SUMMARY
Sex differences are pervasive in human health and disease. One major key to sex-biased differences lies in the sex chromosomes. Although the functions of the X-chromosome proteins are well appreciated, how they compare with their Y-chromosome homologs remains elusive. Herein, using ensemble and single-molecule techniques, we report that the sex chromosome-encoded RNA helicases DDX3X and DDX3Y are distinct in their propensities for liquid-liquid phase separation (LLPS), dissolution, and translation repression. We demonstrate that the N-terminal intrinsically disordered region of DDX3Y more strongly promotes LLPS than the corresponding region of DDX3X and that the weaker ATPase activity of DDX3Y compared to DDX3X contributes to the slower disassembly dynamics of DDX3Y-positive condensates. Interestingly, DDX3Y-dependent LLPS represses mRNA translation and enhances aggregation of FUS more strongly than DDX3X-dependent LLPS. Our study provides a platform for future comparisons of sex chromosome-encoded protein homologs, providing insights into sex differences in RNA metabolism and human disease.
Keywords: Sex chromosome homolog proteins, DDX3X, DDX3Y, RNA helicase, liquid-liquid phase separation, condensates, ATPase activity, translation repression
eTOC Blurb:
Shen et al. report that the Y chromosome-encoded RNA helicase DDX3Y has stronger propensity for liquid-liquid phase separation compared to its X chromosome-encoded homolog DDX3X, which results in the stronger repression of mRNA translation and promotion of FUS aggregation under stress conditions.
Graphical Abstract
INTRODUCTION
Many human disorders manifest in a sex-biased manner, yet the molecular mechanisms responsible for these biases are not fully understood (Mauvais-Jarvis et al., 2020). On a genetic level, the most significant differences between males and females lie in the sex chromosomes. Although the X and Y chromosomes are, by and large, not homologous, a handful of X chromosome-encoded proteins have Y chromosome-encoded homologs (Bellott et al., 2014). These Y chromosome-encoded homologs were historically thought to only be expressed and to function in the reproductive system. However, a growing body of evidence suggests that Y chromosome-encoded homologs are not only expressed throughout the body at the transcript and protein levels but are also evolutionarily conserved (Godfrey et al., 2020). However, the functional differences between the protein homologs encoded on the X and Y chromosomes have not been thoroughly investigated. Emerging evidence is starting to reveal that Y chromosome-encoded proteins may function differently from their X chromosome-encoded homologs (Gozdecka et al., 2018; Nguyen et al., 2020; Shi et al., 2021).
One pair of sex chromosome-encoded proteins are the RNA helicases DDX3X and DDX3Y. In females (specified as XX individuals), DDX3X escapes X-inactivation, and two copies of the protein are expressed, whereas in males (specified as XY individuals), one copy of each of the X- and Y-linked proteins is expressed (Cotton et al., 2015; Ditton et al., 2004). The DDX3X gene (Xp11.4) encodes a DEAD-box RNA helicase (Sharma and Jankowsky, 2014), which is evolutionarily conserved in C. elegans (LAF-1), yeast (Ded1p), Drosophila (Belle), and humans (Sharma and Jankowsky, 2014). The functions of DDX3X are more well studied than those of DDX3Y, and include its activity as a translation-initiation factor (Lai et al., 2008; Lee et al., 2008) for a set of mRNAs with highly structured 5’-UTRs (Ku et al., 2019; Lai et al., 2008; Lee et al., 2008; Phung et al., 2019; Soto-Rifo et al., 2012), and at repeat-associated non-AUG (RAN) start sites (Cheng et al., 2019; Linsalata et al., 2019). In addition, DDX3X (and its homologs) undergoes liquid-liquid phase separation (LLPS) and is a conserved component of stress granules (SGs) (Beckham et al., 2008; Elbaum-Garfinkle et al., 2015; Iserman et al., 2020; Shih et al., 2012; Valentin-Vega et al., 2016). SGs, which are composed of RNAs and proteins and form as part of the stress response, are correlated with changes to mRNA metabolism, including translational repression (Kimball et al., 2003; Moon et al., 2019). Dysregulation of SGs has been implicated in a wide range of human disorders, including cardiomyopathy, Alzheimer’s disease, and amyotrophic lateral sclerosis (ALS), many of which present with a varying degree of sex-biased incidences, progressions, and outcomes (Ash et al., 2014; Baradaran-Heravi et al., 2020; Ferretti et al., 2018; Manjaly et al., 2010; McCombe and Henderson, 2010; Meyer et al., 2014; Schneider et al., 2020; Watkins et al., 2020). Additionally, DDX3X mutations can lead to DDX3X syndrome, a disorder which accounts for 1–3% of intellectual disability cases and is more prevalent in females than in males (Iossifov et al., 2014; Lennox et al., 2020; Ruzzo et al., 2019; Scala et al., 2019; Snijders Blok et al., 2015; Wang et al., 2018b).
Notably, the presence of the DDX3Y gene can only sometimes compensate for the loss of the DDX3X gene (Chen et al., 2016). For example, Ddx3y cannot fully compensate for the loss of Ddx3x during embryonic and neuronal development in mice (Chen et al., 2016; Patmore et al., 2020). Furthermore, male mice, but not female mice, survive conditional knockout of Ddx3x in the bone marrow, but these male mice present with distinct deficiencies in innate antimicrobial immunity compared to Ddx3x conditional knockout female mice (Szappanos et al., 2018). These findings highlight the fact that DDX3X and DDX3Y have roles beyond the reproductive system and suggest that Ddx3x and Ddx3y possibly have distinct functions in immune cells (Szappanos et al., 2018). Indeed, recently compiled proteomic databases show that both DDX3X and DDX3Y proteins are expressed in the human immune system, including in T-cells, B-cells, and NK-cells (Bryk and Wisniewski, 2017; Joshi et al., 2019). Moreover, previous studies revealed that the DDX3Y protein is present in the enteric nervous system and the human heart (Cardinal et al., 2020; Godfrey et al., 2020; Vakilian et al., 2015). Still, potential functional differences between DDX3X and DDX3Y and their contributions to sex-biased human diseases are largely unknown.
Like other DEAD-box helicases, DDX3X and DDX3Y contain a helicase core composed of two RecA-like domains and one intrinsically disordered region (IDR) on each of the N- and C-termini. IDRs are frequently involved in the process of LLPS, driven by weak multivalent interactions (Figure 1A). Although DDX3X and DDX3Y share 92% amino acid sequence identity overall, the N-terminal IDRs (IDR1) are more divergent, accounting for 60% difference between DDX3X and DDX3Y (Figure S1A and S1B). IDR1 of DDX3X specifically is known to be essential for its LLPS in vitro and inside cells (Saito et al., 2019; Shih et al., 2012). Since many of the differences between the sequences of DDX3X and DDX3Y are concentrated in IDR1, we wondered whether DDX3X and DDX3Y differ in their propensity to LLPS and consequently differ in responding to cellular stress.
Figure 1. DDX3Y has a stronger LLPS propensity compared to DDX3X in vitro.
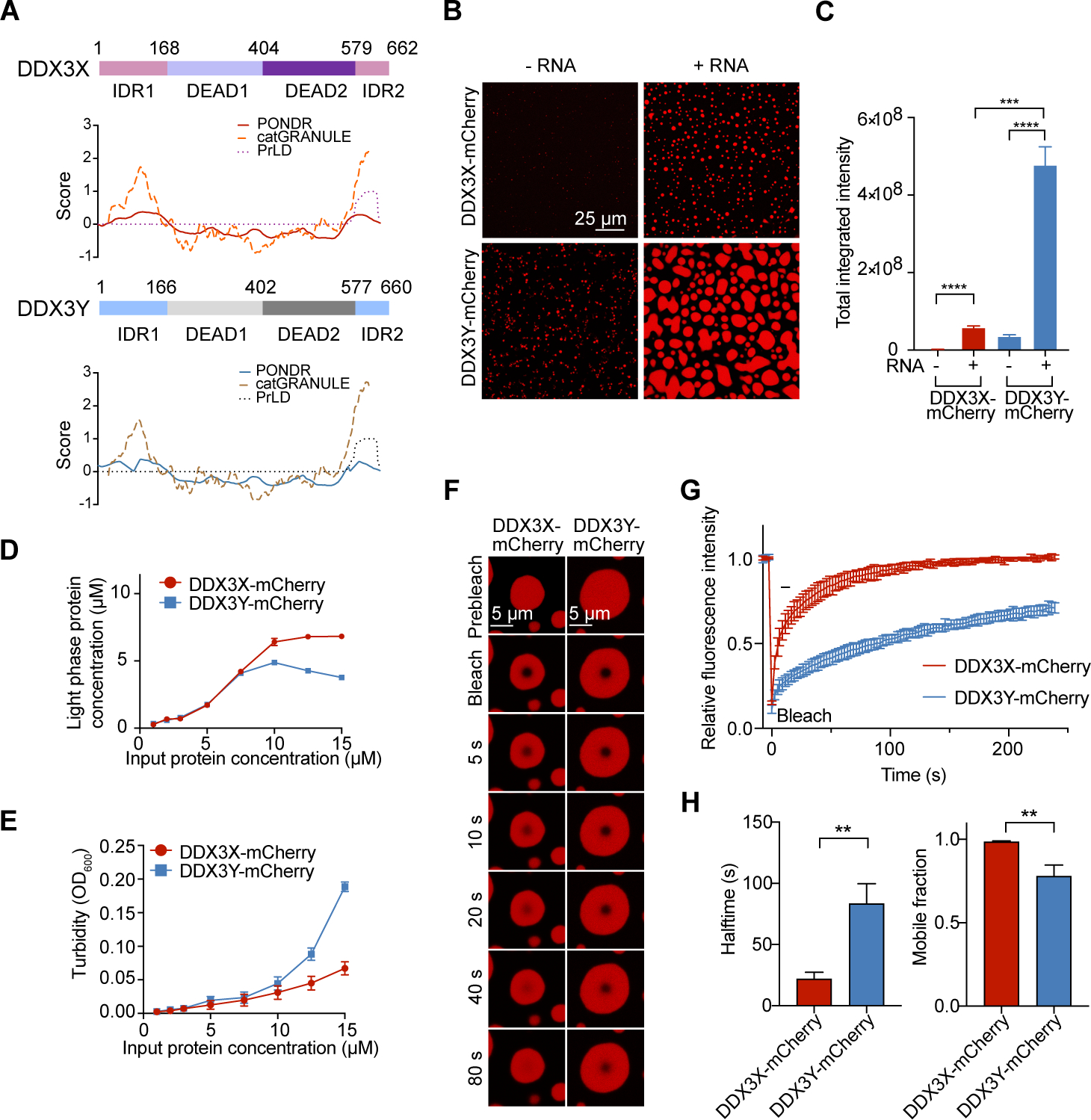
(A) Structural prediction of DDX3X and DDX3Y using PONDR (natural disordered regions), PLAAC (prion-like amino acid regions), and catGRANULE (LLPS propensity).
(B) In vitro droplet formation of 10 μM recombinant DDX3X-mCherry or DDX3Y-mCherry in the absence or presence of 200 ng/μL poly(U)-RNA. Scale bar, 25 μm.
(C) Quantification of the total integrated intensity of DDX3X condensation and DDX3Y condensation in Figure 1B. A two-tailed t-test was used to calculate the p-value. ***p < 0.001, **** p < 0.0001.
(D) Concentrations of DDX3 proteins in the light phase (supernatant after centrifugation) vs. input protein concentrations.
(E) Turbidity (absorbance at 600 nm) of DDX3X-mCherry and DDX3Y-mCherry LLPS. The mean value of turbidity and protein concentration for each condition from three separate protein purifications and three technical repeats were plotted in Figure 1D and 1E.
(F) Time-lapse images of in vitro FRAP experiments. The FRAP experiments were performed identically for DDX3X-mCherry and DDX3Y-mCherry droplets.
(G) FRAP curves for in vitro droplets of DDX3X-mCherry (red) and DDX3Y-mCherry (blue). The traces of the FRAP data represent mean ± s.e.m (n = 3, from three independent experiments).
(H) Halftime and mobile fractions from Figure 1G. A two-tailed t-test was used to calculate the p-value. **p < 0.01.
See also Figure S1.
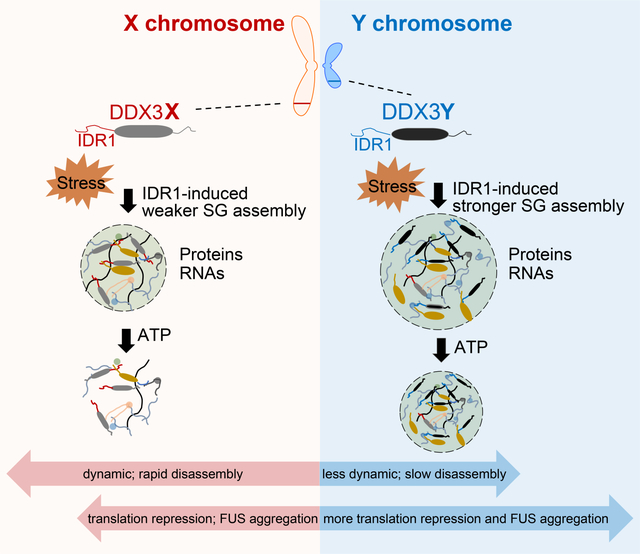
Here, we show that DDX3Y has a greater LLPS propensity than DDX3X. DDX3Y-positive SGs are less dynamic (less able to exchange particles with the light phase) than DDX3X-positive SGs. Furthermore, we demonstrate that while DDX3X-positive and DDX3Y-positive SGs share a large overlap of RNA constituents, there are also RNA components that are unique to either SG. We also show that the condensation of either DDX3X or DDX3Y represses the translation of RNAs, with DDX3Y condensation showing a significantly stronger inhibitory effect. Additionally, we show that both helicases specifically augment the aggregation of FUS in vitro and in cells, with DDX3Y having a more profound effect. Collectively, our results suggest that these sexually dimorphic RNA helicases differentially regulate RNA metabolism through their distinct biochemical and biophysical properties, which might contribute to sex bias in human diseases.
RESULTS
DDX3Y has a stronger propensity than DDX3X for in vitro and cellular phase separation
Given that DDX3X is known to undergo LLPS in vitro, we wanted to establish whether DDX3Y could also phase separate, and how it is compared to DDX3X. Thus, we purified mCherry-tagged full-length DDX3X and DDX3Y to near homogeneity (Figure S1C and S1D). Full-length mCherry-tagged DDX3X and DDX3Y formed noticeable droplets in vitro, and the addition of poly(U)-RNA greatly stimulated this process (Figure 1B and 1C). Strikingly, DDX3Y phase separation was more strongly enhanced by the addition of poly(U)-RNA than DDX3X in vitro; the average integrated fluorescence intensity of DDX3Y droplets was ~10-fold higher than an equal amount of DDX3X (Figure 1B and 1C). As shown in Figure S1E, RNase treatment had no influence on the in vitro droplet formation, indicating negligible RNA carry-over during protein purification. Furthermore, we determined the saturation concentration (Csat) of DDX3X and DDX3Y using sedimentation analysis. The Csat of DDX3X was ~5 μM whereas DDX3Y was ~3 μM, indicating that DDX3Y undergoes LLPS at lower concentrations than DDX3X (Figure 1D). In parallel, we performed a turbidity assay; the results consistently showed that DDX3Y gave substantially higher turbidity at the same total protein concentrations compared to DDX3X (Figure 1E), suggesting a greater degree of phase separation.
We next studied the dynamics of DDX3X and DDX3Y droplets using fluorescent recovery after photobleaching (FRAP). The recovery halftime for DDX3X droplets was ~21.9 seconds, which was significantly faster than the ~83.4 seconds for DDX3Y droplets (Figures 1F, 1G, S1F, and Movies S1 – S4). Also, we observed a larger mobile fraction for DDX3X droplets (~99%) than for DDX3Y droplets (~78%) (Figure 1H). These results demonstrate that DDX3X is less prone to phase separation and that DDX3X droplets are more dynamic than DDX3Y droplets in vitro.
To assess if DDX3Y enters cellular SGs, we expressed DDX3X or DDX3Y in several mammalian cell types, each of which lacks endogenous DDX3Y (HeLa, N2a, and HEK 293T cells). Before expressing either protein, we performed a transient knockdown of endogenous DDX3X with >80% knockdown efficiency (Figure S2A). Exogenous expressions of DDX3X and DDX3Y were at a similar level (Figure S2B and S2C). DDX3X and DDX3Y were diffuse throughout the cytoplasm in unstressed HeLa cells (Figure S2D). Upon arsenite treatment (a commonly used oxidative-stress inducer (Markmiller et al., 2018; Protter and Parker, 2016)), both DDX3X and DDX3Y colocalized with the stress granule marker, G3BP1 (Markmiller et al., 2018) (Figure 2A). Strikingly, the total area of DDX3Y-positive SGs was larger than DDX3X-positive SGs in HeLa cells (1.6-fold), N2a cells (1.3-fold), and HEK293T cells (1.7-fold) (Figure 2B). To control for possible differences in protein concentrations, we expressed DDX3X and DDX3Y across a range of concentrations in HeLa cells with endogenous DDX3X depleted. The results consistently showed that DDX3Y-positive granules were significantly larger than DDX3X-positive granules at similar expressed concentrations (Figure S2E – S2G). Additionally, we quantified the protein half-life of DDX3X and DDX3Y in HeLa cells using cycloheximide (CHX) chase experiments. As shown in Figure 2C and 2D, the cellular half-life of DDX3X is indistinguishable from DDX3Y. FRAP experiments performed on live HeLa cells expressing DDX3X-EGFP or DDX3Y-EGFP (Figure S2H) reveal that the average recovery halftime of DDX3X-positive granules was ~7.6 s, which was significantly faster than the ~10.2 s measured for DDX3Y-positive SGs (Figure 2E – 2G, Movies S5 and S6). Additionally, a larger mobile fraction was observed in DDX3X-positive SGs (86%) than in DDX3Y-positive SGs (77%) (Figure 2G), consistent with our in vitro FRAP experiments. These results together suggest that DDX3X-positive SGs are more dynamic than DDX3Y-positive SGs.
Figure 2. DDX3Y has a stronger LLPS propensity compared to DDX3X in cells.
