Abstract
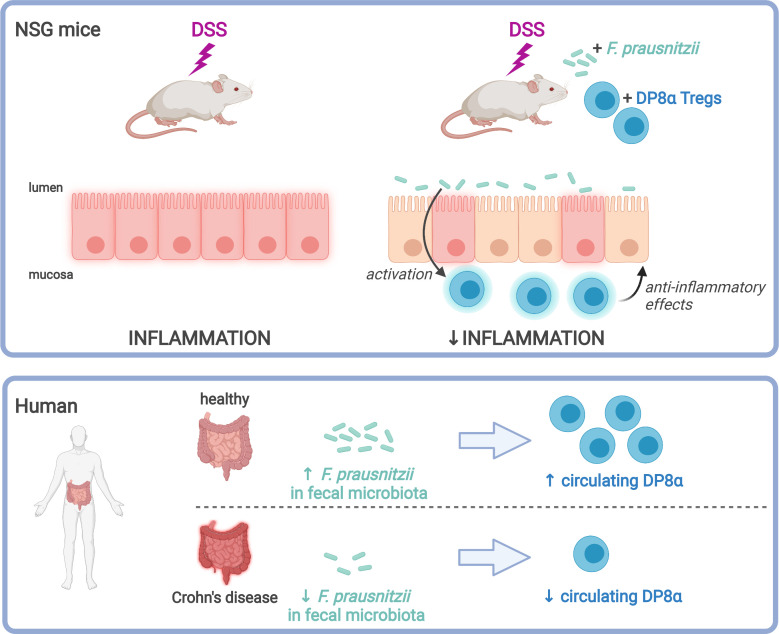
Abundance of Faecalibacterium prausnitzii, a dominant bacterium of the human microbiota that exhibits antiinflammatory effects, is decreased in patients with inflammatory bowel diseases (IBD). In humans, colonic lamina propria contains IL-10–secreting, Foxp3– Tregs characterized by a double expression of CD4 and CD8α (DP8α) and a specificity for F. prausnitzii. This Treg subset is decreased in IBD. The in vivo effect of DP8α cells has not been evaluated yet to our knowledge. Here, using a humanized model of a NSG immunodeficient mouse strain that expresses the HLA D–related allele HLA-DR*0401 but not murine class II (NSG-Ab° DR4) molecules, we demonstrated a protective effect of a HLA-DR*0401–restricted DP8α Treg clone combined with F. prausnitzii administration in a colitis model. In a cohort of patients with IBD, we showed an independent association between the frequency of circulating DP8α cells and disease activity. Finally, we pointed out a positive correlation between F. prausnitzii–specific DP8α Tregs and the amount of F. prausnitzii in fecal microbiota in healthy individuals and patients with ileal Crohn’s disease.
Keywords: Gastroenterology
Keywords: Inflammatory bowel disease, Mouse models, T cells
Introduction
Inflammatory bowel diseases (IBD), including Crohn’s disease (CD) and ulcerative colitis (UC), are characterized by chronic and relapsing inflammation of the intestine. Their incidence rates and prevalence are high and rising in Western countries. Identifying markers to predict relapse or complications and new treatments are much needed and would have large socioeconomic effects. The exact pathogenesis of IBD remains to be deciphered. However, it involves dysregulated immune responses to commensal bacteria (1) and genetic factors as implied by GWAS and mouse models (2–5). In addition, environmental factors and life habits are also involved, and many studies have highlighted the role of gut microbiota alterations in the pathogenesis (6, 7). One of the strongest signals repeatedly identified in the IBD-associated microbiota alteration is the decreased abundance of Faecalibacterium prausnitzii, a dominant bacterium of the Clostridium IV group (8–10). F. prausnitzii is highly prevalent in the normal human gut microbiota and represents by itself around 5% of the total bacterial population in healthy individuals (11). We and others showed that it exerts antiinflammatory properties both in vitro and in vivo in different colitis mouse models (12–14). More than a simple marker of inflammation, the role of the gut microbiota as an actor in IBD pathogenesis is now demonstrated. Among the strongest arguments, one can cite the proinflammatory effect of the IBD-associated microbiota, demonstrated by microbiota transfer from human to mice (15), and the clinical efficacy of fecal microbiota transplantation as a treatment of IBD (16). In mice, Clostridium IVa and XIV bacteria play a role in inducing IL-10–secreting forkhead box P3–positive (Foxp3+) Tregs in the colonic lamina propria (LP), which attenuate colitis (17). In humans, the proportion of Foxp3+ Tregs is low in the colonic LP, as compared with mice (5% and 25% respectively) (18, 19), and, despite the extensive interest in Foxp3+ Tregs as key players in the intestinal immune homeostasis, there is little evidence for the role of these Tregs in IBD. No defect in the abundance or function of these cells was observed in patients with IBD (19, 20), and no polymorphisms in FOXP3 or other genes related to Foxp3+ Treg differentiation was detected by GWAS. In addition, individuals with immune dysregulation, polyendocrinopathy, enteropathy, X-linked (IPEX) syndrome, who thus lack functional Foxp3+ Tregs, do not necessarily develop colitis (21), supporting the hypothesis that Foxp3+ Tregs are not essential in the development of IBD. In this context, we have recently characterized, in the human colonic LP, a subset of Foxp3– IL-10–producing Tregs responding to F. prausnitzii in a MHC class II and T cell receptor–dependent manner (22, 23). These cells share most in vitro regulatory functions with Foxp3+ Tregs and coexpress CD4 and low levels of CD8α in vivo; hence, they were named double-positive CD8α (DP8α). We found that DP8α Tregs are abundant in the healthy colonic LP (up to 13% of CD4+ T cells). They are present also in blood, where CCR6+CXCR6+ DP8α cells react to F. prausnitzii, in contrast to DP8α cells lacking one or both receptors (22); the role for this remains unknown. Interestingly, both colonic and circulating DP8α Tregs are drastically reduced in patients with IBD, as compared with healthy participants (22). Along with these findings, we have recently revealed the ability of F. prausnitzii to induce tolerogenic dendritic cells that favor IL-10–producing CD4+ T cell priming in vitro (24). Altogether, these data argue that F. prausnitzii contributes to the induction of IL-10–producing DP8α Tregs in a manner similar to the induction of IL-10+Foxp3+ Tregs by murine Clostridia (25). This also suggests the existence of a link between reduced levels of F. prausnitzii and decreased activity of F. prausnitzii–specific DP8α Tregs in patients with IBD, potentially resulting in disturbed gastrointestinal homeostasis. Consequently, this suggests that DP8α Tregs could play a role in the control of IBD. Here, we aimed at assessing the role of DP8α Tregs in vivo with a preclinical model. We used immunodeficient mice, humanized for the DRb1*0401 HLA class II allele and injected them with human HLA-DRb1*0401+ CD4+ T cells in the absence or in the presence of human HLA-DRb1*0401–restricted DP8α Tregs, before triggering dextran sulfate sodium–induced (DSS-induced) colitis. In addition, we analyzed the phenotype and frequency of DP8α cells and the abundance of F. prausnitzii in the fecal microbiota in a large cohort of healthy controls and patients with IBD.
Here, we demonstrated the capacity of DP8α Tregs to attenuate colitis severity in a humanized mouse model, supporting their importance in the maintenance of gastrointestinal homeostasis and in IBD prevention. In patients with IBD, we showed that a low abundance of blood DP8α cells was independently associated with several clinical parameters of disease activity, such as flare and elevated C-reactive protein (CRP). Compared with healthy controls, we also confirmed that patients with IBD exhibited a decreased abundance of CCR6+CXCR6+ DP8α Tregs and of F. prausnitzii in their blood and fecal microbiota, respectively. Interestingly, both parameters were positively correlated in patients with CD with ileal involvement, suggesting that F. prausnitzii could induce DP8α Tregs in humans, in the same manner that Clostridia induces Foxp3+ Tregs in mice. These data extend the pathologic relevance of the decreased abundance of DP8α cells and suggest that DP8α Treg abundance may both serve as an indicator of intestinal inflammation and represent a therapeutic target in IBD.
