Summary:
The design of an animal’s body plan is encoded in the genome and execution of this program is a mechanical progression involving coordinated movement of proteins, cells, and whole tissues. Thus, a challenge to understanding morphogenesis is connecting events that occur across various length scales. Here, we describe how a poorly characterized adhesion effector, Arvcf catenin, controls Xenopus head-to-tail axis extension. We find that Arvcf is required for axis extension within the intact organism but not within isolated tissues. We show that the organism scale phenotype results from a defect in tissue scale force production. Finally, we determine that the force defect results from dampening of the pulsatile recruitment of cell adhesion and cytoskeletal proteins to membranes. These results provide a comprehensive understanding of Arvcf function during axis extension and produce insight into how a cellular scale defect in adhesion results in an organism scale failure of development.
Introduction:
Head-to-tail axis extension is an essential step of animal development (Keller, 2002), and genes that govern axis extension are directly linked to human birth defects, in particular neural tube closure defects (Wallingford et al., 2013). While the plan for axis extension is encoded in the genome, execution of this program is a mechanical process involving the highly reproducible and tightly coordinated movement of cell collectives (Mongera et al., 2019). Several collective cell movements contribute to axis extension, including convergent extension (CE), in which a group of cells move towards each other along one axis resulting in extension of the perpendicular axis (Keller, 2002; Tada and Heisenberg, 2012). CE is deeply conserved and has been studied in organisms ranging from nematodes to vertebrates (Huebner and Wallingford, 2018).
Biomechanical analyses have become critical to developmental biology, especially for understanding morphogenetic processes such as collective cell migration (Barriga et al., 2018), neuronal guidance (Thompson et al., 2019), and organogenesis (Tao et al., 2019). Because axis extension requires tissue scale force to push the animal’s anterior (presumptive head) away from its posterior (presumptive tail), the biomechanics of both elongation generally and CE specifically have been explored in recent years (Keller, 2012; Mongera et al., 2018; Shook et al., 2018; Xiong et al., 2018). These forces originate from actomyosin contractility within individual cells and the cellular scale forces must be integrated and transduced across the cell collective by cell-cell adhesions (Lecuit et al., 2011). Understanding how adhesion links cellular scale force production to tissue scale biomechanics, especially in vertebrates, is an ongoing challenge.
Classical cadherins are key adhesion molecules that connect cell-cell contacts to the actomyosin cytoskeleton (Charras and Yap, 2018). Extensive studies have characterized the proteins connecting cadherin-based adhesions to cellular force production machinery, but these data primarily apply to cultured cells or epithelial tissues (Charras and Yap, 2018). Critically, however, cadherin dependent CE in early vertebrate embryos also occurs in mesenchymal tissues and much less is known about cadherin function during collective movement of mesenchymal cells (Theveneau and Mayor, 2012; Walck-Shannon and Hardin, 2014). Thus, to understand early vertebrate development it is essential to improve our comprehension of cadherin-based cell adhesion in mesenchymal tissues.
Cadherins have been shown to control biomechanics across multiple length scales during CE (Huebner et al., 2020; Kale et al., 2018), but cadherin function absolutely requires an array of effector proteins, and less is known about the effector proteins controlling biomechanics in mesenchymal tissues. One poorly characterized cadherin effector, the catenin Armadillo Repeat protein deleted in Velo-Cardio-Facial syndrome (Arvcf) is required for multiple developmental processes, including head-to-tail axis extension (Cho et al., 2011; Fang et al., 2004), but the biomechanical underpinnings of this defect are unclear. At the molecular level, Arvcf is most closely related to p120-catenin and similarly is thought to control cadherin trafficking (McCrea and Park, 2007). Arvcf is also associated with Rho and Rac activity and thus has a role in signaling to the actomyosin cytoskeleton (Fang et al., 2004).
Here, using classic embryology, biomechanical measurements, and cell biology, we found that Arvcf specifically modulates the force generated by CE but interestingly not the ability of a tissue to converge and extend, per se. Rather, this embryonic phenotype results from a failure of CE to generate sufficient force to push an animal’s anterior away from its posterior. We further show that this force generation defect is associated with a modest reduction in cell adhesion and a more pronounced dampening of the oscillatory recruitment of cadherin and actin to cell membranes. These results not only provide a deeper understanding of a poorly defined catenin during an essential biological process, but also illustrates that a modest change in cell behavior can result in an organism scale failure of development.
Results:
Identification of tissue and stage specific Cdh3 protein interactions in Xenopus convergent extension.
The classical cadherin Cdh3 (aka C-cadherin; aka mammalian P-cadherin) plays a crucial role in morphogenesis of the dorsal marginal zone (DMZ) mesoderm of the Xenopus gastrula (Fagotto et al., 2013; Huebner et al., 2021; Lee and Gumbiner, 1995; Pfister et al., 2016), a powerful and deeply studied paradigm for understanding CE (Keller and Sutherland, 2020). Cadherins control numerous cellular processes including proliferation, cell migration, cell polarity, and mechanotransduction and to achieve this assortment of cellular tasks, cadherins interact with an array of structural and signaling molecules (Arslan et al., 2021). However, we currently lack a comprehensive roster of proteins interacting with Cdh3 in the Xenopus DMZ. We therefore used a combination of classic embryology and mass spectrometry to determine the protein interactome for Cdh3.
Details of our affinity purification mass spectrometry (AP-MS) approach can be found in the Methods Section, but briefly, we used mRNA injection to express Cdh3-GFP or GFP alone in the Xenopus dorsal mesoderm and then isolated DMZ (Keller) explants at the onset of gastrulation (~st.10.25)(Keller, 2012)(Fig. 1A). Explants were allowed to undergo CE ex vivo until mid-gastrulation (~st.12) and were lysed. Cdh3-GFP or GFP alone were purified by GFP pulldown and associated proteins were identified by mass spectrometry (Fig. 1A), using the GFP-only pulldown as a negative control. This experiment was performed in two biological replicates with ~800 explants per condition, per replicate.
Figure 1: Tissue and stage-specific Cdh3 affinity purification mass spectrometry results in a highly specific Cdh3 protein interaction dataset.
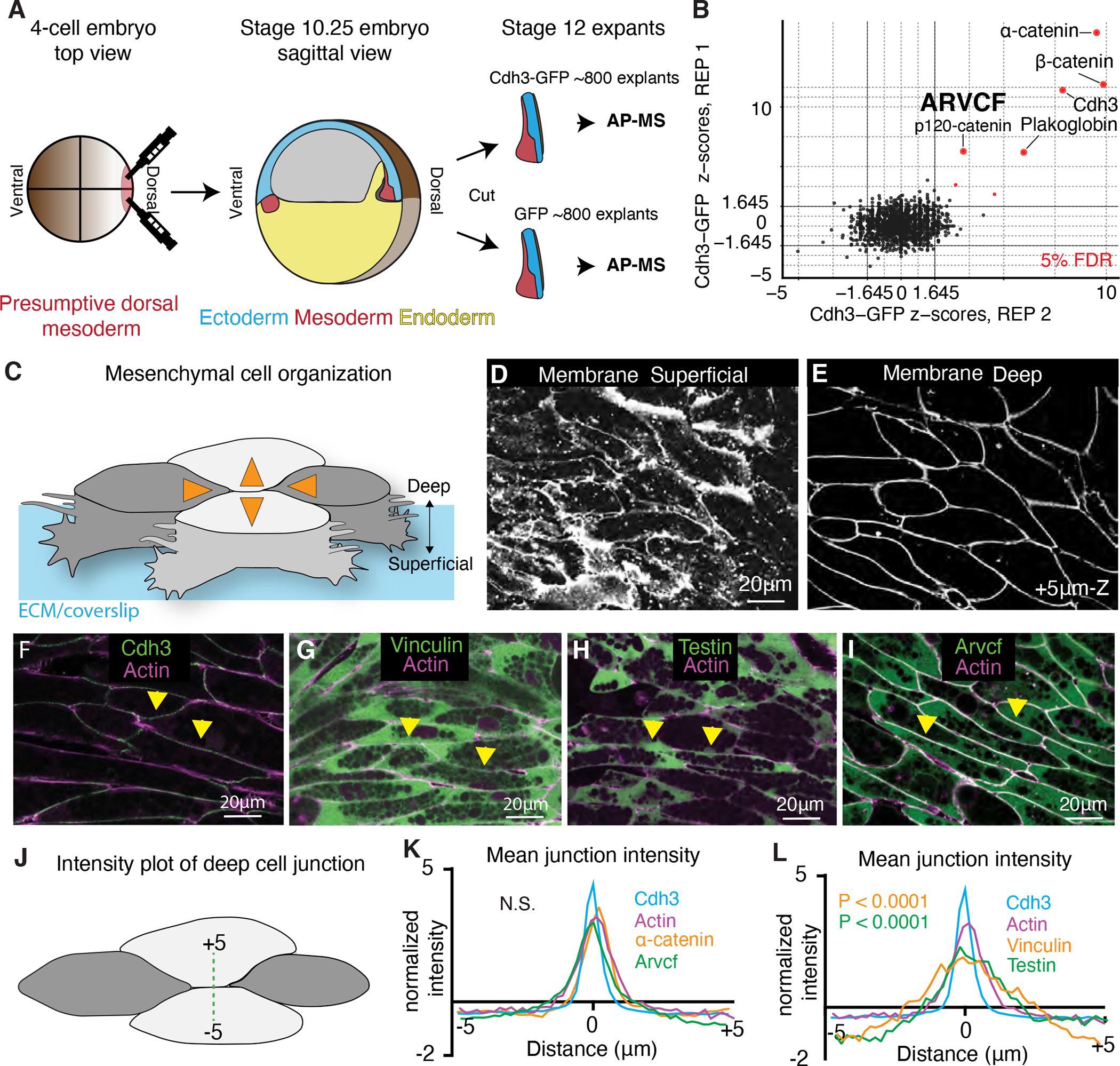
A. Schematic depicting the method used for tissue and stage-specific affinity purification mass spectrometry (APMS) of Cdh3. B. Graph showing the relative protein orthogroup enrichment (see methods) from two replicates of the Cdh3 AP-MS experiment. Here we plot the z-scores from replicate 1 on the y-axis and the z-scores from replicate 2 on the x-axis. Each dot represents a protein identified in the Cdh3 AP-MS dataset and red dots represent proteins that fall below a 5% FDR threshold. C. Cartoon depicting Xenopus mesenchymal cells during convergent extension. Here the mediolateral cells, dark gray, move to each other resulting in displacement of the anterior-posterior cells, light gray. Orange arrows show the cell movements. Mesenchymal cells display apparent structural differences along the superficial (cell-ECM interface) to deep (cell-cell interface) axis. Here polarized lamellar-like structures are observed at the superficial surface. Movement deeper into the cell reveals cell-cell interfaces and actin-based protrusions which extend between neighboring cells. D. Image of the superficial surface of converging and extending Xenopus mesenchymal cells. Cells are labeled with a membrane marker which primarily shows lamellar-like protrusions at the cell-ECM interface. E. Image of the deep surface of the same cells shown in Fig. 1D. Here the membrane marker largely highlights the cell-cell junctions. F. Image of Cdh3 (green) and actin (magenta) at deep cell-cell contacts. Protein localization was visualized by expressing tagged fusion proteins. Yellow arrowheads point to cell junctions. G. Image of vinculin (green) and actin (magenta) at deep cell structures. Protein localization was visualized by expressing tagged fusion proteins. Yellow arrowheads point to cell junctions. H. Image of testin (green) and actin (magenta) at deep cell-cell junctions. Protein localization was visualized by expressing tagged fusion proteins. Yellow arrowheads point to cell junctions. I. Image of the Arvcf (green) and actin (magenta) at deep cell-cell junctions. Protein localization was visualized by expressing tagged fusion proteins. Yellow arrowheads point to cell junctions. J. Schematic displaying the method used to measure fluorescent intensities at cell-cell interfaces. K. Intensity plots of Cdh3 (blue), actin (magenta) α-catenin (orange), and Arvcf (green). Here zero is set at the center of the cell-cell junction and each protein shows a clear peak at the cell junction. Each line is the average over dozens of line plots from a minimum of three replicates. Distributions were statistically compared using a KS test. L. Intensity plots of Cdh3 (blue), actin (magenta), vinculin (orange), and testin (green) at cell-cell junctions. Vinculin and testin lack peaks at the cell-cell junction and the distributions of vinculin and testin were statistically different from Cdh3 and actin as compared by a KS test. Each line represents the average of dozens of line plots over a minimum of three replicates. See also figure S1 and figure S2.
