Abstract
Background
Human rhinoviruses (HRVs) are a common cause of influenza-like illness, with the ability to infect the upper and lower respiratory tracts. In this study we aim to describe the clinical and molecular features of HRV infection in Mexican children and adults.
Methods
We performed a hospital-based, 4-year multicenter prospective observational cohort study of patients with influenza-like illness. Participants who tested positive for HRV were included. We described demographic, clinical, and laboratory characteristics and the association between HRV types, illness severity, and clinical outcomes.
Results
Of the 5662 subjects recruited, 1473 (26%) had HRV; of those, 988 (67.1%) were adults (≥18 years) and 485 (32.9%) were children. One hundred sixty-seven (11.33%) samples were sequenced; 101 (60.5%) were rhinovirus species A (HRV-A), 22 (13.2%) were rhinovirus species B (HRV-B), and 44 (26.3%) were rhinovirus species C (HRV-C). Among children and adults, 30.5% and 23.5%, respectively, were hospitalized (non–intensive care unit [ICU]). The odds of HRV-C are higher than HRV-A for participants in the ICU (compared to outpatient) and when platelets, lymphocytes, white blood cells, and lactate dehydrogenase are increased. The odds of HRV-C are higher than HRV-A and HRV-B with shortness of breath. The odds of HRV-A are higher than HRV-B, and the odds of HRV-B are higher than HRV-C, when mild symptoms like muscle ache and headache occur.
Conclusions
Rhinoviruses are a common cause of influenza-like illness. It is necessary to improve the surveillance, testing, and species identification for these viruses to understand different clinical presentations and risk factors associated with worse outcomes.
Clinical Trials Registration. NCT01418287.
Keywords: clinical presentation, HRV species, influenza-like illness, rhinovirus, severity
This study demonstrated the importance of human rhinovirus (HRV) as a causal agent of influenza-like illness associated with upper and lower respiratory tract symptoms in children and adults. HRV-C was associated with more severe disease and may cause worse outcomes.
Human rhinoviruses (HRVs) have been described as one of the most prevalent human respiratory viruses and a common cause for influenza-like illness (ILI) worldwide [1, 2]. HRVs are responsible for one-half of all acute upper respiratory illnesses and infect billions of people annually [3–5]. Furthermore, it is estimated that HRV infections represent a significant economic burden annually in terms of medical costs, work absenteeism, and inappropriate use of antibiotics [6, 7].
The most common clinical characteristics in patients with HRVs include rhinorrhea, cough, sore throat, headache, diffuse malaise, and nasal congestion. However, they may cause more serious manifestations including bronchiolitis and exacerbations of chronic pulmonary diseases, such as asthma [8, 9], particularly in children, the immunocompromised, and the elderly [10, 11].
HRVs are included in the Picornaviridae family, in the Enterovirus genus, and are specific to humans. HRVs are genetically very diverse and over 150 genotypes have been identified. They are classified into 3 main HRV species (HRV-A, HRV-B, and HRV-C) according to their distinctive genome, phylogenetic sequences, and molecular features [12]. Most HRV respiratory infections are due to HRV-A and HRV-C, and different pathways of epidemiology, transmission, and disease outcomes among the 3 species have been described [13, 14].
Some studies suggest that progression to serious respiratory illness is more likely associated with HRV-A and HRV-C rather than with HRV-B. HRV-C is associated with an increased number of hospitalizations among children [15], especially in those with asthma [16] and cystic fibrosis [17].
Less is known about the epidemiology and clinical characteristics of HRVs in adults. Moreover, in Mexico and other Latin American countries, there is limited information regarding the clinical and molecular characteristics of HRVs as a cause of respiratory illness. Previously, our group described clinical characteristics and outcomes of ILI in an early cohort of 1065 subjects tested for different pathogens. We found that HRV was the most frequently isolated pathogen, infecting 15.3% of enrolled participants; 51% of the population infected with HRVs required hospitalization (62% of adults and 48% of children) [18]. The complete cohort enrolled a total of 5662 participants between 7 April 2010 and 4 April 2014, and HRV continued to be the most common cause of ILI.
In this study, we describe the clinical and molecular features of HRV infection in the Mexican population; we also explored the association between HRV type and more serious illness and worse clinical outcomes.
METHODS
Study Design and Settings
ILI002 was a hospital-based, multicenter prospective observational cohort study of ILI by the Mexican Emerging Infectious Disease Clinical Research Network (LaRed) between 7 April 2010 and 4 April 2014. The study was conducted at 6 hospitals, 5 of them in Mexico City and 1 in San Luis Potosí. Participating hospitals included 2 general hospitals (1 in Mexico City and 1 in San Luis Potosí), 2 tertiary care pediatric hospitals, and 2 tertiary care hospitals (1 is dedicated to the treatment of respiratory disorders; the other provides medical care in a wide range of medical specialties). The study was approved by the ethics committee of each institution (Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán, reference number 116; Instituto Nacional de Enfermedades Respiratorias Ref C12-10; Hospital General Manuel Gea González, reference number 36-50-2010; Instituto Nacional de Pediatría, reference number INP 014/2010; Hospital Infantil de México Federico Gómez, reference number HIM/2010/074; Hospital Regional Dr Ignacio Morones Prieto, reference number 88-12).
Study Population and Definitions
This study analyzed data from participants aged 1 month or older with ILI who sought medical attention and tested positive for HRV in nasopharyngeal swabs or nasal aspirates by real time polymerase chain reaction (rt-PCR). ILI was defined according to a modified case definition criteria from the World Health Organization [19] as having at least 1 respiratory symptom (ie, shortness of breath, cough) and either fever or 1 or more nonrespiratory symptom (ie, malaise, headache), with illness onset within the past 48 hours. By definition, those subjects seeking care for ILI and needing hospitalization are considered to have severe acute respiratory infection (SARI).
Study Procedures
All patients seeking care for ILI who met the inclusion and exclusion criteria at the emergency department or at the intensive care unit (ICU), depending on severity, were invited to participate in the study. After informed assent and consent forms were obtained, sociodemographic and symptom data were collected using previously standardized questionaries, and a complete physical examination was performed. A nasopharyngeal swab, or a nasal aspirate in children, was obtained at baseline for PCR detection of respiratory pathogens in all participants, and a blood sample for complete blood counts and chemistry analysis was collected. The final clinical decision regarding whether the subject would remain in the emergency department, require hospitalization, or receive treatment as an outpatient was made by the attending physician in charge. Follow-up information (symptoms, chronic medical conditions, previous treatment, impact on daily function, hospitalizations, and death) was obtained by phone at day 14 and 60 and at an in-person visit at day 28 through standardized case report forms.
