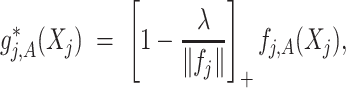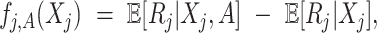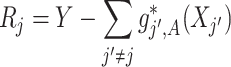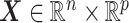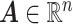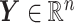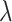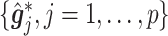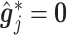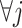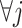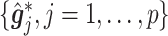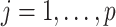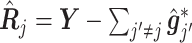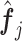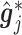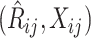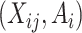Summary
Sparse additive modeling is a class of effective methods for performing high-dimensional nonparametric regression. This article develops a sparse additive model focused on estimation of treatment effect modification with simultaneous treatment effect-modifier selection. We propose a version of the sparse additive model uniquely constrained to estimate the interaction effects between treatment and pretreatment covariates, while leaving the main effects of the pretreatment covariates unspecified. The proposed regression model can effectively identify treatment effect-modifiers that exhibit possibly nonlinear interactions with the treatment variable that are relevant for making optimal treatment decisions. A set of simulation experiments and an application to a dataset from a randomized clinical trial are presented to demonstrate the method.
Keywords: Individualized treatment rules, Sparse additive models, Treatment effect-modifiers, Biomarkers
1. Introduction
Identification of patient characteristics influencing treatment responses, which are often termed treatment effect-modifiers or treatment effect-moderators, is a top research priority in precision medicine. In this article, we develop a flexible yet simple and intuitive regression approach to identifying treatment effect-modifiers from a potentially large number of pretreatment patient characteristics. In particular, we utilize a sparse additive model (Ravikumar and others, 2009) to conduct effective treatment effect-modifier selection.
The clinical motivation behind the development of a high-dimensional additive regression model that can handle a large number of pretreatment covariates, specifically designed to model treatment effect modification and variable selection, is a randomized clinical trial (Trivedi and others, 2016) for treatment of major depressive disorder. A large number of baseline patient characterisitics were collected from each participant prior to randomization and treatment allocation. The primary goal of this study, and thus the goal of our proposed method, is to discover biosignatures of heterogeneous treatment response, and to develop an individualized treatment rule (ITR; e.g., Qian and Murphy 2011; Zhao and others 2012; Laber and Zhao 2015; Kosorok and Laber 2019; Park and others 2020) for future patients based on biosignatures of the heterogeneous treatment response, a key aspect of precision medicine (e.g., Ashley 2015; Fernandes and others 2017, among others). In particular, discovering and identifying which pretreatment patient characteristics influence treatment effects have the potential to significantly enhance clinical reasoning in practice (see, e.g., Royston and Sauerbrei, 2008).
The major challenge in efficiently modeling treatment effect modification from a clinical trial study is that variability due to treatment effect modification (i.e., the treatment-by-covariates interaction effects on outcomes), that is essential for developing ITRs (Qian and Murphy, 2011), is typically dwarfed by a relatively large degree of non-treatment-related variability (i.e., the main effects of the pretreatment covariates on treatment outcomes). In particular, in regression, due to potential confounding between the main effect of the covariates and the treatment-by-covariates interaction effect, misspecification of the covariate main effect may significantly influence estimation of the treatment-by-covariates interaction effect.
A simple and elegant linear model-based approach to modeling the treatment-by-covariates interactions, termed the modified covariate (MC) method, that is robust against model misspecification of the covariates’ main effects was developed by Tian and others (2014). The method utilizes a simple parameterization and bypasses the need to model the main effects of the covariates. See also Murphy (2003); Lu and others (2011), Shi and others (2016), and Jeng and others (2018) for the similar linear model-based approaches to estimating the treatment-by-covariates interactions. However, these approaches assume a stringent linear model to specify the treatment-by-covariates interaction effects and are limited to the binary-valued treatment variable case only. In this work, we extend the optimization framework of Tian and others (2014) for modeling the treatment-by-covariates interactions using a more flexible regression setting based on additive models (Hastie and Tibshirani, 1999) that can accommodate more than two treatment conditions, while allowing an unspecified main effect of the covariates. Additionally, via an appropriate regularization, the proposed approach simultaneously achieves treatment effect-modifier selection in estimation.
In Section 2, we introduce an additive model that has both the unspecified main effect of the covariates and the treatment-by-covariates interaction effect additive components. Then, we develop an optimization framework specifically targeting the interaction effect additive components of the model, with a sparsity-inducing regularization parameter to encourage sparsity in the set of component functions. In Section 3, we develop a coordinate decent algorithm to estimate the interaction effect part of the model and consider an optimization strategy for ITRs. In Section 4, we illustrate the performance of the method in terms of treatment effect-modifier selection and estimation of ITRs through simulation examples. Section 5 provides an illustrative application from a depression clinical trial, and the article concludes with discussion in Section 6.
2. Models
Let 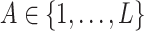 denote a treatment variable assigned with associated probabilities
denote a treatment variable assigned with associated probabilities 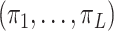 ,
, 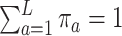 , and
, and 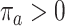 , and let
, and let 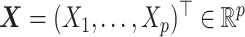 denote pretreatment covariates, independent of
denote pretreatment covariates, independent of  (as in the case of a randomized trial). If the treatment assignment depends on the covariates, then the probabilities
(as in the case of a randomized trial). If the treatment assignment depends on the covariates, then the probabilities  can be replaced by propensity scores
can be replaced by propensity scores 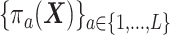 which would typically need to be estimated from data. We let
which would typically need to be estimated from data. We let 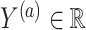 (a = 1,…,L) be the potential outcome if the patient received treatment
(a = 1,…,L) be the potential outcome if the patient received treatment 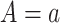
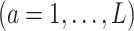 ; we only observe
; we only observe 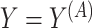 ,
, 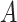 and
and 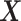 . Throughout the article, without loss of generality, we assume that a larger value of the treatment outcome
. Throughout the article, without loss of generality, we assume that a larger value of the treatment outcome 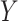 is desirable, and that
is desirable, and that 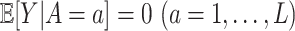 , i.e., the main effect of
, i.e., the main effect of 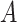 on the outcome is centered at
on the outcome is centered at 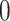 ; this is only to suppress the treatment
; this is only to suppress the treatment 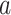 -specific intercepts in regression models in order to simplify the exposition, and can be achieved by removing the treatment level
-specific intercepts in regression models in order to simplify the exposition, and can be achieved by removing the treatment level 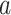 -specific means from
-specific means from 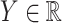 . We model the treatment outcome
. We model the treatment outcome 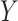 by the following additive model:
by the following additive model:
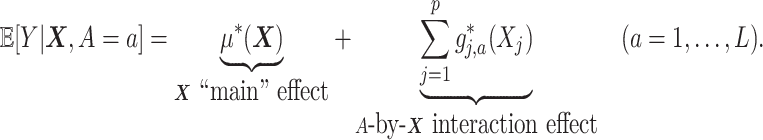 |
(2.1) |
In model (2.1), the first term 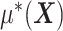 , which in fact will not need to be specified in our exposition, does not depend on the treatment variable
, which in fact will not need to be specified in our exposition, does not depend on the treatment variable 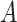 and thus the
and thus the 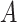 -by-
-by-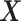 interaction effects are determined only by the second component
interaction effects are determined only by the second component 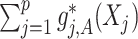 . In terms of modeling treatment effect modification, the term
. In terms of modeling treatment effect modification, the term 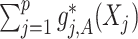 in (2.1) corresponds to the “signal” component, whereas the term
in (2.1) corresponds to the “signal” component, whereas the term 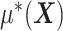 corresponds to a “nuisance” component. In particular, the optimal ITR, under model (2.1), can be shown to satisfy
corresponds to a “nuisance” component. In particular, the optimal ITR, under model (2.1), can be shown to satisfy 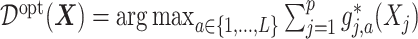 , which does not involve the term
, which does not involve the term 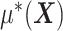 . Therefore, the
. Therefore, the 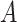 -by-
-by-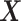 interaction effect term
interaction effect term 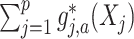 shall be the primary estimation target of model (2.1).
shall be the primary estimation target of model (2.1).
In (2.1), for each individual covariate 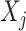 , we utilize a treatment
, we utilize a treatment 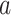 -specific smooth
-specific smooth 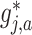 separately for each treatment condition
separately for each treatment condition 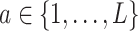 . However, it is useful to treat the set of treatment-specific smooths for
. However, it is useful to treat the set of treatment-specific smooths for 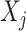 as a single unit, i.e., a single component function
as a single unit, i.e., a single component function 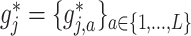 , for the purpose of treatment effect-modifier variable selection.
, for the purpose of treatment effect-modifier variable selection.
In model (2.1), to separate 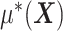 from the component
from the component 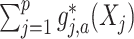 and obtain an identifiable representation, without loss of generality, we assume that the set of treatment
and obtain an identifiable representation, without loss of generality, we assume that the set of treatment 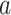 -specific smooths
-specific smooths 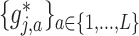 of the
of the 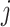 th component function
th component function 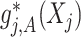 satisfies a condition (almost surely):
satisfies a condition (almost surely):
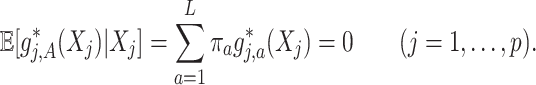 |
(2.2) |
Condition (2.2) implies 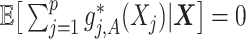 and separates the
and separates the 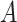 -by-
-by-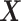 interaction effect component,
interaction effect component, 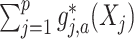 , from the
, from the 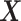 “main” effect component,
“main” effect component, 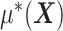 , in model (2.1). For model (2.1), we assume an additive noise structure
, in model (2.1). For model (2.1), we assume an additive noise structure 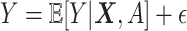 , where
, where 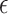 is a zero-mean noise with a finite second moment.
is a zero-mean noise with a finite second moment.