Because of the duplication in the X. laevis genome (Session et al., 2016), we identified enriched proteins by collapsing homeologs and highly related entries into eggNOG vertebrate-level orthologous groups, as outlined in the Methods. Z-scores and confidence intervals for each protein orthogroup were then calculated based on the ratio of peptide counts in the experimental (Cdh3-GFP) compared to negative control (GFP alone) and the standard error from the null distribution based on the global peptide counts. (For a complete list of proteins identified, peptide counts, Log2 fold change over GFP alone, z-scores, p-values, and confidence intervals, see Data Table 1.)
Crucially, our analysis identified Cdh3 itself and the well-known cadherin interactors α-catenin and β-catenin as the most highly enriched proteins in our dataset (Fig. 1B). Using the same parameters (joint z-score over both replicates with a multiple hypothesis-corrected p-value <= 0.05), we also identified less well known but important Cdh3 interactors, including Plakoglobin (aka JUP) (Aktary et al., 2017) and the orthogroup containing the related catenin proteins Arvcf and p120 (McCrea and Park, 2007)(Fig. 1B)(Supp. Fig.1). Taken together, these results demonstrate that our AP-MS dataset effectively identified Cdh3 interacting proteins in the Xenopus gastrula DMZ.
Localization of Cdh3-interaction partners reveal similarities but also differences from the E-cadherin/Cdh1 paradigm
Cells engaged in CE in the Xenopus gastrula DMZ display a unique organization, and previous work has identified two sites of Cdh3 action. Unlike more familiar epithelial cells, which have clear apico-basal polarity, these mesenchymal cells lack a well-defined molecular asymmetry along the axis spanning the superficial cell-extracellular matrix (ECM) interface and the deeper cell-cell junctions (Green and Davidson, 2007)(Fig. 1C). The superficial surface is visually dominated by large actin based lamellar-like structures that become polarized to the mediolateral cell edges during CE (Wallingford et al., 2000)(Fig. 1C, D), and Cdh3 localizes to foci in these protrusions (Pfister et al., 2016)(Supp. Fig. 2A). Observation deeper into the tissue (3–5μm) reveals clear cell-cell junctions (Fig. 1C, E), though extensive actin-based protrusions between neighboring cells at these deep cell-cell interfaces are also observed (Green and Davidson, 2007; Weng et al., 2021); Cdh3 localizes to dynamic foci at deeper cell-cell interfaces (Fagotto et al., 2013; Huebner et al., 2021). Because of the key role of these deeper cell-cell interfaces in CE (Huebner et al., 2021; Shindo et al., 2019; Shindo and Wallingford, 2014; Weng et al., 2021), we used live imaging to identify Cdh3-interacting proteins that localize to these sites.
As mentioned, Cdh3 localizes strongly to the deep cell-cell junctions, where it co-localized with actin, as expected (Fig. 1F; quantified in Fig. 1K–L). Given our interest in biomechanics, we next examined the localization of the mechanosensitive protein α-catenin, a very strong Cdh3 interactor in our AP-MS data (Fig. 1B). α-catenin displayed a similar localization at deep cell-cell junctions (Fig. 1K Supp. Fig. 2B). This result is consistent with the requirement of α-catenin in zebrafish CE (Han et al., 2016).
A surprising result was that the mechanosensitive protein vinculin was only very weakly enriched in our Cdh3 AP-MS dataset from the Xenopus DMZ (Data Table 1). Vinculin is a key component of Cdh1/E-cadherin cell-cell and is specifically associated with cell junctions under tension, including during CE in Drosophila (Huveneers et al., 2012; Kale et al., 2018), so we expected to find vinculin at the deep mesenchymal cell-cell junctions. Nonetheless, vinculin-GFP was not co-localized with Cdh3 and α-catenin at deep cell-cell junctions (Fig. 1G, L). On the other hand, vinculin-GFP was localized to foci specifically in superficially positioned lamellipodia (Supp. Fig. 2C), where Cdh3 is also known to act (Pfister et al., 2016).
This result motivated us to examine another mechanosensitive protein, Testin, which is required for Xenopus CE and is a known tension-dependent effector of cadherin adhesion (Dingwell and Smith, 2006; Oldenburg et al., 2015). Like Vinculin, Testin was present but not highly enriched in our AP-MS dataset (Data Table 1), and moreover, Testin-GFP also did not localize to deep cell-cell junctions (Fig. 1H, L) and was enriched only in foci in the superficial lamella (Supp. Fig. 2D).
Finally, amongst the most highly enriched orthogroups in our Cdh3 AP-MS dataset was that containing the closely related Arvcf and p120 catenins (Data table 1)(Fig. 1B)(Supp. Fig. 1). By examining individual peptides in this orthogroup, we found that while both proteins interact with Cdh3, Arvcf had both the larger peptide count and fold change (Supp. Fig. 1). Arvcf is required for Xenopus CE (Fang et al., 2004; Paulson et al., 2000), yet little is known of its function, and even its localization during CE has not been reported. Using a functional Arvcf-GFP fusion, we found that Arvcf was present in the superficial lamellar structures (Supp. Fig. 2E, white arrows) but moreover was prominently enriched in the deep cell-cell junctions, where it co-localized strongly with Cdh3 (Fig. 1I, yellow arrows, Fig. 1K).
Together, these localization patterns indicate that the Cdh3-dependent mesenchymal cell-cell junctions in Xenopus DMZ cells share some features of the more-thoroughly characterized E-cadherin-dependent adhesions in epithelial cells. However, the absence of vinculin and testin from these junctions suggests important differences as well. These data motivated us to explore cell adhesion during Xenopus CE in more detail, and we chose to focus on the Arvcf catenin.
Arvcf is required for head-to-tail axis extension in the embryo but is dispensable for CE in isolated tissues
The very high confidence interaction of Arvcf with Cdh3 in the Xenopus DMZ (Fig. 1B), its requirement for CE, and its connection to Rho and Rac signaling (Fang et al., 2004; Reintsch et al., 2008) made it a strong candidate for further investigation. We therefore used a previously characterized morpholino oligonucleotide to disrupt Arvcf function (Fang et al., 2004). We recapitulated the described axis elongation defect after Arvcf knockdown, and moreover, this defect was rescued with Arvcf-GFP (Fig. 2A–D).
Figure 2: Arvcf is required for embryonic axis extension.
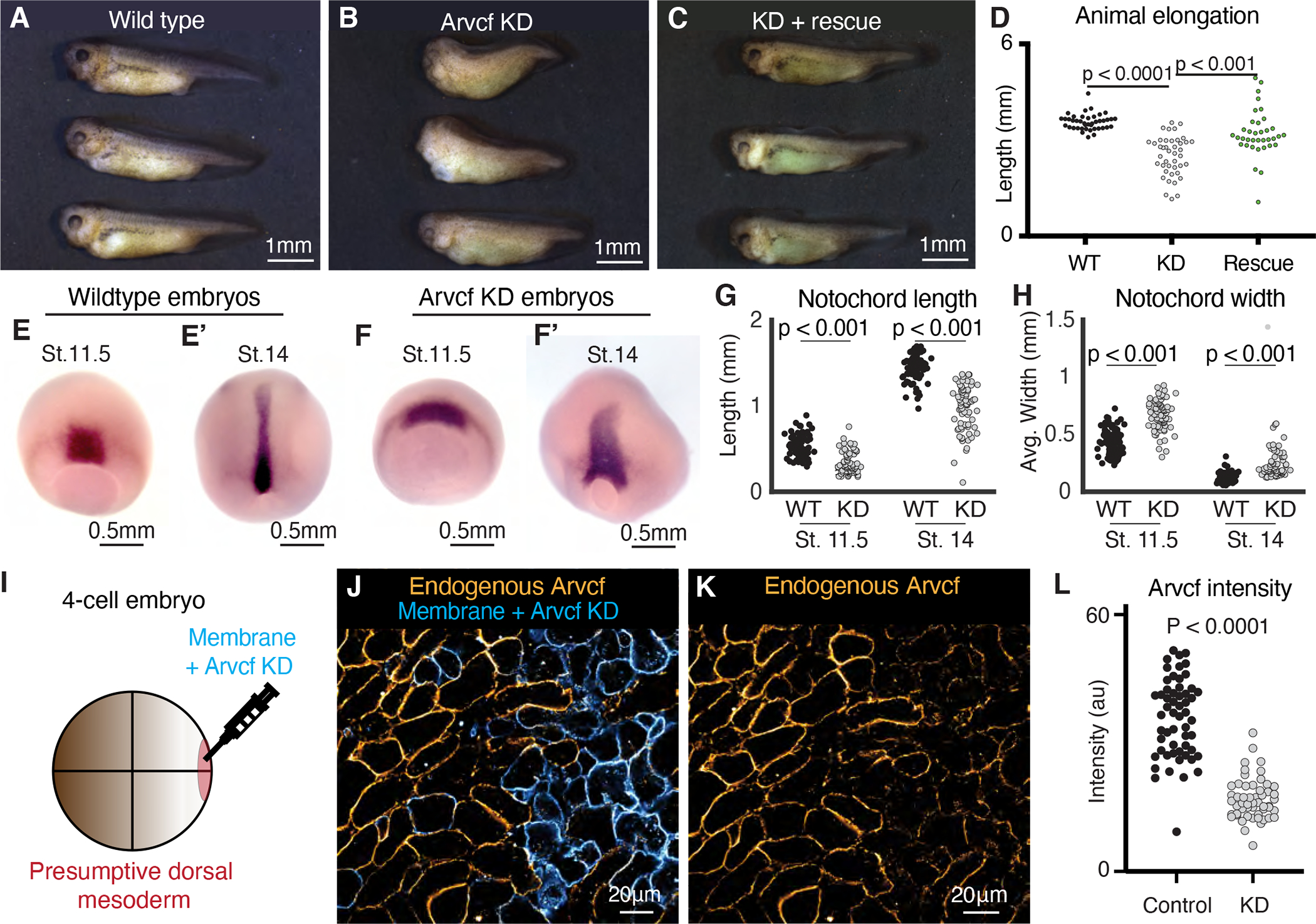
A. Wild type tadpoles (~st.40). B. Sibling embryos to those shown in Fig. 2A in which Arvcf was knocked down in the dorsal mesodermal cells. The Arvcf depleted embryos have a shortened head-to-tail axis and display a characteristic dorsal bend suggesting a CE defect. C. Sibling embryos in which Arvcf was knocked down and then rescued with exogenous expression of Arvcf-GFP. D. Plot showing tadpole (~st.40) head-to-tail length for the wildtype, ARVDF knockdown, and Arvcf rescue conditions. Embryo lengths were statistically compared using an ANOVA test. Each dot represents a single embryo and data was collected from a minimum of three experiments. E. Stage 11.5 wildtype embryos stained by in situ hybridization for the notochord probe Xnot. E’. Stage 14 wildtype embryos stained by in situ hybridization for the notochord probe Xnot. F. Stage 11.5 Arvcf knockdown embryos stained by in situ hybridization for the notochord probe Xnot. F’. Stage 14 Arvcf knockdown embryos stained by in situ hybridization for the notochord prob Xnot. G. Comparison of the total notochord length in wildtype or Arvcf knockdown embryos at stage 11.5 and stage 14. Conditions were statistically compared using a Mann-Whitney test. H. Comparison of the notochord width for wildtype or Arvcf knockdown embryos at stage 11.5 and stage 14. Conditions were statically compared using a Mann-Whitney test. I. Cartoon depicting the microinjection method used to generate mosaic animals. J. Immuno-staining for Arvcf (orange) in an embryo in which Arvcf has been mosaically knocked down (blue cells). K. The same image shown in Fig. 2J except the membrane marker has been removed to better visualize the Arvcf immunostaining. L. Quantification of endogenous Arvcf protein levels from the immunostaining performed on embryos with mosaic Arvcf knockdown. Each dot represents the average ARVCF intensity at the membrane of a single cell and data was collected from a minimum of three replicates. Conditions were statistically compared using a Mann-Whitney test.