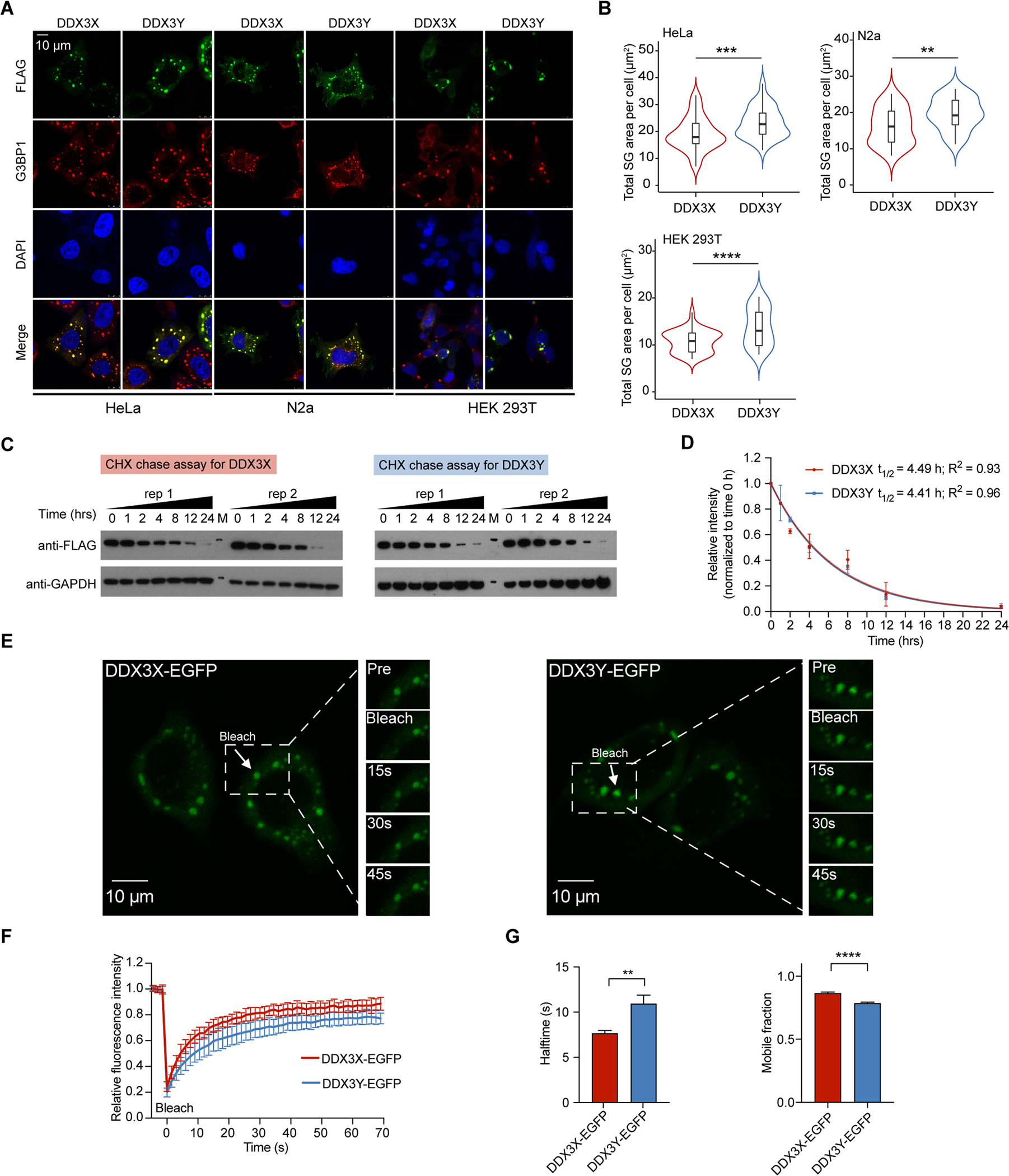
(A) Representative images of co-localization of DDX3X and DDX3Y with G3BP1 in HeLa, N2a, and HEK 293T cells with the endogenous DDX3X knocked down upon arsenite treatment (500 μM, 1 hr). Scale bar, 10 μm.
(B) Violin plots of the total SG area of DDX3X- or DDX3Y-positive SGs per cell across 50 cells upon arsenite treatment (500 μM, 1 hr) in endogenous DDX3X-depleted HeLa, N2a, or HEK 293T cells (n = 50 cells in total, from 3 biologically independent experiments). p values were determined by a two-tailed t-test; **p < 0.01, ***p < 0.001, **** p < 0.0001.
(C) Cycloheximide (CHX) chase assay to determine the cellular half-life of DDX3X and DDX3Y in HeLa cells. Two biological replicates for DDX3X and for DDX3Y were performed. “M” represents the protein ladder on the Western blot membranes (markers: upper 75 kDa, lower 37 kDa).
(D) Quantification of the protein levels in Figure 2C. The intensity of each DDX3X and DDX3Y band was normalized to the corresponding GAPDH intensity before being normalized to the intensity at the corresponding 0 hr time point. The half-lives of DDX3X and DDX3Y were 4.49 hrs and 4.41 hrs, respectively.
(E) Time-lapse images of photobleached SGs in HeLa cells expressing DDX3X-EGFP (left) or DDX3Y-EGFP (right) from in-cell FRAP experiments. The photobleaching events and fluorescence recovery by DDX3X-EGFP- and DDX3Y-EGFP-positive SGs are highlighted by the arrow in each outlined box.
(F) FRAP curves for DDX3X-EGFP (red) and DDX3Y-EGFP (blue) in HeLa cells. The trace of the FRAP data represents mean ± s.e.m. (n = 20 independent measurements, from 3 biologically independent experiments).
(G) The halftime and mobile fractions in Figure 2E & 2F. A two-tailed t-test was used to calculate the p-value. **p < 0.01, **** p < 0.0001.
See also Figure S2.
IDR1 is a major contributor to the higher phase separation propensity of DDX3Y
We constructed several truncated variants of DDX3X and DDX3Y to study the effect of each domain on SG partitioning (Figure S3A and S3B). Deletion of IDR1 from DDX3X (DDX3XΔIDR1) and DDX3Y (DDX3YΔIDR1) led to the sequestration of the variants into cell nuclei and thus completely prevented either protein from entering cytoplasmic SGs, which is consistent with previous work showing that IDR1 of DDX3X contains a nuclear export sequence (Yedavalli et al., 2004) (Figures S1A and S3C). Both DDX3XΔIDR1 and DDX3YΔIDR1 were diffuse in the nucleus, even with arsenite treatment (Figure S3C), so we hypothesized that the high nuclear RNA concentrations prevented these truncations from forming condensates, since this phenomenon has been seen with other RNA-binding proteins with IDRs (Maharana et al., 2018). To test this, we treated cells expressing DDX3XΔIDR1 or DDX3YΔIDR1 with actinomycin D (ActD), which inhibits transcription and thus decreases RNA levels. ActD treatment led to the formation of DDX3X and DDX3Y puncta inside the nucleus, consistent with an RNA-dependent LLPS buffering mechanism (Figure S3D).
Deleting any individual domain in DDX3X and DDX3Y dramatically decreased SG area compared to the full-length (Figures 3A, 3B, S3A, and S3B). Consistent with previous results, IDR1 of either DDX3X or DDX3Y as a standalone protein entered SGs and colocalized with G3BP1. Furthermore, a noticeable fraction of IDR1 from either protein remained diffuse throughout the cytoplasm upon arsenite treatment, indicating that IDR1 alone is less prone to enter SGs. For DDX3X, the deletion of IDR2 (DDX3XΔIDR2) significantly decreased the total SG area of DDX3XΔIDR2-positive SGs per cell (by ~1.4 fold) compared to the wild-type-DDX3X SGs. Deletion of the helicase domains (both DEAD1 and DEAD2) from DDX3X also decreased the total area of DDX3XΔhelicase-SGs (~1.2 fold) per cell, but to a lesser extent than the deletion of IDR1 or IDR2. Similarly, for DDX3Y, deletion of IDR2 or the helicase domains both lead to a ~1.2-fold decrease in the size of DDX3YΔIDR2-positive or DDX3YΔhelicase-positive SGs compared to the full-length DDX3Y SGs. Notably, the total SG area for each DDX3Y truncation was still larger than their DDX3X truncation counterparts (~1.3 to 1.85-fold), suggesting that subtle sequence variation throughout the protein contributes to the distinct behaviors of DDX3X and DDX3Y (Figure 3A and 3B).
Figure 3. IDR1 of DDX3Y more strongly promotes phase separation than IDR1 of DDX3X.
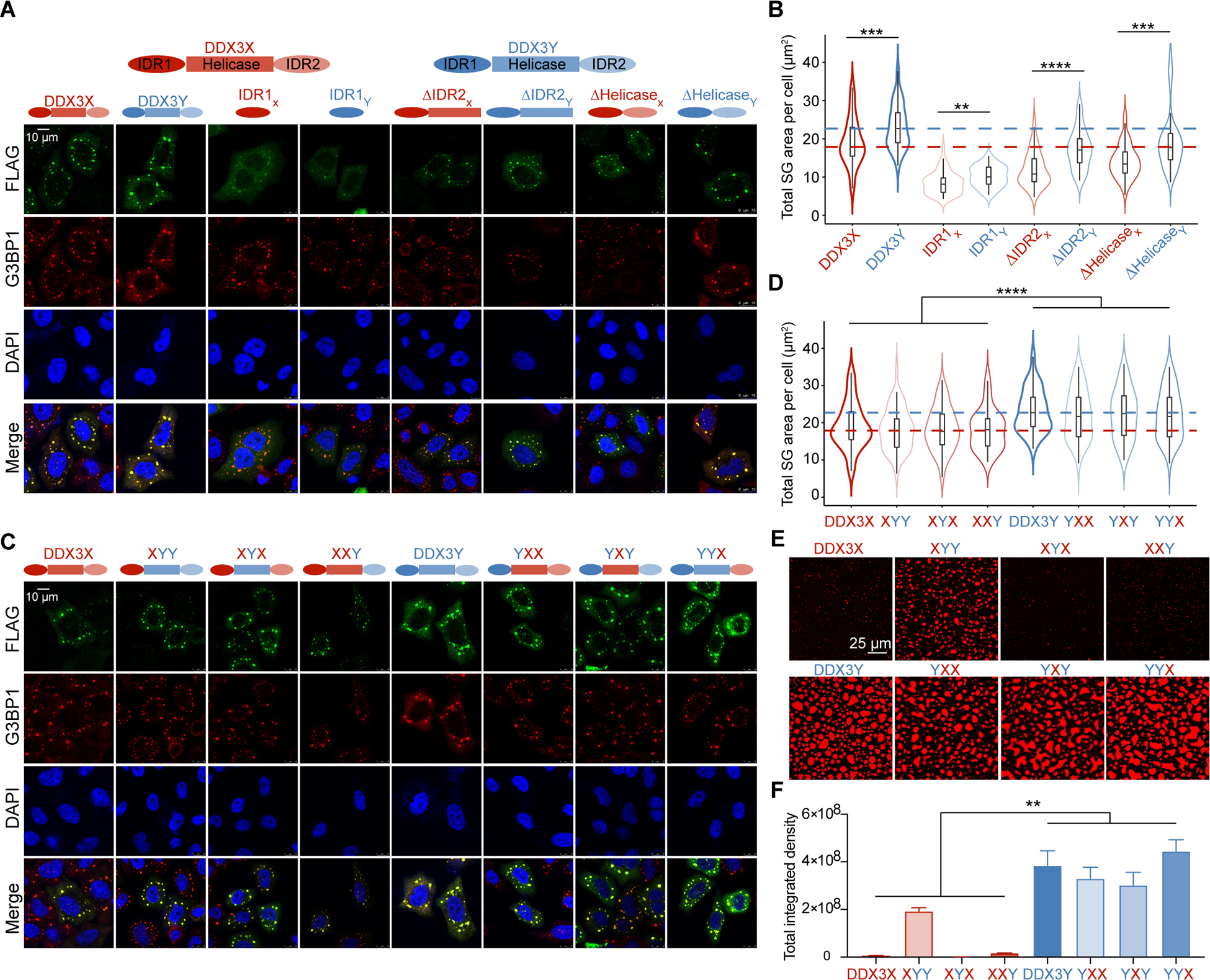
(A) Co-localization of DDX3X or DDX3Y domain truncation variants in HeLa cells with G3BP1 upon 500 μM arsenite treatment for 1 hr. Scale bar, 10 μm.
(B) Violin plots of the total SG area of truncation SGs per cell (n = 50 cells in total, from three biologically independent experiments). The median of the total SG areas per cell of wild-type DDX3X-SGs and DDX3Y-SGs are indicated by red and blue dashed lines, respectively. p values were determined by a two-tailed t-test; **p < 0.01, ***p < 0.001, ****p < 0.0001.
(C) Co-localization of DDX3X or DDX3Y domain swap variants in HeLa cells with G3BP1 upon 500 μM arsenite treatment for 1 hr. Scale bar, 10 μm.
(D) Violin plots of the total SG area of DDX3X or DDX3Y domain swap SGs per cell (50 cells in total from three biologically independent experiments). The median sum of SG areas per cell of wild-type DDX3X-SGs and DDX3Y-SGs is indicated by red and blue dashed lines, respectively. p values were determined by nested t-test to compare all domain-swapped variants with XIDR1 versus all domain-swapped variants with YIDR1; ****p < 0.0001.
(E) In vitro droplet formation of 7.5 μM recombinant DDX3X-mCherry, DDX3Y-mCherry, or domain swap variants of DDX3X and DDX3Y in the presence of 200 ng/μL poly(U)-RNA. Scale bar, 25 μm.
(F) Quantification of the total integrated intensity of the different conditions shown in Figure 3E. Error bars represent s.d. from three repeats at each condition. p values were determined by nested t-test to compare all domain-swapped variants with XIDR1 versus all domain-swapped variants with YIDR1; **p < 0.01.
See also Figure S3.
When we swapped IDRs between DDX3X and DDX3Y, we observed that any domain-swapped variants containing IDR1 of DDX3Y formed significantly larger SGs than the hybrid variants that did not contain IDR1 of DDX3Y, whereas swapping helicase domains or IDR2 did not significantly affect SG sizes (Figures 3C, 3D, S3E, and S3F). When we performed in vitro droplet formation assays with wild-type and domain-swapped variants (which were purified to a similar level of homogeneity, Figure S3G and S3H), our results recapitulated the finding that any domain-swapped variants that contained IDR1 of DDX3Y more readily phase-separated in the presence of RNA than variants which did not contain IDR1 of DDX3Y (Figure 3E and 3F). Our results also suggest that the IDR2 and helicase domains may also contribute to the differences in propensity to undergo LLPS, as the XIDR1YhelicaseYIDR2 variant more readily formed droplets compared to wild-type DDX3X, XIDR1YhelicaseXIDR2, and XIDR1XhelicaseYIDR2 (Figure 3D and 3F). To dissect the contributions of RNA binding and IDRs in phase separation of the proteins, we next performed the in vitro droplet assays with wild-type and domain-swapped variants in the absence of poly(U)-RNA. As shown in Figure S3I, proteins containing IDR1 of DDX3Y formed larger droplets than proteins containing IDR1 of DDX3X. These results suggest that although each of the domains in DDX3 proteins plays a distinct role in facilitating LLPS and in their accumulation into SGs, the sequence of IDR1 is a significant factor in determining the relative droplet sizes.
The weaker ATPase activity of DDX3Y compared to DDX3X weakens its condensate disassembly
DDX3X and DDX3Y harness the energy of ATP hydrolysis to control RNA binding, unwinding, and release (Hondele et al., 2019). Because LLPS droplets assemble through many multivalent, weak interactions between RNAs and proteins (Wang et al., 2018a), we stimulated the ATPase activity of DDX3X and DDX3Y to see if this could induce condensation disassembly by affecting RNA-protein interactions. As shown in Figure 4A and 4B, the addition of 4 mM ATP to protein-RNA droplets induced the disassembly of both DDX3X and DDX3Y condensates. To control for the hydrotropic effect of ATP (Patel et al., 2017) in dissolving the droplets, we repeated the droplet assay using UTP (which is not hydrolyzed by DDX3 and thus could only act as a hydrotrope in this context (Patel et al., 2017)). The addition of UTP had no significant effect on the droplets formed by DDX3X and DDX3Y with RNA (Figure 4A and 4B). These results suggest that the ATPase activity of DDX3X and DDX3Y is a major factor in dissolving DDX3-RNA condensation, possibly by breaking the multivalent protein-RNA interactions.
Figure 4. The weaker ATPase activity of DDX3Y compared to DDX3X weakens its condensate disassembly.
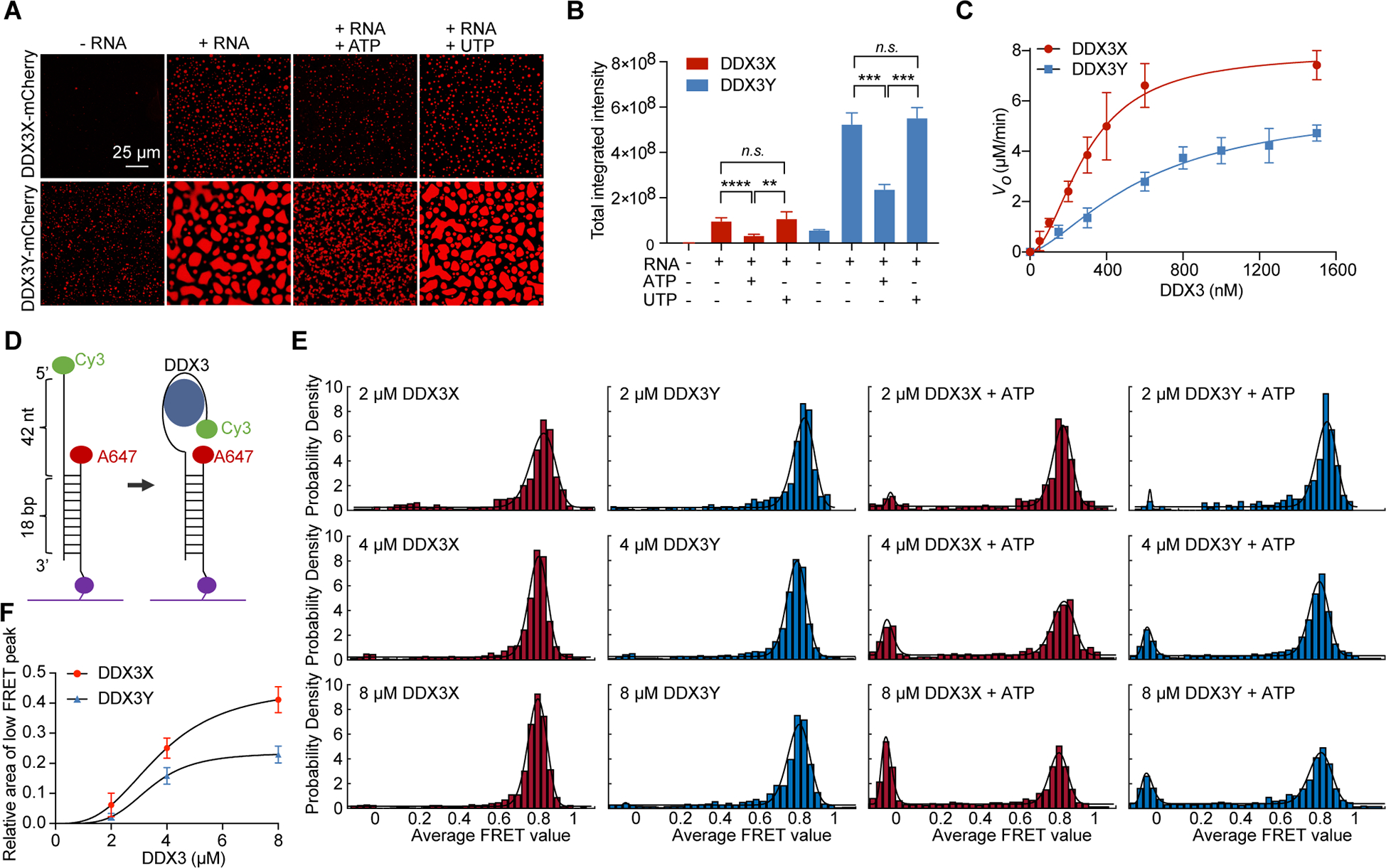
A) In vitro droplet formation of 10 μM recombinant DDX3X-mCherry and DDX3Y-mCherry in the absence and presence of 200 ng/μL poly(U)-RNA with and without the addition of 4 mM ATP or UTP. Scale bar, 25 μm.
(B) Quantification of the total integrated intensity of different groups of condensates shown in Figure 4A. A two-tailed t-test was used to calculate the p-value. n.s. means p > 0.05, **p < 0.01, ***p < 0.001, and **** p < 0.0001.