Notation: For a single component 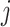 and general component functions
and general component functions 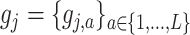 , we define the
, we define the 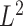 norm of
norm of 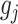 as
as 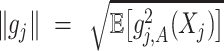 , where expectation is taken with respect to the joint distribution of
, where expectation is taken with respect to the joint distribution of 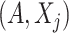 . For a set of random variables
. For a set of random variables 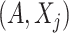 , let
, let 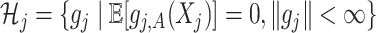 with inner product on the space defined as
with inner product on the space defined as 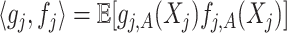 . Sometimes we also write
. Sometimes we also write 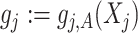 for the notational simplicity.
for the notational simplicity.
Under model (2.1) subject to (2.2), the component functions 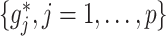 associated with the
associated with the 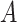 -by-
-by-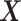 interaction effect can be viewed as the solution to the constrained optimization:
interaction effect can be viewed as the solution to the constrained optimization:
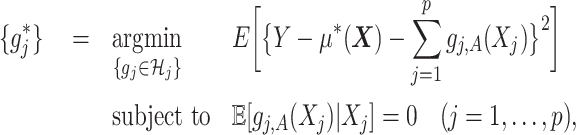 |
(2.3) |
where 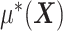 is given from the assumed model (2.1) (and is considered as fixed in (2.3)). Since the minimization in (2.3) is in terms of
is given from the assumed model (2.1) (and is considered as fixed in (2.3)). Since the minimization in (2.3) is in terms of 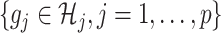 , the objective function part of the right-hand side of (2.3) can be reduced to:
, the objective function part of the right-hand side of (2.3) can be reduced to:
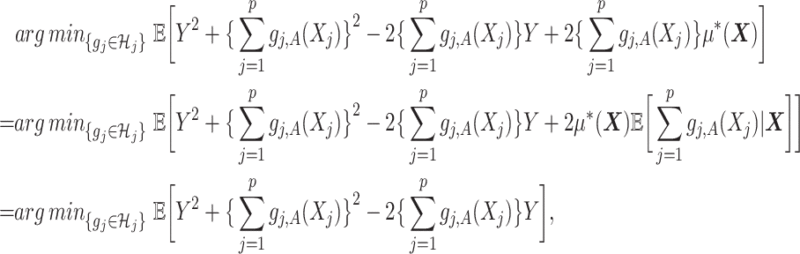 |
where the second line is from an application of the iterated expectation rule conditioning on 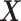 and the third line follows from the constraint
and the third line follows from the constraint 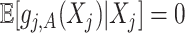
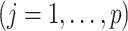 on the right-hand side of (2.3). Therefore, representation (2.3) can be simplified to:
on the right-hand side of (2.3). Therefore, representation (2.3) can be simplified to:
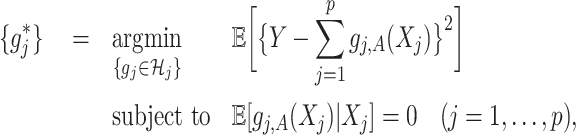 |
(2.4) |
Representation (2.4) of the component functions 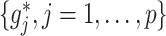 of the underlying model (2.1) is particularly useful when the (high-dimensional) “nuisance” function
of the underlying model (2.1) is particularly useful when the (high-dimensional) “nuisance” function 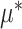 in (2.1) is complicated and prone to specification error. Note,
in (2.1) is complicated and prone to specification error. Note,
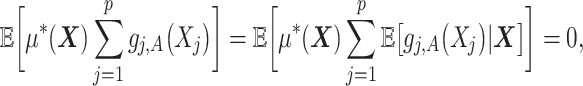 |
(2.5) |
indicating that the component 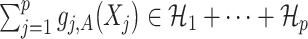 (subject to the identifiability constraint
(subject to the identifiability constraint 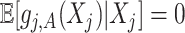 ) is structured to be orthogonal to the
) is structured to be orthogonal to the 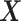 main effect
main effect 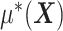 . This orthogonality property is useful for estimating the additive model
. This orthogonality property is useful for estimating the additive model 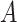 -by-
-by-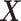 interaction effect
interaction effect 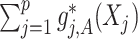 , in the presence of the unspecified
, in the presence of the unspecified 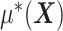 of model (2.1).
of model (2.1).
Under model (2.1), the potential treatment effect-modifiers among 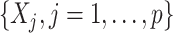 enter the model only through the interaction effect term
enter the model only through the interaction effect term 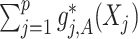 that associates the treatment
that associates the treatment 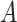 to the treatment outcome
to the treatment outcome 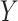 . Ravikumar and others (2009) proposed sparse additive modeling (SAM) for component selection in high-dimensional additive models with a large
. Ravikumar and others (2009) proposed sparse additive modeling (SAM) for component selection in high-dimensional additive models with a large 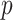 . As in SAM, to deal with a large
. As in SAM, to deal with a large 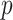 and to achieve treatment effect-modifier selection, we impose sparsity on the set of component functions
and to achieve treatment effect-modifier selection, we impose sparsity on the set of component functions 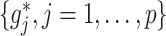 associated with the interaction effect term of model (2.1), under the often practical and reasonable assumption that most covariates are irrelevant as treatment effect-modifiers. This sparsity structure on the index set
associated with the interaction effect term of model (2.1), under the often practical and reasonable assumption that most covariates are irrelevant as treatment effect-modifiers. This sparsity structure on the index set 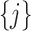 for the nonzero component functions
for the nonzero component functions 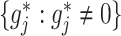 can be usefully incorporated into the optimization-based criterion (2.4) in representing
can be usefully incorporated into the optimization-based criterion (2.4) in representing 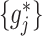 :
:
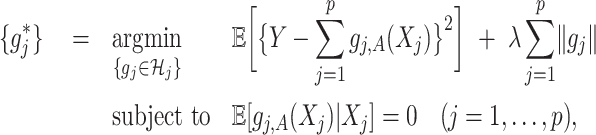 |
(2.6) |
for a sparsity-inducing parameter 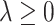 . The term
. The term 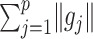 in (2.6) behaves like an
in (2.6) behaves like an 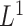 ball across different components
ball across different components 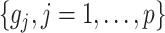 to encourage sparsity in the set of component functions. For example, a large value of
to encourage sparsity in the set of component functions. For example, a large value of 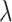 on the right-hand side of (2.6) will generate a sparse solution with many component functions
on the right-hand side of (2.6) will generate a sparse solution with many component functions 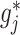 on the left-hand side set exactly to zero.
on the left-hand side set exactly to zero.
3. Estimation
3.1. Model estimation
For each 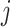 , the minimizer
, the minimizer 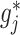 of the optimization problem (2.6) has a component-wise closed-form expression.
of the optimization problem (2.6) has a component-wise closed-form expression.
Theorem 1
Given
, the minimizer
of (2.6) satisfies (almost surely):
(3.7) where
(3.8) in which
(3.9) represents the
th partial residual. In (3.7),
represents the positive part of
.
Note that the 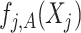 correspond to the projections of
correspond to the projections of 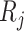 onto
onto 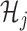 subject to the constraint in (2.6). The proof of Theorem 1 is in the Supplementary Materials available at Biostatistics online.
subject to the constraint in (2.6). The proof of Theorem 1 is in the Supplementary Materials available at Biostatistics online.
The component-wise expression (3.7) for 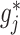 suggests that we can employ a coordinate descent algorithm (e.g., Tseng, 2001) to solve (2.6). Given a sparsity parameter
suggests that we can employ a coordinate descent algorithm (e.g., Tseng, 2001) to solve (2.6). Given a sparsity parameter 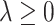 , we can use a standard backfitting algorithm used in fitting additive models (Hastie and Tibshirani, 1999) that fixes the set of current approximates for
, we can use a standard backfitting algorithm used in fitting additive models (Hastie and Tibshirani, 1999) that fixes the set of current approximates for 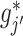 at all
at all 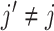 , and obtains a new approximate of
, and obtains a new approximate of 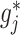 by equation (3.7), and iterates through all
by equation (3.7), and iterates through all 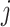 until convergence. A sample version of the algorithm can be obtained by inserting sample estimates into the population expressions (3.9), (3.8), and (3.7) for each coordinate
until convergence. A sample version of the algorithm can be obtained by inserting sample estimates into the population expressions (3.9), (3.8), and (3.7) for each coordinate 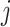 , which we briefly describe next.
, which we briefly describe next.
Given data 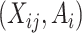
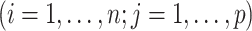 , for each
, for each 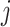 , let
, let 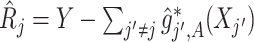 , corresponding to the data-version of the
, corresponding to the data-version of the 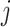 th partial residual
th partial residual 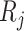 in (3.9), where
in (3.9), where 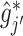 represents a current estimate for
represents a current estimate for 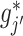 . We estimate
. We estimate 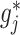 in (3.7) in two steps: (i) estimate the function
in (3.7) in two steps: (i) estimate the function 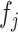 in (3.8); (ii) plug the estimate of
in (3.8); (ii) plug the estimate of 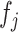 into
into 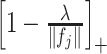 in (3.7), to obtain the soft-thresholded estimate
in (3.7), to obtain the soft-thresholded estimate 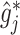 .
.
