Abstract
Major histocompatibility complex (MHC)-associated peptides generated and displayed by antigen-presenting cells in the thymus are essential for the generation of functional and self-tolerant T cells that protect our body from various pathogens. The peptides displayed by cortical thymic epithelial cells (cTECs) are generated by unique enzymatic machineries including the thymoproteasomes, and are involved in the positive selection of self-protective T cells. On the other hand, the peptides displayed by medullary thymic epithelial cells (mTECs) and thymic dendritic cells (DCs) are involved in further selection to establish self-tolerance in T cells. Although the biochemical nature of the peptide repertoire displayed in the thymus remains unclear, many studies have suggested a thymus-specific mechanism for the generation of MHC-associated peptides in the thymus. In this review, we summarize basic knowledge and recent advances in MHC-associated thymic peptides, focusing on the generation and function of thymoproteasome-dependent peptides specifically displayed by cTECs.
Keywords: thymus, T cell, thymic cortex, thymic medulla, thymic selection, thymoproteasome
1. Introduction
The thymus is a primary lymphoid organ that generates self-protective yet self-tolerant T cells. Peptides generated in the thymus and presented by thymic antigen-presenting cells are important for the selection of functionally competent T cells that express the αβ form of the T cell receptor (TCR), during T cell development in the thymus. The developmental stage of T cells in the thymus is identified by the expression of co-receptors CD4 and CD8. Hematopoietic stem cell-derived T progenitor cells that migrate to the thymus express neither CD4 nor CD8, and are thereby termed double negative (DN) thymocytes. The DN thymocytes differentiate into CD4+CD8+ (double positive, DP) thymocytes, and then to either CD4+CD8− (CD4 single positive, CD4SP) or CD4−CD8+ (CD8 single positive, CD8SP) thymocytes. During the development from DN to DP thymocytes, a large variety of antigen-recognition specificities of the TCRs are formed by the V(D)J rearrangement of TCRα and TCRβ genes. The rearrangement of TCRβ gene and the successful expression of in-frame TCRβ protein as a component of the pre-TCR complex in DN thymocytes trigger the rearrangement of TCRα gene and the generation of a large pool of DP thymocytes expressing individual specificities of αβTCRs on their cell surfaces [1, 2]. The antigen-recognition specificities of the αβTCRs expressed by the newly generated DP thymocytes are solely generated within the nucleus and thus inevitably include functionally useless and pathologically self-reactive specificities, in addition to functionally useful and self-protective specificities. In this regard, developing thymocytes need to undergo the process of TCR repertoire selection to generate a functionally competent and useful repertoire of TCR specificities in mature T cells exported from the thymus to the circulation. The thymic cortex is a microenvironment where DP thymocytes are newly generated and positively selected for T cells that are capable of recognizing self-peptides bound to major histocompatibility complex (MHC) molecules. The newly generated DP thymocytes expressing TCRs that interact at a low affinity with self-peptide-MHC complexes presented by cortical thymic epithelial cells (cTECs) receive the signals for survival and differentiation into SP thymocytes as well as for migration to the thymic medulla in a CCL21-CCR7 chemokine-dependent manner [3–5]. The thymic medulla is a microenvironment where medullary thymic epithelial cells (mTECs) express a diverse set of genes including tissue-restricted self-antigens, which is responsible for the establishment of self-tolerance in T cells [6]. Among positively selected thymocytes, those expressing self-reactive TCRs that interact at a high affinity with self-peptide-MHC complexes presented by mTECs and/or dendritic cells (DCs) in the thymic medulla undergo negative selection, eliminating self-reactive T cells by apoptotic cell death [3, 7, 8]. A fraction of self-reactive thymocytes escape the apoptosis and differentiate into regulatory T cells, which are a subpopulation of CD4+ T cells that effectively suppress immune response [9, 10]. Through these processes of thymic selection, mature T cells form a pool of immunocompetent and self-tolerant TCR repertoire.
The TCR engagement with self-peptides associated with either MHC class I (MHC-I) or MHC class II (MHC-II) molecules induces the differentiation of DP thymocytes into CD8SP or CD4SP thymocytes, respectively. The binding of peptides to either MHC-I or MHC-II molecules is largely dependent on the intracellular location of the peptides. Peptides generated in the cytoplasm and the nuclei are produced by the proteasomal digestion of ubiquitinated proteins, transported to the endoplasmic reticulum (ER) through TAP transporter, and loaded onto MHC-I molecules, whereas proteins in endosomal vesicles are degraded by lysosomal endopeptidases and loaded onto MHC-II molecules [11, 12]. The proteolytic enzymes that contribute to the generation of these MHC-associated peptides are different between the thymic cortex and the thymic medulla. In this review, we summarize basic knowledge and recent advances in the generation and function of MHC-associated thymic peptides, focusing on the thymoproteasome-dependent self-peptides specifically displayed by cTECs that induce positive selection of CD8+ T cells.
2. Interaction of peptides with MHC molecules
The interaction of peptides with MHC molecules stabilizes peptide-MHC complexes that are transported to the cell surface to present the peptide antigens to TCRs expressed by T cells [13, 14]. The peptides are an essential part of the cell-surface MHC molecules to fulfill the function of peptide-MHC complexes in the antigen presentation to T cells.
The MHC-I molecule is a heterodimer composed of MHC-I-encoded heavy chain and β2-microgrobulin. The heavy chain consists of three extracellular domains (α1, α2, and α3), a transmembrane domain, and an intracellular domain. The α1 and α2 domains of the heavy chain form a highly polymorphic peptide-binding groove with six pockets (pockets A to F) that accommodate binding to the peptides [15, 16]. The peptides that bind to MHC-I molecule are typically restricted to eight to ten amino acids in length due to the closed structure of the peptide-binding groove at both ends. The peptides have anchor residues that directly interact with the pockets in the peptide-binding groove of MHC-I molecule. Generally, the anchoring at the second and/or the fifth amino acid residues from the N-terminus in addition to the C-terminal amino acid residue of the peptides is important for the binding of the peptides to the peptide-binding groove of MHC-I heavy chain [17–19]. Other amino acid residues of the peptides primarily contribute to the interaction with complementarity-determining regions 3 (CDR3s) of TCRα and TCRβ chains, which are the most variable regions in the TCR molecules [20].
Similar to MHC-I molecule, MHC-II molecule is a heterodimer composed of α and β chains. Each chain has two extracellular domains (α1 and α2 domains in the α chain, and β1 and β2 domains in the β chain) followed by a transmembrane domain and an intracellular domain. The α1 and β1 domains form a highly polymorphic peptide-binding groove. In contrast to the peptide-binding groove in MHC-I molecule, the peptide-binding groove in MHC-II molecule has an open structure at either end so that MHC-II-associated peptides can protrude from the groove, allowing MHC-II molecule to bind to peptides typically ranging from 13 to 25 amino acid residues in length, much longer than the MHC-I-binding peptides whose length is typically restricted to eight to ten residues [21]. The peptides interact with the peptide-binding groove of MHC-II molecule by anchoring at the first, fourth, sixth, and ninth amino acid residues from the N-terminus of the peptides [22, 23]. Other amino acid residues, typically the second, fifth, and eighth residues from the N-terminus, project out from the peptide-binding groove and participate in the interaction with the CDR3s of TCRs [24, 25].
