Abstract
Biscationic quaternary phosphonium compounds (bisQPCs) represent a promising class of antimicrobials, displaying potent activity against both Gram-negative and Gram-positive bacteria. In this study, we explored the effects of structural rigidity on the antimicrobial activity of QPC structures bearing a two-carbon linker between phosphonium groups, testing against a panel of six bacteria, including multiple strains harboring known disinfectant resistance mechanisms. Using simple alkylation reactions, 21 novel compounds were prepared, although alkene isomerization as well as an alkyne reduction were observed during the respective syntheses. The resulting bisQPC compounds showed strong biological activity, but were hampered by diminished solubility of their iodide salts. One compound (P2P-10,10 I) showed single-digit micromolar activity against the entire panel of bacteria. Overall, intriguing biological activity was observed, with less rigid structures displaying better efficacy against Gram-negative strains and more rigid structures demonstrating slightly increased efficacy against S. aureus strains.
Keywords: Antimicrobial, Amphiphiles, Disinfectants, Phosphorus, QPCs
Graphical Abstract

The COVID-19 pandemic has increased the use of already-ubiquitous quaternary ammonium compounds (QACs), but QAC-resistance mechanisms are increasingly prevalent in bacteria. Our groups are developing improved disinfectants of distinctly different structures, and herein we expand our work in an entirely new class of molecules, the quaternary phosphonium compounds (QPCs), to explore rigidity-activity relationships in biscationic structures (bisQPCs).
Introduction
It is becoming increasingly clear that modern society remains highly susceptible to communicable pathogens. With a dwindling number of effective antibiotics, an increase in the regularity of epidemics and pandemics,[1] and a rise in bacterial resistance to our antimicrobial countermeasures, the need for novel antimicrobials has never been more urgent.[2] Disinfectants (for hard surfaces) and antiseptics (for skin) represent first lines of defense against pathogens. One recent report suggested that the use of antiseptics reduces the medical costs of hospital-acquired infections by up to $4.25B annually in the US alone.[3] A current review noted that quaternary ammonium compounds (QACs) have seen a major increase in usage since the beginning of the COVID-19 pandemic, which draws concern for the increased spread of disinfectant resistance through sub-lethal exposure of QACs to microbes.[4] In fact, it was noted that in a recent investigation, 90% of collected dust samples contained QACs, and that amount was found to have doubled since the beginning of the pandemic.[5] This is only exacerbated by their relative chemical stability, leading to prolonged exposure in the environment. Bacterial resistance to the activity of amphiphilic antimicrobials, once thought to be impossible, has become a present and pressing reality.[6]
Our research groups have spent a decade on the construction of over 700 novel cationic disinfectant structures, endeavoring to identify structure-activity relationships (SAR) that can guide the development of novel, safe, and effective antimicrobials.[7–8] Recently, to broaden the structural variety of amphiphilic antiseptics more significantly, we expanded our interests to the production of trivalent sulfonium compounds (TSCs)[9–12] and to quaternary phosphonium compounds (QPCs),[13–17] hypothesizing that expanding beyond positively charged nitrogen species would afford novelty to challenge bacterial resistance mechanisms.[18] A recent publication from our groups reported that select QPC structures demonstrate significant broad-spectrum antimicrobial activity, and in some circumstances, surpass some of the most potent commercial QAC compounds while showing no capitulation to bacterial resistance mechanisms.[13] For example, P6P-10,10 is a biscationic structure (bisQPC) with a simple 1-step synthesis and single-digit micromolar MIC values against a panel of bacteria bearing QAC-resistance mechanisms.
Since little SAR knowledge has been reported for QPCs,[16–17, 19–20] we aimed to mimic previous investigations that focused on modifying the rigidity of a biscationic amphiphile core. Our reports from 2019 and 2020 documented variations in bisQAC rigidity through the addition of linking methylene groups (Figure 1A) and pi-bond modification (Figure 1B), respectively.[21–22] Each of these investigations led to the elucidation of highly potent antimicrobials, although the more rigid TMEDA-based structures in Figure 1A[21] showed superior activity, while the more flexible alkane-linked bispyridinium compounds in Figure 1B[22] were more active than their unsaturated counterparts. We posited that cation proximity might play a role in this pair of observations.
Figure 1.
