SUMMARY:
Gene therapy offers great promise in addressing neuropathologies associated with the central and peripheral nervous systems (CNS and PNS). However, genetic access remains difficult, reflecting the critical need for development of effective and non-invasive gene delivery vectors across species. To that end, we evolved adeno-associated virus serotype 9 (AAV9) capsid in mice, and validated two capsids, AAV-MaCPNS1 and AAV-MaCPNS2, across rodent species (mice and rats) and non-human primate (NHP) species (marmosets and rhesus macaques). Intravenous administration of either AAV efficiently transduced the PNS in rodents, and both the PNS and CNS in NHPs. Furthermore, we used AAV-MaCPNS1 in mice to systemically deliver: (1) the neuronal sensor jGCaMP8s to record calcium signal dynamics in nodose ganglia, and (2) the neuronal actuator DREADD to dorsal root ganglia to mediate pain. This conclusively demonstrates the translatability of these two systemic AAVs across four species, and their functional utility through proof-of-concept studies in mice.
Graphical Abstract
In brief:
Chen et al. evolved a family of AAV capsids variants, including MaCPNS1 and MaCPNS2, that efficiently transduced the PNS in rodents following systemic administration, enabling functional readout and non-invasive modulation of PNS. Both vectors could also enable efficient gene delivery to both PNS and CNS in NHPs.
INTRODUCTION:
Gene delivery to the central and peripheral nervous systems (CNS and PNS) has greatly accelerated neuroscience research in the last decade, and has the potential to translate this research into novel therapies for neurological disorders. However, the lack of potent vectors enabling non-invasive gene delivery across species is a significant bottleneck that can hinder fast progress (Ingusci et al. 2019; Pena et al. 2020; Piguet et al. 2021).
The success of gene delivery relies on a safe and efficient vector, and for this reason, most in vivo applications use adeno-associated viral (AAV) vectors. AAVs offer several advantages, including stable, long-term transgene expression and low immunogenicity (Bedbrook, Deverman, and Gradinaru 2018; O’Carroll, Cook, and Young 2021). The natural serotypes of AAV have demonstrated considerable success in targeting different cell populations within the nervous system through direct routes of gene delivery, such as intracranial (Golebiowski et al. 2017; GuhaSarkar et al. 2017), intracerebroventricular (ICV) (Bey et al. 2020), intrathecal (Hirai et al. 2012), intraganglionic (Yu et al. 2013), intrasciatic (Homs et al. 2011) and intracolonic (Gore et al. 2019). These direct delivery routes suffer from limitations, however, including the need for invasive surgery. In addition, anatomical barriers may restrict surgical access (such as for nodose ganglia (NG) or dorsal root ganglia (DRG)). Finally, localized delivery can lead to incomplete coverage of a large complex system such as the enteric nervous system (ENS) or CNS, and multiple direct interventions may be needed to increase coverage (Gray, Woodard, and Samulski 2010).
An alternative, non-invasive, intravenous (IV) route circumvents these limitations. Some natural serotypes, including AAV9, can target the CNS or PNS systemically. However, lack of specificity towards the target and low efficiency, necessitating high vector load, both potentially lead to toxicity issues (Bourdenx et al. 2014; B. Yang et al. 2014; Vogt et al. 2015; Gombash et al. 2017). Years of capsid engineering efforts have now yielded a toolbox of improved CNS capsids for rodents (Deverman et al. 2016; Körbelin et al. 2016; Ojala et al. 2018; Ravindra Kumar et al. 2020; Nonnenmacher et al. 2021). These include the potent vector AAV-PHP.eB (engineered using CREATE) for the CNS, but its application is restricted to select mice strains (Deverman et al. 2016; Chan et al. 2017; Hordeaux et al. 2018; Matsuzaki et al. 2018; Hinderer et al. 2018). Unlike PHP.B/eB, the recently engineered AAV-F (engineered using iTransduce) and AAV-PHP.Cs (engineered using Multiplexed-CREATE or M-CREATE) for the CNS work across mouse strains (Hanlon et al. 2019; Ravindra Kumar et al. 2020). However, the heterogeneity of the blood-brain barrier (BBB) depending on genetic background has posed a significant challenge for developing capsids that have the potential to translate across species (Hordeaux et al. 2019; Q. Huang et al. 2019; Batista et al. 2020). This issue is particularly acute for non-human primates (NHPs), which are commonly used as pre-clinical research models for gene therapy (Kimura and Harashima 2020; Piguet et al. 2021). With several CNS&PNS-based therapies in the pipeline, there is therefore a high demand for next-generation systemic AAV vectors with potent neurotropic behavior in order to achieve efficient and safe gene delivery for translational applications (Deverman et al. 2018; Tosolini and Smith 2018; Hudry and Vandenberghe 2019; W. Chen et al. 2021; L. Huang et al. 2021; Challis et al. 2022).
Building on our success selecting improved CNS-targeting capsids using M-CREATE (Ravindra Kumar et al. 2020; Goertsen et al. 2021), we decided to test the potential of this method for selecting improved PNS-targeting capsids outperforming the prior engineered variant, AAV-PHP.S, which requires a high dose to exhibit its potent PNS tropism via IV delivery (Chan et al. 2017). Compared to the CNS, the PNS is a more challenging AAV engineering target. Cell populations are sparser, and there is no strong source of selection pressure across targets (akin to the BBB for the CNS). M-CREATE is uniquely well suited to this problem as it capitalizes on deep recovery of capsid libraries across cell-types/organs to select capsids enriched in areas of interest, and a customized analysis pipeline that incorporates positive and negative selections to help identify variants with desired properties.
In this study, we used M-CREATE to identify a family of 7-mer containing AAV9 capsids that appeared to be biased towards PNS areas, and detailed the properties of two such selected AAVs, AAV-MaCPNS1 and AAV-MaCPNS2, in rodents and NHPs. We provide in vivo validation of their tropism in mice, and demonstrate their potential applications with proof-of-concept studies for functional readout and modulation of sensory ganglia. In addition to finding improved PNS-targeting capsids, we sought to address the fundamental question of the translatability of capsids selected in mouse models. To this end, we examined these capsids across four species commonly used in basic through pre-clinical applications: mice, rats, marmosets, and rhesus macaques. The variants discussed in this study show improved efficiency and specificity towards the PNS, and translate their potent behavior across mammalian species. Interestingly, and potentially due to the heterogeneity of the BBB, these variants also show efficient crossing of the BBB to infect the CNS in NHPs (Table S1).
RESULTS:
AAV capsid selection in mice identifies two AAV variants with PNS specificity.
As a starting point for capsid engineering, we chose AAV9 due to its broad tropism when delivered systemically, including for the nervous system (both CNS and PNS) (Foust et al. 2009; Bevan et al. 2011). We diversified the AAV9 capsid by inserting a randomized 7-mer peptide between positions 588 and 589 (Fig. 1A), and intravenously injected the resulting virus library into adult mice of three Cre-transgenic lines: SNAP-Cre (for neurons), GFAP-Cre (for glia), and Tek-Cre (for endothelial cells forming the blood-organ barrier). Two weeks post-injection, we processed peripheral tissues (such as heart, small and large intestines, and DRG) and selectively extracted viral genomes from Cre+ cells using Cre-dependent PCR (see Methods, Fig. 1A–2).
Figure 1: Multiplexed-CREATE selection for AAV capsids targeting the nervous system across species.
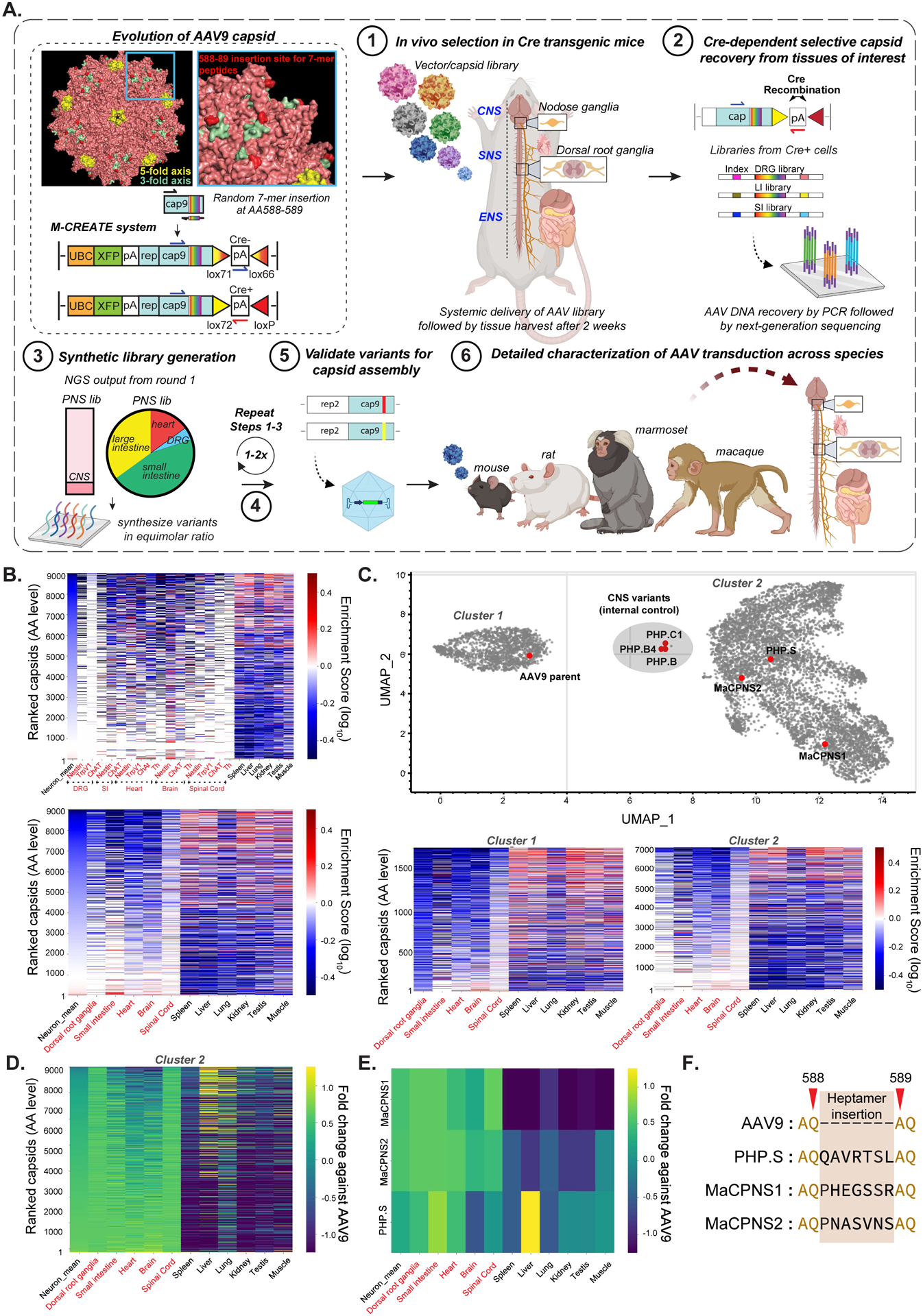
A. An overview of the capsid selection method, Multiplexed-CREATE, and characterization of selected capsids across species. Top left panel illustrates evolution of the AAV9 capsid (PDB 3UX1) with a zoom-in (blue) of a three-fold axis and the 7-mer-i library insertion site between residues 588–589 highlighted in red. The diagrams below demonstrate the arrangement of the acceptor vector in the absence (top) or presence (bottom) of Cre, with the corresponding orientations of the forward/reverse primers used for Cre+ selective recovery. The selection workflow involves four key steps: (1) generation of capsid library and intravenous (IV) delivery into transgenic mouse lines where Cre is restricted to cell types of interest (SNAP-Cre, GFAP-Cre, Tek-Cre, n=2 mice per Cre line). (2) Two weeks post-injection, viral DNA is recovered across cell-types/tissues using Cre-dependent PCR and Illumina next-generation sequencing (NGS), and (3) fed into synthetic pool library production. (4) This library then goes through a second round of in vivo selection. Following this selection process, identified variants are validated for (5) virus production and (6) in vivo transduction across species. B. Heatmaps of capsid variants’ mean enrichment by Cre-dependent recovery across tissues of interest (red text) and Cre-independent recovery across off-targets (black text) after two rounds of selection. Cre lines are plotted separately (top panel, n=3 mice per organ) or grouped by organs (bottom panel). The y-axis represents capsids unique at the amino acid (AA) level, ranked by ‘neuron mean’, which is the mean of the enrichment of all targets of interest. C. UMAP cluster representation of ~9,000 variants that were recovered after two rounds of selection (UMAP parameters: n_neighbors=15, min_dist=0.1, n_components=2, random_state=42, metric=‘correlation’, verbose=3). Three separable clusters are shown along with the positions of known capsids (AAV9, PHP.S, PHP.B, PHP.B4, PHP.C1) and new capsids (MaCPNS1 and MaCPNS2). Heatmaps (below) show enrichment of representative capsids from Clusters-1 and -2 across organs. D. Heatmap of enrichment fold changes against parental AAV9 across organs. E. Heatmap of enrichment fold changes against parental AAV9 of variants MaCPNS1 and MaCPNS2 and previously-engineered variant PHP.S across organs. F. Identity of hepta-AA peptides inserted between positions 588–589 of AAV9 for PHP.S, MaCPNS1 and MaCPNS2 capsids.
Figure 2: Engineered AAVs can efficiently target the peripheral nervous system in mice following systemic delivery.