Although any linear smoothers can be utilized to obtain the estimators 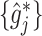 as described in Remark 1 at the end of this section, in this article, we shall focus on regression spline-type estimators which are particularly simple and computationally efficient to implement. Specifically, for each
as described in Remark 1 at the end of this section, in this article, we shall focus on regression spline-type estimators which are particularly simple and computationally efficient to implement. Specifically, for each 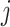 , the function
, the function 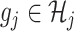 on the right-hand side of (2.6) will be represented by:
on the right-hand side of (2.6) will be represented by:
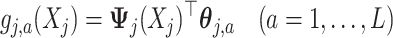 |
(3.10) |
for some prespecified 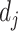 -dimensional basis
-dimensional basis 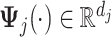 (e.g.,
(e.g., 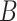 -spline basis on evenly spaced knots on a bounded range for
-spline basis on evenly spaced knots on a bounded range for 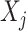 ) and a set of unknown treatment
) and a set of unknown treatment 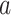 -specific basis coefficients
-specific basis coefficients 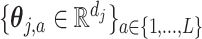 . Given representation (3.10) for the component function
. Given representation (3.10) for the component function 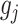 , the constraint
, the constraint 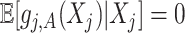 in (2.6) can be simplified to
in (2.6) can be simplified to 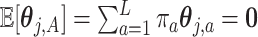 . This constraint can be written succinctly in a matrix form as
. This constraint can be written succinctly in a matrix form as
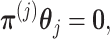 |
(3.11) |
where 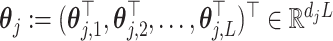 is the vectorized version of the basis coefficients
is the vectorized version of the basis coefficients 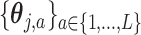 in (3.10), and the
in (3.10), and the 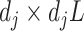 matrix
matrix 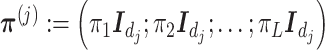 where
where 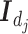 denotes the
denotes the 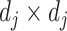 identity matrix.
identity matrix.
Let the 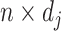 matrices
matrices 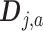
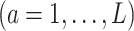 denote the evaluation matrices of the basis function
denote the evaluation matrices of the basis function 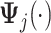 on
on 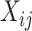
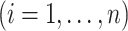 specific to the treatment
specific to the treatment 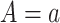
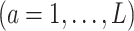 , whose
, whose 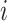 th row is the
th row is the 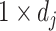 vector
vector 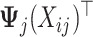 if
if 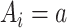 , and a row of zeros
, and a row of zeros 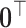 if
if 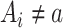 . Then the column-wise concatenation of the design matrices
. Then the column-wise concatenation of the design matrices 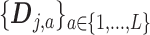 , i.e., the
, i.e., the 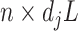 matrix
matrix 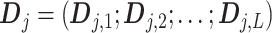 , defines the model matrix associated with the vectorized model coefficient
, defines the model matrix associated with the vectorized model coefficient 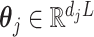 , vectorized across
, vectorized across 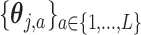 in representation (3.10). Then, we can represent the function
in representation (3.10). Then, we can represent the function 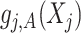 in (3.10), based on the sample data, by the length-
in (3.10), based on the sample data, by the length-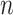 vector:
vector:
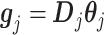 |
(3.12) |
subject to the linear constraint (3.11).
The linear constraint (3.11) on 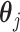 can be conveniently absorbed into the model matrix
can be conveniently absorbed into the model matrix 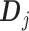 in (3.12) by reparametrization, as we describe next. We can find a
in (3.12) by reparametrization, as we describe next. We can find a 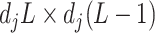 basis matrix
basis matrix 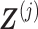 , such that if we set
, such that if we set 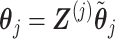 for any arbitrary vector
for any arbitrary vector 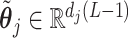 , then the vector
, then the vector 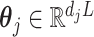 automatically satisfies the constraint (3.11)
automatically satisfies the constraint (3.11) 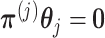 . Such a basis matrix
. Such a basis matrix 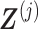 can be constructed by a QR decomposition of the matrix
can be constructed by a QR decomposition of the matrix 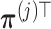 . Then representation (3.12) can be reparametrized, in terms of the unconstrained vector
. Then representation (3.12) can be reparametrized, in terms of the unconstrained vector 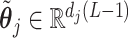 , by replacing
, by replacing 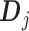 in (3.12) with the reparametrized model matrix
in (3.12) with the reparametrized model matrix 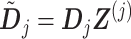 :
:
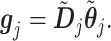 |
(3.13) |
Theorem 1 and Section 2 of Supplementary Materials available at Biostatistics online indicate that the coordinate-wise minimizer 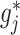 of (2.6) can be estimated based on the sample by
of (2.6) can be estimated based on the sample by
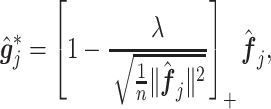 |
(3.14) |
where
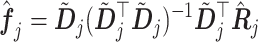 |
(3.15) |
in which 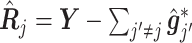 is the estimated
is the estimated 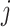 th partial residual vector. In (3.14), the norm
th partial residual vector. In (3.14), the norm 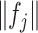 of (3.7) is estimated by the vector norm
of (3.7) is estimated by the vector norm 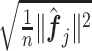 , and the shrinkage factor
, and the shrinkage factor 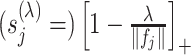 of (3.7) is estimated by
of (3.7) is estimated by 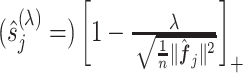 .
.
Based on the sample counterpart (3.14) of the coordinate-wise solution (3.7), a highly efficient coordinate descent algorithm can be conducted to simultaneously estimate all the component functions 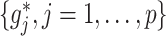 in (2.6). At convergence of the coordinate descent, we have a basis coefficient estimate associated with the representation (3.13),
in (2.6). At convergence of the coordinate descent, we have a basis coefficient estimate associated with the representation (3.13),
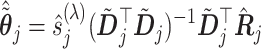 |
(3.16) |
which in turn implies an estimate
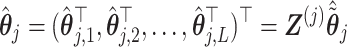 |
for the basis coefficient associated with the representation (3.12). This gives an estimate of the treatment 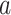 -specific function
-specific function 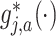
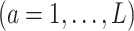 in model (2.1):
in model (2.1):
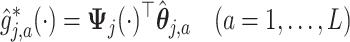 |
(3.17) |
estimated within the class of functions (3.10) for a given tuning parameter 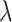 , which controls the shrinkage factor
, which controls the shrinkage factor 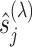 in (3.16). We summarize the computational procedure for the coordinate descent in Algorithm 1.
in (3.16). We summarize the computational procedure for the coordinate descent in Algorithm 1.
Algorithm 1 Coordinate descent
In Algorithm 1, the projection matrices 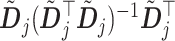
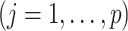 only need to be computed once and therefore the coordinate descent can be performed efficiently. In (3.14), if the shrinkage factor
only need to be computed once and therefore the coordinate descent can be performed efficiently. In (3.14), if the shrinkage factor 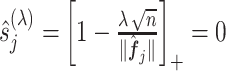 , the associated
, the associated 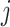 th covariate is absent from the model. The tuning parameter
th covariate is absent from the model. The tuning parameter 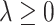 for treatment effect-modifier selection can be chosen to minimize an estimate of the expected squared error of the fitted models,
for treatment effect-modifier selection can be chosen to minimize an estimate of the expected squared error of the fitted models, 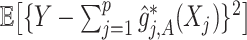 , over a dense grid of
, over a dense grid of 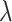 ’s, estimated, for example, by cross-validation. Alternatively, one can utilize the network information criterion (Murata and Amari, 1994) which is a generalization of the Akaike information criterion in approximating the prediction error, for the case where the true underling model, i.e., model (2.1), is not necessarily in the class of candidate models. Throughout the article,
’s, estimated, for example, by cross-validation. Alternatively, one can utilize the network information criterion (Murata and Amari, 1994) which is a generalization of the Akaike information criterion in approximating the prediction error, for the case where the true underling model, i.e., model (2.1), is not necessarily in the class of candidate models. Throughout the article, 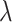 is chosen to minimize
is chosen to minimize 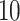 -fold cross-validated prediction error of the fitted models.
-fold cross-validated prediction error of the fitted models.
Remark 1
For coordinate descent, any linear smoothers can be utilized to obtain the sample counterpart (3.14) of the coordinate-wise solution (3.7), i.e., the method is not restricted to regression splines. To estimate the function
in (3.8), we can estimate the first term
in (3.8), using a 1D nonparametric smoother for each treatment level
separately, based on the data
corresponding to the data from the
th treatment condition; we can also estimate the second term
in (3.8) based on the data
which corresponds to the set of data from all treatment conditions, using a 1D nonparametric smoother. Adding these two estimates evaluated at the
observed values of
gives an estimate
in (3.14). Then, we can compute the associated estimate
, which allows implementation of the coordinate descent in Algorithm 1.
3.2. Individualized treatment rule estimation
For a single time decision point, an ITR, which we denote by 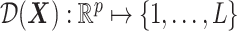 , maps an individual with pretreatment characteristics
, maps an individual with pretreatment characteristics 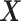 to one of the
to one of the 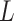 available treatment options. One natural measure for the effectiveness of an ITR
available treatment options. One natural measure for the effectiveness of an ITR 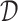 in precision medicine is the so-called “value” (
in precision medicine is the so-called “value” (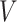 ) function (Murphy, 2005):
) function (Murphy, 2005):
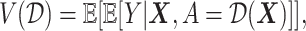 |
(3.18) |
which is the expected treatment response under a given ITR 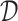 . The optimal ITR
. The optimal ITR 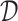 , which we write as
, which we write as 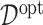 , can be naturally defined to be the rule that maximizes the value
, can be naturally defined to be the rule that maximizes the value 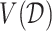 (3.18). Such an optimal rule
(3.18). Such an optimal rule 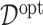 satisfies:
satisfies:
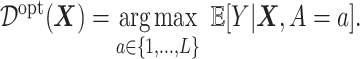 |
(3.19) |
Much work has been carried out to develop methods for estimating the optimal ITR (3.19) using data from randomized clinical trials. Machine learning approaches to estimating (3.19) are often framed in the context of a (weighted) classification problem (Zhang and others, 2012; Zhao and others, 2019), where the function 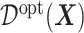 in (3.19) is regarded as the optimal classification rule given
in (3.19) is regarded as the optimal classification rule given 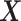 for the treatment assignment with respect to the objective function (3.18). These classification-based approaches to optimizing ITRs include the outcome-weighted learning (OWL) (e.g., Zhao and others, 2012; 2015; Song and others, 2015; Liu and others, 2018) based on support vector machines (SVMs), tree-based classification (e.g., Laber and Zhao, 2015), and adaptive boosting (Kang and others, 2014), among others.
for the treatment assignment with respect to the objective function (3.18). These classification-based approaches to optimizing ITRs include the outcome-weighted learning (OWL) (e.g., Zhao and others, 2012; 2015; Song and others, 2015; Liu and others, 2018) based on support vector machines (SVMs), tree-based classification (e.g., Laber and Zhao, 2015), and adaptive boosting (Kang and others, 2014), among others.
