Abstract
To reduce PCR bias derived from a primer mismatch, the effect of the annealing temperature on the product ratio was investigated by denaturing gradient gel electrophoresis analysis of PCR products from a mixture of perfect-match and one-mismatch templates. These templates were generated by PCR from Pediococcus acidilactici for one mismatch and Micrococcus luteus for the perfect match. PCRs showed that the bias was reduced at lower temperatures. An environmental sample was also examined.
Mixed-template PCRs often cause bias in the final products. This situation is a serious problem in molecular ecological methods, such as denaturing gradient gel electrophoresis (DGGE) analysis used to quantify microbial communities, because band intensities on DGGE gels do not accurately reflect quantitative information for the original microbial populations. Four types of PCR bias have been reported. (i) The ratios of PCR products to cells in multitemplate PCRs are often different among organisms because of different small-subunit ribosomal DNA (rDNA) copy numbers and genome sizes (2, 4, 6). Independent of these differences, reverse transcription-PCR can monitor the relative activities of microbial populations (3). (ii) Sequences with high G+C contents are difficult to amplify because of the low efficiency of template dissociation (11). The use of a denaturant such as acetamide in PCRs has been suggested as a countermeasure (1, 11). (iii) Frequent cycling of template reannealing causes PCR bias (13). Template reannealing could be prevented by running a low number of PCR cycles (13). (iv) Different primer binding energies cause PCR bias (10). The bias could be slightly reduced by running a low number of PCR cycles, but this process would not be an essential countermeasure.
Of these reported types of bias, ii, iii, and iv are characteristics of the PCR itself; therefore, these types of bias are included even in reverse transcription-PCR and might be reduced by modification of the PCR protocol. As deduced from the oligonucleotide dissociation temperature curve, the kinetics of binding between a complementary single oligodeoxynucleotide, such as a primer or a probe, and targeted DNA vary depending on temperature (14). At lower hybridization temperatures, oligonucleotides with a perfect match or one mismatch to the target site hybridize at similar rates. Thus, PCR bias caused by primer binding energies must be reduced by running PCR cycles at low annealing temperatures. The effect of annealing temperatures on PCR bias, however, has not been investigated.
The bacterial 16S rDNA targeted primer pair consisting of 341F (5′-CCTACGGGAGGCAGCAG-3′) with a GC clamp (5′-CGCCCGCCGCGCCCCGCGCCCGTCCCGCCGCCCCCGCCCG-3′) and 907R (5′-CCGTCAATTCCTTTRAGTTT-3′) has been widely used for DGGE analysis of bacterial communities (9). However, in an analysis of probe matches in the ARB software environment (12), only 11,429 and 8,611 organisms have a perfect match and 11,725 and 12,024 organisms have one mismatch with 341F and 907R, respectively, with a database consisting of 1,173 archaean and 15,104 bacterial sequences (ssu_Prok.arb; released on 1 September 2000 by Ribosomal Database Project II [7]). For example, 16S rDNA of Micrococcus luteus perfectly matches 907R, whereas that of Pediococcus acidilactici has one mismatch (Table 1). Therefore, we examined the effect of annealing temperatures using these primers and organisms.
TABLE 1.
Alignment of the sequences of universal reverse primer 907R and its target regions on 16S rDNA
| Sample | Sequencea |
|---|---|
| Primer 907R | 3′-TTTGA(A/G)TTTCCTTAACTGCC-5′ |
| P. acidilactici 16S rDNA | 5′-AAACT C AAAAGAATTGACGG-3′ |
| M. luteus 16S rDNA | 5′-AAACT C AAAGGAATTGACGG-3′ |
| S. marcescens 16S rDNA | 5′-AAACT C AAATGAATTGACGG-3′ |
| B. cereus 16S rDNA | 5′-AAACT C AAAGGAATTGACGG-3′ |
(A/G), A or G.
To simplify the experiment, primer 907R used in this study had no degeneracy (907Rnd: 5′-CCGTCAATTCCTTTAAGTTT-3′). M. luteus IAM1056 and P. acidilactici IAM1233 were obtained from the IAM Culture Collection (Center for Cellular and Molecular Research, Institute of Molecular and Cellular Biosciences, University of Tokyo, Tokyo, Japan) and were cultivated at 30°C for 24 h with nutrient broth (Oxoid, Basingstoke, United Kingdom) and lactobacillus MRS broth (BD, Franklin Lakes, N.J.), respectively. Templates for subsequent PCR experiments were generated by PCRs of nucleic acids extracted from M. luteus and P. acidilactici amplified with 341FGC and 907Rnd (thus, the templates were called M907 and P907, respectively) and with 341FGC and 1100R (5′-TTGCGCTCGTTGCGGGACT-3′) (templates M1100 and P1100, respectively). To remove the primers from the templates, each product was purified with a CONCERT Rapid PCR Purification System (Invitrogen Japan K.K., Tokyo, Japan). In P907 and M907, the original sequences at the 907Rnd priming site were interchanged with the complementary sequence of 907Rnd through the generating reaction. In M1100 and P1100, the original nucleotide sequences at the 907Rnd priming site were conserved. Thus, only P1100 had a mismatch at the 907Rnd priming site. All templates perfectly matched 341FGC. The nucleotide sequences at the 907Rnd priming site of these templates were confirmed by sequence analysis. Sequencing reactions were carried out with an ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Foster City, Calif.) according to the manufacturer's instructions. The products were analyzed with an automatic sequencer (model 377A; Applied Biosystems).