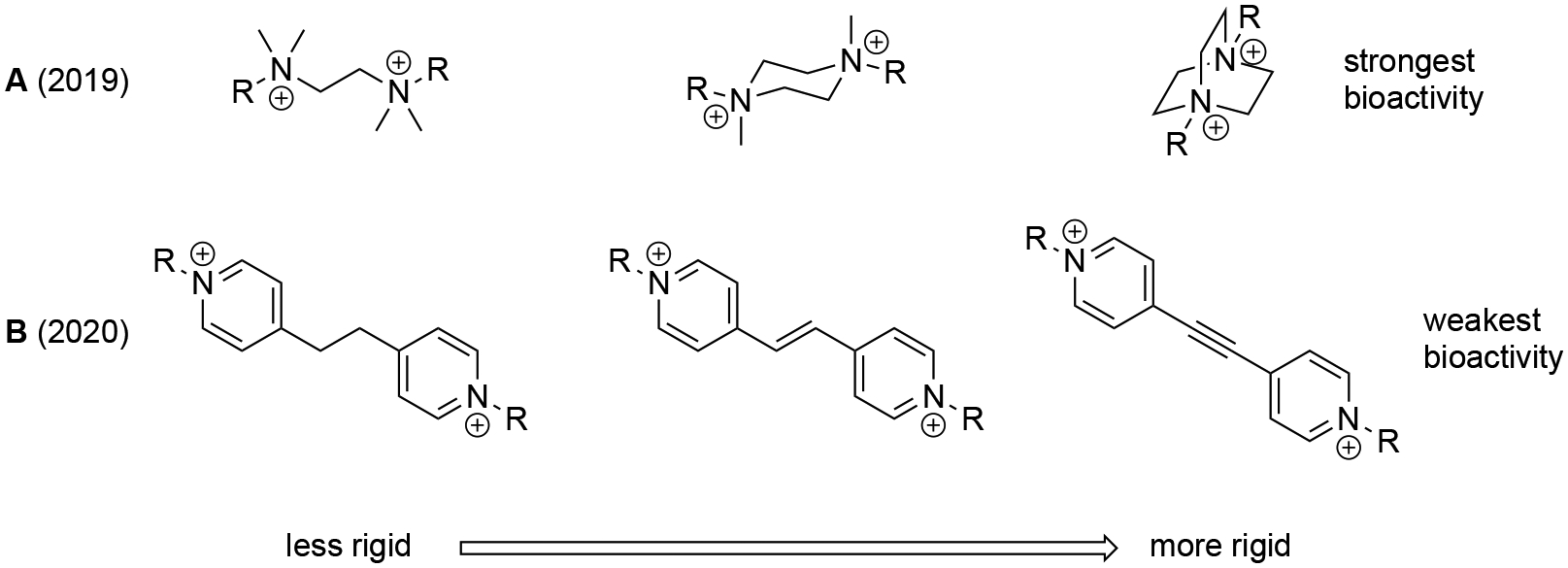
Previous efforts on rigidity-activity relationships in bisQACs.
Therefore, we became optimistic that the commercial availability of alkane, alkene, and alkyne linkers in an ethylene di(bisphenyl)phosphine backbone would open the door to a highly analogous set of bisQPCs with cations separated by a 2-carbon linker of varying rigidity, which we hypothesized may display differentiated antimicrobial activity, perhaps favoring the flexible alkyl linker. As illustrated in Figure 2, we envisioned synthesizing a set of novel bisQPC antimicrobial compounds featuring E-alkene, Z-alkene, and alkyne linkers and assessing the bioactivities of these structures to elucidate the relationship between bisQPC linker rigidity and antimicrobial activity.
Figure 2.