Laboratory Diagnosis
Respiratory samples were tested with the RespiFinder 19 (April 2010 to May 2012) and RespiFinder 22 (previously RespiFinder Plus, June 2012 to March 2014) from PathoFinder BV, Maastricht, the Netherlands. RespiFinder 19 (multiplex rt-PCR test) detects and differentiate 15 viruses (coronaviruses NL63, OC43, and 229E; human metapneumovirus; influenza A, A/H5N1, and B; parainfluenza virus types 1–4; RSV A and B; picornavirus [enterovirus and HRV]; and adenovirus), as well as 4 bacteria (Bordetella pertussis, Chlamydophila pneumoniae, Legionella pneumophila, and Mycoplasma pneumoniae). The RespiFinder 22 removed influenza H5N1 and added bocavirus (type 1), coronavirus HKU1, influenza A/H1N1v, and picornavirus (enterovirus and HRV). Sequenced samples were selected by convenience, clinician judgment, and resource availability to determine HRV species.
RNA Extraction
Total nucleic acids were extracted from 500 µL of nasopharyngeal swabs using the NucliSENS easyMAG system (bioMérieux, Netherlands) and eluted in 110 µL, according to the manufacturer’s instructions.
Sequencing and Phylogenetic Analysis
To determine HRV type, a 549 bp fragment of the VP4/VP2 genomic region was amplified by RT-PCR and sequenced. RT-PCR was performed using 10 µL of nucleic acids with SuperScript III One-Step RT-PCR System with Platinum Taq DNA Polymerase (Invitrogen, ThermoFisher Scientific, Carlsbad, California) with primers VP42F (GGGACCAACTACTTTGGGTGTCCGTGT) and VP42R (GCATCIGGYARYTTCCACCACCANCC) [20]. The amplification program was as follows: RT reaction at 50°C for 60 minutes, initial denaturation at 95°C for 2 minutes, 40 cycles of 95°C for 30 seconds, 55°C for 1 minute, 68°C for 1 minute, and the final extension at 72°C for 5 minutes. RT-PCR products were analyzed in horizontal 2% agarose gel stained with SYBR Safe (Invitrogen, ThermoFisher Scientific, Kiryat Shmona, Israel). All RT-PCR products were purified using the MinElute PCR Purification Kit (Qiagen, Hilden, Germany); the concentration was determined by the Qubit fluorometer using the Qubit ds HS Assay kit (Invitrogen, ThermoFisher Scientific, Eugene, Oregon) and sequenced using the BigDye Terminator v3.1 Cycle Sequencing Kit and the ABI 3130xl genetic analyzer (Applied Biosystems, Foster City, California). The sequences obtained were aligned to reported sequences of HRV in GenBank.
Bioinformatic Analysis
To examine the associations between samples, sequences obtained were manually edited with Kodon software v2.04 (Applied Maths, Sint-Martens-Latem, Belgium). Alignment was performed using the ClustalW program, and phylogenetic relationships were constructed by the neighbor-joining method with bootstrap of 1000 replicates using the MEGA v6 software [21].
Viral Load
To measure viral load, a universal HRV quantitative RT-PCR assay was used. Primers qR447f (5′-GGCCCCTGAATGYGGCTAA-3′), qR561r (5′-GAAACACGGACACCCAAAGTAG-3′), and probe R529 (FAM 5′-AYGGRACCRACTACTTTG-3′ MGB) were used to amplify a 114 bp fragment of the highly conserved region of the 5′ untranslated region as described previously [22]. RNA was amplified in an ABI Prism 7500 SDS Real-Time cycler with the following cycling program: RT reaction at 48°C for 30 minutes, initial denaturation at 95°C for 2 minutes, 45 cycles of 95°C for 15 seconds, and 60°C for 32 seconds. Serial dilution (1 × 101 to 1 × 107 copies/µL) of a plasmid containing the amplified fragment was used to construct a standard curve. All samples were tested in triplicates and viral load was determined by plotting cycle threshold values in the standard curve as viral copies per microliter of extracted RNA.
Statistical Analysis
Categorical variables were summarized by number and frequency and continuous variables by mean and standard deviation. A multinomial regression model was performed, restricted to participants who were sequenced and HRV-A, HRV-B, or HRV-C (with the dependent variable being disease group), to evaluate the univariate relationship of each clinical feature and virus type. Laboratory values were standardized for ease of comparison. A Kruskal-Wallis test was used to compare log10-transformed viral load between groups. To compare between participants who were sequenced and HRV-A, HRV-B, and HRV-C and all others, t tests were used for continuous variables and χ2 tests were used for categorical variables. All analyses were done in R version 3.6.3 software; P < .05 was considered statistically significant. No corrections for multiple comparisons were performed.
RESULTS
A total of 5662 participants were in enrolled in the full cohort, of whom 1473 (26%) tested positive for HRV; the demographic and medical history data for these 1473 participants are described in Table 1. Nine hundred eighty-eight (67.1%) participants were adults (aged ≥18 years) and 485 (32.9%) were children; 69.4% of adults and 35.5% of children were outpatients. Of children and adults, 30.5% and 23.5%, respectively, were hospitalized (non-ICU); 15.3% of children and 6.1% of adults were in the emergency room (Supplementary Figure 1); and 18.8% of children and 1% of adults were in the ICU. One hundred thirty-seven (28.2%) children and 164 (16.6%) adults had a coinfection.
Table 1.