(C) ATPase activity of DDX3X and DDX3Y, as measured by release of free phosphate, in μM/min. Error bars represent ± s.d. from 6 individual replicates. The background value (initial rate when no protein was added) was subtracted from each point before plotting and curve fitting to the Hill equation. DDX3X: Vmax = 7.9 ± 0.4 μM/min, H = 1.9 ± 0.2, K1/2 = 297.2 ± 22.6 nM. DDX3Y: Vmax = 5.9 ± 0.7 μM/min, H = 1.5 ± 0.2, K1/2 = 617.9 ± 123.6 nM. All fitting parameter uncertainties are ± s.e.m.
(D) Schematic of smFRET RNA probe.
(E) FRET efficiency distributions with increasing protein concentrations (2, 4, and 8 μM) of DDX3X and DDX3Y in the presence or absence of ATP. FRET values were collected from over 1000 molecules to build the histograms.
(F) Fitting of the Hill equation to relative values of the low FRET peak area (with background subtracted) in Figure 4E for DDX3X and DDX3Y.
See also Figure S4.
We next studied whether there are intrinsic differences in ATPase activity between DDX3X and DDX3Y, as this could explain the differences observed in their condensate disassembly in response to ATP. Thus, we measured the ATPase activities of both DDX3X and DDX3Y using a malachite green ATPase assay. In this assay, we used MBP-tagged proteins purified to near homogeneity (Figure S4A and S4B). Both of the MBP-tagged DDX3X and DDX3Y displayed the same melting temperatures in differential scanning fluorimetry measurements, and the melting curves are indicative of well-folded proteins devoid of aggregates (Figure S4C and S4D) (Gao et al., 2020).
To ensure that any potential RNA carry-over would not confound our measurements, we performed the malachite green ATPase assays in the absence of additional RNA or with 100 ng/μL total HeLa RNA. In reactions with no added RNA but with 2 mM ATP, DDX3X and DDX3Y reactions had nearly identical amounts of free phosphate after a 30-minute reaction time (Figure S4E). This phosphate is likely due to spontaneous hydrolysis of ATP, as it is also present in the controls with buffer only. When RNA was added, DDX3X reactions produced more free phosphates (indicative of more ATP hydrolysis) than DDX3Y reactions. The malachite green ATPase assays were also performed using RNase treated and untreated MBP-tagged DDX3X and DDX3Y respectively. As shown in Figure S4F, RNase treatment has no noticeable effects on the ATPase activities. Collectively, these results suggest that DDX3X is a more robust ATPase than DDX3Y, and that the possible trace amount of RNA carry-over in the purified MBP-tagged DDX3X and DDX3Y was not responsible for the ATPase activity differences.
To obtain a more complete picture of the ATPase differences between DDX3X and DDX3Y, we performed a continuous ATPase assay as previously reported using the MBP-tagged DDX3X and DDX3Y constructs (Song and Ji, 2019). DDX3X reached a Vmax of 7.9 μM/min while DDX3Y only achieved a Vmax of 5.9 μM/min (Figure 4C). Both DDX3X and DDX3Y hydrolyzed ATP cooperatively (Hill coefficient ≈ 2), as has been previously reported for DDX3X (Song and Ji, 2019). While the ATPase activity of DDX3Y has not been previously investigated, our Vmax values for the full-length DDX3X were higher than those reported for a truncation lacking both IDRs (amino acids 132 – 607) (7.9 vs. 3.1 μM/min, 2.54-fold higher) (Song and Ji, 2019). Our data suggest that ATP hydrolysis activity of DDX3Y is significantly slower than that of DDX3X (Figure 4C).
DDX3X is a non-processive helicase: it binds its dsRNA substrate, binds ATP, unwinds approximately 13 – 19 bp of dsRNA, and releases the two RNA strands upon ATP hydrolysis (Song and Ji, 2019). Given that ATP hydrolysis is a crucial step in this catalytic cycle and that our ATPase data suggest that DDX3X is a more efficient ATPase than DDX3Y, we next investigated how the differences in ATPase activities may affect the dynamics of these enzymes. To this end, we employed smFRET assays with MBP-DDX3X and MBP-DDX3Y to compare their RNA binding abilities and the dynamics of their ATP-dependent interactions with RNA as described previously (Figure 4D) (Kim and Myong, 2016). At protein concentrations of 2, 4, or 8 μM, DDX3X and DDX3Y tightly bound the RNA substrate, resulting in high FRET, with a peak FRET efficiency (E) of 0.81 – 0.85 (Figures 4E, S4G, and Table S1). This binding is specific to DDX3X and DDX3Y, as MBP (a non-RNA interacting protein) gave low FRET, like RNA alone (Figure S4H and S4I).
The addition of ATP to DDX3X and DDX3Y led to a low FRET population that was larger for DDX3X than it was for DDX3Y (Figures 4E, S4J, S4K, and Table S1). Of note, this low-FRET population of RNA molecules does not represent full strand separation of the RNA duplex because alternating direct illumination of the Cy3 and Alexa Fluor 647 probes (lowest traces in Figure S4J and S4K) showed that both labeled strands are present in each individual high- and low-FRET particles. This change in FRET from high to low efficiency likely represents partial unwinding activity. As shown in Figure 4F, the relative areas of the low FRET peak induced by addition of ATP increased along with increasing protein concentrations which exemplifies the cooperativity of the DDX3X- and DDX3Y-catalyzed reactions. Importantly, the smFRET histograms suggest that DDX3X had higher (partial) unwinding activity than DDX3Y, consistent with the kinetics data (Figure 4C). DDX3X also showed a larger proportion of dynamic FRET recordings upon addition of ATP than DDX3Y (Figure S4L). Together, the kinetics and smFRET data support the conclusion that DDX3Y has slower ATPase and (partial) unwinding activities, leading to the less dynamic characteristics of RNA-DDX3Y complexes. Decreased dynamics may contribute to the weaker disassembly of DDX3Y condensates compared to those of DDX3X, which in turn contributes to the observation that DDX3Y condensates persist in the presence of ATP to a greater degree than DDX3X condensates (Figure 4A).
Translation is differentially modulated by DDX3X and DDX3Y
Given that SGs are formed concomitant with translational repression and are thought to harbor proteins that regulate translation (Kimball et al., 2003), we employed a reticulocyte in vitro translation assay to investigate how phase-separated DDX3X and DDX3Y influence the translation of a luciferase reporter (Figure 5A). Upon titration of 4 – 10 μM mCherry-DDX3X or mCherry-DDX3Y into the lysate, there was a dramatic, dose-dependent decrease of luciferase signal with increased concentrations of DDX3X or DDX3Y, indicating a decrease in the in vitro translation (Figure 5B). Indeed, we observed a concentration-dependent increase in protein condensation of DDX3X or DDX3Y within the lysate (Figure 5C and 5D). These results indicate that the condensation of DDX3X and DDX3Y is correlated with repressed mRNA translation. Of note, at each concentration tested, DDX3Y induced a more pronounced decrease in luciferase signal compared to DDX3X (Figure 5B). To ensure that the observed effects were specific to DDX3X and DDX3Y, we also titrated truncated versions of DDX3X and DDX3Y, which only contained the minimally active helicase domain (both RecA-like domains and the N- and C-terminal extensions) (Floor et al., 2016; Song and Ji, 2019) to serve as negative controls. As shown in Figure S5A, the luciferase signal remained steady across all concentrations of the truncated versions of DDX3X and DDX3Y, likely because these constructs lack IDRs. We next studied whether adding ATP could alleviate translation repression by disassembling the DDX3X and DDX3Y condensates in the in vitro translation assays. To this end, the assays were repeated with the addition of 1 mM ATP. Upon ATP addition, in vitro translation was partially restored in both DDX3X and DDX3Y reactions; however, DDX3Y reactions remained less translationally active than the corresponding DDX3X reactions (Figure S5B). These findings support the notion that the decreased ATPase activity of DDX3Y decelerates the dispersal of DDX3Y-containing condensates, and thus ATP does not fully restore in vitro translation.
Figure 5. DDX3X and DDX3Y condensation inhibit the translation of luciferase RNA, and DDX3X- and DDX3Y-positive SGs have shared and unique RNA constituents in cells.
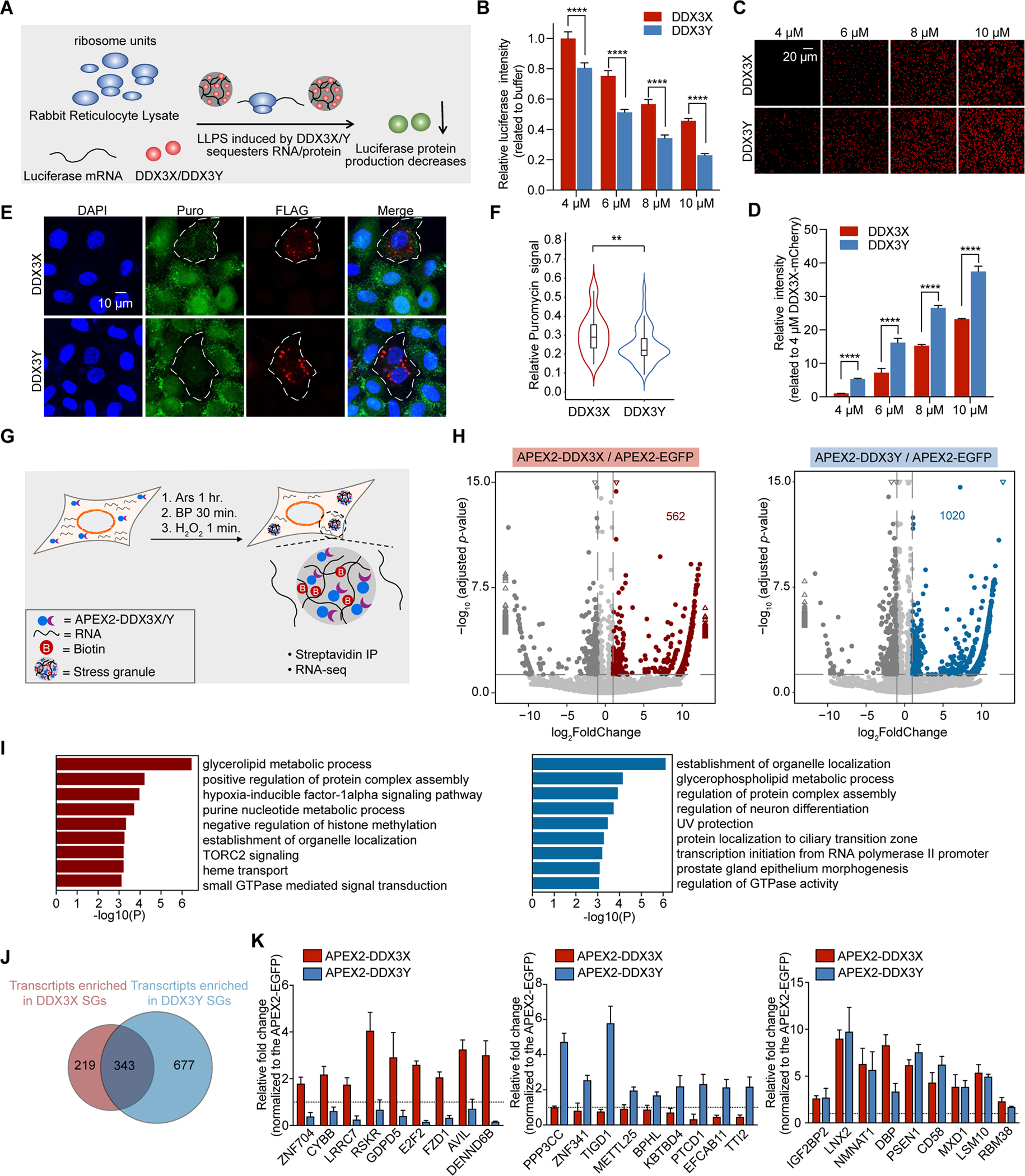
(A) Schematic illustration of the in vitro translation assay.
(B) In vitro translation inhibition at the indicated concentrations of DDX3X-mCherry or DDX3Y-mCherry. p values were determined by two-tailed t-test; ****p < 0.0001.
(C) DDX3X-mCherry and DDX3Y-mCherry phase separation in the reticulocyte assay at each indicated concentration; scale bar, 20 μm.
(D) Quantification of the total integrated intensity of different condensates shown in Figure 5C.
(E) Puromycin incorporation assay to determine the extent of translation repression in cells with DDX3X-positive SGs and DDX3Y-positive SGs. The white outlines indicate the cells which express exogenous DDX3X or DDX3Y.
(F) Quantification of puromycin signal in Figure 5E. Only cells expressing exogenous DDX3X or DDX3Y were selected, and the total puromycin signal in each cell was quantified. The total puromycin signals in neighboring cells not expressing exogenous DDX3X or DDX3Y were quantified similarly and used to normalize the data. p-value was determined by two-tailed t-test; **p < 0.01.
(G) Schematic illustration of APEX2-mediated proximity labeling reaction.
(H) Volcano plots showing differential RNA enrichment in streptavidin pull-downs from APEX2-DDX3X (left) and APEX2-DDX3Y (right) expressing cells compared to APEX2-EGFP expressing cells after 500 μM arsenite treatment for 1 hr. Differentially expressed genes are shown in red and blue for APEX2-DDX3X-enriched and APEX2-DDX3Y-enriched RNAs respectively (adjusted p < 0.05, log2 fold change > 1) and dark gray for APEX2-DDX3X-depleted and APEX2-DDX3Y-depleted RNAs (adjusted p < 0.05, log2 fold change < −1). The rest of the RNAs are shown in light gray for both DDX3X and DDX3Y. The triangles represent the transcripts with log2 fold change > 13 or log2 fold change < −13 in the X-axis; −log10 adjusted p >15 in the Y-axis.
(I) Gene Ontology (GO) analysis of the differentially enriched RNA groups in Figure 5E; red: APEX2-DDX3X-enriched RNAs; and blue: APEX2-DDX3Y-enriched mRNAs.
(J) Venn diagram quantifying the overlapping and distinct RNA clients enriched by APEX2-DDX3X and APEX2-DDX3Y after arsenite treatment.
(K) Relative fold change of select RNAs used to conduct RT-qPCR validation of the sequencing results.
See also Figure S5.
Next, we investigated whether DDX3Y condensation had a stronger translation repression impact than DDX3Y condensation in cells using a puromycin incorporation assay in HeLa cells after transient transfection of FLAG-DDX3X or FLAG-DDX3Y with endogenous DDX3X depleted. Puromycin was incorporated into newly synthesized proteins, and, as shown in Figure 5E and 5F, the formation of DDX3X-positive SGs and DDX3Y-positive SGs significantly reduced puromycin signals compared to the neighboring cells which were not transfected and thus lacked DDX3X-positive or DDX3Y-positive SGs. Notably, the puromycin signals were weaker in cells with DDX3Y-positive SGs than in cells with DDX3X-positive SGs (Figure 5F). These results suggest a potential mechanism by which DDX3Y more effectively inhibit translation through its unique biophysical properties. Phase separation can inhibit translation by sequestering translational machinery and/or mRNA transcripts into the phase-separated compartment (Kim et al., 2019). DDX3Y-containing condensates and SGs are larger and less mobile (Figures 1 and 2), which may result in stronger translation inhibition than the smaller and more dynamic DDX3X condensates which exchange material more rapidly.
APEX-seq captures the protein-RNA interaction patterns of DDX3X and DDX3Y
SGs formed under different stress conditions contain distinct proteins and RNA constituents (Markmiller et al., 2018), which raises the possibility that SG contents vary based on the type of stress presented to a cell. Neither the RNA composition nor the differential effects DDX3X or DDX3Y may exert on resident SG RNAs is known. Because SGs are largely thought to be a mechanism of regulating mRNA metabolism (Buchan and Parker, 2009; Jain et al., 2016; Khong et al., 2017), we sought to define the RNA content of DDX3X- and DDX3Y-positive SGs.
To this end, we employed an adapted ascorbate-peroxidase (APEX2)-based proximity labeling method (Fazal et al., 2019; Marmor-Kollet et al., 2020; Padron et al., 2019) (Figure S5C). First, to examine the specificity of APEX2-catalyzed RNA biotinylation, we generated a mito-APEX2 fusion protein consisting of APEX2 fused to a mitochondrial matrix localization signal (Figure S5D) (Fazal et al., 2019; Mercer et al., 2011). Using a previously described protocol (Fazal et al., 2019), followed by RT-qPCR analysis, we could reliably identify mitochondrial-specific RNAs (ND1 and ND2) labeled by mito-APEX2 (Figure S5E – S5G).
Next, using this validated approach, we expressed APEX2-DDX3X, APEX2-DDX3Y, or APEX2-EGFP (control) in DDX3X knockdown HeLa cells. Cells were then treated with arsenite or DMSO for 1 hr to generate three experimental cell populations. We consistently observed that the total area of APEX2-DDX3Y-positive SGs was significantly larger (1.5-fold) than APEX2-DDX3X-positive SGs (Figure S5H and S5I). The results suggest that the fusion of APEX2 to DDX3X or DDX3Y did not significantly interfere with the ability of either protein to colocalize with SGs in cells upon arsenite treatment.
The above cells were then incubated with biotin-phenol for 30 min, followed by H2O2 treatment for 1 min to activate the APEX2 enzyme and covalently link biotin to RNAs (Figure 5G). To confirm biotinylating, small aliquots of cell lysate for each condition were blotted using a streptavidin antibody; this antibody detected multiple protein bands (consistent with the previous results (Fazal et al., 2019)), confirming that the APEX2 enzyme was active (Figure S5J and S5K). Subsequently, we performed biotin pulldown and poly(dT) extraction to enrich biotinylated poly(A)-RNAs. Enriched RNAs were subjected to next-generation high-throughput RNA-seq. The RNA-seq data from the biological replicates in each group correlated well (Figure S5L).
To determine whether DDX3X- and DDX3Y-positive SGs harbor unique mRNAs, we compared the levels of transcripts between APEX2-DDX3X or APEX2-DDX3Y libraries to APEX2-EGFP libraries. Any transcript for which expression in either the APEX2-DDX3X or APEX2-DDX3Y libraries was at least 2-fold higher than in the APEX2-EGFP dataset (log2 -fold enrichment > 1) was defined as “enriched.” We found that DDX3X-positive SGs enriched 562 RNAs (Figure 5H and Table S2), and Gene Ontology (GO) analysis indicated the encoded proteins were mainly involved in the regulation of glycolipid and nucleic acid metabolism (Figure 5I). DDX3Y-positive SGs enriched 1020 RNAs (Figure 5H); GO analysis suggested that some of the encoded proteins were also involved in the regulation of glycolipid metabolism, while others play roles in transcriptional regulation (Figure 5I). Interestingly, while there was a large pool of RNAs enriched in both DDX3X- and DDX3Y-positive SGs (where 61% of DDX3X- and 34% of DDX3Y-positive SG enriched RNAs are shared), there are also RNA targets that were specific to each helicase (Figure 5J). In the absence of arsenite treatment, there was a small number of transcripts with fold change log2 > 1 or log2 < −1 in APEX2-DDX3X or APEX2-DDX3Y compared to APEX2-EGFP (Figure S5H and S5M). We validated a list of RNA targets specifically enriched by either APEX2-DDX3X or APEX2-DDX3Y, ranging from lower to upper enrichment, using RT-qPCR (Figure 5K). These results indicate that DDX3X- and DDX3Y-positive SGs regulate distinct mRNA targets. However, DDX3X and DDX3Y may act on shared transcripts in divergent ways as these two enzymes inhibit translation to different degrees (Figure 5A – 5F).
