Abstract
Background:
N6-methyladenosine (m6A) modification plays crucial roles in cancers. However, its alteration in colorectal cancer (CRC) is still poorly described. The purpose of this study is to explore the change of m6A modification and the function of m6A binding protein YTHDC2 in CRC.
Methods:
The global level of m6A modification was detected by mass spectrometry and dot blotting assay. The expression of YTHDC2 was investigated using The Cancer Genome Atlas and using real-time polymerase chain reaction (RT-qPCR), western blotting, and immunohistochemistry based on CRC tissues. Kaplan–Meier analysis and Cox proportional hazards regression were performed to analyze the prognostic value of YTHDC2. RNA immunoprecipitation (RIP)-seq and m6A immunoprecipitation (MeRIP)-seq were used to explore the direct targets of YTHDC2. Gene oncology (GO) and Gene Set Enrichment Analysis (GSEA) were used to explore the pathways that could be influenced by YTHDC2.
Results:
No significant difference was observed in the global level of m6A modification on total RNA or mRNA between CRC and adjacent nontumor tissues. We further found a significant decreasing of YTHDC2 in CRC tissues. Kaplan–Meier analysis indicated that lower expression of YTHDC2 was related to the worse disease-free survival and overall survival. In addition, lower expression of YTHDC2 was an independent worse prognostic factor in univariate and multivariate Cox regression analysis. Using YTHDC2-RIP-seq and MeRIP-seq, we identified that YTHDC2 could participate in several important biological signal pathways.
Conclusions:
In summary, this study suggested that the global level of m6A did not change in CRC and identified that lower YTHDC2 as a prognostic marker for worse survival of CRC.
Keywords: Colorectal cancer, N6-methyladenosine, RNA modification, YTHDC2, prognostic biomarker
Introduction
Colorectal cancer (CRC) is the third most common cancer and the second leading cause of cancer-related deaths worldwide. 1 Despite the tremendous progress in the diagnostic and therapeutic strategies of CRC,2,3 the prognosis is still bleak resulting from rapid progression or early metastasis. 4 Therefore, in the era of molecular medicine, identification of diagnostic and therapeutic biomarkers of CRC is warranted.
N6-methyladenosine (m6A) modification, as the most common epigenetic modification of eukaryotic mRNA, 5 plays a key role in various tumor cellular processes. 6 N6-methyladenosine modification is dynamically modulated by methyltransferases (Writers) 7 and demethylases (Erasers)8,9 and recognized by different binding proteins (Readers) 10 to play various biological roles in mRNA splicing, stability, and translocation. 11
The YT521-B homology (YTH) family with RNA-binding domains is the first of “Readers” found to directly recognize the m6A sites of RNA, 12 including YTHDF1-3 13 and YTHDC1-2. 14 YTHDC2 has been previously reported to play a key role in liver steatosis, 15 mammalian spermatogenesis, 16 tumorigenesis, 17 and progression of tumor. 18 In the previous studies, YTHDC2 was identified to be a tumor suppressor in most types of cancer.18,19 However, it is still controversial whether YTHDC2 promotes or suppresses CRC,20-23 which needs to be explored further.
In this study, we verified the levels of m6A and m6A-related regulators in CRC tissues and found the potential role of YTHDC2 in the prognosis of CRC. To identify the clinical prognostic significance, the association between the expression of YTHDC2 and clinicopathological features of CRC patients was further investigated.
Materials & Methods
Study populations
Two hundred and fifteen CRC patients who underwent radical resections in Zhongshan Hospital from 2009 to 2012 were enrolled, and the corresponding tumor tissues were used for tissue microarray (TMA). In addition, CRC tissues and paired-adjacent nontumor tissues were collected from another 24 patients for the paraffin section and the isolation of RNA and protein. The characterization of the study cohort is presented in Supplementary Table 1. Our study was approved by the Clinical Research Ethics Committee of Zhongshan Hospital, Fudan University (protocol ID B2018-038R, approval data 02.05.2018) and was conducted in accordance with the Declaration of Helsinki.
The Cancer Genome Atlas (TCGA) data sets (https://portal.gdc.cancer.gov/) with 638 CRC tissues and 51 adjacent nontumor tissues were used for analyzing the level of m6A-related regulators and Gene Set Enrichment Analysis (GSEA). The Human Protein Atlas data set was used to analyze the overall survival of different YTHDC2 levels.
Cell culture
RKO cell line was purchased from American Type Culture Collection (ATCC, USA) and was cultured in Dulbecco's Modified Eagle Medium (DMEM; SH30243.01, Hyclone Laboratories Inc) with 10% fetal bovine serum (FBS; 16000-044, Gibco, Carlsbad, CA, USA) and 1% penicillin/streptomycin (15140122, Gibco).
