Abstract
Bacteria use trans-translation to rescue stalled ribosomes and target incomplete proteins for proteolysis. Despite similarities between tRNAs and transfer-messenger RNA (tmRNA), the key molecule for trans-translation, new structural and biochemical data show important differences between translation and trans-translation at most steps of the pathways. tmRNA and its binding partner, SmpB, bind in the A site of the ribosome but do not trigger the same movements of nucleotides in the rRNA that are required for codon recognition by tRNA. tmRNA-SmpB moves from the A site to the P site of the ribosome without subunit rotation to generate hybrid states, and moves from the P site to a site outside the ribosome instead of to the E site. During catalysis, transpeptidation to tmRNA appears to require the ribosomal protein bL27, which is dispensable for translation, suggesting that this protein may be conserved in bacteria due to trans-translation. These differences provide insights into the fundamental nature of trans-translation, and provide targets for new antibiotics that may have decrease cross-reactivity with eukaryotic ribosomes.
Keywords: trans-translation, translation, ribosome, tRNA, tmRNA, protein synthesis, antibiotics
Introduction: Ribosome stalling and ribosome rescue
Despite the high fidelity of transcription and translation, ribosomes are frequently trapped on mRNA and need to be rescued. In Escherichia coli, for example, 5% of translation initiations do not produce a complete protein1. Because E. coli ribosomes translate ~50 proteins per cell division cycle, the average ribosome will stall 2–3 times each generation. If these ribosomes could not be rescued, the cell would rapidly lose its protein synthesis capacity and would die1.
In bacteria, ribosomes regularly stall at the 3’ end of mRNAs lacking an in-frame stop codon because of nuclease activity, mRNA damage or premature transcription termination2. When ribosomes stall in the middle of an mRNA, 3’−5’ exonucleases remove the sequence downstream of the ribosome, including the stop codon3. There are also toxins, such as RelE, that cut the mRNA in the A site of the ribosome4. Because multiple ribosomes can initiate translation on the same mRNA, if one ribosome stalls, all the following ribosomes will also be blocked from completing translation and collide. These colliding ribosomes are released by a specialized nuclease, SrmB, that cuts the mRNA between collided ribosomes5. In all these cases, the ribosomes cannot terminate translation normally because there is no stop codon in the mRNA. These “non-stop” ribosomes must be rescued by releasing the peptidyl-tRNA, so the ribosomal subunits can be recycled for productive protein synthesis.
The main mechanism in bacteria to rescue non-stop ribosomes is trans-translation (Figure 1). Non-stop ribosomes are recognized by transfer-messenger RNA (tmRNA) and small protein B (SmpB), which bind to the empty aminoacyl (A) site on the small 30S subunit (Figure 1A). tmRNA contains a tRNA-like domain (TLD) that is aminoacylated with alanine and a reading frame that encodes a proteolysis tag (Figure 2). Binding of tmRNA-SmpB allows translation to restart using tmRNA as a message, thereby adding the proteolysis tag to the nascent polypeptide (Figures 1B–1D)6.Translation terminates at a stop codon encoded on tmRNA6, allowing recycling of the ribosomal subunits, the non-stop mRNA is degraded by RNases and the nascent chain containing the tag is proteolyzed (Figures 1B, 1E, and 1F)7–9. Trans-translation components have been identified in almost every sequenced bacterial genome and are not present in higher eukaryotes10. In many bacteria, including the pathogens Neisseria gonorrhoeae and Mycobacterium tuberculosis, trans-translation is essential for viability11,12. Other bacterial species encode one or more alternative ribosome rescue factors in addition to trans-translation: ArfA, ArfT, or BrfA, which recruit a release factor (RF1 or RF2) to non-stop ribosomes, or ArfB, which can hydrolyze peptidyl-tRNA on the ribosome independently of release factors13–17. These alternative ribosome rescue factors can serve as a backup system for non-stop ribosome rescue when trans-translation is inactivated or overwhelmed13–17. Non-stop ribosomes are rescued by a different pathway in eukaryotes, so antibiotics that target trans-translation may not have the same toxicity and cross-reactivity in eukaryotic cells as antibiotics that target translation, which can also inhibit eukaryotic ribosomes18.
Figure 1. trans-translation relieves stalling at non-stop ribosomes.
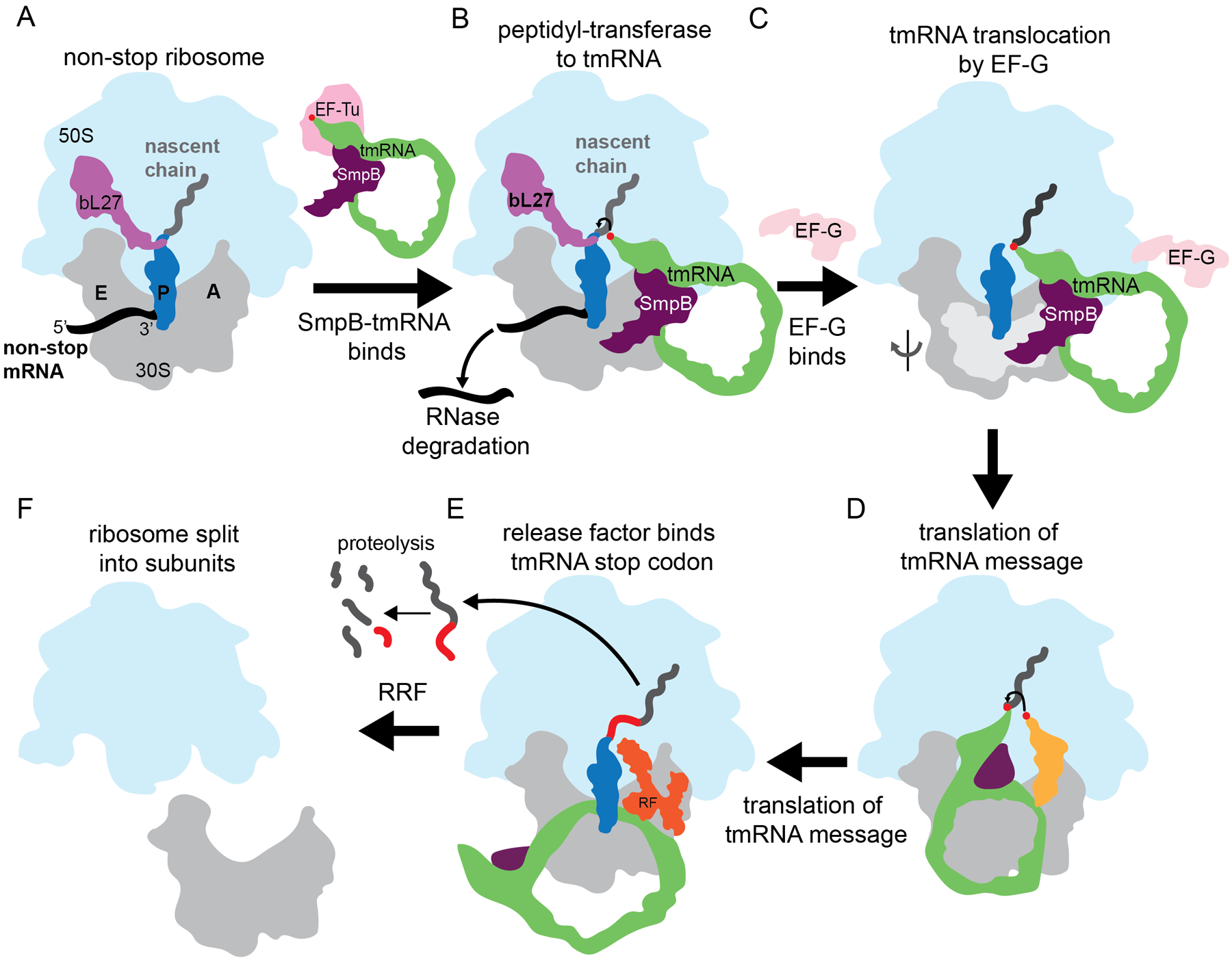
A. A non-stop ribosome arises when the ribosome translates a non-stop mRNA (black), resulting in peptidyl-tRNA (blue) trapped in the P site, and an empty A site. EF-Tu (pink) brings alanyl-tmRNA-SmpB (green and purple) to non-stop complexes. B. SmpB-tmRNA binds to the empty A site, and peptidyl-transfer occurs. C. The 30S ribosomal subunit head rotates relative to the 50S and tilts (light grey), as EF-G (light pink) binds to induce ribosomal translocation of the tRNAs on the 30S. D. Aminoacyl-tRNA (gold) binds next on the tmRNA message. E. Translation proceeds, decoding the protease tag until the tmRNA message terminates at a stop codon. RF1 or RF2 (orange) binds the stop codon and releases the tagged protein. F. The tagged protein is proteolyzed and ribosome recycling factor (RRF) splits the ribosome into its two subunits.
