Abstract
Background
Various machine learning and artificial intelligence methods have been used to predict outcomes of hospitalized COVID-19 patients. However, process mining has not yet been used for COVID-19 prediction. We developed a process mining/deep learning approach to predict mortality among COVID-19 patients and updated the prediction in 6-h intervals during the first 72 h after hospital admission.
Methods
The process mining/deep learning model produced temporal information related to the variables and incorporated demographic and clinical data to predict mortality. The mortality prediction was updated in 6-h intervals during the first 72 h after hospital admission. Moreover, the performance of the model was compared with published and self-developed traditional machine learning models that did not use time as a variable. The performance was compared using the Area Under the Receiver Operator Curve (AUROC), accuracy, sensitivity, and specificity.
Results
The proposed process mining/deep learning model outperformed the comparison models in almost all time intervals with a robust AUROC above 80% on a dataset that was imbalanced.
Conclusions
Our proposed process mining/deep learning model performed significantly better than commonly used machine learning approaches that ignore time information. Thus, time information should be incorporated in models to predict outcomes more accurately.
Keywords: Mortality prediction, Process mining, Deep learning, COVID-19 prediction, Machine learning, SARS-CoV-2
Background
Throughout the COVID-19 pandemic, machine learning and artificial intelligence (AI) methods have been used to understand and predict virus spread, the potential impact of vaccines, morbidity, mortality, and resource allocation [1]. Modeling of COVID-19 morbidity and mortality has yielded insights into disease progression [2, 3], which have been informative for health systems to anticipate resource needs and effective interventions [4]. However, with the emergence of COVID-19 variants and rapid advances in COVID-19 treatment, prevention, and vaccination, 1-time modeling is likely ineffective for understanding how to provide optimal care from the patient, health system, and public health perspectives [4].
Process mining techniques assist in analyzing and optimizing systems using sequences of observations. Process mining approaches have been shown to be valuable in the healthcare industry by enhancing healthcare processes [5, 6]. However, process mining has not yet been used to predict mortality after hospital admission for COVID-19 patients [7, 8] though providing significant advantages over static models. In general, process mining algorithms take a sequential perspective on data points that have been observed over time to derive a single semantic-rich graph structure like a Petri Net. In the context of COVID-19, each patient follows a distinct path throughout such a derived Petri net while being in one state at any point of time. The states naturally embed information of the sequence of observations that lead to this state and of potential future observations leading to subsequent states. This means that process mining algorithms allow to explicitly incorporate the timing and sequence of healthcare events into the modeling process by leveraging the states of a Petri Net.
One significant advantage of process mining techniques over static models is their ability to explicitly incorporate the timing and sequence of healthcare events into the modeling process. For example, let’s assume that a machine learning model uses two specific inputs of blood pressure and blood sugar to predict the mortality of a patient. In this case, a static machine learning model is indifferent to the sequence by which the values of blood pressure and blood sugar were obtained from the patient. Also, the model does not consider when these values were collected (the occurrence times of the events associated with collecting blood pressure and blood sugar values are ignored by the model) in predicting the mortality of the patients. In contrast, for this example, a process mining model uses not only the values of blood pressure and blood sugar, but by leveraging Petri net states, also their collection sequence, and timing in calculating the mortality of the patient. It can be shown that by incorporating the time and sequence information, one can usually generate better prediction models [9]. Therefore, we aimed to utilize a combined process mining and deep learning modeling approach for prediction.
Methodology
University of illinois hospital (UIH) cohort and variables
UIH is a tertiary, academic teaching hospital in Chicago. The University of Illinois at Chicago (UIC) Institutional Review Board approved this study. All admissions to UIH for COVID-19 positive patients were reviewed for the time of the first COVID-19 positive test and the date of admission. If the first positive COVID-19 test was performed greater than 14 days prior to admission or greater than 48 h after admission, the patient was excluded. Patients transferred from another institution were reviewed for prior COVID-19 testing. The patient was excluded if the most recent COVID-19 test has been performed longer than 14 days prior to the transfer. If the transfer was not related to any possible COVID-19 symptoms, the patient was excluded. Symptomatic patients for COVID-19 were included in this cohort, as verified by manual chart review or claim data.
If a patient had multiple hospital admissions at UIH related to COVID-19, each admission encounter was categorized with a final outcome of as death or discharge. All admissions were categorized as intensive care unit (ICU) or Non-ICU.
We partitioned our data into training, validation, and test cohorts using a 60/20/20 split ratio, respectively. Consequently, each admission encounter belonged to a unique cohort.
Variable selection was based on literature review and expert opinion [10]. The variables selected are shown in Table 6, in the appendix section, where demographics, vital signs, laboratory data, and clinical characteristics (comorbidities, diagnosis codes, problem list, clinic notes, procedure reports, location within the hospital) were assessed.
Table 6.
Detailed variables were used as inputs to the proposed model
| Variables related to specific category | Variables | Variables values (if applicable) |
|---|---|---|
| Demographics | Age | |
| Demographics | Gender | |
| Demographics | Race | |
| Process mining | EventCount | |
| Process mining | TokenCount | |
| Process mining | Marking | |
| Process mining | LinearDecay | |
| Process mining | LinearDecay_mean | |
| Process mining | ExpDecay_max | |
| Process mining | LogDecay_mean | |
| Comorbidities | Hypertension | |
| Comorbidities | Diabetes | |
