Introduction
It is clear that regenerative medicine therapies will dominate the future landscape, particularly for genetic disorders and other diseases that have proven intractable with current therapies. As we explore unique combinations of rapidly emerging fields—from computational biology to genome editing to molecular therapeutics—the past goal of ameliorating symptoms of disease is rapidly giving way to the goal of restoring a patient to health and normal function.
Our bodies are adept at managing a state of permanent crisis, as multiple biological functions are constantly disrupted by internal and external forces. In response, molecular and cellular reparative mechanisms are engaged and—unless overwhelmed—restore, repair, or replace damaged tissues. Although we take this process for granted as we recover from a virus or heal from a cut or scrape, when we encounter a condition that is beyond our body’s ability to manage, we become all too aware of the need for medical intervention. New regenerative therapies offer an improved future for patients with untreatable or incurable diseases, particularly genetic disorders.
Regenerative medicine options change as our biomedical understanding changes. Notable examples that have contributed to the development of regenerative solutions are:
Scholarly autopsy: human body reduced to individual organs (15th–17th centuries).
Microscopy and histology: aided by chemical stains that gave unique colors to different cell types (18th–19th centuries).
Cellular biology and genetics (19th–20th centuries).
Molecular biology: structure of proteins and nucleic acids (20th century).
Bioengineering and functional imaging: from optical imaging/optogenetics at one end of the spectrum to functional MRI and brain connectome on the other (21st century).
Even though the increased understanding on each plane—gene, cell, tissue, organism—has not yet led to the ability to link these layers in a system that can reconstruct the original, it is advantageous to consider how induced regeneration on each of these levels operates in monogenic disorders (Figure 1).
Figure 1: Multilayered regeneration in biological systems.
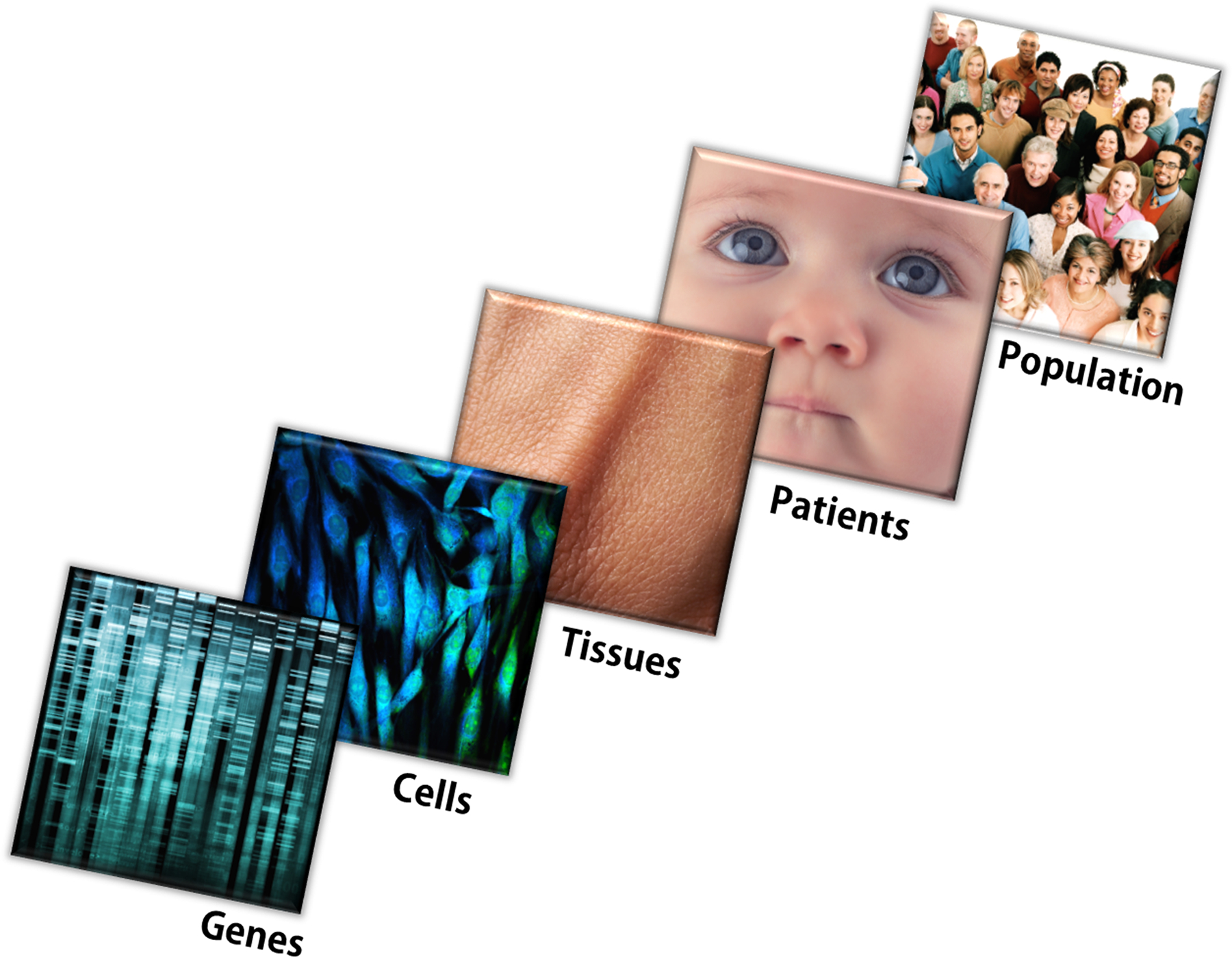
Cells, tissues, organs, organisms and populations are all abstractions of genomes, but they differ in complexity of biological organization and relationships (emerging properties) that define their function. They also use different mechanisms of repair and regeneration. Image credits, L-R: Kheng ho Toh/Hemera, vshivkova/iStock, Dimarik/iStock, valio84s/iStock, Digital Vision, all from Thinkstock.