Under model (2.1), 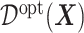 in (3.19) is:
in (3.19) is: 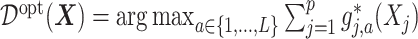 , which can be estimated by:
, which can be estimated by: 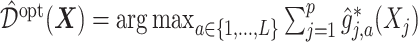 , where
, where 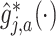 is given in (3.17) at the convergence of the Algorithm 1. This estimator can be viewed as a regression-based approach to estimating (3.19), that approximates the conditional expectations
is given in (3.17) at the convergence of the Algorithm 1. This estimator can be viewed as a regression-based approach to estimating (3.19), that approximates the conditional expectations 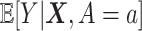
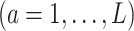 based on the additive model (2.1), while maintaining robustness with respect to model misspecification of the “nuisance” function
based on the additive model (2.1), while maintaining robustness with respect to model misspecification of the “nuisance” function 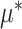 in (2.1) via representation (2.6) for the “signal” components
in (2.1) via representation (2.6) for the “signal” components 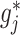 . We illustrate the performance of this ITR estimator
. We illustrate the performance of this ITR estimator 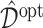 with respect to the value function (3.18), through a set of simulation studies in Section 4.2.
with respect to the value function (3.18), through a set of simulation studies in Section 4.2.
3.3. Feature selection and transformation for individualized treatment rules
Although machine learning approaches that attempt to directly maximize (3.18) without assuming some specific structure on 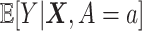
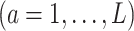 (unlike most of the regression-based approaches) are highly appealing, common machine learning approaches used in optimizing ITRs, including SVMs utilized in the OWL, are often hard to scale to large datasets, due to their taxing computational time. In particular, SVMs are viewed as “shallow” approaches (as opposed to a “deep” learning method that utilizes a learning model with many representational layers) and successful applications of SVMs often require first extracting useful representations for their input data manually or through some data-driven feature transformation (a step called feature engineering) (see, e.g., Kuhn and Johnson, 2019) to have more discriminatory power. Generally, selection and transformation of relevant features can increase the performance, scale, and speed of a machine learning procedure.
(unlike most of the regression-based approaches) are highly appealing, common machine learning approaches used in optimizing ITRs, including SVMs utilized in the OWL, are often hard to scale to large datasets, due to their taxing computational time. In particular, SVMs are viewed as “shallow” approaches (as opposed to a “deep” learning method that utilizes a learning model with many representational layers) and successful applications of SVMs often require first extracting useful representations for their input data manually or through some data-driven feature transformation (a step called feature engineering) (see, e.g., Kuhn and Johnson, 2019) to have more discriminatory power. Generally, selection and transformation of relevant features can increase the performance, scale, and speed of a machine learning procedure.
As an added value, the proposed regression (2.6) based on model (2.1) provides a practical feature selection and transformation learning technique for optimizing ITRs. The set of component functions 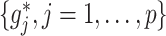 in (2.6) can be used to define data-driven feature transformation functions for the original features
in (2.6) can be used to define data-driven feature transformation functions for the original features 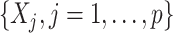 . The resulting transformed features can be used as inputs to a machine learning algorithm for optimizing ITRs and can lead to good results in practical situations.
. The resulting transformed features can be used as inputs to a machine learning algorithm for optimizing ITRs and can lead to good results in practical situations.
In particular, we note that for each 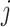 , the component function
, the component function 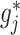 in (2.6) is defined separately from the
in (2.6) is defined separately from the 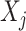 main effect function
main effect function 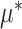 in (2.1). Therefore, the corresponding transformed feature variable
in (2.1). Therefore, the corresponding transformed feature variable 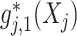 , which represents the
, which represents the 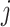 th feature
th feature 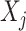 in the new space, highlights only the “signal” nonlinear effect of
in the new space, highlights only the “signal” nonlinear effect of 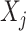 associated with the
associated with the 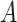 -by-
-by-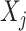 interactions on their effects on the outcome that is relevant to estimating
interactions on their effects on the outcome that is relevant to estimating 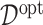 , while excluding the
, while excluding the 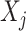 main effect that is irrelevant to the ITR development. This “de-noising” procedure for each variable
main effect that is irrelevant to the ITR development. This “de-noising” procedure for each variable 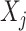 can be very appealing, since irrelevant or partially relevant features can negatively impact the performance of a machine learning algorithm. Moreover, a relatively large value of the tuning parameter
can be very appealing, since irrelevant or partially relevant features can negatively impact the performance of a machine learning algorithm. Moreover, a relatively large value of the tuning parameter 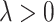 in (2.6) would imply a set of sparse component functions
in (2.6) would imply a set of sparse component functions 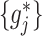 , providing a means of feature selection for ITRs. For the most common case of
, providing a means of feature selection for ITRs. For the most common case of 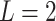 (binary treatment), we have
(binary treatment), we have 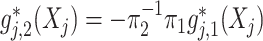 implied by the constraint (2.2) that we impose, which is simply a scalar-scaling of the function
implied by the constraint (2.2) that we impose, which is simply a scalar-scaling of the function 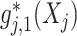 ; this implies that, for each
; this implies that, for each 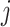 , the mapping
, the mapping 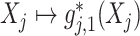 specifies the feature transformation of
specifies the feature transformation of 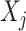 . We demonstrate the utility of this feature selection/transformation, which we use as an input to the OWL approach to optimizing ITRs, through a set of simulation studies in Section 4.2 and a real data application in Section 5.
. We demonstrate the utility of this feature selection/transformation, which we use as an input to the OWL approach to optimizing ITRs, through a set of simulation studies in Section 4.2 and a real data application in Section 5.
4. Simulation study
4.1. Treatment effect-modifier selection performance
In this section, we will report simulation results illustrating the performance of the treatment effect-modifier selection. The complexity of the model for studying the 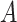 -by-
-by-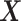 interactions can be summarized in terms of the size of the index set for the component functions
interactions can be summarized in terms of the size of the index set for the component functions 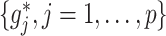 that are not identically zero. We can ascertain the performance of a treatment effect-modifier selection method in terms of these component functions correctly or incorrectly estimated as nonzero. To generate the data, we use the following model:
that are not identically zero. We can ascertain the performance of a treatment effect-modifier selection method in terms of these component functions correctly or incorrectly estimated as nonzero. To generate the data, we use the following model:
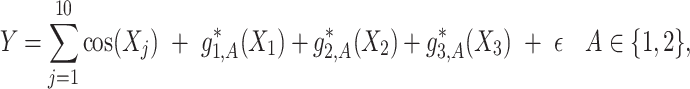 |
(4.20) |
where 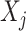
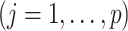 ,
, 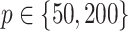 are generated from independent
are generated from independent 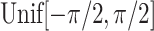 , and the treatment variable
, and the treatment variable 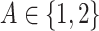 is generated independently of
is generated independently of  and the error term
and the error term 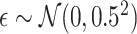 , such that
, such that 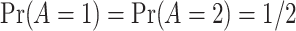 . We set
. We set 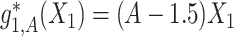 ,
, 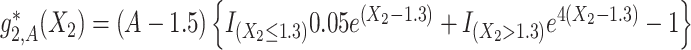 and
and 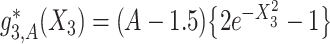 . The graphs of these functions are displayed in Figure S.1 of Supplementary Materials available at Biostatistics online.
. The graphs of these functions are displayed in Figure S.1 of Supplementary Materials available at Biostatistics online.
Under model (4.20), there are only three true treatment effect-modifiers, 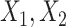 , and
, and 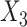 . The other
. The other 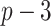 covariates are “noise” covariates, that are not consequential for optimizing ITRs. Also, in (4.20), there are
covariates are “noise” covariates, that are not consequential for optimizing ITRs. Also, in (4.20), there are 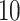 covariates
covariates 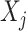
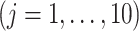 , among the
, among the 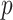 covariates, associated with the
covariates, associated with the 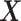 main effects. Under the setting (4.20), the contribution to the variance of the outcome from the
main effects. Under the setting (4.20), the contribution to the variance of the outcome from the 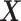 main effect component was about
main effect component was about 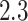 times greater than that from the
times greater than that from the 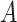 -by-
-by-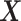 interaction effect component.
interaction effect component.
We consider two approaches to treatment effect-modifier selection: (i) the proposed additive regression approach (2.6) that specifies a sparse set of functions 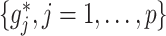 , estimated via Algorithm 1, with the dimension of the cubic
, estimated via Algorithm 1, with the dimension of the cubic  -spline basis
-spline basis 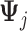 in (3.10) set to be
in (3.10) set to be 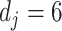
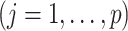 ; and (ii) the linear regression (MC) approach of Tian and others (2014),
; and (ii) the linear regression (MC) approach of Tian and others (2014),
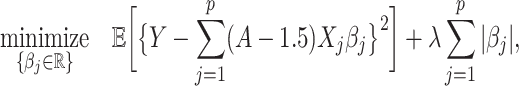 |
(4.21) |
which specifies a sparse vector 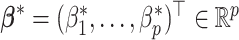 . Given each simulated dataset, the tuning parameter
. Given each simulated dataset, the tuning parameter 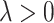 is chosen to minimize a
is chosen to minimize a 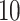 -fold cross-validated prediction error.
