Abstract
The dissimilatory iron-reducing bacterium Geobacter metallireducens was found to require iron at a concentration in excess of 50 μM for continuous cultivation on nitrate. Growth yield (∼3-fold), cytochrome c content (∼7-fold), and nitrate (∼4.5-fold) and nitrite (∼70-fold) reductase activities were all increased significantly when the growth medium was amended with 500 μM iron.
Geobacter metallireducens is a dissimilatory iron-reducing bacterium that also has the ability to respire nitrate to ammonia (7). It has been known for some time that, although G. metallireducens can be transferred indefinitely on freshwater-acetate (FWA) medium with 50 mM Fe(III), it rarely survives repeated transfers on FWA-nitrate medium as prepared according to the method of Lovley and Phillips (7). Biochemical characterization of the components involved in nitrate respiration has shown that c-type cytochromes appear to be overexpressed in nitrate-grown cells (10). Recently, a cytochrome c-containing enzyme complex that exhibits both nitrate and nitrite reductase activities has been described (9). Thus, a possible explanation for the limited number of transfers is that growth on nitrate requires more iron than is supplied in the trace element mixture. Preliminary results indicated that amending FWA-nitrate medium with Fe(III) not only allowed continual culture on nitrate but also improved cell yields (N. MacLennan and J. F. Stolz, unpublished data). Thus, we were interested in determining what effect amending FWA-nitrate medium with iron has on cell yield, cytochrome content, and nitrate and nitrite reductase activities.
(This work was conducted by J. M. Senko in partial fulfillment of an M.S. degree from Duquesne University, Pittsburgh, Pa., 2000.)
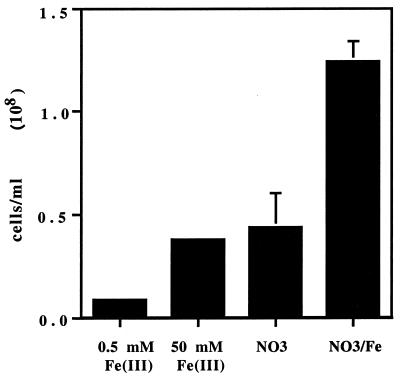
Cultures of G. metallireducens (ATCC 55774) were maintained on FWA-Fe [50 mM Fe(III)] and FWA-nitrate (30 mM sodium nitrate) media prepared according to the method of Lovley and Phillips (7). FWA-nitrate medium was amended with 10, 50, 250, or 500 μM Fe(III) using a stock solution of 50 mM ferric citrate. FWA-Fe medium was also prepared with only 0.5 mM Fe(III) as a control. All media were inoculated using the Hungate method (4), and cultures were transferred every 3 days to fresh media. For cell yield comparison, serum bottles (125 ml) containing 100 ml of medium were inoculated with 3.67 × 106 cells (for a final concentration of 3.56 × 104 cells/ml) from a stock culture grown on nitrate amended with 0.5 mM Fe(III). After 3 days, direct cell counts were performed by fluorescence microscopy using acridine orange (12). The cell yields on the standard FWA-Fe [with 50 mM Fe(III)] and FWA-nitrate media were similar, on the order of 4 × 107 cells/ml (Fig. 1). The cell yield on FWA-nitrate medium amended with 0.5 mM iron, however, was almost threefold greater (Fig. 1). This increase in cell yield could not be attributed to dissimilatory growth on the iron amendment. Minimal growth was observed in cultures grown on FWA-Fe medium that contained only 0. 5 mM iron (Fig. 1). Furthermore, cell yield predictions based on thermodynamic calculations revealed that acetate and nitrate and acetate and Fe are nearly identical in their numbers of moles of cell carbon per mole of substrate carbon; thus, even if the 0.5 mM iron was respired, the amount would be insufficient to account for the observed increase (J. Van Briesen, personal communication). In a separate set of experiments, we found no significant increase in Fe(II) concentration in the medium after 48 h of growth, as determined by the ferrozine assay (7), suggesting that indeed the iron was being assimilated by the cells (data not shown). Increased concentrations of iron were found to be essential for growth with nitrate as the sole source of nitrogen and increased the number of passages. The cultures could be maintained indefinitely on FWA nitrate medium (with ammonium chloride as the nitrogen source) amended with at least 50 μM iron.
FIG. 1.
Cell counts from cultures after 72 h of growth in FWA media with 500 μM Fe(III), 50 mM Fe(III), 30 mM nitrate, or 30 mM nitrate amended with 500 μM Fe(III).