To examine the effect of annealing temperature on the bias caused by primer mismatches, two sets of templates (mixtures of P907 and M907 and of P1100 and M1100) were amplified with the same primer pair, 341FGC-907Rnd, at annealing temperatures of 45, 50, 55, and 60°C. DGGE analyses of the differences between the two sets can reveal the effect of annealing temperature on mismatches. PCR amplifications were performed with 50-μl volumes containing 10 pg of template DNA, 1× EX Taq buffer (TaKaRa, Kyoto, Japan), 200 μM each deoxynucleoside triphosphate, 25 pmol of each primer, and 1.25 U of Taq polymerase (TaKaRa). All reactions were run in triplicate, and mixtures were overlaid with mineral oil. For replicate PCR amplifications, aliquots were taken from a single master mixture. PCR cycling was performed with PCR Express (Thermo Hybaid, Middlesex, United Kingdom) with the following temperature program: 18 cycles at 94°C for 1 min; 45, 50, 55, or 60°C for 1 min; and 72°C for 2 min. This low number of PCR cycles yielded clear bands on DGGE gels (data not shown).
The PCR products were analyzed by DGGE as described by Muyzer et al. (8). After staining with ethidium bromide, nucleic acids in a gel were visualized by UV light (302 nm); digitized images were obtained with a charge-coupled device camera (Image Saver; ATTO, Tokyo, Japan). The band fluorescent intensities were analyzed with NIH-Image (developed at the National Institutes of Health, Bethesda, Md., and available on the Internet at http://rsb.info.nih.gov/nih-image/) and calibrated with Gray Scale (Kodak, Rochester, N.Y.).
In PCRs with the template mixture P907-M907, both templates were expected to be amplified to the same extents, because both templates perfectly match 907Rnd. However, DGGE analysis of the PCR products showed that P907 was preferentially amplified irrespective of the annealing temperature (Table 2). This bias might have been caused by efficient dissociation of template P907, because P907 has a lower G+C content (54.02%, including the GC clamp) than M907 (60.13%). In such a situation, the bias might be reduced by enhancing the efficiency of template dissociation (1, 11). On the contrary, in the analysis of template mixture P1100 (one mismatch with 907Rnd)-M1100 (perfect match), M1100 was selectively amplified at higher annealing temperatures, even though M1100 has a higher GC content (59.29%) than P1100 (53.67%) (Table 2). This selective amplification of M1100 at higher annealing temperatures showed the bias caused by the difference in primer binding energies. The product ratios for P907-M907 and P1100-M1100 were significantly different (P < 0.0001 at 55°C and P < 0.01 at 50°C). Statistical variance analysis of three replicates of each sample was performed using statistical software implemented in Microsoft Excel 98. At an annealing temperature of 45°C, the product ratio for P1100-M1100 was increased to the same extent as the product ratio for P907-M907 (Table 2). This result shows that the bias caused by the difference in primer binding energies was reduced by a lower annealing temperature. An experiment using other organisms, Serratia marcescens (IAM12142; one mismatch with 907Rnd) and Bacillus cereus (IAM 12605; no mismatch with 907Rnd) gave the same results (data not shown). S. marcescens and P. acidilactici have one mismatch in the middle of the 907R priming site (Table 1). If there are organisms that have mismatches at positions different from those used here, the annealing temperature needed for the organisms might be different from those in this study. In any event, the bias caused by the difference in primer binding energies was reduced when the annealing temperature was low.
TABLE 2.
Effect of annealing temperatures on PCR product ratios
| Template | Template ratioa | Mean ± SD product ratio at the following annealing temp (°C)b:
|
|||
|---|---|---|---|---|---|
| 45 | 50 | 55 | 60 | ||
| P907-M907 | 1.07 | 1.61 ± 0.31 | 1.70 ± 0.26 | 2.12 ± 0.12 | BDL |
| P1100-M1100 | 1.12 | 1.47 ± 0.21 | 0.87 ± 0.07 | 0.17 ± 0.08 | BDL |
P907/M907 or P1100/M1100.
Data are P. acidilactici/M. luteus product ratios calculated from three replicates. BDL, below detection limit.
However, Polz and Cavanaugh (10) showed that bias toward a primer with high binding energies was observed at a relatively low annealing temperature, 50°C. One explanation for the disagreement between our results and theirs is the differences in the PCR conditions, such as the addition of 5% acetamide and the larger reaction volume in their study. A reaction with acetamide might require a lower annealing temperature than one without acetamide (11). Generally, larger PCR volumes require lower or longer annealing temperature programs, because the temperature in larger volumes of reaction mixtures changes more slowly than that in smaller volumes.
