Abstract
Snake envenomation is a common but neglected disease that affects millions of people around the world annually. Among venomous snake species in Brazil, the tropical rattlesnake (Crotalus durissus terrificus) accounts for the highest number of fatal envenomations and is responsible for the second highest number of bites. Snake venoms are complex secretions which, upon injection, trigger diverse physiological effects that can cause significant injury or death. The components of C. d. terrificus venom exhibit neurotoxic, myotoxic, hemotoxic, nephrotoxic, and cardiotoxic properties which present clinically as alteration of central nervous system function, motor paralysis, seizures, eyelid ptosis, ophthalmoplesia, blurred vision, coagulation disorders, rhabdomyolysis, myoglobinuria, and cardiorespiratory arrest. In this study, we focused on proteomic characterization of the cardiotoxic effects of C. d. terrificus venom in mouse models. We injected venom at half the lethal dose (LD50) into the gastrocnemius muscle. Mouse hearts were removed at set time points after venom injection (1 h, 6 h, 12 h, or 24 h) and subjected to trypsin digestion prior to high-resolution mass spectrometry. We analyzed the proteomic profiles of >1,300 proteins and observed that several proteins showed noteworthy changes in their quantitative profiles, likely reflecting the toxic activity of venom components. Among the affected proteins were several associated with cellular deregulation and tissue damage. Changes in heart protein abundance offer insights into how they may work synergistically upon envenomation.
Keywords: Crotalus durissus terrificus, snake venom, mass spectrometry-based proteomics, mouse heart, cardiotoxicity, envenomation
Graphical Abstract
1. Introduction
Snakebite is a serious but understudied global health concern, classified by the World Health Organization as a neglected tropical disease. The impacts on public health are especially pronounced in developing countries. About 2.7 million snake envenomations occur around the world annually, resulting in up to 138,000 fatalities and 414,000 permanent disabilities [1, 2].
Brazil is home to four genera of medically significant venomous snakes: Bothrops, Lachesis, Crotalus (Viperidae family) and the Micrurus (Elapidae family) [3]. Crotalus species are distributed from southern Canada to central Argentina. Available epidemiological data, however, are largely limited to bites occurring in Brazil and the United States. According to the Notifiable Diseases Information System (SINAN) of the Health Surveillance Secretariat of the Brazilian Ministry of Health, 11,825 Crotalus envenomations occurred in Brazil, resulting in 108 deaths from 2015 to 2020. Crotalus accounted for the highest number of fatal bites [4] and the second highest number of bites by venomous snakes in the country [5]. The American Association of Poison Control Center’s National Poison Data System (NPDS) indicates that 4,674 venomous snakebites occurred in the United States, of which 426 resulted in major outcomes and six bites led to patient death from 2014 to 2018 (https://aapcc.org/annual-reports).
Advancing our understanding of animal toxins’ physiological activities is critical for improving clinical care, developing more effective antivenoms, and identifying candidate molecules for potential therapeutic applications [6].
Crotalus venom is composed of five major proteins: crotoxin, crotapotin, crotamine, gyroxin, and convulxin. Crotoxin is a neurotoxin that makes up about 65% of the dry weight of the venom [7, 8], and is responsible for observed neurotoxicity [9, 10]. Crotapotin is the acidic subunit of crotoxin and functions as a chaperone to potentiate the action of the basic phospholipase A2 (PLA2) subunit [11, 12]. Crotamine is a myotoxin, which, together with crotoxin, can cause systemic lesions in skeletal muscle tissue [13, 14]. Gyroxin is a neurotoxin that causes aberrant muscle behavior often manifested by severe characteristic barrel rotation of the body [15, 16]. Convulxin is a hemotoxin in the C-type lectin family that can contribute to seizures and respiratory disturbance [17]. Investigations of whole-venom toxicity have also documented coagulation disorders, hemolytic action, nephrotoxicity [18, 19] and cardiotoxicity [20–23].
Proteomic, transcriptomic, immunological, and biochemical analyses have been used to describe whole venom composition, and to study the effects of individual toxins in the venoms of numerous snake species [24–27]. However, few studies have described the proteomic effects of envenomation within cells [28, 29], tissues, or isolated organs [30].
The mammal heart has been the target of countless studies investigating the development of cardiovascular diseases [31, 32]. Numerous substances including industrial chemicals, heavy metals, pharmaceuticals, and biotoxins [33] exhibit cardiotoxicity but snake venom cardiotoxins are often especially remarkable for their specificity and potency [20–22, 34–37]. Molecular analysis of cardiac tissue is an efficient strategy to search for candidate molecules to be targeted in the treatment of heart disease [38]. Mass spectrometry-based technologies can be used to probe the pathology of heart conditions, as well as the effects of drugs and toxins, at the protein level [39].
In this study, we use a proteomic approach to assess the effect of C. d. terrificus venom on cardiac tissue in Swiss mice and explore the mechanisms of action and specific molecular targets of the venom.
2. Experimental section
2.1. Crotalus durissus terrificus venom
Crotalus durissus terrificus venom was extracted and lyophilized at the Butantan Institute Herpetology Department and supplied to our lab through the Strategic Core of Venom and Anti-Venom (NEVAS). The sample used in this study was a pool of venom from 256 individual snakes collected in the states of São Paulo, Goiás, Minas Gerais, Mato Grosso do Sul, and Paraná. The LD50 of this lot of venom (01/14–2) was determined to be 0.71 μg/animal (Karen de Morais Zani and Anita Mitico Tanaka Azevedo of the Butantan Institute Herpetology Department). The venom was diluted with 0.9% NaCl to a concentration of 1.0 mg/ mL, aliquoted, and stored at −80° C until use.
2.2. Animals
We used 64 adult Swiss male mice weighing between 18 and 22 g. These animals were provided by the Central Animal Facility of the Butantan Institute and were approved for experimental use by the Animal Use Ethics Committee of the Butantan Institute under the certification CEUAIB: 2283181019 (2019).
The animals to be used for proteomic analysis (n = 40) were placed in polycarbonate boxes coated with a Millipore filter, containing five mice in each box. For histological analysis (n = 24), three animals were placed in each box. Mice were kept at room temperature (22 °C) and constant humidity (45 to 60%), positive pressure, 12-hour light/dark cycle, and had free access to water and food. The methodology applied in this study followed the standards established by the National Council for the Control of Animal Experimentation (CONCEA) and all applicable legislation.
2.3. Treatment of mice with C. d. terrificus venom
Mice were treated with 0.5 LD50 of the venom (0.355 μg/ animal). Venom was diluted in 50 μL of saline solution (0.9% NaCl) and injected into the gastrocnemius muscle. After 1 h, 6 h, 12 h, or 24 h of the treatment, mice were euthanized and the heart was removed for mass spectrometry analysis and histology analysis. Experiments were performed in quintuplicate for mass spectrometry analysis and in triplicate for histological analysis. Control mice were injected with the vehicle (saline) only and euthanized at the same time periods as mice injected with venom.
2.4. Sample processing for histology analysis
After treatment, animals were anesthetized with isoflurane and hearts were immediately placed in a solution of cadmium chloride (100 mM) for 30 seconds, then fixed in 4% PFA (pH 6.9) for 24 hours and stored in 70% ethanol. Paraffin blocks were assembled and cut at 30 μm thickness and stained with hematoxylin-eosin (H-E) for microscopy analysis. Cardiac tissue slides were stained with H-E according to the protocol by Fischer et al. (2008) [40] and analyzed using objectives of 4x, 10x, 25x, and 40x magnification in a upright Olympus microscope (Shinjuku City, Tokyo, Japan).
2.5. Sample processing for mass spectrometry analysis
To analyze a broad spectrum of proteins from across the entire heart rather than only sampling a limited region of the heart, we chose to use formalin-fixed paraffin-embedded (FFPE) tissue section samples for histological analysis and prepare a separate group of samples specifically for mass spectrometry analysis by processing the entire heart. Heart tissue was dissected and washed with saline solution to remove excess blood, samples were weighed and immediately frozen in liquid nitrogen until further processing for mass spectrometry analysis. The hearts were lysed with phase-transfer surfactant (PTS) lysis buffer as previously described [41], supplemented with protease and phosphatase inhibitors (Halt, Thermo Fisher Scientific, IL, USA). Lysis was performed on ice using a rate of 6 x heart volume-to-weight ratio as previously described [30]. Immediately after lysis, heart tissue was homogenized using a tissue homogenizer (Precellys 24, Bertin Instruments, Montigny-le-Bretonneux, France) at 6,800 g for 30 seconds. Samples were then heated at 95 °C for 5 minutes, sonicated on ice for 20 minutes, then centrifuged at 14,000 g at 4 °C for 30 min. The supernatant was collected and stored at −80 °C until use. Protein concentration was determined using a BCA Protein Assay Kit (Thermo Fisher Scientific, IL, USA) following the manufactureŕs protocol. A 10 μg aliquot from each sample was run on a 10% SDS-PAGE to evaluate total extract.
Protein digestion was performed with trypsin (Sigma-Aldrich, MO, USA) at a 1:50 ratio using a modified FASP protocol, originally described by Wiśniewski and colleagues [42]. Briefly, 200 μg of proteins from the total extract were reduced with 0.02 mM dithiothreitol (DTT) at room temperature, then alkylated with 0.05 mM iodoacetamide on a Microcon YM-10 MWCO 10 KDa filter (Merck Millipore Ltd., County Cork, Ireland) at room temperature while protected from light. The pH of each sample was checked and, if necessary, adjusted to pH 8.0 with 0.1 M HCl or 0.1 M NaOH. Further incubation was done at 37 °C for 18 hours and digestion was terminated with the addition of 2 μL of 10% TFA. Tryptic peptides were labeled through dimethylation, performed by incubating the peptide extract with 20 μL of the light label mixture for the control condition (500 μL of 50 mM TEAB, 2.8 μL of 37% CH2O and 25 μL of 0.6 NaBH3CN) and 20 μL of the heavy marker mixture for samples treated with venom (500 μL of 50 mM TEAB, 5 μL of 20% 13CD2O, 25 μL of 0.6 M NaBD3CN) for 2 hours at room temperature. The reaction was stopped with 5 μL of 1% ammonia and incubated for 30 min at 35 °C. The extracts treated with saline and venom were combined in a 1:1 ratio. Desalting was carried out following the method described by Rappsilber et al. 2007 [43] with some modifications. Briefly, 30 μg of tryptic peptides were desalted on Stage-Tips on 200 μL pipettor tips with three layers of SDB-XC membrane (styrene-divinylbenzene, Empore, 3M, Royersford, PA, USA). The membranes were initially conditioned with 100% methanol, washed with solution A (5% acetonitrile, 0.1% TFA), loaded with samples, and subsequently washed with solution A. The peptides retained on the membranes were eluted three times with 20 μL of solution B composed of 80% acetonitrile, 0.1% TFA and combined. Before proceeding to mass spectrometry analysis, all tryptic peptides were analyzed on an SDS-PAGE stained with silver nitrate in order to verify the digestion quality.
2.6. Mass spectrometry-based proteomic analysis
Light and heavy dimethylated tryptic peptide samples were analyzed on the Q-Exactive HF quadrupole hybrid mass spectrometer (Thermo Scientific, Bremen, Germany) coupled to a nano LC UltiMate 3000 system (Thermo Scientific) using the shotgun/bottom-up approach in positive ion mode. One microgram of peptides from each sample was injected at a gradient of 5 to 50% solvent B (90% acetonitrile, 0.1% TFA) in 60 min at a flow rate of 200 nL/ min. The electrospray source was operated at 2.2kV, and the peptide mixture was analyzed by acquiring full scan mode spectra with a resolution of 120,000 for the determination of MS1 with a maximum injection time of 60ms in the range of 375 to 1500 m/z. The dependent data acquisition (DDA) was carried out by automatically selecting the 20 most intense peaks for the subsequent acquisition of product ion spectra with MS/MS (MS2) with a resolution of 15,000 with a maximum injection time of 40 ms over a range of 200 to 2,000 m/z with a dynamic exclusion of 15 seconds.
