivanova E., Corona C., Eleftheriou C.G., Bianchimano P., Sagdullaev B.T. Retina-specific targeting of pericytes reveals structural diversity and enables control of capillary blood flow. J. Comp. Neurol. 2021;529:1121–1134. doi: 10.1002/cne.25011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juette M.F., Gould T.J., Lessard M.D., Mlodzianoski M.J., Nagpure B.S., Bennett B.T., Hess S.T., Bewersdorf J. Three-dimensional sub-100 nm resolution fluorescence microscopy of thick samples. Nat. Methods. 2008;5:527–529. doi: 10.1038/nmeth.1211. [DOI] [PubMed] [Google Scholar]
- Klein T., Proppert S., Sauer M. Eight years of single-molecule localization microscopy. Histochem. Cell Biol. 2014;141:561–575. doi: 10.1007/s00418-014-1184-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koulen P., Fletcher E.L., Craven S.E., Bredt D.S., Wässle H. Immunocytochemical localization of the postsynaptic density protein PSD-95 in the mammalian retina. J. Neurosci. 1998;18:10136–10149. doi: 10.1523/jneurosci.18-23-10136.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku T., Swaney J., Park J.Y., Albanese A., Murray E., Cho J.H., Park Y.G., Mangena V., Chen J., Chung K. Multiplexed and scalable super-resolution imaging of three-dimensional protein localization in size-adjustable tissues. Nat. Biotechnol. 2016;34:973–981. doi: 10.1038/nbt.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leterrier C., Potier J., Caillol G., Debarnot C., Rueda Boroni F., Dargent B. Nanoscale Architecture of the axon initial segment reveals an organized and robust scaffold. Cell Rep. 2015;13:2781–2793. doi: 10.1016/j.celrep.2015.11.051. [DOI] [PubMed] [Google Scholar]
- Li S., Mitchell J., Briggs D.J., Young J.K., Long S.S., Fuerst P.G. Morphological diversity of the rod spherule: a study of serially reconstructed electron micrographs. PLoS One. 2016;11:e0150024. doi: 10.1371/journal.pone.0150024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes-Jorge L., Llombart C., Ramos D., López-Luppo M., Valença A., Nacher V., Navarro M., Carretero A., Méndez-Ferrer S., Rodriguez-Baeza A., Ruberte J. Intercapillary bridging cells: immunocytochemical characteristics of cells that connect blood vessels in the retina. Exp. Eye Res. 2012;98:79–87. doi: 10.1016/j.exer.2012.03.010. [DOI] [PubMed] [Google Scholar]
- Micheva K.D., Smith S.J. Array tomography: a new tool for imaging the molecular architecture and ultrastructure of neural circuits. Neuron. 2007;55:824. doi: 10.1016/j.neuron.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhaylova M., Cloin B.M.C., Finan K., Van Den Berg R., Teeuw J., Kijanka M.M., Sokolowski M., Katrukha E.A., Maidorn M., Opazo F., et al. Resolving bundled microtubules using anti-tubulin nanobodies. Nat. Commun. 2015;6:7933. doi: 10.1038/ncomms8933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlodzianoski M.J., Cheng-Hathaway P.J., Bemiller S.M., Mccray T.J., Liu S., Miller D.A., Lamb B.T., Landreth G.E., Huang F. Active PSF shaping and adaptive optics enable volumetric localization microscopy through brain sections. Nat. Methods. 2018;15:583–586. doi: 10.1038/s41592-018-0053-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mönkemöller V., Oie C., Hübner W., Hubner W., Huser T., Mccourt P. Multimodal super-resolution optical microscopy visualizes the close connection between membrane and the cytoskeleton in liver sinusoidal endothelial cell fenestrations. Sci. Rep. 2015;5:16279. doi: 10.1038/srep16279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser T., Grabner C.P., Schmitz F. Sensory processing at ribbon synapses in the retina and the cochlea. Physiol. Rev. 2020;100:103–144. doi: 10.1152/physrev.00026.2018. [DOI] [PubMed] [Google Scholar]
