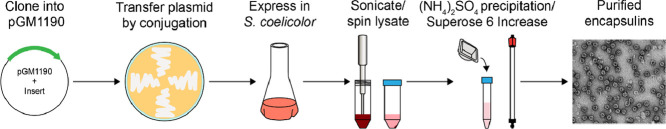
Abstract
In recent years a large number of encapsulin nanocompartment-encoding operons have been identified in bacterial and archaeal genomes. Encapsulin-encoding genes and operons from GC-rich Gram-positive bacteria, particularly of the phylum Actinobacteria, are often difficult to overexpress and purify in a soluble form using standard Escherichia coli expression systems. Here, we present a protocol to heterologously overexpress encapsulin nanocompartments and encapsulin-containing operons in Streptomyces coelicolor. Successful encapsulin production begins with the transfer of a Streptomyces expression plasmid, encoding the gene(s) of interest, via conjugation to the model actinobacterium S. coelicolor. After growing the conjugated S. coelicolor culture to the optimal optical density, protein production is induced by the addition of the inducer thiostrepton, followed by expression in liquid culture for 1–3 days. Cells are lysed and encapsulin proteins purified using ammonium sulfate precipitation and size exclusion chromatography. The method outlined in this protocol can be utilized to improve cargo loading and overall soluble expression of encapsulin systems when compared to expression in E. coli.
-
•
Clone an encapsulin-encoding gene or operon into a Streptomyces expression vector.
-
•
Transfer the Streptomyces expression vector to S. coelicolor via conjugation.
-
•
Heterologously express and purify empty or cargo-loaded encapsulins from S. coelicolor.
Keywords: Encapsulin, Nanocompartment, Streptomyces, Protein production, Protein purification
Graphical abstract
Specification table
| Subject area | Biochemistry, Genetics and Molecular Biology |
| More specific subject area | Protein expression and purification |
| Name of your method | Heterologous Expression and Purification of Encapsulins in Streptomyces coelicolor |
| Name and reference of original method | N/A |
| Resource availability | Reagents and Equipment are listed in the Materials and Reagents section |
Method details
Background
Encapsulin nanocompartments are a diverse class of protein-based compartments found in bacteria and archaea [1]. To date, over 6000 encapsulin-containing operons have been discovered in bacterial and archaeal genomes via genome-mining searches [2,3]. Encapsulins form icosahedral protein shells that self-assemble from HK97-fold protomers [1]. They are defined by their ability to specifically encapsulate dedicated cargo proteins. Encapsulation is mediated by N- or C-terminal targeting peptides or domains present in all cargo proteins which directly interact with the encapsulin shell during self-assembly [1,2,4]. Encapsulins can form T1 (60 subunits, 24 nm) [5], [6], [7], T3 (180 subunits, 32 nm) [8,9], and T4 (240 subunits, 42 nm) [10] shells and have been classified into four main families based on sequence homology and genome neighborhood analysis [2]. Thousands of Family 2B and 3 encapsulins have been identified in operons with putative cargos associated with secondary metabolism [2]. Expressing these complete operons can prove challenging in E. coli, as many of them originate from GC-rich organisms – primarily of the phylum Actinobacteria. Additionally, homogenous cargo loading can be exceedingly difficult when using E. coli as the expression host [5]. To overcome these challenges, we have developed a protocol to heterologously express and purify encapsulins and encapsulin-containing operons in Streptomyces coelicolor. While other Streptomyces species, such as Streptomyces lividans, have been used to heterologously express and purify proteins, we chose to focus on S. coelicolor because it is one of the most well-documented species of Streptomyces – used both for protein expression and heterologous natural product production – and has been used as a standard conjugation host in our laboratory. The methodology presented in this protocol can likely be easily adapted to express and purify proteins from other commonly used Streptomyces species. In developing this protocol, we found it particularly challenging to find coherent and integrated methodologies that begin with the cloning of expression plasmids and follow all the way through to Streptomyces protein production and purification. In this protocol, we have included all steps necessary to clone, conjugate, overexpress, and purify cargo-loaded encapsulin nanocompartments in Streptomyces coelicolor. This protocol will be useful for those seeking to heterologously express and purify soluble cargo-loaded encapsulins originating from high-GC actinobacterial genomes.
Materials and reagents
-
(1)
ElectroMAX DH10B electrocompetent E. coli cells (Invitrogen, catalog number: 18290-015).
-
(2)
E. coli ET12567 (pUZ8002) cells (Life Science Market, catalog number: S0052).
-
(3)
Streptomyces violaceoruber (formerly Streptomyces coelicolor A3(2) (John Innes Center M145)) (ATCC, catalog number: BAA-471).
-
(4)
pGM1190 vector (Created by Muth [11] and available on Addgene, catalog number: 69994).
-
(5)
Monarch DNA gel extraction kit (New England Biolabs, catalog number: T1020S).
-
(6)
Phusion High Fidelity DNA Polymerase (Fisher Scientific, catalog number: F530S).
-
(7)
Gibson Assembly Master Mix (Fisher Scientific, catalog number: NC0260576).
-
(8)
Agarose (Fisher Scientific, catalog number: BP160-100).
-
(9)
0.2 mL Strip tubes (Fisher Scientific, catalog number: AB-0264).
-
(10)
Electroporation cuvettes, 1 mm (Fisher Scientific, catalog number: FB101).
-
(11)
FastDigest NdeI restriction enzyme (ThermoFisher Scientific, catalog number: FD0583).
-
(12)
FastDigest BamHI restriction enzyme (ThermoFisher Scientific, catalog number: FD0054).
-
(13)
10 mM dNTP Mix (Invitrogen, catalog number 18427013).
-
(14)
SOC outgrowth medium (New England Biolabs, catalog number: B9020S).
-
(15)
LB agar, Miller powder (Fisher Scientific, catalog number: BP1425-2).
