Abstract
Three particulate methane monooxygenase PCR primer sets (A189-A682, A189-A650, and A189-mb661) were investigated for their ability to assess methanotroph diversity in soils from three sites, i.e., heath, oak, and sitka, each of which was capable of oxidizing atmospheric concentrations of methane. Each PCR primer set was used to construct a library containing 50 clones from each soil type. The clones from each library were grouped by restriction fragment length polymorphism, and representatives from each group were sequenced and analyzed. Libraries constructed with the A189-A682 PCR primer set were dominated by amoA-related sequences or nonspecific PCR products with nonsense open reading frames. The primer set could not be used to assess methanotroph diversity in these soils. A new pmoA-specific primer, A650, was designed in this study. The A189-A650 primer set demonstrated distinct biases both in clone library analysis and when incorporated into denaturing gradient gel electrophoresis analysis. The A189-mb661 PCR primer set demonstrated the largest retrieval of methanotroph diversity of all of the primer sets. However, this primer set did not retrieve sequences linked with novel high-affinity methane oxidizers from the soil libraries, which were detected using the A189-A650 primer set. A combination of all three primer sets appears to be required to examine both methanotroph diversity and the presence of novel methane monooxygenase sequences.
Methanotrophs are a unique group of organisms which can use methane as a sole source of carbon and energy. The ability of methanotrophs to oxidize methane is due to the possession of the enzyme methane monooxygenase. There are two distinct forms of this enzyme, the cytoplasmic soluble methane monooxygenase and the membrane-bound particulate methane monooxygenase (pMMO) (reviewed in references 21 and 22). Only the pMMO is found universally in methanotrophs and can therefore be used as a functional marker for these organisms. No genetic or structural homology is found between these two enzyme systems despite their similar functions. However, the pMMO enzyme complex shares many similarities with the ammonia monooxygenase (AMO) enzyme complex found in ammonia-oxidizing bacteria (15). These similarities include a high degree of amino acid sequence identity, similar protein complex structures, and broadly similar substrate and inhibition profiles, while each play a crucial role in cell metabolism (6, 11, 29). Methanotrophs and ammonia-oxidizing bacteria can oxidize both methane and ammonia; however, they can obtain energy only from the oxidation of methane and ammonium, respectively (3).
Oligonucleotide primers (A189f and A682r) have been designed to amplify internal fragments of the genes encoding the pMMO and AMO enzyme complexes (11). The sequence information obtained from theses genes encoding pMMO (pmoA) and AMO (amoA) has been used as phylogenetic markers for identification of methanotrophs and ammonia oxidizers (12, 19). The phylogeny of these functional genes closely reflects the 16S rRNA phylogeny of the organisms from which the gene sequences were retrieved. Therefore, retrieval of pmoA and amoA gene sequence information provides information on the diversity of these organisms in different environments (12, 19).
The A189 and A682 primers have been used extensively in environmental studies to provide a molecular profile of the methane-oxidizing community (10, 13, 20). Recently a new reverse pmoA-specific primer, mb661, used in conjunction with the A189 primer, was designed and demonstrated specificity to amplify pmoA sequences while not detecting amoA sequences (4). This new primer was used alongside 16S ribosomal DNA phylogenetic probes to determine in situ populations of methanotrophs in freshwater environments (4). The use of PCR primers to amplify the amoA genes from ammonia oxidizers has recently been critically evaluated (24), with the amoA primer set of Rotthauwe et al. (26) being recommended as the most suitable. The soluble methane monooxygenase genes have also been used to detect methanotrophs in the environment (2, 17, 18); however, this procedure detects only the subgroup of methanotrophs that contain this enzyme.
The pmoA PCR primer set A189-A682 has been adapted for denaturing gradient gel electrophoresis (DGGE) analysis as a means to study pmoA gene diversity (5, 9). The use of degenerate primers in DGGE analysis may cause the appearance of multiple bands for individual organisms, which in complex environments may cause confusion in interpretation of results. The A682r primer has four redundancies within its sequence, which is suspected to cause multiple-banding problems in DGGE analysis. This study attempted to design a new pmoA primer set containing no redundancies which could subsequently be applied in DGGE analysis of pmoA.
Aerobic soils, such as forest soils, play an important role in the global methane cycle by acting as a major sink for atmospheric methane in the atmosphere (1, 14, 30). To investigate methanotroph populations involved in methane oxidation in forest soils, pmoA clone libraries were constructed from three Danish soils, i.e., heath, oak, and sitka, and compared. The soils were obtained from a site at Hjelm Hede, Denmark. The soil types are similar. All were originally heath, but part was planted with sitka spruce over 60 years ago and part has been gradually colonized by oak woodland. The change in vegetation has effected a change in the atmospheric oxidation potentials of the soils. A plant succession from heather to oak vegetation increased methane uptake sixfold, while the introduction of sitka spruce doubled methane uptake rates relative to those of the native heathland. A detailed analysis of the physiochemical properties of the soils and their relative methane uptake potentials will be presented elsewhere (I. R. McDonald et al., unpublished data; N. Høegh et al., unpublished data). However, the major aims of this study on these heathland and forest soils were twofold: to evaluate different pmoA primer sets as tools for investigating methanotroph diversity and to investigate the methanotroph diversity in these soils, which demonstrate novel atmospheric methane oxidation potentials.
MATERIALS AND METHODS
Microbial strains and template DNA.
The microorganisms used in this study were obtained from a culture collection of methanotrophs maintained at the University of Warwick and from the National Collection of Industrial and Marine Bacteria (Aberdeen, United Kingdom). Cultures were grown in nitrate mineral salts medium with the addition of excess methane (20% [vol/vol] in air) as the sole carbon substrate as described previously (32). DNA was extracted from cultures using the methods of Marmur (16).
Sample collection and DNA extraction.