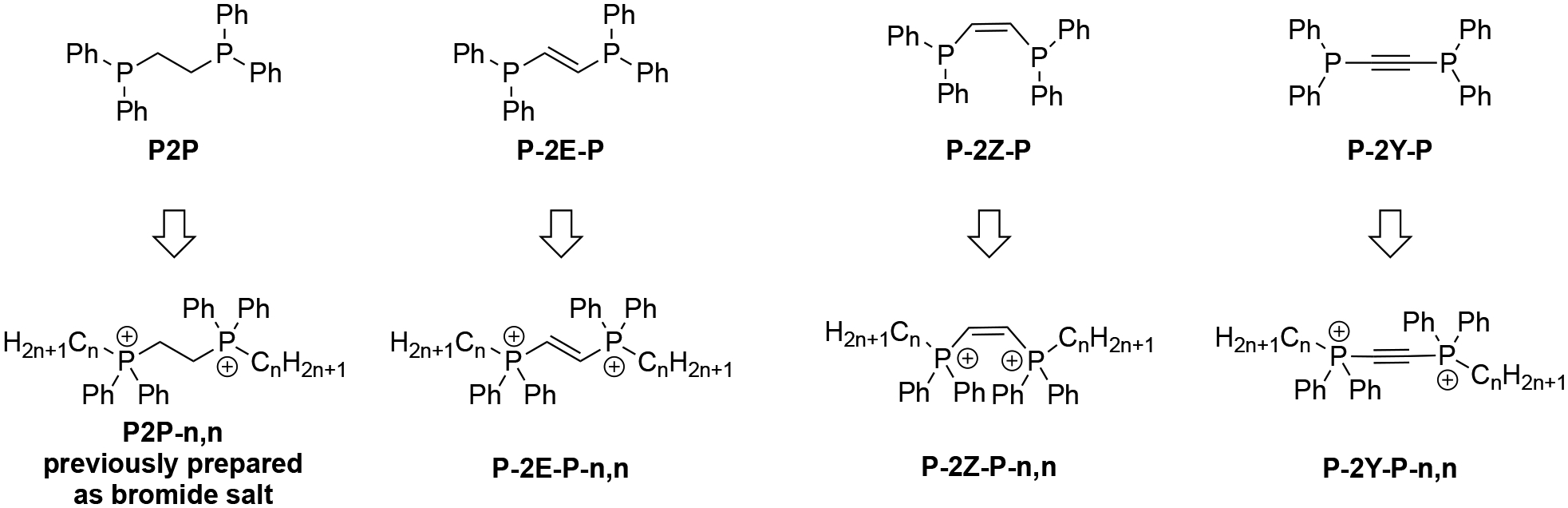
Anticipated preparation of a series of bisQPCs with varying linker rigidity.
Results and Discussion
Towards the synthesis of the rigid bisQPC library, we were initially disappointed to find that mimics of the alkylation conditions we had used in the past for the synthesis of P2P-n,n scaffolds (2.2 equiv. corresponding alkyl bromide, CH3CN, Δ, 24h) were unsuitable for bisphosphines linked by a pi-system, as sluggish alkylation reactions competed with phosphorus oxidation. Instead, a regimen of exposure to excess alkyl iodide in either DMF or acetonitrile, under a blanket of argon, led to varying degrees of success in these conjugated systems. Thus, we adopted this strategy to prepare an analogous set of iodide-containing salts. Accordingly, alkylation of P2P with varying alkyl iodides was, unsurprisingly, high yielding and clean, furnishing P2P-n,n compounds with iodide counterions in 71–95% yields, as illustrated in Scheme 1A. All compounds were purified with trituration, and complete compound characterization is provided in the Supporting Information. Alkylation of the analogous E-alkene (P-2E-P) was similarly effective, leading to comparable yields (57–87%) in similar reaction times (Scheme 1B). Analogous bisalkylation of the Z-alkene P-2Z-P, however, was not successfully accomplished, likely due to the significant steric hindrance of the four phenyl groups in close proximity. This was circumvented by exposure to the desired alkyl bromide, which led to clean monoalkylation, followed by methylation with iodomethane to produce the corresponding bisQPC structures; this was accomplished for three compounds (Scheme 1C). Inspection of 31P, 13C, and 1H NMR spectra of these P-2Z-P derived compounds indicated that the resulting products had isomerized to the E-alkenes; all 31P resonances were observed at precisely 23.8 ppm, matching the P-2E-P derived bisQPCs. This isomerization would seemingly minimize the significant steric strain imposed by the four phenyl groups.
Scheme 1.

Synthesis of P2P-n,n I series, P-2E-P-n,n I series; P-2E-P-n,1 series.
Finally, we found that the bis-alkylation of the alkyne structure proved challenging, showing little reactivity under standard conditions. Screening efforts elucidated a regime of exposure to 10 equiv. of alkyl iodide in DMF at reflux, followed by column chromatography; this led to what appeared to be a bisQPC, albeit in very low yields (≤16%; Scheme 2). Unfortunately, these harsh conditions led to a known side reaction first reported in 1991 by Peter Stang and coworkers;[23] not only was the alkyne reduced to the E-alkene as the literature indicated, the counterion was also edited to the triiodide (I3−) ion. Alkyne reduction was proposed[23] to be the result of participation by a second phosphorus equivalent, which presumably originates from another starting bisphosphine. We were successfully able to support our structural assertion through single crystal X-ray crystallography, as shown in Figure 3; the crystal structure of P2P-8,8 I is shown for comparison.