Convergent extension begins in Xenopus at the onset of gastrulation and continues into tailbud stages (Shih and Keller, 1992), so to better understand the timing of the axis elongation defect, we visualized the notochord at mid-gastrula and early neurula stages (NF Stg. 11.5 and 14, respectively) embryos using in situ hybridization of the notochord specific probe Xnot (von Dassow et al., 1993). We found that the notochord was already slightly shorter and wider by stage 11. 5 and that the defect became more exaggerated by stage 14 (Fig. 2E–H). We then used targeted injections to generate mosaic Arvcf knockdown embryos and used immunostaining for endogenous Arvcf protein at late gastrula stages to confirm deletion of the protein by the morpholino (Fig. 2I–L). Together, these data demonstrate the efficacy and specificity of our approach for Arvcf loss of function. We next sought to better define the nature of the axis elongation defects elicited by Arvcf loss.
The failure of axis extension starting at gastrula stages and the dorsal flexion together strongly suggest a defect in CE. As a direct test, we used a traditional explant assay in which converging and extending tissue is excised from the embryo and allowed to extend ex vivo (Fig. 3A, A’). For this assay, the entire dorsal region of the embryo was excised (dorsal isolates) at late gastrula stage (~st.12) and allowed to elongate unrestrained until early neurulation (~st.14). To our surprise, we observed no difference in the total extent of elongation when comparing the Arvcf knockdown dorsal explants with control (Fig. 3B–C).
Figure 3: External constraint of Arvcf deficient explants recapitulates the embryonic axis extension defect.
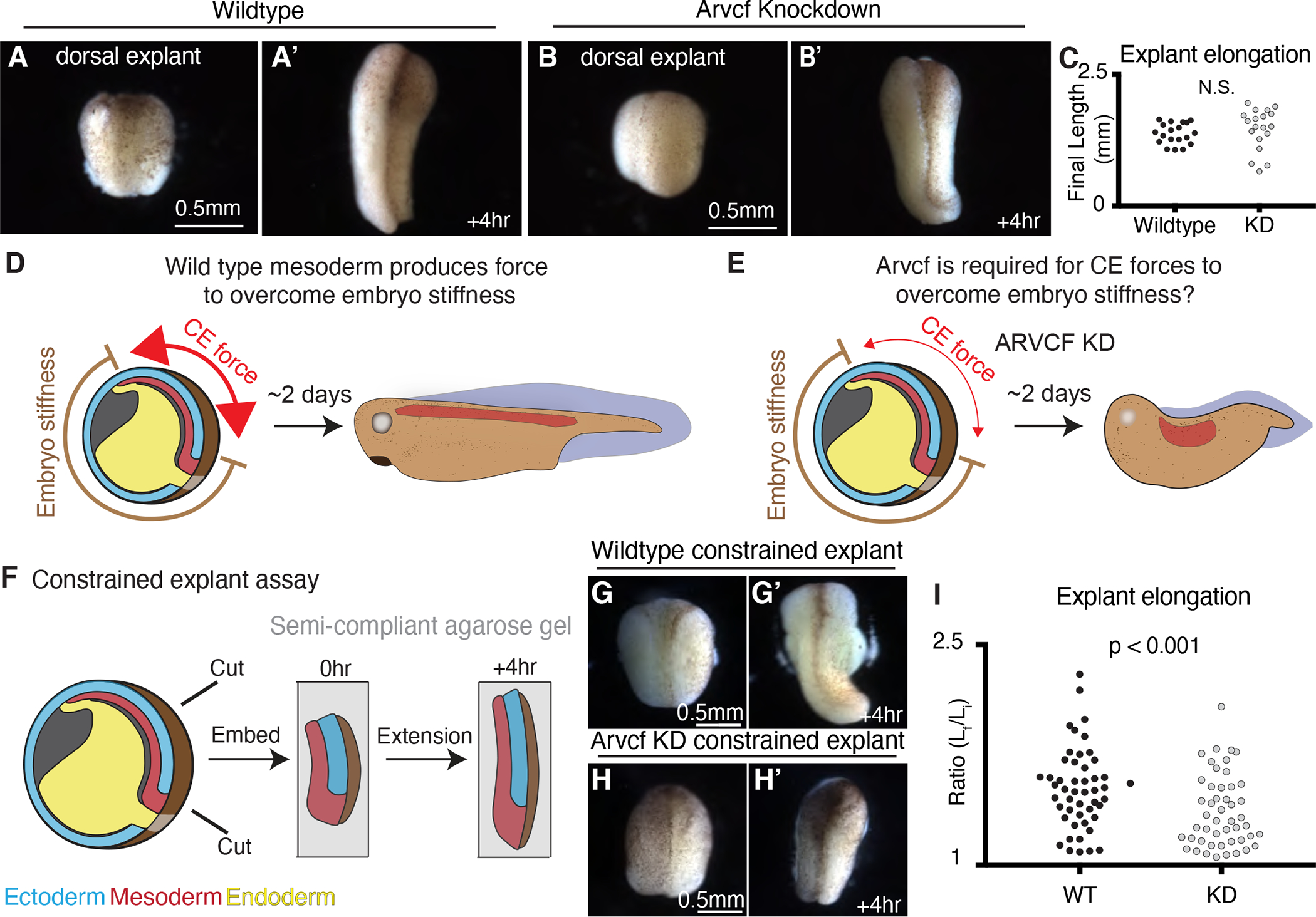
A. Image of a wildtype dorsal isolate. A’. Image of a wildtype dorsal isolate after a four-hour period of extension. B. Image of a dorsal isolate in which Arvcf has been knocked down in the mesodermal cells. B’. Image of an Arvcf depleted dorsal isolate after a four-hour period of extension. C. Graph showing the final dorsal isolate length, after a four-hour elongation, for wildtype and Arvcf depleted embryos. Each dot represents the length of a single explant and conditions were statistically compared using a Mann-Whitney test. D. Cartoon depicting the forces involved in Xenopus axis extension. Here the dorsal mesoderm (red) and the overlaying neural ectoderm (blue, above red) converge and extend generating force to push against the stiff embryo. In the case of wildtype embryos, the CE generated force is sufficiently large (red arrows) to overcome the embryo stiffness and the resulting animals have elongated head-to-tail axis. E. We hypothesize that Arvcf is required for CE generated force and that depletion of Arvcf reduces the tissue level force produced by CE. In this case the reduced CE force is insufficient to push the stiff surrounding embryo and axis extension fails. F. Schematic depicting the constrained explant assay used to mimic the mechanical environment experienced within the embryo. G. Image of a dorsal isolate after embedding in gel. G’. Image of the same dorsal isolate shown in Fig. 3G after 4 hours of elongation. H. Image of an Arvcf deficient dorsal isolate after embedding. H’. Image of the same dorsal isolate in Fig. 3H after a 4-hour interval of extension. I. Graph showing the extent of dorsal isolate elongation during the constrained explant assay for wildtype and Arvcf knockdown dorsal isolates. The y-axis shows the ratio of the final explant length over the final explant length. Each dot represents a single dorsal isolate and conditions were statistically compared using a Mann-Whitney test. See also figure S3.
To explain this surprising result, we considered that axis elongation in Xenopus embryos requires increasing force generation of the converging and extending dorsal tissues to overcome the stiffness of surrounding embryo (Moore et al., 1995; Zhou et al., 2009; Zhou et al., 2015). We therefore hypothesized that Arvcf was not in fact required for CE per se, but instead was required for sufficient CE-mediated force generation to overcome resistance from the surrounding embryo (Fig. 3D, E). To test this idea, dorsal explants were isolated at late gastrula stages and embedded in a semi-compliant 0.3% agarose gel, then incubated until st. 14 (Fig. 3F). In this semi-constrained condition, control explants still elongated effectively (Fig. 3G, G’, I), but under the same conditions, we observed a significant reduction in the extension of explants deficient for Arvcf (Fig. 3H–I).
Thus, by simply adding a subtle mechanical challenge to the Arvcf deficient explants, we were able to recapitulate the embryonic Arvcf knockdown phenotype. This led us to more carefully examine the dynamics of the elongation of unrestrained Arvcf-KD explants, and this revealed a subtle but significant delay in elongation during gastrula stages, even though explants still achieved full length by early neurulation (Supp. Fig. 3). These results therefore suggest a subtle role for Arvcf in CE, specifically in the production of stronger extension forces.
Arvcf is required for tissue-scale force production and stiffening during convergent extension
We next sought to directly quantify the reduced force output of Arvcf deficient dorsal isolates. Following the methods developed by (Zhou et al., 2015), we embedded explants dissected from late gastrula embryos (~ st.12) in semi-rigid, 0.6% agarose gels containing evenly dispersed fluorescent beads (Fig. 4A). Within this semi-rigid gel, wildtype explants converged and produced an extending force that deformed the gel (Fig. 4B, B’). We also observed tissue buckling at ~3 hours when a sudden bend appeared (Supp. Fig. 4A, B, arrow). The embedded fluorescent beads provided fiducial markers for tracking gel deformation, allowing us to observe compression in the gels at the AP poles of the explant (Fig. 4B’). In contrast, Arvcf KD explants also converged and buckled (Fig. 4C), but gel deformation by CE was nearly unobservable (Fig. 4C’).
Figure 4: Arvcf controls force production during vertebrate convergent extension.
