Abstract
The pentraxin-related PTX3, commonly produced by myeloid and endothelial cells, is a humoral pattern-recognition protein of the innate immune system. Because CLL patients’ PTX3 plasma levels are high and most circulating cells in patients with CLL are CLL cells, we reasoned that CLL cells produce PTX3. Western immunoblotting revealed that low-density cells from 7 out of 7 CLL patients produce high levels of PTX3, flow cytometry analysis revealed that the PTX3-producing cells are B lymphocytes co-expressing CD19 and CD5, and confocal microscopy showed that PTX3 is present in the cytoplasm of CLL cells. Because signal transducer and activator of transcription (STAT)-3 is constitutively activated in CLL cells and we identified putative STAT3 binding sites within the PTX3 gene promoter, we postulated that phosphorylated STAT3 triggers transcriptional activation of PTX3. Immunoprecipitation analysis of CLL cells’ chromatin fragments showed that STAT3 antibodies precipitated PTX3 DNA. STAT3 knockdown induced a marked reduction in PTX3 expression, indicating a STAT3-induced transcriptional activation of the PTX3 gene in CLL cells. Using an electromobility shift assay we established and using a dual-reporter luciferase assay we confirmed that STAT3 binds the PTX3 gene promoter. Downregulation of PTX3 enhanced apoptosis of CLL cells, suggesting that inhibition of PTX3 might benefit patients with CLL.
Introduction
The normal B-cell lineage in chronic lymphocytic leukemia (CLL) is gradually replaced by neoplastic B lymphocytes (1) whose differentiation into antibody-producing cells is significantly impaired. As a result, low immunoglobulin serum levels characterize patients with CLL. However, despite their progressive hypogammaglobulinemia, neutropenia and impaired T-cell function (2), untreated CLL patients experience a relatively low incidence of severe bacterial infections (3) and increased rates of autoimmune disorders such as hemolytic anemia, immune thrombocytopenia, paraneoplastic pemphigus, and acquired angioedema (4, 5). Unlike the adaptive immune system whose role in these conditions has been extensively studied (3–5), the role of the evolutionary conserved innate immune system in patients with CLL is comparatively scant.
Pentraxin-related protein 3 (PTX3), C-reactive protein (CRP) and serum amyloid P (SAP) are members of the pentraxin superfamily of inflammatory proteins, characterized by a cyclic pentameric structure (6). Like CRP and SAP, PTX3 is a major component of the humoral arm of the innate immune response system (7). As such, PTX3 exerts antibody-like functions and enhances microbe opsonization, glycosylation-dependent regulation of inflammation and complement activation, thus contributing to facilitation of bacteria clearance (8). Conversely, PTX3 plays a significant role in the development of several autoimmune disorders (9). PTX3 also plays a pleiotropic role in the pathobiology of a variety of cancers. For example, high levels of PTX3 are associated with an inferior outcome in ovarian (10), prostate (11), and head and neck cancers (12). In hepatocellular carcinoma PTX3 accelerates metastasis (13) and in glioblastoma overexpression of PTX3 is associated with high-grade tumor burden and disease severity (14).
Given that PTX3 exerts anti-bacterial functions and induces autoimmunity, and because in patients with CLL serum levels of CRP are high (15), we wondered whether CLL patients harbor high levels of PTX3 and whether, like circulating myeloid cells (16), CLL cells produce PTX3. Furthermore, because in glioblastoma, like in CLL (17), the signal transducer and activator of transcription-3 (STAT3) is continuously phosphorylated on 727 residues and is constantly activated (18, 19), we hypothesized that STAT3 promotes the transcriptional activity of the PTX3 gene in CLL cells. In addition, bearing in mind the role of PTX3 in occurrence and development of various neoplasms (6), we aimed to uncover what role PTX3 plays in the pathogenesis of CLL.
Materials and Methods
CLL patients’ and healthy donors’ peripheral blood sample processing
Heparinized peripheral blood (PB) samples from 30 treatment-naive CLL patients followed at The University of Texas MD Anderson Cancer Center Leukemia Clinic and from 10 healthy individuals were analyzed. Informed consent was obtained under a protocol approved by the Institutional Review Board. Supplemental Table SI depicts the patients’ clinical characteristics. All PB samples were transferred to 15 ml tubes to allow cell sedimentation, plasma was removed and stored at −20°C, and low-density cells (LDCs) were isolated based on a Ficoll-Hypaque 1077 density gradient (Sigma-Aldrich, St. Louis, MO). LDC lymphocytes from the studied CLL patients overwhelmingly expressed cell surface CD19 antigens (>90%), as determined by flow cytometry using an upgraded FACSCalibur flow cytometer (Becton Dickinson, Franklin Lakes, NJ). Normal B lymphocytes or CLL patients’ CD19+ cells were isolated from healthy individuals’ or CLL patients’ LDCs using paramagnetic MACS-nanoparticles and ferromagnetic separation system (Miltenyi Biotec, Auburn, CA). CD19 antigen expression ≥98% was confirmed by flow cytometry.
Enzyme-linked immunosorbent assay (ELISA)
To assess the plasma levels of PTX3, SAP and CRP, we performed ELISA using commercially available kits (Origene Technologies, Rockville, MD; Abnova, Taipei, Taiwan; Invitrogen, Waltham, MA). Briefly, patients’ and healthy individuals’ plasma was incubated in microplate wells in duplicate for 2 h at 37°C. After plasma removal, the wells were washed with PBS (Invitrogen) and incubated with PTX3 antibodies for 2 h. After washing with PBS, a horseradish peroxidase-conjugated detection antibody was added to the microplate wells to detect bound IgGs and color development was measured at 490 nm filter using an EL309 automated microtiter reader (BioTek Instruments, Winooski, VT).
RNA extraction and reverse transcription PCR
To extract total RNA from CLL cells we used TRIzol solution (Invitrogen). RNA quality and concentration were determined using ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE). Total RNA (1 μg) was converted into cDNA using SuperScript First-Strand synthesis system (Invitrogen). PCR reactions were performed using primers listed in Supplemental Table SII and hot-start TaKaRa Taq DNA polymerase (Takara Bio USA, San Jose, CA). PCR products were separated by electrophoresis on an ethidium bromide-infused 2% agarose gel with a GeneRuler 1 kb Plus DNA ladder (Thermo Scientific, Waltham, MA) as reference.
Quantitative real-time PCR (qRT-PCR) analyses
To quantitate mRNA levels of STAT3-target genes we used PCR primers with FAM dye-labeled probes (Supplemental Table SII) and TaqMan Universal PCR master mix (Applied Biosystems, Waltham, MA, USA). qRT-PCR analysis was performed in triplicate on QuantStudio 7 Flex system (Applied Biosystems). Relative gene expression was calculated using delta-delta cycle threshold (Ct) method with GAPDH as endogenous control.
To quantitate expression of genes associated with cell death and survival we used 84-plex SYBR green-based RT2 Profiler PCR array (Human Cell Death PathwayFinder; Qiagen, Germantown, MD). A total of 5 μg of RNA was converted into cDNA using RT2 First Strand kit (Qiagen) and qRT-PCR analysis was performed in duplicate on QuantStudio 7 Flex platform. Relative gene expression was calculated using delta-delta Ct method where Ct value of each gene was normalized to the mean Ct value of 5 housekeeping genes (ACTB, B2M, GAPDH, HPRT1, and RPLP0).
Western immunoblotting
Protein blotting was conducted according to the method we reported before (20). Briefly, by using the Pierce BCA protein assay reagent (Thermo Fisher Scientific, Waltham, MA), cell-derived protein levels were adjusted so that the same protein concentration was used in each experiment. After mixing with 4× Laemmli sample buffer, cell lysates were denatured by heat, electrophoretically separated, transferred to nitrocellulose membranes, and incubated at 4°C overnight. The membranes stained with Ponceaus S stain to verify equal protein loading were blocked, incubated with primary antibodies and horseradish peroxidase-conjugated secondary antibodies (GE Healthcare, Chicago, IL), and probed using chemiluminescence. Antibodies used in the western immunoblotting are listed in Supplemental Table SII.
Flow cytometry
Live PB LDCs were analyzed by flow cytometry. Before staining, cells were triple washed with 2% FBS (Invitrogen) in PBS. To detect cell surface CD19, CD5, and PTX3, unpermeabilized cells were incubated with fluorescently labeled antibodies (Supplemental Table SII) or their corresponding isotype controls. To detect intracellular PTX3 and phosphoserine STAT3, cells were fixed in 2% paraformaldehyde for 10 min at 37°C and permeabilized overnight at −20°C. Cells were then incubated with fluorescently labeled antibodies (Supplemental Table SII) or their corresponding isotype controls. Fluorescence signals were detected using an upgraded FACSCalibur flow cytometer (Becton Dickinson) and data analysis was performed using CellQuest software (BD Biosciences, Franklin Lakes, NJ). Graphic presentations were prepared using CellQuest (BD Biosciences) and WinList (Verity Software House, Topsham, ME) software.