Demographical Characteristics and Medical History of Enrolled Participants Among Children and Adults
| Characteristic | Overall | Children | Adults |
|---|---|---|---|
| No. | 1473 | 485 | 988 |
| Status | |||
| Emergency room | 134 (9.1) | 74 (15.3) | 60 (6.1) |
| Hospitalized (non-ICU) | 380 (25.8) | 148 (30.5) | 232 (23.5) |
| ICU | 101 (6.9) | 91 (18.8) | 10 (1.0) |
| Outpatient | 858 (58.2) | 172 (35.5) | 686 (69.4) |
| Age, y | |||
| Mean (SD) | 26.66 (21.34) | 3.34 (4.1) | 38.11 (16.51) |
| Median (IQR) | 24.00 (5.00–41.00) | 2.00 (0.00–5.00) | 34.00 (24.00–49.00) |
| Categorical age, y | |||
| 0–5 | 383 (26.0) | 383 (79.0) | 0 (0.0) |
| 6–11 | 64 (4.3) | 64 (13.2) | 0 (0.0) |
| 12–17 | 38 (2.6) | 38 (7.8) | 0 (0.0) |
| 18–59 | 868 (58.9) | 0 (0.0) | 868 (87.9) |
| ≥60 | 120 (8.1) | 0 (0.0) | 120 (12.1) |
| Sex | |||
| Female | 895 (60.8) | 220 (45.4) | 675 (68.3) |
| Male | 578 (39.2) | 265 (54.6) | 313 (31.7) |
| Smoking history | |||
| Never smoked | 923 (62.7) | 361 (74.4) | 562 (56.9) |
| Passive smoker | 168 (11.4) | 120 (24.7) | 48 (4.9) |
| Current smoker | 211 (14.3) | 2 (0.4) | 209 (21.2) |
| Former smoker | 171 (11.6) | 2 (0.4) | 169 (17.1) |
| Coinfection | 301 (20.4) | 137 (28.2) | 164 (16.6) |
| Influenza | 55 (3.7) | 20 (4.1) | 35 (3.5) |
| Coronavirus | 79 (5.4) | 18 (3.7) | 61 (6.2) |
| Parainfluenza virus | 49 (3.3) | 27 (5.6) | 22 (2.2) |
| Respiratory syncytial virus | 61 (4.1) | 35 (7.2) | 26 (2.6) |
| Adenovirus | 44 (3.0) | 24 (4.9) | 20 (2.0) |
| Bordetella pertussis | 8 (0.5) | 7 (1.4) | 1 (0.1) |
| Metapneumovirus | 26 (1.8) | 10 (2.1) | 16 (1.6) |
| Mycoplasma pneumoniae | 7 (0.5) | 3 (0.6) | 4 (0.4) |
| Comorbidities | |||
| Asthma | 165 (11.2) | 45 (9.3) | 120 (12.1) |
| Cardiovascular disorders | 148 (10.0) | 19 (3.9) | 129 (13.1) |
| Congenital syndromes | 92 (6.2) | 89 (18.4) | 3 (0.3) |
| Received influenza vaccine | 131 (8.9) | 40 (8.2) | 91 (9.2) |
| Received pneumonia vaccine | 473 (32.1) | 332 (68.5) | 141 (14.3) |
| Current medications | |||
| Antibiotics | 669 (45.4) | 262 (54.0) | 407 (41.2) |
| Antihistamines | 364 (24.7) | 89 (18.4) | 275 (27.8) |
| Antipyretics | 386 (26.2) | 157 (32.4) | 229 (23.2) |
| Systemic steroids | 303 (20.6) | 86 (17.7) | 217 (22.0) |
| Death | 29 (2.0) | 6 (1.2) | 23 (2.3) |
Data are presented as No. (%) unless otherwise indicated.
Abbreviations: ICU, intensive care unit; IQR, interquartile range; SD, standard deviation.
In children, 79% (383) of cases occurred in those ≤5 years of age. In adults, 868 (87.9%) cases were in those 18–59 years old, and 120 (12.1%) were in those ≥60 years old. The most common comorbidities in children were congenital syndromes, occurring in 89 (18.4%) children. In adults, common comorbidities were asthma (12.1%) and cardiovascular disorders (13.1%). Other reported comorbidities included chronic lung disease, human immunodeficiency virus infection, and immunodeficiencies, among others (Supplementary Table 1).
Antibiotic use was common, occurring in 262 (54%) children and 407 (41.2%) adults. Other medication usage included antihistamines (18.4% of children and 27.8% of adults), antipyretics (32.4% of children and 23.2% of adults), and systemic steroids due to an autoimmune or oncologic disease (17.7% of children and 22% of adults). Other medications included cough medicine, decongestants, and inhaled steroids (Supplementary Table 1).
One hundred sixty-seven (15.1%) samples were sequenced, of which 101 (60.5%) were HRV-A, 22 (13.2%) were HRV-B, and 44 (26.3%) were HRV-C. Fifty-seven children (34.1%) had sequencing data available; 26 children (45.6%) had HRV-A and HRV-C, whereas 5 (8.8%) had HRV-B. One hundred ten adults (65.9%) had sequencing data available. Most adults (75 [68.2%]) with sequencing data available had HRV-A, whereas 17 (15.5%) and 18 (16.4%) adults had HRV-B and HRV-C, respectively.
Table 2 presents demographic and medical history data for the 167 cases of HRV that were sequenced and HRV-A, HRV-B, or HRV-C. Thirty-five of these subjects were admitted to the ICU, of whom 33 (94.3%) were children, and 35 subjects were hospitalized (non-ICU), of whom 24 (68.5%) were adults. Ten subjects were in the emergency room, of whom 6 (60%) were children; of the 87 subjects who were outpatients, 80 (92%) were adults.
Table 2.