-fold cross-validated prediction error.
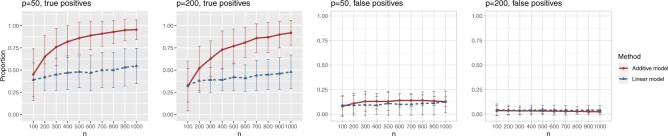
Figure 1 summarizes the results of the treatment effect-modifier selection performance with respect to the true/false positive rates (the left/right two panels, respectively), comparing the proposed additive regression to the linear regression approach, which are reported as the averages (and 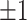 standard deviations) across the
standard deviations) across the 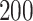 simulation replications. Figure 1 illustrates that, for the both
simulation replications. Figure 1 illustrates that, for the both 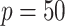 and
and 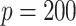 cases, the proportion of the correctly selecting treatment effect-modifiers (i.e., the “true positive”; the left two panels) of the additive regression method (the red solid curves) tends to
cases, the proportion of the correctly selecting treatment effect-modifiers (i.e., the “true positive”; the left two panels) of the additive regression method (the red solid curves) tends to 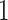 , with
, with 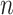 increasing from
increasing from 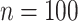 to
to 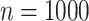 , while the proportion of incorrectly selecting treatment effect-modifiers (i.e., the “false positive”; the right two panels) tends to be bounded above by a small number. On the other hand, the proportion of correctly selecting treatment effect-modifiers for the linear regression method (the blue dotted curves) tends to be only around
, while the proportion of incorrectly selecting treatment effect-modifiers (i.e., the “false positive”; the right two panels) tends to be bounded above by a small number. On the other hand, the proportion of correctly selecting treatment effect-modifiers for the linear regression method (the blue dotted curves) tends to be only around 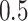 for both choices of
for both choices of 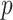 . In Figure S.2 of Supplementary Materials available at Biostatistics online, we further examine the true positive rates reported in Figure 1, by separately displaying the true positive rates associated with selection of
. In Figure S.2 of Supplementary Materials available at Biostatistics online, we further examine the true positive rates reported in Figure 1, by separately displaying the true positive rates associated with selection of 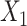 ,
, 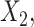 and
and 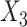 respectively. The more flexible additive regression significantly outperforms the linear regression in terms of selecting the covariates
respectively. The more flexible additive regression significantly outperforms the linear regression in terms of selecting the covariates 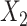 and
and 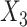 , i.e., the ones that have the nonlinear interaction effects (see Figure S.1 of Supplementary Materials available at Biostatistics online for the functions associated with the interaction effects) with
, i.e., the ones that have the nonlinear interaction effects (see Figure S.1 of Supplementary Materials available at Biostatistics online for the functions associated with the interaction effects) with 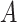 , while the both methods perform at a similar level in selection of
, while the both methods perform at a similar level in selection of 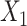 which has a linear interaction effect with
which has a linear interaction effect with 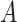 .
.
Figure 1.
The proportion of the three relevant covariates (i.e., 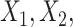 and
and 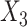 ) correctly selected (the “true positives”; the two gray panels), and the
) correctly selected (the “true positives”; the two gray panels), and the 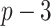 irrelevant covariates (i.e.,
irrelevant covariates (i.e., 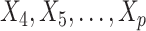 ) incorrectly selected (the “false positives”; the two white panels), respectively (and
) incorrectly selected (the “false positives”; the two white panels), respectively (and 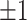 standard deviation), as the sample size
standard deviation), as the sample size 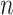 varies from
varies from 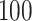 to
to 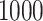 , for each
, for each 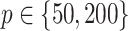 .
.
4.2. Individualized treatment rule estimation performance
In this subsection, we assess the optimal ITR estimation performance of the proposed method based on simulations. We generate a vector of covariates 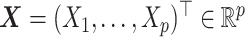 ,
, 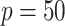 (a
(a 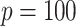 case is considered in Section S.5.1 of Supplementary Materials available at Biostatistics online) based on a multivariate normal distribution with each component having the marginal distribution
case is considered in Section S.5.1 of Supplementary Materials available at Biostatistics online) based on a multivariate normal distribution with each component having the marginal distribution 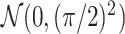 with the correlation between the components
with the correlation between the components 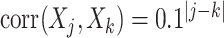 . Responses were generated, for (i) “highly nonlinear”
. Responses were generated, for (i) “highly nonlinear”  -by-
-by- interactions:
interactions:
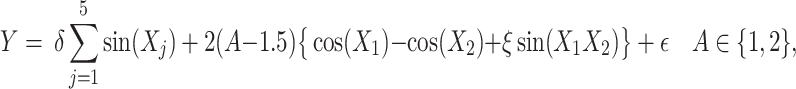 |
(4.22) |
and for (ii) “moderately nonlinear” 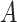 -by-
-by-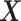 interactions:
interactions:
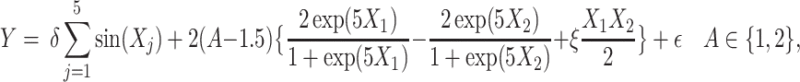 |
(4.23) |
where the treatment variable 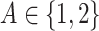 is generated independently from the covariates
is generated independently from the covariates 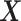 and the error term
and the error term 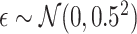 , such that
, such that 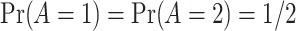 . Models (4.22) and (4.23) are indexed by a pair
. Models (4.22) and (4.23) are indexed by a pair 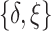 . First, the parameter
. First, the parameter 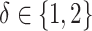 controls the proportion of the variance of the response
controls the proportion of the variance of the response 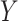 attributable to the
attributable to the 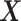 “main” effect:
“main” effect: 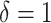 corresponds to a moderate
corresponds to a moderate 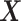 main effect contribution;
main effect contribution; 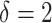 corresponds to a large
corresponds to a large 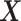 main effect contribution. Estimation of the interaction effect becomes more difficult with a larger
main effect contribution. Estimation of the interaction effect becomes more difficult with a larger 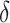 . Second, the parameter
. Second, the parameter 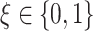 determines whether the
determines whether the 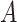 -by-
-by-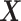 interaction effect term has an exact additive regression structure
interaction effect term has an exact additive regression structure  or whether it deviates from an additive structure
or whether it deviates from an additive structure  . In the case of
. In the case of  , the proposed model (2.1) is correctly specified, whereas, for the case of
, the proposed model (2.1) is correctly specified, whereas, for the case of 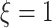 , it is misspecified. For each scenario, we consider the following four approaches to estimating the optimal ITR
, it is misspecified. For each scenario, we consider the following four approaches to estimating the optimal ITR 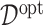 in (3.19).
in (3.19).
The proposed additive regression approach (2.6), estimated via Algorithm 1. The dimension of the basis function
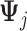 in (3.10) is taken to be
in (3.10) is taken to be 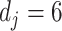
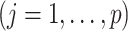 . Given estimates
. Given estimates 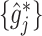 , the estimate of
, the estimate of 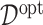 in (3.19) is
in (3.19) is 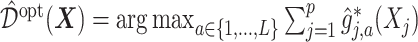 .
.The linear regression (MC) approach (4.21) of Tian and others (2014), implemented through the R-package glmnet, with the sparsity tuning parameter
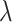 selected by minimizing a 10-fold cross-validated prediction error. Given an estimate
selected by minimizing a 10-fold cross-validated prediction error. Given an estimate 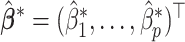 , the corresponding estimate of
, the corresponding estimate of 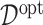 in (3.19) is
in (3.19) is 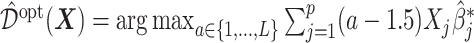 .
.The OWL method (Zhao and others, 2012) based on a Gaussian radial kernel, implemented in the R-package DTRlearn, with a set of feature transformed (FT) covariates
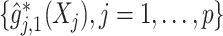 used as an input to the OWL method, in which the functions
used as an input to the OWL method, in which the functions 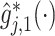
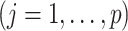 are obtained from the approach in 1. To improve the efficiency of the OWL, we employ the augmented OWL approach of Liu and others (2018). The inverse bandwidth parameter
are obtained from the approach in 1. To improve the efficiency of the OWL, we employ the augmented OWL approach of Liu and others (2018). The inverse bandwidth parameter 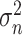 and the tuning parameter
and the tuning parameter 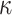 in Zhao and others (2012) are chosen from the grid of
in Zhao and others (2012) are chosen from the grid of 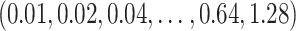 and that of
and that of 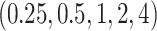 (the default setting of DTRlearn), respectively, based on a
(the default setting of DTRlearn), respectively, based on a 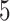 -fold cross-validation.
-fold cross-validation.The same (OWL) approach as in 3 but based on the original features
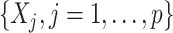 .
.
For each simulation run, we estimate 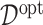 from each of the four methods based on a training set (of size
from each of the four methods based on a training set (of size 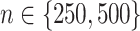 ), and for evaluation of these methods, we compute the value
), and for evaluation of these methods, we compute the value 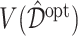 in (3.18) for each estimate
in (3.18) for each estimate 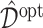 , using a Monte Carlo approximation based on a random sample of size
, using a Monte Carlo approximation based on a random sample of size 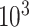 . Since we know the true data generating model in simulation studies, the optimal
. Since we know the true data generating model in simulation studies, the optimal 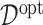 can be determined for each simulation run. Given each estimate
can be determined for each simulation run. Given each estimate 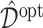 of
of 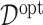 , we report
, we report 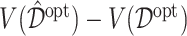 , as the performance measure of
, as the performance measure of 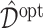 . A larger value (i.e., a smaller difference from the optimal value) of the measure indicates better performance.
