Summary
Alveolar formation increases the surface area for gas exchange. A molecular understanding of alveologenesis remains incomplete. Here we show that the autonomic nerve and alveolar myofibroblast form a functional unit in mice. Myofibroblasts secrete neurotrophins to promote neurite extension/survival while neurotransmitters released from autonomic terminals are necessary for myofibroblast proliferation and migration, a key step in alveologenesis. This establishes a functional link between autonomic innervation and alveolar formation. We also discover that planar cell polarity (PCP) signaling employs a Wnt–Fz/Ror–Vangl cascade to regulate the cytoskeleton and neurotransmitter trafficking/release from the terminals of autonomic nerves. This represents a new aspect of PCP signaling in conferring cellular properties. Together, these studies offer molecular insight into how autonomic activity controls alveolar formation. Our work also illustrates the fundamental principle of how two tissues (e.g., nerves and lungs) interact to build alveoli at the organismal level.
eTOC Blurb
Zhang et al. demonstrate that autonomic nerves and alveolar myofibroblasts form a functional unit during alveolar formation. These nerves secrete neurotransmitters to promote proliferation, contraction and migration of alveolar myofibroblasts for proper secondary septation. Alveolar myofibroblasts secrete neurotrophins to affect autonomic nerve function.
Introduction
The alveolus, the unit of gas exchange, is produced during the last phase of lung development (Burri, 2006; Chao et al., 2016; Pan et al., 2019; Whitsett and Weaver, 2015). Lung branching morphogenesis is followed by the construction of primary saccules, the primitive oxygen exchange apparatus located at the terminal ends of branched airways. The smooth wall of primary saccules is further modified by the generation of secondary crests or septa, which divide the saccules into alveoli. Secondary septation is the most important step during alveologenesis. In this process, alveolar epithelial cells (type I and type II), myofibroblasts and endothelial cells/pericytes undergo coordinated movement to form secondary septa within saccules. As a result, secondary septa consist of an overlying layer of alveolar epithelial cells that ensheathes a core of myofibroblasts and endothelial cells/pericytes. Alveolar myofibroblasts are largely derived from alveolar fibroblasts, which undergo extensive proliferation and differentiation to generate alveolar myofibroblasts at around postnatal (P) day 2 and 3 in mice. Formation of alveoli greatly increases the surface area and efficiency of gas exchange. This information is essential to revealing the pathogenesis of lung diseases caused by immaturity or loss of alveoli. This knowledge is also critical for designing new strategies to promote alveolar formation in disease conditions (Rodriguez-Castillo et al., 2018). The prominent examples include bronchopulmonary dysplasia (BPD), lung infections, including Coronavirus disease 2019 (COVID-19), and chronic obstructive pulmonary disease (COPD). BPD develops in preterm babies whose alveolar development is blocked due to ventilation-induced hyperoxia and lung injury (Silva et al., 2015). COPD is a common disorder that ranks third in mortality in the US. COPD is characterized by inflammation and destruction of alveoli (Patel et al., 2019). No effective treatment is available for COPD.
The nervous system plays a key role in controlling the physiological function of the lungs. Movement of the diaphragm and intercostal muscles during the inspiration-expiration cycle is controlled by motor neurons derived from the ventral neuroectoderm during embryonic development. Contraction of smooth muscles in the trachea and bronchi, glandular secretion and vascular tone regulation are attributed to autonomic innervation. Bronchioles are also innervated. By contrast, the literature is scant in innervation of saccules and alveoli (Watanabe et al., 2018). Nevertheless, a very small number of papers reported the presence of nerve fibers in the alveoli of adult rodent and human lungs by electron microscopy (EM) (Fox et al., 1980; Hung et al., 1972, 1973; Meyrick and Reid, 1971). There were no follow-up studies and no information is available on innervation of saccules and alveoli during lung development. Importantly, whether lung innervation affects alveolar formation is unknown.
Through a combination of mouse genetics, cell-based assays and transcriptomics, we delineate a conceptual framework of alveologenesis. In essence, autonomic nerves and mesenchymal myofibroblasts exhibit reciprocal signaling and form a functional unit. Myofibroblasts secrete neurotrophins to promote neurite extension and survival while neurotransmitters released from autonomic terminals are necessary for myofibroblast proliferation and migration. This is a new function of the autonomic nervous system. Our work also illustrates the fundamental principle of how two tissues (in this case, nerves and lungs) interact to build alveoli at the organismal level.
Results
Lung saccules are innervated by autonomic nerves during postnatal lung development to form alveoli
As a first step toward exploring the relationship between lung innervation and alveolar formation, we performed immunostaining on serial lung tissues from wild-type mice using anti-neurofilament (NF-M) (2H3) (Dodd et al., 1988), anti-Synaptophysin (SYP) or anti-PGP9.5 antibodies specific to nerve fibers. We found that nerve fibers extended and contacted lung saccules at postnatal (P) day 1 to 5 and alveoli derived from saccules (Figures 1A–1C). We sectioned the entire mouse lung along the longitudinal axis and identified two routes through which the nerve fibers reached the saccules. Most nerve fibers (~85%) followed the conducting airways to the bronchoalveolar duct junctions (BADJs) and extended into saccules close to respiratory bronchioles (Figure 1D). Due to the dense packing and folding/looping of bronchioles and saccules in a confined physical space, saccules at the end of one respiratory bronchiole contacted nerves from another respiratory bronchiole (Figure 1D). This would resolve the apparent conundrum in which autonomic nerves do not spread far beyond the BADJ and yet can contact saccules. Quantification revealed that ~40% of these non-surface saccules touched nerve fibers (Figure 1F–1H). A smaller number of nerve fibers (~15%) reached the saccules by passing through the surface of the lungs (Figure 1E). These fibers innervated saccules located more distally to the respiratory bronchioles; ~15% of these surface saccules adjoined nerve fibers (Figure 1F). Myofibroblasts within saccules send out cellular extensions to form a network (Branchfield et al., 2016; Zhang et al., 2020). Thus, we speculate that multiple saccules could form a functional unit, within which the effects of innervated saccules are propagated to those not directly innervated.
Figure 1. Mouse saccules are innervated by neural crest descendants.
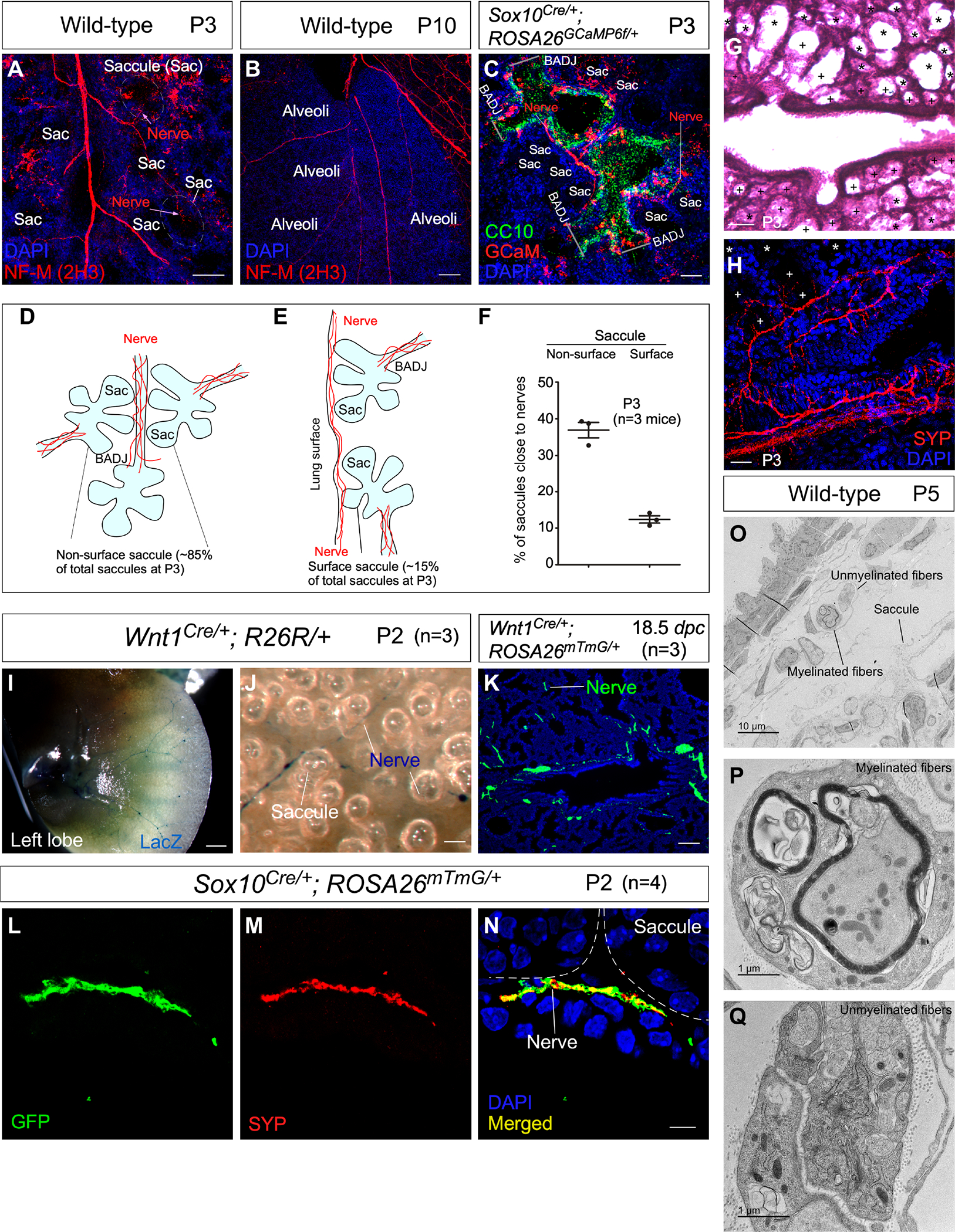
(A-C) Immunofluorescence of mouse lung sections stained with anti-NF-M (2H3) to trace autonomic nerves at postnatal (P) day 3 and 10. Saccules (Sac) and alveoli adjacent to nerves were identified by their morphology. Autonomic nerves were also visualized by GCaM expression in Sox10Cre/+; ROSA26GCaMP6f/+ lungs at P3, in which GCaM was activated from ROSA26GCaMP6f in neural crest descendants (SOX10+) after their migration. The airway epithelium was marked by CC10 expression in club cells, aiding the identification of the bronchoalveolar duct junction (BADJ) that separated the airways and saccules. The addition of a clearing procedure facilitated visualization of nerve fibers.
(D, E) Schematic diagram of the passage of autonomic nerves to reach both surface and non-surface saccules.
(F) Quantification of the % of saccules that are adjacent to autonomic nerves of wild-type mouse lungs at P3. Saccules were divided into the surface and non-surface groups as shown in (D, E).
(G) Hematoxylin-and-eosin (H&E) staining of lung sections from control mice at P3. Saccules marked by (+) were counted as saccules close to nerves whereas saccules marked by (*) were counted as saccules not close to nerves.
(H) Immunofluorescence of lung sections from control mice stained with anti-SYP. Saccules marked by (+) were counted as saccules close to nerves whereas saccules marked by (*) were counted as saccules not close to nerves.
(I, J) Surface view of LacZ-stained left lobes of mouse lungs from Wnt1Cre/+; R26R/+ mice (n=3) at P2. LacZ expression was induced from R26R in neural crest descendants (WNT1+) prior to their migration.
(K) Immunofluorescence of lung sections from Wnt1Cre/+; ROSA26mTmG/+ mice (n=3) at 18.5 days post coitus (dpc) stained with anti-GFP. GFP expression was generated from ROSA26mTmG in WNT1+ cells. Nerve fibers (GFP+) were in close proximity to the developing saccules.
(L-N) Immunofluorescence of lung sections from Sox10Cre/+; ROSA26mTmG/+ mice (n=4) at P2 stained with anti-GFP and anti-SYP. GFP expression was produced from ROSA26mTmG in SOX10+ cells. Autonomic nerve fibers (GFP+SYP+) adjoined developing saccules.
(O-Q) Transmission electron microscopy (TEM) of wild-type lungs to visualize autonomic nerve fibers and their relationship to saccules.
Scale bars, 100 μm (A), 100 μm (B), 50 μm (C), 100 μm (G), 25 μm (H), 1 mm (I), 25 μm (J), 100 μm (K), 5 μm (L-N).
We also generated Wnt1Cre/+; ROSA26mTmG/+ and Wnt1Cre/+; R26R/+ mice to trace innervation of saccules and alveoli. Wnt1Cre is expressed in neural crest progenitors prior to their migration (Danielian et al., 1998; Lewis et al., 2013). Thus, autonomic nerves derived from the neural crest are lineage-labeled by Wnt1Cre and reporter mice. In this setting, Cre recombinase in Wnt1+ cells induced the production of GFP from the ROSA26mTmG allele (Muzumdar et al., 2007) and LacZ (β-galactosidase) from the R26R allele (Soriano, 1999), respectively. We found that labeled autonomic nerve fibers extended to the immediate vicinity of lung saccules (Figures 1I–1K). To confirm this finding, we produced Sox10Cre/+; ROSA26mTmG/+ mice. Sox10Cre (Matsuoka et al., 2005) is expressed in neural crest progenitors after their migration. Similarly, labeled autonomic nerve fibers spread to abut lung saccules (Figures 1L–1N). Both myelinated and unmyelinated fibers could be detected in the vicinity of saccules by transmission electron microscopy (Figures 1O–1Q). Taken together, our results represent a comprehensive study to reveal innervation of saccules by autonomic nerves during postnatal lung development.
Autonomic nerve function and activity are essential for alveolar formation
The close contact of autonomic nerves with saccules prompted us to determine whether autonomic nerves control alveolar formation. We expect that neurotransmitters released by autonomic nerves will be received by cells in the vicinity. To test this idea, we manipulated the activity of TrkA and TrkB in neural crest progenitors. TrkA and TrkB encode receptors for neurotrophins and are essential for neural development. TRKA, the high-affinity neurotrophin receptor for nerve growth factor (NGF), is absolutely necessary for the survival of sympathetic neurons and for proper innervation of their targets (Lu et al., 2005). TRKB, the high-affinity neurotrophin receptor for brain-derived neurotrophic factor (BDNF), is highly expressed in parasympathetic neurons in the brainstem and is also detected in embryonic sympathetic ganglions (Ernsberger, 2009). Information on how TrkA and TrkB may affect alveologenesis is sparse.
To study the role of neurotrophin signaling in alveologenesis, we selectively inactivated TrkA or TrkB in neural crest progenitors using Sox10Cre to convert floxed alleles of TrkA (TrkAf) (Sanchez-Ortiz et al., 2012) and TrkB (TrkBf) (Xu et al., 2000) into null alleles. TrkAf/f; Sox10Cre/+ and TrkBf/f; Sox10Cre/+ mice could not be distinguished from their wild-type littermates by P10 and P7, respectively. No defects were found in saccules, airway smooth muscles and the diaphragm, nor was surfactant production affected in the mutant lungs. Analysis of alveolar development in TrkAf/f; Sox10Cre/+ (Figures 2A and 2B) and TrkBf/f; Sox10Cre/+ (Figures 2G and 2H) mice revealed loss of secondary septa in many regions where an increased mean linear intercept (MLI, a measure of air space size) was noted (Figure 2C and 2I).
Figure 2. Neurotrophin signaling in autonomic nerves regulates alveolar formation.