Confocal microscopy
CLL cells were fixed with 2% paraformaldehyde at 37°C, permeabilized and kept overnight at −20°C. After being washed with PBS supplemented with 2% FBS (Invitrogen), the cells were incubated for 1 h in the presence of rabbit anti-PTX3 and mouse anti-STAT3 antibodies. After washing with PBS, the cells were incubated with Alexa Fluor 488-labeled anti-rabbit and Alexa Fluor 647-labeled anti-mouse antibodies (Supplemental Table SII). After being washed with PBS, the cells were suspended in DAPI dye (Invitrogen) for 15 min and then washed in PBS to remove the unbound dye. Finally, the cells were transferred onto µ-slide VI chamber slides (Ibidi, Fitchburg, WI), imaged using an upgraded FV1000 confocal microscope, and visualized using the FluoView software (Olympus, Tokyo, Japan).
Chromatin immunoprecipitation (ChIP)
Consistent with our previously described ChIP protocol (20), we utilized the SimpleChIP Enzymatic Chromatin IP kit (Cell Signaling Technology, Danvers, MA). Cell proteins were crosslinked to DNA in the presence of 1% formaldehyde and exposed to a lysis buffer on ice. Following nuclease-mediated digestion and ultrasonication, sheared chromatin was incubated with STAT3 antibodies overnight at 4°C (Supplemental Table SII). IgG was used as a negative control. Thereafter, protein-DNA complexes were separated using protein G beads, and the isolated DNA was amplified using PCR. The sequences of the STAT3-target gene primers used in the ChIP analysis are depicted in Supplemental Table SIII.
RNA interference assays
To study effects of STAT3 and PTX3 RNA interference we transfected the cells with short hairpin RNA (shRNA) and small interfering RNA (siRNA), respectively. STAT3-shRNA conjugated with green fluorescent protein was transfected using a lentiviral system as previously described (20). Specifically, pCMV delta R8.2 and pMDG plasmids were used for virion packaging and SuperFect reagents (Qiagen) were used for transfection into 293T cells. The empty lentiviral vector served as a negative control. Concentrated infection medium was obtained by centrifugal filtration of 293T cell supernatant using Amicon Ultra device (EMD Millipore, Burlington, MA) with addition of polybrene (10 ng/ml) and incubated with CLL cells in 10% FBS-supplemented DMEM (Thermo Fisher Scientific).
PTX3-siRNA was transfected into CLL cells using electroporation. Briefly, CLL cells were incubated for 1 h with siRNA and siPORT NeoFX reagent suspended in Opti-MEM reduced serum medium (Thermo Fisher Scientific). Then, cells were submitted to electrical pulses using Gene Pulser Xcell electroporation system (Bio-Rad Laboratories) and incubated in 10% FBS-supplemented RPMI 1640 medium for 24 h. Scrambled siRNA and GAPDH were used as internal controls. Transfection efficiency in both systems was assessed after 48 h by PI/GFP flow cytometry using an upgraded FACSCalibur flow cytometer (Becton Dickinson).
Electromobility shift assay (EMSA)
To process cell specimens for EMSA we used Pierce NE-PER extraction kit (Thermo Fisher Scientific) and incubated nuclear extracts with 4 biotinylated DNA probes targeting sequence of the PTX3 gene promoter. Each probe included one gamma (γ)-interferon activated sequence (GAS)-like element (Supplemental Table SIII) synthesized by Sigma-Genosys Biotechnologies (The Woodlands, TX). The CLL cell nuclear extracts were incubated with PTX3 promoter probes for 30 min at 4°C and separated by polyacrylamide gel electrophoresis. Following membrane transfer, DNA fragments were fixed via ultraviolet light-mediated crosslinking. The biotinylated probes were visualized using streptavidin-horseradish peroxidase (Gel-Shift Kit; Panomics, Fremont, CA). To verify binding specificity, an unlabeled cold probe was used at 7-fold excess concentration. The STAT3-induced effect was assessed by adding anti-STAT3 antibodies or mouse IgG (BD Biosciences) to the nuclear extracts, as described previously (20).
Dual-reporter luciferase assay
To assess effect of STAT3 activation on PTX3 gene transcription we electroporatically transfected MM1 cells with 4 different PTX3 promoter fragments using the Gene Pulser Xcell Electroporation System (Bio-Rad Laboratories, Hercules, CA). The constructs used for transfection consisted of a firefly luciferase reporter sequence and a fragment of PTX3 gene promoter region with 1 to 4 GAS-like elements depicted in Supplemental Table SIII. Following transfection MM1 cells were stimulated by IL-6 or left unstimulated and the luciferase activity was assessed post 24 h using a Dual-Luciferase Reporter Assay System (Promega, Madison, WI) and a BD MonolightTM 3010 luminometer (BD Biosciences). To calculate the reporter activity of individual fragments, the firefly luciferase activity produced by each construct was normalized to Renilla luciferase activity generated by pRL-SV40 control plasmid. STR-profiled and mycoplasma-free MM1 cell line was acquired from ATCC (Manassas, VA).
Quantitation of the cellular apoptosis rate
To assess the levels of apoptotic cell death in CLL cells we utilized annexin V and propidium iodide (PI) flow cytometry. After washing the cells in PBS the specimens were incubated with Cy5-conjugated annexin V and PI solution in the dark and submitted to flow cytometry analysis using an upgraded FACSCalibur flow cytometer (Becton Dickinson). Percent of annexin V+ cells was used to calculate the apoptotic rate of CLL cells.
Statistical analysis
The sample size was chosen to obtain sufficient power (≥80%) of a two-sided test with a level of significance α=0.05. Intergroup difference was detected using t-test or analysis of variance (ANOVA) followed by multiple comparisons by Tukey’s or Dunnett’s method, as appropriate. Calculations were performed and data were visualized using Prism software v8.0 (GraphPad Software, San Diego, CA). The alpha value that was considered to be statistically significant was P<0.05.
Results
CLL cells produce PTX3 and CLL patients’ plasma harbors high PTX3
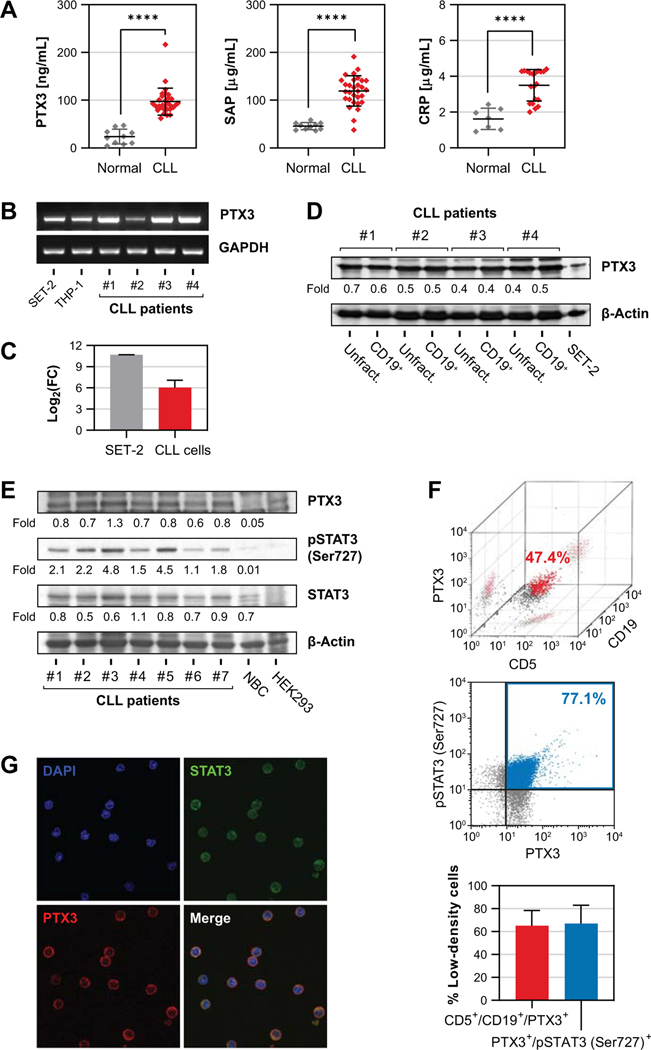
Because levels of the pentraxin CRP were found to be high in patients with CLL (15), we wondered whether CLL patients’ PTX3 levels are high as well. We obtained plasma samples from 30 previously untreated CLL patients and 10 age matched donors with no hematological disease and, by using ELISA, assessed the levels of the pentraxins PTX3, CRP and SAP. We found that levels of all three pentraxins were significantly higher in CLL patients’ plasma than in healthy donors’ plasma (Fig. 1A). Whereas liver cells are the main source of CRP, circulating cells such as monocytes and granulocytes produce PTX3 (21). Because the vast majority of circulating nucleated cells in CLL are CLL cells, we postulated that CLL cells produce PTX3. To test this hypothesis, we first sought to determine whether CLL cells express PTX3. We fractionated (98% to 99% purity as assessed by flow cytometry) CD19+ cells from the PB of 4 randomly selected patients with CLL. Using PCR we found that CLL patients’ CD19+ PB cells expressed PTX3 (Fig. 1B, C) and that PTX3 protein levels were similar in unfractionated and fractionated PB cells of all four patients as assessed by western immunoblot analysis (Fig. 1D). Then, we analyzed CLL cell lysates of 7 different CLL patients by immunoblotting and found high PTX3 protein levels in cell lysates obtained from all patients with CLL but not in the lysates obtained from healthy donors’ CD19+ B lymphocytes. In addition, as commonly found in CLL cells (20), we detected phosphoserine STAT3 and total STAT3 in CLL-cell lysates of those patients’ samples (Fig. 1E). To confirm that CLL cells produce PTX3 we used flow cytometry and confocal microscopy. Flow cytometric analysis gated on small, non-granulated cells revealed that 65% ± 13.4% (n = 4) of CD5- and CD19- expressing CLL cells co-expressed PTX3 and that 67.1% ± 16% (n = 4) of the cells expressed both phosphoserine STAT3 and PTX3 (Fig. 1F). Furthermore, confocal microscopy detected cytoplasmic PTX3 co-localizing with cytoplasmic STAT3 in CLL cells (Fig. 1G), indicating that PTX3, detected on cell surface, was also present in the cytoplasm of CLL cells. Together, these data suggest that CLL cells express, produce and likely release PTX3.