| Comorbidities | Heart Disease | |
| Comorbidities | COPD | |
| Comorbidities | Stroke | |
| Comorbidities | Cerebrovascular Disease | |
| Comorbidities | Cancer | |
| Comorbidities | Respiratory Problems | |
| Comorbidities | Chronic Kidney Disease | |
| Comorbidities | Tuberculosis | |
| Location | COVID-4 | |
| Location | COVID-2 | |
| Location | MEDICAL INTENSIVE | |
| Location | FAMILYMEDICINE | |
| Location | MICU-2 | |
| Location | MED SERVICE A | |
| Location | MED SERVICE D | |
| Location | MED SERVICE C | |
| Location | MED SERVICE B | |
| Location | MiCU-1 | |
| Location | MED SERVICE E | |
| Location | COVID-5 | |
| Location | COVID MICU-3 | |
| Location | MED HEMATOLOGY | |
| Location | MED HEPATOLOGY/LIVER | |
| Location | MED SICKLE CELL | |
| Location | COVID MICU-5 | |
| Location | ORGAN TRANSPLANT | |
| Location | MED ONCOLOGY | |
| Location | COVID MICU-4 | |
| Location | STEM CELL TRANSPLANT | |
| Location | PED PREADMIT ONLY | |
| Location | COVID-6 | |
| Location | SURGERY GENERAL | |
| Location | NEUROSURGERY | |
| Location | MED CARDIO | |
| Location | CORONARY CARE UNIT | |
| Location | NEUROLOGY | |
| Location | MED PREAD ONLY | |
| Location | MED GI | |
| Encounters | Inpatient | |
| Encounters | UIH ER | |
| Encounters | death | |
| Encounters | PREADMIT | |
| Encounters | ER OB | |
| Encounters | 5 W PEDS | |
| Encounters | disch | |
| Procedure reports | RADRPT | |
| Procedure reports | ECG Measurements and Interpretation | |
| Procedure reports | Echo Transthoracic | |
| Procedure reports | Pathology Report | |
| Procedure reports | Echo Transesophageal | |
| Lab | (1,3)-BETA-D-GLUCAN | Normal |
| Lab | (1,3)-BETA-D-GLUCAN INTERPRETATION | Normal |
| Lab | % BASOPHIL | Normal |
| Lab | % EOSINOPHIL | Normal |
| Lab | % LYMPHOCYTE | Normal |
| Lab | % MONOCYTE | Normal |
| Lab | % NEUTROPHIL | Normal |
| Lab | % TRANSFERRIN SAT | Normal, LOW, HI |
| Lab | A. GALACTOMANNAN AG | Normal |
| Lab | A. GALACTOMANNAN INDEX | Normal |
| Lab | A1ANTITRYP | Normal |
| Lab | ABO/RH(D) | No flag |
| Lab | ABS CD19 | Normal, LOW |
| Lab | ABS CD3 | Normal, LOW |
| Lab | ABS CD3/CD4 | LOW |
| Lab | ABS CD3/CD8 | Normal,LOW |
| Lab | ABS CD56 | Normal,LOW,HI |
| Lab | Abs Retic | Normal,HI |
| Lab | ABSOLUTE BAND NEUTROPHIL (MANUAL DIFF) | Normal |
| Lab | ABSOLUTE BASOPHIL (MANUAL DIFF) | HI |
| Lab | ABSOLUTE EOSINOPHIL (MANUAL DIFF) | Normal, HI |
| Lab | ABSOLUTE LYMPHOCYTE (MANUAL DIFF) | Normal, LOW, HI |
| Lab | ABSOLUTE MONOCYTE (MANUAL DIFF) | Normal, LOW, HI |
| Lab | ABSOLUTE NEUTROPHILS (MANUAL DIFF) | Normal, HI |
| Lab | ACETAMINOPHEN | LOW |
| Lab | ACT BICARB | Normal, LOW, HI |
| Lab | ADAMTS13 | LOW |
| Lab | ADDITIONAL TESTING | Normal |
| Lab | ADENOVIRUS | Normal |
| Lab | ADENOVIRUS QUANT BY PCR | Normal |
| Lab | AEROMONAS/PLEISOMONAS SCREEN | Normal |
| Lab | ALB CONC | Normal |
| Lab | ALBUMIN | Normal, LOW |
| Lab | Alcohol, Urn Screen | Normal |
| Lab | ALK PHOS | Normal, LOW, HI |
| Lab | ALT(SGPT) | Normal, LOW, HI |
| Lab | amd | LOW |
| Lab | AMMONIA | HI |
| Lab | AMORPHOUS | Normal |
| Lab | AMPHETAMINES-UR | Normal |
| Lab | Amphetamines, Urn Screen | Normal |
| Lab | AMYLASE | HI |
| Lab | ANION GAP | Normal, HI |
| Lab | ANISOCYTOSIS | Normal |
| Lab | ANTI NUCLEAR AB | Normal |
| Lab | ANTI-HB CORE IGM | Normal |
| Lab | ANTI-MITOCHONDRIAL IGG | Normal |
| Lab | ANTI-SMOOTHMUSCLE | Normal |
| Lab | ANTIBODY SCREEN | No flag |
| Lab | ASPERGILLUS AB BY ID | Normal |
| Lab | AST(SGOT) | Normal, LOW, HI |
| Lab | ATYPICAL BACTERIAL PNEUMONIA | Normal |
| Lab | B-NATRIURETIC PEPTIDE | Normal, HI |
| Lab | BAND NEUTROPHIL | Normal |
| Lab | BARBITURATES-UR | Normal |
| Lab | Barbiturates, Urn Screen | Normal |
| Lab | BASE EXCESS | Normal |
| Lab | BASO | Normal |
| Lab | BASOPHILS | Normal, HI |
| Lab | Benzodiazepines, Urn Screen | Normal |
| Lab | BENZODIAZPINE-UR | Normal |
| Lab | BETAHYDROXYBUTYRIC ACID | Normal, HI |
| Lab | BF ALBUMIN | Normal |
| Lab | BF BILIRUBIN | Normal |
| Lab | BF GLUCOSE | Normal |
| Lab | BF LDH | Normal |
| Lab | BF LYMPH | Normal |
| Lab | BF MACROS/MONOS | Normal |
| Lab | BF MESO | Normal |
| Lab | BF NEUT | Normal |
| Lab | BF TOTAL PROTEIN | Normal |
| Lab | BF-RBC | Normal, HI |
| Lab | BF-WBC | Normal |
| Lab | BILIRUBIN, DIRECT | Normal, HI |
| Lab | BILIRUBIN,TOTAL | Normal, HI |
| Lab | BKV QUANT BY PCR | Normal |
| Lab | BKV RT SPECIMEN | Normal |
| Lab | Blastomyces AB | Normal |
| Lab | BLASTOMYCES INTERPRETATION | Normal |
| Lab | BLASTOMYCES RESULT | Normal |
| Lab | BLASTOMYCES SPECIMEN | Normal |
| Lab | Bordetella parapertussis | Normal |
| Lab | BORDETELLA PERTUSSIS | Normal |
| Lab | BRPR | ABN |
| Lab | BUDDING YEAST | Normal |
| Lab | BUN | Normal, LOW, HI |
| Lab | BUN/CREAT RATIO | Normal, LOW, HI |
| Lab | BURR CELLS | Normal |
| Lab | C DIFFICILE RT PCR | Normal |
| Lab | C-REACTIVE PROTEIN | Normal, HI |
| Lab | CALCIUM | Normal, LOW, HI |
| Lab | CALPROTECTIN, FECAL | HI |
| Lab | CAMPYLOBACTER GROUP BY PCR | Normal |
| Lab | CARBMAZPNE, UNBOUND | Normal |
| Lab | CD19%, TOTAL B CELLS | Normal, HI |
| Lab | CD3/CD4%, HELPER T | Normal, LOW |
| Lab | CD3/CD8%, SUP T CELLS | Normal, HI |
| Lab | CD3%, TOTAL T CELLS | Normal, LOW |
| Lab | CD4 COMMENT | Normal |
| Lab | CD56% | Normal, HI |
| Lab | CDASU 9A Comments | Normal |
| Lab | CEA | HI |
| Lab | CERULOPLASMIN | LOW |
| Lab | CHK | No flag |
| Lab | CHLAMYDIA PNEUMONIAE | Normal |
| Lab | CHLORIDE | Normal, LOW, HI |
| Lab | CHOLESTEROL | Normal, HI |
| Lab | CK MACRO TYPE I | Normal |
| Lab | CK MACRO TYPE II | Normal |
| Lab | CK TOTAL | Normal |
| Lab | CK-BB | Normal |
| Lab | CK-MB | Normal |
| Lab | CK-MM | Normal |
| Lab | CLARITY | Normal |
| Lab | CLUMPED PLATELETS | Normal |
| Lab | CMV QUANT BY PCR | Normal |
| Lab | CO2 CONTENT | Normal, LOW, HI |
| Lab | COCAINE-URINE | Normal |
| Lab | Cocaine, Urn Screen | Normal |
| Lab | COLOR | Normal |
| Lab | COMPLEMENT C3 | LOW |
| Lab | COMPLEMENT C4 | Normal |
| Lab | COPPER | HI |
| Lab | Coronavirus 19 | Normal, ABN |
| Lab | CORONAVIRUS 229E | Normal |
| Lab | CORONAVIRUS HKU1 | Normal |
| Lab | CORONAVIRUS NL63 | Normal |
| Lab | CORONAVIRUS OC43 | Normal |
| Lab | CPK | Normal, LOW, HI |
| Lab | CREAT CONC | Normal |
| Lab | CREATININE | Normal, LOW, HI |
| Lab | Creatinine, Urn Screen | Normal |
| Lab | CROSSMATCH | No flag |
| Lab | CYTOPLASMIC STAINING | Normal |
| Lab | D-DIMER | Normal, HI, CRIT |
| Lab | DIFF METHOD | Normal |
| Lab | DIFFERENTIAL METHOD | Normal |
| Lab | DOHLE BODIES | Normal |
| Lab | EBV QUANT BY PCR | Normal, ABN |
| Lab | EOS | Normal, HI |
| Lab | EOSINOPHIL | Normal, HI |
| Lab | Estimated Creat Clearance | No flag, LOW |
| Lab | Estimated GFR | No flag |
| Lab | ETHANOL | Normal |
| Lab | FENTANYL QUANT URINE | Normal |
| Lab | FERRITIN | Normal, LOW, HI |