In both MHC-I and MHC-II molecules, the highly polymorphic structure of the peptide-binding groove affects the spectrum of MHC-associated peptides among different individuals with different polymorphisms within the same species.
3. Self-peptides bind to MHC class I in the thymus
Self-peptides that are generated through the processing of cytoplasmic and nucleic proteins by the proteasomes are delivered by TAP transporter to the ER lumen. ER aminopeptidases trim the N-terminal amino acid residues of the peptides. The successful formation of peptide-MHC-I complexes through the binding of the peptides to the peptide-binding groove of MHC-I molecules results in the transport of the peptide-MHC-I complexes to the cell surface through the Golgi apparatus. In the thymus, self-peptides bound to MHC-I molecules displayed by cTECs are primarily generated by thymoproteasomes, whereas those displayed by other cells, including mTECs and DCs, are chiefly generated by immunoproteasomes and constitutive proteasomes.
3.1. Thymoproteasome-dependent generation of MHC class I-associated peptides
The thymoproteasome is a form of proteasome whose expression is detected only in cTECs but not in any other cells in the body [26, 27]. The 20S core particle of proteasome responsible for the proteolytic activity is composed of 28 subunits, which are two sets of seven different α subunits (α1– α7) and seven different β subunits (β1– β7). Among these subunits, β1, β2, and β5 subunits are responsible for catalytic activities carrying caspase-like, trypsin-like, and chymotrypsin-like specificities, respectively [28, 29]. In contrast to constitutively expressed proteasomes containing β1, β2, and β5 catalytic subunits, thymoproteasomes expressed by cTECs contain β1i, β2i, and β5t catalytic subunits (Table 1). The β1i and β2i subunits are also assembled with the β5i subunit to shape another form of proteasome, the immunoproteasome, which is expressed in other antigen-presenting cells, including mTECs and DCs in the thymus and antigen-presenting cells exposed to inflammatory cytokines during immune response. On the other hand, β5t is a cTEC-specific subunit and is responsible for cTEC-specific expression of the thymoproteasome [26, 27]. Like many other tissue-specific genes, β5t expression is also detectable in a small (1–5%) population of mTECs [30, 31]. This expression of β5t in mTECs is enriched in MHC-IIhigh mTEC subpopulation and is dependent on Aire [30–32], suggesting that mTEC expression of β5t reflects the promiscuous gene expression of self-genomic components in mTECs, a key process to establish self-tolerance in newly generated T cells in the thymus.
Table 1.
Enzymes differently expressed between cTECs and mTECs in proteomic profiling (P < 0.05)
| Abundant in cTECs than mTECs | Abundant in mTECs than cTECs |
|---|---|
| Lysosomal/endosomal peptidases | Lysosomal/endosomal peptidases |
| Cathepsin B (Ctsb) | Cathepsin A (Ctsa) |
| Cathepsin D (Ctsd) | Cathepsin C (Ctsc) |
| Cathepsin L (Ctsl) | Cathepsin H (Ctsh) |
| TSSP (Prss16) | Cathepsin S (Ctss) |
| Cathepsin Z (Ctsz) | |
| Intracellular peptidases | |
| Calpain 1 (Capn1) | |
| Calpain 2 (Capn2) | |
| Proteasomal catalytic subunits | Proteasomal catalytic subunits |
| β1i (Psmb9) | β1 (Psmb1) |
| β2i (Psmb10) | β2 (Psmb2) |
| β5t (Psmb11) | β5 (Psmb5) |
| β5i (Psmb8) |
The amino acids that compose the S1 pocket, which mainly determine the substrate specificity of proteasome catalytic subunits, are enriched with hydrophilic residues in the β5t subunit, in contrast to the amino acids in the S1 pocket of the β5 and β5i subunits, which are enriched with hydrophobic residues [26, 33]. Indeed, proteasomal chymotrypsin-like activity, which coincides with the hydrophobicity of the S1 pocket amino acids, and which contributes to the generation of hydrophobic C-terminal residues in the produced peptides, is reduced in β5t-containing thymoproteasomes in comparison with the other forms of proteasomes containing β5 or β5i [26]. Consequently, peptides generated by the thymoproteasome are distinct from those generated by the immunoproteasome [34]. A recent study further showed that human thymoproteasomes and immunoproteasomes differ in their catalytic activity with regard to not only substrate specificity but also peptide transport kinetics [35].
Peptides generated by proteasomes bind to MHC-I molecules via anchor residues including the C-terminal anchor enriched with hydrophobic residues [36]. A mass spectrometry analysis of the amino acid sequences of MHC-I-associated peptides in mouse embryonic fibroblasts (MEFs) that predominantly express thymoproteasomes showed that the C-terminal residues are still enriched with hydrophobic amino acids in the peptides eluted from H-2Db- and H-2Kb-peptide complexes, similar to the MHC-I-bound peptides in immunoproteasome-expressing MEFs [34]. In addition, the central anchor residues are enriched with asparagine in H-2Db-associated peptides and with phenylalanine and tyrosine in H-2Kb-associated peptides in either thymoproteasome-expressing or immunoproteasome-expressing MEFs [34]. On the contrary, amino acid residues at the 6th position from the N-terminus of the H-2Db- and H-2Kb-associated peptides are enriched with proline in thymoproteasome-expressing MEFs rather than immunoproteasome-expressing MEFs [34]. Furthermore, amino acid residues at the 7th position from the N-terminus of the H-2Db- and H-2Kb-associated peptides appear to be enriched with acidic residues in immunoproteasome-expressing MEFs [34]. These 6th and 7th amino acid residues of MHC-I-associated peptides often contribute to the interaction with TCRs, so that the distinct preference in amino acid usage at these positions may be related to the biological function of thymoproteasome-expressing cTECs in T cell development and selection.
3.2. Thymoproteasomes optimize positive selection of CD8+ T cells in the thymus
Thymoproteasomes are essential for the optimal generation of CD8+ T cells. The generation of CD8SP thymocytes and peripheral CD8+ T cells is impaired in mice deficient in β5t, encoded by Psmb11 [26, 37]. The antigen responsiveness of CD8+ T cells generated in β5t-deficient mice is also impaired [38]. The impairment in CD8+ T cell development caused by the loss of β5t in the thymus varies among different TCR transgenic mice [37–39], whereas β5t deficiency in the mouse has a highly specific impact on proteasomal components, rather than a pervasive alteration in transcriptome or proteome, in cTECs [40, 41], suggesting the possibility that the thymoproteasome optimizes CD8+ T cell development through the control of TCR repertoire selection via thymoproteasome-dependent MHC-I-associated peptides. Indeed, our recent deep-sequencing analysis detected a difference in TCRα and TCRβ variable region sequences, and thereby an alteration in the TCR repertoire, in CD8+ T cells between control and β5t-deficient mice [42].