Scheme 2.
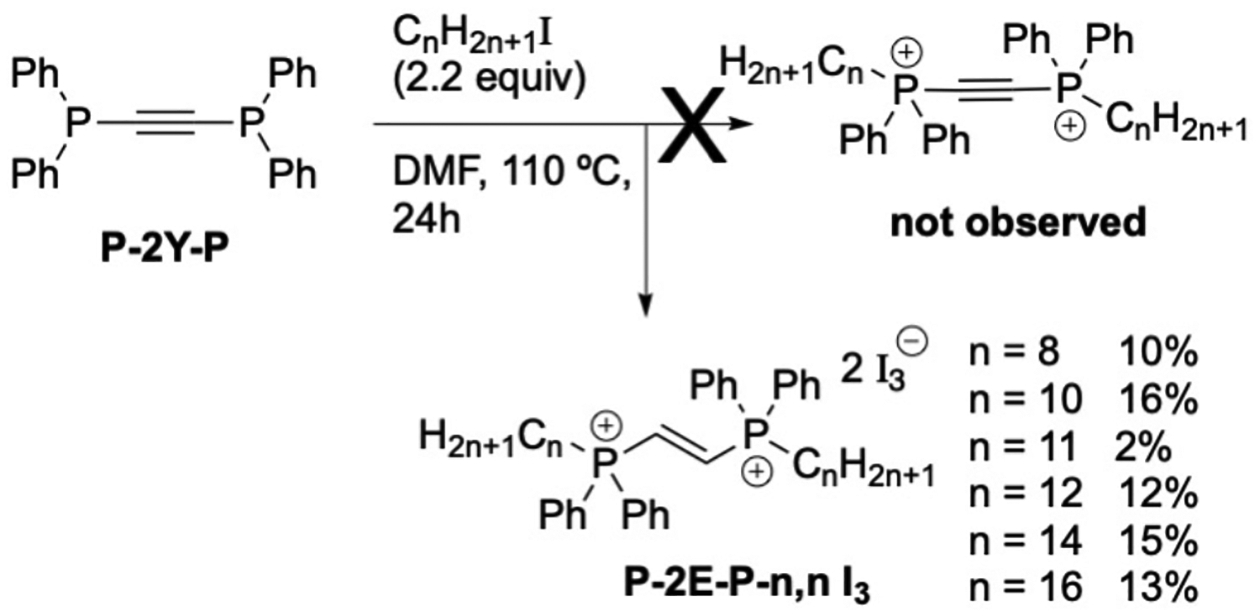
Attempted synthesis of P-2Y-P-n,n.
Figure 3.
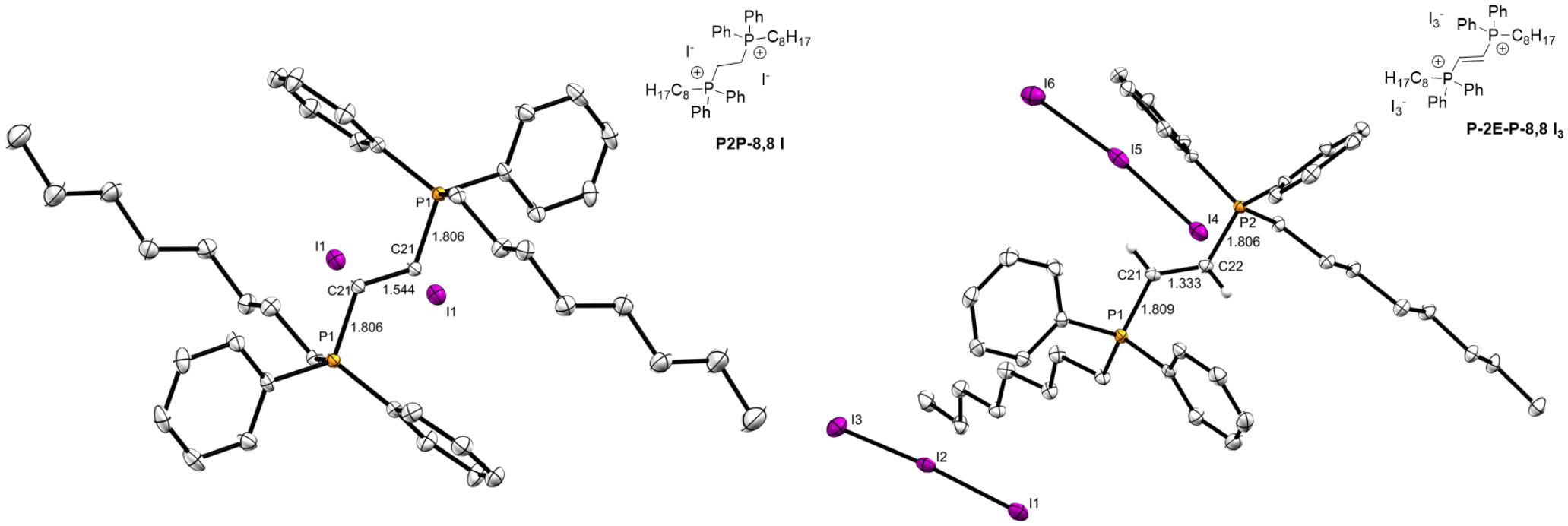
ORTEP diagrams for P2P-8,8 I (left) and P-2E-P-8,8 I3 (right), as a result of alkyne reduction. Bond lengths for the bridging P-C and C-C bonds are labeled in Å. Thermal ellipsoids are shown at the 50% probability level. Except for the hydrogens on the alkene, all other hydrogen atoms are omitted for clarity.
With 21 bisQPCs of varying structure and rigidity in hand, we advanced to assessment of biological activity. Solubility issues became apparent, with this group of compounds showing generally worse solubility as compared to the previously reported bromide counterparts. The bioactivities of the compounds were assessed via minimum inhibitory concentration (MIC) and hemolysis (lysis20) assays, wherein the latter was used as a proxy for cytotoxicity. In addition to the 21 bisQPC compounds, three commercial QACs were utilized for comparison: benzalkonium chloride (BAC; 70% benzyldimethyldodecylammonium chloride and 30% benzyldimethyltetradecylammonium chloride), cetylpyridinium chloride (CPC), and didecyldimethylammonium chloride (DDAC). To determine the MIC values, the compounds were each screened against a panel of six bacteria, including three Gram-positive strains (methicillin-susceptible Staphylococcus aureus [MSSA; SH1000], community-acquired methicillin-resistant S. aureus [CA-MRSA; USA 300–0114], hospital-acquired