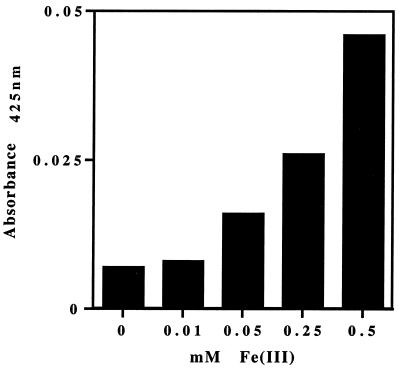
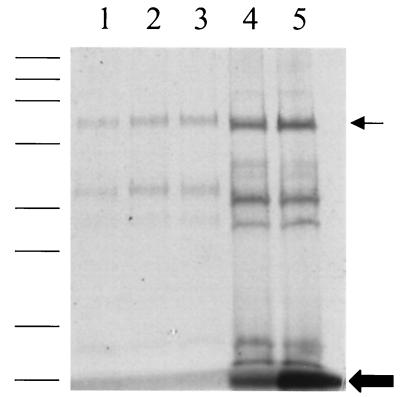
The cytochrome content of cell lysates was determined by difference spectra and sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis. For this set of experiments, cultures were maintained on 10 ml of medium in 22-ml septum-topped tubes. FWA-nitrate medium was amended with 10, 50, 250, or 500 μM Fe(III), with unamended medium being used as a control. After the eighth transfer, cells were harvested by centrifugation, resuspended in 1 ml of bicarbonate buffer (2.5 g/liter, pH 6.8), and lysed by sonication (9). The protein concentration was determined by the Bradford method, using bovine serum albumin as the standard (1). Heme content was compared using difference spectra (Fig. 2). The spectra were determined on a Cary 3E UV–visible-light spectrophotometer at both 425 nm (Fig. 2) and 550 nm (data not shown) using the alkaline pyridine hemochrome method (15). Total heme content increased as the concentration of iron increased. The highest concentration of heme was detected in cells from FWA-nitrate medium amended with 500 μM Fe(III) (Fig. 2). Further evidence that the increase in iron was affecting heme content was provided by heme activity assays in denaturing polyacrylamide gels (Fig. 3). Equal amounts in protein (25 μg/lane) were loaded onto precast SDS-polyacrylamide gels (4% stacker gel–12% resolving gel; Bio-Rad, Hercules, Calif.) (5). After electrophoresis, the gels were stained with Coomassie brilliant blue to visualize proteins or were developed for peroxidase activity with dimethoxybenzidene to detect covalently bound heme (2). A marked increase in peroxidase activity, indicating a greater amount of covalently bound heme, was seen in all the cytochrome species as iron concentration increased (Fig. 3). These results suggest that not enough iron was available for heme synthesis at the lower iron concentrations (i.e., <50 μM), although total protein content was the same. At the highest iron concentrations, however, free heme was detected at the bottom of the heme-stained gels (Fig. 3). This result suggests that, under these conditions, insufficient quantities of apoprotein were being synthesized or that the process of heme ligation was limiting. The increase in free heme C content (Fig. 3) is indeed striking. Heme biosynthesis is carefully regulated in both prokaryotes and eukaryotes due in part to the cytotoxicity of porphyrin (3, 8).
FIG. 2.
Reduced minus oxidized absorbance at 425 nm of cell lysates grown on FWA-nitrate medium amended with 0, 10, 50, 250, and 500 μM ferric citrate.
FIG. 3.
SDS-polyacrylamide gel (4-to-12% gradient) stained for covalently bound heme of cell lysates harvested after eight transfers on FWA-nitrate medium amended with iron. Lane 1, 0 μM iron; lane 2, 10 μM iron; lane 3, 50 μM iron; lane 4, 250 μM iron; lane 5, 500 μM iron. Molecular-mass standards are 208, 115, 79.5, 49.5, 34.8, 28.3, 20.4, and 7.2 kDa. Large arrow, free heme; small arrow, 62-kDa cytochrome c nitrite reductase.
These same cell lysates were used to measure the rates of nitrate and nitrite reduction. Nitrate and nitrite reductase activities, as determined by the methyl viologen assay (16), also increased with increasing iron concentration (Table 1). Nitrate reductase activity was increased over 4.5-fold in cells grown in medium amended with either 250 or 500 μM iron. The increase in nitrite reductase activity was most dramatic with an almost 70-fold increase in activity in cells grown in medium amended with 500 μM iron (Table 1). The latter results are not surprising, as nitrite reduction is catalyzed by a multiheme cytochrome c nitrite reductase (9). The increase in nitrate reductase activity may be attributed to the tight coupling of nitrate and nitrite reduction in G. metallireducens (9). The functional enzyme complex which includes a multiheme cytochrome c exhibits both nitrate and nitrite reductase activities (9). Nitrite does not accumulate in the medium (7) and is not detectable as an intermediate in the nitrate reductase assay (9). This tight coupling is not observed in the closely related species Desulfovibrio desulfuricans (6), Sulfurospirillum deleyianum (13, 14), and Sulfurospirillum barnesii (11), which reduce nitrate and nitrite in separate steps. Interestingly, nitrite reduction is the rate-limiting step in assimilatory nitrate reduction in the marine alga Thalassiosira weissflogii under iron-limited growth conditions (A. Milligan, http://www.ocgy.ubc.ca/∼pjhlab/milligan.html). While it is well-known that iron is an important trace element, the high concentrations needed by G. metallireducens for growth on nitrate is apparently due to the essential involvement of c-type cytochromes. The overproduction of free heme C with increasing iron may reflect novel regulatory aspects of heme biosynthesis.