Figure 2. Comparison of tmRNA to tRNA structure.
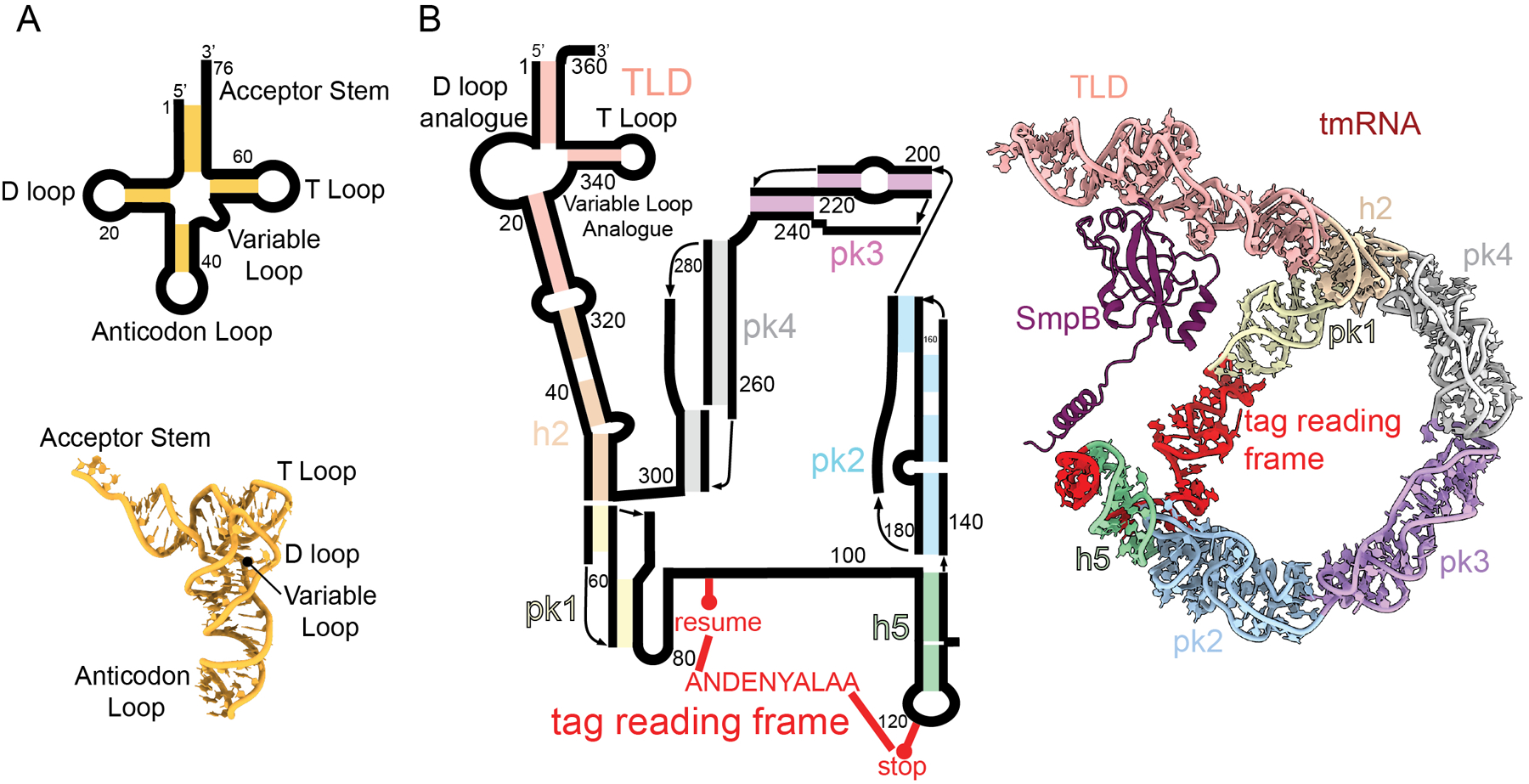
A. (top) Secondary structure of a tRNA with stem regions colored in gold. The three-dimensional structure of a tRNA with regions indicated (bottom; PDB code 4V5D). B. Secondary structure of tmRNA with stems in colored blocks: TLD (red), h2 (orange), pk1 (yellow), h5 (green), pk2 (blue), pk3 (purple), pk4 (grey) and the tag reading frame (red) (left). The three-dimensional structure of tmRNA with SmpB bound with the same color scheme as in panel B (right; PDB code 7AC7).
