Abstract
Escherichia coli O157:H7 and O157 nonmotile isolates (E. coli O157) previously were recovered from feces, hides, and carcasses at four large Midwestern beef processing plants (R. O. Elder, J. E. Keen, G. R. Siragusa, G. A. Barkocy-Gallagher, M. Koohmaraie, and W. W. Laegreid, Proc. Natl. Acad. Sci. USA 97:2999–3003, 2000). The study implied relationships between cattle infection and carcass contamination within single-source lots as well as between preevisceration and postprocessing carcass contamination, based on prevalence. These relationships now have been verified based on identification of isolates by genomic fingerprinting. E. coli O157 isolates from all positive samples were analyzed by pulsed-field gel electrophoresis of genomic DNA after digestion with XbaI. Seventy-seven individual subtypes (fingerprint patterns) grouping into 47 types were discerned among 343 isolates. Comparison of the fingerprint patterns revealed three clusters of isolates, two of which were closely related to each other. Remarkably, isolates carrying both Shiga toxin genes and nonmotile isolates largely fell into specific clusters. Within lots analyzed, 68.2% of the postharvest (carcass) isolates matched preharvest (animal) isolates. For individual carcasses, 65.3 and 66.7% of the isolates recovered postevisceration and in the cooler, respectively, matched those recovered preevisceration. Multiple isolates were analyzed from some carcass samples and were found to include strains with different genotypes. This study suggests that most E. coli O157 carcass contamination originates from animals within the same lot and not from cross-contamination between lots. In addition, the data demonstrate that most carcass contamination occurs very early during processing.
Escherichia coli O157:H7 or O157 nonmotile (both referred to herein as E. coli O157) are classified as enterohemorrhagic E. coli and can cause diseases ranging in severity from nonbloody diarrhea (46) to hemolytic-uremic syndrome and death. Several factors have been associated with E. coli O157 virulence, including production of at least one of two Shiga toxins, intimin, and enterohemolysin (37, 41). E. coli O157:H7 has been declared an adulterant in ground beef due to frequent association of disease with consumption of undercooked hamburgers (26, 49). The organism commonly is present in cattle feces, suggesting that the animal is the source of beef contamination.
Studies clarifying the direct role of animal infection in subsequent carcass contamination, as well as the frequency of cross-contamination, have been limited (12, 13). A few reports have suggested that hides are a significant source of bacterial carcass contamination (6, 10, 34, 44). Most reports have approached the problem by examining the potential for carcass contamination at critical processing steps by following changes in total aerobic, coliform, or generic E. coli counts or some combination of these (6, 17–19). These measurements can imply causality but lack the ability to directly link sources with the introduction of specific organisms (contamination events). Pulsed-field gel electrophoresis (PFGE) genotyping has been used to track sources of Listeria contamination (20), and commonly is used by the Centers for Disease Control and Prevention and others to track sources of E. coli O157 outbreaks (e.g., see references 2, 5, 7, 24, and 42). However, prior to this study it had not been used to track E. coli O157 contamination of carcasses.
Previously, we reported the preharvest and postharvest prevalence of E. coli O157 at four large, Midwestern processing plants during July and August (13). Samples were taken from animals (preharvest) and carcasses (postharvest) within the same lot, but not necessarily from the same animals. A correlation was noted between the prevalence of E. coli O157 found preharvest and postharvest. Furthermore, carcasses found contaminated in the cooler also were found to be contaminated preevisceration. These correlations suggested relationships (i) between isolates entering the plant with animals and those that appear on carcasses within the same lot and (ii) between isolates found on carcasses in the cooler and those found on the carcass earlier in processing. However, in the absence of specific identification of the isolates these relationships could not be confirmed. The recovered E. coli O157 isolates have now been characterized by XbaI PFGE genotyping. The data have been used to track and confirm the sources of carcass contamination throughout processing and to examine isolate relatedness along with genotypic variability.
MATERIALS AND METHODS
E. coli O157 isolates.
The recovery of E. coli O157 isolates from cattle feces, hides, and carcasses has been described previously (13). Within each single-source lot, different samples were not necessarily taken from the same animals, but each individual carcass was tracked and sampled preevisceration, postevisceration, and postprocessing (final sampling, in the cooler). Lots ranged in size from 35 to 85 animals, and 20% of each lot was sampled. The characterization of one isolate per positive sample was described in regards to biochemical and immunological analyses, the presence of toxin and other virulence genes, and motility (13). At the time, up to three additional E. coli O157 isolates from postharvest samples also were recovered and stored from additional, randomly chosen, morphologically correct colonies picked from selective plates after enrichment and immunomagnetic separation. Some of these secondary isolates have now been characterized by the same methods.
Genomic fingerprint analyses.
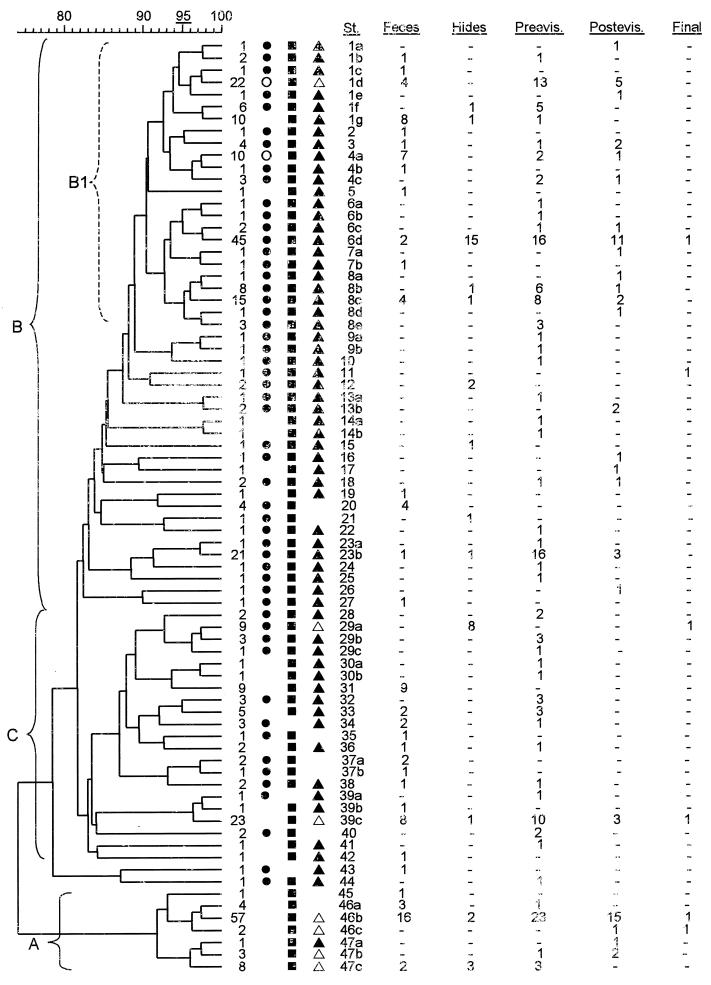
E. coli. O157 isolate fingerprints generated and analyzed in this study were based on PFGE separation of XbaI-digested genomic DNA as previously described (11); this is the method used by members of PulseNet (http://www.cdc.gov/ncidod/dbmd/pulsenet/pulsenet.htm). Pulsed-field gel certified agarose was obtained from Bio-Rad (Hercules, Calif.); Tris-borate-EDTA running buffer and lysozyme were purchased from Sigma (St. Louis, Mo.). XbaI and RsaI were purchased from New England Biolabs (Beverly, Mass.), and Taq polymerase was purchased from Promega (Madison, Wis.). Lambda concatemers (Bio-Rad) were used as size markers. E. coli O157 strain G5244 (Bio-Rad) was used as a control and for standardization of gels. Banding patterns were analyzed and comparisons made using Molecular Analyst Fingerprinting software (Bio-Rad), employing the Dice similarity coefficient in conjunction with the unweighted pair group method using arithmetic averages (UPGMA) for clustering. Isolates were grouped into types that likely had the same origin based on fingerprint pattern similarities. Types were defined strictly as isolates that grouped together and had one; one and two; or one, two, and three band differences among their fingerprints (approximately >95% Dice similarity) (Fig. 1). Isolates with two or three band differences in their fingerprints, but not grouping with isolates that had a one-band difference, were classified as distinct types. Subtypes were defined as isolates with identical fingerprint patterns.