| Lab | FIBRINOGEN | Normal, HI |
| Lab | FINE GRAN CAST | HI |
| Lab | FK506/TACROLIMUS | Normal |
| Lab | Flu A (POCT) | Normal |
| Lab | FLU A H1 SEASONAL | Normal |
| Lab | FLU A H1N1 2009 | Normal, ABN |
| Lab | FLU B | Normal |
| Lab | Flu B (POCT) | Normal |
| Lab | FOLATE | Normal |
| Lab | FREE T4 | Normal, LOW |
| Lab | GLUCOSE | Normal, LOW, HI, CRIT |
| Lab | GLUCOSE (POCT) | Normal, LOW, HI, CRIT |
| Lab | HAPTOGLOBIN | Normal, HI |
| Lab | HCT | Normal, LOW, HI |
| Lab | HCV REAL TIME PCR | Normal |
| Lab | HDL | Normal, LOW |
| Lab | HELP/SUPP RATIO | Normal |
| Lab | Hemoglobin—POCT | LOW |
| Lab | HEMOGLOBIN A2 | Normal |
| Lab | HEMOGLOBIN F | Normal, HI |
| Lab | HEP A IGM AB | Normal |
| Lab | HEP B CORE AB,TOTAL | Normal |
| Lab | HEP B SURF AB,QUANT | Normal |
| Lab | HEP B SURFACE AG | Normal |
| Lab | HEP C ANTIBODY | Normal, ABN |
| Lab | HGB | Normal, LOW, HI |
| Lab | HGB A | Normal |
| Lab | HGB A1C | Normal, HI |
| Lab | HGB C | Normal |
| Lab | HGB S | Normal |
| Lab | HISTOPLASMA INTERPRETATION | Normal |
| Lab | HISTOPLASMA RESULT | Normal |
| Lab | HISTOPLASMA SPECIMEN | Normal |
| Lab | HIV 1 Antibody | Normal |
| Lab | HIV 1 Antigen | Normal |
| Lab | HIV 2 Antibody | Normal |
| Lab | HIV Antigen and Antibody Screen NC | Normal |
| Lab | HIV1AB | Normal |
| Lab | HIV1AG | Normal |
| Lab | HIV2AB | Normal |
| Lab | HOWELL JOLLY | Normal |
| Lab | HSV TYPE I | Normal |
| Lab | HSV TYPE II | Normal |
| Lab | HUMAN METAPNEUMOVIRUS | Normal |
| Lab | HUMAN RHINOVIRUS/ENTEROVIRUS | Normal |
| Lab | HVABAG | Normal |
| Lab | HYALINE CAST | Normal |
| Lab | HYPOCHROMASIA | Normal |
| Lab | IGA | Normal, LOW, HI |
| Lab | IGG | Normal, LOW |
| Lab | IGM | Normal, LOW, HI |
| Lab | IMMUNOFIX SERUM | Normal |
| Lab | Influenza A Equivocal (Inconclusive) | Normal |
| Lab | INFLUENZA A, H3 SUBTYPE | Normal |
| Lab | Influenza A, No Subtype Detected | Normal |
| Lab | INR | Normal, HI, CRIT |
| Lab | INTERLEUKIN 6 | Normal, HI |
| Lab | INTERPRETATION | Normal |
| Lab | IONIZED CALCIUM | Normal, LOW |
| Lab | IRON | Normal, LOW, HI |
| Lab | Issue Date/Time | No flag |
| Lab | LACTIC ACID | Normal, LOW, HI, CRIT |
| Lab | LARGE PLATELETS | Normal |
| Lab | LDH | Normal, HI |
| Lab | LDL, CALCULATED | Normal, HI |
| Lab | LEGIONELLA AG, UR | Normal |
| Lab | LEUK ESTERASE | Normal, ABN |
| Lab | LEVETIRACETAM LEVEL | LOW |
| Lab | LIPASE | Normal, LOW, HI |
| Lab | LITHIUM | Normal |
| Lab | LYMPH | Normal, LOW, HI |
| Lab | LYMPHOCYTE | Normal, LOW, HI |
| Lab | MACROCYTOSIS | Normal |
| Lab | MAGNESIUM | Normal, LOW ,HI |
| Lab | MARIJUANA-URINE | Normal, ABN |
| Lab | Marijuana, Urn Screen (THC, Urn, Screen) | Normal |
| Lab | MCH | Normal, LOW, HI |
| Lab | MCHC | Normal, LOW |
| Lab | MCV | Normal, LOW, HI |
| Lab | MEAS O2 SAT-MV | Normal, LOW, HI |
| Lab | META | HI |
| Lab | Methadone, Urn Screen | Normal |
| Lab | METHANOL | Normal |
| Lab | MICROALB/CREAT RATIO | HI |
| Lab | MICROCYTOSIS | Normal |
| Lab | MITOGEN MINUS NIL | Normal |
| Lab | MONO | Normal, LOW, HI |
| Lab | MONOCYTE | Normal, LOW, HI |
| Lab | MPV | Normal, LOW, HI |
| Lab | MRSA Transcribed Result | No flag |
| Lab | MUCUS | Normal |
| Lab | MYELO | HI |
| Lab | NEUT | Normal, LOW, HI |
| Lab | NEUTROPHIL | Normal, LOW, HI |
| Lab | NIL (NEGATIVE CONTROL) | Normal |
| Lab | NITRITE | Normal, ABN |
| Lab | NON FENTANYL URINE | Normal |
| Lab | Non-HDL Chol | No flag |
| Lab | NOROVIRUS GI/GII BY PCR | Normal |
| Lab | NUCLEATED RBC'S | Normal |
| Lab | O2 SAT | Normal, LOW, HI |
| Lab | O2 SAT MEASURED | Normal, LOW |
| Lab | OPIATE HYDROCODONE | Normal |
| Lab | OPIATE ACETYL MORPHINE | Normal |
| Lab | OPIATE CODEINE | Normal |
| Lab | OPIATE HYDROMORPHONE | Normal |
| Lab | OPIATE MORPHINE | Normal |
| Lab | OPIATE OXYCODONE | Normal |
| Lab | OPIATE OXYMORPHONE | Normal |
| Lab | OPIATES NORHYDROCODONE | Normal |
| Lab | OPIATES NOROXYCODONE | Normal |
| Lab | OPIATES NOROXYMORPHONE | Normal |
| Lab | OPIATES-URINE | Normal, ABN |
| Lab | Opiates, Urn Screen | Normal |
| Lab | OVA AND PARASITES EXAM | Normal |
| Lab | OVALOCYTES | Normal |
| Lab | PARA1 | Normal |
| Lab | PARA2 | Normal |
| Lab | PARA3 | Normal |
| Lab | PARA4 | Normal |
| Lab | PARVOVIRUS QUANT BY PCR | Normal |
| Lab | PCO2 | Normal, LOW, HI, CRIT |
| Lab | PCT FREE CARB | Normal |
| Lab | PERFORMING LAB | Normal |
| Lab | PH | Normal, LOW, HI |
| Lab | PHENCYCLIDINE UR | Normal |
| Lab | Phencyclidine, Urn Screen | Normal |
| Lab | PHENYTOIN FREE | Normal |
| Lab | PHENYTOIN TOTAL | Normal, LOW |
| Lab | PHOSPHORUS | Normal, LOW, HI, CRIT |
| Lab | PLT | Normal, LOW, HI, CRIT |
| Lab | PLT ESTIMATE | Normal |
| Lab | PO2 | Normal, LOW, HI |
| Lab | POIKILOCYTOSIS | Normal |
| Lab | POLYCHROMASIA | Normal |
| Lab | POTASSIUM | Normal, LOW,HI, CRIT |
| Lab | PRO BNP,NT | Normal, HI |
| Lab | PROCALCITONIN | Normal |
| Lab | Product Code | No flag |
| Lab | Product Identification | No flag |
| Lab | PROLACTIN | Normal |
| Lab | Propoxyphene, Urn Screen | Normal |
| Lab | PROT/CREAT RATIO | Normal |
| Lab | PROTHROMBIN TIME | Normal, HI |
| Lab | PTH-INTACT | HI |
| Lab | PTT | Normal, LOW, HI, CRIT |
| Lab | QTBG INTERPRETATION | Normal |
| Lab | QUANTIFERON TB RESULT | Normal |
| Lab | RBC | Normal, LOW, HI |
| Lab | RDW | Normal, HI |
| Lab | REACTIVE LYMPHS | Normal |
| Lab | RESPIRATORY PCR PANEL SPECIMEN SOURCE | Normal |
| Lab | RESPIRATORY SYNCYTIAL VIRUS | Normal |
| Lab | RETIC COUNT | Normal, HI |
| Lab | ROTAVIRUS A BY PCR | Normal |
| Lab | SALICYLATE | Normal |
| Lab | SALMONELLA SPECIES BY PCR | Normal |
| Lab | SARS-CoV-2 IGG AB | Normal, ABN |
| Lab | SCHISTOCYTES | Normal |
| Lab | SED RATE-WEST | Normal, HI |
| Lab | SEND OUT RESULT: | Normal |
| Lab | SEND OUT TEST: | Normal |
| Lab | SERUM ALB ELECT | Normal |
| Lab | SERUM ALPHA 1 | Normal |
| Lab | SERUM ALPHA 2 | Normal |
| Lab | SERUM BETA | Normal |
| Lab | SERUM GAMMA | Normal |
| Lab | SERUM HCG | Normal |
| Lab | SERUM OSMOLALITY | Normal, LOW, HI, CRIT |
| Lab | SERUM TOTAL PROTEIN | Normal |
| Lab | SFIX ENHANCED REPORT | Normal |
| Lab | SHIGA TOXIN 1 BY PCR | Normal |
| Lab | SHIGA TOXIN 2 BY PCR | Normal |
| Lab | SHIGELLA SPECIES BY PCR | Normal |
| Lab | SICKLE CELLS | Normal |
| Lab | SODIUM | Normal, LOW, HI |
| Lab | SPECIMEN SOURCE | Normal |
| Lab | SPECIMEN TYPE | Normal |
| Lab | SPHEROCYTES | Normal |
| Lab | SQUAMOUS EPI'S | Normal, HI |
| Lab | Status Information | No flag |
| Lab | STREPTOCOCCUS PNEUMONIAE AG, URINE | Normal |
| Lab | SYPHILIS FOLLOW UP, RPR QUANT | Normal |
| Lab | TARGET CELLS | Normal |
| Lab | TB AG MINUS NIL | Normal |
| Lab | TB SCR COMMENT | Normal |
| Lab | TB2 AG MINUS NIL | Normal |
| Lab | TEARDROPS | Normal |
| Lab | TOTAL CARB | Normal |
| Lab | TOTAL IRON BINDING | Normal, LOW, HI |