Because trans-translation requires all the general translation factors, it was long assumed that tmRNA-SmpB interactions with the ribosome largely mimicked those of tRNAs. However, recent structural and biochemical studies show that there are remarkable differences at almost every step of the pathway. First, trans-translation relies on SmpB binding to tmRNA (Figure 2B)19, which are two macromolecules unique to trans-translation and do not play a role in canonical translation. Second, there are differences in how tmRNA-SmpB binds to the A site as compared to normal tRNAs (Figure 2A). For example, during normal translation, the tRNA anticodon binds a cognate mRNA codon, while in trans-translation tmRNA lacks an equivalent anticodon and instead SmpB binds to the empty A site of the ribosome rather than to mRNA20–22. Third, structural data suggests differences in how tmRNA translocates through the ribosome21,22. And finally, despite similarities between the acceptor stem of both tmRNA and tRNA to position the 3’ end of the activated aminoacyl, structural and biochemical studies suggests a crucial role of the ribosomal protein bL27 (“b” designates a bacterial specific ribosomal protein23) in trans-translation, particularly the positioning or flexibility of the bL27 N terminus. These differences are potential targets for antibiotic development and may explain evolutionary conservation of some features of the ribosome and general translation factors.
tRNA and tmRNA Delivery to the A site
Addition of each amino acid to the nascent polypeptide during translation elongation begins when a ternary complex of aminoacyl-tRNA•EF-Tu•GTP binds at the A site. For the ribosome to select the correct tRNA, the three-nucleotide tRNA anticodon base pairs with the three-nucleotide mRNA codon on the 30S subunit in the A-site decoding center (Figure 3A). Cognate tRNA binding to an mRNA codon induces 16S rRNA nucleotides A1492 and A1493, and G530 to rearrange to monitor the presence of Watson-Crick pairing between the codon and anticodon24. Nucleotides A1492 and A1493 interact with the first and second base pairs of the codon-anticodon interaction, respectively, and G530 interacts with the anticodon second nucleotide and codon third nucleotide (Figure 3A)24–26. While Watson-Crick pairing between the codon and the anticodon is essential for the fidelity of tRNA selection, the ribosome employs both kinetic and induced fit mechanisms to ensure correct tRNA selection27. The 30S head domain closes around the tRNA, triggering GTP hydrolysis by EF-Tu on the large 50S subunit, almost 80 Å distant. If a non-cognate tRNA is present, GTP hydrolysis still occurs, but at a slower rate, providing an opportunity for the tRNA to dissociate28,29. EF-Tu•GDP dissociation permits full accommodation of tRNA into the A site on the 50S subunit and specifically, in the peptidyl transferase center (PTC). This accommodation positions the tRNA acceptor stem containing the activated aminoacyl group for rapid catalysis (Figure 3A).
Figure 3. SmpB-tmRNA forms distinct A-site interactions as compared to tRNA.
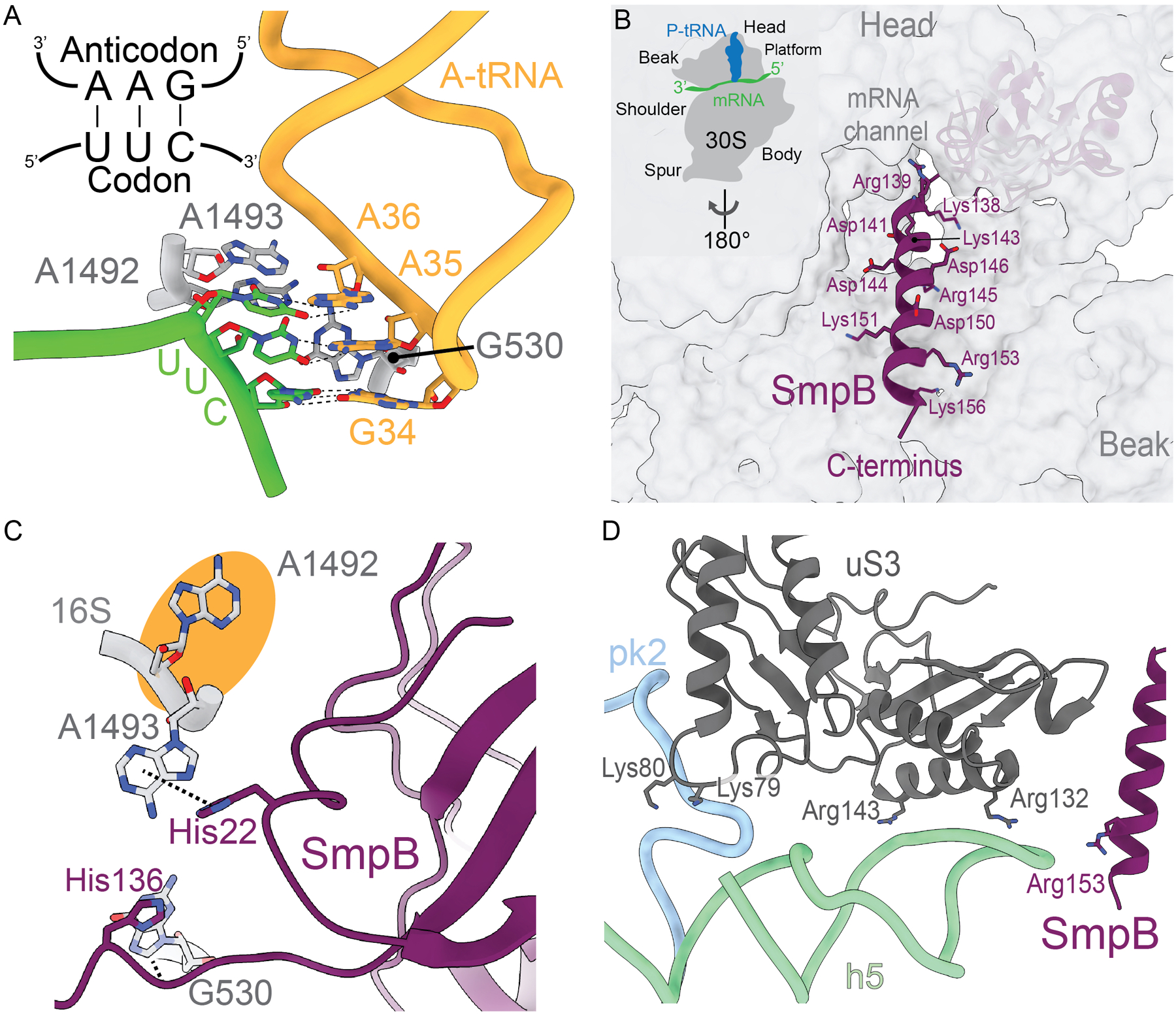
A. An A-site tRNA (gold) anticodon nucleotides (34, 35, 36) interact with the cognate mRNA codon (shown as UUC; lime green). When there are Watson-Crick base-pairs formed, signaling a correct tRNA, 16S rRNA nucleotides A1492, A1493, and G530 (grey) flip from their rRNA helices to monitor the pairing. PDB code 4V5D. B. The positively charged C-terminal tail of SmpB (purple) inserts into the mRNA channel on the 30S, forming electrostatic interactions with the phosphate backbone of 16S rRNA nucleotides (grey). PDB code 6Q97. C. SmpB residues His22 and His136 (purple) form π-stacking interactions with 16S rRNA nucleotides A1493 and G530 (grey), such that they flip out into a conformation similar to that observed during decoding of a mRNA-tRNA pair. 16S rRNA A1492 does not flip out (orange circle). PDB code 6Q97. D. The phosphate backbone of tmRNA pk2 (blue) and h5 (green) form electrostatic interactions with positively charged residues Lys79, Lys80, Arg132, and Arg143 of ribosomal protein uS3 (dark grey) to mediate tmRNA binding to the ribosome. SmpB residue Arg153 (purple) also forms electrostatic interactions with tmRNA h5. PDB code 6Q97.