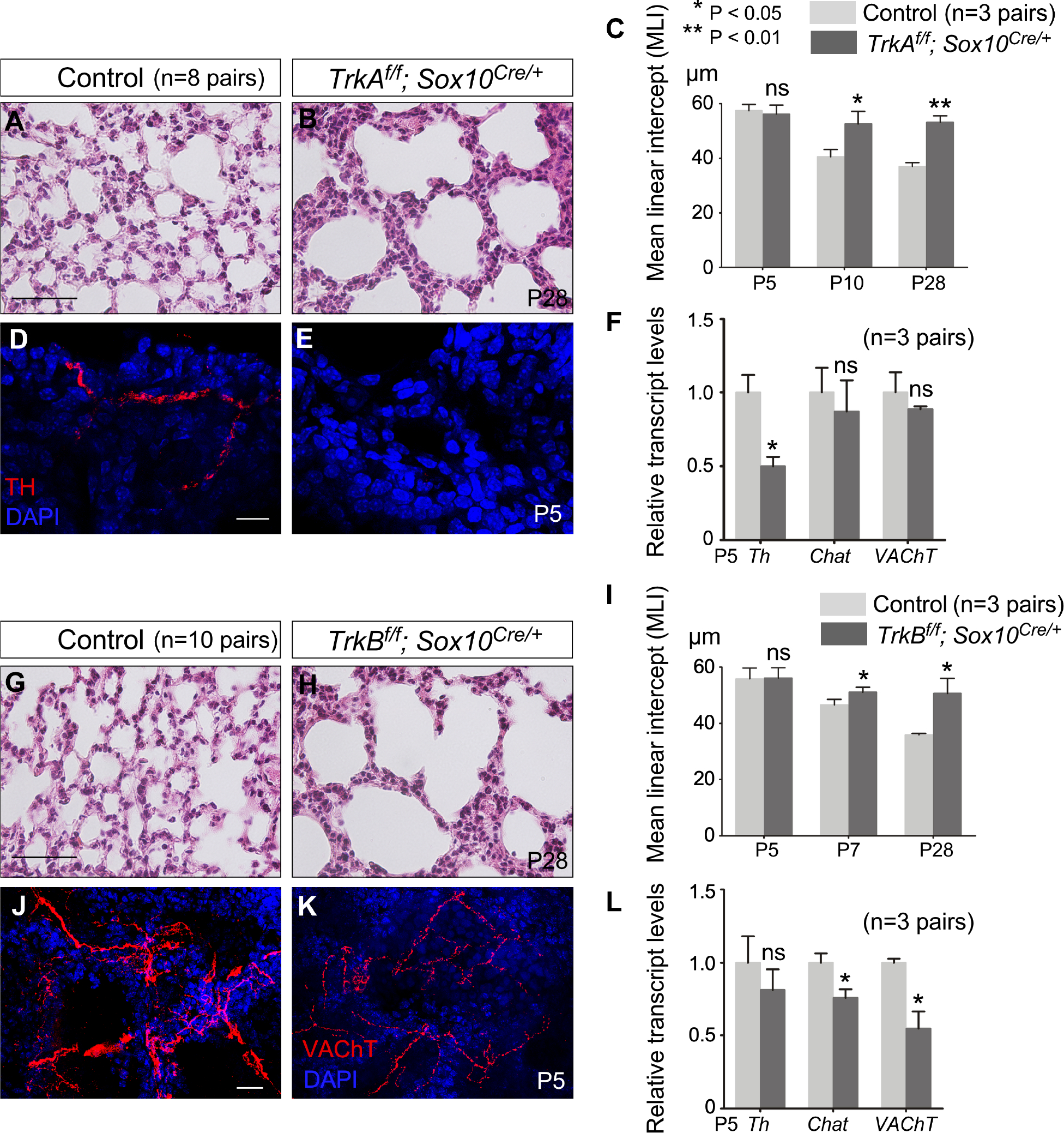
(A, B) Hematoxylin-and-eosin (H&E) staining of lung sections from control and TrkAf/f; Sox10Cre/+ mice (n=8 pairs) at postnatal (P) day 28.
(C) Measurement of the mean linear intercept (MLI) of control and TrkAf/f; Sox10Cre/+ lungs (n=3 pairs) at P5, P10 and P28.
(D, E) Immunofluorescence of lung sections from control and TrkAf/f; Sox10Cre/+ mice (n=3 pairs) stained with anti-TH (tyrosine hydroxylase) to label sympathetic nerve fibers at P5.
(F) qPCR analysis of transcript levels in control and TrkAf/f; Sox10Cre/+ lungs (n=3 pairs) at P5. Th expression levels were reduced in TrkAf/f; Sox10Cre/+ lungs compared to controls while the expression levels of Chat and VAChT were unaltered.
(G, H) H&E staining of lung sections from control and TrkBf/f; Sox10Cre/+ mice (n=10 pairs) at P28.
(I) Measurement of the MLI in control and TrkBf/f; Sox10Cre/+ lungs (n=3 pairs) at P5, P7 and P28.
(J, K) Immunofluorescence of lung sections from control and TrkBf/f; Sox10Cre/+ mice (n=5 pairs) stained with anti-VAChT to label parasympathetic nerve fibers at P5.
(L) qPCR analysis of transcript levels in control and TrkBf/f; Sox10Cre/+ lungs (n=3 pairs) at P5. The expression levels of Chat and VAChT were reduced in TrkBf/f; Sox10Cre/+ lungs compared to controls whereas the expression levels of Th were unaltered.
Scale bars, 100 μm (A, B, G, H), 10 μm (D, E), 25 μm (J, K). All values are mean ± SEM. (*) p<0.05; (**) p<0.01; ns, not significant (unpaired Student’s t-test).
Sympathetic nerves labeled by tyrosine hydroxylase (TH) were barely detectable in TrkAf/f; Sox10Cre/+ lungs (Figures 2D–2F). Likewise, parasympathetic nerves distinguished by vesicular acetylcholine transporter (VAChT) were significantly reduced in TrkBf/f; Sox10Cre/+ lungs (Figures 2J–2L). These results suggest that the alveolar defects in TrkAf/f; Sox10Cre/+ and TrkBf/f; Sox10Cre/+ mice were caused by loss of autonomic function due to defective neurotrophin signaling. Removal of individual neurotrophin receptors using Wnt1Cre (Lewis et al., 2013) yielded similar alveolar phenotypes.
To further probe the function of autonomic innervation in alveolar formation, we abolished autonomic nerve activity through the production of Sox10Cre/+; RC::Ptox/+ mice. Cre activity in Sox10+ cells induced the generation of a tetanus toxin light chain-GFP fusion (referred to as Ptox) from the RC::Ptox locus (Kim et al., 2009) (abbreviated as Ptox in the figures). As a result, neurotransmission in autonomic nerves was blocked by tetanus toxin. As expected, the distribution and density of autonomic nerves and neurotransmitter production were largely unperturbed in Sox10Cre/+; RC::Ptox/+ lungs (Figures 3F, 3G and Figure S1). We found that loss of autonomic activity led to alveolar phenotypes with an increased MLI (Figures 3A–3E). Regional alveolar defects (without apparent defects in the airways or other organs) were observed in Sox10iCreER/+; RC::Ptox/+ lungs in which Ptox was induced by tamoxifen (Figures 3H–3J). The alveolar defect in Sox10Cre/+; RC::Ptox/+ lungs was more severe than that in TrkAf/f; Sox10Cre/+ or TrkBf/f; Sox10Cre/+ lungs, suggesting that both TrkA and TrkB regulate autonomic nerve function in the lung. Together, these studies establish a functional role of autonomic innervation in alveolar formation.
Figure 3. Autonomic nerve activity regulates alveolar formation.
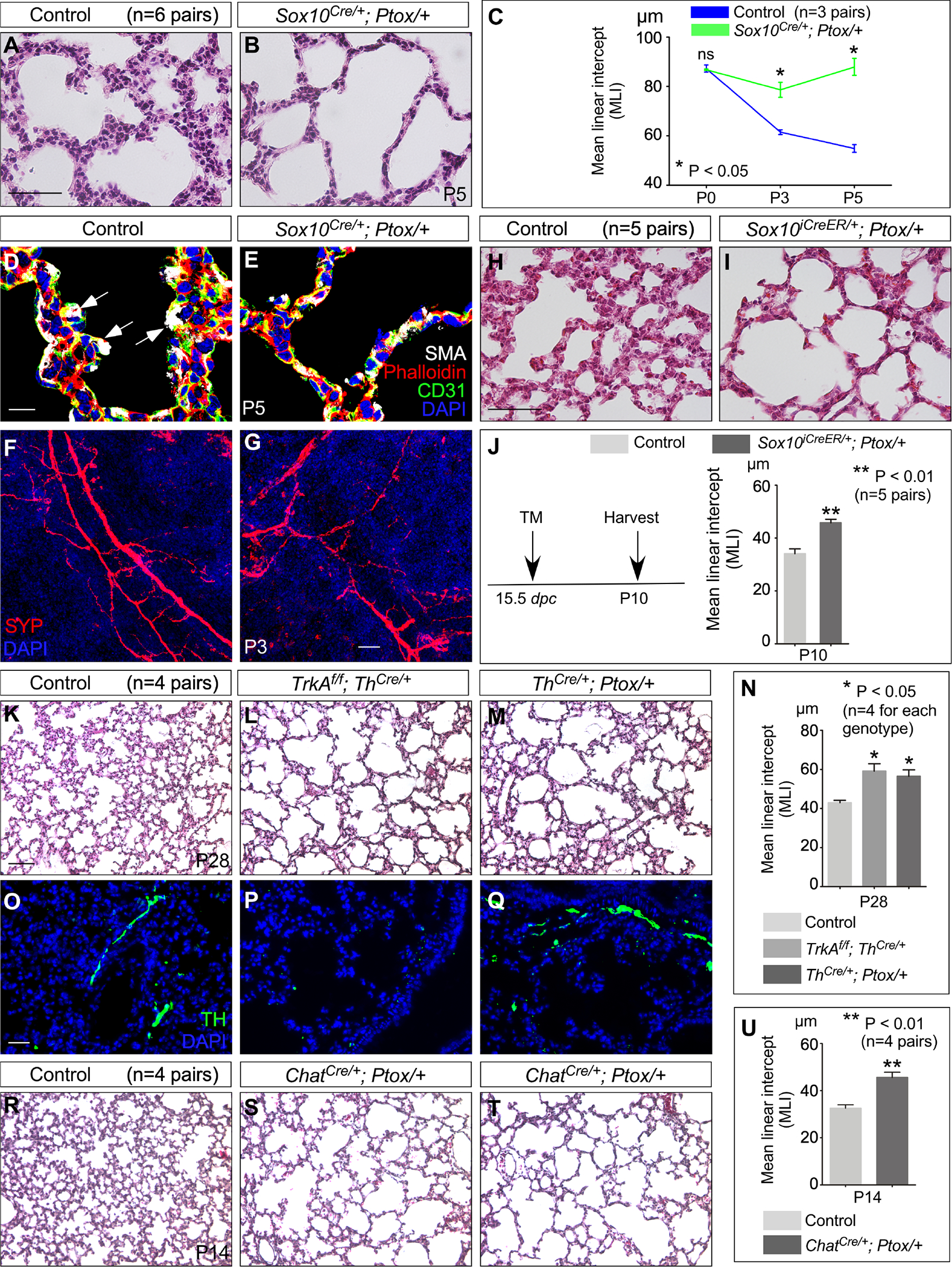
(A, B) H&E staining of lung sections from control and Sox10Cre/+; Ptox/+ mice (n=6 pairs) at P5.
(C) Measurement of the MLI in control and Sox10Cre/+; Ptox/+ lungs (n=3 pairs) at P0, P3 and P5.
(D, E) Immunofluorescence of lung sections from control and Sox10Cre/+; Ptox/+ mice (n=3 pairs) stained with anti-SMA (smooth muscle actin), anti-CD31 and phalloidin at P5. CD31 labels endothelial cells and phalloidin binds to F-actin. Arrows point to secondary septation.
(F, G) Immunohistochemical analysis of control and Sox10Cre/+; Ptox/+ mouse lungs at P3. Immunoreactivity of SYP, which marked nerve fibers, showed no apparent difference between control and mutant lungs.
(H, I) H&E staining of lung sections from control and Ptox-mutant mice (conditional induction by tamoxifen).
(J) Measurement of the MLI in control and Sox10iCreER/+; Ptox/+ lungs (n=5 pairs) at P10. Sox10iCreER/+; Ptox/+ mice were injected with tamoxifen at 15.5 dpc and lungs were collected at P10.
(K-M) H&E staining of lung sections from control, TrkAf/f; ThCre/+ and ThCre/+; Ptox/+ mice at P28.
(N) Measurement of the MLI in control, TrkAf/f; ThCre/+ and ThCre/+; Ptox/+ lungs at P28 (n=4 for each genotype).
(O-Q) Immunohistochemical analysis of control, TrkAf/f; ThCre/+ and ThCre/+; Ptox/+ mouse lungs at P28. Immunoreactivity of TH, which marked sympathetic nerves, was reduced in TrkAf/f; ThCre/+ lungs but not in ThCre/+; Ptox/+ lungs.
(R-T) H&E staining of lung sections from control and ChatCre/+; Ptox/+ mice at P14.
(U) Measurement of the MLI in control and ChatCre/+; Ptox/+ lungs at P14 (n=4 pairs).
Scale bars, 100 μm (A, B), 10 μm (D, E), 50 μm (F, G), 100 μm (H, I), 100 μm (K-M, R-T), 25 μm (O-Q). All values are mean ± SEM. (*) p<0.05; (**) p<0.01; ns, not significant (unpaired Student’s t-test and one-way ANOVA).
Wnt1Cre and Sox10Cre mark neural crest lineages, which give rise to cell types other than the autonomic nervous system. This could complicate the interpretation of alveolar phenotypes in our studies. To address this issue, we utilized Th-Cre and Chat-Cre mice, in which Cre recombinase is expressed in the sympathetic nervous system, labeled by tyrosine hydroxylase (TH), and the parasympathetic nervous system, marked by choline acetyltransferase (ChAT), respectively. We generated TrkAf/f; ThCre/+ and ThCre/+; RC::Ptox/+ mice to perturb sympathetic function or activity, and ChatCre/+; RC::Ptox/+ mice to disrupt parasympathetic activity. Analysis of alveolar development in TrkAf/f; ThCre/+ and ThCre/+; RC::Ptox/+ (Figures 3K–3Q) and ChatCre/+; RC::Ptox/+ (Figures 3R–3U) mice revealed loss of secondary septa in many regions where an increased MLI was noted (Figure 3N and 3U). These studies further bolster our model in which autonomic innervation controls alveologenesis.
Planar cell polarity (PCP) signaling in neural crest progenitors controls alveolar formation
We analyzed the expression profiles of signaling pathways in neural crest progenitors (Soldatov et al., 2019) to gain mechanistic insights into how autonomic innervation regulates alveolar formation. Many components of the PCP (a non-canonical Wnt) pathway are present in neural crest progenitors, suggesting a potential role in controlling autonomic function. For instance, Vangl1, Vangl2, Ror1, Ror2, Fzd2, Fzd3 are expressed in neural crest progenitors and their derivatives (e.g., nerves) (Figure S2).
The PCP pathway is a fundamental, conserved mechanism for polarizing a field of cells within the plane of an epithelial cell sheet and is essential in many tissues (Campanale et al., 2017; Davey and Moens, 2017; Zallen, 2007). PCP signaling is initiated by the binding of non-canonical Wnt ligands to the Frizzled (Fz) receptors and Ror (receptor tyrosine kinase-like orphan receptor 2) coreceptors (Wang et al., 2016). The signal is relayed by a set of core PCP components that include cytoplasmic and membrane proteins. The outcome of PCP signaling is an altered actomyosin cytoskeleton that is local and is associated with polarized cellular function in a field of cells. Interestingly, previous studies reported the involvement of PCP signaling in axon sprouting (Zou, 2012) or postsynaptic compartmentalization (Nagaoka and Kishi, 2016). However, a functional role of PCP signaling in controlling autonomic function and alveolar formation in the lung has not been reported.
To investigate the relationship between PCP signaling, autonomic function and alveolar formation, we disrupted PCP signaling in neural crest progenitors. To this end, we focused on Vangl1 and Vangl2, mammalian homologs of Drosophila van gogh (vangl). Vangl1 and Vangl2 encode four-pass transmembrane proteins and are critical for PCP signaling. We manipulated VANGL1/2 activity to control PCP signaling and used Wnt1Cre and Sox10Cre to convert a floxed allele of Vangl2 (Vangl2f) (Song et al., 2010) into a null allele in neural crest progenitors. Vangl1 plays a minor role in this process since Vangl1gt/gt mice, homozygous for the Vangl1gt null allele (Torban et al., 2008), are fully viable and fertile. Thus, our study has focused on Vangl2. Vangl2f/f; Sox10Cre/+ mice were born at the expected Mendelian frequency and could not be distinguished from their wild-type littermates at birth by their outer appearance or activity. Most Vangl2f/f; Sox10Cre/+ mice succumbed before P30; lethality was observed at different stages of alveolar development. Analysis of Vangl2f/f; Sox10Cre/+ lungs at different postnatal stages revealed enlarged saccules with an increased MLI; many regions lacked rudimentary secondary septation (Figures 4A–4O and Figure S3). These results support a critical role of PCP signaling in autonomic function and subsequently alveolar formation. Likewise, Vangl2f/f; Wnt1Cre/+ (Danielian et al., 1998; Lewis et al., 2013) mice displayed alveolar phenotypes (Figure S2), resembling those of Vangl2f/f; Sox10Cre/+ mice. Interestingly, most Vangl2f/f; Wnt1Cre/+ mice survived to P30.
Figure 4. A signaling cascade of planar cell polarity controls alveolar formation.