RNA isolation and mRNA isolation
Colorectal cancer tissues and adjacent nontumor tissues were stored at RNAlater (R0901, Sigma-Aldrich, Daemstadt, Germany) storage and frozen immediately, then stored at −80°C until RNA isolation. Total RNA was extracted using the TRIzol® reagent (Ambion, Life technologies, Carlsbad, CA, USA) and dissolved in RNase-free water. Poly(A) + mRNA was isolated twice using the Magnetic mRNA Isolation Kit (S1550S, New England Biolabs Inc, Nebraska, USA) according to the manufacturer’s instructions. RNAs were quantified by NanoDrop 2000 spectrophotometer (Thermo Scientific, Waltham, MA, USA).
RNA digestion and LC-MS/MS analysis
Liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis was performed as previous reported. 24 In brief, a total of 300 ng RNA sample was fully digested by Nucleoside Digestion Mix (M0649S, New England Biolabs Inc) according to the manufacturer’s instructions. The LC-MS/MS analysis was performed on Agilent6410 QQQ triple-quadrupole mass spectrometer. Nucleosides were quantified using retention time and nucleoside to base ion mass transitions of 284 to 152 (G), 282.1 to 150.1 (m6A), and 268 to 136 (A).
Dot blotting
Dot blotting assay was performed as previously reported. 25 In brief, equal amounts total RNA and mRNA were denatured at 95°C for 10 min, followed by chill on ice immediately. RNAs (1 µL) were dropped onto the nitrocellulose membrane and ultraviolet (UV) cross-linking was performed. The membranes were blocked with 3% bovine serum albumin (BSA) in 0.1% TBS-Tween (TBST) and incubated with primary antibody for overnight (4°C), anti-m6A (202003, Synaptic Systems, Germany, 1:1000). The membranes were washed with TBST and incubated with secondary antibody goat antirabbit IgG-HRP (L3012, Signalway Antibody, 1:5000) for 1 hour at room temperature. The dots were visualized applying ECL kit (Applygen Technologies Inc, Beijing, China). The images of dots were observed using the ChemiDoc Touch Imaging system (Bio-Rad, Hercules, CA, USA).
Quantitative real-time polymerase chain reaction (RT-PCR) analysis
Total RNA was reverse transcribed into cDNA using a PrimeScript™ RT Reagent Kit with gDNA Eraser (Takara Bio Inc, Japan) according to the manufacturer’s protocol. The cDNA was used for RT-PCR using the SYBR Green Master Mix (Roche) on a Roche LightCycler 480 II system. Primer sequences were listed in Table 1.
Table 1.
Primer sequences of target genes in this study.
| Target gene | Primer sequence (5ʹ→3ʹ) |
|---|---|
| GAPDH | F: AGGTCGGAGTCAACGGATTT R: TGACGGTGCCATGGAATTTG |
| METTL3 | F: CTTGCATGGATTCTGAGGCC R: GTCAGCCATCACAACTGCAA |
| METTL14 | F: AATCGCCTCCTCCCAAATCT R: CCACCTCTTTCTCCTCGGAA |
| WTAP | F: TCAGTGCGGGGTATGAAAGT R: ACCTTTCCCACTCACTGCTT |
| ALKBH5 | F: TCATCAACGACTACCAGCCC R: GGCTTGAACTGGAACTTGCA |
| FTO | F: AGACACCTGGTTTGGCGATA R: CCAAGGTTCCTGTTGAGCAC |
| YTHDF1 | F: GCAACTCTCCTGGAAACGTC R: TTGATGATGAACACACGCCC |
| YTHDF2 | F: GTGTTGGAGAAGCTTCGGTC R: AGCAGCATCCAGTCTCTTGT |
| YTHDF3 | F: CTGCCAAACCTCAACCGAAA R: GAGCCTTTACCACTGACCCT |
| YTHDC1 | F: TTTCCTTCGTCGCACACAAG R: CGGTCTCTGTCTCGATCACA |
| YTHDC2 | F: TCTGAGAATTGGGCTGTCGT R: TTCTCCTTTGGCCCTGTCAA |
Western blotting
Western blotting was performed following standard protocols. In brief, total proteins were extracted from CRC tissues and adjacent nontumor tissues using SDS lysis buffer (P0013G, Beyotime Biotechnology, Shanghai, China). The protein concentration was measured with BCA Protein Assay Kit (P0010S, Beyotime Biotechnology). The total proteins were separated by 10% SDS-PAGE (sodium dodecyl sulfate polyacrylamide gel electrophoresis) and then transferred to nitrocellulose membrane. The membranes were blocked by 5% skimmed milk for 1 hour, then incubated overnight at 4°C using the primary antibody against human YTHDC2 (35440, Cell Signaling Technology, 1:1000), β-actin (66009-1, Proteintech, 1:2000). The membranes were further incubated using secondary antibody goat anti-rabbit IgG-HRP (L3012, Signalway antibody, 1:5000) and secondary antibody goat antimouse IgG-HRP (L3032, Signalway antibody, 1:5000) for 1 hour. β-Actin was used as the loading control. Protein bands were detected with ECL kit and ChemiDoc Touch Imaging System (Bio-Rad).