DDX3X and DDX3Y co-phase separate into SGs
While our study up to this point has focused on either DDX3X or DDX3Y individually, XY individuals express both proteins simultaneously (Cotton et al., 2015; Ditton et al., 2004; Godfrey et al., 2020). To examine whether DDX3X and DDX3Y go into the same SGs, we expressed DDX3X and DDX3Y together in cells with endogenous DDX3X transiently knocked down. We found that all the antibodies we tested were highly cross-reactive (Figure S6A), likely due to the high similarity of these two proteins. Thus, we used proteins with different tags (FLAG vs. HA), and tagged homologs were expressed to a similar extent (Figure S6B).
Given that SG size and composition are sensitive to different types of stressors (Fujimura et al., 2012; Markmiller et al., 2018; Saito et al., 2019; Szaflarski et al., 2016), we studied SGs triggered by a range of stressors, including energy depletion (CCCP), osmotic stress (sorbitol), translation inhibition (puromycin), proteasome inhibition (MG132), and ER stress (thapsigargin). Under these conditions, MG132 and thapsigargin did not induce SG formation (Figure S6C). However, when CCCP, sorbitol, or puromycin were used to stress cells, DDX3X, DDX3Y, and G3BP1 colocalize, although the fluorescence intensity of FLAG-DDX3X was much lower than HA-DDX3Y (Figure 6A). To ensure that our observations were not due to differential recognition by the anti-FLAG and anti-HA antibodies, we repeated the experiments using HA-DDX3X and FLAG-DDX3Y. We observed the same lower intensity of DDX3X, even though total protein levels for each DDX3 homolog were similar and were not affected by either tag (Figures S6D). These results suggest that, while DDX3X and DDX3Y phase separate to SGs together, DDX3Y has a stronger propensity to go into SGs than DDX3X, in line with our previous observations.
Figure 6. A combination of DDX3X and DDX3Y shows a stronger propensity of LLPS and translation repression than DDX3X alone; and DDX3Y enhances FUS aggregation and accelerates TDP-43 aggregation more than DDX3X.
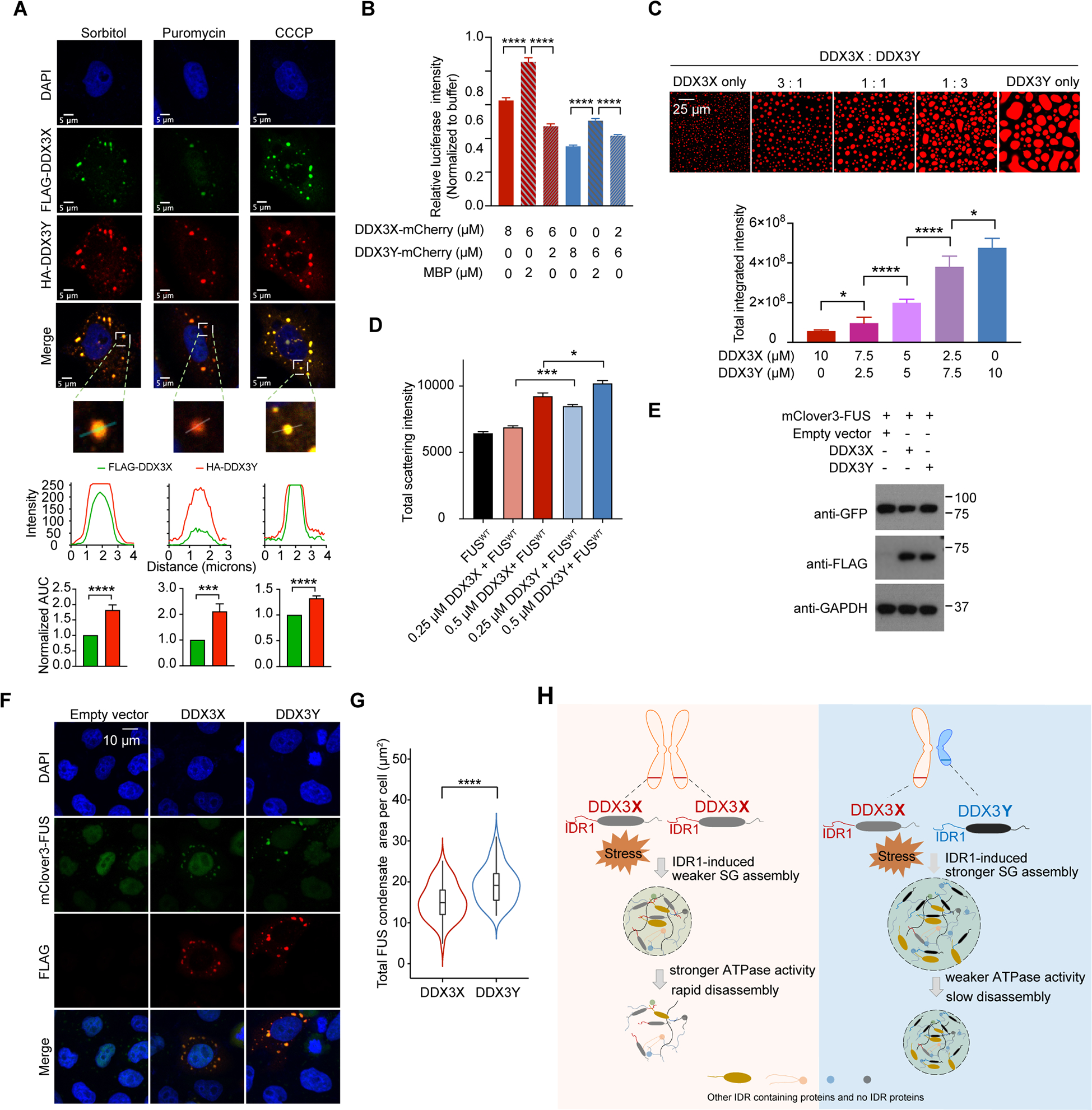
(A) Immunofluorescence images of SGs containing both FLAG-DDX3X and HA-DDX3Y in HeLa cells treated with sorbitol, puromycin, or CCCP. Below each image, traces of fluorescence intensity profiles through positions denoted by the white lines in the merged images. The area under the curve (AUC) normalized to that of FLAG-DDX3X is plotted for each intensity profile and shows that the signal from HA-DDX3Y is consistently higher than the signal from FLAG-DDX3X.
(B) A mixture of DDX3X-mCherry and DDX3Y-mCherry at the annotated concentrations differentially repress in vitro translation of luciferase RNA. Overall, DDX3Y represses translation more than DDX3X. p values were determined by two-tailed t-test; ****p < 0.0001.
(C) In vitro droplet formation of recombinant DDX3X-mCherry and DDX3Y-mCherry at the annotated ratios in the presence of 200 ng/μL poly(U)-RNA (top panel). Scale bar, 25 μm. Quantification of the total integrated intensity of each type of condensate (bottom panel). p values were determined by two-tailed t-test; *p < 0.05, ****p < 0.0001.
(D) DDX3Y more strongly promotes FUS aggregation than DDX3X in vitro. The area under the curve for each time course of light scattering at 395 nm in Figure S6E was used to compare the extent of aggregation for each condition. p values were determined by two-tailed t-test; *p < 0.05, ***p < 0.001.
(E) Western blots showing the expression of DDX3X and DDX3Y in DOX-inducible stable cell lines expressing mClover3-FUS.
(F) Representative images showing the localization of Flag-DDX3X, Flag-DDX3Y, and mClover-FUS upon arsenite treatment (500 μm, 1 hr). Scale bar, 10 μm.
(G) The total area of granules containing mClover3-FUS and Flag-DDX3X or granules containing mClover3-FUS and Flag-DDX3Y per cell was quantified (n = 50 cells in total, from three biologically independent experiments). p values were determined by a two-tailed t-test; ****p < 0.0001.
(H) Schematic illustration of how sexually dimorphic RNA helicases DDX3X and DDX3Y differentially regulate RNA translation through phase separation.
See also Figure S6.
Given that DDX3X and DDX3Y can co-phase separate into SGs, we wondered how mixtures of DDX3X and DDX3Y affected translation compared to DDX3X or DDX3Y alone. As shown in Figure 6B, a 3:1 ratio of DDX3X (6 μM) and DDX3Y (2 μM), which is close to the physiological ratios between DDX3X and DDX3Y (Godfrey et al., 2020), led to a 1.4-fold more robust repression of translation compared to DDX3X (8 μM) alone, and a 3.6-fold more robust repression of translation compared to a mixture of DDX3X (6 μM) and MBP (2 μM). Moreover, a mixture of DDX3X (2 μM) and DDX3Y (6 μM) less efficiently repressed translation relative to DDX3Y (8 μM) alone. Furthermore, the in vitro LLPS assays were performed with different ratios of DDX3X and DDX3Y in the presence of RNA. The addition of DDX3Y to DDX3X stimulated droplet formation, and condensation was positively correlated with the relative amount of DDX3Y (Figure 6C).
DDX3Y more strongly promotes FUS aggregation than DDX3X
Dysregulation of SGs can promote FUS aggregation, leading to cell death (Bentmann et al., 2012; Guo et al., 2018; Kamelgarn et al., 2016; Silva et al., 2019). Thus, we studied how DDX3X and DDX3Y influence in vitro FUS fibrillization. When we added 1 μM wild-type FUS to either DDX3X or DDX3Y, we saw that the aggregation of FUS, detected by light scattering at 395 nm, was enhanced by both helicases but was more extensively aggravated in the presence of DDX3Y (Figure S6E). To quantify the effect of DDX3X and DDX3Y on FUS aggregation over time, we measured the area under the curve (AUC) for each scattering time course. Both DDX3X and DDX3Y enhance FUS aggregation but that DDX3Y had a stronger enhancement, even at high concentrations of either protein (Figures 6D and S6E). We also studied the colocalization of FUS and DDX3X or DDX3Y in HeLa cells upon arsenite treatment. We constructed a DOX-inducible stable cell line that expressed FUS. As shown in Figure 6E – 6F, while wild-type FUS was mainly located in the nucleus with transfection of empty vector, a portion of FUS formed puncta which colocalize with DDX3X-positive granules and DDX3Y-positive granules in the cytoplasm with transfection of DDX3X and DDX3Y expression plasmids. Furthermore, DDX3Y-FUS puncta were significantly larger than DDX3X-FUS puncta (Figure 6G). As XY individuals are at a higher risk for developing ALS (Manjaly et al., 2010), these data not only suggest that DDX3X and DDX3Y affect FUS aggregation but also suggest that the enhanced propensity to promote LLPS by DDX3Y might lead to a XY-specific increase of FUS aggregation.
DDX3Y more strongly accelerates TDP-43 aggregation than DDX3X
Finally, we tested whether DDX3X or DDX3Y might also stimulate the aggregation of TDP-43, another prominent RNA-binding protein connected to ALS and FTD (Portz et al., 2021; Tan et al., 2017). We found that DDX3X or DDX3Y did not enhance the total amount of TDP-43 aggregation (Figure S6F and S6G). However, both helicases accelerated TDP-43 aggregation (Figure S6F and S6H). The halftime t1/2 (i.e. the time at which 50% TDP-43 aggregation had occurred) was reduced from ~6.9 hrs in the absence of helicase to ~5.7 hrs in the presence of DDX3X and ~ 4.4 hrs in the presence of DDX3Y (Figure S6F and S6H). These findings suggest that DDX3Y can accelerate TDP-43 aggregation more potently than DDX3X.
DISCUSSION
Herein, we report the first comparative study of the sexually dimorphic RNA helicases DDX3X and DDX3Y in the regulation of translation via distinct phase separation behaviors (Figure 6H). Importantly, we reveal the molecular mechanism underpinning the higher propensity of DDX3Y to phase separate and its lower propensity to disassemble once condensed. Firstly, we find that the condensation propensity differences of DDX3X and DDX3Y are most likely due to the sequence composition of both IDR1s. Overall, the percentages of both negatively and positively charged amino acids in YIDR1 (17.3% and 16.1% respectively) are higher than in XIDR1(16.7% and 14.9% respectively), whereas the percentage of charged amino acids is similar between the other domains of DDX3X and DDX3Y (Figure S6I). This finding suggests that YIDR1 can form more charge-charge interactions to support phase separation. In addition to these electrostatic interactions, cation-π and π-π (Vernon et al., 2018) interactions are known to facilitate LLPS (Qamar et al., 2018). As such, Tyr to Phe and Arg to Lys mutations in IDRs dramatically impair phase separation (Schuster et al., 2020). In line with this, Phe84 and Lys118 in XIDR1 correspond to Tyr83 and Arg116 in YIDR1, suggesting that YIDR1 is capable of more cation-π and π-π interactions than XIDR1, which might result in the stronger phase separation of DDX3Y found in these studies (Figure S1A). Indeed, Lys118 in DDX3X is a known site of post-translational acetylation (Saito et al., 2019). Acetylation at this site decreases the phase separation of DDX3X in vitro and inside of cells through the disruption of cation-π interactions. Given that the analogous position in DDX3Y is an arginine (Arg116), which is often thought of as a “non-acetyl” mimetic, this position is likely a key source of difference between DDX3X and DDX3Y and merits future study.
Second, our data suggest that the distinct dynamics of DDX3X- and DDX3Y-positive SGs may also be related to differences in their ATPase-driven SG remodeling activity (Jain et al., 2016; Tauber et al., 2020) (Figure 4). Furthermore, the differences in dynamics of DDX3X and DDX3Y condensates may also explain why DDX3Y more strongly repressed translation than DDX3X. DDX3X-positive SGs and DDX3Y-positive SGs sequester distinct mRNAs in addition to a shared pool of transcripts, suggesting that the differences in ATPase activity and translational repression may have functional consequences, especially under stress (Figure 5G – 5K). The results shared here suggest that DDX3X and DDX3Y might influence the translation of the overlapping mRNA targets to different degrees in addition to exerting differential regulation of distinct RNA components. They might also differentially sequester other translational components.
One important question that remains to be answered is the degree to which DDX3X and DDX3Y overlap functionally. Studies by others have suggested that DDX3X and DDX3Y are redundant in protein synthesis under unstressed conditions (Venkataramanan et al., 2021). Our data suggest that their divergent roles may not be apparent until they are driven to phase separate during the stress response.
Stress leads to various human disorders, many of which display sex-biased features. For instance, ALS is ~20% more common in males than females (Manjaly et al., 2010). We showed that DDX3Y more strongly promotes FUS self-assembly in vitro and formed larger FUS granules in cells (Figure 6D – 6G), possibly through its stronger phase separation propensity compared to DDX3X. Furthermore, DDX3Y more strongly accelerated TDP-43 aggregation than DDX3X (Figure S6F – S6H). Other known SG resident proteins such as TIA-1, hnRNPA1, and hnRNPA2 have been previously indicated in ALS (Fernandes et al., 2020; Gilks et al., 2004; Harrison and Shorter, 2017; Khalfallah et al., 2018; Molliex et al., 2015). Future investigation on the scope and extent of DDX3X’s and DDX3Y’s impact on these proteins in sex specificity in neurodegenerative diseases will be exciting.
Limitations of the Study
HeLa cells may not be perfectly suited to the task of uncovering the biological targets in DDX3Y-specific SGs, because they lack a Y chromosome and thus may also lack some of the specific transcripts that may be targeted by DDX3Y. However, we feel that reconstituting HeLa cells (with endogenous DDX3X depleted) with ectopically expressed DDX3X or DDX3Y provided a clean system to reveal the different RNA sequestration impacts of DDX3X and DDX3Y due to their intrinsic differences in phase separation. Thus, we believe our findings are an excellent proof of principle. Additionally, while our recombinant proteins were purified to apparent homogeneity and are not expected to have unequal carryover of RNA, there could be some effect of potential impurities on the magnitude of the differences we measure. However, we believe that the difference itself is a true observation about these proteins. The apparent unwinding activity observed in the smFRET assays is not an indication of complete unwinding by DDX3X/DDX3Y and this assay is not a conventional assay to study helicase activity. Both the donor and acceptor fluorophores being present in all molecules compiled in the smFRET analysis indicates only partial unwinding of the RNA substrate. We believe our smFRET assay is mainly reflecting the ATPase and dynamic activity of the proteins with RNA and give insight into the differences between DDX3X and DDX3Y in these features. Our experiments should serve as a crucial first step towards understanding the role of sexually dimorphic proteins in disease.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Kathy Fange Liu (liufg@pennmedicine.upenn.edu).
Materials availability
All materials generated in this study are available on request to Lead Contact.