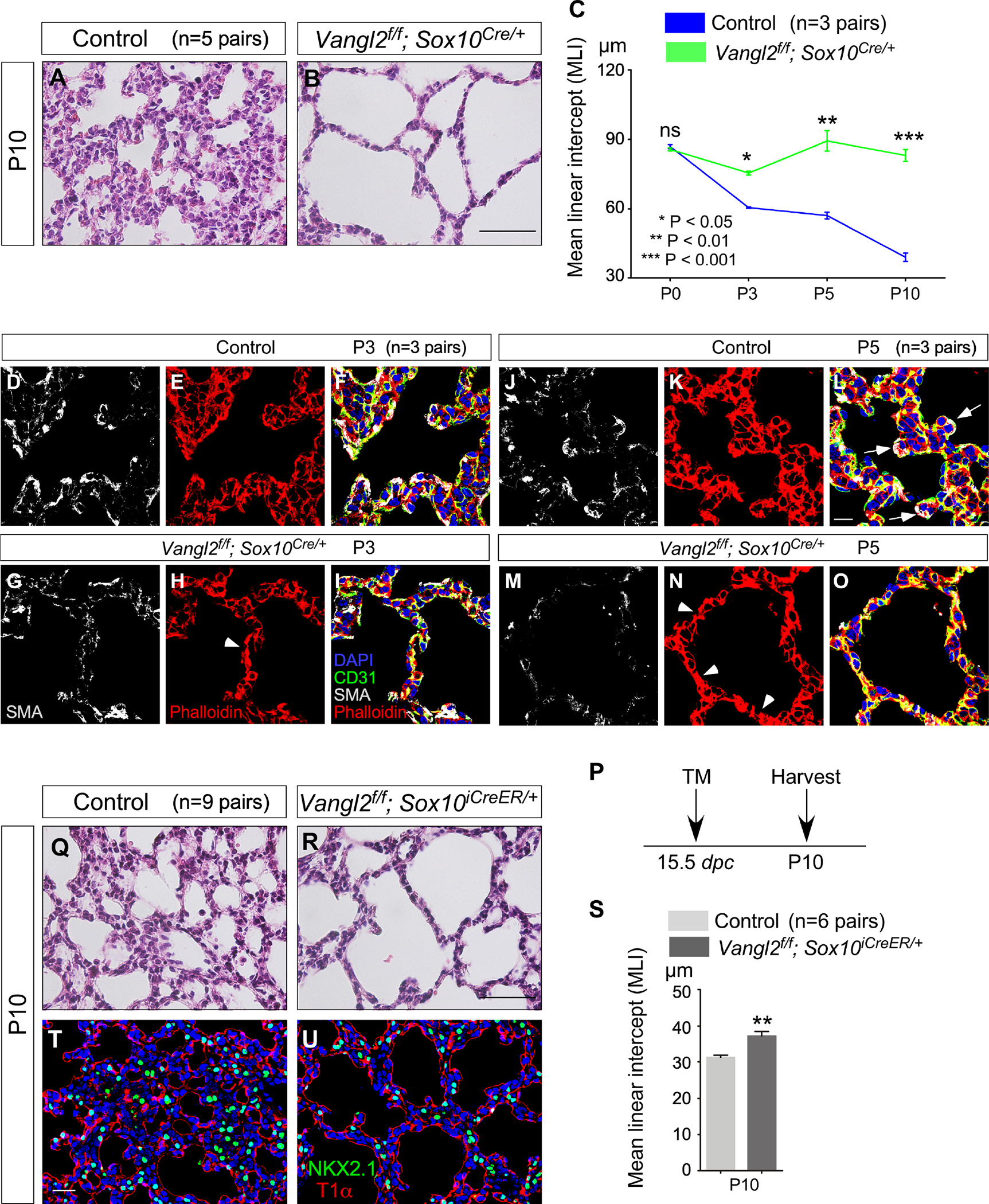
(A, B) Hematoxylin-and-eosin (H&E) staining of lung sections from control and Vangl2f/f; Sox10Cre/+ mice (n=5 pairs) at postnatal (P) day 10.
(C) Measurement of the mean linear intercept (MLI) in control and Vangl2f/f; Sox10Cre/+ lungs (n=3 pairs) at P0, P3, P5 and P10. The early phenotypes suggest that the requisite expansion and migration of myofibroblasts prior to alveologenesis are disrupted in the absence of PCP signaling in autonomic nerves.
(D-O) Immunofluorescence of lung sections from control and Vangl2f/f; Sox10Cre/+ (n=3 pairs for each stage) mice stained with anti-SMA (smooth muscle actin), anti-CD31 and phalloidin at P3 and P5. CD31 labels endothelial cells and phalloidin binds to F-actin. Arrows point to sites of secondary septation. Arrowheads indicate disorganized cytoskeleton in the mutant lungs.
(P) Schematic diagram of the time course of tamoxifen administration and tissue harvest for control and Vangl2f/f; Sox10iCreER/+ mice.
(Q, R) H&E staining of lung sections from control and Vangl2f/f; Sox10iCreER/+ mice (n=9 pairs) at P10.
(S) Measurement of the MLI in control and Vangl2f/f; Sox10iCreER/+ lungs (n=6 pairs) at P10.
(T, U) Immunofluorescence of lung sections from control and Vangl2f/f; Sox10iCreER/+ mice (n=3 pairs) stained with anti-NKX2.1 and T1α at P10. NKX2.1 labeled epithelial cells while T1α marked alveolar type I cells.
Scale bars, 100 μm (A, B), 10 μm (D-O), 100 μm (Q, R), 25 μm (T, U). All values are mean ± SEM. (*) p<0.05; (**) p<0.01; ns, not significant (unpaired Student’s t-test).
As an alternative approach, we produced Vangl2f/f; Sox10iCreER/+ (McKenzie et al., 2014) and administered tamoxifen to eliminate Vangl2. Vangl2f/f; Sox10iCreER/+ mice that had received tamoxifen at 15.5 days post coitus (dpc) exhibited regional alveolar defects (Figures 4P–4U) (without apparent defects in the airways or other organs), suggesting a direct effect of PCP signaling on local alveolar formation.
We set out to reveal the signaling cascade that controls Vangl2 function in neural crest progenitors. We tested the function of Ror2 in neural crest progenitors using a floxed allele of Ror2 (Ror2f) (Ho et al., 2012) and inspected Ror2f/f; Wnt1Cre/+ and Ror2f/f; Sox10Cre/+ mice. Unfortunately, both died soon after birth due to craniofacial defects. To overcome neonatal lethality, we produced Ror2f/f; Sox10iCreER/+ and administered tamoxifen to eliminate Ror2. Ror2f/f; Sox10iCreER/+ mice that had received tamoxifen at 15.5 dpc developed regional alveolar defects (Figures 5A–5F) (without apparent defects in the airways or other organs). Together, these studies establish a Ror2–Vangl1/2 signaling cascade, which functions in neural crest progenitors to regulate alveologenesis. Wnt5a mRNA is primarily expressed in myofibroblasts (Zhang et al., 2020) by PLISH (proximity ligation in situ hybridization) (Nagendran et al., 2018), consistent with the available data from single cell analysis (Guo et al., 2019). WNT5A has multiple targets in the lung (Zhang et al., 2020). Our findings suggest that WNT5A released from myofibroblasts could trigger a Fz/Ror2–Vangl1/2 cascade in neural crest progenitors and control autonomic function.
Figure 5. Planar cell polarity signaling controls neurotransmitter trafficking/release from autonomic nerve terminals and alveolar formation.
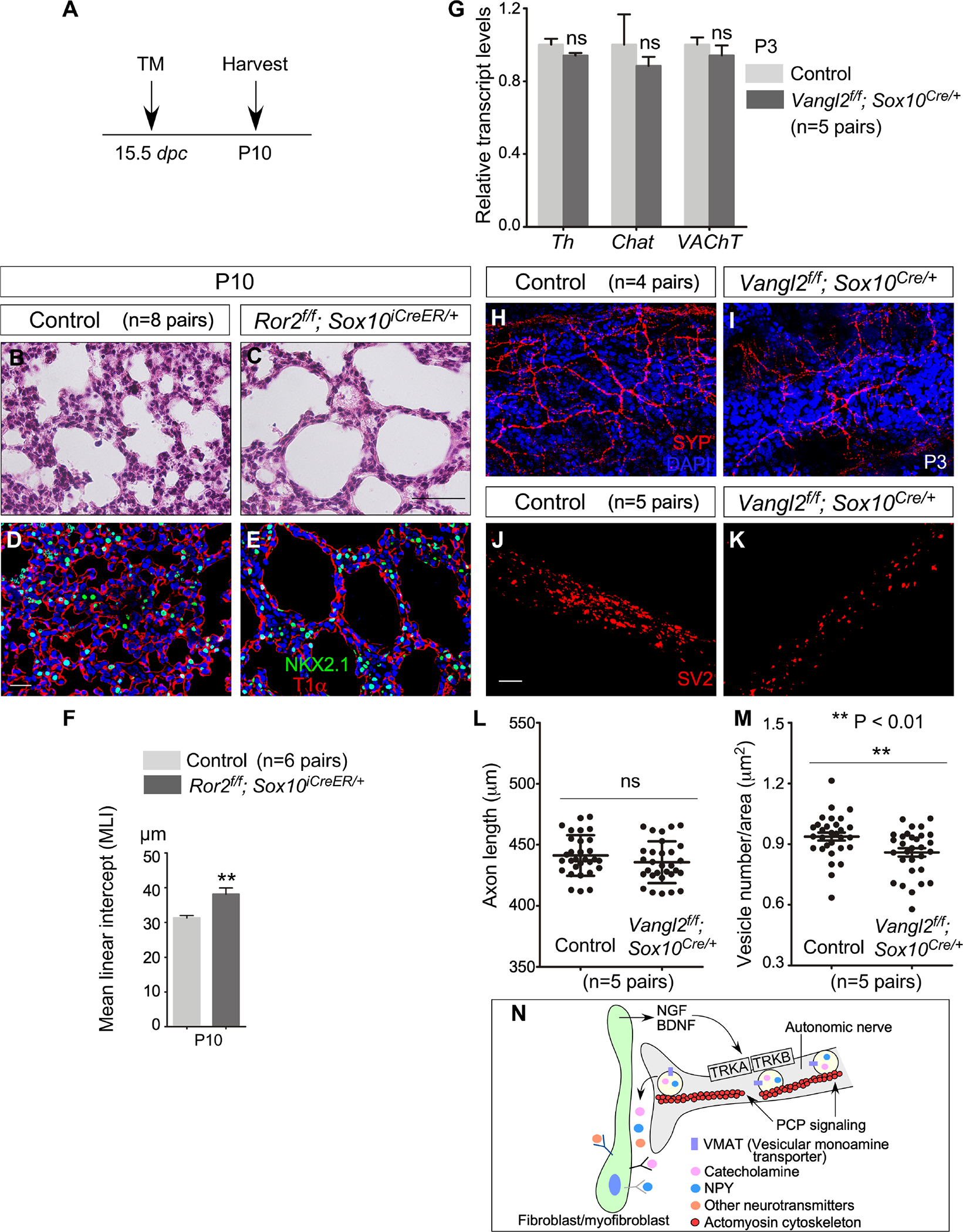
(A) Schematic diagram of the time course of tamoxifen administration and tissue harvest for control and Ror2f/f; Sox10iCreER/+ mice.
(B, C) H&E staining of lung sections from control and Ror2f/f; Sox10iCreER/+ mice (n=8 pairs) at P10.
(D, E) Immunofluorescence of lung sections from control and Ror2f/f; Sox10iCreER/+ mice (n=3 pairs) stained with anti-NKX2.1 and T1α at P10.
(F) Measurement of the MLI in control and Ror2f/f; Sox10iCreER/+ lungs (n=6 pairs) at P10.
(G) qPCR analysis of transcript levels in control and Vangl2f/f; Sox10Cre/+ lungs (n=5 pairs) at P3. The expression levels of Th, Chat and VAChT were unaltered in Vangl2f/f; Sox10Cre/+ lungs compared to controls.
(H, I) Immunofluorescence of lung sections from control and Vangl2f/f; Sox10Cre/+ mice (n=4 pairs) stained with anti-SYP at P3.
(J, K) Immunofluorescence of cultured neurons derived from the superior cervical ganglion (SCG) of control and Vangl2f/f; Sox10Cre/+ mice (n=5 pairs) stained with anti-SV2 to detect vesicles.
(L) Quantification of axon length of cultured neurons of SCG from control and Vangl2f/f; Sox10Cre/+ mice (n=5 pairs).
(M) Quantification of vesicle number of cultured neurons of SCG from control and Vangl2f/f; Sox10Cre/+ mice (n=5 pairs).
(N) Schematic diagram of regulation of neurotransmitter trafficking and release at the nerve terminals by PCP signaling.
Scale bars, 100 μm (B, C), 25 μm (D, E, H, I), 5 μm (J, K). All values are mean ± SEM. (*) p<0.05; (**) p<0.01; ns, not significant (unpaired Student’s t-test).
Neurotransmitters fail to be released from autonomic nerves when PCP signaling is eliminated
The distribution and density of autonomic nerves and neurotransmitter synthesis were mostly unperturbed in Vangl2f/f; Sox10Cre/+ lungs (Figures 5G–5I). We thus speculate that loss of PCP signaling impacts neurotransmitter trafficking and release from autonomic nerve terminals, and consequently autonomic function in the lung. In support of this idea, we recently reported disruption of intracellular trafficking and release of PDGF from alveolar epithelial cells due to loss of PCP signaling (Zhang et al., 2020). To test our hypothesis, neurons were dissociated from the superior cervical ganglions (SCG) of control and Vangl2f/f; Sox10Cre/+ mice and cultured following the established procedure (Amendola et al., 2015; He and Baas, 2003; Jackson and Tourtellotte, 2014). The axon length was indistinguishable between control and mutant neurons (Figures 5J–5L). This is consistent with the observation that the spatial distribution of nerves and expression of neurotransmitters did not appear to be perturbed in Vangl2f/f; Sox10Cre/+ lungs (Figures 5H and 5I). However, the number of vesicles (labeled by SV2) was reduced in axons of SCG neurons derived from Vangl2f/f; Sox10Cre/+ mice compared to controls (Figures 5J, 5K and 5M). These results favor a model in which PCP signaling is required for neurotransmitter trafficking and release from the autonomic nerves (Figure 5N).
Myofibroblasts are a target of autonomic innervation in the lungs
Autonomic nerves that extend to the vicinity of developing saccules could influence the function of one or several components of the secondary septa during alveolar formation. To examine lung cell types in the neighborhood of autonomic nerves, we employed PdgfraCreER/+; ROSA26mTmG/+ and PdgfraCre/+; ROSA26mTmG/+ mice (Kang et al., 2010; Roesch et al., 2008) to label fibroblasts/myofibroblasts (PDGFRA+) with GFP (Figures 6A–6H). Myofibroblasts express PDGFRA, the receptor for platelet-derived growth factor (PDGF), and smooth muscle actin (SMA), and proliferate and migrate in response to PDGF signaling. Myofibroblasts play an active role in promoting secondary septation. PdgfraCreER offered the advantage of labeling sparse, individual myofibroblasts by tamoxifen administration to detect their cellular extensions while PdgfraCre enabled visualization of the myofibroblast network. In lungs from PdgfraCreER/+; ROSA26mTmG/+ and PdgfraCre/+; ROSA26mTmG/+ mice harvested at P3 and P5, myofibroblasts within saccules sent out cytoplasmic extensions (Figures 6A and 6B), which formed a network (Figures 6C–6H), and were in close proximity to or in contact with nerve fibers labeled by NF-M (2H3) or SYP (Figures 6I–6K). This raised the possibility that neurotransmitters released by autonomic nerves affect myofibroblast function such as its proliferation and migration prior to/during alveologenesis.
Figure 6. Alveolar myofibroblasts are targets of autonomic innervation.
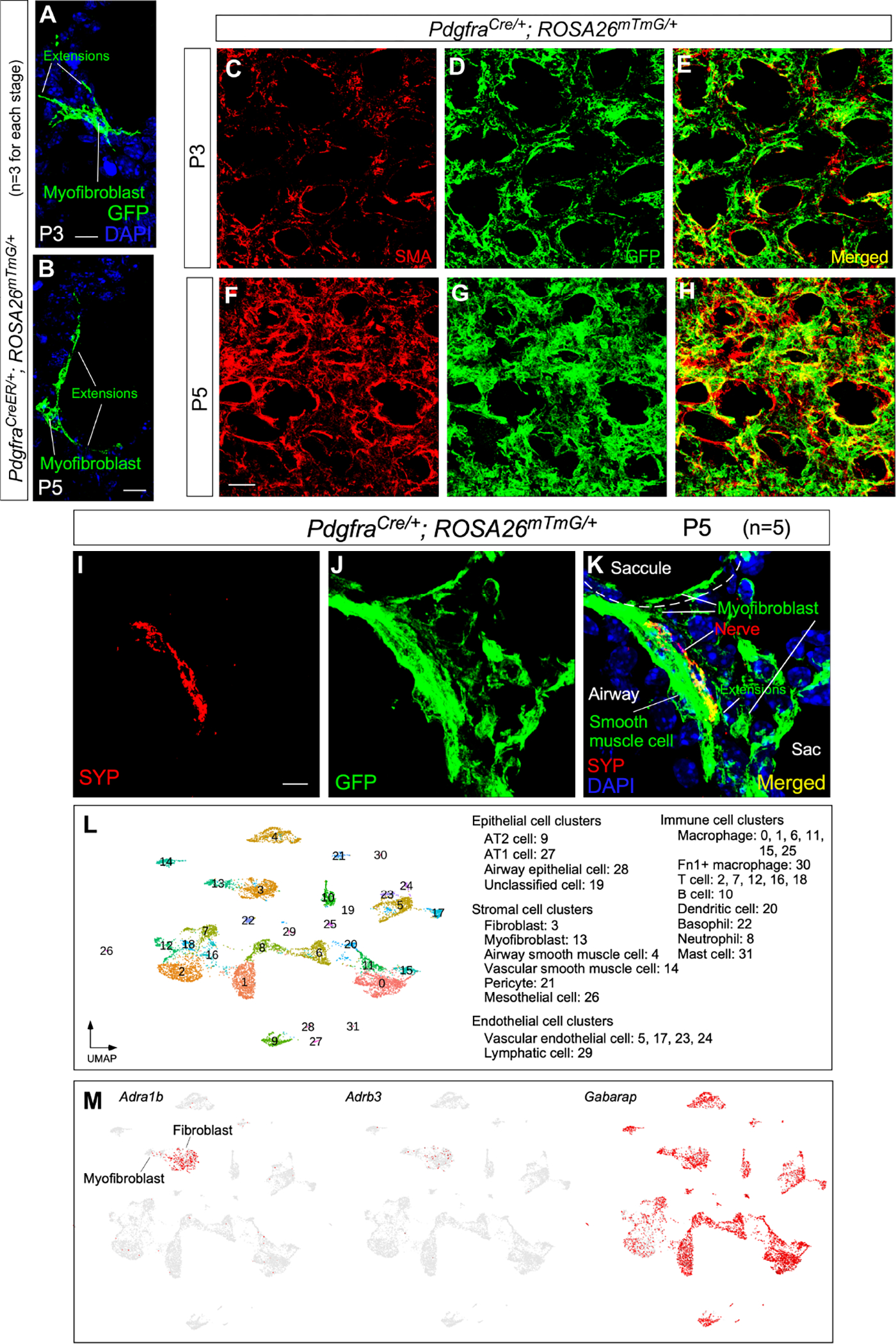
(A, B) Immunofluorescence of lung sections from PdgfraCreER/+; ROSA26mTmG/+ mice (n=3) at P3 and P5 stained with anti-GFP. Leaky expression of Pdgfra-CreER labeled single myofibroblasts with GFP induced from ROSA26mTmG. Myofibroblasts sent out cellular extensions.