TMA and immunohistochemistry (IHC)
The TMA slides of CRC specimens were obtained from Zhongshan Hospital. Immunohistochemistry (IHC) staining was performed as described previously. 26 In brief, TMA slides were incubated with YTHDC2 antibody (ab220160, Abcam, Cambridge, UK, 1:500) overnight at 4°C. After washing with phosphate-buffered saline (PBS), TMA slides were subjected to the secondary antibody for 1 hour at room temperature, stained with diaminobenzidine and counterstained with hematoxylin. Staining intensity was assessed by 2 independent pathologists who were blinded to the clinical data. A 4-point scale (0, undetectable; 1, weak; 2, moderate; 3, strong) was used and the percentage of positively stained cells was expressed as 4 categories (1: 0%-25%, 2: 26%-50%, 3: 51%-75%, and 4: 76%-100%). The 2 scores were multiplied together to yield the final score. The patients with a greater than or equal to 8 final score were defined as high expression group.
RIP-Seq AND MeRIP-Seq
The RNA immunoprecipitation (RIP) assay was performed according to the instruction of the EZ-Magna RIP Kit (17-701, Merck Millipore, USA) with anti-YTHDC2 (ab22016, Abcam) and corresponding control rabbit IgG. The m6A immunoprecipitation (MeRIP) procedure was performed according to the instruction of the Gen Seq m6A-MeRIP Kit (GS-T-001, Gen Seq). Transcriptome sequencing and bioinformatics analysis were all done by Cloud-Seq Biotech (Shanghai, China).
Statistical analysis
Statistical differences between groups were determined by using a chi-squared test. Survival analysis was calculated by Kaplan–Meier analysis and compared using the log-rank test. All the results are presented as the mean ± standard deviation. P < .05 was considered to indicate statistically different. Statistical analysis was performed using SPSS software version 22.0.
Results
Global level of m6A modification in CRC patients
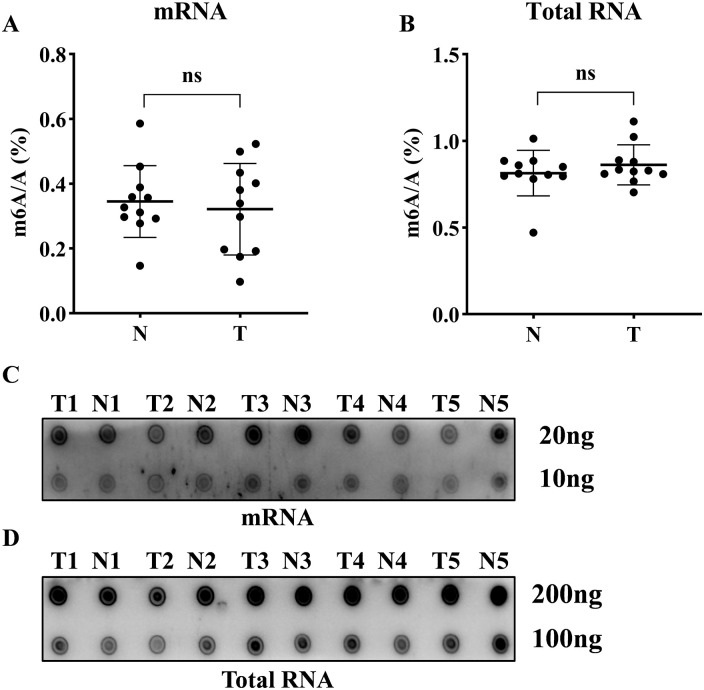
The percentage ratio of m6A to A was determined in 11 CRC patients using the LC-MS/MS method. No significant difference was observed in the amount of m6A between tumor tissues and adjacent nontumor tissues in mRNA or total RNA samples (Figure 1A and B). Dot blot assay also revealed that there was no significant difference in m6A levels between the tumor and adjacent nontumor tissues (Figure 1C and D). These results suggested that the alteration of global m6A level might not be essential in CRC carcinogenesis.
Figure 1.
The global levels of m6A RNA modification in CRC patients: (A-B) The percentage of m6A in relation to adenosine in mRNA and total RNA derived from 11 randomly selected CRC patients by LC-MS/MS method. (C-D) Global m6A level in mRNA and total RNA of CRC tissues and paired-adjacent nontumor tissues were shown as 5 randomly selected samples by dot blot assay. A: adenosine; N: adjacent nontumor tissues; T: tumor tissues; the data in (A-B) are presented as the means ± SD; ns: no significant difference.