Results
Validation of the mouse model and adoptive transfer of human cells.
To evaluate the effect of DP8α Tregs on intestinal inflammation in vivo, we used a model of DSS-induced colitis in nonobese diabetic Prkdcscid.Il2rg–/– (NSG) mice that lack murine MHC class II and instead can express a HLA allele of interest under the control of the murine MHC class II promoter, as described previously (26). NSG mice lack murine T cells, B cells, and NK cells and can be reconstituted with human CD4+ T cells from allelically matched HLA class II donors. We used NSG mice expressing the frequently expressed DRb1*0401 allele (NSG-Ab°DR4 mice), reconstituted with purified HLA-DRb1*0401–expressing CD4+ T cells. Then, to assess the role of DP8α Tregs in intestinal inflammation, we set up a colitis model in these mice and generated a DP8α Treg clone responding to HLA-DRb1*0401–expressing mouse antigen-presenting cells loaded with F. prausnitzii. As a negative control, a HLA-DR–restricted DP8α Treg clone also reactive to F. prausnitzii, but not restricted by the specific HLA-DRb1*0401, was used.
To select such clones, we used PBMCs from 2 donors from a blood bank (Etablissement Français du Sang [EFS], Pays de Loire, France), one typed homozygous for the relevant DRb1*0401 allele and the second, used as a control, from a DRb1*0408/DRb1*1396 donor. CD4+ T cells were magnetically sorted, and DP8α Tregs were FACS sorted and cloned simultaneously before being expanded using polyclonal stimulation as we described previously (22). We first selected DP8α Treg clone best responders to F. prausnitzii–loaded autologous monocytes and then, among these, a clone from each donor whose response was specifically inhibited by a blocking anti–HLA-DR antibody (Supplemental Figure 1, A–F; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.154722DS1). Finally, we confirmed that murine bone marrow–derived dendritic cells (BM-DCs) from NSG-Ab°DRb1*0401+ mice loaded with F. prausnitzii induced a potent cytokine response (IL-10 and IFN-γ) from the selected DRb1*0401 homozygous clone but not from the DRb1*0408/DRb1*1396 one (Supplemental Figure 1, A–D), validating not only the HLA-DRb1*0401–restricted response to F. prausnitzii of the first clone, but also, importantly, its ability to respond as well to BM-DCs from NSG-Ab°DR4 mice.
We set up the DSS-induced colitis model in a preliminary experiment (data not shown), in which mice received i.p. injection of human allelically matched effector CD4+ T cells 10 days before DSS administration. We determined that the dose of 1% DSS in drinking water was suitable to induce weight loss and colitis without causing too much mortality and to allow recovery upon DSS removal.
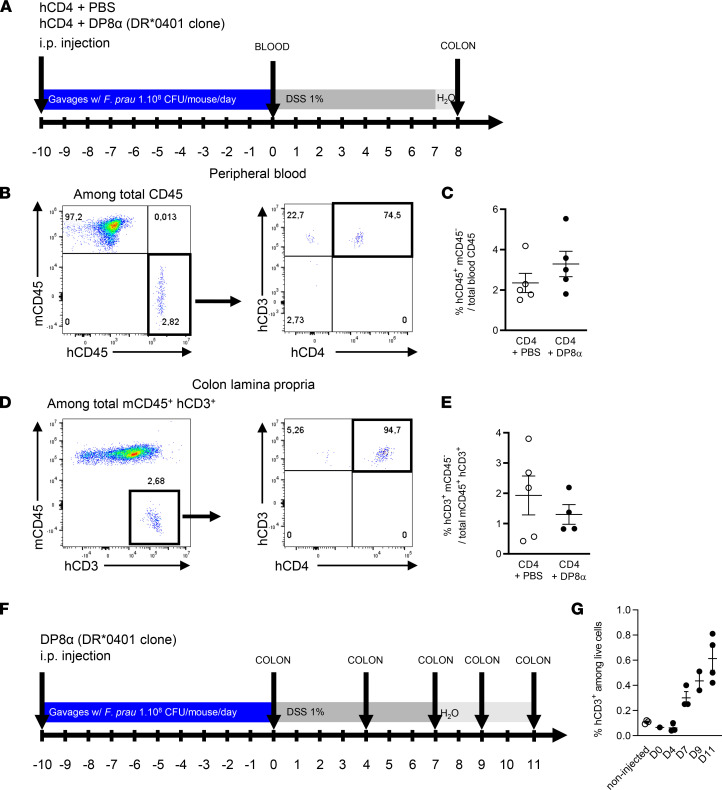
We used flow cytometry to evaluate the presence of human cells in NSG-Ab°DR4 mice after i.p. injection of CD4+ effector T cells in combination or not with allelically matched DRb1*0401-restricted DP8α Tregs. This was evaluated in the blood 10 days after T cell injection, before the start of DSS-induced colitis (day 0), and, in the colonic LP, 10 days after T cell injection and 8 days after the start of DSS treatment (day 8) to ensure T cell recruitment to the intestinal mucosa of mice (Figure 1A). Cells were gated according to side scatter (SSC-A) and forward scatter (FSC-A) after doublet discrimination (data not shown). The frequency of human cells was then measured in mouse blood as the fraction of human CD45+ cells among total, mouse and human, CD45+ cells. Most human cells were CD3+ T cells (Figure 1B), and abundance of these circulating human T cells was similar in mice injected with CD4+ T cells only and in those coinjected with the DP8α Treg clone (Figure 1C), consistent with the fact that DP8α Tregs rapidly and massively migrate into tissues and do not stay in the blood. In the colon LP, we measured the fraction of human CD3+ cells (Figure 1D). Human cells were virtually all CD4+ (Figure 1E). As observed in blood, the frequency of human T cells in the colon LP was not significantly different between CD4+PBS and CD4+DP8α groups (Figure 1E). We could not distinguish CD4+ effector T cells from DP8α clones, as most DP8α clones lose CD8α expression after in vitro expansion, without any effect on their in vitro regulatory functions (our unpublished observations). To address this issue and confirm the ability of the DP8α clone to colonize the colonic mucosa in NSG-AB°DR4 mice, we injected a group of mice with DP8α cells only and compared them with vehicle-treated mice. After 10 days of gavage with F. prausnitzii, the mice received 1% DSS in drinking water for 7 days (Figure 1F), and the frequency of human CD3+ cells in the colon LP was quantified by flow cytometry at different time points (Figure 1G). Data show an engraftment of the human DP8α clone in the colonic mucosa, which increased over time (Figure 1G). Altogether, we hereby demonstrated the presence of i.p. infused human T cells in the blood of NSG-AB°DR4 mice before DSS treatment (1.5%–5.5% of human CD45+ cells among total mouse and human CD45+ cells) and in the colon LP following DSS treatment (0.57%–3.8% of human CD3+ T cells), thus validating “humanization” of our mouse model and allowing further investigations.
Figure 1. Detection of human cells in whole blood and the colon lamina propria of NSG-Ab°DR4 mice.
(A) The presence of human cells after injection of CD4+ effector T cells in presence or absence of DP8α Tregs was assessed by flow cytometry in 2 independent experiments in the peripheral blood 10 days after injection, before the start of DSS treatment (day 0) and in the colon lamina propria 8 days after the start of DSS treatment (day 8). (B) After doublet discrimination and exclusion of debris, human CD3+ and human CD4+ cells were analyzed among human CD45+ (hCD45+) and murine CD45– (mCD45–) cells. (C) Frequency of human CD45+ and mouse CD45– cells among total CD45+ cells in CD4+PBS (n = 5, empty dots) and CD4+DP8α (n = 5, black dots) groups. (D) Detection of human cells in the colon lamina propria was determined by the expression of human CD3 and the absence of expression of mouse CD45 among total mCD45+ and human CD3+ cells. (E) Frequency of human CD3+ and mouse CD45– cells among total colonic mouse CD45+ and human CD3+ cells in CD4+PBS (n = 5, empty dots) and CD4+DP8α (n = 4, black dots) groups. Original magnification, ×4. (F and G) DP8α clone was injected i.p. and analyzed by flow cytometry in the colon lamina propria of the mice after 10 days of daily gavage with F. prausnitzii followed by 0, 4, 7, 9, or 11 days of treatment with DSS. “Non-injected” indicates the control group without cells injected. Results are presented as the mean ± SEM.
