Abstract
Microcin 24 is an antimicrobial peptide secreted by uropathogenic Escherichia coli. Secretion of microcin 24 provides an antibacterial defense mechanism for E. coli. In a plasmid-based system using transformed Salmonella enterica, we found that resistance to microcin 24 could be seen in concert with a multiple-antibiotic resistance phenotype. This multidrug-resistant phenotype appeared when Salmonella was exposed to an E. coli strain expressing microcin 24. Therefore, it appears that multidrug-resistant Salmonella can arise as a result of an insult from other pathogenic bacteria.
Microcins are antimicrobial peptides secreted by bacteria as a means of disabling neighboring bacteria (1). Microcin 24 (Mcc24) is a colicin secreted by a strain of uropathogenic Escherichia coli (18). A previous study demonstrated that Mcc24 has activity against Salmonella enterica and most E. coli strains but not against Campylobacter or Listeria strains (18). We also found that Mcc24 does not have activity against multiple-antibiotic-resistant Klebsiella pneumoniae (ATCC MCV37) (data not shown). In the present study, we evaluated the ability of S. enterica serotype Typhimurium to develop resistance to Mcc24 and how the development of Mcc24 resistance related to resistance to antibiotics. This study was undertaken to determine if the use of Mcc24 in probiotic bacteria, as described previously (23), could lead to antibiotic resistance in Salmonella.
We pursued the multiple-antibiotic resistance (mar) operon as a potential component of Mcc24 resistance since this operon regulates resistance to a variety of foreign substances in Salmonella (7). The mar operon regulates an efflux system that facilitates the expulsion of antibiotics and organic solvents (6). The mar operon is regulated by MarR, a repressor protein that prevents the transcription of marAB (15). Transcription of marAB occurs when MarR is absent due to an inhibition of translation of marR (20), altered as a result of mutagenesis of marR (20) or sequestration by salicylate (5). As part of this study we used a plasmid-based system to attenuate the Mar phenotype in a manner similar to that of White et al. (22), who used antisense DNA inhibition to block the expression of marA, a regulator of an efflux system (16). S. enterica serotype Typhimurium strain SL1344 (24) was transformed with pBAD, a high-copy-number plasmid with a pBM1 replicon (Invitrogen) containing the PCR-derived marR gene. The marR gene was cloned from SL1344 DNA using PCR conditions described previously (3) with 5′-ATGAAAAGCACCAGTGATCTGTTC-3′ and 5′-CCTACGGCAGATT-TTTCTTGAGCAA-3′ as forward and reverse primers, respectively. The expression of marR in pBAD is under the control of arabinose via the araBAD promoter (12, 14).
SL1344 was also transformed with another plasmid containing the CFP (cyan derivative of green fluorescent protein [8]; Clontech) gene cloned into pCRII Blunt, a high-copy-number plasmid with a pBMI replicon and the Lac promoter for transcription of the cloned gene (Invitrogen), as previously described (10). This plasmid was included to visually distinguish S. enterica serotype Typhimurium from an Mcc24-producing E. coli strain (ampicillin-resistant strain MC4100pGOB18 [23]). SL1344 cotransformants, designated SL1344/CFP/marR, were propagated in Lennox L broth (GIBCO-BRL) containing 100 μg of ampicillin per ml and 64 μg of kanamycin per ml. Ampicillin resistance is conferred by a beta-lactamase encoded by pBAD, while kanamycin resistance is conferred by a phosphotransferase encoded by pCRII Blunt.
To evaluate Mcc24 resistance in S. enterica serotype Typhimurium in a system that mimics the in vivo commingling of Salmonella and E. coli, we flooded SL1344/CFP/marR (approximately 2 × 108 bacteria) on Lennox L agar. Next we soaked a filter disk (Bacto concentration disks, sterile blanks; Difco) in Lennox L broth containing Mcc24-producing E. coli MC4100pGOB18 (109 organisms/ml) and placed this disk in the middle of the SL1344/CFP/marR-laden agar plate. Bacteria were then grown together on Lennox L agar plates containing ampicillin at 37°C overnight.