-
(16)
LB broth, Miller mix (Fisher Scientific, catalog number: BP97235).
-
(17)
ISP Medium No. 1 (Himedia, catalog number: M356-500G).
-
(18)
2X-YT Mix (RPI, catalog number X-15640-1000.0).
-
(19)
Mueller-Hinton Agar (Fluka Analytical, catalog number: 70191-500G).
-
(20)
d-(-)-Mannitol (Fisher Scientific, catalog number: AAA140300B).
-
(21)
Soya flour, 32 ounces (OlivNation Soy Flour purchased from Amazon).
-
(22)
Apramycin sulfate (Gold Biotechnology, catalog number: A-600-5).
-
(23)
Kanamycin sulfate (Fisher Scientific, catalog number: BP906-5).
-
(24)
Chloramphenicol (Fisher Scientific, BP-904-100).
-
(25)
Thiostrepton (Gold Biotechnology, catalog number: T-300-1).
-
(26)
Dimethyl sulfoxide (Sigma, catalog number: D8418-100mL).
-
(27)
Sodium hydroxide (Alfa Aesar, catalog number: 1823-500G).
-
(28)
d-sucrose (Fisher Scientific, catalog number: BP220-10).
-
(29)
Microbiology malt extract (Millipore Sigma, catalog number: 1.05391.0500).
-
(30)
Dextrose (Fisher Scientific, catalog number: D16-1).
-
(31)
Acid casein peptone (Fisher Scientific, catalog number: BP1424-500).
-
(32)
Sodium chloride (Fisher Scientific, catalog number: S271-500).
-
(33)
Tris base (EMD Millipore, catalog number: 648311-1KG).
-
(34)
37% Hydrochloric acid (Acros Organic, catalog number: 42379-5000).
-
(35)
SIGMAFAST protease inhibitor cocktail tablets, EDTA-Free (Sigma-Aldrich, catalog number: S8830-20TAB).
-
(36)
Lysozyme, from hen egg white (Sigma-Aldrich, catalog number; 10837059001).
-
(37)
Magnesium chloride hexahydrate (Fisher Scientific, catalog number: M33-500).
-
(38)
Benzonase nuclease (Sigma-Aldrich, catalog number: E1014-25KU).
-
(39)
14% SDS-PAGE gels (Invitrogen, catalog number: XP00145BOX).
Equipment
-
(1)
Floor centrifuge (Beckman Coulter Avanti JXN-26 with JA-12 rotor) or equivalent.
-
(2)
Tabletop centrifuge (Eppendorf 5425 with FA24 × 2 rotor) or equivalent.
-
(3)
Benchtop refrigerated centrifuge (Eppendorf 5810R with A-4-81 rotor) or equivalent.
-
(4)
Orbital shaker/incubator (Infors HT Multitron Standard orbital shaker/incubator) or equivalent.
-
(5)
Incubator with grate (VWR gravity convection incubator, catalog number: 89511-422) or equivalent.
-
(6)
Electroporator (Bio-Rad MicroPulser Electroporator, catalog number: 1652100) or equivalent.
-
(7)
Pipettes (Gilson Pipetman L P2L, P20L, P200L, and P100L) or equivalent.
-
(8)
Pipette gun (Eppendorf Easypet 3, catalog number: 4430000018) or equivalent.
-
(9)
FPLC (Cytiva ÄKTA pure) or equivalent.
-
(10)
Superose 6 Increase 10/300 GL column (Cytiva, catalog number: 29-0915-96).
-
(11)
Laboratory rocker (Chemglass Life Sciences 3D shaker/rocker, catalog number: CLS-4029-100).
-
(12)
Agarose gel power supply (BioRad PowerPac Basic, catalog number: 1645050) or equivalent.
-
(13)
Horizontal electrophoresis chamber for agarose gels (Biorad Mini-Sub Cell GT, catalog number 1704487EDU) or equivalent.
-
(14)
125 mL baffle flasks (Kimax, catalog number: 25630).
-
(15)
SDS-PAGE power supply (Invitrogen PowerEase 90 W power supply, catalog number: PS0090) or equivalent.
-
(16)
SDS-PAGE gel chamber (Invitrogen Xcell SureLockTM Mini-Cell, catalog number: EI0001) or equivalent.
-
(17)
Sonicator equipped with 1/8 in probe (FisherbrandTM Model 120 Sonic Dismembrator, catalog number: FB120110) or equivalent.
-
(18)
Gel imaging system (BioRad ChemiDocTM Imaging System, catalog number:12003153) or equivalent.
-
(19)
Ultrapure water source (Milipore MilliQ Reference Water Purification System) or equivalent.
Software
-
(1)
GE Life Sciences Unicorn 7.3 or equivalent.
Recipes
Modified YEME medium adapted from Kieser et al. [12]
-
•
Yeast extract – 3 g.
-
•
Acid casein peptone - 5 g.
-
•
Oxoid malt extract - 3 g.
-
•
Glucose - 10 g.
-
•
Sucrose - 340 g (34% w/v final).
-
•
Distilled water up to 1000 mL.
-
•
After autoclaving, add 5 mM MgCl2.
-
•
Add sterile glycine to 0.5% w/v.
The YEME medium outlined in Kieser et al. calls for 5 grams of Difco Bacto-peptone. We have, however, found acceptable growth and expression using acid casein peptone (Fisher sciences, BP1424-500) and routinely use it in place of Difco Bacto-peptone. If expression or growth issues occur using our modified YEME recipe, try using Difco Bacto-peptone.
Mannitol Soya flour medium (MS) adapted from Kieser et al. [12]
-
•
Agar - 10 g.
-
•
Mannitol - 10 g.
-
•
Soya Flour - 10 g.
-
•
Tap water - 500 mL.
Mix the agar, mannitol, and soya flour in 2 L Erlenmeyer flask or baffle flask. Cover the lid with foil and autoclave twice. Some flour may form clumps during the preparation despite thorough mixing. We used plates with small clumps and have not encountered any issues.