methicillin-resistant S. aureus [HA-MRSA; ATCC 33591]) and three Gram-negative strains (Enterococcus faecalis [OG1RF], Escherichia coli [MC4100], Acinetobacter baumannii [ATCC 17498]). To evaluate the hemolysis activities, the compound concentrations resulting in 20% red blood cell lysis (lysis20) were determined. The results of the MIC and hemolysis assays are reported below in Table 1.
Table 1.
Antimicrobial activity (MIC) and hemolysis (lysis20) of the prepared saturated and unsaturated bisQPCs compared to their saturated counterparts and commercially available QACs against Gram-positive strains methicillin-susceptible S. aureus (MSSA), community-acquired methicillin-resistant S. aureus (CA-MRSA) and hospital-acquired methicillin-resistant S. aureus (HA-MRSA) and Gram-negative strains E. faecalis, E. coli, and A. baumannii.

|
Previously published data for comparison. See Reference 13.
A few insights of structure-activity relationships became quickly apparent. Of first note, the theme continued that bisQPCs are promising antibacterial agents, with one new compound (P2P-10,10 I) showing single-digit micromolar activity against the entire panel of bacteria tested; other compounds such as P2P-11,11 I and P-2E-P 10,10 I showed comparable activity. With very few exceptions, methicillin-resistant bacteria (HA-MRSA and CA-MRSA) had bioactivity essentially equal to MSSA, supporting the observation that existing resistance mechanisms are not effective against this bisQPC species.