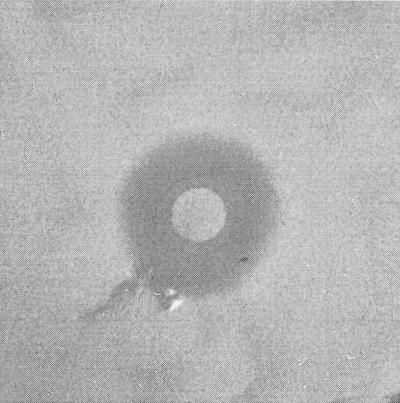
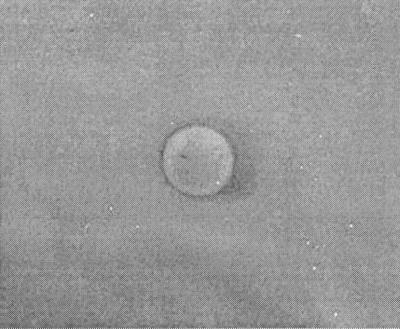
As shown in Fig. 1, a zone of inhibition was observed around the disk soaked in E. coli strain MC4100pGOB18. Table 1 reveals that this zone was due to a secreted protein since the inhibition was present if trichloroacetic acid (TCA)-precipitated supernatants from E. coli strain MC4100pGOB18 were used instead of broth containing the bacteria. The zone also disappeared for SL1344 transformed with pCRXL, a high-copy-number plasmid with a ColE1 replicon, the Lac promoter for transcription of the cloned gene (Invitrogen), and the Mcc24 immunity gene (mtfl; GenBank accession number U47048). The mtfl gene was amplified by PCR from the pGOB18 plasmid. Additionally, the zone of inhibition disappeared if TCA-precipitated supernatants from E. coli strain MC4100pGOB18 were treated with 1 mg of pronase E (Sigma) per ml. As shown in Fig. 2, the zone was also absent if SL1344/CFP/marR or SL1344 was grown in Lennox L broth and incubated on Lennox L agar, both containing 3.5 mM salicylate (Sigma). Salicylate can activate the Mar phenotype by physically binding to MarR (5), thus competitively eliminating the repressor effect of MarR on the mar operon (15). Therefore, it appears that resistance to Mcc24 can occur through the mar response. This is also apparent since the zone was absent for S. enterica serotype Typhimurium strain 8431 (Table 1), a mutant exhibiting a Mar-like phenotype (4). Additionally, the salicylate-mediated induction of Mcc24 resistance was prevented by 0.2%-arabinose-induced episomal expression of MarR from pBAD. That is, repression of marA transcription occurred as a result of competitive antagonism of salicylate-mediated derepression by episomally derived MarR. Chromosomal and episomal marR DNA sequences were not changed by the salicylate or arabinose treatment. Sequencing was performed using pBAD-specific primers for episomal marR, open reading frame 221, and marA flanking sequences for chromosomal marR (GenBank accession number U54468; data not shown).
FIG. 1.
Mcc24-mediated zone of inhibition of SL1344/CFP/marR. SL1344/CFP/marR and E. coli strain MC4100pGOB18 (center of plate) were incubated together as described in the text.
TABLE 1.
Changes in MICs in relationship to Mcc24 resistance
| Treatment | Strain | MIC (μg/ml) ofa:
|
Zone of inhibition around MC4100pGOB18 disk | |||
|---|---|---|---|---|---|---|
| CIP | TET | CHL | RIF | |||
| Exposure to disk soaked in MC4100pGOB18 | SL1344 | 0.125 | 4 | 2 | 12.5 | Yes |
| Exposure to TCA-precipitated supernatants from MC4100pGOB18 | SL1344 | NDb | ND | ND | ND | Yes |
| Exposure to pronase-treated TCA-precipitated supernatants from MC4100pGOB18 | SL1344 | ND | ND | ND | ND | No |
| 3.5 mM salicylate | SL1344 | 4 | 16 | 32 | 50 | No |
| Exposure to disk soaked in MC4100pGOB18 | SL1344/mtfI-pCRXL | ND | ND | ND | ND | No |
| None | 8431 | 4 | 32 | 64 | 50 | ND |
| Exposure to disk soaked in MC4100pGOB18 | 8431 | 4 | 32 | 64 | 50 | No |
| None | SL1344/CFP/marR | 0.125 | 4 | 2 | 12.5 | ND |
| Exposure to disk soaked in MC4100pGOB18 | SL1344/CFP/marR | 0.125 | 4 | 2 | 12.5 | Yes |
| Exposure to disk soaked in MC4100pGOB18 in the presence of 3.5 mM salicylate | SL1344/CFP/marR | 4 | 16 | 32 | 50 | No |
| Exposure to disk soaked in MC4100pGOB18 in the presence of 3.5 mM salicylate plus 0.2% arabinose | SL1344/CFP/marR | 0.5 | 8 | 8 | 12.5 | Yes |
| Exposure to disk soaked in MC4100pGOB18 in the presence of 3.5 mM salicylate plus two exposures to 0.2% arabinose | SL1344/CFP/marR | 0.25 | 4 | 4 | 12.5 | Yes |
Concentrations of antibiotics were as follows: 8, 4, 2, 1, 0.5, 0.25, and 0.125 μg/ml for ciprofloxacin (CIP); 64, 32, 16, 8, 4, 2, and 1 μg/ml for tetracycline (TET) and for chloramphenicol (CHL); and 100, 50, 25, 12.5, 6.3, 3.2, and 1.6 μg/ml for rifampin (RIF).