Core soil samples were taken from three sites located at Hjelm Hede in Northern Jutland, Denmark. The sampling sites were (i) native heathland (heath), (ii) established oak (oak), and (iii) sitka spruce (sitka). Core soil samples (15-cm diameter, 35-cm depth) were obtained from each of the three vegetation sites using the method of Hall et al. (7). The oak and sitka cores were extruded from the sample tube and sectioned into 5-cm sections before being placed in airtight collection bags. The heath core was sectioned into 5-cm sections down to 15 cm and then into 2-cm sections (15 to 27 cm). The soil samples were sieved (4-mm mesh) to remove stone and roots and to homogenize the soil. Total DNA was extracted from 2 g of each core section using the methods of McDonald et al. (17). High-molecular-mass DNA was excised from a 1% (wt/vol) agarose gel and purified (Geneclean II kit; Bio 101) to remove humic compounds which interfered with PCR amplification. This method yielded consistently high-quality DNA, which could be easily digested with restriction endonucleases and was suitable as a template in PCR amplification experiments. In this study, only DNA extracted from the core sections demonstrating the highest methane oxidation potentials was used (heath, 21 to 23 cm; oak, 5 to 10 cm; sitka, 20 to 25 cm) (Høegh et al., unpublished data)
Design of a new pmoA-specific primer, A650.
The pmoA and amoA sequences of methanotrophs and nitrifiers presently available from the GenBank database were aligned and then scanned for conserved regions within the pmoA gene which could provide a suitable primer target site. From this analysis, a reverse primer, A650r (5′ ACGTCCTTACCGAAGGT 3′), was designed. No unique region at the start of the pmoA sequence could be identified as being suitable for a new forward primer. The A650 primer was used in conjunction with the A189f primer to amplify a 478-bp internal section of the pmoA gene. The primer was tested against a range of methanotrophs and nitrifiers, including Methylococcus capsulatus (Bath), Methylococcus capsulatus (strain M), Methylomicrobium agile (A30), Methylobacter whittenburyi, Methylocaldum tepidum (LK6), Methylomonas methanica (S1), Methylomicrobium album (BG8), Methylosinus trichosporium (OB3b), Methylocystis parvus (OBBP), Methylosphaera hansonii, Nitrosomonas europaea (NCIMB 11850), Nitrosospira sp. (Np22), Nitrosococcus oceanus (NCIMB 11848), Nitrosomonas eutropha, and Nitrosospira multiformis (NCIMB 11849).
PCR amplification.
PCR amplification reactions were performed in 50-μl (total volume) reaction mixtures in 0.5-ml Microfuge tubes using a DNA thermal cycler with a hot lid (Touchdown model; Hybaid, Teddington, Middlesex, United Kingdom). All PCR amplifications of the pmoA gene used the A189f primer in combination with either the A682, A650, or mb661 primer. Individual reagents and their concentrations or amounts were as follows: 1× PCR buffer, 1.5 mM MgCl2, 0.05% W-1 (supplied with the Taq DNA polymerase), 20 μg of bovine serum albumin (Boehringer Mannheim), 200 μmol of each deoxynucleoside triphosphate, 20 pmol of each primer, 1 μl of template DNA (approximately 5 to 50 ng), and 5 U of Taq polymerase (Life Technologies). Each primer set used the same thermal profile. Taq polymerase was added after the initial denaturation step of 96°C for 5 min, followed by 30 cycles of 94°C for 1 min, 56°C for 1 min, and 72°C for 1 min. A final extension period of 5 min at 72°C was included (11).
Construction of clone banks and restriction fragment length polymorphism (RFLP) analysis.
The size and purity of each PCR product were checked on 1% (wt/vol) agarose gels (27), and the products were then ligated into the pCR 2.1 vector supplied with the TA cloning kit (Invitrogen, San Diego, Calif.) according to the manufacturer's instructions. Individual colonies containing inserts were suspended in 3 ml of nutrient broth containing ampicillin (50 μg/ml) and grown overnight at 37°C. Small-scale preparations of plasmids were performed using the methods of Saunders and Burke (28). Plasmids were digested with the restriction enzyme combinations of EcoRI-RsaI and EcoRI-PvuII-HincII. Digests were resolved on 2% (wt/vol) agarose gels and grouped manually, based on the restriction pattern obtained.
DNA sequencing and analysis.
Small-scale preparations of clones from libraries were done by the method of Saunders and Burke (28), and DNA for direct sequencing of DGGE bands was prepared by purification of PCR products using a Wizard PCR purification kit (Promega, Southampton, United Kingdom). DNA sequencing reactions were carried out by cycle sequencing with the Dye Termination kit of PE Applied Biosystems (Warrington, Cheshire, United Kingdom). Phylogenetic analyses of the DNA and deduced amino acid sequences were carried out using the ARB program (http://www.mikro.biologie.tu-muenchen.de). Sequences were manually aligned with the pmoA and amoA sequences obtained from the GenBank database. Regions of sequence ambiguity and incomplete data were excluded from the analyses. Results were depicted as a consensus tree, combining the results of evolutionary distance (Dayhoff percentage of acceptable point mutations model), maximum-parsimony, and maximum-likelihood analyses of the data sets. Multifurcations indicate points where the branching order was not supported by all three methods.
DGGE analysis of the pmoA gene.
A GC clamp (23) was attached to the 5′ end of the pmoA-specific A650 primer. PCR amplification was performed with the GC-A650 primer and the A189 primer. A touchdown PCR program was optimized and consisted of an initial denaturation step of 5 min at 94°C, followed by 20 touchdown cycles (65 to 55°C) and 10 further cycles at 55°C for 1 min, followed by 72°C for 1 min and a final extension of 72°C for 5 min. PCR amplification with the GC-A189–A682 primer set was performed as outlined by Henckel et al. (9). PCR products were analyzed as described above.