Figure 1.
CLL cells express PTX3. (A) PB was harvested from CLL patients and healthy donors and plasma levels of PTX3 (left panel), CRP (middle panel) and SAP (right panel) were measured by ELISA. Error bars denote mean with standard deviation. Comparisons between groups were made using unpaired t-test. ****, P<0.0001. (B and C) PTX3 transcript levels in PB LDCs from CLL patients were analyzed using reverse transcription PCR. As shown in panel B, PTX3 mRNA levels detected in specimens of 4 CLL patients were comparable to levels detected in the leukemic cell lines SET-2 (JAK2+ megakaryoblastic) and THP-1 (monocytic). Panel C depicts comparison of PTX3 gene expression between cells from CLL patients (n = 4) and SET-2 cells as assessed by qRT-PCR. mRNA levels were calculated using delta-delta Ct method with GAPDH as internal control and normalized to THP-1 cells. (D) Western immunoblotting analysis of unfractionated (Unfract.) and CD19+ fractionated cells obtained from 4 CLL patients. As shown, levels of PTX3 protein in total LDCs are comparable to levels in CD19-purified population. SET-2 cells served as positive control and β-actin was used as loading control. (E) Western blot analysis of CLL patients (n = 7) and B lymphocytes from a healthy donor. PTX3, STAT3 and serine 727-phosphorylated STAT3 (pSTAT3) proteins were observed in samples from all 7 patients. However, PTX3 and pSTAT3 were not observed in normal B lymphocytes (NBC). HEK293 cells were used as control. (F) Immunophenotype analysis of CLL cells by flow cytometry. Upper panel: Representative 3D dot-plot from a single CLL patient depicting 47.4% of the LDCs co-expressing CD5, CD19 and PTX3. Middle panel: Representative 2D dot-plot from a single CLL patient depicting 77.1% of the LDCs co-expressing phosphoserine STAT3 and PTX3. Lower panel: Summary of data from 4 different patients. As shown, 65% ± 13.4% of CD5- and CD19- expressing LDCs were also expressors of PTX3 and 67.1% ± 16% of the cells expressed both phosphorylated STAT3 and PTX3. Bars denote mean and error bars indicate standard deviation. (G) Fluorescence micrographs of permeabilized CLL cells by confocal microscopy. Depicted are STAT3 and PTX3 proteins in the cytoplasm of the B lymphocytes from a CLL patient. Original magnification, ×100.
STAT3 antibodies co-immunoprecipitate STAT3 protein and PTX3 DNA, and STAT3 silencing downregulates PTX3 levels
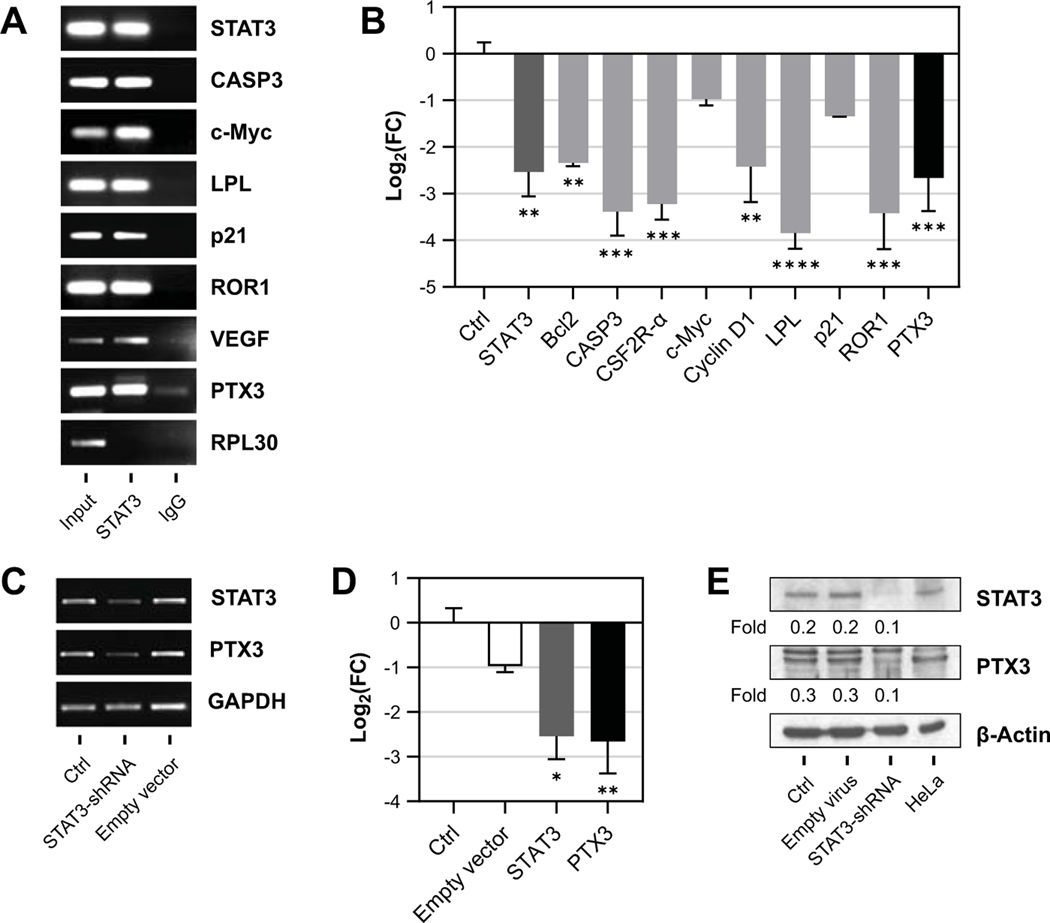
We and other investigators have previously demonstrated that STAT3 in CLL cells is constitutively activated (17, 20, 22) and, as a key regulator of transcription (20, 23–28), provides CLL cells with survival advantage (22). Because CLL patients’ plasma harbors high levels of PTX3 and since CLL cells co-express PTX3 and phosphoserine STAT3 (Fig. 1), we wondered whether expression of PTX3 is induced by constitutively activated STAT3. First, we asked whether STAT3 binds DNA containing the PTX3 gene. We isolated CLL cell extracts and, using ChIP, we found that STAT3 antibodies precipitated STAT3 protein as well as PTX3 DNA and the DNA containing other genes induced by STAT3 – caspase 3 (CASP3) (29), c-Myc, lipoprotein kinase (LPL) (26), p21, tyrosine kinase like orphan receptor-1 (ROR1) (30), STAT3, and vascular endothelial growth factor (VEGF) (31) (Fig. 2A).