2.7. Data processing and statistical methods
Raw data files were analyzed using PEAKS Studio version X plus software (Bioinformatics Solutions Inc, Toronto, Canada) using de novo sequencing tools and Search DB for the classic search of sequences against the Mus musculus databank, downloaded from UniProtKB/ Swiss-Prot in July 2020. The parameters used were: carbamidomethylation of cysteine as fixed modification; methionine oxidation and deamination of asparagine and glutamine as variable modifications with mass error tolerance ±10 ppm for MS and ±0.02 Da for MS/ MS. Trypsin was selected as the enzyme used in protein digestion and two lost cleavages per peptide and up to three post-translational modifications were allowed. Protein decoy was enabled and false discovery rate (FDR) using the Benjamini-Hochberg method was set to 0.1% and the ALC (average local confidence) was set to 80%. The maximum allowable retention time window was three minutes. Protein quantification was performed by dimethylation labeled as light (+28 Da) for control and heavy (+36 Da) for venom treated samples [44]. The identification of protein groups, normalization of the ion spectra areas, and determination of the significance of the protein groups were performed using the algorithm included in the PEAKS X plus software. Protein identification comparison was analyzed using an online Venn diagram generator (http://bioinformatics.psb.ugent.be/webtools/Venn/). Unique proteins identified in the venom-treated mice for each time point were obtained through a screening analysis, in which only proteins present in each of the venom treated-condition (exclusive proteins) were analyzed. Data from all time points were analyzed through the functional enrichment analysis available in the Webgestalt tool (http://www.webgestalt.org/) as described by Wang et al. (2017) [45]. Overrepresentation Enrichment Analysis (ORA) was created for the Gene Ontology (GO) of biological process, cellular component, and molecular function. The FDR was set to 0.05 using the Benjamini Rochberg method, which was used in the statistical tests to determine the enrichment when comparing among all tested time points.
Semi-quantitative analysis values from the raw data table generated by the PEAKS X plus software were filtered according to the minimum occurrence of at least three of the five valid values of the normalized ratio in each experimental condition. Afterward, the data were transformed using log2 and z-scored using Perseus software version 1.6.5.0 [46] and submitted to a hierarchical grouping based on Euclidean distance. Clusters were analyzed performing an ORA analysis for Mammalian phenotype ontology in order to verify the behavior of the proteins grouped in each cluster. Also, the same protein set was analyzed using STRING [47], keeping only proteins that were involved in cardiomyopathy and formed a protein-protein interaction (PPI) network. The network was uploaded to Cytoscape [48, 49] to generate a pathway overview. Principal component analysis (PCA) used the normalized protein quantification values to find which linear combinations would explain most of the variability of the differentially expressed proteins for the different assays. The PCA studies were performed using the R-statistics FactoMineR [50] and Factoextra (http://www.sthda.com/english/rpkgs/factoextra) for graphical visualization.
All of the raw data were uploaded and stored at the Center for Computational Mass Spectrometry of the University of California, San Diego, MassIVE website and can be downloaded from: ftp://massive.ucsd.edu/MSV000087836/
3. Results
3.1. Unique proteins identified in venom-treated mice
Hearts were dissected, lysed and total protein was quantified using the BCA protein assay kit. Cardiac tissue yield ranged from 5.72 to 8.18 mg (Supplemental Table 1). Each sample was lysed using PTS buffer, reduced with DTT, alkylated with IAA, and digested with trypsin. Among all treatment and control samples, mass spectrometry analysis of trypsin-digested tissue samples identified a total of 20,146 spectra corresponding to 9,767 peptides and 1,341 distinct proteins (Supplemental Table 2). We identified 1,047 unique proteins at the 1 h time point, 747 at 6 h, 860 at 12 h, and 1,027 at 24 h (Supplemental Figure 1A and Supplemental Table 3). To identify proteins unique to the venom treatment groups, we compared control (saline) groups against the venomtreated samples from each time point. In the venom treatment samples, we identified 73 unique proteins at the 1 h time point, 5 at 6 h, 11 at 12 h, and 40 at 24 h (Supplemental Figure 1A and Supplemental Table 3). Functional enrichment analysis of differentially abundant proteins at each time point revealed that after 1 h of venom treatment, proteins involved in “cell adhesion mediator activity” and “mitochondrial gene expression” showed the highest and the lowest enrichment rates, respectively (Supplemental Figure 1B). In the 6 h post-treatment group, “deaminase activity” and “foam cell differentiation” showing the highest enrichment rates and “platelet-derived growth factor receptor signaling pathway” showed the lowest enrichment rate (Supplemental Figure 1C). At the 12 h venom-treatment time, the GO category “peroxisome organization” showed an FDR ≤ 0.05 with the highest enrichment rate, while “somatic diversification of immune receptors” showed the lowest enrichment rate (Supplemental Figure 1D). At 24 h venom treatment, the “retromer complex” GO function presented the highest enrichment rate, while “signal transduction in response to DNA damage” showed the lowest enrichment rate (Supplemental Figure 1E).
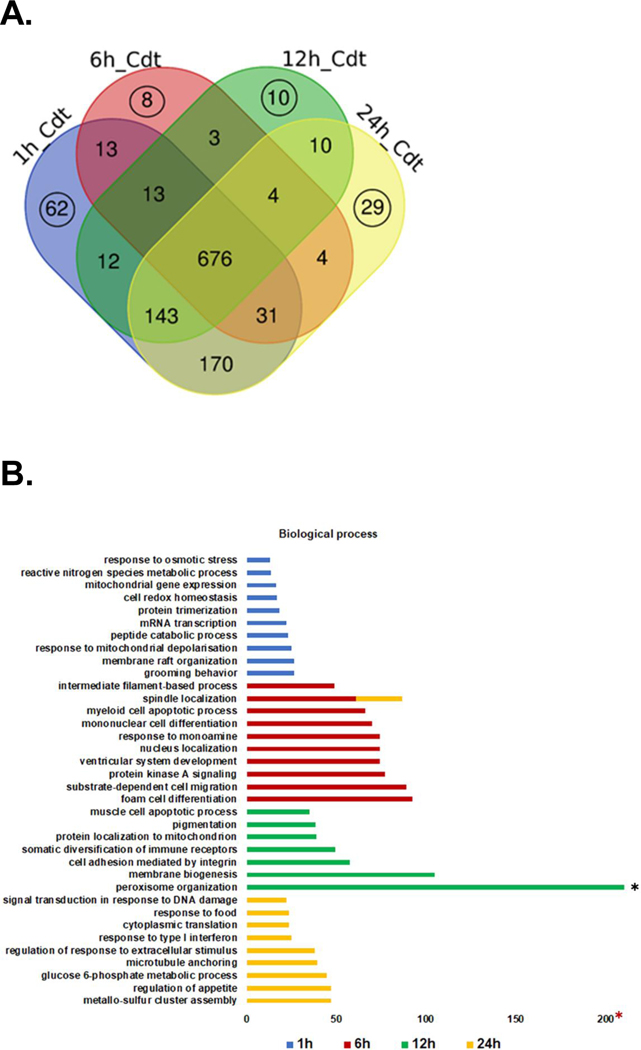
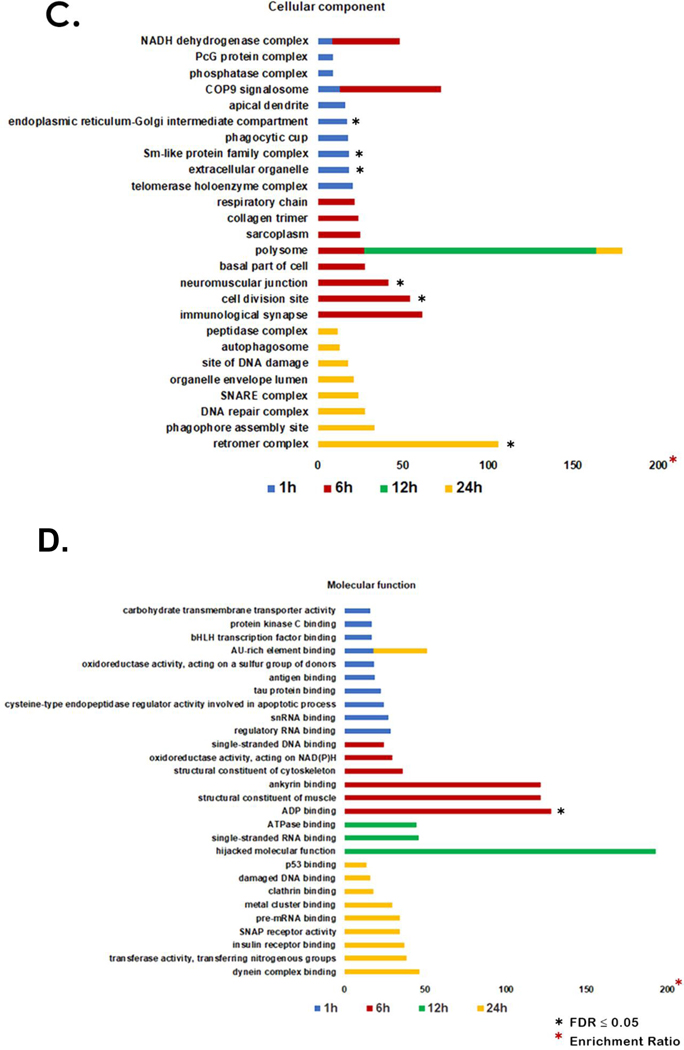
Among the 676 proteins identified only in venom-treated samples, 62 were exclusively present in the 1 h group, 8 in the 6 h group, 10 in the 12 h group, and 29 in the 24 h group (Figure 1A and Supplemental Table 4). We used the Webgestalt tool to perform functional enrichment and classify these exclusive proteins based on their biological process (Figure 1B), cellular components (Figure 1C), and molecular function (Figure 1D) at each time point.
Figure 1.
Identification of proteins in the venom treatment groups. A. Venn diagram comparing proteins exclusively found in the venom treatment group at each time point is represented by black circles. These were submitted to subsequent functional analysis. B. Enrichment for GO biological process. C. Enrichment for GO cellular component. D. Enrichment for GO molecular function.
A total of 36 GO categories were identified and enriched in order to classify identified proteins according to which biological processes they are involved in. Many of the GO categories were only represented at certain time points. However, “spindle localization” was enriched in both the 6 h and 24 h sample groups. “Peroxisome organization,” present in the 12 h group, exhibited the highest enrichment rate (FDR ≤ 0.05) of any GO category (Figure 1B).
Cellular component analysis showed 26 enriched GO categories. “Endoplasmic reticulum-Golgi intermediate compartment”, “Sm-like protein family complex”, and “extracellular organelle” showed enrichment with FDR ≤ 0.05 in the 1 h venom treatment group. In the 6 h treatment group, “neuromuscular junction” and “cell division site” showed enrichment with FDR ≤ 0.05. In the 24 h venom treatment group, “retromeric complex” showed the highest enrichment rate with FDR ≤ 0.05. The “polysome” GO category showed significant enrichment rates after 6 h, 12 h and 24 h venom treatment while “NADH dehydrogenase complex” and “COP9 signalosome” were enriched at 1 h and 6 h after venom treatment (Figure 1C).
Among the 28 GO categories enriched and identified in the molecular function analysis, “AU-rich element binding” were enriched at 1 h and 24 h after venom treatment. “ADP binding” was the only category to show an enrichment rate with FDR ≤ 0.05 at 6 h venom treatment. “Hijacked molecular function” was observed at 12 h after venom treatment and showed the highest enrichment rate of any of the GO categories (Figure 1D).