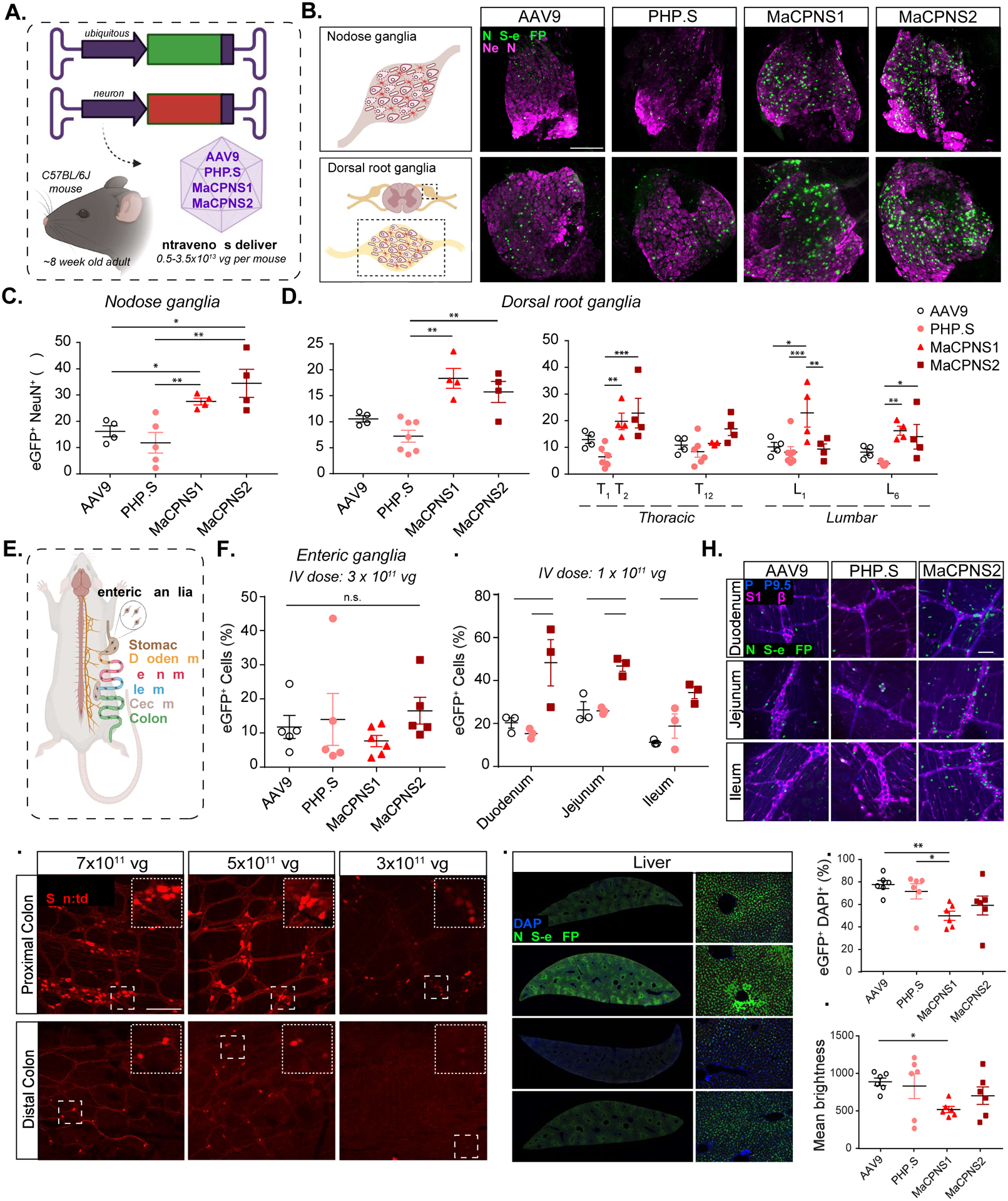
A. Illustration demonstrating the IV administration of AAV capsids packaged with ssAAV:CAG-2xNLS-eGFP and ssAAV:hSyn-tdTomato genome in a mouse model (~8 week-old young C57BL/6J adults). B. (Top) An illustration of the nodose ganglia (NG) (left), and representative images of AAV9, PHP.S, MaCPNS1 and MaCPNS2 vector-mediated expression of nuclear-localized (NLS) eGFP (green) in NG (right). (Bottom) An illustration of the dorsal root ganglia (DRG) in the spinal cord (left) and (right) representative images of AAV vector-mediated expression of NLS-eGFP (green) in DRG across segment T2 (Thoracic) of the spinal cord (n≥4 per group, ~8 week-old C57BL/6J males, 3×1011 vg IV dose per mouse, 3 weeks of expression). Magenta: αNeuN antibody staining for neurons. Images are matched in fluorescence intensity to the respective AAV9 control. Scale bar: 200 μm. C. Percentage of AAV-mediated eGFP expression overlapping with the αNeuN marker in NG. One-way ANOVA non-parametric Kruskal-Wallis test (exact P=0.0003), and follow-up multiple comparisons with uncorrected Dunn’s test are reported (P=0.0499 for AAV9 versus MaCPNS1, P=0.0251 for AAV9 versus MaCPNS2, P=0.0094 for PHP.S versus MaCPNS1, P=0.0038 for PHP.S versus MaCPNS2). *P ≤ 0.05, **P ≤ 0.01 are shown, P > 0.05 is not shown; n≥4 per group, same experimental parameters as B. Each data point represents 1–2 nodose ganglia per mouse comprising >700 cells, mean ± s.e.m is plotted. D. Percentage of eGFP expression overlapping with the αNeuN marker in DRG (left) where each data point shows the mean per mouse across select DRGs within thoracic and lumbar segments of the spinal cord. A one-way ANOVA, non-parametric Kruskal-Wallis test (exact P=0.0005), and follow-up multiple comparisons with uncorrected Dunn’s test are reported (P=0.0018 for PHP.S versus MaCPNS1, P=0.0087 for PHP.S versus MaCPNS2). *P ≤ 0.05, **P ≤ 0.01 are shown, P > 0.05 is not shown; n≥4 per group, same experimental parameters as B. Each data point shows mean ± s.e.m of DRGs across different areas of each mouse, comprising a mean of 1–2 DRGs per area with >200 αNeuN+ cells per DRG. (Right) Percentage of eGFP expression overlapping with the αNeuN marker in DRG across spinal cord areas in individual mice. A two-way ANOVA and Tukey’s multiple comparisons tests with adjusted P values are reported (*P ≤ 0.05, **P ≤ 0.01 and ***P ≤ 0.001 are shown, P > 0.05 is not shown). Each data point shows the mean ± s.e.m of 1–2 DRG per mouse comprising >200 αNeuN+ cells per DRG. E. Illustration of the adult mouse gastrointestinal (GI) tract, highlighting the enteric ganglia (zoom-in) that are spread across the different segments of the GI tract. F. Percentage of cells expressing NLS-eGFP delivered by AAV vectors in the myenteric plexus across the GI tract: stomach, duodenum, jejunum, ileum, proximal colon and cecum (ssAAV:CAG-2xNLS-eGFP genome, n≥5 per group, ~8 weeks old C57BL/6J males, 3×1011 vg IV dose per mouse, 3 weeks of expression). A one-way ANOVA non-parametric Kruskal-Wallis test (approximate P=0.2985), and follow-up multiple comparisons using uncorrected Dunn’s test are reported (individual P > 0.05, n.s.). Each data point shows the mean ± s.e.m of >100 enteric ganglia per intestinal segment per mouse). G. Percentage of cells expressing NLS-eGFP (green in H) in the myenteric plexus of small intestinal segments: duodenum, jejunum and ileum delivered by AAV vectors: AAV9, PHP.S and MaCPNS2 (ssAAV:CAG-2xNLS-eGFP genome, n=3 per group, 1×1011 vg IV dose per mouse, 3 weeks of expression). Two-way ANOVA, Tukey’s multiple comparisons tests with adjusted P values are reported (*P ≤ 0.05, **P ≤ 0.01 and ***P ≤ 0.001 are shown, P > 0.05 is not shown). Each data point shows the mean ± s.e.m of ≥ 2 images per mouse comprising >100 enteric ganglia. H. shows representative images of AAV-mediated eGFP expression across the individual intestinal segments analyzed in G. Scale bar: 50 μm. The tissues were co-stained with αS100β (magenta) antibody for glia and αPGP9.5 (blue) for neurons. The images are matched in fluorescence intensity to the respective AAV9 control. I. MaCPNS2 vector-mediated expression of tdTomato (red) from ssAAV:hSyn-tdTomato in the proximal and distal segments of the colon at three different IV doses per mouse: 7×1011 vg, 5×1011 vg, 3×1011 vg (3 weeks of expression, n=3 per group, scale bar: 200 μm, dotted white box inset represents a zoomed-in view of the indicated area in each image). Images in the distal and proximal colon are matched in fluorescence intensity. J. Representative images of AAV vector-mediated expression of NLS-eGFP (green) from ssAAV:CAG-2xNLS-eGFP in the liver (n=6 per group, 3×1011 vg IV dose/mouse, 3 weeks of expression, blue: DAPI staining of nuclei, scale bar: 1 mm (left, full cross-sectional image), and 50 μm (right, higher magnification view). The images are matched in fluorescence intensity to the respective AAV9 control. K. Percentage of eGFP+ cells overlapping with the DAPI marker in the liver. A non-parametric Kruskal-Wallis test (approximate P=0.0168), and follow-up multiple comparisons with uncorrected Dunn’s test (P=0.0035 for AAV9 versus MaCPNS1 and P=0.0189 for PHP.S versus MaCPNS1) are reported (*P ≤ 0.05, **P ≤ 0.01 are shown, P > 0.05 is not shown; same experimental parameters as J., each data point shows the mean ± s.e.m of 4 images per mouse comprising >780 DAPI+ cells per image). L. Mean brightness of the eGFP+ cells quantified in K. A non-parametric Kruskal-Wallis test (approximate P=0.1698), and follow-up multiple comparisons with uncorrected Dunn’s test (P=0.0433 for AAV9 versus MaCPNS1) is reported (*P ≤ 0.05 is shown, P > 0.05 is not shown; each data point shows the mean ± s.e.m of 4 images per mouse comprising >780 DAPI+ cells per image).
After round-1 (R1) selection, we recovered a total of ~9,000 variants from the Cre lines in the PNS tissues of interest: heart, DRG, small and large intestine. Of these, ~10% overlapped with the CNS libraries (Fig. 1A–3). We then synthesized these variants in an equimolar ratio to create a synthetic oligopool library for round-2 (R2) PNS selection. This synthetic pool also included a spike-in library of previously-validated internal controls (see Methods). We intravenously injected the R2 virus library into different Cre lines: Nestin-Cre (for neurons), CHAT-Cre (for cholinergic neurons), TRPV1-Cre (for primary afferent thermosensitive neurons), and Tek-Cre (for endothelial cells, providing a negative selection). Two weeks post-injection, we selectively recovered capsids by Cre-dependent PCR from tissues of interest (DRG, heart, small and large intestine, brain, and spinal cord (SC)), and other targets (spleen, liver, lung, kidney, testis and muscle) by performing Cre-independent PCR (see Methods, Fig. 1A–4, 1B).
Figure 3: Systemic delivery of GCaMP sensor and excitatory DREADD actuator using AAV-MaCPNS1 enable functional characterization of DRG neurons in mouse models.
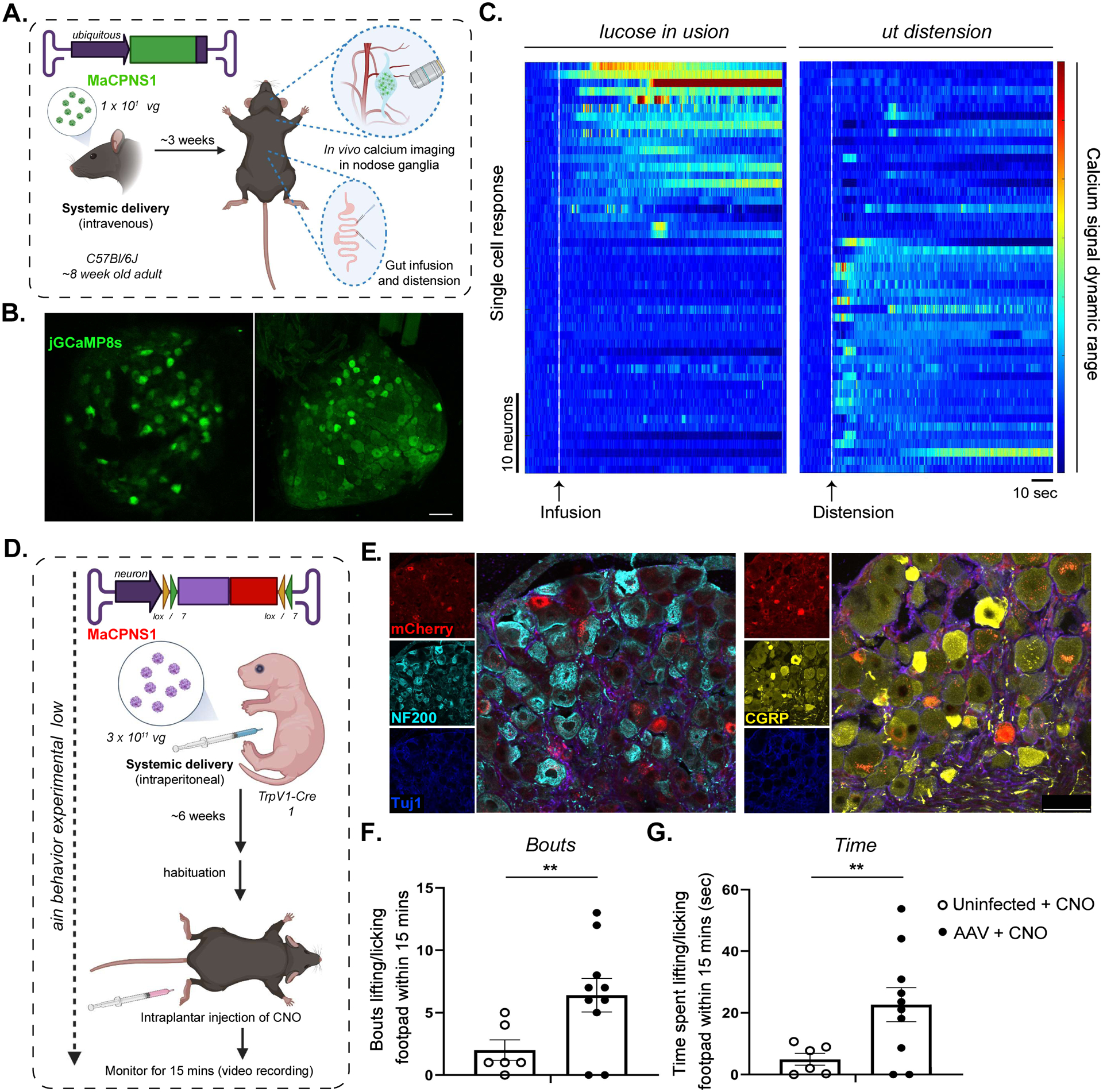
A. Illustration of IV administration of MaCPNS1 capsid packaged with ssAAV:CAG-jGCaMP8s genome in mice (~8 week-old young adults, C57BL/6J males, 1×1012 vg IV dose/mouse, n=4). Three weeks post-expression, the mice were anesthetized and subjected to in vivo calcium imaging in nodose ganglia (NG, top zoom-in), and glucose infusion and distension in the gut (bottom zoom-in). B. Representative images of MaCPNS1 vector-mediated expression of jGCaMP8s (green) in NG in vivo (left), and in post-hoc fixed tissue (right) (scale bar: 100 μm). C. Single-cell activity response measured by calcium signal dynamics in the NG (data pooled from 4 experimental mice). Left panel: NG neuronal response to glucose infusion (white dotted line). Right panel: NG neuronal response to gut distension (white dotted line). D. Illustration of the pain induction experimental workflow. MaCPNS1 vector with ssAAV:hSyn-DIO-hM3D(Gq)-mCherry was intraperitoneally administered to a TrpV1-Cre mouse model (postnatal stage 1 (P1), males, 3×1011 vg IV dose/mouse). After six weeks of expression, the mice (AAV injected or untreated) were habituated and subjected to intraplantar injection with the agonist CNO, after which the mice were monitored for nocifensive lifting/licking behaviors. E. Representative images of DRG sections showing MaCPNS1 vector-mediated mCherry (red) expression. The tissues were co-stained with αNF200 (cyan), αTuj1 (blue) and αCGRP (yellow) markers. F. Total bouts of, and G total time spent lifting or licking the footpad within 15 minutes of injection in uninfected (n=6) and MaCPNS1-infected (n=10) mice treated with CNO. **P ≤ 0.01 by unpaired t-test. Mean+/− sem are shown.
Figure 4: Engineered AAVs can efficiently target the peripheral nervous system in adult rats following systemic administration.
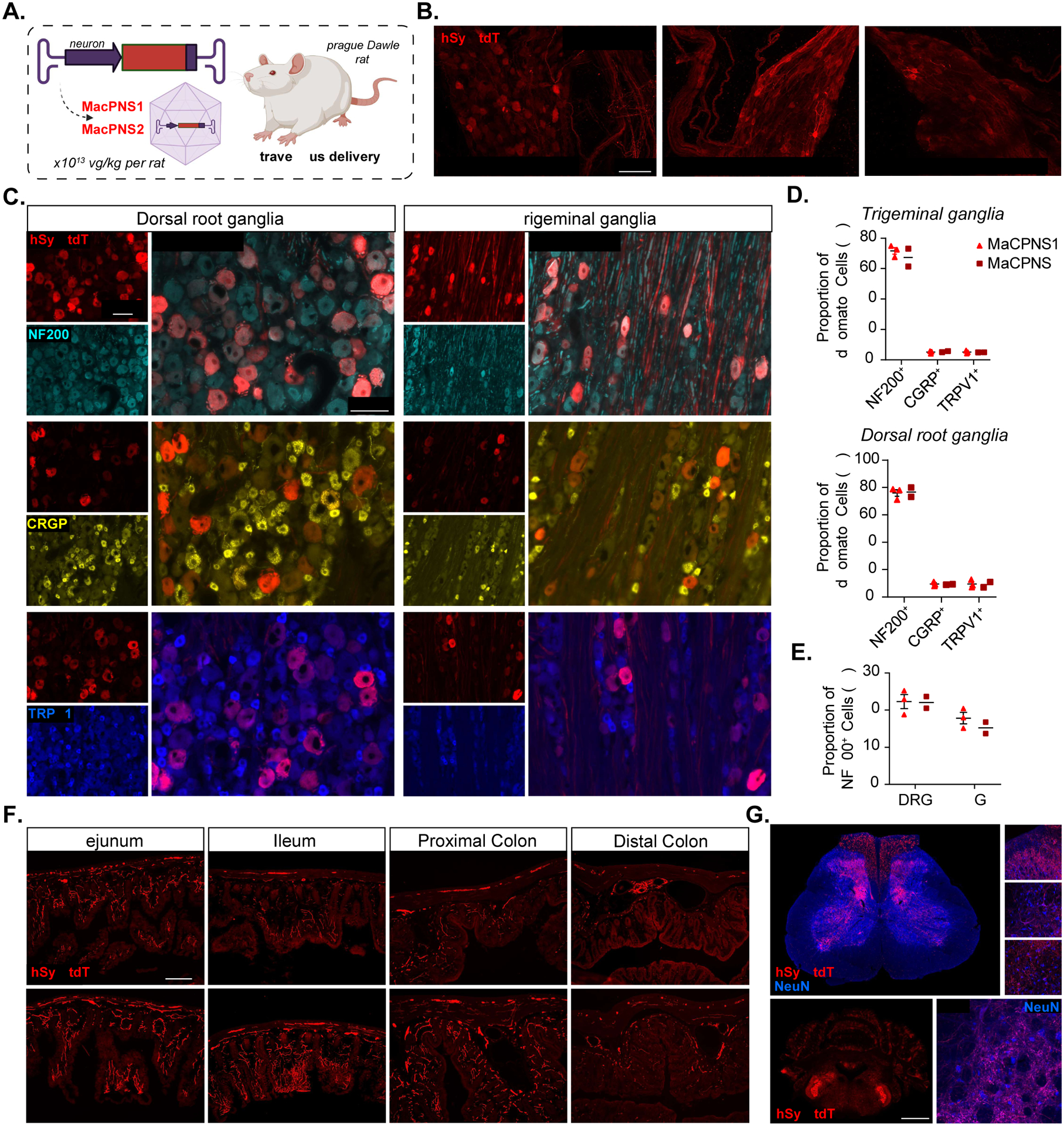
A. Illustration of IV administration of AAV capsids MaCPNS1 and MaCPNS2 packaged with ssAAV:hSyn-tdTomato genome in a rat model (young adults, Sprague Dawley, male, 2×1013 vg/kg per rat). The rat tissues were stained with αDsRed (red) antibody against tdTomato B. Representative images of MaCPNS1 vector-mediated tdTomato (red) expression in major pelvic ganglia (left), sympathetic chain ganglia (middle) and inferior mesenteric ganglia (right) in adult rats 3 weeks post expression (n≥2 per group, scale bar: 100 μm). C. Representative images of MaCPNS1 vector-mediated tdTomato (red) expression in DRG (left) and TG (right). The tissues were co-stained with either αNF200 (cyan), αCGRP (yellow), or αTRPV1 (blue) markers (scale bars: 100 μm). D. Quantification of proportion of AAV-mediated tdTomato expressing cells that overlap with αNF200, αCGRP and αTRPV1 markers in TG (above) and DRG (below), and E. proportion of αNF200 marker+ cells that overlap with the AAV-mediated tdTomato expressing cells in DRG and TG (n≥2 per group, each data point represents the average of at least 3 images from each rat, mean ± s.e.m is plotted for n>2, mean is plotted for n=2). F. Representative images of MaCPNS1 and MaCPNS2 vector-mediated tdTomato expression across different segments of the GI tract: jejunum, ileum, proximal colon and distal colon (scale bar: 200 μm). G. Representative images of MaCPNS1 vector-mediated tdTomato expression in the spinal cord (above, scale bar: 500 μm) with zoomed-in views of selected areas (white boxes, right, scale bar: 100 μm), and hindbrain (below, scale bar: 2 mm) with zoomed-in view of Sp5O region (scale bar: 100 μm). The tissues were co-stained with αNeuN (blue) antibodies.