PCR products were separated using a Dcode system (Bio-Rad, Munich, Germany) on 1-mm-thick polyacrylamide gels (7.5% [wt/vol] acrylamide-bisacrylamide [37.5:1]) (Bio-Rad) prepared with and electrophoresed in 0.5× TAE (0.02 M Tris base, 0.01 M sodium acetic acid, 0.5 mM EDTA, pH 7.4) at 60°C and a constant voltage. PCR products amplified with the A189–A650-GC primer set were run on a gradient of 55 to 65% (65% corresponds to 7.5% [wt/vol] acrylamide, 4.55 M urea, and 26% [vol/vol] deionized formamide) at a constant voltage of 150 V for 6 h. PCR products amplified with the GC-A189–A682 primers were run according to the procedures of Henckel et al. (9). Gels were stained with 1:50,000 (vol/vol) SYBR-Gold (Molecular Probes) for 30 min before being photographed. Distinct DNA bands were excised and suspended in 100 μl of water overnight to elute DNA. The bands were reamplified and run again on the DGGE system to ensure purity and correct mobility within the gels. Direct sequencing of the DNA bands was performed as described above, and the analysis of derived sequences was also performed as described above.
Nucleotide sequence accession numbers.
The environmental clone sequences have been deposited in the GenBank database under accession numbers AF368354 to AF368374.
RESULTS
Clone library construction.
Three different pmoA-specific primers sets (A189-A682, A189-A650, and A189-mb661) were used to construct clone libraries from three soil samples (heath, oak, and sitka). PCR amplification products of the predicted size were obtained from each of the soils using the three primer sets. Libraries of approximately 50 clones were constructed for each soil and from each pmoA primer set (nine libraries in total). The libraries were subjected to RFLP analysis and grouped based on their representative RFLP patterns. This use of methanotroph-specific primers and operational taxonomic units (OTUs) has been shown to be effective in screening environmental clone libraries and providing an indication of methanotroph diversity (4, 12). In this study, random sequencing within classified OTU groups always demonstrated identical clone sequences, suggesting that the clone groupings based on restriction analysis were robust.
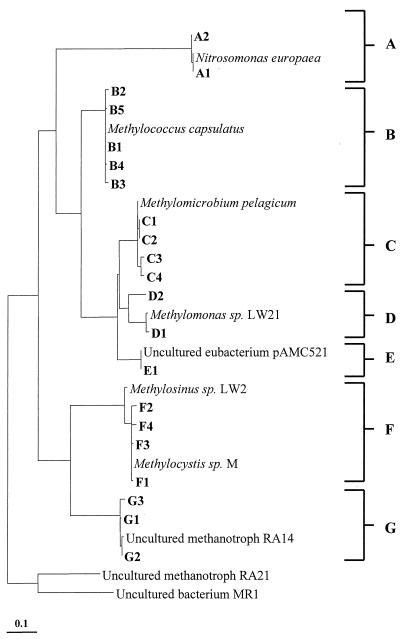
Phylogenetic analysis of sequences of representative clones from each OTU group of each library was performed. All pmoA sequences obtained were affiliated closely with established pmoA groupings and were subsequently assigned to broad phylogenetic groups, designated A through G. Table 1 provides an overview of the proportion of OTU-grouped clones found in each library, while Fig. 1 indicates the phylogenetic affiliations of these clone sequences. The group A sequences (A1 and A2) were related to the amoA genes of Nitrosomonas. Group B sequences (B1, B2, B3, B4, and B5) were related to the pmoA genes of the genus Methylococcus. Group C sequences (C1, C2, C3, and C4) include relatives of the pmoA genes from uncultured bacteria (presumed methanotrophs). Group D sequences (D1 and D2) are affiliated with the pmoA genes of Methylomonas. Group E (E1) includes sequences also affiliated with pmoA sequences from uncultured bacteria (presumed methanotrophs). Group F (F1, F2, F3, and F4) contains sequences related to the pmoA genes of the type II methanotrophs Methylosinus and Methylocystis. Finally, group G sequences (G1, G2, and G3) were related to the recently described environmental pmoA clone RA14.
TABLE 1.
Representative proportions of clones in each OTU group for each clone library
| Soil | Resultsa with primer set
|
|||||
|---|---|---|---|---|---|---|
| A189-A682
|
A189-A650
|
A189-mb661
|
||||
| Clone type | % of library | Clone type | % of library | Clone type | % of library | |
| Heath | A1 | 59 | B2 | 98 | B1 | 46 |
| A2 | 2 | F4 | 2 | C1 | 4 | |
| No match | 29 | C2 | 2 | |||
| No sequence | 10 | D1 | 6 | |||
| E1 | 4 | |||||
| F1 | 28 | |||||
| F3 | 2 | |||||
| No sequence | 8 | |||||
| Oak | A1 | 4 | B1 | 87 | B4 | 24 |
| B1 | 4 | G1 | 9 | C1 | 52 | |
| Miscellaneous | 34 | No sequence | 4 | C4 | 2 | |
| No match | 45 | D2 | 12 | |||
| No sequence | 13 | F2 | 10 | |||
| Sitka | A1 | 66 | B1 | 82 | B1 | 68 |
| Miscellaneous | 19 | B3 | 2 | B5 | 9 | |
| No match | 15 | G2 | 9 | F1 | 23 | |
| G3 | 7 | |||||
No sequence, OTU groups for which no representatives were sequenced; no match, OTU groups for which sequence information was not valid or for which no discernible open reading frame could be identified; miscellaneous, OTU groups containing individual clones which have not been characterized by sequencing.
FIG. 1.
Phylogenetic analysis of the derived amino acid sequences encoded by pmoA and amoA genes retrieved from the Danish soils. Bar, 10 inferred substitutions per 100 amino acid positions. Retrieved sequences and their relationships with known pmoA sequences are grouped into established phylogenetic families A, B, C, D, E, F, and G. Uncultured methanotroph pmoA sequences RA21 and MR1 were used as the outgroup.
A189-A682 clone libraries.