Because tmRNA does not have an anticodon, SmpB-mediated interactions with the ribosome are largely responsible for driving EF-Tu dissociation and A-site accommodation during trans-translation. EF-Tu•GTP delivers an Ala-tmRNA-SmpB to the ribosome and SmpB forms interactions with the decoding center of the ribosome to facilitate tmRNA accommodation30. SmpB interacts with the tRNA-like domain (TLD) and helix 5 of tmRNA, resulting in placement of SmpB in the general position of the anticodon stem of a tRNA (Figure 2B)30. Upon binding the A site, the C-terminal tail of SmpB, which contains highly conserved positively charged residues required for trans-translation, makes electrostatic contacts with the 16S rRNA backbone (Figure 3B)20–22. tmRNA also makes more extensive contacts with the ribosome than tRNAs due to its large size and complex structure. tmRNA is ~360 nucleotides in length and is highly structured, composed of several helices and pseudoknots (Figure 2B)31. The TLD acceptor and T-loop interact with EF-Tu, while the D and variable loops interact with SmpB. The proteolysis tag reading frame, helix 5 (H5), and four pseudoknots form a ring around the 30S, engaging the head domain22. Pseudoknot 2 (PK2) and H5 bind the solvent side of non-stop ribosomes adjacent to the mRNA entrance channel21. Specifically, PK2 nucleotides C183-A184-A185 interact with ribosomal protein uS3 residues Arg72, Pro73, and Ile77 to anchor tmRNA to the 30S subunit (Figure 3D). Similar to how uS3 interacts with mRNA, uS3 residues Lys79 and Lys80 make electrostatic interactions with the PK2 backbone, and Arg132 and Arg143 with the H5 backbone21(Figure 3D).
tmRNA-SmpB allow for communication between the mRNA entrance tunnel, the 30S head domain and EF-Tu in a manner distinct from ternary complex binding. The SmpB C-terminal tail, in particular residues Gly132 to Arg139, is important for activation of GTP hydrolysis on EF-Tu32. SmpB residue His136 π-stacks with decoding center nucleotide G530 and is thought to play a role similar to tRNA in stimulating GTP hydrolysis on EF-Tu32 (Figure 3C). SmpB residue His22 also π-stacks with A1493, inducing G530 and A1493 into similar conformations that occur upon cognate tRNA binding to the A site (Figure 3C)21,22. One notable structural difference between tmRNA-SmpB-30S and tRNA-30S interactions is that tmRNA-SmpB binding does not cause A1492 to flip out of 16S rRNA helix 44 (h44), instead mimicking the decoding center orientation when the A site is empty (Figures 3A and 3C). This may be because there is no anticodon recognition requirement, or a difference in requirements of tmRNA-SmpB interaction on the ribosome.
Structural differences in the decoding center upon tmRNA-SmpB accommodation are supported by biochemical data. During normal translation, mutation of A1492, A1493 or G530 results in dominant lethality33, a 30- to 60-fold decrease in EF-Tu GTP hydrolysis rates and up to a 20-fold decrease in peptide bond formation rates, likely due to a reduction in A-site tRNA accomodation33. Mutational studies of A1492, A1493 or G530 demonstrate that these nucleotides are not essential for tmRNA-SmpB binding and EF-Tu GTP hydrolysis20. Ribosomes with a point mutation in nucleotides A1492, A1493, or G530 do not have a decrease in EF-Tu GTP hydrolysis rates when tmRNA-SmpB is delivered20. Ribosomes with mutations in decoding center nucleotides show only a 2-fold decrease in peptide bond formation rate between a P-site tRNA and the A-site tmRNA20, suggesting that there are less defects in tmRNA-SmpB binding and accommodation. So while A1492, A1493, and G530 play an essential role in canonical decoding, activation, and accommodation32, these nucleotides appear to play a limited role in accommodation of tmRNA-SmpB20,33.
Translocation differences
The movement of A-site, P-site and E-site tRNAs occurs independently on each ribosomal subunit34. Immediately after peptidyl transfer, the ribosome is in a ‘pretranslocational state’ with a deacylated tRNA in the P site and the peptidyl-tRNA in the A site. The chemical moieties located at the CCA ends of tRNAs have low affinity for these sites, causing the P- and A-tRNAs to spontaneously translocate to the E and P sites on the 50S, adopting A/P and P/E tRNA “hybrid states” (Figure 4A), resulting in intersubunit ratcheting35. In this hybrid position, the anticodon ends remain fixed on the 30S and requires EF-G to translocate to the next tRNA binding site and the ribosomal subunits rachet relative to each other. Translocation of both tRNAs to the E and P sites on the 30S is facilitated by binding and GTP hydrolysis by EF-G, resulting in a ‘posttranslocational state’. EF-G•GTP binding facilitates a ~20° rotation of the 30S and ~3° 30S head tilt, triggering GTP hydrolysis. Translocation is accompanied by the head domain of the 30S undergoing a ~7–10° counterclockwise rotation relative to the 50S subunit36 (Figure 4B)37. These collective changes leave the A site empty, peptidyl-tRNA in the P site, and a deacylated tRNA in the E site, coupled with the movement of mRNA by one codon38.
Figure 4. tmRNA does not induce a 30S rotational state upon formation of A/P and E/P hybrid states after peptidyl transfer.