YTHDC2 was down-regulated in CRC tissues
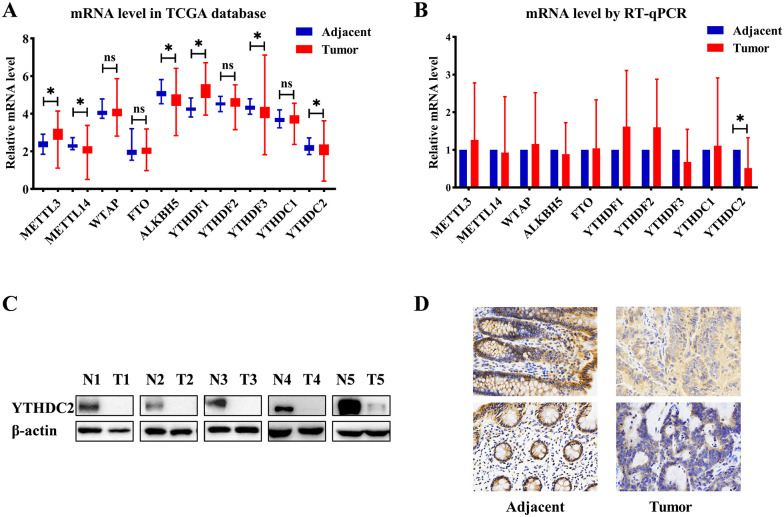
We further explore the expression of m6A regulators (Writers: METTL3, METTL14, WTAP; Erasers: FTO, ALKBH5; Readers: YTHDF1-3, YTHDC1-2). The mRNA levels of m6A regulators were analyzed using TCGA database. Compared with adjacent nontumor tissues, the mRNA levels of METTL3 and YTHDF1 were increased in tumor tissues, and METTL14, ALKBH5, YTHDF3, and YTHDC2 were deceased in tumor tissues (Figure 2A). Moreover, we performed RT-qPCR to verify the result in tumor and adjacent nontumor tissues from 24 CRC patients. However, we have not observed a statistically significant difference in the expression of m6A regulators, except YTHDC2 (P = .005, Figure 2B and Supplementary Figure 1).
Figure 2.
The expression of m6A-associated proteins in CRC patients: (A) mRNA levels of m6A-related regulators in TCGA database, (B) mRNA levels of m6A-related regulators in CRC patients (n = 24), (C) the protein level of YTHDC2 in representative CRC patients by western blotting, and (D) the protein level of YTHDC2 in representative CRC by IHC. The data in (A-B) are presented as the means ± SD; ns: no significant difference; * P < .05.
We next verified that YTHDC2 was significantly decreased in CRC tumor tissues from Zhongshan Hospital by western blot and immunohistochemical staining of primary CRC lesions and adjacent nontumor tissues (Figure 2C and D). The significant low expression of YTHDC2 in tumors prompted us to investigate the clinical and functional consequences of YTHDC2 in CRC.
Low expression of YTHDC2 was associated with poor prognosis
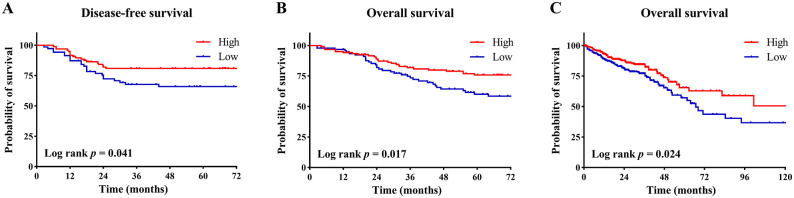
To investigate the clinical significance of YTHDC2 in CRC, we performed IHC staining for YTHDC2 in CRC TMA of Zhongshan cohort. The patients with low YTHDC2 expression had significant worse disease-free survival (DFS) and overall survival (OS; P = .041; P = .017, as shown in Figure 3A and B), which was consistent with the results from The Protein Atlas data set (Figure 3C). Multivariable Cox regression analysis showed that low YTHDC2 expression was an independent risk factor in DFS and OS (Tables 2 and 3). However, we have observed that no clinicopathological factor was significantly associated with YTHDC2 expression (Supplementary Table 2).
Figure 3.
Survival curves based on the expression of YTHDC2: (A) disease-free survival analysis in CRC patients from TMA, (B) overall survival analysis in CRC patients from TMA, (C) overall survival analysis in CRC patients from The Human Protein Atlas database. The median expression value was taken as the cut-off value and divided the patients into high YTHDC2 group and low YTHDC2 group.
Table 2.