The clone libraries constructed with the A189-A682 primer set resulted in limited successful retrieval of methanotroph pmoA sequences. The heath and sitka libraries were dominated by the clone sequence A1 (59 and 66% of clone libraries, respectively), which demonstrated high identity (>98%) to the amoA gene of Nitrosomonas europaea. Only 4% of clones were affiliated with this sequence in the oak library. The libraries consisted of a high proportion of clone types which contained no open reading frames of significant length and which failed to show homology to other sequences after database analysis (heath, 29%; oak, 45%; and sitka, 15%). The primers appeared to retrieve a large number of hybrid nonsense amplified bands of the correct insert size, although the sequences were not identified as chimeric. A small number of clones in the libraries also failed to provide any sequence data, as indicated in Table 1 (heath, 10%; oak, 13%).
The only clones recovered from the libraries that were affiliated with methanotrophic sequences were found in the oak library. The retrieved sequence (B1) showed high sequence identity (>99%) with the pmoA gene of Methylococcus capsulatus. However, this clone type constituted only 4% of the clone library (Table 1). A total of 34% of clones within the oak library, when analyzed by RFLP, were found to be single clones representing different OTU groups. Sequence information was not recovered from these individual OTU patterns.
From these results it can be concluded that the primer set A189-A682 was inadequate to assess methanotroph diversity in these soils, and therefore the use of other primer sets was investigated.
Design of primer A650.
All available pmoA sequences were aligned, and a new pmoA-specific reverse primer, A650, was designed for use in both clone library construction and DGGE analysis. To facilitate DGGE analysis, the A650 primer was designed without redundancies. Designing a primer specific to all pmoA sequences was problematic; therefore, mismatches with some pmoA sequences occur. A selection of pmoA and amoA sequences showing both matching and mismatching regions compared to all three reverse primers used in this study is shown in Table 2. The A650 primer did not match any ammonia monooxygenase amoA gene sequences.
TABLE 2.
pmoA and amoA sequence alignments of reference methanotrophs and ammonia oxidizers, showing target regions for the pmoA-and amoA-specific reverse primer A682 and the pmoA-specific reverse primers A650 and mb661
| Primer or target | Sequencea with pmoA reverse primer:
|
||
|---|---|---|---|
| A682 | A650 | mb661 | |
| Primer (3′-5′) | C G S A A G A A G A G N C G S A A G | T G G A A G C C A T T C C T G C A | C C A T T Y C T G C A A C G M G G C C |
| Target (5′-3′) | g c g t t c t t c t c g g c g t t c | a c c t t c g g t a a g g a c g t | g g t a a g g a c g t t g c t c c g g |
| c c c | a g | ||
| a | |||
| t | |||
| Target strains | |||
| Type I pmoA sequences | |||
| Methylococcus capsulatus (Bath) | . . a . . . . . . . . . . . . . . . | . . . . . . . . . . . . . . . . . | . . . . . . . . . . . g . . . . . . . |
| Methylobacter sp. strain BB5. 1 | . . t . . . . . . . . a . . . . . | . . . . . . . . . . . . . . . . . . . | |
| Uncultured methanotroph pAMC521 | . . t . . . . . . . . . . . . . . | . . . . . . . . . . . . . . . . . . . | |
| Uncultured proteobacterium U | . . t . . . . . . . . a . . . . . | . . . . . . . . . . . . . n . . . a . | |
| Methylomonas methanica (S1) | . . . . . . . . . . . a . . . . . | . . . . . . . . . . . . . . . . . t . | |
| Methylomonas sp. strain LW21 | . . t . . . . . . . . a . . . . . | . . . . . . . . . . . . . . . . . . . | |
| Methylomicrobium album (BG8) | . . t . . . . . . . . . . . . . . | . . . . . . . . . . . . . . . . . a . | |
| Methylocaldum tepidum (LK6) | . . . . . . . . . . . . . . . . . . | . . . . . . . . . . . a . . . . | . . . . . . . . . . . . . g . . . . . |
| Type II pmoA sequences | |||
| Methylosinus trichosporium (OB3b) | . . . . . . . . . . . . . g . . . . | . . . . . . . . . . . . . . . . . | . . . . . . . . . . . . . . . . . . . |
| Methylosinus sp. strain LW2 | . . g . . . . . . . . a . . . . . | . . . . . . . . . . . . . . . . . . n | |
| Methyocystis paryus (OBBP) | . . . . . . . . . . . . . . . . . | . . . . . . . . . . . . . t . . . . . | |
| Methylocystis sp. strain M | . . . . . . . . . . . . . g . . . . | . . . . . . . . . . . . . . . . . | . . . . . . . . . . . . . t . . . . . |
| High-affinity pmoA sequences | |||
| Uncultured methanotroph RA14 | . . . . . . . . . . . . . . t . . | . . . . . . . . t . . c . t . g g c . | |
| Uncultured methanotroph RA21 | . . . . . . . . g g g c c . t t c | . . g g g c c . t t c c . . . g t t t | |
| Ammonia oxidizer amoA sequences | |||
| Nitrosomonas europaea (NCIMB 11850) | . . a . . . . . . . . . . . . . . . | . . . . . t . . . g g t c . t a c | . . . g g t c . t a c c a . a g t t a |
| Nitrosospira sp. strain Np22 | . . . . . . . . . . . . . . . . . . | . . . . . t . . . g g t c . . a c | . . . g g t c . . a c c a . . g t . a |
| Nitrosococcus oceanus (NC1MB 11848) | . . t . . . . . . . . . . g t . . t | . . g . . t . . . . . . . . . . . | . . . . . . . . . . . g . t c . . . . |
| Nitrosomonas eutropha | . . a . . . . . . g . . . . . . . t | . . . . . t . . . g g t c . . a c | . . . g g t c . . a c c a . a g t t a |
Nucleotides identified for mispairings and pairings are indicated by dots. The bases showing mismatches refer to the sequence of the target pmoA or amoA gene and not the probe.