After two rounds of in vivo selection, among all the variants that we included in the R2 library, 6,300 variants showed a bias towards one or more of the PNS tissues (Fig. 1B, Supplementary Fig. 1A). We next sought to further classify the recovered variants based on distinct tropisms. We used the Uniform Manifold Approximation and Projection (UMAP) algorithm (McInnes, Healy, and Melville 2020), which takes into account differences among variants’ enrichments across organs, to identify clusters of variants representing distinct tropisms (Fig. 1C, top panel). Members of Cluster-1 (1,846 variants), which includes AAV9, show relatively higher enrichment in off-target tissues such as the liver (Fig. 1C, bottom left panel). Cluster-2 members (7,148 variants), including our previously engineered variant PHP.S, exhibit relatively higher enrichment in the PNS and lower enrichment in off-target tissues (Fig. 1C, bottom right panel). Cluster-3 members (5 variants), including variants from the internal control, were highly enriched in the CNS.
Based on this analysis, we reasoned that Cluster-2 might contain promising variants distinct from the parental AAV9, which we indeed observed by comparing their tropism directly to that of AAV9 (Fig. 1D). We identified two new capsids from Cluster-2 that exhibited low off-target transduction compared to AAV9 and PHP.S (Fig. 1E) and that could package viral genomes with similar efficiency to AAV9 (see Methods, Supplementary Fig. 1B). The first capsid, with a 7-mer peptide insertion of PHEGSSR between the 588–89 residues of the AAV9 parent, we will henceforth refer to as AAV-MaCPNS1. The second, with an insertion of PNASVNS, we will refer to as AAV-MaCPNS2 (Fig. 1F).
IV-delivered AAV-MaCPNS1/2 efficiently transduces sensory and enteric ganglia in mice, with low liver transduction.
To characterize the transduction capability of AAV-MaCPNS1 and AAV-MaCPNS2 variants in vivo, we packaged these variants with a single-stranded (ss) AAV genome carrying a strong ubiquitous promoter, CAG, driving expression of nuclear-localized eGFP reporter and intravenously injected them into adult mice at 3×1011 vg per animal (Fig. 2A–H,J–L; see Methods). By quantifying expression in nodose ganglia (NG) that overlapped with the NeuN neuronal marker, we found that MaCPNS1 and MaCPNS2 had mean transduction efficiencies of ~28% and ~35%, respectively. By comparison, the efficiency of AAV9 was ~16%, and AAV-PHP.S ~12% (Fig. 2B–C). Thus, MaCPNS1/2 capsids exhibit about 2-fold higher transduction of NG than previously available vectors.
Next, we investigated the efficiency of DRG transduction from selected spinal levels (thoracic and lumbar). The MaCPNS1 vector demonstrated a mean transduction efficiency of ~18%, and MaCPNS2 ~16%, compared to ~11% for AAV9 and ~7% for AAV-PHP.S (Fig. 2B bottom panel, 2D, left), with some variability in transduction across different segments of the SC (Fig. 2D, right). Thus, compared to known capsids, MaCPNS1/2 can achieve about 2-fold higher transduction of DRG via IV delivery. Increasing the vector dose to 1×1012 vg, we observed that PHP.S had higher transduction in DRG compared to AAV9 while MaCPNS1 exhibited even higher transduction (Supplementary Fig. 2A–C), further confirming that MaCPNS1/2 capsids transduce DRG in adult mice more efficiently than either AAV9 or PHP.S.
To investigate the transduction efficiency of the new vectors in the ENS, we assessed AAV-mediated eGFP expression in the enteric ganglia of the myenteric plexus across different areas of the GI tract – stomach, duodenum, jejunum, ileum, proximal colon and cecum (Fig. 2E–F). We observed a mean transduction efficiency of ~8% by MaCPNS1, ~17% by MaCPNS2, ~13% by PHP.S and ~12% by AAV9 (Fig. 2F). By analyzing individual GI segments, we noticed variability in transduction of enteric ganglia, with a bias of MaCPNS2 transduction towards small intestinal (SI) segments (duodenum, jejunum and ileum) over other areas, indicating a modest improvement in SI ENS transduction by MaCPNS2 compared to other vectors. To more closely examine this SI bias, we slightly lowered the vector dose to 1×1011 vg and observed a 2-fold increase in transduction of the SI by MaCPNS2 compared to AAV9 (Fig. 2G–H). Next, we investigated if the transduction efficiency across large intestinal (LI) segments could be improved by a higher dose (Fig. 2I, Supplementary Fig. 2D). Compared to the initial dose of 3×1011 vg, we observed that a MaCPNS2 dose of either 5×1011 vg or 7×1011 vg achieved robust and uniform transduction of the enteric ganglia of the myenteric plexus in the proximal colon as well as sparser transduction in the distal colon. Thus intravenously-administered MaCPNS2 offers improved transduction of enteric ganglia in the SI, with the option of additional targeting of LI segments at higher doses.
To investigate the liver transduction of MaCPNS1/2 capsids, we assessed eGFP expression in vivo (Fig. 2J–L). MaCPNS1 exhibited 1.5-fold lower transduction compared to the same dose of AAV9 or PHP.S (Fig. 2K). Quantifying the mean brightness of the eGFP fluorescence signal revealed that the new vectors also exhibited 1.3- to 2.1-fold lower signal compared to AAV9 or PHP.S (Fig. 2L), suggesting reduced transgene expression per cell. Collectively these data suggest that MaCPNS1 exhibits lower transduction of hepatocytes and lower transgene expression per liver cell compared to other vectors used for PNS. In addition to the liver, we also investigated other tissues, including the CNS, given AAV9’s ability to cross the BBB. At a modest IV dose of 3×1011 vg, CNS transduction was low for all vectors we tested, including AAV9 (Supplementary Fig. 2E). Even when we increased the dose of MaCPNS2 to 5–7×1011 vg, we did not observe CNS transduction in mice (Supplementary Fig. 2F). In addition, MaCPNS1/2 transduction of cardiac muscle was similar to that of AAV9 (Supplementary Fig. 2E).
All of the above experiments were conducted in adult mice. However, many potential applications require early age intervention (Gray 2016; Privolizzi et al. 2021; Ali et al. 2021). We therefore investigated MaCPNS1/2 transduction in DRG in neonates using an alternative delivery route. We introduced MaCPNS1/2 capsids packaged with ssAAV:hSyn-tdTomato into postnatal stage 1 (P1) mice via intraperitoneal (IP) injection, with a dose of 3×1011 vg per mouse. After 6 weeks of expression, MaCPNS1 transduced ~39% of neurons (marked with Tuj1) in DRG and MaCPNS2 transduced ~35%, including ~55% of all CGRP+ neurons and ~19% of all NF200+ neurons (Supplementary Fig. 2G–I). Examining targeting preference, we found that the capsids were predominantly targeting two types of neurons: about 60% of transduced neurons were CGRP+ and ~11% NF200+ (Supplementary Fig. 2J).
In summary, we found that systemic delivery of MaCPNS1 and MaCPNS2 vectors in mice can efficiently transduce sensory ganglia (such as NG and DRG) compared to AAV9, with MaCPNS2 distinguished from MaCPNS1 by enhanced transduction of the ENS. Further, MaCPNS1 and MaCPNS2 exhibited improved specificity for the PNS, with relatively lower transduction of AAV9’s primary target, the liver.
Systemic MaCPNS1-mediated sensor and actuator expression enable functional characterization of neurons in nodose ganglia and dorsal root ganglia.
Functional readout of PNS activity during physiological conditions is key to understanding the interaction between the brain and peripheral system (R. Chen, Canales, and Anikeeva 2017; Zanos et al. 2018; Walters et al. 2019; Jiman et al. 2020). However, commonly-used imaging with genetically encoded calcium indicators (GECIs) in the CNS has been challenging in the PNS due to the low efficiency of delivering GECI to PNS targets. To test the applicability of MaCPNS1/2 capsids to this problem, we intravenously delivered MaCPNS1 capsid packaged with a recently-developed GECI, jGCaMP8s (Zhang et al. 2020), to adult mice. After three weeks of expression, we recorded calcium signals in vivo during procedures on the gut (Fig. 3A–C). We infused the gut with either glucose solution or saline (to induce distension) while recording neuronal activity in the NG of the anesthetized mice and observed distinguishable neuronal responses in NG (Fig. 3A–C). No significant expression was observed in the CNS or liver of these mice.
After verifying the new vectors’ potential for enabling functional readout of the PNS, we took the further step of seeing whether we could achieve functional modulation with the vectors. We sought to construct a mouse pain-induction system with improved temporal control, a critical tool for understanding and potentially managing pain. To activate DRG TrpV1 neurons, we packaged Cre-dependent excitatory Designer Receptors Exclusively Activated by Designer Drugs (DREADD) (Krashes et al. 2011) into MaCPNS1 and intraperitoneally injected the vector into P1 TRPV1-Cre pups (Fig. 3D). After 6 weeks of expression (Fig. 3E), we gave mice an intraplantar injection of the DREADD agonist clozapine N-oxide (CNO) and evaluated resulting pain-like behaviors (see Methods). We observed an increase in the number of bouts and overall time spent lifting or licking the injected footpad in the experimental AAV-administered group compared to the control group, indicating nocifensive pain-like behavior (Fig. 3F–G).
Together, these results provide a proof of concept for the application of these new vectors to a wide range of studies involving the monitoring and modulation of sensory processes, including pain.
IV-administered AAV-MaCPNS1/2 efficiently transduces the PNS in adult rats.
Having validated the new variants’ transduction profiles in mouse models, we next investigated their efficacy in another common research model system, rats. Systemic delivery of MaCPNS1/2 capsids packaged with ssAAV:hSyn-tdTomato in Sprague Dawley adults (Fig. 1A) transduced sensory ganglia (DRG and trigeminal ganglia (TG)), sympathetic chain ganglia (SCG), parasympathetic ganglia (inferior mesenteric ganglia (IMG)), mixed sympathetic-parasympathetic ganglia (the major pelvic ganglion (MPG)), and enteric ganglia across the SI and LI (Fig. 4A–F, Supplementary Fig. 3A–B). The majority of cells transduced in both DRG and TG were NF200+ (about 70–75%), with only 5% either CGRP+ or TRPV1+ (Fig. 4C–D). In terms of efficiency, both capsids transduced ~22% of the NF200+ cells in DRG, and 15%-17% of the NF200+ cells in TG (Fig. 4E). Interestingly, this cell type bias contrasts with what we observed in neonatal mice, where the vectors showed a bias towards CGRP+ neurons (Supplementary Fig. 2I–J). This profile could result from differences in species or age/time of injection, as well as the different systemic routes of vector delivery (similar tropism shifts have been reported elsewhere (Foust et al. 2009)). The tropism diversity highlights the necessity of considering all relevant experimental variables to achieve the desired transduction profile.
The vectors’ efficient labeling of sensory ganglia prompted us to look further into the ENS (Fig. 4F, Supplementary Fig. 3C). Analyzing segments of the SI and LI, we found that MaCPNS1 and MaCPNS2 were comparable at transducing the myenteric plexus and the submucosal plexus, including the vascular and periglandular plexuses. Next, we investigated projections to the SC and brainstem (Fig. 4G, Supplementary Fig. 3D). Sensory nerve fibers entering the dorsal horn of the SC were densely labeled, and the ascending afferent tracts in the dorsal column were also strongly transduced by both virus vectors. In the brainstem, fibers were densely labeled in the spinal trigeminal nucleus oralis (Sp5O), which potentially projected from the TG. In addition to the transduction in the PNS and the nerve projections labeling in the CNS, we also observed some labeling of neuronal cell bodies across regions in the brain (Supplementary Fig. 3D). Furthermore, we observed no expression in the liver (Supplementary Fig. 3E).
Together, these results show that the potent PNS tropism of the MaCPNS1/2 vectors is conserved across rodent models tested.
IV-delivered AAV-MaCPNS1/2 transduces the adult marmoset CNS and PNS more efficiently than AAV9.
Novel capsids selected in mice do not always translate to NHPs (Hordeaux et al. 2018; Matsuzaki et al. 2018). After validating the MaCPNS1/2 capsids in mice and rats, we therefore decided to assess their performance in NHPs. We first chose the marmoset, a New World monkey and an emerging animal model for translational research. Owing to the limited availability of these animals, in each adult animal we tested two viral capsids (AAV9, MaCPNS1 or MaCPNS2) packaging different fluorescent reporters (either ssAAV:CAG-eGFP or ssAAV:CAG-tdTomato) (Fig. 5A). Compared to the parent AAV9, intravenously-delivered MaCPNS1/2 capsids carrying either ssAAV:CAG-eGFP or ssAAV:CAG-tdTomato genome showed robust transduction of the PNS and the CNS. In the PNS, we observed enhanced transduction of DRG, the SI and the ascending fiber tracts in the dorsal column of the SC (Fig. 5B–D), similar to what we observed in rodents. In the CNS, robust brain-wide transduction by MaCPNS1/2 capsids, but not AAV9, was evident in regions including the cortex, thalamus, globus pallidus, cerebellum and brainstem (Fig. 5E–F). Further antibody staining and quantification revealed that capsids mainly transduce neurons and astrocytes in the marmoset brain. In the marmoset cortex, MaCPNS2 displayed a ~5.5-fold increase in neuronal transduction over AAV9 while MaCPNS1 displayed a ~4-fold increase over AAV9 (Supplementary Fig. 4A–C). Among the neurons transduced, few overlapped with the PV marker, indicating the vectors’ potential bias to excitatory population (Supplementary Fig. 4D). For astrocyte transduction, MaCPNS2 displayed a ~25-fold increase over AAV9 in cortex (Supplementary Fig. 4B). Similar improvement in both neuronal and astrocytic transduction was also observed in the thalamus (Supplementary Fig. 4C).
Figure 5: Engineered AAVs can efficiently transduce the central and peripheral nervous system in marmoset.