(C-H) Immunofluorescence of lung sections from PdgfraCre/+; ROSA26mTmG/+ mice (n=3 for each stage) at P3 and P5 stained with anti-GFP. Myofibroblasts expressed Pdgfra-Cre and were labeled by GFP produced from ROSA26mTmG. Cellular extensions of myofibroblasts were interconnected to form a network, which was apparent by P5.
(I-K) Immunofluorescence of lung sections from PdgfraCre/+; ROSA26mTmG/+ mice (n=5) at P5 stained with anti-GFP and anti-SYP. Nerve fibers (SYP+) could be within 0.1 μm of cellular extensions of myofibroblasts (GFP+) located in saccules.
(L) Single-cell RNA-seq (scRNA-seq) analysis of mouse distal lung cells. Cell clusters of distal lung cells were displayed on UMAP.
(M) UMAP plots of cell clusters that expressed Adra1b, Adrb3 (receptors) and Gabarap across all cell clusters, darker red indicating higher relative expression.
Scale bars, 10 μm (A, B), 25 μm (C-H), 5 μm (I-K).
To further test this hypothesis, we dissected distal murine lungs at P3 and sorted lung cells for single-cell RNA-seq (scRNA-seq) analysis (Mizikova and Thebaud, 2021) (Figure 7A). Receptors for neurotransmitters were present in myofibroblasts, supporting our model in which myofibroblasts are a target of autonomic innervation in the lungs (Figures 6L and 6M and Figure S4).
Figure 7. A functional interaction between alveolar fibroblasts/myofibroblasts and autonomic nerves is essential for alveologenesis.
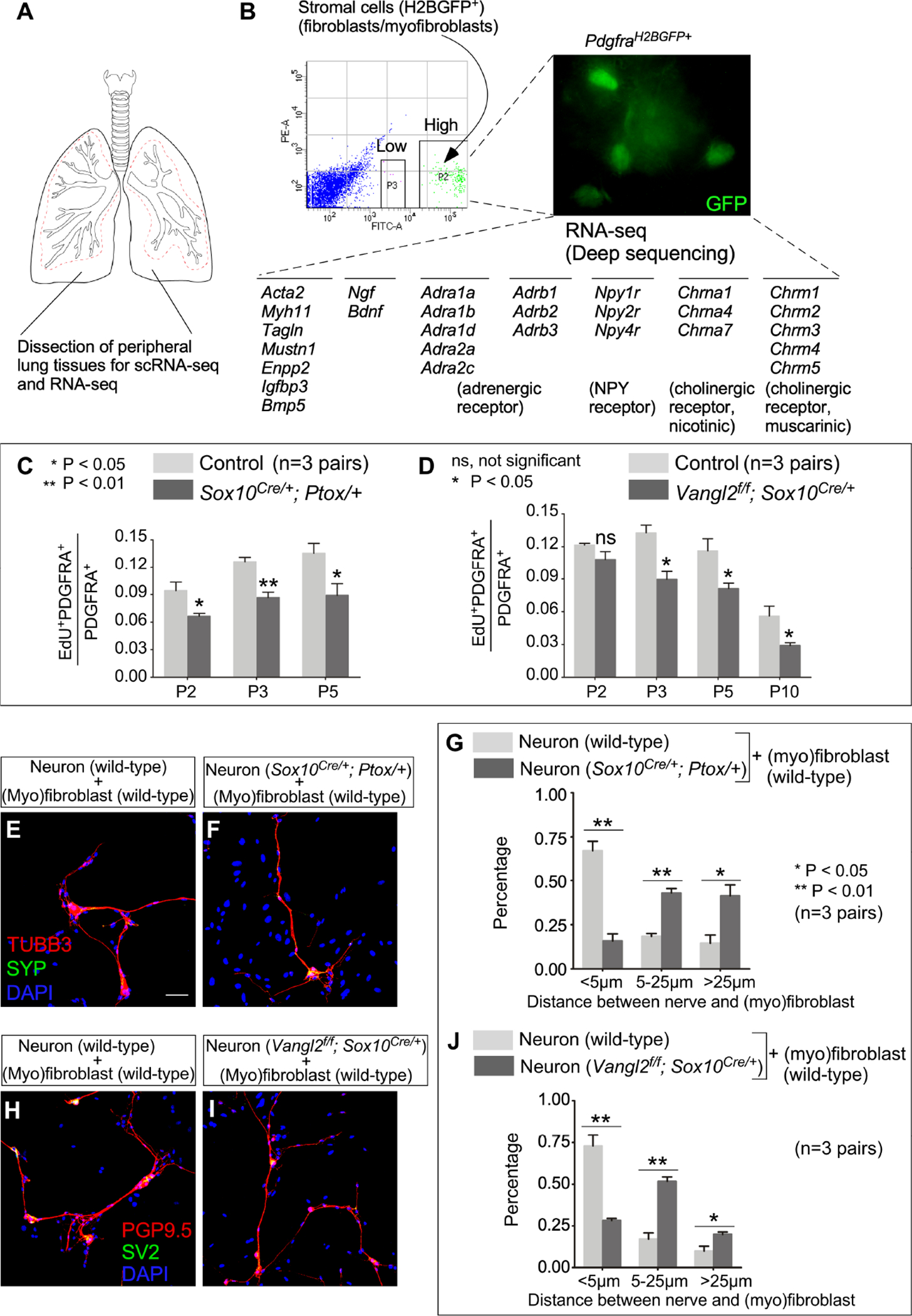
(A) Schematic diagram of dissection of peripheral lung tissues for scRNA-seq and RNA-seq.
(B) RNA-seq analysis of sorted murine myofibroblasts. (Myo)fibroblasts (GFP+) were isolated from the lungs of PdgfraH2BGFP/+ mice at postnatal (P) day 3 by fluorescence-activated cell sorting (FACS). A partial list of genes involved in neurotrophin signaling and synaptic transmission in sympathetic and parasympathetic nerves were shown.
(C) Quantification of the ratio of proliferating (myo)fibroblasts (EdU+PDGFRA+) to (myo)fibroblasts (PDGFRA+) in control and Sox10Cre/+; Ptox/+ lungs (n=3 pairs) at P2, P3 and P5.
(D) Quantification of the ratio of proliferating (myo)fibroblasts (EdU+PDGFRA+) to (myo)fibroblasts (PDGFRA+) in control and Vangl2f/f; Sox10Cre/+ lungs (n=3 pairs) at P2, P3, P5 and P10.
(E, F) Immunofluorescence of cultured neurons and (myo)fibroblasts.
(G) Quantification of the distance between nerves and (myo)fibroblasts, which was measured as the shortest distance between the nuclei of (myo)fibroblasts and the nerve fibers.
(H, I) Immunofluorescence of cultured neurons and (myo)fibroblasts.
(J) Quantification of the distance between nerves and (myo)fibroblasts, which was measured as the shortest distance between the nuclei of (myo)fibroblasts and the nerve fibers.
Scale bars, 100 μm (E, F, H, I). All values are mean ± SEM. (*) p<0.05; (**) p<0.01; ns, not significant (unpaired Student’s t-test).
Loss of autonomic nerve function or activity leads to impaired proliferation and migration of (myo)fibroblasts
scRNA-seq of lung cells (Figures 6L and 6M) and RNA-Seq (Figure 7B) of isolated myofibroblasts uncovered expression of receptors for neurotransmitters in myofibroblasts, characteristic of targets of autonomic innervation. Indeed, loss of adrenergic receptors (Rohrer et al., 1999; Susulic et al., 1995) resulted in alveolar defects (Figure S5). To assess if autonomic innervation impacts fibroblast/myofibroblast [abbreviated as (myo)fibroblast] function, we analyzed (myo)fibroblast proliferation by EdU incorporation in multiple mouse lines with disrupted autonomic innervation or function. (Myo)fibroblast proliferation was reduced in Vangl2f/f; Sox10Cre/+ and Sox10Cre/+; RC::Ptox/+ lungs (Figures 7C and 7D and Figure S6). Moreover, myofibroblasts failed to migrate to the prospective sites of secondary septation in lungs deficient in neurotrophin signaling (Figure S7). In support of this observation, the rate of (myo)fibroblast migration in wound healing assays in response to conditioned media derived from PC12 (pheochromocytoma) cells (Zerby and Ewing, 1996) was enhanced when the conditioned media was treated with KCl to release neurotransmitters, mainly catecholamines (Figure S7). Together, these results suggest that neurotransmitters from autonomic nerves can promote (myo)fibroblast proliferation and migration.
Fibroblasts/Myofibroblasts aggregate around autonomic nerve fibers in coculture
Our results support a functional interaction between autonomic nerves and (myo)fibroblasts. To further test this idea, we performed coculture of SCG neurons and fibroblasts/myofibroblasts derived from the distal lungs of wild-type mice. Fibroblasts/myofibroblasts were found to accumulate along the nerve fibers (Figure S8). By contrast, aggregation of wild-type fibroblasts/myofibroblasts failed to occur when cocultured with SCG neurons derived from Sox10Cre/+; RC::Ptox/+ (Figures 7E–7G) or Vangl2f/f; Sox10Cre/+ (Figures 7H–7J) mice. Our results suggest that neurotransmitter release was impaired in the mutant neurons. Together, these results support a role of neurotransmitters in regulating fibroblasts/myofibroblasts migration.
Neurotrophins secreted from fibroblasts/myofibroblasts control autonomic nerve function and alveolar formation
As a target of autonomic innervation, myofibroblasts likely produce neurotrophic factors for nerve growth and survival. The trophic interaction between neurons and their targets is exemplified by the neuromuscular junction. Indeed, our scRNA-seq and RNA-Seq analysis in this study disclosed the presence of Bdnf and Ngf in myofibroblasts (Figure 8A).
Figure 8. Alveolar fibroblasts/myofibroblasts are essential for neural function during alveologenesis.
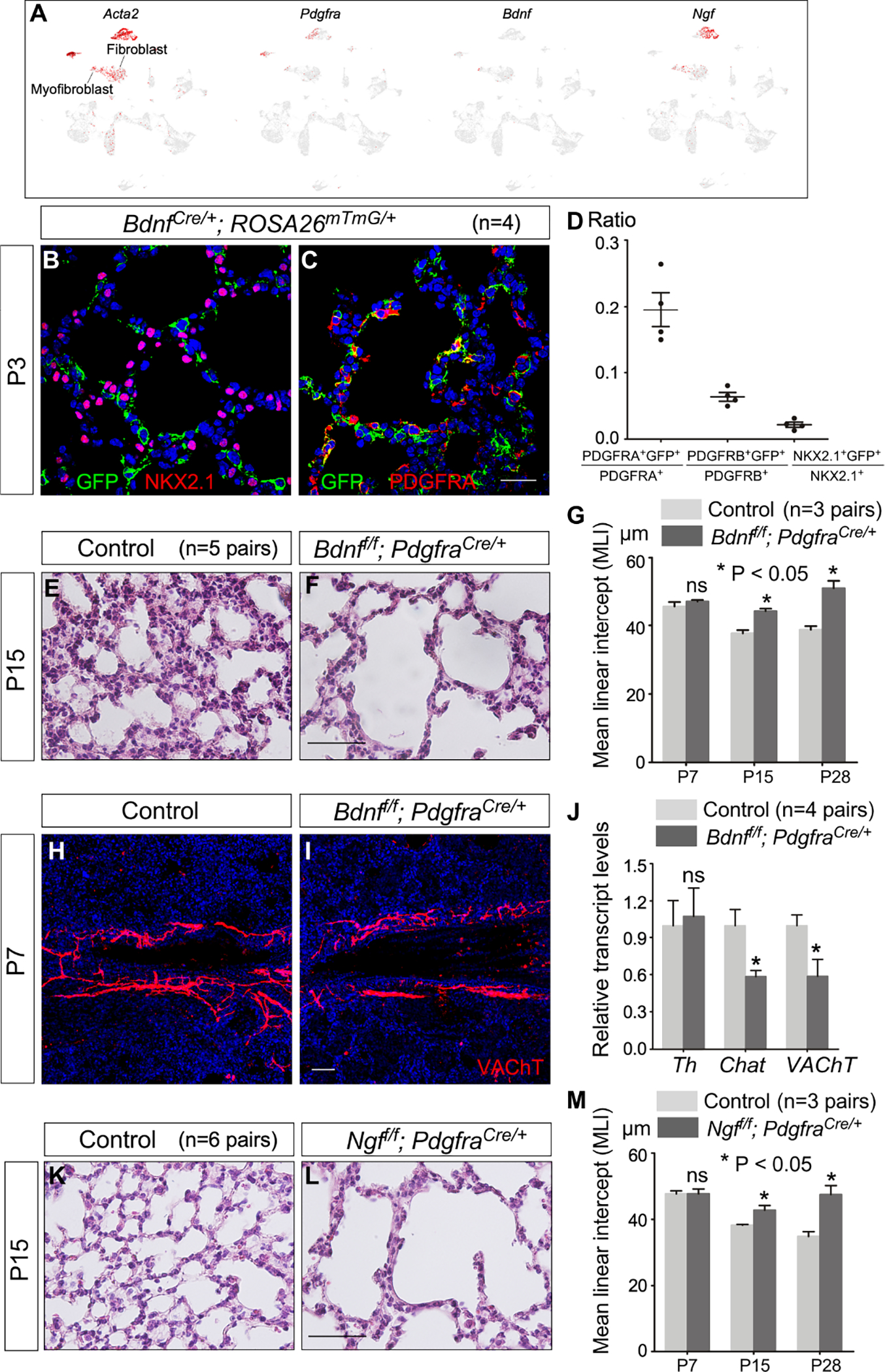
(A) Single-cell RNA-seq (scRNA-seq) analysis of mouse distal lung cells. UMAP plots of cell clusters that expressed Acta2, Pdgfra, Bdnf and Ngf across all cell clusters, darker red indicating higher relative expression. Bdnf and Ngf were also expressed in airway smooth muscle and vascular smooth muscle cells, two known sites of neurotrophin production. We also noted the incomplete nature of the existing scRNA-seq data. For instance, Ngf is not detected in any lung cell types by the publicly available scRNA-seq (LungMAP). Interestingly, Ngf-LacZ signal in the lungs of NgfLacZ/+ mice was high in pulmonary neuroendocrine cells (PNECs).
(B, C) Immunofluorescence of lung sections from BdnfCre/+; ROSA26mTmG/+ mice (n=4 pairs) stained with anti-GFP, anti-NKX2.1 and anti-PDGFRA at P3. NKX2.1 labeled epithelial cells, PDGFRA marked fibroblasts/myofibroblasts and PDGFRB detected pericytes. Of note, removal of Bdnf by Sox9Cre in the lung epithelium did not lead to alveolar defects.
(D) Quantification of the percentage of (myo)fibroblasts, pericytes and epithelial cells labeled by Bdnf-Cre, respectively. Expression of Bdnf-Cre induced GFP expression from ROSA26mTmG/+.
(E, F) Hematoxylin-and-eosin (H&E) staining of lung sections from control and Bdnff/f; PdgfraCre/+ mice (n=5 pairs) at P15.
(G) Measurement of the mean linear intercept (MLI) in control and Bdnff/f; PdgfraCre/+ mice (n=3 pairs) at P7, P15 and P28.
(H, I) Immunofluorescence of lung sections from Bdnff/f; PdgfraCre/+ mice (n=3 pairs) stained with anti-VAChT at P7. VAChT marked parasympathetic nerves.
(J) qPCR analysis of transcript levels in control and Bdnff/f; PdgfraCre/+ lungs (n=4 pairs) at P7. The expression levels of Chat and VAChT were reduced in Bdnff/f; PdgfraCre/+ lungs compared to controls.