| Lab | TOTAL PROTEIN | Normal, LOW, HI |
| Lab | Total Syphilis Antibody IGG and IGM | ABN |
| Lab | TOXIC VACUOLIZATION | Normal |
| Lab | TRANS EPI CELLS | Normal, HI |
| Lab | TRANSFERRIN | Normal, LOW |
| Lab | Treponema pallidum Antibody by TP-PA | Normal |
| Lab | TRIGLYCERIDE | Normal, HI |
| Lab | TROPONIN I | Normal, HI, CRIT |
| Lab | TSH | Normal, LOW, HI |
| Lab | Unit Blood Type | No flag |
| Lab | Unit Number | No flag |
| Lab | UR CHLORIDE-RANDOM | Normal |
| Lab | UR CREATININE | Normal |
| Lab | UR OSMOLALITY | Normal, LOW, HI |
| Lab | UR PH | Normal |
| Lab | UR POTASSIUM-RANDOM | Normal |
| Lab | UR SODIUM-RANDOM | Normal |
| Lab | UR TOTAL PROTEIN | Normal |
| Lab | UR UREA N-RANDOM | Normal |
| Lab | URIC ACID | Normal, LOW, HI |
| Lab | Urine bacteria | ABN |
| Lab | URINE BILIRUB | Normal |
| Lab | URINE BLOOD | Normal,ABN |
| Lab | URINE CLARITY | Normal |
| Lab | URINE COLOR | Normal |
| Lab | URINE GLUCOSE | Normal,ABN |
| Lab | URINE HCG | Normal |
| Lab | URINE KETONES | Normal,ABN |
| Lab | Urine pregnancy test—POCT | No flag |
| Lab | URINE PROTEIN | Normal,ABN |
| Lab | Urine RBC's | Normal,HI |
| Lab | URINE SP GRAV | Normal,HI |
| Lab | Urine WBC's | Normal,HI |
| Lab | UROBILINOGEN | Normal,HI |
| Lab | VANCOMYCIN-RANDOM | Normal |
| Lab | VIBRIO GROUP BY PCR | Normal |
| Lab | VITAMIN B1 | Normal |
| Lab | VITAMIN B12 | Normal,HI |
| Lab | VITAMIN D (25 OH) | LOW |
| Lab | Volume | No flag |
| Lab | WAXY CAST | Normal |
| Lab | WBC | Normal,LOW,HI |
| Lab | WBC CLUMPS | Normal |
| Lab | WHOLE BLOOD GLUC | Normal,HI,CRIT |
| Lab | WHOLE BLOOD HGB | Normal,LOW |
| Lab | WHOLE BLOOD K | Normal,LOW,HI,CRIT |
| Lab | WHOLE BLOOD NA | Normal,LOW,HI |
| Lab | YERSINIA ENTEROCOLITICA BY PCR | Normal |
| Lab | ZINC, BLOOD | Normal |
| Vit | BMI | ok |
| Vit | BP diastolic | ok |
| Vit | BP systolic | ok |
| Vit | Pulse rate | ok |
| Vit | Respiratory rate | ok |
| Vit | SPO2 | ok,crit |
| Vit | Temp (DegC) | ok,crit |
Converting electronic health records (EHRs) to an event log
Process mining algorithms utilize event logs as their input. Event logs consist of a sequence of events with a name describing the observed action and its corresponding timestamp (i.e., when the event occurred). The temporally ordered sequence of such events is called a trace. Commonly, a trace contains only events that belong to the same context. In this paper, the observations of a specific COVID-19 admission formed a trace. This can also be understood as a trajectory. The set of all traces (i.e., all COVID-19 admissions in the dataset) comprised an event log.
The extracted traces of the event log were performed at 6 h, 12 h, 18 h, 24 h, 30 h, 36 h, 42 h, 48 h, 54 h, 60 h, 66 h, and 72 h of the hospital admission. Patients that had died or been discharged before a given time of the prediction were excluded from contributing date to times after discharge or death.
For each admission, static features were extracted that did not change over the course of the hospital encounter (i.e. demographic information, comorbidities). The patient-centric trajectory of the hospital encounter was then represented as a trace. A trace started with the first occurrence of an event related to the hospital encounter and ended with the occurrence of an outcome event: either discharge or death. Each event was associated with the timestamp of observation. In this way, the state of the patient can be reconstructed at each point of time. Events can be either location-based, vital signs, lab measurements, report-based, encounter-based, or ICU-based.
Location-based events represented that a patient moved to a particular location. For example: the emergency room, ICU, non-ICU inpatient teams, among others. Vital sign events represented the observation of a particular vital sign, which were subsequently recorded as either “ok” or “critical”. Laboratory measurements were flagged as either normal or abnormal to create the laboratory events. Report-based events corresponded to procedure reports (e.g. electrocardiograms or radiological testing). Report-based events correspond to a performed procedure without considering individual findings or outcomes within the reports. Encounter-based events represented specific highlights (admission, observation status, discharge, or death) during the hospital stay. ICU-based events were based on the admission or not to the ICU, therefore, there were ICU-in and ICU-out events recorded.
After the conversion of the EHR data, a set of traces (i.e., an event log) was obtained. Each set of traces corresponded to one hospital admission and used the events to describe the health trajectory of the patient from admission to either discharge or death. Due to the definition of events and the sequential structure of traces, the traces could be used to create subtraces, such that a subtrace contained only events from, e.g., admission time to 24 h into the hospital encounter.
Process mining/deep learning model development
A process mining/deep learning model was developed to predict the likelihood of mortality every 6-h within the first 72 h of hospital admission. Our approach is a combination of both process mining and deep learning modeling. The process mining modeling output were used as the input to the deep learning model for the prediction. The patient trajectories were used to extract a process graph model using a process mining discovery algorithm [11]. The resulting process model and the patient trajectories from admission to the time of prediction were fed to the Decay Replay Mining (DREAM) algorithm [12]. The DREAM algorithm enhances the process model with functions that parameterize time using the patient trajectories. As an output, the DREAM algorithm provides a state of the process model for each patient that contains time information. Hence, the outputs of the DREAM algorithm are called timed state samples (TSS). The TSS corresponds to the health condition of a patient up to the time of prediction and contains information on the observed events and process states, and their interarrival times. Comorbidities and demographic information were used as independent variables. The generated TSS, together with demographic information and comorbidities, were then fed to a Neural Network (NN) model to predict mortality for each 6-h interval within the first 72 h. The same process model was used for all time intervals, and the architecture of the NN is shown in Fig. 1. Also, Table 1 provides more details about the deep learning modeling parameters. Figure 2 illustrates the complete overview of our proposed approach. The corresponding source code is publicly available on our Github repository. Descriptive statistics, model development, and statistical analysis were conducted using Python, version 3.6.