Data and code availability
RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Original western blot images have been deposited at Mendeley and are publicly available as of the date of publication. The DOI is listed in the key resources table. Microscopy data reposted in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key resources table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-G3BP1 | Proteintech | Cat# 13057-2-AP, RRID:AB_2232034 |
| Rabbit polyclonal anti-HA | Abcam | Cat# ab9110, RRID:AB_307019 |
| Mouse monoclonal anti-FLAG | Sigma-Aldrich | Cat# F3165, RRID:AB_259529 |
| Rat monoclonal anti-FLAG | Invitrogen | Cat# MA1-142, RRID:AB_2536846 |
| HRP conjugated anti-FLAG | Invitrogen | Cat# MA1-91878-HRP, RRID:AB_2537626 |
| Rabbit polyclonal anti-GAPDH | Invitrogen | Cat# PA1-16777, RRID:AB_568552 |
| Mouse monoclonal anti-GFP | Santa Cruz | Cat# sc-996, RRID:AB_2187785 |
| Mouse anti-puromycin, clone 12D10, Alexa Fluor 488 conjugated antibody | Sigma-Aldrich | Cat# MABE343-AF488, RRID:AB_2736875 |
| Goat anti-mouse IgG (H&L) (HRP) | Abcam | Cat# ab6789, RRID:AB_955439 |
| Mouse monoclonal anti-ATP5A | Abcam | Cat# ab14748, RRID:AB_301447 |
| Goat anti-Mouse IgG H&L (Alex Fluor® 647) | Abcam | Cat# ab150115, RRID:AB_2687948 |
| Goat anti-Rabbit IgG H&L (Alexa Fluor® 594) | Abcam | Cat# ab150080, RRID:AB_2650602 |
| Goat anti-Mouse IgG H&L (Alexa Fluor® 488) | Abcam | Cat# ab150113, RRID:AB_2576208 |
| Goat Anti-Rat IgG H&L (Alexa Fluor® 488) | Abcam | Cat# ab150157, RRID:AB_2722511 |
| Mouse monoclonal anti-DDX3 | Abcam | Cat# ab196032 |
| Rabbit polyclonal anti-DDX3Y | Invitrogen | Cat# PA5-90055, RRID:AB_2805908 |
| Bacterial and Virus Strains | ||
| One Shot™ TOP10 Chemically Competent E. coli | Invitrogen | Cat# 404003 |
| BL21(DE3)-RIL Competent E. coli | Agilent | Cat# 230204 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Streptavidin-HRP | Cell Signaling Technology | Cat# 3999, RRID:AB_10830897 |
| DDX3X-mCherry recombinant protein | This paper | N/A |
| DDX3Y-mCherry recombinant protein | This paper | N/A |
| MBP-DDX3X recombinant protein | This paper | N/A |
| MBP-DDX3Y recombinant protein | This paper | N/A |
| N-Lauroylsarcosine sodium salt solution | Sigma-Aldrich | Cat# L7414-10ML |
| SUPERase·In™ RNase Inhibitor | Invitrogen | Cat# AM2694 |
| Proteinase K solution (20 mg/mL) | Life Technologies | Cat# AM2548 |
| Lipofectamine 2000 | Thermo Fisher | Cat# 11668019 |
| Pierce streptavidin magnetic beads | Thermo Fisher | Cat# 88816 |
| Agencourt AMPure XP | Beckman Coulter | Cat# A63881 |
| TRIzol | Invitrogen | Cat# 15596026 |
| IPTG | Thermo Fisher | Cat# 34060 |
| DAPI | Sigma-Aldrich | Cat# D9542 |
| Fisher BioReagents™ Bovine Serum Albumin, Heat Shock Treated | Fisher Scientific | Cat# BP1600-100 |
| Polyuridylic acid potassium salt | Sigma-Aldrich | Cat# P9528-10MG |
| Sodium arsenite | Spectrum Chemical | Cat# S-222 |
| Sorbitol solution | Spectrum Chemical | Cat# S-1525 |
| Thapsigargin | ACROS Organics | Cat# AC328570010 |
| CCCP | Alfa Aesar | Cat# AAL06932MC |
| Puromycin | Takara Bio USA | Cat# 631305 |
| MG132 | Sigma-Aldrich | Cat# 474790-1 MG |
| Hydrogen peroxide solution | Sigma-Aldrich | Cat# H1009 |
| Biotin-phenol | Iris Biotech | Cat# LS-3500.1000 |
| Sodium azide | Sigma-Aldrich | Cat# S2002 |
| Sodium ascorbate | Sigma-Aldrich | Cat# A7631 |
| Trolox | Sigma-Aldrich | Cat# 238813 |
| Cycloheximide | Sigma-Aldrich | Cat# C7698 |
| Critical Commercial Assays | ||
| Malachite Green Phosphate Assay Kit | Sigma-Aldrich | Cat# MAK307-1KT |
| Flexi® Rabbit Reticulocyte Lysate System | Promega | Cat# L4540 |
| Luna® Universal One-Step RT-qPCR Kit | NEB | Cat# M3005L |
| Truseq Stranded mRNA Library Prep | Illumina | Cat# 20020594 |
| Dynabeads™ mRNA Purification Kit (for mRNA purification from total RNA preps) | Invitrogen | Cat# 61006 |
| EnzChek™ Phosphate Assay Kit | Thermo Fisher | Cat# E6646 |
| RNA Clean and Concentrator-5 | Zymo Research | Cat# R1016 |
| Deposited Data | ||
| APEX-seq with stress treatment | This paper | GEO: GSE171792 |
| APEX-seq without stress treatment | This paper | GEO: GSE193783 |
| Raw data | This paper; Mendeley Data | https://dx.doi.org/10.17632/9hs5d4fvgd.1 |
| Experimental Models: Cell Lines | ||
| Human: HeLa | ATCC | CCL-2, RRID:CVCL_0030 |
| Human: HEK 293T | ATCC | CRL-3216, RRID:CVCL_0063 |
| Mouse: N2a | ATCC | CCL-131, RRID:CVCL_0470 |
| Oligonucleotides | ||
| Primers for all the recombinant DNA | This paper | Table S3 |
| Recombinant DNA | ||
| pPB DDX3X | This paper | N/A |
| pPB DDX3Y | This paper | N/A |
| pPB DDX3X-IDR1 | This paper | N/A |
| pPB DDX3Y-IDR1 | This paper | N/A |
| pPB DDX3X-Helicae | This paper | N/A |
| pPB DDX3Y-Helicae | This paper | N/A |
| pPB DDX3X-IDR2 | This paper | N/A |
| pPB DDX3Y-IDR2 | This paper | N/A |
| pPB DDX3X-ΔIDR1 | This paper | N/A |
| pPB DDX3Y-ΔIDR1 | This paper | N/A |
| pPB DDX3X-ΔIDR2 | This paper | N/A |
| pPB DDX3Y-ΔIDR2 | This paper | N/A |
| pPB DDX3X-ΔHelicase | This paper | N/A |
| pPB DDX3Y-ΔHelicase | This paper | N/A |
| pPB DDX3XIDR1-DDX3YHelicae-DDX3YIDR2 | This paper | N/A |
| pPB DDX3YIDR1-DDX3XHelicae-DDX3XIDR2 | This paper | N/A |
| pPB DDX3XIDR1-DDX3Xhelicae-DDX3YIDR2 | This paper | N/A |
| pPB DDX3YIDR1-DDX3Yhelicae-DDX3XIDR2 | This paper | N/A |
| pPB DDX3XIDR1-DDX3Yhelicae-DDX3XIDR2 | This paper | N/A |
| pPB DDX3YIDR1-DDX3Xhelicae-DDX3YIDR2 | This paper | N/A |
| pcDNA3 mito-APEX2 | This paper | N/A |
| pPB DDX3X-EGFP | This paper | N/A |
| pPB DDX3Y-EGFP | This paper | N/A |
| pcDNA3 FLAG-DDX3X | This paper | N/A |
| pcDNA3 FLAG-DDX3Y | This paper | N/A |
| pcDNA3 HA-DDX3X | This paper | N/A |
| pcDNA3 HA-DDX3Y | This paper | N/A |
| pET-MCN_His-TEV_V5-DDX3X-mCherry | (Hondele et al., 2019) | N/A |
| pET-MCN_His-TEV_V5-DDX3Y-mCherry | This paper | N/A |
| pET-MCN_His-TEV_V5-DDX3XIDR1-DDX3YHelicae-DDX3YIDR2-mCherry | This paper | N/A |
| pET-MCN_His-TEV_V5-DDX3YIDR1-DDX3XHelicae-DDX3XIDR2-mCherry | This paper | N/A |
| pET-MCN_His-TEV_V5-DDX3XIDR1-DDX3Xhelicae-DDX3YIDR2-mCherry | This paper | N/A |
| pET-MCN_His-TEV_V5-DDX3YIDR1-DDX3Yhelicae-DDX3XIDR2-mCherry | This paper | N/A |
| pET-MCN_His-TEV_V5-DDX3XIDR1-DDX3Yhelicae-DDX3XIDR2-mCherry | This paper | N/A |
| pET-MCN_His-TEV_V5-DDX3YIDR1-DDX3Xhelicae-DDX3YIDR2-mCherry | This paper | N/A |
| pMAL-c2X | (Walker et al., 2010) | Addgene Plasmid #75286, RRID:Addgene_75286 |
| pMAL-c2X-DDX3X | This paper | N/A |
| pMAL-c2X-DDX3Y | This paper | N/A |
| pcDNA5/FRT/TO APEX2-GFP | (Padron et al., 2019) | Addgene Plasmid #129640, RRID:Addgene_129640 |
| pJ4M/TDP-43 | (Hallegger et al., 2021) | Addgene plasmid #104480, RRID:Addgene_104480 |
| pPB APEX2-DDX3X | This paper | N/A |
| pPB APEX2-DDX3Y | This paper | N/A |
| pPB APEX2-EGFP | This paper | N/A |
| Software and Algorithms | ||
| HISAT2 | (Kim et al., 2015) | https://ccb.jhu.edu/software/hisat2/index.shtml, RRID:SCR_015530 |
| DESeq2 | (Love et al., 2014) | https://bioconductor.org/packages/release/bioc/html/DESeq2.html, RRID:SCR_015687 |
| featureCounts | (Liao et al., 2014) | http://bioinf.wehi.edu.au/featureCounts/, RRID:SCR_012919 |
| trimmomatic | (Bolger et al., 2014) | http://www.usadellab.org/cms/index.php?page=trimmomatic, RRID:SCR_011848 |
| R | https://www.r-project.org | https://www.r-project.org, RRID:SCR_000036 |
| Fiji | (Schindelin et al., 2012) | https://imagej.net/Fiji, RRID:SCR_002285 |
| GraphPad Prism | GraphPad Software | https://graphpad.com/scientificsoft, RRID:SCR_002798 |
| Other | ||
| 384-well microscopy plates | Brooks Life Sciences | Cat#4ti-0203 |
| Amicon ultra-15 | EMD Millipore | Cat# UFC901024 |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell culture, transfection and Escherichia coli strains
HeLa, HEK293T, and N2a cells were cultured in DMEM + GlutaMAX (GIBCO) with 10% FBS (GIBCO) and 1% Pen/Strep (Corning) in a humidified incubator with 5% CO2 at 37°C.
For bacterial cell culture, TOP10 and BL21(DE3)-RIL chemically competent bacterial strains grew in lysogeny broth containing the corresponding antibiotics at 200 rpm, 37°C.
The negative control siRNA from Ambion (AM4611) was used as a control siRNA in the knockdown experiments. DDX3X siRNA was purchased from Ambion (Assay ID 145803). Lipofectamine 2000 (Invitrogen) and Lipofectamine RNAiMax (Invitrogen) from Invitrogen were used for plasmids and siRNA transfection, respectively. It took 48 hrs for siRNA knockdown and 24 hrs for plasmid expression.
METHOD DETAILS
Constructs
For recombinant MBP-DDX3X and MBP-DDX3Y protein expression: DNA fragments encoding human DDX3X were PCR-amplified from the HeLa cDNA library, and the human DDX3Y coding sequence was PCR-amplified from pCMV6-DDX3Y (Origene RC226072). These DNA fragments were then inserted into the pMAL-c2x vector (Walker et al., 2010) (Addgene Plasmid #75286) using restriction enzymes BamHI and SalI. A TEV enzyme digestion sequence (protein sequence ENLYFQG; DNA sequence GAAAACCTGTACTTCCAGGGA) was added to the forward primers, and a His-tag sequence (protein sequence HHHHHH; DNA sequence CATCATCACCATCACCAC) was added to the reverse primers. For in vitro LLPS experiments, the pETMCN_His-TEV_V5-DDX3Y-mCherry construct was made by swapping the DDX3Y with the DDX3X in the pETMCN_His-TEV_V5-DDX3X-mCherry (Hondele et al., 2019) using NdeI and BamHI restriction enzymes. For expression of DDX3X and DDX3Y in mammalian cells, DNA fragments encoding the full-length DDX3X and full-length DDX3Y were inserted into the pPB vector with FLAG (DYKDDDDK) and HA (YPYDVPDYA) tags before the N-terminus of the proteins using MfeI and SalI (XhoI was used for digesting the pPB vector) restriction enzymes. The plasmids expressing FLAG or HA single-tagged DDX3X and DDX3Y were made by inserting the DNA fragments encoding full-length DDX3X or full-length DDX3Y into the modified pcDNA3 vector with FLAG or HA tag in frame at the N-terminus.
For the truncation variants of DDX3X and DDX3Y, the following truncations were put into the pPB vector (domain prediction based on PONDR (Xue et al., 2010)): IDR1 of DDX3X (amino acids 1–168); IDR1 of DDX3Y (amino acids 1 – 164); IDR2 of DDX3X (amino acids 580 – 662); IDR2 of DDX3Y (amino acids 579 – 660); helicase domain of DDX3X (amino acids 169 – 579); helicase domain of DDX3Y (amino acids 165 – 578); ΔIDR1 truncation variant of DDX3X (amino acid 169 – 662); ΔIDR1 truncation variant of DDX3Y (amino acids 165 – 660); ΔIDR2 truncation variant of DDX3X (amino acids 1 – 579); ΔIDR2 truncation variant of DDX3Y (amino acids 1 – 578); ΔHelicase truncation variant of DDX3X (amino acids Δ169 – 579); ΔHelicase truncation variant of DDX3Y (amino acids Δ165 – 578). For domain-swap variants of DDX3X and DDX3Y, the following constructs were inserted into the pPB vector: DDX3XIDR1-DDX3YHelicae-DDX3YIDR2; DDX3YIDR1-DDX3XHelicae-DDX3XIDR2; DDX3XIDR1-DDX3Xhelicae-DDX3YIDR2; DDX3YIDR1-DDX3Yhelicae-DDX3XIDR2; DDX3XIDR1-DDX3Yhelicae-DDX3XIDR2; DDX3YIDR1-DDX3Xhelicae-DDX3YIDR2. DDX3X-EGFP, DDX3X-mCherry, DDX3Y-EGFP, and DDX3Y-mCherry with a linker sequence (amino acids GlyGlySerGly) inserted between DDX3X/DDX3Y and EGFP/mCherry were inserted into the pPB vector using MfeI and SalI restriction enzymes. Those domain-swap variants were cloned into the pETMCN_His-TEV_V5-mCherry vector to express mCherry-tagged proteins in E.coli as well.
For APEX2-seq experiments, the DNA fragment encoding APEX2 was PCR amplified from pcDNA5/FRT/TO APEX2-GFP (Addgene, 129640) and fused to DDX3X and DDX3Y using fusion PCR. APEX2-DDX3X and APEX2-DDX3Y were inserted into the pPB vector using MfeI and Sal I restriction enzymes, and APEX2-EGFP was inserted into the pPB vector using MfeI and XhoI restriction enzymes. To validate the biotin labeling efficiency of APEX2, the plasmid pPB mito-APEX2 was constructed. The DNA fragment encoding a mitochondria matrix localization sequence (amino acids MLATRVFSLVGKRAISTSVCVRAH) derived from COX4 was added to the APEX2 forward primer for the PCR reaction. The PCR product was subsequently inserted into the pcDNA3 vector by using BamHI and XhoI restriction enzymes.
All the sequences of the primers used for these clones are summarized in Table S3, and each plasmid was validated by Sanger sequencing.
Protein purification
The pMAL-c2X-DDX3X, pMAL-c2X-DDX3Y plasmids were transformed into Escherichia coli strain BL21-RIL to express the MBP-tagged recombinant proteins. The pETMCN_His-TEV_V5-DDX3X-mCherry (Hondele et al., 2019), pETMCN_His-TEV_V5-DDX3Y-mCherry and other domain-swap variants in the pETMCN_His-TEV_V5-mCherry vectors were transformed into E. coli strain BL21-RIL to express the mCherry-tagged recombinant proteins. The bacteria were cultured in lysogeny broth at 37°C till OD600 nm = 0.8 before administration of 1 mM IPTG at 16°C for 16 hrs. The pellets from 2 L bacterial culture were resuspended with 80 mL binding buffer (25 mM Tris-HCl, pH 7.5, 500 mM NaCl) and sonicated. After centrifuging at 12,000 rpm for 30 min to remove the cell debris, the supernatant was loaded to a Ni-NTA column. Next, 10 column volumes of the binding buffer supplemented with 50 mM imidazole was used as buffer A to wash away the non-specific binding proteins. Another 10 column volumes of a high salt buffer (25 mM Tris-HCl, pH 7.5, and 2 M NaCl) was used to decrease the amount of bound RNAs from DDX3X or DDX3Y. Finally, four column volumes of the binding buffer supplemented with 500 mM imidazole was used to elute the bound proteins. The DDX3X-mCherry and DDX3Y-mCherry recombinant proteins were dialyzed into the storage buffer (25 mM Tris-HCl, pH 7.5, 500 mM NaCl, 10% glycerol, and 2 mM DTT) and the His-tag was cleaved using TEV enzyme simultaneously with the dialysis. Then, the proteins were concentrated using Amicon Ultra-15 (Millipore) tubes before loading to a Superdex 200 column, with buffer (25 mM Tris-HCl, pH 7.5, and 500 mM NaCl) for mCherry tagged proteins and buffer (25 mM Tris-HCl, pH 7.5, 200 mM NaCl) for MBP tagged proteins. MBP-tagged proteins were purified at 4°C, while mCherry-tagged proteins were purified at room temperature. The purity of the proteins was analyzed by SDS-PAGE. Purified proteins were aliquoted, snap-frozen in liquid nitrogen and stored at −80°C. Once thawed, aliquots were never refrozen.
GST-TEV-FUSWT was purified as described (Sun et al., 2011). Briefly, E. coli cells were lysed by sonication on ice in PBS with protease inhibitors (cOmplete, EDTA-free, Roche Applied Science). The protein was purified over Glutathione Sepharose 4 Fast Flow (GE Healthcare) and eluted from the beads using 50 mM Tris-HCl pH 8, 20 mM trehalose, and 20 mM glutathione. Purified protein was snap-frozen in liquid nitrogen and stored at −80°C.
pJ4M/TDP-43 was a gift from Nicolas Fawzi (Addgene plasmid # 104480). TDP-43 was purified as previously described (Hallegger et al., 2021). Briefly, the plasmid was transformed into BL21(DE3) RIL E. coli. Cells were harvested by centrifugation and lysed by lysozyme (1 mg/mL) and sonication in wash buffer (20 mM Tris-HCl pH 8.0, 1 M NaCl, 10 mM imidazole, 10% glycerol, 1 mM DTT, 5 μM Pepstatin A, 100 μM PMSF, and cOmplete, EDTA-free, Roche Applied Science protease inhibitors). The protein was purified over Ni-NTA agarose beads (QIAGEN) and eluted from the beads using elution buffer (wash buffer except with 300 mM imidazole rather than 10 mM imidazole). The protein was further purified over amylose resin (NEB) and eluted with 20 mM Tris-HCl pH 8.0, 1 M NaCl, 10 mM imidazole, 10% glycerol, 1 mM DTT, 5 μM Pepstatin A, 100 μM PMSF, and 10 mM maltose. The protein was concentrated, snap-frozen in liquid nitrogen, and stored at −80°C.
Differential scanning fluorimetry assay
Purified MBP-DDX3X and MBP-DDX3Y proteins were diluted to 0.25 mg/mL. 19 μL of each protein was transferred to a well of a 384-well plate, and 1 μL of 5-fold SYPRO Orange (Thermo Fisher) was added to each well. The plate was sealed and spun at 3,600 g for 2 min. The fluorescent signal at 570 nm was collected using a RT-qPCR machine with the temperature ramping from 20 to 95°C. The data were analyzed using DSF World (Wu et al., 2020).
In vitro LLPS assay
The in vitro LLPS was set up at room temperature with total volume of 20 μL in PCR tube. Proteins were diluted to 100 μM (DDX3X-mCherry and DDX3Y-mCherry) or 75 μM (DDX3X-mCherry, DDX3Y-mCherry and all the domain-swap mutants) using the storage buffer. 10 μL buffer (25 mM Tris-HCl, pH 7.5, 500 mM NaCl) and 2 μL diluted protein were transferred to the PCR tube. Then, 8 μL water was added to the tube to observe the LLPS of protein alone; 2 μL polyU-RNA (2 mg/mL, dissolved in water) and 6 μL water were added to the tube to observe the LLPS of protein with RNA; 2 μL polyU-RNA (2 mg/mL, dissolved in water), 2 μL ATP buffer (40 mM ATP and 50 mM MgCl2) or UTP buffer (40 mM UTP and 50 mM MgCl2) and 4 μL water were added to the tube to observe the LLPS of protein with RNA and ATP or UTP. To observe the LLPS of different combinations of DDX3X-mCherry and DDX3Y-mCherry, the proteins were diluted to the proper concentrations. Then 10 μL buffer, 2 μL diluted protein and 8 μL water were transferred to a PCR tube. The mixtures were mixed by pipetting and transferred to 384-well glass-bottomed plate. After incubating at room temperature for 1 hr, the plate was spun at 100 g for 1 min. Then the images were taken using a Zeiss LSM 880 confocal microscope under a 63 × oil lens.
