Abstract
Background:
Radiographs are widely used to evaluate radiographic progression with modified stoke ankylosing spondylitis spinal score (mSASSS).
Objective:
This pilot study aimed to develop a deep learning model for grading the corners of the cervical and lumbar vertebral bodies for computer-aided detection of mSASSS in patients with ankylosing spondylitis (AS).
Methods:
Digital radiographic examination of the spine was performed using Discovery XR656 (GE Healthcare) and Digital Diagnost (Philips). The disk points were detected between the bodies using a key-point detection deep learning model from the image obtained in DICOM (digital imaging and communications in medicine) format from the cervical and lumbar spinal radiographs. After cropping the vertebral regions around the disk point, the lower and upper corners of the vertebral bodies were classified as grade 3 (total bony bridges) or grades 0, 1, or 2 (non-bridges). We trained a convolutional neural network model to predict the grades in the lower and upper corners of the vertebral bodies. The performance of the model was evaluated in a validation set, which was separate from the training set.
Results:
Among 1280 patients with AS for whom mSASSS data were available, 5,083 cervical and 5245 lumbar lateral radiographs were reviewed. The total number of corners where mSASSS was measured in the cervical and lumbar vertebrae, including the upper and lower corners, was 119,414. Among them, the number of corners in the training and validation sets was 110,088 and 9326, respectively. The mean accuracy, sensitivity, and specificity for mSASSS scoring in one corner of the vertebral body were 0.91604, 0.80288, and 0.94244, respectively.
Conclusion:
A high-performance deep learning model for grading the corners of the vertebral bodies was developed for the first time. This model must be improved and further validated to develop a computer-aided tool for assessing mSASSS in the future.
Keywords: ankylosing spondylitis, artificial intelligence, deep learning, mSASSS, radiographic progression
Introduction
Ankylosing spondylitis (AS) is a chronic inflammatory disease characterized by the inflammation and ankylosis of the spinal joint.1,2 In some patients, progression of structural changes in the spinal joint is noted, which results in irreversible limitation of motion and reduced quality of life. Therefore, it is necessary to quantify radiographic progression to confirm the disease state and the prognosis of the patient.
Regarding the assessment of radiographic progression, magnetic resonance imaging (MRI) and computed tomography (CT) are useful methods for detecting structural changes in the spine of patients with AS; however, increased cost and radiation exposure pose a problem.3–5 Therefore, radiographs are widely used to evaluate radiographic progression with modified stoke ankylosing spondylitis spinal score (mSASSS) in both cohort and clinical trials. Although radiographs are less expensive and widely available compared with MRI or CT, it is difficult to distinguish the small changes in the vertebral structures and requires 2 years or more to confirm radiographic progression. In addition, the validation of radiographic progression is time-consuming and increases the workload for radiologists.4,6
With the recent advances in artificial intelligence (AI) technology, the analysis of medical images using deep learning has shown promising results in the quantification, detection, and diagnosis of various diseases.7–9 A recent study recently reported the use of deep learning in the field of rheumatic diseases to detect radiographic sacroiliitis in axial spondyloarthritis. 10 It exhibited excellent performance in the detection of definite radiographic sacroiliitis with areas under the curve of 0.97 and 0.94 for the validation and test datasets, respectively, suggesting that these models can be utilized as classification tools in research and diagnostic aid in clinical practice.
With the detection of sacroiliitis, the quantification of the spinal radiographic progression is also necessary to assess the disease status of the patients. Computer-aided assessment of the structural damage of the spine using deep learning algorithms may also be useful in evaluating the radiographic progression in clinical trials or clinical practice for patients with AS. The purpose of this study was to develop a computer-aided detection algorithm using deep learning for grading the structural damage by training the vertebral corners of the spinal radiographs in patients with AS.
Methods
Patients and data collection
The data of 1280 patients who were diagnosed with AS between January 2001 and December 2018 at a single center according to the modified New York criteria 11 were reviewed. To evaluate the patient’s radiographic progression, radiographs were taken approximately every 2 years, and mSASSS was assessed.12,13 Digital radiographic examination of the spine was performed using Discovery XR656 (GE Healthcare) and Digital Diagnost (Philips).