3.2. Semi-quantitative analysis
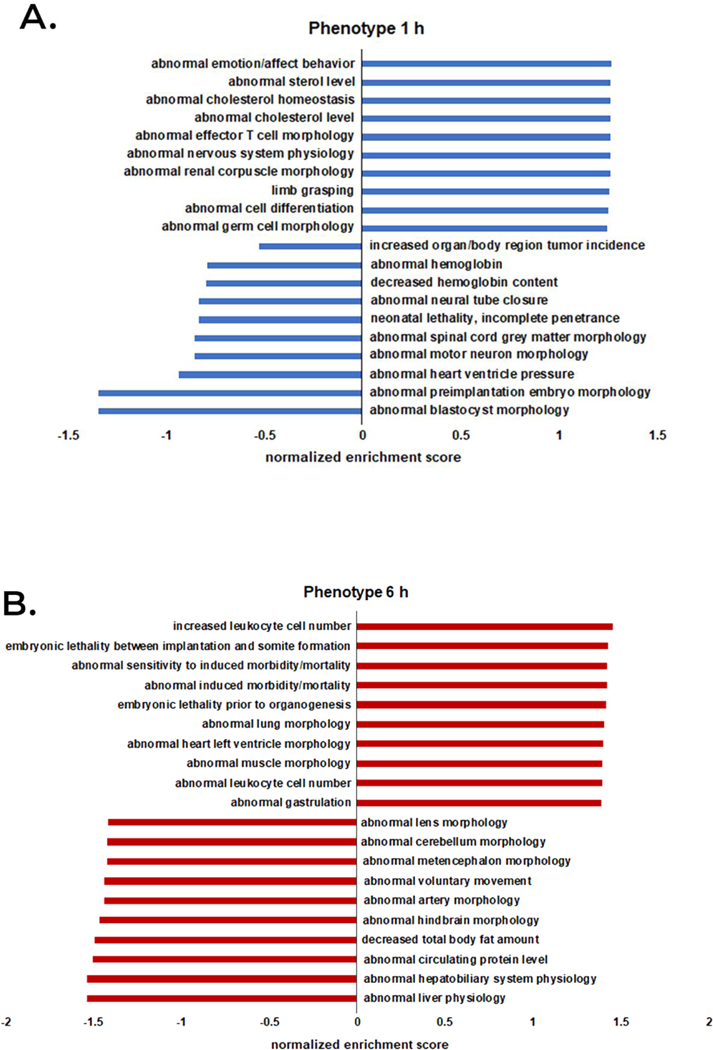
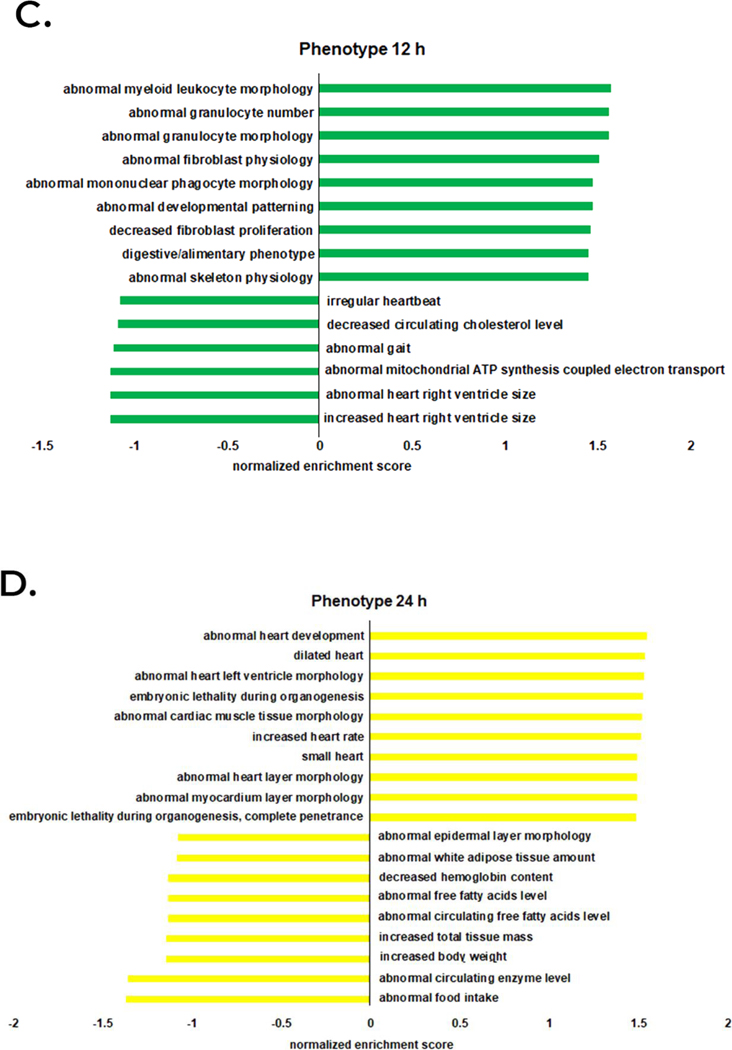
Quantitative mass spectrometry analysis was performed on a total of 1,108 proteins identified in either control and venom-treated samples (Supplemental Table 5). Quantitative enrichment analysis was performed using gene set enrichment analysis (GSEA) supported by the Webgestalt tool. At the 1 h time point, the venom-treated group showed enrichment of several GO categories including “abnormal cholesterol homeostasis”, “abnormal cholesterol level”, “abnormal nervous system physiology”, and “abnormal cell differentiation” and a decrease in “abnormal motor neuron morphology” and “abnormal heart ventricle pressure” categories (Figure 2A). At 6 h, the venom-treated group showed increased enrichment for “abnormal sensitivity to induced morbidity/mortality”, “abnormal induced morbidity/mortality”, “abnormal heart left ventricle morphology”, “abnormal muscle morphology”, and “abnormal leukocyte cell number” and decreased enrichment was observed for “abnormal artery morphology”, “decrease total body fat amount” and “abnormal circulating protein level” (Figure 2B). At 12 h, the venom treatment group showed an increase in enrichment for “abnormal fibroblast physiology”, “abnormal developmental patterning”, “decreased fibroblast proliferation”, and “digestive/alimentary phenotype” and decreased enrichment for “irregular heartbeat”, “decrease circulating cholesterol level”, “abnormal mitochondrial ATP synthesis coupled electron transport”, “abnormal heart right ventricle size”, and “increase heart right ventricle size” (Figure 2C). At 24 h, the venom treatment group showed increased enrichment for “abnormal heart development”, “dilated heart”, “abnormal heart left ventricle morphology”, “abnormal cardiac muscle tissue morphology”, “increased heart rate”, “small heart”, “abnormal heart layer morphology”, and “abnormal myocardium layer morphology” and decreased enrichment for “abnormal white adipose tissue amount”, “abnormal circulating free fatty acids level”, “increase total tissue mass”, “increase body weight”, “abnormal circulating enzyme level”, and “abnormal food intake” (Figure 2D).
Figure 2.
Semi-quantitative functional analysis for mammalian phenotype enrichment. All protein intensity values identified by mass spectrometry quantification analysis were averaged and submitted to functional analysis. A-D denote 1 h to 24 h time points, respectively.
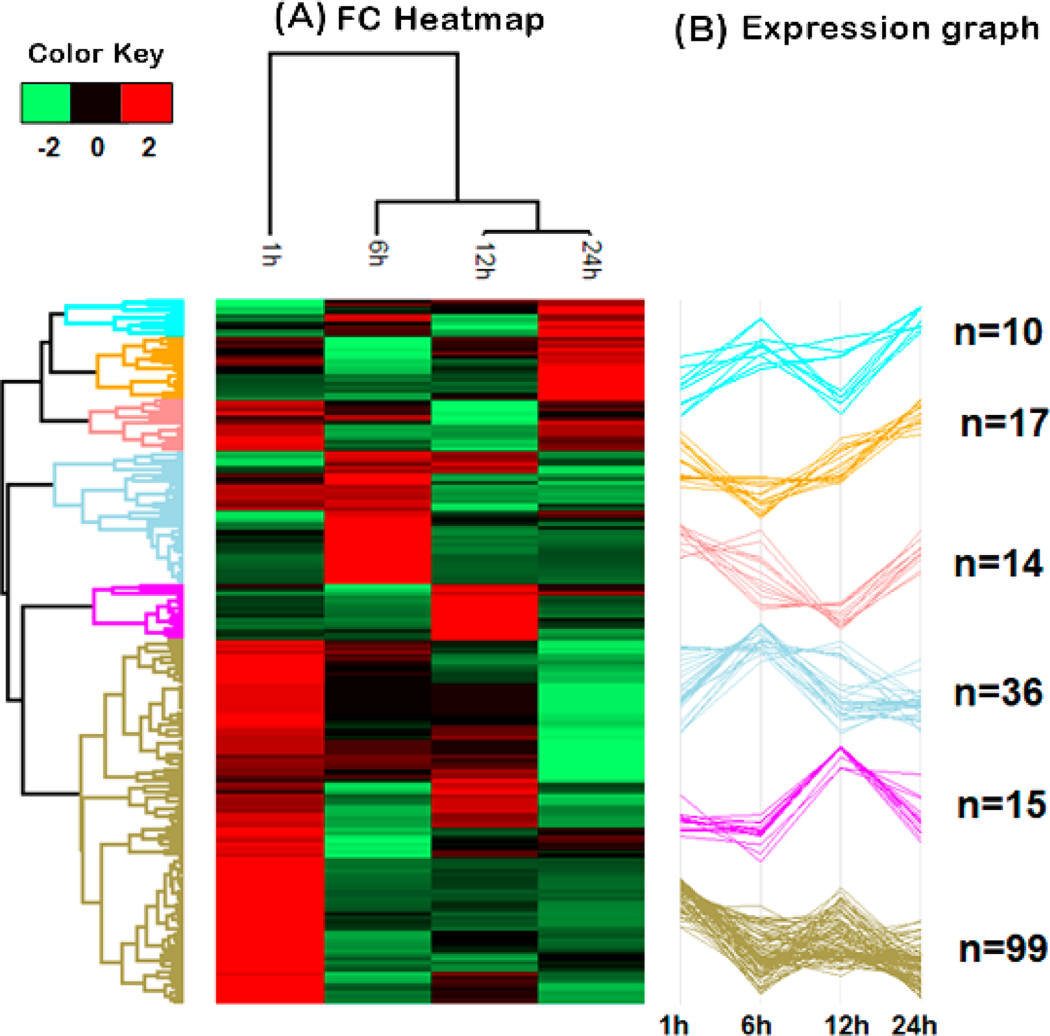
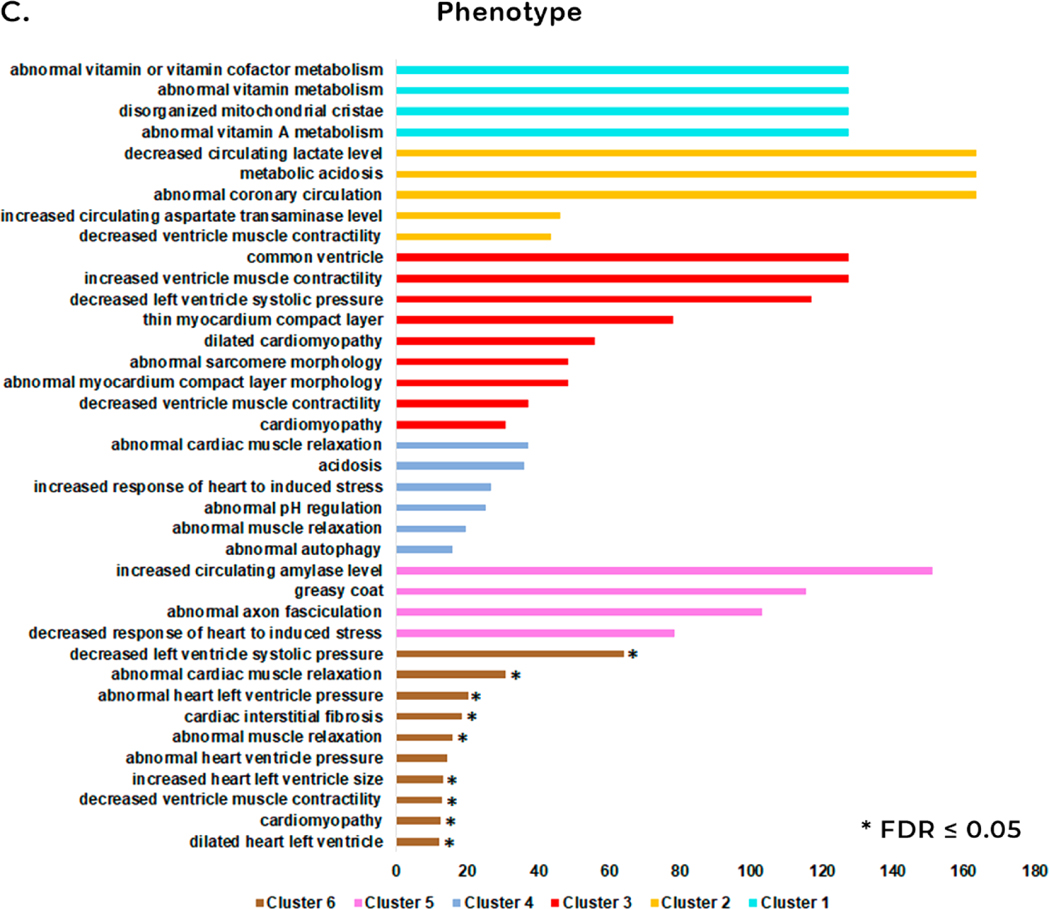
Analysis of changes in relative abundance identified 191 unique proteins among all time points in both the control and venom-treated samples (Supplemental Table 6). Hierarchical clustering analysis clustered venom treatment proteins into six clusters (Figure 3A). Based on these results, we performed an Overrepresentation Enrichment Analysis (ORA) for phenotype (Mammalian Phenotype Ontology) using the Webgestalt tool for each cluster (Figure 3C).