Immunofluorescence cell staining
Cells were passaged to a 6-well plate with a coverslip in each well and cultured overnight. The cells were washed once in PBS and then fixed using 4% paraformaldehyde in PBST (PBS with 0.05% Tween-20) at room temperature for 15 min. Then, the cells were washed twice by PBST and permeabilized by 0.5% Triton at room temperature for 20 min. After being washed once by PBST, the cells were blocked with 1% BSA in PBST at room temperature for 30 min. Then, the blocking solution was replaced with 1 mL blocking solution supplemented with desired primary antibodies (at 1:1000 dilution) and incubated at room temperature for 1 hr or 4 °C overnight. After 4 washes with PBST, the corresponding Alexa Fluor-conjugated secondary antibodies were applied (1:1,000 diluted in the blocking solution) and incubated at room temperature for 1 hr. After being washed three times by PBST, the cells were incubated with 0.5 μg/mL DAPI for 1 min. After 4 times PBST washes, an antifade reagent (Invitrogen) was used to mount the slides. The images were taken using a Leica TCS SP8 confocal microscope. The “analyze particles tool” in Fiji (Schindelin et al., 2012) was utilized to quantify the sizes of the stress granules in mammalian cells in about 50 different cells per condition. The sizes of the stress granule in cells were analyzed by two researchers independently.
Fluorescence recovery after photobleaching
The FRAP assays were conducted using the bleaching module of the Zeiss LSM 880 confocal microscope for DDX3X and DDX3Y droplets individually. The 488 nm laser was used to bleach the EGFP signal, and the 561 nm laser was used to bleach the mCherry signal. Bleaching was focused on a circular region of interest (ROI) using 100% laser power, and time-lapse images were collected afterward. A same-sized circular area away from the bleaching point was selected as an unbleached control. The fluorescence intensity was directly measured in the Zen software. The values were reported as relative to pre-bleaching time points. GraphPad Prism was used to plot the data. The halftime for each replicate was calculated using the following formula: y=a•(1-exp(−b•x)) + c, in which a is the slow recovery fraction, c is the rapid diffusion fraction, and b is the recovery rate. The halftime is ln2 / b, and a mobile fraction is a + c. The two-tail t-test was used to calculate the p-values. For FRAP of the live cells, cells expressing DDX3X-EGFP or DDX3Y-EGFP were cultured in 35 mm poly-D-lysine coated glass-bottomed dishes (Mattek). Before taking the images, cells were treated with 500 μM sodium arsenite for 1 hr in the FluoroBrite DMEM medium with 10% FBS. A 20 × lens was used at zoom scale 6. For FRAP of in vitro LLPS, a 63 × oil lens was used.
Formation of stress granules in cells
Cells were seeded in a 6-well plate and cultured overnight. The following stressors: 500 μM sodium arsenite (1 hr), 1 M sorbitol (1 hr), 50 μM thapsigargin (1 hr), 40 μg/mL puromycin (3 hrs), and 10 μM MG132 (3 hrs) were added to the cell culture media (DMEM + 10% FBS) each in a separate well. For the stress condition of 60 μM CCCP, CCCP was added to glucose-free DMEM with 10% FBS. After the treatment, cells were fixed and subjected to the immunofluorescence imaging procedure detailed above.
Sequence alignment
The amino acid sequences of DDX3X and DDX3Y were downloaded from CCDS Database and aligned using Clustal Omega. The alignment results were redrawn using ESPript 3.0 (Robert and Gouet, 2014). The sequences were used to predict the natural disordered regions by PONDR (Xue et al., 2010), prion-like amino acid regions by PLAAC (Lancaster et al., 2014) and LLPS propensity by catGRANULE (Mitchell et al., 2013).
Turbidity assay
DDX3X-mCherry and DDX3Y-mCherry proteins were diluted to 10, 20, 30, 50, 75, 100, 125 and 150 μM using the storage buffer. 15 μL buffer (25 mM Tris-HCl, pH 7.5, 200 mM NaCl), 3 μL diluted protein were transferred to the PCR tube, 3 μL polyU-RNA (2 mg/mL, dissolved in water), and 9 μL water were mixed in a PCR tube. After incubating for 20 min at room temperature, the mixtures were transferred to a 384-well black plate with a clear flat bottom. The turbidity was measured by using a Tecan plate reader at OD600 nm. Then, the solution was transferred to a clean Eppendorf tube, and spun at 16,000 g for 2 min. The supernatant was used to measure protein concentration using Bradford method.
Construction of DOX-inducible cell lines
5 μg of the lentiviral vectors expressing mClover3-FUS, 2.5 μg VSVG plasmid, and 3.75 μg pPAX2 plasmid were co-transfected to 100% confluent HEK 293T cells in 6-well plate with 40 μL polyethylenimine. The medium was changed after 6 to 8 hrs. 500 μL medium containing the virus was collected twice a day and 500 μL fresh medium was replenished at each time. The virus was collected in a total three-day period, and then spun down at 3000 rpm for 5 min at room temperature. The HeLa cells were infected with lentivirus with 8 μg/mL polybrene. After induction with 100 ng/mL of doxycycline (DOX) for two days, the cells were subjected to cell sorting. Western blots were performed to validate the expression of mClover3-FUS by using anti-GFP antibody.
Colocalization of DDX3X/Y with FUS in cells
The DOX-inducible HeLa cells expressing mClover3-FUS were seeded in a 6-well plate with 100 ng/mL of DOX and a coverslip in the well. 1 μg of empty vector, DDX3X-pPB, or DDX3Y-pPB were transfected to those cells respectively using lipofectamine 2000. 24 hrs later, sodium arsenite was used to each well at a final concentration of 500 μM for 1 hr. The cells were then subjected to the immunofluorescence cell stain protocol as discussed above.
Cycloheximide chase assay
The cycloheximide (CHX) chase assay was performed to explore the half-life of DDX3X and DDX3Y proteins in cells. HeLa cells were seeded in 6-well plate and transfected with 1 μg of DDX3X-pPB and DDX3Y-pPB respectively. 24 hrs later, CHX was added to the cells at a final concentration of 100 μg/mL and incubated for different time intervals. Then, the cells were collected and lysed with RIPA buffer (25 mM Tris-HCl, pH 7.4, 150 mM NaCl, 2 mM EDTA, and 1% NP-40). The protein concentrations were measured using a Bradford assay and the same amount of cell lysate was used in Western blot analyses. The intensity for each band was quantified using Fiji. The intensity for each DDX3X and DDX3Y band was normalized to the corresponding GAPDH intensity firstly, and then for each replicate at different time points were normalized to time 0 hr. The data were plotted in Prism and an exponential decay formula was used to determine the half-life of DDX3X and DDX3Y. The degradation rate Kdecay was estimated by ln(At/A0) = − Kdecay t where At and A0 stand for the quantity at time t and time 0. Thus, the half-life (t1/2), when 50% of the protein is decayed is described by t1/2 = ln2 / Kdecay.
Puromycin incorporation assay
HeLa cells were seeded in a 6-well plate with a coverslip in it, and then transfected with 1 μg of pPB-DDX3X and pPB-DDX3Y respectively. After 24 hrs, a final concentration of 500 μM sodium arsenite was added to the DMEM and incubated for 5 min to trigger stress granule formation. Then, the medium was replaced by fresh DMEM medium with puromycin (1 μg/mL). After incubating for 30 min in a humidified incubator with 5% CO2 at 37°C, the cells were then washed with 1 × PBS and subjected to the immunofluorescence cell staining protocol as described above using anti-puromycin and anti-FLAG antibodies.
Malachite green ATPase assay
ATPase measurements were taken using the Malachite Green Phosphate Assay Kit according to the manufacturer’s instructions. Briefly, 1.0 μM MPB-DDX3X or MBP-DDX3Y was incubated with 100 ng/μL total RNA extracted from HeLa cells for 15 min in the reaction buffer (25 mM Tris-HCl, pH 8, 200 mM NaCl, 1 mM DTT, and 2 mM MgCl2) before the addition of 2 mM ATP. The reaction was incubated at room temperature for 30 min. Then the reaction was quenched by the addition of malachite green mixture and left for an additional 30 min to develop the color. The samples were then loaded into a clear-bottom 384-well plate and absorbance at OD600 nm was measured. Values were converted from absorbance units to μM free phosphate using a standard curve generated with the kit’s phosphate standard. Background values (free phosphates detected from reactions lacking RNA) were subtracted from the value from reactions with RNA. Data were plotted in Prism. Significance was calculated using Student’s two-tailed t test.
Continuous ATPase assay
ATPase measurements were taken using the EnzChek Phosphate Assay Kit (ThermoFisher) as previously described (Song and Ji, 2019). Briefly, MBP-DDX3X or MBP-DDX3Y was titrated at the indicated concentrations into 500 μL reactions containing 40 mM Tris-HCl pH 7.5, 50 mM NaCl, 0.5 mM MgCl2, 2 mM DTT, 0.5 units Super RNaseIN, 0.01% NP-40 substitute and 100 nM annealed duplex RNA (sequence in Table S3). After 10 min of incubation at room temperature, the reaction was started by adding 2 mM magnesium ATP. The reaction progress was monitored over five min at 30 sec intervals as absorbance at 360 nm. Absorbance values were the converted to μM/min using a standard curve and initial rates were plotted and fit to Hill kinetics using Prism.
smFRET
PEG-passivated slides were prepared according to previously published protocols with minor modifications (Jamiolkowski et al., 2017). Briefly, glass coverslips and slides were sonicated at 40°C twice with acetone for 15 min followed by sonication with methanol (25 min), 1 M KOH (40 min), and ethanol (15 min). Plasma cleaning was then used to remove any remaining contaminants from the surfaces. Plasma cleaned slides and coverslips were then incubated in a solution composed of 1.4 mL of 3-aminopropyltriethoxysilane (APTES), 2.3 mL of glacial acetic acid, and 46 mL of methanol overnight at room temperature. Silicated slides and coverslips were then incubated with polyethylene glycol (PEG, Laysan Bio, Inc., containing 20% (w/w) mPEG succinimidyl valerate, MW 2000 and 1% biotin-PEG-SC, MW 2000) in 0.1 M sodium bicarbonate (pH 8.3) for four hrs followed by an additional incubation overnight in a humidifying chamber. Slides and coverslips were then washed with MilliQ water and used to construct flow chambers for single molecule experiments with double-sided sticky tape. Double stranded RNA with a 5’ overhang was obtained as follows: 5’-biotin/ACCGCUGCCGUCGCUCCG/AlexF647N/−3’; and 5’-/Cy3/UUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUCGGAGCGACGGCAGCGGU-3’ were ordered from IDT; the two strands were annealed by heating the sample at 65°C for 5 min, then slowly cooling the sample to room temperature over 3 hrs. 2 mM protocatechuic acid (PCA), 0.05 uM protocatechuate-3,4,dioxygenase, 2.45 mM 6-hydroxy-2,5,7,8- tetramethylchromane-2-carboxylic (Trolox), 1 mM cyclooctatetraene (COT), and 1 mM 4-nitrobenzyl alcohol (NBA) were added to the imaging buffer (125 mM NaCl and 50 mM Tris-HCl pH 7.5). 70 pM biotinylated, fluorescent dsRNA was added to the flow cell and incubated for 6 min. Unbound RNA was washed out and recording either commenced or DDX3X or DDX3Y were first added as indicated in the text. The TIRF microscope, optics and camera were used to record smFRET as in previous report (Jamiolkowski et al., 2017). All FRET measurements were carried out at room temperature (~23°C) using a frame interval of 100 ms with alternating 532 nm/640 nm laser excitation (ALEX; effective frame interval 200 ms). Data analyses were carried out using custom scripts written in either Python or Java. Background-corrected fluorescence intensities and FRET distributions were corrected for differential quantum yield, differential detector sensitivity, direct excitation of the A647 by the 532 nm laser and leakage of Cy3 fluorescence into the acceptor detector channel as in previous report (Hellenkamp et al., 2018; Jamiolkowski et al., 2017). Only recordings exhibiting a single step photobleaching event in the direct acceptor excitation (ALEX) channel were further analyzed and included in FRET distributions.
RNA isolation
A Direct-zol RNA MicroPrep kit (Zymo Research) was used to isolate total RNA following the manufacturer’s instructions. DNA digestion was performed on the column at room temperature for 15 min to remove the DNA contamination of the extracted RNA. For a larger scale of RNA purification, total RNA was purified by TRIzol reagent (Invitrogen), following the manufacturer’s instruction.
RT-qPCR
RT-qPCR was performed using a Luna Universal One-Step RT-qPCR kit (NEB) to assess the relative abundance of RNA in each sample. All samples used for RT-qPCR were treated with DNase I to remove possible DNA contamination. The primers used for RT-qPCR are listed in Table S3.
Cellular APEX labeling
To enable APEX labeling in mammalian cells, we followed a previously established protocol (Fazal et al., 2019). Briefly, the cells were treated with or without 500 μM sodium arsenite in DMEM media for 30 min. Then, the DMEM media with 500 μM sodium arsenite was replaced with DMEM media supplemented with 500 μM sodium arsenite and 500 mM biotin-phenol for another 30 min. Next, H2O2 (Sigma-Aldrich) was added to each cell culture dish at 1 mM final concentration for exactly 1 min with gentle agitation. To stop the labeling, the culture media was removed, and the quenching solution (10 mM sodium ascorbate, 10 mM sodium azide, and 5 mM Trolox in PBS) was immediately used to wash the cells three times. Finally, 1 mL quenching solution was applied to cover the cells. Cells were then collected with a cell scraper. The unlabeled control samples were prepared in parallel under the same procedure as aforementioned, without the addition of the H2O2.
Validation of APEX labeling
To validate APEX labeling in cells, after the cellular labeling reaction, the cells were lysed using RIPA buffer (25 mM Tris-HCl, pH 7.4, 150 mM NaCl, 2 mM EDTA, and 1% NP-40) supplemented with 10 mM sodium ascorbate, 10 mM sodium azide, 5 mM Trolox, and proteinase inhibitor on ice for 15 min. After centrifugation, the supernatant of the cell lysate was loaded on an SDS-PAGE gel and transferred to a PVDF membrane (Millipore) by a semi-dry transfer instrument. The membrane was blocked with 5% BSA in PBST at 4 °C overnight. 1:20,000 diluted streptavidin-HRP antibody (cell signaling) in 5% BSA was used at room temperature for 1 hr to detect the biotinylated proteins. After washing three times with PBST, the membrane was visualized by ECL Western Blotting Detection Kit (Thermo Fisher). The endogenous biotinylated proteins were also visible at 130, 75, and 72 kDa in both the labeled and unlabeled cells (as expected).
Purification and sequencing biotinylated RNA
After the cellular APEX-labeling, total RNA was extracted using TRIzol following the manufacturer’s instruction. To enrich biotinylated RNAs, Pierce streptavidin magnetic beads (Thermo Fisher) were used (20 μL beads per 50 μg RNA). The beads were washed 3 times with 500 μL B&W buffer (5 mM Tris-HCl, pH 7.5, 0.5 mM EDTA, 1 M NaCl, and 0.1% TWEEN 20), followed by 2 times with 500 μL solution A (0.1 M NaOH and 50 mM NaCl), and once with 500 μL solution B (100 mM NaCl). The beads were then suspended in 125 μL solution B. 50 μg RNA diluted in 125 μL water was mixed with the beads and incubated at 4 °C for 2 hrs with rotation. Then, the beads were placed on a magnetic stand to remove the solution and washed 3 times with 500 μL B&W buffer. Finally, the beads were resuspended in 54 μL water. To release the biotinylated RNA, 33 μL proteinase buffer (3 × PBS, 6% N-Lauryl sarcosine sodium solution (Sigma-Aldrich), 30 μM EDTA, and 15 μM DTT) was added to the beads with 10 μL proteinase K (Thermo Fisher) and 5 μL RNase inhibitor. The beads were then incubated at 42 °C for 1 hr and 55°C for 1 hr on a shaker at 600 rpm. The RNA was then purified using a RNA Clean and Concentrator 5 kit (Zymo Research) following the manufacturer’s instruction. The resulting RNA was then used for RNA-seq library construction using a TruSeq Stranded mRNA Kit (Illumina). The concentrations for all the libraries were determined by KAPA Library Quantification Kit (KAPA Biosystems) following the manufacturer’s protocol and subjected to Next-generation high-throughput sequencing using an Illumina NextSeq 550 with a single-end 75-bp read length.
Validation of the APEX labeling of RNA
To test the specificity of APEX2 labeling, APEX2 was fused with a mitochondrial localization signal (MLATRVFSLVGKRAISTSVCVRAH, derived from COX4) at its N terminus. The localization of the fused protein was validated by immunofluorescence. Total RNA was first extracted from the labeled and unlabeled cells, which was then followed by enrichment of biotinylated RNA using the streptavidin magnetic beads as described above. To test for the RNA enrichment, primers against mitochondria-translated transcripts ND1 and ND2, and cytoplasm translated transcripts GAPDH and TRMT10A were designed and listed in Table S3. RT-qPCR was performed in the labeled and unlabeled controls. The ratios of RNA recovered in the labeled samples relative to unlabeled controls were calculated.
High throughput data analysis
The high throughput sequencing reads were adaptor and quality trimmed using Trimmomatic (Bolger et al., 2014) with the following command: trimmomatc SE -phred33 ILLUMINACLIP:TruSeq3-SE.fa:2:30:10 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:30. Then, the reads were aligned to the GRCh38 human genome reference using HISAT2(Kim et al., 2015). The default parameters were used. Aligned reads were quantified using featureCounts (Liao et al., 2014). Read counts were further analyzed using DESeq2 (Love et al., 2014). Gene ontology (GO) analysis was carried out with the Metascape (Zhou et al., 2019).
In Vitro translation
In vitro translation assays were performed using the Flexi Rabbit Reticulocyte Lysate System (Promega). Each reaction (25 μL) contains 10 μL rabbit reticulocyte lysate, 0.25 μL amino acid mixture minus leucine (1 mM), 0.25 μL of amino acid mixture minus methionine (1 mM), 1 μL Mg(OAc)2, 0.25 μL luciferase mRNA (1 mg/mL), 0.5 μL RNase inhibitor, 0.25 μL DTT (1 M), and 12.5 μL of different concentrations of DDX3X-mCherry, DDX3Y-mCherry, or buffer (25 mM HEPES-KOH, pH 7.4, 150 mM KCl, and 2 mM DTT). Assembled reactions were incubated at 30°C for 90 mins. A standard reaction containing 75 μL of the luciferase substrate mixed with 5 μL of the unpurified translation mixture in a white 96-well plate. The luminescence was measured using a luminometer (Promega). To observe any LLPS in the lysate, the lysate from the translation assays was transferred to 384-well glass-bottomed plate. Then, the plate was spun at 100 g for 1 min. The images were taken using a Zeiss LSM 880 confocal microscope under a 63 × oil lens.
FUS aggregation assay
First, 0, 0.25, or 0.5 μM MBP-TEV-DDX3X or MBP-TEV-DDX3Y was incubated with 1 μg TEV protease for 30 min at room temperature. Then, 1 μM GST-TEV-FUSWT (or an equal volume of elution buffer) was added to the reaction and turbidity was used to assess aggregation by measuring absorbance at 395 nm in a Tecan plate reader. Readings from DDX3X or DDX3Y alone were subtracted from the appropriate FUS conditions. Area under the curve was used to compare the extent of aggregation for each condition (GraphPad Prism).
TDP-43 aggregation assay
Firstly, 0, 0.25, or 0.5 μM MBP-TEV-DDX3X or MBP-TEV-DDX3Y in buffer (200 mM NaCl, 25 mM Tris-HCl pH 8.0) was incubated in TDP-43 assay buffer (150 mM NaCl, 20 mM HEPES-NaOH pH 7.0, 1mM DTT) with 0.5 μg TEV protease for 30 minutes at room temperature. Turbidity was assessed by measuring absorbance at 395 nm in a Tecan plate reader. TDP-43 was buffer exchanged into TDP-43 assay buffer (Bio-Rad Micro Bio-Spin Chromatography Columns, following manufacturer’s instructions) and concentration was determined via NanoDrop. Turbidity measurements were paused after 30 minutes in order to add 4.0 μM TDP-43 (or an equal volume of TDP-43 assay buffer) to the reaction. Then, turbidity measurements were resumed for an additional 16 hours. The data was standardized by subtracting out the initial reading at t = 1 min. from each respective condition. Values from conditions with DDX3X or DDX3Y alone were then subtracted from the appropriate conditions with TDP-43. Area under the curve analysis was used to compare the extent of aggregation for each condition. The t1/2 of aggregation was determined by performing a nonlinear regression (asymmetric sigmoidal) on the data starting after TDP-43 addition (t = 31 min.) (GraphPad Prism).