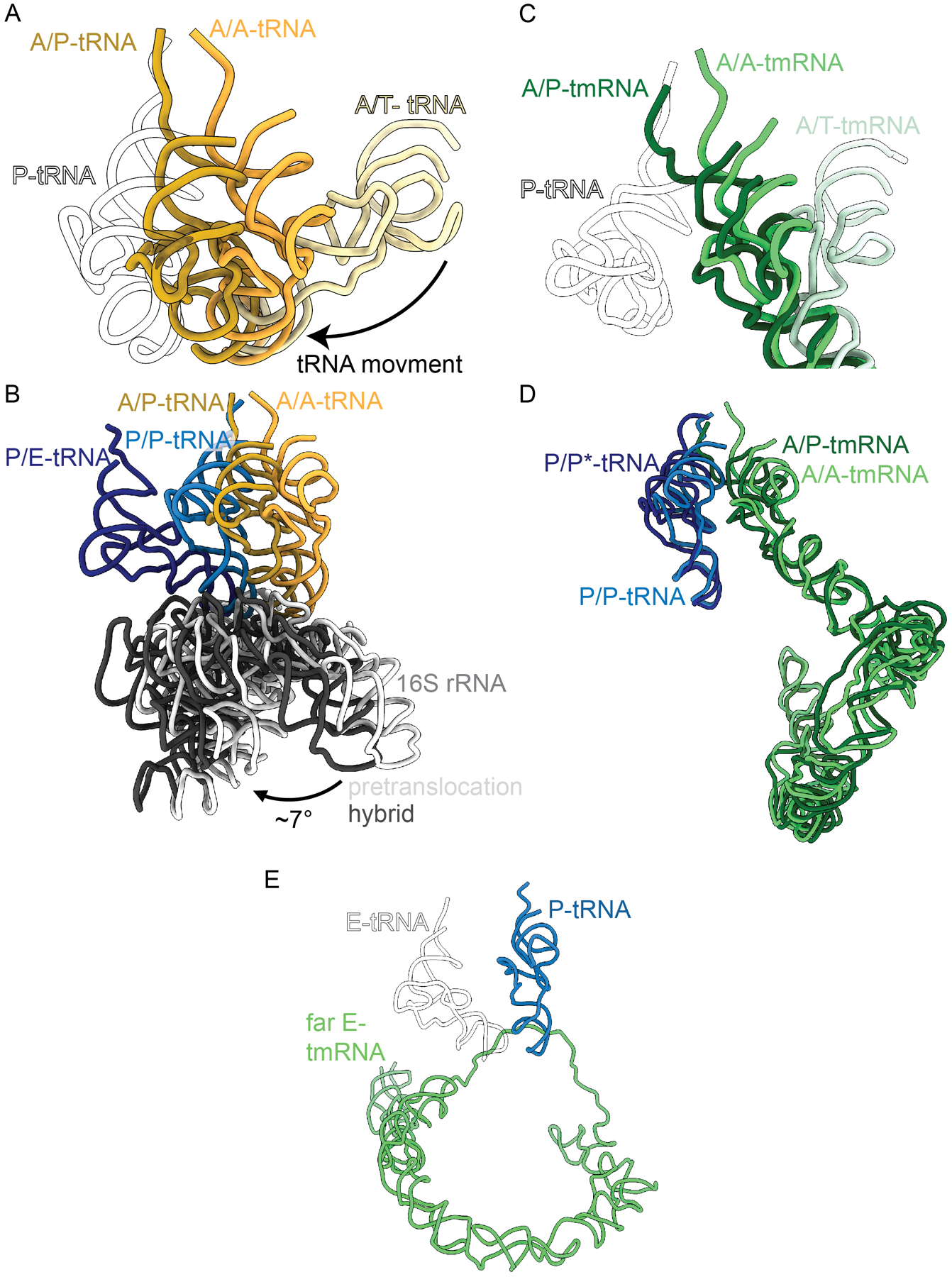
A. tRNA transitions from a pre-accommodated A/T tRNA state (pale yellow), when EF-Tu brings tRNAs to the ribosome, to an A/A tRNA state once a tRNA has been accommodated in the 50S A site (gold), and finally to an A/P tRNA state after peptidyl transfer and spontaneous movement to the P site but only on the 50S (dark yellow). P-site tRNA (white outline) is provided as reference. PDB codes 6WD2, 6WDE, 6WDG. B. While the tRNAs undergo translocation from A/A to A/P (yellow), and P/P to P/E (blue) the small ribosomal subunit head (grey) undergoes a rotation (light to dark grey) in relation to the 50S subunit. PDB codes 6WDE, 6WDG. C. Transition from an A/T state (mint) when the A-tmRNA is bound to EF-Tu, to an A/A state once a tmRNA has been accommodated in the A site (green), to an A/P (dark green). P-tRNA (white outline) provided as reference. PDB codes 7ABZ, 7AC7, 6Q97. D. The transition from A/A to A/P state of tmRNA (green) occurs without ratcheting. The P-tRNA (blue) goes from a P/P state to a P/P* transition state, where the P/P* acceptor stem (dark blue) is slightly offset from its original position. PDB codes 7AC7, 6Q97. E. Position of the acceptor stem of tmRNA (green) when it enters a ‘far E’ state, with a canonical E-tRNA (black outline of white tRNA) for reference. PDB codes 6Q9A, 4V5D.
During trans-translation, tmRNA-SmpB also needs to translocate through the ribosome to translate the tmRNA-encoded proteolysis tag. Recent ribosome structures have elucidated differences in translocation of tmRNA-SmpB from the A to the P site21,22. The tmRNA TLD acceptor stem forms an A/P hybrid state after peptidyl transfer without intersubunit ratcheting (Figure 4D)21. The A/P-tRNA acceptor end also moves beyond the P site, ~9.8 Å closer towards the E site (Figure 4B). This may be to accommodate the movement of the tmRNA, without fully adopting the P/E conformation. The movement of tmRNA into an A/P hybrid state without any ratcheting of the subunits reflects a major difference between the two processes and is likely a result of the size of tmRNA, and the additional interactions that tmRNA makes with the ribosome as compared to a tRNA. There are additional conformational differences that appear to be unique for tmRNA translocation. During tmRNA-mediated translocation from the A to the P site, the 30S head rotates ~14° with a 12° head tilt, angles greater than in translation (i.e., ~7–10° rotation and ~5–6° tilt)22,35,39,40. This increase may be explained by the additional RNA structural elements that tmRNA has including H2 and PK1. tmRNA H2 is positioned between the 30S and 50S during translocation and helps mediate tmRNA movement across the intersubunit bridges into the E site22. At the same time, the PK ring rotates along with the 30S head, possibly mediating the increase in tilt angle, allowing tmRNA to translocate effectively22.
After translocation of tmRNA to the P site, the first codon of the tmRNA proteolysis tag reading frame is in the A site and base-pairs with an incoming aminoacyl-tRNA. After peptidyl transfer, the P-site tmRNA and A-site tRNA need to be translocated to the E and P sites, respectively. This movement places the second codon of the proteolysis tag reading frame in the A site for decoding7. It was previously speculated that the TLD of P-site tmRNA translocates to the E site and the A-site peptidyl-tRNA to the P site, analogous to normal translation7. However, biochemical evidence or a structure of tmRNA in the E site have been elusive21,22. Instead, recent structures show that rather than deacylated tmRNA-SmpB occupying the E site in a similar manner as deacylated tRNA, deacylated tmRNA-SmpB transits past the E site, occupying a ‘far E-site state’ on the surface of the ribosome21,22 (Figure 4E). It is thought that tmRNA adopts this far E-site state because otherwise its large size and extended RNA helices would clash with regions of the ribosome21,22. Another reason tmRNA-SmpB may bind on the ribosomal surface adjacent to the E site instead of occupying the E site is because movement the tmRNA TLD is likely to be constrained during translation of the tag reading frame. While this hypothesis makes biological sense, there is no structure of tmRNA past the ‘far E site’, as it translates the remainder of the proteolysis tag21, so it remains unknown how tmRNA continues to associate with the ribosome. However, it is agreed that the large size of tmRNA likely imposes severe constraints on ribosome dynamics such as the 30S head rotation and tilt required for normal tRNA translocation21,22. Further investigation is needed to fully assess differences in the tmRNA translocation process.
Peptidyl transferase activity and a unique role for bL27 in trans-translation
After aminoacyl-tRNA or tmRNA is accommodated on the 50S with the acceptor stem located in the PTC, the α-amino group of the aminoacyl-tRNA spontaneously attacks the terminal carbonyl group of the peptidyl-tRNA to form a new peptide bond and transfer the nascent polypeptide to the A-site tRNA (Figure 1B), with several 23S rRNA nucleotides mediating this interaction (Figures 5A and 5E)41. Translation and trans-translocation appear to have the same rRNA nucleotide requirements for transpeptidation activity (Figures 5A–5F).