I. DNA regeneration
Genomic integrity is maintained by robust repair mechanisms that can be co-opted for targeted gene correction. The extraordinary combination of basic DNA repair science with the ability to create artificial molecules capable of DNA cleavage at a chosen genomic site has enabled precision gene correction, termed gene editing (1).
Changing the DNA sequence in genomic context was long thought impossible. A significant breakthrough came with the elegant repurposing of naturally occurring DNA-binding proteins and associated endonuclease for targeted DNA cleavage. Zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) have been the most prominent customizable DNA-binding proteins. Cells respond to double-stranded DNA breaks either by non-homologous end-joining (NHEJ) or—in actively dividing cells and in the presence of a target sequence-complementary donor template—by homology-driven repair (HDR). NHEJ typically disrupts the target sequence by local accumulation of insertions and deletions (indels), and has been useful in disabling genes, e.g., for generating rodent models of human disease and in clinical trials with inactivation of genes operational in human cancers and infections. HDR can (in the presence of a wild-type template) rewrite a specific genomic site, restoring the normal sequence and function of the affected gene. Gene editing offers the additional advantages that:
The gene restored to function remains under the physiological control of the native promoter.
Gene-editing molecules can be multiplexed to modify several molecular targets at once.
When paired with activation or repression, they can be used to regulate gene expression.
Nevertheless, the need to construct a unique ZFN or TALEN protein for each target sequence is costly in effort and supplies. When clustered regularly interspaced palindromic repeats (CRISPRs) appeared (1)—with the RNA-DNA complementarity between the target site and guide RNA and Cas9 nuclease—the field acquired the most powerful gene engineering tool yet developed. As ZFN, TALEN, and CRISPR nucleases can be programmed to almost any DNA sequence, gene editing has been used successfully in numerous monogenic disorders (2).