FIG. 1.
Relatedness of E. coli O157 isolates. The dendrogram was generated using the Dice coefficient and UPGMA analysis (see Materials and Methods). The scale at the top of the dendrogram indicates the level of similarity between isolates; types include isolates connected at approximately >95% similarity. One isolate of each distinct genomic pattern (subtype) was included in the dendrogram; thus, a final branch may represent multiple isolates as indicated at the end of each branch. The presence of stx1 is indicated by solid circles; the presence of stx2 is indicated by solid squares. Subtypes that include isolates that carry stx1 and stx2 as well as isolates that carry only stx2 are indicated by open circles and solid squares. The presence of only motile isolates is indicated by filled triangles. The presence of nonmotile and motile isolates is indicated by open triangles. The absence of a symbol indicates the absence of that characteristic from all isolates within the subtype. The numbers of isolates within each subtype that were recovered at each sampling site are indicated. Subtype designations follow the symbols. Abbreviations: St., subtype designations; Preevis., preevisceration; Postevis., postevisceration.
Genomic fingerprint stability.
Fourteen isolates were selected for analysis of genome stability. These isolates included pairs from types 23, 4, 6, and 1, each with fingerprints differing by one band (Fig. 1). Also included were three isolates each from types 39 and 46; each group included two identical isolates from large subtypes and one isolate with a one-band difference. Cells were recovered from frozen (−70°C) stocks and heavily streaked onto Trypticase soy agar (TSA) (Difco Laboratories, Detroit, Mich.), followed by overnight incubation at 37°C. This was considered the day 1 culture. An isolated colony was subsequently subcultured (passaged) daily, alternating TSA and sorbitol MacConkey's agar supplemented with cefixime (0.5 mg/liter) and potassium tellurite (2.5 mg/liter; Dynal, Lake Success, N.Y.) (ctSMAC). At days 1, 5, 10, and 15 genomic DNA fingerprints were prepared as described above. In addition, cells from the same isolated colony subcultured from TSA to ctSMAC on day 1 were transferred to 3 ml of brilliant green bile broth (Difco) and passaged daily at 37°C. Genomic DNA fingerprints were prepared from these broth cultures at day 3.
PCR-restriction fragment length polymorphism analysis.
The presence of the H7 gene was detected in the nonmotile isolates by the method of Fields et al. (15). In brief, approximately 1.8 kb of the fliC gene was amplified by PCR. Annealing temperatures were adjusted from 60 to 45°C as necessary. RsaI digests of the PCR products were examined by agarose gel electrophoresis for the characteristic H7 banding pattern.
Statistical analyses.
Chi-squared analyses were performed to compare frequencies of types per group using the general linear module of SAS (SAS Institute, Inc., Cary, N.C.). Results were considered significant at P ≤ 0.5.
RESULTS
Relatedness and distribution of E. coli O157 isolates.
One randomly selected E. coli O157 isolate from each positive sample recovered during a study in beef processing plants (13) was examined by XbaI PFGE genomic DNA fingerprinting. A total of 77 different patterns, or subtypes, were identified. The isolates divided into three main clusters (see Materials and Methods and Fig. 1). Cluster A included 76 isolates recovered from five lots during two trips to the same plant (Fig. 1 and Table 1). Clusters B and C were derived from one branch of the dendrogram and included 191 and 74 isolates, respectively (Fig. 1). A smaller group of tightly related strains within cluster B included 142 isolates (cluster B1 [Fig. 1]). Two isolates fell outside of the clusters but were more closely related to clusters B and C (types 43 and 44 [Fig. 1]).
TABLE 1.
Distribution of subtypes among lots
| Lota | Subtype(s)b recovered within cluster:
|
Nonclustered subtype | ||||
|---|---|---|---|---|---|---|
| B1 | B (not B1) | C | A | |||
| A1-1 | 1d, 1g, 4a | 23b | 43 | |||
| A1-2 | 14a, 14b, 23b | |||||
| A1-3 | 23a, 23b | 39c | ||||
| A2-1 | 20 | |||||
| A2-2 | 4c | |||||
| A2-3 | 4c | 33 | ||||
| A2-4 | 1d | 23b, 27 | ||||
| B1-1 | ||||||
| B1-2 | 25 | |||||
| B1-3 | 1d, 8a, 8b, 8e | |||||
| B1-4 | 1d, 7b, 8b | 31 | ||||
| B2-1 | 5, 6a, 6d | 29a | ||||
| B2-2 | 1b, 1c, 6c, 6d, 8c | 21 | 29a, 38 | |||
| B2-3 | 6b, 6d | 10, 19, 26 | 29a | |||
| B2-4 | 6d | 12, 15 | 30a | 44 | ||
| C1-1 | 1d, 1e, 1f, 4b, 8d, 8e | 11 | 46b | |||
| C1-2 | 33, 41 | 45, 46a, 46b, 46c | ||||
| C1-3 | 1d, 1f, 7a | 46b, 47a | ||||
| C1-4 | 1d, 4a | 16 | 35 | 46b, 47b, 47c | ||
| C2-1 | 8c | 46a, 46b, 46c, 47b | ||||
| C2-2 | 3 | 9a, 13b, 22 | 37a, 37b | |||
| C2-3 | 1a, 3 | 13b | 29b, 39c | |||
| C2-4 | 9b, 13a, 24 | 29c, 28, 34, 36 | ||||
| 39a, 39b, 39c | ||||||
| D1-1 | 1d | 32 | ||||
| D1-2 | 1b | 32 | ||||
| D1-3 | ||||||
| D1-4 | 17 | 42 | ||||
| D2-1 | 39c | |||||
| D2-2 | 1f, 2, 8c | 18 | 33, 40 | |||
| D2-3 | 1f, 8c | 30b, 31 | ||||
For lot designations, the letter indicates the plant, the first number indicates the trip, and the last number indicates the lot.