Demographical Characteristics and Medical History of the 176 Cases for Rhinovirus That Were Sequenced According to Virus Type and Age Group
| Characteristic | Overall | Children | Adults | ||||
|---|---|---|---|---|---|---|---|
| A | B | C | A | B | C | ||
| No. of participants | 167 | 26 | 5 | 26 | 75 | 17 | 18 |
| Status | |||||||
| Emergency room | 10 (6.0) | 4 (15.4) | 1 (20.0) | 1 (3.8) | 2 (2.7) | 0 (0.0) | 2 (11.1) |
| Hospitalized (non-ICU) | 35 (21.0) | 3 (11.5) | 0 (0.0) | 8 (30.8) | 18 (24.0) | 1 (5.9) | 5 (27.8) |
| ICU | 35 (21.0) | 16 (61.5) | 4 (80.0) | 13 (50.0) | 2 (2.7) | 0 (0.0) | 0 (0.0) |
| Outpatient | 87 (52.1) | 3 (11.5) | 0 (0.0) | 4 (15.4) | 53 (70.7) | 16 (94.1) | 11 (61.1) |
| Age, y | |||||||
| Mean (SD) | 25.19 (20.13) | 3 (3.69) | 0.8 (1.3) | 3.31 (5.08) | 38.69 (15.07) | 34.18 (15.4) | 30.89 (10.36) |
| Median (IQR) | 23.00 (4.00–41.00) | 1.00 (0.00–4.75) | 0.00 (0.00–1.00) | 0.50 (0.00–5.50) | 38.00 (24.00–50.00) | 28.00 (23.00–47.00) | 27.00 (22.00–37.75) |
| Categorical age, y | |||||||
| 0–5 | 44 (26.3) | 20 (76.9) | 5 (100.0) | 19 (73.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| 6–11 | 8 (4.8) | 4 (15.4) | 0 (0.0) | 4 (15.4) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| 12–17 | 5 (3.0) | 2 (7.7) | 0 (0.0) | 3 (11.5) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| 18–59 | 102 (61.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 69 (92.0) | 15 (88.2) | 18 (100.0) |
| ≥60 | 8 (4.8) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 6 (8.0) | 2 (11.8) | 0 (0.0) |
| Sex | |||||||
| Female | 97 (58.1) | 5 (19.2) | 1 (20.0) | 9 (34.6) | 55 (73.3) | 14 (82.4) | 13 (72.2) |
| Male | 70 (41.9) | 21 (80.8) | 4 (80.0) | 17 (65.4) | 20 (26.7) | 3 (17.6) | 5 (27.8) |
| Smoking history | |||||||
| Never smoked | 116 (69.5) | 18 (69.2) | 4 (80.0) | 22 (84.6) | 56 (74.7) | 9 (52.9) | 7 (38.9) |
| Passive smoker | 20 (12.0) | 8 (30.8) | 1 (20.0) | 4 (15.4) | 3 (4.0) | 1 (5.9) | 3 (16.7) |
| Current smoker | 15 (9.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 7 (9.3) | 5 (29.4) | 3 (16.7) |
| Former smoker | 16 (9.6) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 9 (12.0) | 2 (11.8) | 5 (27.8) |
| Coinfection | 13 (7.8) | 6 (23.1) | 1 (20.0) | 5 (19.2) | 1 (1.3) | 0 (0.0) | 0 (0.0) |
| Influenza | 4 (2.4) | 1 (3.8) | 0 (0.0) | 2 (7.7) | 1 (1.3) | 0 (0.0) | 0 (0.0) |
| Coronavirus | 1 (0.6) | 1 (3.8) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Parainfluenza virus | 4 (2.4) | 2 (7.7) | 1 (20.0) | 1 (3.8) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Respiratory syncytial virus | 1 (0.6) | 0 (0.0) | 0 (0.0) | 1 (3.8) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Adenovirus | 2 (1.2) | 2 (7.7) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Bordetella pertussis | 1 (0.6) | 0 (0.0) | 0 (0.0) | 1 (3.8) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Metapneumovirus | 1 (0.6) | 0 (0.0) | 0 (0.0) | 1 (3.8) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Mycoplasma pneumoniae | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Comorbidities | |||||||
| Asthma | 20 (12.0) | 1 (3.8) | 0 (0.0) | 2 (7.7) | 12 (16.0) | 0 (0.0) | 5 (27.8) |
| Cardiovascular disorders | 22 (13.2) | 1 (3.8) | 1 (20.0) | 3 (11.5) | 12 (16.0) | 3 (17.6) | 2 (11.1) |
| Congenital syndromes | 9 (5.4) | 5 (19.2) | 2 (40.0) | 2 (7.7) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Received flu vaccine | 13 (7.8) | 1 (3.8) | 0 (0.0) | 3 (11.5) | 6 (8.0) | 0 (0.0) | 3 (16.7) |
| Received pneumonia vaccine | 50 (29.9) | 18 (69.2) | 4 (80.0) | 12 (46.2) | 10 (13.3) | 1 (5.9) | 5 (27.8) |
| Current medications | |||||||
| Antibiotics | 78 (46.7) | 18 (69.2) | 4 (80.0) | 18 (69.2) | 24 (32.0) | 2 (11.8) | 12 (66.7) |
| Antihistamines | 35 (21.0) | 3 (11.5) | 0 (0.0) | 3 (11.5) | 17 (22.7) | 5 (29.4) | 7 (38.9) |
| Antipyretics | 40 (24.0) | 3 (11.5) | 4 (80.0) | 9 (34.6) | 17 (22.7) | 2 (11.8) | 5 (27.8) |
| Systemic steroids | 44 (26.3) | 10 (38.5) | 1 (20.0) | 7 (26.9) | 18 (24.0) | 2 (11.8) | 6 (33.3) |
| Death | 4 (2.4) | 2 (7.7) | 0 (0.0) | 0 (0.0) | 2 (2.7) | 0 (0.0) | 0 (0.0) |
Data are presented as No. (%) unless otherwise indicated.
Abbreviations: ICU, intensive care unit; IQR, interquartile range; SD, standard deviation.
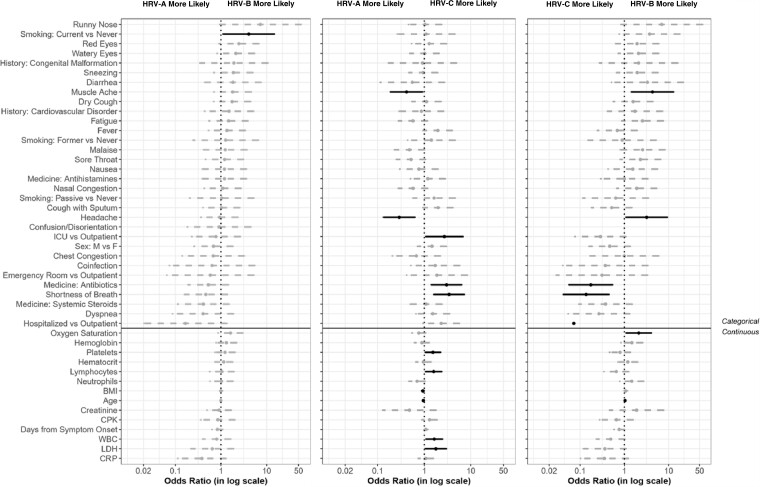
Of the 26 children with HRV-A, 16 (61.5%) were admitted to the ICU, 4 (15.4%) were in the emergency room, and 3 (11.5%) each were hospitalized (non-ICU) and outpatient. Of the 5 children with HRV-B, 4 (80%) were in the ICU and 1 (20%) was in the emergency room. Fifty percent of the 26 children with HRV-C were in the ICU; only 1 (3.8%) was in the emergency room, 8 (30.8%) were hospitalized (non-ICU), and 4 (15.4%) were outpatient. Of the 75 adults with HRV-A, most were outpatient (70.7%); 2 (2.7%) each were in the emergency room and in the ICU, and 18 (24%) were hospitalized (non-ICU). Similarly, 16 of the 17 (94.1%) adults with HRV-B were outpatient and 1 (5.9%) was hospitalized. Finally, of the 18 adults with HRV-C, 11 (61.1%) were outpatient, 5 (27.8%) were hospitalized, and 2 (11.1%) were in the emergency room. Hospitalization status overall was significantly different between the 167 sequenced participants with HRV-A, HRV-B, and HRV-C and the remaining 1306 participants (P < .001) (Supplementary Table 3). Of the 167 sequenced participants with HRV-A, HRV-B, and HRV-C, only 1 adult (HRV-A) had a coinfection, whereas 12 children had a coinfection (6 HRV-A, 1 HRV-B, and 5 HRV-C). Significantly fewer participants who were sequenced and HRV-A, HRV-B, and HRV-C had a coinfection compared to others (7.8% and 22.1%, respectively; P < .001). There was a significant difference in the distribution of enrollment year between the sequenced participants with HRV-A, HRV-B, and HRV-C and others; specifically, there were fewer sequenced subjects enrolled in 2014. There were no other differences between the group of participants that was sequenced and HRV-A, HRV-B, and HRV-C and all others in the cohort (Supplementary Table 3). Of the sequenced participants, 30.6% had comorbidities; 12% had asthma, 22% had cardiovascular diseases, and 5.4% had congenital syndromes (Supplementary Table 2). There were no statistical differences between HRV types for any of the comorbidities (data not shown). Figure 1 presents the odds ratios and 95% confidence intervals for the associations of each clinical feature and the HRV types from the univariate logistic regression model. For laboratory variables, the odds ratio reflects the effect of an increase of 1 standard deviation. Supplementary Tables 4 and 5 show the full data of the laboratory values of the participants with HRV infection by age, as well as the laboratory values for the participants who were sequenced according to the viral type.