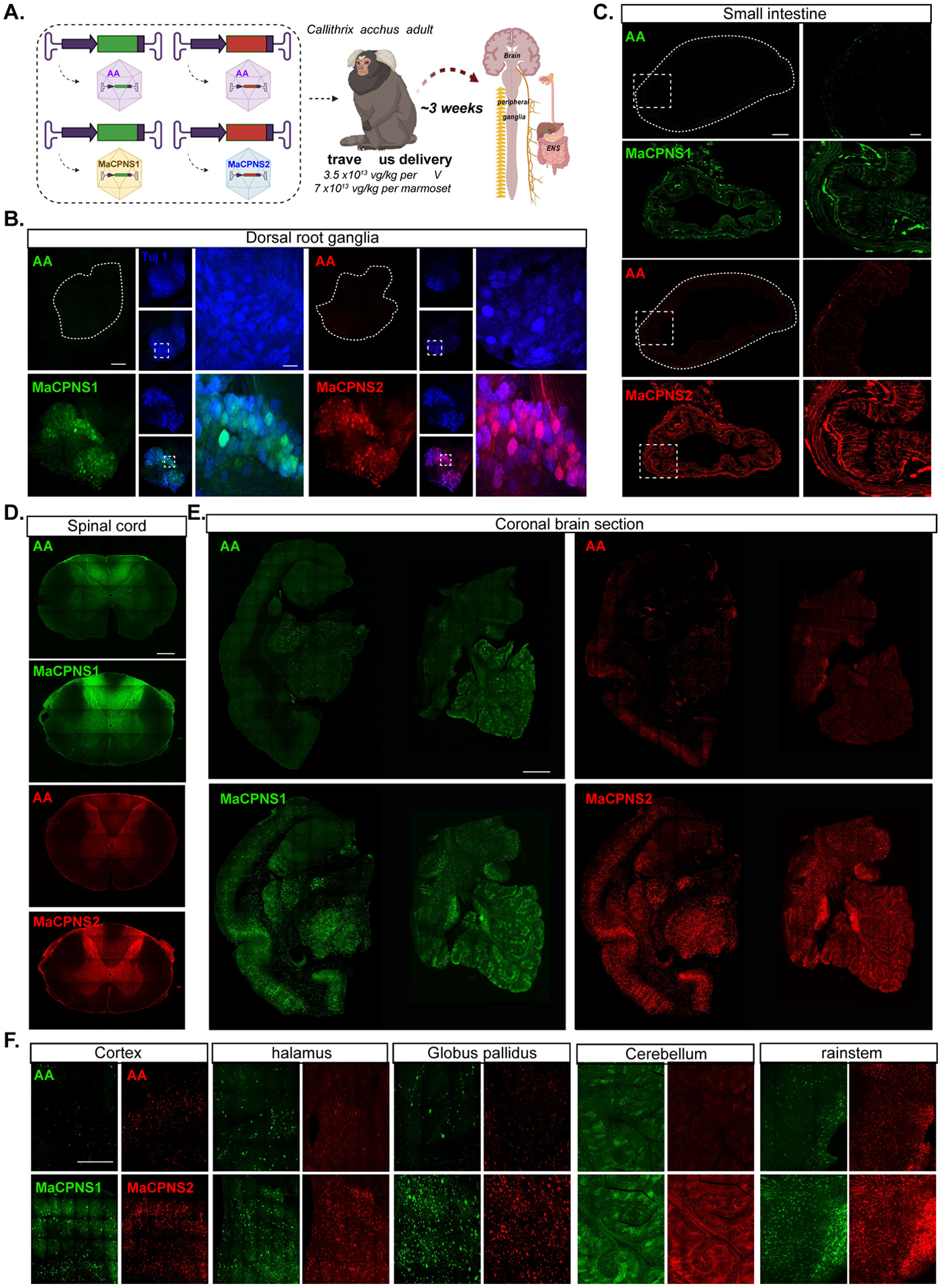
A. Illustration of AAV vector delivery to adult marmoset to study transduction across the CNS and PNS after 3 weeks of expression. The capsids (AAV9/MaCPNS1/MaCPNS2) and their corresponding genomes (ssAAV:CAG-eGFP/tdTomato) are shown on the left. Two AAV vectors packaged with colored fluorescent reporters were mixed and intravenously delivered at a total dose of 7×1013 vg/kg per adult marmoset (16 month old Callithrix jacchus, i.e. 3.5×1013 vg/kg per AAV). Representative images of marmoset B., DRGs (scale bar: 200 μm, left and 500 μm, right), C., small intestine (scale bar: 500 μm, left and 100 μm, right), D., fibers in the dorsal column of the spinal cord (scale bar: 500 μm), E., coronal brain sections of midbrain (left) and hindbrain (right) (scale bar: 500 μm), and F., select brain areas: cortex, thalamus, globus pallidus, cerebellum and brainstem (scale bar: 400 μm), showing AAV9 vector-mediated expression of eGFP (green) or tdTomato (red), MaCPNS1-mediated expression of eGFP (green) and MaCPNS2-mediated expression of tdTomato (red). The images are matched in fluorescence intensity to the respective AAV9 control. Zoomed-in views of selected areas (dotted white boxes) are shown on the right in B. and C. In B. the zoomed-in view shows the overlap of MaCPNS1 and MaCPNS2-mediated expression with the neuronal marker Tuj1 (blue).
These results demonstrate that the MaCPNS1/2 capsids can efficiently cross the BBB in adult marmosets, while still exhibiting the enhanced PNS tropism observed in rodents.
IV-delivered AAV-MaCPNS1/2 transduce the infant rhesus macaque CNS and PNS more efficiently than AAV9.
Encouraged by the tropism of MaCPNS1/2 capsids in marmosets, we further assessed their transduction potential in another NHP, the rhesus macaque, an Old World monkey and a common pre-clinical research model for AAV gene therapy. We employed the same strategy as in marmosets, intravenously administering AAV9, MaCPNS1 and MaCPNS2 capsids packaging either ssAAV:CAG-eGFP or ssAAV:CAG-tdTomato to an infant rhesus macaque (Fig. 6A). Similar to our results in marmosets, we observed that MaCPNS1/2 efficiently targeted both the CNS and PNS compared to the parent AAV9 after three weeks of expression. We saw enhanced transduction in the SC, DRG and GI tract, including the esophagus, colon and SI (Fig. 6B–D), as observed in rodents and marmosets. In the SC, there was strong labeling of both ascending fiber tracts in the dorsal column as well as the peripheral fiber tracks coming into the dorsal horn. Within the grey matter of the lumbar SC, MaCPNS1 displayed a ~25-fold increase in neuronal transduction over AAV9. For non-neuronal cells, MaCPNS2 displayed a ~130-fold increase in transduction over AAV9 (Supplementary Fig. 5A–B). Similar improvement in transduction was also observed in the thoracic SC (Supplementary Fig. 5C–D). In the lumbar and thoracic DRG, MaCPNS2 displayed ~77-fold and ~44-fold increases in neuronal transduction over AAV9, respectively. MaCPNS1 also showed ~37-fold and ~22-fold increases in neuronal targeting in lumbar and thoracic DRG. Within lumbar DRG (Supplementary Fig. 5E), MaCPNS2 was highly specific to neurons, with ~94% of all transduced cells being neurons. In thoracic DRG, both vectors displayed high specificity in targeting neurons (Supplementary Fig. 5F). In the CNS, MaCPNS1/2 capsids mediated enhanced brain-wide transduction including in areas such as the cortex, hippocampus, putamen and brainstem (Fig. 6E–F, Supplementary Fig. 6, Video S1–4), similar to our observations in marmosets. Both vectors transduced neurons and astrocytes but not oligodendrocytes or endothelial cells (Supplementary Fig. 7A–B). In the macaque cortex, neuron transduction by MaCPNS1 and MaCPNS2 was ~6-fold and ~11-fold higher, respectively, than AAV9 (Supplementary Fig. 7B). Most neurons transduced by both vectors were PV-, indicating a potential bias to excitatory populations (Supplementary Fig. 7D). MaCPNS2 also displayed a ~44-fold increase in astrocyte transduction over AAV9 in cortex (Supplementary Fig. 7B). A similar improvement in neuronal and astrocytic transduction was also observed in the thalamus (Supplementary Fig. 7C). Consistent with our observations in marmoset, MaCPNS1’s transduction was more biased towards neurons while MaCPNS2 showed increased transduction of both neurons and astrocytes. No significant difference in liver transduction was observed between MaCPNS1, MaCPNS2 and AAV9 (Supplementary Fig. 6).
Figure 6: Engineered variants efficiently target the central and peripheral nervous system in macaque following systemic delivery.
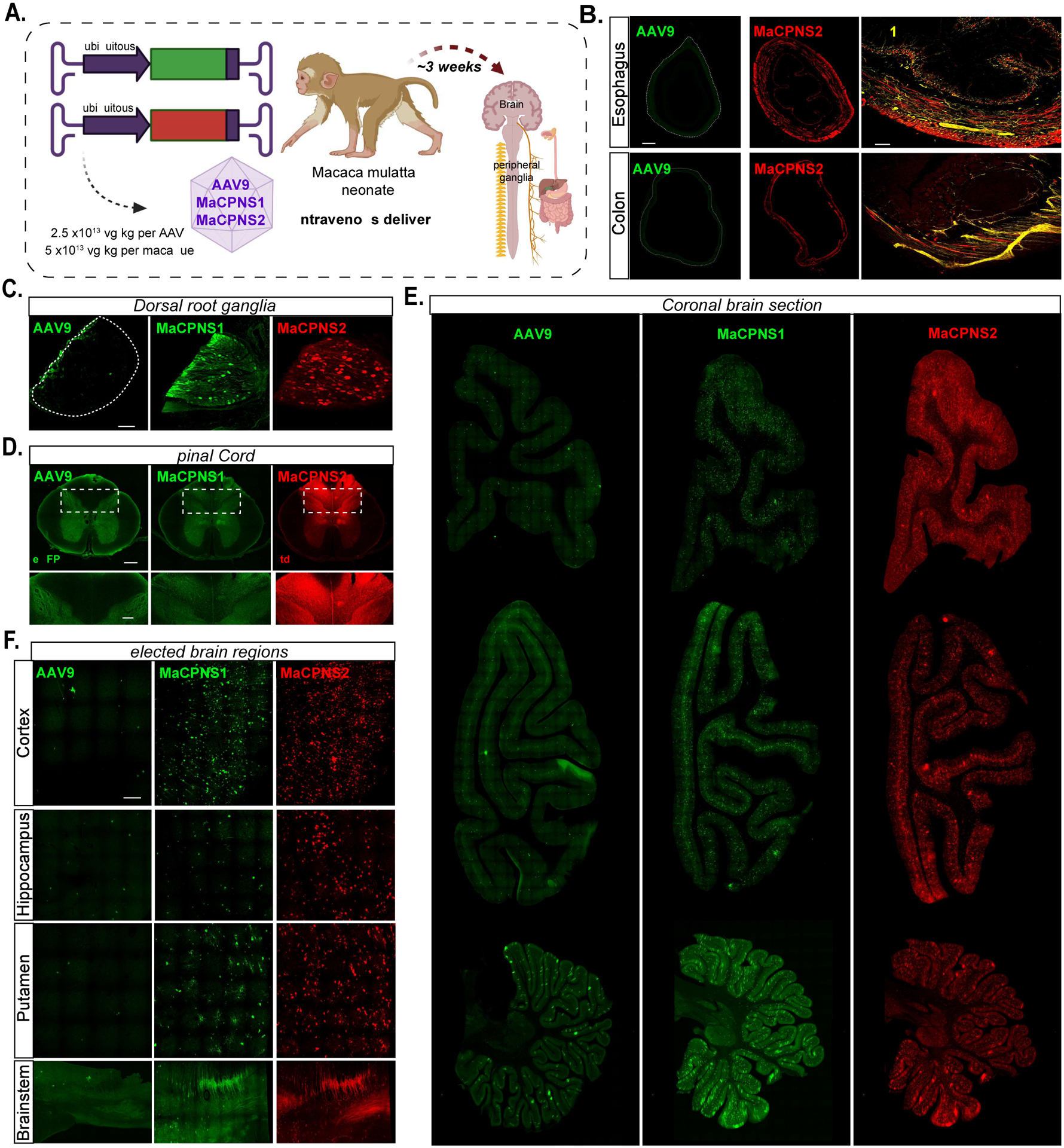
A. Illustration of AAV vector delivery to macaque to study transduction across the CNS and PNS after 4 weeks of expression. The capsids (AAV9/MaCPNS1/MaCPNS2) and their corresponding genomes (ssAAV:CAG-eGFP/tdTomato) are shown on the left. Two AAVs packaged with different fluorescent proteins were mixed and intravenously injected at a dose of 5×1013 vg/kg per macaque (Macaca mulatta, injected within 10 days of birth, female, i.e. 2.5×1013 vg/kg per AAV). Representative images of macaque B., GI regions of esophagus (top panel) and colon (bottom panel) (scale bar: 500 μm, left and 200 μm, right), C., DRGs (scale bar: 200 μm), D., spinal cord (scale bar: 500 μm, top and 200 μm, bottom), E., coronal sections of forebrain, hindbrain and cerebellum (scale bar: 2 mm), and F., selected brain areas: cortex, hippocampus, putamen, and brainstem (scale bar: 200 μm, top and 500 μm, bottom), showing AAV9 vector-mediated expression of eGFP (green), MaCPNS1-mediated expression of eGFP (green) and MaCPNS2-mediated expression of tdTomato (red). The eGFP images of MaCPNS1 are matched in fluorescence intensity to the AAV9 control. The zoomed-in views in B (right panels) show the overlap of MaCPNS2-mediated expression of tdTomato (red) with the neuronal marker Tuj1 (yellow). The zoomed-in views in D (bottom panels) show AAV-mediated expression of eGFP (green) and tdTomato (red) in the fibers in the dorsal column (white dashed boxes).
These experiments demonstrate that the new capsids, MaCPNS1/2, can efficiently transduce the PNS and CNS in both New and Old World monkeys, making them useful vectors for translational research across the nervous system.
DISCUSSION:
In this study, we describe systemic AAVs which address the pressing need for efficient gene delivery vectors to target the nervous system across species. By in vivo selection and data analysis, we identified a library of capsids with divergent tropism compared to their parent, AAV9. Two variants, MaCPNS1 and MaCPNS2, were noteworthy for their potent neurotropic behavior in the mouse model in which they were selected. In contrast to our previously-engineered PNS-targeting variant, AAV-PHP.S, which requires a high dose to be potent, intravenous delivery of a modest dose of the new variants in adult mice showed about 2-fold improvement in the transduction of NG and DRG compared to AAV9. In addition to improved sensory ganglia transduction, the MaCPNS2 capsid showed improved transduction of the SI of the ENS. These MaCPNS1/2 capsids also stood out from AAV9 and PHP.S in their specificity for the PNS, with lower transduction in the liver. In addition to their performance in adult mice, the new vectors efficiently transduced DRG when delivered at the P1 neonatal stage in mice, via a technically easy intraperitoneal injection. In neonatal mice, both vectors showed a significant bias towards transducing CGRP+ neurons.
Experiments involving functional readout or modulation in the sensory system require a high copy-number of functional proteins. This can be challenging to achieve with systemic delivery, and is thought to require a combination of both an efficient vector and an engineered genetic indicator/probe (Bedbrook et al. 2019; Michelson, Vanni, and Murphy 2019; Grødem et al. 2021). In this study, we demonstrate that it is possible to use the new MaCPNS1 capsid systemically for both monitoring and modulating neuronal function. By systemically delivering a recently engineered GECI, jGCaMP8s, we were able to visualize vagal neuron calcium dynamics in response to gut glucose infusion and distension. By demonstrating the use of viral vector-mediated GECI delivery for imaging in wild-type mice, we highlight the possibility of performing similar imaging studies in species where transgenic models may not be available. Following our success with GECI sensor delivery, we extended the application of the MaCPNS1 vector to systemic delivery of a DREADD actuator to a TrpV1-Cre neonate, enabling pain induction by chemogenetic modulation of TrpV1+ neurons in DRG, which mediate thermosensation and pain (Cavanaugh et al. 2011; Mishra et al. 2011; Pogorzala, Mishra, and Hoon 2013). These proof-of-concept experiments demonstrate the potential of MaCPNS1/2 for modulating different sensory modalities with higher temporal resolution. With the rapid development of ultra-sensitive opsins (Bedbrook et al. 2019; Gong et al. 2020) and a wireless light source (Y. Yang et al. 2021), these newly-developed AAV variants could potentially open the door to less invasive modulation of hard-to-access peripheral ganglia with precise temporal control.
MaCPNS1 and MaCPNS2 also show promise in rats, another commonly used research model for PNS applications (Ravagli et al. 2020; Draxler et al. 2021). Both capsids translated their potent PNS tropism across rodents, showing efficient transduction of sensory ganglia, sympathetic ganglia, parasympathetic ganglia, and enteric neurons. Detailed cell-type characterization in DRG and TG of adult rats showed that the vectors were biased towards transducing NF200+ neurons, in contrast to the bias towards CGRP+ neurons we observed in DRG in mice injected intraperitoneally at the neonate P1 stage. Such tropism shifts have been previously noted with other AAV serotypes, and emphasize the importance of considering the roles various experimental conditions play in determining a vector’s tropism in a given animal model (Foust et al. 2009; Matsuzaki et al. 2018).