- Nanguneri S., Flottmann B., Horstmann H., Heilemann M., Kuner T. Three-dimensional, tomographic super-resolution fluorescence imaging of serially sectioned thick samples. PLoS One. 2012;7:e38098. doi: 10.1371/journal.pone.0038098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehme E., Freedman D., Gordon R., Ferdman B., Weiss L.E., Alalouf O., Naor T., Orange R., Michaeli T., Shechtman Y. DeepSTORM3D: dense 3D localization microscopy and PSF design by deep learning. Nat. Methods. 2020;17:734–740. doi: 10.1038/s41592-020-0853-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwenhuizen R.P.J., Lidke K.A., Bates M., Puig D.L., Grünwald D., Stallinga S., Rieger B. Measuring image resolution in optical nanoscopy. Nat. Methods. 2013;10:557–562. doi: 10.1038/nmeth.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Sullivan M.L., Puñal V.M., Kerstein P.C., Brzezinski J.A.T., Glaser T., Wright K.M., Kay J.N. Astrocytes follow ganglion cell axons to establish an angiogenic template during retinal development. Glia. 2017;65:1697–1716. doi: 10.1002/glia.23189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier N., Keller D., Gönczy P., Manley S. Resolution doubling in 3D-STORM imaging through improved buffers. PLoS One. 2013;8:e69004. doi: 10.1371/journal.pone.0069004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavani S.R.P., Thompson M.A., Biteen J.S., Lord S.J., Liu N., Twieg R.J., Piestun R., Moerner W.E. Three-dimensional, single-molecule fluorescence imaging beyond the diffraction limit by using a double-helix point spread function. Proc. Natl. Acad. Sci. USA. 2009;106:2995–2999. doi: 10.1073/pnas.0900245106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postma M., Goedhart J. PlotsOfData-A web app for visualizing data together with their summaries. PLoS Biol. 2019;17:e3000202. doi: 10.1371/journal.pbio.3000202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punge A., Rizzoli S.O., Jahn R., Wildanger J.D., Meyer L., Schönle A., Kastrup L., Hell S.W. 3D reconstruction of high-resolution STED microscope images. Microsc. Res. Tech. 2008;71:644–650. doi: 10.1002/jemt.20602. [DOI] [PubMed] [Google Scholar]
- Puthussery T., Gayet-Primo J., Taylor W.R. Localization of the calcium-binding protein secretagogin in cone bipolar cells of the mammalian retina. J. Comp. Neurol. 2010;518:513–525. doi: 10.1002/cne.22234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasnik I., Mckinney S.A., Ha T. Nonblinking and long-lasting single-molecule fluorescence imaging. Nat. Methods. 2006;3:891–893. doi: 10.1038/nmeth934. [DOI] [PubMed] [Google Scholar]
- Robichaux M.A., Potter V.L., Zhang Z., He F., Liu J., Schmid M.F., Wensel T.G. Defining the layers of a sensory cilium with STORM and cryoelectron nanoscopy. Proc. Natl. Acad. Sci. USA. 2019;116:23562–23572. doi: 10.1073/pnas.1902003116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanes J.R., Zipursky S.L. Design principles of insect and vertebrate visual systems. Neuron. 2010;66:15–36. doi: 10.1016/j.neuron.2010.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarthy V. Collagen IV mRNA expression during development of the mouse retina: an in situ hybridization study. Invest. Ophthalmol. Vis. Sci. 1993;34:145–152. [PubMed] [Google Scholar]
- Saxton W.O., Baumeister W. The correlation averaging of a regularly arranged bacterial cell envelope protein. J. Microsc. 1982;127:127–138. doi: 10.1111/j.1365-2818.1982.tb00405.x. [DOI] [PubMed] [Google Scholar]
- Schmitz F., Drenckhahn D. Dystrophin in the retina. Prog. Neurobiol. 1997;53:547–560. doi: 10.1016/s0301-0082(97)00047-6. [DOI] [PubMed] [Google Scholar]
- Sigal Y.M., Speer C.M., Babcock H.P., Zhuang X. Mapping synaptic input fields of neurons with super-resolution imaging. Cell. 2015;163:493–505. doi: 10.1016/j.cell.2015.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suleiman H., Zhang L., Roth R., Heuser J.E., Miner J.H., Shaw A.S., Dani A. Nanoscale protein architecture of the kidney glomerular basement membrane. Elife. 2013;2:e01149. doi: 10.7554/elife.01149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao C., Zhang X. Retinal proteoglycans act as cellular receptors for basement membrane assembly to control astrocyte migration and angiogenesis. Cell Rep. 2016;17:1832–1844. doi: 10.1016/j.celrep.2016.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillberg P.W., Chen F., Piatkevich K.D., Zhao Y., Yu C.C., English B.P., Gao L., Martorell A., Suk H.J., Yoshida F., et al. Protein-retention expansion microscopy of cells and tissues labeled using standard fluorescent proteins and antibodies. Nat. Biotechnol. 2016;34:987–992. doi: 10.1038/nbt.3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uesugi R., Yamada M., Mizuguchi M., Baimbridge K.G., Kim S.U. Calbindin D-28k and parvalbumin immunohistochemistry in developing rat retina. Exp. Eye Res. 1992;54:491–499. doi: 10.1016/0014-4835(92)90127-e. [DOI] [PubMed] [Google Scholar]