Protein quantification and Western blot
Protein concentrations of the samples were calculated using the Bradford Assay (Bio-Rad). Protein samples were boiled at 95°C in Laemmli sample buffer for 10 min. After brief centrifugation, the samples were loaded onto SDS-PAGE gels. After running at 180 V for 1 hr, the gels were transferred to the PVDF membranes (Millipore) by semi-dry transfer apparatus at 20 V for 50 min. Then, the PVDF membranes were blocked with 5% milk or BSA in 1 × PBST for 30 mins at room temperature or 4°C overnight. The membranes were then incubated in 3% milk or BSA in 1 × PBST containing the corresponding primary antibodies overnight at 4°C. After washing three times with 1 × PBST, the horseradish peroxidase (HRP)-conjugated secondary antibodies (1:20,000) in 1% of milk were applied and incubated at room temperature for 1 hr. After washing three times with 1 × PBST, the membranes were visualized using ECL Western Blotting Detection Kit (Thermo Fisher).
QUANTIFICATION AND STATISTICAL ANALYSIS
Images were analyzed using Fiji. All data are presented as the mean ± standard error of mean (s.e.m.) or standard deviation (s.d.) from the independent determinations. The statistical analyses were performed using the GraphPad Prism (GraphPad Software, Inc,; La Jolla, CA, USA). Differences of means were tested for statistical significance with unpaired two-tailed Student’s t-test. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; n.s. means p > 0.05.
Supplementary Material
Table S2. Summary of DDX3X-specific, DDX3Y-specific, and overlapped transcripts from APEX-seq, related to Figure 5.
Table S3. Summary of the cloning and RT-qPCR primers used in this study, related to Figures 1, 2, 3, 4, 5 and 6.
Movie S1. Time-lapse imaging video of FRAP experiment with DDX3X-mCherry LLPS, related to Figure 1.
Movie S2. Time-lapse imaging video of FRAP experiment with DDX3Y-mCherry LLPS, related to Figure 1.
Movie S3. A biological replicate of time-lapse imaging video of FRAP experiment with DDX3X-mCherry LLPS, related to Figures 1 and S1.
Movie S4. A biological replicate of time-lapse imaging video of FRAP experiment with DDX3Y-mCherry LLPS, related to Figures 1 and S1.
Movie S5. Time-lapse imaging video of FRAP experiments with DDX3X-EGFP Stress Granule in HeLa Cells, related to Figure 2.
Movie S6. Time-lapse imaging video of FRAP experiments with DDX3Y-EGFP Stress Granule in HeLa Cells, related to Figure 2.
Highlights:
The N-terminal IDR of DDX3Y more strongly promotes condensation than DDX3X.
Slower ATPase activity of DDX3Y contributes to weaker dissolution of condensates.
Stronger phase separation of DDX3Y than DDX3X leads to more translation repression.
DDX3Y condensates enhance aggregation of FUS more strongly than DDX3X.
ACKNOWLEDGEMENTS
This work was supported by the National Institutes of Health (R35GM133721 to K.F.L., R35GM133721–03S1 to A.Y., T32GM132039 to A.Y. and M.C.O., T32GM008275 to K.E.C. and C.M.F., F31NS111870. to C.M.F., R01GM099836, R21AG061784, and R21AG065854 to J.S., and R35GM118139 to Y.E.G.). K.F.L. is supported by the American Cancer Society (RSG-22–064-01-RMC). J.S. is supported by grants from Target ALS, the Amyotrophic Lateral Sclerosis Association, the Office of the Assistant Secretary of Defense for Health Affairs through the Amyotrophic Lateral Sclerosis Research Program (W81XWH-20–1-0242), the G. Harold and Leila Y. Mathers Foundation, and Sanofi. We thank Dr. Bede Portz for the pMAL-c2X-DDX3X plasmid, Kollin Schultz for his help with DSF experiments, Dr. Matthew Kayser for sharing the Leica SP8 confocal microscope, Dr. Brain Capell for sharing the Nextseq550 sequencer, Dr. Karsten Weis for sharing the pKW4557 (DDX3X-mCherry) plasmid, and Dr. Ophir Shalem for sharing the plasmid expressing mClover3-FUS and mClover3-TDP-43 with us. We thank Drs. Ronen Marmorstein, Hillary Nelson, Gregory Van Duyne, and Kristen Lynch for their constructive discussions and for editing this manuscript.
Footnotes
DECLARATION of INTERESTS
J.S. is a consultant for Dewpoint Therapeutics, Vivid Sciences, Korro Bio, Neumora, and ADRx.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ash PE, Vanderweyde TE, Youmans KL, Apicco DJ, and Wolozin B (2014). Pathological stress granules in Alzheimer’s disease. Brain Res. 1584, 52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baradaran-Heravi Y, Van Broeckhoven C, and van der Zee J (2020). Stress granule mediated protein aggregation and underlying gene defects in the FTD-ALS spectrum. Neurobiol. Dis. 134, 104639. [DOI] [PubMed] [Google Scholar]
- Beckham C, Hilliker A, Cziko AM, Noueiry A, Ramaswami M, and Parker R (2008). The DEAD-box RNA helicase Ded1p affects and accumulates in Saccharomyces cerevisiae P-bodies. Mol. Biol. Cell 19, 984–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellott DW, Hughes JF, Skaletsky H, Brown LG, Pyntikova T, Cho TJ, Koutseva N, Zaghlul S, Graves T, Rock S, et al. (2014). Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature 508, 494–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentmann E, Neumann M, Tahirovic S, Rodde R, Dormann D, and Haass C (2012). Requirements for stress granule recruitment of fused in sarcoma (FUS) and TAR DNA-binding protein of 43 kDa (TDP-43). J. Biol. Chem. 287, 23079–23094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, and Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryk AH, and Wisniewski JR (2017). Quantitative analysis of human red blood cell proteome. J. Proteome Res. 16, 2752–2761. [DOI] [PubMed] [Google Scholar]
- Buchan JR, and Parker R (2009). Eukaryotic stress granules: the ins and outs of translation. Mol. Cell 36, 932–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinal T, Bergeron KF, Soret R, Souchkova O, Faure C, Guillon A, and Pilon N (2020). Male-biased aganglionic megacolon in the TashT mouse model of Hirschsprung disease involves upregulation of p53 protein activity and Ddx3y gene expression. PLoS Genet. 16, e1009008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Chan CH, Chen CM, Tsai YS, Tsai TY, Wu Lee YH, and You LR (2016). Targeted inactivation of murine Ddx3x: essential roles of Ddx3x in placentation and embryogenesis. Hum. Mol. Genet. 25, 2905–2922. [DOI] [PubMed] [Google Scholar]
- Cheng W, Wang S, Zhang Z, Morgens DW, Hayes LR, Lee S, Portz B, Xie Y, Nguyen BV, Haney MS, et al. (2019). CRISPR-Cas9 screens identify the RNA helicase DDX3X as a repressor of C9ORF72 (GGGGCC)n repeat-associated non-AUG translation. Neuron 104, 885–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotton AM, Price EM, Jones MJ, Balaton BP, Kobor MS, and Brown CJ (2015). Landscape of DNA methylation on the X chromosome reflects CpG density, functional chromatin state and X-chromosome inactivation. Hum. Mol. Genet. 24, 1528–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditton HJ, Zimmer J, Rajpert-De Meyts E, and Vogt PH (2004). The AZFa gene DBY (DDX3Y) is widely transcribed but the protein is limited to the male germ cells by translation control. Hum. Mol. Genet. 13, 2333–2341. [DOI] [PubMed] [Google Scholar]
- Elbaum-Garfinkle S, Kim Y, Szczepaniak K, Chen CC, Eckmann CR, Myong S, and Brangwynne CP (2015). The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc. Natl. Acad. Sci. U.S.A. 112, 7189–7194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazal FM, Han S, Parker KR, Kaewsapsak P, Xu J, Boettiger AN, Chang HY, and Ting AY (2019). Atlas of subcellular RNA localization revealed by APEX-Seq. Cell 178, 473–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes N, Nero L, Lyons SM, Ivanov P, Mittelmeier TM, Bolger TA, and Buchan JR (2020). Stress granule assembly can facilitate but is not required for TDP-43 cytoplasmic aggregation. Biomolecules 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti MT, Iulita MF, Cavedo E, Chiesa PA, Schumacher Dimech A, Santuccione Chadha A, Baracchi F, Girouard H, Misoch S, Giacobini E, et al. (2018). Sex differences in Alzheimer disease - the gateway to precision medicine. Nat. Rev. Neurol. 14, 457–469. [DOI] [PubMed] [Google Scholar]
- Floor SN, Condon KJ, Sharma D, Jankowsky E, and Doudna JA (2016). Autoinhibitory interdomain interactions and subfamily-specific extensions redefine the catalytic core of the human DEAD-box protein DDX3. J. Biol. Chem. 291, 2412–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura K, Sasaki AT, and Anderson P (2012). Selenite targets eIF4E-binding protein-1 to inhibit translation initiation and induce the assembly of non-canonical stress granules. Nucleic Acids Res. 40, 8099–8110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao K, Oerlemans R, and Groves MR (2020). Theory and applications of differential scanning fluorimetry in early-stage drug discovery. Biophys. Rev. 12, 85–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, and Anderson P (2004). Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell 15, 5383–5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey AK, Naqvi S, Chmatal L, Chick JM, Mitchell RN, Gygi SP, Skaletsky H, and Page DC (2020). Quantitative analysis of Y-Chromosome gene expression across 36 human tissues. Genome Res. 30, 860–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozdecka M, Meduri E, Mazan M, Tzelepis K, Dudek M, Knights AJ, Pardo M, Yu L, Choudhary JS, Metzakopian E, et al. (2018). UTX-mediated enhancer and chromatin remodeling suppresses myeloid leukemogenesis through noncatalytic inverse regulation of ETS and GATA programs. Nat. Genet. 50, 883–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Kim HJ, Wang H, Monaghan J, Freyermuth F, Sung JC, O’Donovan K, Fare CM, Diaz Z, Singh N, et al. (2018). Nuclear-import receptors reverse aberrant phase transitions of RNA-binding proteins with prion-like domains. Cell 173, 677–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallegger M, Chakrabarti AM, Lee FCY, Lee BL, Amalietti AG, Odeh HM, Copley KE, Rubien JD, Portz B, Kuret K, et al. (2021). TDP-43 condensation properties specify its RNA-binding and regulatory repertoire. Cell 184, 4680–4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison AF, and Shorter J (2017). RNA-binding proteins with prion-like domains in health and disease. Biochem. J. 474, 1417–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellenkamp B, Schmid S, Doroshenko O, Opanasyuk O, Kuhnemuth R, Rezaei Adariani S, Ambrose B, Aznauryan M, Barth A, Birkedal V, et al. (2018). Precision and accuracy of single-molecule FRET measurements-a multi-laboratory benchmark study. Nat. Methods 15, 669–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hondele M, Sachdev R, Heinrich S, Wang J, Vallotton P, Fontoura BMA, and Weis K (2019). DEAD-box ATPases are global regulators of phase-separated organelles. Nature 573, 144–148. [DOI] [PubMed] [Google Scholar]
- Iossifov I, O’Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, Stessman HA, Witherspoon KT, Vives L, Patterson KE, et al. (2014). The contribution of de novo coding mutations to autism spectrum disorder. Nature 515, 216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iserman C, Desroches Altamirano C, Jegers C, Friedrich U, Zarin T, Fritsch AW, Mittasch M, Domingues A, Hersemann L, Jahnel M, et al. (2020). Condensation of Ded1p promotes a translational switch from housekeeping to stress protein production. Cell 181, 818–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Wheeler JR, Walters RW, Agrawal A, Barsic A, and Parker R (2016). ATPase-modulated stress granules contain a diverse proteome and substructure. Cell 164, 487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamiolkowski RM, Chen C, Cooperman BS, and Goldman YE (2017). tRNA fluctuations observed on stalled ribosomes are suppressed during ongoing protein synthesis. Biophys. J. 113, 2326–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi RN, Stadler C, Lehmann R, Lehtio J, Tegner J, Schmidt A, and Vesterlund M (2019). TcellSubC: an atlas of the subcellular proteome of human T cells. Front. Immunol. 10, 2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamelgarn M, Chen J, Kuang L, Arenas A, Zhai J, Zhu H, and Gal J (2016). Proteomic analysis of FUS interacting proteins provides insights into FUS function and its role in ALS. Biochim. Biophys. Acta 1862, 2004–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalfallah Y, Kuta R, Grasmuck C, Prat A, Durham HD, and Velde CV (2018). TDP-43 regulation of stress granule dynamics in neurodegenerative disease-relevant cell types. Sci. Rep. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khong A, Matheny T, Jain S, Mitchell SF, Wheeler JR, and Parker R (2017). The stress granule transcriptome reveals principles of mRNA accumulation in stress granules. Mol. Cell. 68, 808–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, and Salzberg SL (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Tsang B, Vernon RM, Sonenberg N, Kay LE, and Forman-Kay JD (2019). Phospho-dependent phase separation of FMRP and CAPRIN1 recapitulates regulation of translation and deadenylation. Science 365, 825–829. [DOI] [PubMed] [Google Scholar]
- Kim Y, and Myong S (2016). RNA remodeling activity of DEAD box proteins tuned by protein concentration, RNA length, and ATP. Mol. Cell 63, 865–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimball SR, Horetsky RL, Ron D, Jefferson LS, and Harding HP (2003). Mammalian stress granules represent sites of accumulation of stalled translation initiation complexes. Am. J. Physiol. Cell Physiol. 284, C273–C284. [DOI] [PubMed] [Google Scholar]
- Ku YC, Lai MH, Lo CC, Cheng YC, Qiu JT, Tarn WY, and Lai MC (2019). DDX3 Participates in Translational Control of Inflammation Induced by Infections and Injuries. Mol. Cell Biol. 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai MC, Lee YH, and Tarn WY (2008). The DEAD-box RNA helicase DDX3 associates with export messenger ribonucleoproteins as well as tip-associated protein and participates in translational control. Mol. Biol. Cell 19, 3847–3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster AK, Nutter-Upham A, Lindquist S, and King OD (2014). PLAAC: a web and command-line application to identify proteins with prion-like amino acid composition. Bioinformatics 30, 2501–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CS, Dias AP, Jedrychowski M, Patel AH, Hsu JL, and Reed R (2008). Human DDX3 functions in translation and interacts with the translation initiation factor eIF3. Nucleic Acids Res. 36, 4708–4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennox AL, Hoye ML, Jiang R, Johnson-Kerner BL, Suit LA, Venkataramanan S, Sheehan CJ, Alsina FC, Fregeau B, Aldinger KA, et al. (2020). Pathogenic DDX3X mutations impair RNA metabolism and neurogenesis during fetal cortical development. Neuron 106, 404–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. [DOI] [PubMed] [Google Scholar]
- Linsalata AE, He F, Malik AM, Glineburg MR, Green KM, Natla S, Flores BN, Krans A, Archbold HC, Fedak SJ, et al. (2019). DDX3X and specific initiation factors modulate FMR1 repeat-associated non-AUG-initiated translation. EMBO Rep. 20, e47498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maharana S, Wang J, Papadopoulos DK, Richter D, Pozniakovsky A, Poser I, Bickle M, Rizk S, Guillen-Boixet J, Franzmann TM, et al. (2018). RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science 360, 918–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjaly ZR, Scott KM, Abhinav K, Wijesekera L, Ganesalingam J, Goldstein LH, Janssen A, Dougherty A, Willey E, Stanton BR, et al. (2010). The sex ratio in amyotrophic lateral sclerosis: A population based study. Amyotroph. Lateral Scler. 11, 439–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markmiller S, Soltanieh S, Server KL, Mak R, Jin W, Fang MY, Luo EC, Krach F, Yang D, Sen A, et al. (2018). Context-dependent and disease-specific diversity in protein interactions within stress granules. Cell 172, 590–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmor-Kollet H, Siany A, Kedersha N, Knafo N, Rivkin N, Danino YM, Moens TG, Olender T, Sheban D, Cohen N, et al. (2020). Spatiotemporal proteomic analysis of stress granule disassembly using APEX reveals regulation by SUMOylation and links to ALS pathogenesis. Mol. Cell 80, 876–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauvais-Jarvis F, Bairey Merz N, Barnes PJ, Brinton RD, Carrero JJ, DeMeo DL, De Vries GJ, Epperson CN, Govindan R, Klein SL, et al. (2020). Sex and gender: modifiers of health, disease, and medicine. Lancet 396, 565–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCombe PA, and Henderson RD (2010). Effects of gender in amyotrophic lateral sclerosis. Gend. Med. 7, 557–570. [DOI] [PubMed] [Google Scholar]
- Mercer TR, Neph S, Dinger ME, Crawford J, Smith MA, Shearwood AM, Haugen E, Bracken CP, Rackham O, Stamatoyannopoulos JA, et al. (2011). The human mitochondrial transcriptome. Cell 146, 645–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer S, van der Meer P, van Tintelen JP, and van den Berg MP (2014). Sex differences in cardiomyopathies. Eur. J. Heart Fail 16, 238–247. [DOI] [PubMed] [Google Scholar]