ND, not determined.
FIG. 2.
Induction of the Mar phenotype in SL1344/CFP/marR. SL1344/CFP/marR and E. coli strain MC4100pGOB18 (center of plate) were incubated together in the presence of 3.5 mM salicylate.
To evaluate the potential relationship between resistance to Mcc24 and multiple-antibiotic resistance, we determined the MICs of ciprofloxacin, chloramphenicol, tetracycline, and rifampin. These four antibiotics were chosen since they represent part of the Mar phenotype (6). MICs were determined by inoculating 106 bacteria into 1-ml aliquots of Mueller-Hinton broth (Difco) containing serial dilutions of antibiotics per NCCLS guidelines. Bacteria were grown aerobically, and MICs were ascribed based on the lowest concentration of antibiotic that inhibited growth. Table 1 reveals that salicylate-mediated Mcc24 resistance occurred in concert with an increase in ciprofloxacin MICs. Additionally, SL1344/CFP/marR exhibited increases in the MICs of chloramphenicol, tetracycline, and rifampin. Simultaneous exposure to arabinose prevented the salicylate-mediated change in ciprofloxacin MICs.
To evaluate the mar response in relationship to Mcc24 resistance in Salmonella, we repeatedly selected SL1344/CFP/marR colonies that bordered the zone of inhibition (Table 2). Salmonella colonies were expanded by vigorous shaking for 4 to 6 h at 37°C in Lennox L broth containing ampicillin and kanamycin. Broth cultures (approximately 2 × 108 bacteria) were then replated with the disk soaked in E. coli strain MC4100pGOB18 on Lennox L agar containing ampicillin. The zone disappeared after 9, 11, or 18 expansions and replatings in three separate experiments. As a control, SL1344/CFP/marR colonies from the edge of the plate were also expanded and replated. SL1344/CFP/marR colonies from the edge of the plate remained sensitive to Mcc24. We have been unable to isolate Mcc24-resistant SL1344/CFP/marR colonies from the edge of the plate. In a representative experiment in which 18 expansion-replating procedures were required to induce Mcc24 resistance, we found that resistance to Mcc24 and increased MICs of ciprofloxacin, tetracycline, chloramphenicol, and rifampin could be prevented by episomal expression of MarR. Mcc24-sensitive microbes became Mcc24 resistant, however, after removal of the arabinose. Chromosomal and episomal marR DNA sequences were not changed by the expansion processes or by the salicylate or arabinose treatment. Thus, Mcc24 exposure-mediated resistance was apparently due to activation of the mar system. While a previous study suggests that resistance to microcin MccB17 can be due to an export of the peptide in E. coli (11), this is the first study to document mar-mediated resistance to a microcin.
TABLE 2.