Figure 3.
Hierarchical clustering of protein quantification at 1 h, 6 h, 12 h, and 24 h post venom treatment. A. Hierarchical cluster of the most significant proteins for each treatment showing changes in protein abundance over time. The fold change (FC) in protein abundance of venom treated samples over the control samples is represented on a log2 scale. (B) Expression graphs of each cluster and the numbers of proteins composing each cluster. The protein quantification values were normalized by Z-score in rows. (C) The GO mammalian phenotype enrichment for all clusters.
The expression plot in cluster 1 shows an increase in the relative abundance of proteins at 6 h and 24 h and a decrease in abundance at 1 h and 12 h after venom treatment (Figure 3B). Enrichment analysis for phenotype of this set of proteins showed an increase of four GO categories: “abnormal vitamin or vitamin cofactor metabolism”, “abnormal vitamin metabolism”, “disorganized mitochondrial cristae” and “abnormal vitamin A metabolism” (Figure 3C, shown in light blue). Cluster 2 showed a decrease in the relative abundance of proteins at 6 h and an increase at 1 h, 12 h and 24 h (Figure 3B). The total number of GO categories identified with the highest enrichment rate were: “decreased circulating lactate level”, “metabolic acidosis” and “abnormal coronary circulation” (Figure 3C, shown in orange). Cluster 3 showed a decrease in the relative abundance of proteins at 12 h and an increase in relative abundance at 1 h, 6 h, and 24 h after venom treatment (Figure 3B). The enrichment analysis for the phenotype of these proteins were related to phenomena that occur in the heart. Among them, we highlight “common ventricle” and “increased ventricle muscle contractility” with the highest enrichment rate (Figure 3C, shown in red). Cluster 4 showed an increase in relative abundance of proteins at 6 h and a decrease in abundance at 1 h, 12 h and 24 h after venom treatment (Figure 3B). Phenotype enrichment analysis revealed several GO categories, of which “abnormal cardiac muscle relaxation” and “acidosis” showed the highest enrichment rates (Figure 3C, shown in dark blue). Relative expression analysis of proteins in cluster 5 grouped proteins that increased in the relative abundance at 12 h and a decrease in this abundance at 1 h, 6 h, and 24 h after envenomation (Figure 3B). Phenotype enrichment analysis of proteins in this cluster yielded four GO categories, including “decreased response of heart to induced stress”. (Figure 3C, shown in pink). Cluster 6 showed decreased protein abundance from 6 h to 24 h (Figure 3B). Functional enrichment for phenotype showed 10 GO categories with potential relevance to pathology in the heart, including “dilated heart left ventricle”, “cardiomyopathy”, “decreased ventricle muscle contractility”, “increased heart left ventricle size”, “abnormal heart ventricle pressure”, “abnormal muscle relaxation”, “cardiac interstitial fibrosis”, “abnormal heart left ventricle pressure”, “abnormal cardiac muscle relaxation”, and “decreased left ventricle systolic pressure” (Figure 3C, shown in brown).
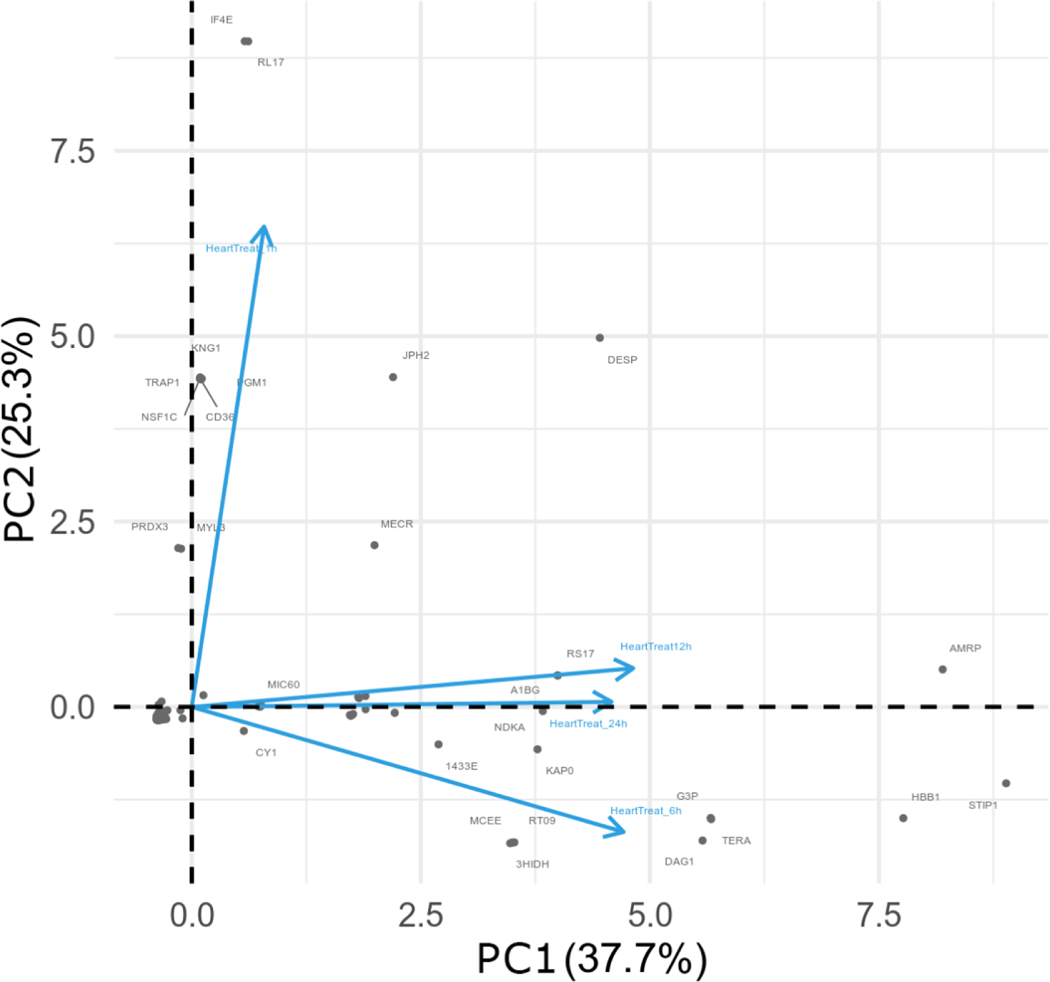
3.3. PCA analysis
For all treatment groups, proteins present in at least three replicates were subjected to Principal Component Analysis (PCA) and time points were projected into new component space to visualize differences at each time point. The orthogonal projection between the vectors for the 1h and 6h time points highlight key proteins at each time point potentially involved in physiological activity in heart tissue resulting from venom injection. The first two components (PC1 and PC2) explain 63% of the total variability in protein expression (Figure 4).
Figure 4.
Comparison of protein log2 fold change (FC) profiles across different time points upon venom treatment. The first two components, PC1 and PC2, explain 63% of the protein variability observed among all time points, as shown in the 2D graph. Note that 12 h and 24 h data graph closer to each other, representing higher correlation between their respective protein profiles. The 1 h and 6 h venom treatment vectors are orthogonal in terms of the observed proteome and indicate that those proteins, and their respective profiles, are very divergent.
At 6h after envenomation, an intermediary time point for the systemic response, there was a negative correlation based on PC2 to the other time points, corroborating the clustering analysis. This is especially evident in Cluster 4, where many proteins increased in abundance. Notably included in this increase are MYOZ2 and NDUS6, which are related to “increased response of heart to induced stress” and “abnormal cardiac muscle relaxation”, respectively (Figure 3C). A strong positive correlation between PC1 and PC2 presented at the 12h and 24h time points, again corroborating the clustering analysis (Figure 3C). This is evident in Cluster 2, where many proteins decreased in abundance. Notably, though, some proteins increased in abundance, including SODM and MYG, which are both related to “decreased ventricle muscle contractility”. At the 1h time point, a strong positive correlation presented in the PC2, significantly diverging from the other time points. This is due to a set of exclusive recruited proteins associated with the early response to the venom treatment, as evidenced by the clustering analysis (Figure 3C). Cluster 6, for example, includes PDLI5 and MYH6 related to “dilated heart left ventricle”, “cardiomyopathy” and “abnormal heart left ventricle pressure”.