Figure 5. Trans-translation induces little to no changes in the peptidyl transferase center (PTC) except to bL27.
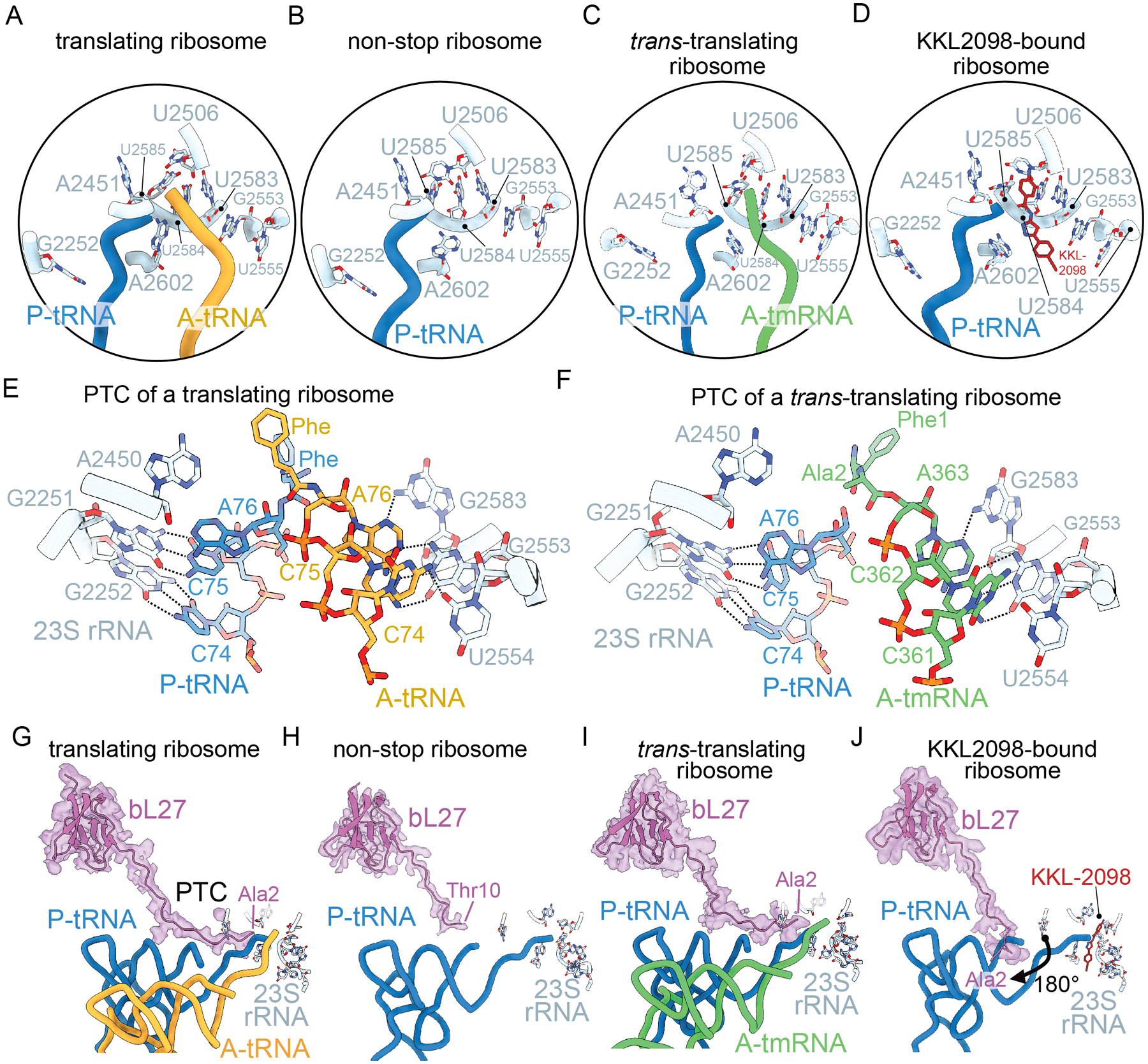
A-D PTC nucleotides (light blue) adopts similar conformations across all structures shown: A. 70S ribosome with nonhydrolyzable Phe-tRNAPhe A- (blue) and P-site tRNAs (gold) (PDB code 4V5D); B. non-stop 70S ribosome with a non-hydrolyzable fMet-tRNAfMet in the P site (blue) (PDB code 5MDZ); C. non-stop 70S ribosome with tRNAPhe in the P site (blue) and a peptidyl-tmRNA in the A site (green) (PDB code 7AC7); and D. non-stop 70S ribosome with KKL-2098 (red) bound (PDB code 6OM6). E. Base pairing and hydrogen bonding interactions between the 23S rRNA (light blue) and the P-site tRNA (blue) and A-site tRNA (gold) CCA ends (PDB code 4V5D). 23S rRNA nucleotides A2450 and G2583 interacts with the A-site tRNA A76 nucleotide, G2553 helps position the A-site tRNA, and U2554 interacts with the A-site tRNA C74 nucleotide. G2251 and G2252 base pair with C74 and C75 of the P-site tRNA, respectively. F. Base pairing and hydrogen bonding interactions between the 23S rRNA (light blue) and the P-site tRNA (blue) and tmRNA (green) CCA ends (PDB code 7AC7). The interactions between 23S rRNA and tRNA are conserved with tmRNA. G. bL27 extends into the PTC (23S rRNA nucleotides depicted as light blue) when an A-site tRNA containing a nonhydrolyzable aminoacyl group (gold) is bound (PDB code 4V5D). bL27 is fully resolved fully to the N-terminus residue Ala2 (map is shown in purple). H. In a non-stop ribosome, the N-terminus of bL27 is not resolved past Thr10 (PDB code 5MDZ). I. When tmRNA (green) is fully accommodated in the 50S A site, the N-terminus of bL27 is fully resolved to Ala2 (PDB code 7AC7). J. When trans-translation inhibitor KKL-2098 (red) is bound at the PTC, bL27 is fully resolved to Ala2, but rotates ~180° away from the PTC of the ribosome (PDB code 6OM6).
While the reaction is mediated by the 23S rRNA, ribosomal protein bL27 also plays a role in peptidyl-transferase activity18. As with most ribosomal proteins, bL27 has a globular structure which interacts with L33 and H80 and H81 of 23S rRNA but also has a long N-terminus that extends into the PTC (Figure 5G). In most ribosome structures using deacylated tRNAs, the first 20 residues of bL27 are unresolvable, and therefore considered to be flexible. Consistent with this assumption, when the 3’ ends of the A-site and P-site tRNAs are stabilized with proper chemical groups, most of the N-terminus of bL27 can be modeled and is within 4 Å of 23S rRNA nucleotide A2501 (Figures 5G and 5I). In a non-stop ribosome using a nonhydrolyzable fMet-tRNAfMet in the P site, the bL27 N-terminus cannot be resolved before residue Thr10 (Figure 5H). N-terminal mutational and deletion studies of bL27 reveal a slight growth defect but these bL27 variants are readily incorporated into ribosomes42,43. However, peptidyl transferase activity is reduced even when only the first three N-terminal amino acids are deleted42,43. These data support a model whereby the bL27 N-terminus may influence the orientation of 23S rRNA nucleotides 2506 and 2585 or the positioning tRNAs at the PTC42–44.