The primer was tested for specificity via PCR against target and nontarget organisms at moderate stringency (see Materials and Methods). All methanotrophs tested (Methylococcus capsulatus [Bath], Methylococcus capsulatus [strain M], Methylomicrobium agile [A30], Methylobacter whittenburyi, Methylosphaera hansonii, Methylocaldum tepidum [LK6], Methylomonas methanica [S1], Methylomicrobium album [BG8], Methylosinus trichosporium [OB3b], and Methylocystis parvus [OBBP]) gave positive amplification of pmoA products, with the exceptions of Methylomicrobium album (BG8) and Methylosphaera hansonii. Table 2 shows a 1-bp mismatch with the primer and Methylomicrobium album (BG8). Despite 1-bp mismatches for Methylomonas methanica and Methylocaldum tepidum, positive amplification with this primer was observed under the PCR conditions used. The pmoA gene sequences from the organisms Methylomicrobium agile (A30), Methylobacter whittenburyi, and Methylosphaera hansonii have not been determined and are therefore not presented in Table 2. No PCR products were produced with DNA from ammonia-oxidizing nitrifiers (Nitrosomonas europaea [NCIMB 11850], Nitrosospira sp. [Np22], Nitrosococcus oceanus [NCIMB 11848], Nitrosomonas eutropha, and Nitrosospira multiformis [NCIMB 11849]).
A189-A650 clone libraries.
Clone libraries constructed with the A189-A650 primer set were dominated by pmoA sequences B1 and B2, showing high identity (>98%) to Methylococcus capsulatus (Bath) pmoA sequences (heath, 98%; oak, 87%; and sitka, 82% of the libraries). Limited methanotroph diversity was detected within the soils with this primer set. For example, only two OTU groups were identified in the heath and oak libraries.
The oak and sitka libraries contained a group of clones (G1, G2, and G3) closely affiliated with the pmoA clone RA14, presumably from an unknown organism. These types of sequences have been found in a range of soils that oxidize atmospheric methane (10, 12). Within the oak library the sequences contributed 9% of clones (G1), while within the sitka library they represented 16% of clones (G2 and G3). This primer set also retrieved a pmoA sequence from the heath library affiliated with the pmoA gene from the type II methanotroph Methylocystis sp. strain M (clone type F4, >98% sequence identity).
The specificity of the A189-A650 primer set for subgroups of methanotrophs was confirmed by the retrieval of only methanotroph pmoA sequences. No amoA sequences from ammonia-oxidizing bacteria were retrieved. Also, only two ambiguous sequences, where sequence information could not be obtained, were found in the oak library. However, due to the targeting of the primers to subgroups of methanotroph pmoA sequences, a pronounced bias was observed within the libraries in recovering Methylococcus capsulatus pmoA sequences. No pmoA sequences from other type I organisms were identified, notably no Methylomonas- or Methylomicrobium-associated pmoA sequences. The alignment of sequences in Table 2 demonstrates that the A650 primer has one or two mismatches to these organisms.
mb661 clone libraries.
Costello and Lidstrom (4) designed a new reverse pmoA primer (mb661) to specifically amplify pmoA sequences and not amoA sequences. The A189-mb661 primer set was found to be more useful for studying methanotroph diversity in freshwater environments.
When used in this study, clone libraries constructed with the A189-mb661 primer set contained pmoA sequences affiliated with both type I and type II methanotroph pmoA sequences (Table 1 and Fig. 1). Methylococcus capsulatus-affiliated pmoA sequences (>98% sequence identity) again represented a large proportion of clones within the libraries (heath, clone type B1 = 46%, oak, clone type B4 = 24%; and sitka, clone types B1 and B5 = 77% of the libraries). The largest representative of clones in the oak library (52%) was clone type C1, which is related to the pmoA sequence of an uncultured proteobacterium affiliated with Methylomicrobium, a type I methanotroph. Clones showing similar high identities to this uncultured organism were found in the heath library (C1 and C2), although together they constituted only 6% of the clones recovered. Other type I-related pmoA sequences found in the libraries included Methylomonas-affiliated pmoA sequences D1 (6% of heath library) and D2 (12% of oak library). However, clone type D2 demonstrated only 94% homology to the pmoA sequence of Methylomonas sp. strain LW21, the closest database match. Clone type E1 (100% identity with uncultured eubacterium pAMC521) was only a minor representative of the heath library (4%); however, it was another pmoA sequence which demonstrated the application of the A189-mb661 primer set for retrieval of a wide diversity of type I pmoA sequences from the environment.
A significant proportion of clones identified in the libraries were affiliated with the pmoA sequences of type II methanotrophs. For example, clone type F1 constituted 28% of clones in the heath library and 23% in the sitka library, while clone type F2 constituted 10% of the oak library. All type II pmoA clone types F1, F2, F3, and F4 possessed high identity (>97%) with the pmoA gene of Methylocystis sp. strain M.
DGGE analysis of soils using pmoA
PCR-amplified pmoA gene fragments from a selection of control methanotroph cultures and the soils investigated in this study were analyzed by DGGE. Two different pmoA-specific primer sets were used in this study, both of which incorporated a GC clamp (GC-A189–A682 and A189–A650-GC) (23). The A650 pmoA primer was designed for application in DGGE analysis; hence, no redundancies were incorporated when designing the primer.