50 mg/mL apramycin stock
-
•
Apramycin sulfate 500 mg.
-
•
Sterile water 10 mL.
Add apramycin sulfate and water to sterile 15 mL conical tube. Vortex until fully suspended. Sterile filter into a sterile 15 mL conical tube using a 0.2 μm filter. Make 500 μL aliquots in sterile 1.5 mL microcentrifuge tubes. Store at -20°C for no longer than 1 year.
25 mg/mL nalidixic acid stock in 0.3 M sodium hydroxide
-
•
250 mg nalidixic acid.
-
•
3 mL of 1 M sodium hydroxide.
-
•
Fill to 10 mL with sterile water.
Weigh out nalidixic acid into a sterile 15 mL conical tube. Add 3 mL of 1 M sodium hydroxide. Fill to 10 mL with water. Vortex until fully solubilized. Sterile filter using a 0.2 μm filter into a sterile 15 mL conical tube. Make 500 μL aliquots in sterile 1.5 mL microcentrifuge tubes. Store at -20°C for no longer than 1 year.
50 mg/mL thiostrepton stock
-
•
500 mg thiostrepton.
-
•
10 mL 100% DMSO.
Add 500 mg thiostrepton to 15 mL conical tube. Fill to 10 mL with DMSO, vortexing in between. Make 0.5 mL aliquots in sterile 1.5 mL microcentrifuge tubes. Store at -20°C for no longer than a year.
SEC buffer – 150 mM sodium chloride, 20 mM Tris pH 7.5
-
•
20 mL Tris pH 7.5.
-
•
37.5 mL 4M sodium chloride.
-
•
Fill to 1 L with MilliQ water.
Add the Tris and sodium chloride to a 1 L graduated cylinder. Fill the cylinder to 1 L with MilliQ water with mixing. Filter and degas using 0.2 um vacuum filter. Store in 1 L bottle at 4°C.
Luria Broth (Miller)
-
•
12.5 grams of granulated Luria broth.
-
•
0.5 L of deionized water.
Mix the granulated Luria broth and deionized water together in 1 L bottle. Autoclave the bottle with the lid loosely screwed on. Store at room temperature or at 4°C.
2X-YT
-
•
1 capsule of 2X-YT mix.
-
•
0.5 L of deionized water.
Fill a 1 L bottle with 0.5 L of deionized water. Add 1 capsule of powdered 2X-YT mix to the bottle. Autoclave with the bottle lid loosely screwed on. Store at room temperature or at 4°C.
Procedure
(A) Gibson cloning of the encapsulin gene or operon into the pGM1190 vector
Section A outlines the steps required to PCR amplify and insert a single gene or a complete operon into the pGM1190 vector [11] which will be used as the S. coelicolor expression vector.
-
(1)PCR amplify insert using forward primer with overhang 5’-GTCAGAGAAGGGAGCGGACATATG…3’ and reverse primer with overhang 5′-ATTCGAGCTCGGTACCCGGGGGATCCCTA…3’. The overhang for the forward primer contains the start methionine, while the overhang for the reverse primer contains the stop codon, both shown in bold. These primers are designed to anneal to a pGM1190 vector that has been cut with NdeI and BamHI restriction enzymes during Gibson cloning, while also maintaining the NdeI/BamHI sites to directly restriction clone into the pGM1190 vector if necessary. The NdeI cut site is underlined in the forward primer and the BamHI cut site is underlined in the reverse primer. Maintaining the NdeI/BamHI sites also allows for plasmid screening by restriction digest.
-
(a)Note: PCR amplification from GC-rich templates such as Streptomyces genomic DNA can be challenging. We have found that the addition of 2–6% DMSO or use of the GC Buffer provided with Phusion High Fidelity DNA Polymerase kit can improve PCR efficiency. Additionally, annealing temperatures between 68 and 72°C have been found to be optimal. We have also seen higher rates of success using genomic DNA that has been partially fragmented via several freeze-thaw cycles. In some instances, we have also had improved success using the Q5 High-Fidelity DNA Polymerase kit (NEB, catalog number: M0491S) with the provided buffer for GC-rich targets.
-
(a)
-
(2)Cut pGM1190 vector (2 µg) with BamHI and NdeI restriction enzymes using manufacturer's recommended protocol.
-
(a)Note: 2 µg of pGM1190 vector should be sufficient for performing at least five Gibson assembly reactions in the later steps and compensate for low yields that commonly occur when purifying DNA by agarose gel extraction.
-
(a)
-
(3)
Run the digested pGM1190 vector from Step A.2. and PCR-amplified insert from Step A.1. over 1% agarose gel at 125 V for approximately 20–25 min, or until dye front reaches the end of the agarose gel.
-
(4)
Extract and purify DNA from Step 3 using NEB Monarch DNA gel extraction kit according to the manufacturer's recommended protocol.
-
(5)
Gibson assemble the insert and vector using 2X Gibson Master Mix according to the manufacturer's protocol. Our standard protocol is to prepare the mix with a 3:1 molar ratio of insert to template and incubate at 50°C for 15 min. Longer incubation times may be used when multiple fragments are being assembled in the same reaction.
-
(6)
Transform electrocompetent E. coli DH10B with 1 μL of the Gibson-assembled vector by electroporation using a 1 mm gap electroporation cuvette and 1.8 keV.
-
(7)
Resuspend the electroporated cells in 300 μL of SOC outgrowth media and pipette into a sterile 1.5 mL microcentrifuge tube. Shake the tubes at 200 rpm and 37°C for 45 min.
-
(8)Plate 100 μL of the transformants on LB-agar plates containing 50 μg/mL apramycin. Incubate the plates for 18 h at 37°C.
-
(a)Note: It may be useful to also plate 100 μL of a 1:10 dilution as competency can vary among batches and suppliers.