When comparing the more-flexible P2P compounds bearing an alkyl linker with those bearing a rigidified alkene linker, the differentiation was somewhat nuanced. While overall activity against S. aureus (SA) is roughly comparable, several alkene-bearing QPCs showed improved activity. For example, the P-2EP-n,n series with chain lengths of n = 10, 11, 12, and 14 all showed a two-to-eight-fold increase in activity (MIC values of ≤4 μM) against MSSA and CA-MRSA strains tested compared to their alkyl QPC counterparts. Conversely, examination of the data for the Gram-negative strains, the alkyl bridged P2P series (e.g., P2P-10,10 I and P2P-11,11 I) displayed superior activity compared to the rigidified structures. It is thus difficult to declare a superior broad-spectrum bisQPC; ease in preparation and higher yields suggest that the alkyl-linked QPCs would be better antimicrobial candidates.
Asymmetric compounds, particularly P-2E-P-14,1 I, showed low micromolar activity against four of the six strains tested, except for Gram-negative species E. coli and A. baumannii. The triiodide compounds, however, fared significantly worse in the biological assays, showing instability as well as insolubility during the course of the tests. As a result, all these compounds were essentially inactive against the Gram-negative strains.
The cytotoxicity of these compounds, using red blood cell lysis as a proxy, was moderate, correlating with antimicrobial activity. This aligns with previous reports of the ability of cationic amphiphilic molecules to lyse eukaryotic lipid membranes in addition to bacterial cell membranes.[24–25]
Finally, it is noteworthy that these compounds, all bearing iodide counterions in order to best assemble an analogous set of structures bearing varied rigidity, fared worse overall than their bromide counterparts. We believe this to be primarily a function of solubility, which was uniformly worse for the iodide salts, akin to previous observations from our group in multiQAC investigations.[26]
Conclusion
In sum, this report has shown that bisQPC structures continue to be promising novel antimicrobial compounds. The incorporation of rigidity elements within their architecture did not lead to substantially differentiated bioactivity, though the diminished solubility of iodide salts led to decreased antimicrobial activity in an absolute sense. While pi-bond-derived rigidity was observed to be only a modest factor in these SAR analyses, it is still worth considering the addition of rigidity from alternative hydrocarbon linkers, which led to improved activity in TMEDA-derived bisQACs.
Experimental Section
Synthesis of QPC library.
Reagents and solvents were used from Sigma-Aldrich, City Chemical LLC, Alfa Aesar, and Fisher Chemical, without further purification. All reactions were carried out under an argon atmosphere unless otherwise noted, with reagent grade solvents and magnetic stirring. All yields refer to spectroscopically pure compounds. 1H, 13C, and 31P NMR spectra were measured with a 400MHz or 500MHz JEOL spectrophotometer, and chemical shifts were reported on a δ-scale (ppm) downfield from TMS or 85% H3PO4. Coupling constants were calculated in Hertz. The solvent used was chloroform-d (CDCl3), with an internal reference of 7.26 ppm for 1H NMR and 77.16 ppm for 13C NMR. Accurate mass spectrometry data was acquired on an AB Sciex 5600 TripleTOF using electrospray ionization in positive mode. Detailed synthetic procedures and characterization data are provided in the Supporting Information.
X-ray Crystallography.
X-ray intensity data for P2P-8,8 I (CCCD reference number 2156703) were collected on a Rigaku XtaLAB Synergy-S instrument equipped with a Dectris Pilatus3 R 200K HPC area detector. X-ray intensity data for P-2E-P-8,8 I3 (CCCD reference number 2156704) were collected on a Rigaku XtaLAB Synergy-S instrument with a HyPix-6000HE HPC area detector. All data were collected at a temperature of 100 K using Mo-Kα radiation (λ=0.71073 Å). Intensity data was integrated using CrysAlisPro[27] which produced a listing of unaveraged F2 and σ(F2) values. The structures were solved using SHELXT and refined using SHELXL.[28] The placement of hydrogen atoms was defined based upon calculated positions with respect to the parent atoms using a riding model and thermal parameters were refined isotropically. The thermal parameters for all non-hydrogen atoms were refined anisotropically. Detailed collection data for the complexes are available in the Supporting Information. Supplementary crystallographic data for all crystal structures can be obtained free of charge via http://www.ccdc.cam.ac.uk/conts/retrieving.html, or from the Cambridge Crystallographic Data Centre, 12 Union Road, Cambridge CB2 1EZ, UK; fax: (+44) 1223-336-033; or deposit@ccdc.cam.ac.uk.
Biological Assays.