Univariate and multivariate Cox regression analysis for disease-free survival (n = 173).
| Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | ||
| Age (years) | >65/⩽65 | 0.985 (0.538-1.806) | 0.962 | / | / |
| Sex | Male/female | 1.598 (0.861-2.965) | 0.137 | / | / |
| CEA (ng/mL) | >5/⩽5 | 2.605 (1.430-4.746) | 0.002 | 1.488 (0.773-2.864) | 0.234 |
| CA199 (U/mL) | >37/⩽37 | 4.453 (2.344-8.458) | <0.000 | 3.313 (1.685-3.673) | 0.001 |
| Tumor location | Left-side/right-side | 0.995 (0.519-1.908) | 0.988 | / | / |
| Tumor size (cm) | >5.0/⩽5.0 | 2.062 (1.075-3.953) | 0.029 | 0.992 (0.473-2.080) | 0.983 |
| Histological type | Mucinous/adenocarcinoma | 0.624 (0.277-1.402) | 0.253 | / | / |
| Primary differentiation | Poor/well | 1.997 (1.089-3.661) | 0.025 | 1.551 (0.829-2.902) | 0.17 |
| T-stage | T3-4/T1-2 | 4.222 (1.508-11.817 | 0.006 | 2.653 (0.824-8.542) | 0.102 |
| N-stage | N1-2/N0 | 2.442 (1.341-4.449) | 0.004 | 1.891 (1.021-3.500) | 0.043 |
| YTHDC2 level | Low/high | 1.807 (0.990-3.300) | 0.054 | 1.996 (1.085-3.673) | 0.026 |
Table 3.
Univariate and multivariate Cox regression analysis for overall survival (n = 215).
| Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | ||
| Age (years) | >65/⩽65 | 1.158 (0.703-1.907) | 0.565 | / | / |
| Sex | Male/female | 2.204 (1.263-3.847) | 0.005 | 1.462 (0.806-2.650) | 0.211 |
| CEA (ng/mL) | >5/⩽5 | 2.821 (1.696-4.694) | <0.000 | 1.580 (0.911-2.739) | 0.103 |
| CA199 (U/mL) | >37/⩽37 | 3.864 (2.337-6.391) | <0.000 | 2.025 (1.172-3.500) | 0.011 |
| Tumor location | Left-side/right-side | 0.777 (0.465-1.298) | 0.336 | / | / |
| Tumor size (cm) | >5.0/⩽5.0 | 2.463 (1.396-4.345) | 0.002 | 2.560 (1.340-4.891) | 0.004 |
| Histological type | Mucinous/adenocarcinoma | 1.002 (0.457-2.199) | 0.996 | / | / |
| Primary differentiation | Poor/well | 1.919 (1.167-3.157) | 0.01 | 1.648 (0.976-2.783) | 0.061 |
| T-stage | T3-4/T1-2 | 5.106 (1.854-14.062 | 0.002 | 1.257 (0.405-3.900) | 0.692 |
| N-stage | N1-2/N0 | 2.966 (1.766-4.982) | <0.000 | 1.644 (0.939-2.880) | 0.082 |
| M-stage | M1/M0 | 7.874 (4.754-13.053) | <0.000 | 4.798 (2.722-8.455) | <0.000 |
| YTHDC2 level | Low/high | 1.812 (1.102-2.979) | 0.019 | 1.959 (1.157-3.315) | 0.012 |
Signal pathways and cellular processes associated with YTHDC2
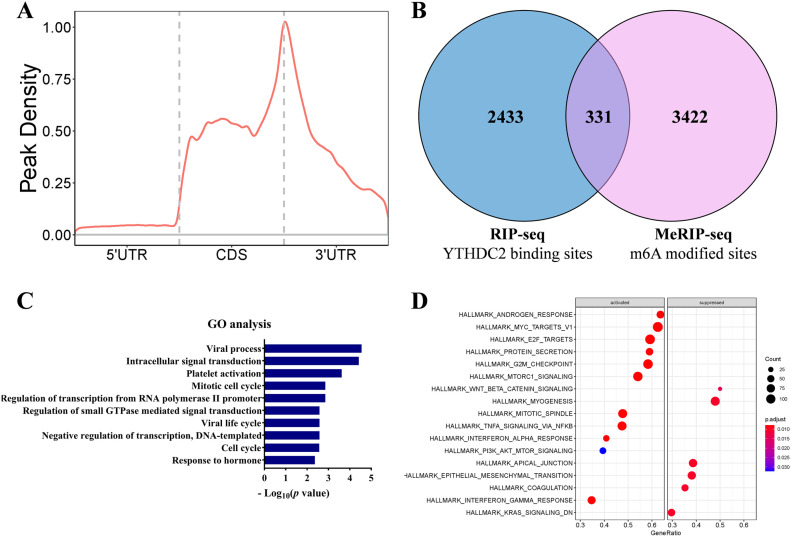
Furthermore, we performed MeRIP-seq and RIP-seq in RKO cell to investigate the effects of YTHDC2 on signal pathways and cellular processes. As a result, the MeRIP-Seq identified an amount of methylation peaks enriched in the region of CDS and 3ʹUTR in mRNA (Figure 4A). A total of 2764 mRNAs were identified as the possible direct targets of YTHDC2 by the RIP-seq, of which 331 mRNAs are m6A-modified (Figure 4B). Pathway analysis revealed that the enriched gene sets of the coding genes corresponding to the 331 mRNAs involved cell cycle and mitotic cell cycle (Figure 4C). In addition, we turned to GSEA to inspect the signal pathways involved in the high and low YTHDC2 expression groups based on TCGA database. We found that the YTHDC2 was related to the following biological signal pathways, including MYC targets, E2F targets, G2M checkpoint (Figure 4D), which were all associated with cell cycle. These results suggested that YTHDC2 may be associated with tumor progression by regulating these pathways.