Figure 2.
STAT3 antibodies co-immunoprecipitate STAT3 protein and PTX3 DNA, and STAT3-shRNA abrogates PTX3 expression in CLL cells. (A) Chromatin immunoprecipitation of DNA from CLL cell extracts was performed to assess STAT3 binding to the PTX3 gene. After incubation with STAT3 antibodies, chromatin fragments were precipitated and submitted to DNA extraction. Ribosomal protein L-30 (RPL30) was used as negative control. Preprocessed samples that were not submitted to immunoprecipitation are denoted as input and IgG represents isotype control (negative control). (B) CLL cells transfected with STAT3-shRNA were compared to CLL cells transfected with an empty vector (Ctrl). mRNA levels of STAT3-target genes were quantified using qRT-PCR, including STAT3, BCL2, CASP3, GMCSFR2, CyclinD1, LPL, p21, ROR1, and PTX3. The mRNA levels in CLL cells transfected with STAT3-shRNA were calculated using delta-delta cycle threshold method. (C-E) mRNA levels of PTX3 transcripts from CLL cells transfected with STAT3-shRNA as assessed by qRT-PCR. Empty lentiviral vector was used as a negative control. As shown in panel C, levels of PTX3 transcript were markedly reduced compared to non-transfected cells or cells transfected with an empty vector. GAPDH was used as internal control. As shown in panel D, similar results were obtained using qRT-PCR and, as shown in panel E, PTX3 protein levels, assessed by western immunoblotting, were significantly lower in STAT3-shRNA-treated CLL cells. In contrast, the transfected CLL cells exhibited 3-fold lower levels of PTX3 compared with control, non-transfected cells, or with cells transfected with an empty vector. In this experiment, the positive control was HeLa cell line and the loading control was β-actin. Bars denote mean and error bars indicate standard deviation. Differences in gene expression were determined using one-way ANOVA and Dunnett’s multiple comparison test. FC, fold change. *, P<0.05; **, P<0.01; ***, P<0.001; ****, P<0.0001.
Because these data suggested that STAT3 binds the PTX3 gene, we sought to determine whether STAT3 induces the expression of PTX3. We transfected CLL cells with STAT3-shRNA and found that knockdown of STAT3 downregulated PTX3 transcript levels, but also transcript levels of other STAT3-activated genes such as B-cell lymphoma-2 (Bcl2), CASP3 (29), colony stimulating factor-2 alpha receptor (CSF2R-α) (27), c-Myc, cyclin-D1, LPL (26), p21, and ROR1 (30) (Fig. 2B). Furthermore, we found that unlike the lentiviral empty vector, STAT3-shRNA significantly reduced STAT3 and PTX3 gene expression and protein levels (Fig. 2C-E), indicating that STAT3 transcriptionally activates the PTX3 gene.
The PTX3 gene promoter is activated by STAT3
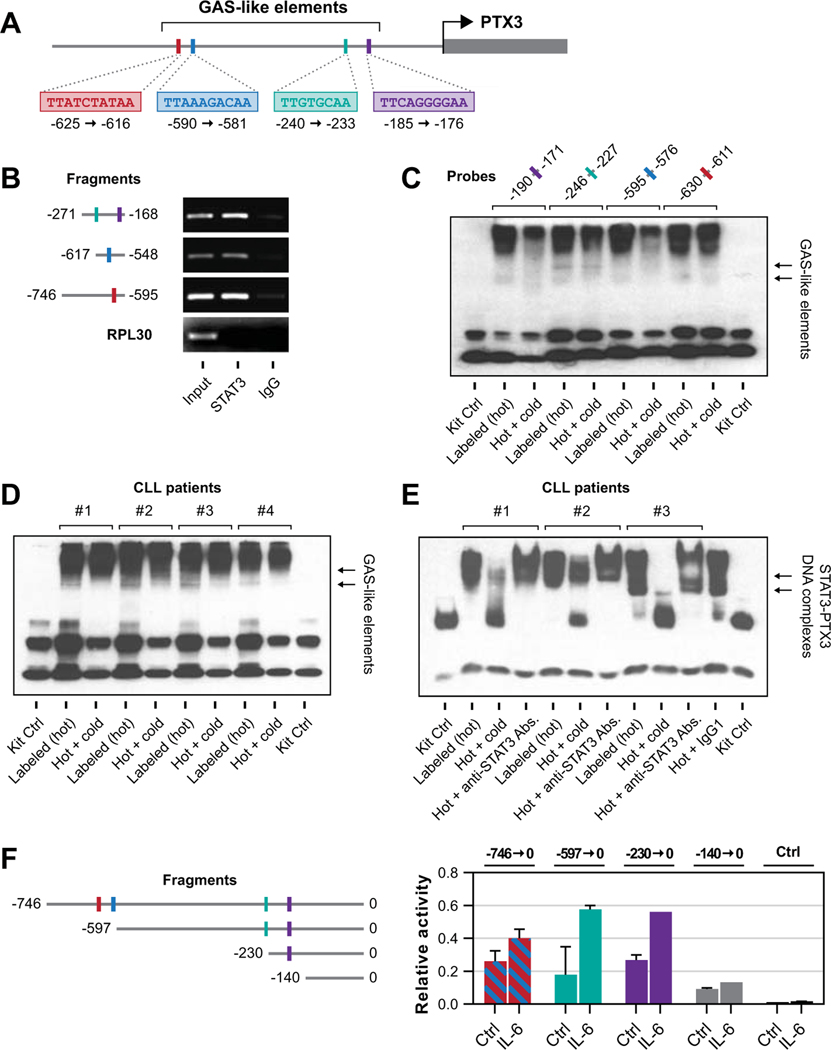
To delineate these findings and to determine whether the PTX3 gene promoter region harbors STAT3-binding sites, we performed sequence analysis of the PTX3 gene. Using TFSEARCH online tool we detected four GAS-like elements in the promoter region constituting putative STAT3 binding sites (Fig. 3A). To test if any of these GAS-like elements are bound by STAT3, we designed three pairs of primers to amplify truncated segments harboring putative STAT3 binding site regions. Primers 1 and 2 spanned one STAT3 binding site whereas primer 3 spanned two proximal sites. Then, using ChIP, we demonstrated that STAT3 antibodies precipitated DNA of each of those 3 truncated PTX3 gene promoter regions (Fig. 3B), suggesting STAT3 binds the PTX3 gene promoter at these genetic loci.
Figure 3.
STAT3 binds the PTX3 promoter and promotes transcription of the PTX3 gene. (A) Results of the analysis of the PTX3 promoter genomic sequence. The arrow denotes the PTX3 gene start codon. As shown, we identified 4 GAS-like elements upstream of the PTX3 gene that represent putative binding sites for STAT3. (B) ChIP analysis revealed that STAT3 protein and DNA fragments containing PTX3 gene promoter from CLL cells were co-immunoprecipitated by anti-STAT3 antibodies (Input). In contrast, isotype control (IgG) did not precipitate CLL-cell chromatin fragments. The first fragment, spanning from −271 bp to −168 bp, harbors 2 putative STAT3-binding sites whereas each one of the two other fragments harbors one putative STAT3 binding site. RPL30 served as negative control. (C) CLL cell nuclear extracts were incubated with biotinylated (hot) probes targeting GAS-like elements within the PTX3 gene promoter that corresponded to putative STAT3 binding sites. Probes included individual putative STAT3-binding sites according to the sequence analysis of the PTX3 promoter as shown in panel A. Using EMSA we detected binding of CLL nuclear extracts in all 4 sites, whereas the incubation with a non-biotinylated (cold) probe reduced the binding. (D) Nuclear extracts obtained from CLL cells of 4 patients were incubated with biotinylated (hot) probes designed to contain one of the putative STAT3 binding sites. Shown are results of EMSA performed on CLL nuclear extracts from all 4 patients. Whereas STAT3 antibodies bound the putative binding site, the unlabeled (cold) probe attenuated the STAT3 binding. (E) Similar to the previous experiment nuclear extract from 3 different CLL patients were analyzed using EMSA. As shown, the unlabeled (cold) probe or anti-STAT3 antibodies (Abs.), but not their isotype control (IgG1), significantly reduced the binding of the labeled (hot) probe, suggesting that STAT3 binds to the PTX3 gene promoter. (F) The luciferase assay confirmed that STAT3 induces the expression of PTX3. Left panel: Schematic diagram of the PTX3 gene promoter fragments analyzed in these experiments. Right panel: Reporter construct consisting of luciferase gene and fragments of the PTX3 gene promoter harboring putative STAT3-binding sites that were used for transfection into MM1 cells (legend in the left panel). IL-6, known to phosphorylate STAT3, was added and relative luciferase activity was assessed. As shown, no luciferase activity was detected in the short fragment lacking STAT3 binding site whereas luciferase activity, enhanced by IL-6 was detected by transfecting fragments harboring STAT3 binding site into MM1 cells. Data from 3 different experiments are depicted. Bars denote mean and error bars indicate standard deviation.