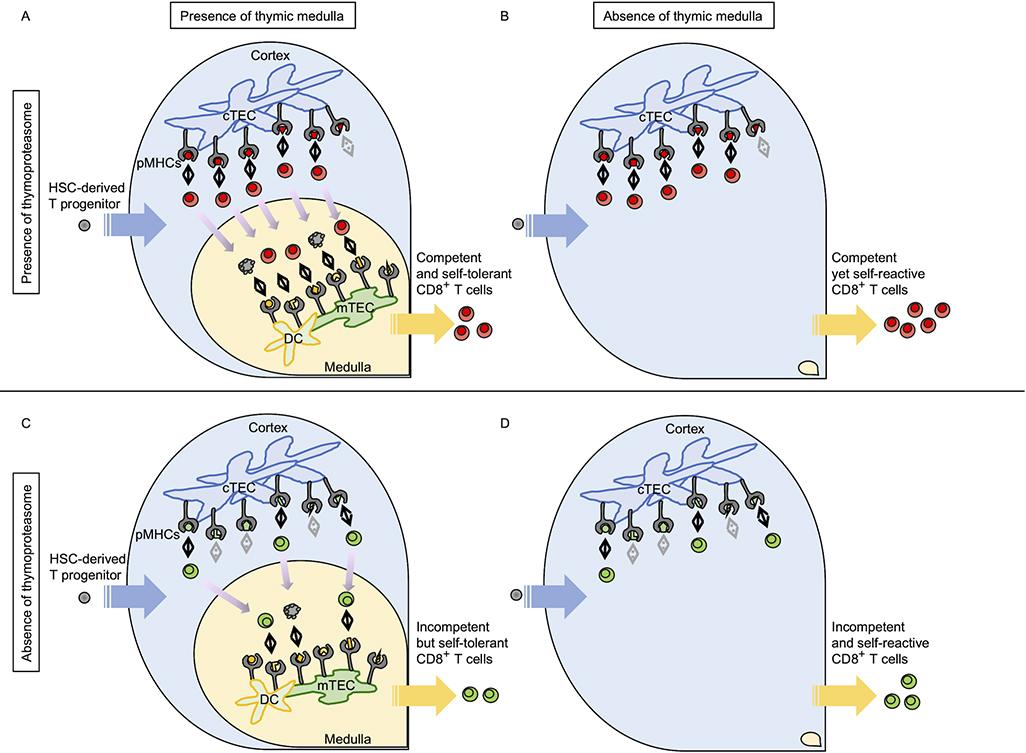
In β5t-deficient mice, β5 and β5i are compensatively expressed in cTECs so that cTECs maintain proteasome-dependent proteolysis and an undiminished cell number [37, 40]. Interestingly, we recently found that the impaired generation of CD8+ T cells caused by β5t deficiency is equivalently detected in the presence and the absence of the thymic medulla [42]. It was also found that β5t deficiency affects CD8+ T cell development in the thymus where MHC-I molecules are expressed only in cTECs but not in other antigen-presenting cells, including mTECs and DCs [42]. These results indicate that the thymoproteasome optimizes the generation of CD8+ T cells in the thymic cortex, independent of the thymic medulla or any additional antigen-presenting cells other than cTECs (Figure 1). Furthermore, we found that β5t deficiency impairs the development of CD69+CCR7− cortical DP thymocytes in the absence of MHC-II molecules, even in the presence of the transgenic overexpression of anti-apoptotic molecule Bcl2 [42], indicating that the thymoproteasome optimizes the positive selection of CD8+ T cells in the thymic cortex independent of thymocyte negative selection (Figure 1). Whether and how thymoproteasome-dependent MHC-I-associated peptides expressed by cTECs actually contribute to the positive selection of CD8+ T cells have yet to be determined. A recent study of RNA sequencing analysis reported that functions of β5t extended beyond its role in self-peptide production [43], although their results have been under controversy [40, 41, 44].
Figure 1.
Thymoproteasomes optimize the generation of CD8+ T cells in the thymic cortex, independent of antigen presentation by cells other than cTECs.
Hematopoietic stem cell (HSC)-derived T progenitor cells (in gray) migrate to the thymus, proliferate, and differentiate into DP thymocytes. DP thymocytes undergo positive selection in the thymic cortex. (A, B) Thymoproteasomes in cTECs produce a unique set of MHC-I-associated peptides (in red) that efficiently induce positive selection of CD8SP thymocytes (in red). (A) Positively selected cortical thymocytes migrate into the medulla to be screened by mTECs and DCs for negative selection to establish self-tolerance, so that functionally competent and self-tolerant CD8+ T cells are exported from the thymus to the circulation. (B) In the absence of the thymic medulla, thymoproteasome-mediated positively selected thymocytes are exported from the thymus without medullary establishment of self-tolerance. (C, D) In β5t-deficient thymus, cTECs produce thymoproteasome-independent MHC-I-associated peptides (in green). CD8SP thymocytes (in green) positively selected without β5t are reduced in number and suboptimal in TCR responsiveness, and are therefore functionally incompetent, regardless of the presence (C) or absence (D) of the medulla. (C) Positively selected thymocytes are still screened in the medulla to establish self-tolerance. CD8+ T cells exported from the thymus are self-tolerant but impaired in antigen responsiveness. (D) Abnormally positively selected and therefore functionally incompetent T cells are exported from the thymus without medullary establishment of self-tolerance.
3.3. Peptide switch hypothesis and proteasome switch
It was hypothesized in the 1980s that MHC-associated peptides presented by cTECs are different from those presented by other antigen-presenting cells so that positively selected thymocytes in the thymic cortex can escape from cell death induced by negative selection [45, 46]. This hypothesis was re-advocated recently as the “peptide switch” hypothesis, in which the difference in MHC-associated peptides for positive selection and negative selection is important to form the TCR repertoire.
This hypothesis was indirectly examined for CD8+ T cells by the analyses of β1i/β2i/β5i/β5t-deficient (4KO) mice and β5i-deficient β5t-transgenic (β5i-KO β5t-Tg) mice. All cells in the thymus of the 4KO mice are expected to be solely the constitutive form of the proteasomes due to the lack of both immunoproteasomes and thymoproteasomes [47], whereas all cells in the thymus of the β5i-KO β5t-Tg mice are expected to express thymoproteasome-specific subunit β5t while lacking immunoproteasome-specific subunit β5i [48]. In both of these mutant mice, the number of CD8SP thymocytes was markedly reduced, although the number of cortical thymocytes positively selected to the CD8+ lineage was not reduced, and negative selection of their CD8+ lineage thymocytes was enhanced [47, 48], suggesting the contribution of the “proteasome switch” to the successful generation of CD8+ T cells in the thymus.
However, as described in section 3.2, our analysis of β5t-deficient mice showed that thymoproteasomes optimize the cortical positive selection of CD8+ T cells independent of thymocyte negative selection [42]. An independent study analyzed mice in which the β5i-encoding sequence was knocked into β5t locus and endogenous β5i was deficient (β5tβ5i β5i-KO) [39]. In these mice, cTECs are expected to express immunoproteasomes, and other antigen-presenting cells in the thymus are expected to express constitutive proteasomes. It was shown that in these mice, the loss of β5t still impaired positive selection of CD8+ lineage thymocytes without the enhancement of thymocyte negative selection, even in the presence of the “proteasome switch” [39]. Thus, the contribution of β5t and thymoproteasomes to CD8+ T cell development is independent of the “proteasome switch” [49].