Administration of DP8α cells with F. prausnitzii protects against DSS-induced colitis.
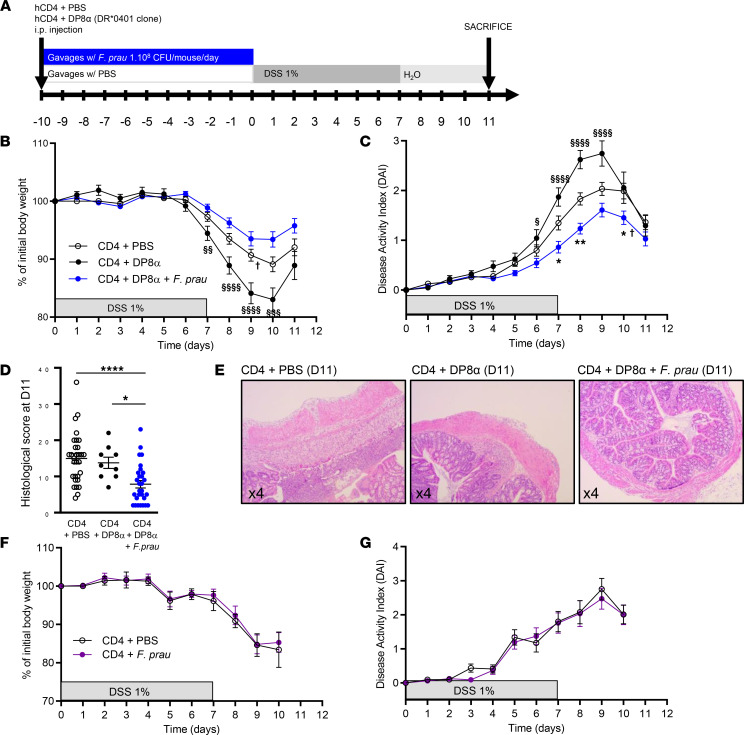
Next, we studied the ability of human DRb1*0401-restricted DP8α Tregs, alone or in combination with F. prausnitzii intragastric gavage, to alleviate DSS-induced colitis in NSG-Ab°DR4 mice. The mice were injected i.p. with human CD4+ peripheral T cells and PBS or DP8α clone and received PBS or F. prausnitzii by gavage for 10 days before the start of DSS treatment for 7 days, followed by a recovery phase of 4 days with regular water (Figure 2A). As shown by assessing body weight loss and disease activity index (DAI), DP8α Tregs combined with F. prausnitzii exhibited protective effects, while the administration of DP8α Tregs alone did not (Figure 2, B and C). The histological score, evaluating the severity, spread of inflammation, and crypt damage, was significantly lower in the CD4+DP8α Tregs+F. prausnitzii group compared with the CD4+PBS and CD4+DP8α cell group (Figure 2, D and E), confirming a protective effect of activated DP8α Tregs.
Figure 2. Administration of DP8α Tregs and F. prausnitzii, but not that of DP8α or F. prausnitzii alone, protects NSG-Ab°DR4 mice from DSS-induced colitis.
(A) Experimental outline: NSG-Ab°DR4 female mice were injected i.p. with PBS or human peripheral CD4 effector T cells with either a human DRb1*0401-restricted DP8α Treg clone or PBS and received daily gavage with 1 × 108 CFU of F. prausnitzii or 200 μL vehicle 1X PBS for 10 days before 1% DSS supplementation in drinking water for 7 days, followed by 4 days of regular drinking water. (B) Body weight and (C) disease activity index (DAI) were evaluated in the CD4+PBS (n = 44), CD4+DP8α (n = 20), and CD4+DP8α+F. prausnitzii (n = 45) groups in 5 independent experiments. (D and E) Histological score was assessed in the CD4+PBS (n = 30), CD4+DP8α (n = 9), and CD4+DP8α+F. prausnitzii (n = 29) groups in 3 independent experiments. (F) Body weight and (G) DAI were also evaluated in the CD4+PBS group (n = 10, open circles), as compared with the CD4+F. prausnitzii group (n = 11, purple circles) in another series of 2 independent experiments. Body weight and DAI are presented as the mean ± SEM. For comparison between multiple groups, 1-way ANOVA was performed, and P values of less than 0.05 were considered significant (*P < 0.05, **P < 0.01, ****P < 0.0001, §P < 0.05, §§P < 0.01, §§§P < 0.001, §§§§P < 0.0001). Only significant statistical results after adjustment for false discovery rate are shown (Benjamini-Hochberg, q < 0.1). The symbol † indicates the beginning of mortality.
Importantly, the intragastric administration of F. prausnitzii alone, without DP8α cells, did not have any protective effect, ruling out an independent effect of the bacterium (Figure 2, F and G). Moreover, there was no protective effect of DP8α Tregs combined with F. prausnitzii when using the DR-restricted DRb1*0408/DRb1*1396-expressing DP8α clone (Supplemental Figure 2, A–C), which did not recognize F. prausnitzii on BM-DCs from NSG-Ab°DR4 mice (Supplemental Figure 1, D and F).
DP8α cells administered with F. prausnitzii decrease gut inflammation and ameliorate histological score in DSS-induced colitis in mice.
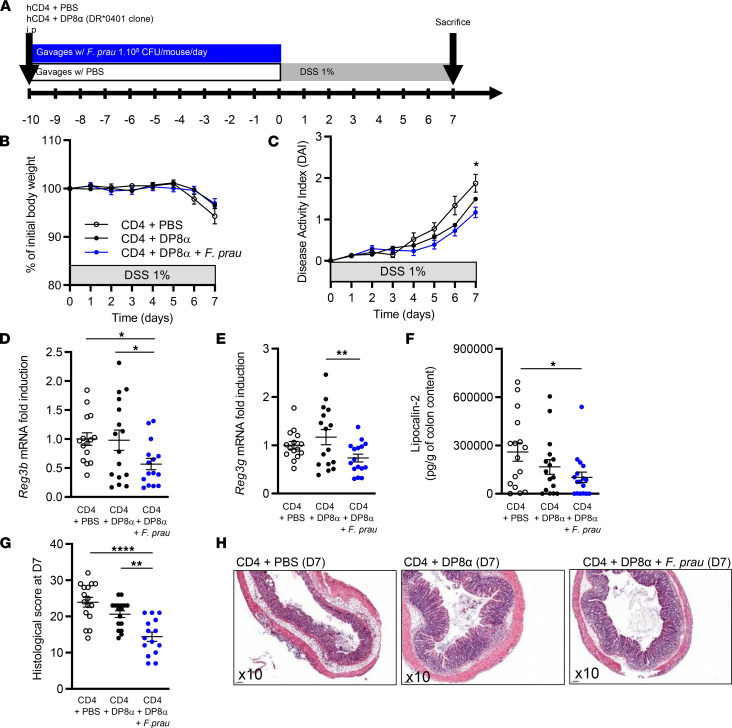
To further compare mice receiving CD4+DP8α Tregs+F. prausnitzii versus CD4+PBS control mice, we sacrificed mice (n = 16–17 per group) earlier, at day 7 after DSS initiation, and collected colon samples to evaluate inflammation severity (Figure 3A).
Figure 3. Administration of DP8α Tregs and F. prausnitzii lowers inflammation induced by DSS-induced colitis in NSG-Ab°DR4 mice.