Figure 4.
The biological pathways influenced by YTHDC2: (A) distribution of m6A peaks across the length of mRNAs in RKO, (B) Venn diagram showing overlap of the target genes of RIP-seq and MeRIP-seq (triple repetition), (C) GO analysis of the overlap targets using David (https://david.ncifcrf.gov/), and (D) GSEA based on TCGA database. The median expression value was taken as the cut-off value and divided the patients into high YTHDC2 group and low YTHDC2 group.
Discussion
Accumulating evidences indicate that the disturbance of m6A modification may contribute to the initiation, progression, and metastasis of cancer. 11 However, the association between m6A modification and CRC is still unclear. In this study, we first quantified the m6A modification in total RNA or mRNA of CRC patients. In addition, we identified m6A reader YTHDC2 as a markedly downregulated protein in CRC. Further analysis suggested that YTHDC2 was associated with the prognosis of CRC and several important signal pathways.
To our knowledge, this is the first study to quantify the m6A/A in both total RNA and mRNA of CRC, as m6A is the most common chemical modification of mRNA. Unfortunately, no difference was observed in m6A quantity in either total RNA or mRNA, which however seemed reasonable. First, the mechanism of m6A modification in physiological and pathological process is so complex 27 that its function cannot be fully explained by only global level change. Similar results were also found in ovarian cancer and head and neck squamous cell carcinoma studies.28,29 Second, we also detected the expression levels of “Writers” and “Erasers” which affects the global level of m6A modification.7-9 Corresponding to the no difference of global level of m6A modification, we also found no significant differences in the “Writers” and “Erasers” of m6A. Based on the results, we speculated that m6A modification might play a biological role in CRC through the disturbances in specific mRNA sites or the change of different “Readers.”
Pan et al 30 reported that m6A levels in RNA were substantially elevated in both adenoma and CRC, which was opposite to our results. However, the detection methods of m6A levels in RNA were totally different. The LC-MS/MS method was used in our study, which could be more direct and accurate. In addition, we also performed repeated identification at the mRNA level. RNA m6A quantification kit and IHC were used by Pan et al, and the accuracy could be affected by the specificity of antibody. The another reason for the difference may be related to tumor heterogeneity. Given the opposite results, it is valuable to further explore the level and function of m6A in larger sample size studies using more accurate method.
Notably, YTHDC2, the “Reader” of m6A, was significantly lower expressed in CRC tissues. This study initially explored the expression of YTHDC2 in CRC at the levels of RNA and protein, and the results was similar with previous TCGA-based studies.20-22 In addition, low expression of YTHDC2 was reported in other types of cancers, such as lung adenocarcinoma, 17 non–small-cell lung cancer, 18 and head and neck squamous cell carcinoma, 19 which suggested that YTHDC2 probably functions as a tumor suppressor. The survival analysis in our study also showed that YTHDC2 was an independent factor for better prognosis in CRC based on TMA. Therefore, the lower expression of YTHDC2 may play a key role in CRC through m6A modification. Tanabe et al 23 reported that YTHDC2 promoted the metastasis of CRC cell lines in vitro and in vivo. However, Tanabe et al did not further explore the m6A binding function of YTHDC2, which could be important to clarify the biological function of YTHDC2. In addition, we detected the level of YTHDC2 in tissues and public data sets, which could be more reliable. Based on these results, the biological functions of YTHDC2 should be explored further.
As a reader of m6A modification, YTHDC2 may play a role by influencing the metabolism of m6A mRNA. To explore this hypothesis, RKO cell was used to detect the m6A-modified mRNAs bound by YTHDC2. Our results showed that YTHDC2 might regulate several important pathways, such as cell cycle. This is the first study to report this function of YTHDC2 in CRC cell, which can provide the evidence for further YTHDC2 researches.