Nevertheless, it should be emphasized that it remains unclear whether or not the switch in MHC-associated peptides actually contributes to T cell development. To our knowledge, no studies have directly clarified the structure and function of thymoproteasome-dependent MHC-I-associated peptides in CD8+ T cell development. It is also unclear whether the “proteasome switch” reflects a “peptide switch”, and whether the loss of the “proteasome switch” results in the loss of the “peptide switch”. Transcriptomes and proteomes in cTECs and mTECs are highly different [40], so that MHC-associated peptides involved in positive and negative selection in the cortex and the medulla may be substantially distinct even in the absence of any “proteasome switch”.
3.4. Role of immunoproteasomes in the generation of CD8+ T cells
In contrast to positive selection-inducing cTECs that express thymoproteasomes, many other thymic antigen-presenting cells, including mTECs and DCs, express immunoproteasomes and constitutive proteasomes [40, 50] (Table 1). Although the generation of CD8+ T cells is not reduced in mice deficient in immunoproteasome-specific component β5i [51], the development of CD8+ T cells that express either transgenic TCR specific for f-actin capping protein CPα192–99 or TCR specific for viral glycoprotein GP118–125 is impaired in β5i-deficient mice, probably due to the alteration in the thymic repertoire selection [52, 53]. In addition, mice that lack all of immunoproteasome catalytic subunits β1i, β2i, and β5i exhibit the reductions in the surface expression of MHC-I complexes and in the number of CD8+ T cells [54]. It is also shown that in mice deficient in β1i, β2i, and β5i, processing-dependent antigen presentation by DCs is impaired in inducing in vitro T cell response [54]. Thus, the immunoproteasome and its components contribute to the generation of MHC-I-associated peptides and the development and selection of CD8+ T cells. It is important to note that β1i and β2i are catalytic components of the thymoproteasome, so that the defective CD8+ T cell development in mice deficient in β1i, and/or β2i may be due in part to the defects in the thymoproteasome.
4. Self-peptides bind to MHC class II in the thymus
Self-peptides that are associated with MHC-II molecules are generated in the endosomal and lysosomal vesicles. The peptide-binding groove of MHC-II molecules newly generated in the ER is associated with invariant chain (Ii) [55]. The Ii-associated MHC-II molecules are transported from the ER through the Golgi apparatus to the endosomes. In the late endosomes, Ii undergoes proteolytic degradation to leave a short fragment termed CLIP (class II associated Ii chain peptide) in the peptide-binding groove of MHC-II molecules. However, MHC-II-like molecule H-2M in mouse (or HLA-DM in human) facilitates the replacement of CLIP with peptides available in the endosomes, and the resultant peptide-MHC-II complexes are transported to the cell surface for antigen presentation to T cells [55]. Autophagy and endosomal lysosomal endopeptidases, including cathepsins and thymus-specific serine protease (TSSP), participate in the generation of MHC-II-associated self-peptides in the thymus.
4.1. Autophagy
In DCs, peptides bound to MHC-II molecules are often derived from exogenous proteins captured by endocytosis. Compared with DCs, TECs are less active in endocytosis to present exogenous peptides [56]. However, TECs are highly active in autophagy, which delivers cytosolic molecules into the endosomal and lysosomal vesicles and provides cytoplasm-derived peptides to the MHC-II antigen presentation system [57–59]. Unlike other cells, the high activity in autophagy in TECs is constitutive, independent of starvation, and detected in both cTECs and mTECs [58, 59]. The development of CD4+ T cells that express MHC-II-restricted transgenic TCRs is impaired in the thymus deficient in Atg5, an essential component of the autophagosome, suggesting the role of autophagy in T cell positive selection by cTECs [59]. Additionally, the transplantation of Atg5-deficient thymus into athymic mice resulted in T cell-mediated autoimmune inflammation in the host mice, suggesting that autophagy is also involved in the mTEC-mediated self-tolerance in T cells [59]. A reduction in autophagy in TECs was also detected by knocking down autoimmunity-associated gene CLEC16A [60]. Thus, autophagy contributes to T cell selection in the thymus, possibly via the provision of MHC-II-associated self-peptides in TECs.
4.2. Cathepsins
Cathepsin L (Ctsl) is a lysosomal endopeptidase abundantly detected in cTECs, whereas cathepsin S (Ctss) is preferably expressed in other antigen-presenting cells including mTECs (Table 1). Ctsl degrades Ii chain that is associated with the peptide-binding groove of MHC-II molecules, and Ctsl-deficient mice are defective in the generation of CD4SP thymocytes and CD4+ T cells, which is partly due to the inefficient Ii chain degradation [61]. It has been additionally shown that Ctsl regulates CD4+ T cell selection independently of its effect on Ii chain, suggesting the possibility that Ctsl plays a role in the generation of positively selecting MHC-II-associated peptide ligands [62].
Like Ctsl, Ctss is involved in Ii chain degradation [63]. Unlike Ctsl, Ctss is more abundant in mTECs than cTECs (Table 1). However, the role of Ctss in the thymus is unclear because normal T cell development is reported in Ctss-deficient mice [63, 64].
In addition to Ctsl and Ctss, we have detected the abundant expression of other cathepsin family members in TECs in the proteomic profiling of cTECs and mTECs in mouse [40]. Namely, cathepsins B (Ctsb) and D (Ctsd), which are involved in exogenous antigen processing [65], are abundant in cTECs compared with mTECs [40] (Table 1). Interestingly, Ctsb and Ctsd are detected in abundance in human thymic epithelial tumors [66]. On the other hand, cathepsins A, C, H, and Z are more abundant in mTECs than cTECs [40] (Table 1). It is interesting to find out whether and how these cathepsins play a role in the thymus, including the generation of self-peptides that induce T cell repertoire selection.
4.3. Thymus-specific serine protease
TSSP, encoded by Prss16, is an endosomal peptidase that is more abundant in cTECs than mTECs (Table 1). Prss16 transcript is detectable in other cells including thymic DCs [67]. In contrast to Ctsl deficiency, the generation of CD4+ T cells is not globally affected in TSSP-deficient mice [68]. However, studies using MHC-II-restricted TCR transgenic T cells have shown that the generation of CD4+ T cells specific for certain antigens, including Dby male antigen, OVA323–339, and islet amyloid polypeptide, is impaired in the thymus of TSSP-deficient mice, suggesting that TSSP in cTECs affects the generation of self-peptides that induce positive selection of those CD4+ T cells [68, 69]. The role of TSSP in the thymic establishment of self-tolerance in T cells has also been suggested [67, 70].
4.4. Calpains
Calpains are cytoplasmic Ca2+-dependent and non-lysosomal proteases consisting of 17 isoforms. We have shown in our proteomic profiling of cTECs and mTECs that calpain 1 and calpain 2 are more abundant in cTECs than mTECs [40] (Table 1). It has been speculated that endogenous peptides generated by cytoplasmic protease, including proteasomes, may leak from the MHC-I antigen presentation pathway into the MHC-II presentation pathway [3, 71]. Calpain 1 is another candidate cytoplasmic protease that is possibly involved in the generation of endogenous peptides for the MHC-II pathway. One study has shown that the inhibition of calpain 1 reduces MHC-II-associated presentation of a peptide derived from cytoplasmic glutamate decarboxylase (GAD), but not derived exogenously from endocytosed GAD, to MHC-II-restricted GAD-specific T cells [72]. Cytoplasmic peptides processed by cytoplasmic proteases, including calpains, may be further trimmed by endosomal peptidases for MHC-II-associated presentation, because the inhibition of Ctsb reduces the MHC-II-dependent presentation of cytoplasmic GAD [72]. These results suggest the role of calpain 1 in the processing of MHC-II-associated endogenous peptides. So far, the role of calpains in the thymus is unknown.