To further confirm that STAT3 binds the PTX3 gene promoter and identify which of the four GAS-like elements bind STAT3, we utilized an EMSA. Using biotin-labeled DNA probes (Supplemental Table SIII) targeting each of the 4 putative STAT3 binding sites within the PTX3 gene promoter, we found that all 4 DNA probes aggregated with CLL cell nuclear fraction. The addition of unbiotinylated (cold) probes partially reversed the binding (Fig. 3C), confirming adequate specificity and suggesting that all putative binding sites within the PTX3 promoter bind CLL-cell-derived nuclear protein. To validate that these findings are common in CLL cells, we obtained nuclear extracts from CLL cells of 4 different randomly selected CLL patients and, using probe 2, confirmed that CLL-cell nuclear protein binds the PTX3 promoter of 4 different CLL patients (Fig. 3D). Then, using STAT3 antibodies, we established that amongst the CLL cell nuclear extract proteins STAT3 is the protein that binds the PTX3 gene promoter. Using EMSA with probe 2 and nuclear extracts from 3 different randomly selected CLL patients we found that the addition of STAT3 antibodies induced a super-shift or reduced the protein-to-DNA binding (Fig. 3E), confirming that STAT3 binds to promoter of the PTX3 gene.
To delineate these findings and to test whether STAT3 activates the PTX3 gene, we used MM1 cells. In MM1 cells, STAT3 is transcriptionally inactive and extracellular ligands, such as IL-6, phosphorylate and activate STAT3 (32). Although IL-6-induced STAT3 activation in MM1 cells is mediated by phosphorylation on tyrosine 705, as opposed to the serine-727 phosphorylation in CLL cells, we have adapted this system as a model of STAT3 activation as previously reported (33). We transfected MM1 cells with a luciferase reporter gene attached to 5 different PTX3 promoter fragments, one of which did not harbor a STAT3 binding site. Importantly, transfected MM1 cells that lacked any putative GAS-like element did not produce any luciferase activity (Fig. 3F). In sharp contrast, a high additive luciferase activity was observed in cells that were transfected with 1 or 2 PTX3 promoter fragments harboring putative STAT3 binding site, particularly after stimulation with IL-6. Luciferase activity was similar in fragments that included 2, 3 or 4 putative STAT3 binding sites. It is important to note that IL-6 to minor extent also activates STAT1 through their common receptor subunit gp130 (32). However, STAT1 is recruited to the more restricted consensus GAS sites (34). Therefore stimulation of the PTX3 promoter is likely mediated primarily by STAT3, as is the case with most acute-phase protein-encoding genes (35).
PTX3 protects CLL cells from apoptosis
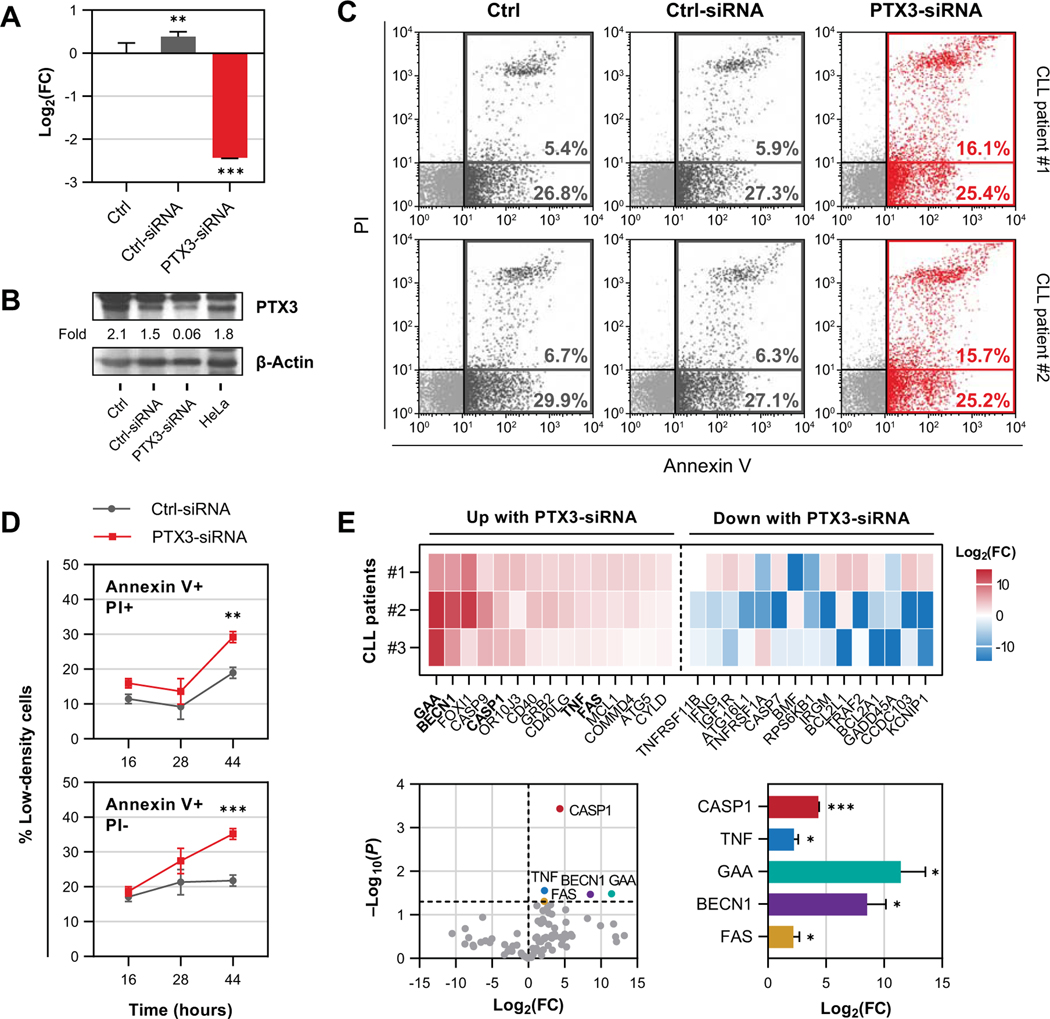
Because PTX3 expression is induced by STAT3, and because in solid tumors PTX3 provides neoplastic cells with a survival advantage (7, 14, 36), we wondered whether, similar to STAT3, PTX3 protects CLL cells from apoptosis. To answer this question, we transfected CLL cells with PTX3-siRNA. Using qRT-PCR and western immunoblotting we found that PTX3-mRNA, but not Ctrl (scrambled)-siRNA significantly reduced PTX3 expression and protein levels (Fig. 4A, B). Then, using annexin V/PI flow cytometry, we found that transfection with PTX3-siRNA induced apoptosis of CLL cells (Fig. 4C, D). As shown in Fig. 4C, CLL cells 16 h following transfection with PTX3-siRNA exhibited a higher spontaneous apoptosis rates than CLL cells transfected with Ctrl (scrambled)-siRNA. Although this effect was initially observed only in late (annexin V+/PI+) and not in early apoptotic cells (annexin V+/PI-), both apoptotic fractions increased over time (Fig. 4D).
Figure 4.
PTX3 induces anti-apoptotic effect in CLL cells. (A and B) Apoptotic rates of CLL cells were assessed after transfection with PTX3-siRNA or Ctrl (scrambled)-siRNA. Untreated CLL cells served as negative control (Ctrl). As shown in panel A, transfection with PTX3-siRNA significantly attenuated PTX3 mRNA expression as assessed by qRT-PCR. Similarly, transfection with PTX3-siRNA significantly reduced PTX3 protein expression as assessed by western immunoblotting. These effects were not observed after transfecting CLL cells with scrambled siRNA. Differences in gene expression were determined using one-way ANOVA and Tukey’s multiple comparison test. (C and D) Apoptosis rates of CLL cells after PTX3 silencing was assessed using annexin V and PI flow cytometry. Shown in panel C are representative dot-plots from 2 different CLL patients. Transfection with PTX3-siRNA, but not with Ctrl (scrambled)-siRNA, induced higher rates of apoptosis as assessed after 16 h in annexin V+ gate. Non-transfected CLL cells were used as a negative control (Ctrl). Panel D summarizes time dependent changes in early (annexin V+/PI-; lower facet) and late apoptotic cell fractions (annexin V+/PI+; upper facet) apoptosis rates of LDCs from different CLL patients (n = 3) 16, 28, and 44 h following PTX3-siRNA transfection. As shown, apoptotic rates of PTX3-silenced CLL cells increased over time in both fractions and the most significant effect occurred 44 h after transfection. (E) Differential expression of genes associated with cell death and survival in CLL cells transfected with PTX3-siRNA compared to Ctrl (scrambled)-siRNA-transfected cells. RNA from CLL patients (n = 3) was analyzed using 84-plex qRT-PCR array. Upper panel: Heatmap displaying 15 most upregulated and 15 most downregulated genes following PTX3 silencing. Lower left panel: Volcano plot showing differentially expressed genes (in color). Lower right panel: Relative levels of differentially expressed genes. Bars or dots represent mean and error bars indicate standard deviation. Comparisons were made between siRNA-transfected groups using paired t-test. FC, fold change. *, P<0.05; **, P<0.01; ***, P<0.001.