-
(a)
-
(9)
Screen colonies for insert by colony PCR or by restriction digest.
-
(10)
Pick 2–4 colonies and prepare overnight cultures in 5 mL LB containing 50 μg/mL apramycin.
-
(11)
Purify plasmids from the overnight growths using Qiagen miniprep plasmid extraction kit following the manufacturer's protocols.
-
(12)
Sequence the plasmids by Sanger sequencing.
-
(13)
After the plasmid sequence has been verified by Sanger sequencing, transform E. coli ET12567 (pUZ8002) chemically competent cells with the assembled plasmid by adding at least 50 ng of plasmid to a 40 μL aliquot of chemically competent ET12567 (pUZ8002) cells on ice. ET12567 is a methylation-deficient strain carrying the tra-containing plasmid pUZ8002 to optimize conjugation efficiency to Streptomyces hosts. Heat shock the cells in a 37°C water bath for 90 s, then put on ice for 3 min. Add 300 μL of SOC outgrowth media and shake the transformed cells at 37°C for 45 min at 200 rpm. Plate 50–100 μL of the transformed cells on LB-agar plates containing 50 μg/mL apramycin, 30 μg/mL chloramphenicol, and 50 μg/mL kanamycin. Incubate the plates overnight at 37°C. Colony-containing plates can be stored at 4°C for 1 month if necessary.
(B) Preparing the Streptomyces coelicolor spore stock for conjugation
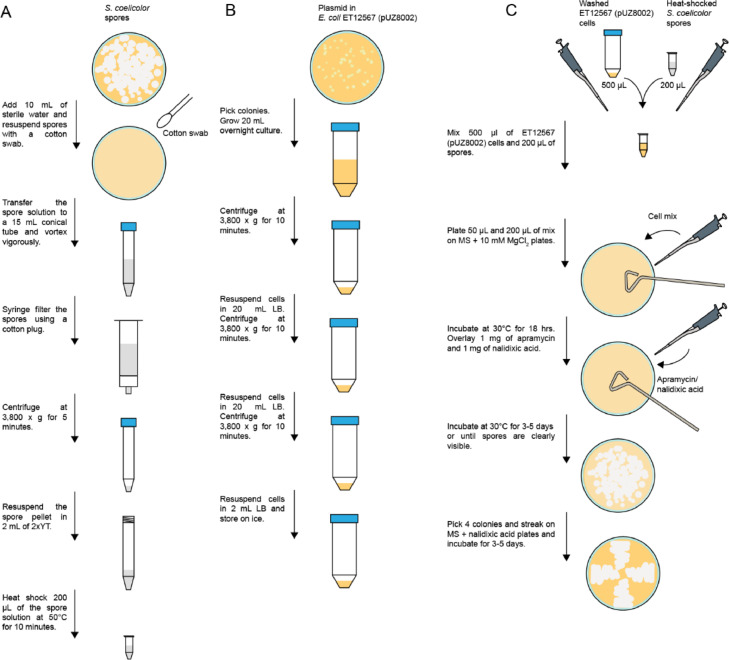
Section B addresses handling the S. coelicolor stock and creating a spore stock to be used for conjugation with E. coli ET12567 (pUZ8002). The steps outlined in Sections B and C very closely follow the protocols suggested by Tong et. al. [13] and Kieser et. al. [12]. All steps require the use of sterile media, containers, and pipette tips. It is critical that the S. coelicolor stocks do not get contaminated during handling. Fig. 1A illustrates the steps required to prepare a spore stock for conjugation as presented in Section B.
-
(1)In a laminar hood, resuspend the desiccated S. coelicolor stock from ATCC in 1 mL ISP medium #1. Pipette 100 μL of resuspended stock into a sterile centrifuge tube and prepare 1:10, 1:100, 1:1000, and 1:10,000 serial dilutions in ISP medium #1. Plate 100 μL of each serial dilution on fresh MS-agar plates, taking care to maintain proper sterile technique.
-
(a)Note: if a laminar hood is unavailable, Step B.1. and all subsequent steps that require handling Streptomyces during conjugation can be performed close to an active Bunsen burner on a benchtop surface.
-
(a)
-
(2)Take 500 μL of the remaining resuspended stock and add 500 μL of sterile 50% glycerol. Mix, and store at -80°C as a stock for future use.
-
(a)Note: Steps B.1. and B.2. assume the reader does not already have a viable Streptomyces coelicolor glycerol stock and is starting from the desiccated cells shipped from ATCC. If the reader already has a pre-existing glycerol stock, then scrape a small amount of the glycerol stock using a sterile pipette tip or sterile toothpick, resuspend in 200 μL of 2X-YT media, and plate on MS-agar plates using dilutions as in B.1.
-
(a)
-
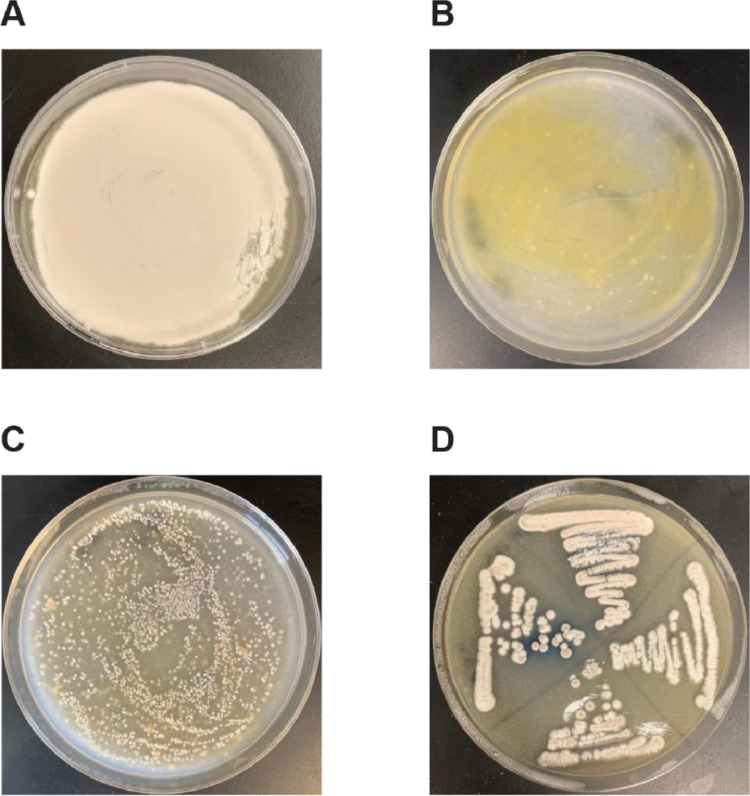
(3)Store plates inverted at 30°C for 2–5 days until plates are rich with visible spores. Refer to Fig. 2A for an example of a plate dense with spores.