Assessment of radiographic progression
Two radiologists (Seunghun Lee and Kyung Bin Joo) independently assessed the radiographs according to the mSASSS (0–72). They evaluated mSASSS without EMR information. K. Joo re-evaluated mSASSS without referring to the previous assessments 4 months after interpreting all the radiographs. The intra-observer reliability with consistency (one-way model) and inter-observer reliability with agreement (two-way model) were calculated. The intra-observer reliability with consistency [intraclass coefficient (ICC) = 0.978, 95% CI, 0.976–0.979] and inter-observer reliability with agreement between the two readers were excellent (ICC = 0.946, 95% CI, 0.941–0.950).12–14 The mSASSS was evaluated using the lateral view of the cervical and lumbar radiographs of the spine. The radiologist evaluated 12 anterior corners from the lower corner of the body of the second cervical vertebra to the upper corner of the body of the first thoracic vertebra and 12 anterior corners from the lower corner of the body of the 12th thoracic vertebra to the upper corner of the sacrum. Each lower and upper anterior corner was labeled as grade 0 (normal), 1 (erosions, sclerosis, and/or squaring), 2 (syndesmophytes), or 3 (total body bridges).
Training with deep learning
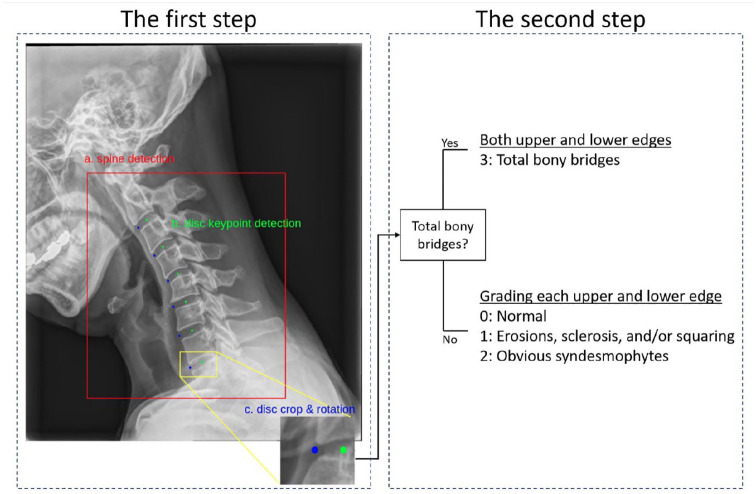
Among the radiographs, the lateral views of the cervical and lumbar spine were selected. Since we were interested in grading the anterior corners of the vertebral body, the modeling process consisted of two steps to analyze per radiograph (Figure 1). The first step was to automatically detect each vertebral region for analysis, and the second step was to automatically grade each corner of the vertebral body in the detected region. All the images converted from DICOM format to PNG (portable network graphic) format were used for training and validation. The contrast was automatically adjusted during this automatic conversion process.
Figure 1.
The two steps of the modeling process for training. The first step is the automatic detection of the (a) spinal region and (b) disk point; two points (green and blue) are detected per disk space. (c) The region is cropped around the center point. The second step grades the upper and lower corners.
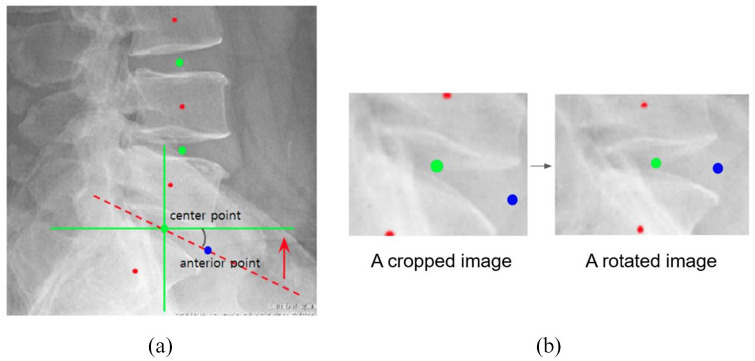
Initially, we built an algorithm to detect the regions of the vertebra in the lateral view of the radiograph. This algorithm finds two disk points per disk space from the second cervical to the first thoracic vertebral body of the detected region. Two disk points were located—one at the center of the disk and the other, anterior to the disk. Therefore, six disks were selected from each of the cervical and lumbar vertebrae, and 12 disk points were located. The two disk points formed one region for grading the upper and lower corners of one vertebra. Therefore, six grading regions, each on the cervical and lumbar vertebrae, were cropped around the center point as a better performance was obtained with the center point on the vertebral body than with the anterior point on the marginal bone. To increase the accuracy of the grading, the anterior and center points of the disk were rotated horizontally. After finding the angle between the horizontal line and the straight line connecting the center and the anterior point, the center point was rotated by that angle to make all spinal bodies horizontal (Figure 2).