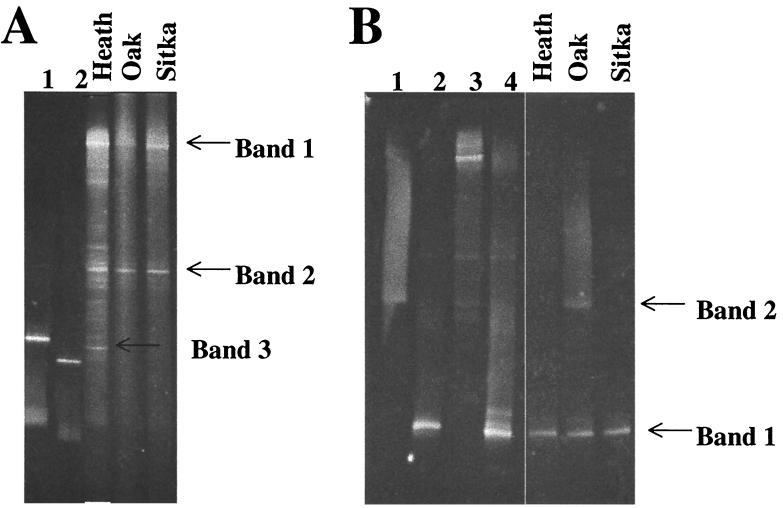
The DGGE profiles of the heath, oak, and sitka soils amplified with the GC-A189–A682 primer sets are shown in Fig. 2A. The heath soil represented the most complex profile, indicating a more diverse pmoA-amoA population. The profiles of the oak and sitka soils were dominated by two bands (bands 1 and 2 in Fig. 2A), which were also present in the heath profile. Bands 1 and 2 (Fig. 2A) were excised from the gel, reamplified by PCR, and run on an identical DGGE gel to confirm their positions relative to those in the original sample. Band 1 was successfully sequenced and possessed 100% identity to amoA from Nitrosomonas europaea. Band 2 was recalcitrant to reamplification and sequencing. Band 3, excised from the heath profile, produced a valid DNA sequence which did not have any closely matched sequences after database analysis.
FIG. 2.
DGGE profiles of control strains and Danish soils obtained with pmoA primers sets. (A) A189-A682 primer set. Lane 1, Methylocystis strain M; lane 2, Methylosinus trichosporium OB3b. (B) A189-A650 primer set. Lane 1, Methylococcus capsulatus (Bath); lane 2, Methylobacter whittenburyi; lane 3, Methylocystis strain M; lane 4, Methylosinus trichosporium OB3b.
The results from DGGE analysis of the soils with the A189-A682 primer set were similar to those found with the same primer set during the clone library analysis. First, amoA sequences affiliated with Nitrosomonas europaea amoA sequences dominated. Second, the difficulty in obtaining valid sequence from amplified bands in the profile was similar to that for the nonspecific amplified products obtained with these primers in the clone libraries.
DGGE analysis of amplified pmoA sequences obtained from the soils using the A189–A650-GC primer set is shown in Fig. 2B. The DGGE profile demonstrated that one major band (band 1) appeared in all the soils. Excision of the band, PCR reamplification, and sequencing indicated that the band sequence was closely related to the pmoA sequence of Methylosinus trichosporium OB3b (≈97% identity). This band, when amplified from the soils, migrated in the DGGE gel the same distance as the pmoA PCR-amplified product obtained with DNA from Methylosinus trichosporium OB3b (Fig. 2B). Repeated attempts to obtain valid sequence from band 2 in the oak soil (Fig. 2B) failed. The A189–A682-GC primer set recovered little diversity of pmoA genes from the soils. Multiple bands seen with control organisms may be due to the presence of multiple copies of pmoA in methanotrophs.
The retrieval of a pmoA sequence related to the Methylosinus trichosporium OB3b sequence is contradictory to the library analysis of soils, since they were dominated by pmoA sequences from Methylococcus capsulatus. It is suspected that a bias caused by the GC clamp on the A650 primer may have an effect on PCR amplification.
DISCUSSION
In this study, we have compared three pmoA primer sets to assess their potentials for investigating methanotroph diversity and subsequently comparing this diversity between three soil samples with contrasting cover vegetation, i.e., heath, oak, and sitka. The pmoA gene sequences retrieved in this study were used to infer methanotroph diversity, since the phylogeny of the pmoA genes reflects that of the 16S rRNAs of methanotrophs (4, 19)
Comparison of primer sets for assessing methanotroph diversity.
Costello and Lidstrom (4) highlighted the disadvantage of the A189-A682 PCR primer set, in that it amplifies both amoA and pmoA sequences (11, 24). In this study the A189-A682 PCR could not be used to assess methanotroph diversity. Most sequences recovered from the libraries were homologous to the amoA gene of Nitrosomonas europaea. Also, a large number of clones in each library were nonspecific amplified PCR products exhibiting no sensible open reading frame.
The frequencies of PCR-derived rRNA gene clones within libraries cannot be claimed with confidence to represent the relative abundances of different components of the microbial community (8). In molecular studies, however, it is likely that more abundant sequences are preferentially amplified, while less abundant sequences are discriminated against (31). Since the dominant clone type in the A189-A682 libraries demonstrated a high degree of homology to the amoA gene of Nitrosomonas europaea, it is likely that the amoA genes are more abundant than the corresponding pmoA sequences of methanotrophs and hence are preferentially amplified, thus dominating the libraries.
The A650 primer was designed in this study principally for incorporation into clone library and DGGE analysis. While the clone libraries were dominated by pmoA sequences closely related to the pmoA gene of Methylococcus capsulatus, DGGE profiles exhibited a dominant band which when sequenced revealed a pmoA sequence closely related to that of Methylosinus trichosporium. The GC clamp on the primer is suspected to have a large effect on hybridization specificity and sequence amplification. The use of the A650 primer for DGGE analysis therefore appears to be limited, as it does not reflect true diversity of pmoA sequences in these soils. However, this does dramatically demonstrate the role that a particular primer can play in PCR bias and suggests that it is not possible to assign dominance of a strain in an environment based upon the number of similar clones in a clone library.
An inherent disadvantage of PCR-based molecular techniques is the possible bias in selected amplification of some sequences, which can affect the measure of diversity observed. Within the A650 clone libraries, a possible large bias towards amplification of pmoA sequences related to Methylococcus capsulatus is observed. A direct comparison between the A189-A650 and A189-mb661 libraries shows that the clone types C, D, and F are not detected in the A189-A650 library. Table 2 demonstrates that the A650 primer set has one or two base pair mismatches with the pmoA gene from type I methanotrophs affiliated with the type C, D, and F sequences. Similarly, the type G clones affiliated with RA14-like pmoA sequences which are linked with possible novel, high-affinity methanotrophs are not found in the A189-mb661 libraries. The mb661 primer has many mismatches with these RA14-type pmoA sequences (Table 2).