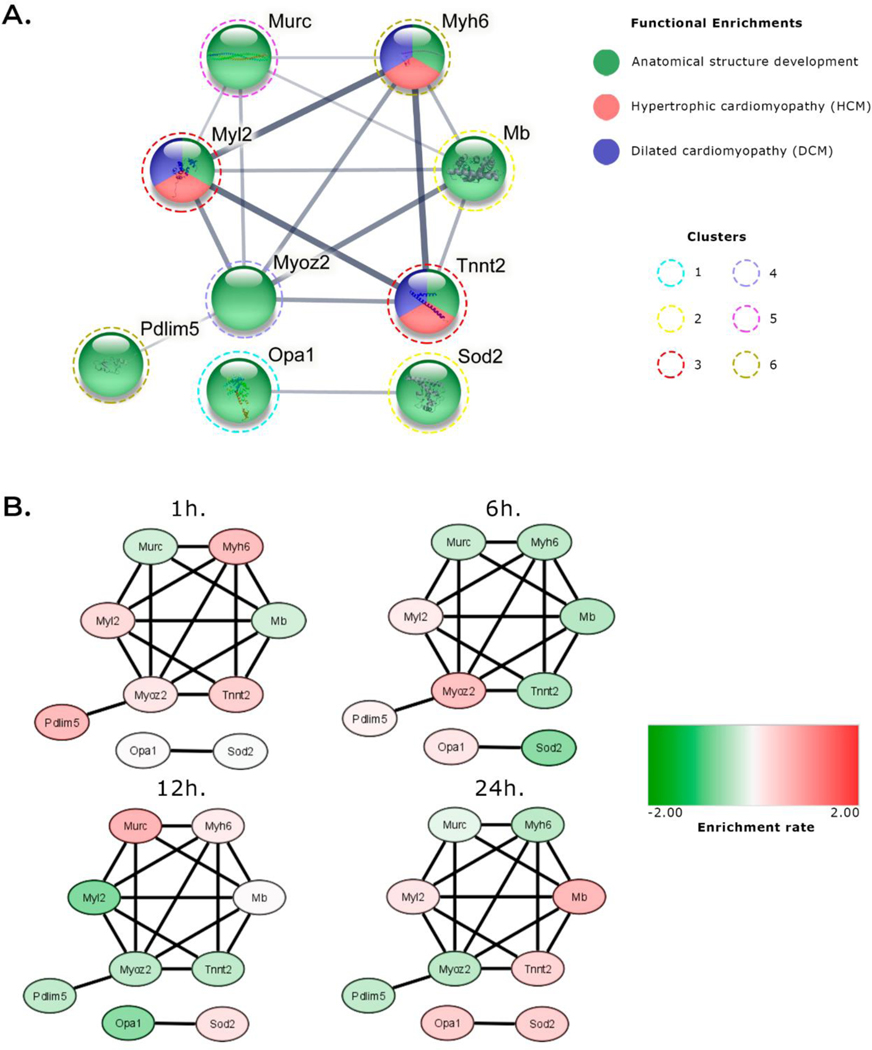
3.4. Network analysis
Network analysis (Figure 5) found nine proteins with important roles in cardiac tissue that fell into two clusters. The first cluster contained Murc, Myh6, Mb, Tnnt2, Myoz2, Pdim5, and Myl2, and the second cluster contained Opa1 and Sod2 (Figure 5A). All of these proteins are involved in heart structure development, and Myh6, Tnnt2 and Myl2 are also involved in Hypertrophic cardiomyopathy (HCM) and Dilated cardiomyopathy (DCM) (Figure 5A). All of these proteins showed increased abundance at 1h. Only Myl2 was upregulated at the 6 h time point, only Myh6 was upregulated at 12 h, and at 24 h both Myl2 and Tnnt2 were upregulated (Figure 5B).
Figure 5.
Network visualization of semi-quantitative highlighted proteins. A. STRING PPI analysis. The edge width represents the confidence found for the given interaction. The color of each node is represented by its functional assignment or related disease. The dashed colored line around nodes represents the cluster of origin. B. Network analysis combining quantitative results using Cytoscape. This result shows individual patterns of up- or down-regulation (represented as red and green, respectively) of these proteins. Minimum required interaction score: medium confidence (0.400).
3.5. Histology analysis
Examination by microscopy of H-E stained heart tissue sections did not show major changes in gross histopathology during the acute phase of venom challenge. The heart tissues collected at 1 h, 6 h, 12 h and 24 h after venom treatment did not show apparent increase of necrosis or inflammatory infiltration when compared to the control groups. There were signs of blood vasculature leakage in some of animals 12 h or 24 h after venom treatment, but the alteration was subtle and could be detected in only half of the venom treated animals at the 12 h to 24 h time points (Supplemental Figure 2). No such changes were observed at the 1hr or 6hr time points (Chi Square p=0.045, considering the observation of both 12 h and 24 h).
4. Discussion
Crotalus d. terrificus venom is known to have myotoxic, nephrotoxic, neurotoxic, and cardiotoxic activities which result in a variety of clinical symptoms including alteration of central nervous system function, motor paralysis, eyelid ptosis, ophthalmoplesia, blurred vision, rhabdomyolysis, myoglobinuria and cardiorespiratory arrest, and in the most severe cases, death. Clinical effects of C. d. terrificus venom on patients’ have been previously described, as have the physiological effects of both whole venom and isolated venom components in animal models. Siqueira and colleagues (1990) observed extensive and severe myocardial injury in a patient bitten by C. d. terrificus. Electrocardiogram alterations included low QRS voltage, diffuse changes in ventricular repolarization, and increased serum enzymes such as total CK, CK-MB, HDL cholesterol, aspartate (AST), and alanine aminotransferases (ALT) which peaked 48 h after hospitalization [22]. In a clinical review of 21 cases of children envenomated by C. d. terrificus, Cupo and colleagues (1993) describe tachycardia, serum CK, lactate dehydrogenase (LD), isoenzymes, CK-MB, and LD1–5 levels similar to those observed in acute myocardial infarction [20]. Santos and colleagues (1990) analyzed the effect of crotoxin, isolated from C. d. terrificus venom, perfused into guinea pig hearts for 90 min. They noted decreased contraction force and negative inotropic effect without changes in heart rate. They also reported increased CK activity related to the reduced force of contraction [21]. In an earlier study, Breithaupt (1976) perfused phospholipase A2, crotaponin, or a combination of the two into rat and rabbit hearts. They observed no alterations on heart rate, contractility, or coronary flow with any of these conditions [51].
More recently, Simoes and colleagues described the effect of C. d. cascavella venom in the heart of Wistar rats. They observed that 30 μg/ mL venom induced significant negative inotropic effects due to reduced contractile force with no changes in rhythmicity. This suggested the absence venom action on the sinoatrial node and participation of the NO/cGMP/PKG pathway. The venom induced hypotension followed by bradycardia. Neither total-CK or CK-MB levels showed any change, suggesting that the venom is not myotoxic in cardiac tissue. Interestingly, electron microscopy analysis of cardiac tissue showed that the negative inotropic effect is not related to cardiac tissue damage or cell death but may be associated with focal injury to the cell ultrastructure by inducing mitochondrial dysfunction [34].
Our characterization of heart tissue after exposure to C. d. terrificus venom found changes in abundance of several noteworthy proteins at various time periods following after treatment. Functional analysis of the identified proteins based on the GO phenotype analysis found affects on several proteins associated with mitochondria at different time points after venom injection, corroborating the electron micrograph analysis performed by Simoes and colleagues [34].
At 1 h (Figure 2A) after venom treatment, we observed upregulation of proteins related to abnormalities of the renal corpuscle. Kidney disorders following Crotalid envenomation have been previously reported. Acute kidney injury (AKI) and renal failure was the most severe complication reported; proteinuria, and hematuria, which are the most common manifestations of Crotalid snakebites [37, 52–55].
Crotoxin exhibits strong neurotoxic action, impacting both the nervous system and the conduction pathways of cardiac tissue [56]. Interestingly, we detected enrichment of several GO protein categories related to lipid disorders associated with abnormal levels of sterols and cholesterols. Our data support Marinovic and coworkers, who found that crotamine reduces the level of low-density cholesterol and triglycerides, and increases levels of high-density cholesterol [57]. Our observation of downregulation of proteins related to tumorigenicity also supports previous reports of antitumoral activity by crotamine [58].
The 6 h post-venom time point approximately corresponds to the average time from envenomation before patients receive medical assistance (personal communication with doctors at Hospital Vital Brazil (HVB), Butantan Institute São Paulo, specialized in the care of patients bitten by venomous animals). This is the time point when we observed a pronounced increase in GO categories related to processes contributing to ultimate mortality, suggesting that the venom causing critical disturbances in the heart and in the organism as a whole. Protein GO categories related to ventricular abnormalities were observed as at other time points but at 6 h, we also observed an increase in left ventricle pressure. This may signal a change in the scenario observed at 1 h post-venom time point, although we cannot explain the exact mechanism for this occurrence. We also observed changes in proteins belonging to GO categories related to embryonic development and lung abnormalities. Proteins downregulated at the 6 h time point mostly belong to GO categories related to impaired liver function, abnormal voluntary movement, and altered brain morphology. Interestingly, we previously reported changes in several proteins, including proteins related to brain morphology, in the cerebellum of mice treated with C. d. terrificus venom [30].
At the 12 h time point (Figure 2C), we observed an increase in a protein GO category related to the abnormal number of granulocytes, which may indicate an activation of local immune activity against the venom. An increase in proteins belonging to GO categories related to myeloid leukocyte morphology and abnormal morphology of mononuclear phagocytes and granulocytes also indicate implications for these immune cell types. Cruz et al. (2005) have previously observed that C. d. terrificus venom contributed to morphological, functional, and biochemical alterations of macrophages in BALB/c mice [59]. We observed an increased enrichment for the GO categories and function related to abnormal fibroblast physiology and decreased fibroblast proliferation. In contrast to earlier time points, we observed a reduction of GO categories related to arrhythmia, abnormal right ventricle size, and increased right ventricle size at 12 h. We also saw a reduction of proteins in the GO category involved in decreased circulating cholesterol, in stark contrast to the 1 h time point. Winkler and colleagues (1993) noted a decrease in circulating cholesterol in patients and rabbits after Vipera palaestinae envenomation which negatively correlated with overall case severity [60]. The authors suggested that this phenomenon was likely due to transcapillary lipoprotein leakage as well as changes in lipoprotein transport and metabolism caused by venom components such as PLA2.
At the 24 h time point (Figure 2D), we observed an increase in several GO categories and function involved in cardiac cell damage and altered heart morphology, suggesting possible damage to the cardiac tissue in mice. These data corroborate previous reports of similar phenotypes, such as bilateral ventricle damage and infiltration of immune defense cells[23]. Protein GO categories associated with white adipose tissue were reduced at the 24 h time point, as was body mass. This supports similar observations presented by Morinovic et al. [57].
Hierarchical clustering analysis allowed us to observe changes in protein abundance among post venom treatment time points (Figure 3 and Supplemental Table 6). Among the identified proteins, we highlight OPA1 (dynamin-like 120 kDa protein) in cluster 1, SODM (superoxide dismutase) and MYG (myoglobin) in cluster 2, MLRV (Myosin Regulatory Light Chain) and TNNT2 (Troponin T2) proteins in cluster 3, MYOZ2 (myozenin 2) in cluster 4, CAVN4 (caveolae-associated protein 4) in cluster 5, and PDLI5 (PDZ and LIM domain protein 5) from cluster 6 (Table 1). All these proteins are related to dysfunctions and damage in the heart and in other organs. SODM is an antioxidant enzyme, which is associated with oxidative stress. Interestingly, one of the enzyme families present in C. d. terrificus venom is the L-amino acid oxidases (LAAOs). The LAAOs are flavoenzymes that catalyze the stereospecific oxidative deamination of L-amino acid substrates to yield an alpha-keto acid, ammonia, and hydrogen peroxide (H2O2). Although LAAÓs physiological role in venom is still unknown, it is speculated that they may be related to venom conservation and stabilization of the venom glands due to their antibacterial properties. L-amino acid oxidases also cause plasma clotting and platelet aggregation. Some authors suggest that these hemolytic activities may be related to the H2O2 produced during the chemical reaction catalyzed by LAAOs, which may cause oxidative stress to the targeted cells and tissues. Although no references are found relating the oxidative stress and free radical pathways to snake venom activity in heart tissue, oxidative stress following Crotalus envenomation has been noted in other mammalian organs, such as liver and kidney [61–65].
Table 1.