(K, L) H&E staining of lung sections from control and Ngff/f; PdgfraCre/+ mice (n=6 pairs) at P15.
(M) Measurement of the MLI in control and Ngff/f; PdgfraCre/+ mice (n=3 pairs) at P7, P15 and P28.
Scale bars, 25 μm (B, C), 100 μm (E, F, K, L), 50 μm (H, I). All values are mean ± SEM. (*) p<0.05; ns, not significant (unpaired Student’s t-test).
We directly examined the expression of Bdnf in the lungs, taking advantage of BdnfCre (Tan et al., 2016). In BdnfCre/+; ROSA26mTmG/+ mice, GFP was induced by Cre in Bdnf-expressing cells. We found that ~20% of (myo)fibroblasts were labeled by GFP while GFP signal was barely detected in alveolar epithelial cells (NKX2.1+) (Figures 8B–8D and Figure S9). In addition, scRNA-seq data at P3 showed that 13.5% of alveolar myofibroblasts, 5.4% of alveolar fibroblasts, 2.6% of alveolar epithelial cells, 5.9% of pericytes, 8% of airway/vascular smooth muscle cells, and 2.5% of endothelial cells express Bdnf. Thus, alveolar fibroblasts/myofibroblasts are the major BDNF-producing cells during alveolar development.
To evaluate whether BDNF secretion from (myo)fibroblasts could promote neurite extension and survival, we selectively inactivated Bdnf in (myo)fibroblasts using PdgfraCre (Roesch et al., 2008) to convert a floxed allele of Bdnf (Bdnff) (Rios et al., 2001) to a null allele. Autonomic function was disrupted in Bdnff/f; PdgfraCre/+ mice (Figures 8H–8J), which developed alveolar defects (Figures 8E–8G).
We then investigated the functional role of NGF produced in (myo)fibroblasts. SCG neurons in culture are dependent on NGF for their survival and maintenance of neuronal properties (e.g., production of vesicles) (Figure S10). Importantly, Ngff/f; PdgfraCre/+ mice, in which a floxed allele of Ngf (Ngff) (Muller et al., 2012) was converted to a null allele in (myo)fibroblasts, exhibited alveolar defects (Figures 8K–8M). These results indicate a trophic role of fibroblasts/myofibroblasts on autonomic nerves in the lung. Of note, few alveolar epithelial cells or (myo)fibroblasts express TrkA/TrkB by scRNA-seq at postnatal day 3. Interestingly, alveolar type II (AT2) cells exhibited increased growth in response to BDNF signaling during alveolar repair (Paris et al., 2020). This implies that alveolar epithelial cells could be a target of NGF/BDNF signaling during alveologenesis.
We did not observe uniform innervation of saccules and (myo)fibroblasts. Nevertheless, myofibroblasts within and across saccules are connected through cellular extensions. Thus, we anticipate that the effect of autonomic innervation will be amplified through a network of myofibroblasts and diffusion of neurotransmitters within and across saccules. Together, our work has identified a reciprocal trophic relationship between the autonomic nerves and fibroblasts/myofibroblasts. This functional unit mediates the effect of autonomic nerves on controlling alveolar formation.
Discussion
The discovery of autonomic control of alveolar formation through (myo)fibroblast innervation has opened up a new area of investigation and changed our fundamental understanding of alveologenesis. Our work provides several lines of evidence to support a direct effect of autonomic innervation on alveolar formation and delineates a conceptual framework of alveologenesis. They include the proximity of nerve fibers to (myo)fibroblasts, a functional consequence of disrupting autonomic innervation and activity, a signaling cascade that regulates neurotransmitter trafficking and release, and a reciprocal trophic relationship between (myo)fibroblasts and autonomic nerves. These studies not only offer mechanistic insights into alveologenesis but also shed light on understanding the disease mechanisms of COPD and its treatment. If alveolar repair/regeneration reactivates programs for alveologenesis, we anticipate that autonomic nerve function is also essential in the process of repairing/regenerating damaged alveoli.
scRNA-seq is a powerful tool to identify gene expression in individual cell clusters. However, the functional roles of these genes in animals, regardless of their widespread or restricted expression, can only be revealed through in vivo studies. For alveolar formation, there is no organoid model that recapitulates the production of the alveolus. Our genetic analysis in mice enabled us to pinpoint the key functions of neurotrophin and PCP signaling in neural crest progenitors and (myo)fibroblasts to control alveologenesis despite their broad expression. In this study, we did not test all the components of neurotrophin and PCP signaling. For instance, TrkC (a high-affinity neurotrophin receptor and p75 a (a low-affinity neurotrophin receptor) could play a minor role in nerve development in the lung. Similarly, we have initially focused on a Ror2–Vangl1/2 axis in controlling neural development, but cannot rule out the possibility that many Fz receptors and Ror1 play a role in neural development in the lung. Notwithstanding, a combinatorial use of select members of signaling cascades in diverse tissues seems to be a common feature and confers unique properties of a given tissue.
It is interesting to note the effects of sympathectomy or vagotomy on postnatal lungs have been reported (Dotta and Mortola, 1992; Mortola et al., 1987; Wong et al., 1998). These experiments did not examine alveolar development nor was molecular analysis performed. It is also unclear how denervation hypersensitivity of target tissues and collateral sprouting of nerves have contributed to target tissue function in these studies. To reexamine the issue of innervation and alveologenesis, we have used modern genetic and molecular tools in this study to selectively manipulate the signals and receptions in distinct cell populations. Our work has revealed a autonomic nerve–(myo)fibroblast circuit that regulates alveologenesis.
The nature of nerve endings in lung saccules is unknown. It is possible that saccules are innervated by free nerve endings, i.e., the presence of pre-synaptic clear core vesicles (classical neurotransmitters) or dense core vesicles (neuropeptides) without post-synaptic densities. In this case, neurotransmitters will reach the targets likely by diffusion. It is also possible that the nerve endings form specialized structures on the targets (such as fibroblasts/myofibroblasts). It remains to be determined if a autonomic nerve–(myo)fibroblast circuit is functionally analogous to the neuromuscular junction (NMJ).
We did not observe uniform innervation of saccules and (myo)fibroblasts. Nevertheless, myofibroblasts within and across saccules are connected through cellular extensions. Thus, we anticipate that the effect of autonomic innervation will be amplified through a network of myofibroblasts and diffusion of neurotransmitters within and across saccules.
Our analysis has revealed defects in myofibroblast migration and aggregation when autonomic nerve function is disrupted. We speculate that neurotransmitters regulate myofibroblast contraction, an important property for myofibroblast migration and aggregation. We propose that neurotransmitters regulate the actomyosin cytoskeleton in myofibroblasts, which is essential for their contraction and migration. However, it is possible that various neurotransmitters released from sympathetic and parasympathetic nerves regulate other aspects of myofibroblast function.
Our investigation has identified myofibroblasts as a main target of autonomic innervation in saccules and alveoli. Whether alveolar epithelial or endothelial cells/pericytes are also targets of autonomic nerves requires future genetic and molecular investigation that utilizes a similar approach. We cannot rule out the possibility that the effects of autonomic nerves on alveologenesis are indirect. They could be mediated by mechanisms such as surfactant homeostasis, alveolar fluid clearance, peristalsis of airways via airway smooth muscle cells and others.
Taken together, the functional unit of autonomic nerves and alveolar myofibroblasts represents a new mode of cell-cell interactions. In conjunction with signaling cascades, the autonomic-myofibroblast unit confers cellular properties essential for alveolar formation. It also significantly expands our understanding of how nerves interact with tissues. Our work illustrates the fundamental principle of how two tissues (e.g., nerves and lungs) interact to build alveoli at the organismal level. They also establish the foundation for treating lung diseases caused by loss of alveoli.
Our results raise the question of whether studies in mice recapitulate human lung diseases such as COPD/emphysema. We previously documented a reduction in the expression levels of WNT5A and VANGL2 in COPD/emphysema lungs (Zhang et al., 2020), suggesting a link between PCP signaling and alveolar loss/repair in these patients. We found that expression of several neurotransmitters or enzymes involved in neurotransmitter synthesis was perturbed in COPD/emphysema lungs (Figure S11). These findings set the stage for future investigations into the role of autonomic function in alveolar repair in mammals.
Limitations of the study
Many of the Cre lines used in this study label multiple cell populations in several organs, posing a general problem for interpreting the phenotypes. Attribution of the phenotypes to a given lineage cannot be absolutely ascertained. For instance, Wnt1Cre and Sox10Cre are expressed in neural crest progenitors that give rise to multiple lineages including the autonomic nerves. Similarly, PdgfraCre is expressed in subsets of fibroblasts and myofibroblasts. Moreover, due to Cre expression outside the lungs, we cannot rule out the possibility that their effects on other organs have contributed to the alveolar phenotypes. Bdnf and Ngf are broadly expressed and we have only functionally tested their activity in fibroblasts/myofibroblasts in this study. We also cannot rule out the contribution of systemic factors from the adrenal gland (especially the adrenal medulla) to alveolar formation. It is difficult to discern the contributions of myofibroblast migration and contraction to secondary septation. A collective interpretation of the expression and functional data from multiple Cre lines offers new insight and a model to which one single piece of data cannot lead.
STAR Methods
RESOURCES AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed and will be fulfilled by the lead contact, Pao-Tien Chuang (pao-tien.chuang@ucsf.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
RNA-seq and scRNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Any additional information relating to the data reported in this paper is available from the lead contact upon request. This paper does not report original code.
Key resources table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-Acetylated tubulin | MilliporeSigma | Cat# T6793; RRID:AB_477585 |
| Mouse monoclonal anti-ACTA2 | Thermo Scientific Lab Vision | Cat# MS-113-P0; RRID:AB_64001 |
| Goat polyclonal anti-CC10 (S-20) | Santa Cruz Biotechnology | Cat# sc-9773; RRID:AB_2183391 |
| Chicken polyclonal anti-GFP | Aves Labs | Cat# GFP-1010; RRID:AB_2307313 |
| Mouse monoclonal anti-HOP (E-1) | Santa Cruz Biotechnology | Cat# sc-398703; RRID:AB_2687966 |
| Mouse monoclonal anti-Neurofilament (NF-M) | Developmental Studies Hybridoma Bank | Cat# 2H3; RRID:AB_531793 |
| Rabbit monoclonal anti-NKX2.1 | Epitomics | Cat# 2044-1; RRID:AB_1267367 |
| Rabbit polyclonal anti-PDGF receptor alpha | Cell Signaling Technology | Cat# 3164; RRID:AB_2162351 |
| Rabbit monoclonal anti-Phospho-PDGF Receptor α (Tyr754) (23B2) | Cell Signaling Technology | Cat# 2992; RRID:AB_390728 |
| Goat polyclonal anti-PDGF R beta | R&D Systems | Cat# AF1042; RRID:AB_2162633 |
| Rat monoclonal anti-PECAM-1 (MEC 13.3) (CD31) | Santa Cruz Biotechnology | Cat# sc-18916; RRID:AB_627028 |
| Rabbit polyclonal anti-PGP9.5 | AnaSpec | Cat# 53772; RRID:AB_2272470 |
| Rabbit polyclonal anti-prosurfactant protein C (proSP-C) | MilliporeSigma | Cat# AB3786; RRID:AB_91588 |
| Mouse monoclonal anti-SV2 | Developmental StudiesHybridoma Bank | Cat# SV2; RRID:AB_2315387 |
| Rabbit monoclonal anti-Synaptophysin (SYP) | Invitrogen | Cat# 180130; RRID:AB_10836766 |
| Hamster monoclonal anti-T1α | Developmental Studies Hybridoma Bank | Cat# 8.1.1; RRID:AB_531893 |
| Mouse monoclonal anti-TUBB3 | Developmental Studies Hybridoma Bank | Cat# 6G7; RRID:AB_528497 |
| Rabbit polyclonal anti-Tyrosine hydroxylase | Synaptic Systems | Cat# 213 102; RRID:AB_2619896 |
| Rabbit polyclonal anti-VAChT | Synaptic Systems | Cat# 139 103; RRID:AB_887864 |
| Rat monoclonal anti-VANGL2 clone 2G4 | MilliporeSigma | Cat# MABN750; RRID:AB_2721170 |
| Bacterial and virus strains | ||
| Biological samples | ||
| Human emphysema/COPD patient tissues | Paul Wolters, University of California, San Francisco | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| 5-Bromo-4-Chloro-3-Indolyl-D-Galactoside (X-gal) | Research Products International | Cat# B71800-1.0 |
| Collagenase B | Roche | Cat# 11088815001 |
| 2’-Deoxy-5-ethynyluridine (EdU) | BiosynthCarbosynth | Cat# 61135-33-9 |
| Deoxyribonuclease I from bovine pancreas | MilliporeSigma | Cat# 9003-98-9 |
| Dispase | BD Biosciences | Cat# 354235 |
| DMEM | Mediatech | Cat# 10-013-CV |
| Fetal Bovine Serum (FBS) | Gibco | Cat# 10437-028 |
| Fibronectin | Corning | Cat# 354008 |
| Glutaraldehyde, 8% aqueous solution, EM grade | Electron Microscopy Sciences | Cat# 16000 |
| 4-Hydroxytamoxifen (4-OHT) | Toronto Research Chemicals | Cat# H954705 |
| Nerve Growth Factor (NGF 2.5S) | Gibco | Cat# 13257-019 |
| Paraformaldehyde | MilliporeSigma | Cat# P6148 |
| Paraformaldehyde, 16% solution, EM grade | Electron Microscopy Sciences | Cat# 15700 |
| Penicillin/Streptomycin | Gibco | Cat# 15070-063 |
| 1X RBC Lysis Buffer | Invitrogen | Cat# 00-4333-57 |
| Rhodamine-conjugated phalloidin | Molecular Probes | Cat# R415 |
| RNase-free DNase | QIAGEN | Cat# 79254 |
| Tamoxifen | Toronto Research Chemicals | Cat# T006000 |
| TRIzol® Reagent | Ambion | Cat# 15596018 |
| Critical commercial assays | ||
| Click-iT™ EdU Alexa Fluor™ 488 Imaging Kit | Thermo Fisher | Cat# C10337 |
| Maxima First Strand cDNA Synthesis Kit | Thermo Scientific | Cat# K1641 |
| RNeasy Mini Kit | QIAGEN | Cat# 74104 |
| RNeasy Plus Mini Kit | QIAGEN | Cat# 74134 |
| RNeasy Plus Micro Kit | QIAGEN | Cat# 74034 |
| TSA Plus Cyanine 3 (Cy3) Fluorescein detection kit | Perkin-Elmer | Cat# NEL753001KT |
| Two-well culture insert | Ibidi | Cat# 80209 |
| Deposited data | ||
| Bulk RNA-Seq data for peripheral lungs of PdgfraH2BGFP mice at P3 | This paper | GSE189466 |
| scRNA-Seq data for peripheral lungs of control mice at P3 | This paper | GSE189466 |
| Experimental models: Cell lines | ||
| Primary fibroblasts derived from peripheral lungs of control mice at P3 | This paper | N/A |
| Primary neurons derived from the superior cervical ganglion (SCG) of Ptox; Sox10Cre mice at P3 | This paper | N/A |
| Primary neurons derived from the superior cervical ganglion (SCG) of Sox10Cre mice at P3 | This paper | N/A |
| Primary neurons derived from the superior cervical ganglion (SCG) of Vangl2f/f; Sox10Cre mice at P3 | This paper | N/A |
| Rat: PC12 cells | ATCC | CRL-1721 |
| Experimental models: Organisms/strains | ||