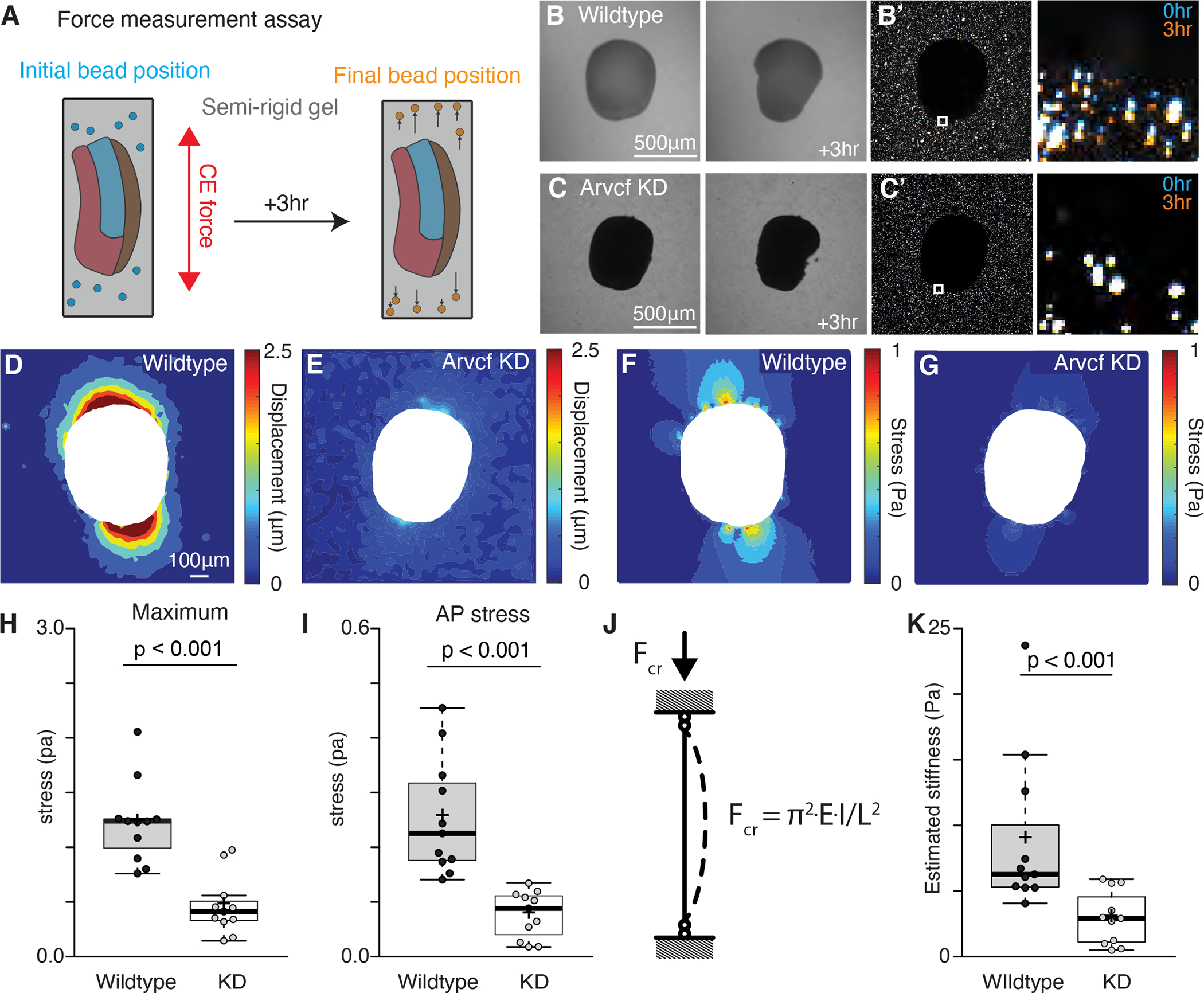
A. Schematic depicting the assay used to measure the CE force production. Dorsal explants were excised from late gastrula embryos (~st.12) and embedded in semi-rigid 0.6% agarose gels with known mechanical properties. Fluorescent beads were also embedded in gel to allow visualization of the gel deformation. Explants were then incubated for three hours to allow CE. Then with the known mechanical properties of the gel and the displacement field of the beads we calculated the stress fields generated by each explant. B. Images of a wildtype explant embedded in a semi-rigid agarose gel and then a second image of the same explant after a three-hour incubation. White arrow points to the direction of the out-of-plane explant buckling. B’. Image of the same explant and gel shown in Fig. 4B but here we are visualizing the beads embedded in the gel and surrounding the explant. The inset focuses on the beads adjacent to the explant and the zoomed in image shows both the initial bead position (blue) and the final bead position (orange). C. Image of an Arvcf depleted explant embedded in a semi-rigid agarose gel and a second image of the same explant after a three-hour incubation. C’. Image of the beads surrounding the explant shown in Fig. 4C. The inset focuses on a subset of beads next to the explant and the zoomed image shows the initial bead position (blue) and the final bead position (orange). D. Displacement field measured by PIV from the beads in Fig. 4B’. E. Displacement field measured by PIV from the beads in Fig. 4C’. F. Von Mises stress field estimated using finite element method in the gel shown in Fig. 4B. G. Von Mises stress field estimated using finite element method in the gel shown in Fig. 4C. H. Graph showing the maximum compressive stress along the explant-gel interface. Conditions were statistically compared using a Mann-Whitney test and Arvcf depleted explants applied a significantly lowered force on the gel. I. Graph showing the average compressive stress along the AP axis. Conditions were statistically compared using a Mann-Whitney test and Arvcf depleted explants applied a significantly lowered extending force along the AP axis. J. Schematic depicting the buckling model to estimate tissue stiffness. Explant was modeled as a column with a rectangular cross-section. When it converged and extended in a semi-rigid gel, the reactive force applied a uniform longitudinal load that caused an out-of-plane tissue buckling, K. Graph showing the estimated tissue stiffness using a simplified buckling model. Conditions were statistically compared using a Mann-Whitney test and Arvcf depleted explants were significantly softer. See also figure S4.
To quantify the compressive force generated by elongating explants, we used particle image velocimetry (PIV) (Thielicke and Stamhuis, 2014) to quantify the movement of fluorescent beads, and thus gel deformation (Fig. 4D, E). Using finite element analysis to estimate the stress field (Fig. 4F, G), we compared the maximum tissue force before the explants buckled (~3hr). Wildtype explants generated maximum compressive stress of 1 Pa at the AP poles and average stress of 0.2 Pa along the AP axis (Fig. 4F, H, I). These values are comparable with prior assessment of CE forces in Xenopus (Zhou et al., 2015). By contrast, explants deficient for Arvcf generated significantly less force, displaying a roughly 60% reduction in both maximum and mean stress (Fig. 4G, H, I).
Finally, we calculated the tissue stiffness using the critical buckling load, an effective estimation of biomechanical stiffness across length scales, including during tissue morphogenesis (Badel et al., 2013; Kikumoto et al., 2006; Trushko et al., 2020). We treated explants as a columns with a rectangular cross-section, on which tissue scale extending force applied a uniform longitudinal load (Fig. 4J). This estimation also revealed a significant reduction in the tissue stiffness of explants deficient for Arvcf (Fig. 4K). These data demonstrate that Arvcf is not only required for tissue scale force production but also for tissue stiffening during CE.
Arvcf knockdown disrupts cell adhesion during convergent extension
We next sought to understand the cell biological basis for reduced force production in the absence of Arvcf. A previous study showed that Arvcf loss results in subtly reduced bulk levels of Cdh3 protein (Fang et al., 2004), but how it affects Cdh3 membrane localization is not known. We therefore used targeted microinjection to knockdown Arvcf in one half of the dorsal mesoderm and used immuno-staining to measure endogenous Cdh3 protein levels (Fig. 5A, B). We measured Cdh3 intensity and found a modest but significant reduction in endogenous Cdh3 protein levels at cell-cell junctions after Arvcf KD (Fig. 5A–C).
Figure 5: Arvcf KD reduces cell adhesion but only has a modest effect on cell intercalation.
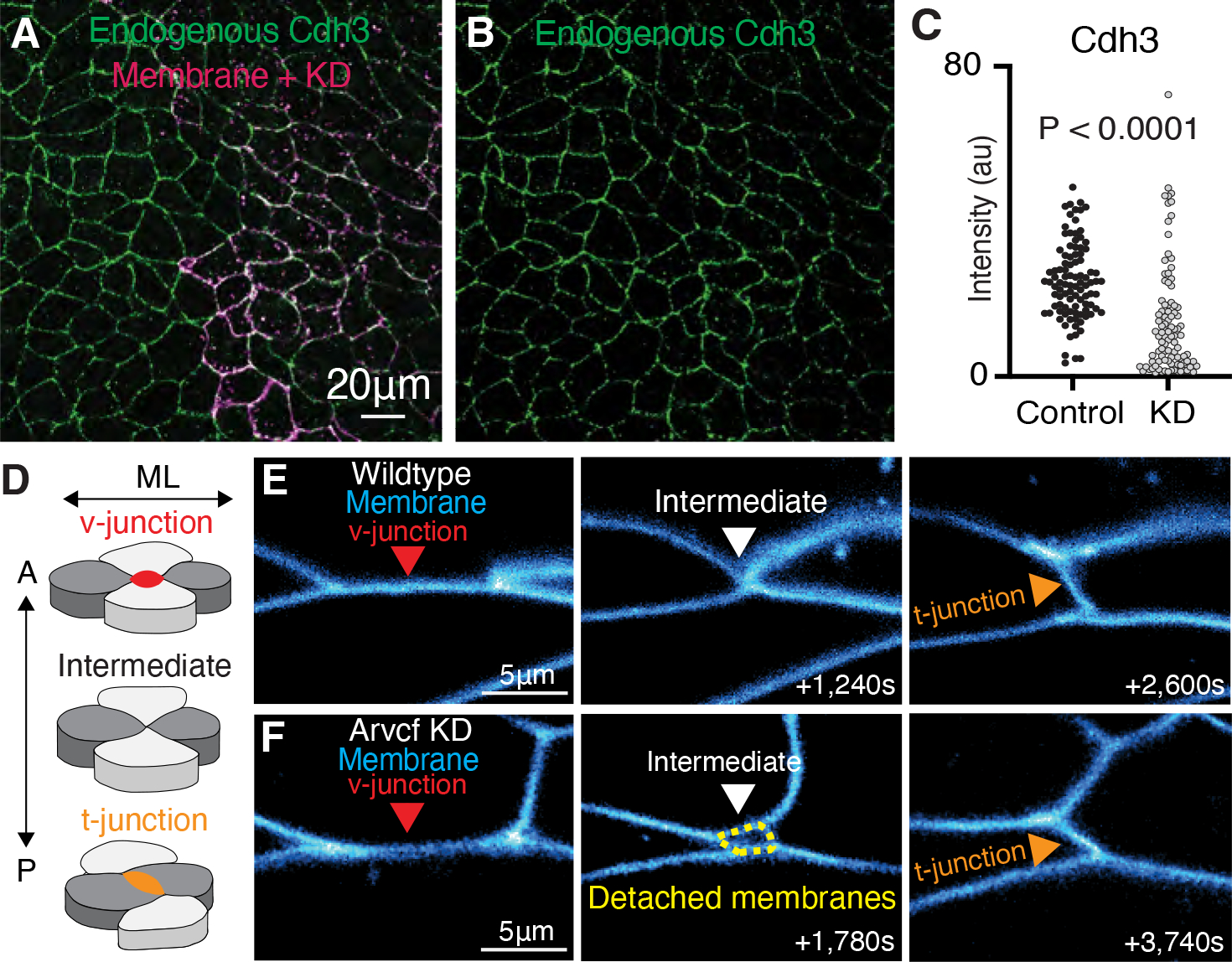
A. Immunostaining for endogenous Cdh3 (green) in a field of cells where Arvcf had been mosaically knocked down. A membrane marker (magenta) was used as a tracer for the Arvcf morpholino. B. Image showing the isolated Cdh3 channel from Fig. 5A. C. Graph displaying the measurement of endogenous Cdh3 intensity from wildtype or Arvcf depleted cells. Each dot represents the average cdh3 intensity of a single cell and conditions were statistically compared using a Mann-Whitney test. D. Cartoon depiction of the cell movements that drive CE with emphasis on the cell-cell junctions. Initially there is a cell-cell junction between the anterior-posterior cells (light gray) termed a v-junction (red). The cells then intercalate bringing the mediolateral cells (dark gray) together. The mediolateral cells then form a new cell-cell contact (t-junction; orange) pushing the anterior posterior cells apart. E. Still frames from a time-lapse movie of wildtype cells intercalating. Cell membranes are labeled blue, and the cell intercalation can be visualized as the v-junction is replaced by a t-junction. F. Frames from a time-lapse movie showing one example of Arvcf depleted cells intercalating. Here we initially observe a v-junction which shortens, forms a 4-cell intermediate, which then resolves to form a new t-junction. One feature that was unique to the intercalation of the Arvcf depleted cells was that there were often gaps (yellow dashed lines) between the membranes at the intermediate state. Despite these gaps, cells were able to intercalate after ARVCF-KD.
To ask if this reduction of cortical Cdh3 was accompanied by changes in cell-cell adhesion, we made time-lapse movies of CE. For this analysis, we simplify CE to the rearrangements of groups of four cells in which anterior-posterior neighbors are initially connected via a cell junction (v-junction) which shortens by during intercalation, resulting in formation of a new cell contact (t-junction)(Fig. 5D, E). In time-lapse movies, Arvcf-depleted cells were motile and underwent largely normal cell intercalation events, but these were characterized by transient detachment of cell-cell junctions, leaving extra-cellular spaces (Fig. 5F). These detachments were always transient but were particularly pronounced as v-junctions were replaced by t-junctions (Fig. 5F). Moreover, in a companion paper, we show that the rate of junction shortening was modestly, but significantly reduced in Arvcf depleted cells (Weng et al., 2021). Thus, v-junctions in Arvcf-depleted cells were slow to shorten and displayed transient detachment from neighboring cells but ultimately succeed in intercalating.
Loss of Arvcf leads to reduced amplitude of cortical actin oscillations and reduced cortex tension at v-junctions.
In addition to its role in cell adhesion, Arvcf functionally interacts with the small GTPases Rho and Rac during CE (Fang et al., 2004), but again the cell biological impact for these interactions is not known. These small GTPases control actin assembly and actomyosin contraction, so we examined the effect of Arvcf loss on the enrichment of actomyosin at v-junctions using biosensors for actin (lifeact-RFP) and contractile myosin (myosin light chain 9, Myl9-GFP). We found that the normal cortical enrichment of actomyosin at v-junction during Xenopus CE (Shindo et al., 2019; Shindo and Wallingford, 2014) remained normal after loss of Arvcf (Fig. 6A–D), suggesting that Arvcf is not required for the steady state polarization of the contractile actomyosin machinery at v-junctions.