5. Future perspective
Despite the importance of MHC-associated thymic peptides for the generation of immunocompetent and self-tolerant T cells, the identities of positively selecting peptides expressed by cTECs and negatively selecting peptides expressed by mTECs remain to be determined. Peptides bound to MHC molecules in whole human thymus have been identified [73, 74]. Furthermore, neuroendocrine-related peptides, including neurotensin, have been detectable in cultured human TEC lines in an MHC-I associated manner [75, 76]. However, MHC-associated peptides displayed by individual thymic antigen-presenting cells, including freshly prepared cTECs and mTECs, have never been identified in mouse or human. As cTECs and mTECs have their unique spectrum in the expression of peptide-producing enzymes, such as thymoproteasomes, cathepsins, and TSSP, it is speculated that MHC-associated peptides expressed by cTECs and mTECs carry unique characteristics for inducing positive selection and negative selection, respectively, of developing thymocytes. Thus, it is important to identify MHC-associated peptides expressed by cTECs and mTECs to further improve our understanding of the mechanism for TCR repertoire formation in the thymus.
The identification of amino acid sequences of MHC-associated peptides is most frequently performed by the immunoprecipitation of MHC molecules, the elution of associated peptides, and the mass spectrometry analysis of the peptides. However, current mass spectrometry-based analysis of MHC-associated peptides relies on a large number of cells, typically 108 cells or more, per analysis [77], which is quite a high hurdle for the analysis with TECs, because only up to 104 cTECs and up to 105 mTECs can be isolated from one normal mouse by the enzymatic digestion of the thymus [78]. Even though cTECs and mTECs of order 106 are present in the thymus in situ in one normal adult mouse [78], the tight adhesion of TECs, in particular cTECs, with neighboring cells including thymocytes makes highly inefficient to isolate TECs, especially cTECs, from the thymus [79].
Interestingly, our recent analysis has indicated that keratin 5 promoter-driven cyclin D1 transgenic (K5D1) mouse, which carries a massively enlarged yet functionally capable thymus [40, 80], would be a useful tool to obtain a large number of mouse TECs. We have reported that cTECs and mTECs isolated from K5D1 mouse retain transcriptomic profiles that characterize cTECs and mTECs in normal mouse, and the thymus from K5D1 mouse is capable of producing functionally competent T cells in a thymoproteasome-dependent manner and in establishing self-tolerance in T cells in a mTEC-dependent manner [40]. Importantly, approximately 2 × 105 cTECs and approximately 2 × 105 mTECs could be isolated from one K5D1 mouse, which would enable various biochemical analyses for studies of cTECs and mTECs [40].
Through the use of K5D1 mouse thymus in combination with newly improved methods to determine amino acid sequences from small amounts of proteins [81–83], we hope to be able to identify MHC-associated peptides isolated from cTECs and mTECs in genetically manipulated mice, for example, by comparing MHC-I-associated peptides expressed by cTECs obtained from control and thymoproteasome-deficient mice. Such an analysis is expected to be beneficial towards advancing our understanding of the mechanism for positive and negative selection of T cells in the thymus.
Highlights.
MHC-associated peptides displayed in the thymus form the T cell repertoire.
The thymoproteasome generates MHC class I-associated peptides in cTECs.
Thymoproteasome-dependent MHC-associated peptides optimally induce positive selection of self-protective CD8+ T cells
Acknowledgements
We thank Dr. Hidetaka Kosako for providing information on new technology for proteomic analyses. This work was supported by grants from MEXT-JSPS (I.O.) and the Intramural Research Program of the US National Institutes of Health, the National Cancer Institute, and the Center for Cancer Research (Y.T.).
Abbreviations:
- cTECs
cortical thymic epithelial cells
- mTECs
medullary thymic epithelial cells
- DCs
dendritic cells
- MHC
major histocompatibility complex
Footnotes
Declaration of Competing Interest
The authors report no competing interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Fehling HJ, Krotkova A, Saint-Ruf C, von Boehmer H. Crucial role of the pre-T-cell receptor alpha gene in development of alpha beta but not gamma delta T cells. Nature. 1995;375:795–8. doi: 10.1038/375795a0. [DOI] [PubMed] [Google Scholar]
- [2].Irving BA, Alt FW, Killeen N. Thymocyte development in the absence of pre-T cell receptor extracellular immunoglobulin domains. Science. 1998;289:905–8. doi: 10.1126/science.280.5365.905 [DOI] [PubMed] [Google Scholar]
- [3].Klein L, Kyewski B, Allen PM, Hogquist KA. Positive and negative selection of the T cell repertoire: what thymocytes see (and don’t see). Nat Rev Immunol. 2014;14:377–91. doi: 10.1038/nri3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ueno T, Saito F, Gray DH, Kuse S, Hieshima K, Nakano H, et al. CCR7 signals are essential for cortex-medulla migration of developing thymocytes. J Exp Med. 2004;200:493–505. doi: 10.1084/jem.20040643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kozai M, Kubo Y, Katakai T, Kondo H, Kiyonari H, Schaeuble K, et al. Essential role of CCL21 in establishment of central self-tolerance in T cells. J Exp Med. 2017;214:1925–35. doi: 10.1084/jem.20161864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kyewski B, Derbinski J. Self-representation in the thymus: an extended view. Nat Rev Immunol. 2004;4:688–98. doi: 10.1038/nri1436 [DOI] [PubMed] [Google Scholar]
- [7].Gallegos AM, Bevan MJ. Central tolerance to tissue-specific antigens mediated by direct and indirect antigen presentation. J Exp Med. 2004;200:1039–49. doi: 10.1084/jem.20041457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Koble C, Kyewski B. The thymic medulla: a unique microenvironment for intercellular self-antigen transfer. J Exp Med. 2009;206:1505–13. doi: 10.1084/jem.20082449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Jordan MS, Boesteanu A, Reed AJ, Petrone AL, Holenbeck AE, Lerman MA, et al. Thymic selection of CD4+CD25+ regulatory T cells induced by an agonist self-peptide. Nat Immunol 2001;2:301–6. doi: 10.1038/86302 [DOI] [PubMed] [Google Scholar]