Description of proteins identified and highlighted from each cluster 1–6 shown in Fig. 3.
| Accession # | Protein | Protein name | Cluster | Related GO category | Description | Fold change | Reference | |||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
|
|
|
|||
| 1 h | 6 h | 12 h | 24 h | |||||||
| P58281 | OPA1 | Dynamin-like 120 kDa protein | 1 | disorganized mitochondrial cristae | Maintain the normal structure and function of mitochondrial crests, and preserves the structure of the inner membrane to protect cells from apoptosis. The reduction of OPA1 may lead to cell death and contribute to the progression of heart failure. | 0.03 | 0.43 | −1,39 | 0.93 | [74,75] |
| P09671 | SODM | Superoxide dismutase | 2 | decreased ventricle muscle contractility | Play an important role in inducing the mitochondrial pathway of apoptosis in the heart, arising mainly through the mitochondrial permeability transition. | −0.04 | −1.39 | 0.53 | 0.89 | [76–80] |
| P04247 | MYG | Myoglobin | 2 | decreased ventricle muscle contractility | Present in the cytoplasm of skeletal and cardiac muscles, and can be detected in high plasma concentrations as one of the first biomarkers in the first hours after an acute myocardial infarction | −0.54 | −0.89 | 0.04 | 1.39 | [81–83] |
| P51667 | MRLV | Myosin regulatory light chain | 3 | cardiomyopathy, decreased ventricle muscle contractility, abnormal sarcomere morphology and dilated cardiomyopathy | Play an important role in the structure and function of the cardiac muscle, Mutations found in the gene encoding MLRV have been reported to be associated with left ventricular middle chamber hypertrophic cardiomyopathy (HCM) and a role in the development and progression of chronic heart failure | 0.68 | 0.34 | −1.48 | 0.46 | [66–69] |
| P50752 | TNNT2 | Troponin T2 | 3 | cardiomyopathy, decreased ventricle muscle contractility, abnormal sarcomere morphology and dilated cardiomyopathy | Present a physiological activity in calcium regulation of the actin thin filament function, which ultimately becomes essential for the cardiac muscles contraction. It has also been reported that mutations in the gene encoding TNNT2 could result in dilated cardiomyopathy | 0.91 | −0.95 | −0.78 | 0.81 | [70,72,84] |
| Q9JJW5 | MYOZ2 | Myozenin 2 | 4 | Increased response of heart to induced stress | Related to hypertrophic cardiomyopathy (HCM) due to mutations in its encoding gene | 0.43 | 1.21 | −0.81 | −0.83 | [85,86] |
| P52503 | NDUS6 | NADH:Ubiquinone Oxidoreductase Subunit S protein | 4 | abnormal cardiac muscle relaxation | Relate to possible specific cardiomyopathy due to mitochondrial complex I deficiency, which may increase the risk of heart failure and death | −0.34 | 1.49 | −0.61 | −0.54 | [87–89] |
| A2AMM0 | CAVN4 | Caveolae-associated protein 4 | 5 | decreased response of heart to induced stress | Mutations in the MURC/Cavin-4 gene of the CAVN4 protein can be observed in individuals with dilated heart disease and a possible gene deletion could reduce ischemic-reperfusion injury in mice | −0.56 | −0.65 | 1.48 | −0.28 | [90] |
| Q8CI51 | PDLI5 | PDZ and LIM domain protein 5 | 6 | dilated heart left ventricle, cardiomyopathy, abnormal heart left ventricle pressure | Responsible for maintaining cellular homeostasis when the heart is subjected to biomechanical stress and thus regulates the activity of the cardiac calcium channel, preserving cardiac contractility | 1.35 | 0.17 | −0.75 | −0.77 | [91,92] |
| Q02566 | MYH6 | Myosin-6 | 6 | dilated heart left ventricle, cardiomyopathy, abnormal heart left ventricle pressure | Mutations in the gene encoding MYH6 may be related to diseases such as hypertrophic cardiomyopathy CMH and dilated CMD | 1.26 | −0.78 | 0.34 | −0.83 | [71–73] |
Some of the proteins noted above were also subjected to PCA analysis. Network and PPI analysis of the highlighted proteins revealed that all of these proteins play distinct roles in heart function: Myh6 is involved in muscle contraction, Tnnt2 serves as the tropomyosin-binding subunit of troponin, and Myl2 plays key roles in heart development and function (Figure 5A). These proteins have been reported to be involved in cardiomyopathies [66–73]. In the present study, these proteins showed upregulation at 1 h, which might suggest that they are involved in an early response to venom damage (Figure 5B), although we were not able to observe a consistent pattern through the entire post venom treatment process.
Histological analysis showed no significant sign of morphological alterations after C. d. terrificus venom treatment when compared to control. This is in line with recent observations by Simoes and colleagues (2021) who injected C. d. cascavellas venom in Wistar rats [34]. However, when De Paola and Rossi (1993) injected high dose of C. d. terrificus venom (80 μg in 0.2 mL of saline) intraperitoneally in Wistar rats, they observed swollen muscle fibers, contraction bands, and myocytolytic necrosis with a large number of mast cells corresponding to a mild to moderate edema in the interstitial space and infiltration of mononuclear cells 4 days after treatment [23]. In our experiments, we also observed signs of blood vasculature leakage in some of the animals at 12 h and 24 h after venom injection. Although not significant, these data corroborate the molecular changes observed in the mass spectrometry-based proteomic analysis.
Our observations shed new light on the clinical outcome of C. d. terrificus snakebite. At each time point after venom treatment, we noted altered profiles of proteins involved in classic Crotalid envenomation symptoms such as impaired kidney function, nervous system disruption, and damaged cardiac tissue. Furthermore, the modulations we observed may indicate proteins that could be incorporated into the production of improved antivenoms, facilitate improvements in clinical treatment strategies, or be candidates for novel therapeutic interventions to reduce the debilitating effects of Crotalid snakebite.
5. Conclusion
In this study, we analyzed the proteomic profile of >1,300 proteins present in mouse cardiac tissue 1 h, 6 h, 12 h, and 24 h after injection of 0.5 LD Crotalus durissus terrificus venom into the gastrocnemius muscle. We found substantial alterations of proteins related to heart tissue damage at both early and later time points following venom treatment. Venom induced changes in several cardiac tissue proteins that work synergistically through distinct functions in the heart tissue, and thus trigger a variety of immunological and biochemical effects leading to cellular damage in the early time points and, later, to structural damage.
Supplementary Material
Significance.
Venom of the tropical rattlesnake (Crotalus durissus terririficus) is known to be neurotoxic, myotoxic, nephrotoxic and cardiotoxic. Although there are several studies describing the biochemical effects of this venom, no work has yet described its proteomic effects in the cardiac tissue of mice. In this work, we describe the changes in several mouse cardiac proteins upon venom treatment. Our data shed new light on the clinical outcome of the envenomation by C. d. terrificus, as well as candidate proteins that could be investigated in efforts to improve current treatment approaches or in the development of novel therapeutic interventions in order to reduce mortality and morbidity resulting from envenomation.
Highlights.
C. d. terrificus rattlesnake venom treatment of mice showed changes in cardiac protein profile
C. d. terrificus venom modulated proteins involved in biological and cellular process, molecular function, and phenotypic profile
C. d. terrificus venom modulated GO categories related to cardiac cardiac tissue damage
Network analysis showed disturbances of proteins related to cardiac tissue damage
6. Acknowledgements
We thank Dr. Solange M.T. Serrano, Prof Hugo A. Armelin, Prof Graziella E. Ronsein, and Dr Eduardo S. Kitano for insightful discussion.
7. Funding
This work was supported by grants 2013/07467-1, 2016/04000-3, and 2017/17943-6 from the São Paulo Research Foundation (FAPESP). ESF was supported by an Erna and Jakob Michael “Visiting Professorship”, Department of Biological Regulation, Weizmann Institute of Science, Rehovot, Israel and grant 302809/2016-3 from the Brazilian National Council for Scientific and Technological Development, CNPq, Brazil. WSS was supported by the Coordination for the Improvement of Higher Education Personnel (CAPES, Brazil) institutional fellowship #88882.442313/2019-01 (Butantan Institute, Brazil), and SSSA was supported by CNPq institutional fellowship #131408/2019-4 (University of São Paulo, Brazil). CYK was supported by FAPESP master’s degree program fellowship 2017/06496-9. ZC was supported in part by a grant from the National Cancer Institute (NCI) of National Institutes of Health (NIH) R01CA245673.
Footnotes
No conflict of interest statement
The authors declare that there is no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].World Health Organization, Snakebite Envenoming, Available online: https://www.who.int/news-room/fact-sheets/detail/snakebite-envenoming, 2021. (accessed on 8 January, 2022).
- [2].Jadon RS, Sood R, Bauddh NK, Ray A, Soneja M, Agarwal P, Wig N, Ambispective study of clinical picture, management practices and outcome of snake bite patients at tertiary care Centre in Northern India, J. Family Med. Prim. Care 10 (2) (2021) 933–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Azevedo-Marques MM, Cupo P, Hering SE, Accidents by venomous animals: venomous snakes (Portuguese), Medicina (Ribeirao Preto. Online) 36 (2/4) (2003) 480–489. [Google Scholar]
- [4].Brazilian Ministry of Health, Accidents by Venomous Animals: What to do and How to Avoid Them (Portuguese). Available online: https://antigo.saude.gov.br/saude-de-a-z/acidentes-por-animais-peconhentos, 2019. (accessed 12 August, 2021).
- [5].SINAN - Notifiable Diseases Information System (Sistema de Informação deAgravos de Notificação), Notification of Health and Diseases, – present: accident by poisonous animals (2020) (Portuguese). Available online: http://tabnet.datasus.gov.br/cgi/deftohtm.exe?sinannet/cnv/animaisbr.def, 2007. (accessed 12 August, 2021).