. A larger value (i.e., a smaller difference from the optimal value) of the measure indicates better performance.
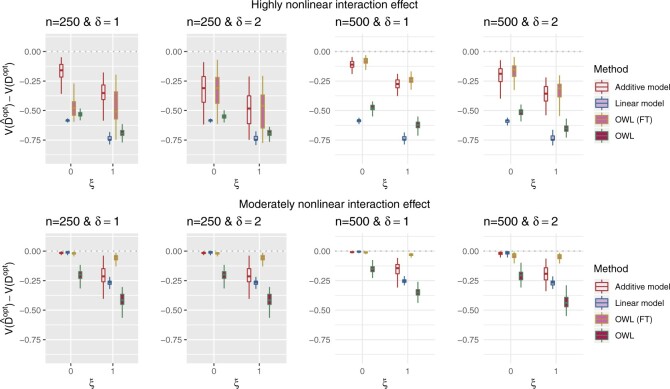
In Figure 2, we present the boxplots, obtained from 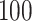 simulation runs, of the normalized values
simulation runs, of the normalized values 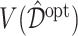 (normalized by the optimal values
(normalized by the optimal values 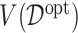 ) of the decision rules
) of the decision rules 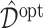 estimated from the four approaches, for each combination of
estimated from the four approaches, for each combination of 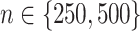 ,
, 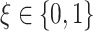 (corresponding to correctly specified or misspecified additive interaction effect models, respectively) and
(corresponding to correctly specified or misspecified additive interaction effect models, respectively) and 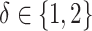 (corresponding to moderate or large main effects, respectively), for the highly nonlinear and moderately nonlinear
(corresponding to moderate or large main effects, respectively), for the highly nonlinear and moderately nonlinear 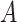 -by-
-by-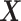 interaction effect scenarios, in the top and bottom panels, respectively. The proposed additive regression clearly outperforms the OWL (without feature transformation) method in all scenarios (both the top and bottom panels) and also the linear regression approach in all of the highly nonlinear
interaction effect scenarios, in the top and bottom panels, respectively. The proposed additive regression clearly outperforms the OWL (without feature transformation) method in all scenarios (both the top and bottom panels) and also the linear regression approach in all of the highly nonlinear 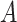 -by-
-by-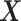 interaction effect scenarios (the top panels). For the moderately nonlinear
interaction effect scenarios (the top panels). For the moderately nonlinear 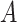 -by-
-by-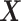 interaction effect scenarios (the bottom panels), when
interaction effect scenarios (the bottom panels), when  , all the methods except the OWL perform at a near-optimal level. On the other hand, when
, all the methods except the OWL perform at a near-optimal level. On the other hand, when 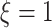 (i.e., when the underlying
(i.e., when the underlying 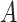 -by-
-by-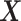 interaction effect model deviates from the additive structure), the more flexible additive model significantly outperforms the linear model. We have also considered a linear
interaction effect model deviates from the additive structure), the more flexible additive model significantly outperforms the linear model. We have also considered a linear 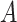 -by-
-by-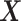 interaction effect case in Section S.5.2 of Supplementary Materials available at Biostatistics online, in which the linear regression outperforms the additive regression, but only slightly, whereas if the underlying model deviates from the exact linear structure and
interaction effect case in Section S.5.2 of Supplementary Materials available at Biostatistics online, in which the linear regression outperforms the additive regression, but only slightly, whereas if the underlying model deviates from the exact linear structure and 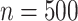 , the more flexible additive regression tends to outperform the linear model. This suggests that, in the absence of prior knowledge about the form of the interaction effect, employing the proposed additive regression is more suitable for optimizing ITRs than the linear regression. Comparing the two OWL methods (OWL (FT) and OWL), Figure 2 illustrates that the feature transformation based on the estimated component functions
, the more flexible additive regression tends to outperform the linear model. This suggests that, in the absence of prior knowledge about the form of the interaction effect, employing the proposed additive regression is more suitable for optimizing ITRs than the linear regression. Comparing the two OWL methods (OWL (FT) and OWL), Figure 2 illustrates that the feature transformation based on the estimated component functions 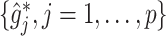 provides a considerable benefit in their performance. This suggests the utility of the proposed model (2.1) as a potential feature transformation and selection tool for a machine learning algorithm for optimizing ITRs. In Figure 2, comparing the cases with
provides a considerable benefit in their performance. This suggests the utility of the proposed model (2.1) as a potential feature transformation and selection tool for a machine learning algorithm for optimizing ITRs. In Figure 2, comparing the cases with 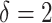 to those with
to those with 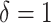 , the increased magnitude of the main effect generally dampens the performance of all approaches, as the “noise” variability in the data generation model increases.
, the increased magnitude of the main effect generally dampens the performance of all approaches, as the “noise” variability in the data generation model increases.
Figure 2.
Boxplots based on 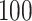 simulation runs, comparing the four approaches to estimating
simulation runs, comparing the four approaches to estimating 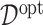 , given each scenario indexed by
, given each scenario indexed by 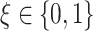 ,
, 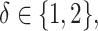 and
and 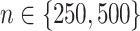 , for the highly nonlinear
, for the highly nonlinear 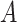 -by-
-by-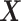 interaction effect case in the top panels, and the moderately nonlinear
interaction effect case in the top panels, and the moderately nonlinear 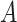 -by-
-by-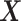 interaction effect case in the bottom panels. The dotted horizontal line represents the optimal value corresponding to
interaction effect case in the bottom panels. The dotted horizontal line represents the optimal value corresponding to 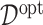 .
.
5. Application to data from a depression clinical trial
In this section, we illustrate the utility of the proposed additive regression for estimating treatment effect modification and optimizing individualized treatment rules, using data from a depression clinical trial study, comparing an antidepressant and placebo for treating major depressive disorder (Trivedi and others, 2016). The goal of the study is to identify baseline characteristics that are associated with differential response to the antidepressant versus placebo and to use those characteristics to guide treatment decisions when a patient presents for treatment.
Study participants (a total of 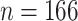 participants) were randomized to either placebo (
participants) were randomized to either placebo (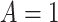 ;
; 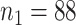 ) or an antidepressant (sertraline) (
) or an antidepressant (sertraline) (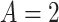 ;
; 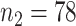 ). Subjects were monitored for 8 weeks after initiation of treatment, and the primary endpoint of interest was the Hamilton Rating Scale for Depression (HRSD) score at week 8. The outcome
). Subjects were monitored for 8 weeks after initiation of treatment, and the primary endpoint of interest was the Hamilton Rating Scale for Depression (HRSD) score at week 8. The outcome 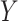 was taken to be the improvement in symptoms severity from baseline to week
was taken to be the improvement in symptoms severity from baseline to week 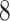 , taken as the difference, i.e., we take: week 0 HRSD score–week 8 HRSD score. (Larger values of the outcome
, taken as the difference, i.e., we take: week 0 HRSD score–week 8 HRSD score. (Larger values of the outcome 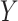 are considered desirable.) The study collected baseline patient clinical data, prior to treatment assignment. These pretreatment clinical data
are considered desirable.) The study collected baseline patient clinical data, prior to treatment assignment. These pretreatment clinical data 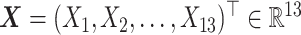 include:
include: 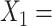 Age at evaluation;
Age at evaluation; 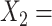 Severity of depressive symptoms measured by the HRSD at baseline;
Severity of depressive symptoms measured by the HRSD at baseline; 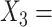 Logarithm of duration (in month) of the current major depressive episode; and
Logarithm of duration (in month) of the current major depressive episode; and 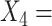 Age of onset of the first major depressive episode. In addition to these standard clinical assessments, patients underwent neuropsychiatric testing at baseline to assess psychomotor slowing, working memory, reaction time (RT), and cognitive control (e.g., post-error recovery), as these behavioral characteristics are believed to correspond to biological phenotypes related to response to antidepressants (Petkova and others, 2017) and are considered as potential modifiers of the treatment effect. These neuropsychiatric baseline test measures include:
Age of onset of the first major depressive episode. In addition to these standard clinical assessments, patients underwent neuropsychiatric testing at baseline to assess psychomotor slowing, working memory, reaction time (RT), and cognitive control (e.g., post-error recovery), as these behavioral characteristics are believed to correspond to biological phenotypes related to response to antidepressants (Petkova and others, 2017) and are considered as potential modifiers of the treatment effect. These neuropsychiatric baseline test measures include: 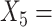 (A not B) RT-negative;
(A not B) RT-negative; 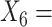 (A not B) RT-non-negative;
(A not B) RT-non-negative; 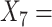 (A not B) RT-all;
(A not B) RT-all; 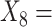 (A not B) RT-total correct;
(A not B) RT-total correct; 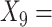 Median choice RT;
Median choice RT; 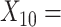 Word fluency;
Word fluency; 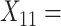 Flanker accuracy;
Flanker accuracy; 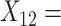 Flanker RT;
Flanker RT; 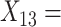 Post-conflict adjustment.
Post-conflict adjustment.