| Mouse: Adrb1− [FVB.129-Adrb1tm1Bkk/J] | The Jackson Laboratory | Stock# 031490 |
| Mouse: Adrb2− [FVB.129-Adrb2tm1Bkk/J] | The Jackson Laboratory | Stock# 031496 |
| Mouse: Adrb3− [FVB/N-Adrb3tm1Lowl/J] | The Jackson Laboratory | Stock# 006402 |
| Mouse: BdnfCre [B6.FVB-Bdnfem1(cre)Zak/J] | The Jackson Laboratory | Stock# 030189 |
| Mouse: Bdnff [STOCK Bdnftm3Jae/J] | The Jackson Laboratory | Stock# 004339 |
| Mouse: ChatCre [B6;129S6-Chattm2(cre)Lowl/J] | The Jackson Laboratory | Stock# 006410 |
| Mouse: GCaMP6f [B6;129S-Gt(ROSA)26Sortm95.1(CAG-GCaMP6f)Hze/J] | The Jackson Laboratory | Stock# 024105 |
| Mouse: Ngff | Dr. Liliana Minichiello | N/A |
| Mouse: NgfLacZ [Ngftm1Ddg] | Dr. David Ginty | N/A |
| Mouse: PDGFRaCre [C57BL/6-Tg(Pdgfra-cre)1Clc/J] | The Jackson Laboratory | Stock# 013148 |
| Mouse: PDGFRaCreER [B6N.Cg-Tg(Pdgfra-cre/ERT)467Dbe/J] | The Jackson Laboratory | Stock# 018280 |
| Mouse: PDGFRaH2BGFP [B6.129S4-Pdgfratm11(EGFP)Sor/J] | The Jackson Laboratory | Stock# 007669 |
| Mouse: RC::PFtox [B6;129S6-Gt(ROSA)26Sortm8(CAG-mCherry,-EGFP)Dym/J] | The Jackson Laboratory | Stock# 029486 |
| Mouse: Ror2f [B6;129S4-Ror2tm1.1Meg/J] | The Jackson Laboratory | Stock# 018354 |
| Mouse: ROSA26mTmG [B6.129(Cg)-Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J] | The Jackson Laboratory | Stock# 007676 |
| Mouse: R26R [B6.129S4-Gt(ROSA)26Sortm1Sor/J] | The Jackson Laboratory | Stock# 003474 |
| Mouse: ROSA26tdTomato [B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J] | The Jackson Laboratory | Stock# 007914 |
| Mouse: Sox10Cre [B6;CBA-Tg(Sox10-cre)1Wdr/J] | The Jackson Laboratory | Stock# 025807 |
| Mouse: Sox10iCreER [CBA;B6-Tg(Sox10-icre/ERT2)388Wdr/J] | The Jackson Laboratory | Stock# 027651 |
| Mouse: ThCre [B6;129X1-Thtm1(cre)Te/Kieg] | Dr. Ted Ebendal | N/A |
| Mouse: TrkAf [B6.129P2(SJL)-Ntrk1tm1Ddg/J] | The Jackson Laboratory | Stock# 022362 |
| Mouse: TrkBf [Ntrk2tm1.1Lfr] | Dr. Baoji Xu | N/A |
| Mouse: Vangl1gt [Vangl1GT(XL802)Byg] | Dr. Philippe Gros | N/A |
| Mouse: Vangl2f [Vangl2tm1.1Yy] | Dr. Yingzi Yang | N/A |
| Mouse: Wnt1Cre [H2az2Tg(Wnt1-Cre)11Rth] | The Jackson Laboratory | Stock# 003829 |
| Mouse: Wnt1Cre2 [129S4.Cg-E2f1Tg(Wnt1-cre)2Sor/J] | The Jackson Laboratory | Stock# 022137 |
| Oligonucleotides | ||
| qPCR primer: Mus musculus Chat (fwd): 5’-GCCATTGTGAAGCGGTTTG-3’ | This paper | N/A |
| qPCR primer: Mus musculus Chat (rev): 5’-TCATTCAGCCAGTATTCAGAGAC-3’ | This paper | N/A |
| qPCR primer: Mus musculus Gapdh (fwd): 5’-AGGTTGTCTCCTGCGACTTCA-3’ | This paper | N/A |
| qPCR primer: Mus musculus Gapdh (rev): 5’-CCAGGAAATGAGCTTGACAAAGTT-3’ | This paper | N/A |
| qPCR primer: Mus musculus Th (fwd): 5’-AAGATCAAACCTACCAGCCG-3’ | This paper | N/A |
| qPCR primer: Mus musculus Th (rev): 5’-TACGGGTCAAACTTCACAGAG-3’ | This paper | N/A |
| qPCR primer: Mus musculus VAChT (fwd): 5’-CGTATCAGTCTATGGCAGTGTC-3’ | This paper | N/A |
| qPCR primer: Mus musculus VAChT (rev): 5’-TGAGTGAACGATATGGCCTG-3’ | This paper | N/A |
| qPCR primer: Homo sapiens CHAT (fwd): 5’-GCCCTGCCGTGATCTTTG-3’ | This paper | N/A |
| qPCR primer: Homo sapiens CHAT (rev): 5’-GCCTTGTAGCTGAGTACACC-3’ | This paper | N/A |
| qPCR primer: Homo sapiens DBH (fwd): 5’-ACGTACTGGTGCTACATTAAGG-3’ | This paper | N/A |
| qPCR primer: Homo sapiens DBH (rev): 5’-TTGCCCTTGGTGACGATG-3’ | This paper | N/A |
| qPCR primer: Homo sapiens GAPDH (fwd): 5’-CTGACTTCAACAGCGACACC-3’ | This paper | N/A |
| qPCR primer: Homo sapiens GAPDH (rev): 5’-TAGCCAAATTCGTTGTCATACC-3’ | This paper | N/A |
| qPCR primer: Homo sapiens NPY (fwd): 5’-AAAGCACAGAAAATGTTCCCAG-3’ | This paper | N/A |
| qPCR primer: Homo sapiens NPY (rev): 5’-GCTGAAAATAGGAAAAGGCCAG-3’ | This paper | N/A |
| qPCR primer: Homo sapiens TH (fwd): 5’-CCGTGCTAAACCTGCTCTTC-3’ | This paper | N/A |
| qPCR primer: Homo sapiens TH (rev): 5’-GGTGGATTTTGGCTTCAAACG-3’ | This paper | N/A |
| qPCR primer: Homo sapiens VIP (fwd): 5’-TTCTCACAGACTTCGGCATG-3’ | This paper | N/A |
| qPCR primer: Homo sapiens VIP (rev): 5’-TCAGGTTCATTTGCTCCCTC-3’ | This paper | N/A |
| Recombinant DNA | ||
| Software and algorithms | ||
| ImageJ | N/A | https://imagej.nih.gov/ij/ |
| Prism 5.0 | GraphPad | https://www.graphpad.com/ |
| RStudio | RStudio® | https://www.rstudio.com/ |
| Other | ||
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
All mouse experiments in this study were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee (IACUC) of the University of California, San Francisco (UCSF). Matings were set up to obtain mice with the corresponding genotypes described in this study. Information of mouse ages, numbers and genotypes were included in the main text and figures. N pairs in the figure legends mean n controls and n mutants, which were collected from as many litters as needed to reach that number. All the controls used in this study are littermate controls. Their genotypes would vary depending on the exact crosses used in a given experiment. The mouse strains used in this study are listed below. TrkAf [B6.129P2(SJL)-Ntrk1tm1Ddg/J] mice, TrkBf [Ntrk2tm1.1Lfr] mice (provided by Dr. Baoji Xu), Vangl1gt [Vangl1GT(XL802)Byg] and Vangl2f [Vangl2tm1.1Yy] mice (provided by Dr. Yingzi Yang), Ror2f mice [B6;129S4-Ror2tm1.1Meg/J], Bdnff mice [STOCK Bdnftm3Jae/J], Ngff mice, Adrb1− mice [FVB.129-Adrb1tm1Bkk/J], Adrb2− mice [FVB.129-Adrb2 tm1Bkk/J], Adrb3− mice [FVB/N-Adrb3tm1Lowl/J], NgfLacZ mice [Ngftm1Ddg] (provided by Dr. David Ginty), ROSA26mTmG mice [STOCK Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J], ROSA26tdTomato [B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J], R26R mice [B6;129S4-Gt(ROSA)26Sortm1Sor/J], GCaMP6f mice [B6;129S-Gt(ROSA)26Sortm95.1(CAG-GCaMP6f)Hze/J], RC::Ptox mice (derived from RC::PFtox mice [Gt(ROSA)26Sortm7(CAG-mCherry,-EGFP/tetX)Dym] provided by Dr. Susan Dymecki), Wnt1Cre [Tg(Wnt1-Cre)11Rth] mice (provided by Dr. Andy McMahon), Wnt1Cre (line2) or Wnt1Cre2 mice [129S4.Cg-E2f1Tg(Wnt1-cre)2Sor/J] (provided by Dr. Jeffrey Bush), Sox10Cre mice [B6;CBA-Tg(Sox10-cre)1Wdr/J], Sox10iCreER mice [CBA;B6-Tg(Sox10-icre/ERT2)388Wdr/J], ThCre mice [B6;129X1-Thtm1(cre)Te/Kieg], ChatCre mice [B6;129S6-Chattm2(cre)Lowl/J], BdnfCre mice [B6.FVB-Bdnfem1(cre)Zak/J], PdgfraCre mice [C57BL/6-Tg(Pdgfra-cre)1Clc/J] and PdgfraCreER mice [B6N.Cg-Tg(Pdgfra-cre/ERT)467Dbe/J] were obtained from Jackson Laboratory or the investigators indicated. Of note, we observed recombination of floxed alleles in germ cells when the male mice harbored a Sox10Cre allele. Thus, in this study, Sox10Cre was transmitted only through female mice. Germline recombination of floxed alleles was also occasionally observed for PdgfraCre and Wnt1Cre.
Mouse neurons
Neurons from the mouse superior cervical ganglion (SCG) were isolated and cultured as previously described (Jackson and Tourtellotte, 2014). Briefly, SCGs from wild-type and Vangl2f/f; ROSA26mTmG/+ mice at postnatal (P) day 3 were dissected, rinsed with 100% ethanol, and washed extensively with sterilized PBS for 5 min. Neurons were dissociated through serial enzymatic digestions; incubation in 500 μl collagenase B (1 mg/ml) solution for 30 min at 37°C followed by 500 μl 0.25% trypsin for another 30 min. Cells were rinsed with SCG culture medium (DMEM containing 10% FBS, 1× Penicillin/streptomycin, 1× L-glutamine and 100 ng/ml Nerve growth factor (NGF)) and pipetted 50 times in SCG culture medium to release single cells. Dissociated cells from SCGs were plated onto poly-D-lysine coated coverslips in a 24-well plate, and media were replaced every two days.
Mouse lung cells and fibroblasts/myofibroblasts
Lungs were collected from wild-type and PdgfraH2BGFP/+ mice, and inflated with the digestion solution (1.2 U/ml dispase, 0.5mg/ml collagenase B and 50 U/ml DNase I) through the trachea. Lungs devoid of the trachea were minced into small pieces, placed in the digestion solution and rocked for 30 min at 37°C. The tissues were pipetted several times and digested for another 20–30 min at 37°C. The samples were placed on ice after tissues had dissolved. The samples were mixed with twice the volume of the sorting buffer (phenol red free DMEM with 2% FBS, 2× Penicillin/streptomycin and 100 U/ml DNase I), filtered through 70 μm cell strainers, and spun at 600g for 5 min at 4°C. The cells were resuspended in 2 ml of red blood cell (RBC) lysis buffer for 3 min, and then mixed with 5 ml of sorting buffer to quench the reaction. The samples were refiltered through 40 μm cell strainers. After spinning at 600g for 5 min at 4°C, the cells were resuspended in 1 ml of sorting buffer and purified by fluorescence-activated cell sorting (FACS).
Human lung tissues
Lung samples were obtained at the time of lung transplantation performed for severe emphysema (Global Initiative for Chronic Obstructive Lung Disease Criteria, stages III or IV) during the study period 2015–2019. Control lung tissues were obtained from donor lungs not utilized for lung transplantation. Our studies indicate that these lungs are physiologically and pathologically normal (Ware et al., 2002). Written informed consent was obtained from all subjects and the study was approved by the University of California, San Francisco Institutional Review Board (IRB approval # 13-10738).
METHODS DETAILS
Tamoxifen administration
4-Hydroxytamoxifen (4-OHT) solution was prepared by dissolving powder in ethanol to make a stock solution at a concentration of 20 mg/ml, which was then diluted 1:10 in corn oil (MilliporeSigma) to make a working solution at a final concentration of 2 mg/ml. 4-OHT solution was stored at −20°C. 4-OHT was administered to pregnant female mice carrying Vangl2f/f; Sox10iCreER and Ror2 f/f; Sox10iCreER embryos and their littermates at 15.5 days post coitus (dpc) by delivering 100–150 μL of 4-OHT (2mg/ml) per mouse to their stomach through a feeding needle. Three days post-4-OHT treatment, pregnant females were transferred to cages that housed healthy nursing females as foster mothers. Lungs from the pups were harvested at postnatal (P) day 10, when the majority of secondary septa were being generated.
Mean linear intercept (MLI)
Measurement of the MLI was performed as previously described (Zhang et al., 2022; Zhang et al., 2020).
Histology, immunofluorescence and immunohistochemistry
For histological analysis of alveolar development, mouse lungs at the indicated time points were dissected and fixed with 4% paraformaldehyde (PFA) in PBS for 1 hr at 4°C. The samples were embedded in paraffin wax and sectioned at 7 μm. Haematoxylin-and-eosin staining was performed as previously described (Lin et al., 2017; Zhang et al., 2020). Images were taken using a SPOT 2.3 CCD camera connected to a Nikon Eclipse E1000 microscope.
LacZ staining was performed to visualize pulmonary neurons and nerves. Lungs from NgfLacZ/+ and Wnt1Cre/+; R26R/+ mice at P3 were collected and fixed in 4% PFA on ice for 30 min. The lungs were washed in 0.02% NP-40 in PBS for 2 hr, immersed in X-gal staining solution (5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6, 2 mM MgCl2, 0.01% sodium deoxycholate, 0.02% NP-40 and 1 mg/ml X-gal in PBS), and rocked at 37°C for 3 days. Images for LacZ-stained lungs were taken using a Nikon SMZ1500 microscope.
Whole mount immunofluorescence was also performed to determine the distribution of pulmonary neurons and nerves. Lungs from Wnt1Cre/+; ROSA26mTmG/+ or Sox10Cre/+; ROSA26mTmG/+ mice were collected and sectioned at 200–300 μm. Whole mount immunostaining was performed by incubating samples with mouse anti-NF-M (2H3) or chicken anti-GFP antibodies at 4°C overnight. After washes with PBS for 5 hr, donkey anti-mouse Alexa Fluor 594 or donkey anti-chicken Alexa Fluor 488 were added, and samples were incubated at 4°C overnight.
Immunofluorescence was performed as previously described (Zhang et al., 2020). For frozen sections, fixed lungs were placed in 30% sucrose until they sank, embedded in OCT and sectioned at 7 μm. The primary antibodies we used include: rabbit anti-PGP9.5 (1:150, AnaSpec), rabbit anti-Synaptophysin (SYP) (1:200, Invitrogen), mouse anti-neurofilament (NF-M) (1:200, Developmental Studies Hybridoma Bank), mouse anti-SV2 (1:200, Developmental Studies Hybridoma Bank), mouse anti-TUBB3 (1:200, Developmental Studies Hybridoma Bank), rabbit anti-VAChT (1:200, Synaptic Systems), rabbit anti-Tyrosine hydroxylase (TH) (1:200, Synaptic Systems), rabbit anti-NKX2.1 (1:100, Epitomics), chicken anti-GFP (1:200, Aves Labs), rabbit anti-calcitonin gene-related peptide (CGRP) (1:400; MilliporeSigma), rabbit anti-prosurfactant protein C (proSP-C) (1:200, MilliporeSigma), hamster anti-T1α (1:200, Developmental Studies Hybridoma Bank), mouse anti-HOPX (1:100, Santa Cruz Biotechnology), goat anti-CC10 (1:200, Santa Cruz Biotechnology), mouse anti-acetylated tubulin (1:200, MilliporeSigma), mouse anti-ACTA2 (1:200, Thermo Scientific Lab Vision), rat anti-PECAM-1 (CD31) (1:150, Santa Cruz Biotechnology), rabbit anti-PDGFRA (1:150, Cell Signaling Technology), rabbit anti-phospho-PDGFRA (Tyr754) (1:100, Cell Signaling Technology), goat anti-PDGFRB (1:200, R&D Systems) and rat anti-VANGL2 (1:150, MilliporeSigma). Secondary antibodies and conjugates used were donkey anti-rabbit Alexa Fluor 488 or 594 (1:1000, Life Technologies), donkey anti-chicken Alexa Fluor 488 or 647 (1:1000, Life Technologies), donkey anti-mouse Alexa Fluor 488 or 594 (1:1000, Life Technologies), and donkey anti-rat Alexa Fluor 594 (1:1000, Life Technologies). The biotinylated secondary antibodies used were goat anti-hamster (1:1000, Jackson ImmunoResearch Laboratories), donkey anti-rabbit (1:1000, Jackson ImmunoResearch Laboratories), donkey anti-rat (1:1000, Jackson ImmunoResearch Laboratories) and horse anti-mouse (1:1000, Jackson ImmunoResearch Laboratories). The signal was detected using streptavidin-conjugated Alexa Fluor 488, 594, or 647 (1:1000, Life Technologies) or HRP-conjugated streptavidin (1:1000, Perkin-Elmer) coupled with fluorogenic substrate Alexa Fluor 594 tyramide for 30 s (1:200, TSA kit; Perkin Elmer). F-actin was stained with rhodamine-conjugated phalloidin (1:200; MilliporeSigma).
Confocal images were captured on a Leica SPE laser-scanning confocal microscope. Adjustment of red/green/blue/grey histograms and channel merges was performed using LAS AF Lite (Leica Microsystems).
Quantification of saccule innervation
Mouse lungs at postnatal day 3 were collected and fixed in 4% PFA for 1 hr. The samples were processed, paraffin embedded and serially sectioned at 50 μm along the coronal plane of the entire lung. Quantification was performed on hematoxylin-and-eosin stained sections; the percentage of saccules innervated inside the lung was calculated as % = Ni/Nt, where Ni is the number of saccules in the immediate vicinity of airways, which are innervated and Nt is the total number of saccules inside the lung.
For counting the number of innervated saccules on the lung surface, lungs from Wnt1Cre/+; R26R/+ mice were collected and stained with LacZ. The percentage of saccules innervated on the lung surface was calculated as the ratio of saccules in the immediate vicinity of nerves on the lung surface to the total number of saccules on the lung surface. Most of the surface saccules are located at the far end from the BADJs and are at the periphery of the lung.