- [6].Fox JW, Serrano SMT, Approaching the golden age of natural product pharmaceuticals from venom libraries: an overview of toxins and toxin-derivatives currently involved in therapeutic or diagnostic applications, Curr. Pharm. Des. 13 (28) (2007) 2927–2934. [DOI] [PubMed] [Google Scholar]
- [7].Slotta KH, Fraenkel-Conrat H, Two active proteins from rattlesnake venom, Nature 142 (3587) (1938) 213. [Google Scholar]
- [8].Gonçalves JM, Purification and properties of crotamine, in: Buckley EE, Porges N (Eds.), American Association for the Advancemet of Science, Washington, DC, 1956, pp. 261–273. [Google Scholar]
- [9].Brazil OV, Pharmacology of crystalline crotoxin. II. Neuromuscular blocking action, Mem. Inst. Butantan 33 (3) (1966) 981–992. [PubMed] [Google Scholar]
- [10].Lomeo RDS, Gonçalves APDF, da Silva CN, de Paula AT, Costa Santos DO, Fortes-Dias CL, Gomes DA, de Lima ME, Crotoxin from Crotalus durissus terrificus snake venom induces the release of glutamate from cerebrocortical synaptosomes via N and P/Q calcium channels, Toxicon: Off. J. Int. Soc. Toxinol. 85 (2014) 5–16. [DOI] [PubMed] [Google Scholar]
- [11].Fortes-Dias CL, Lin Y, Ewell J, Diniz CR, Liu TY, A phospholipase A2 inhibitor from the plasma of the south American rattlesnake (Crotalus durissus terrificus). Protein structure, genomic structure, and mechanism of action, J. Biolo. Chem. 269 (22) (1994) 15646–15651. [PubMed] [Google Scholar]
- [12].Fernandes CAH, Pazin WM, Dreyer TR, Bicev RN, Cavalcante WLG, Fortes-Dias C, Ito AS, Oliveira CLP, Fernandez RM, Fontes MRM, Biophysical studies suggest a new structural arrangement of crotoxin and provide insights into its toxic mechanism, Sci. Rep. 7 (1) (2017) 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bieber AL, Nedelkov D, Structural, biological and biochemical studies of myotoxin a and homologous myotoxins, J. Toxicol.Toxin Rev. 16 (1–2) (1Ɛ997) 33–52. [Google Scholar]
- [14].Oguiura N, Boni-Mitake M, Rádis-Baptista G, New view on crotamine, a small ´ basic polypeptide myotoxin from South American rattlesnake venom, Toxicon 46 (4) (2005) 363–370. [DOI] [PubMed] [Google Scholar]
- [15].Barrabin H, Martiarena JL, Vidal JC, Barrio A, in: Rosemberg P (Ed.), Isolation and characterization of gyroxin from Crotalus durissus terrificus venom, Pergamon Press, New York, 1978, pp. 113–133. [Google Scholar]
- [16].Barrio A, Gyroxin, a new neurotoxin of Crotalus durissus terrificus venom, Assn Latinoamer Cienc Fisiol, Serrano 665, 1414 Buenos Aires, Argentina (1961) 224–232. [Google Scholar]
- [17].Prado-Franceschi J, O., Vital Brazil, Convulxin, a new toxin from the venom of the south American rattlesnake Crotalus durissus terrificus, Toxicon 19 (6) (1981) 875–887. [DOI] [PubMed] [Google Scholar]
- [18].Hadler WA, Brazil OV, Pharmacology of crystalline crotoxin. IV. Nephrotoxicity, Mem. Inst. Butantan 33 (3) (1966) 1001–1008. [PubMed] [Google Scholar]
- [19].Tokarnia CH, Brito MF, Barbosa JD, Dobereiner J, Clinicopathological features ¨ of snakebite envenomation by Crotalus durissus terrificus and Bothrops spp. in production animals (Portuguese), Pesqui. Vet. Bras. 34 (4) (2014) 301–312. [Google Scholar]
- [20].Cupo P, Azevedo-Marques MM, Hering SE, Acute myocardial infarction-like enzyme profile in human victims of Crotalus durissus terrificus envenoming, Trans. R. Soc. Trop. Med. Hyg. 84 (3) (1990) 447–451. [DOI] [PubMed] [Google Scholar]
- [21].Santos PEB, Souza SD, Freire-Maia L, Almeida AP, Effects of crotoxin on the isolated guinea pig heart, Toxicon 28 (2) (1990) 215–224. [DOI] [PubMed] [Google Scholar]
- [22].de Siqueira JE, Higuchi MDL, Nabut N, Lose A, Souza JK, Nakashima M, Myocardial lesions after snake bites by the Crotalus durissus terrificus species (rattlesnake). A case report, Arq. Bras. Cardiol. 54 (5) (1990) 323–325. [PubMed] [Google Scholar]
- [23].de Paola F, Rossi MA, Myocardial damage induced by tropical rattlesnake (Crotalus durissus terrificus) venom in rats, Cardiovasc. Pathol. 2 (1) (1993) 77–81. [DOI] [PubMed] [Google Scholar]
- [24].Gonçalves-Machado L, Pla D, Sanz L, Jorge RJB, Leitão-De-Araújo M, Alves ML, Alvares DJ, De Miranda J, Nowatzki J, de Morais-Zani K, Fernandes W, Tanaka-Azevedo AM, Fernandez J, Zingali RB, Gutierrez JM, Corrêa-Netto C, Calvete JJ, Combined venomics, venom gland transcriptomics, bioactivities, and antivenomics of two Bothrops jararaca populations from geographic isolated regions within the Brazilian Atlantic rainforest, J. Proteome 135 (2016) 73–89. [DOI] [PubMed] [Google Scholar]
- [25].Nicolau CA, Carvalho PC, Junqueira-de-Azevedo ILM, Teixeira-Ferreira A, Junqueira M, Perales J, Neves-Ferreira AGC, Valente RH, An in-depth snake venom proteopeptidome characterization: benchmarking Bothrops jararaca, J. Proteome 151 (2017) 214–231. [DOI] [PubMed] [Google Scholar]
- [26].Reeks T, Lavergne V, Sunagar K, Jones A, Undheim E, Dunstan N, Fry B, Alewood PF, Deep venomics of the Pseudonaja genus reveals inter- and intra- specific variation, J. Proteome 133 (2016) 20–32. [DOI] [PubMed] [Google Scholar]
- [27].Oliveira ISD, Cardoso IA, Bordon KDCF, Carone SEI, Boldrini-França J, Pucca MB, Zoccal KF, Faccioli LH, Sampaio SV, Rosa JC, Arantes EC, Global proteomic and functional analysis of Crotalus durissus collilineatus individual venom variation and its impact on envenoming, J. Proteome 191 (2019) 153–165. [DOI] [PubMed] [Google Scholar]
- [28].Menezes MC, Kitano ES, Bauer VC, Oliveira AK, Cararo-Lopes E, Nishiyama MY, Zelanis A, Serrano SMT, Early response of C2C12 myotubes to a sub-cytotoxic dose of hemorrhagic metalloproteinase HF3 from Bothrops jararaca venom, J. Proteome 198 (2019) 163–176. [DOI] [PubMed] [Google Scholar]
- [29].Kisaki CY, Arcos SSS, Montoni F, da Silva Santos W, Calacina HM, Lima IF, Cajado-Carvalho D, Ferro ES, Nishiyama-Jr MY, Iwai LK, Bothrops Jararaca snake venom modulates key cancer-related proteins in breast tumor cell lines, Toxins 13 (8) (2021) 1–28, 519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Montoni F, Andreotti DZ, Eichler RADS, Santos WDS, Kisaki CY, Arcos SSS, Lima IF, Soares MAM, Nishiyama-Jr MY, Nava-Rodrigues D, Ferro ES, Carvalho VM, Iwai LK, The impact of rattlesnake venom on mice cerebellum proteomics points to synaptic inhibition and tissue damage, J. Proteomics 221 (2020) 1–12. [DOI] [PubMed] [Google Scholar]
- [31].Teston EF, Cecilio HPM, Santos AL, Arruda GOD, Radovanovic CAT, Marcon SS, Factors associated with cardiovascular diseases in adults, Medicina (Ribeirao Preto. Online) 49 (2) (2016) 95–102. [Google Scholar]
- [32].Radovanovic CAT, Santos LAD, Carvalho MDDB, Marcon SS, Arterial hypertension and other risk factors associated with cardiovascular diseases among adults, Rev. Latino-Am. Enfermagem 22 (4) (2014) 547–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Bharadwaj LA, in: Ballantyne B, Marrs TC, Syversen T, Casciano DA, Sahu SC (Eds.), Cardiac Toxicology, John Wiley & Sons, Ltd, Chichester, UK, 2009, pp. 1–22. [Google Scholar]
- [34].Simoes LO, Alves QL, Camargo SB, Araujo FA, Hora VRS, Jesus RLC, Barreto BC, Macambira SG, Soares MBP, Meira CS, Aguiar MC, Couto RD, Lomonte B, Menezes-Filho JE, Cruz JS, Vannier-Santos MA, Casais ESLL, Silva DF, Cardiac effect induced by Crotalus durissus cascavella venom: Morphofunctional evidence and mechanism of action, Toxicol. Lett. 337 (2021) 121–133. [DOI] [PubMed] [Google Scholar]
- [35].Wollberg Z, Shabo-Shina R, Intrator N, Bdolah A, Kochva E, Shavit G, Oron Y, Vidne BA, Gitter S, A novel cardiotoxic polypeptide from the venom of Atractaspis engaddensis (burrowing asp): cardiac effects in mice and isolated rat and human heart preparations, Toxicon: Off. J. Int. Soc. Toxinol. 26 (6) (1988) 525–534. [DOI] [PubMed] [Google Scholar]
- [36].Cupo P, Azevedo-Marques MM, Hering SE, Clinical and laboratory features of south American rattlesnake (Crotalus durissus terrificus) envenomation in children, Trans. R. Soc. Trop. Med. Hyg. 82 (6) (1988) 924–929. [DOI] [PubMed] [Google Scholar]
- [37].Azevedo-Marques MM, Cupo P, Coimbra TM, Hering SE, Rossi MA, Laure CJ, Myonecrosis, myoglobinuria and acute renal failure induced by south American rattlesnake (Crotalus durissus terrificus) envenomation in Brazil, Toxicon: Off. J. Int. Soc. Toxinol. 23 (4) (1985) 631–636. [DOI] [PubMed] [Google Scholar]
- [38].Celermajer DS, Chow CK, Marijon E, Anstey NM, Woo KS, Cardiovascular disease in the developing world: prevalences, patterns, and the potential of early disease detection, J. Am. Coll. Cardiol. 60 (14) (2012) 1207–1216. [DOI] [PubMed] [Google Scholar]
- [39].Stanley BA, Gundry RL, Cotter RJ, Van Eyk JE, Heart disease, clinical proteomics and mass spectrometry, Dis. Markers 20 (3) (2004) 167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Fischer AH, Jacobson KA, Rose J, Zeller R, Hematoxylin and eosin staining of tissue and cell sections, CSH Protocols 3 (5) (2008) pdb.prot4986-pdb.prot4986. [DOI] [PubMed] [Google Scholar]
- [41].Masuda T, Tomita M, Ishihama Y, Phase transfer surfactant-aided trypsin digestion for membrane proteome analysis, J. Proteome Res. 7 (2) (2008) 731–740. [DOI] [PubMed] [Google Scholar]
- [42].Wisniewski JR, Zougman A, Nagaraj N, Mann M, Universal sample preparation method for proteome analysis, Nat. Methods 6 (5) (2009) 359–362. [DOI] [PubMed] [Google Scholar]
- [43].Rappsilber J, Mann M, Ishihama Y, Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips, Nat. Protoc. 2 (8) (2007) 1896–1906. [DOI] [PubMed] [Google Scholar]
- [44].Boersema PJ, Raijmakers R, Lemeer S, Mohammed S, Heck AJR, Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics, Nat. Protoc. 4 (4) (2009) 484–494. [DOI] [PubMed] [Google Scholar]
- [45].Wang J, Vasaikar S, Shi Z, Greer M, Zhang B, WebGestalt 2017: a more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit, Nucleic Acids Res. 45 (W1) (2017) W130–W137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, Cox J, The Perseus computational platform for comprehensive analysis of (prote) omics data, Nat. Methods 13 (9) (2016) 731–740. [DOI] [PubMed] [Google Scholar]
- [47].Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C, STRING v10: protein-protein interaction networks, integrated over the tree of life, Nucleic Acids Res. 43 (Database issue) (2015) D447–D452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T, Cytoscape: a software environment for integrated models of biomolecular interaction networks, Genome Res. 13 (11) (2003) 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Doncheva NT, Morris JH, Gorodkin J, Jensen LJ, Cytoscape StringApp: network analysis and visualization of proteomics data, J. Proteome Res. 18 (2) (2019) 623–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Lê S, Josse J, Husson F, FactoMineR: an R package for multivariate analysis, J. Stat. Softw. 25 (1) (2008) 1–18. [Google Scholar]
- [51].Breithaupt H, Neurotoxic and myotoxic effects of crotalus phospholipase a and its complex with crotapotin, Naunyn Schmiedeberg’s Arch. Pharmacol. 292 (3) (1976) 271–278. [DOI] [PubMed] [Google Scholar]
- [52].Chugh KS, Sakhuja V, in: Tu AT(Ed.), Renal Disease Caused by Snake Venom, Routledge, Abingdon, England, United Kingdom, 2017, pp. 471–493. [Google Scholar]