- [10].Sakaguchi S Naturally arising CD4+ regulatory T cells for immunologic self-tolerance and negative control of immune responses. Ann Rev Immunol 2004;22:531–62. doi: 10.1146/annurev.immunol.21.120601.141122 [DOI] [PubMed] [Google Scholar]
- [11].van Bleek GM, Nathenson SG. Presentation of antigenic peptides by MHC class I molecules Trends Cell Biol. 1992;2:202–7. doi: 10.1016/0962-8924(92)90247-k [DOI] [PubMed] [Google Scholar]
- [12].Pieters J MHC class II-restricted antigen processing and presentation. Adv Immunol. 2000;75:159–208. doi: 10.1016/s0065-2776(00)75004-8 [DOI] [PubMed] [Google Scholar]
- [13].Germain RN, Margulies DH. The biochemistry and cell biology of antigen processing and presentation. Anne Rev Immunol. 1993;11:403–50. doi: 10.1146/annurev.iy.11.040193.002155. [DOI] [PubMed] [Google Scholar]
- [14].Reich Z, Altman JD, Boniface JJ, Lyons DS, Kozono H, Ogg G, et al. Stability of empty and peptide-loaded class II major histocompatibility complex molecules at neutral and endosomal pH : Comparison to class I proteins. Proc Natl Acad Sci USA. 1997;94:2495–500. doi: 10.1073/pnas.94.6.2495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Saper MA, Bjorkman PJ, Wiley DC. Refined structure of the human histocompatibility antigen HLA-A2 at 2.6A resplution. J Mol Biol. 1991;219:277–319. doi: 10.1016/0022-2836(91)90567-p. [DOI] [PubMed] [Google Scholar]
- [16].Matsui M, Hioe CR, Frelinger JA. Roles of the six peptide-binding pockets of the HLA-A2 molecule in allorecognition by human cytotoxic T-cell clones. Proc Natl Acad Sci U S A. 1993;90:674–78. doi: 10.1073/pnas.90.2.674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Falk K, Rotzschke O, Stevanovic S, Jung G, Rammensee HG. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature. 1991;351:290–6. doi: 10.1038/351290a0 [DOI] [PubMed] [Google Scholar]
- [18].Hunt DF, Henderson RA, Shabanowitz J, Sakaguchi K, Michel H, Sevilir N, et al. Characterization of peptides bound to the class I MHC molecule HLA-A2.1 by mass spectrometry. Science. 1992;255:1261–3. doi: 10.1126/science.1546328 [DOI] [PubMed] [Google Scholar]
- [19].Wieczorek M, Abualrous ET, Sticht J, Álvaro-Benito M, Stolzenberg S, Noé F, Freund C. Major histocompatibility complex (MHC) class I and MHC class II proteins: conformational plasticity in antigen presentation. Front Immunol. 2017;8:292. doi: 10.3389/fimmu.2017.00292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Garboczi DN, Biddison WE. Shapes of MHC restriction. Immunity. 1999;10:1–7. doi: 10.1016/s1074-7613(00)80001-1 [DOI] [PubMed] [Google Scholar]
- [21].Chicz RM, Urban RG, Lane WS, Gorga JC, Stern LJ, Vignali DA, Strominger JL. Predominant naturally processed peptides bound to HLA-DR1 are derived from MHC-related molecules and are heterogeneous in size. Nature. 1992;358:764–8. doi: 10.1038/358764a0 [DOI] [PubMed] [Google Scholar]
- [22].Stern LJ, Brown JH, Jardetzky TS, Gorga JC, Urban RG, Strominger JL, Wiley DC. Crystal structure of the human class II MHC protein HLA-DR1 complexed with an influenza virus peptide. Nature. 1994;368:215–21. doi: 10.1038/368215A0 [DOI] [PubMed] [Google Scholar]
- [23].Jones EY, Fugger L, Strominger JL, Siebold C. MHC class II proteins and diseases: a structural perspective. Nat Rev Immunol. 2006;6:271–82. doi: 10.1038/nri1805 [DOI] [PubMed] [Google Scholar]
- [24].Arnold PY, Gruta NLL, Miller T, Vignali KM, Adams PS, Woodland DL, Vignali DAA. The majority of immunogenic epitopes generate CD4+ T cells that are dependent on MHC class II-bound peptide-flanking residues. J Immunol. 2002;169:739–49. doi: 10.4049/jimmunol.169.2.739 [DOI] [PubMed] [Google Scholar]
- [25].Liu J, Gao GF. Major histocompatibility complex: interaction with peptides. In: eLS. John Wiley & Sons, Ltd: Chichester, 2011. doi: 10.1002/9780470015902.a0000922.pub2 [DOI] [Google Scholar]
- [26].Murata S, Sasaki K, Kishimoto T, Niwa S, Hayashi H, Takahama Y, Tanaka, K. Regulation of CD8+ T cell development by thymus-specific proteasomes. Science. 2007;316:1337–53. doi: 10.1126/science.1141915 [DOI] [PubMed] [Google Scholar]
- [27].Ripen AM, Nitta T, Murata S, Tanaka K, Takahama Y. Ontogeny of thymic cortical epithelial cells expressing the thymoproteasome subunit beta5t. Eur J Immunol. 2011;41:1278–87. doi: 10.1002/eji.201041375 [DOI] [PubMed] [Google Scholar]
- [28].Dick TP, Nussbaum AK, Deeg M, Heinemeyer W, Groll M, Schirle M, et al. Contribution of proteasomal beta-subunits to the cleavage of peptide substrates analyzed with yeast mutants. J Bio Chem. 1998;273:25637–46. doi: 10.1074/jbc.273.40.25637 [DOI] [PubMed] [Google Scholar]
- [29].Kisselev AF, Akopian TN, Castillo V, Goldberg AL. Proteasome active sites allosterically regulate each other, suggesting a cyclical bite-chew mechanism for protein breakdown. Mol Cell. 1999;4:395–402. doi: 10.1016/s1097-2765(00)80341-x [DOI] [PubMed] [Google Scholar]
- [30].Ohigashi I, Zuklys S, Sakata M, Mayer CE, Hamazaki Y, Minato N, et al. Adult thymic medullary epithelium is maintained and regenerated by lineage-restricted cells rather than bipotent progenitors. Cell Rep. 2015;13:1432–43. doi: 10.1016/j.celrep.2015.10.012 [DOI] [PubMed] [Google Scholar]
- [31].Mayer CE, Žuklys S, Zhanybekova S, Ohigashi I, Teh HY, Sansom SN, et al. Dynamic spatio-temporal contribution of single β5t+ cortical epithelial precursors to the thymus medulla. Eur J Immunol. 2016;46:846–56. doi: 10.1002/eji.201545995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Sansom SN, Shikama-Dorn N, Zhanybekova S, Nusspaumer G, Macaulay IC, Deadman ME, et al. Population and single-cell genomics reveal the Aire dependency, relief from Polycomb silencing, and distribution of self-antigen expression in thymic epithelia. Genome Res. 2014;24:1918–31. doi: 10.1101/gr.171645.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Murata S, Takahama Y, Kasahara M, Tanaka K. The immunoproteasome and thymoproteasome: functions, evolution and human disease. Nat Immunol. 2018;19:923–31. doi: 10.1038/s41590-018-0186-z. [DOI] [PubMed] [Google Scholar]