The conservation of the vectors’ potent PNS tropism across rodents prompted us to test their performance in NHPs. We first tested the new AAVs in a New World monkey, the marmoset, which has recently been gaining attention in the neuroscience community as a promising animal model for biomedical research (Marx 2016; Miller et al. 2016; Jennings et al. 2016). Intravenous delivery of MaCPNS1 and MaCPNS2 to adult marmosets showed potent PNS tropism. However, the vectors also efficiently crossed the BBB to transduce the CNS, making them potent vectors across the nervous system. We further validated the vectors’ tropism in an Old World monkey, the rhesus macaque, which is more closely related to humans and is widely used as an animal model for pre-clinical research, including gene therapy (Jennings et al. 2016; Hudry and Vandenberghe 2019; Bey et al. 2020). As in marmosets, intravenously delivered MaCPNS1 and MaCPNS2 efficiently transduced both the PNS and CNS in an infant rhesus macaque. The enhanced CNS tropism we observed in NHPs may be explained by the heterogeneity of the BBB across species.
The conservation of these AAV variants’ potent PNS tropism across species validates the usefulness of selecting capsids in mouse models, a preferred model among capsid engineers due to the relatively fewer challenges implementing iterations of in vivo selection or capsid evolution given animal availability. However, the question of translatability for CNS tropism requires further investigation. Prior CNS-specific selections have yielded capsids which may or may not be translatable across species (Matsuzaki et al. 2018; Goertsen et al. 2021) or whose potential has yet to be tested.
In summary, here we introduce new AAVs to address some significant challenges in the field of gene delivery vectors for the nervous system (Table S1). Non-invasive delivery of transgenes across the nervous system can be transformative for many applications, including basic science, as demonstrated with our previously-engineered AAV vectors (Chakrabarti et al. 2020; Marvaldi et al. 2020; Reynaud-Dulaurier and Decressac 2020; Levy et al. 2020; Takano et al. 2020; Asencor et al. 2021; Duan et al. 2021; Monteys et al. 2021). With several therapeutic candidates now in the pipeline for various neurological disorders (Deverman et al. 2018; Hudry and Vandenberghe 2019; Sevin and Deiva 2021; Privolizzi et al. 2021), the new systemic AAV vectors described in this study, AAV-MaCPNS1 and AAV-MaCPNS2, offer hope to accelerate translational research as well.
STAR METHODS:
RESOURCE AVAILABILITY
Lead contact:
Further information and requests for resources should be directed to the Lead Contact, Viviana Gradinaru (viviana@caltech.edu).
Materials availability:
All mouse strains used in this study are available from Jackson Laboratories. All plasmids and viral vectors generated for the study have been made available from Addgene or can be obtained from the lead contact upon request. Accession numbers are listed in the key resources table.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-NeuN antibody | Abcam | Cat #ab177487; RRID:AB_2532109 |
| Chicken anti-PGP9.5 antibody | Invitrogen | Cat #PA1–10011; RRID:AB_1088162 |
| Goat anti-chicken Alexa647 antibody | Invitrogen | Cat #A32933; RRID: AB_2762845 |
| Donkey anti-rabbit Alexa555 antibody | Invitrogen | Cat #A21432; RRID: AB_2535853 |
| Rabbit anti-CGRP antibody | Millipore | Cat #PC205L; RRID: AB_2068524 |
| Rabbit anti-Neurofilament 200 antibody | Sigma | Cat #N4142; RRID: AB_477272 |
| Mouse anti-Tuj1antibody | Abcam | Cat #ab7751; RRID: AB_306045 |
| Donkey anti-rabbit DyLight488 antibody | Abcam | Cat #ab96919; RRID: AB_10679362 |
| Donkey anti-mouse Alexa647 antibody | Abcam | Cat #ab150107; RRID: AB_2890037 |
| Rabbit anti-dsRed antibody | Takara Bio | Cat #632496; RRID: AB_10013483 |
| Mouse anti-Neurofilament 200 antibody | Sigma-Aldrich | Cat #N0142; RRID: AB_477257 |
| Goat anti-calcitonin gene-related peptide antibody | Bio-Rad | Cat #1720–9007; RRID: AB_2290729 |
| Goat anti-TRPV1 antibody | Neuromics | Cat #GT15129; RRID: AB_1624144 |
| Donkey anti-rabbit AF594 antibody | Jackson Immuno | Cat #711-585-152; RRID: AB_2340621 |
| Donkey anti-mouse AF488 antibody | Jackson Immuno | Cat #715-545-150; RRID: AB_2340846 |
| Donkey anti-goat AF488 antibody | Jackson Immuno | Cat #705-545-147; RRID: AB_2336933 |
| Mouse anti-NeuN antibody | Chemicon | Cat #MAB377; RRID: AB_2298772 |
| Rabbit anti-S100 beta antibody | Abcam | Cat #ab52642; RRID: AB_882426 |
| Rabbit anti-Parvalbumin antibody | Abcam | Cat #ab181086; RRID: N/A |
| Rabbit anti-Olig2 antibody | Abcam | Cat #ab109186; RRID: AB_10861310 |
| Rabbit anti-GLUT1 antibody | Millipore | Cat #07–1401; RRID: AB_11212210 |
| Goat anti-rabbit Alexa647 antibody | Invitrogen | Cat # A21245, RRID: AB_141775 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6J | Jackson Laboratory | Cat #000664 |
| Mouse: Tek-Cre | Jackson Laboratory | Cat #008862 |
| Mouse: ChAT-IRES-Cre | Jackson Laboratory | Cat #006410 |
| Mouse: Nestin-Cre | Jackson Laboratory | Cat #003771 |
| Mouse: TRPV1-Cre | Jackson Laboratory | Cat #017769 |
| Mouse: TH-Cre | European Mouse Mutant Archive | Cat #EM::00254 |
| Sprague-Dawley rat | University of Melbourne | N/A |
| Marmoset | University of California San Diego | N/A |
| Rhesus Macaque | University of California Davis | N/A |
| Recombinant DNA | ||
| pUCmini-iCAP-AAV.MaCPNS1 | In house | Deposited at Addgene #185136 |
| pUCmini-iCAP-AAV.MaCPNS2 | In house | Deposited at Addgene #185137 |
| pAAV:hSyn1-tdTomato plasmid | Addgene | #51506 |
| pAAV:CAG-tdTomato plasmid | Addgene | #59462 |
| pAAV:hSyn-DIO-hM3D(Gq)-mCherry plasmid | Addgene | #44361 |
| pGP-AAV-CAG-FLEX-jGCaMP8s-WPRE plasmid | Addgene | #162380 |
| Software and algorithms | ||
| Graphpad Prism v9 | Graphpad Prism Inc | https://www.graphpad.com/scientific-software/prism/ |
| Keyence BZ-X Analyzer | Keyence | https://www.keyence.com/ |
| Zen | Zeiss | https://www.zeiss.com/microscopy/us/products/microscope-software/zen.html |
| Imaris | Oxford Instruments | https://imaris.oxinst.com/ |
| Biorender | Biorender | https://biorender.com/ |
| Adobe Illustrator | Adobe | https://www.adobe.com/products/illustrator/ |
| UMAP analysis code | McInnes, Healy, and Melville 2020 | https://github.com/lmcinnes/umap |
| CaImAn analysis code | Giovannucci et al., 2019 | https://github.com/flatironinstitute/CaImAn-MATLAB |
| Next generation sequencing analysis code | Ravindra Kumar et al., 2020 | https://github.com/GradinaruLab/mCREATE |
Data and code availability
This paper did not report original code. All data are available upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All animal procedures in mice that were carried out in this study were approved by the California Institute of Technology Institutional Animal Care and Use Committee (IACUC), and Harvard Medical School Institutional Animal Care and Use Committee (IACUC). C57BL/6J (000664), Tek-Cre (008863) (Kisanuki et al. 2001), ChAT-IRES-Cre (006410) (Rossi et al. 2011), Nestin-Cre (003771) (Giusti et al. 2014), and TRPV1-Cre (017769) (Cavanaugh et al. 2011) mice were purchased from the Jackson Laboratory (JAX). TH-Cre mice were obtained from the European Mouse Mutant Archive (EM::00254) and crossed with wild-type C57BL/6N mice. Heterozygous TH-Cre mice were used. For capsid selection experiments, 6–8 week old male and female mice were used. For in vivo validation studies of AAV capsid variants, 6–8 weeks old male mice were used.
All procedures performed on rats in this study were approved by the Animal Ethics Committee of the University of Melbourne (Ethics Number 1814639) and complied with the Australian Code for the Care and Use of Animals for Scientific Purposes (National Health and Medical Research Council of Australia). Six male Sprague-Dawley rats (Biomedical Sciences Animal Facility, University of Melbourne) aged 7 weeks were used in this study. Rats were housed in groups of 3 with environmental enrichment under a 12-hour (h) light-dark cycle with ad libitum access to food and water. Six male animals were used in this study and received tail vein injections of AAVs.
All experimental procedures performed on marmosets were approved by the University of California, San Diego, Institutional Animal Care and Use Committee (IACUC) and in accordance with National Institutes of Health and the American Veterinary Medical Association guidelines. 2 female animals and 1 male animal were used in this study and received intravenous injections of AAVs.
All experimental procedures performed on rhesus macaques were approved by the Institutional Animal Care and Use Committee at the University of California, Davis and the California National Primate Research Center (CNPRC). Two infant female animals were used in this study and received intravenous injections of AAVs.
For all the experiments performed in this study, the animals were randomly assigned, and the experimenters were not blinded while performing the experiments in this study unless indicated otherwise.
METHOD DETAILS
Library plasmid preparation
The plasmids used for AAV library preparation were described previously (Ravindra Kumar et al, 2020, Deverman et al, 2016) (plasmids available from Caltech CLOVER Center upon request). Briefly, plasmid rAAV-ΔCap-in-cis-Lox2 (Fig. 1A) was used for building the heptamer insertion (7-mer-i) AAV library. Plasmid pCRII-9Cap-XE was used as a PCR template for the DNA library generation. Plasmid AAV2/9-REP-AAP-ΔCap was used to supplement the AAV library during virus production.
Plasmid preparation for capsid characterization
The AAV capsid variants such as AAV-MaCPNS1 (Addgene plasmid # 185136) and AAV-MaCPNS2 (Addgene plasmid # 185137) capsids were built by inserting 7-mer peptides between AAs 588–589 of the AAV9 cap gene in the pUCmini-iCAP-PHP.B backbone ((Deverman et al. 2016), Addgene plasmid # 103002). The AAV-PHP.S capsid was described previously ((Chan et al. 2017), Addgene plasmid # 103006).
For in vivo validation of AAV capsids, we packaged the vectors with a single-stranded (ss) rAAV genome: pAAV:CAG-2xNLS-EGFP (Deverman et al. 2016) (available from Caltech CLOVER Center upon request, a similar version with 1xNLS is in Addgene, plasmid # 104061), pAAV:CAG-EGFP, pAAV:hSyn1-tdTomato (a gift from Hongkui Zeng, Addgene plasmid # 51506, (Oh et al. 2014)), pAAV:CAG-tdTomato (a gift from Edward Boyden, Addgene plasmid # 59462), pAAV:hSyn-DIO-hM3D(Gq)-mCherry (a gift from Bryan Roth, Addgene plasmid # 44361, (Krashes et al. 2011)). To make the pAAV:CAG-jGCaMP8s plasmid, jGCaMP8s was synthesized as a gBlocks Gene Fragment (IDT) based on the sequence in the plasmid pGP-AAV-CAG-FLEX-jGCaMP8s-WPRE (Addgene plasmid # 162380) and subcloned into the plasmid pAAV:CAG-EGFP by replacing the EGFP gene.
AAV capsid library generation
The round-1 (R1) and round-2 (R2) libraries were generated as described previously (Ravindra Kumar et al, 2020). Briefly, the R1 library involved a randomized 21-nucleotide (7xNNK mutagenesis) insertion between AAs 588–589 of AAV9 capsid. The R2 library was built using a synthetic pool method (Ravindra Kumar et al. 2020). The R2 library was composed of an equimolar ratio of ~9000 variants that were recovered from the tissues of interest in R1 (DRG, heart, small and large intestine). The Spike-in variants as part of the synthetic pool library consisted of previously validated variants such as AAV-PHP.B, AAV-PHP.B4, AAV-PHP.C1 (CNS variants), AAV-PHP.S (PNS variant) and AAV9 parent.
In vivo selection and capsid library recovery
For capsid selection in vivo, the virus library was intravenously administered to male and female mice of various Cre transgenic lines (n=2–3 per Cre line) at 2×1011 vg per mouse in R1 selection, and at 1×1012 vg per mouse in R2 selection. Two weeks post injection, mice were euthanized, the organs of interest were harvested and snap-frozen on dry ice. The tissues were stored at −80°C for long-term. To recover capsids from the tissue, the tissues were processed using Trizol, and the rAAV genomes were recovered by Cre-dependent PCR or Cre-independent PCR as previously described (Ravindra Kumar et al. 2020). The AAV DNA library, virus library and the libraries recovered from tissue post in vivo selection were processed for NGS as also described previously (Ravindra Kumar et al, 2020).
AAV vector production for in vivo characterization
The AAV vectors were produced using an optimized vector production protocol (Challis et al, 2019). The average yield was ~ 1×1012 vg per plate.
AAV vector administration and tissue harvest in Mice
For the intravenous injection procedures in mice, the AAV vectors were injected intravenously via the retro-orbital route into 6–8 week old adult mice at a dose of 0.1–1×1012 vg per mouse. The retro-orbital injections were performed as described previously (Yardeni et al. 2011; Challis et al. 2019). The expression times were ~3 weeks from the time of injection. The dosage and expression time were kept consistent across different experimental groups unless noted otherwise.
For the intraperitoneal injection procedures in mice, neonatal pups at postnatal stage 1 (P1) were intraperitoneally injected with the AAV vectors at a dose of 3×1011 or 1×1012 vg per mouse. Six weeks after AAV administration, tissue collections were performed.
To harvest the tissues of interest, the mice were anesthetized with Euthasol (pentobarbital sodium and phenytoin sodium solution, Virbac AH) and transcardially perfused using 30 – 50 mL of 0.1 M phosphate buffered saline (PBS) (pH 7.4), followed by 30–50 mL of 4% paraformaldehyde (PFA) in 0.1 M PBS. The organs were collected and post-fixed 24–48 h in 4% PFA at 4°C. Following this, the tissues were washed with 0.1 M PBS twice and stored in fresh PBS-azide (0.1 M PBS containing 0.05% sodium azide) at 4°C.
AAV vector administration and tissue harvest in Rat
Six 7-week-old male Sprague Dawley rats received lateral tail vein injections of either MaCPNS1 or MaCPNS2 (2×1013 vg/kg – 3 rats/group). After a 3-week incubation period, animals were transcardially perfused with saline followed by 4% PFA, as per a published protocol (dx.doi.org/10.17504/protocols.io.bahzib76). Tissues were then dissected out and post-fixed in 4% PFA for 1 h before being washed in 0.1 M PBS (3 × 30 min). Tissues were stored in PBS-azide until processing. In one rat (MaCPNS2 injection), the perfusion fixation process was unsuccessful, so tissues were fixed only by immersion (18–24 h), prior to washing and storage as described above.
AAV vector administration and tissue harvest in Marmoset
Marmoset monkeys were anesthetized using an intramuscular Ketamine (20 mg/kg) and Acepromazine (0.5 mg/kg) injection. An intravenous catheter was placed in the saphenous vein of the hind leg and flushed with ~2 mL of LRS (Lactated Ringer’s solution) for 2 min. Viruses were pooled together in a single syringe (~500–900 μL) and infused at a rate of 200 μL/min into the catheter. Following the infusion, the catheter was flushed with ~3 mL of LRS for 2 min and removed. The animal was then returned to a recovery cage.
Following an incubation period of 4–6 weeks post viral injection, the animals were euthanized by injecting pentobarbital intraperitoneally. Two researchers worked in parallel to harvest the tissue as quickly as possible to limit degradation. Each organ – brain, lungs, kidneys, etc - was removed and separated into two parts. One half of the tissue was flash-frozen in 2-methylbutane that was chilled with dry ice to preserve mRNA and DNA in the harvested tissues. The other half of the tissue was fixed in 4% PFA solution for estimation of protein expression. Flash-frozen tissue samples were transferred to a −80°C freezer, while PFA-fixed tissue samples were stored in a 4°C fridge.
AAV vector administration and tissue harvest in Rhesus Macaque
Two female rhesus macaques were injected within 10 days of birth. Prior to injection, animals were anesthetized with ketamine (0.1 mL) and the skin over the saphenous vein was shaved and sanitized. AAVs (2.5×1013 vg/kg) were slowly infused into the saphenous vein for ~1 min in < 0.75 mL of 0.1 M PBS. Animals were monitored while they recovered from anesthesia in their home enclosure, and daily for the remainder of the study. Monkeys were individually housed within sight and sound of conspecifics.