The subtypes of E. coli O157 recovered from samples within each lot are indicated. Subtypes are grouped according to overall clusters (see Fig. 1).
Isolates also were typed or categorized into closely related groups as described in Materials and Methods. Forty-seven types were identified. Clusters A, B, and C included 3, 27, and 15 types, respectively (Fig. 1). Thirty-seven types included a small number of isolates and were predominately found in cluster C and in cluster B outside of B1 (65 isolates, one to five per type [Fig. 1]). Types 1, 6, and 46 accounted for 155 of the 343 isolates analyzed (45.2% [Fig. 1]). Isolates of type 1 were recovered from samples taken at all of the plants (Table 1). Isolates of types 6 and 46 were recovered from several lots sampled at individual plants (Table 1). These data do not indicate that the isolates are endemic in the plants, because they were recovered from preharvest samples in addition to postharvest samples (Fig. 1). Instead, they may represent geographically predominant strains.
The data were examined to determine if specific characteristics were associated with closely related isolates versus isolates distributed throughout the XbaI PFGE clusters. Isolates with different Shiga toxin profiles were distributed unevenly among the clusters; strains carrying only one stx gene were predominantly found among clusters A and C. These clusters included 67% of the subtypes (56% of the types) with isolates carrying only one stx gene, even though they accounted for just 36% of the total number of subtypes (38% of the types). These clusters also accounted for only 19% of the subtypes (22% of the types) with isolates carrying both stx genes. Overall, clusters A and C accounted for 81.6% of the stx2 isolates, while cluster B included 85.9% of the stx1 stx2 isolates (Fig. 1). Furthermore, cluster A included 52 of the 73 nonmotile isolates (71.2%), but at least one motile isolate was recovered for five of the seven subtypes in this cluster (Fig. 1). Only one nonmotile isolate grouped into cluster B1 (Fig. 1). The presence of the H7 fliC gene in all of the nonmotile isolates was confirmed by PCR-restriction fragment length polymorphism analysis (data not shown).
The data were further examined by chi-square analyses to determine if various characteristics were associated specifically with isolates recovered from particular types of samples (see Materials and Methods). Some bias was found between isolates from various sample sites in the XbaI PFGE genotype cluster, Shiga toxin profile, and motility characteristics (Table 2). Fecal isolates were underrepresented in cluster B relative to isolates from other sample sites. Compared to isolates from other sample sites, postevisceration isolates were relatively less prevalent in cluster C and more prevalent in cluster A. Also, a substantially smaller proportion of fecal isolates than carcass isolates carried both stx1 and stx2. The converse was true for isolates carrying only stx2. Finally, a larger proportion of preharvest isolates than postharvest isolates were nonmotile.
TABLE 2.
Distribution of phenotypic and genotypic characteristics based on subtypes, types, and sample sites
| No. (%) of:
|
No. (%) of isolates recovered from sample siteb or time
|
No. (%) of isolates overall | ||||||
|---|---|---|---|---|---|---|---|---|
| Subtypesa | Typesa | Feces | Hide | Preevis. | Postevis. | Final | ||
| Cluster | ||||||||
| B | 47 (61) | 27 (57) | 39 (43) | 24 (63) | 88 (59) | 38 (63) | 2 (33) | 191 (56) |
| C | 21 (27) | 15 (32) | 29 (32) | 9 (24) | 31 (21) | 3 (5) | 2 (33) | 74 (22) |
| A | 7 (9) | 3 (6) | 22 (24) | 5 (13) | 28 (19) | 19 (32) | 2 (33) | 76 (22) |
| stx genotype | ||||||||
| stx1 | 3 (4) | 3 (6) | 3 (3) | 0 (0) | 2 (1) | 0 (0) | 0 (0) | 5 (1) |
| stx2 | 24 (31) | 16 (34) | 58 (64) | 7 (18) | 54 (36) | 25 (42) | 3 (50) | 147 (43) |
| stx1 stx2 | 52 (68) | 31 (66) | 30 (33) | 31 (82) | 92 (62) | 35 (58) | 3 (50) | 191 (56) |
| Motility | ||||||||
| Motile | 69 (90) | 41 (87) | 63 (69) | 24 (63) | 128 (86) | 50 (83) | 5 (83) | 270 (79) |
| Nonmotile | 15 (19) | 11 (23) | 28 (31) | 14 (37) | 20 (14) | 10 (17) | 1 (17) | 73 (21) |
The sum of the number of subtypes and types carrying specified stx genes or classified by motility is greater than the total numbers of each because some subtypes and types include isolates with more than one Shiga toxin profile or include both motile and nonmotile isolates (see Fig. 1).
Abbreviations: Preevis., preevisceration; Postevis., postevisceration.
Genomic variation among preharvest and postharvest E. coli O157 isolates overall.
The variation in genomic fingerprints of isolates recovered both preharvest and postharvest was examined; fingerprints of both types of isolates varied substantially (Table 3). Across all lots with at least one positive sample, there was an average of one new type per 4.6 preharvest isolates and per 6.5 postharvest isolates. Thirty-two types included only preharvest or only postharvest isolates. Twenty of these types included only one isolate, and none included more than nine isolates (Fig. 1).
TABLE 3.
Variation in E. coli O157 recovered at packing plantsa
| Collection time and/or site | No. of isolates | No. of types | % Uniqueb |
|---|---|---|---|
| Preharvest | 129 | 28 | 14.7 |
| Feces | 91 | 23 | 16.5 |
| Hide | 38 | 12 | 10.5 |
| Postharvest | 214 | 34 | 13.6 |
| Preevisceration | 148 | 28 | 13.5 |
| Postevisceration | 60 | 17 | 13.6 |
| Final | 6 | 5 | 16.7 |
| Overall | 343 | 47 | 14.0 |
Isolates were typed based on relatedness of XbaI PFGE genomic fingerprint patterns (see Materials and Methods).
Percentage of unique isolates, or isolates unlike any other within the same lot.
In order to discern the potential for the presence of multiple isolate types in the samples, genomic fingerprints were generated for a group of 153 supplemental E. coli O157 isolates. This group was comprised of one to three additional isolates per sample (depending on availability) from 61 of the postharvest samples. The isolates were from samples in lots C1-3 and B2-2 or were from samples with an initial isolate that was (i) of a different genomic type than all preharvest isolates in the same lot or (ii) of a different genotype than another isolate(s) from a sample(s) of the same carcass taken at a different processing point(s). One or more of the additional isolates from 36 samples had a genomic fingerprint distinct from that of the initial sample isolate, although in many cases the change was by one band (data not shown). As many as three genomically distinct isolates were found within a sample. Seven new types were identified among the additional E. coli O157 isolates.
Genomic variation among preharvest and postharvest E. coli O157 isolates within lots.