Figure 2.
A vertebral disk rotated horizontally using the anterior and center points. After finding the angle (red arrow) between the horizontal line (green line) and the line connecting the center and the anterior point (red dotted line). (a) The image is rotated by that angle (red arrow) around the point at the center to make all spinal bodies horizontal. (b) The red dot is for illustration purposes only to indicate the center of the vertebral body and is not used in the algorithm.
The modified HRNet model, a key-point detection deep learning model, was used for detecting the region of interest for the anterior and center points of the disk in the spine. The HRNet model learns various resolutions by changing the scale of the image being trained. Compared with the previous image learning by downsampling the original image, HRNet maintains the resolution of the original image while downsampling; thus, the image information is not lost. 15 In addition, it also infers key-points by averaging the heatmaps generated at various resolutions during image testing; hence, the key-points of various scales can be inferred well. HRNet has high accuracy because various image resolutions continuously integrate information between the networks in parallel. In this study, the original image was reduced in resolution (256 × 192) to fit this model. Although the resolution was reduced, it is possible to achieve accurate results by detecting the target regardless of the resolution.
Using Pytorch version 1.1, this model was trained with 1080 radiograph images for the training set and 120 radiograph images for the validation set. Training was conducted for 30 h using the graphic card GTX 3090; the batch size was 32, the learning rate was 0.001, and the optimizer was Adam. In addition, transfer learning of the human pose estimate pretrained model was used for retraining.
Subsequently, the region consisting of a bridge between the upper and lower corners was graded as 3 for both corners. In the absence of a bridge between the upper and lower corners, the grades were classified as 0, 1, or 2. An algorithm for grading the upper and lower corners of the vertebral body in the selected region was developed using a convolutional neural network (CNN) model of residual network (ResNet) 152. 16
ResNet has a very deep network of up to 152 layers, characterized by introducing residual connections to solve the problem of vanishing gradients. 16 In CNN, the performance could be improved by stacking layers deeply, but since actual gradient vanishing/exploding takes place, the performance decreases and training errors increase. ResNet has improved the performance of image analysis by solving this phenomenon. The model was trained using Tensorflow version 2.4. The model was trained for 48 h using the graphics card GTX 3090. The batch size was 3, the learning rate was 0.000001, and the optimizer was Adam. The phenomenon of the loss not converging was encountered when a high learning rate was used; therefore, an appropriate learning rate was selected. Data augmentation techniques, such as rotation, flip, shift, and CLAHE, were used. A learning rate scheduler was not used; the learning rate decay provided by the Adam optimizer was used.
The ImageNet pretrained model was retrained by transfer learning. To facilitate the learning, the learning was carried out by shuffling the order of the images every epoch. The number of epochs during training was 10,000, and validation was performed for each epoch. The weight values of the final model were used as the value of the epoch with the best validation result. In addition, since the number of image data for each mSASSS grade was unbalanced, the image data of grades 0, 1, and 3 were downsampled to match the number of image data of grade 2 with the smallest number. The number of all grades was set to 300 of grade 2, and the downsampled dataset was changed every epoch. For example, out of 10,000 vertebral body data, the data set from 0 to 300 was used in the first epoch, and the data set from 301 to 600 was used in the second epoch to ensure that all data was used.
The evaluation of the model performance
Since the number of grades was unbalanced, it was difficult to divide the data into training, validation, and test sets. Therefore, we performed an analysis by dividing the data into training and validation sets. The radiographs were divided into training and validation sets in a ratio of 8:2. We reduced a high percentage of grade 0 compared with other grades to reduce the imbalance between the classes during training and validation of the grade of each corner. The performance of the model was evaluated in a validation set and separated from the trained set. The actual and predicted values of the grades for each corner were compared using a confusion matrix. In addition, the sensitivity, specificity, positive predictive value, F1 score, and accuracy of predicting the grades were evaluated. All analyses were performed using Python (Python Software Foundation, version 3).