(A) Experimental outline: NSG-Ab°DR4 female mice were injected i.p. with 2 × 106 human peripheral CD4 effector T cells alone or in combination with 2 × 106 human DRb1*0401-restricted DP8α clones and received daily gavage with 200 μL 1X PBS or 1 × 108 CFU of F. prausnitzii, respectively, for 10 days before 1% DSS supplementation in drinking water for 7 days. Mice were sacrificed at day 7. (B) Body weight and (C) disease activity index were assessed during the protocol in all groups of mice. (D and E) mRNA levels of Reg3b and Reg3g were analyzed by RT-qPCR in the proximal colon at day 7. (F) Lipocalin-2 secretion (pg/g of colon content) was measured by ELISA in the colon content at day 7. (G and H) Histological score was obtained from the colons of mice at day 7. Original magnification, ×10. Results are represented as the mean ± SEM. For comparison between multiple groups, 1-way ANOVA was performed, and P values of less than 0.05 were considered significant (*P < 0.05, **P < 0.01, ****P < 0.0001). Only significant statistical results after adjustment for false discovery rate are shown (Benjamini-Hochberg, q < 0.1). Each figure is representative of n = 3 independent experiments (CD4+PBS, n = 17; CD4+DP8α, n = 16; CD4+DP8α+F. prausnitzii, n = 17).
At day 7, no statistically significant difference was observed regarding body weight loss (Figure 3B). However, statistically significant protection was observed regarding DAI in the CD4+DP8α Tregs+F. prausnitzii group (Figure 3C). Due to the important immunodeficiency of NSG mice, the numerous cytokines we explored (namely murine and human IL10, IL1B, TNFA, IL-17A, and IL-22) were either weakly expressed and not significantly different between the study groups or not detected (data not shown). Murine Cxcl1, Cxcl2, and Cxcl5, expressed by innate immune cells, were detected by RT-qPCR in the distal and proximal colon but no statistically significant difference was observed between the CD4+DP8α Tregs+F. prausnitzii and CD4+PBS groups (data not shown). However, we showed that mRNA expression of antimicrobial C-type lectin molecules regenerating islet-derived protein 3 β (Reg3b) and 3 γ (Reg3g) were significantly decreased in the proximal colons of CD4+DP8α+F. prausnitzii mice when compared with either the CD4+DP8α group or the CD4+PBS group for Reg3b (Figure 3, D and E). At the protein level, a lower abundance of the intestinal inflammation marker lipocalin-2 (Lcn-2) was observed in the total colon content of mice treated with DP8α+F. prausnitzii, as compared with that in controls (Figure 3F). The histological score was significantly lower in the CD4+DP8α Tregs+F. prausnitzii group, confirming a protective effect of activated DP8α Tregs (the combination of DP8α and F. prausnitzii) but not DP8α Tregs alone (Figure 3, G and H).
Altogether, these data show that the administration of DP8α Tregs together with their cognate antigen, F. prausnitzii, ameliorates DSS-induced colitis in humanized mice, emphasizing the role of F. prausnitzii–activated DP8α cells in protecting against intestinal inflammation.
The number of circulating CCR6+CXCR6+ DP8α Tregs and the quantity of F. prausnitzii in fecal microbiota are positively correlated in healthy controls and patients with CD with ileal involvement.
To evaluate the translational relevance of our findings in mice, we took advantage of a large cohort of 250 patients with IBD (185 with CD and 65 with UC; clinical characteristics are described in Supplemental Tables 1 and 2) and 73 healthy controls (HCs). Human PBMCs were isolated and analyzed by flow cytometry to evaluate the frequency and phenotype of blood DP8α cells. DNA from stool samples was extracted in a subcohort of 52 patients with IBD and 10 HCs to quantify F. prausnitzii in the fecal microbiota.
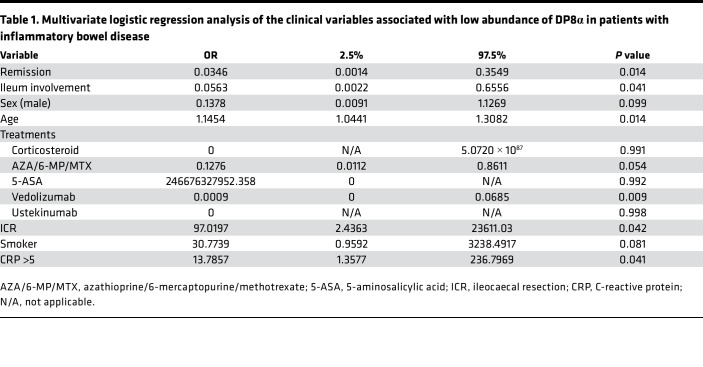
Multivariate logistic regression identified several clinical parameters independently associated with the abundance of DP8α cells in peripheral blood (Table 1). Interestingly, low frequency (less than median) of DP8α cells was associated with parameters of disease activity, including flare and elevated CRP (>5), supporting a potential antiinflammatory role of these cells. The other factors associated with a low frequency of DP8α cells included previous ileal resection and the absence of ileal involvement.
Table 1. Multivariate logistic regression analysis of the clinical variables associated with low abundance of DP8α in patients with inflammatory bowel disease.
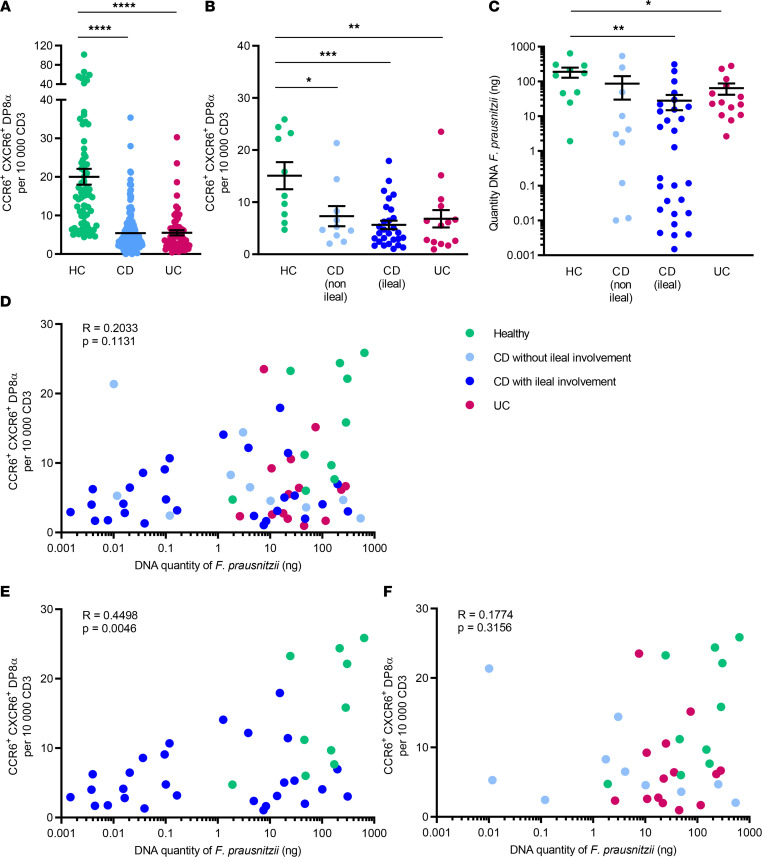
As previously demonstrated (22), we confirmed that the frequency of CCR6+CXCR6+ DP8α cells per 10,000 CD3+ T cells was decreased in patients with CD and UC, as compared with healthy controls in the whole cohort (Figure 4A) and also in the subcohort of patients with available fecal microbiota data (Figure 4B). This decrease was seen in both UC and CD, independently of ileal involvement. The quantity of fecal F. prausnitzii was also significantly decreased in patients with ileal CD and UC, as compared with controls, although the signal was stronger in patients with CD and ileal involvement (Figure 4C), consistent with previous findings (27). Interestingly, we observed that the number of circulating CCR6+CXCR6+ DP8α cells, previously described as specific for F. prausnitzii (22), was positively correlated with the abundance of F. prausnitzii in the fecal microbiota of HCs and patients with CD with ileal involvement but not in patients with purely colonic CD and UC (Figure 4, D–F).