To confirm that PTX3 silencing indeed induced apoptosis and not necrosis of CLL cells, we profiled expression of 84 genes associated with different aspects of cell death using a qRT-PCR array (Fig. 4E). Remarkably, we found that PTX3 silencing significantly induced upregulation of 3 canonical pro-apoptotic genes – caspase 1 (CASP1), tumor necrosis factor (TNF), and Fas receptor (FAS). In addition, we found significantly high levels of 2 genes involved in autophagy – α-glucosidase (GAA) and beclin-1 (BECN1). No changes in analyzed necrosis-associated genes (n = 28) or anti-apoptotic genes (n = 11) were observed. Taken together, our data suggest that PTX3 selectively protects CLL cells from programmed cell death by apoptosis.
Discussion
PTX3 is an innate immune protein secreted by myeloid and endothelial cells. Here we show that despite CLL patients’ substantial neutropenia (2), their PTX3 plasma levels are significantly higher than those of healthy individuals. We also demonstrate that CLL cells produce PTX3, that constitutively activated STAT3 induces the transcriptional activation of the PTX3 gene, and that PTX3 provides CLL cells with survival advantage.
The response to invading pathogens is carried out by the innate and the adaptive immune systems. Both systems recruits cellular and humoral immune response mechanisms that often interact with each other (37). While the humoral response of the adaptive immune system consists of production and release of antibodies, the humoral response of the innate immune system consists of humoral pattern recognition proteins including the pentraxins CRP, SAP and PTX3 (38). A strict distinction between cells that produce antibodies and cells that produce antibody-like proteins, such as PTX3, typifies all mammals. Under normal physiological conditions, B lymphocyte-derived cells produce antibodies whereas monocytes, monocyte-derived cells, neutrophils and endothelial cells produce and release PTX3 (39). Here we show that in CLL this strict dichotomy no longer exists. CLL cells are B cell-derived neoplastic cells that maintain the ability to produce and secrete antibodies. Yet, as we demonstrated in this study, they aberrantly produce PTX3.
We found that the expression of PTX3 in CLL cells is driven by STAT3, a transcription factor that is ubiquitously expressed in human cells (40). STAT3 regulates the activity of a wide range of cellular pathways. During embryonic life, STAT3 regulates cells growth and maintenance of pluripotency (41, 42). In adult life, STAT3 acts as a master activator of vital pathways including activation of cell survival programs (43), cell-cycle progression (44), and highly orchestrated processes such as angiogenesis (45). Normally, the activation of STAT3 depends on extracellular stimulation. However, in CLL cells STAT3 is constitutively activated and activates the transcription of a plethora of STAT3-target genes (17, 20). Here we demonstrate that the transcriptional promoter of the PTX3 gene harbors STAT3 binding sites. Upon binding to those sites, STAT3 activates the transcription of the PTX3 gene in CLL cells. Remarkably, STAT3 plays a dual role in activating the innate immune system. In addition to inducing the expression of the humoral pattern recognition molecule PTX3, STAT3 also activates cellular component of the innate system in CLL cells. STAT3 induces the expression ROR1 (30), Wnt5a (24), and GLI1 (23), members of the Wnt and Hedgehog signaling pathway. Both the Wnt and Hedgehog signaling pathways were implicated in activating the cellular component of the innate immune system (46, 47).
Similar to several inflammatory cytokines (48), including cytokines driven by STAT3 (49, 50), PTX3 protects CLL cells from undergoing spontaneous apoptosis. This effect is not unique to CLL cells. In agreement with our findings, several investigators found that PTX3 provides normal and neoplastic cells with survival advantage (7, 51–53). However, in CLL the effect of PTX3 is not restricted to CLL cell function. Along with directly inducing changes in CLL cells, PTX3 exerts several systemic manifestations. PTX3 exerts antibody-like anti-bacterial functions and complement activation (8, 54). In addition, PTX3 plays a role in the induction of autoimmunity (9). Whether PTX3 is directly involved in driving autoimmune disorders such as hemolytic anemia or immune thrombocytopenia, commonly found in patients with CLL, has not yet been determined.
PTX3 binds to the cellular Fϲγ receptor and plays a role in tissue remodeling. As such, PTX3 promotes the differentiation of CD14+ monocytes into fibrocytes that participate in the induction of tissue fibrosis (55). Increased PTX3 plasma levels were detected in patients with myelofibrosis (56), and PTX3 levels correlated with disease burden and clinical outcome (57). Reticulin fibrosis is commonly detected in the bone marrow of most CLL patients and the degree of reticulin fibrosis correlates with CLL patients’ survival (58). Because PTX3 plasma levels are increased in CLL patients, it is likely that PTX3 induces BM fibrosis and affects the clinical outcome of patients with CLL.
Given that PTX3 exerts pleiotropic effects that benefit neoplastic cells, several investigators suggested that inhibition of PTX3 would benefit patients with different neoplasms (9, 59, 60). Whether inhibition of PTX3 would mitigate disease activity or improve outcomes of patients affected by CLL remains to be determined.
Supplementary Material
Key Points.
Constitutively activated STAT3 induces the expression of PTX3 in CLL cells.
PTX3 attenuates spontaneous apoptosis rate of CLL cells.
Acknowledgements
We thank Dr. Giorgio Inghirami, Department of Pathology, University of Torino (Torino, Italy), for providing pCMV delta R8.2 and pMDG plasmids.
This work was supported by a grant from the CLL Global Research Foundation and by the Cancer Center Support Grant from the National Cancer Institute (NCI), National Institutes of Health (NIH) P30 CA016672.