A few limitations must be considered when interpreting these results. First, we had just explored the change of the global level of m6A modification and failed to clarify its certain role in CRC. Second, the biological function and detail mechanism of YTHDC2 was not verified by experiments, which was necessary to further understand the function of YTHDC2 in CRC. These questions need to be researched by higher-quality study in the future.
Conclusions
Our study provides the evidence that there is no significant difference in global level of m6A between CRC and paired adjacent nontumor tissues, which also suggests that only the alteration of m6A in specific mRNA may produce effect in CRC. In addition, YTHDC2 is significantly low-expressed and correlated with the prognosis of CRC.
Supplemental Material
Supplemental material, sj-docx-1-onc-10.1177_11795549221104441 for The m6A RNA Modification Quantity and the Prognostic Effect of Reader YTHDC2 in Colorectal Cancer by Tianyu Liu, Wentao Tang, Yijiao Chen, Yu Liu, Donghao Xu, Yudong Jiang, Shizhao Zhou, Xiaorui Qin, Li Ren, Wenju Chang and Jianmin Xu in Clinical Medicine Insights: Oncology
Supplemental material, sj-docx-2-onc-10.1177_11795549221104441 for The m6A RNA Modification Quantity and the Prognostic Effect of Reader YTHDC2 in Colorectal Cancer by Tianyu Liu, Wentao Tang, Yijiao Chen, Yu Liu, Donghao Xu, Yudong Jiang, Shizhao Zhou, Xiaorui Qin, Li Ren, Wenju Chang and Jianmin Xu in Clinical Medicine Insights: Oncology
Acknowledgments
The results related to the public data sets shown in our study are in part based on data generated by the TCGA Research Network: https://www.cancer.gov/tcga.
Footnotes
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This research was funded by The National Natural Science Foundation of China (grant nos. 82072678, 81602035, and 81472228); Shanghai Science and Technology Committee Project (grant no. 19511121301); and Clinical Research Plan of SHDC (grant nos. SHDC2020CR5006, SHDC2020CR1033B, and SHDC2020CR3037B).
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: Conceptualization, Tianyu Liu, Wentao Tang, Yijiao Chen and Yu Liu; Methodology, Tianyu Liu, Wentao Tang, Yijiao Chen and Wenju Chang; Bioinformatics Analysis, Donghao Xu and Yudong Jiang; Investigation, Shizhao Zhou and Xiaorui Qin; Writing -Original Draft, Tianyu Liu; Writing -Review & Editing, Wenju Chang and Li Ren; Funding Acquisition, Jianmin Xu; Supervision, Jianmin Xu and Wenju Chang. All authors read and approved the final manuscript.
ORCID iDs: Tianyu Liu  https://orcid.org/0000-0001-5174-2539
https://orcid.org/0000-0001-5174-2539
Yudong Jiang  https://orcid.org/0000-0002-9405-4100
https://orcid.org/0000-0002-9405-4100
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209-249. [DOI] [PubMed] [Google Scholar]
- 2. Tang W, Ren L, Liu T, et al. Bevacizumab plus mFOLFOX6 versus mFOLFOX6 alone as first-line treatment for RAS mutant unresectable colorectal liver-limited metastases: the become randomized controlled trial. J Clin Oncol. 2020;38:3175-3184. [DOI] [PubMed] [Google Scholar]
- 3. Ye LC, Liu TS, Ren L, et al. Randomized controlled trial of cetuximab plus chemotherapy for patients with KRAS wild-type unresectable colorectal liver-limited metastases. J Clin Oncol. 2013;31:1931-1938. [DOI] [PubMed] [Google Scholar]
- 4. Guan X. Cancer metastases: challenges and opportunities. Acta Pharm Sin B. 2015;5:402-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169:1187-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. He L, Li H, Wu A, Peng Y, Shu G, Yin G. Functions of N6-methyladenosine and its role in cancer. Mol Cancer. 2019;18:176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu J, Yue Y, Han D, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10:93-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jia G, Fu Y, Zhao X, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zheng G, Dahl JA, Niu Y, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Haussmann IU, Bodi Z, Sanchez-Moran E, et al. M(6)A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature. 2016;540:301-304. [DOI] [PubMed] [Google Scholar]
- 11. Chen XY, Zhang J, Zhu JS. The role of m(6)A RNA methylation in human cancer. Mol Cancer. 2019;18:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Stoilov P, Rafalska I, Stamm S. YTH: a new domain in nuclear proteins. Trends Biochem Sci. 2002;27:495-497. [DOI] [PubMed] [Google Scholar]