Figure 4. The frequency of circulating CCR6+CXCR6+ DP8α Tregs and the quantity of F. prausnitzii DNA in fecal microbiota are both decreased in patients with IBD and are positively correlated in patients with Crohn’s disease with ileal involvement.
(A) Frequency of circulating CCR6+CXCR6+ DP8α cells per 10,000 CD3+ T cells in the whole cohort of healthy controls (HC, n = 73), patients with Crohn’s disease (CD, n = 185), and patients with ulcerative colitis (UC, n = 65) or (B) according to localization (with or without ileal involvement) in a subcohort of HCs (n = 10) and patients with IBD (n = 10 CD without ileal involvement, n = 28 CD with ileal involvement, and n = 14 patients with UC). (C) Quantity of F. prausnitzii DNA in stool samples in HCs and patients with IBD. (D–F) Spearman’s correlation of the frequency of circulating CCR6+CXCR6+ DP8α cells per 10,000 CD3+ T cells and the quantity of F. prausnitzii DNA in fecal microbiota in the different groups. Results in A–C are represented as the mean ± SEM, For comparison between multiple groups, 1-way ANOVA was performed, and P values of less than 0.05 were considered significant (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). Only significant statistical results after adjustment for false discovery rate are shown (Benjamini-Hochberg, q < 0.1).
Discussion
In this study, we demonstrated the protective role of DP8α Tregs in vivo in the development of DSS-induced colitis in a model of immunodeficient humanized mice. In presence of CD4+ effector T cells, the administration of DR*0401-restricted DP8α Tregs in combination with F. prausnitzii intragastric gavage attenuated colitis severity, as assessed by the decrease of the DAI, body weight loss, Lcn-2 concentration in colon content, and RNA expression of the antimicrobial molecules Reg3b and Reg3g in the colons of NSGAb°DR*0401 mice, while the administration of DP8α Tregs or F. prausnitzii alone had no protective effect. This underlines the importance of F. prausnitzii stimulation to activate DP8α Tregs to exert their antiinflammatory role and suggests that these Tregs could play a role in intestinal homeostasis and in the control of intestinal inflammation. We had previously demonstrated the protective effect of F. prausnitzii and its supernatant in a trinitrobenzene sulfonic acid–induced acute colitis model (13) and in a model of chronic colitis using dinitrobenzene sulfonic acid (12). However, it is difficult to compare those studies with the present one, as the colitis model and mouse strains are different. In other models of colitis, the effect of F. prausnitzii was not clearly demonstrated. Kawade et al. showed that live but not inactivated F. prausnitzii was able to prevent the effect of DSS-induced colitis in a model of BALB/c mice (28), and another study showed that it is the administration of F. prausnitzii supernatant that ameliorates DSS colitis in C57BL/6J mice and that this effect is mediated by Th17 cells (29). In the present study, we used F. prausnitzii, in particularly immunodeficient mice with defects in innate immunity and lacking adaptive immune cells, in which the only adaptive immune cells present are, therefore, the human CD4 effector T cells and DP8α Tregs administered i.p. So, the system we set up is designed to specifically test the effect of DP8α Tregs. In humans, it is likely that F. prausnitzii acts through different mechanisms and targets, including DP8α cells but also epithelial cells (30, 31) and antigen-presenting cells likely responsible for DP8α Treg induction/priming (24).
To extend our knowledge regarding the clinical relevance of DP8α abundance and its link with F. prausnitzii, we took advantage of a large cohort of 250 patients with IBD. Interestingly, we observed that a low frequency of circulating total DP8α cells was associated with several parameters of disease activity (including flare status and elevated CRP) and also with previous ileocecal resection and the absence of ileal involvement. These findings suggest that patients with CD with severe terminal ileum involvement who required ileocecal resection and thus displayed a more severe phenotype have fewer DP8α cells. Although we observed that F. prausnitzii abundance was decreased in patients with ileal CD and UC, patients with both CD and ileal involvement exhibited the lowest levels of F. prausnitzii, consistent with previous findings (27). In these patients, we also observed a high heterogeneity in the quantity of F. prausnitzii, with two distinct groups of patients (high and low F. prausnitzii), that is not entirely explained by localization of the disease (ileal or ileocolonic), previous surgery, or activity of the disease. In 2008, we showed that a low proportion of F. prausnitzii on resected ileal Crohn mucosa constituted a risk factor for endoscopic recurrence at 6 months (13). Low abundance of F. prausnitzii has been confirmed in numerous CD populations ever since, but some studies indicate that this mostly applies to patients with ileal CD (27, 32).
To evaluate the potential link between F. prausnitzii and the induction of DP8α cells in humans, we analyzed the correlation between the number of F. prausnitzii–specific DP8α T cells, identified by the surface expression of CCR6 and CXCR6 (22), and the amount of fecal F. prausnitzii. We observed a statistically significant positive correlation between both parameters only in healthy individuals and patients with CD with ileal involvement but not in patients with purely colonic CD or UC. This might be due to the largest decrease in F. prausnitzii being found in patients with CD with ileal involvement, which allowed us to detect more distinctly correlations with DP8α cell abundance.
A main limitation of the present study relies in the rather low strength of the association between circulating DP8α Tregs and fecal F. prausnitzii (r = 0.45). The relationship between both variables is probably highly complex and depends on multiple different factors. This correlation might be strengthened by increasing the number of patients; unfortunately, we were able to collect stool samples from only 20.8% of the initial cohort of 250 patients with IBD. It would also be of major interest to study the association between ileocolonic LP DP8α and F. prausnitzii in patients with IBD, because circulating DP8α cells do not necessarily represent their intestinal counterparts. However, our data support the protective role of DP8α Tregs observed in vivo in DSS-induced colitis and highlight the potential role of F. prausnitzii as a direct inducer of mucosal DP8α cells in humans.
In conclusion, we showed that DP8α cells activated by F. prausnitzii protect against intestinal inflammation in vivo. The detailed underlying mechanisms remain to be deciphered. The association among low circulating levels of DP8α cells, disease activity, and inflammation supports the antiinflammatory role of these cells in humans. Although it does not prove any causal relationship, the association between blood F. prausnitzii–specific CCR6+CXCR6+ DP8α cells and the abundance of fecal F. prausnitzii in ileal CD supports the role of F. prausnitzii in inducing DP8α Tregs in humans with a potential role in gut homeostasis. Such a role is also supported both by the T cell receptor specificity of DP8α Tregs for this bacterium (22, 23) and mechanistically by the ability of F. prausnitzii to induce IL-10– and IL-27–secreting dendritic cells (24), which are well-known inducers of IL-10–secreting Tregs (33, 34). Supplementing the gut microbiota of patients with IBD with F. prausnitzii, notably to induce DP8α cells, could therefore represent a promising therapeutic strategy.
Methods
Further information can be found in Supplemental Methods.
Participants.
Peripheral blood and stool samples were obtained from healthy individuals and individuals with IBD prospectively recruited between 2018 and 2020 in the Gastroenterology Department of the Saint Antoine Hospital (AP-HP). Clinical characteristics and treatments of patients recruited to the study can be found in Supplemental Tables 1 and 2. Human PBMCs derived from healthy volunteers, who gave informed consent, at the EFS. PBMCs were isolated by Ficoll gradient centrifugation and used to sort polyclonal CD4+ T cells or generate F. prausnitzii–specific DP8α Treg clones, as detailed below, and were also used as HCs for patients with IBD.
Culture of F. prausnitzii A2-165.