The proposed approach (2.6) to estimating the 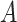 -by-
-by-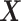 interaction effect part
interaction effect part 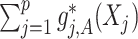 of model (2.1), estimated via Algorithm 1, simultaneously selected
of model (2.1), estimated via Algorithm 1, simultaneously selected 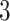 pretreatment covariates as treatment effect-modifiers:
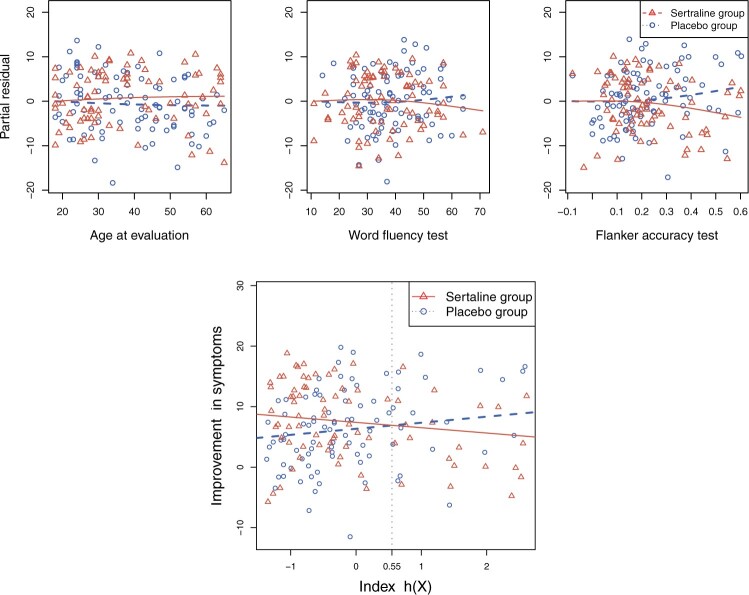
pretreatment covariates as treatment effect-modifiers: 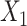 (“Age at evaluation”),
(“Age at evaluation”), 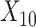 (“Word fluency test”), and
(“Word fluency test”), and 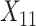 (“Flanker accuracy test”). The top panels in Figure 3 illustrate the estimated non-zero component functions
(“Flanker accuracy test”). The top panels in Figure 3 illustrate the estimated non-zero component functions 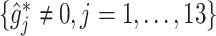 (i.e., the component functions corresponding to the selected covariates
(i.e., the component functions corresponding to the selected covariates 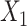 ,
, 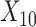 , and
, and 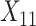 ) and the associated partial residuals. The linear regression approach (4.21) to estimating the
) and the associated partial residuals. The linear regression approach (4.21) to estimating the 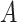 -by-
-by-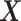 interactions selected a single covariate,
interactions selected a single covariate, 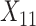 , as a treatment effect-modifier.
, as a treatment effect-modifier.
Figure 3.
The top panels: Scatterplots of partial residuals vs. the covariates associated with estimated non-zero component functions 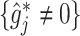 for placebo (blue circles) and the active drug (red triangles) treated participants; for each panel, the blue dashed curve represents
for placebo (blue circles) and the active drug (red triangles) treated participants; for each panel, the blue dashed curve represents 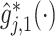 , corresponding to the placebo (
, corresponding to the placebo (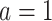 ), and the red solid curve represents
), and the red solid curve represents 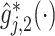 , corresponding to the active drug (
, corresponding to the active drug (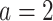 ). The bottom panel: A scatterplot of the treatment response
). The bottom panel: A scatterplot of the treatment response 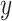 versus the index
versus the index 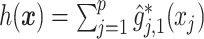 , for the drug (red triangles) and the placebo (blue circles) groups; the blue dashed line is
, for the drug (red triangles) and the placebo (blue circles) groups; the blue dashed line is 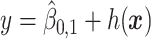 and the red solid line is
and the red solid line is 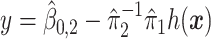 ; a gray dashed vertical line indicates the threshold value
; a gray dashed vertical line indicates the threshold value 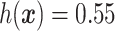 associated with the ITR
associated with the ITR 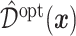 .
.
Note, in the binary (i.e., 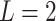 ) treatment case, any ITR
) treatment case, any ITR 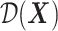 partitions the domain of
partitions the domain of 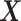 ,
, 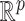 , into two regions:
, into two regions: 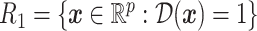 and
and 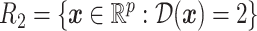 . Let
. Let 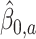
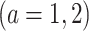 represent the treatment
represent the treatment 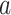 -specific intercept estimates. Then the proposed estimator
-specific intercept estimates. Then the proposed estimator 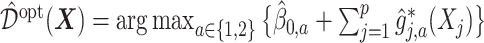 , is equivalent to:
, is equivalent to:
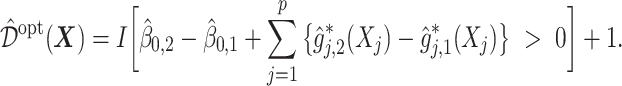 |
(5.24) |
In this dataset, 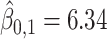 (corresponding to the placebo) and
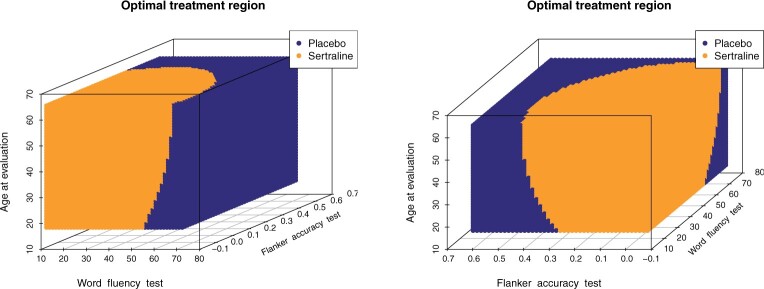
(corresponding to the placebo) and 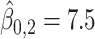 (corresponding to the drug). The optimal treatment region in the
(corresponding to the drug). The optimal treatment region in the 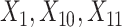 space, implied by the ITR (5.24), is illustrated in Figure 4, where we have utilized
space, implied by the ITR (5.24), is illustrated in Figure 4, where we have utilized 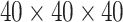 equally spaced grid points (i.e.,
equally spaced grid points (i.e., 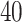 for each axis) for visualization of the regions
for each axis) for visualization of the regions 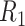 and
and 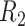 .
.
Figure 4.
The decision regions 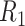 (corresponding to the placebo, dark blue) and
(corresponding to the placebo, dark blue) and 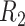 (corresponding to the active drug, orange) displayed in the 3D cube of the three covariates
(corresponding to the active drug, orange) displayed in the 3D cube of the three covariates 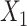 (age at evaluation),
(age at evaluation), 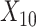 (word fluency test), and
(word fluency test), and 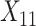 (Flanker accuracy test), evaluated at
(Flanker accuracy test), evaluated at 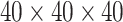 equally spaced grid points (
equally spaced grid points (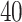 for each axis), shown from two different angles (left and right panels).
for each axis), shown from two different angles (left and right panels).
For an alternative way of visualizing the ITR (5.24), let us define a 1D index 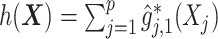 . By the constraint (2.2), we have the relationship
. By the constraint (2.2), we have the relationship 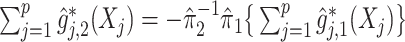 . Therefore, the term
. Therefore, the term 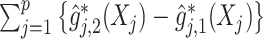 in the decision rule (5.24) can be reparametrized, with respect to
in the decision rule (5.24) can be reparametrized, with respect to 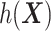 , as
, as 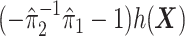 . In this dataset,
. In this dataset, 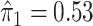 and
and 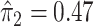 , and thus the ITR (5.24) can be re-written as:
, and thus the ITR (5.24) can be re-written as: 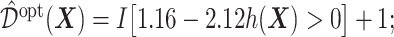 this ITR indicates that, for a patient with pretreatment characteristics
this ITR indicates that, for a patient with pretreatment characteristics 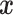 , if
, if 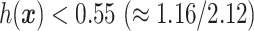 , then he/she would be recommended the active drug (
, then he/she would be recommended the active drug (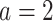 ) and the placebo (
) and the placebo (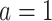 ), otherwise. For example, for a patient with
), otherwise. For example, for a patient with 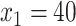 ,
, 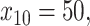 and
and  , the index
, the index 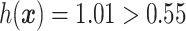 (see the bottom panel in Figure 3 for a visualization) and thus the patient would be recommended the placebo.
(see the bottom panel in Figure 3 for a visualization) and thus the patient would be recommended the placebo.
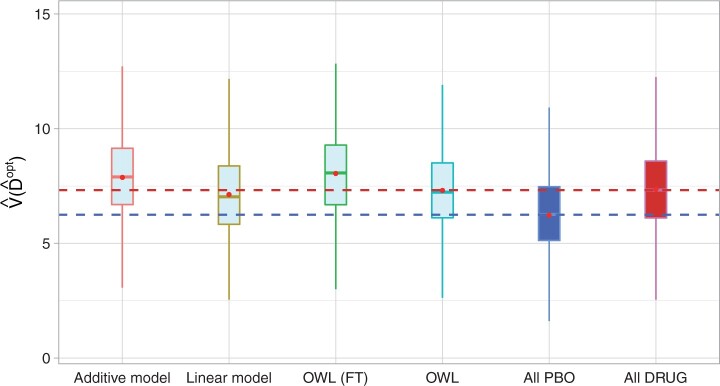
To evaluate the performance of the ITRs (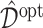 ) obtained from the four different approaches described in Section 4.2, we randomly split the data into a training set and a testing set (of size
) obtained from the four different approaches described in Section 4.2, we randomly split the data into a training set and a testing set (of size 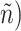 using a ratio of
using a ratio of 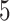 to
to 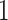 , replicated
, replicated 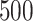 times, each time computing an ITR
times, each time computing an ITR 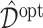 based on the training set, then estimating its value
based on the training set, then estimating its value 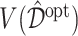 in (3.18) by an inverse probability weighted estimator (Murphy, 2005):
in (3.18) by an inverse probability weighted estimator (Murphy, 2005): 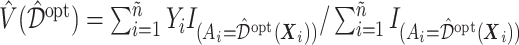 , computed based on the testing set of size
, computed based on the testing set of size 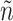 . For comparison, we also include two naïve rules: treating all patients with placebo (“All PBO”) and treating all patients with the active drug (“All DRUG”), each regardless of the individual patient’s characteristics
. For comparison, we also include two naïve rules: treating all patients with placebo (“All PBO”) and treating all patients with the active drug (“All DRUG”), each regardless of the individual patient’s characteristics 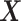 . The resulting boxplots obtained from the
. The resulting boxplots obtained from the 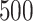 random splits are illustrated in Figure 5. A larger value of the measure indicates better performance.
random splits are illustrated in Figure 5. A larger value of the measure indicates better performance.