II. Cellular regeneration
Cellular replacement maintains tissue homeostasis in the process of regular wear and tear, as well as in response to acute or chronic injury. Hematopoietic diseases (Fanconi anemia, dyskeratosis congenita, sickle cell anemia) have been cured by regeneration of donor hematopoietic stem cells (HSCs). Maturation and refinement of our understanding of cell renewal has made induction of pluripotent stem cells from differentiated cells possible. We have recently seen the first evidence of engraftment of true stem cells outside the hematopoietic system in the generation of skin from patient’s own gene-corrected induced pluripotent stem cells (iPSCs; Hirsch et a., in press). Monogenic disorders, difficult to study due to limited specimens and genotypic (and phenotypic) heterogeneity, have also benefited tremendously from the discovery of induced pluripotency. Disease modeling (for example by differentiating patient-specific iPSCs to organoids), drug testing using disease-appropriate iPSC-progeny cells, and potentially using gene-corrected iPSCs for therapy—are the foundation of clinically meaningful regenerative interventions (3). Additionally, successful interventions in regenerative medicine will likely build upon the 3D structure of tissues. Advances in bioengineering—bioprinting of heart valves, blood vessels, etc.—have already made this clear.
The challenges of using stem cells are either intrinsic (possibility of aberrant growth from stem cells, either benign—mesenchymal stem cells [MSCs] injected into eyelids for cosmetic indication differentiated into bone—or malignant—theoretical because no reports to date in human trials reported MSCs have led to cancer, although there are reports of donor-derived leukemias in the field of hematopoietic cell transplantation ([HCT]) or extrinsic (such as the possibility of immune response to the injected cells from an allogeneic donor, which could lead to rejection of stem cell grafts, or the acceptance of the graft with autoimmune consequences.
III. Tissue regeneration
Regeneration of lymphohematopoietic system from allogeneic HSCs opened the field of blood and marrow transplantation (BMT) in 1968, and it has become standard of care in many malignant and nonmalignant disorders for more than 50,000 patients worldwide every year.
In addition to leukemia and lymphoma, allogeneic BMT has been used for enzymopathies (mucopolysaccharidosis type I, alpha-manosidosis), hemoglobinopathies (thalassemia, sickle cell disease), chromosome sensitivity disorders (Fanconi anemia, dyskeratosis congenita), channelopathies (osteopetrosis, adrenoleukodystrophy), and extra-cellular matrix disorders (epidermolysis bullosa). Thus allogeneic BMT was the first gene therapy (delivering the wild-type gene in transplanted donor cells) and the first regenerative solution for inherited diseases (4).
IV. Organismal regeneration
Earlier this year, gene engineering of dominant negative MYBCP3 mutations that lead to hypertrophic cardiomyopathy in adults was accomplished in human embryos (not intended for implantation) (5). This proof-of-concept study showed improved gene editing efficiency (gene-editing reagents were injected directly into embryonic cells) and undetectable (with current techniques) unwanted side effects on genome integrity (Cas9 protein degrades rapidly, so the time it can act on off-target sites is shorter compared to its delivery as DNA).
V. 4th dimension: time
Monogenic disorders are triggered by DNA lesions in a single gene (one or both alleles of one gene). However, the consequences of each mutation change over time. Typically, the cellular, tissue, and organ damage accumulates (with progression of clinical signs and symptoms), and the other organ systems react to the malfunction that was initially confined to a single cell type or tissue. Dominant among these reactions are inflammatory and immune responses that frequently amplify the single gene mutation into a disease cascade. With accumulated organ damage resulting from overwhelmed or unproductive tissue repair, the disease phenotype changes over time. As a result, the disease of an infant, a teenager, and an adult with the same monogenic DNA lesion is different (Figure 2). It is critical to choose treatment modalities appropriate to disease progression. The same formulation of gene/cell therapy that has favorable and lasting impact in the dynamic and resilient state of infancy will have different efficacy in an adult counterpart with the accumulated degenerated cells and exhausted mechanisms of tissue repair.