Figure 6: Depletion of Arvcf resulted in a dampening of the oscillatory temporal dynamics of Cdh3 and actin.
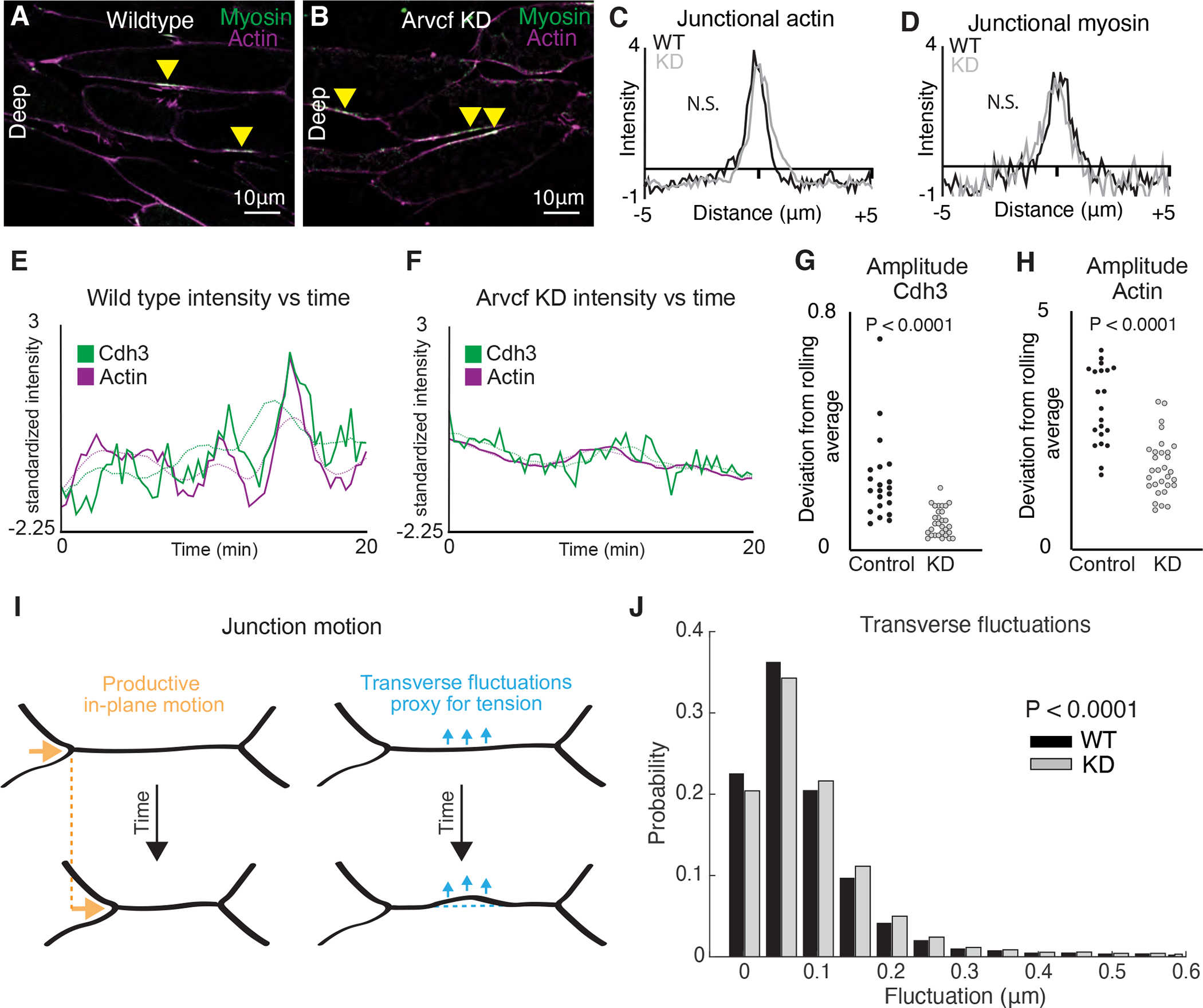
A. Image of deep cell-cell interfaces showing actin in magenta and myosin in green. Yellow arrowheads point to myosin accumulations at v-junctions. B. Image of deep cell-cell interfaces after Arvcf knockdown. Yellow arrows point to myosin enrichments at v-junctions. C. Intensity plots showing the mean intensity of actin at v-junctions. Wildtype is shown in black and Arvcf knockdown cells are shown in gray. Conditions were statistically compared using a KS test. D. Intensity plots showing mean intensity of myosin at deep v-junctions. Black shows wildtype and gray shows Arvcf depleted cells. Conditions were statistically compared using a KS test. E. Cdh3 (green) and actin (magenta) intensity plotted over time from a wildtype shortening v-junction. Dashed lines show the rolling average (over 3.33 min) for either Cdh3 or actin. F. Plot showing Cdh3 (green) and actin (magenta) intensity over time from an Arvcf depleted shortening v-junction. Dashed lines show the rolling average. G. We quantified the oscillation amplitude as the deviation of a protein’s intensity plot from that same protein’s rolling average. This graph shows the average Cdh3 amplitude for wildtype and Arvcf knockdown v-junctions. Each dot is the average amplitude from a single shortening v-junction. Conditions were compared using a Mann-Whitney test. H. The same quantification shown in Fig. 6J but looking at actin oscillation amplitude. Again, each dot represents the average amplitude at a single shortening v-junction and conditions were compared using a Mann-Whitney test. I. Schematic showing the transverse fluctuations which we use as a proxy for local junction tension. Orange shows the productive in-plane motion of junction shortening and blue shows the transverse fluctuations that are opposed to in-plane motion. Dashed lines represent the initial junction position. J. Quantification of the probability that a wildtype (black) or Arvcf knockdown (gray) junction will undergo a transverse fluctuation of a given size. The Arvcf knockdown junctions had significantly larger transverse fluctuations. Conditions were compared using a Mann-Whitney test.
Cell intercalation in Xenopus involves not only polarized steady state enrichment of actomyosin at v-junctions (Shindo and Wallingford, 2014), but also oscillations that correlate with intercalation speed and efficiency (Shindo et al., 2019). Despite the normal bulk levels, we observed a dramatic reduction in the amplitude of actomyosin pulses is Arvcf depleted junctions (Fig. 6E, F). To quantify this result, we measured the rolling average actin intensity (Fig. 6E, F) dashed lines) and then defined the oscillation amplitude for each junction as the mean deviation between the junction intensity and the rolling average. Using this method, we found a significant difference in the amplitude of actin oscillation in Arvcf-KD cells (Fig. 6G, H).
Finally, such a reduction in actin oscillation amplitude at v-junctions may be expected to reduce the force generated by individual v-junctions, which may explain the tissue level phenotype described above. To test this idea, we employed our method for estimating tension in shortening junctions (Huebner et al., 2021). This method estimates tension by quantifying the non-productive fluctuations of the cell cortex transverse to the axis of junction shortening in time-lapse movies (Fig. 6I). Because it is non-invasive, the method overcomes two limitations of traditional laser-cutting; it rules out any possible contribution of the wounding response to observed cortex dynamics, and moreover enables continuous assessment of tension in single junctions over time. Arvcf depleted junctions displayed a significantly higher probability of undergoing large transverse fluctuations (Fig. 6J) indicating that junctions lacking Arvcf experience lower cortical tension as compared to controls. Thus, our data suggest that in the absence of Arvcf, modestly reduced cell adhesion and dampened actin dynamics result in reduced cortex tension along v-junctions; this in turn results in slower intercalation and an overall reduction in elongation force for the tissue.
Discussion:
Our over-arching goal here was to explore the role of cadherin-based cell adhesions specifically in the collective movement of mesenchymal cells in a vertebrate embryo. Here, proteomic, biomechanical, and cell biological approaches revealed a role for Arvcf catenin in tuning the multiscale biomechanics of CE during Xenopus gastrulation. Specifically, we showed that Arvcf disrupts embryo elongation, but does not disrupt the elongation of isolated explants of axial tissues. In this way, the phenotype elicited by Arvcf loss is distinct from that observed for other regulators of CE, as disruption of PCP proteins, cadherins, Rho GTPases, or actin regulators all elicit parallel elongation defects in embryos and also explants.
Moreover, Arvcf was not required for the cell movements that drive CE per se, resulting in only a modest alterations of cell behaviors that drive intercalation but instead is required to achieve the normal levels of extension force. We further show that loss of Arvcf leads to a modest reduction in cell adhesion, dampening of cortical actin oscillation, and reduced cortex tension in v-junctions. We conclude that the reduced oscillatory recruitment of this actomyosin machinery reduces force production at the cellular level which results in the observed tissue-scale force defect. We feel that several aspects of this study are of note.
First, our results also provide insights into molecular mechanisms of cadherin-based cell adhesion, specifically in mesenchymal cells. Unlike the well-characterized E-cadherin-based junctions in epithelial cells (Rodriguez-Boulan and Macara, 2014), mesenchymal cell junctions are amorphous, lacking the more ordered features such as adherens junctions and tight junctions (Ewald et al., 2012). Nonetheless, mesenchymal cell collectives maintain cohesion during collective movement (Theveneau and Mayor, 2012), including during CE (Walck-Shannon and Hardin, 2014). In this context, it is significant that our APMS indicates that Cdh3-mediated junctions in Xenopus mesenchyme share some but not all features of the canonical Cdh1 (E-cadherin) junctions in epithelia. For example, while α- and β-catenin were strongly bound to Cdh3 and enriched in cell-cell junction, vinculin was not. This result is surprising as vinculin is a key mechanosensity component of Cdh1/epithelial junctions (le Duc et al., 2010), including during Drosophila CE (Huveneers et al., 2012; Kale et al., 2018), and its absence represents a key difference between Cdh3/mesenchymal junctions and Cdh1/epithelial junctions. It worth noting, therefore, that mammalian Cdh3 (aka P-cadherin) is implicated in collective cell movements in the skin and mammary gland (Cetera et al., 2018; Radice et al., 1997), so further exploration of the composition of cadherin adhesion complexes in other cell and tissue types should be informative.
In addition, our Cdh3 AP-MS dataset led us to re-examine the Arvcf catenin, which has known roles in vertebrate development (Cho et al., 2011; Fang et al., 2004) but has not been re-examined in the past decade and its cell biological mechanisms of action remain unclear. Our finding that Arvcf specifically controls the magnitude of tissue-scale CE force generation is interesting, as nether Arvcf or its closest homolog p120 catenin have been implicated in force production or force sensitivity (McCrea and Park, 2007). Moreover, the finding that loss of Arvcf elicits a slight reduction in cell and tissue forces is also important for providing a molecular insight into the progressive tissue stiffening that has been well-characterized in amphibian embryos (Keller and Sutherland, 2020; Moore et al., 1995; Shook et al., 2018; Zhou et al., 2009), as well as in other vertebrates (Mongera et al., 2019). Perhaps most significantly in this light, our data suggest that Arvcf-mediated control of actomyosin and cell adhesion is a central feature of embryos’ “mechanical accommodation” abilities that allow for consistently normal development despite the wide range of mechanical variation between individual embryos (Zhou et al., 2015).
The exact mechanism of Arvcf action remains unclear, but our work here provides several clues. First, we observed a modest reduction in cell adhesion, likely stemming from a modest loss of junctional Cdh3 (Fig. 5), consistent with previous work in Xenopus (Fang et al., 2004). Second, and more significant in our view, was the reduction of the amplitude of actin oscillations and the reduced cell cortex tension at v-junctions (Fig. 6). These results provide an advance in their own right, as the cell biological basis of Arvcf-associated CE defects have not been explored, but the data become more significant in light of results from our companion paper.