-
(a)Note: Once sporulated, plates can be stored at 4°C for 1-2 months. However, it is best practice to use fresh spores for conjugation.
-
(a)
-
(4)
When ready to prepare a spore stock for conjugation, pipette 10 mL of 2X-YT onto the plate. Refer to Fig. 1A for a visual workflow of Steps B.4. to B.10.
-
(5)
Use a sterile cotton swab to gently remove the spores from the surface of the plate. Be careful not to dig into the MS-agar.
-
(6)
After the spores are completely resuspended, pipette them into a sterile 15 mL conical tube and vortex vigorously to break apart spore chains.
-
(7)
Pour over a sterile syringe containing a sterile cotton plug. Collect the flowthrough in a sterile conical tube.
-
(8)
Centrifuge the tube for 10 min at 3800 x g at 4°C to pellet spores.
-
(9)
Carefully discard the clarified supernatant.
-
(10)
Resuspend the spore pellet in 2 mL 2X-YT. This spore stock can be used for downstream conjugations and can be stored in the refrigerator for up to 2 weeks.
a. Note: The spore stock at this stage can also be stored in 20% glycerol at -80°C instead of 2X-YT. Spores from the 20% glycerol stock can also be used in the subsequent conjugation steps in Section C, however, we have observed significantly lower conjugation efficiencies from the 20% glycerol stocks. If a glycerol stock is used for conjugation, we recommend centrifuging the thawed glycerol stock for 10 min at 3800 x g to pellet the spores, then resuspend the spores in 2X-YT to be used in Section C.
Fig. 1.
Flowchart for transferring plasmids into S. coelicolor by conjugation. (A) Flowchart for preparing S. coelicolor spore stocks ready for conjugation according to Section B. (B) Flowchart for preparing E. coli ET12567 (pUZ8002) cells according to Section C. (C) Flowchart for conjugating plasmids into S. coelicolor as in Section C.
Fig. 2.
Images of plates for a successful conjugation. (A) Lawn of spores as should be expected in Step B.1. (B) A typical plate after an overnight incubation prior to antibiotic overlay as in Step C.9. (C) A plate containing S. coelicolor conjugates after 5 days after adding antibiotics as in Step C.11. (D) A succesful streak of 4 spore colonies onto MS-agar containing apramycin and nalidixic acid as in Step C.14.
(C) Conjugation of Streptomyces coelicolor
Section C provides the necessary steps to transfer the modified pGM1190 vector into S. coelicolor via conjugation with E. coli ET12567 (pUZ8002) cells. Steps C.1. to C.6. are illustrated in Fig. 1B. Steps C.7. to C.13. are illustrated in Fig. 1C.
-
(1)
The day before the conjugation, prepare an overnight culture of 20 mL LB containing 50 μg/mL apramycin, 30 μg/mL chloramphenicol, and 50 μg/mL kanamycin. Inoculate the 20 mL LB overnight culture with a colony of E. coli ET12567 (pUZ8002) containing the desired plasmid. Grow overnight with shaking at 37°C.
-
(2)
The next day, centrifuge the E. coli ET12567(pUZ8002) cells for 10 min at 3800 x g at 4°C.
-
(3)
Pipette 200 μL of the prepared spore stock from Step B.10. into a sterile microcentrifuge tube and incubate at 50°C for 10 min to activate the spores. After 10 min, the tube containing the spores can be stored at room temperature to briefly cool prior to conjugation.
-
(4)
Dispose of the supernatant from Step C.2., resuspend the E. coli ET12567 (pUZ8002) cell pellet in 20 mL LB, and centrifuge again for 10 min at 3800 x g at 4°C.
-
(5)
Repeat Step C.4.
-
(6)
After the E. coli ET12567 (pUZ8002) cells have been washed twice with LB, resuspend the final cell pellet in 2 mL LB, and store the tube containing the cells on ice.
-
(7)
Mix 500 μL of the E. coli ET12567 (pUZ8002) suspension with 200 μL of the activated S. coelicolor spores in a sterile tube.
-
(8)
Pipette 200 μL, 100 μL, and 50 μL of the conjugation mix on separate MS-Agar plates containing 10 mM MgCl2. Incubate the plates at 30°C for 18 h. See Fig. 2B for an example of a plate to be overlaid after an overnight incubation.
-
(9)
The following day prepare a solution containing 1 mg/mL nalidixic acid (from a stock in 300 mM NaOH) and 1 mg/mL apramycin in sterile water. Prepare 1 mL of this mixture for each plate to be overlayed.
-
(10)
Pipette 1 mL of the antibiotic overlay mixture to each plate. Spread the antibiotic over the plate by gently rocking. A spreader may be used to help spread the overlay, but it is not necessary and must be used very gently.
-
(11)
Incubate the plates at 30°C for 3–5 days until spores are visible. See Fig. 2C for an example of a plate with visible spores from a successful conjugation.