Abbreviations used in this article:
- ChIP
chromatin immunoprecipitation
- CLL
chronic lymphocytic leukemia
- CRP
C-reactive protein
- Ct
cycle threshold
- GAS
gamma (γ)-interferon activated sequence
- LDC
low-density cell
- PB
peripheral blood
- PI
propidium iodide
- PTX3
pentraxin-related protein 3
- qRT-PCR
quantitative real time PCR
- SAP
serum amyloid P
- shRNA
short hairpin RNA
- siRNA
small interfering RNA
- STAT3
signal transducer and activator of transcription-3
Footnotes
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Rozovski U, Hazan-Halevy I, Keating MJ, and Estrov Z. 2014. Personalized medicine in CLL: Current status and future perspectives. Cancer Lett. 352: 4–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsang M, and Parikh SA. 2017. A Concise Review of Autoimmune Cytopenias in Chronic Lymphocytic Leukemia. Curr. Hematol. Malig. Rep. 12: 29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsiodras S, Samonis G, Keating MJ, and Kontoyiannis DP. 2000. Infection and immunity in chronic lymphocytic leukemia. Mayo Clin. Proc. 75: 1039–1054. [DOI] [PubMed] [Google Scholar]
- 4.Strati P, and Caligaris-Cappio F. 2011. A matter of debate in chronic lymphocytic leukemia: is the occurrence of autoimmune disorders an indicator of chronic lymphocytic leukemia therapy? Curr. Opin. Oncol. 23: 455–460. [DOI] [PubMed] [Google Scholar]
- 5.Hamblin TJ 2006. Autoimmune complications of chronic lymphocytic leukemia. Semin. Oncol. 33: 230–239. [DOI] [PubMed] [Google Scholar]
- 6.Wang Z, Wang X, Zou H, Dai Z, Feng S, Zhang M, Xiao G, Liu Z, and Cheng Q. 2020. The Basic Characteristics of the Pentraxin Family and Their Functions in Tumor Progression. Front. Immunol. 11: 1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doni A, Stravalaci M, Inforzato A, Magrini E, Mantovani A, Garlanda C, and Bottazzi B. 2019. The Long Pentraxin PTX3 as a Link Between Innate Immunity, Tissue Remodeling, and Cancer. Front. Immunol. 10: 712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deban L, Russo RC, Sironi M, Moalli F, Scanziani M, Zambelli V, Cuccovillo I, Bastone A, Gobbi M, Valentino S, Doni A, Garlanda C, Danese S, Salvatori G, Sassano M, Evangelista V, Rossi B, Zenaro E, Constantin G, Laudanna C, Bottazzi B, and Mantovani A. 2010. Regulation of leukocyte recruitment by the long pentraxin PTX3. Nat. Immunol. 11: 328–334. [DOI] [PubMed] [Google Scholar]
- 9.Wu Q, Cao F, Tao J, Li X, Zheng SG, and Pan H-F. 2020. Pentraxin 3: A promising therapeutic target for autoimmune diseases. Autoimmun. Rev. 19: 102584. [DOI] [PubMed] [Google Scholar]
- 10.Chang X, Li D, Liu C, Zhang Z, and Wang T. 2021. Pentraxin 3 is a diagnostic and prognostic marker for ovarian epithelial cancer patients based on comprehensive bioinformatics and experiments. Cancer Cell Int. 21: 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stallone G, Cormio L, Netti GS, Infante B, Selvaggio O, Di Fino G, Ranieri E, Bruno F, Prattichizzo C, Sanguedolce F, Tortorella S, Bufo P, Grandaliano G, and Carrieri G. 2014. Pentraxin 3: A Novel Biomarker for Predicting Progression from Prostatic Inflammation to Prostate Cancer. Cancer Res. 74: 4230–4238. [DOI] [PubMed] [Google Scholar]
- 12.Zhou H, He Y, Li L, Wu C, and Hu G. 2021. Identification novel prognostic signatures for Head and Neck Squamous Cell Carcinoma based on ceRNA network construction and immune infiltration analysis. Int. J. Med. Sci. 18: 1297–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song T, Wang C, Guo C, Liu Q, and Zheng X. 2018. Pentraxin 3 overexpression accelerated tumor metastasis and indicated poor prognosis in hepatocellular carcinoma via driving epithelial-mesenchymal transition. J. Cancer 9: 2650–2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Locatelli M, Ferrero S, Boneschi FM, Boiocchi L, Zavanone M, Gaini SM, Bello L, Valentino S, Barbati E, Nebuloni M, Mantovani A, and Garlanda C. 2013. The long pentraxin PTX3 as a correlate of cancer-related inflammation and prognosis of malignancy in gliomas. J. Neuroimmunol. 260: 99–106. [DOI] [PubMed] [Google Scholar]
- 15.Herishanu Y, Polliack A, Shenhar-Tsarfaty S, Weinberger R, Gelman R, Ziv-Baran T, Zeltser D, Shapira I, Berliner S, and Rogowski O. 2017. Increased serum C-reactive protein levels are associated with shorter survival and development of second cancers in chronic lymphocytic leukemia. Ann. Med. 49: 75–82. [DOI] [PubMed] [Google Scholar]
- 16.Bottazzi B, Doni A, Garlanda C, and Mantovani A. 2010. An Integrated View of Humoral Innate Immunity: Pentraxins as a Paradigm. Annu. Rev. Immunol. 28: 157–183. [DOI] [PubMed] [Google Scholar]
- 17.Frank DA, Mahajan S, and Ritz J. 1997. B lymphocytes from patients with chronic lymphocytic leukemia contain signal transducer and activator of transcription (STAT)1 and STAT3 constitutively phosphorylated on serine residues. J. Clin. Invest. 100: 3140–3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin G-S, Chen Y-P, Lin Z-X, Wang X-F, Zheng Z-Q, and Chen L. 2014. STAT3 serine 727 phosphorylation influences clinical outcome in glioblastoma. Int. J. Clin. Exp. Pathol. 7: 3141–3149. [PMC free article] [PubMed] [Google Scholar]
- 19.Ganguly D, Sims M, Cai C, Fan MY, and Pfeffer LM. 2018. Chromatin Remodeling Factor BRG1 Regulates Stemness and Chemosensitivity of Glioma Initiating Cells. Stem Cells 36: 1804–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hazan-Halevy I, Harris D, Liu Z, Liu J, Li P, Chen X, Shanker S, Ferrajoli A, Keating MJ, and Estrov Z. 2010. STAT3 is constitutively phosphorylated on serine 727 residues, binds DNA, and activates transcription in CLL cells. Blood 115: 2852–2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bottazzi B, Inforzato A, Messa M, Barbagallo M, Magrini E, Garlanda C, and Mantovani A. 2016. The pentraxins PTX3 and SAP in innate immunity, regulation of inflammation and tissue remodelling. J. Hepatol. 64: 1416–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishdorj G, Johnston JB, and Gibson SB. 2010. Inhibition of Constitutive Activation of STAT3 by Curcurbitacin-I (JSI-124) Sensitized Human B-Leukemia Cells to Apoptosis. Mol. Cancer Ther. 9: 3302–3314. [DOI] [PubMed] [Google Scholar]
- 23.Rozovski U, Harris DM, Li P, Liu Z, Jain P, Manshouri T, Veletic I, Ferrajoli A, Bose P, Thompson P, Jain N, Verstovsek S, Wierda W, Keating MJ, and Estrov Z. 2021. STAT3 induces the expression of GLI1 in chronic lymphocytic leukemia cells. Oncotarget 12: 401–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rozovski U, Harris DM, Li P, Liu Z, Jain P, Ferrajoli A, Burger JA, Bose P, Thompson PA, Jain N, Wierda WG, Uziel O, Keating MJ, and Estrov Z. 2019. STAT3-Induced Wnt5a Provides Chronic Lymphocytic Leukemia Cells with Survival Advantage. J. Immunol. 203: 3078–3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rozovski U, Harris DM, Li P, Liu Z, Jain P, Ferrajoli A, Burger J, Thompson P, Jain N, Wierda W, Keating MJ, and Estrov Z. 2018. STAT3-activated CD36 facilitates fatty acid uptake in chronic lymphocytic leukemia cells. Oncotarget 9: 21268–21280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rozovski U, Grgurevic S, Bueso-Ramos C, Harris DM, Li P, Liu Z, Wu JY, Jain P, Wierda W, Burger J, O’Brien S, Jain N, Ferrajoli A, Keating MJ, and Estrov Z. 2015. Aberrant LPL Expression, Driven by STAT3, Mediates Free Fatty Acid Metabolism in CLL Cells. Mol. Cancer Res. 13: 944–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li P, Harris D, Liu Z, Rozovski U, Ferrajoli A, Wang Y, Bueso-Ramos C, Hazan-Halevy I, Grgurevic S, Wierda W, Burger J, O’Brien S, Faderl S, Keating M, and Estrov Z. 2014. STAT3-Activated GM-CSFR alpha Translocates to the Nucleus and Protects CLL Cells from Apoptosis. Mol. Cancer Res. 12: 1267–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li P, Grgurevic S, Liu Z, Harris D, Rozovski U, Calin GA, Keating MJ, and Estrov Z. 2013. Signal Transducer and Activator of Transcription-3 Induces MicroRNA-155 Expression in Chronic Lymphocytic Leukemia. PLoS One 8: e64678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rozovski U, Harris DM, Li P, Liu Z, Wu JY, Grgurevic S, Faderl S, Ferrajoli A, Wierda WG, Martinez M, Verstovsek S, Keating MJ, and Estrov Z. 2016. At High Levels, Constitutively Activated STAT3 Induces Apoptosis of Chronic Lymphocytic Leukemia Cells. J. Immunol. 