Changes in MICs in relationship to Mcc24 exposure in a representative experiment
| Treatment | Straina | MIC (μg/ml) ofb:
|
Zone of inhibition around MC4100pGOB18 disk | |||
|---|---|---|---|---|---|---|
| CIP | TET | CHL | RIF | |||
| Two exposures to disks soaked in MC4100pGOB18 | SL1344/CFP/marR border colonies | 0.125 | 4 | 2 | 12.5 | Yes |
| 17 exposures to disks soaked in MC4100pGOB18 | SL1344/CFP/marR border colonies | 0.25 | ND | ND | ND | Yes |
| 18 exposures to disks soaked in MC4100pGOB18 | SL1344/CFP/marR border colonies | 2 | 16 | 32 | 50 | No |
| 17 exposures to disks soaked in MC4100pGOB18 followed by another exposure in the presence of 0.2% arabinose | SL1344/CFP/marR border colonies | 0.5 | ND | ND | ND | Yes |
| 17 exposures to disks soaked in MC4100pGOB18 and then two more exposures in the presence of 0.2% arabinose | SL1344/CFP/marR border colonies | 0.25 | ND | ND | ND | Yes |
| 17 exposures to disks soaked in MC4100pGOB18 one exposure to 0.2% arabinose, and then 2 to 3 additional exposures to disks soaked in MC4100pGOB18 | SL1344/CFP/marR border colonies | 2 | 16 | 32 | 50 | No |
| 18 expansions without exposure to disks soaked in MC4100pGOB18 | SL1344/CFP/marR edge colonies | 0.125 | 4 | 2 | 12.5 | Yes |
| 50 expansions without exposure to disks soaked in MC4100pGOB18 | SL1344/CFP/marR edge colonies | 0.125 | 4 | 2 | 12.5 | Yes |
“Border” indicates that colonies were selected from the border of the zone of inhibition, and “edge” indicates that colonies were selected from the edge of the agar plate.
Concentrations of antibiotics were as described in Table 1, note a. ND, not determined.
Although the mechanism of action for Mcc24 is currently unknown, other microcins provide antimicrobial action by inhibiting DNA gyrase, by inhibiting protein expression (17, 21), or by forming pores in target bacteria (9). Since marR DNA sequences were not altered and since ancillary MarR may prevent derepression, it is feasible that Mcc24 can inhibit translation. The Mcc24-mediated inhibition of translation may decrease MarR levels below a threshold that normally maintains repression of mar. Derepression then ensues when this threshold is breached. That is, Mcc24 indirectly mediates resistance to itself.
In this study, we found that Salmonella can develop resistance to an E. coli-derived microcin. Extrapolation of our results indicates that exposure of Salmonella to Mcc24 can result in multidrug-resistant Salmonella. Therefore, the ongoing problem with multidrug-resistant Salmonella may be due to the activation of innate pathways and the acquisition of genetic determinants of antibiotic resistance such as integrons (2, 13, 19). Since ciprofloxacin resistance in Salmonella is uncommon, the events described in this study are probably a rare situation in vivo. Nonetheless, it unfortunately appears that microcin production gives an advantage to pathogenic E. coli while also potentially selecting for multidrug-resistant Salmonella.
Acknowledgments
We thank Ruth Willson for technical assistance, Sandy Johnson for secretarial assistance, Chuck Greiner for photography, and Kristi Anderson, Kim Brogden, and Bonita Glatz for reviewing the manuscript. S.A.C. and T.S.F. contributed equally to this work.
REFERENCES
- 1.Baquero F, Bouanchaud D, Martinez-Perez M, Fernandez C. Microcin plasmids: a group of extrachromosomal elements coding for low-molecular-weight antibiotics in Escherichia coli. J Bacteriol. 1978;135:342–347. doi: 10.1128/jb.135.2.342-347.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Briggs C E, Fratamico P M. Molecular characterization of an antibiotic resistance gene cluster of Salmonella typhimurium DT104. Antimicrob Agents Chemother. 1999;43:846–849. doi: 10.1128/aac.43.4.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carlson S A, Browning M, Ferris K, Jones B D. Identification of diminished tissue culture invasiveness among multiple antibiotic resistant Salmonella typhimurium DT104. Microb Pathog. 2000;28:37–44. doi: 10.1006/mpat.1999.0322. [DOI] [PubMed] [Google Scholar]
- 4.Carlson S A, Ferris K E. Augmentation of antibiotic resistance in multiple antibiotic resistant Salmonella typhimurium DT104 following exposure to penicillin derivatives. Vet Microbiol. 2000;73:25–35. doi: 10.1016/s0378-1135(00)00154-1. [DOI] [PubMed] [Google Scholar]