F. prausnitzii strain A2-165 (DSMZ collection, Braunschweig, Germany; DSM no. 17677) was grown overnight at 37°C in anaerobic conditions (90% N2, 5% CO2, and 5% H2) in LyBHI (brain-heart infusion medium supplemented with 0.5% yeast extract [Difco Laboratories] and 5 mg/L hemin) supplemented with cellobiose (1 mg/mL; MilliporeSigma), maltose (1 mg/mL, MilliporeSigma), and cysteine (0.5 mg/mL, MilliporeSigma). A2-165 was pelleted, resuspended in PBS, and stored at –80°C. A2-165 was oxygen-inactivated before administration to mice.
Mouse model.
NOD.Cg-Prkdcscid Il2rgtm1Wjl H2-Ab1tm1Gru Tg(HLA-DRB1)31Dmz/SzJ (NSG-Ab°DR4) mice are immunodeficient mice that lack murine T cells, B cells and NK cells, humanized for the expression of a human MHC class II allele (HLA-DR*0401) in absence of murine MHC II. The animals were purchased from The Jackson Laboratories and housed and bred under specific pathogen– and opportunistic pathogen–free conditions at the Saint-Antoine Research Center (Plateforme d’Hébergement et d’expérimentation animale; Centre De Recherche scientifique Saint-Antoine; INSERM UMR_S938) and fed ad libitum. Ten- to twelve-week-old female mice were assigned to the experimental groups after matching age and weight, and the animals were group housed in the same cage.
Human cell isolation, cloning, and expansion.
Human PBMCs were isolated by Ficoll gradient centrifugation from healthy donor blood (EFS) and HLA typing was performed (EFS). CD4 T cells from HLA-DRb1*04+ donors were magnetically purified using CD4 microbeads according to the supplier’s instructions (Miltenyi Biotec), while CD3+CD4+CD8αloCCR6+CXCR6+ F. prausnitzii–specific DP8α Tregs were sorted and cloned using a BD FACSAria III. CD4+ T cells and DP8α Treg clones were expanded on feeder cells, as we previously described (22).
DSS-induced colitis in humanized mice.
The 10- to 12-week-old NSG-Ab°DR4 mice were injected i.p. with 2 × 106 human peripheral CD4 T cells isolated from a HLA DR*0401+ healthy donor, alone or in combination with 2 × 106 cells from F. prausnitzii–reactive DP8α Treg clones (either the HLA-DR*0401–restricted clone, obtained from a homozygous DR*0401 donor, or the HLA-DR*0408–restricted one). Following the injection of human cells, the mice received daily intragastric gavages with either 200 μL of PBS alone or 200 μL of the A2-165 F. prausnitzii strain in PBS (approximately 1 × 108 CFU/mouse/d) for 10 days, before the start of colitis induction. To induce colitis, mice were administered DSS (36,000–50,000 Da; MP Biomedicals) at 1%–1.5% in drinking water ad libitum for 7 days and were then sacrificed or allowed to recover by drinking unsupplemented water for 3 to 4 days. In all experiments, body weight, blood in the stool, and stool consistency were analyzed daily. The severity of colitis was assessed using the DAI, with the modified method of Cooper and colleagues (35). Diarrhea was scored as follows: 0, normal; 2, loose stools; 4, watery diarrhea. Blood in stool was scored as follows: 0, normal; 2, slight bleeding; 4, gross bleeding. Weight loss was scored as follows: 0, none; 1, 1–5%; 2, 5%–10%; 3, 10%–15%; 4, <15%. DAI was the average of these scores.
Whole blood staining, isolation of LP cells, and flow cytometry staining in mice.
Whole blood from NSG-Ab°DR4 mice was collected from the submandibular vein before the start of DSS treatment. 100 μL of whole blood was stained with APC-labeled anti-mouse CD45, APC-Vio770–labeled anti-human CD45, PE-Vio770–labeled anti-human CD3, and Vioblue-labeled anti-human CD4 (all from Miltenyi Biotec) for 10 minutes at room temperature and then incubated with red blood cell lysis solution (Miltenyi Biotec) for 20 minutes and fixed in 1% paraformaldehyde solution for 20 minutes. Finally, the cells were washed and resuspended in PBS 0.5% BSA, 2 mM EDTA before acquisition. For isolation of LP immune cells, the colon was washed with cold PBS and minced into 0.5 cm long pieces before incubation in chelating buffer HBSS (MilliporeSigma) with 5% FBS (PAA Laboratories), 5 mM EDTA (MilliporeSigma), and 1 mM 1,4-Dithiothreitol (MilliporeSigma) for 20 minutes at 37°C under agitation. The LP was cleaned of epithelial cells after washes with PBS and filtration through 100 μm mesh cell strainer. Cells of the LP were isolated from fibrous matter using collagenase IV (680 UI/mL, Roche) and DNAse I (1 mg/mL, MilliporeSigma) digestion for 30 minutes at 37°C under agitation. Cells were washed in RPMI 1640 medium (Gibco) supplemented with 10% FBS, 1% penicillin streptomycin (Gibco), 10 mM HEPES (Gibco), and 50 μM β-mercaptoethanol (MilliporeSigma). LP immune cells were enriched at the interface of a 40%:80% Percoll gradient (GE Healthcare Life Sciences), washed, and stained for flow cytometry with the same antibodies used in whole blood, with addition of eFluor 506 fixable viability dye (Thermo Fisher Scientific) to assess cell death. Data were acquired on a CytoFLEX (Beckman Coulter) flow cytometer, and data were analyzed on FlowJo V10 (Tree Star).
qRT-PCR in murine colon.
Proximal colon tissues from NSG-Ab°DR4 mice were harvested after 7 days of DSS treatment and frozen in liquid nitrogen. Tissues were transferred in Lysing Matrix D tubes (MP Biomedicals) and homogenized on a FastPrep (MP Biomedicals) bead beating machine. RNA was extracted from homogenized tissues using an RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. RNA samples were reversed transcribed using the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). qPCR were performed using SYBR Green PCR Master Mix (Applied Biosystems) in a StepOnePlus apparatus (Applied Biosystems). The oligonucleotides used were as follows: Rplp0 — sense, 5′-AGATTCGGGATATGCTGTTGGC-3′; antisense, 5′-TCGGGTCCTAGACCAGTGTTC-3′; Reg3b — sense, 5′-ATGCTGCTCTCCTGCCTGATG-3′; antisense, 5′-CTAATGCGTGCGGAGGGTATATTC-3′; Reg3g — sense, 5′-TTCCTGTCCTCCATGATCAAAA-3′; antisense, 5′-CATCCACCTCTGTTGGGTTCA-3′. The 2−ΔΔCt quantification method was used for the analyses with mouse Rplp0 as an endogenous control and normalization to CD4+PBS-treated mice.
Quantification of colon Lcn-2 levels.
During sacrifice and sample collection, colon contents were collected from NSG-Ab°DR4 mice and frozen for further analyses. Frozen colon contents were weighted and suspended in PBS. Samples were vortexed for 20 minutes to get a homogenous suspension and centrifuged for 10 minutes at 10,000g at 4°C. Clear supernatants were collected ad stored at –80°C until analysis. Lcn-2 levels were measured using a Duoset murine Lcn-2 ELISA Kit (R&D Systems) according to the manufacturer’s instructions, and results are expressed as pg/g of colon content.
Histological score.
Colon fragments taken from the midpart of the colon were fixed in 4% paraformaldehyde solution and embedded in paraffin, and 5 mm sections were stained with hematoxylin and eosin for histological scoring. Tissues were scored blindly using established methods for DSS colitis as previously described (4). Inflammation severity, inflammation extent, and crypt damage were graded. For each feature, the product of the grade and the percentage involvement was established. The histological score was obtained by adding the subscore of each feature.
Flow cytometry to track circulating DP8α Tregs in patients and HCs.