196: 4400–4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li P, Harris D, Liu Z, Liu J, Keating M, and Estrov Z. 2010. Stat3 Activates the Receptor Tyrosine Kinase Like Orphan Receptor-1 Gene in Chronic Lymphocytic Leukemia Cells. PLoS One 5: e11859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Badoux X, Bueso-Ramos C, Harris D, Li P, Liu Z, Burger J, O’Brien S, Ferrajoli A, Keating MJ, and Estrov Z. 2011. Cross-talk between chronic lymphocytic leukemia cells arid bone marrow endothelial cells: role of signal transducer and activator of transcription 3. Hum. Pathol. 42: 1989–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heinrich PC, Behrmann I, Muller-Newen G, Schaper F, and Graeve L. 1998. Interleukin-6-type cytokine signalling through the gp130/Jak/STAT pathway. Biochem. J. 334: 297–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rozovski U, Wu JY, Harris DM, Liu Z, Li P, Hazan-Halevy I, Ferrajoli A, Burger JA, O’Brien S, Jain N, Verstovsek S, Wierda WG, Keating MJ, and Estrov Z. 2014. Stimulation of the B-cell receptor activates the JAK2/STAT3 signaling pathway in chronic lymphocytic leukemia cells. Blood 123: 3797–3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gerhartz C, Heesel B, Sasse J, Hemmann U, Landgraf C, SchneiderMergener J, Horn F, Heinrich PC, and Graeve L. 1996. Differential activation of acute phase response factor/STAT3 and STAT1 via the cytoplasmic domain of the interleukin 6 signal transducer gp130 .1. Definition of a novel phosphotyrosine motif mediating STAT1 activation. J. Biol. Chem. 271: 12991–12998. [DOI] [PubMed] [Google Scholar]
- 35.Schmitz J, Dahmen H, Grimm C, Gendo C, Muller-Newen G, Heinrich PC, and Schaper F. 2000. The cytoplasmic tyrosine motifs is full-length glycoprotein 130 have different roles in IL-6 signal transduction. J. Immunol. 164: 848–854. [DOI] [PubMed] [Google Scholar]
- 36.Rathore M, Girard C, Ohanna M, Tichet M, Ben Jouira R, Garcia E, Larbret F, Gesson M, Audebert S, Lacour JP, Montaudie H, Prod’Homme V, Tartare-Deckert S, and Deckert M. 2019. Cancer cell-derived long pentraxin 3 (PTX3) promotes melanoma migration through a toll-like receptor 4 (TLR4)/NF-kappa B signaling pathway. Oncogene 38: 5873–5889. [DOI] [PubMed] [Google Scholar]
- 37.Haseeb M, Anwar MA, and Choi S. 2018. Molecular Interactions Between Innate and Adaptive Immune Cells in Chronic Lymphocytic Leukemia and Their Therapeutic Implications. Front. Immunol. 9: 2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Razvina O, Jiang S, Matsubara K, Ohashi R, Hasegawa G, Aoyama T, Daigo K, Kodama T, Hamakubo T, and Naito M. 2015. Differential expression of pentraxin 3 in neutrophils. Exp. Mol. Pathol. 98: 33–40. [DOI] [PubMed] [Google Scholar]
- 39.Porte R, Davoudian S, Asgari F, Parente R, Mantovani A, Garlanda C, and Bottazzi B. 2019. The Long Pentraxin PTX3 as a Humoral Innate Immunity Functional Player and Biomarker of Infections and Sepsis. Front. Immunol. 10: 794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Demaria M, Camporeale A, and Poli V. 2014. STAT3 and metabolism: How many ways to use a single molecule? Int. J. Cancer 135: 1997–2003. [DOI] [PubMed] [Google Scholar]
- 41.Hirano T, Ishihara K, and Hibi M. 2000. Roles of STAT3 in mediating the cell growth, differentiation and survival signals relayed through the IL-6 family of cytokine receptors. Oncogene 19: 2548–2556. [DOI] [PubMed] [Google Scholar]
- 42.Humphrey RK, Beattie GM, Lopez AD, Bucay N, King CC, Firpo MT, Rose-John S, and Hayek A. 2004. Maintenance of pluripotency in human embryonic stem cells is STAT3 independent. Stem Cells 22: 522–530. [DOI] [PubMed] [Google Scholar]
- 43.Akira S 2000. Roles of STAT3 defined by tissue-specific gene targeting. Oncogene 19: 2607–2611. [DOI] [PubMed] [Google Scholar]
- 44.Fukada T, Ohtani T, Yoshida Y, Shirogane T, Nishida K, Nakajima K, Hibi M, and Hirano T. 1998. STAT3 orchestrates contradictory signals in cytokine-induced G(1) to S cell-cycle transition. EMBO J. 17: 6670–6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao P, Niu N, Wei T, Tozawa H, Chen X, Zhang C, Zhang J, Wada Y, Kapron CM, and Liu J. 2017. The roles of signal transducer and activator of transcription factor 3 in tumor angiogenesis. Oncotarget 8: 69139–69161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kling JC, and Blumenthal A. 2017. Roles of WNT, NOTCH, and Hedgehog signaling in the differentiation and function of innate and innate-like lymphocytes. J. Leukoc. Biol. 101: 827–840. [DOI] [PubMed] [Google Scholar]
- 47.Jridi I, Cante-Barrett K, Pike-Overzet K, and Staal FJT. 2021. Inflammation and Wnt Signaling: Target for Immunomodulatory Therapy? Front. Cell Dev. Biol. 8: 615131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rozovski U, Keating MJ, and Estrov Z. 2013. Targeting inflammatory pathways in chronic lymphocytic leukemia. Crit. Rev. Oncol. Hematol. 88: 655–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lian C, Huang Q, Zhong X, He Z, Liu B, Zeng H, Xu N, Yang Z, Liao C, Fu Z, and Guo H. 2021. Pentraxin 3 secreted by human adipose-derived stem cells promotes dopaminergic neuron repair in Parkinson’s disease via the inhibition of apoptosis. FASEB J. 35: e21748. [DOI] [PubMed] [Google Scholar]
- 50.Qiu L, Xu R, Wang S, Li S, Sheng H, Wu J, and Qu Y. 2015. Honokiol ameliorates endothelial dysfunction through suppression of PTX3 expression, a key mediator of IKK/I kappa B/NF-kappa B, in atherosclerotic cell model. Exp. Mol. Med. 47: e171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Daigo K, Nakakido M, Ohashi R, Fukuda R, Matsubara K, Minami T, Yamaguchi N, Inoue K, Jiang S, Naito M, Tsumoto K, and Hamakubo T. 2014. Protective effect of the long pentraxin PTX3 against histone-mediated endothelial cell cytotoxicity in sepsis. Sci. Signal. 7: ra88. [DOI] [PubMed] [Google Scholar]
- 52.Liu F, Yang L, Zhou X, Sheng W, Cai S, Liu L, Nan P, and Xu Y. 2014. Clinicopathological and genetic features of Chinese hereditary nonpolyposis colorectal cancer (HNPCC). Med. Oncol. 31: 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Choi B, Lee E-J, Song D-H, Yoon S-C, Chung Y-H, Jang Y, Kim S-M, Song Y, Kang S-W, Yoon S-Y, and Chang E-J. 2014. Elevated Pentraxin 3 in bone metastatic breast cancer is correlated with osteolytic function. Oncotarget 5: 481–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Deban L, Jaillon S, Garlanda C, Bottazzi B, and Mantovani A. 2011. Pentraxins in innate immunity: lessons from PTX3. Cell Tissue Res. 343: 237–249. [DOI] [PubMed] [Google Scholar]
- 55.Pilling D, Cox N, Vakil V, Verbeek JS, and Gomer RH. 2015. The Long Pentraxin PTX3 Promotes Fibrocyte Differentiation. PLoS One 10: e0119709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Verstovsek S, Manshouri T, Pilling D, Bueso-Ramos CE, Newberry KJ, Prijic S, Knez L, Bozinovic K, Harris DM, Spaeth EL, Post SM, Multani AS, Rampal RK, Ahn J, Levine RL, Creighton CJ, Kantarjian HM, and Estrov Z. 2016. Role of neoplastic monocyte-derived fibrocytes in primary myelofibrosis. J. Exp. Med. 213: 1723–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Veletic I, Manshouri T, Newberry KJ, Garnett J, Verstovsek S, and Estrov Z. 2019. Pentraxin-3 plasma levels correlate with tumour burden and overall survival in patients with primary myelofibrosis. Br. J. Haematol. 185: 382–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tadmor T, Shvidel L, Aviv A, Ruchlemer R, Bairey O, Yuklea M, Herishanu Y, Braester A, Levene N, Vernea F, Ben-Ezra J, Bejar J, Polliack A, and Israeli CLLSG. 2013. Significance of bone marrow reticulin fibrosis in chronic lymphocytic leukemia at diagnosis A Study of 176 Patients With Prognostic Implications. Cancer 119: 1853–1859. [DOI] [PubMed] [Google Scholar]
- 59.Chi J-Y, Hsiao Y-W, Li C-F, Lo Y-C, Lin Z-Y, Hong J-Y, Liu Y-M, Han X, Wang S-M, Chen B-K, Tsai KK, and Wang J-M. 2015. Targeting chemotherapy-induced PTX3 in tumor stroma to prevent the progression of drug-resistant cancers. Oncotarget 6: 23987–24001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tung J-N, Ko C-P, Yang S-F, Cheng C-W, Chen P-N, Chang C-Y, Lin C-L, Yang T-F, Hsieh Y-H, and Chen K-C. 2016. Inhibition of pentraxin 3 in glioma cells impairs proliferation and invasion in vitro and in vivo. J. Neurooncol. 129: 201–209. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.