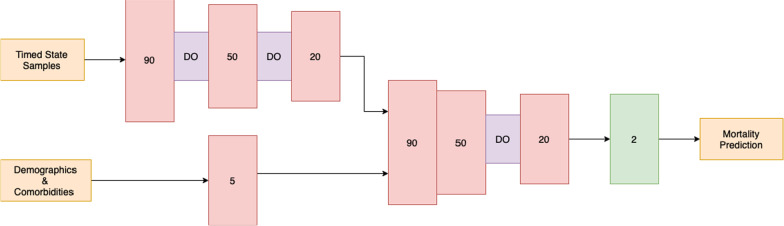
Fig. 1.
Architecture of Neural Network (NN). This Figure shows the details of the NN architecture. The timed state samples, demographics information and comorbidities were fed separately to two branches which first branch contains three hidden layers with 90, 50 and 20 neurons respectively. After the first and after the second hidden layers, there is a dropout layer with a rate of 20%. Moreover, the second branch contains one hidden layer with 5 neurons. The two branches were then concatenated to a branch with three hidden layers, containing 90, 50, and 20 neurons respectively. There is a dropout layer after the second concatenated hidden layer with the rate of 30%. At the end, the output layer included softmax activation function to predict mortality of the COVID- 19 patients
Table 1.
Deep learning model parameters
| Hours | Epoch | Batch size | Dropout rate | Activation function | Learning rate | optimizer |
|---|---|---|---|---|---|---|
| 6,12, 18, 30, 42, 54, 60, 66, 72 | 350 | 12 | 0.5 | Relu | 5e-4 | Adam |
| 24, 36 | 350 | 12 | 0.7 | Relu | 5e-4 | Adam |
| 48 | 350 | 8 | 0.7 | Relu | 5e-4 | Adam |
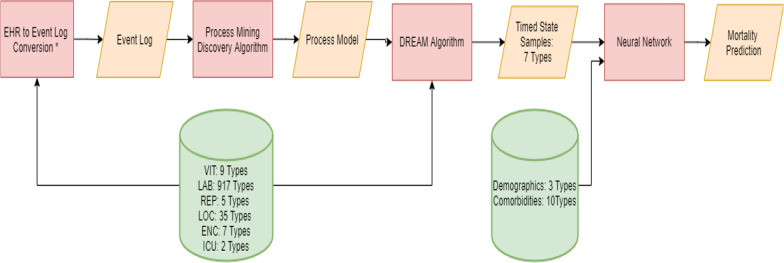
Fig. 2.
Process Mining/Deep Learning Model Development: The orange parallelograms represent the input/ output data. Four different algorithms were used in this methodology which is shown in red rectangles. The green cylinders represent the variable types that were coming directly from the database and were used as the inputs to the algorithms. *Refer to Section Converting Electronic Health Records (EHRs) to an Event Log for more details
Machine learning models
We compared the results of the process mining approach with results of a published model and self-developed models using machine learning algorithms that did not directly utilize time information.
The first model was a Logistic Regression (LR) model developed using data from 305 patients in China [13]. Core features in this model were age, Lactate dehydrogenase (LDH), and C-reactive protein (CRP).
The self-developed model was trained using the UIH data cohorts to explore other machine learning algorithms for the time interval modeling task. The development of these models utilized the variables described above. However, the data were kept in the original tabular format, as opposed to the event log format. The time component of the data was implicitly added to the training process by splitting a single training instance into multiple instances based on the time interval. This conversion allowed the developed models to witness instances from low time intervals that had limited information and from high intervals with more complete information. A variety of popular machine learning algorithms were evaluated to classify mortality at each 6- hour time interval within 72 h of admission. These algorithms included Logistic Regression (LR) [14], Decision Trees [15], Support Vector Machine (SVM) [16], Random Forest [17], XGBoost [18], LightGBM [19], and CatBoost [20]. The training process of these models included both a forward step feature selection and a grid search of model parameters. This search process aimed to find the best model with the fewest input features. The best model was determined based on the Average Area Under the Receiver Operating Characteristic Curve (AUROC) [21] of the validation cohort at each time interval.
Model evaluation
The primary evaluation metric for model development and selection was the AUROC. We used Delong’s test to calculate 95% confidence intervals (CI) of the AUROCs and compare AUROC CIs between models [22]. In addition, we calculated the accuracy, sensitivity and specificity of models across the time intervals [22], with 95% CIs.
Analysis of contribution of process mining unique variables
Shapley value analysis [23] was conducted on the testing cohort to find out the impact of each variable in the process mining model prediction and to identify variables associated with the mortality prediction of the COVID-19 patients in the 6-h intervals within the first 72 h, and to compare it to the self-developed machine learning and Chinese LR [13] models.
Results
UIH cohort characteristics
Table 2 shows the demographics, clinical characteristics, and medical conditions of the study population per encounter. There was a total of 508 encounters of 481 unique patients. The training cohort included 303 encounters (60%), the validation and testing cohorts the remaining 101 (20%) and 104 (20%) encounters, respectively. Given the size of the data, more traditional machine learning models have an advantage over deep learning based models. With the emergence of more COVID-19 data these models have the potential to be updated with more information. In the current state, data augmentation methods have the potential to be implemented with the goal of increasing overall performance. In this study, we do not implement any data augmentation, as the purpose of this work is to focus on the utilization of time information through the process mining algorithms.
Table 2.
Encounter characteristics of the training, validation, and testing cohorts
| Characteristics | Training cohort (N = 303) |
Validation cohort (N = 101) |
Testing cohort (N = 104) |
p-value train versus Test* | p-value validation versus test* | p-value train + validation versus test* |
|---|---|---|---|---|---|---|
| Number of unique patients N (%) | 288 (95.0) | 96 (95.0) | 97 (93.3) | |||
| Primary outcome (N, (%)) | ||||||
| Mortality | 43 (14.2) | 6 (5.9) | 11 (10.6) | 0.18 | 0.12 | < 0.0001 |
| Demographics | ||||||
| Age in years Mean (std) | 56.6 (16.6) | 56.6 (15.6) | 53.4 (14.2) | 0.012 | 0.028 | 0.009 |
| Female N (%) | 147 (48.5) | 50 (49.5) | 56 (53.8) | 0.18 | 0.27 | 0.18 |
| Race/ethnicity (N, (%)) | 0.63 | 0.95 | 0.76 | |||
| Black | 137 (45.2) | 51 (50.5) | 49 (47.1) | |||
| Hispanic | 36 (11.9) | 13 (12.9) | 16 (15.4) | |||
| Other, non- hispanic | 112 (37.0) | 30 (29.7) | 32 (30.7) | |||
| White | 18 (5.9) | 7 (6.9) | 7 (6.7) | |||
| Mean (std) of the number of laboratory measurements per encounter | ||||||
| 636 (786) | 510 (663) | 531 (972) | 0.078 | 0.228 | 0.090 | |
| Mean (std) vital signs measurements per encounter | ||||||
| 999 (1540) | 765 (1344) | 802 (1971) | 0.026 | 0.12 | 0.030 | |
| Comorbidities | 0.81 | 0.69 | 0.81 | |||
| Mean (std) comorbidities per encounter | 1.0 (1.1) | 1.0 (1.1) | 0.9 (0.9) | |||
| Hypertension N (%) | 128 (42.2) | 43 (42.6) | 37 (35.6) | |||
| Diabetes N (%) | 89 (29.4) | 32 (31.7) | 30 (28.8) | |||
| Heart disease N (%) | 12 (3.9) | 1 (1.0) | 2 (1.9) | |||
| COPD N (%) | 3 (1.0) | 0 (0.0) | 1 (1.0) | |||
| Stroke N (%) | 1 (0.3) | 0 (0.0) | 0 (0.0) | |||
| Cerebrovascular disease N (%) | 0 (0.0) | 2 (2.0) | 0 (0.0) | |||
| Cancer N (%) | 4 (1.3) | 2 (2.0) | 1 (1.0) | |||
| Respiratory problems N (%) | 44 (14.5) | 12 (11.9) | 15 (14.4) | |||
| Chronic kidney disease N (%) | 28 (9.2) | 11 (10.9) | 6 (5.7) | |||
| Tuberculosis N (%) | 3 (1.0) | 1 (1.0) | 3 (2.9) | |||
Bold indicates p-value < 0.05
Significance was set at 0.05
Patients older than 89 have been clipped to age 90
*Continuous variables were compared using a t-test and categorical variables were compared using a Chi-square test
The testing cohort was slightly younger than the training and validation cohorts (mean 53.4 vs. 56.6 years, p = 0.009). Though the distribution of race was not significantly different between the cohorts, the proportion of self-described Black patients was slightly higher in the validation (50.5%) and testing (47.1%) cohorts compared to the training cohort (45.2%). There were no statistically significant differences in the number of comorbidities per encounter in each cohort.