- 5.Cohen S P, Hachler H, Levy S B. Genetic and functional analysis of the multiple antibiotic resistance (mar) locus in Escherichia coli. J Bacteriol. 1993;175:1484–1492. doi: 10.1128/jb.175.5.1484-1492.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen S P, McMurry L M, Hooper D C, Wolfson J S, Levy S B. Cross-resistance to fluoroquinolones in multiple-antibiotic-resistant (Mar) Escherichia coli selected by tetracycline or chloramphenicol: decreased drug accumulation associated with membrane changes in addition to OmpF reduction. Antimicrob Agents Chemother. 1989;33:1318–1325. doi: 10.1128/aac.33.8.1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen S P, Yan W, Levy S B. A multidrug resistance regulatory chromosomal locus is widespread among enteric bacteria. J Infect Dis. 1993;168:484–488. doi: 10.1093/infdis/168.2.484. [DOI] [PubMed] [Google Scholar]
- 8.Cormack B, Valdivia R, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 9.de Lorenzo V, Pugsley A. Microcin E492, a low-molecular-weight peptide antibiotic which causes depolarization of the Escherichia coli cytoplasmic membrane. Antimicrob Agents Chemother. 1985;27:666–669. doi: 10.1128/aac.27.4.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frana T S, Carlson S A. Development and use of a plasmid encoding green fluorescence protein in multiple antibiotic resistant Salmonella. BioTechniques. 2001;30:28–32. doi: 10.2144/01301bm03. [DOI] [PubMed] [Google Scholar]
- 11.Garrido M, Herrero M, Kolter R, Moreno F. The export of the DNA replication inhibitor microcin B17 provides immunity for the host cell. EMBO J. 1988;7:1853–1862. doi: 10.1002/j.1460-2075.1988.tb03018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guzman L-M, Belin D, Carson M J, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hall R M, Stokes H W. Integrons: novel DNA elements which capture genes by site-specific recombination. Genetica. 1993;90:115–132. doi: 10.1007/BF01435034. [DOI] [PubMed] [Google Scholar]
- 14.Lee N, Francklyn C, Hamilton E P. Arabinose-induced binding of AraC protein to araI2 activates the araBAD operon promoter. Proc Natl Acad Sci USA. 1987;81:8814–8818. doi: 10.1073/pnas.84.24.8814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin R G, Rosner J L. Binding of purified multiple antibiotic-resistance repressor protein (MarR) to mar operator sequences. Proc Natl Acad Sci USA. 1995;92:5456–5460. doi: 10.1073/pnas.92.12.5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McDermott P F, White D G, Podglajen I, Alekshun M N, Levy S B. Multidrug resistance following expression of the Escherichia coli marA gene in Mycobacterium smegmatis. J Bacteriol. 1998;180:2995–2998. doi: 10.1128/jb.180.11.2995-2998.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moreno F, San Millan J L, Hernandez-Chico C, Kolter R. Genetics and biochemistry of antibiotic production. Boston, Mass: Butterworth-Heimann; 1995. [Google Scholar]
- 18.O'Brien G, Mahanty H. Colicin 24, a new plasmid-borne colicin from a uropathogenic strain of Escherichia coli. Plasmid. 1994;31:288–296. doi: 10.1006/plas.1994.1030. [DOI] [PubMed] [Google Scholar]
- 19.Ridley A, Threlfall E J. Molecular epidemiology of antibiotic resistance genes in multiresistant epidemic Salmonella typhimurium DT 104. Microb Drug Resist. 1998;4:113–118. doi: 10.1089/mdr.1998.4.113. [DOI] [PubMed] [Google Scholar]
- 20.Sulavik M C, Dazer M, Miller P F. The Salmonella typhimurium mar locus: molecular and genetic analyses and assessment of its role in virulence. J Bacteriol. 1997;179:1857–1866. doi: 10.1128/jb.179.6.1857-1866.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vizan J, Hernandez-Chico C, del Castillo I, Moreno F. The peptide antibiotic microcin B17 induces double-strand cleavage of DNA mediated by E. coli DNA gyrase. EMBO J. 1991;10:467–476. doi: 10.1002/j.1460-2075.1991.tb07969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.White D G, Maneewannakul K, von Hofe E, Zillman M, Eisenberg W, Field A K, Levy S B. Inhibition of the multiple antibiotic resistance (mar) operon in Escherichia coli by antisense DNA analogs. Antimicrob Agents Chemother. 1997;41:2699–2704. doi: 10.1128/aac.41.12.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wooley R, Gibbs P, Shotts E J. Inhibition of Salmonella typhimurium in the chicken intestinal tract by a transformed avirulent avian Escherichia coli. Avian Dis. 1999;432:45–50. [PubMed] [Google Scholar]
- 24.Wray C, Sojka W J. Experimental Salmonella typhimurium in calves. Res Vet Sci. 1978;25:139–143. [PubMed] [Google Scholar]