- van den Dries K., Schwartz S.L., Byars J., Meddens M.B., Bolomini-Vittori M., Lidke D.S., Figdor C.G., Lidke K.A., Cambi A. Dual-color superresolution microscopy reveals nanoscale organization of mechanosensory podosomes. Mol. Biol. Cell. 2013;24:2112–2123. doi: 10.1091/mbc.e12-12-0856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecino E., Rodriguez F.D., Ruzafa N., Pereiro X., Sharma S.C. Glia-neuron interactions in the mammalian retina. Prog. Retin. Eye Res. 2016;51:1–40. doi: 10.1016/j.preteyeres.2015.06.003. [DOI] [PubMed] [Google Scholar]
- Xu J., Tehrani K.F., Kner P. Multicolor 3D super-resolution imaging by quantum dot stochastic optical reconstruction microscopy. ACS Nano. 2015;9:2917–2925. doi: 10.1021/nn506952g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tissue was prepared using a standard preparatory method (see supplemental Table 1, materials and methods) to image calbindin-labeled horizontal cells across 10 μm. The image is color coded by depth, with colors indicating the axial position throughout the stack (purple 0 μm to yellow 10 μm). Synapses appear jagged and incomplete, with little structural detail connecting them back to the cell body.
Tissue was prepared using our optimized RAIN-STORM protocol, and calbindin-labeled horizontal cells were imaged across 10 μm. The image is color coded by depth, with colors indicating the axial position throughout the stack (purple 0 μm to yellow 10 μm). Fine neurite structural details are observed and individual synapses are clearly visible.
Tissue was prepared using our optimized RAIN-STORM protocol, and collagen IV-labeled vessels were imaged across 10 μm. The image is color coded by depth, with colors indicating the axial position throughout the stack (purple 0 μm to yellow 10 μm). The structure and morphology of hollow vessels are well preserved, and cross-vessel nanotube connections are apparent.
Tissue was prepared using our optimized RAIN-STORM protocol, and GFAP-labeled astrocytes were imaged across 10 μm. The image is color coded by depth, with colors indicating the axial position throughout the stack (purple 0 μm to yellow 10 μm). The structure and morphology of astrocyte filamentous fibers were observed, which appear to form a highly branched network.
Tissue was prepared using our optimized RAIN-STORM protocol, and mouse bipolar neurons in the outer plexiform layer were labeled with PKCα (magenta) and presynaptic ribbons were labeled with RIBEYE (cyan). The image is colored by channel but covers a full 10 μm depth through the tissue slice. Individual neurite tips can clearly be observed invaginating individual presynaptic ribbons.
Human retina was prepared using our optimized RAIN-STORM protocol, and human bipolar neurons in the outer plexiform layer were labeled with PKCα (magenta) and presynaptic ribbons were labeled with RIBEYE (cyan). The image is colored by channel but covers a full 10 μm depth through the tissue slice. As in mouse samples, individual human bipolar neurite tips are directly apposed to invaginating individual presynaptic ribbons. While fewer synapses were observed in the human samples relative to the mouse, this decrease is likely due to the inherent delay in human postmortem sample collection rather than a biologically relevant difference.
Data Availability Statement
-
•
All data reported in this paper will be shared by the Lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the Lead contact upon request.