Results
Training and validation sets
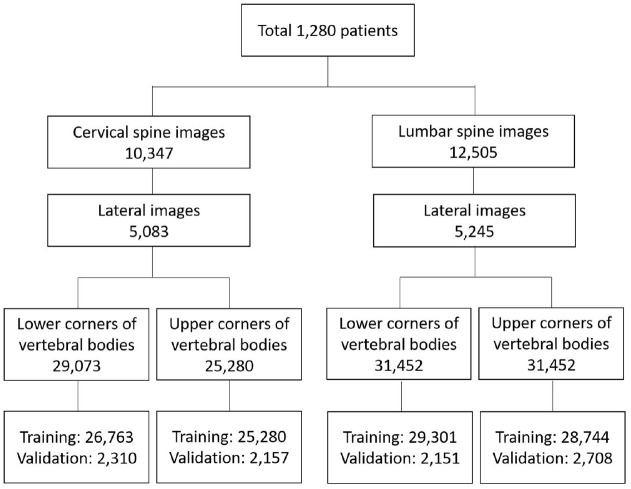
Among 1280 patients with AS with radiographs, 5083 cervical and 5245 lumbar lateral radiographs labeled with mSASSS were reviewed (Figure 3). The total number of corners, including the upper and lower corners on the cervical and lumbar spine radiographs, was 119,414, which were divided into 110,088 training and 9326 validation sets.
Figure 3.
Flow chart for the patients and radiographs.
Prediction of the grade
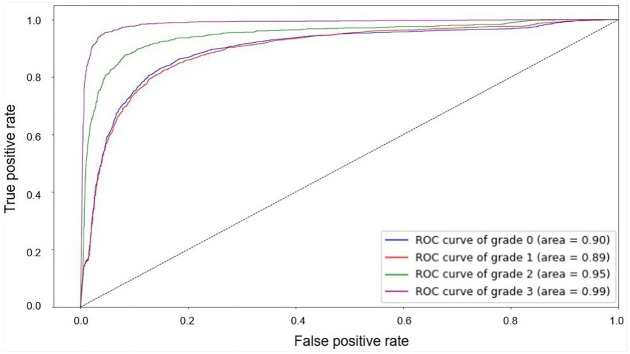
The actual and predicted values for each grade were presented in a confusion matrix (Table 1). The performance for each grade is listed in Table 2. The sensitivity and accuracy for grade 3 were relatively high compared with those for the other grades (0.93652 and 0.95743, respectively). For grade 2, the sensitivity was low but the specificity was high, compared with the values for the other grades (0.63869 and 0.97266, respectively). The accuracy was relatively low for grade 1 (0.87958). The mean accuracy, sensitivity, and specificity for all grades were 0.91604, 0.80288, and 0.94244, respectively. The receiver operating characteristic curve for each grade is shown in Figure 4.
Table 1.
Matrix of the actual and predicted grade in the validation set.
| Grade | Predicted class | |||||
|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | Total | ||
| Actual class | 0 | 2250 | 295 | 54 | 19 | 2618 |
| 1 | 502 | 2169 | 75 | 46 | 2792 | |
| 2 | 96 | 147 | 700 | 153 | 1096 | |
| 3 | 25 | 58 | 96 | 2641 | 2820 | |
| Total | 2873 | 2669 | 925 | 2859 | 9326 | |
Table 2.
Performance for each scoring in the validation set.
| Grade | True positive | True negative | False positive | False negative | Sensitivity | Specificity | Positive predictive value | F1 score | Accuracy |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 2250 | 6085 | 623 | 368 | 0.859 | 0.907 | 0.783 | 0.820 | 0.894 |
| 1 | 2169 | 6034 | 500 | 623 | 0.777 | 0.923 | 0.813 | 0.794 | 0.880 |
| 2 | 700 | 8005 | 225 | 396 | 0.639 | 0.973 | 0.757 | 0.693 | 0.933 |
| 3 | 2641 | 6288 | 218 | 179 | 0.937 | 0.966 | 0.924 | 0.930 | 0.957 |
Figure 4.
The receiver operating characteristic curve for each grade.