In that paper, we show that Xenopus CE is driven by the spatial and temporal integration of cell crawling and junction contraction (Weng et al., 2021), two mechanisms that are physically separated in epithelial CE (Huebner and Wallingford, 2018; Sun et al., 2017; Williams et al., 2014). We further show that the integration of the two modes is crucial for high efficiency cell intercalation, and that loss of Arvcf results in reduced cell intercalation velocities, which parallels the delayed tissue elongation reported here. Strikingly, mosaic labelling experiments in that paper further suggest that the slower intercalation results from a specific defect in lamellipodia-mediated cell crawling (Weng et al., 2021), and this effect likely explains the dampened amplitude of overall actin oscillations at v-junctions observed here (Fig. 6). This result is curious: On the one hand, it is consistent with the previous finding that Arvcf is required for normal Rac activation during Xenopus CE (Fang et al., 2004), and Rac is generally considered to be essential for lamellipodial protrusions (Nobes and Hall, 1995). On the other hand, other work has suggested that disruption of Rac is not sufficient to disrupt lamellipodia formation in Xenopus (Tahinci and Symes, 2003).
Clearly, further exploration will be required, but the data from the two papers together suggest the following model: In the absence of Arvcf, reduced cell adhesion and reduced protrusive activity elicit in an overall reduction in cell cortex tension along v-junctions. This in turn slows cell intercalation and limits the amount of tissue-level force that can be produced by the tissue. Thus, in the context of the embryo depleted of Arvcf, CE is not sufficiently powerful to overcome the load created by other attached tissues, resulting in stunted axis elongation. Thus, our study here of the subtle effects of Arvcf loss provide a complement to previous studies of essential regulators such as Cdh3 itself or the PCP proteins in CE, and also inform our understanding of mechanical robustness of morphogenesis generally (e.g. (Davidson, 2017; von Dassow and Davidson, 2009).
Limitations of the study
In this study we investigated Arvcf function exclusively during CE of the Xenopus DMZ. Moving forward it would be exciting to test if Arvcf has a similar role in other converging and extending tissues such as the Xenopus neural plate. As well it would be good to test Arvcf function during CE in other organisms such as Drosophila or mouse. Similarly, we performed the Cdh3 AP-MS experiment exclusively in the DMZ. Testing the Cdh3 protein interactions in other mesenchymal tissues would allow us to better understand if the observed Cdh3 protein interactions are specific to the DMZ or more general to mesenchymal tissues. A final limitation is that we use a knockdown approach to investigate Arvcf function. While our knockdown is efficient (Fig. 2J–L) and specific (Fig. 2A–D) it would be insightful to see if a complete knockout would exaggerate the knockdown phenotype.
STAR Methods:
Resource availability
Lead contact
Requests for resources and reagents should be directed to and will be made available by the lead contact, John B. Wallingford, Wallingford@austin.utexas.edu.
Materials availability
Unique materials (eg. plasmids) generated by this study will be made available by the lead contact upon request.
Data and code availability
Proteomics data has been deposited into MassIVE which in turn was passed to ProteomeXchange. The MassIVE accession # is MSV000087312. The ProteomeXchange # is PXD025665 (http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD025665-1).
No new code was generated during this study.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and subject details
Xenopus laevis used in this study were acquired from Nasco which has since stopped selling live animals. However, the Nasco Xenopus colony has been moved to and are available from Xenopus1. All Xenopus research performed for this study was approved by IACUC. A more thorough description of Xenopus handling is described below in the method detail section.
Method details
Xenopus embryo manipulations
Adult Xenopus were maintained in a recirculating aquatic system and female Xenopus were ovulated at a maximum of once every three months to ensure high quality embryos. Ovulation is induced by injection of 600 units of human gonadotropin and ovulating females are incubated at 16°C overnight. Following incubation ovulating females produced eggs for ~8 hours. Eggs were acquired by gently “squeezing” ovulating females and then immediately fertilized using crushed testes. Embryos were reared in 1/3X Marc’s Modified Ringer’s (MMR) solution unless otherwise noted. Eggs were dejellied in 3% cysteine (pH 8) for 10 minutes and washed in 1/3X MMR prior to further manipulation.
For microinjection embryos were placed in 2% ficoll in 1/3X MMR 10 minutes prior to injection and then returned to 1/3X MMR 30 minutes after microinjection. Embryos were injected using a Parker’s Picospritizer III with an MK1 manipulator. Embryos were injected in dorsal blastomeres to target the presumptive mesoderm and were injected at the 4- or 32-cell stage as noted in the manuscript.
Microdissections were performed in Steinberg’s solution and explants were maintained in Steinberg’s after dissection. Dissections were performed under a stereoscope using eyelash knives and hair loops. Embryos were dissected at st.~10.25 (Keller explants (Sive et al., 2007)) for mass spec experiments and for imaging experiments. Embryos were dissected at ~st.12 (dorsal explants (Zhou et al., 2015)) for biomechanical assays and the later stage explants were used here because earlier stage explants were not sufficiently stiff for biomechanical manipulations and measurements.
Morpholino, antibodies, plasmids, and cloning
The Arvcf morpholino was previously characterized (Cho et al., 2011; Fang et al., 2004) and was ordered from Gene Tools. The Arvcf polyclonal antibody was also previously developed and characterized (Paulson et al., 2000) and was gifted from the laboratory of Pierre D. McCrea. Cdh3 antibody was ordered from the Developmental Studies Hybridoma Bank (catalog number 6B6). The α-catenin antibody was purchased from Sigma (catalog number C2081). Cdh3-GFP (Pfister et al., 2016), lifeact-RFP, membrane-GFP, and membrane-BFP were made in pCS105. Vinculin-GFP was made in pCS108 and was a gift from the Paris Skourides lab. Myl9-GFP was made in pCS107 (Shindo et al., 2019). The Xenopus Arvcf and testin sequences were obtained from www.xenbase.org and the open reading frames were amplified from a Xenopus cDNA library. The Arvcf and testin ORFs were then inserted into the pCS10R-GFP vector.
Arvcf MO: 5’- ACACTGGCAGACCTGAGCCTATGGC -3’
Morpholino and mRNA microinjections
Capped mRNA was produced using Thermo Fisher SP6 mMessage mMachine kit (catalog number AM1340). mRNAs were injected at the following concentrations vinculin-GFP (50pg), testin-GFP (50pg), Arvcf for imaging and rescue (50pg), lifeact-RFP (100pg), membrane-GFP for imaging and mass-spec (100pg), membrane-BFP (100pg), Myl9-GFP (50pg), Cdh3-GFP for imaging and mass-spec (50pg). Arvcf morpholino was injected at a concentration of 30ng.
Immunoprecipitation of Xenopus Keller explants for mass-spectrometry
Embryos were injected in the presumptive dorsal mesoderm at the 4-cell stage with either Cdh3-GFP or GFP alone. Keller explants were then excised from st.10.25 embryos and explants elongated to ~st.12. Explant immunoprecipitation (IP) was then performed using a GFP-Trap Agarose Kit (ChromoTek, catalog number gtak-20) and proteins were eluted in 2X sample buffer. The IP experiment was performed in 2 replicates with approximately 800 explants per condition per replicate.
Affinity purification-mass spectrometry
Immunoprecipitated proteins were prepared for mass spectrometry as described in (Lee et al., 2020). Two mass spectrometry injections were used for each biological replicate. Mass spectrometry was performed on a Thermo Orbitrap Fusion for the first and second injections of the first biological replicate of Cdh3-GFP and on a Themo Orbitrap Fusion Lumos Tribrid for the third and fourth injections. Both injections of the second biological replicate of Cdh3-GFP were performed on a Thermo Orbitrap Fusion. In all cases, peptides were separated using revere phase chromatography on a Dionex Ultimate 3000 RSLCnano UHPLC system (Thermo Scientific) with a C18 trap to Acclaim C18 PepMap RSLC column (Dionex; Thermo Scientific) configuration and eluted using a 3% to 45% gradient over 60 min. with direct injection into the mass spectrometer using nano-electrospray. For the second biological replicate and the second two injections of the first biological replicate, data were collected using a data-dependent high energy-induced dissociation (HCD) method. The first and second injections of the first biological replicate were collected using a collision-induced dissociation (CID) method. Full precursor ion scans (MS1) were collected at 120,000 m/z resolution, and monoisotopic precursor selection and charge-state screening were enabled using Advanced Peak Determination (APD), with ions of charge ≥ +2 selected for HCD with stepped collision energy of 30% +/− 3% (Lumos) or 31% +/− 4% (Orbitrap Fusion) or 35% collision energy for CID.
Protein interaction analysis
Raw MS/MS spectra were processed using Proteome Discoverer (v2.3). We used the Percolator node in Proteome Discoverer to assign unique peptide spectral matches (PSMs) at FDR < 5% to the composite form of the X. laevis reference proteome described in (Drew et al., 2020) which comprises both genome-derived Xenbase JGI v9.1 + GenBank X. laevis protein sets, but with homeologs and highly related entries combined into eggNOG vertebrate-level orthology groups (Huerta-Cepas et al., 2017), based on the method developed in (McWhite et al., 2020). To identify proteins statistically significantly associated with Cdh3, we calculated both a log2 fold-change and a Z-score for each prey protein based on the observed PSMs in the bait versus control pulldown. Calculations for the fold-change and Z-score were performed as described in (Lee et al., 2020) and (Lu et al., 2007). We determined significance by calculating p-values for each Z-score using the pnorm distribution function available in the R Stats Package (v3.6.1). We corrected for multiple comparisons by computing the Benjamini-Hochberg false discovery rate using the p.adjust function, also from the R Stats Package (v3.6.1). Probability values and false discovery rates are provided in data table 1.
Cellular scale imaging of Xenopus mesodermal cells
For a detailed description of high magnification Xenopus imaging please see (Kieserman et al., 2010). Briefly, Keller explants were mounted on fibronectin coated coverslips, mesoderm facing the coverslip, and held in place with a glass “chip” and vacuum grease. Explants were incubated at room temperature for 4 hours or at 16°C overnight. Confocal images were acquired using either a Nikon A1R or a Zeiss LSM700 (40x or 60x lens). Time-lapse movies were acquired with a 20 second time interval. Images were either acquired at the superficial surface (at the coverslip/cell interface) or at the deep cell surface (5μm above the coverslip).
Measurement of protein intensities at deep cell junctions
Image analysis was performed using the open-source image analysis software Fiji (Schindelin et al., 2012). Images were first processed using a 50-pixel rolling ball radius background subtraction. Then a 10μm straight line was drawn across each cell-cell junction with the line centered on the junction. The line was then converted to a region of interest and the Multi Plot tool was used to extract intensity values across the region of interest. Intensity plots were then statistically compared using a Kolmogorov-Smirnov test.
Imaging and measurement of embryos and dorsal explants
Whole embryos and dorsal explants images were acquired using a Zeiss AXIO Zoom stereoscope. Embryos, for the knockdown and rescue experiment, were kept at room temperature until stage 40 and then fixed with MEMFA in glass vials for 1 hour. Post fixation samples were washed three times in 1X PBS and images were acquired. The embryo anterior-posterior length was then measured using the line and measurement tools in Fiji and statistically compared using an ANOVA test. Dorsal explants were dissected from late gastrula embryos (~st.12), allowed to heal for 30 minutes, and pre-CE images were acquired. Explants were then incubated for 4 hours at room temperature and post-elongation images were acquired. Explant lengths were measured using Fiji and statistically compared using a Mann-Whitney test. The time-lapse movies of unconstrained explant elongation were performed as described above except images were acquired every 20 minuets during the 4-hour period of elongation. The explant elongation rate curves were statistically compared using a Wilcoxon test.