Figure 5.
Boxplots of the estimated values of the treatment rules 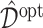 estimated from
estimated from 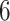 approaches, obtained from
approaches, obtained from 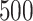 randomly split testing sets. Higher values are preferred.
randomly split testing sets. Higher values are preferred.
The results in Figure 5 demonstrate that the proposed additive regression approach, which allows nonlinear flexibility in developing ITRs, tends to outperform the linear regression approach, in terms of the estimated value. The additive regression approach also shows some superiority over the method OWL (without feature transformation). In comparison to the OWL methods, the proposed additive regression, in addition to its superior computational efficiency, provides a means of simultaneously selecting treatment effect-modifiers and allows a visualization for the heterogeneous effects attributable to each estimated treatment effect-modifier as in Figure 3, which is an appealing feature in practice. Moreover, the estimated component functions 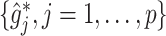 of the proposed regression provide an effective means of performing feature transformation for
of the proposed regression provide an effective means of performing feature transformation for 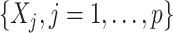 . As in Section 4.2, the FT OWL approach appears to have a considerable improvement over the OWL that bases on the original untransformed covariates.
. As in Section 4.2, the FT OWL approach appears to have a considerable improvement over the OWL that bases on the original untransformed covariates.
6. Discussion
In this article, we have developed a sparse additive model, via a structural constraint, specifically geared to identify and model treatment effect-modifiers. The approach utilizes an efficient back-fitting algorithm for model estimation and variable selection. The proposed sparse additive model for treatment effect modification extends existing linear model-based regression methods by providing nonlinear flexibility to modeling treatment-by-covariate interactions. Encouraged by our simulation results and the application, future work will investigate the asymptotic properties related to treatment effect-modifier selection and estimation consistency, in addition to developing hypothesis testing procedures for treatment-by-covariates interaction effects.
Modern advances in biotechnology, using measures of brain structure and function obtained from neuroimaging modalities (e.g., magnetic resonance imaging (MRI), functional MRI, and electroencephalography), show the promise of discovering potential biomarkers for heterogeneous treatment effects. These high-dimensional data modalities are often in the form of curves or images and can be viewed as functional data (e.g., Ramsay and Silverman, 1997). Future work will also extend the additive model approach to the context of functional additive regression (e.g., Fan and others, 2014, 2015). The goal of these extensions will be to handle a large number of functional-valued covariates while achieving simultaneous variable selection, which will extend current functional linear model-based methods for precision medicine (McKeague and Qian, 2014; Ciarleglio and others, 2015; 2018) to a more flexible functional regression setting, as well as to longitudinally observed functional data (e.g., Park and Lee 2019).
7. Software
R-package samTEMsel (Sparse Additive Models for Treatment Effect-Modifier Selection) contains R-codes to perform the methods proposed in the article, and is publicly available on GitHub (github.com/syhyunpark/samTEMsel).
Supplementary Material
Acknowledgments
We are grateful to the editors, the associate editor, and two referees for their insightful comments and suggestions. Conflict of Interest: None declared.
Supplementary material
Supplementary material is available at http://biostatistics.oxfordjournals.org
Funding
National Institute of Mental Health (NIMH) (5 R01 MH099003).
References
- Ashley, E. (2015). The precision medicine initiative: a new national effort. The Journal of the American Medical Association 313, 2117. [DOI] [PubMed] [Google Scholar]
- Ciarleglio, A., Petkova, E., Ogden, R. T. and Tarpey, T. (2015). Treatment decisions based on scalar and functional baseline covariate decisions based on scalar and functional baseline covariates. Biometrics 71(4), 884–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciarleglio, A., Petkova, E., Ogden, R. T. and Tarpey, T. (2018). Constructing treatment decision rules based on scalar and functional predictors when moderators of treatment effect are unknown. Journal of Royal Statistical Society: Series C 67, 1331–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, Y., Foutz, N., James, G. M. and Jank, W. (2014). Functional response additive model estimation with online virtual stock markets. The Annals of Applied Statistics 8, 2435–2460. [Google Scholar]
- Fan, Y., James, G. M. and Radchanko, P. (2015). Functional additive regression. The Annals of Statistics 43, 2296–2325. [Google Scholar]
- Fernandes, B., Williams, L., Steiner, J., Leboyer, M., Carvalho, A. and Berk, M. (2017). The new field of ‘precision psychiatry’. BMC Medicine 15, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie, T. and Tibshirani, R. (1999). Generalized Additive Models. London: Chapman and Hall/CRC. [DOI] [PubMed] [Google Scholar]
- Jeng, X., Lu, W. and Peng, H. (2018). High-dimensional inference for personalized treatment decision. Electronic Journal of Statistics 12, 2074–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, C., Janes, H. and Huang, Y. (2014). Combining biomarkers to optimize patient treatment recommendations. Biometrics 70, 696–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosorok, M. R. and Laber, E. B. (2019). Precision medicine. Annual Review of Statistics and Its Application 6(1), 263–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn, M. and Johnson, K. (2019). Feature Engineering and Selection: A Practical Approach for Predictive Models. London: Chapman and Hall/CRC. [Google Scholar]
- Laber, E. B. and Zhao, Y. (2015). Tree-based methods for individualized treatment regimes. Biometrika 102, 501–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., Wang, Y., Kosorok, M. R., Zhao, Y. and Zeng, D. (2018). Augmented outcome⣳weighted learning for estimating optimal dynamic treatment regimens. Statistics in Medicine 37, 3776–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, W., Zhang, H. and Zeng, D. (2011). Variable selection for optimal treatment decision. Statistical Methods in Medical Research 22, 493–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeague, I. and Qian, M. (2014). Estimation of treatment policies based on functional predictors. Statistica Sinica 24, 1461–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata, N. and Amari, S. (1994). Network Information Criterion-determining the number of hidden units for an artificial neural network model. IEEE Transactions on Neural Networks 5(6), 865–872. [DOI] [PubMed] [Google Scholar]
- Murphy, S. A. (2003). Optimal dynamic treatment regimes. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 65, 331–355. [Google Scholar]
- Murphy, S. A. (2005). A generalization error for q-learning. Journal of Machine Learning 6, 1073–1097. [PMC free article] [PubMed] [Google Scholar]
- Park, H. and Lee, S. (2019). Logistic regression error-in-covariate models for longitudinal high-dimensional covariates. Stat 8, e246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, H., Petkova, E., Tarpey, T. and Ogden, R. T. (2020). A constrained single-index regression for estimating interactions between a treatment and covariates. Biometrics, 1–13. 10.1111/biom.13320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkova, E., Ogden, R.T., Tarpey, T., Ciarleglio, A., Jiang, B., Su, Z., Carmody, T., Adams, P., Kraemer, H., Grannemann, B.. and others. (2017). Statistical analysis plan for stage 1 embarc (establishing moderators and biosignatures of antidepressant response for clinical care) study. Contemporary Clinical Trials Communications 6, 22–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian, M. and Murphy, S. A. (2011). Performance guarantees for individualized treatment rules. The Annals of Statistics 39(2), 1180–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay, J. O. and Silverman, B. W. (1997). Functional Data Analysis. New York: Springer. [Google Scholar]
- Ravikumar, P., Lafferty, J., Liu, H. and Wasserman, L. (2009). Sparse additive models. Journal of Royal Statistical Society: Series B 71, 1009–1030. [Google Scholar]
- Royston, P. and Sauerbrei, W. (2008). Interactions between treatment and continuous covariates: a step toward individualizing therapy. Journal of Clinical Oncology 26(9), 1397–1399. [DOI] [PubMed] [Google Scholar]
- Shi, C., Song, R. and Lu, W. (2016). Robust learning for optimal treatment decision with np-dimensionality. Electronic Journal of Statistics 10, 2894–2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, R., Kosorok, M., Zeng, D., Zhao, Y., Laber, E. B. and Yuan, M. (2015). On sparse representation for optimal individualized treatment selection with penalized outcome weighted learning. Stat 4, 59–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, L., Alizadeh, A., Gentles, A. and Tibshrani, R. (2014). A simple method for estimating interactions between a treatment and a large number of covariates. Journal of the American Statistical Association 109(508), 1517–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi, M., McGrath, P., Fava, M., Parsey, R., Kurian, B., Phillips, M., Pquendo, M., Bruder, G., Pizzagalli, D., Toups, M., and others. (2016). Establishing moderators and biosignatures of antidepressant response in clinical care (embarc): rationale and design. Journal of Psychiatric Research 78, 11–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng, P. (2001). Convergence of block coordinate descent method for nondifferentiable maximation. Journal of Optimization Theory and Applications 109, 474–494. [Google Scholar]
- Zhang, B., Tsiatis, A. A., Davidian, M., Zhang, M. and Laber, E. (2012). Estimating optimal treatment regimes from classification perspective. Stat 1, 103–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y., Laber, E., Ning, Y., Saha, S. and Sands, B. (2019). Efficient augmentation and relaxation learning for individualized treatment rules using observational data. Journal of Machine Learning Research 20, 1–23. [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y., Zeng, D., Rush, A. J. and Kosorok, M. R. (2012). Estimating individualized treatment rules using outcome weighted learning. Journal of the American Statistical Association 107, 1106–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y., Zheng, D., Laber, E. B. and Kosorok M. R. (2015). New statistical learning methods for estimating optimal dynamic treatment regimes. Journal of the American Statistical Association 110, 583–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.