Human PBMCs were isolated by Ficoll gradient centrifugation. After isolation, PBMCs were stained for 45 minutes at 4°C in PBS 0.1% bovine serum albumin with the following antibodies: anti-human CD3-PE-Cy7 (clone UCHT1, BD), anti-human CD4-FITC (clone 13B8.2, Beckman Coulter), anti-human CD8a-BV421 (clone RPA-T8, BD), anti-human CCR6-PE (clone G034E3, Biolegend), and anti-human CXCR6-APC (clone K041E5, Biolegend). Fluorescence was measured on a BD LSR II flow cytometer (BD Biosciences) and analyzed using FlowJo or DIVA softwares. DP8α Tregs (CD3+CD4+CD8αloCCR6+CXCR6+ cells) were then quantified among total CD3+ cells, as described previously (22).
DNA extraction and bacterial quantification in fecal microbiota of patients and HCs.
Fecal genomic DNA was extracted from the stool samples using a previously described method (4), which is based on the Godon DNA extraction method (36). More precisely, the feces samples were weighed and then resuspended for 10 minutes at room temperature in 250 μL 4 M guanidine thiocyanate in 0.1 M Tris-HCl (pH 7.5) (MilliporeSigma) and 40 μL of 10% N-lauroyl sarcosine (MilliporeSigma). After the addition of 500 μL of 5% N-lauroyl sarcosine in 0.1 M phosphate buffer (pH 8.0), the 2 mL tubes were incubated at 70°C for 1 hour. One volume (750 mL) of 0.1 mm–diameter silica beads (MilliporeSigma) (previously sterilized by autoclaving) was added, and the tube was shaken at 6.5 m/s 3 times for 30 seconds each in a FastPrep apparatus (MP Biomedicals). 15 mg of polyvinylpolypyrrolidone (MilliporeSigma) was added to the tube, which was then vortexed and centrifuged for 5 minutes at 20,000g at 4°C. After recovery of the supernatant, the pellets were washed with 500 μL TENP (50 mM Tris [pH 8], 20 mM EDTA [pH 8], 100 mM NaCl, 1% polyvinylpolypyrrolidone) and centrifuged for 5 minutes at 20,000g at 4°C, and the new supernatant was added to the first supernatant. The washing step was repeated 2 times. The pooled supernatant (about 2 mL) was briefly centrifuged to remove particles and then split into two 2 mL tubes. Nucleic acids were precipitated by the addition of 1 volume of isopropanol for 10 minutes at room temperature and centrifugation for 10 minutes at 20,000g at 4°C. Pellets were resuspended and pooled in 450 μL 100 mM phosphate buffer, pH 8, and 50 μL 5 M potassium acetate. The tube was placed at 4°C overnight and centrifuged at 20,000g for 30 minutes. The supernatant was then transferred to a new tube containing 20 μL RNase (1 mg/mL) and incubated at 37°C for 30 minutes. Nucleic acids were precipitated by the addition of 50 μL 3 M sodium acetate and 1 mL absolute ethanol. The tube was incubated for 10 minutes at room temperature, and the nucleic acids were recovered by centrifugation at 20,000g for 15 minutes. The DNA pellet was finally washed with 70% ethanol, dried, and resuspended in 100 μL Tris–EDTA buffer. DNA suspensions were stored at −20°C for real-time quantitative PCR (qPCR) analysis. Quantifications of F. prausnitzii were performed by qPCR using SYBR Green PCR Master Mix (Applied Biosystems) in a StepOnePlus apparatus (Applied Biosystems). Each reaction was done in duplicate in a final volume of 20 μL with 0.2 μM of each primer and 5 μL of the appropriate dilution of DNA. F. prausnitzii was quantified using specific primers: sense, 5′-CCATGAATTGCCTTCAAAACTGTT-3′, and antisense, 5′-GAGCCTCAGCGTCAGTTGGT-3′ (13). Amplifications were performed with the following conditions: denaturation step, 10 minutes at 95°C; 40 cycles of 95°C denaturation for 15 seconds; 55°C extension for 1 minutes, followed by a standard melting curve program. We used quantified F. prausnitzii DNA as reference standard to quantify F. prausnitzii DNA in each sample.
Statistics.
GraphPad Prism version 6.0 was used for statistical analysis in mouse experiments, comparisons of DP8α frequencies or abundance of F. prausnitzii between healthy donors and patients, and for all graphs representations. For data displayed in graphs, the results are presented as the mean ± SEM. For comparison between multiple groups, 1-way ANOVA was performed, and P values of less than 0.05 were considered significant. Only significant statistical results after adjustment for false discovery rate are shown (Benjamini-Hochberg, q < 0.1). Spearman’s correlations were used for correlations between DP8α frequencies and abundance of fecal F. prausnitzii in HCs and patients. Statistical analysis of the patient cohort was performed in the R statistical environment (version 3.6.2). For multivariate logistic regression with stepwise variable selection, the variables taken into consideration were: sex, age, disease (UC/CD), activity (flare/remission), topography (ileum, colon), treatment, history of surgery (ileocecal resection, colectomy, ileostomy), smoking (active or not), and CRP (elevated or normal).
Study approval.
All human samples were collected with informed consent. Approval for human studies was obtained from the local ethics committee (Comité de Protection de Personnes Ile de France IV, IRB 00003835 Suivitheque study; registration no. 2012/05NICB). For isolation of PBMCs to purify and clone CD4+ T cells and DP8α Treg clones, blood samples were obtained from EFS (convention no. CPDL-PLER-2017 21). All animal experiments were carried out in strict accordance with the French national and European laws and conformed to the Council Directive on the approximation of laws, regulations, and administrative provisions of the Member States regarding the protection of animals used for experimental and other scientific purposes (86/609/Eec). All animal experiments were conducted according to the institutional guidelines approved by the Comité d’éthique en expérimentation animale no. 5 Charles Darwin, Paris, France (reference no. APAFIS#8231-2016121612402727 v2).
Author contributions
FJ, FA, and HS conceptualized the study. ST, EG, and NR provided methodology. ST, EG, NR, CD, CO, MS, CG, LB, IAS, TL, and SC provided investigation. ST and HS provided formal analysis. ST, NR, and HS wrote the original draft. ST, EG, NR, FJ, FA, and HS reviewed and edited the manuscript. FA, FJ, and HS acquired funding. TL provided mice. JMC and PL provided bacteria. HS provided human data and samples. FJ, FA, and HS supervised the study. The order of first authorship was determined randomly.
Supplementary Material
Acknowledgments
The authors thank the patients recruited in the Gastroenterology Department of Saint-Antoine Hospital (AP-HP) and the nurses, and the members of the animal core facility (Plateforme d’Hébergement et d’expérimentation animale, Saint-Antoine Research Center). The authors also thank B. Solhonne (Saint-Antoine Research Center) for her contribution for histology staining, M. Mohty and B. Gaugler (Saint-Antoine Research Center) for the use of the CytoFLEX flow cytometer, and M-L Michel (INRAE) for advice on murine LP isolation. The graphical abstract was created with Biorender.com.
Version 1. 05/10/2022
In-Press Preview
Version 2. 06/22/2022
Electronic publication
Footnotes
Conflict of interest: HS has received unrestricted study grants from Danone, Biocodex, and Enterome; board membership, consultancy, or lecture fees from Carenity, Abbvie, Astellas, Danone, Ferring, Mayoly Spindler, MSD, Novartis, Roche, Tillotts, Enterome, Maat, BiomX, Biose, Novartis, and Takeda; and is a cofounder of Exeliom Biosciences.
Copyright: © 2022, Touch et al. This is an open access article published under the terms of the Creative Commons Attribution 4.0 International License.
Reference information: JCI Insight. 2022;7(12):e154722.https://doi.org/10.1172/jci.insight.154722.
Contributor Information
Sothea Touch, Email: sothea.touch2@gmail.com.
Emmanuelle Godefroy, Email: emmanuelle.godefroy@univ-nantes.fr.
Nathalie Rolhion, Email: nathalie.rolhion@inserm.fr.
Camille Danne, Email: camille.danne@inrae.fr.
Cyriane Oeuvray, Email: cyriane.oeuvray@gmail.com.