For all biological assays, laboratory strains of methicillin-susceptible Staphylococcus aureus MSSA (SH1000), Staphylococcus aureus CA-MRSA (USA300–0114), Staphylococcus aureus HA-MRSA (ATCC 33591), Escherichia coli (MC4100), Enterococcus faecalis (OG1RF), Acinetobacter baumannii (ATCC 17978), were grown with shaking at 37 °C overnight from freezer stocks in 5 mL of the indicated media: SH1000, OG1RF, MC4100, USA300–0114, and ATCC 17978 were grown in BD™ Mueller-Hinton broth (MHB), whereas ATCC 33591 was grown in BD™ tryptic soy broth (TSB). The inoculum were then streaked onto Mueller-Hinton agar (MHA) plates and grown overnight at 37 °C. From the plates, single colonies were selected, diluted into 50 mL of fresh media and grown overnight at 37 °C with shaking for use in the MIC assays. Optical density (OD) measurements were obtained using a SpectraMax iD3 plate reader (Molecular Devices, United States).
Minimum Inhibitory Concentration (MIC) Assays.
Compounds were serially diluted two-fold from stock solutions (1.0 mM) to yield twelve 100 μL test concentrations, wherein the starting concentration of DMSO was 2.5%. Overnight S. aureus, E. faecalis, E. coli, A. baumannii, USA300–0114 (CA-MRSA), and ATCC 33591 (HA-MRSA) cultures were diluted to ca. 106 CFU/mL in MHB or TSB and regrown to mid-exponential phase, as determined by optical density recorded at 600 nm (OD600). All cultures were then diluted again to ca. 106 CFU/mL and 100 μL were inoculated into each well of a U-bottom 96-well plate (Corning, 351177) containing 100 μL of compound solution. Plates were incubated statically at 37 °C for 72 hours upon which wells were evaluated visually for bacterial growth. The MIC was determined as the lowest concentration of compound resulting in no bacterial growth visible to the naked eye, based on the highest value in three independent experiments. Aqueous DMSO controls were conducted as appropriate for each compound.
Red Blood Cell (RBC) Hemolysis Assay (Lysis20)
RBC lysis assays were performed on mechanically defibrinated sheep blood (Hemostat Labs: DSB030). An aliquot of 1.5 mL blood was placed into a microcentrifuge tube and centrifuged at 3,800 rpm for ten minutes. The supernatant was removed, and the cells were resuspended with 1 mL of phosphate-buffered saline (PBS). The suspension was centrifuged as described above, the supernatant was removed, and cells were resuspended 4 additional times in 1 mL PBS. The final cell suspension was diluted twentyfold with PBS. Compounds were serially diluted with PBS two-fold from stock solutions (1.0 mM) to yield 100 μL of twelve test concentrations on a flat-bottom 96-well plate (Corning, 351172), wherein the starting concentration of DMSO was 2.5%. To each of the wells, 100 μL of the twentyfold suspension dilution was then inoculated. TritonX (1% by volume) served as a positive control (100% lysis marker) and sterile PBS served as a negative control (0% lysis marker). Samples were then placed in an incubator at 37 °C and shaken at 200 rpm. After 1 hour, the samples were centrifuged at 3,800 rpm for ten minutes. The absorbance of the supernatant was measured with a UV spectrometer at a 540 nm wavelength. The concentration inducing 20% RBC lysis was then calculated for each compound based upon the absorbances of the TritonX and PBS controls. Aqueous DMSO controls were conducted as appropriate for each compound. This procedure was adapted from Peng et al.[29]
Supplementary Material
Acknowledgements
The authors thank Robert Carden for his assistance in evaluating earlier synthetic routes and Samantha Brayton for her help in obtaining mass spectrometric data.
This work was funded by the National Institute of General Medical Sciences (R35 GM119426 to W.M.W.; T32 GM008602 to C.A.S.) and Major Research Instrumentation grants from the National Science Foundation (CHE-1827930 and CHE-2018399 to K.P.C.M.).