- [53].Martins AMC, Toyama MH, Havt A, Novello JC, Marangoni S, Fonteles MC, Monteiro HSA, Determination of Crotalus durissus cascavella venom components that induce renal toxicity in isolated rat kidneys, Toxicon 40 (8) (2002) 1165–1171. [DOI] [PubMed] [Google Scholar]
- [54].Danzig LE, Hemodialysis of acute renal failure following rattlesnake bite, with recovery, JAMA 175 (2) (1961) 136–137. [DOI] [PubMed] [Google Scholar]
- [55].Albuquerque PLMM, Jacinto CN, Silva Junior GB, Lima JB, Veras MDSB, Daher EF, Acute kidney injury caused by Crotalus and Bothrops snake venom: a review of epidemiology, clinical manifestations and treatment, Rev. Inst. Med. Trop. Sao Paulo 55 (5) (2013) 295–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Hernández M, Scannone H, Finol HJ, Pineda ME, Fernández I, Vargas AM, Girón M´E, Aguilar I, Rodríguez-Acosta A, Alterations in the ultrastructure of ´ cardiac autonomic nervous system triggered by crotoxin from rattlesnake (Crotalus durissus cumanensis) venom, Exp. Tox. Pathol. 59 (2) (2007) 129–137. [DOI] [PubMed] [Google Scholar]
- [57].Marinovic MP, Campeiro JD, Lima SC, Rocha AL, Nering MB, Oliveira EB, Mori MA, Hayashi MAF, Crotamine induces browning of adipose tissue and increases energy expenditure in mice, Sci. Rep. 8 (1) (2018) 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].dos Santos IGC, Fortes-Dias CL, dos Santos MC, Pharmacological applications of Brazilian snake venoms with emphasis in Crotalus durissus terrificus and Crotalus durissus ruruima, Sci. Amaz. 6 (1) (2017) 42–53. [Google Scholar]
- [59].Cruz AH, Mendonca RZ, Petricevich VL, Crotalus durissus terrificus venom interferes with morphological, functional, and biochemical changes in murine macrophage, Mediat. Inflamm. 2005 (6) (2005) 349–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Winkler E, Chovers M, Almog S, Pri-Chen S, Rotenberg M, Tirosh M, Ezra D, Halkin H, Decreased serum cholesterol level after snake bite (Vipera palaestinae) as a marker of severity of envenomation, J. Lab. Clin. Med. 121 (6) (1993) 774–778. [PubMed] [Google Scholar]
- [61].Silva JGD, Soley BDS, Gris V, Pires ADRA, Caderia SMSC, Eler GJ, Hermoso APM, Bracht A, Dalsenter PR, Acco A, Effects of the Crotalus durissus terrificus snake venom on hepatic metabolism and oxidative stress, J. Biochem. Mol. Toxicol. 25 (3) (2011) 195–203. [DOI] [PubMed] [Google Scholar]
- [62].Yamasaki SC, Villarroel JS, Barone JM, Zambotti-Villela L, Silveira PF, Aminopeptidase activities, oxidative stress and renal function in Crotalus durissus terrificus envenomation in mice, Toxicon 52 (3) (2008) 445–454. [DOI] [PubMed] [Google Scholar]
- [63].Frezzatti R, Silveira PF, Allopurinol reduces the lethality associated with acute renal failure induced by Crotalus durissus terrificus snake venom: comparison with probenecid, PLoS Negl. Trop. Dis. 5 (9) (2011), e1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Costa CRC, Belchor MN, Rodrigues CFB, Toyama DO, de Oliveira MA, Novaes DP, Toyama MH, Edema induced by a Crotalus durissus terrificus venom serine protease (Cdtsp 2) involves the PAR pathway and PKC and PLC activation, Int. J. Mol. Sci. 19 (8) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Melendez-Martinez D, Munoz JM, Barraza-Garza G, Cruz-Perez MS, Gatica- Colima A, Alvarez-Parrilla E, Plenge-Tellechea LF, Rattlesnake Crotalus molossus nigrescens venom induces oxidative stress on human erythrocytes, J. Venom Anim. Toxins. Incl. Trop. Dis. 23 (2017) 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Sheikh F, Lyon RC, Chen J, Functions of myosin light chain-2 (MYL2) in cardiac muscle and disease, Gene 569 (1) (2015) 14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Li Y, Wu G, Tang Q, Huang C, Jiang H, Shi L, Tu X, Huang J, Zhu X, Wang H, Slow cardiac myosin regulatory light chain 2 (MYL2) was down-expressed in chronic heart failure patients, Clin. Cardiol. 34 (1) (2011) 30–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Flavigny J, Richard P, Isnard R, Carrier L, Charron P, Bonne G, Forissier Le JF, Desnos M, Dubourg O, Komajda M, Schwartz K, Hainque B, Identification of two novel mutations in the ventricular regulatory myosin light chain gene (MYL2) associated with familial and classical forms of hypertrophic cardiomyopathy, J. Mol. Med. (Berlin, Germany) 76 (34) (1998) 208–214. [DOI] [PubMed] [Google Scholar]
- [69].Kabaeva ZT, Perrot A, Wolter B, Dietz R, Cardim N, Correia JM, Schulte HD, Aldashev AA, Mirrakhimov MM, Osterziel KJ, Systematic analysis of the regulatory and essential myosin light chain genes: genetic variants and mutations in hypertrophic cardiomyopathy, Eur. J. Human Gene. 10 (11) (2002) 741–748. [DOI] [PubMed] [Google Scholar]
- [70].Hershberger RE, Pinto JR, Parks SB, Kushner JD, Li D, Ludwigsen S, Cowan J, Morales A, Parvatiyar MS, Potter JD, Clinical and functional characterization of TNNT2 mutations identified in patients with dilated cardiomyopathy, Circ. Cardiovasc. Genet. 2 (4) (2009) 306–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Carniel E, Taylor MRG, Sinagra G, Di Lenarda A, Ku L, Fain PR, Boucek MM, Cavanaugh J, Miocic S, Slavov D, Graw SL, Feiger J, Zhu XZ, Dao D, Ferguson DA, Bristow MR, Mestroni L, Alpha-myosin heavy chain: a sarcomeric gene associated with dilated and hypertrophic phenotypes of cardiomyopathy, Circulation 112 (1) (2005) 54–59. [DOI] [PubMed] [Google Scholar]
- [72].Hershberger RE, Norton N, Morales A, Li D, Siegfried JD, Gonzalez- Quintana J, Coding sequence rare variants identified in MYBPC3, MYH6, TPM1, TNNC1, and TNNI3 from 312 patients with familial or idiopathic dilated cardiomyopathy, Circ. Cardiovasc. Genet. 3 (2) (2010) 155–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Niimura H, Patton KK, McKenna WJ, Soults J, Maron BJ, Seidman JG, Seidman CE, Sarcomere protein gene mutations in hypertrophic cardiomyopathy of the elderly, Circulation 105 (4) (2002) 446–451. [DOI] [PubMed] [Google Scholar]
- [74].Olichon A, Baricault L, Gas N, Guillou E, Valette A, Belenguer P, Lenaers G, Loss of OPA1 perturbates the mitochondrial inner membrane structure and integrity, leading to cytochrome c release and apoptosis, J. Biol. Chem. 278 (10) (2003) 7743–7746, 10.1074/jbc.C200677200, [DOI] [PubMed] [Google Scholar]
- [75].Chen L, Gong Q, Stice JP, Knowlton AA, Mitochondrial OPA1, apoptosis, and heart failure, Cardiovasc. Res. 84 (1) (2009) 91–99, 10.1093/cvr/cvp181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Van Remmen H, Williams MD, Guo Z, Estlack L, Yang H, Carlson EJ, Epstein CJ, Huang TT, Richardson A, Knockout mice heterozygous for Sod2 show alterations in cardiac mitochondrial function and apoptosis, Am. J. Physiol. Heart Circ. Physiol. 281 (3) (2001) H1422–H1432, 10.1152/ajpheart.2001.281.3.H1422. [DOI] [PubMed] [Google Scholar]
- [77].Fukai T, Folz RJ, Landmesser U, Harrison DG, Extracellular superoxide dismutase and cardiovascular disease, Cardiovasc. Res. 55 (2) (2002) 239–249, 10.1016/s0008-6363(02)00328-0. [DOI] [PubMed] [Google Scholar]
- [78].Li X, Lin Y, Wang S, Zhou S, Ju J, Wang X, Chen Y, Xia M, Extracellular Superoxide Dismutase Is Associated With Left Ventricular Geometry and Heart Failure in Patients With Cardiovascular Disease, J. Am. Heart Assoc. 9 (15) (2020) 1–19, 10.1161/JAHA.120.016862, e016862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Romuk E, Jache Wć, Kozielska-Nowalany E, Birkner E, Zemła-Woszek A, Wojciechowska C, Superoxidedismutase activity as a predictor of adverse outcomes in patients with nonischemic dilated cardiomyopathy, Cell Stress Chaperones 24 (3) (2019) 661–673, 10.1007/s12192-019-00991-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Winnik S, Auwerx J, Sinclair DA, Matter CM, Protective effects of sirtuins in cardiovascular diseases: from bench to bedside, Eur. Heart J. 36 (48) (2015) 3404–3412, 10.1093/eurheartj/ehv290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Ohman EM, Casey C, Bengtson JR, Pryor D, Tormey W, Horgan JH, Early detection of acute myocardial infarction: additional diagnostic information from serum concentrations of myoglobin in patients without ST elevation, Br. Heart J. 63 (6) (1990) 335–338, 10.1136/hrt.63.6.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Parikh SV, de Lemos JA, Biomarkers in cardiovascular disease: integrating pathophysiology into clinical practice, Am. J. Med. Sci. 332 (4) (2006) 186–197. [DOI] [PubMed] [Google Scholar]
- [83].Störk TV, Wu AH, Müller-Bardorff M, Gareis R, Müller R, Hombach V, Katus H, Mockel M, North-Wurttemberg Infarction Study (NOWIS) Group, ¨ Diagnostic and prognostic role of myoglobin in patients with suspected acute coronary syndrome. North-Württemberg Infarction Study (NOWIS) Group, Am. J. Cardiol. 86 (12) (2000) 1371–1374, 10.1016/s0002-9149(00)01246-7. [DOI] [PubMed] [Google Scholar]
- [84].Wei B, Jin JP, TNNT1, TNNT2, and TNNT3: Isoform genes, regulation, and structure-function relationships, Gene 581 (1) (2016) 1–13, 10.1016/j.gene.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Osio A, Tan L, Chen SN, Lombardi R, Nagueh SF, Shete S, Roberts R, Willerson JT, Marian AJ, Myozenin 2 is a novel gene for human hypertrophic cardiomyopathy, Circ. Res. 100 (6) (2007) 766–768, 10.1161/01.RES.0000263008.66799.aa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Marian AJ, Genetic determinants of cardiac hypertrophy, Curr. Opin. Cardiol. 23 (3) (2008) 199–205, 10.1097/HCO.0b013e3282fc27d9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Ke BX, Pepe S, Grubb DR, Komen JC, Laskowski A, Rodda FA, Hardman BM, Pitt JJ, Tyan MT, Lazarou M, Koleff J, Cheung MMH, Smolich JJ, Thorburn DR, Tissue-specific splicing of an Ndufs6 gene-trap insertion generates a mitochondrial complex I deficiency-specific cardiomyopathy, Proc. Natl. Acad. Sci. USA 109 (16) (2012) 6165–6170, 10.1073/pnas.1113987109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Loeffen JL, Smeitink JA, Trijbels JM, Janssen AJ, Triepels RH, Sengers RC, van den Heuvel LP, Isolated complex I deficiency in children: clinical, biochemical and genetic aspects, Hum. Mutat. 15 (2) (2000) 123–134, . [DOI] [PubMed] [Google Scholar]
- [89].Spiegel R, Shaag A, Mandel H, Reich D, Penyakov M, Hujeirat Y, Saada A, Elpeleg O, Shalev SA, Mutated NDUFS6 is the cause of fatal neonatal lactic acidemia in Caucasus Jews, Eur. J. Hum. Genet. 17 (9) (2009) 1200–1203, 10.1038/ejhg.2009.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Nishi M, Ogata T, Cannistraci CV, Ciucci S, Nakanishi N, Higuchi Y, Sakamoto A, Tsuji Y, Mizushima K, Matoba S, Systems Network Genomic Analysis Reveals Cardioprotective Effect of MURC/Cavin-4 Deletion Against Ischemia/Reperfusion Injury, J. Am. Heart Assoc. 8 (15) (2019) 1–106, 10.1161/JAHA.119.012047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Cheng H, Kimura K, Peter AK, Cui Li Ouyang K. Shen T, Liu Y, Gu Y, Dalton ND, Evans SM Knowlton KU, Peterson KL, Chen J, Loss of enigma homolog protein results in dilated cardiomyopathy, Circ. Res. 107 (3) (2010) 348–356, 10.1161/CIRCRESAHA.110.218735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Li A, Ponten F, dos Remedios CG, The interactome of LIM domain proteins: the contributions of LIM domain proteins to heart failure and heart development, Proteomics 12 (2) (2012) 203–225. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.