The diversity of E. coli O157 isolates within each lot was investigated. A surprisingly high number of E. coli O157 genomic types were recovered within each lot: as many as six types for 11 preharvest isolates and up to eight types for 15 postharvest isolates (Table 1 and data not shown). Within lots that had at least one positive sample, an average of one new type was recovered per 2.6 preharvest isolates and per 2.9 postharvest isolates. Larger lots did not necessarily include more types than smaller lots. Some of the most distantly related isolates were recovered from the same lots (e.g., lot C1-1 [Table 1]). Twenty-one lots included at least one isolate of a unique type within that lot, i.e., an isolate that was unlike any others recovered within that lot. Unique isolates were recovered proportionately from preharvest samples as well as postharvest samples (Table 2).
In order to discern a link between carriage by live animals and carcass contamination, the 17 lots with at least two preharvest and two postharvest isolates were examined by lot for a correlation between preharvest and postharvest isolate types (Table 4). Isolates of the same type were considered “a match,” or “matching.” Overall, within these 17 lots 68.2% (120 of 176) of the isolates recovered postharvest matched at least one preharvest isolate from the same lot. Statistical analyses of these data were not possible due to the large number of unknown variables, such as the total potential number of isolate types per sample.
TABLE 4.
E. coli O157 isolate matches
| Lota | No. of postharvest matching preharvestb | Total no. of postharvest samplesc | No. of postevisceration matching preeviscerationd | Total no. of carcass samplese |
|---|---|---|---|---|
| A1-1 | 13 | 13 | 3 | 3 |
| A1-2 | ||||
| A1-3 | 5 | 9 | ||
| A2-1 | ||||
| A2-2 | ||||
| A2-3 | ||||
| A2-4 | ||||
| B1-1 | ||||
| B1-2 | ||||
| B1-3 | 1 | 2 | ||
| B1-4 | 4 | 7 | ||
| B2-1 | 11 | 12 | 4 | 4 |
| B2-2 | 16 | 17 | 5 | 7 |
| B2-3 | 5 | 7 | 0 | 1 |
| B2-4 | 1 | 3 | ||
| C1-1 | 0 | 8 | 1 | 2 |
| C1-2 | 28 | 29 | 10 | 11 |
| C1-3 | 5 | 14 | 2 | 6 |
| C1-4 | 8 | 14 | 1 | 5 |
| C2-1 | 3 | 5 | 1 | 1 |
| C2-2 | 1 | 4 | 0 | 1 |
| C2-3 | 0 | 1 | ||
| C2-4 | 9 | 15 | 1 | 2 |
| D1-1 | ||||
| D1-2 | ||||
| D1-3 | ||||
| D1-4 | ||||
| D2-1 | 2 | 2 | ||
| D2-2 | 9 | 13 | 3 | 3 |
| D2-3 | 0 | 4 |
For lot designations, the letter indicates the plant, the first number indicates the trip, and the last number indicates the lot. Lots indicated in boldface type included preharvest isolates unlike any postharvest isolates.
The number of postharvest samples with isolates of the same type as preharvest sample isolates. Only the 17 lots including at least two preharvest and two postharvest E. coli O157 positive samples were analyzed.
The total number of postharvest samples testing E. coli O157 positive. Only the 17 lots including at least two preharvest and two postharvest E. coli O157 positive samples were included.
The number of carcasses with matching postevisceration and preevisceration isolates. Only lots with carcasses testing positive at both sites were included.
The total number of carcasses testing E. coli O157 positive both preevisceration and postevisceration. Only lots with carcasses testing positive at both sites were included.
Only one isolate was examined from each positive sample, leaving open the possibility that a different isolate from the same sample would match within the lot. One to three additional isolates (depending on availability) were examined from 42 postharvest samples for which the initial isolate did not match any preharvest isolate within the same lot. Isolates with different genomic types were recovered from 24 of these samples. At least one additional isolate matched a preharvest isolate from the same lot for eight of the samples, slightly increasing, to 72.7% (128 of 176), the proportion of identified matches between postharvest and preharvest isolates. Seven of the additional isolates were of unique types, i.e., unlike any other within the same lot.
Genomic variation among postharvest E. coli O157 isolates by carcass.
The fingerprints of isolates from carcass samples taken throughout processing were compared, in order to discern if the carcass contamination found later in processing corresponded to that which was on the same carcass early in processing. The initial preevisceration and postevisceration isolates examined from 32 of the 49 carcasses contaminated at both processing points (65.3%) were of the same genomic type, i.e., matched (Table 4). Additional preevisceration and/or postevisceration isolates from 15 of the carcasses were examined that did not have matching initial isolates. Matches were revealed between preevisceration and postevisceration isolates from seven more carcasses. Thus, the detected proportion of carcasses with postevisceration isolates traceable to preevisceration isolates increased to 79.6% (39 of 49). Again, statistical analyses were impossible.
Upon first analysis, four of the isolates from the six final samples positive for E. coli O157 matched those recovered preevisceration from the same carcass. For the remaining two carcasses, additional isolates from the positive carcass samples also did not match. However, based on the information above, it is possible that additional matches were present and simply remained undetected.
Stability of E. coli O157 genomic fingerprints.
In order to determine the stability of the E. coli O157 fingerprints, 14 isolates were passaged (subcultured) as described in Materials and Methods. There was no change in the XbaI genomic fingerprint patterns for 12 of these isolates during passaging (data not shown).
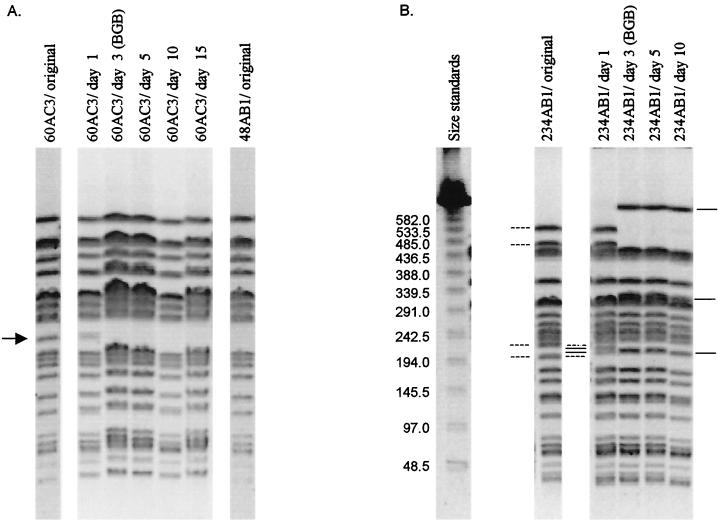
A new XbaI PFGE pattern appeared during passaging of isolate 60AC3. A single band of approximately 212 kb was lost after a 3-day broth passaging or by day 5 of passaging on plates (Fig. 2A). This isolate originally was the only one of subtype 23a. The new pattern derived from 60AC3 after passaging was the same as the pattern of subtype 23b including 21 isolates. The XbaI PFGE pattern of isolate 48AB1, of subtype 23b, did not change during passaging (data not shown).