References
- [1].Mehand MS, Al-Shorbaji F, Millett P, Murgue B, Antiviral Res 2018, 159, 63–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Gupta R, Science 2020, 370, 913–914. [DOI] [PubMed] [Google Scholar]
- [3].Schmier JK, Hulme-Lowe CK, Semenova S, Klenk JA, DeLeo PC, Sedlak R, Carlson PA, Clin. Outcomes Res 2016, 8, 197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Vereshchagin AN, Frolov NA, Egorova KS, Seitkalieva MM, Ananikov VP, Int. J. Mol. Sci 2021, 22, 6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Zheng G, Filippelli GM, Salamova A, Environ. Sci. Technol. Lett 2020, 7, 760–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Jennings MC, Minbiole KPC, Wuest WM, ACS Infect. Dis 2015, 1, 288–303. [DOI] [PubMed] [Google Scholar]
- [7].Morrison KR, Allen RA, Minbiole KPC, Wuest WM, Tetrahedron Lett 2019, 60, 150935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Minbiole KPC, Jennings MC, Ator LE, Black JW, Grenier MC, LaDow JE, Caran KL, Seifert K, Wuest WM, Tetrahedron 2016, 72, 3559–3566. [Google Scholar]
- [9].Feliciano JA, Leitgeb AJ, Schrank CL, Allen RA, Minbiole KPC, Wuest WM, Carden RG, Bioorg. Med. Chem. Lett 2021, 37, 127809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Nikolaev AE, Semenov VÉ, Voloshina AD, Kulik NV, Reznik VS, Pharm. Chem. J 2010, 44, 130–133. [Google Scholar]
- [11].Li L, Jia D, Wang H, Chang C, Yan J, Zhao ZK, New J. Chem 2020, 44, 303–307. [Google Scholar]
- [12].Hirayama M, Biocontrol Sci 2012, 17, 27–35. [DOI] [PubMed] [Google Scholar]
- [13].Sommers KJ, Michaud ME, Hogue CE, Scharnow AM, Amoo LE, Petersen AA, Carden RG, Minbiole KPC, Wuest WM, ACS Infect. Dis 2022, 8, 387–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Xue Y, Pan Y, Xiao H, Zhao Y, RSC Adv 2014, 4, 46887–46895. [Google Scholar]
- [15].Xue Y, Xiao H, Zhang Y, Int. J. Mol. Sci 2015, 16, 3626–3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kanazawa A, Ikeda T, Endo T, Antimicrob. Agents and Chemother 1994, 38, 945–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Terekhova NV, Tatarinov DA, Shaihutdinova ZM, Pashirova TN, Lyubina AP, Voloshina AD, Sapunova AS, Zakharova LY, Mironov VF, Bioorg. Med. Chem. Lett 2020, 30, 127234. [DOI] [PubMed] [Google Scholar]
- [18].Carden RG, Sommers KJ, Schrank CL, Leitgeb AJ, Feliciano JA, Wuest WM, Minbiole KPC, ChemMedChem 2020, 15, 1974–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kanazawa A, Ikeda T, Endo T, J. Polym. Sci. Part A 1994, 32, 1997–2001. [Google Scholar]
- [20].Lukáč M, Devínsky F, Pisárčik M, Papapetropoulou A, Bukovský M, Horváth B, J. Surfactants Deterg 2017, 20, 159–171. [Google Scholar]
- [21].Kontos RC, Schallenhammer SA, Bentley BS, Morrison KR, Feliciano JA, Tasca JA, Kaplan AR, Bezpalko MW, Kassel WS, Wuest WM, Minbiole KPC, ChemMedChem 2019, 14, 83–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Leitgeb AJ, Feliciano JA, Sanchez HA, Allen RA, Morrison KR, Sommers KJ, Carden RG, Wuest WM, Minbiole KPC, ChemMedChem 2020, 15, 667–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Stang PJ, Zhdankin VV, J. Am. Chem. Soc 1991, 113, 4571–4576. [Google Scholar]
- [24].Groothuis FA, Timmer N, Opsahl E, Nicol B, Droge STJ, Blaauboer BJ, Kramer NI, Chem. Res. Toxicol 2019, 32, 1103–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Yousif LF, Stewart KM, Kelley SO, ChemBioChem 2009, 10, 1939–1950. [DOI] [PubMed] [Google Scholar]
- [26].Grenier MC, Davis RW, Wilson-Henjum KL, LaDow JE, Black JW, Caran KL, Seifert K, Minbiole KPC, Bioorg. Med. Chem. Lett 2012, 22, 4055–4058. [DOI] [PubMed] [Google Scholar]
- [27].CrysAlisPro 1.171.41.122a: Rigaku Oxford Diffraction, Rigaku Corporation, Oxford, UK. (2021). [Google Scholar]
- [28].Sheldrick G, Acta Crystallogr. A 2015, 71, 3–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Peng L, DeSousa J, Su Z, Novak BM, Nevzorov AA, Garland ER, Melander C, Chem. Commun 2011, 47, 4896–4898. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