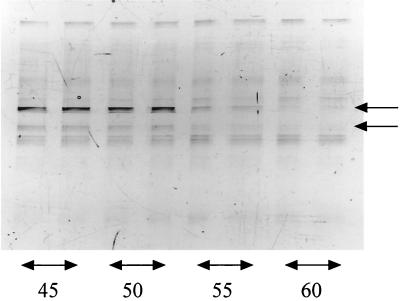
Furthermore, we examined a composting sample for which the nucleic acid extraction method was described elsewhere (5). PCRs were performed for 21 cycles with 31.7 ng of template. These PCR conditions were designed to amplify as much template as possible. Even the major bands on the DGGE profiles at lower annealing temperatures were not observed at higher temperatures (Fig. 1). This result suggested that PCRs at lower annealing temperatures can be used for larger numbers of organisms than can those at higher temperatures. These results indicate that annealing temperature is an important parameter in PCR for molecular ecological research.
FIG. 1.
Effect of annealing temperature on the DGGE profiles of an environmental sample. The numbers under the lanes show the annealing temperatures. PCRs for each annealing temperature were run in duplicate. The arrows indicate the bands that appeared only at low annealing temperatures.
Acknowledgments
We are grateful to Susumu Takii for incisive comments on the manuscript and to Hiroya Shinozaki for advice and discussions.
This work was supported by a grant from the Ministry of Education, Culture, Sports, Science and Technology (12440219).
REFERENCES
- 1.Dutton C M, Paynton C, Sommer S S. General method for amplifying regions of very high G + C content. Nucleic Acids Res. 1993;21:2953–2954. doi: 10.1093/nar/21.12.2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farrelly V, Rainey F A, Stackebrandt E. Effect of genome size and rrn gene copy number on PCR amplification of 16S rRNA genes from a mixture of bacterial species. Appl Environ Microbiol. 1995;61:2798–2801. doi: 10.1128/aem.61.7.2798-2801.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Felske A, Akkermans A D L, De Vos W M. Quantification of 16S rRNAs in complex bacterial communities by multiple competitive reverse transcription-PCR in temperature gradient gel electrophoresis fingerprints. Appl Environ Microbiol. 1998;64:4581–4587. doi: 10.1128/aem.64.11.4581-4587.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fogel G B, Collins C R, Li J, Brunk C F. Prokaryotic genome size and SSU rDNA copy number: estimation of microbial relative abundance from a mixed population. Microb Ecol. 1999;38:93–113. doi: 10.1007/s002489900162. [DOI] [PubMed] [Google Scholar]
- 5.Ishii K, Fukui M, Takii S. Microbial succession during a composting process as evaluated by denaturing gradient gel electrophoresis analysis. J Appl Microbiol. 2000;89:768–777. doi: 10.1046/j.1365-2672.2000.01177.x. [DOI] [PubMed] [Google Scholar]
- 6.Klappenbach J A, Dunbar J M, Schmidt T M. rRNA operon copy number reflects ecological strategies of bacteria. Appl Environ Microbiol. 2000;66:1328–1333. doi: 10.1128/aem.66.4.1328-1333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maidak B L, Cole J R, Parker C T, Jr, Garrity G M, Larsen N, Li B, Lilburn T G, McCaughey M J, Olsen G J, Overbeek R, Pramanik S, Schmidt T M, Tiedje J M, Woese C R. A new version of the RDP (Ribosomal Database Project) Nucleic Acids Res. 1999;27:171–173. doi: 10.1093/nar/27.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muyzer G, Brinkhoff T, Nübel U, Santegoeds C, Schäfer H, Wawer C. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology. In: Akkermans A D L, Elsas J D V, Bruijn F J D, editors. Molecular microbial ecology manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1997. pp. 1–27. F. J.. [Google Scholar]
- 9.Muyzer G, de Waal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Polz M F, Cavanaugh C M. Bias in template-to-product ratios in multitemplate PCR. Appl Environ Microbiol. 1998;64:3724–3730. doi: 10.1128/aem.64.10.3724-3730.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reysenbach A-L, Giver L J, Wickham G S, Pace N R. Differential amplification of rRNA genes by polymerase chain reaction. Appl Environ Microbiol. 1992;58:3417–3418. doi: 10.1128/aem.58.10.3417-3418.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strunk O, Ludwig W, Gross O, Reichel B, Stuckmann N, May M, Nonhoff B, Lenke M, Ginhart T, Vilbig A, Westran R. ARB—a software environment for sequence data. Munich, Germany: Technische Universität München; 1998. [Google Scholar]
- 13.Suzuki M T, Giovannoni S J. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng D, Alm E W, Stahl D A, Raskin L. Characterization of universal small-subunit rRNA hybridization probes for quantitative molecular microbial ecology studies. Appl Environ Microbiol. 1996;62:4504–4513. doi: 10.1128/aem.62.12.4504-4513.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]