TABLE 1.
Nitrate and nitrite reductase activities of cell lysates grown on FWA medium amended with iron with nitrate as the terminal electron acceptor
| Fe amendment (μM)a | Reductase activity (nmol min−1 mg−1)
|
|
|---|---|---|
| Nitrate | Nitrite | |
| 0 | 19 | 90 |
| 50 | 63 | 100 |
| 250 | 89 | 2,900 |
| 500 | 89 | 6,100 |
The unamended medium contained 4 μM iron.
Acknowledgments
We thank P. Basu for helpful discussion, J. Van Briesen for cell yield calculations, and J. Bergeron for the ferrozine assays.
This work was supported in part by grant MCB 9305399 from the National Science Foundation and Duquesne University.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seiman J G, Smith J A, Struhl K. Short protocols in molecular biology. New York, N.Y: Wiley and Sons; 1989. [Google Scholar]
- 2.Francis R T, Becker R R. Specific indication of hemoproteins in polyacrylamide gels using a double staining process. Anal Chem. 1984;136:504–514. doi: 10.1016/0003-2697(84)90253-7. [DOI] [PubMed] [Google Scholar]
- 3.Hamza I, Chauhan S, Hassett R, O'Brian M R. The bacterial Irr protein is required for the coordination of heme biosynthesis with iron availability. J Biol Chem. 1998;273:21669–21674. doi: 10.1074/jbc.273.34.21669. [DOI] [PubMed] [Google Scholar]
- 4.Hungate R E. A roll tube method for the cultivation of strict anaerobes. Methods Microbiol. 1969;3B:117–132. [Google Scholar]
- 5.Laemmli U. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 6.Liu M-C, Peck H D., Jr The isolation of a hexaheme cytochrome from Desulfovibrio desulfuricans and its identification as a new type of nitrite reductase. J Biol Chem. 1981;256:13159–13164. [PubMed] [Google Scholar]
- 7.Lovley D R, Phillips E J. Novel mode of energy metabolism: organic carbon oxidation coupled to dissimilatory reduction of iron or manganese. Appl Environ Microbiol. 1988;54:1472–1480. doi: 10.1128/aem.54.6.1472-1480.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.May B K, Dogra S C, Sadlon T J, Bhasker C R, Cox T C, Bottomley S S. Molecular regulation of heme biosynthesis in higher vertebrates. Prog Nucleic Acid Res Mol Biol. 1995;51:1–52. doi: 10.1016/s0079-6603(08)60875-2. [DOI] [PubMed] [Google Scholar]
- 9.Murillo F M, Gugliuzza T, Senko J, Basu P, Stolz J. A heme C-containing enzyme complex that exhibits nitrate and nitrite reductase activity from the dissimilatory iron reducing bacterium Geobacter metallireducens. Arch Microbiol. 1999;172:313–320. doi: 10.1007/s002030050785. [DOI] [PubMed] [Google Scholar]
- 10.Naik R R, Murillo F M, Stolz J F. Evidence for a novel nitrate reductase in the dissimilatory iron-reducing bacterium Geobacter metallireducens. FEMS Microbiol Lett. 1992;106:53–58. [Google Scholar]
- 11.Oremland R S, Switzer Blum J, Burns Bindi A, Dowdle P R, Herbel M, Stolz J F. Simultaneous reduction of nitrate and selenate by cell suspensions of selenium-respiring bacteria. Appl Environ Microbiol. 1999;65:4385–4392. doi: 10.1128/aem.65.10.4385-4392.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parsons T R, Maita Y, Lalli C M. Manual of chemical and biological methods for seawater analysis. New York, N.Y: Pergamon Press; 1984. Direct counting of bacteria by fluorescence microscopy; pp. 123–126. [Google Scholar]
- 13.Schumacher W, Kroneck P M H. Anaerobic energy metabolism of the sulfur-reducing bacterium “Spirillum” 5175 during dissimilatory nitrate reduction to ammonia. Arch Microbiol. 1992;157:464–470. [Google Scholar]
- 14.Schumacher W, Hole U, Kroneck P M H. Ammonia-forming cytochrome c nitrite reductase from Sulfurospirillum deleyianum is a tetraheme protein: new aspects of molecular composition and spectroscopic properties. Biochem Biophys Res Commun. 1994;205:911–916. doi: 10.1006/bbrc.1994.2751. [DOI] [PubMed] [Google Scholar]
- 15.Smith K. Porphyrins and metalloporphyrins. Oxford, United Kingdom: Elsevier Scientific; 1975. [Google Scholar]
- 16.Stolz J F, Gugliuzza T, Blum J S, Oremland R, Murillo F M. Differential cytochrome content and reductase activity in Geospirillum barnesii strain SeS3. Arch Microbiol. 1997;167:1–5. doi: 10.1007/s002030050408. [DOI] [PubMed] [Google Scholar]