Figure 1.
Odds ratios and associated 95% confidence intervals for the rhinovirus types for each characteristic in a univariate model. Characteristics below the solid black line are laboratory parameters, body mass index, and age. Abbreviations: BMI, body mass index; CPK, creatine phosphokinase; CRP, C-reactive protein; F, female; HRV, human rhinovirus; ICU, intensive care unit; LDH, lactate dehydrogenase; M, male; WBC, white blood cell.
The Figure 1 shows that the odds of HRV-A are higher than HRV-B in the presence of muscle ache, headache, and increases in body mass index and age. The odds of HRV-B are higher than HRV-A in participants who are current smokers (compared to those who have never smoked), and higher than HRV-C when displaying muscle ache, headache, and higher oxygen saturation. The odds of HRV-C are higher than both HRV-A and HRV-B when antibiotics are used and when shortness of breath is present. The odds of HRV-C are higher than HRV-A for participants in the ICU (compared to outpatient) and in the presence of increases in platelets, lymphocytes, white blood cells (WBCs), and lactate dehydrogenase (LDH). The odds of HRV-C are higher than HRV-B for hospitalized (non-ICU) participants.
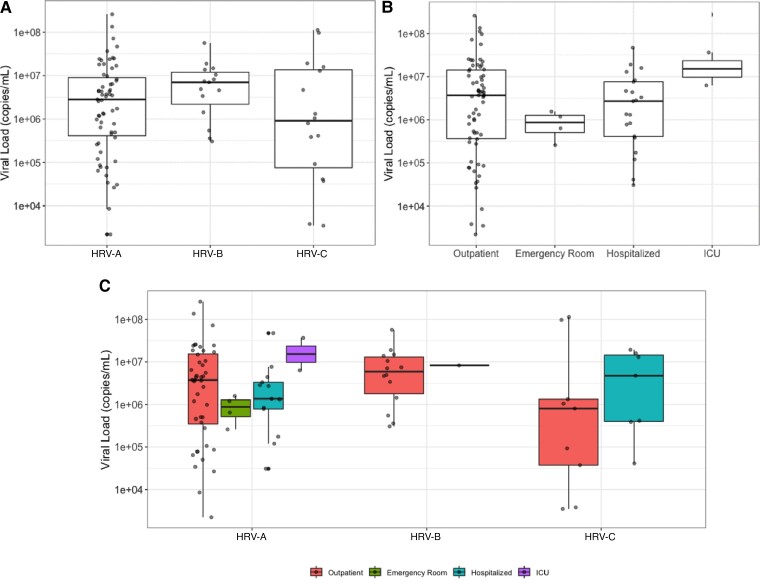
Of the 167 subjects with positive HRV tests who were sequenced, 94 (56.3%) had viral load information available. There were no significant differences in viral load between the HRV types or between hospitalization statuses (Figure 2).
Figure 2.
Viral load (copies/mL) by human rhinovirus (HRV) type and hospitalization status at baseline. No statistically significant differences associated with HRV type (P = .243) (A) or hospitalization status (P = .318) (B). P values are from a Kruskal-Wallis test for comparison between groups. Abbreviations: HRV, human rhinovirus; ICU, intensive care unit.
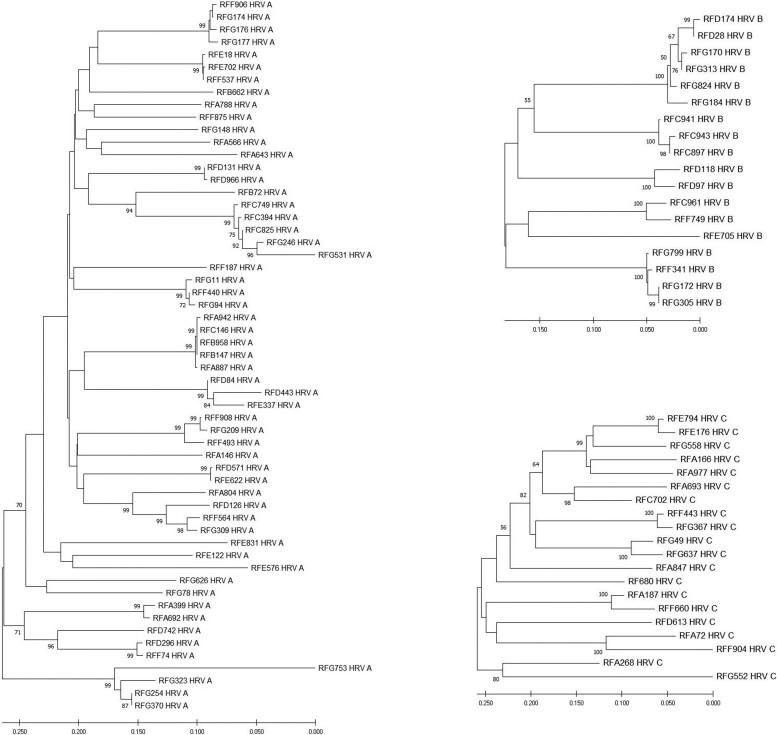
Figure 3 presents the phylogenetic analysis of the sequenced samples. Phylogenetic analysis shows a high diversity of sequences in circulation among Mexican HRV species, and we could not find any relationship between the clades and seasonality or outbreaks.