FIG. 2.
Genomic fingerprints from passaged isolates. (A) Fingerprints of strain 60AC3 during passaging. The isolate and passaging day are indicated for each lane. The arrow indicates the band that was lost during passaging. (B) Fingerprints of strain 234AB1 during passaging. Size standards are lambda concatemers (Bio-Rad); sizes are given in kilobases. The isolate and passaging day are indicated for each lane. Dashed lines indicate bands lost from the original fingerprint, solid lines indicate bands gained in comparison to the original fingerprint. Bands marked between lanes 1 and 2 pertain to changes in the day 1 pattern, bands marked outside of lanes 1 and 5 pertain to changes between the original fingerprint and the day 3, 5, and 10 fingerprints. BGB, brilliant green bile broth.
After 1 day of passaging, the fingerprint from isolate 234AB1 (subtype 6c) had a four-band difference from the original pattern for that isolate; slight changes in size were noted for two bands (Fig. 2B). After further passaging in broth or on plates, a seven-band difference altogether occurred in the isolate 234AB1 fingerprints. One of the two previously altered bands was lost, as were two large bands. In addition, new bands appeared. This resulted in an apparent net loss of approximately 227 kb of DNA. The new pattern was unlike any others observed in this study and therefore constituted a new, closely related type.
DISCUSSION
E. coli O157 genomic variability.
Many genomic types and subtypes of E. coli O157 have been identified by XbaI PFGE fingerprinting in this and other studies (3, 7, 14, 21, 23, 31, 43). The nature and significance of these genomic differences remain unclear. Bacteriophage have been implicated as a causative agent, and the genomic variations have been suggested to be related to the direct or indirect ability to cause disease (30). A study using multilocus enzyme electrophoresis analysis of housekeeping genes detected little difference among E. coli O157 strains (51), which argues that the diversity in PFGE patterns is due in large part to nonevolutionary events such as horizontal DNA transfer. However, inversions, translocations, and point mutations could have caused some of the genotypic variation (9, 47). For example, the fingerprint pattern changes that resulted from repeated subculturing of isolate 234AB1 were suggestive of inversion or translocation events, as well as a loss of large amounts of DNA. Changes in genome size may not be uncommon in Enterobacteriaceae (8, 50) and can be the result of duplications, deletions, and horizontal DNA transfer events associated with elements such as conjugative transposons, insertion elements, and lysogenic bacteriophage (9, 47). Although duplications may not be unlikely in E. coli (22), numerous studies have suggested that horizontal DNA transfer by a variety of mechanisms occurs in and across many bacterial species in vivo (16, 25, 27, 35, 38–40, 48). Rode et al. (45) found that a sepsis-associated strain of E. coli and a uropathogenic strain of E. coli had distinct deletion and insertion events associated with novel DNA in comparison to E. coli K-12 rather than genomic rearrangements, which suggests horizontal DNA transfer events had occurred. DNA transfer by the E. coli O157 Shiga toxin phages in vivo has been reported (1), and even the E. coli K-12 genome contains evidence of substantial horizontal or lateral DNA transfer (8, 32).
This study is the first to report that motility as well as carriage of stx genes corresponded to specific genotypic clusters. The only similar observation was made by Karch et al. (28), who reported a group of distinct patterns for sorbitol-fermenting E. coli O157:H− isolates. For the most part, the nonmotile isolates recovered in this study did have XbaI PFGE genotypes identical to those of some motile, H7+ isolates. These nonmotile isolates may have undetected mutations or may simply prefer different conditions to stimulate expression of the H7 antigen and motility, such as passaging in semisolid media. Reports differ on the correlation of E. coli O157 Shiga toxin profiles with subtypes (33, 43). The data reported here demonstrated a strong association between Shiga toxin profile and XbaI PFGE subtype, although two subtypes included both stx2 and stx1 stx2 isolates (subtypes 1d and 4a [Fig. 1]). The absence of stx1 in the stx2 isolates was confirmed by colony blotting and an additional PCR procedure (data not shown). Murase et al. (36) noted by a different PFGE technique that loss of an ∼70-kb band corresponded to loss of either stx gene. It is possible that the presence or absence of a band this size was not clearly distinguished in these analyses. Alternatively, other phage may be present in the strains lacking stx1, such that their size and position masked the absence of an stx1 phage.
Two or three main E. coli O157 relatedness clusters have been found in this and other studies by various genomic analyses (30, 33, 51). It has been suggested that genomic variation is related to the ability of the organism to cause disease (30). It is possible that each cluster consists of one or a few core genotypes that are primarily altered by independent horizontal DNA transfer events resulting in the multitude of subtypes and types. The detection of a few predominant genotypes and many less populous genotypes in this and other studies is in keeping with this hypothesis (31, 43). The derivation of several genotypes from a single genotype during in vivo passaging of E. coli O157 has been observed, although the causes of the alterations were not determined (4, 29). The derivation of two new genotypes from a single E. coli O157 isolate was also observed during in vitro passaging in this study.
Up to four types or subtypes of E. coli O157 were found per lot in cattle feces (data not shown). This observation is similar to the results of previous studies (31, 43). Rice et al. also (43) reported no relationship between the number of subtypes and the number of samples per farm. The number of preharvest E. coli O157 XbaI PFGE types recovered per lot increased to as many as six when hide isolates were added to the analysis. Therefore, the cattle may have actually carried or been exposed to more types of E. coli O157 than those recovered from feces. Exposure of hides to E. coli O157 in the feces of wild animals, potential difficulties recovering all possible types from different sources (i.e., feces or hides), or the possible inability of all E. coli O157 types to survive under various conditions could account for the extra variation.
Tracking of E. coli O157 carcass contamination.
Chapman et al. (12) studied E. coli O157 contamination of carcasses at a South Yorkshire abattoir and found that 30% of the carcasses (seven carcasses) from cattle with feces positive for E. coli O157 were contaminated, and 8% of the adjacent carcasses (two carcasses) also were contaminated. Direct contamination and cross-contamination were implicated by phage typing and plasmid profiles of the strains. In addition, Byrne et al. (10) showed that spreading E. coli O157:H7-inoculated feces onto hides resulted in contamination of the carcass and workers' hands and knives. Because of the relatively low number of E. coli O157-positive hides detected, the data from this study were insufficient to provide evidence that either hides or feces were more likely to be the direct source of E. coli O157 on the carcasses. The data did clearly demonstrate a strong relationship between preharvest and postharvest isolates within a lot, corroborating the previous observation of an overall positive correlation between preharvest and postharvest prevalence by lot (13). The isolate in vitro-passaging data suggest that more of the carcass isolates could have originated from preharvest isolates within the same lot, but the matches were not identified because of genomic alterations between individual sample isolations that were sufficient to change the designated XbaI PFGE type. Expanding the typing limitations may have revealed additional valid matches but likely also would have misidentified matches not reflective of the actual source of contamination. In addition, the ratios of types to numbers of isolates suggest that if additional preharvest isolates had been recovered, more potentially matching types might have been found.