-
(12)Using sterile pipette tips or toothpicks, pick 4 spore colonies from Step C.11. and streak the spore colonies onto an MS-agar plate containing 50 μg/mL apramycin and 25 μg/mL nalidixic acid.
-
aNote: This step helps to enrich the S. coelicolor and further screen against S. coelicolor cells that do not contain the desired plasmid.
-
a
-
(13)
Incubate the struck plate at 30°C for 3–5 days until spores are visible. See Fig. 2C for an example of a well-struck plate with all positive conjugates.
(D) Small-scale expression in Streptomyces coelicolor
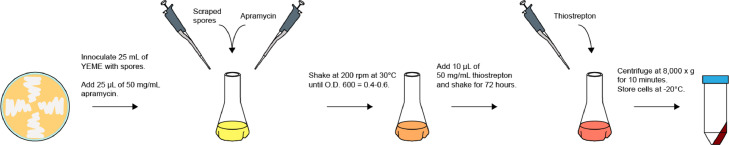
Section D provides the necessary steps for expressing an encapsulin or encapsulin-containing operon that has been inserted in the pGM1190 vector. The steps for Section D are illustrated in Fig. 3.
-
(1)
Prepare a sterile 125 mL baffled flask by adding 3–6 mm glass beads and autoclaving on dry cycle.
-
(2)
Prepare 500 mL YEME media stock.
-
(3)
Add 25 mL YEME to autoclaved 125 mL baffle flask.
-
(4)
Add 25 μL of 50 mg/mL apramycin to the flask.
-
(5)
Using a pipette tip, scrape off approximately ¼ of the spores from a single struck colony in Step C.13. Use this to inoculate the YEME media.
-
(6)
Shake at 30°C for ca. 72 h, or until O.D. 600 reaches approximately 0.4–0.8. The culture may start to appear red due to production of secondary metabolites.
-
(7)When the O.D. 600 reaches 0.4–0.8, induce protein expression by adding 10 μL of 50 mg/mL thiostrepton in DMSO to bring the thiostrepton concentration to 20 μg/mL.
-
aNote: The amount of thiostrepton can be modified to modulate expression levels if needed. Refer to Fig. S1 in the Further Notes section for the effect of different thiostrepton concentrations on overall expression levels.
-
a
-
(8)
Express the cells with shaking at 30°C for 24–72 h. The media will often appear dark red or dark blue due to the production of secondary metabolites.
-
(9)
After cells have been expressed for the desired amount of time, centrifuge cells for 10 min at 8000 x g. Discard the supernatant, and store cell pellet at -20°C. For longer term storage, store cell pellet at -80°C.
Fig. 3.
Flowchart for encapsulin expression in S. coelicolor. Outline of the steps required to produce proteins in S. coelicolor as presented in Section D.
(E) Protein purification from Streptomyces coelicolor
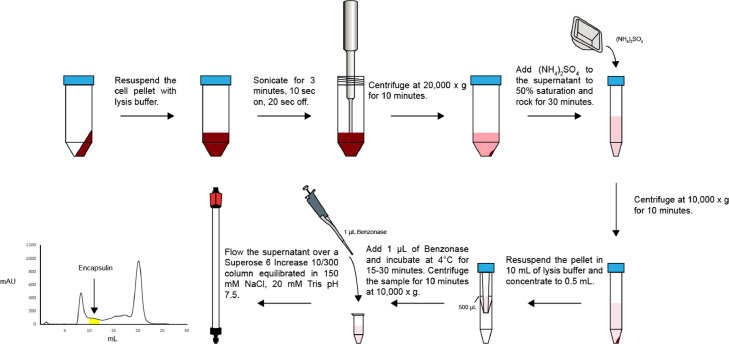
Section E outlines the steps required to purify overexpressed encapsulins from S. coelicolor. The steps for Section E are illustrated in Fig. 4. This method is suitable for purifying encapsulins from 1 and 2 grams of frozen S. coelicolor cell pellets.
-
(1)
Prepare 1 L of lysis buffer containing 150 mM NaCl, 20 mM Tris pH 7.5, 2 mM MgCl2.
-
(2)
Weigh the frozen cell pellet from Step D.9. Add cold lysis buffer at a ratio of 5 mL / 1 gram of cell pellet. Keep the lysate on ice as much as possible for the remainder of the purification.
-
(3)
Add 1 mg/mL of lysozyme to the lysis buffer.
-
(4)
Add 1 μL of Benzonase nuclease to the lysis buffer.
-
(5)
Add SIGMAFAST protease inhibitor cocktail. For small scale purifications, it may be helpful to prepare a 10X concentrated stock of SIGMAFAST protease inhibitor cocktail in lysis buffer and store at -80°C until use.
-
(6)
Sonicate the cell pellet using a 1/8-inch diameter tip with the following parameters: 80% power (18 watts), 10 s on, 20 s off, 3 min total sonication time. Keep the tube containing the lysate on ice the entire time.
-
(7)
Centrifuge the cell lysate for 10 min, 20,000 x g at 4°C in order to remove cell debris.
-
(8)Carefully collect supernatant into a 15 mL conical tube and place at 4°C for 30 min to improve Benzonase digestion of contaminating nucleic acids.
-
(a)Note: If nucleic acid contamination is not a concern, Step E.8. is not necessary.
-
(a)
-
(9)Add ammonium sulfate to the lysate to 50% saturation. For 1 mL of lysate, add 314 mg of ammonium sulfate. This can be scaled according to the volume of the lysate.
-
(a)Note: Ammonium sulfate at 50% saturation is a general recommendation for encapsulins. However, the amount of ammonium sulfate added may have to be adjusted depending on the surface charge distribution of the encapsulin protein being purified.
-
(a)
-
(10)Gently rock at 4°C for at least 30 min to precipitate protein.
-
(a)Note: Longer incubations in ammonium sulfate can often increase yield.
-
(a)
-
(11)
Centrifuge sample for 10 min at 10,000 x g, 4°C to precipitate the protein sample.