Figure 2: Same genotype results in different clinical presentations over time.
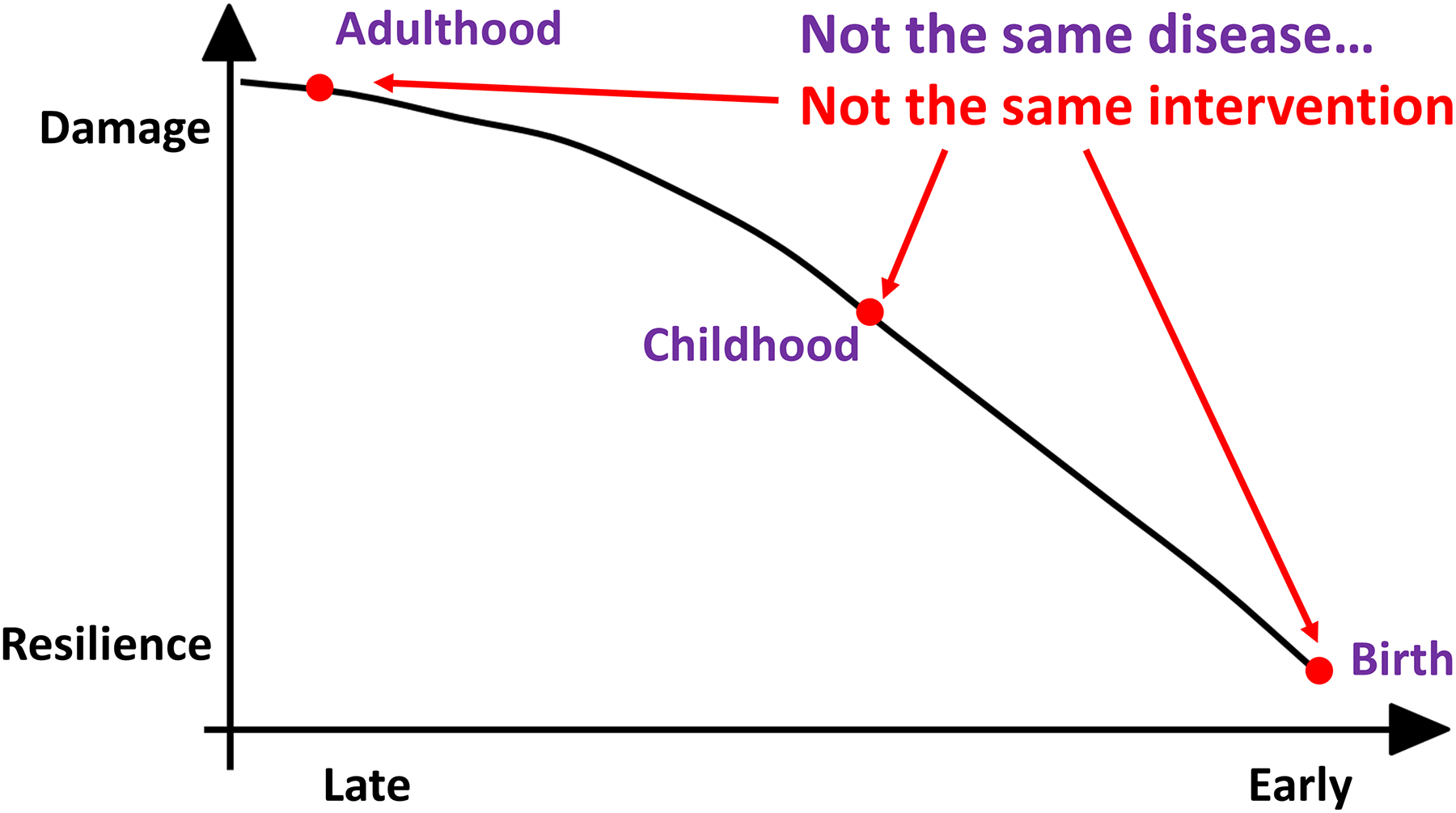
Even though genetic disorders originate from discrete DNA mutation(s) their clinical manifestation changes over time. There is more disease burden, more reactive changes (for example, immunological), and, typically, less capacity to respond to regenerative medicine interventions. This is obvious but frequently forgotten. It is critical, however, as different treatments are relevant to patients of different ages with the same genotype.