Immunostaining Keller explants and quantification of endogenous Arvcf and Cdh3 in knockdown cells
Embryos were injected at the 4-cells stage in a single dorsal blastomere with Arvcf morpholino and membrane BFP to generate animals with a mosaic Arvcf knockdown. Keller explants were then dissected from early gastrula embryos (~st.10.25) and mounted on fibronectin coated coverslips. Samples were incubated at room temperature for four hours or overnight at 16°C. Explants were then fixed in 4% paraformaldehyde for 1 hour and washed 3 times in PBS to remove fixative. Next samples were permeabilized with 0.05% Triton X-100 in PBS for 30 minutes and blocked in 1% Normal Goat Serum (NGS) in PBS for 2 hours at room temperature. The primary antibody, for both Arvcf and Cdh3, was diluted 1:100 in 1% NGS/PBS and explants were incubated in primary antibody overnight at 4°C. Samples were then blocked a second time at room temperature for 1 hour and then washed twice with fresh blocking solution. Secondary antibody (goat anti-Mouse 488, #A32723) was diluted 1:500 and samples were incubated at 4°C overnight. Finally, samples were washed three times in 1X PBS and imaged. Images were processed using a 50-pixel rolling ball radius background subtraction. Then the Fiji segmented line tool was used to create a region of interest circling the plasma membrane of wildtype or Arvcf knockdown cells. The protein mean intensity was then measured and samples were statistically compared using a Mann-Whitney test.
Constrained explant elongation assay
We used semi-compliant agarose gel to provide external mechanical constraint to CE. We used 0.3% low-melting-point (LMP) agarose (Promega, Madison, WI) gel freshly made in the Steinberg’s solution and kept at 37 °C to remain liquid. Explants were dissected from late gastrula embryos (~st.12) and allowed to heal and clear debris for 30 min. Explants were then rinsed twice with liquid gel cooled down to RT and transferred to individual culture wells casted with 2% agarose gel and pre-filled with liquid LMP agarose gel. Once explants were positioned and oriented, culture wells were moved to a 13.5 °C incubator for 20 min to solidify the LMP agarose gel, then moved back to RT. Explants were then incubated for 4 hours to allow CE.
In situ hybridization
Whole-mount in situ hybridization was performed as descried previously using a DIG-labeled single-strand RNA probe against a partial sequence of Xnot (Monsoro-Burq, 2007; Sive et al., 2000). This antisense probe has been well characterized, which shows Xnot expression in the prenotochordal region about the dorsal lip at stage 10.5 and along the dorsal midline exclusive to the notochord up to stage 16 (von Dassow et al., 1993). Bright field images were captured with a Zeiss Axio Zoom. V16 stereo microscope with Carl Zeiss Axiocam HRc color microscope camera or a Leica stereo microscope MDG41 with AmScope microscope digital camera WF200.
Tissue scale force measurement and tissue stiffness estimation
We adapted a previously reported method for the biomechanical testing of the explants during CE (Zhou et al., 2015). We embedded explants dissected from late gastrula embryos (~st.12) in 0.6% LMP agarose gel instead. 0.6% LMP agarose gel was the softest one that limited the induced maximum strain below 10% within 3-hour incubation, which enabled a linear viscoelastic finite element model for force estimation. 0.5 μm red fluorescent latex beads (Sigma) were evenly dispersed in the liquid LMP agarose gel as markers for tracking gel deformation. After gel solidified, the fluorescent beads were scanned during CE with a confocal microscope (Nikon A1R or Zeiss LSM 700, 10x objective). Z-stacks were collected over 40 μm near the dorsal-ventral mid-plane every 20 min.
We observed both wildtype and KD explants embedded in 0.6% agarose gel buckled at the end of the 3-hour incubation. A sudden bend of an explant at the AP poles (“buckling”) was also associated with reduced gel deformation and redirected explant elongation. We took the last frame before explant buckling to quantify the maximum explant elongation force. We first created maximum z-projections of the beads image and aligned it to the one from the first time point using ImageJ (NIH) and analyzed bead displacement using PIVlab (Thielicke and Stamhuis, 2014; Zhou et al., 2015). We then applied finite element analysis (FEA) to estimate the explant elongation force using customized MATLAB scripts. We assumed that all forces on the gel were applied at the gel-tissue interface and were compressive. The gel was modeled as an isotropic, linear viscoelastic material with bulk elastic modulus of 30 Pa and Poisson’s ratio of 0.5 (Normand et al., 2000; Zhou et al., 2015). Forces were estimated by iterations that minimized the difference between a simulated displacement field and the measured displacement filed from bead tracking. The Von Mises stresses were then calculated for display. The maximum compressive (2nd principle) stresses and average stress along the AP axis were calculated for statistics.
We next used the maximum elongation force, which was also the critical buckling force to estimate tissue stiffness. We simplified explant as an isotropic, linear viscoelastic column with a rectangular cross section and assumed the reactive tissue extending force applied a uniform load along the AP axis. Euler’s buckling load give the formula Fcr = π2EI/L2, where Fcr is Euler’s critical load, E is the Young’s modulus of the explant, I is the minimum area moment of inertia of the cross section, and L is the length of the explant in the AP axis. We can also express I as I = t3w/12, where w is the width of the explant, and t is the thickness. With the assumption of uniform loading, Fcr = Fst/ts, where ts is the scanned thickness and Fs is the estimated force from FEA. Thus, the tissue stiffness can be expressed as E = 12FsL2/π2wt2ts.
Measurement of cell polarization
Embryos were injected and Keller explants were prepared as described above. We then used the Fiji straight line tool to measure the mediolateral length and anterior-posterior width of individual cells. We then report the cell polarization as the ratio of the mediolateral length over the anterior-posterior width. Samples were statistically compared using a Mann-Whitney test.
Measurement of Cdh3 and actin oscillations
Images were processed with 50-pixel rolling ball radius background subtraction. The Fiji segmented line tool, with width set to the thickness of the junction (~16 pixels), was used to set a line of interest (LOI) across the length of the cell junction. Next the multi-plot tool was used to extract cdh3 intensity values across the length of the cell junction and the measure tool was used to collect data such as junction length and mean intensity values. The Fiji Time Lapse plugin Line Interpolator Tool was used to make successive measurements for movies. Here a segmented line LOI was drawn every 10–30 frames, the line interpolator tool was then used to fill in the LOIs between the manually drawn LOIs allowing rapid semi-manual segmentation. The multi-plot tool and measure tool were then used to extract data for each time-point of the movie.
Transverse fluctuation analysis
Transverse fluctuation was analyzed along the shortening v-junctions in wildtype and Arvcf depleted explants using MATLAB. Briefly, embryos were injected at the 4-cell stage in both dorsal blastomeres with membrane BFP with or without Arvcf morpholino. Keller explants were then dissected from early gastrula embryos, mounted on fibronectin coated coverslips, and incubated at room temperature for four hours. Live-cell imaging and image analysis including cell segmentation and junction detection has been described previously (Weng et al., 2021). Then, we defined the baseline position of a junction at any time point as the moving average over 2 μm along the junction length following by averaging over 20 sec on time. The transverse fluctuation is the transverse deviation from a baseline, which is measured the shortest distance from any point of the junction to the baseline.
Quantification and statistical analysis
Statistical analysis was performed using MATLAB or Prism software. Statistical significance was assessed using either a Mann-Whitney test, Analysis of Variance (ANOVA) test, or a Kolmogorov-Smirnov (KS) test and the type of test used for each analysis is reported in the figure legends. P-values are reported directly in the figures and N values are reported either in the figure or the figure legends.
Supplementary Material
Acknowledgements
We would like to acknowledge Dr. Daniel J. Dickinson for critical reading of this manuscript. Work in the JBW lab was supported by grants from the NICHD (RO1 HD099191; R21 HD103882). E.M.M. additionally support from the Welch Foundation (F1515) and the NIGMS (R35 GM122480).
Footnotes
Declaration of interests:
J.B.W. is a member of the Developmental Cell advisory board.
References
- Aktary Z, Alaee M, Pasdar M, 2017. Beyond cell-cell adhesion: Plakoglobin and the regulation of tumorigenesis and metastasis. Oncotarget 8, 32270–32291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arslan FN, Eckert J, Schmidt T, Heisenberg C-P, 2021. Holding it together: when cadherin meets cadherin. Biophysical Journal. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badel P, Rohan CP, Avril S, 2013. Finite Element simulation of buckling-induced vein tortuosity and influence of the wall constitutive properties. J Mech Behav Biomed Mater 26, 119–126. [DOI] [PubMed] [Google Scholar]
- Barriga EH, Franze K, Charras G, Mayor R, 2018. Tissue stiffening coordinates morphogenesis by triggering collective cell migration in vivo. Nature 554, 523–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cetera M, Leybova L, Joyce B, Devenport D, 2018. Counter-rotational cell flows drive morphological and cell fate asymmetries in mammalian hair follicles. Nature Cell Biology 20, 541–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charras G, Yap AS, 2018. Tensile Forces and Mechanotransduction at Cell–Cell Junctions. Current Biology 28, R445–R457. [DOI] [PubMed] [Google Scholar]
- Cho K, Lee M, Gu D, Munoz WA, Ji H, Kloc M, McCrea PD, 2011. Kazrin, and its binding partners ARVCF- and delta-catenin, are required for Xenopus laevis craniofacial development. Developmental Dynamics 240, 2601–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson LA, 2017. Mechanical design in embryos: mechanical signalling, robustness and developmental defects. Philosophical Transactions of the Royal Society B: Biological Sciences 372, 20150516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingwell KS, Smith JC, 2006. Tes regulates neural crest migration and axial elongation in Xenopus. Developmental Biology 293, 252–267. [DOI] [PubMed] [Google Scholar]
- Drew K, Lee C, Cox RM, Dang V, Devitt CC, McWhite CD, Papoulas O, Huizar RL, Marcotte EM, Wallingford JB, 2020. A systematic, label-free method for identifying RNA-associated proteins in vivo provides insights into vertebrate ciliary beating machinery. Dev Biol 467, 108–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald AJ, Huebner RJ, Palsdottir H, Lee JK, Perez MJ, Jorgens DM, Tauscher AN, Cheung KJ, Werb Z, Auer M, 2012. Mammary collective cell migration involves transient loss of epithelial features and individual cell migration within the epithelium. Journal of Cell Science 125, 2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagotto F, Rohani N, Touret A-S, Li R, 2013. A Molecular Base for Cell Sorting at Embryonic Boundaries: Contact Inhibition of Cadherin Adhesion by Ephrin/Eph-Dependent Contractility. Developmental Cell 27, 72–87. [DOI] [PubMed] [Google Scholar]
- Fang X, Ji H, Kim S-W, Park J-I, Vaught TG, Anastasiadis PZ, Ciesiolka M, McCrea PD, 2004. Vertebrate development requires ARVCF and p120 catenins and their interplay with RhoA and Rac. The Journal of Cell Biology 165, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green JBA, Davidson LA, 2007. Convergent extension and the hexahedral cell. Nature Cell Biology 9, 1010–1015. [DOI] [PubMed] [Google Scholar]