-
(12)
Resuspend the protein pellet immediately in 10 mL of 150 mM NaCl, 20 mM Tris pH 7.5, 2 mM MgCl2 and concentrate to at least 0.5 mL in a 100 kDa cutoff Amicon concentrator. This step helps to reduce the concentration of ammonium sulfate in the protein sample and can prevent downstream precipitation of protein. Additionally, removing ammonium sulfate can improve the activity of Benzonase.
-
(13)
Add 0.5 μL of Benzonase nuclease to the sample. Let the protein sample incubate at 4°C for 15–30 min.
-
(14)
Centrifuge the protein sample for 10 min, 10,000 x g, 4°C. There will likely be a deep red pellet consisting mostly of insoluble material. The supernatant may also appear red.
-
(15)
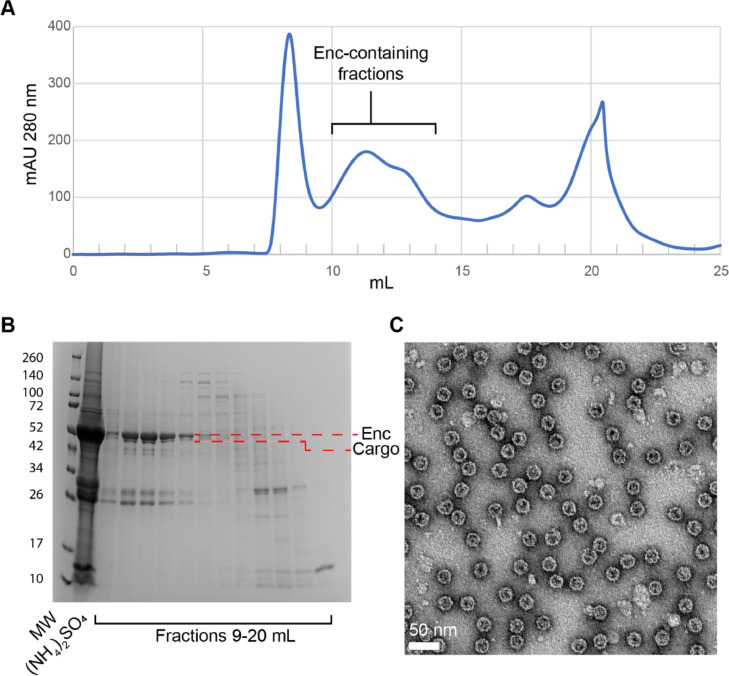
Run the clarified supernatant over a Superose 6 Increase 10/300 GL column pre-equilibrated in 150 mM NaCl, 20 mM Tris pH 7.5 at a flow rate of 0.5 mL/min and collect 1 mL fractions. T1 encapsulins typically elute between 10 and 13 mL. See Fig. 5A for a typical elution profile when running on a Superose 6 Increase 10/300 GL column.
-
(16)
Verify the purity of encapsulin-containing fractions by running the protein-containing fractions on an SDS-PAGE gel. See Fig. 5B for an example SDS-PAGE gel from an encapsulin purification.
-
(17)
Store the protein sample at 4°C.
-
(18)
The protein sample should be clean enough after this step for analysis by negative stain transmission electron microscopy (TEM) and even cryo-EM. See Fig. 5C for an example negative stain TEM micrograph of encapsulins according to the steps provided in Section E. If higher purity is required, it may be necessary to further purify the protein by ion exchange chromatography.
Fig. 4.
Outline of encapsulin purification from S. coelicolor. Steps required to purify an overexpressed encapsulin from S. coelicolor according to Section E.
Fig. 5.
Successful purification of an encapsulin from S. coelicolor. (A) Size exclusion chromatography (Superose 6 Increase 10/300 GL) of an encapsulin (Q54255) and its cargo protein (A0A2 × 2LWM5) according to Section E. T1 encapsulin samples generally elute between 10 and 13 mL. Enc: encapsulin. (B) SDS-PAGE gel of the encapsulin sample purified in panel A. Enc and the cargo protein bands were unambiguously identified via tryptic digest and mass spectrometry identification. The bands around the 26 and 34 kDa marker bands represent the following often co-purifying ribosomal proteins as identified via tryptic digest and mass spectrometry: TsnR family 23S rRNA methyltransferase, 29 kDa (A0A6M9XPU2), 30S ribosomal protein S3, 30 kDa (Q9L0D4), 30S ribosomal protein S4, 24 kDa (Q9KXP5). The encapsulin protein is most enriched in fractions 10–12 and is less enriched in fraction 13. MW: molecular weight marker, (NH4)2SO4: ammonium sulfate pellet. (C) Negative stain TEM micrograph of fraction 10 from panels A and B. The micrograph was collected using an FEI Morgagni 100 kV electron microscope. The sample concentration: 0.1 mg/mL, stain: 0.2% uranyl formate, grid: freshly glow-discharged formvar-reinforced carbon grid (EMS, FCF200-AU-EC).
Method validation and further notes
Effect of thiostrepton on expression levels
We have observed relatively consistent expression levels between 5 and 20 μg/mL thiostrepton. Fig. S1 illustrates the different levels of expression of encapsulin and cargo protein when induced at different concentrations of thiostrepton at different time points. We chose 20 μg/mL thiostrepton for 72 h for our protocol due to the reduced background and increased mass of cell pellet compared to 24 h of induction.
S. coelicolor expression improves cargo loading and solubility
While yields can vary depending on the protein being expressed, we have found the protocol presented here to provide a more homogenous sample with significantly improved cargo-loading and solubility compared with E. coli expression. To demonstrate this, we initially co-expressed a Family 2B encapsulin and its cargo protein from S. griseus in E. coli and observed that only 34% of encapsulins contained apparent cargo by 2D class averages of extracted particles from a cryo-EM dataset as shown in Fig. S2A and S2C. When the same proteins were expressed in S. coelicolor, we observed cargo in almost 96% of the particles from based on a cryo-EM dataset as shown in Fig. S2B and S2C.