There were statistically more events in the training cohort (516.0 ± 3,882.3), compared to the testing (186.8 ± 1,217.4) and validation (176.6 ± 1,133.4) cohorts (P = 0.014). Conversely, there were no statistically significant differences across encounter types by cohort (P = 0.96); laboratory events were the most frequent (94%, 94%, and 93% in the training, testing, and validation cohorts, respectively), followed by location (3.6%, 3.3% and 4.3% in the training, testing and validation cohorts, respectively) and vital signs events (0.9%, 1.2% and 1.2% in the training, testing and validation cohorts, respectively).
Evaluation metrics and proposed and baseline model performance
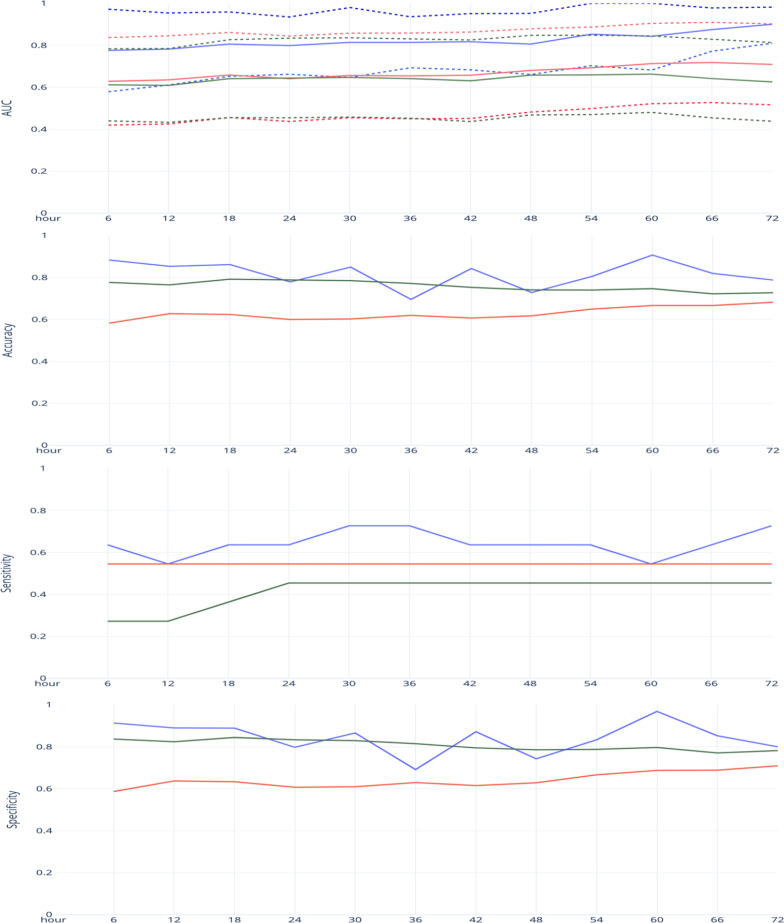
The process mining/ deep learning approach surmounted all of the time intervals in terms of AUROC compared to both the best baseline model and the best existing model in the literature. Also, in terms of specificity and accuracy, the proposed approach yielded the highest results in 9 intervals out of 12. Lastly, comparing the sensitivity metric results, our proposed model resulted in the best results in 10 intervals. The summary of the evaluation metrics for both the proposed approach and the baseline models is illustrated in Fig. 3 (detailed numbers in Table 3). Moreover, Table 4 shows an evaluation of the sensitivity and specificity for the three models. Hence, the experimental results indicate that our approach outperformed all evaluation metrics in most time intervals. A t-test of means is performed to test the stated null and alternative hypothesis for both the sensitivity and specificity over the 72-h time range with a threshold of 0.5. This analysis shows that the PM model outperformed both the RF and LR models.
Fig. 3.
Statistical metrics for all 6-h intervals within the first 72 h on the testing cohort. Blue indicators the Process Mining Model. Green indicators the Random Forest Model. Red indicators the Logistical Regression Model. Dashed lines indicate the upper and lower 95% confidence interval of the model’s AUROC
Table 3.
Detailed results on the testing cohort
| Hour | Confusion matrix | AUROC | Specificity | Sensitivity | Accuracy | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PM | RF | LR | PM | RF | LR | PM | RF | LR | PM | RF | LR | PM | RF | LR | ||
| 6 |
84;8 4;7 |
54;38 5;6 |
77;15 8;3 |
0.776 | 0.628 | 0.611 | 0.913 | 0.587 | 0.837 | 0.636 | 0.545 | 0.273 | 0.883 | 0.583 | 0.776 | |
| 12 |
81;10 5;6 |
58;33 5;6 |
75;16 8;3 |
0.782 | 0.635 | 0.608 | 0.890 | 0.637 | 0.824 | 0.545 | 0.545 | 0.273 | 0.853 | 0.627 | 0.765 | |
| 18 |
80;10 4;7 |
57;33 5;6 |
76;14 7;4 |
0.806 | 0.658 | 0.640 | 0.889 | 0.633 | 0.844 | 0.636 | 0.545 | 0.364 | 0.861 | 0.624 | 0.792 | |
| 24 |
67;17 4;7 |
51;33 5;6 |
70;14 6;5 |
0.799 | 0.640 | 0.644 | 0.798 | 0.607 | 0.833 | 0.636 | 0.545 | 0.455 | 0.779 | 0.600 | 0.789 | |
| 30 |
71;11 3;8 |
50;32 5;6 |
68;14 6;5 |
0.814 | 0.656 | 0.646 | 0.866 | 0.610 | 0.829 | 0.727 | 0.545 | 0.455 | 0.849 | 0.602 | 0.785 | |
| 36 |
56;25 3;8 |
51;30 5;6 |
66;15 6;5 |
0.814 | 0.654 | 0.641 | 0.691 | 0.630 | 0.815 | 0.727 | 0.545 | 0.455 | 0.696 | 0.619 | 0.771 | |
| 42 |
68;10 4;7 |
48;30 5;6 |
62;16 6;5 |
0.817 | 0.657 | 0.631 | 0.872 | 0.615 | 0.795 | 0.636 | 0.545 | 0.455 | 0.843 | 0.606 | 0.752 | |
| 48 |
52;18 4;7 |
44;26 5;6 |
55;15 6;5 |
0.806 | 0.680 | 0.657 | 0.743 | 0.629 | 0.786 | 0.636 | 0.545 | 0.455 | 0.728 | 0.617 | 0.740 | |
| 54 |
55;11 4;7 |
44;22 5;6 |
52;14 6;5 |
0.853 | 0.692 | 0.659 | 0.833 | 0.667 | 0.788 | 0.636 | 0.545 | 0.455 | 0.805 | 0.649 | 0.740 | |
| 60 |
62;2 5;6 |
44;20 5;6 |
51;13 6;5 |
0.843 | 0.713 | 0.662 | 0.969 | 0.688 | 0.797 | 0.545 | 0.545 | 0.455 | 0.907 | 0.667 | 0.746 | |
| 66 |
52;9 4;7 |
42;19 5;6 |
47;14 6;5 |
0.875 | 0.718 | 0.641 | 0.852 | 0.689 | 0.770 | 0.636 | 0.545 | 0.455 | 0.819 | 0.667 | 0.722 | |
| 72 |
44;11 3;8 |
39;16 5;6 |
43;12 6;5 |
0.9 | 0.709 | 0.625 | 0.800 | 0.709 | 0.782 | 0.727 | 0.545 | 0.455 | 0.788 | 0.681 | 0.727 | |
Table 4.