- 13. Shi H, Wang X, Lu Z, et al. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 2017;27:315-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wojtas MN, Pandey RR, Mendel M, Homolka D, Sachidanandam R, Pillai RS. Regulation of m(6)A transcripts by the 3’→5’ RNA helicase YTHDC2 is essential for a successful meiotic program in the mammalian germline. Mol Cell. 2017;68:374.e12-387.e12. [DOI] [PubMed] [Google Scholar]
- 15. Zhou B, Liu C, Xu L, et al. N(6)-methyladenosine reader protein YT521-B homology domain-containing 2 suppresses liver steatosis by regulation of mRNA stability of lipogenic genes. Hepatology. 2021;73:91-103. [DOI] [PubMed] [Google Scholar]
- 16. Hsu PJ, Zhu Y, Ma H, et al. Ythdc2 is an N(6)-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017;27:1115-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ma L, Chen T, Zhang X, et al. The m(6)A reader YTHDC2 inhibits lung adenocarcinoma tumorigenesis by suppressing SLC7A11-dependent antioxidant function. Redox Biol. 2021;38:101801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sun S, Han Q, Liang M, Zhang Q, Zhang J, Cao J. Downregulation of m(6) A reader YTHDC2 promotes tumor progression and predicts poor prognosis in non-small cell lung cancer. Thorac Cancer. 2020;11:3269-3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li Y, Zheng JN, Wang EH, Gong CJ, Lan KF, Ding X. The m6A reader protein YTHDC2 is a potential biomarker and associated with immune infiltration in head and neck squamous cell carcinoma. PeerJ. 2020;8:e10385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhuang J, Lin C, Ye J. M(6) A RNA methylation regulators contribute to malignant progression in rectal cancer. J Cell Physiol. 2020;235:6300-6306. [DOI] [PubMed] [Google Scholar]
- 21. Chen Y, Wang S, Cho WC, Zhou X, Zhang Z. Prognostic implication of the m(6)A RNA methylation regulators in rectal cancer. Front Genet. 2021;12:604229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ji L, Chen S, Gu L, Zhang X. Exploration of potential roles of m6A regulators in colorectal cancer prognosis. Front Oncol. 2020;10:768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tanabe A, Tanikawa K, Tsunetomi M, et al. RNA helicase YTHDC2 promotes cancer metastasis via the enhancement of the efficiency by which HIF-1α mRNA is translated. Cancer Lett. 2016;376:34-42. [DOI] [PubMed] [Google Scholar]
- 24. Ma H, Wang X, Cai J, et al. N(6-)Methyladenosine methyltransferase ZCCHC4 mediates ribosomal RNA methylation. Nat Chem Biol. 2019;15:88-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Si W, Li Y, Ye S, et al. Methyltransferase 3 mediated miRNA m6A methylation promotes stress granule formation in the early stage of acute ischemic stroke. Front Mol Neurosci. 2020;13:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mao Y, Feng Q, Zheng P, et al. Low tumor infiltrating mast cell density confers prognostic benefit and reflects immunoactivation in colorectal cancer. Int J Cancer. 2018;143:2271-2280. [DOI] [PubMed] [Google Scholar]
- 27. Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m-A RNA methylation. Nat Rev Genet. 2014;15:293-306. [DOI] [PubMed] [Google Scholar]
- 28. Xu F, Li J, Ni M, et al. FBW7 suppresses ovarian cancer development by targeting the N(6)-methyladenosine binding protein YTHDF2. Mol Cancer. 2021;20:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Romanowska K, Rawłuszko-Wieczorek AA, Marczak Kosińska ŁA, Suchorska WM, Golusiński W. The m(6)A RNA modification quantity and mRNA expression level of RNA methylation-related genes in head and neck squamous cell carcinoma cell lines and patients. Biomolecules. 2021;11:908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pan J, Liu F, Xiao X, et al. METTL3 promotes colorectal carcinoma progression by regulating the m6A-CRB3-Hippo axis. J Exp Clin Cancer Res. 2022;41:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-onc-10.1177_11795549221104441 for The m6A RNA Modification Quantity and the Prognostic Effect of Reader YTHDC2 in Colorectal Cancer by Tianyu Liu, Wentao Tang, Yijiao Chen, Yu Liu, Donghao Xu, Yudong Jiang, Shizhao Zhou, Xiaorui Qin, Li Ren, Wenju Chang and Jianmin Xu in Clinical Medicine Insights: Oncology
Supplemental material, sj-docx-2-onc-10.1177_11795549221104441 for The m6A RNA Modification Quantity and the Prognostic Effect of Reader YTHDC2 in Colorectal Cancer by Tianyu Liu, Wentao Tang, Yijiao Chen, Yu Liu, Donghao Xu, Yudong Jiang, Shizhao Zhou, Xiaorui Qin, Li Ren, Wenju Chang and Jianmin Xu in Clinical Medicine Insights: Oncology