VI. Regeneration of environment
All cells operate in concert with other cells. Early cell therapy, made possible by the understanding of immune tolerance in the 1950s and by HCT in the 1960s, primarily used allogeneic donors. These therapies created numerous side effects, both from the chemotherapy and radiation used in recipient preparation, and in the immune side effects ranging from profound transient immune suppression to graft-versus-host disease. Immune reaction that proved beneficial in some cases (graft-versus-leukemia effect in myelogenous cancers), was often devastating to patients with genetic disorders (DNA repair-deficiency disorder Fanconi anemia). It seems obvious that for inherited diseases, by definition both systemic and constitutional, gene therapy of multiple cell types, or “stem cell niche therapy,” will grow in significance and impact.
VII. Restoration of health
The new ambition of medicine is changing from focus on disease to focus on health. This conceptually different direction is only possible because of regenerative medicine. Drugs and biologicals, although life-changing in numerous disorders, typically only alleviate symptoms of disease or partially restore organ function. In contrast, regenerative approaches aim at full repair or replacement of diseased tissues and shift the therapeutic goal from disease control to restoration of full function, i.e., healthy organ and organism.
The origin is also different: the power of regenerative medicine is derived from the abundance of natural processes (awakening our “inner salamander”), not the near-chaotic abundance of the current pharmacopoeia.
The discovery of genes revolutionized our understanding of the biological world. Now the ability to edit genes has re-defined the way we think about medicine. Recent advances in molecular science include development of self-inactivating retroviruses and lentiviruses, adeno-associated viral vectors, and transposons. Gene editing is based on the concepts of nucleic acids transmitting information via a digital code and the natural mutating variance of nucleic acids as a substrate for evolutionary selection. Human genetic disorders are clinical representations of these variations. Gene editing offers—for the first time—a technology whereby the disease-causing mutations are eliminated by re-coding the affected gene. Synthetic molecules with the dual functions of identifying and cutting the DNA at a specific genomic location proximal to the targeted mutation are one of the most elegant examples of current biology’s emerging ability to engineer clinically relevant interventions at the site of the problem, i.e., at a molecular level. Recent experience from phase I/II clinical trials (transduced autologous cells for severe immune deficiencies and demyelinating disorders, and particularly chimeric antigen receptor-assisted T-cell therapy [CAR-T]) have made gene-corrected therapy of genetic and other disorders a realistic—inevitable—next step. The licensing of CAR-T (tisagelecleucel) in August of 2017 can be seen as the turning point in the history of gene cell therapy and a harbinger of a future where regenerative medicine will govern our practice of medicine and experience of life.
Funding Support:
NIH R01 AR063070, NIH R01 AR059947, NIH/NCI P01 CA065493, NIH P01 CA111412.
Footnotes
Conflict of Interest: The author declares no conflicts.
References
- (1).Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA & Charpentier E A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–21 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Osborn MJ, Belanto JJ, Tolar J & Voytas DF Gene editing and its application for hematological diseases. Int J Hematol 104, 18–28 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Tolar J Translating genome engineering to better clinical outcomes. Transl Res 161, 199–204 (2013). [DOI] [PubMed] [Google Scholar]
- (4).Tolar J, Sodani P & Symons H Alternative donor transplant of benign primary hematologic disorders. Bone Marrow Transplant 50, 619–27 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Tang L et al. CRISPR/Cas9-mediated gene editing in human zygotes using Cas9 protein. Mol Genet Genomics 292, 525–33 (2017). [DOI] [PubMed] [Google Scholar]