- Mitchell SF, Jain S, She M, and Parker R (2013). Global analysis of yeast mRNPs. Nat. Struct. Mol. Biol. 20, 127–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, and Taylor JP (2015). Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon SL, Morisaki T, Khong A, Lyon K, Parker R, and Stasevich TJ (2019). Multicolour single-molecule tracking of mRNA interactions with RNP granules. Nat. Cell Biol. 21, 162–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TA, Wu K, Pandey S, Lehr AW, Li Y, Bemben MA, Badger JD 2nd, Lauzon JL, Wang T, Zaghloul KA, et al. (2020). A cluster of autism-associated variants on X-linked NLGN4X functionally resemble NLGN4Y. Neuron 106, 759–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padron A, Iwasaki S, and Ingolia NT (2019). Proximity RNA labeling by APEX-seq reveals the organization of translation initiation complexes and repressive RNA granules. Mol. Cell 75, 875–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A, Malinovska L, Saha S, Wang J, Alberti S, Krishnan Y, and Hyman AA (2017). ATP as a biological hydrotrope. Science 356, 753–756. [DOI] [PubMed] [Google Scholar]
- Patmore DM, Jassim A, Nathan E, Gilbertson RJ, Tahan D, Hoffmann N, Tong Y, Smith KS, Kanneganti TD, Suzuki H, et al. (2020). DDX3X suppresses the susceptibility of hindbrain lineages to medulloblastoma. Dev. Cell 54, 455–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phung B, Ciesla M, Sanna A, Guzzi N, Beneventi G, Cao Thi Ngoc P, Lauss M, Cabrita R, Cordero E, Bosch A, et al. (2019). The X-linked DDX3X RNA helicase dictates translation reprogramming and metastasis in melanoma. Cell Rep. 27, 3573–3586. [DOI] [PubMed] [Google Scholar]
- Portz B, Lee BL, and Shorter J (2021). FUS and TDP-43 phases in health and disease. Trends Biochem. Sci. 46, 550–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protter DSW, and Parker R (2016). Principles and properties of stress granules. Trends Cell Biol. 26, 668–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qamar S, Wang GZ, Randle SJ, Ruggeri FS, Varela JA, Lin JQ, Phillips EC, Miyashita A, Williams D, Strohl F, et al. (2018). FUS phase separation is modulated by a molecular chaperone and methylation of arginine cation-pi interactions. Cell 173, 720–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert X, and Gouet P (2014). Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzzo EK, Perez-Cano L, Jung JY, Wang LK, Kashef-Haghighi D, Hartl C, Singh C, Xu J, Hoekstra JN, Leventhal O, et al. (2019). Inherited and de novo genetic risk for autism impacts shared networks. Cell 178, 850–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito M, Hess D, Eglinger J, Fritsch AW, Kreysing M, Weinert BT, Choudhary C, and Matthias P (2019). Acetylation of intrinsically disordered regions regulates phase separation. Nat. Chem. Biol. 15, 51–61. [DOI] [PubMed] [Google Scholar]
- Scala M, Torella A, Severino M, Morana G, Castello R, Accogli A, Verrico A, Vari MS, Cappuccio G, Pinelli M, et al. (2019). Three de novo DDX3X variants associated with distinctive brain developmental abnormalities and brain tumor in intellectually disabled females. Eur. J. Hum. Genet. 27, 1254–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider JW, Oommen S, Qureshi MY, Goetsch SC, Pease DR, Sundsbak RS, Guo W, Sun M, Sun H, Kuroyanagi H, et al. (2020). Dysregulated ribonucleoprotein granules promote cardiomyopathy in RBM20 gene-edited pigs. Nat. Med. 26, 1788–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster BS, Dignon GL, Tang WS, Kelley FM, Ranganath AK, Jahnke CN, Simpkins AG, Regy RM, Hammer DA, Good MC, et al. (2020). Identifying sequence perturbations to an intrinsically disordered protein that determine its phase-separation behavior. Proc. Natl. Acad. Sci. U.S.A. 117, 11421–11431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D, and Jankowsky E (2014). The Ded1/DDX3 subfamily of DEAD-box RNA helicases. Crit. Rev. Biochem. Mol. 49, 343–360. [DOI] [PubMed] [Google Scholar]
- Shi B, Li W, Song YS, Wang ZJ, Ju R, Ulman A, Hu J, Palomba F, Zhao YF, Le JP, et al. (2021). UTX condensation underlies its tumour-suppressive activity. Nature 597, 726–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih JW, Wang WT, Tsai TY, Kuo CY, Li HK, and Wu Lee YH (2012). Critical roles of RNA helicase DDX3 and its interactions with eIF4E/PABP1 in stress granule assembly and stress response. Biochem. J. 441, 119–129. [DOI] [PubMed] [Google Scholar]
- Silva JM, Rodrigues S, Sampaio-Marques B, Gomes P, Neves-Carvalho A, Dioli C, Soares-Cunha C, Mazuik BF, Takashima A, Ludovico P, et al. (2019). Dysregulation of autophagy and stress granule-related proteins in stress-driven Tau pathology. Cell Death Differ. 26, 1411–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijders Blok L, Madsen E, Juusola J, Gilissen C, Baralle D, Reijnders MR, Venselaar H, Helsmoortel C, Cho MT, Hoischen A, et al. (2015). Mutations in DDX3X are a common cause of unexplained intellectual disability with gender-specific effects on Wnt signaling. Am. J. Hum. Genet. 97, 343–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, and Ji X (2019). The mechanism of RNA duplex recognition and unwinding by DEAD-box helicase DDX3X. Nat. Commun. 10, 3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto-Rifo R, Rubilar PS, Limousin T, de Breyne S, Decimo D, and Ohlmann T (2012). DEAD-box protein DDX3 associates with eIF4F to promote translation of selected mRNAs. EMBO J. 31, 3745–3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Diaz Z, Fang X, Hart MP, Chesi A, Shorter J, and Gitler AD (2011). Molecular determinants and genetic modifiers of aggregation and toxicity for the ALS disease protein FUS/TLS. PLoS Biol. 9, e1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szaflarski W, Fay MM, Kedersha N, Zabel M, Anderson P, and Ivanov P (2016). Vinca alkaloid drugs promote stress-induced translational repression and stress granule formation. Oncotarget 7, 30307–30322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szappanos D, Tschismarov R, Perlot T, Westermayer S, Fischer K, Platanitis E, Kallinger F, Novatchkova M, Lassnig C, Muller M, et al. (2018). The RNA helicase DDX3X is an essential mediator of innate antimicrobial immunity. Plos Pathog. 14, e1007397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan RH, Ke YD, Ittner LM, and Halliday GM (2017). ALS/FTLD: experimental models and reality. Acta Neuropathol. 133, 177–196. [DOI] [PubMed] [Google Scholar]
- Tauber D, Tauber G, Khong A, Van Treeck B, Pelletier J, and Parker R (2020). Modulation of RNA condensation by the DEAD-box protein eIF4A. Cell 180, 411–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakilian H, Mirzaei M, Tabar MS, Pooyan P, Rezaee LH, Parker L, Haynes PA, Gourabi H, Baharvand H, and Salekdeh GH (2015). DDX3Y, a male-specific region of Y chromosome gene, may modulate neuronal differentiation. J. Proteome Res. 14, 3474–3483. [DOI] [PubMed] [Google Scholar]
- Valentin-Vega YA, Wang YD, Parker M, Patmore DM, Kanagaraj A, Moore J, Rusch M, Finkelstein D, Ellison DW, Gilbertson RJ, et al. (2016). Cancer-associated DDX3X mutations drive stress granule assembly and impair global translation. Sci. Rep. 6, 25996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataramanan S, Gadek M, Calviello L, Wilkins K, and Floor SN (2021). DDX3X and DDX3Y are redundant in protein synthesis. RNA 27, 1577–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernon RM, Chong PA, Tsang B, Kim TH, Bah A, Farber P, Lin H, and Forman-Kay JD (2018). Pi-Pi contacts are an overlooked protein feature relevant to phase separation. Elife 7, e31486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker IH, Hsieh PC, and Riggs PD (2010). Mutations in maltose-binding protein that alter affinity and solubility properties. Appl. Microbiol. Biotechnol. 88, 187–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Choi JM, Holehouse AS, Lee HO, Zhang X, Jahnel M, Maharana S, Lemaitre R, Pozniakovsky A, Drechsel D, et al. (2018a). A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell 174, 688–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Posey JE, Rosenfeld JA, Bacino CA, Scaglia F, Immken L, Harris JM, Hickey SE, Mosher TM, Slavotinek A, et al. (2018b). Phenotypic expansion in DDX3X - a common cause of intellectual disability in females. Ann. Clin. Transl. Neurol. 5, 1277–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins J, Ghosh A, Keerie AFA, Alix JJP, Mead RJ, and Sreedharan J (2020). Female sex mitigates motor and behavioural phenotypes in TDP-43(Q331K) knock-in mice. Sci. Rep. 10, 19220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Yu J, Gale-Day Z, Woo A, Suresh A, Hornsby M, and Gestwicki JE (2020). Three essential resources to improve differential scanning fluorimetry (DSF) experiments. bioRxiv. [Google Scholar]
- Xue B, Dunbrack RL, Williams RW, Dunker AK, and Uversky VN (2010). PONDR-FIT: a meta-predictor of intrinsically disordered amino acids. Biochim. Biophys. Acta. 1804, 996–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yedavalli VSRK, Neuveut C, Chi YH, Kleiman L, and Jeang KT (2004). Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell 119, 381–392. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, and Chanda SK (2019). Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S2. Summary of DDX3X-specific, DDX3Y-specific, and overlapped transcripts from APEX-seq, related to Figure 5.
Table S3. Summary of the cloning and RT-qPCR primers used in this study, related to Figures 1, 2, 3, 4, 5 and 6.
Movie S1. Time-lapse imaging video of FRAP experiment with DDX3X-mCherry LLPS, related to Figure 1.
Movie S2. Time-lapse imaging video of FRAP experiment with DDX3Y-mCherry LLPS, related to Figure 1.
Movie S3. A biological replicate of time-lapse imaging video of FRAP experiment with DDX3X-mCherry LLPS, related to Figures 1 and S1.
Movie S4. A biological replicate of time-lapse imaging video of FRAP experiment with DDX3Y-mCherry LLPS, related to Figures 1 and S1.
Movie S5. Time-lapse imaging video of FRAP experiments with DDX3X-EGFP Stress Granule in HeLa Cells, related to Figure 2.
Movie S6. Time-lapse imaging video of FRAP experiments with DDX3Y-EGFP Stress Granule in HeLa Cells, related to Figure 2.
Data Availability Statement
RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Original western blot images have been deposited at Mendeley and are publicly available as of the date of publication. The DOI is listed in the key resources table. Microscopy data reposted in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key resources table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-G3BP1 | Proteintech | Cat# 13057-2-AP, RRID:AB_2232034 |
| Rabbit polyclonal anti-HA | Abcam | Cat# ab9110, RRID:AB_307019 |
| Mouse monoclonal anti-FLAG | Sigma-Aldrich | Cat# F3165, RRID:AB_259529 |
| Rat monoclonal anti-FLAG | Invitrogen | Cat# MA1-142, RRID:AB_2536846 |
| HRP conjugated anti-FLAG | Invitrogen | Cat# MA1-91878-HRP, RRID:AB_2537626 |
| Rabbit polyclonal anti-GAPDH | Invitrogen | Cat# PA1-16777, RRID:AB_568552 |
| Mouse monoclonal anti-GFP | Santa Cruz | Cat# sc-996, RRID:AB_2187785 |
| Mouse anti-puromycin, clone 12D10, Alexa Fluor 488 conjugated antibody | Sigma-Aldrich | Cat# MABE343-AF488, RRID:AB_2736875 |
| Goat anti-mouse IgG (H&L) (HRP) | Abcam | Cat# ab6789, RRID:AB_955439 |
| Mouse monoclonal anti-ATP5A | Abcam | Cat# ab14748, RRID:AB_301447 |
| Goat anti-Mouse IgG H&L (Alex Fluor® 647) | Abcam | Cat# ab150115, RRID:AB_2687948 |
| Goat anti-Rabbit IgG H&L (Alexa Fluor® 594) | Abcam | Cat# ab150080, RRID:AB_2650602 |
| Goat anti-Mouse IgG H&L (Alexa Fluor® 488) | Abcam | Cat# ab150113, RRID:AB_2576208 |
| Goat Anti-Rat IgG H&L (Alexa Fluor® 488) | Abcam | Cat# ab150157, RRID:AB_2722511 |
| Mouse monoclonal anti-DDX3 | Abcam | Cat# ab196032 |
| Rabbit polyclonal anti-DDX3Y | Invitrogen | Cat# PA5-90055, RRID:AB_2805908 |
| Bacterial and Virus Strains | ||
| One Shot™ TOP10 Chemically Competent E. coli | Invitrogen | Cat# 404003 |
| BL21(DE3)-RIL Competent E. coli | Agilent | Cat# 230204 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Streptavidin-HRP | Cell Signaling Technology | Cat# 3999, RRID:AB_10830897 |
| DDX3X-mCherry recombinant protein | This paper | N/A |
| DDX3Y-mCherry recombinant protein | This paper | N/A |
| MBP-DDX3X recombinant protein | This paper | N/A |
| MBP-DDX3Y recombinant protein | This paper | N/A |
| N-Lauroylsarcosine sodium salt solution | Sigma-Aldrich | Cat# L7414-10ML |
| SUPERase·In™ RNase Inhibitor | Invitrogen | Cat# AM2694 |
| Proteinase K solution (20 mg/mL) | Life Technologies | Cat# AM2548 |
| Lipofectamine 2000 | Thermo Fisher | Cat# 11668019 |
| Pierce streptavidin magnetic beads | Thermo Fisher | Cat# 88816 |
| Agencourt AMPure XP | Beckman Coulter | Cat# A63881 |
| TRIzol | Invitrogen | Cat# 15596026 |
| IPTG | Thermo Fisher | Cat# 34060 |
| DAPI | Sigma-Aldrich | Cat# D9542 |
| Fisher BioReagents™ Bovine Serum Albumin, Heat Shock Treated | Fisher Scientific | Cat# BP1600-100 |
| Polyuridylic acid potassium salt | Sigma-Aldrich | Cat# P9528-10MG |
| Sodium arsenite | Spectrum Chemical | Cat# S-222 |
| Sorbitol solution | Spectrum Chemical | Cat# S-1525 |
| Thapsigargin | ACROS Organics | Cat# AC328570010 |
| CCCP | Alfa Aesar | Cat# AAL06932MC |
| Puromycin | Takara Bio USA | Cat# 631305 |
| MG132 | Sigma-Aldrich | Cat# 474790-1 MG |
| Hydrogen peroxide solution | Sigma-Aldrich | Cat# H1009 |
| Biotin-phenol | Iris Biotech | Cat# LS-3500.1000 |
| Sodium azide | Sigma-Aldrich | Cat# S2002 |
| Sodium ascorbate | Sigma-Aldrich | Cat# A7631 |
| Trolox | Sigma-Aldrich | Cat# 238813 |
| Cycloheximide | Sigma-Aldrich | Cat# C7698 |
| Critical Commercial Assays | ||
| Malachite Green Phosphate Assay Kit | Sigma-Aldrich | Cat# MAK307-1KT |
| Flexi® Rabbit Reticulocyte Lysate System | Promega | Cat# L4540 |
| Luna® Universal One-Step RT-qPCR Kit | NEB | Cat# M3005L |
| Truseq Stranded mRNA Library Prep | Illumina | Cat# 20020594 |
| Dynabeads™ mRNA Purification Kit (for mRNA purification from total RNA preps) | Invitrogen | Cat# 61006 |
| EnzChek™ Phosphate Assay Kit | Thermo Fisher | Cat# E6646 |
| RNA Clean and Concentrator-5 | Zymo Research | Cat# R1016 |
| Deposited Data | ||
| APEX-seq with stress treatment | This paper | GEO: GSE171792 |
| APEX-seq without stress treatment | This paper | GEO: GSE193783 |
| Raw data | This paper; Mendeley Data | https://dx.doi.org/10.17632/9hs5d4fvgd.1 |
| Experimental Models: Cell Lines | ||
| Human: HeLa | ATCC | CCL-2, RRID:CVCL_0030 |
| Human: HEK 293T | ATCC | CRL-3216, RRID:CVCL_0063 |
| Mouse: N2a | ATCC | CCL-131, RRID:CVCL_0470 |
| Oligonucleotides | ||
| Primers for all the recombinant DNA | This paper | Table S3 |
| Recombinant DNA | ||
| pPB DDX3X | This paper | N/A |
| pPB DDX3Y | This paper | N/A |
| pPB DDX3X-IDR1 | This paper | N/A |
| pPB DDX3Y-IDR1 | This paper | N/A |
| pPB DDX3X-Helicae | This paper | N/A |
| pPB DDX3Y-Helicae | This paper | N/A |
| pPB DDX3X-IDR2 | This paper | N/A |
| pPB DDX3Y-IDR2 | This paper | N/A |
| pPB DDX3X-ΔIDR1 | This paper | N/A |
| pPB DDX3Y-ΔIDR1 | This paper | N/A |
| pPB DDX3X-ΔIDR2 | This paper | N/A |
| pPB DDX3Y-ΔIDR2 | This paper | N/A |
| pPB DDX3X-ΔHelicase | This paper | N/A |
| pPB DDX3Y-ΔHelicase | This paper | N/A |
| pPB DDX3XIDR1-DDX3YHelicae-DDX3YIDR2 | This paper | N/A |
| pPB DDX3YIDR1-DDX3XHelicae-DDX3XIDR2 | This paper | N/A |
| pPB DDX3XIDR1-DDX3Xhelicae-DDX3YIDR2 | This paper | N/A |
| pPB DDX3YIDR1-DDX3Yhelicae-DDX3XIDR2 | This paper | N/A |
| pPB DDX3XIDR1-DDX3Yhelicae-DDX3XIDR2 | This paper | N/A |
| pPB DDX3YIDR1-DDX3Xhelicae-DDX3YIDR2 | This paper | N/A |
| pcDNA3 mito-APEX2 | This paper | N/A |
| pPB DDX3X-EGFP | This paper | N/A |
| pPB DDX3Y-EGFP | This paper | N/A |
| pcDNA3 FLAG-DDX3X | This paper | N/A |
| pcDNA3 FLAG-DDX3Y | This paper | N/A |
| pcDNA3 HA-DDX3X | This paper | N/A |
| pcDNA3 HA-DDX3Y | This paper | N/A |
| pET-MCN_His-TEV_V5-DDX3X-mCherry | (Hondele et al., 2019) | N/A |
| pET-MCN_His-TEV_V5-DDX3Y-mCherry | This paper | N/A |
| pET-MCN_His-TEV_V5-DDX3XIDR1-DDX3YHelicae-DDX3YIDR2-mCherry | This paper | N/A |
| pET-MCN_His-TEV_V5-DDX3YIDR1-DDX3XHelicae-DDX3XIDR2-mCherry | This paper | N/A |
| pET-MCN_His-TEV_V5-DDX3XIDR1-DDX3Xhelicae-DDX3YIDR2-mCherry | This paper | N/A |
| pET-MCN_His-TEV_V5-DDX3YIDR1-DDX3Yhelicae-DDX3XIDR2-mCherry | This paper | N/A |
| pET-MCN_His-TEV_V5-DDX3XIDR1-DDX3Yhelicae-DDX3XIDR2-mCherry | This paper | N/A |
| pET-MCN_His-TEV_V5-DDX3YIDR1-DDX3Xhelicae-DDX3YIDR2-mCherry | This paper | N/A |
| pMAL-c2X | (Walker et al., 2010) | Addgene Plasmid #75286, RRID:Addgene_75286 |
| pMAL-c2X-DDX3X | This paper | N/A |
| pMAL-c2X-DDX3Y | This paper | N/A |
| pcDNA5/FRT/TO APEX2-GFP | (Padron et al., 2019) | Addgene Plasmid #129640, RRID:Addgene_129640 |
| pJ4M/TDP-43 | (Hallegger et al., 2021) | Addgene plasmid #104480, RRID:Addgene_104480 |
| pPB APEX2-DDX3X | This paper | N/A |
| pPB APEX2-DDX3Y | This paper | N/A |
| pPB APEX2-EGFP | This paper | N/A |
| Software and Algorithms | ||
| HISAT2 | (Kim et al., 2015) | https://ccb.jhu.edu/software/hisat2/index.shtml, RRID:SCR_015530 |
| DESeq2 | (Love et al., 2014) | https://bioconductor.org/packages/release/bioc/html/DESeq2.html, RRID:SCR_015687 |
| featureCounts | (Liao et al., 2014) | http://bioinf.wehi.edu.au/featureCounts/, RRID:SCR_012919 |
| trimmomatic | (Bolger et al., 2014) | http://www.usadellab.org/cms/index.php?page=trimmomatic, RRID:SCR_011848 |
| R | https://www.r-project.org | https://www.r-project.org, RRID:SCR_000036 |
| Fiji | (Schindelin et al., 2012) | https://imagej.net/Fiji, RRID:SCR_002285 |
| GraphPad Prism | GraphPad Software | https://graphpad.com/scientificsoft, RRID:SCR_002798 |
| Other | ||
| 384-well microscopy plates | Brooks Life Sciences | Cat#4ti-0203 |
| Amicon ultra-15 | EMD Millipore | Cat# UFC901024 |