Tissues were collected 4 weeks post AAV administration. Animals were deeply anesthetized and euthanized using sodium pentobarbital in accordance with guidelines for humane euthanasia of animals at the CNPRC. The whole body was perfused with ice cold RNase-free 0.1 M PBS. Brains were removed from the skull and blocked into 4 mm thick slabs in the coronal plane. Brain slabs and organs were subsequently post-fixed in 4% PFA for 48 h. One hemisphere of each animal was cryoprotected in 10%,15%, and 30% sucrose in 0.1 M PBS.
Immunohistochemistry on tissues
In mice and NHP experiments, tissue sections, typically 100-μm thick, were first incubated in blocking buffer (10% normal donkey serum (NDS), 0.1% Triton X-100, and 0.01% sodium azide in 0.1 M PBS, pH 7.4) with primary antibodies (rabbit anti-NeuN (Abcam ab177487, 1:500), chicken anti-PGP9.5 (Invitrogen PA1–10011, 1:500), rabbit anti-S100 beta (Abcam ab52642, 1:200), rabbit anti-Parvalbumin (Abcam 181086, 1:200), rabbit anti-Olig2 (Abcam ab109186, 1:200), rabbit anti-GLUT1 (Millipore 07–1401, 1:200)) at appropriate dilutions for 24 h at room temperature (RT) on a rocker. After primary antibody incubation, the tissues were washed 1–3 times with wash buffer 1 (0.1% Triton X-100 in 0.1 M PBS buffer, pH 7.4) over a period of 5–6 h in total. The tissues were then incubated in blocking buffer with secondary antibodies (goat anti-chicken Alexa647 (Invitrogen, A32933, 1:1000), donkey anti-rabbit Alexa555 (Invitrogen, A21432, 1:1000), goat anti-rabbit Alexa647 (Invitrogen, A21245, 1:1000) at appropriate dilutions for 12–24h at RT and then washed 3 times in 0.1 M PBS over a total duration of 5–6 h. When performing DNA staining, the tissues were incubated with 4′,6-Diamidine-2′-phenylindole dihydrochloride (DAPI) (Sigma Aldrich, 10236276001, 1:1000) in 0.1 M PBS for 15 min followed by a single wash for 10 min in 0.1 M PBS. The DAPI and/or antibody-stained tissue sections were mounted with ProLong Diamond Antifade Mountant (ThermoFisher Scientific, P36970) before imaging under the microscope.
For immunostaining of DRG in mice neonates and the pain induction experiment, DRGs from spinal cord segments: thoracic levels 11–13 and lumbar levels 1–5 or 1–6 were dissected, fixed for 2 h in 4% PFA (diluted in 0.1 M PBS) at 4°C, incubated overnight at 4°C in 30% sucrose solution (diluted in 0.1 M PBS), and embedded in OCT (Tissue-Tek®). Sections of 14 μm thickness were cut and blocked with blocking buffer-1 (10% NDS, in PBST solution containing 0.3% Triton X-100 and 0.1 M PBS, pH 7.4) for 2 h at RT. Tissues were stained with primary antibodies (rabbit anti-CGRP (Millipore PC205L, 1:500), rabbit anti-Neurofilament 200 (Sigma N4142, 1:500), mouse anti-Tuj1 (Abcam ab7751, 1:1,000)) that were diluted in blocking buffer-2 (2% NDS in PBST solution) and incubated overnight at 4°C. Sections were washed in PBST buffer, then stained with secondary antibodies (donkey anti-rabbit DyLight488 (Abcam ab96919, 1:1000), donkey anti-mouse Alexa647 (Abcam ab150107, 1:500)) diluted in blocking buffer-2 for 2 h at RT. After washing with 0.1 M PBS, sections were mounted in VectaShield (Vector Labs), and imaged in an Olympus Fluoview 1000 confocal microscope with 20X magnification at HMS Microscopy Resources on the North Quad (MicRoN) Core. Five DRG images per mouse were used for quantification.
To immunostain rat organs and sensory ganglia, cryosections (14 μm) mounted onto gelatinized slides were washed in PBS (1 × 10 min) followed by 1 h incubation in blocking solution (PBS containing 0.1% Triton X-100 and 10% horse serum). Sections were incubated with primary antibody (hypertonic PBS, 18–24 h in a humid chamber), washed in PBS (1 × 15 min) and incubated with secondary antibody (2 h). Sections were again washed in PBS (1 × 15 min) and then coverslipped using buffered glycerol. This method is described in detail in (dx.doi.org/10.17504/protocols.io.w3ffgjn). DRG and trigeminal tissue were labelled for DsRed and each of the 3 markers (NF-200, CGRP, TRPV1) while liver tissue was labelled for DsRed and DAPI. Primary antibodies were: rabbit anti-dsRed (Takara Bio, 632496, 1:5000); mouse antineurofilament 200 (Sigma-Aldrich, N0142, 1:4000); goat anti-calcitonin gene-related peptide, Bio-Rad,1720-9007,1:5000); goat anti-TRPV1 (Neuromics, GT15129, 1:1000). Secondary antibodies were: donkey anti-rabbit AF594 (Jackson ImmunoResearch Laboratories, 711-585-152, 1:1000); donkey anti-mouse AF488 (Jackson, 715-545-150, 1:2000); donkey anti-goat AF488 (Jackson, 705-545-147, 1:1000).
To immunostain rat CNS, cryosections (40 μm) of brain (coronal) and L4-L5 spinal cord (transverse) were collected in a 1 in 4 series and stored in PBS-azide until use. Sections were washed in PBS (3 × 10 min) followed by 2 h incubation in blocking solution (PBS containing 0.5% Triton X-100 and 10% horse serum), washed in PBS (3 × 10 min) then incubated for 48 h with primary antibody (0.1% PBS-azide containing 0.5% Triton X-100 and 2% horse serum). Tissues were then washed in PBS (3 × 10 min) followed by 4 h incubation with secondary antibody. Tissues were washed in PBS (3 × 10 min) and then mounted and coverslipped using buffered glycerol. This method is described in detail in (dx.doi.org/10.17504/protocols.io.bakkicuw). Primary antibodies used were: rabbit anti-dsRed (Takara Bio, 632496, 1:5000) and mouse anti-NeuN (Chemicon, MAB377, 1:2000). Secondary antibodies are described above.
To immunostain whole mounts of autonomic ganglia (IMG, MPG, sympathetic chain) and intestine, tissues were washed in PBS (3 × 30 min) followed by 2 h incubation in blocking solution (PBS containing 0.5% Triton X-100 and 10% horse serum). Tissues were again washed in PBS (3 × 30 min) before 72 h incubation with primary antibody (0.1% PBS-Azide containing 0.5% Triton X-100 and 2% horse serum). Tissues were then washed in PBS (3 × 30 min) followed by 24 h incubation with secondary antibody. Tissues were washed in PBS (3 × 30 min) before being cleared using ethyl cinnamate (Masselink et al. 2019). Briefly, tissues were washed in methanol (100%, 3 × 30 min) followed by dichloromethane (1 × 30 min) and then ethyl cinnamate (2 × 30 min). Tissues were then mounted on glass slides and coverslipped using ethyl cinnamate.
Optical clearing for thick tissues
For imaging tissues >100 microns in thickness such as gut, optical clearing was carried out by incubating small pieces (0.5–1 cm length) of tissue in 1–2 mL of ScaleS4(0) (Hama et al., 2015) solution overnight at RT with gentle agitation. The tissues were then mounted in fresh ScaleS4(0) solution with spacers (0.1–1 mm thick) on glass slides and imaged under the microscope.
Tissue imaging and image processing
The images used in this study were acquired with a Zeiss LSM 880 confocal microscope using the following objectives: Plan-Apochromat 10X 0.45 M27 (working distance 2.0 mm), and Plan-Apochromat 25X 0.8 Imm Corr DIC M27 multi-immersion. The liver images were acquired with a Keyence BZ-X700 microscope using a 20X objective. The images were then processed in the following image processing software: Zen Black 2.3 SP1 (for Zeiss confocal images) and BZ-X Analyzer (for Keyence images).
Image collection and analysis for cryosections of rat sensory ganglia and organs was performed using wide-field fluorescence microscopy with an ApoTome attachment (Zeiss AxioImager M2). Quantification of AAV+ and AAV+/Marker+ neurons was performed for DRG and trigeminal ganglia on 4 non-sequential sections with only nucleated neuronal profiles counted. Counts were performed manually while viewing sections under the microscope. For CNS sections and whole mounts of intestine and ganglia, image collection and analysis was performed using wide-field fluorescence microscopy with an ApoTome attachment (Zeiss AxioImager M2).
Nodose calcium imaging experiment
In vivo vagal ganglion imaging was conducted as previously described (Williams et al. 2016). Briefly, animals were anesthetized with pentobarbital at a dose of 100 mg/kg body weight, injected intraperitoneally (i.p.). A skin incision was made to expose the abdominal region. For intestinal glucose infusion and distension, an input tubing (HelixMark Silicone Tubing 60-011-04) was inserted in the jejunum and the output tubing (HelixMark Silicone Tubing 60-011-09) was set at the ileum. Surgical thread was used to fasten and secure tubing sites. During the abdominal surgery, isotonic saline was applied occasionally to prevent tissue drying. After suturing the abdominal region, a midline neck incision was made. Muscles were separated and retracted laterally to expose the trachea and vagus trunk. To free the vagal ganglion, the carotid artery was retracted aside, and the vagus nerve was transected superior to the jugular ganglion. The vagal ganglion was then placed on a custom 5-mm diameter glass coverslip (72290-01, Electron Microscopy Sciences), and immediately immersed in silicon adhesive (KWIK-SIL, World Precision Instruments). Imaging was conducted with a Leica SP8 confocal microscope, with a frame rate of 1 Hz. For glucose stimulus, 300 mM glucose solution was infused at a flow rate of 800 μL/min for 2 min. Intestinal distension stimulus was achieved by closing the exit port for 1 min while infusing saline.
Pain behavior experiment
Trpv1-Cre neonatal pups at postnatal stage 1 (P1) were intraperitoneally injected with 10 μL of MaCPNS1:Syn-DIO-hM3D(Gq)-mCherry virus (3×1013 vg/ml). Six weeks after injection, mice were subjected to a pain test in an infrared Behavior Observation Box (iBOB) (Harvard Apparatus) which allowed video recording of mice in the dark and independent of an observer. Prior to Clozapine-N-oxide (CNO) injection, mice were habituated for 30 min in individual chambers of the iBOB. Mice were then given intraplantar injection of 10 μL of CNO or vehicle (5% Dimethyl sulfoxide (DMSO) in 0.1 M PBS). Injections were performed under light restraint without anesthesia. Mice were immediately placed in the iBOB and recorded. Total bouts of lifting/licking of the footpad and total time spent licking/lifting the footpad for the first 15 minutes after CNO injection were quantified.
Analysis of pain behavior
The 20-min videos recorded in the pain-induction experiment were scored by a blinded observer to quantify the total bouts and time spent licking and shaking the injected paw within 15 min. GraphPad Prism was used for statistical comparison and plotting.
Quantification of AAV transduction in vivo
The quantification of AAV transduction across NG, DRG and GI tract was carried out by manually counting fluorescent expression resulting from the AAV genome. The Adobe Photoshop CC 2018 Count Tool was used for this purpose. To quantify expression in the liver, we used Keyence Analyzer automated cell count software. The efficiency was determined by the percentage of cells expressing EGFP or tdTomato relative to a specific cell marker, namely, NeuN, PGP9.5, DAPI, CGRP, NF200, TRPV1, Tuj1, GLUT1, Olig2 or S100β.
NGS data alignment, processing and analysis
The raw fastq DNA files were aligned to the AAV9 capsid template using custom alignment software as described previously (Ravindra Kumar et al. 2020) (https://github.com/GradinaruLab/mCREATE). The NGS data analysis was carried out using a custom data-processing pipeline with scripts written in Python https://github.com/GradinaruLab/mCREATE) and plotting software such as Plotly, Seaborn, and GraphPad PRISM 7.05. The AAV9 capsid structure model was produced with PyMOL.
The enrichment score for a variant was determined using the following formula:
Where N is the total number of variants in a library.
UMAP Clustering analysis
For profiling the overall tropism of the AAV library post R2 selection, a non-linear algorithm, UMAP (McInnes, Healy, and Melville 2020), was used for dimension reduction and visualization with a script adapted from https://github.com/lmcinnes/umap. After the M-CREATE data-processing pipeline, the R2 capsid library yielded enrichment scores across 22 targets (tissues, Fig. 1B). These enrichment scores served as 22 dimensions that were fed into the UMAP algorithm, and the parameters considered were n_neighbors (15), n_components (2), random_state (42), distance_metric (‘correlation’), verbose (3).
Data analysis for calcium imaging
Imaging data was analyzed using CaImAn-MATLAB (Giovannucci et al. 2019; Corder et al. 2019) with modified MATLAB code adapted from https://github.com/flatironinstitute/CaImAn-MATLAB. Basically, imaging frames from the same animal were first registered to correct for motion. A non-negative matrix factorization (CNMF) algorithm was applied to recognize individual cells and to extract fluorescence activities. A 30 s window before the stimulus onset was used as baseline signal. To quantify signals, the mean (μ) and standard deviation (σ) of F0(t) over the baseline period were computed as F(t) = (F0(t) − μ)/σ. Cells were defined as responsive if the average ΔF/F(σ) value during the stimulus period was more than 3 s.d. above the baseline mean activity.
Supplementary Material
Video S1. Marmoset brain injected with MaCPNS1 and MaCPNS2, related to Figure 5. MaCPNS1 packaging ssAAV:CAG-eGFP and MaCPNS2 packaging ssAAV:CAG- tdTomato were mixed and intravenously delivered at a total dose of 7×1013 vg/kg per adult marmoset (16 month old Callithrix jacchus, i.e. 3.5×1013 vg/kg per AAV). Representative overview of marmoset midbrain.
Videos S2. Macaque midbrain injected with MaCPNS1 and MaCPNS2, related to Figure 6. MaCPNS1 packaging ssAAV:CAG-eGFP and MaCPNS2 packaging ssAAV:CAG- tdTomato were mixed and intravenously injected at a dose of 5×1013 vg/kg per macaque (Macaca mulatta, injected within 10 days of birth, female, i.e. 2.5×1013 vg/kg per AAV). Representative overview of macaque midbrain.
Videos S3. Macaque hindbrain injected with MaCPNS1 and MaCPNS2, related to Figure 6. MaCPNS1 packaging ssAAV:CAG-eGFP and MaCPNS2 packaging ssAAV:CAG- tdTomato were mixed and intravenously injected at a dose of 5×1013 vg/kg per macaque (Macaca mulatta, injected within 10 days of birth, female, i.e. 2.5×1013 vg/kg per AAV). Representative overview of macaque hindbrain.
Videos S4. Macaque cerebellum injected with MaCPNS1 and MaCPNS2, related to Figure 6. MaCPNS1 packaging ssAAV:CAG-eGFP and MaCPNS2 packaging ssAAV:CAG- tdTomato were mixed and intravenously injected at a dose of 5×1013 vg/kg per macaque (Macaca mulatta, injected within 10 days of birth, female, i.e. 2.5×1013 vg/kg per AAV). Representative overview of macaque cerebellum.
Highlights:
Direct evolution identified AAV vectors targeting peripheral nervous system (PNS)
Systemically administrated MaCPNS1 and MaCPNS2 efficiently target PNS in rodents
Systemic MaCPNS1 is used for functional readout and non-invasive modulation of PNS
Systemic MaCPNS1 and MaCPNS2 transduce both PNS and CNS in macaque and marmoset
ACKNOWLEDGMENTS:
We thank members of the Gradinaru group for their assistance in this study: Yaping Lei for help with virus production, Elisha Mackey for mouse colony management, Tim Miles, Zhe Qu and Erin Sullivan for lab management, Miguel Chuapoco for discussion on NHP experiments, Patricia Anguiano for administrative assistance and the entire Gradinaru group for discussions. We thank I. Antoshechkin and the Millard and Muriel Jacobs Genetics and Genomics Laboratory at Caltech for providing sequencing service. We thank Cassandra Tang-Wing and Chris Chamberlain at UCSD for the injection of the marmoset. We thank Michael Metke and Vikram Pal Singh at UCSD for the tissue collection from marmoset. We thank the staff at the California National Primate Research Center for support of the macaque experiment. We thank Catherine Oikonomou for help with manuscript editing. Diagrams in figures were created with BioRender.com.