We further expressed the same Family 2B encapsulin without cargo in both S. coelicolor and E. coli and found that the S. coelicolor overexpression dramatically improved the solubility of the encapsulin compared to expression in E. coli, as seen in Fig. S2D. The protein purified from S. coelicolor additionally had improved purity over protein purified from E. coli following the same purification protocol.
Ethics statement
CRediT authorship contribution statement
Michael P. Andreas: Conceptualization, Methodology, Investigation, Writing – original draft, Writing – review & editing, Visualization. Tobias W. Giessen: Conceptualization, Writing – original draft, Writing – review & editing, Visualization, Supervision.
Declaration Competing of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
We gratefully acknowledg funding from the NIH (R35GM133325). Research reported in this publication was supported by the University of Michigan Cryo-EM Facility (U-M Cryo-EM). U-M Cryo-EM is grateful for support from the U-M Life Sciences Institute and the U-M Biosciences Initiative.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.mex.2022.101787.
Appendix. Supplementary materials
Data availability
Data will be made available on request.
References
- 1.Giessen T.W. Encapsulins. Annu. Rev. Biochem. 2022;91:353–380. doi: 10.1146/annurev-biochem-040320-102858. https://www.ncbi.nlm.nih.gov/pubmed/35303791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andreas M.P., Giessen T.W. Large-scale computational discovery and analysis of virus-derived microbial nanocompartments. Nat. Commun. 2021;12(1):4748. doi: 10.1038/s41467-021-25071-y. https://www.ncbi.nlm.nih.gov/pubmed/34362927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giessen T.W., Silver P.A. Widespread distribution of encapsulin nanocompartments reveals functional diversity. Nat. Microbiol. 2017;2:17029. doi: 10.1038/nmicrobiol.2017.29. https://www.nature.com/articles/nmicrobiol201729 [DOI] [PubMed] [Google Scholar]
- 4.Altenburg W.J., Rollins N., Silver P.A., Giessen T.W. Exploring targeting peptide-shell interactions in encapsulin nanocompartments. Sci. Rep. 2021;11(1):4951. doi: 10.1038/s41598-021-84329-z. https://www.ncbi.nlm.nih.gov/pubmed/33654191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nichols R.J., LaFrance B., Phillips N.R., Radford D.R., Oltrogge L.M., Valentin-Alvarado L.E., Bischoff A.J., Nogales E., Savage D.F. Discovery and characterization of a novel family of prokaryotic nanocompartments involved in sulfur metabolism. Elife. 2021;10:e59288 doi: 10.7554/eLife.59288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sutter M., Boehringer D., Gutmann S., Gunther S., Prangishvili D., Loessner M.J., Stetter K.O., Weber-Ban E., Ban N. Structural basis of enzyme encapsulation into a bacterial nanocompartment. Nat. Struct. Mol. Biol. 2008;15(9):939–947. doi: 10.1038/nsmb.1473. https://www.ncbi.nlm.nih.gov/pubmed/19172747 [DOI] [PubMed] [Google Scholar]
- 7.Tang Y., Mu A., Zhang Y., Zhou S., Wang W., Lai Y., Zhou X., Liu F., Yang X., Gong H., Wang Q., Rao Z. Cryo-EM structure of Mycobacterium smegmatis DyP-loaded encapsulin. Proc. Natl. Acad. Sci. U.S.A. 2021;118(16) doi: 10.1073/pnas.2025658118. e2025658118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akita F., Chong K.T., Tanaka H., Yamashita E., Miyazaki N., Nakaishi Y., Suzuki M., Namba K., Ono Y., Tsukihara T., Nakagawa A. The crystal structure of a virus-like particle from the hyperthermophilic archaeon Pyrococcus furiosus provides insight into the evolution of viruses. J. Mol. Biol. 2007;368(5):1469–1483. doi: 10.1016/j.jmb.2007.02.075. https://www.ncbi.nlm.nih.gov/pubmed/17397865 [DOI] [PubMed] [Google Scholar]
- 9.McHugh C.A., Fontana J., Nemecek D., Cheng N.Q., Aksyuk A.A., Heymann J.B., Winkler D.C., Lam A.S., Wall J.S., Steven A.C., Hoiczyk E. A virus capsid-like nanocompartment that stores iron and protects bacteria from oxidative stress. EMBO J. 2014;33(17):1896–1911. doi: 10.15252/embj.201488566. <Go to ISI>://WOS:000341839500008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giessen T.W., Orlando B.J., Verdegaal A.A., Chambers M.G., Gardener J., Bell D.C., Birrane G., Liao M., Silver P.A. Large protein organelles form a new iron sequestration system with high storage capacity. Elife. 2019;8:e46070 doi: 10.7554/eLife.46070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muth G. The pSG5-based thermosensitive vector family for genome editing and gene expression in actinomycetes. Appl. Microbiol. Biotechnol. 2018;102(21):9067–9080. doi: 10.1007/s00253-018-9334-5. https://www.ncbi.nlm.nih.gov/pubmed/30191290 [DOI] [PubMed] [Google Scholar]
- 12.Kieser T., Bibb M.J., Buttener M.J., Chater K.F., Hopwood D.A. John Innes Foundation; 2000. Practical Streptomyces Genetics. ISBN: 0-7084-0623-8. [Google Scholar]
- 13.Tong Y., Whitford C.M., Blin K., Jorgensen T.S., Weber T., Lee S.Y. CRISPR-Cas9, CRISPRi and CRISPR-BEST-mediated genetic manipulation in streptomycetes. Nat. Protoc. 2020;15(8):2470–2502. doi: 10.1038/s41596-020-0339-z. https://www.ncbi.nlm.nih.gov/pubmed/32651565 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.