Additional differences between normal and trans-translation in transpeptidation are elucidated from experiments with small molecules that inhibit trans-translation but not normal translation. When trans-translation inhibitor KKL-2098 is bound to a non-stop ribosome, the N-terminus bL27 is resolved from Ala2, but in position never seen previously (Figure 5I). The first eight N-terminal residues rotate ~180° away from the PTC at Gly818. This alternative orientation of bL27 when trans-translation is inhibited but normal translation is not, suggests that bL27 plays a unique role in mediating the peptidyl transferase activity of tmRNA not required for a tRNA. This observation is interesting, because tRNA, tmRNA and 23S rRNA PTC nucleotides adopt similar positions in the A site (Figures 5E–5F). These results strongly suggest that there is an association between trans-translation and bL27 stabilization that is not needed for translation. One attractive model is that the acceptor arm of tmRNA is more flexible in the A site than the acceptor arm of tRNAs, and bL27 is required to properly align tmRNA for transpeptidation. If this model is correct, trans-translation should be dramatically slower on ribosomes lacking bL27. This model would also explain why no A/P hybrid state is formed during transpeptidation to tmRNA, if tmRNA is flexible enough to reach the PTC without subunit rotation.
Antibiotics and trans-translation
Targeting trans-translation is an emerging strategy that shows promise in combating the antibiotic resistance global health crisis18,45. There is no clinically approved antibiotic that targets trans-translation. Investigation of specific trans-translation inhibitors has yielded a family of acylaminooxadiazoles with potent and broad-spectrum antibiotic activity45. How these inhibitors confer specificity for trans-translation but not translation is not yet known, but there are several possibilities due to the innate differences in the two processes explored in this review. trans-translation relies on SmpB binding to tmRNA, which are two moieties completely unique to trans-translation and do not play a role in canonical translation. And finally, structural data suggests differences in how tmRNA translocates through the ribosome21,22. The previously unseen conformation of bL27 in addition to the increase in trans-translation inhibitor potency with bL27 truncations that is not seen with any other ribosome targeting antibiotic suggests a different mechanism between the two processes that make trans-translation a druggable target.
Conclusions
Despite the similarity in processes between translation and trans-translation, there are several key differences throughout the different steps of trans-translation that are important to decipher to fully understand trans-translation biology. The differences provide a foundation for why trans-translation can be such a specific and effective target for antibiotic development.
Abbreviations:
- PTC
peptidyl transferase center
- TLD
tRNA-like domain
Footnotes
Conflict of Interest. KCK is an inventor on a patent covering the acylaminooxadiazole molecules described above.
Data Sharing Statement.
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
References
- 1.Keiler KC Mechanisms of ribosome rescue in bacteria. Nature Reviews Microbiology 13, 285–297, doi: 10.1038/nrmicro3438 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Keiler KC & Feaga HA Resolving nonstop translation complexes is a matter of life or death. Journal of Bacteriology 196, 2123–2130, doi: 10.1128/JB.01490-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hui MP, Foley PL & Belasco JG Messenger RNA degradation in bacterial cells. Annual Review of Genetics 48, 537–559, doi: 10.1146/annurev-genet-120213-092340 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pedersen K, Zavialov AV, Pavlov MY, Elf J, Gerdes K & Ehrenberg M The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell 112, 131–140, doi: 10.1016/s0092-8674(02)01248-5] (2003). [DOI] [PubMed] [Google Scholar]
- 5.Saito K, Kratzat H, Campbell A, Buschauer R, Burroughs AM, Berninghausen O, Aravind L, Green R, Beckmann R & Buskirk AR Ribosome collisions induce mRNA cleavage and ribosome rescue in bacteria. Nature 603, 503–508, doi: 10.1038/s41586-022-04416-7 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keiler KC Biology of trans-translation. Annual Review of Microbiology 62, 133–151, doi: 10.1146/annurev.micro.62.081307.162948 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Keiler KC, Waller PR & Sauer RT Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271, 990–993, doi: 10.1126/science.271.5251.990 (1996). [DOI] [PubMed] [Google Scholar]
- 8.Richards J, Mehta P & Karzai AW RNase R degrades non-stop mRNAs selectively in an SmpB-tmRNA-dependent manner. Molecular Microbiology 62, 1700–1712, doi: 10.1111/j.1365-2958.2006.05472.x (2006). [DOI] [PubMed] [Google Scholar]
- 9.Yamamoto Y, Sunohara T, Jojima K, Inada T & Aiba H SsrA-mediated trans-translation plays a role in mRNA quality control by facilitating degradation of truncated mRNAs. RNA 9, 408–418, doi: 10.1261/rna.2174803 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hudson CM, Lau BY & Williams KP Ends of the line for tmRNA-SmpB. Frontiers in Microbiology 5, 421, doi: 10.3389/fmicb.2014.00421 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang C, Wolfgang MC, Withey J, Koomey M & Friedman DI Charged tmRNA but not tmRNA-mediated proteolysis is essential for Neisseria gonorrhoeae viability. The EMBO Journal 19, 1098–1107, doi: 10.1093/emboj/19.5.1098 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alumasa JN, Manzanillo PS, Peterson ND, Lundrigan T, Baughn AD, Cox JS & Keiler KC Ribosome Rescue Inhibitors Kill Actively Growing and Nonreplicating Persister Mycobacterium tuberculosis Cells. ACS Infectious Diseases 3, 634–644, doi: 10.1021/acsinfecdis.7b00028 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chadani Y et al. Ribosome rescue by Escherichia coli ArfA (YhdL) in the absence of trans-translation system. Mol Microbiol 78, 796–808, doi: 10.1111/j.1365-2958.2010.07375.x (2010). [DOI] [PubMed] [Google Scholar]
- 14.Chadani Y, Ono K, Kutsukake K & Abo T Escherichia coli YaeJ protein mediates a novel ribosome-rescue pathway distinct from SsrA- and ArfA-mediated pathways. Molecular Microbiology 80, 772–785, doi: 10.1111/j.1365-2958.2011.07607.x (2011). [DOI] [PubMed] [Google Scholar]