All of the primer sets used in this study provide some valid information on methanotroph diversity. The A189-A682 primer set is useful for investigating both amoA diversity of ammonia-oxidizing bacteria and pmoA diversity of methanotrophs. However, this primer set may be limited to environments where methanotroph populations are high, such as peat (19), and for the detection of novel sequences such as RA14 and RA21 (12). Reay et al. (25) noted, however, that this primer set may not be able to amplify all type I methanotrophs. The A650 primer is limited due to suspected bias and a lack of complete coverage of all methanotroph genera, although the addition of degeneracies might increase the diversity of the methanotrophs that it detects by PCR. The advantage of the A189-A650 primer set, however, is that it retrieves pmoA sequences linked to novel uncultured organisms which maybe involved in atmospheric methane oxidation (10, 12). The A189-mb661 primer set demonstrated the greatest recovery of pmoA diversity in this study. For studying environments and investigating pmoA diversity of type I and type II methanotrophs, while not amplifying amoA sequences, this primer set appears to be the best. However a combination of all three primer sets will provide the most information on pmoA-amoA diversity in the environment.
Comparison of methanotroph diversity between the soil samples.
A comparison of the diversity of the soils with the different primer sets is difficult, which is not surprising given the associated biases and problems caused by PCR. Also, the fact that only 50 clones were screened per clone library means that some representatives may have been missed due to the small sample size.
All three soil samples were investigated initially due to their ability to oxidize atmospheric concentrations of methane and therefore their potential to possess high-affinity methanotrophs with unique pmoA sequences. However, all pmoA sequences retrieved from the soils grouped with established phylogenetic affiliations of pmoA genes.
RA14 pmoA sequences linked with possible high-affinity methanotrophs (10, 12) were found in both the oak and sitka libraries constructed with the A189-A650 primer set. However, these sequences were not found in the corresponding library of the heath soil, despite this soil having the ability to oxidize the lowest level of methane (unpublished data). It is possible that other high-affinity methanotrophs containing pmoA genes which cannot be amplified by the A189-A650 primer set occur in this soil.
The pmoA library of the heath soil demonstrated the highest diversity of methanotrophs with the A189-mb661 PCR primer set (seven OTU groups), although it exhibited the lowest diversity with the A189-A650 PCR primer set (98% B2 clone type). Given the problems with the A650 primer discussed above, the A189-mb661 library probably reflects a more realistic picture of the diversity of the type I and type II methanotrophs. If we use only the A189-mb661 primer set to determine established type I and type II methanotroph diversity, then the heath library possessed the greatest diversity, followed by the oak library and then the sitka library.
Although the libraries constructed with the A189 and A682 primers cannot be used to assess methanotroph diversity between the soils, they do provide some information. A total of 34% of clones within the oak library constructed with the A189-A682 primer set were single OTU groups, representatives of which were not sequenced. Given the problems of nonspecific amplification with this primer set, many of the OTU groups may have been nonsense sequences. The low retrieval of amoA sequences from this soil library, in contrast to the heath and sitka libraries, may suggest that there is a low diversity of ammonium oxidizers, that the soil is not dominated by these organisms, or that this primer set does not amplify amoA from all ammonia oxidizers (24). The A189-A682 DGGE also showed the most complex profile with the heath soil, again indicating possible highest amoA-pmoA diversity with this soil.
This study highlighted some of the problems associated with using molecular techniques to analyze methanotroph diversity in environmental samples. The distinct biases between pmoA-specific primer sets have been shown and should be taken into consideration when designing experiments to investigate methanotroph diversity in the environment.
ACKNOWLEDGMENT
This work was supported under the European Community RTD Biotechnology Programme (BIO 4 CT 960419).
REFERENCES
- 1.Amaral J A, Ren T, Knowles R. Atmospheric methane consumption by forest soils and extracted bacteria at different pH values. Appl Environ Microbiol. 1998;64:2397–2402. doi: 10.1128/aem.64.7.2397-2402.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Auman A J, Stolyar S, Costello A M, Lidstrom M E. Molecular characterization of methanotrophic isolates from freshwater lake sediment. Appl Environ Microbiol. 2000;66:5259–5266. doi: 10.1128/aem.66.12.5259-5266.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bedard C, Knowles R. Physiology, biochemistry, and specific inhibitors of CH4, NH4+, and CO oxidation by methanotrophs and nitrifiers. Microbiol Rev. 1989;53:68–84. doi: 10.1128/mr.53.1.68-84.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costello A M, Lidstrom M E. Molecular characterization of functional and phylogenetic genes from natural populations of methanotrophs in lake sediments. Appl Environ Microbiol. 1999;65:5066–5074. doi: 10.1128/aem.65.11.5066-5074.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunfield P F, Liesack W, Henckel T, Knowles R, Conrad R. High-affinity methane oxidation by a soil enrichment culture containing a type II methanotroph. Appl Environ Microbiol. 1999;65:1009–1014. doi: 10.1128/aem.65.3.1009-1014.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilbert B, McDonald I R, Finch R, Stafford G P, Nielsen A K, Murrell J C. Molecular analysis of the pmo (particulate methane monooxygenase) operons from two type II methanotrophs. Appl Environ Microbiol. 2000;66:966–975. doi: 10.1128/aem.66.3.966-975.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hall G H, Simon B M, Pickup R W. CH4 production in blanket bog peat: a procedure for sampling, sectioning and incubating samples whilst maintaining anaerobic conditions. Soil Biol Biochem. 1996;28:9–15. [Google Scholar]