This work was primarily supported by grants from the National Institutes of Health (NIH) to V.G.: NIH BRAIN R01MH117069, NIH Pioneer DP1OD025535 and SPARC 1OT2OD024899. Additional funding includes: (to V.G.) the Vallee Foundation, the Moore Foundation, the CZI Neurodegeneration Challenge Network, and the NSF NeuroNex Technology Hub grant 1707316, a grant from the Pew Charitable Trusts, the Heritage Medical Research Institute and the Beckman Institute for CLARITY, Optogenetics and Vector Engineering Research (CLOVER) for technology development and dissemination; NIH R01 DK127257 (to I.M.C.); CZI Neurodegeneration Challenge Network (to I.M.C.); NIH R01 DC012087 (to C.T.M.); NIH P51 OD011107 (to A.S.F.); BRAIN Initiative Armamentarium UF1MH128336 (to V.G., A.S.F., C.T.M.); Department of Defense grant W81XWH-17-1-0588 (to S.K.M. and V.G.); and NIH SPARC 3OT2OD023872 (to J.R.K.).
DECLARATION OF INTERESTS
The California Institute of Technology has filed and licensed patent applications for some of the work described in this manuscript, with X.C., S.R.K. and V.G. listed as inventors. V.G. is a cofounder and board member of Capsida Biotherapeutics, a fully integrated AAV engineering and gene therapy company. V.G. is a member of the Neuron advisory board. The remaining authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this paper.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES:
- Ali Hossamaldein Gaber, Ibrahim Khalid, Elsaid Mahmoud Fawzi, Mohamed Reem Babiker, Abeidah Mahmoud I. A., Al Rawwas Azhar Othman, Elshafey Khaled, et al. 2021. “Gene Therapy for Spinal Muscular Atrophy: The Qatari Experience.” Gene Therapy, July, 1–5. 10.1038/s41434-021-00273-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asencor Andoni I., Dvoryanchikov Gennady, Tsoulfas Pantelis, and Chaudhari Nirupa. 2021. “AAV-PHP.S-Mediated Delivery of Reporters to Cranial Ganglion Sensory Neurons.” 10.1101/2021.09.14.460327. [DOI] [Google Scholar]
- Batista Ana Rita, King Oliver D., Reardon Christopher P., Davis Crystal, Shankaracharya null, Philip Vivek, Gray-Edwards Heather, et al. 2020. “Ly6a Differential Expression in Blood-Brain Barrier Is Responsible for Strain Specific Central Nervous System Transduction Profile of AAV-PHP.B.” Human Gene Therapy 31 (1–2): 90–102. 10.1089/hum.2019.186. [DOI] [PubMed] [Google Scholar]
- Bedbrook Claire N., Deverman Benjamin E., and Gradinaru Viviana. 2018. “Viral Strategies for Targeting the Central and Peripheral Nervous Systems.” Annual Review of Neuroscience 41 (1): 323–48. 10.1146/annurev-neuro-080317-062048. [DOI] [PubMed] [Google Scholar]
- Bedbrook Claire N., Yang Kevin K., Robinson J. Elliott, Mackey Elisha D., Gradinaru Viviana, and Arnold Frances H.. 2019. “Machine Learning-Guided Channelrhodopsin Engineering Enables Minimally-Invasive Optogenetics.” Nature Methods 16 (11): 1176–84. 10.1038/s41592-019-0583-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan Adam K., Duque Sandra, Foust Kevin D., Morales Pablo R., Braun Lyndsey, Schmelzer Leah, Chan Curtis M., et al. 2011. “Systemic Gene Delivery in Large Species for Targeting Spinal Cord, Brain, and Peripheral Tissues for Pediatric Disorders.” Molecular Therapy: The Journal of the American Society of Gene Therapy 19 (11): 1971–80. 10.1038/mt.2011.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bey Karim, Deniaud Johan, Dubreil Laurence, Joussemet Béatrice, Cristini Joseph, Ciron Carine, Hordeaux Juliette, et al. 2020. “Intra-CSF AAV9 and AAVrh10 Administration in Nonhuman Primates: Promising Routes and Vectors for Which Neurological Diseases?” Molecular Therapy. Methods & Clinical Development 17 (June): 771–84. 10.1016/j.omtm.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdenx Mathieu, Dutheil Nathalie, Bezard Erwan, and Dehay Benjamin. 2014. “Systemic Gene Delivery to the Central Nervous System Using Adeno-Associated Virus.” Frontiers in Molecular Neuroscience 7: 50. 10.3389/fnmol.2014.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanaugh Daniel J., Chesler Alexander T., Jackson Alexander C., Sigal Yaron M., Yamanaka Hiroki, Grant Rebecca, O’Donnell Dajan, et al. 2011. “Trpv1 Reporter Mice Reveal Highly Restricted Brain Distribution and Functional Expression in Arteriolar Smooth Muscle Cells.” The Journal of Neuroscience: The Official Journal of the Society for Neuroscience 31 (13): 5067–77. 10.1523/JNEUROSCI.6451-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti Sampurna, Pattison Luke A., Doleschall Balint, Rickman Rebecca H., Blake Helen, Callejo Gerard, Heppenstall Paul A., and St Ewan. Smith John. 2020. “Intraarticular Adeno-Associated Virus Serotype AAV-PHP.S–Mediated Chemogenetic Targeting of Knee-Innervating Dorsal Root Ganglion Neurons Alleviates Inflammatory Pain in Mice.” Arthritis & Rheumatology 72 (10): 1749–58. 10.1002/art.41314. [DOI] [PubMed] [Google Scholar]
- Challis Rosemary C., Kumar Sripriya Ravindra, Chan Ken Y., Challis Collin, Beadle Keith, Jang Min J., Kim Hyun Min, et al. 2019. “Systemic AAV Vectors for Widespread and Targeted Gene Delivery in Rodents.” Nature Protocols 14 (2): 379. 10.1038/s41596-018-0097-3. [DOI] [PubMed] [Google Scholar]
- Challis Rosemary C., Kumar Sripriya Ravindra, Chen Xinhong, Goertsen David, Coughlin Gerard M., Hori Acacia M., Chuapoco Miguel R., Otis Thomas S., et al. 2022. “Adeno-Associated Virus Toolkit to Target Diverse Brain Cells.” Annual Review of Neuroscience 45:447–69. 10.1146/annurev-neuro-111020-100834 [DOI] [PubMed] [Google Scholar]
- Chan Ken Y., Jang Min J., Yoo Bryan B., Greenbaum Alon, Ravi Namita, Wu Wei-Li, Sánchez-Guardado Luis, et al. 2017. “Engineered AAVs for Efficient Noninvasive Gene Delivery to the Central and Peripheral Nervous Systems.” Nature Neuroscience 20 (8): 1172–79. 10.1038/nn.4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Ritchie, Canales Andres, and Anikeeva Polina. 2017. “Neural Recording and Modulation Technologies.” Nature Reviews. Materials 2 (2): 16093. 10.1038/natrevmats.2016.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Wenli, Yao Shun, Wan Jie, Tian Yu, Huang Lan, Wang Shanshan, Akter Farhana, Wu Yinqiu, Yao Yizheng, and Zhang Xiaochun. 2021. “BBB-Crossing Adeno-Associated Virus Vector: An Excellent Gene Delivery Tool for CNS Disease Treatment.” Journal of Controlled Release: Official Journal of the Controlled Release Society 333 (May): 129–38. 10.1016/j.jconrel.2021.03.029. [DOI] [PubMed] [Google Scholar]
- Corder Gregory, Ahanonu Biafra, Grewe Benjamin F., Wang Dong, Schnitzer Mark J., and Scherrer Grégory. 2019. “An Amygdalar Neural Ensemble That Encodes the Unpleasantness of Pain.” Science (New York, N.Y.) 363 (6424): 276–81. 10.1126/science.aap8586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deverman Benjamin E., Pravdo Piers L., Simpson Bryan P., Kumar Sripriya Ravindra, Chan Ken Y., Banerjee Abhik, Wu Wei-Li, et al. 2016. “Cre-Dependent Selection Yields AAV Variants for Widespread Gene Transfer to the Adult Brain.” Nature Biotechnology 34 (2): 204–9. 10.1038/nbt.3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deverman Benjamin E., Ravina Bernard M., Bankiewicz Krystof S., Paul Steven M., and Sah Dinah W. Y.. 2018. “Gene Therapy for Neurological Disorders: Progress and Prospects.” Nature Reviews Drug Discovery 17 (9): 641–59. 10.1038/nrd.2018.110. [DOI] [PubMed] [Google Scholar]
- Draxler Peter, Moen Aurora, Galek Karolina, Boghos Ani, Ramazanova Dariga, and Sandkühler Jürgen. 2021. “Spontaneous, Voluntary, and Affective Behaviours in Rat Models of Pathological Pain.” Frontiers in Pain Research 2: 23. 10.3389/fpain.2021.672711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Yangyang, Ye Tao, Qu Zhe, Chen Yuewen, Miranda Abigail, Zhou Xiaopu, Lok Ka-Chun, et al. 2021. “Brain-Wide Cas9-Mediated Cleavage of a Gene Causing Familial Alzheimer’s Disease Alleviates Amyloid-Related Pathologies in Mice.” Nature Biomedical Engineering, July, 1–13. 10.1038/s41551-021-00759-0. [DOI] [PubMed] [Google Scholar]
- Foust Kevin D., Nurre Emily, Montgomery Chrystal L., Hernandez Anna, Chan Curtis M., and Kaspar Brian K.. 2009. “Intravascular AAV9 Preferentially Targets Neonatal Neurons and Adult Astrocytes.” Nature Biotechnology 27 (1): 59–65. 10.1038/nbt.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannucci Andrea, Friedrich Johannes, Gunn Pat, Kalfon Jérémie, Brown Brandon L., Koay Sue Ann, Taxidis Jiannis, et al. 2019. “CaImAn an Open Source Tool for Scalable Calcium Imaging Data Analysis.” ELife 8 (January): e38173. 10.7554/eLife.38173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giusti Sebastian A., Vercelli Claudia A., Vogl Annette M., Kolarz Adam W., Pino Natalia S., Deussing Jan M., and Refojo Damian. 2014. “Behavioral Phenotyping of Nestin-Cre Mice: Implications for Genetic Mouse Models of Psychiatric Disorders.” Journal of Psychiatric Research 55 (August): 87–95. 10.1016/j.jpsychires.2014.04.002. [DOI] [PubMed] [Google Scholar]
- Goertsen David, Flytzanis Nicholas C., Goeden Nick, Chuapoco Miguel R., Cummins Alexander, Chen Yijing, Fan Yingying, et al. 2021. “AAV Capsid Variants with Brain-Wide Transgene Expression and Decreased Liver Targeting after Intravenous Delivery in Mouse and Marmoset.” Nature Neuroscience, December. 10.1038/s41593-021-00969-4. [DOI] [PubMed] [Google Scholar]
- Golebiowski Diane, van der Bom Imramsjah M. J., Kwon Churl-Su, Miller Andrew D., Petrosky Keiko, Bradbury Allison M., Maitland Stacy, et al. 2017. “Direct Intracranial Injection of AAVrh8 Encoding Monkey β-N-Acetylhexosaminidase Causes Neurotoxicity in the Primate Brain.” Human Gene Therapy 28 (6): 510–22. 10.1089/hum.2016.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gombash SE, Cowley CJ, Fitzgerald JA, Lepak CA, Neides MG, Hook K, Todd LJ, et al. 2017. “Systemic Gene Delivery Transduces the Enteric Nervous System of Guinea Pigs and Cynomolgus Macaques.” Gene Therapy 24 (10): 640–48. 10.1038/gt.2017.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Xin, Mendoza-Halliday Diego, Ting Jonathan T., Kaiser Tobias, Sun Xuyun, Bastos André M., Wimmer Ralf D., et al. 2020. “An Ultra-Sensitive Step-Function Opsin for Minimally Invasive Optogenetic Stimulation in Mice and Macaques.” Neuron 107 (1): 38–51.e8. 10.1016/j.neuron.2020.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore Reshma, Riedl Maureen S., Kitto Kelley F., Fairbanks Carolyn A., and Vulchanova Lucy. 2019. “AAV-Mediated Gene Delivery to the Enteric Nervous System by Intracolonic Injection.” Methods in Molecular Biology (Clifton, N.J.) 1950: 407–15. 10.1007/978-1-4939-9139-6_24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray Steven J. 2016. “Timing of Gene Therapy Interventions: The Earlier, the Better.” Molecular Therapy 24 (6): 1017–18. 10.1038/mt.2016.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray Steven J, Woodard Kenton T, and Samulski R Jude. 2010. “Viral Vectors and Delivery Strategies for CNS Gene Therapy.” Therapeutic Delivery 1 (4): 517–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grødem Sverre, Nymoen Ingeborg, Guro Helén Vatne Valgerdur Björnsdottir, Lensjø Kristian Kinden, and Fyhn Marianne. 2021. “An Updated Suite of Viral Vectors for in Vivo Calcium Imaging Using Local and Retro-Orbital Injections.” BioRxiv, January, 2021.05.14.443815. 10.1101/2021.05.14.443815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GuhaSarkar Dwijit, Neiswender James, Su Qin, Gao Guangping, and Sena-Esteves Miguel. 2017. “Intracranial AAV-IFN-β Gene Therapy Eliminates Invasive Xenograft Glioblastoma and Improves Survival in Orthotopic Syngeneic Murine Model.” Molecular Oncology 11 (2): 180–93. 10.1002/1878-0261.12020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanlon Killian S., Meltzer Jonah C., Buzhdygan Tetyana, Cheng Ming J., Sena-Esteves Miguel, Bennett Rachel E., Sullivan Timothy P., et al. 2019. “Selection of an Efficient AAV Vector for Robust CNS Transgene Expression.” Molecular Therapy. Methods & Clinical Development 15 (December): 320–32. 10.1016/j.omtm.2019.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinderer Christian, Katz Nathan, Buza Elizabeth L., Dyer Cecilia, Goode Tamara, Bell Peter, Richman Laura K., and Wilson James M.. 2018. “Severe Toxicity in Nonhuman Primates and Piglets Following High-Dose Intravenous Administration of an Adeno-Associated Virus Vector Expressing Human SMN.” Human Gene Therapy 29 (3): 285–98. 10.1089/hum.2018.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai Takashi, Enomoto Mitsuhiro, Machida Akira, Yamamoto Mariko, Kuwahara Hiroya, Tajiri Mio, Hirai Yukihiko, et al. 2012. “Intrathecal ShRNA-AAV9 Inhibits Target Protein Expression in the Spinal Cord and Dorsal Root Ganglia of Adult Mice.” Human Gene Therapy Methods 23 (2): 119–27. 10.1089/hgtb.2012.035. [DOI] [PubMed] [Google Scholar]
- Homs J, Ariza L, Pagès G, Udina E, Navarro X, Chillón M, and Bosch A. 2011. “Schwann Cell Targeting via Intrasciatic Injection of AAV8 as Gene Therapy Strategy for Peripheral Nerve Regeneration.” Gene Therapy 18 (6): 622–30. 10.1038/gt.2011.7. [DOI] [PubMed] [Google Scholar]
- Hordeaux Juliette, Wang Qiang, Katz Nathan, Buza Elizabeth L., Bell Peter, and Wilson James M.. 2018. “The Neurotropic Properties of AAV-PHP.B Are Limited to C57BL/6J Mice.” Molecular Therapy 26 (3): 664–68. 10.1016/j.ymthe.2018.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hordeaux Juliette, Yuan Yuan, Clark Peter M., Wang Qiang, Martino R. Alexander, Sims Joshua J., Bell Peter, Raymond Angela, Stanford William L., and Wilson James M.. 2019. “The GPI-Linked Protein LY6A Drives AAV-PHP.B Transport across the Blood-Brain Barrier.” Molecular Therapy: The Journal of the American Society of Gene Therapy 27 (5): 912–21. 10.1016/j.ymthe.2019.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Lan, Wan Jie, Wu Yinqiu, Tian Yu, Yao Yizheng, Yao Shun, Ji Xiaoyun, Wang Shengjun, Su Zhaoliang, and Xu Huaxi. 2021. “Challenges in Adeno-Associated Virus-Based Treatment of Central Nervous System Diseases through Systemic Injection.” Life Sciences 270 (April): 119142. 10.1016/j.lfs.2021.119142. [DOI] [PubMed] [Google Scholar]
- Huang Qin, Chan Ken Y., Tobey Isabelle G., Chan Yujia Alina, Poterba Tim, Boutros Christine L., Balazs Alejandro B., et al. 2019. “Delivering Genes across the Blood-Brain Barrier: LY6A, a Novel Cellular Receptor for AAV-PHP.B Capsids.” PloS One 14 (11): e0225206. 10.1371/journal.pone.0225206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudry Eloise, and Vandenberghe Luk H.. 2019. “Therapeutic AAV Gene Transfer to the Nervous System: A Clinical Reality.” Neuron 101 (5): 839–62. 10.1016/j.neuron.2019.02.017. [DOI] [PubMed] [Google Scholar]