Figure 3.
Phylogenetic analysis of human rhinovirus (HRV). The 558-bp fragment VP4/VP2 genomic region was analyzed by MEGA version 6 software. Associations between samples were done by the neighbor-joining method with bootstrap analysis of 1000 replicates for each HRV A, B, and C type.
DISCUSSION
Our study analyzed data from ILI002, where participants with ILI/SARI were prospectively included for 4 years. We identified enterovirus/HRV infection in 26% of the ILI002 participants (1473/5662) between 2010 and 2014, the most common pathogen isolated overall. Of these, 988 (67.1%) were adults and 485 (32.9%) were children. Sequencing and HRV species determination was performed in 167 samples.
The prevalence of enterovirus/HRV found in our study is similar to what has been previously observed. García et al studied a large sample of children and young adults in several Central and South American countries. They reported that 16% of ILI cases were positive for HRV [23].
Clinically, HRV is characterized by symptoms compatible with ILI, and it can cause more severe disease, including pneumonia and bronchiolitis [24]. In Mexico, there is only 1 study of HRV in 306 children, characterizing HRV as an important cause of lower and upper respiratory tract infection [1]. In our study, the most affected group of children was those ≤5 years old (79%). Of note, 64.5% of the children who tested positive for HRV were either in the emergency room, hospitalized (non-ICU), or required ICU support. Only 28.2% had coinfection with another virus, supporting the hypothesis of the pathogenic role of HRV.
Worldwide, little has been described regarding HRV infection and severe disease in adults. Recently, Chiu et al found that 21.5% of adult patients with respiratory tract infections who visited Taiwan hospitals were positive for HRV [25]. In our study, most adults with HRV were outpatients, although 23.5% required hospitalization and 1 required the ICU. Golke et al found that chronic pulmonary conditions such as asthma and chronic obstructive pulmonary disease were comorbidities for HRV infection in both inpatient and outpatient adults [26]. In more critical patients, Fica et al found that chronic pulmonary conditions and heart disease were the most common comorbidities for HRV-positive subjects with SARI, along with diabetes mellitus and some neurologic diseases [27]. In our study, the most common comorbidities among adult HRV-positive cases were asthma (12.1%) and cardiovascular disorders (13.1%).
It has been reported that HRV-A and HRV-C may be associated with more severe disease compared with HRV-B in children [4]. More recently, it has been described that HRV-C was more frequently isolated in children aged <2 years [28]. In our cohort, from the genotyped samples, the most common species among children <5 years old were HRV-A and HRV-C.
Some studies have yielded divergent results regarding HRV severity [14, 26, 29]. This may be influenced by the nature of the study (prospective vs retrospective) or population type (children vs adults). For example, McCulloch et al did not find an association between hospitalized adults and genotype [29]. We assessed whether there was a univariate association between symptoms, disease severity, and HRV species among the whole cohort (children and adults). We found that the odds of laboratory abnormalities (increases in platelets, lymphocytes, WBC, and LDH) increased with HRV-C infection when compared with HRV-A. We also found that HRV-C was associated with increased odds of being in the ICU. Interestingly, the odds of having shortness of breath were higher in subjects with HRV-C, compared to subjects with either HRV-A or HRV-B. The odds of muscle ache and headache were higher in subjects infected with HRV-A and HRV-B compared to HRV-C. These results support the hypothesis that HRV-C is associated with a more severe disease. Of note, there is a higher rate of ICU admissions in the sequenced group (21%) compared to all other participants (5.1%). This suggests that the sequenced subgroup represents a sicker group of patients (Supplementary Table 3).
Overall, we found a high frequency of antibiotic use in the treatment of HRV infection (41.2% of infected adults and 54% of children). Antibiotic use was more common in patients with HRV-C compared to those with HRV-B. The overuse of antibiotics is a major health problem worldwide and exposes patients to potential harm for no benefit. The use of antibiotics in viral infections have been also reported by other authors: Li et al reported a 26.8% of antibiotic use in hospitalized patients with SARI [30], and Fica et al reported 90% antibiotic use in hospitalized adult patients with HRV infection [27].
We did not find any associations between viral load and hospitalization status nor viral load and HRV species, suggesting that there could be a pathogenic mechanism intrinsic to HRV-C, or host-response differences, that cause more severe disease and are not related to viral load. This hypothesis is supported by evidence that HRVs modulate and induce different immune responses [31–33], and it has been demonstrated that HRV-C can effectively evade and suppress the innate immune response [34].
This study is the first to describe the clinical and molecular features of HRV infection in the Mexican population, including both children and adults. The data were collected in a prospective manner, using previously standardized tools. Limitations of this study include the observational nature of the study and that most bacterial infections were not evaluated. In addition, RespiFinder multiplex PCR only detects enterovirus/HRV infection; however, after sequencing the available samples, HRV was confirmed as the causative agent.
Although our study took place from 2010 to 2014, the topic is still of interest because the role of HRVs as a cause of lower respiratory tract infections has been underestimated [35]. Moreover, recent data demonstrate the importance of studying and performing HRV epidemiological surveillance [35, 36]. It has been proposed that HRV delayed the development of the 2009 influenza A/H1N1 pandemic through viral interference, and interestingly during the coronavirus disease 2019 pandemic, the prevalence of most respiratory pathogens was unusually low. However, HRV remained the main virus co-circulating with SARS-CoV-2 [36].
This hospital-based study included patients seeking medical care by ILI/SARI, and this potentially would select for patients with more comorbidities and potentially more serious illness than the entire population. However, these results still show how HRV may cause severe disease in children and adults and that, in particular, HRV-C infection may cause worse outcomes.
In conclusion, this study highlights the frequency and importance of HRV as a cause of ILI/SARI and reflects the need to improve surveillance and testing for HRV. Testing may also favor appropriate use of antibiotics. Species identification will help to improve the understanding of the wide range of disease severity caused by different HRV species and subtypes and risk factors for outcomes in various types of patients.
Supplementary Material
Contributor Information
Arturo Galindo-Fraga, Departamento de Epidemiología, Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán, Mexico City, Mexico.
Paola del Carmen Guerra-de-Blas, Mexican Emerging Infectious Diseases Clinical Research Network, Mexico City, Mexico.