Lung clearing
Lung tissue was cleared using the CUBIC method as previously described (Gomez-Gaviro et al., 2017; Susaki et al., 2015) with minor modifications. In brief, 300 μm lung slices were immersed in CUBIC reagent-1 (25% Urea, 25% Quadrol and 15% Triton X-100 dissolved in ddH2O), rocked at 37°C for overnight, washed 3 times with PBST (0.1% Tween-20 in PBS), and incubated with the primary antibodies at 4°C for 3 days. After washing with PBST, the lung tissues were incubated with secondary antibodies at 4°C for 1 day. The samples were then immersed in CUBIC reagent-2 (25% Urea, 50% sucrose and 10% triethanolamine dissolved in ddH2O) for 2 hr. The lung slices were mounted with VECTASHIELD containing 10 μm/ml DAPI.
Cell proliferation assays
The rate of cell proliferation was determined through a short pulse of EdU incorporation as previously reported (Zhang et al., 2020). 0.25 mg of EdU (5 mg/ml solution) per mouse was administered intraperitoneally to pups at postnatal day 2, 3, 5 and 7 one hour prior to tissue collection; 0.375 mg of EdU per mouse was used for mice at postnatal day 10 and 12. The Click-iT EdU Alexa Fluor 488 Imaging Kit (Life Technologies) was used to assess EdU incorporation on paraffin-embedded lung sections. Alveolar fibroblasts and myofibroblasts were identified by antibodies against PDGFRA on lung sections. The proliferation rate of fibroblasts/myofibroblasts was calculated as the ratio of (EdU+PDGFRA+ cells)/(PDGFRA+ cells).
In vitro myofibroblast migration assay
Myofibroblast migration was examined using the Culture-Insert 2 Well system (ibidi). In brief, mouse lungs at postnatal day 3 were dissected, minced into small pieces, placed in digestion solution (1.2 U/ml dispase, 0.5mg/ml collagenase B and 50 U/ml DNase I), and rocked at 37°C for 2 hr to release single cells. The samples were mixed with an equal volume of culture medium (DMEM with 10% FBS, 2× penicillin/streptomycin and 1× L-glutamine) and filtered through 40 μm cell strainers. After centrifugation at 600g for 10 min, the dissociated cells were resuspended in 200 μl culture medium and plated into individual wells (100 μl per well). Lung myofibroblasts quickly attached to fibronectin–coated plates in 2–3 hr. Culture medium was replaced to remove non-adherent cells and cells were incubated until they reached 100% confluence. Confluent myofibroblasts were switched to starvation medium (DMEM with 0.5% FBS and 1× penicillin/streptomycin) for 16 hr. Meanwhile, PC12 cells were plated and cultured until they reached 100% confluence. PC12 cells were maintained in starvation medium containing 15 mM KCl for 30 min to generate conditioned medium. The PC12-conditioned and control media were added to starved lung myofibroblasts after removal of the 2-well culture insert. Migration of myofibroblasts into the denuded area was allowed to proceed for another 36–48 hr.
RNA-Seq
RNA-Seq was performed as previously described (Zhang et al., 2020) with minor modifications. Briefly, peripheral lung tissues were manually dissected from the lungs of PdgfraH2BGFP/+ mice at postnatal (P) day 3 and digested to release lung cells as described above. GFP-labeled myofibroblasts were sorted by FACS into TRIzol (Life Technologies). RNA was extracted using the RNeasy Micro Plus Kit (Qiagen) following the manufacturer’s instructions. The quality of extracted RNA was evaluated by an Agilent 2100 Bioanalyzer. The samples were sequenced on an Illumina HiSeq 2000 or HiSeq 4000. RNA-Seq data have been deposited in Gene Expression Omnibus (GSE189466).
scRNA-seq
scRNA-seq of distal mouse lung cells was performed following the standard procedures. Peripheral lung tissues were manually dissected from mouse lungs at postnatal (P) day 3 and processed as described above. FACS-sorted single cells were loaded onto 3′ library chips according to the manufacturer’s protocol for Chromium 10X Single Cell 3′ Library v3 (10X Genomics). Sample quality control, library preparation and sequencing were performed according to the manufacturer’s protocol.
The 10X Cell Ranger software version 5.0 was used to align reads to the mouse genome and generate feature-barcode matrices. The R package Seurat (version 3.2.2) was used for single cell transcriptome analysis. The SCTransform normalization method was used to control for between-sample sequencing depth variation. Cells were removed according to the following thresholds: <300 genes/cell or >5000 genes per cell, >20000 UMIs/cell, 15% mitochondrial content, <0.01% hemoglobin content. Among the cells retained, the effects of mitochondrial and ribosomal content were regressed out prior to clustering. Genes were excluded from the final dataset if they were expressed in fewer than 5 cells. For differential expression analysis between clusters, genes were detected if they are expressed in at least 10% of cells in a cluster with a log fold change of at least 0.25. For DEG analysis between comparison groups, genes were detected if expressed in at least 5% of cells in the group with a log fold change of at least 0.25. scRNA-seq data have been deposited in Gene Expression Omnibus (GSE189466).
qPCR analysis
RNA was prepared from the mouse lungs. In brief, the cranial lobe of lungs from mice of the indicated genotypes was homogenized in 1 ml TRIzol (Life Technologies). The homogenates were mixed with 200 μl chloroform and centrifuged for 15 min at 4°C. The upper aqueous layer was removed and mixed with an equal volume of 70% ethanol. RNA was extracted with the RNeasy Mini Kit (Qiagen) following the manufacturer’s instructions. RNA was then reverse-transcribed with the Maxima First Strand cDNA Synthesis Kit (Thermo Scientific). Quantitative PCR (qPCR) was performed on the ABI Prism 7900HT Sequence Detection System. Primers for qPCR are listed as follows: mouse Th forward, 5’-AAGATCAAACCTACCAGCCG-3’; reverse, 5’-TACGGGTCAAACTTCACAGAG-3’, mouse Chat forward, 5’-GCCATTGTGAAGCGGTTTG-3’; reverse, 5’-TCATTCAGCCAGTATTCAGAGAC-3’, mouse VAChT forward, 5’-CGTATCAGTCTATGGCAGTGTC-3’; reverse, 5’-TGAGTGAACGATATGGCCTG-3’, mouse Gapdh forward, 5’-AGGTTGTCTCCTGCGACTTCA-3’; reverse, 5’-CCAGGAAATGAGCTTGACAAAGTT-3’.
RNA extraction and qPCR analysis of human lungs were performed as described above. The following primers were used: human TH forward, 5’-CCGTGCTAAACCTGCTCTTC-3’; reverse, 5’-GGTGGATTTTGGCTTCAAACG-3’, human DBH forward, 5’-ACGTACTGGTGCTACATTAAGG-3’; reverse, 5’-TTGCCCTTGGTGACGATG-3’, human CHAT forward, 5’-GCCCTGCCGTGATCTTTG-3’; reverse, 5’-GCCTTGTAGCTGAGTACACC-3’, human NPY forward, 5’-AAAGCACAGAAAATGTTCCCAG-3’; reverse, 5’-GCTGAAAATAGGAAAAGGCCAG-3’, human VIP forward, 5’-TTCTCACAGACTTCGGCATG-3’; reverse, 5’-TCAGGTTCATTTGCTCCCTC-3’, human GAPDH forward, 5’-CTGACTTCAACAGCGACACC-3’; reverse, 5’-TAGCCAAATTCGTTGTCATACC-3’.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical comparisons between two or more groups were presented as mean ± SEM. The P values were calculated by two-tailed Student’s t-tests and one-way ANOVA. Statistical significance was assessed as *P < 0.05, **P < 0.01 and *** P < 0.001. Detailed biological replicates (n numbers) were indicated in the figure legends.
Supplementary Material
Highlights.
Mouse lung air sacs are innervated by autonomic nerves during alveolar development.
Vangl2 regulates neurotransmitter release from autonomic nerves to control alveologenesis.
Alveolar myofibroblasts are a target of autonomic innervation during secondary septation.
Alveolar myofibroblasts secrete neurotrophins to regulate autonomic nerve function.
Acknowledgements
We thank Evelyn Chuang for figure preparation and Allan Basbaum, Andy Chang and Ross Metzger for critical reading of the manuscript. Some data for this study were acquired at the Nikon Imaging Center at CVRI and the UCSF Laboratory for Cell Analysis. This work was supported by grants (R01 HL142876) from the National Institutes of Health to P.T.C.
Footnotes
Declaration of interests
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amendola J, Boumedine N, Sangiardi M, and El Far O (2015). Optimization of neuronal cultures from rat superior cervical ganglia for dual patch recording. Sci Rep 5, 14455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branchfield K, Li R, Lungova V, Verheyden JM, McCulley D, and Sun X (2016). A three-dimensional study of alveologenesis in mouse lung. Dev Biol 409, 429–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burri PH (2006). Structural aspects of postnatal lung development - alveolar formation and growth. Biol Neonate 89, 313–322. [DOI] [PubMed] [Google Scholar]
- Campanale JP, Sun TY, and Montell DJ (2017). Development and dynamics of cell polarity at a glance. J Cell Sci 130, 1201–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao CM, Moiseenko A, Zimmer KP, and Bellusci S (2016). Alveologenesis: key cellular players and fibroblast growth factor 10 signaling. Mol Cell Pediatr 3, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielian PS, Muccino D, Rowitch DH, Michael SK, and McMahon AP (1998). Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr Biol 8, 1323–1326. [DOI] [PubMed] [Google Scholar]
- Davey CF, and Moens CB (2017). Planar cell polarity in moving cells: think globally, act locally. Development 144, 187–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd J, Morton SB, Karagogeos D, Yamamoto M, and Jessell TM (1988). Spatial regulation of axonal glycoprotein expression on subsets of embryonic spinal neurons. Neuron 1, 105–116. [DOI] [PubMed] [Google Scholar]
- Dotta A, and Mortola JP (1992). Postnatal development of the denervated lung in normoxia, hypoxia, or hyperoxia. J Appl Physiol (1985) 73, 1461–1466. [DOI] [PubMed] [Google Scholar]
- Ernsberger U (2009). Role of neurotrophin signalling in the differentiation of neurons from dorsal root ganglia and sympathetic ganglia. Cell Tissue Res 336, 349–384. [DOI] [PubMed] [Google Scholar]
- Fox B, Bull TB, and Guz A (1980). Innervation of alveolar walls in the human lung: an electron microscopic study. J Anat 131, 683–692. [PMC free article] [PubMed] [Google Scholar]
- Gomez-Gaviro MV, Balaban E, Bocancea D, Lorrio MT, Pompeiano M, Desco M, Ripoll J, and Vaquero JJ (2017). Optimized CUBIC protocol for three-dimensional imaging of chicken embryos at single-cell resolution. Development 144, 2092–2097. [DOI] [PubMed] [Google Scholar]
- Guo M, Du Y, Gokey JJ, Ray S, Bell SM, Adam M, Sudha P, Perl AK, Deshmukh H, Potter SS, et al. (2019). Single cell RNA analysis identifies cellular heterogeneity and adaptive responses of the lung at birth. Nat Commun 10, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, and Baas PW (2003). Growing and working with peripheral neurons. Methods Cell Biol 71, 17–35. [DOI] [PubMed] [Google Scholar]
- Ho HY, Susman MW, Bikoff JB, Ryu YK, Jonas AM, Hu L, Kuruvilla R, and Greenberg ME (2012). Wnt5a-Ror-Dishevelled signaling constitutes a core developmental pathway that controls tissue morphogenesis. Proc Natl Acad Sci U S A 109, 4044–4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung KS, Hertweck MS, Hardy JD, and Loosli CG (1972). Innervation of pulmonary alveoli of the mouse lung: an electron microscopic study. Am J Anat 135, 477–495. [DOI] [PubMed] [Google Scholar]
- Hung KS, Hertweck MS, Hardy JD, and Loosli CG (1973). Electron microscopic observations of nerve endings in the alveolar walls of mouse lungs. Am Rev Respir Dis 108, 328–333. [DOI] [PubMed] [Google Scholar]
- Jackson M, and Tourtellotte W (2014). Neuron Culture from Mouse Superior Cervical Ganglion. Bio Protoc 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SH, Fukaya M, Yang JK, Rothstein JD, and Bergles DE (2010). NG2+ CNS glial progenitors remain committed to the oligodendrocyte lineage in postnatal life and following neurodegeneration. Neuron 68, 668–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JC, Cook MN, Carey MR, Shen C, Regehr WG, and Dymecki SM (2009). Linking genetically defined neurons to behavior through a broadly applicable silencing allele. Neuron 63, 305–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis AE, Vasudevan HN, O’Neill AK, Soriano P, and Bush JO (2013). The widely used Wnt1-Cre transgene causes developmental phenotypes by ectopic activation of Wnt signaling. Dev Biol 379, 229–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Yao E, Zhang K, Jiang X, Croll S, Thompson-Peer K, and Chuang PT (2017). YAP is essential for mechanical force production and epithelial cell proliferation during lung branching morphogenesis. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B, Pang PT, and Woo NH (2005). The yin and yang of neurotrophin action. Nat Rev Neurosci 6, 603–614. [DOI] [PubMed] [Google Scholar]
- Matsuoka T, Ahlberg PE, Kessaris N, Iannarelli P, Dennehy U, Richardson WD, McMahon AP, and Koentges G (2005). Neural crest origins of the neck and shoulder. Nature 436, 347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie IA, Ohayon D, Li H, de Faria JP, Emery B, Tohyama K, and Richardson WD (2014). Motor skill learning requires active central myelination. Science 346, 318–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyrick B, and Reid L (1971). Nerves in rat intra-acinar alveoli: an electron microscopic study. Respir Physiol 11, 367–377. [DOI] [PubMed] [Google Scholar]
- Mizikova I, and Thebaud B (2021). Looking at the developing lung in single-cell resolution. Am J Physiol Lung Cell Mol Physiol 320, L680–L687. [DOI] [PubMed] [Google Scholar]
- Mortola JP, Saetta M, and Bartlett D Jr. (1987). Postnatal development of the lung following denervation. Respir Physiol 67, 137–145. [DOI] [PubMed] [Google Scholar]
- Muller M, Triaca V, Besusso D, Costanzi M, Horn JM, Koudelka J, Geibel M, Cestari V, and Minichiello L (2012). Loss of NGF-TrkA signaling from the CNS is not sufficient to induce cognitive impairments in young adult or intermediate-aged mice. J Neurosci 32, 14885–14898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, and Luo L (2007). A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605. [DOI] [PubMed] [Google Scholar]
- Nagaoka T, and Kishi M (2016). The planar cell polarity protein Vangl2 is involved in postsynaptic compartmentalization. Neurosci Lett 612, 251–255. [DOI] [PubMed] [Google Scholar]
- Nagendran M, Riordan DP, Harbury PB, and Desai TJ (2018). Automated cell-type classification in intact tissues by single-cell molecular profiling. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H, Deutsch GH, Wert SE, Ontology S, and Consortium N.M.A.o.L.D.P. (2019). Comprehensive anatomic ontologies for lung development: A comparison of alveolar formation and maturation within mouse and human lung. J Biomed Semantics 10, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paris AJ, Hayer KE, Oved JH, Avgousti DC, Toulmin SA, Zepp JA, Zacharias WJ, Katzen JB, Basil MC, Kremp MM, et al. (2020). STAT3-BDNF-TrkB signalling promotes alveolar epithelial regeneration after lung injury. Nat Cell Biol 22, 1197–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AR, Patel AR, Singh S, Singh S, and Khawaja I (2019). Global Initiative for Chronic Obstructive Lung Disease: The Changes Made. Cureus 11, e4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios M, Fan G, Fekete C, Kelly J, Bates B, Kuehn R, Lechan RM, and Jaenisch R (2001). Conditional deletion of brain-derived neurotrophic factor in the postnatal brain leads to obesity and hyperactivity. Mol Endocrinol 15, 1748–1757. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Castillo JA, Perez DB, Ntokou A, Seeger W, Morty RE, and Ahlbrecht K (2018). Understanding alveolarization to induce lung regeneration. Respir Res 19, 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesch K, Jadhav AP, Trimarchi JM, Stadler MB, Roska B, Sun BB, and Cepko CL (2008). The transcriptome of retinal Muller glial cells. J Comp Neurol 509, 225–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrer DK, Chruscinski A, Schauble EH, Bernstein D, and Kobilka BK (1999). Cardiovascular and metabolic alterations in mice lacking both beta1- and beta2-adrenergic receptors. J Biol Chem 274, 16701–16708. [DOI] [PubMed] [Google Scholar]
- Sanchez-Ortiz E, Yui D, Song D, Li Y, Rubenstein JL, Reichardt LF, and Parada LF (2012). TrkA gene ablation in basal forebrain results in dysfunction of the cholinergic circuitry. J Neurosci 32, 4065–4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva DM, Nardiello C, Pozarska A, and Morty RE (2015). Recent advances in the mechanisms of lung alveolarization and the pathogenesis of bronchopulmonary dysplasia. Am J Physiol Lung Cell Mol Physiol 309, L1239–1272. [DOI] [PubMed] [Google Scholar]
- Soldatov R, Kaucka M, Kastriti ME, Petersen J, Chontorotzea T, Englmaier L, Akkuratova N, Yang Y, Haring M, Dyachuk V, et al. (2019). Spatiotemporal structure of cell fate decisions in murine neural crest. Science 364. [DOI] [PubMed] [Google Scholar]
- Song H, Hu J, Chen W, Elliott G, Andre P, Gao B, and Yang Y (2010). Planar cell polarity breaks bilateral symmetry by controlling ciliary positioning. Nature 466, 378–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P (1999). Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet 21, 70–71. [DOI] [PubMed] [Google Scholar]
- Susaki EA, Tainaka K, Perrin D, Yukinaga H, Kuno A, and Ueda HR (2015). Advanced CUBIC protocols for whole-brain and whole-body clearing and imaging. Nat Protoc 10, 1709–1727. [DOI] [PubMed] [Google Scholar]
- Susulic VS, Frederich RC, Lawitts J, Tozzo E, Kahn BB, Harper ME, Himms-Hagen J, Flier JS, and Lowell BB (1995). Targeted disruption of the beta 3-adrenergic receptor gene. J Biol Chem 270, 29483–29492. [DOI] [PubMed] [Google Scholar]
- Tan CL, Cooke EK, Leib DE, Lin YC, Daly GE, Zimmerman CA, and Knight ZA (2016). Warm-Sensitive Neurons that Control Body Temperature. Cell 167, 47–59 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torban E, Patenaude AM, Leclerc S, Rakowiecki S, Gauthier S, Andelfinger G, Epstein DJ, and Gros P (2008). Genetic interaction between members of the Vangl family causes neural tube defects in mice. Proc Natl Acad Sci U S A 105, 3449–3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Chang H, Rattner A, and Nathans J (2016). Frizzled Receptors in Development and Disease. Curr Top Dev Biol 117, 113–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware LB, Wang Y, Fang X, Warnock M, Sakuma T, Hall TS, and Matthay M (2002). Assessment of lungs rejected for transplantation and implications for donor selection. Lancet 360, 619–620. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Nakamura R, Takase Y, Susaki EA, Ueda HR, Tadokoro R, and Takahashi Y (2018). Comparison of the 3-D patterns of the parasympathetic nervous system in the lung at late developmental stages between mouse and chicken. Dev Biol 444 Suppl 1, S325–S336. [DOI] [PubMed] [Google Scholar]
- Whitsett JA, and Weaver TE (2015). Alveolar development and disease. Am J Respir Cell Mol Biol 53, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong KA, Bano A, Rigaux A, Wang B, Bharadwaj B, Schurch S, Green F, Remmers JE, and Hasan SU (1998). Pulmonary vagal innervation is required to establish adequate alveolar ventilation in the newborn lamb. J Appl Physiol (1985) 85, 849–859. [DOI] [PubMed] [Google Scholar]
- Xu B, Zang K, Ruff NL, Zhang YA, McConnell SK, Stryker MP, and Reichardt LF (2000). Cortical degeneration in the absence of neurotrophin signaling: dendritic retraction and neuronal loss after removal of the receptor TrkB. Neuron 26, 233–245. [DOI] [PubMed] [Google Scholar]
- Zallen JA (2007). Planar polarity and tissue morphogenesis. Cell 129, 1051–1063. [DOI] [PubMed] [Google Scholar]
- Zerby SE, and Ewing AG (1996). Electrochemical monitoring of individual exocytotic events from the varicosities of differentiated PC12 cells. Brain Res 712, 1–10. [DOI] [PubMed] [Google Scholar]
- Zhang K, Yao E, Chen B, Chuang E, Wong J, Seed RI, Nishimura SL, Wolters PJ, and Chuang PT (2022). Acquisition of cellular properties during alveolar formation requires differential activity and distribution of mitochondria. Elife 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Yao E, Lin C, Chou YT, Wong J, Li J, Wolters PJ, and Chuang PT (2020). A mammalian Wnt5a-Ror2-Vangl2 axis controls the cytoskeleton and confers cellular properties required for alveologenesis. Elife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y (2012). Does planar cell polarity signaling steer growth cones? Curr Top Dev Biol 101, 141–160. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq and scRNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Any additional information relating to the data reported in this paper is available from the lead contact upon request. This paper does not report original code.