- 8.Head I M, Saunders J R, Pickup R W. Microbial evolution, diversity and ecology: a decade of ribosomal RNA analysis of uncultivated microorganisms. Microb Ecol. 1998;35:1–21. doi: 10.1007/s002489900056. [DOI] [PubMed] [Google Scholar]
- 9.Henckel T, Friedrich M, Conrad R. Molecular analyses of the methane-oxidizing microbial community in rice field soil by targeting the genes of the 16S rRNA, particulate methane monooxygenase, and methanol dehydrogenase. Appl Environ Microbiol. 1999;65:1980–1990. doi: 10.1128/aem.65.5.1980-1990.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henckel T, Jäckel U, Schnell S, Conrad R. Molecular analyses of novel methanotrophic communities in forest soil that oxidize atmospheric methane. Appl Environ Microbiol. 2000;66:1801–1808. doi: 10.1128/aem.66.5.1801-1808.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holmes A J, Costello A M, Lidstrom M E, Murrell J C. Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol Lett. 1995;132:203–208. doi: 10.1016/0378-1097(95)00311-r. [DOI] [PubMed] [Google Scholar]
- 12.Holmes A J, Roslev P, McDonald I R, Iversen N, Henriksen K, Murrell J C. Characterization of methanotrophic bacterial populations in soils showing atmospheric methane uptake. Appl Environ Microbiol. 1999;65:3312–3318. doi: 10.1128/aem.65.8.3312-3318.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jensen S, Holmes A J, Olsen R A, Murrell J C. Detection of methane oxidizing bacteria in forest soil by monooxygenase PCR amplification. Microb Ecol. 2000;39:282–289. [PubMed] [Google Scholar]
- 14.King G M. Responses of atmospheric methane consumption by soils to global climate change. Global Change Biol. 1997;3:351–362. [Google Scholar]
- 15.Klotz M G, Norton J M. Multiple copies of ammonia monooxygenase (amo) operons have evolved under biased AT/GC mutational pressure in ammonia-oxidising autotrophic bacteria. FEMS Microbiol Lett. 1998;168:311. doi: 10.1111/j.1574-6968.1998.tb13288.x. [DOI] [PubMed] [Google Scholar]
- 16.Marmur J. A procedure for the isolation of deoxyribonucleic acid from microorganisms. J Mol Biol. 1961;3:208–218. [Google Scholar]
- 17.McDonald I R, Hall G H, Pickup R W, Murrell J C. Methane oxidation potential and preliminary analysis of methanotrophs in blanket bog peat using molecular ecology techniques. FEMS Microbiol Ecol. 1996;21:197–211. [Google Scholar]
- 18.McDonald I R, Kenna E M, Murrell J C. Detection of methanotrophic bacteria in environmental samples with the PCR. Appl Environ Microbiol. 1995;61:116–121. doi: 10.1128/aem.61.1.116-121.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McDonald I R, Murrell J C. The particulate methane monooxygenase gene pmoA and its use as a functional gene probe for methanotrophs. FEMS Microbiol Lett. 1997;156:205–210. doi: 10.1111/j.1574-6968.1997.tb12728.x. [DOI] [PubMed] [Google Scholar]
- 20.McDonald I R, Upton M, Hall G, Pickup R W, Edwards C, Saunders J R, Ritchie D A, Murrell J C. Molecular ecological analysis of methanogens and methanotrophs in blanket bog peat. Microb Ecol. 1999;38:225–233. doi: 10.1007/s002489900172. [DOI] [PubMed] [Google Scholar]
- 21.Murrell J C, Gilbert B, McDonald I R. Molecular biology and regulation of methane monooxygenase. Arch Microbiol. 2000;173:325–332. doi: 10.1007/s002030000158. [DOI] [PubMed] [Google Scholar]
- 22.Murrell J C, McDonald I R, Gilbert B. Regulation of expression of methane monooxygenases by copper ions. Trends Microbiol. 2000;8:221–225. doi: 10.1016/s0966-842x(00)01739-x. [DOI] [PubMed] [Google Scholar]
- 23.Muyzer G, Teske A, Wirsen C O, Jannasch H W. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch Microbiol. 1995;164:165–172. doi: 10.1007/BF02529967. [DOI] [PubMed] [Google Scholar]
- 24.Purkhold U, Pommerening-Roser A, Juretschko S, Schmid M C, Koops H P, Wagner M. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl Environ Microbiol. 2000;66:5368–5382. doi: 10.1128/aem.66.12.5368-5382.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reay, D. S., S. Radajewski, J. C. Murrell, N. McNamara, and D. B. Nedwell. Impact of land-use on the activity and diversity of methane oxidising bacteria in forest soils. Soil Biol. Biochem., in press.
- 26.Rotthauwe J H, Witzel K P, Liesack W. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol. 1997;63:4704–4712. doi: 10.1128/aem.63.12.4704-4712.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 28.Saunders S E, Burke J F. Rapid isolation of miniprep DNA for double strand sequencing. Nucleic Acids Res. 1990;18:4948. doi: 10.1093/nar/18.16.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Semrau J D, Chistoserdov A, Lebron J, Costello A M, Davagnino J, Kenna E M, Holmes A J, Finch R, Murrell J C, Lidstrom M E. Particulate methane monooxygenase genes in methanotrophs. J Bacteriol. 1995;177:3071–3079. doi: 10.1128/jb.177.11.3071-3079.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith K A, Dobbie K E, Ball B C, Bakken L R, Sitaula B K, Hansen S, Brumme R, Borken W, Christensen S, Prieme A, Fowler D, MacDonald J A, Skiba U, Klemedtsson L, Kasimir-Klemedtsson A, Degórska A, Orlanski P. Oxidation of atmospheric methane in Northern European soils, comparison with other ecosystems, and uncertainties in the global terrestrial sink. Global Change Biol. 2000;6:791–803. [Google Scholar]
- 31.Ward D M, Bateson M M, Weller R, Ruff Roberts A L. Ribosomal RNA analysis of microorganisms as they occur in nature. In: Marshall K C, editor. Advances in microbial ecology. New York, N.Y: Plenum Press; 1992. pp. 219–286. [Google Scholar]
- 32.Whittenbury R, Davies S L, Davey J F. Exospores and cysts formed by methane-utilizing bacteria. J Gen Microbiol. 1970;61:219–226. doi: 10.1099/00221287-61-2-219. [DOI] [PubMed] [Google Scholar]