The presence of more types of E. coli O157 in postharvest samples than in preharvest samples suggests that additional types of E. coli O157 were present preharvest and were not identified in this study. Since a proportionate number of unique isolates were recovered preharvest as well as postharvest, and a proportionate number of types were unique to preharvest isolates and postharvest isolates, the data do not suggest cross-contamination. Cross-contamination of the carcasses presumably would be due to animals within lots entering the plant earlier in the day. Cross-contamination previously was suggested as the source of E. coli O157 on carcasses within lots that did not include positive preharvest samples (13). When the XbaI PFGE genotypes were examined with regard to matches between postharvest and preharvest isolates across lots, the data were found to be inconclusive. In some cases preharvest (animal) isolates matched postharvest (carcass) isolates from an earlier lot, and in several instances preharvest isolates from different lots were of the same type (data not shown). Therefore, cross-contamination between lots could not be discerned. However, for each trip to a plant, there was no evidence of cross-contamination worsening over the course of the sampling period. The numbers of matching preharvest and postharvest isolates did not decrease for lots processed later in the day, and carcasses from later lots were not more frequently contaminated than carcasses from lots processed earlier in the day. The latter observations need to be interpreted with caution, though, as the study was not designed to compare the data in this manner. Overall, while cross-contamination probably occurred to a limited extent, some of the prevalence data may have been reflective of a higher difficulty in recovering E. coli O157 from preharvest sources. Competing microflora in fecal and hide samples necessitates the use of more-stringent enrichment conditions for these samples compared to carcass samples (13).
For many postharvest samples, more than one genomic type of E. coli O157 was recovered. These data could suggest that individual carcass contamination originated from multiple sources or that contamination sources (feces and hide) may harbor multiple isolates that can be transferred to the carcass in a single contamination event. Work identifying E. coli O157 of multiple genomic types in individual cattle feces supports the latter interpretation (31). Only one isolate was examined per preharvest sample in this study, so additional types may have been present in hide or fecal samples and remained undetected.
Further studies are needed to detail the potential for carcass-to-carcass cross-contamination. We did not sample adjacent carcasses, so the potential for direct cross-contamination was not discerned. The clear majority of carcass contamination with E. coli O157 occurred prior to any direct contact between carcasses (13), although cross-contamination during very early processing steps via equipment and workers could have occurred (10). Regarding potential cross-contamination later in processing, most of the carcasses found to be contaminated later in processing had been contaminated earlier in processing (49 of 59 [reference 13]). In addition, the PFGE patterns of most isolates from the later processing samples matched those of isolates recovered from the same carcass earlier in processing. Therefore, E. coli O157 contamination found later in processing was due largely to contamination that occurred early in processing and not to carcass-to-carcass contamination.
Several additional and particularly interesting observations were made during this study. First, postevisceration isolates were underrepresented in cluster C. Second, predominant preharvest and postharvest E. coli O157 types within a lot often were not the same (data not shown). For example in lot A1-1, one type included 12 of 15 preharvest isolates and only 2 of 13 postharvest isolates, and a second type included only 2 preharvest isolates but included 11 postharvest isolates. (The third type included one preharvest isolate.) Third, fecal isolates were less likely than isolates from other samples to group into cluster B. Fecal isolates also were less likely to carry both stx genes, being more likely to carry only stx2. Finally, compared to carcass isolates, fecal and hide isolates were slightly more likely to be nonmotile. Further experiments are necessary to examine these phenomena. The data could be evidence that some E. coli O157 genomic types are more successful at passing through processing steps or are more easily transferred to the carcass or could reflect the fact that different types are more easily recovered from the different sample sites based on recovery and/or enrichment methods. If these hypotheses were true, one would expect that a limited number of preevisceration types or preharvest types would be found postevisceration or postharvest, respectively. However, of the preevisceration isolates overall, 67.6% were of the same type as postevisceration isolates, and within the 17 lots used to analyze preharvest/postharvest isolate relatedness, 72.1% of the preharvest isolates were of types also found postharvest.
In summary, this study tracked carcass contamination by E. coli O157 in processing plants within the United States. A strong association between E. coli O157 carried by live animals and on carcasses within the same lot was first demonstrated by prevalence data (13). XbaI PFGE genotyping data have further implied that E. coli O157 found on carcasses is primarily the result of transfer within a lot rather than cross-contamination between lots, although some cross-contamination may occur (13). Furthermore, the tracking data based on XbaI PFGE genotyping confirm that the majority of E. coli O157 found on the carcass is the result of preevisceration contamination, despite a dramatic reduction throughout processing in the number of carcasses contaminated (13). Taken together, these data indicate the need to apply additional in-plant intervention strategies aimed at preventing direct contamination of the carcasses early in processing. In addition, a substantial level of genomic variation was observed among the E. coli O157 isolates recovered during this study. The significance of this divergence remains to be seen, although it has been implied to affect, either directly or indirectly, the ability of the organism to cause disease in humans (30). Work is in progress to investigate the relationships between these genomic differences and various aspects of the organism's ability to eventually cause disease, such as relative virulence or survival during processing, storage, and cooking.
ACKNOWLEDGMENTS
We thank Kim Kucera, Sandy Fryda-Bradley, and Ron Mlejnek for excellent technical assistance and Carol Grummert for excellent secretarial assistance.