- [34].Sasaki K, Takada K, Ohte Y, Kondo H, Sorimachi H, Tanaka K, et al. Thymoproteasomes produce unique peptide motifs for positive selection of CD8+ T cells. Nat Commun. 2015;6:7484. doi: 10.1038/ncomms8484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Kuckelkorn U, Stübler S, Textoris-Taube K, Kilian C, Niewienda A, Henklein P, et al. Proteolytic dynamics of human 20S thymoproteasome. J Bio Chem. 2019;294:7740–54. doi: 10.1074/jbc.RA118.007347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hörig H, Young ACM, Papadopoulos NJ, DiLorenzo TP, Nathenson SG. Binding of longer peptides to the H-2Kb heterodimer is restricted to peptides extended at their C terminus: refinement of the inherent MHC class I peptide binding criteria. J Immunol. 1999;163:4434–41 [PubMed] [Google Scholar]
- [37].Nitta T, Murata S, Sasaki K, Fujii H, Mat Ripen A, Ishimaru N, et al. Thymoproteasome shapes immunocompetent repertoire of CD8+ T cells. Immunity. 2010;32:29–40. doi: 10.1016/j.immuni.2009.10.009 [DOI] [PubMed] [Google Scholar]
- [38].Takada K, Van Laethem F, Xing Y, Akane K, Suzuki H, Murata S., et al. TCR affinity for thymoproteasome-dependent positively selecting peptides conditions antigen responsiveness in CD8+ T cells. Nat Immunol. 2015;16:1069–76. doi: 10.1038/ni.3237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Xing Y, Jameson S, Hogquist K. Thymoproteasome subunit-β5t generates peptide-MHC complexes specialized for positive selection. PNAS. 2013;110:6979–84. doi: 10.1073/pnas.1222244110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Ohigashi I, Tanaka K, Kondo K, Fujimori S, Kondo H, Palin AC, et al. Trans-omics impact of thymoproteasome in cortical thymic epithelial cells. Cell Rep. 2019;29: 2901–16. doi: 10.1016/j.celrep.2019.10.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ohigashi I, Takahama Y. Specific impact of β5t on proteasome subunit composition in cortical thymic epithelial cells. Cell Rep. 2021;36:109657. doi: 10.1016/j.celrep.2021.109657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Ohigashi I, Frantzeskakis M, Jacques A, Fujimori S, Ushio A, Yamashita F, Ishimaru N, et al. Thymoproteasome hardwires TCR repertoire of CD8+ T cells with cortical positive selection independent of negative selection. J Exp Med. 2021;218: e20201904. doi: 10.1084/jem.20201904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Apavaloaei A, Brochu S, Dong M, Rouette A, Hardy MP, Villafano G, et al. PSMB11 orchestrates the development of CD4 and CD8 thymocytes via regulation of gene expression in cortical thymic epithelial cells. J Immunol. 2019;202:966–78. doi: 10.4049/jimmunol.1801288 [DOI] [PubMed] [Google Scholar]
- [44].Apavaloaei A, Laverdure JP, Perreault C. PSMB11 regulates gene expression in cortical thymic epithelial cells. Cell Rep. 2021;36:109546. doi: 10.1016/j.celrep.2021.109546 [DOI] [PubMed] [Google Scholar]
- [45].Kourilsky P, Claverie JM. MHC restriction, alloreactivity, and thymic education: a common link? Cell. 1989;56:327–9. doi: 10.1016/0092-8674(89)90233-x [DOI] [PubMed] [Google Scholar]
- [46].Marrack P, Kappler J. The T cell receptor. Science. 1987;238:1073–9. doi: 10.1126/science.3317824 [DOI] [PubMed] [Google Scholar]
- [47].Kincaid E, Murata S, Tanaka K, Rock K. Specialized proteasome subunits have an essential role in the thymic selection of CD8+ T cells. Nat Immunol. 2016;17:938–46. doi: 10.1038/ni.3480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Tomaru U, Konno S, Miyajima S, Kimoto R, Onodera M, Kiuchi S, et al. Restricted expression of the thymoproteasome is required for thymic selection and peripheral homeostasis of CD8+ T cells. Cell Rep. 2019;26:639–51. doi: 10.1016/j.celrep.2018.12.078 [DOI] [PubMed] [Google Scholar]
- [49].Ohigashi I, Takahama Y. Thymoproteasome optimizes positive selection of CD8+ T cells without contribution of negative selection. Adv Immunol. 2021;149:1–23. doi: 10.1016/bs.ai.2021.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Nil A, Firat E, Sobek V, Eichmann K, Niedermann G. Expression of housekeeping and immunoproteasome subunit gene is differentially regulated in positively and negatively selecting thymic stroma subsets. Eur J Immunol. 2004;34:2681–89. doi: 10.1002/eji.200425032 [DOI] [PubMed] [Google Scholar]
- [51].Fehling H, Swat W, Laplace C, Kühn R, Raiewsky K, Müller U, von Boehmer H. MHC class I expression in mice lacking the proteasome subunit LMP-7. Science. 1994;265:1234–7. doi: 10.1126/science.8066463 [DOI] [PubMed] [Google Scholar]
- [52].Basler M, Mundt S, Groettrup M. The immunoproteasome subunit LMP7 is required in the murine thymus for filling up a hole in the T cell repertoire. Eur J Immunol. 2018;48:419–29. dio: 10.1002/eji.201747282 [DOI] [PubMed] [Google Scholar]
- [53].Osterloh P, Linkemann K, Tenzer S, Rammensee H, Radsak M, Busch D, Schild H. Proteasomes shape the repertoire of T cells participating in antigen-specific immune responses. PNAS. 2006;103:5042–7. doi: 10.1073/pnas.0509256103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Kincaid E, Che J, York I, Escobar H, Reyes-Vargas E, Delgado J, et al. Mice completely lacking immunoproteasomes show major changes in antigen presentation. Nat Immunol. 2012;13:129–35. doi: 10.1038/ni.2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Cresswell P Invariant chain structure and MHC class II function. Science. 1995;85:505–7. doi: 10.1016/S0092-8674(00)81025-9 [DOI] [PubMed] [Google Scholar]
- [56].Klein L, Roettinger B, Kyewski B. Sampling of complementing self-antigen pools by thymic stromal cells maximizes the scope of central T cell tolerance. Eur J Immunol. 2001;31:2476–86. doi: [DOI] [PubMed] [Google Scholar]
- [57].Kasai M, Tanida I, Ueno T, Kominami E, Seki S, Ikeda T, Mizouchi T. Autophagic compartments gain acess to the MHC class II compartments in thymic epithelium. J Immunol. 2009;183:7278–85. doi: 10.4049/jimmunol.0804087 [DOI] [PubMed] [Google Scholar]
- [58].Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–11. doi: 10.1091/mbc.E03-09-0704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Nedjic J, Aichinger M, Emmerich J, Mizushima N, Klein L. Autophagy in thymic epithelium shapes the T-cell repertoire and is essential for tolerance. Nature 2008;455:396–400. doi: 10.1038/nature07208 [DOI] [PubMed] [Google Scholar]