- 15.Feaga HA, Viollier PH & Keiler KC Release of nonstop ribosomes is essential. mBio 5, e01916, doi: 10.1128/mBio.01916-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goralski TDP, Kirimanjeswara GS & Keiler KC A New Mechanism for Ribosome Rescue Can Recruit RF1 or RF2 to Nonstop Ribosomes. mBio 9, doi: 10.1128/mBio.02436-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shimokawa-Chiba N, Muller C, Fujiwara K, Beckert B, Ito K, Wilson DN & Chiba S Release factor-dependent ribosome rescue by BrfA in the Gram-positive bacterium Bacillus subtilis. Nature Communications 10, 5397, doi: 10.1038/s41467-019-13408-7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aron ZD, Mehrani A, Hoffer ED, Connolly KL, Srinivas P, Torhan MC, Alumasa JN, Cabrera M, Hosangadi D, Barbor JS, Cardinale SC, Kwasny SM, Morin LR, Butler MM, Opperman TJ, Bowlin TL, Jerse A, Stagg SM, Dunham CM & Keiler KC trans-Translation inhibitors bind to a novel site on the ribosome and clear Neisseria gonorrhoeae in vivo. Nature Communications 12, 1799, doi: 10.1038/s41467-021-22012-7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karzai AW, Susskind MM & Sauer RT SmpB, a unique RNA-binding protein essential for the peptide-tagging activity of SsrA (tmRNA). The EMBO Journal 18, 3793–3799, doi: 10.1093/emboj/18.13.3793 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller MR, Liu Z, Cazier DJ, Gebhard GM, Herron SR, Zaher HS, Green R & Buskirk AR The role of SmpB and the ribosomal decoding center in licensing tmRNA entry into stalled ribosomes. RNA 17, 1727–1736, doi: 10.1261/rna.2821711 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rae CD, Gordiyenko Y & Ramakrishnan V How a circularized tmRNA moves through the ribosome. Science 363, 740–744, doi: 10.1126/science.aav9370 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guyomar C, D’Urso G, Chat S, Giudice E & Gillet R Structures of tmRNA and SmpB as they transit through the ribosome. Nature Communications 12, 4909, doi: 10.1038/s41467-021-24881-4 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ban N, Beckmann R, Cate JH, Dinman JD, Dragon F, Ellis SR, Lafontaine DL, Lindahl L, Liljas A, Lipton JM, McAlear MA, Moore PB, Noller HF, Ortega J, Panse VG, Ramakrishnan V, Spahn CM, Steitz TA, Tchorzewski M, Tollervey D, Warren AJ, Williamson JR, Wilson D, Yonath A & Yusupov M A new system for naming ribosomal proteins. Current Opinion in Structural Biology 24, 165–169, doi: 10.1016/j.sbi.2014.01.002 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogle JM, Murphy FV, Tarry MJ & Ramakrishnan V Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell 111, 721–732, doi: 10.1016/s0092-8674(02)01086-3 (2002). [DOI] [PubMed] [Google Scholar]
- 25.Demeshkina N, Jenner L, Westhof E, Yusupov M & Yusupova G A new understanding of the decoding principle on the ribosome. Nature 484, 256–259, doi: 10.1038/nature10913 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Loveland AB, Demo G, Grigorieff N & Korostelev AA Ensemble cryo-EM elucidates the mechanism of translation fidelity. Nature 546, 113–117, doi: 10.1038/nature22397 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pape T, Wintermeyer W & Rodnina M Induced fit in initial selection and proofreading of aminoacyl-tRNA on the ribosome. The EMBO Journal 18, 3800–3807, doi: 10.1093/emboj/18.13.3800 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gromadski KB & Rodnina MV Kinetic determinants of high-fidelity tRNA discrimination on the ribosome. Molecular Cell 13, 191–200, doi: 10.1016/s1097-2765(04)00005-x (2004). [DOI] [PubMed] [Google Scholar]
- 29.Daviter T, Gromadski KB & Rodnina MV The ribosome’s response to codon-anticodon mismatches. Biochimie 88, 1001–1011, doi: 10.1016/j.biochi.2006.04.013 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Neubauer C, Gillet R, Kelley AC & Ramakrishnan V Decoding in the absence of a codon by tmRNA and SmpB in the ribosome. Science 335, 1366–1369, doi: 10.1126/science.1217039 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams KP & Bartel DP Phylogenetic analysis of tmRNA secondary structure. RNA 2, 1306–1310 (1996). [PMC free article] [PubMed] [Google Scholar]
- 32.Miller MR & Buskirk AR An unusual mechanism for EF-Tu activation during tmRNA-mediated ribosome rescue. RNA 20, 228–235, doi: 10.1261/rna.042226.113 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cochella L, Brunelle JL & Green R Mutational analysis reveals two independent molecular requirements during transfer RNA selection on the ribosome. Nature Structural & Molecular Biology 14, 30–36, doi: 10.1038/nsmb1183 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Moazed D & Noller HF Intermediate states in the movement of transfer RNA in the ribosome. Nature 342, 142–148, doi: 10.1038/342142a0 (1989). [DOI] [PubMed] [Google Scholar]
- 35.Frank J & Agrawal RK A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature 406, 318–322, doi: 10.1038/35018597 (2000). [DOI] [PubMed] [Google Scholar]
- 36.Ratje AH, Loerke J, Mikolajka A, Brunner M, Hildebrand PW, Starosta AL, Donhofer A, Connell SR, Fucini P, Mielke T, Whitford PC, Onuchic JN, Yu Y, Sanbonmatsu KY, Hartmann RK, Penczek PA, Wilson DN & Spahn CM Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature 468, 713–716, doi: 10.1038/nature09547 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo Z & Noller HF Rotation of the head of the 30S ribosomal subunit during mRNA translocation. Proceedings of the National Academy of Sciences 109, 20391–20394, doi: 10.1073/pnas.1218999109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodnina MV & Wintermeyer W The ribosome as a molecular machine: the mechanism of tRNA-mRNA movement in translocation. Biochemical Society Transactions 39, 658–662, doi: 10.1042/BST0390658 (2011). [DOI] [PubMed] [Google Scholar]
- 39.Valle M, Zavialov A, Sengupta J, Rawat U, Ehrenberg M & Frank J Locking and unlocking of ribosomal motions. Cell 114, 123–134, doi: 10.1016/s0092-8674(03)00476-8 (2003). [DOI] [PubMed] [Google Scholar]
- 40.Nguyen K & Whitford PC Steric interactions lead to collective tilting motion in the ribosome during mRNA-tRNA translocation. Nature Communications 7, 10586, doi: 10.1038/ncomms10586 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rodnina MV The ribosome as a versatile catalyst: reactions at the peptidyl transferase center. Current Opinion in Structural Biology 23, 595–602, doi: 10.1016/j.sbi.2013.04.012 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Wower IK, Wower J & Zimmermann RA Ribosomal protein L27 participates in both 50 S subunit assembly and the peptidyl transferase reaction. Journal of Biological Chemistry 273, 19847–19852, doi: 10.1074/jbc.273.31.19847 (1998). [DOI] [PubMed] [Google Scholar]
- 43.Maguire BA, Beniaminov AD, Ramu H, Mankin AS & Zimmermann RA A protein component at the heart of an RNA machine: the importance of protein l27 for the function of the bacterial ribosome. Molecular Cell 20, 427–435, doi: 10.1016/j.molcel.2005.09.009 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Trobro S & Aqvist J Role of ribosomal protein L27 in peptidyl transfer. Biochemistry 47, 4898–4906, doi: 10.1021/bi8001874 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Ramadoss NS, Alumasa JN, Cheng L, Wang Y, Li S, Chambers BS, Chang H, Chatterjee AK, Brinker A, Engels IH & Keiler KC Small molecule inhibitors of trans-translation have broad-spectrum antibiotic activity. Proceedings of the National Academy of Sciences 110, 10282–10287, doi: 10.1073/pnas.1302816110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.