Discussion
We developed a deep learning model for grading the changes in the corners of the vertebral body with good performance on spinal radiographs in patients with AS. This study suggests that computer-aided detection of marginal bony changes in the spine, reflecting structural damage in AS, is possible. However, there is room for further improvement in the performance, which required improvement for clinical application and research for the assessment of mSASSS. Thus, a better algorithm application, more high-quality data, and external validation are needed.
It is very difficult to quantify the radiographic progression in patients with AS.4,17,18 Although several quantification methods have been developed for imaging, such as CT and MRI. The mSASSS that can be evaluated on a radiograph is the appropriate and preferred scoring method for radiographic progression in clinical trials.4,6 However, it is also less validated in detecting radiographic changes and has been difficult to use routinely in a clinical setting. 19 Therefore, improving the reliability of the scoring methods for assessing the radiographic progression is essential.
For the reliability of mSASSS, two or more experienced radiologists are required to research radiographic progression.4,19 In addition, it is also important to detect the microscopic changes as the structural changes in the spine take a long time to be confirmed through radiography. Rheumatologists need to detect the progression of a patient’s radiographic progression in clinical practice, and patients often wonder how the ankylosis is progressing. MRI can be a good alternative,20,21 but it is time-consuming to acquire and score images. 19 Moreover, MRI is an expensive method to obtain an image of the radiographic changes during every follow-up. Therefore, we suggest that computer-aided detection methods to evaluate mSASSS on radiographs using deep learning will be useful for quantifying the spinal radiographic progression in clinical practice and research.
Various algorithms for image analysis and classification have been developed. Most image analysis algorithms using deep learning have been developed based on CNN. CNN is a structure that extracts features from data and identifies patterns of features. Regarding the deep learning model for image processing in this study, two CNN algorithms were used. First, a modified HRNet derived from CNN was used for the detection of the vertebral body, and 24 disk points were located. HRNet can maintain high-resolution representation throughout processing and showed improved performance compared with the other key-point detection models.15,22 Second, ResNet 152 model was used to train the grade of the corners of the vertebral body. ResNet overcomes the limitation by transferring the features of the previous layer to the subsequent layer to retain the characteristics of the features.16,22,23 Therefore, the model can be deepened by using residual connections; as a result, the accuracy of the model is further increased.
A dataset is an important factor in improving the prediction power of deep learning. Although AS is a rare disease, a large number of spine radiographs have been collected over a long period of time so that the spine changes of many patients can be identified at a single center. Therefore, to improve the prediction performance, we segmented and processed vertebral bodies from cervical and lumbar spinal radiographs to create a training dataset of over 100,000. In addition, this dataset has been labeled with mSASSS in the past, and its excellent reliability has been verified through various studies on radiographic progression.12,13,24
Taken together, the deep learning model detected all regions of the vertebra of interest and successfully graded the upper and lower corners of the vertebral body with good performance. Grading of the corner of the vertebral body grade 3, which is easy for humans to detect, is also found in deep learning models. However, there was a high percentage of misreading in predicting grade 0 as 1, grade 1 as 0, and grade 2 as 1 or 3. In the assessment of mSASSS, including 24 corners of the vertebral bodies on the cervical and lumbar vertebrae, higher accuracy may be required than the grading accuracy on each corner of the vertebral body.
There are some limitations to this study. First, this study used a model based on single-center data. A multi-center test set is required for external validation. Second, grading was performed on the vertebral bodies in patients with scoliosis capable of key-point detection; however, cases with severe malformations or artificial structures were excluded from the data set. It is necessary to address this issue while evaluating the overall mSASSS in the future. Third, because the developed algorithm was not made with a software, clinical verification was not possible in a medical environment, and this verification will be carried out in the next study. Fourth, because this study was performed by splitting the data at the radiograph level, there may be a risk of overfitting.
mSASSS is a representative tool for quantitatively assessing the radiographic progression in patients with AS. This pilot study presented a deep learning-based solution for computer-aided detection of the structural changes in the vertebral body using mSASSS-labeled radiographs. The performance of predicting the grades for changes in the vertebral corners in the lateral view of cervical and lumbar radiographs was promising. In the future, external validation is required, along with performance improvement with additional datasets.
Acknowledgments
This is a collaborative study with CRESCOM Co., Ltd.