Ana M Ortega-Villa, National Institute of Allergy and Infectious Diseases, Bethesda, Maryland, USA.
Allyson Mateja, Clinical Monitoring Research Program Directorate, Frederick National Laboratory for Cancer Research, Frederick, Maryland, USA.
Jesus Arturo Ruiz Quiñones, Departamento de Epidemiología, Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán, Mexico City, Mexico.
Pilar Ramos Cervantes, Departamento de Infectología, Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán, Mexico City, Mexico.
Fernando Ledesma Barrientos, Departamento de Infectología, Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán, Mexico City, Mexico.
Ana A Ortiz-Hernández, División de Desarrollo y Enlace Interinstitucional, Instituto Nacional de Pediatría, Mexico City, Mexico.
Beatriz Llamosas-Gallardo, División de Desarrollo y Enlace Interinstitucional, Instituto Nacional de Pediatría, Mexico City, Mexico.
Alejandra Ramírez-Venegas, Departamento de Investigación en Tabaquismo y en Enfermedades Pulmonares Obstructivas Crónicas, Instituto Nacional de Enfermedades Respiratorias “Ismael Cosío Villegas”, Mexico City, Mexico.
Rafael Valdéz Vázquez, Departamento de Infectología, Hospital General “Dr Manuel Gea González,” Mexico City, Mexico.
Daniel Noyola Chepitel, Microbiology Department, Facultad de Medicina, Universidad Autónoma de San Luis Potosí, San Luis Potosí, Mexico.
Sarbelio Moreno-Espinosa, Departamento de Infectología, Hospital Infantil de México Federico Gómez, Instituto Nacional de Salud, Mexico City, Mexico.
John H Powers, Clinical Research Directorate, Frederick National Laboratory for Cancer Research, Frederick, Maryland, USA.
M Lourdes Guerrero, Departamento de Infectología, Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán, Mexico City, Mexico.
Guillermo M Ruiz-Palacios, Departamento de Infectología, Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán, Mexico City, Mexico.
John H Beigel, National Institute of Allergy and Infectious Diseases, Bethesda, Maryland, USA.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. Investigators and Coordinators: Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán: Guillermo M. Ruiz-Palacios, M. Lourdes Guerrero, Arturo Galindo-Fraga, Bricia Roa, Itzel Cruz-Gaona, Diana Aguilar-Cruz; Instituto Nacional de Enfermedades Respiratorias: Alejandra Ramírez-Venegas, Paulina M. Paulín-Prado, Nora Bautista; Hospital General Dr Gea González: Rafael Valdéz, Irma Jiménez, Lorena Hernández, Patricia Rodríguez, Javier Reyes-Mar, José Alfonso Maya, Ana Laura Corona; Instituto Nacional de Pediatría: Beatriz Llamosas-Gallardo, Juliana Estevez-Jiménez, Ana A. Ortiz-Hernández, Diana Andrade-Platas; Hospital Infantil de México Federico Gómez: Sarbelio Moreno-Espinosa, Ana Estela Gamiño Arroyo, Mónica González Matus, Luis Mendoza Garcés. Central Laboratory at the Department of Infectious Diseases, Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán: Santiago Pérez-Patrigeon, Pilar Ramos-Cervantes, Violeta Ibarra-González, Julia Martínez-López, Luis A. García-Andrade, Fernando Ledesma-Barrientos; Departamento de Microbiología, Facultad de Medicina, Universidad Autónoma de San Luis Potosí. San Luis Potosí: Daniel Ernesto Noyola Cherpitel, Daniel Hernandez; Coordinación de los Institutos Nacionales de Salud y Hospitales de Alta Especialidad, Secretaría de Salud, México: Dr Guillermo Ruíz Palacios y Santos; National Institute of Allergy and Infectious Diseases (NIAID): Mary Smolskis, Christian Yoder, Dean Follmann; SAIC Frederick in support of NIAID: John Beigel, Wenjuan Gu; Clifford Lane. Westat: Laura Freimanis-Hance, Isabel Trejos, Amanda Fournier, Bernadette Tetra; Leidos Biomedical Research (LBR): Gema Souto Adeva; LaRed Director: Justino Regalado Pineda; LaRed Coordinating Center: Abelardo Montenegro Liendo, Juan Francisco Galán, Hugo Arroyo-Figueroa, Nadine Mascareñas, Eli Becerril, Peter Quidgley, Ana Vilardell Dávila. We are indebted to the study participants for their contribution to the study.
Ethical approval. The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the institutional review board (or ethics committee) of each participating institution.
Patient consent. Written informed consent and assent were obtained from all participants involved in the study.
Data availability. The data presented in this study are available on request from the corresponding author. The data are not publicly available because a data-sharing agreement is needed to provided for (1) a commitment to using the data only for research purposes and not to identify any individual participant; (2) a commitment to securing the data using appropriate computer technology; and (3) a commitment to destroying or returning the data after analyses are completed.
Disclaimer. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does the mention of trade names, commercial products, or organizations imply endorsement by the US government.
Financial support. This work was supported by the Mexican Emerging Infectious Diseases Clinical Research Network (LaRed). LaRed is funded by the Mexico Ministry of Health and by the National Institute of Allergy and Infectious Diseases (NIAID) in United States. This project has been funded in part by Consejo Nacional de Ciencia y Tecnología (CONACYT) (Fondo Sectorial Secretaría de Salud (SSA)/Instituto Mexicano del Seguro Social (IMSS)/Instituto de Seguridad y Servicios Sociales de los Trabajadores del Estado (ISSSTE), projects numbers 71260 and 127088) and the NIAID, National Institutes of Health (NIH) through a contract with Westat, Inc (contract number HHSN2722009000031, task order number HHSN27200002). This project has been funded in whole or in part with federal funds from the National Cancer Institute, NIH (contract number 75N91019D00024).