Key resources table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-Acetylated tubulin | MilliporeSigma | Cat# T6793; RRID:AB_477585 |
| Mouse monoclonal anti-ACTA2 | Thermo Scientific Lab Vision | Cat# MS-113-P0; RRID:AB_64001 |
| Goat polyclonal anti-CC10 (S-20) | Santa Cruz Biotechnology | Cat# sc-9773; RRID:AB_2183391 |
| Chicken polyclonal anti-GFP | Aves Labs | Cat# GFP-1010; RRID:AB_2307313 |
| Mouse monoclonal anti-HOP (E-1) | Santa Cruz Biotechnology | Cat# sc-398703; RRID:AB_2687966 |
| Mouse monoclonal anti-Neurofilament (NF-M) | Developmental Studies Hybridoma Bank | Cat# 2H3; RRID:AB_531793 |
| Rabbit monoclonal anti-NKX2.1 | Epitomics | Cat# 2044-1; RRID:AB_1267367 |
| Rabbit polyclonal anti-PDGF receptor alpha | Cell Signaling Technology | Cat# 3164; RRID:AB_2162351 |
| Rabbit monoclonal anti-Phospho-PDGF Receptor α (Tyr754) (23B2) | Cell Signaling Technology | Cat# 2992; RRID:AB_390728 |
| Goat polyclonal anti-PDGF R beta | R&D Systems | Cat# AF1042; RRID:AB_2162633 |
| Rat monoclonal anti-PECAM-1 (MEC 13.3) (CD31) | Santa Cruz Biotechnology | Cat# sc-18916; RRID:AB_627028 |
| Rabbit polyclonal anti-PGP9.5 | AnaSpec | Cat# 53772; RRID:AB_2272470 |
| Rabbit polyclonal anti-prosurfactant protein C (proSP-C) | MilliporeSigma | Cat# AB3786; RRID:AB_91588 |
| Mouse monoclonal anti-SV2 | Developmental StudiesHybridoma Bank | Cat# SV2; RRID:AB_2315387 |
| Rabbit monoclonal anti-Synaptophysin (SYP) | Invitrogen | Cat# 180130; RRID:AB_10836766 |
| Hamster monoclonal anti-T1α | Developmental Studies Hybridoma Bank | Cat# 8.1.1; RRID:AB_531893 |
| Mouse monoclonal anti-TUBB3 | Developmental Studies Hybridoma Bank | Cat# 6G7; RRID:AB_528497 |
| Rabbit polyclonal anti-Tyrosine hydroxylase | Synaptic Systems | Cat# 213 102; RRID:AB_2619896 |
| Rabbit polyclonal anti-VAChT | Synaptic Systems | Cat# 139 103; RRID:AB_887864 |
| Rat monoclonal anti-VANGL2 clone 2G4 | MilliporeSigma | Cat# MABN750; RRID:AB_2721170 |
| Bacterial and virus strains | ||
| Biological samples | ||
| Human emphysema/COPD patient tissues | Paul Wolters, University of California, San Francisco | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| 5-Bromo-4-Chloro-3-Indolyl-D-Galactoside (X-gal) | Research Products International | Cat# B71800-1.0 |
| Collagenase B | Roche | Cat# 11088815001 |
| 2’-Deoxy-5-ethynyluridine (EdU) | BiosynthCarbosynth | Cat# 61135-33-9 |
| Deoxyribonuclease I from bovine pancreas | MilliporeSigma | Cat# 9003-98-9 |
| Dispase | BD Biosciences | Cat# 354235 |
| DMEM | Mediatech | Cat# 10-013-CV |
| Fetal Bovine Serum (FBS) | Gibco | Cat# 10437-028 |
| Fibronectin | Corning | Cat# 354008 |
| Glutaraldehyde, 8% aqueous solution, EM grade | Electron Microscopy Sciences | Cat# 16000 |
| 4-Hydroxytamoxifen (4-OHT) | Toronto Research Chemicals | Cat# H954705 |
| Nerve Growth Factor (NGF 2.5S) | Gibco | Cat# 13257-019 |
| Paraformaldehyde | MilliporeSigma | Cat# P6148 |
| Paraformaldehyde, 16% solution, EM grade | Electron Microscopy Sciences | Cat# 15700 |
| Penicillin/Streptomycin | Gibco | Cat# 15070-063 |
| 1X RBC Lysis Buffer | Invitrogen | Cat# 00-4333-57 |
| Rhodamine-conjugated phalloidin | Molecular Probes | Cat# R415 |
| RNase-free DNase | QIAGEN | Cat# 79254 |
| Tamoxifen | Toronto Research Chemicals | Cat# T006000 |
| TRIzol® Reagent | Ambion | Cat# 15596018 |
| Critical commercial assays | ||
| Click-iT™ EdU Alexa Fluor™ 488 Imaging Kit | Thermo Fisher | Cat# C10337 |
| Maxima First Strand cDNA Synthesis Kit | Thermo Scientific | Cat# K1641 |
| RNeasy Mini Kit | QIAGEN | Cat# 74104 |
| RNeasy Plus Mini Kit | QIAGEN | Cat# 74134 |
| RNeasy Plus Micro Kit | QIAGEN | Cat# 74034 |
| TSA Plus Cyanine 3 (Cy3) Fluorescein detection kit | Perkin-Elmer | Cat# NEL753001KT |
| Two-well culture insert | Ibidi | Cat# 80209 |
| Deposited data | ||
| Bulk RNA-Seq data for peripheral lungs of PdgfraH2BGFP mice at P3 | This paper | GSE189466 |
| scRNA-Seq data for peripheral lungs of control mice at P3 | This paper | GSE189466 |
| Experimental models: Cell lines | ||
| Primary fibroblasts derived from peripheral lungs of control mice at P3 | This paper | N/A |
| Primary neurons derived from the superior cervical ganglion (SCG) of Ptox; Sox10Cre mice at P3 | This paper | N/A |
| Primary neurons derived from the superior cervical ganglion (SCG) of Sox10Cre mice at P3 | This paper | N/A |
| Primary neurons derived from the superior cervical ganglion (SCG) of Vangl2f/f; Sox10Cre mice at P3 | This paper | N/A |
| Rat: PC12 cells | ATCC | CRL-1721 |
| Experimental models: Organisms/strains | ||
| Mouse: Adrb1− [FVB.129-Adrb1tm1Bkk/J] | The Jackson Laboratory | Stock# 031490 |
| Mouse: Adrb2− [FVB.129-Adrb2tm1Bkk/J] | The Jackson Laboratory | Stock# 031496 |
| Mouse: Adrb3− [FVB/N-Adrb3tm1Lowl/J] | The Jackson Laboratory | Stock# 006402 |
| Mouse: BdnfCre [B6.FVB-Bdnfem1(cre)Zak/J] | The Jackson Laboratory | Stock# 030189 |
| Mouse: Bdnff [STOCK Bdnftm3Jae/J] | The Jackson Laboratory | Stock# 004339 |
| Mouse: ChatCre [B6;129S6-Chattm2(cre)Lowl/J] | The Jackson Laboratory | Stock# 006410 |
| Mouse: GCaMP6f [B6;129S-Gt(ROSA)26Sortm95.1(CAG-GCaMP6f)Hze/J] | The Jackson Laboratory | Stock# 024105 |
| Mouse: Ngff | Dr. Liliana Minichiello | N/A |
| Mouse: NgfLacZ [Ngftm1Ddg] | Dr. David Ginty | N/A |
| Mouse: PDGFRaCre [C57BL/6-Tg(Pdgfra-cre)1Clc/J] | The Jackson Laboratory | Stock# 013148 |
| Mouse: PDGFRaCreER [B6N.Cg-Tg(Pdgfra-cre/ERT)467Dbe/J] | The Jackson Laboratory | Stock# 018280 |
| Mouse: PDGFRaH2BGFP [B6.129S4-Pdgfratm11(EGFP)Sor/J] | The Jackson Laboratory | Stock# 007669 |
| Mouse: RC::PFtox [B6;129S6-Gt(ROSA)26Sortm8(CAG-mCherry,-EGFP)Dym/J] | The Jackson Laboratory | Stock# 029486 |
| Mouse: Ror2f [B6;129S4-Ror2tm1.1Meg/J] | The Jackson Laboratory | Stock# 018354 |
| Mouse: ROSA26mTmG [B6.129(Cg)-Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J] | The Jackson Laboratory | Stock# 007676 |
| Mouse: R26R [B6.129S4-Gt(ROSA)26Sortm1Sor/J] | The Jackson Laboratory | Stock# 003474 |
| Mouse: ROSA26tdTomato [B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J] | The Jackson Laboratory | Stock# 007914 |
| Mouse: Sox10Cre [B6;CBA-Tg(Sox10-cre)1Wdr/J] | The Jackson Laboratory | Stock# 025807 |
| Mouse: Sox10iCreER [CBA;B6-Tg(Sox10-icre/ERT2)388Wdr/J] | The Jackson Laboratory | Stock# 027651 |
| Mouse: ThCre [B6;129X1-Thtm1(cre)Te/Kieg] | Dr. Ted Ebendal | N/A |
| Mouse: TrkAf [B6.129P2(SJL)-Ntrk1tm1Ddg/J] | The Jackson Laboratory | Stock# 022362 |
| Mouse: TrkBf [Ntrk2tm1.1Lfr] | Dr. Baoji Xu | N/A |
| Mouse: Vangl1gt [Vangl1GT(XL802)Byg] | Dr. Philippe Gros | N/A |
| Mouse: Vangl2f [Vangl2tm1.1Yy] | Dr. Yingzi Yang | N/A |
| Mouse: Wnt1Cre [H2az2Tg(Wnt1-Cre)11Rth] | The Jackson Laboratory | Stock# 003829 |
| Mouse: Wnt1Cre2 [129S4.Cg-E2f1Tg(Wnt1-cre)2Sor/J] | The Jackson Laboratory | Stock# 022137 |
| Oligonucleotides | ||
| qPCR primer: Mus musculus Chat (fwd): 5’-GCCATTGTGAAGCGGTTTG-3’ | This paper | N/A |
| qPCR primer: Mus musculus Chat (rev): 5’-TCATTCAGCCAGTATTCAGAGAC-3’ | This paper | N/A |
| qPCR primer: Mus musculus Gapdh (fwd): 5’-AGGTTGTCTCCTGCGACTTCA-3’ | This paper | N/A |
| qPCR primer: Mus musculus Gapdh (rev): 5’-CCAGGAAATGAGCTTGACAAAGTT-3’ | This paper | N/A |
| qPCR primer: Mus musculus Th (fwd): 5’-AAGATCAAACCTACCAGCCG-3’ | This paper | N/A |
| qPCR primer: Mus musculus Th (rev): 5’-TACGGGTCAAACTTCACAGAG-3’ | This paper | N/A |
| qPCR primer: Mus musculus VAChT (fwd): 5’-CGTATCAGTCTATGGCAGTGTC-3’ | This paper | N/A |
| qPCR primer: Mus musculus VAChT (rev): 5’-TGAGTGAACGATATGGCCTG-3’ | This paper | N/A |
| qPCR primer: Homo sapiens CHAT (fwd): 5’-GCCCTGCCGTGATCTTTG-3’ | This paper | N/A |
| qPCR primer: Homo sapiens CHAT (rev): 5’-GCCTTGTAGCTGAGTACACC-3’ | This paper | N/A |
| qPCR primer: Homo sapiens DBH (fwd): 5’-ACGTACTGGTGCTACATTAAGG-3’ | This paper | N/A |
| qPCR primer: Homo sapiens DBH (rev): 5’-TTGCCCTTGGTGACGATG-3’ | This paper | N/A |
| qPCR primer: Homo sapiens GAPDH (fwd): 5’-CTGACTTCAACAGCGACACC-3’ | This paper | N/A |
| qPCR primer: Homo sapiens GAPDH (rev): 5’-TAGCCAAATTCGTTGTCATACC-3’ | This paper | N/A |
| qPCR primer: Homo sapiens NPY (fwd): 5’-AAAGCACAGAAAATGTTCCCAG-3’ | This paper | N/A |
| qPCR primer: Homo sapiens NPY (rev): 5’-GCTGAAAATAGGAAAAGGCCAG-3’ | This paper | N/A |
| qPCR primer: Homo sapiens TH (fwd): 5’-CCGTGCTAAACCTGCTCTTC-3’ | This paper | N/A |
| qPCR primer: Homo sapiens TH (rev): 5’-GGTGGATTTTGGCTTCAAACG-3’ | This paper | N/A |
| qPCR primer: Homo sapiens VIP (fwd): 5’-TTCTCACAGACTTCGGCATG-3’ | This paper | N/A |
| qPCR primer: Homo sapiens VIP (rev): 5’-TCAGGTTCATTTGCTCCCTC-3’ | This paper | N/A |
| Recombinant DNA | ||
| Software and algorithms | ||
| ImageJ | N/A | https://imagej.nih.gov/ij/ |
| Prism 5.0 | GraphPad | https://www.graphpad.com/ |
| RStudio | RStudio® | https://www.rstudio.com/ |
| Other | ||