- Ingusci Selene, Verlengia Gianluca, Soukupova Marie, Zucchini Silvia, and Simonato Michele. 2019. “Gene Therapy Tools for Brain Diseases.” Frontiers in Pharmacology 10: 724. 10.3389/fphar.2019.00724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings Charles G., Landman Rogier, Zhou Yang, Sharma Jitendra, Hyman Julia, Movshon J. Anthony, Qiu Zilong, et al. 2016. “Opportunities and Challenges in Modeling Human Brain Disorders in Transgenic Primates.” Nature Neuroscience 19 (9): 1123–30. 10.1038/nn.4362. [DOI] [PubMed] [Google Scholar]
- Jiman Ahmad A., Ratze David C., Welle Elissa J., Patel Paras R., Richie Julianna M., Bottorff Elizabeth C., Seymour John P., Chestek Cynthia A., and Bruns Tim M.. 2020. “Multi-Channel Intraneural Vagus Nerve Recordings with a Novel High-Density Carbon Fiber Microelectrode Array.” Scientific Reports 10 (1): 15501. 10.1038/s41598-020-72512-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Seigo, and Harashima Hideyoshi. 2020. “Current Status and Challenges Associated with CNS-Targeted Gene Delivery across the BBB.” Pharmaceutics 12 (12): 1216. 10.3390/pharmaceutics12121216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisanuki YY, Hammer RE, Miyazaki J, Williams SC, Richardson JA, and Yanagisawa M. 2001. “Tie2-Cre Transgenic Mice: A New Model for Endothelial Cell-Lineage Analysis in Vivo.” Developmental Biology 230 (2): 230–42. 10.1006/dbio.2000.0106. [DOI] [PubMed] [Google Scholar]
- Krashes Michael J., Koda Shuichi, Ye ChianPing, Rogan Sarah C., Adams Andrew C., Cusher Daniel S., Maratos-Flier Eleftheria, Roth Bryan L., and Lowell Bradford B.. 2011. “Rapid, Reversible Activation of AgRP Neurons Drives Feeding Behavior in Mice.” The Journal of Clinical Investigation 121 (4): 1424–28. 10.1172/JCI46229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Körbelin Jakob, Dogbevia Godwin, Michelfelder Stefan, Ridder Dirk A., Hunger Agnes, Wenzel Jan, Seismann Henning, et al. 2016. “A Brain Microvasculature Endothelial Cell-Specific Viral Vector with the Potential to Treat Neurovascular and Neurological Diseases.” EMBO Molecular Medicine 8 (6): 609–25. 10.15252/emmm.201506078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krashes Michael J., Koda Shuichi, Ye ChianPing, Rogan Sarah C., Adams Andrew C., Cusher Daniel S., Maratos-Flie Eleftheria, Roth Bryan L., and Lowell Bradford B.. 2011. “Rapid, Reversible Activation of AgRP Neurons Drives Feeding Behavior in Mice.” The Journal of Clinical Investigation 121 (4): 1424–28. 10.1172/JCI46229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy Jonathan M., Yeh Wei-Hsi, Pendse Nachiket, Davis Jessie R., Hennessey Erin, Butcher Rossano, Koblan Luke W., Comander Jason, Liu Qin, and Liu David R.. 2020. “Cytosine and Adenine Base Editing of the Brain, Liver, Retina, Heart and Skeletal Muscle of Mice via Adeno-Associated Viruses.” Nature Biomedical Engineering 4 (1): 97–110. 10.1038/s41551-019-0501-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvaldi Letizia, Panayotis Nicolas, Alber Stefanie, Dagan Shachar Y., Okladnikov Nataliya, Koppel Indrek, Di Pizio Agostina, et al. 2020. “Importin A3 Regulates Chronic Pain Pathways in Peripheral Sensory Neurons.” Science 369 (6505): 842–46. 10.1126/science.aaz5875. [DOI] [PubMed] [Google Scholar]
- Marx Vivien. 2016. “Neurobiology: Learning from Marmosets.” Nature Methods 13 (11): 911–16. 10.1038/nmeth.4036. [DOI] [PubMed] [Google Scholar]
- Masselink Wouter, Reumann Daniel, Murawala Prayag, Pasierbek Pawel, Taniguchi Yuka, Bonnay François, Meixner Katharina, Knoblich Jürgen A., and Tanaka Elly M.. 2019. “Broad Applicability of a Streamlined Ethyl Cinnamate-Based Clearing Procedure.” Development (Cambridge, England) 146 (3): dev166884. 10.1242/dev.166884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki Yasunori, Konno Ayumu, Mochizuki Ryuta, Shinohara Yoichiro, Nitta Keisuke, Okada Yukihiro, and Hirai Hirokazu. 2018. “Intravenous Administration of the Adeno-Associated Virus-PHP.B Capsid Fails to Upregulate Transduction Efficiency in the Marmoset Brain.” Neuroscience Letters 665: 182–88. 10.1016/j.neulet.2017.11.049. [DOI] [PubMed] [Google Scholar]
- McInnes Leland, Healy John, and Melville James. 2020. “UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction.” ArXiv:1802.03426 [Cs, Stat], September. http://arxiv.org/abs/1802.03426. [Google Scholar]
- Michelson Nicholas J., Vanni Matthieu P., and Murphy Timothy H.. 2019. “Comparison between Transgenic and AAV-PHP.EB-Mediated Expression of GCaMP6s Using in Vivo Wide-Field Functional Imaging of Brain Activity.” Neurophotonics 6 (2): 025014. 10.1117/1.NPh.6.2.025014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller Cory T, Freiwald Winrich A, Leopold David A, Mitchell Jude F, Silva Afonso C, and Wang Xiaoqin. 2016. “Marmosets: A Neuroscientific Model of Human Social Behavior.” Neuron 90 (2): 219–33. 10.1016/j.neuron.2016.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra Santosh K., Tisel Sarah M., Orestes Peihan, Bhangoo Sonia K., and Hoon Mark A.. 2011. “TRPV1-Lineage Neurons Are Required for Thermal Sensation.” The EMBO Journal 30 (3): 582–93. 10.1038/emboj.2010.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteys Alex Mas, Hundley Amiel A., Ranum Paul T., Tecedor Luis, Muehlmatt Amy, Lim Euyn, Lukashev Dmitriy, Sivasankaran Rajeev, and Davidson Beverly L.. 2021. “Regulated Control of Gene Therapies by Drug-Induced Splicing.” Nature 596 (7871): 291–95. 10.1038/s41586-021-03770-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonnenmacher Mathieu, Wang Wei, Child Matthew A., Ren Xiao-Qin, Huang Carol, Amy Zhen Ren Jenna Tocci, et al. 2021. “Rapid Evolution of Blood-Brain-Barrier-Penetrating AAV Capsids by RNA-Driven Biopanning.” Molecular Therapy. Methods & Clinical Development 20 (March): 366–78. 10.1016/j.omtm.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Carroll Simon J., Cook William H., and Young Deborah. 2021. “AAV Targeting of Glial Cell Types in the Central and Peripheral Nervous System and Relevance to Human Gene Therapy.” Frontiers in Molecular Neuroscience 13: 256. 10.3389/fnmol.2020.618020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh Seung Wook, Harris Julie A., Ng Lydia, Winslow Brent, Cain Nicholas, Mihalas Stefan, Wang Quanxin, et al. 2014. “A Mesoscale Connectome of the Mouse Brain.” Nature 508 (7495): 207–14. 10.1038/nature13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojala David S., Sun Sabrina, Santiago-Ortiz Jorge L., Shapiro Mikhail G., Romero Philip A., and Schaffer David V.. 2018. “In Vivo Selection of a Computationally Designed SCHEMA AAV Library Yields a Novel Variant for Infection of Adult Neural Stem Cells in the SVZ.” Molecular Therapy: The Journal of the American Society of Gene Therapy 26 (1): 304–19. 10.1016/j.ymthe.2017.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena Stefanie A., Iyengar Rahul, Eshraghi Rebecca S., Bencie Nicole, Mittal Jeenu, Aljohani Abdulrahman, Mittal Rahul, and Eshraghi Adrien A.. 2020. “Gene Therapy for Neurological Disorders: Challenges and Recent Advancements.” Journal of Drug Targeting 28 (2): 111–28. 10.1080/1061186X.2019.1630415. [DOI] [PubMed] [Google Scholar]
- Piguet Françoise, de Saint Denis Timothée, Audouard Emilie, Beccaria Kevin, André Arthur, Wurtz Guillaume, Schatz Raphael, et al. 2021. “The Challenge of Gene Therapy for Neurological Diseases: Strategies and Tools to Achieve Efficient Delivery to the Central Nervous System.” Human Gene Therapy 32 (7–8): 349–74. 10.1089/hum.2020.105. [DOI] [PubMed] [Google Scholar]
- Pogorzala Leah A., Mishra Santosh K., and Hoon Mark A.. 2013. “The Cellular Code for Mammalian Thermosensation.” The Journal of Neuroscience: The Official Journal of the Society for Neuroscience 33 (13): 5533–41. 10.1523/JNEUROSCI.5788-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Privolizzi Riccardo, Chu Wing Sum, Tijani Maha, and Ng Joanne. 2021. “Viral Gene Therapy for Paediatric Neurological Diseases: Progress to Clinical Reality.” Developmental Medicine & Child Neurology 63 (9): 1019–29. 10.1111/dmcn.14885. [DOI] [PubMed] [Google Scholar]
- Ravagli Enrico, Mastitskaya Svetlana, Thompson Nicole, Iacoviello Francesco, Shearing Paul R., Perkins Justin, Gourine Alexander V., Aristovich Kirill, and Holder David. 2020. “Imaging Fascicular Organization of Rat Sciatic Nerves with Fast Neural Electrical Impedance Tomography.” Nature Communications 11 (1): 6241. 10.1038/s41467-020-20127-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar Ravindra, Sripriya Timothy F. Miles, Chen Xinhong, Brown David, Dobreva Tatyana, Huang Qin, Ding Xiaozhe, et al. 2020. “Multiplexed Cre-Dependent Selection Yields Systemic AAVs for Targeting Distinct Brain Cell Types.” Nature Methods 17 (5): 541–50. 10.1038/s41592-020-0799-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynaud-Dulaurier Robin, and Decressac Michael. 2020. “PHP.B/EB Vectors Bring New Successes to Gene Therapy for Brain Diseases.” Frontiers in Bioengineering and Biotechnology 8: 1137. 10.3389/fbioe.2020.582979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi Jari, Balthasar Nina, Olson David, Scott Michael, Berglund Eric, Lee Charlotte E., Choi Michelle J., Lauzon Danielle, Lowell Bradford B., and Elmquist Joel K.. 2011. “Melanocortin-4 Receptors Expressed by Cholinergic Neurons Regulate Energy Balance and Glucose Homeostasis.” Cell Metabolism 13 (2): 195–204. 10.1016/j.cmet.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevin Caroline, and Deiva Kumaran. 2021. “Clinical Trials for Gene Therapy in Lysosomal Diseases With CNS Involvement.” Frontiers in Molecular Biosciences 8: 624988. 10.3389/fmolb.2021.624988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano Tetsuya, Wallace John T., Baldwin Katherine T., Purkey Alicia M., Uezu Akiyoshi, Courtland Jamie L., Soderblom Erik J., et al. 2020. “Chemico-Genetic Discovery of Astrocytic Control of Inhibition in Vivo.” Nature 588 (7837): 296–302. 10.1038/s41586-020-2926-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosolini Andrew P., and Smith George M.. 2018. “Editorial: Gene Therapy for the Central and Peripheral Nervous System.” Frontiers in Molecular Neuroscience 11: 54. 10.3389/fnmol.2018.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt Christoph C., Bruegmann Tobias, Malan Daniela, Ottersbach Annika, Roell Wilhelm, Fleischmann Bernd K., and Sasse Philipp. 2015. “Systemic Gene Transfer Enables Optogenetic Pacing of Mouse Hearts.” Cardiovascular Research 106 (2): 338–43. 10.1093/cvr/cvv004. [DOI] [PubMed] [Google Scholar]
- Walters Marie C., Sonner Martha J., Myers Jessica H., and Ladle David R.. 2019. “Calcium Imaging of Parvalbumin Neurons in the Dorsal Root Ganglia.” ENeuro 6 (4): ENEURO.0349–18.2019. 10.1523/ENEURO.0349-18.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams Erika K., Chang Rui B., Strochlic David E., Umans Benjamin D., Lowell Bradford B., and Liberles Stephen D.. 2016. “Sensory Neurons That Detect Stretch and Nutrients in the Digestive System.” Cell 166 (1): 209–21. 10.1016/j.cell.2016.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Bin, Li Shaoyong, Wang Hongyan, Guo Yansu, Gessler Dominic J, Cao Chunyan, Su Qin, et al. 2014. “Global CNS Transduction of Adult Mice by Intravenously Delivered RAAVrh.8 and RAAVrh.10 and Nonhuman Primates by RAAVrh.10.” Molecular Therapy 22 (7): 1299–1309. 10.1038/mt.2014.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Yiyuan, Wu Mingzheng, Vázquez-Guardado Abraham, Wegener Amy J., Grajales-Reyes Jose G., Deng Yujun, Wang Taoyi, et al. 2021. “Wireless Multilateral Devices for Optogenetic Studies of Individual and Social Behaviors.” Nature Neuroscience 24 (7): 1035–45. 10.1038/s41593-021-00849-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yardeni Tal, Michael Eckhaus H. Douglas Morris, Marjan Huizing, and Shelley Hoogstraten-Miller. 2011. “Retro-Orbital Injections in Mice.” Lab Animal 40 (5): 155–60. 10.1038/laban0511-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Hongwei, Fischer Gregory, Ferhatovic Lejla, Fan Fan, Light Alan R., Weihrauch Dorothee, Sapunar Damir, Nakai Hiroyuki, Park Frank, and Hogan Quinn H.. 2013. “Intraganglionic AAV6 Results in Efficient and Long-Term Gene Transfer to Peripheral Sensory Nervous System in Adult Rats.” PloS One 8 (4): e61266. 10.1371/journal.pone.0061266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanos Theodoros P., Silverman Harold A., Levy Todd, Tsaava Tea, Battinelli Emily, Lorraine Peter W., Ashe Jeffrey M., Chavan Sangeeta S., Tracey Kevin J., and Bouton Chad E.. 2018. “Identification of Cytokine-Specific Sensory Neural Signals by Decoding Murine Vagus Nerve Activity.” Proceedings of the National Academy of Sciences 115 (21): E4843–52. 10.1073/pnas.1719083115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Yan; Rózsa Márton; Bushey Daniel; Zheng Jihong; Reep Daniel; Broussard Gerard Joey; Tsang Arthur; Tsegaye Getahun; Patel Ronak, et al. 2020. “jGCaMP8 Fast Genetically Encoded Calcium Indicators.” Janelia Research Campus. Online resource. 10.25378/janelia.13148243.v1 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1. Marmoset brain injected with MaCPNS1 and MaCPNS2, related to Figure 5. MaCPNS1 packaging ssAAV:CAG-eGFP and MaCPNS2 packaging ssAAV:CAG- tdTomato were mixed and intravenously delivered at a total dose of 7×1013 vg/kg per adult marmoset (16 month old Callithrix jacchus, i.e. 3.5×1013 vg/kg per AAV). Representative overview of marmoset midbrain.
Videos S2. Macaque midbrain injected with MaCPNS1 and MaCPNS2, related to Figure 6. MaCPNS1 packaging ssAAV:CAG-eGFP and MaCPNS2 packaging ssAAV:CAG- tdTomato were mixed and intravenously injected at a dose of 5×1013 vg/kg per macaque (Macaca mulatta, injected within 10 days of birth, female, i.e. 2.5×1013 vg/kg per AAV). Representative overview of macaque midbrain.
Videos S3. Macaque hindbrain injected with MaCPNS1 and MaCPNS2, related to Figure 6. MaCPNS1 packaging ssAAV:CAG-eGFP and MaCPNS2 packaging ssAAV:CAG- tdTomato were mixed and intravenously injected at a dose of 5×1013 vg/kg per macaque (Macaca mulatta, injected within 10 days of birth, female, i.e. 2.5×1013 vg/kg per AAV). Representative overview of macaque hindbrain.
Videos S4. Macaque cerebellum injected with MaCPNS1 and MaCPNS2, related to Figure 6. MaCPNS1 packaging ssAAV:CAG-eGFP and MaCPNS2 packaging ssAAV:CAG- tdTomato were mixed and intravenously injected at a dose of 5×1013 vg/kg per macaque (Macaca mulatta, injected within 10 days of birth, female, i.e. 2.5×1013 vg/kg per AAV). Representative overview of macaque cerebellum.
Data Availability Statement
This paper did not report original code. All data are available upon request.