Footnotes
ORCID iDs: Bon San Koo  https://orcid.org/0000-0002-4212-2634
https://orcid.org/0000-0002-4212-2634
Kim Tae-Hwan  https://orcid.org/0000-0002-3542-2276
https://orcid.org/0000-0002-3542-2276
Contributor Information
Bon San Koo, Division of Rheumatology, Department of Internal Medicine, Inje University Seoul Paik Hospital, College of Medicine, Inje University, Seoul, Korea.
Jae Joon Lee, CRESCOM, Seongnam-si, Korea.
Jae-Woo Jung, CRESCOM, Seongnam-si, Korea.
Chang Ho Kang, Department of Radiology, Korea University Anam Hospital, Seoul, Korea.
Kyung Bin Joo, Department of Rheumatology, Hanyang University Hospital for Rheumatic Diseases, Seoul, Korea.
Tae-Hwan Kim, Department of Rheumatology, Hanyang University Hospital for Rheumatic Diseases, Seoul, Korea.
Seunghun Lee, Department of Radiology, Hanyang University Hospital for Rheumatic Diseases, 222-1, Wangsimni-ro, Seongdong-gu, Seoul 04763, Korea.
Declarations
Ethics approval and consent to participate: The Hanyang University Seoul Hospital Institutional Review Board approved this study (HYUH 2020-01-011). The need to obtain informed consent was waived as this is an anonymized radiographic image analysis study.
Consent for publication: Not applicable
Author contributions: Bon San Koo: Conceptualization; Investigation; Project administration; Writing – original draft; Writing – review & editing.
Jae Joon Lee: Formal analysis; Investigation; Methodology; Project administration; Software; Validation; Visualization; Writing – review & editing.
Jae-Woo Jung: Formal analysis; Investigation; Methodology; Project administration; Software; Validation; Visualization; Writing – review & editing.
Chang Ho Kang: Formal analysis; Investigation; Methodology; Project administration; Software; Supervision; Validation; Visualization; Writing – review & editing.
Kyung Bin Joo: Investigation; Project administration; Resources; Writing – review & editing.
Tae-Hwan Kim: Conceptualization; Data curation; Investigation; Project administration; Resources; Supervision; Writing – original draft; Writing – review & editing.
Seunghun Lee: Conceptualization; Data curation; Formal analysis; Investigation; Project administration; Resources; Supervision; Validation; Visualization; Writing – original draft; Writing – review & editing
Funding: The authors received no financial support for the research, authorship, and/or publication of this article.
Competing Interests: The authors declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: Seunghun Lee, Tae-Hwan Kim and Chan Ho Kang are CRESCOM Co., Ltd. Stockholders and Jae-Joon Lee is a founder of CRESCOM Co., Ltd., a startup company whose eventual products and services will be related to the subject matter of the article. The authors indicate no other conflicts of interest pertaining to the content of this article.
Availability of data and materials: The disclosure of personally identifiable information of patients, including radiographs, is restricted by the Institutional Review Board.
References
- 1. Inman RD. Axial spondyloarthritis: current advances, future challenges. J Rheum Dis 2021; 28: 55–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ranganathan V, Gracey E, Brown MA, et al. Pathogenesis of ankylosing spondylitis: recent advances and future directions. Nat Rev Rheumatol 2017; 13: 359–367. [DOI] [PubMed] [Google Scholar]
- 3. Tins BJ, Butler R. Imaging in rheumatology: reconciling radiology and rheumatology. Insights Imag 2013; 4: 799–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. van der Heijde D, Landewé R. Selection of a method for scoring radiographs for ankylosing spondylitis clinical trials, by the Assessment in Ankylosing Spondylitis Working Group and OMERACT. J Rheumatol 2005; 32: 2048–2049. [PubMed] [Google Scholar]
- 5. Ostergaard M, Boesen M. Imaging in rheumatoid arthritis: the role of magnetic resonance imaging and computed tomography. Radiol Med 2019; 124: 1128–1141. [DOI] [PubMed] [Google Scholar]
- 6. Wanders AJ, Landewe RB, Spoorenberg A, et al. What is the most appropriate radiologic scoring method for ankylosing spondylitis? A comparison of the available methods based on the Outcome Measures in Rheumatology Clinical Trials filter. Arthritis Rheum 2004; 50: 2622–2632. [DOI] [PubMed] [Google Scholar]