REFERENCES
- 1.Acheson D W K, Reidl J, Zhang X, Keusch G T, Mekalanos J J, Waldor M K. In vivo transduction with Shiga toxin 1-encoding phage. Infect Immun. 1998;66:4496–4498. doi: 10.1128/iai.66.9.4496-4498.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ackman D, Marks S, Mack P, Caldwell M, Root T, Birkhead G. Swimming-associated haemorrhagic colitis due to Escherichia coli O157:H7 infection: evidence of prolonged contamination of a fresh water lake. Epidemiol Infect. 1997;119:1–8. doi: 10.1017/s095026889700770x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akiba M, Masuda T, Sameshima T, Katsuda K, Nakazawa M. Molecular typing of Escherichia coli O157:H7 (H-) isolates from cattle in Japan. Epidemiol Infect. 1999;122:337–341. doi: 10.1017/s0950268899002198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akiba M, Sameshima T, Nakazawa M. Clonal turnover of enterohemorrhagic Escherichia coli O157:H7 in experimentally infected cattle. FEMS Microbiol Lett. 2000;184:79–83. doi: 10.1111/j.1574-6968.2000.tb08994.x. [DOI] [PubMed] [Google Scholar]
- 5.Banatvala N, Magnano A R, Carter M L, Barrett T J, Bibb W F, Vasile L L, Mshar P, Lambert-Fair M A, Green J H, Bean N H, Tauxe R V. Meat grinders and molecular epidemiology: two supermarket outbreaks of Escherichia coli O157:H7 infection. J Infect Dis. 1996;173:480–483. doi: 10.1093/infdis/173.2.480. [DOI] [PubMed] [Google Scholar]
- 6.Bell R G. Distribution and sources of microbial contamination on beef carcasses. J Appl Microbiol. 1997;82:292–300. doi: 10.1046/j.1365-2672.1997.00356.x. [DOI] [PubMed] [Google Scholar]
- 7.Bender J B, Hedberg C W, Besser J M, Boxrud D J, MacDonald K L, Osterholm M T. Surveillance for Escherichia coli O157:H7 infections in Minnesota by molecular subtyping. N Engl J Med. 1997;337:388–394. doi: 10.1056/NEJM199708073370604. [DOI] [PubMed] [Google Scholar]
- 8.Bergthorsson U, Ochman H. Distribution of chromosome length variation in natural isolates of Escherichia coli. Mol Biol Evol. 1998;15:6–16. doi: 10.1093/oxfordjournals.molbev.a025847. [DOI] [PubMed] [Google Scholar]
- 9.Brunder W, Karch H. Genome plasticity in Enterobacteriaceae. Int J Med Microbiol. 2000;290:153–165. doi: 10.1016/S1438-4221(00)80084-3. [DOI] [PubMed] [Google Scholar]
- 10.Byrne C M, Bolton D J, Sheridan J J, McDowell D A, Blair I S. The effects of preslaughter washing on the reduction of Escherichia coli O157:H7 transfer from cattle hides to carcasses during slaughter. Lett Appl Microbiol. 2000;30:142–145. doi: 10.1046/j.1472-765x.2000.00689.x. [DOI] [PubMed] [Google Scholar]
- 11.Centers for Disease Control and Prevention. Standardized molecular subtyping of foodborne bacterial pathogens by pulsed-field gel electrophoresis. CDC training manual. Atlanta, Ga: Foodborne and Diarrheal Diseases Branch, Centers for Disease Control and Prevention; 1998. [Google Scholar]
- 12.Chapman P A, Siddons C A, Wright D J, Norman P, Fox J, Crick E. Cattle as a possible source of verocytotoxin-producing Escherichia coli O157 infections in man. Epidemiol Infect. 1993;111:439–447. doi: 10.1017/s0950268800057162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elder R O, Keen J E, Siragusa G R, Barkocy-Gallagher G A, Koohmaraie M, Laegreid W W. Correlation of enterohemorrhagic Escherichia coli prevalence in feces, hides, and carcasses of beef cattle during processing. Proc Natl Acad Sci USA. 2000;97:2999–3003. doi: 10.1073/pnas.060024897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faith N G, Shere J A, Brosch R, Arnold K W, Ansay S E, Lee M-S, Luchansky J B, Kaspar C W. Prevalence and clonal nature of Escherichia coli O157:H7 on dairy farms in Wisconsin. Appl Environ Microbiol. 1996;62:1519–1525. doi: 10.1128/aem.62.5.1519-1525.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fields P I, Blom K, Hughes H J, Helsel L O, Feng P, Swaminathan B. Molecular characterization of the gene encoding H antigen in Escherichia coli and development of a PCR-restriction fragment length polymorphism test for identification of E. coli O157:H7 and O157:NM. J Clin Microbiol. 1997;35:1066–1070. doi: 10.1128/jcm.35.5.1066-1070.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garrigues-Jeanjean N, Wittmer A, Ouriet M F, Duval-Iflah Y. Transfer of the shuttle vector pRRI207 between Escherichia coli and Bacteroides spp. in vitro and in vivo in the digestive tract of axenic mice and in gnotoxenic mice inoculated with a human microflora. FEMS Microbiol Ecol. 1999;29:33–43. [Google Scholar]
- 17.Gill C O, McGinnis J C. Improvement of the hygienic performance of the hindquarters skinning operations at a beef packing plant. Int J Food Microbiol. 1999;51:123–132. doi: 10.1016/s0168-1605(99)00111-7. [DOI] [PubMed] [Google Scholar]
- 18.Gill C O, McGinnis J C, Badoni M. Assessment of the hygienic characteristics of a beef carcass dressing process. J Food Prot. 1996;59:136–140. doi: 10.4315/0362-028X-59.2.136. [DOI] [PubMed] [Google Scholar]
- 19.Gill C O, McGinnis J C, Badoni M. Use of total or Escherichia coli counts to assess the hygienic characteristics of a beef carcass dressing process. Int J Food Microbiol. 1996;31:181–196. doi: 10.1016/0168-1605(96)00982-8. [DOI] [PubMed] [Google Scholar]
- 20.Giovannacci I, Ragimbeau C, Queguiner S, Salvat G, Vendeuvre J-L, Carlier V, Ermel G. Listeria monocytogenes in pork slaughtering and cutting plants. Use of RAPD, PFGE and PCR-REA for tracing and molecular epidemiology. Int J Food Microbiol. 1999;53:127–140. doi: 10.1016/s0168-1605(99)00141-5. [DOI] [PubMed] [Google Scholar]
- 21.Grif K, Karch H, Schneider C, Daschner F D, Beutin L, Cheasty T, Smith H, Rowe B, Dierich M P, Allerberger F. Comparative study of five different techniques for epidemiological typing of Escherichia coli O157. Diagn Microbiol Infect Dis. 1998;32:165–176. doi: 10.1016/s0732-8893(98)00103-5. [DOI] [PubMed] [Google Scholar]
- 22.Haack K R, Roth J R. Recombination between chromosomal IS200 elements supports frequent duplication formation in Salmonella typhimurium. Genetics. 1995;141:1245–1252. doi: 10.1093/genetics/141.4.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heuvelink A E, Van Den Biggelaar F L A M, De Boer E, Herbes R G, Melchers W J G, Huis in 'T Veld J H J, Monnens L A H. Isolation and characterization of verocytotoxin-producing Escherichia coli O157 strains from Dutch cattle and sheep. J Clin Microbiol. 1998;36:878–882. doi: 10.1128/jcm.36.4.878-882.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hilborn E D, Mermin J H, Mshar P A, Hadler J L, Voetsch A, Wojtkunski C, Swartz M, Mshar R, Lambert-Fair M A, Farrar J A, Glynn M K, Slutsker L. A multistate outbreak of Escherichia coli O157:H7 infections associated with consumption of mesclun lettuce. Arch Intern Med. 1999;159:1758–1764. doi: 10.1001/archinte.159.15.1758. [DOI] [PubMed] [Google Scholar]