Statistical comparison of evaluation metrics
| Hypothesis | AUROC (p-value) |
|
|---|---|---|
| Null | Alternative | |
| PM = LR | PM > LR |
< 0.05 (PM has a significantly better AUROC than LR) |
| PM = LR | LR > PM |
> 0.05 (LR does not have a significantly better AUROC than PM) |
| PM = RF | PM > RF |
< 0.05 (PM has a significantly better AUROC than RF) |
| PM = RF | RF > PM |
> 0.05 (RF does not have a significantly better AUROC than PM) |
| RF = LR | RF > LR |
> 0.05 (RF does not have a significantly better AUROC than LR) |
| RF = LR | LR > RF |
> 0.05 (LR does not have a significantly better AUROC than RF) |
Shapley value analysis
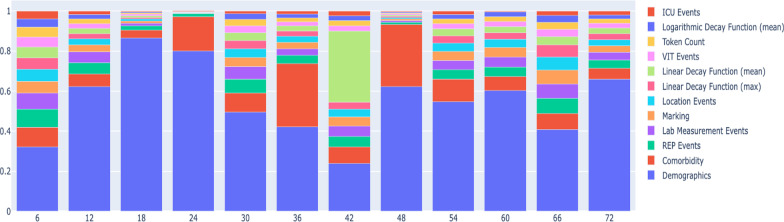
Figure 4 illustrates the results of the Shapley value analysis for all 6-h intervals within the first 72 h of admission. Also, the exact Shapley values are shown in Table 5. In almost all cases, demographic characteristics had the most significant impact on the prediction of mortality, followed by comorbidities. Age was strongly associated with mortality [9]. The impact of other variables varied from one time interval to another and comparing the value of the Shapley analysis for other variables, no consistent order was observed. The Shapley value analysis confirmed that the process mining-related variables–including the time decay function values, markings, and token counts– were consistently important for predicting mortality .
Fig. 4.
illustrates the results of the Shapley value analysis for all 6-h intervals within the first 72 h of COVD-19 patients
Table 5.
Shapley value analysis summary
| Category | Time intervals | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 6 Hr | 12Hr | 18Hr | 24Hr | 30Hr | 36Hr | 42Hr | 48 Hr | 54Hr | 60Hr | 66Hr | 72Hr | |
| Demographics | 0.0144 | 0.0706 | 0.5983 | 1.014 | 0.0657 | 0.0622 | 0.0222 | 0.2034 | 0.0422 | 0.0274 | 0.0199 | 0.0698 |
| Comorbidity | 0.0044 | 0.0071 | 0.0264 | 0.2162 | 0.0126 | 0.0465 | 0.0076 | 0.1012 | 0.0087 | 0.0032 | 0.0039 | 0.0058 |
| REP Events | 0.0041 | 0.0064 | 0.0143 | 0.0201 | 0.0092 | 0.0061 | 0.0049 | 0.0041 | 0.0036 | 0.0022 | 0.0037 | 0.0044 |
| Lab Measurement events | 0.0035 | 0.0062 | 0.0092 | 0.0023 | 0.0083 | 0.0048 | 0.0048 | 0.0026 | 0.0036 | 0.0022 | 0.0035 | 0.0041 |
| marking | 0.0027 | 0.0040 | 0.0079 | 0.0023 | 0.0061 | 0.0048 | 0.0043 | 0.0025 | 0.0034 | 0.0022 | 0.0034 | 0.0033 |
| Location events | 0.0027 | 0.0033 | 0.0068 | 0.0023 | 0.0058 | 0.0044 | 0.0035 | 0.0023 | 0.0032 | 0.0019 | 0.0032 | 0.0033 |
| Linear decay function (max) | 0.0025 | 0.0030 | 0.0058 | 0.0022 | 0.0053 | 0.0039 | 0.0033 | 0.0022 | 0.0028 | 0.0015 | 0.0029 | 0.0032 |
| Linear decay function (mean) | 0.0024 | 0.0030 | 0.0055 | 0.0018 | 0.0052 | 0.0038 | 0.033 | 0.0022 | 0.0028 | 0.0013 | 0.0020 | 0.0029 |
| VIT events | 0.0023 | 0.0027 | 0.0053 | 0.0017 | 0.0046 | 0.0031 | 0.0025 | 0.0019 | 0.0020 | 0.0012 | 0.0018 | 0.0026 |
| Token count | 0.0022 | 0.0027 | 0.0044 | 0.0017 | 0.0042 | 0.0028 | 0.0023 | 0.0016 | 0.0018 | 0.0011 | 0.0017 | 0.0024 |
| Logarithmic decay function (mean) | 0.0018 | 0.0026 | 0.0042 | 0.0016 | 0.0038 | 0.0027 | 0.0023 | 0.0015 | 0.0017 | 0.0011 | 0.0017 | 0.0021 |
| ICU Events | 0.0018 | 0.0019 | 0.0026 | 0.0013 | 0.0018 | 0.0024 | 0.0022 | 0.0014 | 0.0013 | 0.0002 | 0.0011 | 0.0020 |
Discussion
Using a cohort of hospitalized COVID-19 patients from a large medical center in the United States, we developed a process mining model using routine clinical data and the sequence of clinical events to evaluate mortality risk. Process mining performed significantly better than traditional predictive models over 6-h intervals within the first 72 h after hospital admission. Furthermore, we corroborate prior findings indicating that demographic characteristics and comorbidities are strong mortality predictors in COVID-19 [24, 25]. Interestingly, process mining-related variables such as time decay function values, markings, and token counts were found to have a strong predictive value. These findings advance our understanding of COVID-19 mortality prediction and support further studies using process mining for dynamic risk prediction.
Although previous studies have consistently demonstrated the underlying factors associated with COVID-19 mortality [24], our results highlight those traditional models such as logistic regression or random forest might underestimate the mortality prediction. In contrast to more traditional models, process mining leverages time and the sequence of events. Technically, this was realized through the usage of time functions, which activated the observation of events, and which decayed over time [12]. Multiple types of time decay functions were used, such as linear, exponential, and logarithmic. Each of those functions was initialized based on the mean or maximum patient history duration that was observed in the derivation data set.
By following this approach, predictive models can be developed that update outcome probability based on the time of the prediction. Thus, the likelihood of mortality may change over time, even if no further events have been observed.
The time decay functions values at a given time were fed into a NN, along with event features. Ideally, the NN does not just simply learn the impact of the duration of the last event observation on the outcome probability, but models potentially complex time relationships, such as event interarrival times that have an effect on the outcome probability. These complex time relationships could be the durations between specific lab measurements, or the duration from admission to ICU in the interplay of performed procedures. As clinician behavior may affect event timings and sequencing, the clinician behavior itself may be playing a role in the prediction.
Our results suggest that evaluating the clinical course and the sequence of events up until the time of a prediction can improve predictions as compared to only looking at factors present on admission [25]. Our results help reconcile and summarize findings that demographics, clinical events, laboratory data, and comorbidities can help predict mortality in COVID-19 inpatients. To date, work on artificial intelligence modeling in COVID-19 includes several methodologies, the most frequent being LR, XGBoost, support vector machine, RF, among others [7]. Although current artificial intelligence models have exhibited promising mortality predictive ability, it is unclear which of these methodologies might provide a better prediction compared to others. Moreover, available models do not consider the patient time course in addition to baseline covariates [26, 27]. This is crucial since it can promote early identification of COVID-19 patients with high mortality risk, helping improve clinical decision-making and resource allocation.
At a more general level, our findings are consistent with the concurrent evaluation of the clinical course and available clinical data [24]. Therefore, our work highlights the importance of a comprehensive evaluation of COVID-19 inpatients, including the sequence of clinical events.
A second important finding of this study was the added value of TSS on the process mining model development as time passes, which to date has not been used in COVID-19 prediction models [7]. Based on the results of the Shapley analysis, the time decay function values, and the distinct process mining variables such as markings and token counts, consistently demonstrated an important role in the mortality risk. Hence, our findings underscore the importance of carefully modeling mortality risk while taking into account the series of clinical events among hospitalized COVID-19.
Our approach outperformed other published models in terms of the accuracy, specificity, sensitivity, and AUROC values [13], as well as the best baseline internal model.
Study limitations
Our results should be interpreted in the light of several limitations. First, our modeling was performed using data from a single site, and these models may have performed differently in other cohorts; as a result, our process should be repeated externally to validate the value of adding time and sequence information in other data sets. Second, our data reflect the first COVID-19 wave in Chicago, therefore, it may not reflect the impact from COVID-19 variants, developed therapies, or vaccination. Third, our dataset contained only a modest number of patients and validation in larger cohorts is needed. Lastly, data validation for report time versus event occurrence time, were demanding, limiting the evaluation of the process mining model in real-time.
Conclusion
A process mining/deep learning approach using admission data and clinical course of hospitalized COVID-19 patients was able to predict mortality in 6-h intervals within the first 72 h of admission and performed significantly better than the commonly used approach of using only the initial admission results. Our findings underscore the importance of adopting clinical event times and sequencing in the study of COVID-19 mortality, which may help identify underlying characteristics among patients at risk. Since the use of TSS in process mining improved the prediction of COVID-19 mortality, strategies should be considered while identifying those sequential clinical changes, therefore helping to target treatments and resources among those at risk.