Marjolène Straube, Email: marjolene.straube@hotmail.fr.
Chloé Galbert, Email: chloe.galbert.crsa@gmail.com.
Loïc Brot, Email: loic.brot@sorbonne-universite.fr.
Sead Chadi, Email: sead.chadi@inrae.fr.
Tatiana Ledent, Email: tatiana.ledent@inserm.fr.
Jean-Marc Chatel, Email: jean-marc.chatel@inrae.fr.
Philippe Langella, Email: philippe.langella@inra.fr.
Francine Jotereau, Email: francine.jotereau@univ-nantes.fr.
Frédéric Altare, Email: frederic.altare@inserm.fr.
Harry Sokol, Email: harry.sokol@aphp.fr.
References
- 1.Goethel A, et al. The interplay between microbes and the immune response in inflammatory bowel disease. J Physiol. 2018;596(17):3869–3882. doi: 10.1113/JP275396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jostins L, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491(7422):119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khor B, et al. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474(7351):307–317. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lamas B, et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med. 2016;22(6):598–605. doi: 10.1038/nm.4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu JZ, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47(9):979–986. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lavelle A, Sokol H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat Rev Gastroenterol Hepatol. 2020;17(4):223–237. doi: 10.1038/s41575-019-0258-z. [DOI] [PubMed] [Google Scholar]
- 7.Nishida A, et al. Gut microbiota in the pathogenesis of inflammatory bowel disease. Clin J Gastroenterol. 2018;11(1):1–10. doi: 10.1007/s12328-017-0813-5. [DOI] [PubMed] [Google Scholar]
- 8.Gevers D, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15(3):382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lloyd-Price J, et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019;569(7758):655–662. doi: 10.1038/s41586-019-1237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sokol H, et al. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis. 2009;15(8):1183–1189. doi: 10.1002/ibd.20903. [DOI] [PubMed] [Google Scholar]
- 11.Miquel S, et al. Faecalibacterium prausnitzii and human intestinal health. Curr Opin Microbiol. 2013;16(3):255–261. doi: 10.1016/j.mib.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 12.Martín R, et al. The commensal bacterium Faecalibacterium prausnitzii is protective in DNBS-induced chronic moderate and severe colitis models. Inflamm Bowel Dis. 2014;20(3):417–430. doi: 10.1097/01.MIB.0000440815.76627.64. [DOI] [PubMed] [Google Scholar]
- 13.Sokol H, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105(43):16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang M, et al. Faecalibacterium prausnitzii inhibits interleukin-17 to ameliorate colorectal colitis in rats. PLoS One. 2014;9(10):e109146. doi: 10.1371/journal.pone.0109146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Britton GJ, et al. Microbiotas from humans with inflammatory bowel disease alter the balance of gut Th17 and RORγt+ regulatory T cells and exacerbate colitis in mice. Immunity. 2019;50(1):212–224. doi: 10.1016/j.immuni.2018.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benech N, Sokol H. Fecal microbiota transplantation in gastrointestinal disorders: time for precision medicine. Genome Med. 2020;12(1):58. doi: 10.1186/s13073-020-00757-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Atarashi K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500(7461):232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 18.Maynard CL, et al. Regulatory T cells expressing interleukin 10 develop from Foxp3+ and Foxp3- precursor cells in the absence of interleukin 10. Nat Immunol. 2007;8(9):931–941. doi: 10.1038/ni1504. [DOI] [PubMed] [Google Scholar]
- 19.Saruta M, et al. Characterization of FOXP3+CD4+ regulatory T cells in Crohn’s disease. Clin Immunol. 2007;125(3):281–290. doi: 10.1016/j.clim.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 20.Park O, et al. Analysis of the Foxp3/scurfin gene in Crohn’s disease. Ann N Y Acad Sci. 2005;1051(1):218–228. doi: 10.1196/annals.1361.125. [DOI] [PubMed] [Google Scholar]
- 21.Lord JD. Promises and paradoxes of regulatory T cells in inflammatory bowel disease. World J Gastroenterol. 2015;21(40):11236–11245. doi: 10.3748/wjg.v21.i40.11236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Godefroy E, et al. Expression of CCR6 and CXCR6 by gut-derived CD4+/CD8α+ T-regulatory cells, which are decreased in blood samples from patients with inflammatory bowel diseases. Gastroenterology. 2018;155(4):1205–1217. doi: 10.1053/j.gastro.2018.06.078. [DOI] [PubMed] [Google Scholar]
- 23.Sarrabayrouse G, et al. CD4CD8αα lymphocytes, a novel human regulatory T cell subset induced by colonic bacteria and deficient in patients with inflammatory bowel disease. PLoS Biol. 2014;12(4):e1001833. doi: 10.1371/journal.pbio.1001833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alameddine J, et al. Faecalibacterium prausnitzii skews human DC to prime IL10-producing T cells through TLR2/6/JNK signaling and IL-10, IL-27, CD39, and IDO-1 induction. Front Immunol. 2019;10:143. doi: 10.3389/fimmu.2019.00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kashiwagi I, et al. Smad2 and Smad3 inversely regulate TGF-β autoinduction in clostridium butyricum-activated dendritic cells. Immunity. 2015;43(1):65–79. doi: 10.1016/j.immuni.2015.06.010. [DOI] [PubMed] [Google Scholar]
- 26.Goettel JA, et al. Fatal autoimmunity in mice reconstituted with human hematopoietic stem cells encoding defective FOXP3. Blood. 2015;125(25):3886–3895. doi: 10.1182/blood-2014-12-618363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Willing B, et al. Twin studies reveal specific imbalances in the mucosa-associated microbiota of patients with ileal Crohn’s disease. Inflamm Bowel Dis. 2009;15(5):653–660. doi: 10.1002/ibd.20783. [DOI] [PubMed] [Google Scholar]
- 28.Kawade Y, et al. Administration of live, but not inactivated, Faecalibacterium prausnitzii has a preventive effect on dextran sodium sulfate‑induced colitis in mice. Mol Med Rep. 2019;20(1):25–32. doi: 10.3892/mmr.2019.10234. [DOI] [PubMed] [Google Scholar]
- 29.Huang X-L. Faecalibacterium prausnitzii supernatant ameliorates dextran sulfate sodium induced colitis by regulating Th17 cell differentiation. World J Gastroenterol. 2016;22(22):5201–5210. doi: 10.3748/wjg.v22.i22.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lenoir M, et al. Butyrate mediates anti-inflammatory effects of Faecalibacterium prausnitzii in intestinal epithelial cells through Dact3. Gut Microbes. 2020;12(1):1–16. doi: 10.1080/19490976.2020.1826748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quévrain E, et al. The presence of the anti-inflammatory protein MAM, from Faecalibacterium prausnitzii, in the intestinal ecosystem. Gut. 2016;65(5):882. doi: 10.1136/gutjnl-2015-311094. [DOI] [PubMed] [Google Scholar]
- 32.Willing BP, et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology. 2010;139(6):1844–1854. doi: 10.1053/j.gastro.2010.08.049. [DOI] [PubMed] [Google Scholar]
- 33.Comi M, et al. Interleukin-10-producing DC-10 is a unique tool to promote tolerance via antigen-specific T regulatory type 1 cells. Front Immunol. 2018;9:682. doi: 10.3389/fimmu.2018.00682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takenaka MC, Quintana FJ. Tolerogenic dendritic cells. Semin Immunopathol. 2017;39(2):113–120. doi: 10.1007/s00281-016-0587-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cooper HS, et al. Clinicopathologic study of dextran sulfate sodium experimental murine colitis. Lab Invest. 1993;69(2):238–249. [PubMed] [Google Scholar]
- 36.Godon JJ, et al. Molecular microbial diversity of an anaerobic digestor as determined by small-subunit rDNA sequence analysis. Appl Environ Microbiol. 1997;63(7):2802–2813. doi: 10.1128/aem.63.7.2802-2813.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.