Potential conflicts of interest. All authors: The authors declare that they do not have conflicts of interest to disclose.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Aponte FE, Taboada B, Espinoza MA, et al. Rhinovirus is an important pathogen in upper and lower respiratory tract infections in Mexican children. Virol J 2015; 12:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Souty C, Masse S, Valette M, et al. Baseline characteristics and clinical symptoms related to respiratory viruses identified among patients presenting with influenza-like illness in primary care. Clin Microbiol Infect 2019; 25:1147–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Drysdale SB, Mejias A, Ramilo O. Rhinovirus—not just the common cold. J Infect 2017; 74(Suppl 1):S41–6. [DOI] [PubMed] [Google Scholar]
- 4. Lee WM, LemanskeRF, Jr, Evans MD, et al. Human rhinovirus species and season of infection determine illness severity. Am J Respir Crit Care Med 2012; 186:886–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nguyen DNT, Mai LQ, Bryant JE, et al. Epidemiology and etiology of influenza-like-illness in households in Vietnam: it’s not all about the kids!. J Clin Virol 2016; 82:126–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fendrick AM, Monto AS, Nightengale B, Sarnes M. The economic burden of non-influenza-related viral respiratory tract infection in the United States. Arch Intern Med 2003; 163:487–94. [DOI] [PubMed] [Google Scholar]
- 7. Hellgren J, Cervin A, Nordling S, Bergman A, Cardell LO. Allergic rhinitis and the common cold—high cost to society. Allergy 2010; 65:776–83. [DOI] [PubMed] [Google Scholar]
- 8. Hershenson MB. Rhinovirus-induced exacerbations of asthma and COPD. Scientifica (Cairo) 2013; 2013:405876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. To KKW, Yip CCY, Yuen KY. Rhinovirus—from bench to bedside. J Formos Med Assoc 2017; 116:496–504. [DOI] [PubMed] [Google Scholar]
- 10. Ljubin-Sternak S, Meštrović T, Ivković-Jureković I, et al. The emerging role of rhinoviruses in lower respiratory tract infections in children—clinical and molecular epidemiological study from Croatia, 2017–2019. Front Microbiol 2019; 10:2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hung IF, Zhang AJ, To KK, et al. Unexpectedly higher morbidity and mortality of hospitalized elderly patients associated with rhinovirus compared with influenza virus respiratory tract infection. Int J Mol Sci 2017; 18:259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vandini S, Biagi C, Fischer M, Lanari M. Impact of rhinovirus infections in children. Viruses 2019; 11:521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chu HY, Englund JA, Strelitz B, et al. Rhinovirus disease in children seeking care in a tertiary pediatric emergency department. J Pediatr Infect Dis Soc 2016; 5:29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen W-J, Arnold JC, Fairchok MP, et al. Epidemiologic, clinical, and virologic characteristics of human rhinovirus infection among otherwise healthy children and adults: rhinovirus among adults and children. J Clin Virol 2015; 64:74–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cox DW, Khoo SK, Zhang G, et al. Rhinovirus is the most common virus and rhinovirus-C is the most common species in paediatric intensive care respiratory admissions. Eur Respir J 2018; 52:1800207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Calvo C, Casas I, García-García ML, et al. Role of rhinovirus C respiratory infections in sick and healthy children in Spain. Pediatr Infect Dis J 2010; 29:717–20. [DOI] [PubMed] [Google Scholar]
- 17. de Almeida MB, Zerbinati RM, Tateno AF, et al. Rhinovirus C and respiratory exacerbations in children with cystic fibrosis. Emerg Infect Dis 2010; 16:996–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Galindo-Fraga A, Ortiz-Hernández AA, Ramírez-Venegas A, et al. Clinical characteristics and outcomes of influenza and other influenza-like illnesses in Mexico City. Int J Infect Dis 2013; 17:e510–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. World Health Organization . WHO surveillance case definitions for ILI and SARI. https://www.who.int/teams/global-influenza-programme/surveillance-and-monitoring/case-definitions-for-ili-and-sari#:~:text=ILI%20case%20definition,within%20the%20last%2010%20days. Accessed 16 April 2021.
- 20. Jin Y, Yuan XH, Xie ZP, et al. Prevalence and clinical characterization of a newly identified human rhinovirus C species in children with acute respiratory tract infections. J Clin Microbiol 2009; 47:2895–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 2013; 30:2725–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ng KT, Chook JB, Oong XY, et al. Performance of a TaqMan assay for improved detection and quantification of human rhinovirus viral load. Sci Rep 2016; 6:34855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. García J, Espejo V, Nelson M, et al. Human rhinoviruses and enteroviruses in influenza-like illness in Latin America. Virol J 2013; 10:305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bizot E, Bousquet A, Charpié M, et al. Rhinovirus: a narrative review on its genetic characteristics, pediatric clinical presentations, and pathogenesis. Front Pediatr 2021; 9:643219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chiu YT, Tien N, Lin HC, et al. Detection of respiratory pathogens by application of multiplex PCR panel during early period of COVID-19 pandemic in a tertiary hospital in Central Taiwan [manuscript published online ahead of print 6 October 2021]. J Microbiol Immunol Infect 2021. doi: 10.1016/j.jmii.2021.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Golke P, Hönemann M, Bergs S, Liebert UG. Human rhinoviruses in adult patients in a tertiary care hospital in Germany: molecular epidemiology and clinical significance. Viruses 2021; 13:2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fica A, Dabanch J, Andrade W, et al. Clinical relevance of rhinovirus infections among adult hospitalized patients. Braz J Infect Dis 2015; 19:118–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Adam DC, Chen X, Scotch M, MacIntyre CR, Dwyer D, Kok J. The molecular epidemiology and clinical phylogenetics of rhinoviruses among paediatric cases in Sydney, Australia. Int J Infect Dis 2021; 110:69–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McCulloch DJ, Sears MH, Jacob JT, et al. Severity of rhinovirus infection in hospitalized adults is unrelated to genotype. Am J Clin Pathol 2014; 142:165–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li W, Yu B, Zhou J, et al. Genetic diversity and epidemiology of human rhinovirus among children with severe acute respiratory tract infection in Guangzhou, China. Virol J 2021; 18:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kim JH, Jang JY, Jang YJ. Human rhinovirus serotypes induces different immune responses. Virol J 2021; 18:232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kirchberger S, Majdic O, Stöckl J. Modulation of the immune system by human rhinoviruses. Int Arch Allergy Immunol 2007; 142:1–10. [DOI] [PubMed] [Google Scholar]
- 33. Megremis S, Niespodziana K, Cabauatan C, et al. Rhinovirus species-specific antibodies differentially reflect clinical outcomes in health and asthma. Am J Respir Crit Care Med 2018; 198:1490–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pang LL, Yuan XH, Shao CS, et al. The suppression of innate immune response by human rhinovirus C. Biochem Biophys Res Commun 2017; 490:22–8. [DOI] [PubMed] [Google Scholar]
- 35. Esneau C, Duff AC, Bartlett NW. Understanding rhinovirus circulation and impact on illness. Viruses 2022; 14:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kiseleva I, Ksenafontov A. COVID-19 shuts doors to flu but keeps them open to rhinoviruses. Biology (Basel) 2021; 10:733. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.