- [60].Schuster C, Gerold KD, Schober L, Probst L, Boerner K, Kim MJ, et al. The autoimmunity-associated gene CLEC16A modulates thymic epithelial cell autophagy and alters T cell selection. Immunity. 2015;42:942–52. doi: 10.1016/j.immuni.2015.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Nakagawa T, Roth W, Wong P, Nelson A, Farr A, Deussing J, et al. Cathepsin L: critical role in Ii degradation and CD4 T cell selection in the thymus. Science. 1998;280:450–3. doi: 10.1126/science.280.5362.450 [DOI] [PubMed] [Google Scholar]
- [62].Honey K, Nakagawa T, Peter C, Rudensky A. Cathepsin L regulates CD4+ T cell selection independently of its effect on invariant chain : a role in the generation of positively selecting peptide ligands. J Exp Med. 2002;195:1349–58. doi: 10.1084/jem.20011904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Nakagawa TY, Brissette WH, Lira PD, Griffiths RJ, Petrushova N, Stock J, et al. Impaired invariant chain degradation and antigen presentation and diminished collagen-induced arthritis in cathepsin S null mice. Immunity. 1999;10:207–17. doi: 10.1016/s1074-7613(00)80021-7 [DOI] [PubMed] [Google Scholar]
- [64].Shi GP, Villadangos JA, Dranoff G, Small C, Gu L, Haley KJ, et al. Cathepsin S required for normal MHC Class II peptide loading and germinal center development. Immunity. 1999;10:197–206. doi: 10.1016/s1074-7613(00)80020-5 [DOI] [PubMed] [Google Scholar]
- [65].Mizuochi T, Yee S-T, Kasai M, Muno D, Kominami E. Both cathepsin B and cathepsin D are necessary for the processing of ovalbumin as well as for the degradation of class II MHC invariant chain. Immunol Lett. 1994;43:189–93. doi: 10.1016/0165-2478(94)90221-6 [DOI] [PubMed] [Google Scholar]
- [66].Yamaguchi N, Tomaru U, Kiuchi T, Ishizu A, Deguchi T, Otsuka N, et al. Expression of cathepsin B, D and K in thymic epithelial tumours. J Clin Pathol. 2021;74:84–90. doi: 10.1136/jclinpath-2020-206551 [DOI] [PubMed] [Google Scholar]
- [67].Viret C, Leung-Theung-Long S, Serre L, Lamare C, Vignali DAA, Malissen B, et al. Thymus-specific serine protease controls autoreactive CD4 T cell development and autoimmune diabetes in mice. J Clin Invest. 2011;121:1810–21. doi: 10.1172/JCI43314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Gommeaux J, Grégoire C, Nguessan P, Richelme M, Malissen M, Guerder S, et al. Thymus-specific serine protease regulates positive selection of a subset of CD4+ thymocytes. Eur J Immunol. 2009;39:956–64. doi: 10.1002/eji.200839175 [DOI] [PubMed] [Google Scholar]
- [69].Viret C, Mahiddine K, Baker RL, Haskins K, Guerder S. The T Cell Repertoire–Diversifying Enzyme TSSP Contributes to Thymic Selection of Diabetogenic CD4 T Cell Specificities Reactive to ChgA and IAPP Autoantigens. J Immunol. 2015;195:1964–73. doi: 10.4049/jimmunol.1401683 [DOI] [PubMed] [Google Scholar]
- [70].Serre L, Girard M, Ramadan A, Menut P, Rouquié N, Lucca LE, et al. Thymic-specific serine protease limits central tolerance and exacerbates experimental autoimmune encephalomyelitis. J Immunol. 2017;199:3748–3756. doi: 10.4049/jimmunol.1700667 [DOI] [PubMed] [Google Scholar]
- [71].Nedjic J, Aichinger M, Mizushima N, Klein L. Macroautophagy, endogenous MHC II loading and T cell selection: the benefits of breaking the rules. Curr Opin Immunol. 2009;21:92–7. doi: 10.1016/j.coi.2009.01.013 [DOI] [PubMed] [Google Scholar]
- [72].Lich JD, Elliott JF, Blum JS. Cytoplasmic processing is a prerequisite for presentation of an endogenous antigen by major histocompatibility complex class II proteins. J Exp Med. 2000;191:1513–24. doi: 10.1084/jem.191.9.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Adamopoulou E, Tenzer S, Hillen N, Klug P, Rota IA, Tietz S, et al. Exploring the MHC-peptide matrix of central tolerance in the human thymus. Nat Commun. 2013;4:2039. doi: 10.1038/ncomms3039 [DOI] [PubMed] [Google Scholar]
- [74].Collado JA, Alvarez I, Ciudad MT, Espinosa G, Canals F, Pujol-Borrell R, et al. Composition of the HLA-DR-associated human thymus peptidome. Eur J Immunol. 2013;43:2273–82. doi: 10.1002/eji.201243280 [DOI] [PubMed] [Google Scholar]
- [75].Vanneste Y, Thome AN, Vandersmissen E, Charlet C, Franchimont D, Martens H, et al. Identification of neurotensin-related peptides in human thymic epithelial cell membranes and relationship with major histocompatibility complex class I molecules. J Neuroimmunol. 1997;76:161–6. doi: 10.1016/s0165-5728(97)00052-0 [DOI] [PubMed] [Google Scholar]
- [76].Geenen V The thymus and the science of self. Semin Immunopathol. 2021;43:5–14. doi: 10.1007/s00281-020-00831-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Caron E, Kowalewski DJ, Chiek Koh C, Sturm T, Schuster H, Aebersold R. Analysis of Major Histocompatibility Complex (MHC) Immunopeptidomes Using Mass Spectrometry. Mol Cell Proteomics. 2015;14:3105–17. doi: 10.1074/mcp.O115.052431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Sakata M, Ohigashi I, Takahama Y. Cellularity of thymic epithelial cells in the postnatal mouse. J Immunol. 2018;200:1382–88. doi: 10.4049/jimmunol.1701235 [DOI] [PubMed] [Google Scholar]
- [79].Nakagawa Y, Ohigashi I, Nitta T, Sakata M, Tanaka K, Murata S, Kanagawa O, Takahama Y. Thymic nurse cells provide microenvironment for secondary TCRa rearrangement in cortical thymocytes. Proc Natl Acad Sci U S A. 2012;109:20572–77. doi: 10.1073/pnas.1213069109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Robles AI, Larcher F, Whalin RB, Murillas R, Richie E, Gimenez-Conti IB, et al. Expression of cyclin D1 in epithelial tissues of transgenic mice results in epidermal hyperproliferation and severe thymic hyperplasia. Proc Natl Acad Sci USA. 1996;93:7634–38. doi: 10.1073/pnas.93.15.7634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Swaminathan J, Boulgakov AA, Hernandez ET, Bardo AM, Bachman JL, Marotta J, et al. Highly parallel single-molecule identification of proteins in zeptomole-scale mixtures. Nat Biotechnol. 2018;36:1076–82. doi: 10.1038/nbt.4278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Zhu Y, Piehowski PD, Zhao R, Chen J, Shen Y, Moore RJ, et al. Nanodroplet processing platform for deep and quantitative proteome profiling of 10–100 mammalian cells. Nat Commun. 2018;9:882. doi: 10.1038/s41467-018-03367-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Alfaro JA, Bohländer P, Dai M, Filius M, Howard CJ, van Kooten XF, et al. The emerging landscape of single-molecule protein sequencing technologies. Nat Methods. 2021;18,604–17. doi: 10.1038/s41592-021-01143-1 [DOI] [PMC free article] [PubMed] [Google Scholar]