- 25.Jain R, Rivera M C, Lake J A. Horizontal gene transfer among genomes: the complexity hypothesis. Evolution. 1999;96:3801–3806. doi: 10.1073/pnas.96.7.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson R P, Clark R C, Wilson J B, Read S C, Rahn K, Renwick S A, Sandhu K A, Alves D, Karmali M A, Lior H, McEwen S A, Spika J S, Gyles C L. Growing concerns and recent outbreaks involving non-O157:H7 serotypes of verotoxigenic Escherichia coli. J Food Prot. 1996;59:1112–1122. doi: 10.4315/0362-028X-59.10.1112. [DOI] [PubMed] [Google Scholar]
- 27.Karaolis D K R, Somara S, Maneval D R, Jr, Johnson J A, Kaper J B. A bacteriophage encoding a pathogenicity island, a type-IV pilus and a phage receptor in cholera bacteria. Nature. 1999;399:375–379. doi: 10.1038/20715. [DOI] [PubMed] [Google Scholar]
- 28.Karch H, Böhm H, Schmidt H, Gunzer F, Aleksic S, Heesemann J. Clonal structure and pathogenicity of Shiga-like toxin-producing, sorbitol-fermenting Escherichia coli O157:H−. J Clin Microbiol. 1993;31:1200–1205. doi: 10.1128/jcm.31.5.1200-1205.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karch H, Rüssmann H, Schmidt H, Schwarzkopf A, Heesemann J. Long-term shedding and clonal turnover of enterohemorrhagic Escherichia coli O157 in diarrheal diseases. J Clin Microbiol. 1995;33:1602–1605. doi: 10.1128/jcm.33.6.1602-1605.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim J, Nietfeldt J, Benson A K. Octamer-based genome scanning distinguishes a subpopulation of Escherichia coli O157:H7 strains in cattle. Proc Natl Acad Sci USA. 1999;96:13288–13293. doi: 10.1073/pnas.96.23.13288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laegreid W W, Elder R O, Keen J E. Prevalence of Escherichia coli O157:H7 in range beef calves at weaning. Epidemiol Infect. 1999;123:291–298. doi: 10.1017/s0950268899002757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lawrence J G, Ochman H. Molecular archaeology of the Escherichia coli genome. Proc Natl Acad Sci USA. 1998;95:9413–9417. doi: 10.1073/pnas.95.16.9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee M-S, Kaspar C W, Brosch R, Shere J, Luchansky J B. Genomic analysis using pulsed-field gel electrophoresis of Escherichia coli O157:H7 isolated from dairy calves during the United States national dairy heifer evaluation project (1991–1992) Vet Microbiol. 1996;48:223–230. doi: 10.1016/0378-1135(95)00135-2. [DOI] [PubMed] [Google Scholar]
- 34.McEvoy J M, Doherty A M, Finnerty M, Sheridan J J, McGuire L, Blair I S, McDowell D A, Harrington D. The relationship between hide cleanliness and bacterial numbers on beef carcasses at a commercial abattoir. Lett Appl Microbiol. 2000;30:390–395. doi: 10.1046/j.1472-765x.2000.00739.x. [DOI] [PubMed] [Google Scholar]
- 35.Muñoz R, García E, López R. Evidence for horizontal transfer from Streptococcus to Escherichia coli of the kfiD gene encoding the K5-specific UDP-glucose dehydrogenase. J Mol Evol. 1998;46:432–436. doi: 10.1007/pl00006322. [DOI] [PubMed] [Google Scholar]
- 36.Murase T, Yamai S, Watanabe H. Changes in pulsed-field gel electrophoresis patterns in clinical isolates of enterohemorrhagic Escherichia coli O157:H7 associated with loss of Shiga toxin genes. Curr Microbiol. 1999;38:48–50. doi: 10.1007/pl00006771. [DOI] [PubMed] [Google Scholar]
- 37.Nataro J P, Kaper J B. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Netherwood T, Bowden R, Harrison P, O'Donnell A G, Parker D S, Gilbert H J. Gene transfer in the gastrointestinal tract. Appl Environ Microbiol. 1999;65:5139–5141. doi: 10.1128/aem.65.11.5139-5141.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nikolich M P, Hong G, Shoemaker N B, Salyers A A. Evidence for natural horizontal transfer of tetQ between bacteria that normally colonize humans and bacteria that normally colonize livestock. Appl Environ Microbiol. 1994;60:3255–3260. doi: 10.1128/aem.60.9.3255-3260.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ochman H, Lawrence J G, Groisman E A. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 41.Ostroff S M, Tarr P I, Neill M A, Lewis J H, Hargrett-Bean N, Kobayashi J M. Toxin genotypes and plasmid profiles as determinants of systemic sequelae in Escherichia coli O157:H7 infections. J Infect Dis. 1989;160:994–998. doi: 10.1093/infdis/160.6.994. [DOI] [PubMed] [Google Scholar]
- 42.Preston M A, Johnson W, Khakhria R, Borczyk A. Epidemiologic subtyping of Escherichia coli serogroup O157 strains isolated in Ontario by phage typing and pulsed-field gel electrophoresis. J Clin Microbiol. 2000;38:2366–2368. doi: 10.1128/jcm.38.6.2366-2368.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rice D H, McMenamin K M, Pritchett L C, Hancock D D, Besser T E. Genetic subtyping of Escherichia coli O157 isolates from 41 Pacific Northwest USA cattle farms. Epidemiol Infect. 1999;122:479–484. doi: 10.1017/s0950268899002496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ridell J, Korkeala H. Special treatment during slaughtering in Finland of cattle carrying an excessive load of dung; meat hygienic aspects. Meat Sci. 1993;35:223–228. doi: 10.1016/0309-1740(93)90052-J. [DOI] [PubMed] [Google Scholar]
- 45.Rode C K, Melkerson-Watson L J, Johnson A T, Bloch C A. Type-specific contributions to chromosome size differences in Escherichia coli. Infect Immun. 1999;19:230–236. doi: 10.1128/iai.67.1.230-236.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodrigue D C, Mast E E, Greene K D, Davis J P, Hutchinson M A, Wells J G, Barrett T J, Griffin P M. A university outbreak of Escherichia coli O157:H7 infections associated with roast beef and an unusually benign clinical course. J Infect Dis. 1995;172:1122–1125. doi: 10.1093/infdis/172.4.1122. [DOI] [PubMed] [Google Scholar]
- 47.Sanderson K E, Liu S-L. Chromosomal rearrangements in enteric bacteria. Electrophoresis. 1998;19:569–572. doi: 10.1002/elps.1150190417. [DOI] [PubMed] [Google Scholar]
- 48.Shoemaker N B, Wang G-R, Salyers A A. Evidence for natural transfer of a tetracycline resistance gene between bacteria from the human colon and bacteria from the bovine rumen. Appl Environ Microbiol. 1992;58:1313–1320. doi: 10.1128/aem.58.4.1313-1320.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takeda Y. Enterohaemorrhagic Escherichia coli. World Health Stat Q. 1997;50:74–80. [PubMed] [Google Scholar]
- 50.Thong K L, Puthucheary S D, Pang T. Genome size variation among recent human isolates of Salmonella typhi. Res Microbiol. 1997;148:229–235. doi: 10.1016/S0923-2508(97)85243-6. [DOI] [PubMed] [Google Scholar]
- 51.Whittam T S, Wolfe M L, Wachsmuth I K, Ørskov F, Ørskov I, Wilson R A. Clonal relationships among Escherichia coli strains that cause hemorrhagic colitis and infantile diarrhea. Infect Immun. 1993;61:1619–1629. doi: 10.1128/iai.61.5.1619-1629.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]