- Han MKL, Hoijman E, Nöel E, Garric L, Bakkers J, de Rooij J, 2016. αE-catenin-dependent mechanotransduction is essential for proper convergent extension in zebrafish. Biology Open 5, 1461–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huebner RJ, Malmi-Kakkada AN, Sarikaya S, Weng S, Thirumalai D, Wallingford JB, 2020. Mechanical heterogeneity along single cell-cell junctions is driven by lateral clustering of cadherins during vertebrate axis elongation. bioRxiv, 2020.2002.2011.944033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huebner RJ, Malmi-Kakkada AN, Sarıkaya S, Weng S, Thirumalai D, Wallingford JB, 2021. Mechanical heterogeneity along single cell-cell junctions is driven by lateral clustering of cadherins during vertebrate axis elongation. eLife 10, e65390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huebner RJ, Wallingford JB, 2018. Coming to Consensus: A Unifying Model Emerges for Convergent Extension. Developmental Cell 46, 389–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J, Forslund K, Coelho LP, Szklarczyk D, Jensen LJ, von Mering C, Bork P, 2017. Fast Genome-Wide Functional Annotation through Orthology Assignment by eggNOG-Mapper. Mol Biol Evol 34, 2115–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huveneers S, Oldenburg J, Spanjaard E, van der Krogt G, Grigoriev I, Akhmanova A, Rehmann H, de Rooij J, 2012. Vinculin associates with endothelial VE-cadherin junctions to control force-dependent remodeling. J Cell Biol 196, 641–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kale GR, Yang X, Philippe J-M, Mani M, Lenne P-F, Lecuit T, 2018. Distinct contributions of tensile and shear stress on E-cadherin levels during morphogenesis. Nature Communications 9, 5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller R, 2002. Shaping the Vertebrate Body Plan by Polarized Embryonic Cell Movements. Science 298, 1950. [DOI] [PubMed] [Google Scholar]
- Keller R, 2012. Physical biology returns to morphogenesis. Science 338, 201–203. [DOI] [PubMed] [Google Scholar]
- Keller R, Sutherland A, 2020. Chapter Ten - Convergent extension in the amphibian, Xenopus laevis, in: Solnica-Krezel L (Ed.), Current Topics in Developmental Biology. Academic Press, pp. 271–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieserman EK, Lee C, Gray RS, Park TJ, Wallingford JB, 2010. High-Magnification In Vivo Imaging of Xenopus Embryos for Cell and Developmental Biology. Cold Spring Harbor Protocols 2010, pdb.prot5427. [DOI] [PubMed] [Google Scholar]
- Kikumoto M, Kurachi M, Tosa V, Tashiro H, 2006. Flexural rigidity of individual microtubules measured by a buckling force with optical traps. Biophys J 90, 1687–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- le Duc Q, Shi Q, Blonk I, Sonnenberg A, Wang N, Leckband D, de Rooij J, 2010. Vinculin potentiates E-cadherin mechanosensing and is recruited to actin-anchored sites within adherens junctions in a myosin II-dependent manner. J Cell Biol 189, 1107–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuit T, Lenne P-F, Munro E, 2011. Force Generation, Transmission, and Integration during Cell and Tissue Morphogenesis. Annual Review of Cell and Developmental Biology 27, 157–184. [DOI] [PubMed] [Google Scholar]
- Lee C, Cox RM, Papoulas O, Horani A, Drew K, Devitt CC, Brody SL, Marcotte EM, Wallingford JB, 2020. Functional partitioning of a liquid-like organelle during assembly of axonemal dyneins. Elife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C-H, Gumbiner BM, 1995. Disruption of Gastrulation Movements in Xenopus by a Dominant-Negative Mutant for C-cadherin. Developmental Biology 171, 363–373. [DOI] [PubMed] [Google Scholar]
- Lu P, Vogel C, Wang R, Yao X, Marcotte EM, 2007. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat Biotechnol 25, 117–124. [DOI] [PubMed] [Google Scholar]
- McCrea PD, Park J. i., 2007. Developmental functions of the P120-catenin sub-family. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 1773, 17–33. [DOI] [PubMed] [Google Scholar]
- McWhite CD, Papoulas O, Drew K, Cox RM, June V, Dong OX, Kwon T, Wan C, Salmi ML, Roux SJ, Browning KS, Chen ZJ, Ronald PC, Marcotte EM, 2020. A Pan-plant Protein Complex Map Reveals Deep Conservation and Novel Assemblies. Cell 181, 460–474.e414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongera A, Michaut A, Guillot C, Xiong F, Pourquié O, 2019. Mechanics of Anteroposterior Axis Formation in Vertebrates. Annual Review of Cell and Developmental Biology 35, 259–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongera A, Rowghanian P, Gustafson HJ, Shelton E, Kealhofer DA, Carn EK, Serwane F, Lucio AA, Giammona J, Campàs O, 2018. A fluid-to-solid jamming transition underlies vertebrate body axis elongation. Nature 561, 401–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsoro-Burq AH, 2007. A rapid protocol for whole-mount in situ hybridization on Xenopus embryos. CSH Protoc 2007, pdb.prot4809. [DOI] [PubMed] [Google Scholar]
- Moore SW, Keller RE, Koehl MA, 1995. The dorsal involuting marginal zone stiffens anisotropically during its convergent extension in the gastrula of Xenopus laevis. Development 121, 3131–3140. [DOI] [PubMed] [Google Scholar]
- Nobes CD, Hall A, 1995. Rho, Rac, and Cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell 81, 53–62. [DOI] [PubMed] [Google Scholar]
- Normand V, Lootens DL, Amici E, Plucknett KP, Aymard P, 2000. New insight into agarose gel mechanical properties. Biomacromolecules 1, 730–738. [DOI] [PubMed] [Google Scholar]
- Oldenburg J, van der Krogt G, Twiss F, Bongaarts A, Habani Y, Slotman JA, Houtsmuller A, Huveneers S, de Rooij J, 2015. VASP, zyxin and TES are tension-dependent members of Focal Adherens Junctions independent of the α-catenin-vinculin module. Scientific Reports 5, 17225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson AF, Mooney E, Fang X, Ji H, McCrea PD, 2000. Xarvcf, Xenopus member of the p120 catenin subfamily associating with cadherin juxtamembrane region. J Biol Chem 275, 30124–30131. [DOI] [PubMed] [Google Scholar]
- Pfister K, Shook DR, Chang C, Keller R, Skoglund P, 2016. Molecular model for force production and transmission during vertebrate gastrulation. Development 143, 715–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radice GL, Ferreira-Cornwell MC, Robinson SD, Rayburn H, Chodosh LA, Takeichi M, Hynes RO, 1997. Precocious Mammary Gland Development in P-Cadherin–deficient Mice. Journal of Cell Biology 139, 1025–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reintsch WE, Mandato CA, McCrea PD, Fagotto F, 2008. Inhibition of cell adhesion by xARVCF indicates a regulatory function at the plasma membrane. Developmental Dynamics 237, 2328–2341. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Boulan E, Macara IG, 2014. Organization and execution of the epithelial polarity programme. Nature Reviews Molecular Cell Biology 15, 225–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A, 2012. Fiji: an open-source platform for biological-image analysis. Nature Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Session AM, Uno Y, Kwon T, Chapman JA, Toyoda A, Takahashi S, Fukui A, Hikosaka A, Suzuki A, Kondo M, van Heeringen SJ, Quigley I, Heinz S, Ogino H, Ochi H, Hellsten U, Lyons JB, Simakov O, Putnam N, Stites J, Kuroki Y, Tanaka T, Michiue T, Watanabe M, Bogdanovic O, Lister R, Georgiou G, Paranjpe SS, van Kruijsbergen I, Shu S, Carlson J, Kinoshita T, Ohta Y, Mawaribuchi S, Jenkins J, Grimwood J, Schmutz J, Mitros T, Mozaffari SV, Suzuki Y, Haramoto Y, Yamamoto TS, Takagi C, Heald R, Miller K, Haudenschild C, Kitzman J, Nakayama T, Izutsu Y, Robert J, Fortriede J, Burns K, Lotay V, Karimi K, Yasuoka Y, Dichmann DS, Flajnik MF, Houston DW, Shendure J, DuPasquier L, Vize PD, Zorn AM, Ito M, Marcotte EM, Wallingford JB, Ito Y, Asashima M, Ueno N, Matsuda Y, Veenstra GJC, Fujiyama A, Harland RM, Taira M, Rokhsar DS, 2016. Genome evolution in the allotetraploid frog Xenopus laevis. Nature 538, 336–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih J, Keller R, 1992. Patterns of cell motility in the organizer and dorsal mesoderm of Xenopus laevis. Development 116, 915. [DOI] [PubMed] [Google Scholar]
- Shindo A, Inoue Y, Kinoshita M, Wallingford JB, 2019. PCP-dependent transcellular regulation of actomyosin oscillation facilitates convergent extension of vertebrate tissue. Developmental Biology 446, 159–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo A, Wallingford JB, 2014. PCP and septins compartmentalize cortical actomyosin to direct collective cell movement. Science 343, 649–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shook DR, Kasprowicz EM, Davidson LA, Keller R, 2018. Large, long range tensile forces drive convergence during Xenopus blastopore closure and body axis elongation. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sive HL, Grainger RM, Harland RM, 2000. Early development of Xenopus laevis : a laboratory manual. [Google Scholar]
- Sive HL, Grainger RM, Harland RM, 2007. Xenopus laevis Keller Explants. Cold Spring Harbor Protocols 2007, pdb.prot4749. [DOI] [PubMed] [Google Scholar]
- Sun Z, Amourda C, Shagirov M, Hara Y, Saunders TE, Toyama Y, 2017. Basolateral protrusion and apical contraction cooperatively drive Drosophila germ-band extension. Nat Cell Biol 19, 375–383. [DOI] [PubMed] [Google Scholar]
- Tada M, Heisenberg CP, 2012. Convergent extension: using collective cell migration and cell intercalation to shape embryos. Development 139, 3897–3904. [DOI] [PubMed] [Google Scholar]
- Tahinci E, Symes K, 2003. Distinct functions of Rho and Rac are required for convergent extension during Xenopus gastrulation. Developmental Biology 259, 318–335. [DOI] [PubMed] [Google Scholar]
- Tao H, Zhu M, Lau K, Whitley OKW, Samani M, Xiao X, Chen XX, Hahn NA, Liu W, Valencia M, Wu M, Wang X, Fenelon KD, Pasiliao CC, Hu D, Wu J, Spring S, Ferguson J, Karuna EP, Henkelman RM, Dunn A, Huang H, Ho H-YH, Atit R, Goyal S, Sun Y, Hopyan S, 2019. Oscillatory cortical forces promote three dimensional cell intercalations that shape the murine mandibular arch. Nature Communications 10, 1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theveneau E, Mayor R, 2012. Cadherins in collective cell migration of mesenchymal cells. Current Opinion in Cell Biology 24, 677–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thielicke W, Stamhuis EJ, 2014. PIVlab – Towards User-friendly, Affordable and Accurate Digital Particle Image Velocimetry in MATLAB. Journal of Open Research Software 2. [Google Scholar]
- Thompson AJ, Pillai EK, Dimov IB, Foster SK, Holt CE, Franze K, 2019. Rapid changes in tissue mechanics regulate cell behaviour in the developing embryonic brain. eLife 8, e39356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trushko A, Di Meglio I, Merzouki A, Blanch-Mercader C, Abuhattum S, Guck J, Alessandri K, Nassoy P, Kruse K, Chopard B, Roux A, 2020. Buckling of an Epithelium Growing under Spherical Confinement. Developmental Cell 54, 655–668.e656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Dassow G, Schmidt JE, Kimelman D, 1993. Induction of the Xenopus organizer: expression and regulation of Xnot, a novel FGF and activin-regulated homeo box gene. Genes & Development 7, 355–366. [DOI] [PubMed] [Google Scholar]
- von Dassow M, Davidson LA, 2009. Natural variation in embryo mechanics: gastrulation in Xenopus laevis is highly robust to variation in tissue stiffness. Developmental Dynamics 238, 2–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walck-Shannon E, Hardin J, 2014. Cell Intercalation from top to bottom. Nature reviews. Molecular cell biology 15, 34–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallingford JB, Niswander LA, Shaw GM, Finnell RH, 2013. The Continuing Challenge of Understanding, Preventing, and Treating Neural Tube Defects. Science 339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallingford JB, Rowning BA, Vogeli KM, Rothbacher U, Fraser SE, Harland RM, 2000. Dishevelled controls cell polarity during Xenopus gastrulation. Nature 405, 81–85. [DOI] [PubMed] [Google Scholar]
- Weng S, Huebner RJ, Wallingford JB, 2021. Convergent extension requires adhesion-dependent biomechanical integration of cell crawling and junction contraction. Cell Reports, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams M, Yen W, Lu X, Sutherland A, 2014. Distinct apical and basolateral mechanisms drive PCP-dependent convergent extension of the mouse neural plate. Developmental cell 29, 34–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong F, Ma W, Bénazéraf B, Mahadevan L, Pourquié O, 2018. Mechanical Coupling Coordinates the Co-elongation of Axial and Paraxial Tissues in Avian Embryos. bioRxiv, 412866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Kim HY, Davidson LA, 2009. Actomyosin stiffens the vertebrate embryo during crucial stages of elongation and neural tube closure. Development 136, 677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Pal S, Maiti S, Davidson LA, 2015. Force production and mechanical accommodation during convergent extension. Development (Cambridge, England) 142, 692–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Proteomics data has been deposited into MassIVE which in turn was passed to ProteomeXchange. The MassIVE accession # is MSV000087312. The ProteomeXchange # is PXD025665 (http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD025665-1).
No new code was generated during this study.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