- 7. Galbusera F, Casaroli G, Bassani T. Artificial intelligence and machine learning in spine research. JOR Spine 2019; 2: e1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lee JG, Jun S, Cho YW, et al. Deep learning in medical imaging: general overview. Korean J Radiol 2017; 18: 570–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yoon JH, Kim EK. Deep learning-based artificial intelligence for mammography. Korean J Radiol 2021; 22: 1225–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bressem KK, Vahldiek JL, Adams L, et al. Deep learning for detection of radiographic sacroiliitis: achieving expert-level performance. Arthritis Res Ther 2021; 23: 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. van der Linden S, Valkenburg HA, Cats A. Evaluation of diagnostic criteria for ankylosing spondylitis: a proposal for modification of the New York criteria. Arthritis Rheum 1984; 27: 361–368. [DOI] [PubMed] [Google Scholar]
- 12. Koo BS, Oh JS, Park SY, et al. Tumour necrosis factor inhibitors slow radiographic progression in patients with ankylosing spondylitis: 18-year real-world evidence. Ann Rheum Dis 2020; 79: 1327–1332. [DOI] [PubMed] [Google Scholar]
- 13. Lee TH, Koo BS, Nam B, et al. Conventional disease-modifying antirheumatic drugs therapy may not slow spinal radiographic progression in ankylosing spondylitis: results from an 18-year longitudinal dataset. Ther Adv Musculoskelet Dis 2020; 12: 75912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Koo BS, Lee S, Oh JS, et al. Early control of C-reactive protein levels with non-biologics is associated with slow radiographic progression in radiographic axial spondyloarthritis. Int J Rheum Dis 2022; 25: 311–316. [DOI] [PubMed] [Google Scholar]
- 15. Sun K, Xiao B, Liu D, et al. Deep high-resolution representation learning for human pose estimation. In: Proceedings of the 2019 IEEE/CVF conference on computer vision and pattern recognition (CVPR), Long Beach, CA, 15–20 June 2019, pp. 5686–5696. New York: IEEE. [Google Scholar]
- 16. He K, Zhang X, Ren S, et al. Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, Las Vegas, NV, 27–30 June 2016, pp. 770–778. New York: IEEE. [Google Scholar]
- 17. Braun J, Baraliakos X. Imaging of axial spondyloarthritis including ankylosing spondylitis. Ann Rheum Dis 2011; 70: i97–i103. [DOI] [PubMed] [Google Scholar]
- 18. Ramiro S, Heijde D, Sepriano A, et al. Spinal radiographic progression in early axial spondyloarthritis: five-year results from the DESIR cohort. Arthritis Care Res 2018; 71: 1678–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. van der Heijde D, Braun J, Deodhar A, et al. Modified stoke ankylosing spondylitis spinal score as an outcome measure to assess the impact of treatment on structural progression in ankylosing spondylitis. Rheumatology 2019; 58: 388–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mandl P, Navarro-Compán V, Terslev L, et al. EULAR recommendations for the use of imaging in the diagnosis and management of spondyloarthritis in clinical practice. Ann Rheum Dis 2015; 74: 1327–1339. [DOI] [PubMed] [Google Scholar]
- 21. Kucybała I, Urbanik A, Wojciechowski W. Radiologic approach to axial spondyloarthritis: where are we now and where are we heading. Rheumatol Int 2018; 38: 1753–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Alzubaidi L, Zhang J, Humaidi AJ, et al. Review of deep learning: concepts, CNN architectures, challenges, applications, future directions. J Big Data 2021; 8: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. He K, Zhang X, Ren S, et al. Identity mappings in deep residual networks. In: European conference on computer vision. Berlin: Springer, 2016, pp. 630–645. [Google Scholar]
- 24. Koo B, Oh J, Park S, et al. Suppressing inflammation rather than lowering the disease activity score should be targeted during TNF inhibitor treatment to slow radiographic changes in patients with Ankylosing Spondylitis [abstract]. Arthritis Rheumatol 2020; 72 (suppl. 10). https://acrabstracts.org/abstract/suppressing-inflammation-rather-than-lowering-the-disease-activity-score-should-be-targeted-during-tnf-inhibitor-treatment-to-slow-radiographic-changes-in-patients-with-ankylosing-spondylitis/ (accessed 14 July 2022) [Google Scholar]