There are several avenues for future research. First, the resulting DREAM model can be used to discover if the non-observance of future events (such as action to be performed) has a positive or negative impact on the prediction to facilitate decision making. Such research efforts might enable the detection of improved intervention points in time. Second, sensitivity analyses can be performed to investigate the modeled time dependencies to gain new knowledge about COVID-19 care. This also allows us to investigate the robustness of the model to detect weaknesses that can be further improved. Lastly, our modeling can be used on larger and more diverse datasets and could be continued to be applied as new variants are observed and new vaccines and treatments introduced to assess their impact on clinical outcomes.
Acknowledgements
Not applicable
Abbreviations
- AUROC
Average area under the receiver operating characteristic curve
- AI
Artificial intelligence
- COVID-19
Coronavirus disease 2019
- DREAM
Decay replay mining
- LR
Logistic regression
- NN
Neural network
- RF
Random forest
- TSS
Timed state sample
- LDH
Lactate dehydrogenase
- CRP
C-reactive protein
Appendix
Table 6 shows the variables which were used as inputs to the proposed model. These variables are related to one of the following categories: demographics information, process mining, comorbidities, locations, encounters, procedure reports and the lab measurements. Moreover, where applicable, possible values of the variables are shown.
Author contributions
MP, SH, HD, JT: Involved in all aspects of this study. WG, JMR, LC, YZ, AT, KMK, AB: Data acquisition and interpretation, and revision of the manuscript. MP, SH, JT have equal contribution in this paper. All authors read and approved the final manuscript.
Funding
This research has been funded by the University of Illinois at Chicago Center for Clinical and Translational Science (CCTS) Award UL1TR002003. The funding body did not take part in the design of the study and collection, analysis, and interpretation of data and writing the manuscript.
Availability of data and materials
The datasets generated and/or analyzed during the current study are not publicly available due privacy but are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
This study was approved by University of Illinois at Chicago Internal Review Board. Permission from University of Illinois at Chicago Privacy Board and Internal Review Board were required to access the data used in this study. All the experiment protocols involving human data were in accordance with the University of Illinois at Chicago Privacy Board and Internal Review Board guidelines. Our research was provided a waiver of informed consent, parental permission and assent from the University of Illinois at Chicago IRB granted under 45 CFR 46.116(f).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Miotto R, Li L, Kidd BA, Dudley JT. Deep patient: an unsupervised representation to predict the future of patients from the electronic health records. Sci Rep. 2016;6(1):26094. doi: 10.1038/srep26094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Reilly KM, Sandman F, Allen D, Jarvis CI, Gimma A, Douglas A, et al. Predicted norovirus resurgence in 2021–2022 due to the relaxation of nonpharmaceutical interventions associated with COVID-19 restrictions in England: a mathematical modeling study. BMC Med. 2021;19(1):299. doi: 10.1186/s12916-021-02153-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morciano M, Stokes J, Kontopantelis E, Hall I, Turner AJ. Excess mortality for care home residents during the first 23 weeks of the COVID-19 pandemic in England: a national cohort study. BMC Med. 2021;19(1):71. doi: 10.1186/s12916-021-01945-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alballa N, Al-Turaiki I. Machine learning approaches in COVID-19 diagnosis, mortality, and severity risk prediction: a review. Inform Med Unlocked. 2021;24:100564. doi: 10.1016/j.imu.2021.100564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ghasemi M, Amyot D. Process mining in healthcare: a systematised literature review. Int J Electron Healthc. 2016;9:60. doi: 10.1504/IJEH.2016.078745. [DOI] [Google Scholar]
- 6.Theis J, Fau - Galanter W, Galanter W, Fau - Boyd A, Boyd A, Fau - Darabi H, Darabi H. Improving the In-Hospital Mortality Prediction of Diabetes ICU Patients Using a Process Mining/Deep Learning Architecture. LID.2021. 10.1109/JBHI.2021.3092969. (2168–2208 (Electronic)).
- 7.Adamidi ES, Mitsis K, Nikita KS. Artificial intelligence in clinical care amidst COVID-19 pandemic: a systematic review. Comput Struct Biotechnol J. 2021;19:2833–2850. doi: 10.1016/j.csbj.2021.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prediction of unplanned 30-day readmission for ICU patients with heart failure [Internet]. Available from: https://www.medrxiv.org/content/10.1101/2021.10.06.21264643v1. [DOI] [PMC free article] [PubMed]
- 9.Pishgar MRM, Theis J, Darabi H. Process mining model to predict mortality in paralytic ileus patients. In: International Conference on Cyber-physical Social Intelligence. 2021.
- 10.Galanter W, Rodríguez-Fernández JM, Chow K, Harford S, Kochendorfer KM, Pishgar M, et al. Predicting clinical outcomes among hospitalized COVID-19 patients using both local and published models. BMC Med Inform Decis Mak. 2021;21(1):224. doi: 10.1186/s12911-021-01576-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Augusto A, Conforti R, Dumas M, La Rosa M, Polyvyanyy A. Split miner: automated discovery of accurate and simple business process models from event logs. Knowl Inf Syst. 2019;59(2):251–284. doi: 10.1007/s10115-018-1214-x. [DOI] [Google Scholar]
- 12.Theis J, Darabi H. Decay replay mining to predict next process events. IEEE Access Pract Innov Open Solut. 2019;7:119787–119803. [Google Scholar]
- 13.Ma X, Ng M, Xu S, Xu Z, Qiu H, Liu Y, et al. Development and validation of prognosis model of mortality risk in patients with COVID-19. Epidemiol Infect. 2020;148:e168-e. doi: 10.1017/S0950268820001727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wright RE, In L, Grimm G, Yarnold PR. Logistic regression, reading and understanding multivariate statistics. 1995. pp. 217–44.
- 15.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J. Scikit-learn: machine learning in Python. J Mach Learn Res. 2011;12:2825–30. [Google Scholar]
- 16.Fürnkranz J. Decision Tree. In: Sammut C, Webb GI, editors. Encyclopedia of machine learning. Boston: Springer; 2010. pp. 263–267. [Google Scholar]
- 17.Breiman L. Random forests. Mach Learn. 2001;45(1):5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 18.Tianqi Chen CG. XGBoost: A scalable tree boosting system. Association for Computing Machinery. 2016.
- 19.Ke GMQ, Finley T, Wang T, Chen W, Ma W, et al. LightGBM: a highly efficient gradient boosting decision tree. Adv Neural Inf Process Syst. 2017;30:3146–3154. [Google Scholar]
- 20.Ostroumova L, Gusev G, Vorobev A, Dorogush AV, Gulin A. CatBoost: unbiased boosting with categorical features. NeurIPS; 2018.
- 21.Siddiqui MK, Morales-Menendez R, Ahmad S. Application of receiver operating characteristics (roc) on the prediction of obesity. Braz Arch Biol Technol. 2020 doi: 10.1590/1678-4324-2020190736. [DOI] [Google Scholar]
- 22.DeLong ER, DeLong DM, Fau - Clarke-Pearson DL, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. xxxx. (0006–341X (Print)). [PubMed]
- 23.Lundberg SM, Lee SI. A unified approach to interpreting model predictions. 2017.
- 24.Tian W, Jiang W, Yao J, Nicholson CJ, Li RH, Sigurslid HH, et al. Predictors of mortality in hospitalized COVID-19 patients: a systematic review and meta-analysis. J Med Virol. 2020;92(10):1875–1883. doi: 10.1002/jmv.26050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mesas AE, Cavero-Redondo I, Álvarez-Bueno C, Sarriá Cabrera MA, Maffei de Andrade S, Sequí-Dominguez I, et al. Predictors of in-hospital COVID-19 mortality: a comprehensive systematic review and meta-analysis exploring differences by age sex and health conditions. PLoS One. 2020;15(11):e0241742. doi: 10.1371/journal.pone.0241742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Argenziano MG, Bruce SL, Slater CL, Tiao JR, Baldwin MR, Barr RG, et al. Characterization and clinical course of 1000 patients with coronavirus disease 2019 in New York: retrospective case series. BMJ. 2020;369:m1996. doi: 10.1136/bmj.m1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analyzed during the current study are not publicly available due privacy but are available from the corresponding author on reasonable request.